- 1Department of Pharmacology and Clinical Pharmacy, Faculty of Pharmacy, Universitas Padjadjaran, Bandung, Indonesia
- 2Laboratory of Translational Pharmaceutical Research, Faculty of Pharmacy, Universitas Padjadjaran, Bandung, Indonesia
- 3Department of Pharmaceutical Analysis and Medical Chemistry, Faculty of Pharmacy, Universitas Padjadjaran, Bandung, Indonesia
- 4Pharmacy Study Program, Faculty of Science and Technology, Universitas Mandala Waluya, Kendari, Indonesia
- 5Pharmacy Study Program, Faculty of Pharmacy, Universitas Halu Oleo, Kendari, Indonesia
- 6Department of Pharmacy, Faculty of Mathematics and Natural Sciences, Universitas Islam Bandung, Bandung, Indonesia
- 7Research Collaboration Centre for Theranostic Radio Pharmaceuticals, National Research and Innovation Agency (BRIN), Bandung, Indonesia
The Wnt/β-catenin signaling pathway is critically involved in breast cancer progression, particularly in the triple-negative subtype (TNBC). Aberrant activation of this pathway promotes tumor proliferation, with β-catenin functioning as a central effector regulated by GSK-3β-mediated phosphorylation and degradation. Despite its therapeutic significance, no selective Wnt/β-catenin inhibitors have been clinically approved, underscoring the need for alternative strategies. Natural compounds such as α-mangostin have emerged as potential modulators of this pathway. This study investigates the potential of α-mangostin, a natural xanthone compound, to suppress Wnt/β-catenin signaling through complementary in silico approaches examining its interaction with proteins related to the Wnt signaling pathway, followed by in vitro validation using the MDA-MB-231 triple-negative breast cancer cell line (ER-/PR-/HER2-). In parallel, MCF-7 cells (ER+/PR+/HER2-) were used as a comparator to evaluate the differential inhibitory effects on breast cancer cells with distinct hormonal profiles. Molecular docking demonstrated favorable binding of α-mangostin to β-catenin and LRP6, with higher affinity toward LRP6. Molecular dynamics simulations confirmed the stability of these complexes, particularly the α-mangostin-LRP6 complex, which exhibited minimal RMSD and SASA fluctuations. Consistently, MM/PBSA calculations revealed the most favorable binding free energy for α-mangostin with LRP6 (−96.659 kJ/mol). In vitro WST-8 assays revealed that α-mangostin reduced cell viability in both cell lines, with a greater suppressive effect observed in combination with LiCl. Treatment with 10 µM α-mangostin, alone or with LiCl, significantly downregulated the Wnt transcriptional targets CCND1 (5.2-fold) and MYC (3.3-fold) in MDA-MB-231 cells, as determined by RT-qPCR, thereby indicating a potent suppressive effect on the Wnt pathway. Collectively, these findings indicate that α-mangostin exerts anticancer effects by targeting multiple components of the Wnt/β-catenin pathway, with LRP6 emerging as its primary target. Further investigations are warranted to elucidate its impact on β-catenin phosphorylation and to validate its efficacy in vivo.
1 Introduction
According to data from the Global Burden of Cancer Study (GLOBOCAN) by the World Health Organization (WHO) in 2020, breast cancer exhibits a high incidence rate in Asia, accounting for 45.5% of cases and contributing to 50.5% of cancer-related deaths (Sung et al., 2021). In Indonesia specifically, breast cancer ranks highest among new cases, with 65,858 instances, representing 16.6% of the total 396,914 reported cancer cases (Andinata et al., 2023). Among the breast cancer subtypes, triple-negative breast cancer (TNBC) is known for its poor prognosis. TNBC has limited treatment options compared to other breast cancer subtypes (Chaudhuri et al., 2022; Lee, 2023). The aggressive nature of TNBC underscores the urgency for targeted therapies that can effectively manage its progression and improve patient outcomes.
TNBC is characterized by the absence of estrogen receptor, progesterone receptor, and Human Epidermal Receptor-2 (HER-2) expression (Treeck et al., 2020). This type of cancer is more prevalent in individuals with BRCA1/BRCA2 gene mutations, which predispose them to cancer development (Mehrgou and Akouchekian, 2016). Alterations influence cancer progression in multiple signaling pathways, including the Wnt/β-catenin pathway. Various target genes of the Wnt signaling pathway have been identified and are implicated in regulating processes such as cell proliferation, invasion, metastasis, apoptosis, and resistance to chemotherapy (Zhang and Wang, 2020). The Wnt signaling pathway encompasses two main branches: canonical and non-canonical pathways, each playing distinct roles in cellular function and cancer progression (Liu et al., 2022). Understanding the intricate mechanisms of Wnt signaling in TNBC could lead to novel therapeutic strategies targeting this aggressive subtype of breast cancer.
The canonical Wnt pathway initiates with the binding of the Wnt ligand to the receptor complex composed of frizzled (FZD) and low-density lipoprotein receptor-related protein 5/6 (LRP5/6) (Jeong and Jho, 2021). This binding event prevents GSK-3β from phosphorylating β-catenin, thereby inhibiting its ubiquitination and degradation (Lin et al., 2020). Consequently, β-catenin accumulates in the cytoplasm and translocates into the nucleus, where it binds to TCF/LEF transcription factors and stimulates the transcription of target genes such as CCND1 (Cyclin D1), MYC (c-Myc), AXIN2, and BIRC5 (Survivin) (Liu et al., 2022; Pai et al., 2017). Additionally, the Wnt/β-catenin signaling pathway plays a crucial role in TNBC, with β-catenin, Axin, and APC being key players in this pathway. Aberrant activation of this pathway, often observed in TNBC, contributes to cell proliferation, tumor development, progression, and metastasis, making it a potential therapeutic target for this aggressive form of breast cancer. Part of the destruction complex that regulates β-catenin levels. In the absence of Wnt signaling, AXIN, along with APC and GSK-3β, promotes the phosphorylation and degradation of β-catenin. However, when Wnt signaling is activated, AXIN’s function is inhibited, leading to β-catenin stabilization and nuclear translocation. Another critical component of the destruction complex. APC binds to β-catenin and promotes its degradation. Mutations in APC can lead to the stabilization of β-catenin and aberrant Wnt signaling (Jeong and Jho, 2021; Wu et al., 2025). Elucidating these molecular mechanisms is crucial not only for understanding cancer pathogenesis but also for developing targeted therapies that can potentially inhibit Wnt signaling in cancer.
Several synthetic Wnt inhibitors have been developed, each targeting different components of the pathway. For instance, LGK974 blocks the secretion of porcupine, a Wnt-acyltransferase (Liu et al., 2013), while ICG-001 and its derivative PRI-724 disrupt the β-catenin/CBP interaction, thereby inhibiting downstream transcriptional activity in head-and-neck squamous carcinoma and pancreatic cancer cells. These inhibitors are currently undergoing preclinical and clinical evaluation across various cancer types (Kimura et al., 2022). In addition to synthetic compounds, numerous natural products have demonstrated potential in modulating Wnt signaling. For example, curcumin, resveratrol, and epigallocatechin gallate (EGCG) have been reported to inhibit Wnt/β-catenin signaling through mechanisms such as downregulation of β-catenin expression or interference with its nuclear translocation (Ashrafizadeh et al., 2020; Vallée et al., 2019; Yang et al., 2016). Owing to their multitarget effects and relatively low toxicity, these natural agents represent promising adjunctive strategies for Wnt-targeted cancer therapy.
A natural compound recognized for its anti-breast cancer properties is α-mangostin (Muchtaridi et al., 2019; Nalla and Ganta, 2023; Nurhidayah et al., 2023; Sarmoko et al., 2023). Research has shown that α-mangostin effectively inhibits proliferation and induces apoptosis in SKBR3, MCF-7, and MDA-MB-231 cells, with IC50 values of 9.69 μM, 11.37 μM, and 7.46 μM, respectively (Zhu et al., 2021). The Wnt-mediated modulation of α-mangostin’s anticancer activity has been reported in both osteosarcoma and colon cancer cells. A recent study by Yang S. et al. (2021) demonstrated that α-mangostin induces endoplasmic reticulum (ER) stress due to excessive reactive oxygen species (ROS) generation, leading to the inactivation of the Wnt signaling pathway and subsequently triggering apoptosis through caspase cleavage (Yang S. et al., 2021). In addition, an earlier investigation by Yoo et al. (2011) in colon cancer cells found that α-mangostin inhibits TCF/β-catenin transcriptional activity and downregulates β-catenin protein levels. However, the stability of β-catenin remains unaffected (Yoo et al., 2011). Despite these findings, no study to date has evaluated the effect of α-mangostin on the expression of Wnt/β-catenin target genes in TNBC. Therefore, the present study aims to investigate the therapeutic potential of α-mangostin in breast cancer, focusing specifically on its effects on the Wnt signaling pathway through both in vitro and in silico approaches. In vitro experiments investigated the inhibitory effects of α-mangostin on MDA-MB-231 and MCF-7 cells under conditions of Wnt/β-catenin pathway activation. To evaluate the inhibition of breast cancer cell proliferation, MCF-7 cells (ER+/PR+/HER2−, luminal subtype) were used as a comparator against MDA-MB-231 cells (triple-negative, ER−/PR−/HER2−). Gene expression levels of CCND1 and MYC were quantified using RT-qPCR to assess the downstream impact of α-mangostin on Wnt pathway target genes involved in cell proliferation and survival in MDA-MB-231 cells. Concurrently, in silico methods will be employed to elucidate the interaction between α-mangostin and key components of the Wnt/β-catenin signaling pathway, including LRP6 and β-catenin. This integrated approach aims to provide comprehensive insights into the therapeutic potential of α-mangostin in targeting the Wnt/β-catenin pathway and its implications for breast cancer treatment, notably in TNBC.
2 Materials and methods
2.1 In silico approaches
2.1.1 Molecular docking simulation
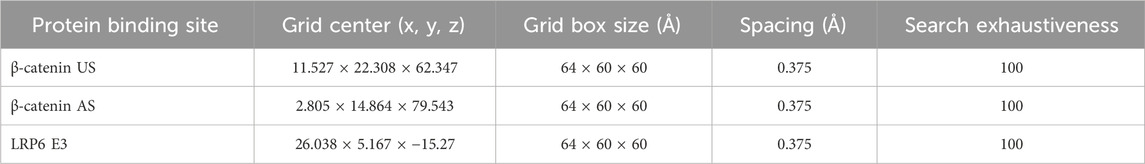
The initial structures for molecular modeling were obtained from the crystal structures of β-catenin (PDB ID: 2GL7, resolution 2.60 Å) and LRP6 (PDB ID: 3S2K, resolution 2.80 Å) (Ahn et al., 2011; Sampietro et al., 2006) (Table 1). Missing loop regions were reconstructed using the auto model and loop model functions of MODELER in Discovery Studio (DS) 2019 Client (Kemmish et al., 2017). Prior to molecular docking simulations, water molecules and ligands were removed, and hydrogen atoms were added to the protein structures. Further preparation of β-catenin and LRP6 was carried out using UCSF Chimera and AutoDockTools 4.2.6, with AD4 atom types and Gasteiger charges assigned to optimize docking accuracy (Forli et al., 2016; Pettersen et al., 2004).
β-Catenin possesses two primary binding sites: the canonical β-catenin union site (β-catenin US), which includes Asp16 from TCF4, Lys435, and His470, with the NZ atom of Lys435 defined as the grid center; and an allosteric site (β-catenin AS), characterized by Pro521, Arg528, and Asp583, with the ND1 atom of His524 designated as the center. For LRP6, the docking grid was centered at Gly227 from DKK1, consistent with previously reported binding interactions. Initial docking poses for subsequent molecular dynamics (MD) simulations were generated using AutoDockTools 4.2.6, with docking parameters optimized according to a previous study (Vélez-Vargas et al., 2023). The best binding poses were selected based on binding affinity and further validated by visual inspection using Discovery Studio (DS) 2019 Client, PLIP, and UCSF Chimera. For comparative analysis, hit derivatives were also evaluated using Schrödinger’s Glide software (version 2019–1, Schrödinger, LLC, New York, NY, United States, 2017) (Durrant and McCammon, 2011). These docking results provided the foundation for further MD simulations, enabling a detailed These docking results served as the basis for MD simulations, allowing detailed investigation of the stability and interactions of α-mangostin and its derivatives with β-catenin and LRP6.
2.1.2 Molecular dynamics (MD) simulation
All-atom molecular dynamics (MD) simulations of the top-ranked docking poses were performed using GROMACS 2016.3 with the PLUMED 2.4 plugin (Abraham et al., 2015). Protein and ligand parameters were generated using the CHARMM36 force field and CGenFF (Wurl and Ferreira, 2023), with systems solvated in a TIP3P water box (10 Å buffer) and neutralized with 0.15 M NaCl. Energy minimization was performed using the steepest descent method until a tolerance of 1000 kJ/mol was achieved, followed by NVT equilibration (25 ps, 303.15 K). NPT production runs for 500 ns at 303.15 K and 1 bar, controlled by the Nosé–Hoover thermostat and Parrinello–Rahman barostat (Shiga et al., 2023). Bond constraints involving hydrogen atoms were applied using the LINCS algorithm (Hess et al., 1997). The production runs employed a 2 fs timestep with trajectories saved every 1 ps; electrostatic interactions were treated with PME, and van der Waals interactions were truncated at 12 Å. An upper-wall restraint (200 kJ/mol·nm-2) was applied when the ligand center of mass exceeded 12 Å from the binding site. Protein–ligand interactions were analyzed over the final 200 ns using GROMACS 2016.3, Discovery Studio 2019 (Kemmish et al., 2017), and VMD 1.9.4 (Humphrey et al., 1996).
2.2 In vitro evaluation
2.2.1 Cell culture
MDA-MB-231 and MCF-7 cell lines were obtained from the European Collection of Authenticated Cell Cultures (ECACC; MDA-MB-231, catalog no. 92020424; MCF-7, catalog no. 86012803) and maintained at the Translational Pharmaceutical Research Laboratory, Faculty of Pharmacy, Universitas Padjadjaran, Sumedang, Indonesia.
MDA-MB-231 cells were maintained at 37 °C in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 2 mM L-glutamine, 100 IU/mL penicillin, and 10 μg/mL streptomycin in a humidified atmosphere containing 5% CO2. The cells reached optimal growth at a density of 1–3 × 10^4 cells/cm2. Once confluent, cultures were harvested by adding 1–2 mL of 0.25% trypsin. The detached cells were transferred to a conical tube, neutralized with fresh DMEM containing FBS to a final volume of 10 mL, and centrifuged at 2000 rpm for 5 min. The supernatant was discarded, and the resulting pellet was resuspended in 1 mL of medium. Viable cells were then counted using a hemocytometer. MCF-7 cells were cultured following the same procedure, except that Eagle’s Minimum Essential Medium (EMEM, EBSS) supplemented with 2 mM L-glutamine, 1% non-essential amino acids (NEAA), and 10% FBS was used.
2.2.2 Cell proliferation inhibitor assay
Cell proliferation was assessed using the WST-8 assay. MDA-MB-231 and MCF-7 cells were seeded into 96-well plates and treated for 24 h with α-mangostin (60, 120, 240, and 480 µM), LiCl (2.5, 5, and 10 mM) or their combinations. After treatment, cells were washed with PBS and incubated with 100 µL of 0.5 mg/mL WST-8 solution at 37 °C for 2–4 h. Absorbance was measured at 450 nm using a Tecan Infinite spectrophotometer, and cell viability was calculated relative to untreated controls.
2.2.3 Expression quantification of CCND1 and MYC
The MDA-MB-231 cell line was seeded into 6-well plates and incubated for 24 h. Cells were then treated with α-mangostin at concentrations of 5 μM and 10 µM or with cisplatin at 1.25 µM and 2.5 µM, either alone or in combination with LiCl (5 mM). RNA was isolated using the GeneZol™ Kit according to the manufacturer’s instructions, and the resulting RNA was resuspended in nuclease-free water (NFW). RT-qPCR was performed with the SensiFAST™ SYBR® No-ROX One-Step Kit in a total reaction volume of 20 μL. The amplification program consisted of an initial denaturation at 95°C for 5 s, followed by 40 cycles of 95 °C for 30 s, 55 °C for 60 s, and 72 °C for 60 s. Primers specific to the target genes are listed in Table 2, with GAPDH used as the housekeeping control gene for normalization (Sun et al., 2015; Xiang et al., 2021). Relative gene expression was calculated using the 2−ΔΔCT method, and all experiments were conducted in biological triplicates (n = 3).
2.2.4 Statistical analyses
Statistical analyses were performed using Microsoft Excel and GraphPad Prism 10.0.0 (GraphPad Software). All data are expressed as mean ± SD. Differences between groups were analyzed using one-way ANOVA followed by Tukey’s post hoc test, with significant p-values indicated in the figures. Significance levels are indicated as follows: P < 0.05; *P < 0.01; and **P < 0.001.
3 Results
3.1 In silico approach
3.1.1 Dynamic molecular behavior and system stability
Previous findings suggest that α-mangostin exerts antiproliferative effects in MDA-MB-231 cells by modulating the transcription of key Wnt-regulated genes involved in proliferation. To further elucidate the molecular basis of this effect, in silico analyses were performed to investigate the interaction of α-mangostin with β-catenin and LRP6. Figure 1 shows the binding profiles of α-mangostin and XAV939 within the β-catenin union site (US), the allosteric site (AS), and the LRP6 receptor. The surface representations highlight the spatial orientation and electrostatic characteristics of the binding pockets, providing context for the stability of interactions. At the β-catenin US, α-mangostin exhibited a docking score of −5.03 kcal/mol with a relatively weak binding affinity (Kd = 206.27 μM). By comparison, XAV939 demonstrated slightly stronger binding at −5.62 kcal/mol with a Kd of 76.33 μM, suggesting better accommodation within the same pocket. The β-catenin AS displayed similar energetics, with α-mangostin docking at −5.03 kcal/mol (Kd = 204.77 μM) and XAV939 at −5.40 kcal/mol (Kd = 109.27 μM), consistent with the more solvent-exposed and structurally relaxed nature of this site. In contrast, α-mangostin exhibited substantially stronger binding to LRP6, with a docking score of −7.23 kcal/mol and a low dissociation constant of 5.02 μM, indicating high affinity. XAV939 also bound LRP6 with favorable energetics (ΔG = −6.63 kcal/mol, Kd = 13.70 μM), but with lower affinity compared to α-mangostin. The binding orientation of α-mangostin in LRP6 was more deeply encapsulated, suggesting enhanced shape complementarity. Electrostatic surface mapping further revealed that LRP6 provides superior charge compatibility, facilitating stronger non-covalent interactions, including hydrogen bonding and π-interactions. Taken together, these results indicate that LRP6 serves as a more selective and energetically favorable receptor for α-mangostin. The improved structural anchoring in LRP6 supports its potential for greater biological efficacy and retention, which may be critical for downstream pharmacological activity (Medina-Barandica et al., 2023).
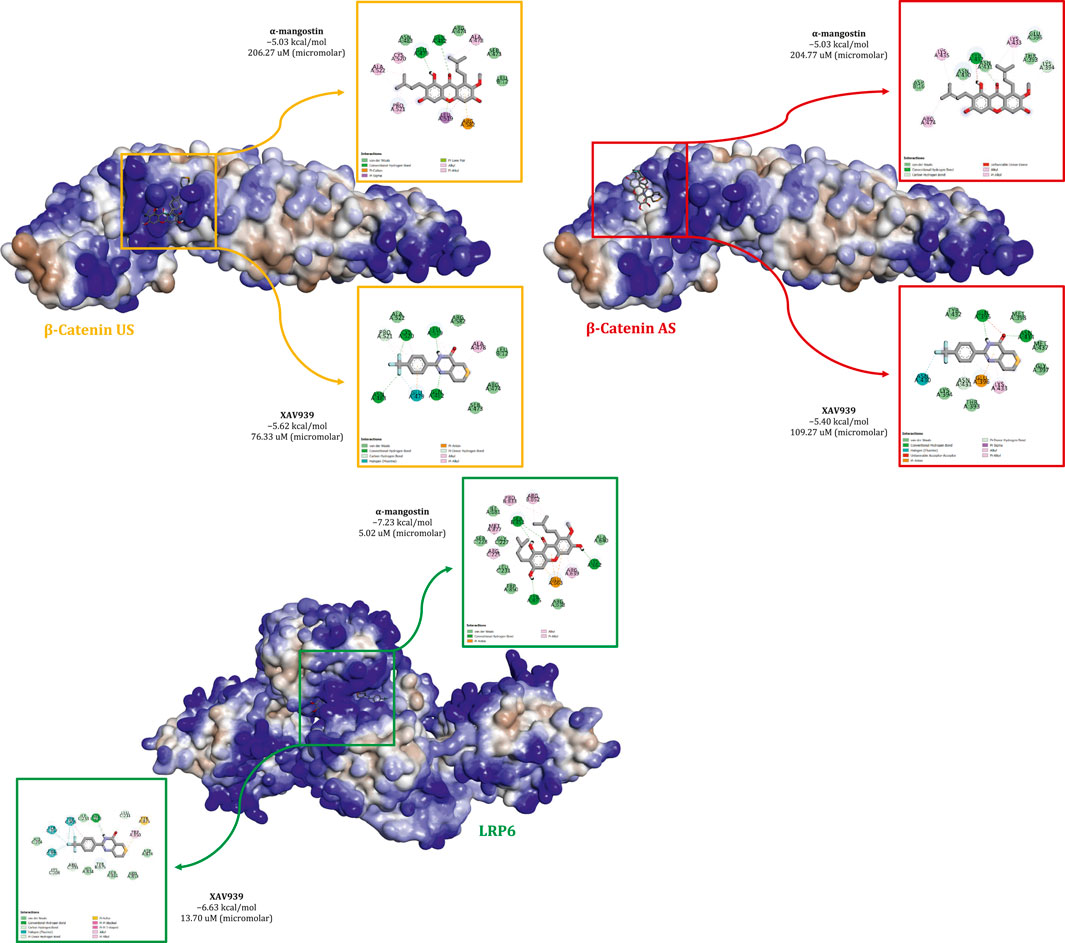
Figure 1. Comparative binding interactions of α-mangostin and XAV939 with β-catenin (union site, US; allosteric site, AS) and LRP6, showing docking affinities, key molecular interactions, and electrostatic surface representations.
Structural stability of the protein–ligand complexes was assessed by root mean square deviation (RMSD) analysis (Figure 2). In the β-catenin US complex, α-mangostin induced a gradual increase in protein RMSD, exceeding 0.35 nm toward the end of the simulation, whereas XAV939 maintained greater stability with values below 0.30 nm. Ligand RMSD plots showed that α-mangostin fluctuated around 0.20–0.22 nm, while XAV939 remained slightly lower at 0.15–0.20 nm. The β-catenin AS complex demonstrated improved stability, with both ligands maintaining consistent protein RMSD values between 0.25 and 0.30 nm. Ligand fluctuations in AS were also reduced compared to US, although α-mangostin still exhibited slightly higher deviation than the reference ligand. By contrast, LRP6 complexes exhibited the highest stability, with protein RMSD values consistently within the range of 0.20–0.30 nm throughout the simulation. Ligand RMSDs in LRP6 remained particularly low, with α-mangostin deviating by less than 0.15 nm. These findings indicate that LRP6 provides a more rigid and stable binding environment compared to β-catenin. The stable RMSD patterns observed in LRP6 suggest minimal structural rearrangements during ligand binding. Overall, these results highlight LRP6 as a conformationally stable and favorable target, supporting its potential for further drug development under physiological conditions (Aulifa et al., 2024).
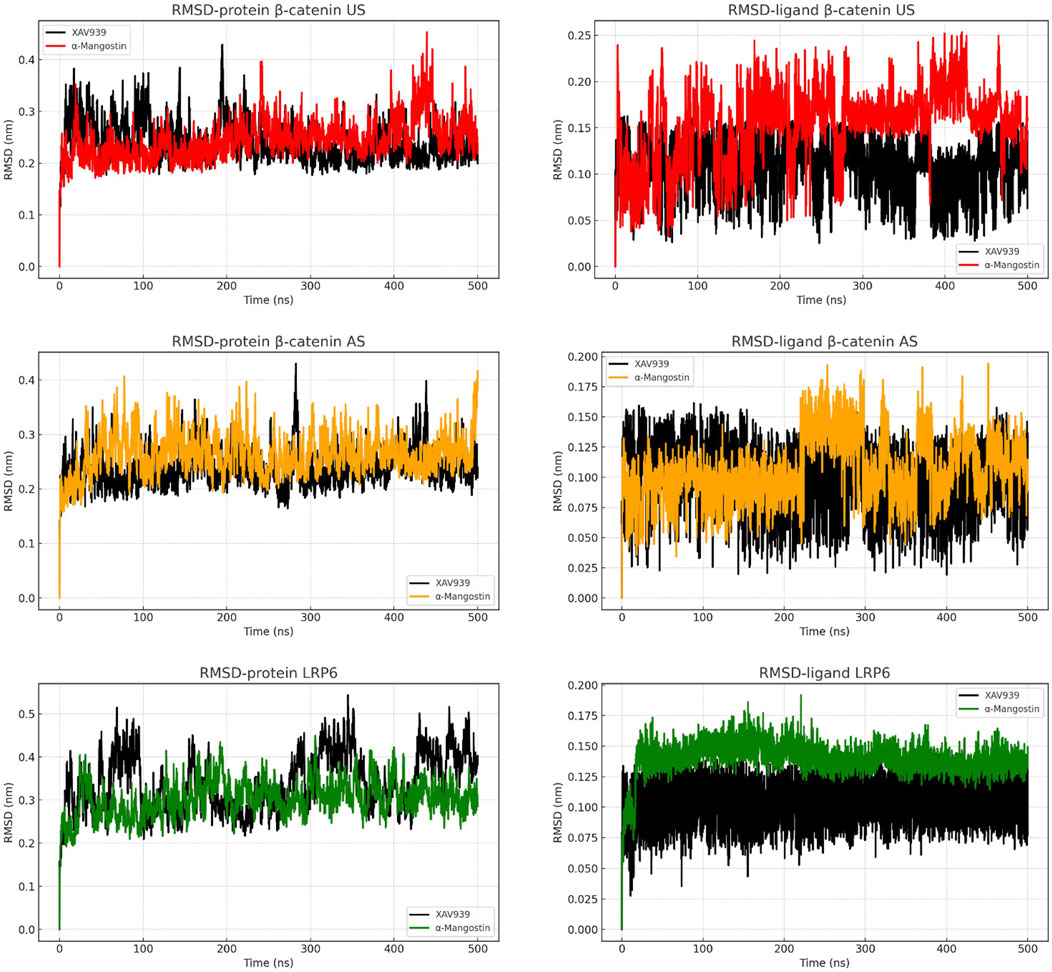
Figure 2. RMSD analysis of backbone atoms and ligand dynamics in β-catenin (union site, US; allosteric site, AS) and LRP6 throughout the simulation time.
To examine residue-level dynamics, root mean square fluctuation (RMSF) analyses were performed, as shown in Figure 3. In the β-catenin US complex, RMSF values for both ligands remained below 0.5 nm, with α-mangostin producing lower fluctuations than XAV939 across several regions. A minor peak was observed near residue index 600 for both ligands, indicating localized flexibility. In the AS configuration, the pattern changed markedly, as XAV939 displayed prominent peaks exceeding 2.0 nm, while α-mangostin exhibited restrained movements, with most residues fluctuating by less than 0.5 nm. These results suggest that α-mangostin stabilizes local dynamics more effectively in the AS. For LRP6, residue fluctuations were even more controlled, particularly in chain A, where RMSF values ranged between 0.2 and 0.4 nm. Chains B and C demonstrated similarly low fluctuations, with α-mangostin yielding slightly smoother profiles and values consistently below 0.3 nm. Across all chains, α-mangostin reduced residue mobility more effectively than the reference ligand. Reduced flexibility at the binding site contributes to enhanced ligand residency and complex stability. Overall, these RMSF trends support the conclusion that LRP6 provides a more rigid and stable interaction interface compared to β-catenin, a property advantageous for the design of ligands targeting dynamic protein systems.
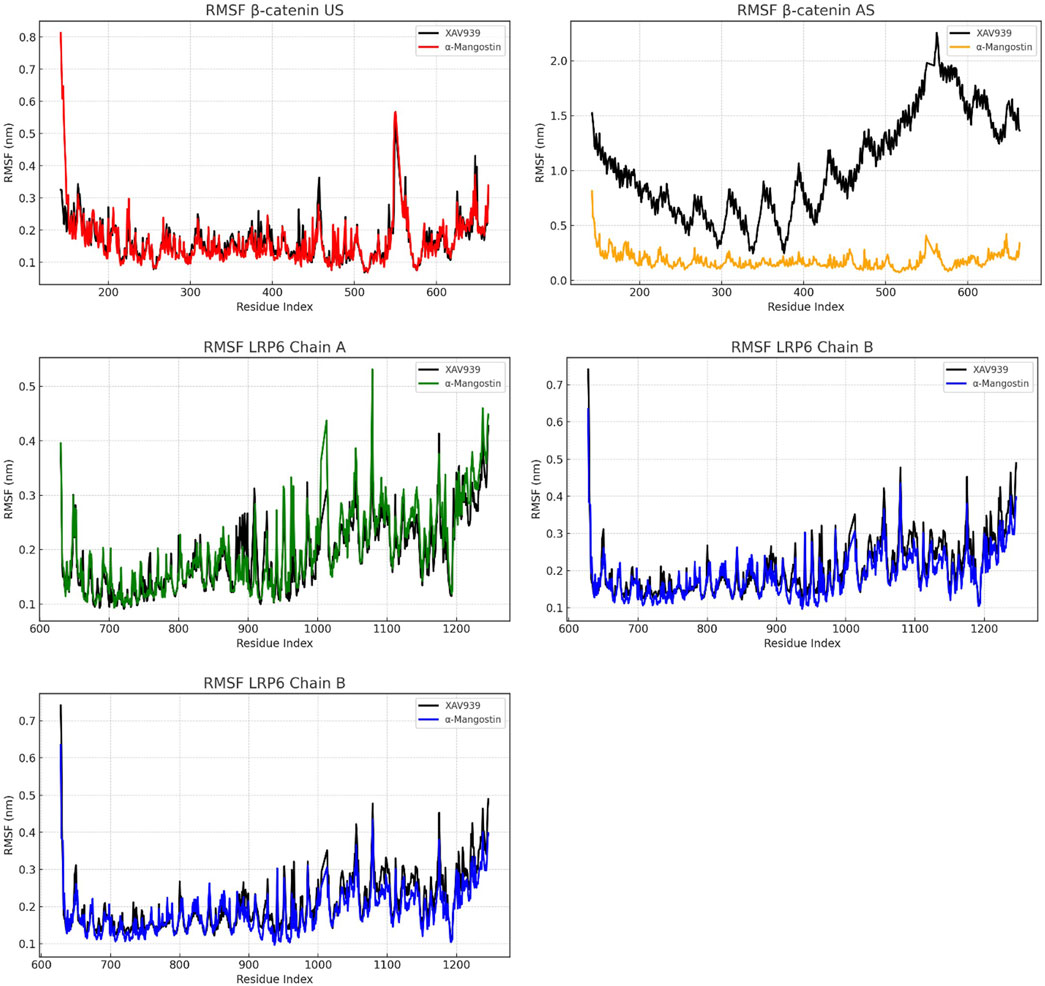
Figure 3. RMSF analysis of β-catenin (union site, US; allosteric site, AS) and LRP6, showing residue-level flexibility and solvent-exposed interactions.
Figure 4 presents the radius of gyration (Rg) and solvent-accessible surface area (SASA) analyses to evaluate protein compactness and surface exposure. In the β-catenin US complex, Rg values for α-mangostin fluctuated between 3.30 and 3.50 nm, slightly higher than those observed with XAV939. SASA plots indicated that α-mangostin increased solvent exposure, reaching approximately 240 nm2. In the AS complex, both ligands exhibited higher Rg values, up to 4.35 nm, while SASA values remained comparable, suggesting moderate surface breathing and reduced folding compactness. By contrast, LRP6 demonstrated tighter Rg distributions (4.10–4.30 nm) and consistently lower SASA values compared with the β-catenin systems. α-Mangostin further reduced SASA in LRP6 relative to XAV939, reflecting deeper ligand embedding within the protein pocket. Compact protein structures are generally associated with enhanced energetic stability and resistance to denaturation or degradation (Buchberger et al., 2010). while reduced solvent exposure favors stronger hydrophobic interactions and improved binding efficiency. Collectively, the Rg and SASA results confirm that LRP6 maintains a more compact and tightly folded conformation during simulation, consistent with earlier findings on backbone and residue-level dynamics. Moreover, α-mangostin contributes to sustaining protein compactness in LRP6, reinforcing its potential as a stable binder.
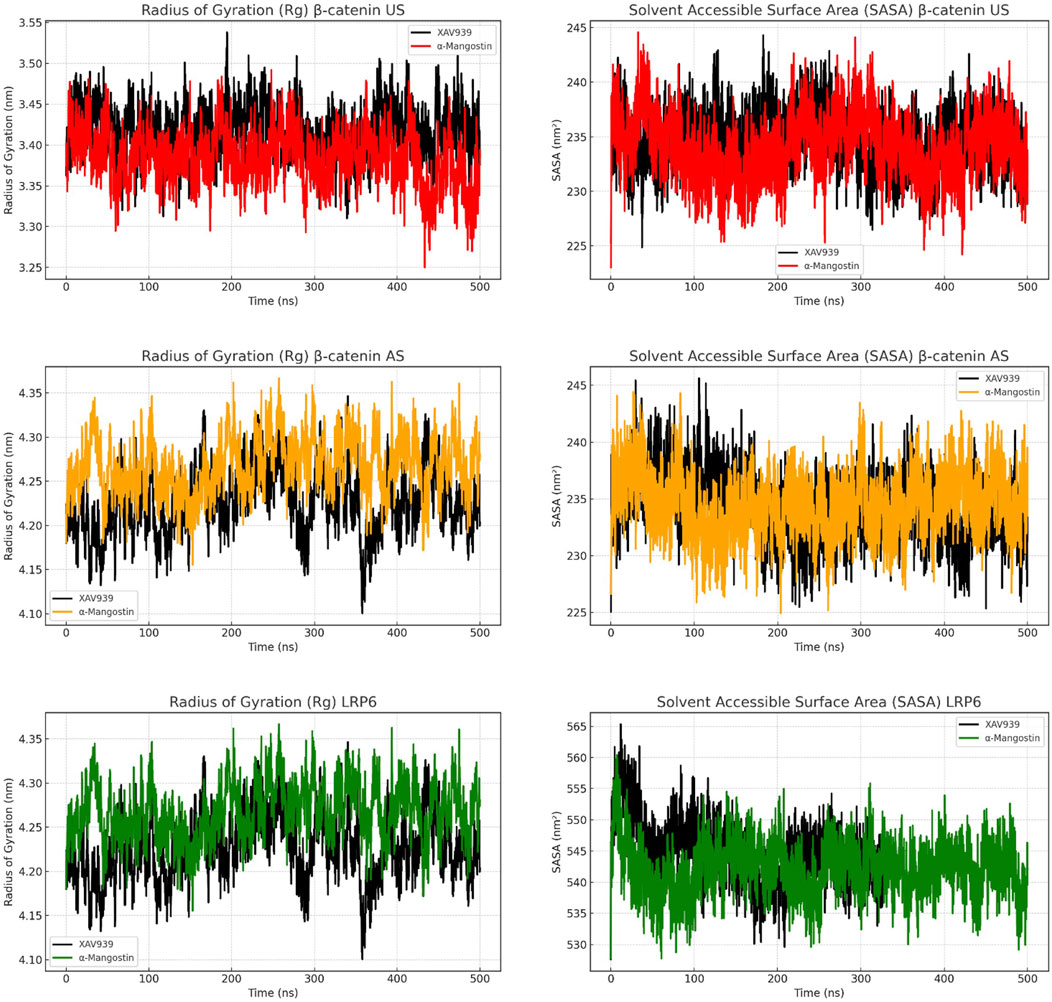
Figure 4. Figure 6. Radius of gyration (Rg) and solvent-accessible surface area (SASA) analyses of β-catenin (union site, US; allosteric site, AS) and LRP6 following molecular dynamics simulations, evaluating structural compactness and solvent exposure over time.
Hydration behavior around the ligand-binding regions was evaluated using radial distribution function (RDF) analysis, as shown in Figure 5. RDF describes the spatial arrangement of solvent molecules in the vicinity of ligands. In the β-catenin US complex, both ligands exhibited broad and less intense RDF peaks, suggesting a disordered solvation shell. XAV939 reached a maximum g(r) value of approximately 13, whereas α-mangostin was slightly lower at around 12. The β-catenin AS complex displayed sharper peaks, although still less structured compared with LRP6. In this system, XAV939 peaked at g(r) = 14, while α-mangostin remained lower, indicating that α-mangostin generates a less disrupted hydration environment. By contrast, LRP6 showed the most structured hydration profile, with sharp peaks centered at 0.5–0.6 nm. Notably, α-mangostin reached a g(r) value close to 25, significantly higher than XAV939. These results suggest that LRP6 forms a compact and stable hydration shell around the ligand. A well-ordered water layer enhances molecular rigidity and contributes favorable enthalpic interactions, thereby influencing ligand stability and retention within the binding pocket. Collectively, the RDF patterns support the conclusion that LRP6 provides a more dynamically favorable environment, consistent with the RMSD and RMSF analyses, which demonstrate the stability of LRP6–ligand complexes.
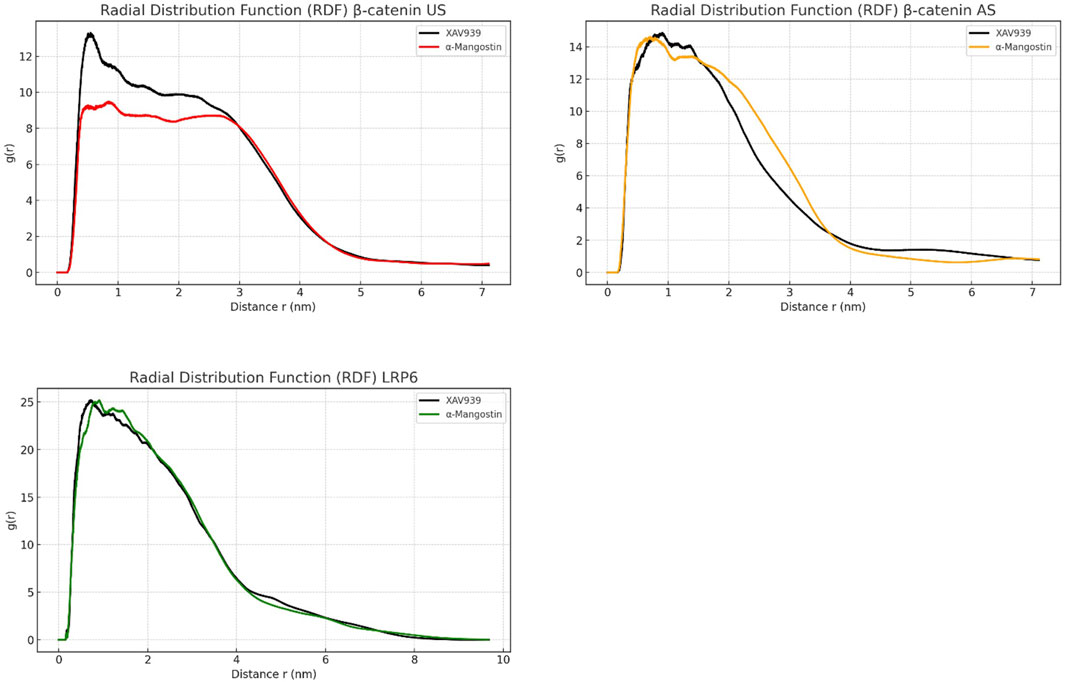
Figure 5. Radial distribution function (RDF) analysis of β-catenin (union site, US; allosteric site, AS) and LRP6 following molecular dynamics simulations, illustrating atomic-level spatial distribution around the ligand-binding regions.
Altogether, the simulation results provide a cohesive view of the structural behavior of β-catenin and LRP6 when bound to XAV939 and α-mangostin. β-Catenin, particularly in its AS binding configuration, exhibited greater flexibility and higher solvent exposure, which may limit its suitability as a stable target for inhibitors (Hankey et al., 2018). Although the US configuration showed somewhat improved stability, it still demonstrated more fluctuations compared with LRP6. Across all structural metrics evaluated (RMSD, RMSF, RDF, Rg, and SASA), LRP6 consistently outperformed β-catenin, indicating that LRP6 is a structurally rigid and solvent-protected receptor well suited for ligand interactions (Raisch et al., 2019). α-Mangostin performed favorably by stabilizing both protein and solvent dynamics, particularly within the LRP6 binding pocket, suggesting its potential role as a multi-site inhibitor of Wnt signaling. Given that β-catenin regulates downstream oncogenic pathways, especially in colorectal and breast cancers. At the same time, LRP6 functions as a co-receptor in the upstream Wnt complex; targeting both proteins could provide synergistic suppression of Wnt signaling. Previous studies have also shown that LRP6-targeted agents reduce cancer proliferation with fewer off-target effects. The consistent molecular stability observed in this study further supports α-mangostin’s candidacy for development as a Wnt pathway inhibitor (Ariyanto et al., 2023). Nevertheless, additional in vitro and in vivo studies are warranted to validate these simulation-based insights.
3.1.2 Mechanism of binding interaction as deduced from calculations of binding free energy
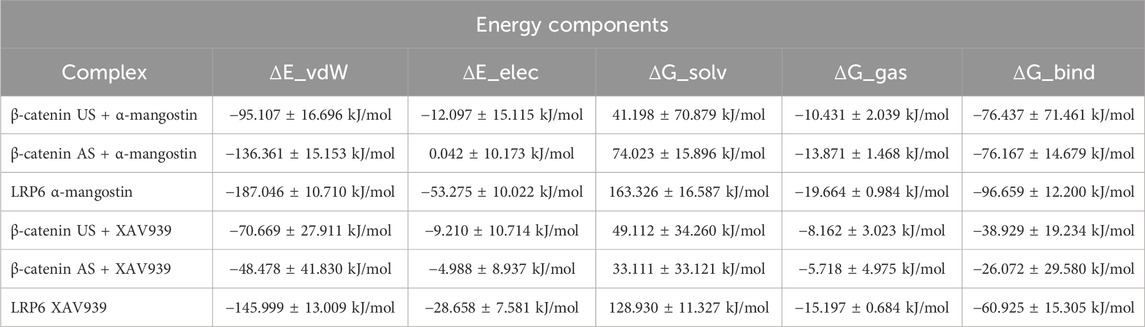
The MM/PBSA method was applied to estimate the binding free energies of α-mangostin and XAV939 with β-catenin (US and AS) and LRP6, providing insights into the stability of the complexes. As shown in Table 3, the calculated ΔG_bind values for α-mangostin were −76.437 kJ/mol at the β-catenin US site, −76.167 kJ/mol at the AS site, and −96.659 kJ/mol with LRP6. These results indicate that LRP6 forms the most energetically favorable complex with α-mangostin. The electrostatic contribution (ΔE_elec) was particularly significant in the LRP6–α-mangostin complex (−53.275 kJ/mol), whereas the AS site showed negligible electrostatics (0.042 kJ/mol). Van der Waals interactions (ΔE_vdW) followed a similar pattern, being strongest in the LRP6 complex (−187.046 kJ/mol). These findings suggest that LRP6 provides a deeply hydrophobic and structurally complementary binding pocket for α-mangostin. The gas-phase interaction energy (ΔG_gas), which combines ΔE_vdW and ΔE_elec, was lowest for the LRP6 complex (−19.664 kJ/mol), further supporting its thermodynamic advantage. Although LRP6 showed the highest desolvation penalty (ΔG_solv = 163.326 kJ/mol), this was offset by its favorable gas-phase and non-covalent interactions. In comparison, the β-catenin US and AS sites demonstrated similar ΔG_bind values, with weaker electrostatic contributions in the AS site balanced by stronger hydrophobic interactions. Collectively, the MM/PBSA results confirm that α-mangostin exhibits a stronger binding affinity for LRP6 than for β-catenin.
A similar trend was observed with the reference ligand XAV939, although its ΔG_bind values were consistently less negative than those of α-mangostin. XAV939 bound to the β-catenin US with a ΔG_bind of −38.929 kJ/mol and to the AS with −26.072 kJ/mol, indicating relatively weak complex formation. Its interaction with LRP6 was more favorable (ΔG_bind = −60.925 kJ/mol) but remained notably less negative than that of the α-mangostin–LRP6 complex. In all comparisons, α-mangostin demonstrated stronger binding, particularly with LRP6, thereby reinforcing its potential as a superior inhibitor scaffold for modulating the Wnt/β-catenin pathway. The strong electrostatic and hydrophobic contributions observed in the α-mangostin–LRP6 complex suggest robust anchoring and reduced dissociation, which may support prolonged biological activity. These findings are consistent with structural dynamics results, including the low RMSF and stable RDF profiles observed in LRP6 simulations. The pronounced ΔG_solv further underscores the role of hydrophobic stabilization within the binding pocket. The comparable ΔG_bind values at the β-catenin US and AS imply that both regions remain accessible targets, albeit thermodynamically less favorable than those at LRP6. Collectively, LRP6’s superior energetic profile highlights it as a more attractive therapeutic target. These binding energy trends provide a rationale for ligand optimization strategies aimed at strengthening van der Waals and electrostatic interactions. Experimental validation using biophysical approaches such as isothermal titration calorimetry (ITC) or surface plasmon resonance (SPR) will be essential to confirm these computational predictions.
3.2 Inhibitory effect of α-mangostin on the proliferation of MDA-MB-231 and MCF-7 breast cancer cells
We evaluated the antiproliferative effects of α-mangostin on two distinct subtypes of breast cancer: triple-negative (MDA-MB-231, ER−/PR−/HER2−) and luminal (MCF-7, ER+/PR+/HER2−). During the experiment, we co-treated the cells with lithium chloride (LiCl), an agonist of the canonical Wnt signaling pathway, which inhibits GSK-3β activity and effectively stabilizes free cytosolic β-catenin in cancer cells (Sun and Liu, 2017). In this study, LiCl was applied at concentrations of 2.5, 5, and 10 mM as a control. Previous studies have shown that low concentrations of LiCl (around 4 mM) can promote mesenchymal stem cell proliferation, whereas higher concentrations (20–40 mM) exert inhibitory effects on cell proliferation (De Boer et al., 2004).
In the proliferation assay with MDA-MB-231 cells, α-mangostin was tested at concentrations ranging from 60 to 480 µM in combination with LiCl to activate Wnt signaling. After 24 h of treatment, α-mangostin markedly suppressed cell viability, reducing survival to 20% even at the lowest concentration (60 µM), thereby demonstrating a strong growth-inhibitory effect. In contrast, treatment with LiCl alone at varying concentrations did not significantly affect cell viability, which remained above 80%. Notably, co-treatment with α-mangostin and LiCl further reduced cell viability (Figure 6A), indicating that α-mangostin effectively suppressed the growth of MDA-MB-231 cells irrespective of Wnt activation status. A similar effect was observed in MCF-7 cells, where α-mangostin displayed even greater potency, markedly reducing viability at the lowest tested concentration (60 µM). The cytotoxic effect was further enhanced when cells were co-treated with LiCl (Figure 6B). However, because MCF-7 cells exhibited extremely low viability under these treatment conditions, further molecular analyses could not be performed on this cell line. Instead, subsequent investigations focused on the TNBC MDA-MB-231 cell line, in which the impact of α-mangostin on Wnt target gene transcription could be more reliably evaluated. Furthermore, since aberrant Wnt signaling is more prominently associated with TNBC (Pohl et al., 2017), this focus on MDA-MB-231 cells was considered well justified.
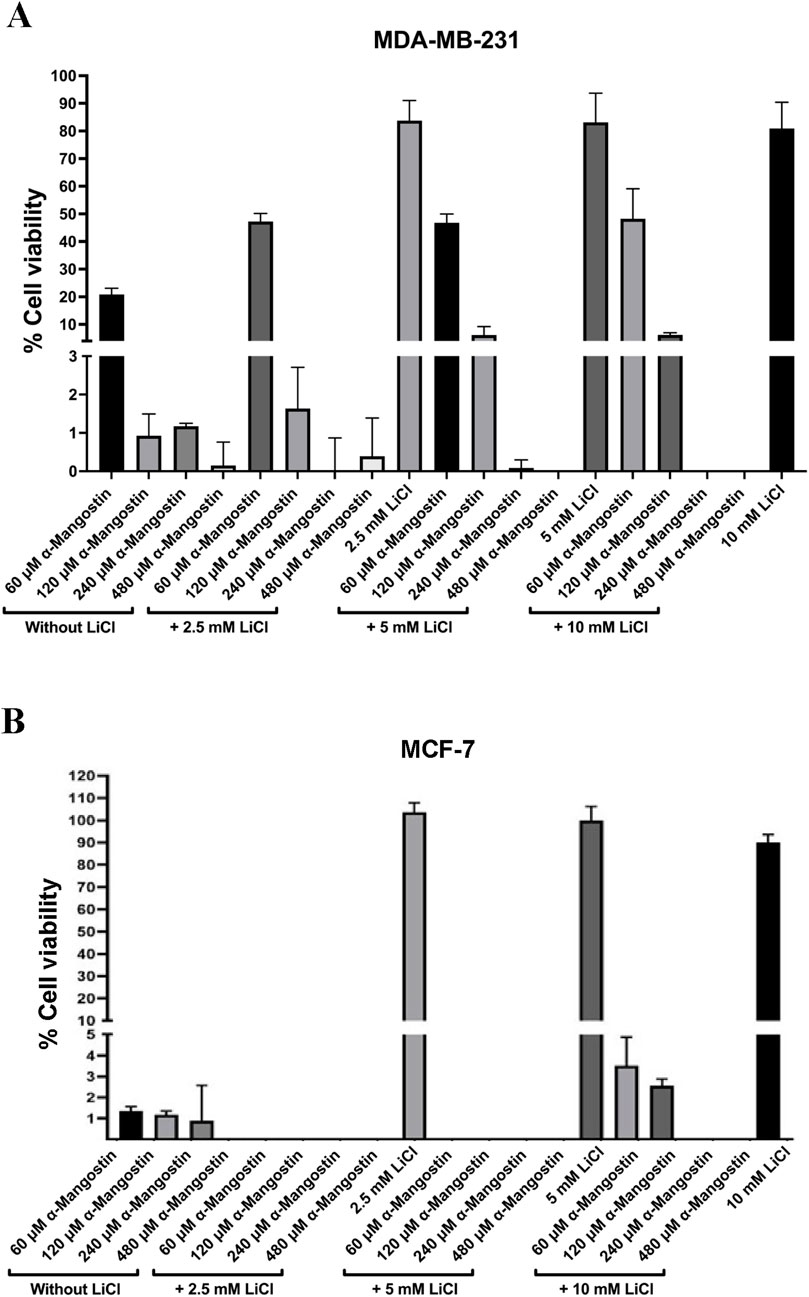
Figure 6. The antiproliferative effect of α-mangostin, both alone and in combination with lithium chloride (LiCl), on (A) MDA-MB-231 and (B) MCF-7 breast cancer cells. Data are presented as the mean of three independent experiments ±SD.
3.3 Antiproliferative effect of α-mangostin correlates with CCND1 and MYC transcription levels in breast cancer cells
Based on in silico affinity test results, α-mangostin exhibited a strong binding affinity for β-catenin, which may contribute to its degradation. Consistently, cell proliferation inhibition assays revealed that α-mangostin significantly reduced the viability of MDA-MB-231 cells. To further clarify the underlying mechanism, we performed subsequent analyses on MDA-MB-231 cells and measured the expression of proliferation-related genes regulated by Wnt signaling, specifically CCND1 and MYC (Sanjari et al., 2020). Wnt signaling was activated using LiCl, which inhibits GSK-3β-mediated phosphorylation of β-catenin (Park et al., 2020). This inhibition prevents β-catenin degradation, allowing it to accumulate in the nucleus and activate transcription factors that promote cell proliferation. Our results revealed that 10 µM α-mangostin significantly suppressed CCND1 transcription in both the absence and presence of LiCl, with a 5.2-fold reduction compared to untreated cells (Figure 7A). For comparison, we also evaluated CCND1 expression following cisplatin treatment, as alterations in this gene are linked to chemoresistance; however, cisplatin did not produce a significant effect on CCND1 transcription. Similarly, α-mangostin (10 µM) reduced MYC mRNA levels by 3.3-fold relative to the untreated group, independent of LiCl treatment (Figure 7B). Collectively, these findings suggest that α-mangostin exerts antiproliferative effects in MDA-MB-231 cells by modulating the transcription of key Wnt-regulated genes involved in proliferation.
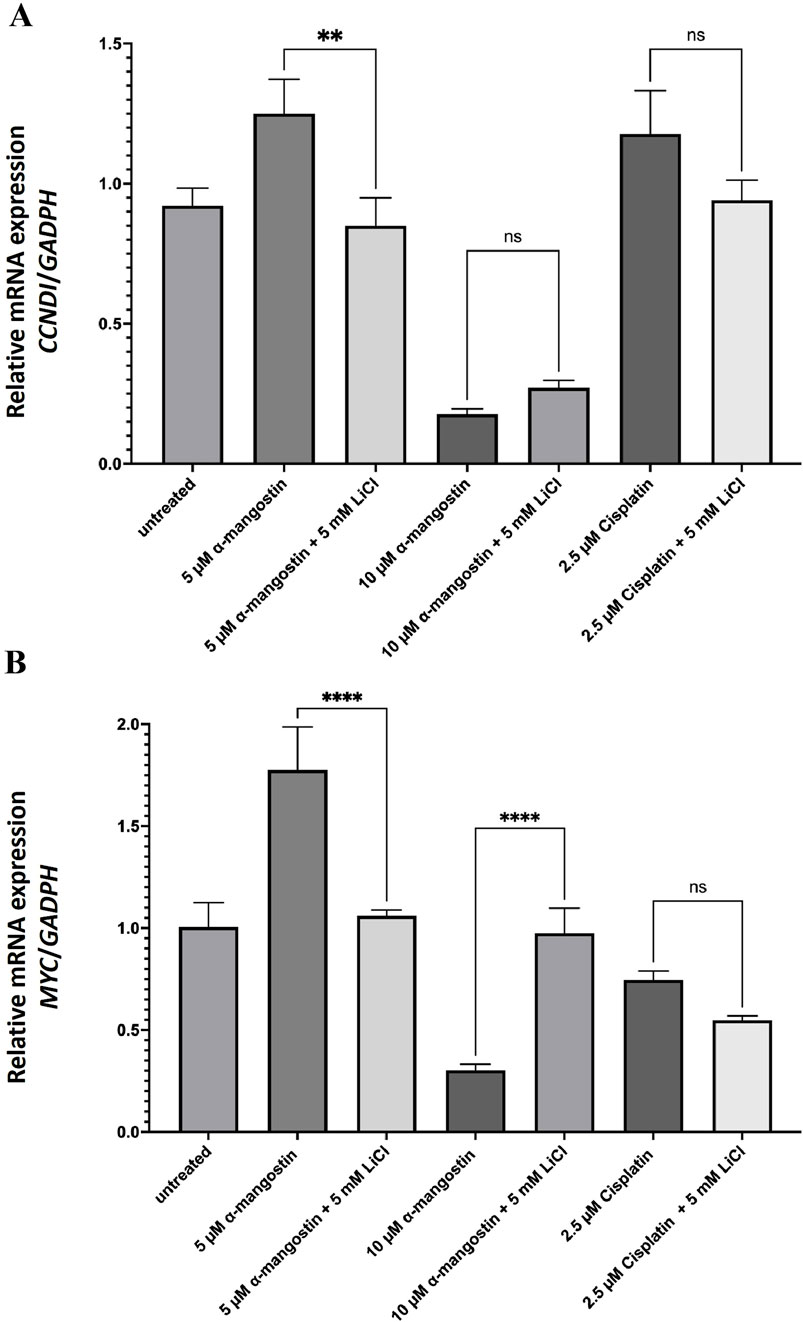
Figure 7. The mRNA expression levels of (A) CCND1 and (B) MYC in LiCl-induced MDA-MB-231 cells following treatment with α-mangostin. Data represent the mean ± standard deviation (SD) of three independent experiments. Statistical significance: **p < 0.01; ***p < 0.001; ns, not significant.
4 Discussions
The Wnt/β-catenin signaling pathway plays a central role in the pathogenesis of multiple cancers, including TNBC, where its aberrant activation promotes tumor progression, metastasis, and resistance to chemotherapy (Pohl et al., 2017; Yang et al., 2022; Yang Z. et al., 2021). Under normal conditions, β-catenin is regulated by glycogen synthase kinase-3β (GSK-3β), which phosphorylates β-catenin and targets it for ubiquitination and subsequent proteasomal degradation (Shang et al., 2017). In TNBC, however, dysregulation of this regulatory mechanism results in β-catenin stabilization and nuclear translocation, where it activates oncogenic targets such as CCND1 (Cyclin D1) and MYC (c-Myc), thereby driving uncontrolled cell proliferation (Dey et al., 2013; Kafri et al., 2016; Xu et al., 2016). Although β-catenin represents an attractive therapeutic target, no clinically approved drugs are currently available that directly and selectively inhibit this pathway (Morris et al., 2022). This limitation highlights the need for alternative therapeutic strategies, including the exploration of natural compounds that have the potential to modulate Wnt/β-catenin signaling.
Among natural compounds, α-mangostin has been reported to exert broad-spectrum anticancer effects; however, its direct impact on the Wnt/β-catenin pathway in TNBC remains insufficiently characterized. The present study provides new evidence showing that α-mangostin significantly suppresses the proliferation of MDA-MB-231 cells, even in the presence of Wnt pathway activation by LiCl. This suggests that α-mangostin exerts cytotoxic effects independently of β-catenin stabilization, distinguishing it from conventional Wnt-targeting agents. Our results are consistent with those of Yoo et al. (2011), in colon cancer cells, α-mangostin disrupted Wnt/β-catenin signaling by reducing β-catenin expression at both the mRNA and protein levels. Notably, this reduction occurred without initiating the canonical degradation process of β-catenin and was independent of β-catenin mutation status. Furthermore, LiCl treatment, which activates Wnt signaling, did not diminish the inhibitory effects of α-mangostin, suggesting that its mechanism of action bypasses the conventional β-catenin degradation pathway. Similarly, in our study, α-mangostin modulated the transcription of downstream Wnt target genes without directly affecting β-catenin degradation, further supporting the hypothesis of an alternative mechanism of pathway suppression.
Additionally, α-mangostin demonstrated greater cytotoxic sensitivity in MCF-7 cells, reinforcing its strong antiproliferative effects in luminal breast cancer. Previous studies have attributed its activity in MCF-7 cells to the induction of mitochondrial-mediated apoptosis through Bax oligomerization, cytochrome c release, and caspase activation (Simon et al., 2022). Consistently, Nalla et al. (2023) reported that α-mangostin suppressed Ki-67 expression, inhibited cell migration by modulating EMT markers such as MMP-2 and PKM-2, and downregulated STAT3 activation (Nalla et al., 2023). Furthermore, both α-mangostin and γ-mangostin were shown to inhibit the migration of MDA-MB-231 cells via transcriptional suppression of CXCR4, underscoring their multi-targeted capacity in attenuating breast cancer progression (Sarmoko et al., 2023).
Molecularly, our results confirm that α-mangostin significantly downregulated CCND1 and MYC expression in MDA-MB-231 cells, irrespective of LiCl-mediated Wnt activation. This observation is consistent with previous reports indicating that α-mangostin inhibits TCF/LEF transcriptional activity by promoting β-catenin degradation, thereby reducing the expression of Wnt target genes (Zhu et al., 2021), as illustrated in Figure 8. Notably, LiCl treatment did not reverse the β-catenin degradation induced by α-mangostin, suggesting that its mechanism may bypass the classical Wnt/β-catenin signaling cascade (Yoo et al., 2011). Furthermore, cisplatin, a widely used chemotherapeutic agent, had no significant effect on CCND1 expression in our study, supporting the notion that prolonged cisplatin exposure contributes to Wnt-mediated chemoresistance in TNBC (Yin et al., 2013).
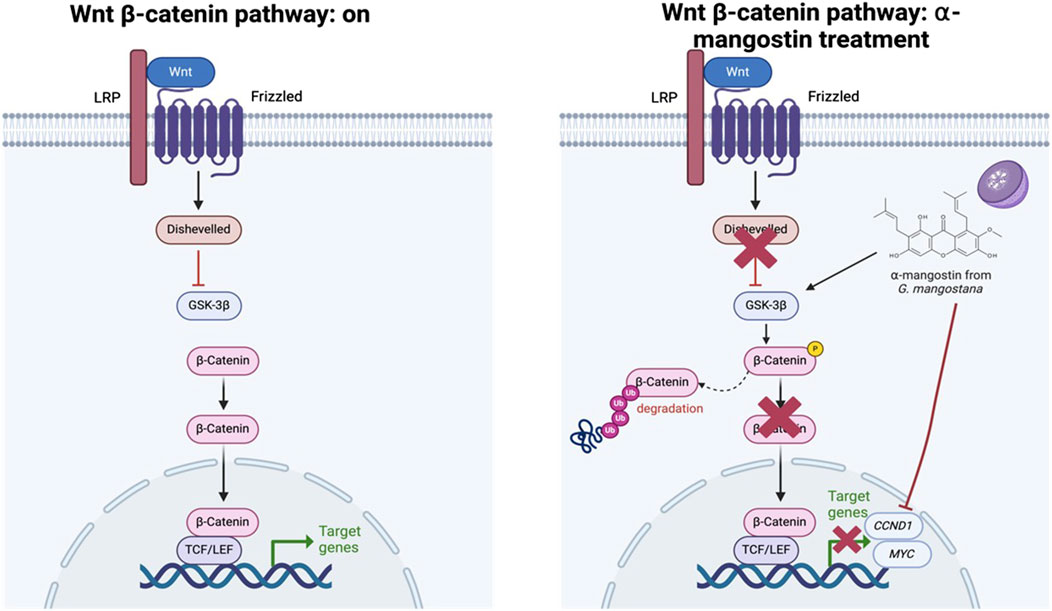
Figure 8. Proposed mechanism of action of α-mangostin in modulating the Wnt/β-catenin signaling pathway.
Beyond its role in cell cycle regulation, α-mangostin has also been shown to modulate intracellular reactive oxygen species (ROS) levels, triggering endoplasmic reticulum (ER) stress in a Wnt-dependent manner. In osteosarcoma cells, α-mangostin inhibited GSK-3β activity, leading to ROS-induced ER stress and blocking β-catenin nuclear translocation (Yang S. et al., 2021). Similarly, an increase in ROS levels was observed in MDA-MB-231 cells treated with α-mangostin (Sarmoko et al., 2023), despite the well-documented antioxidant properties of this compound. This apparent dual role, acting as both pro-oxidant and antioxidant, is reminiscent of polyphenols such as curcumin, which exhibit context-dependent oxidative modulation in cancer cells (Wolnicka-Glubisz and Wisniewska-Becker, 2023). Considering these dual effects, it is important to recognize their implications when developing rational strategies for natural compound-based cancer therapeutics. The ability of α-mangostin to selectively induce oxidative stress in malignant cells while sparing normal cells underscores its therapeutic potential. Harnessing this biphasic behavior could provide a foundation for more effective and targeted anticancer strategies, potentially minimizing drug resistance and improving treatment outcomes.
Our molecular docking and simulation analyses indicate that α-mangostin modulates Wnt/β-catenin signaling by targeting both β-catenin and LRP6. Since β-catenin is central to oncogenic transcription, particularly when exacerbated by APC mutations (Hankey et al., 2018), and LRP6 serves as an upstream Wnt co-receptor, the stronger interaction of α-mangostin with LRP6 suggests its potential to block Wnt signaling at early stages of pathway activation. This dual-inhibition strategy, suppressing signaling at multiple levels (Raisch et al., 2019), could mitigate drug resistance often associated with single-target therapies, thereby improving overall treatment efficacy (Raisch et al., 2019; Ariyanto et al., 2023).
The absence of clinically approved selective Wnt/β-catenin inhibitors, despite the pathway’s involvement through components such as β-catenin, GSK-3β, Axin, APC, and LRP6, underscores the therapeutic relevance of natural compounds like α-mangostin. Our earlier in silico findings suggested that α-mangostin may inhibit GSK-3β by modulating the Wnt/β-catenin pathway, although this had not been validated in vitro. To address this, the present study performed comparative in silico analyses, docking α-mangostin and the reference Wnt inhibitor XAV939 to LRP6, followed by molecular dynamics simulations and MM/PBSA binding energy calculations. Consistently, α-mangostin demonstrated a more favorable interaction profile with LRP6, as reflected by more negative binding free energy and enhanced structural stability. These results strengthen the hypothesis that α-mangostin targets LRP6 to modulate Wnt signaling. While experimental validation using LRP6-specific inhibitors or knockdown approaches was beyond the scope of this study, it remains a critical direction for future work, which could be pursued using techniques such as Western blotting or β-catenin luciferase reporter assays (Lai et al., 2009). Taken together, our findings support α-mangostin as a promising therapeutic candidate for aggressive and therapy-resistant breast cancer subtypes. Unlike conventional Wnt-targeting therapies, α-mangostin exerts its effects independently of β-catenin stabilization, making it a potentially more versatile treatment option.
A similar phenomenon was reported with EGCG in breast cancer cells, where β-catenin expression remained unchanged while MYC transcription was suppressed (Kim et al., 2006). Likewise, the polyphenol curcumin inhibited Wnt signaling by preventing β-catenin nuclear translocation, thereby downregulating downstream targets such as CCND1 and MYC (Prasad et al., 2009). Given its multi-target effects, including regulation of the cell cycle, induction of apoptosis, and inhibition of metastasis, further in vitro and in vivo investigations are warranted to establish α-mangostin as a potential therapeutic agent.
Although drug development targeting the Wnt signaling pathway in cancer has made promising progress, no specific Wnt-targeted therapy has yet been approved for clinical use. This remains a significant challenge in advancing therapeutic strategies targeting the canonical Wnt/β-catenin pathway. Small-molecule inhibitors, both synthetic and naturally derived, often display favorable bioavailability and cellular permeability; however, concerns about off-target effects persist (Yu et al., 2021). These concerns arise primarily from the essential role of the Wnt/β-catenin pathway in normal physiological processes, making the selective targeting of this pathway inherently difficult. In addition, the scarcity of well-defined druggable structures within Wnt/β-catenin signaling components further complicates the development of selective and effective inhibitors, thereby limiting their clinical translation (Cui et al., 2018). Addressing these obstacles will require a comprehensive evaluation of existing compounds and the design of novel strategies, such as combination approaches or synergistic therapeutic regimens, which may ultimately enable the successful development of effective Wnt-targeted cancer treatments.
Future investigations should prioritize evaluating the synergistic potential of α-mangostin in combination with established chemotherapeutic agents to improve treatment outcomes and mitigate resistance. Furthermore, a detailed exploration of the molecular interactions between α-mangostin, GSK-3β, and β-catenin will be essential to clarify its precise role in modulating Wnt signaling. Preclinical validation using TNBC animal models will also be critical to confirm its therapeutic efficacy under physiological conditions. By uncovering a novel mechanism of action, this study provides a foundation for the potential clinical application of α-mangostin as an anti-TNBC agent. With continued research, α-mangostin may emerge as a promising addition to the current repertoire of breast cancer therapeutics, offering new opportunities for patients with limited treatment options.
5 Conclusion
Our study demonstrated that α-mangostin suppresses the transcription of CCND1 (cyclin D1) and MYC (c-Myc) in Wnt-activated MDA-MB-231 TNBC cells. Computational analyses further revealed that α-mangostin binds at sites overlapping with LRP6 inhibitors, suggesting its potential to disrupt the LRP6–β-catenin complex. Together, these findings highlight a novel mechanism by which α-mangostin may modulate the Wnt/β-catenin signaling pathway. Future studies should investigate the synergistic potential of α-mangostin in combination with established chemotherapies to enhance therapeutic outcomes and overcome resistance. In addition, a more profound exploration of its molecular interactions with GSK-3β and β-catenin will be essential to define further its role in regulating this pathway.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
RA: Data curation, Formal Analysis, Investigation, Methodology, Supervision, Validation, Writing – review and editing. CD: Investigation, Methodology, Writing – original draft, Writing – review and editing. AF: Data curation, Methodology, Supervision, Writing – review and editing. TF: Formal Analysis, Investigation, Methodology, Writing – review and editing. MM: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Ministry of Education and Culture of the Republic of Indonesia through Universitas Padjadjaran through Academic Leadership Grants No. No. 1503/UN6.3.1/PT.00/2024.
Acknowledgments
The authors gratefully acknowledge Rector Universitas Padjadjaran and Universitas Udayana, Indonesia, for the use of their facilities for this study. We also thank the 6th ISPST and 15th Annual ISCC 2024 Committee of Universitas Padjadjaran, who have facilitated the preparation of this manuscript, including helping with English proofreading.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25. doi:10.1016/j.softx.2015.06.001
Ahn, V. E., Chu, M. L., Choi, H. J., Tran, D., Abo, A., and Weis, W. I. (2011). Structural basis of Wnt signaling inhibition by dickkopf binding to LRP5/6. Dev. Cell 21, 862–873. doi:10.1016/j.devcel.2011.09.003
Andinata, B., Bachtiar, A., Oktamianti, P., Partahi, J. R., and Dini, M. S. A. (2023). A comparison of cancer incidences between dharmais cancer hospital and GLOBOCAN 2020: a descriptive study of top 10 cancer incidences. 17:4 119–121. doi:10.33371/ijoc.v17i2.982
Ariyanto, E. F., Wirajati, F., Rahman, P. H., Berbudi, A., and Rohmawaty, E. (2023). Mechanism of action of Indonesian medicinal plants in inhibiting 3T3-L1 adipocyte differentiation: a review. J. Appl. Pharm. Sci. doi:10.7324/japs.2023.6711
Ashrafizadeh, M., Ahmadi, Z., Farkhondeh, T., and Samarghandian, S. (2020). Resveratrol targeting the wnt signaling pathway: a focus on therapeutic activities. J. Cell. Physiology 235, 4135–4145. doi:10.1002/jcp.29327
Aulifa, D. L., Al Shofwan, A. A., Megantara, S., Fakih, T. M., and Budiman, A. (2024). Elucidation of molecular interactions between drug-polymer in amorphous solid dispersion by a computational approach using molecular dynamics simulations. Adv. Appl. Bioinform Chem. 17, 1–19. doi:10.2147/AABC.S441628
Buchberger, A., Bukau, B., and Sommer, T. (2010). Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell 40, 238–252. doi:10.1016/j.molcel.2010.10.001
Chaudhuri, A., Kumar, D. N., Shaik, R. A., Eid, B. G., Abdel-Naim, A. B., Md, S., et al. (2022). Lipid-based nanoparticles as a pivotal delivery approach in triple negative breast Cancer (TNBC) therapy. Int. J. Mol. Sci. 23, 10068. doi:10.3390/ijms231710068
Cui, C., Zhou, X., Zhang, W., Qu, Y., and Ke, X. (2018). Is β-Catenin a druggable target for cancer therapy? Trends Biochem. Sci. 43, 623–634. doi:10.1016/j.tibs.2018.06.003
De Boer, J., Wang, H. J., and Van Blitterswijk, C. (2004). Effects of Wnt signaling on proliferation and differentiation of human mesenchymal stem cells. Tissue Eng. Mar-Apr 10, 393–401. doi:10.1089/107632704323061753
Dey, N., Young, B., Abramovitz, M., Bouzyk, M., Barwick, B., De, P., et al. (2013). Differential activation of Wnt-β-Catenin pathway in triple negative breast cancer increases MMP7 in a PTEN dependent manner. PLOS ONE 8, e77425. doi:10.1371/journal.pone.0077425
Durrant, J. D., and McCammon, J. A. (2011). Molecular dynamics simulations and drug discovery. BMC Biol. Oct. 28 (9), 71. doi:10.1186/1741-7007-9-71
Forli, S., Huey, R., Pique, M. E., Sanner, M. F., Goodsell, D. S., and Olson, A. J. (2016). Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 11, 905–919. doi:10.1038/nprot.2016.051
Hankey, W., Frankel, W. L., and Groden, J. (2018). Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: implications for therapeutic targeting. Cancer Metastasis Rev. Mar. 37, 159–172. doi:10.1007/s10555-017-9725-6
Hess, B., Bekker, H., Berendsen, H. J. C., and Fraaije, JGEM (1997). LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472. doi:10.1002/(sici)1096-987x(199709)18:12<1463::aid-jcc4>3.3.co;2-l
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: visual molecular dynamics. J. Mol. Graph 14 (33-38), 33–28. doi:10.1016/0263-7855(96)00018-5
Jeong, W., and Jho, E. H. (2021). Regulation of the low-density lipoprotein receptor-related protein LRP6 and its association with disease: Wnt/β-catenin signaling and beyond. Front. Cell Dev. Biol. 9, 714330. doi:10.3389/fcell.2021.714330
Kafri, P., Hasenson, S. E., Kanter, I., Sheinberger, J., Kinor, N., Yunger, S., et al. (2016). Quantifying β-catenin subcellular dynamics and cyclin D1 mRNA transcription during Wnt signaling in single living cells. eLife 5, e16748. doi:10.7554/eLife.16748
Kemmish, H., Fasnacht, M., and Yan, L. (2017). Fully automated antibody structure prediction using BIOVIA tools: validation study. PLoS One 12 (5), e0177923. doi:10.1371/journal.pone.0177923
Kim, J., Zhang, X., Rieger-Christ, K. M., Summerhayes, I. C., Wazer, D. E., Paulson, K. E., et al. (2006). Suppression of wnt signaling by the green tea Compound (–)-Epigallocatechin 3-Gallate (EGCG) in invasive breast cancer cells: REQUIREMENT OF THE TRANSCRIPTIONAL REPRESSOR HBP1. J. Biol. Chem. 281, 10865–10875. doi:10.1074/jbc.M513378200
Kimura, K., Kanto, T., Shimoda, S., Harada, K., Kimura, M., Nishikawa, K., et al. (2022). Safety, tolerability, and anti-fibrotic efficacy of the CBP/β-catenin inhibitor PRI-724 in patients with hepatitis C and B virus-induced liver cirrhosis: an investigator-initiated, open-label, non-randomised, multicentre, phase 1/2a study. EBioMedicine 80, 104069. doi:10.1016/j.ebiom.2022.104069
Lai, S.-L., Chien, A. J., and Moon, R. T. (2009). Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. 19, 532–545. doi:10.1038/cr.2009.41
Lee, J. (2023). Current treatment landscape for early triple-negative breast cancer (TNBC). J. Clin. Med. 15, 12. doi:10.3390/jcm12041524
Lin, J., Song, T., Li, C., and Mao, W. (2020). GSK-3β in DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy of cancer. Biochim. Biophys. Acta Mol. Cell Res. May 1867, 118659. doi:10.1016/j.bbamcr.2020.118659
Liu, J., Pan, S., Hsieh, M. H., Ng, N., Sun, F., Wang, T., et al. (2013). Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. 110, 20224–20229. doi:10.1073/pnas.1314239110
Liu, J., Xiao, Q., Xiao, J., Niu, C., Li, Y., Zhang, X., et al. (2022). Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 7, 3. doi:10.1038/s41392-021-00762-6
Medina-Barandica, J., Contreras-Puentes, N., Tarón-Dunoyer, A., Durán-Lengua, M., and Alviz-Amador, A. (2023). In-silico study for the identification of potential destabilizers between the spike protein of SARS-CoV-2 and human ACE-2. Inf. Med. Unlocked 40, 101278. doi:10.1016/j.imu.2023.101278
Mehrgou, A., and Akouchekian, M. (2016). The importance of BRCA1 and BRCA2 genes mutations in breast cancer development. Med. J. Islam Repub. Iran. 30, 369. Available online at: https://pubmed.ncbi.nlm.nih.gov/27493913/.
Morris, A., Pagare, P. P., Li, J., and Zhang, Y. (2022). Drug discovery efforts toward inhibitors of canonical Wnt/β-catenin signaling pathway in the treatment of cancer: a composition-of-matter review (2010–2020). Drug Discov. Today 27, 1115–1127. doi:10.1016/j.drudis.2021.11.014
Muchtaridi, M., Yusuf, M., Syahidah, H. N., Subarnas, A., Zamri, A., Bryant, S., et al. (2019). Cytotoxicity of chalcone of eugenia aquea burm F. Leaves against T47D breast cancer cell lines and its prediction as an estrogen receptor antagonist based on pharmacophore-molecular dynamics simulation. Adv. Appl. Bioinforma. Chem. 12, 33–43. doi:10.2147/AABC.S217205
Nalla, S., and Ganta, S. (2023). Defensive impact of kaempferide against neurodegenerative studies: in vitro and in vivo investigations. Chem. Afr. 6, 2483–2493. doi:10.1007/s42250-023-00673-9
Nalla, L. V., Dharavath, A., Behera, S. K., and Khairnar, A. (2023). Alpha mangostin inhibits proliferation, migration, and invasion of human breast cancer cells via STAT3 inhibition. Adv. Cancer Biol. - Metastasis 7, 100089. doi:10.1016/j.adcanc.2023.100089
Nurhidayah, W., Widyasari, E. M., Daruwati, I., Mahendra, I., Subroto, T., Khairul Ikram, N. K., et al. (2023). Radiosynthesis, stability, lipophilicity, and cellular uptake evaluations of [(131)I]Iodine-α-Mangostin for breast cancer diagnosis and therapy. Int. J. Mol. Sci. May 12, 24. doi:10.3390/ijms24108678
Pai, S. G., Carneiro, B. A., Mota, J. M., Costa, R., Leite, C. A., Barroso-Sousa, R., et al. (2017). Wnt/beta-catenin pathway: modulating anticancer immune response. J. Hematol. and Oncol. 10, 101. doi:10.1186/s13045-017-0471-6
Park, H.-B., Kim, J.-W., and Baek, K.-H. (2020). Regulation of wnt signaling through ubiquitination and deubiquitination in cancers. Int. J. Mol. Sci. 21, 3904. doi:10.3390/ijms21113904
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi:10.1002/jcc.20084
Pohl, S.-G., Brook, N., Agostino, M., Arfuso, F., Kumar, A. P., and Dharmarajan, A. (2017). Wnt signaling in triple-negative breast cancer. Oncogenesis 6, e310. doi:10.1038/oncsis.2017.14
Prasad, C. P., Rath, G., Mathur, S., Bhatnagar, D., and Ralhan, R. (2009). Potent growth suppressive activity of curcumin in human breast cancer cells: modulation of Wnt/beta-catenin signaling. Chemico-Biological Interact. 181, 263–271. doi:10.1016/j.cbi.2009.06.012
Raisch, J., Côté-Biron, A., and Rivard, N. (2019). A role for the WNT Co-Receptor LRP6 in pathogenesis and therapy of epithelial cancers. Cancers (Basel). 11, 1162. doi:10.3390/cancers11081162
Sampietro, J., Dahlberg, C. L., Cho, U. S., Hinds, T. R., Kimelman, D., and Xu, W. (2006). Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Mol. Cell 24, 293–300. doi:10.1016/j.molcel.2006.09.001
Sanjari, M., Kordestani, Z., Safavi, M., Mashrouteh, M., FekriSoofiAbadi, M., and Ghaseminejad Tafreshi, A. (2020). Enhanced expression of cyclin D1 and C-myc, a prognostic factor and possible mechanism for recurrence of papillary thyroid carcinoma. Sci. Rep. 10, 5100. doi:10.1038/s41598-020-61985-1
Sarmoko, S., Novitasari, D., Toriyama, M., Fareza, M. S., Choironi, N. A., Itoh, H., et al. (2023). Different modes of mechanism of gamma-mangostin and alpha-mangostin to inhibit cell migration of triple-negative breast cancer cells concerning CXCR4 downregulation and ROS generation. Iran. J. Pharm. Res. 22, e138856. doi:10.5812/ijpr-138856
Shang, S., Hua, F., and Hu, Z.-W. (2017). The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget 8 (20), 33972–33989. doi:10.18632/oncotarget.15687
Shiga, M., Morishita, T., and Sorai, M. (2023). Interfacial tension of carbon dioxide - water under conditions of CO2 geological storage and enhanced geothermal systems: a molecular dynamics study on the effect of temperature. Fuel 337, 127219. doi:10.1016/j.fuel.2022.127219
Simon, S. E., Lim, H. S., Jayakumar, F. A., Tan, E. W., and Tan, K. O. (2022). Alpha-mangostin activates MOAP-1 tumor suppressor and mitochondrial signaling in MCF-7 human breast cancer cells. Evidence-Based Complementary Altern. Med. 2022, 7548191. doi:10.1155/2022/7548191
Sun, X., and Liu, Y. (2017). Activation of the Wnt/β-catenin signaling pathway May contribute to cervical cancer pathogenesis via upregulation of Twist. Oncol. Lett. 14, 4841–4844. doi:10.3892/ol.2017.6754
Sun, W., Qiu, G., Zou, Y., Cai, Z., Wang, P., Lin, X., et al. (2015). Knockdown of TMEM45A inhibits the proliferation, migration and invasion of glioma cells. Int. J. Clin. Exp. Pathol. 8, 12657–12667. doi:10.1111/1440-1681.13220
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Treeck, O., Schuler-Toprak, S., Skrzypczak, M., Weber, F., and Ortmann, O. (2020). Knockdown of PTEN decreases expression of estrogen receptor β and tamoxifen sensitivity of human breast cancer cells. Steroids 156, 108575. doi:10.1016/j.steroids.2019.10857
Vallée, A., Lecarpentier, Y., and Vallée, J.-N. (2019). Curcumin: a therapeutic strategy in cancers by inhibiting the canonical WNT/β-catenin pathway. J. Exp. and Clin. Cancer Res. 38, 323. doi:10.1186/s13046-019-1320-y
Vélez-Vargas, L. C., Santa-González, G. A., Uribe, D., Henao-Castañeda, I. C., and Pedroza-Díaz, J. (2023). In vitro and in silico study on the impact of chlorogenic acid in colorectal cancer cells: proliferation, apoptosis, and interaction with β-Catenin and LRP6. Pharmaceuticals 16, 276. doi:10.3390/ph16020276
Wolnicka-Glubisz, A., and Wisniewska-Becker, A. (2023). Dual action of curcumin as an Anti- and pro-oxidant from a biophysical perspective. Antioxidants (Basel), 12. doi:10.3390/antiox12091725
Wu, X., Que, H., Li, Q., and Wei, X. (2025). Wnt/β-catenin mediated signaling pathways in cancer: recent advances, and applications in cancer therapy. Mol. Cancer 10 (24), 171. doi:10.1186/s12943-025-02363-1
Wurl, A., and Ferreira, M. T. (2023). Atomistic MD simulations of n-alkanes in a phospholipid bilayer: CHARMM36 versus slipids. Macromol. Theory Simulations 32, 2200078. doi:10.1002/mats.202200078
Xiang, J., Zhou, L., He, Y., and Wu, S. (2021). LDH-A inhibitors as remedies to enhance the anticancer effects of PARP inhibitors in ovarian cancer cells. Aging (Albany NY) 13, 25920–25930. doi:10.18632/aging.203780
Xu, J., Chen, Y., Huo, D., Khramtsov, A., Khramtsova, G., Zhang, C., et al. (2016). β-catenin regulates c-Myc and CDKN1A expression in breast cancer cells. Mol. Carcinog. 55, 431–439. doi:10.1002/mc.22292
Yang, C., Du, W., and Yang, D. (2016). Inhibition of green tea polyphenol EGCG((−)-epigallocatechin-3-gallate) on the proliferation of gastric cancer cells by suppressing canonical wnt/β-catenin signalling pathway. Int. J. Food Sci. Nutr. 67, 818–827. doi:10.1080/09637486.2016.1198892
Yang, S., Zhou, F., Dong, Y., and Ren, F. (2021). α-Mangostin induces apoptosis in human osteosarcoma cells through ROS-mediated endoplasmic reticulum stress via the WNT pathway. Cell Transplant. 30, 09636897211035080. doi:10.1177/09636897211035080
Yang, Z., Wang, M., Ren, Y., Li, L., Cao, L., Zhang, W., et al. (2021). Inhibition of Wnt10b/β-catenin signaling alleviates pulmonary fibrogenesis induced by paraquat in vivo and in vitro. Life Sci. 286 (2021/12/01/), 120027. doi:10.1016/j.lfs.2021.120027
Yang, X., Cao, D., Ma, W., Gao, S., Wen, G., and Zhong, J. (2022). Wnt signaling in triple-negative breast cancers: its roles in molecular subtyping and cancer cell stemness and its crosstalk with non-coding RNAs. Life Sci. 300, 120565. doi:10.1016/j.lfs.2022.120565
Yin, S., Xu, L., Bonfil, R. D., Banerjee, S., Sarkar, F. H., Sethi, S., et al. (2013). Tumor-initiating cells and FZD8 play a major role in drug resistance in triple-negative breast cancer. Mol. Cancer Ther. 12, 491–498. doi:10.1158/1535-7163.MCT-12-1090
Yoo, J.-H., Kang, K., Jho, E. H., Chin, Y.-W., Kim, J., and Nho, C. W. (2011). α- and γ-Mangostin inhibit the proliferation of colon cancer cells via β-catenin gene regulation in Wnt/cGMP signalling. Food Chem. 129, 1559–1566. doi:10.1016/j.foodchem.2011.06.007
Yu, F., Yu, C., Li, F., Zuo, Y., Wang, Y., Yao, L., et al. (2021). Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct. Target Ther. 30 (6), 307. doi:10.1038/s41392-021-00701-5
Zhang, Y., and Wang, X. (2020). Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 13, 165. doi:10.1186/s13045-020-00990-3
Keywords: triple negative breast cancer, α-mangostin, Wnt/β-catenin, GSK-3β, CCND1, MYC
Citation: Amalia R, Dewi C, Fristiohady A, Fakih TM and Muchtaridi M (2025) Unraveling the influence of α-mangostin on MDA-MB-231 cell line via WNT/β-catenin signaling pathway: in silico and in vitro approaches. Front. Pharmacol. 16:1600281. doi: 10.3389/fphar.2025.1600281
Received: 26 March 2025; Accepted: 15 September 2025;
Published: 01 October 2025.
Edited by:
Germain Sotoing Taiwe, University of Buea, CameroonReviewed by:
Barathan Muttiah, University of Malaya, MalaysiaArpana Parihar, Advanced Materials and Processes Research Institute (CSIR), India
Tatiana Takahasi Komoto, University of Ribeirão Preto, Brazil
Nagesh Kishan Panchal, The University of Texas Health Science Center at San Antonio, United States
Sümeyra Çetinkaya, Field crops central research institute, Türkiye
Copyright © 2025 Amalia, Dewi, Fristiohady, Fakih and Muchtaridi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muchtaridi Muchtaridi, bXVjaHRhcmlkaUB1bnBhZC5hYy5pZA==
 Riezki Amalia
Riezki Amalia Citra Dewi2,3,4
Citra Dewi2,3,4 Taufik Muhammad Fakih
Taufik Muhammad Fakih Muchtaridi Muchtaridi
Muchtaridi Muchtaridi