- 1Department of Pharmacy Practice, College of Pharmacy, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
- 2Biochemistry Department, Faculty of Pharmacy, Tanta University, Tanta, Egypt
- 3Pharmaceutical Analysis Center, King Abdullah International Medical Research Center (KAIMRC), King Saud bin Abdulaziz University for Health Sciences (KSAU-HS), Ministry of National Guard Health Affairs (MNGHA), Riyadh, Saudi Arabia
- 4Blood and Cancer Research Department, King Abdullah International Medical Research Center (KAIMRC), King Saud bin Abdulaziz University for Health Sciences (KSAU-HS), Ministry of National Guard Health Affairs (MNGHA), Riyadh, Saudi Arabia
- 5Department of Pharmaceutical Care Services, Medical Affairs, King Abdullah Bin Abdulaziz University Hospital, Riyadh, Saudi Arabia
- 6Clinical Pharmacy and Pharmacy Practice Department, Faculty of Pharmacy, Egyptian Russian University, Cairo, Egypt
- 7Department of Clinical Pharmacy and Pharmacy Practice, Faculty of Pharmacy, Damanhour University, Damanhour, Egypt
Background: Hepatocellular carcinoma (HCC), a leading cause of cancer-related mortality, is commonly treated with doxorubicin (DOX). However, its effectiveness varies significantly among patients.
Aim: The present study aimed to identify potential genetic variants affecting the response of HCC patients to DOX.
Methods: 78 patients with HCC who received DOX via transarterial chemoembolization (TACE) technology were selected. DNA was extracted from blood for genome-wide genotyping using the Applied Biosystems™ Axiom™ Precision Medicine Diversity Research™ Array. Genetic data were analysed using Axiom™ Analysis Suite software v5.2.
Results: Six hits in five genes [AK3 (rs378117), TRPM3 (rs1329774 and rs4745058), CDH4 (rs2427043), LINC00504 (rs76228864), and GRIN2D (rs76754767)] were associated with a risk of tumour progression, whereas variants in HPGD (rs45593131) and RC3H2 (rs2792999) were suggested as protective factors. rs8038528 in the PCSK6 gene was categorized as a low-response variant associated with an unsatisfactory reduction in α-fetoprotein (AFP) levels after DOX chemotherapy (P = 6.82 × 10−5). In contrast, three SNPs (rs1998853, rs12440990, and rs4774596) located within two genes (NPAS3 and DMXL2) were identified as predictors of good response rates to the treatment, as AFP levels were reduced by ≥ 20%. Death incidents showed associations with five SNPs that reached p ≤ 5.0 × 10−8; four of these are located within the DENND1B, LOC107986086, TMEM169, and RNF152 genes.
Conclusion: These findings support the incorporation of pharmacogenomic testing into clinical practice for HCC therapy, paving the way for customized treatment methods that may improve therapeutic efficacy and patient outcomes. Future research is needed to replicate these genetic connections.
1 Introduction
According to the International Agency for Research on Cancer (IARC) and partners, hepatocellular carcinoma (HCC) is the most common subtype of primary liver cancer. HCC accounts for 80% of all primary liver cancers globally (Rumgay et al., 2022) and is the third leading cause of cancer-related mortality (Konyn et al., 2021). However, the incidence and risk factors for HCC vary across different regions and countries. The highest rates were reported in eastern Asia, northern Africa, and southeastern Asia (Rumgay et al., 2022). In the Arab world, HCC represents a major health problem, especially in Egypt, which has the highest number of cases (Abudeif, 2019). The main risk factor for HCC in Egypt is infection with hepatitis C virus (HCV), which affects approximately 15% of the population (Abudeif, 2019). In other Arab countries, such as Saudi Arabia, the United Arab Emirates (UAE), and Morocco, hepatitis B virus (HBV) is the predominant cause of HCC (El-Kassas and Elbadry, 2022). Other factors that contribute to HCC in the Arab region include obesity, diabetes, alcohol consumption, and nonalcoholic steatohepatitis (NASH) (Hashim et al., 2021).
The American Association for the Study of Liver Diseases (AASLD) developed evidence-based practice guidelines, including therapeutic and preventive aspects of care. These guidelines consider changing epidemiological patterns, with an increasing proportion of cases related to nonviral aetiologies (Singal et al., 2023). The Barcelona Clinic Liver Cancer Staging and Treatment Allocation (BCLC) categorization divides HCC into five stages: very early, early, intermediate, advanced, and terminal (Li et al., 2023). Early-stage HCC is best managed by curative treatment, which includes surgical resection, ablation, or transplantation. Patients with intermediate-stage disease, particularly unresectable HCC, often receive palliative therapy with locoregional treatment, whereas systemic treatment, including chemotherapy, tyrosine kinase inhibitors, or immunotherapy, is reserved for patients with advanced disease (Singal et al., 2023).
One method of locoregional treatment is transarterial chemoembolization (TACE), which involves injecting a chemotherapy drug (such as doxorubicin [DOX]) mixed with an embolic agent (such as ethiodized oil or microspheres) into the hepatic artery that supplies blood to the tumour. This localized administration causes an infarct and subsequent necrosis of the tumour. This treatment approach is preferred over systemic chemotherapy, as the drug can directly target cancer cells and block their blood supply while minimizing exposure to the rest of the body (Bessar et al., 2021). Additionally, in theory, TACE provides better groundwork for subsequent major surgery and likely prolongs long-term survival if it is initiated at an early stage of HCC. Over the last decade, TACE has been shown to be a promising technique for the treatment of HCC in terms of overall survival (OS) or the tumour response, with minimal adverse events (Chen et al., 2017; Han et al., 2019). However, it is still accompanied by several limitations and challenges, such as determining the optimal dose and selecting the most effective chemotherapy drug (epirubicin, DOX, cisplatin, or adriamycin).
DOX is the most common cytotoxic drug employed in TACE (Bessar et al., 2021; Tam, 2013) as a mono-drug or in combination with chemotherapy (Fan et al., 2017; Liu et al., 2015; Feng et al., 2018) at fixed or variable doses depending on the patient variables (body surface area and weight) and the size of the tumour (Bessar et al., 2021). Although it can be infused systemically, this route is avoided because of its severe side effects, especially cardiac toxicity (van der Zanden et al., 2021). To date, the exact mechanisms by which DOX mediates cancer cell dysfunction or death are not fully understood (Micallef and Baron, 2021). However, they are mostly attributed to the ability of DOX to intercalate into tumour cell DNA, inhibit topoisomerase II, disrupt mitochondrial function, and potentiate free radical generation and oxidative damage, in addition to its immunomodulatory role (Kciuk et al., 2023). Some of these mechanisms are also linked to multiorgan toxicity induced by DOX due to intercalation in the DNA of healthy cells, particularly when it is infused systemically (van der Zanden et al., 2021; Pugazhendhi et al., 2018). Furthermore, cumulative evidence from in vivo and in vitro studies examining the chemical structure and metabolism of DOX has suggested alternative mechanisms of anticancer activity (Bisht et al., 2024).
The Pharmacogenomics Knowledge Base (PharmGKB) database has built integrated complex pharmacokinetic (PK)–pharmacodynamic (PD) pathways illustrating the fate of this drug molecule in the human body (Thorn et al., 2013), which were deemed beneficial for pharmacogenomic (PGx) research attempting to define how genetic variations may explain interindividual and interethnic variability in DOX PKs and PDs and hence affect the clinical response to DOX (Thorn et al., 2013). In summary, DOX is transported into cells by the solute carrier family (SLC) of membrane proteins (e.g., SLC28A3 and SLC22A16). A significant fraction (approximately 50%) of the unchanged drug undergoes efflux from body cells via transporters of the ATP-binding cassette family (ABCB1, ABCC1, ABCG2, and ABCC2) and RA1-binding proteins (RABLP1). The remaining proportion of drugs in cells undergoes metabolism, mainly to doxorubicinol and DOX-semiquinone radicals; both metabolites have lower antineoplastic activity, but they are more prone to induce cardiotoxicity (Kassner et al., 2008). A carbonic anhydrase isoform (CBR1) was found to be its main metabolic carrier to liver cells (Kassner et al., 2008; Gonzalez-Covarrubias et al., 2009), whereas an oxidoreductase isoform (AKR1A) was shown to be its most important carrier into heart tissues (Mordente et al., 2003).
To date, studies of several candidate genes have examined the impact of germline genetic polymorphisms in selected key genes involved directly in DOX PK/PD pathways to determine their individual associations with effectiveness and safety in multiethnic groups of patients with several types of cancer, such as breast neoplasms (Nyangwara et al., 2024; Ruiz-Pinto et al., 2018; Hertz et al., 2016; Lal et al., 2016; Faraji et al., 2016), non-Hodgkin lymphoma (Wojnowski et al., 2005), osteosarcoma (Windsor et al., 2011), soft tissue sarcoma (Gelderblom et al., 2014), or acute lymphoblastic leukaemia (Gándara-Mireles et al., 2021).
However, the associations obtained from these studies are often inconclusive, limited to specific cancer complications (such as cardiotoxicity), and confined to certain ethnogeographic groups (Nyangwara et al., 2024; Ruiz-Pinto et al., 2018; Hertz et al., 2016; Lal et al., 2016; Faraji et al., 2016). Therefore, the identified PGx markers were rated based on the PharmGKB clinical annotation scoring system as level 3, which indicates a lower level of evidence based on a single study or several studies that failed to replicate the association or were based on preliminary evidence (Whirl-Carrillo et al., 2012; Whirl-Carrillo et al., 2021). Therefore, in terms of clinical significance, further replication in larger cohort studies involving diverse ethnic populations is needed. Consequently, these constrained findings have hindered the development and implementation of unified guidelines for routine pharmacogenomic testing in oncology clinical practice employing DOX chemotherapy (Nagy et al., 2020). In routine practice, the pharmacogenetic profile of DOX was rarely examined under the condition its usage via TACE technology in HCC patients (Liang et al., 2016). This finding motivated us to conduct the present study. The current pharmacogenomic study aimed to determine potential variants that predict the effectiveness and outcomes of DOX chemotherapy mediated by TACE in HCC patients.
2 Materials and methods
2.1 Study design and patients
This prospective study was conducted between August 2022 and December 2023 in qualified oncology departments that specialize in the management of hepatocellular carcinoma (HCC) at university hospitals. The Institutional Review Board (IRB) of Princess Nourah University (PNU) in Riyadh, Saudi Arabia, approved all study procedures (IRB Log Number: 23–0177) in compliance with recognized ethical standards.
Initially, 224 consecutive patients with HCC, both male and female, were screened for potential inclusion in the study. Each patient underwent a comprehensive review of their medical history and laboratory results. The eligibility criteria required participants to be aged 18 years or older and to have a confirmed diagnosis of intermediate- or advanced-stage HCC, as verified by histological, radiological, or pathological analysis following the guidelines of AASLD (Singal et al., 2023). Some patients were excluded after staging due to various reasons as described in the Flow chart (Supplementary Figure S1).
Enrolled patients received DOX through TACE technology based on expert opinion, as their tumors were deemed unresectable or they were not candidates for radiofrequency ablation. Patients were excluded from the study if they met any of the following criteria: age over 75 years, presence of portal vein thrombosis, white blood cell count less than 3 × 109/L, platelet count below 50 × 109/L, serum bilirubin level greater than 3 mg/dL, serum creatinine concentration exceeding 1.5 mg/dL, history of co-occurring illnesses, or a diagnosis of other types of cancer.
All patients provided written informed consent before enrollment. Primary data were collected from medical records, including demographic and clinical information, baseline laboratory values, and concurrent medication use. The data were entered anonymously into an electronic annotated data entry program (Harris et al., 2019).
In accordance with a standard TACE treatment protocol, each patient was intra-arterially infused with a combination emulsion containing two medications: ethionized oil and DOX. For tumours with a diameter <5 cm, the ethionized oil dose was <5 mL. Tumours ≥5 cm in size were administered a maximum dose of 10 mL. The DOX dose ranged between 30 and 60 mg/m2, depending on the tumour size, extent, and blood supply.
2.2 Study oversight
This observational study conducted follow-up over 13 months to comprehensively capture clinical outcomes, specifically treatment efficacy. Triple pelvic abdominal computed tomography (CT) scans were conducted prior to and 1 month subsequent to TACE to detect recurrence and assess the necessity for an additional therapeutic cycle. Follow-up appointments were arranged for all patients, including those who attained a complete response (CR), to determine any adverse effects and to evaluate hematologic, renal, and hepatic function.
2.3 Clinical outcome definitions
To assess the efficacy of DOX delivered through TACE, we assertively utilized critical outcome measures: overall survival (OS), progression-free survival (PFS), and α-fetoprotein reduction. These metrics were meticulously defined comprehensively in previous studies (Lencioni and Llovet, 2010; Liu et al., 2019; Masior et al., 2023; Bruix, 2021), reinforcing the rigor of our, evaluation and the significance of our findings.
2.4 Genotyping method and quality control
On the first day of the patients’ visit, blood samples were collected into two 5 mL EDTA-containing tubes for genotyping. Genomic DNA was extracted from whole blood using the QIAsymphony SP automated extraction system and a QIAsymphony® DSP DNA Midi Kit, following the manufacturer’s instructions (Qiagen). A Nanodrop spectrophotometer was utilized to determine the concentration and purity of the extracted DNA.
The extracted genomic DNA samples were then amplified using multiplex PCR with a QIAGEN Multiplex PCR Kit, as per the manufacturer’s instructions (Qiagen). This amplification step allowed for high-resolution genotyping of highly homologous regions of the genome.
Following amplification, genome-wide genotyping of the participants was carried out using the Applied Biosystems™ Axiom™ Precision Medicine Diversity (PMD) Research™ Array technology on the automated Applied Biosystems™ GeneTitan™ Multi-Channel (MC) instrument (Affymetrix Inc., Santa Clara, CA, United States). This Axiom Array solution provides comprehensive coverage of over 850,000 single-nucleotide polymorphisms (SNPs), insertions or deletions (indels), and copy number variants (CNVs), along with dense whole-genome coverage across diverse populations [catalogue identifier: 951961; Thermo Fisher Scientific, https://www.thermofisher.com/order/catalogue/product/951961/] (Thermo Fisher Scientific, 2024). It also includes a thorough analysis of over 5,000 pharmacogenomic (PGx) markers in more than 1,100 core and extended pharmacokinetic/pharmacodynamic (PK/PD) genes, evaluated at clinical annotation levels of evidence 1–4 (as established by PharmGKB). These genes influence the absorption, distribution, metabolism, and excretion (ADME) of commonly prescribed medications.
The generated genotyping profiles were analyzed using the Applied Biosystems™ Axiom™ Analysis Suite software version 5.2 (Thermo Fisher Scientific, Santa Clara, CA, United States). Markers corresponding to candidate genes with a genotyping call rate of less than 95%, minor allele frequency (MAF) lower than 0.01, or Hardy-Weinberg equilibrium (HWE) P-value less than 0.001 were excluded from the association analysis. Additionally, samples with a genotyping call rate below 93% were also excluded.
2.5 Selection of markers of interest
The Ensembl Variant Effect Predictor (VEP) tool was used to annotate and prioritize the identified markers based on their predicted effects (McLaren et al., 2016). The selected variants were located on known genes and characterized by their functional consequences for gene activity.
2.6 Statistical and bioinformatics analyses
PLINK version 1.90p 64-bit (16 April 2021) was used for the statistical analysis (www.cog-genomics.org/plink/1.9/), General Public Licence v3 (Purcell et al., 2007). A threshold P value <5 × 10−8 was used for genome-wide association study (GWAS) analyses as a strict significance point to identify potential loci (Witte et al., 1996). However, some signals with lower association signals (P < 5 × 10−5) were also suggested for variants with known functional consequences. The Kaplan–Meier analysis was performed to examine the long-term significance of the effects of the identified top hit genotypes on PFS and OS. Manhattan plots were generated using the R statistical package (qqman) version R-4.2.2. The association values shown in Manhattan plots are reported as–log10 P values. The haplotype analysis software tool version 1.05, prepared by Eliades and Eliades (2009), was used to determine the common significant haplotypes.
3 Results
3.1 Selection of axiom array markers
Approximately 70,000 variants did not exist in the genotyped cohort. Thus, the remaining 780,166 variants were tested and analysed in 81 HCC patients (59 males, 22 females); however, three samples were removed (Supplementary Figure S1) because of the low sample genotyping call rate (<93%). The average genotyping call rate in the remaining samples was 99.6%. Several genotyping quality control (QC) measures were applied; thus, 45,901 variants were removed because of a low variant genotyping call rate (<95%), 138 variants were removed because of the Hardy‒Weinberg exact test, and 258,664 variants were removed because of the minor allele threshold (<1%). The remaining 475,463 variants and 78 patients passed filters and QC measures (35 patients experienced progressive symptoms, 43 had no progression, 8 patients died, 34 had a good AFP response, and 41 had no response). The genomic inflation factor for all GWAS analyses was <1.05.
3.2 Patient characteristics
Table 1 shows the demographic and clinical data of HCC patients who experienced progression (symptomatic or radiographic) versus patients with no evidence of progression. The majority of the studied patients were males (71.8%), yet the male gender difference between the two tested groups (74.4% with no progression vs 68.6% of patients who experienced progression) was not statistically significant (P = 0.568). The mean age of the studied patients was 62.01 years (61.93 ± 6.47 vs 62.11 ± 6.69 years; P = 0.903). Additionally, baseline laboratory data (including albumin, haemoglobin, bilirubin, alanine transaminase (ALT) and aspartate transaminase (AST) levels) were not different between patients who experienced progression and their counterparts who did not have any evidence of progression. On the other hand, baseline AFP measurements were substantially lower in the nonprogression group than in the tumour progression group (mean = 15.5 ng/mL vs. 123.0 ng/mL, respectively; P = 0.009). The majority of the patients in both groups received TACE once (90.7% vs 94.3%, respectively; P = 0.55). Macroscopic vascular invasion and metastatic extension were more common among patients who experienced progression than among those who did not (P = 0.003 and 0.005, respectively). The other clinical characteristics of the patients were equally distributed, as shown in Table 1.

Table 1. The HCC cases with symptomatic or radiographic progression were compared to those without progression.
A multivariate Cox proportional hazards model with a forward likelihood ratio revealed that the presence of metastatic disease extension was the most significant predictor of shorter OS, with an HR of 8.36 (95% CI 1.1–63.5, P = 0.04), while hepatitis B etiology was the strongest predictor of shorter PFS (HR: 5.04, 95% CI 1.26–20.1, P = 0.022). In addition, combined genetic and nongenetic multivariate association analyses revealed that metastatic disease extension (P = 0.015) was the only non-genetic predictor factor modulating the impact of DOX on OS in conjunction with other genotypic variants.
3.3 Genetic associations with progression and PFS
Table 2 and Figure 1 present the association results and Manhattan plots, respectively, of tumour progression outcomes in patients with advanced HCC receiving DOX. Table 2 shows the top six SNPs considered risk variants for enhanced progression of HCC tumours at P < 5 × 10−5. These SNPs were located within the AK3, TRPM3 (2 SNPs), CDH4, LINC00504, and GRIN2D loci. In contrast, two markers in two genes (HPGD and RC3H2) were demonstrated to protect against disease progression at P < 5 × 10−5.
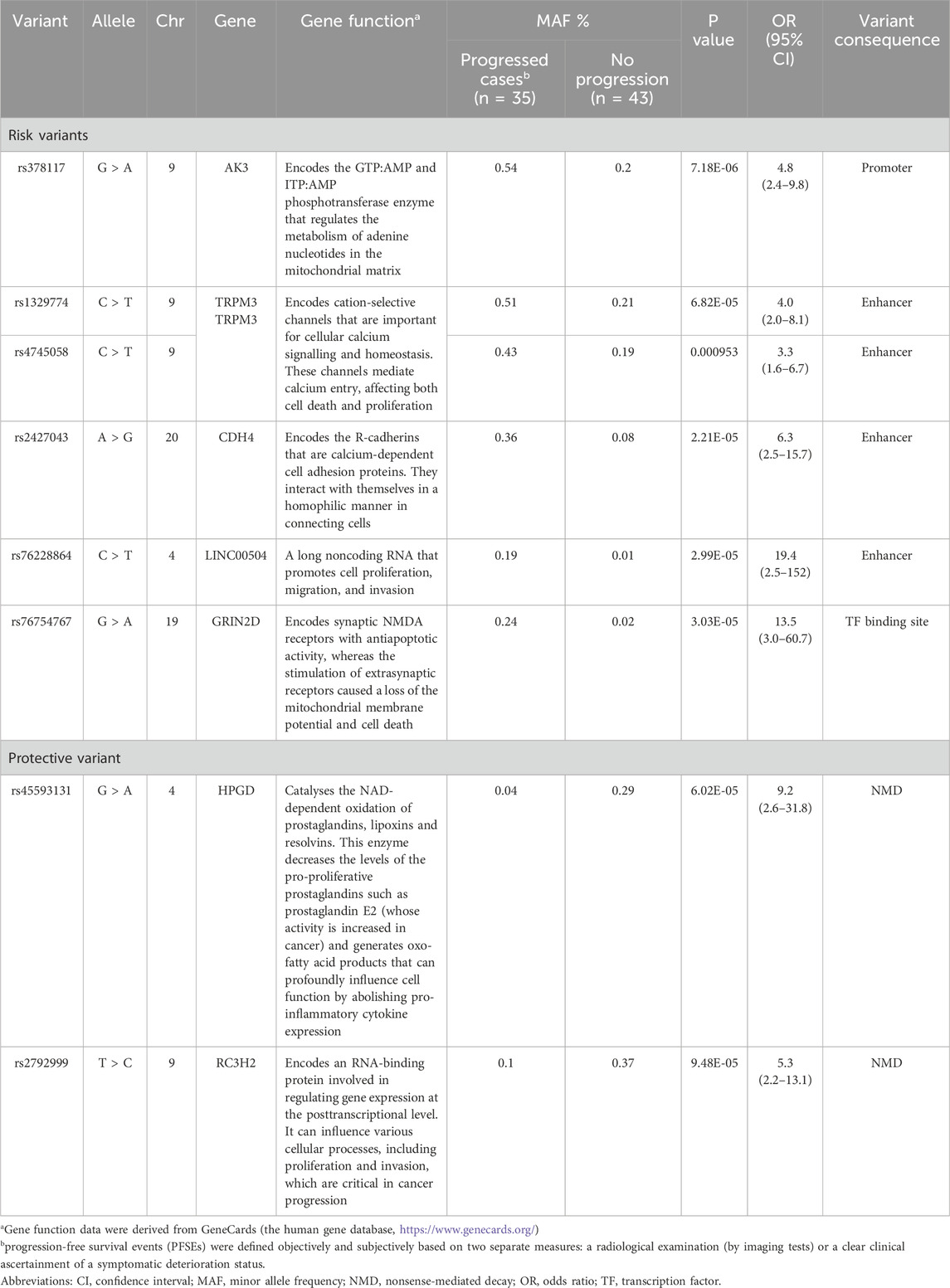
Table 2. Top markers suggested as risk and protective factors for clinical symptomatic and radiological progression among HCC patients receiving DOX chemotherapy.
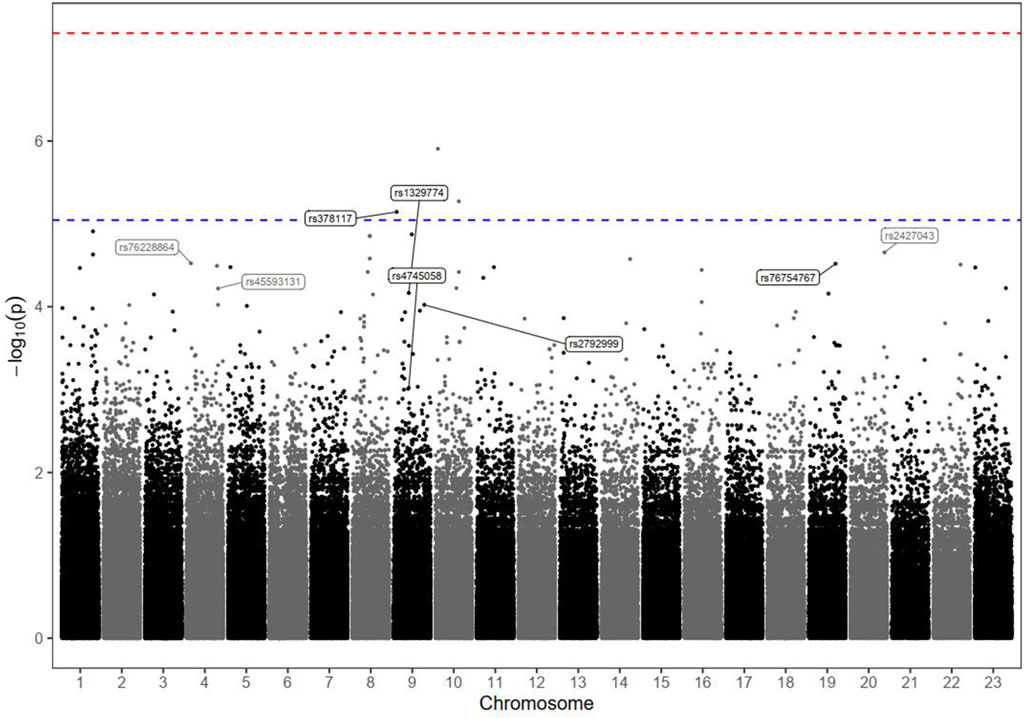
Figure 1. Manhattan plot of tumour progression markers in HCC patients receiving DOX chemotherapy. The plot shows the statistical association between each genomic variant and the outcome. The dots on the plot represent the chromosomal location on the x-axis and their statistical significance with respect to the phenotype of interest according to the logarithmic P value (-log10) on the y-axis. Each dot on the plot represents a single genomic variant. Variants that exceed the threshold line are statistically significant and are worth investigating. The–log10 P values of the detected SNPs were plotted across the 23 chromosomes. None of the SNPs met the genome-wide significance threshold (P < 5 × 10−8), as indicated by the red dashed line. Only the top markers of interest were annotated; the rs ID numbers of some SNPs above and below the blue dashed line (P < 9 × 10−6) are shown.
The results of the Kaplan–Meier analysis, which confirmed the long-term significance of the effects of these identified top hit genotypes on PFS (during the study follow-up period), are described in Supplementary Figure S2. Interestingly, the estimated median PFS time was significantly shorter among HCC patients carrying the risk variant genotypes (in homozygous or heterozygous forms) than among those characterized by the reference status (log-rank P < 0.001). In contrast, the duration of the PFS period tended to be much longer among patients carrying protective variants than among those with major-allele genotypes (log-rank P < 0.001). The variants in linkage disequilibrium (LD) with the identified markers (r2 ≥ 0.6) are shown in Supplementary Excel Sheet 1. In addition, the haplotype analysis revealed no significant associations with any of the commonly detected haplotypes (Supplementary Excel Sheet 2).
3.4 Genetic associations with the biological response
The GWAS analysis results for the tumour biological response in patients with advanced HCC receiving DOX are depicted in Table 3, and Manhattan plots are shown in Figure 2.
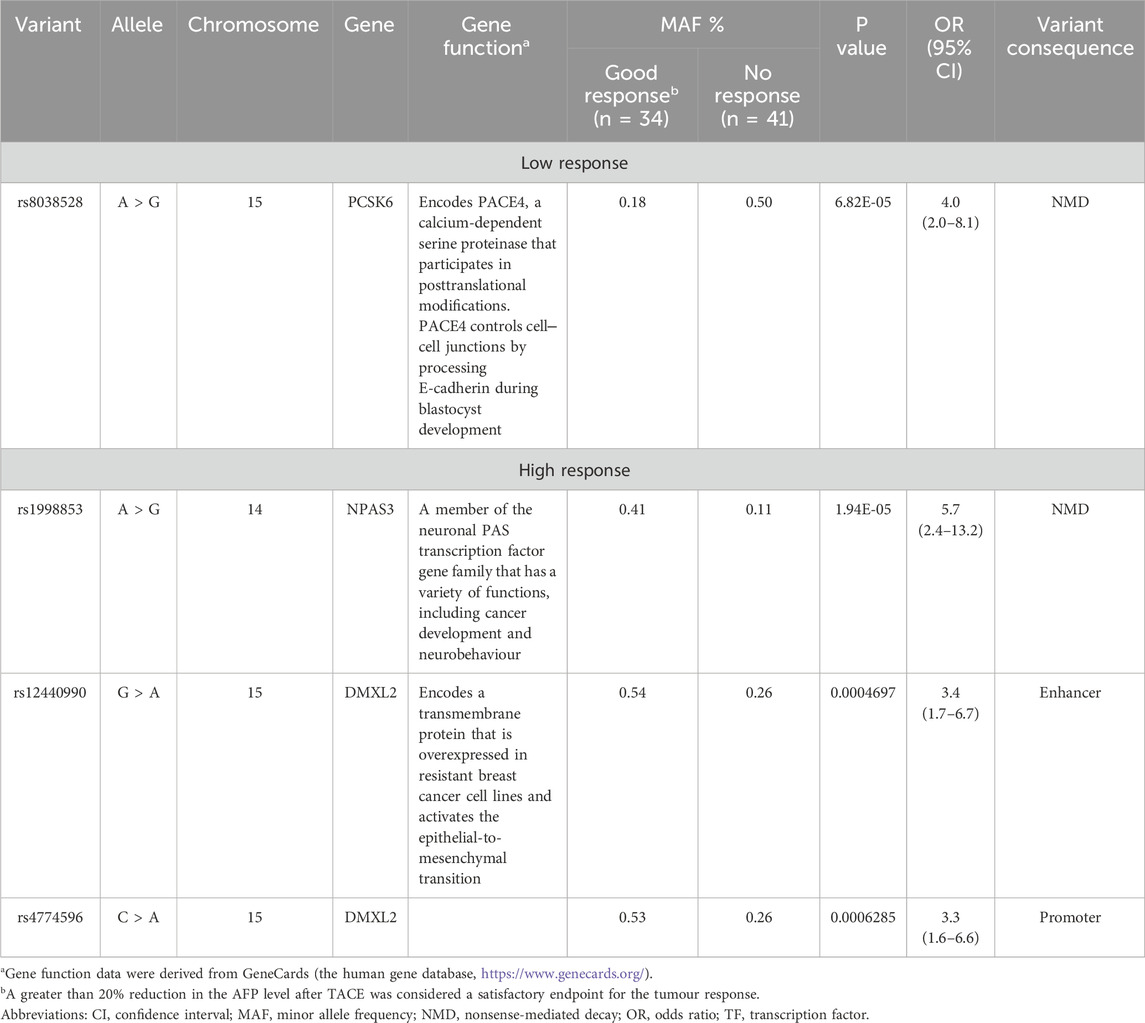
Table 3. Top markers associated with the AFP response among HCC patients receiving DOX chemotherapy.
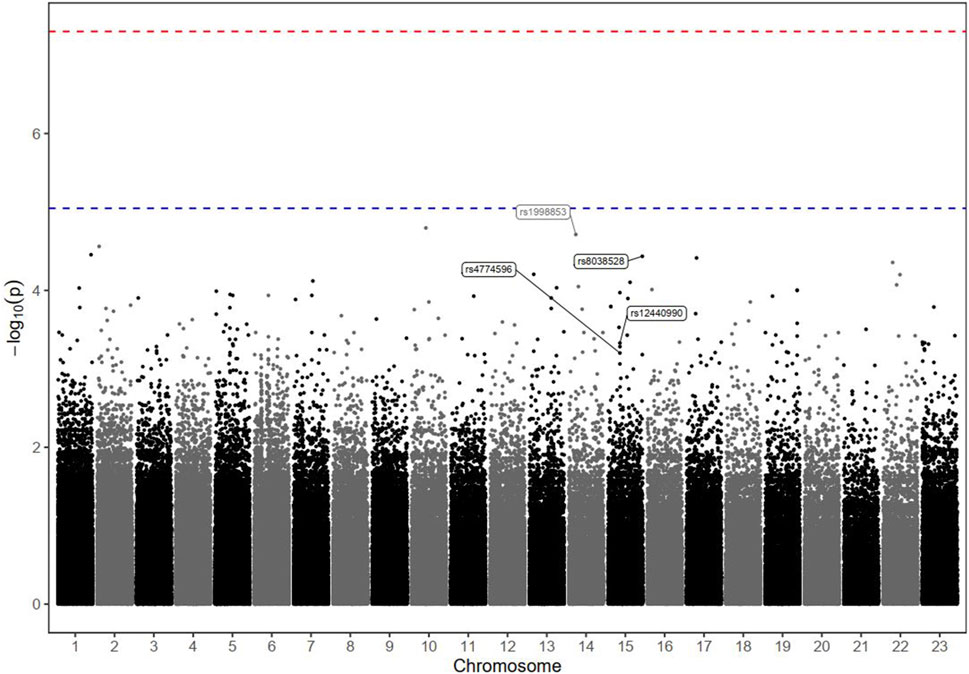
Figure 2. Manhattan plot of markers associated with the biological response (defined by the percentage of AFP reduction) in HCC patients receiving DOX chemotherapy. The–log10 P values of the detected SNPs were plotted across the 23 chromosomes. None of the SNPs met the genome-wide significance threshold (P < 5 × 10−8), as indicated by the red dashed line. Only four markers of interest were annotated; their rs ID numbers are shown below the blue dashed line (P < 9 × 10−6).
Among the screened SNPs, only one polymorphism (rs8038528 in the PCSK6 gene) demonstrated the potential to be associated with a lower AFP response, where patients harbouring the variant (allele A) were (4 times) less likely to respond to DOX treatment based on an assessment of AFP% reduction levels (P = 6.82 × 10−5). In contrast, 3 SNPs (rs1998853, rs12440990, and rs4774596) located within 2 genes (NPAS3 and DMXL2) were suggested as high-response variants, where carriers of the variant alleles exhibited a greater probability of a biological AFP response (OR = 5.7, 3.4, and 3.3, respectively), however, at variable significance levels (Table 3).
LD data for the identified markers with r2 ≥ 0.6 are shown in Supplementary Excel Sheet 1. A haplotype analysis (Supplementary Excel Sheet 2) including the four mentioned SNPs revealed a significant association with the protective haplotype GA AA GG CC (p = 0.019).
3.5 Genetic associations with death and OS
The results of the GWAS analysis for the death incidence in patients with advanced HCC receiving DOX chemotherapy are displayed in Table 4, and the Manhattan plots are shown in Figure 3. As shown in Table 4, 20 markers of different genes had the potential to impact the incidence of death in our HCC cohort. Interestingly, 5 SNPs reached the genome-wide significance level (P < 5.0 × 10−8); 4 of them were located within defined genes (DENND1B, LOC107986086, TMEM169, and RNF152 loci), whereas one SNP, located on chromosome 7, was intergenic. The strongest observed signal was for the rs1582409 variant in the DENND1B (DENN domain containing 1B) locus on chromosome 1 (OR = 63.18, 95% CI = 6.77–589.4, P = 1.78 × 10−9).
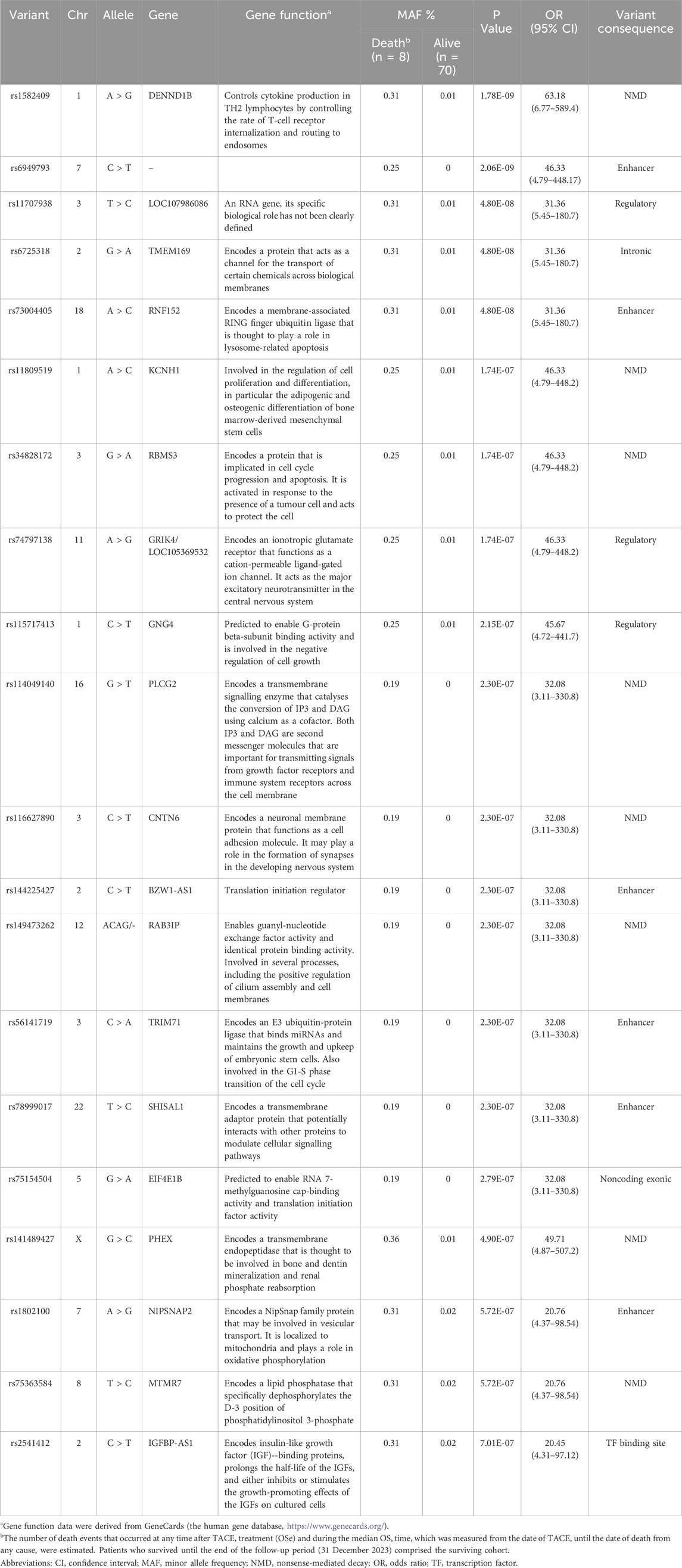
Table 4. Important markers suggested as risk factors for death among HCC patients receiving DOX chemotherapy.
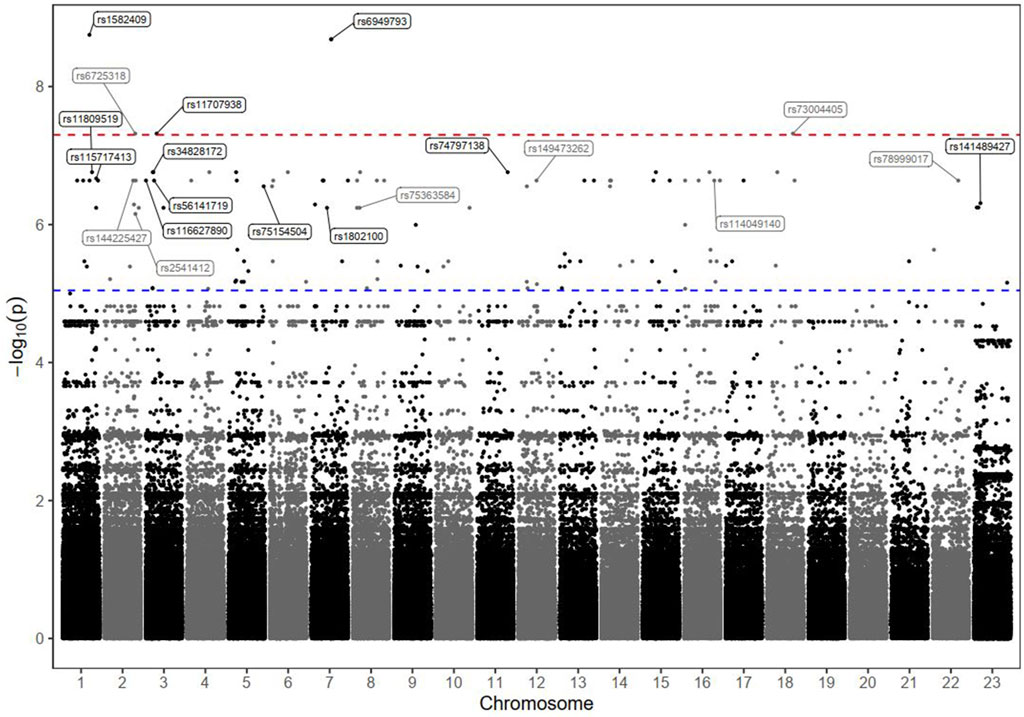
Figure 3. Manhattan plot of death markers in HCC patients receiving DOX chemotherapy. The–log10 P values of the detected SNPs were plotted across the 23 chromosomes. Five SNPs that met the genome-wide significance threshold (P < 5 × 10−8) are shown above the red dashed line. Other markers of interest were also annotated; their rs ID numbers are shown above the blue dashed line (P < 9 × 10−6).
The results of the Kaplan–Meier analysis are shown in Supplementary Figure S3. The estimated median OS time tended to be much shorter among the patients carrying any of the identified risk variants (in homozygous or heterozygous forms) than among those characterized by the reference status (log-rank P < 0.001).
Furthermore, the LD data for the identified markers with r2 ≥ 0.6 are shown in Supplementary Excel Sheet 1. A 10 SNP haplotype analysis (including the top ten associated markers in Supplementary Excel Sheet 2) revealed a significant association with the protective haplotype AA CC TT GG AA AA GG AA GG GG CC (P = 0.00000103).
4 Discussion
This pharmacogenomic study used a GWAS approach to overcome the shortcomings of association findings obtained from previous studies that focused on candidate genes related to DOX therapy (Supplementary Table S1) in various types of cancer, including HCC (Nyangwara et al., 2024; Ruiz-Pinto et al., 2018; Hertz et al., 2016; Lal et al., 2016; Faraji et al., 2016; Wojnowski et al., 2005; Windsor et al., 2011; Gelderblom et al., 2014; Gándara-Mireles et al., 2021; Liang et al., 2016). To our knowledge, this study is the first to examine the ability of a large portion of the genome to modulate the clinical response of HCC patients to DOX therapy. The associations discussed here are confined to the suggested top genetic variants linked to each clinical outcome (tumour progression, tumour response, and death).
Several genes have been suggested to modulate tumour progression and the biological response, although these genes did not reach the genome-wide association significance level of 5.0 × 10−8. In contrast, strong genetic associations were detected with a potential impact on the death outcome in this HCC cohort. Notably, none of the genes associated with death overlapped with the identified genes involved in tumour progression and reduced AFP levels. This finding likely indicates alternative molecular mechanisms underlying each outcome. Consequently, we attempted to interpret the observed associations in terms of the molecular mechanisms underlying gene–HCC and gene–DOX interactions in light of updated research and the scientific literature.
4.1 Progression-related variants in HCC
The AK3 gene encodes the adenylate kinase (AK) 3 protein, which is a GTP:AMP and ITP:AMP phosphotransferase enzyme found in the mitochondrial matrix (Liu et al., 2019). The SNP rs378117 in AK3 is a promoter variant associated with reduced gene activity and probably serves as a predictor of progressive symptoms and radiological deterioration of the tumour in our cohort. The literature revealed contradictory findings of the enhanced metastasis of cancer cells with over- or underexpression of genes encoding AK isoforms and, consequently, enzymatic changes depending on the tissue specificity and different phases (initial vs advanced stages of tumour growth) (Klepinin et al., 2020). Consistent with our observations, some studies have shown that AK3 is downregulated in lung cancer (Balinsky et al., 1984) and hepatoma (Criss et al., 1970). Similarly, an in vitro cell line study showed that AK3 knockout was associated with reduced cell proliferation and increased expression of oxidative stress-related proteins, indicating the importance of this enzyme in maintaining mitochondrial homeostasis (Fujisawa et al., 2022). Moreover, a subsequent study documented significantly lower levels of AK3 expression in thiopurine-resistant human leukaemia cells than in wild-type cells, indicating a role for the AK3 variant in metastasis progression (Karim et al., 2011). Additionally, the suppression of AK3 (as a phosphotransferase and glycolysis-related gene) triggers a cancerous transformation, leading to an uncontrolled cell cycle and growth with a subsequently worse prognosis and shorter survival of patients with breast cancer (Klepinin et al., 2020; Chao et al., 2024). Our study is the first to report the novel impact of AK3 (rs378117) on increasing the progression of human HCC.
The product of the TRPM3 gene belongs to the family of transient receptor potential (TRP) channels. TRP channels are cation-selective channels that are important for cellular calcium signalling and homeostasis. The protein encoded by this gene mediates calcium entry, consequently affecting both cell death and proliferation (Clapham et al., 2005; Gkika and Prevarskaya, 2009). A large-scale database analysis revealed that the level of the TRPM3 transcript was decreased in the majority of cancer tissues. Conversely, the Kaplan–Meier analysis revealed that high TRPM3 mRNA expression levels were significantly associated with an improved prognosis and increased OS of all examined patients with various cancer subtypes (Qin et al., 2020). Similarly, the variants detected within the TRPM3 gene (rs1329774 and rs4745058) in our study are likely linked to downregulated gene expression and thus are significantly associated with tumour progression. These results suggest that targeting ion channels such as TRPM3 to overcome drug resistance in cancer treatment could be an active area for drug discovery and development.
The CDH4 gene, which encodes the R-cadherin protein, has been significantly implicated in the progression and treatment of various cancers. However, the expression level and function of CDH4 in different types of cancer remain controversial (Aacr Project GENIE Consortium, 2017). In some cancers, it acts as an oncogene (bladder cancer), whereas in others, it acts as a tumour suppressor or antioncogene (such as in lung cancer, nasopharyngeal carcinoma, gastric cancer, and colorectal cancer) (Uhlen et al., 2017; Li et al., 2017). This dual role may complicate the development of therapies targeting CDH4 (Xie J.et al., 2023). The specific role and expression of CDH4 in HCC were not previously defined. Interestingly, our data revealed a CDH4 variant (rs2427043) with a potential impact on the progression of tumours in the HCC population, reflecting an increased risk of downregulated expression of the CDH4 protein. Similarly, a previous GWAS revealed that two other CDH4 variants (rs1122269 and rs4925193) were associated with significant trends (although not at the level of the current study) in modifying the OS outcomes of patients receiving gemcitabine treatment for pancreatic cancer (Li et al., 2016). Our results, together with those of gemcitabine studies, imply the importance of further functional and clinical studies to define the role of the CDH4 gene as a PGx biomarker for the individualization of chemotherapy or as a novel target for cancer drug development.
LINC00504 is a long noncoding RNA (lncRNA) that is associated with several types of cancer, including HCC. It is highly expressed in cancer cells compared with normal cells, where it acts as an oncogene associated with an unfavourable prognosis (Mathias et al., 2021; Hou et al., 2023). Its overexpression is associated with aggressive tumour characteristics and correlates with poorer patient outcomes. Elevated levels of LINC00504 correlate with an increased tumour size, advanced stage, and lymph node metastasis, indicating a negative impact on the prognosis (Zhang, 2020). On the other hand, LINC00504 has been shown to influence DOX resistance in HCC cells through various mechanisms, specifically by influencing apoptotic pathways, promoting the epithelial‒mesenchymal transition (EMT), and enhancing metabolic adaptations (Shan and Li, 2019). High levels of LINC00504 are associated with reduced sensitivity to DOX in HCC cells (Ma et al., 2019). Studies indicate that HCC cell lines with elevated LINC00504 expression tend to show greater resistance to DOX treatment than those with lower expression levels (Mathias et al., 2021). Our findings revealed a high allele frequency (19%) of rs76228864 in LINC00504 among patients with tumour progression versus only 1% in the control group. This finding reflects the strong association of the LINC00504 marker with the risk of HCC progression.
GRIN2D, which encodes the GluN2D subunit of N-methyl-D-aspartate receptors (NMDARs), has been identified as a potential oncogene and a novel therapeutic target in certain types of cancer (Wang et al., 2023). For example, in pancreatic and colorectal cancer, GRIN2D was overexpressed in patients’ tissues at both the mRNA and protein levels. Consequently, upregulated GRIN2D could effectively promote tumour growth and liver metastasis by activating subcellular signalling pathways and transcription factors. Furthermore, high expression levels of GRIN2D in HCC are associated with increased infiltration of various immune cells, including T cells and natural killer (NK) cells. This finding suggests that GRIN2D may facilitate a more active immune response within the tumour microenvironment, potentially impacting tumour progression and the patient prognosis (Zhang Y. G. et al., 2022; Xie Z. et al., 2023).
Additionally, high expression levels of GRIN2D are associated with increased resistance to DOX, potentially through its effects on the expression of P-glycoprotein (P-gp), a well-known efflux pump that contributes to multidrug resistance by actively transporting drugs out of cells, thereby reducing efficacy (Zhang Z. et al., 2022). Accordingly, combination therapies that incorporate DOX with agents targeting GRIN2D or related pathways are being explored as strategies to improve therapeutic outcomes in patients with HCC (Duan and Liu, 2018). Consistent with previous reports, our results identified a transcription factor variant in GRIN2D (rs76754767) as a risk factor for HCC tumour progression.
The HPGD gene is recognized as a tumour suppressor in various cancers, including HCC. Its function involves the degradation of prostaglandins, particularly PGE2, which is known to promote inflammation and tumour progression. The downregulation of HPGD is associated with increased levels of PGE2 in the tumour microenvironment, leading to the increased proliferation and survival of hepatocytes, which contributes to the development of HCC from metabolic dysfunction-associated steatohepatitis (Hu et al., 2023). Therefore, restoring HPGD levels or inhibiting PGE2 signalling has been explored as a therapeutic option to mitigate tumour growth and improve the immune response or apoptosis signalling pathways (Ren et al., 2023; Li C. J. et al., 2023). Interestingly, our data revealed a protective relationship between the HPGD variant rs45593131 and HCC progression. RC3H2 (an RNA-binding protein with multiple C3H-type zinc fingers) is involved in regulating gene expression at the posttranscriptional level. It can influence various cellular processes, including proliferation and invasion, which are critical in cancer progression (Zhang et al., 2015).
Studies have shown that RC3H2 is upregulated in HCC tissues compared with normal liver tissues. Its overexpression correlates with increased cell proliferation and invasion capabilities, suggesting that RC3H2 may play a significant role in the aggressiveness of HCC (Zhao et al., 2023).
Consistent with these reports, our data revealed that rs2792999 in RC3H2 is a protective variant with a strong negative association with HCC tumour progression. This variant is likely associated with decreased RC3H2 expression.
4.2 AFP response-related variants
The PCSK6 gene (proprotein convertase subtilisin/kexin type 6), which is located on chromosome 15q26.3, encodes PACE4, a calcium-dependent serine proteinase that participates in posttranslational modifications. PACE4 controls cell‒cell junctions by processing E-cadherin during blastocyst development. PCSK6 is upregulated in pancreatic cancer liver metastases, and its inactivation results in the decreased migratory potential of tumour cells in pancreatic cancer and reprogramming of cell‒cell junctions (He et al., 2022). However, in the liver, PCSK9 binds to and degrades the low-density lipoprotein receptor, which is a major contributor to hypercholesterolemia. Therefore, PCSK6 plays an important role in the cardiovascular system and homeostasis (Suur et al., 2021; Bordicchia et al., 2019). A previous study (using an animal model) showed that the overexpression of PCSK6 protects the heart from the long-term damage caused by DOX, reduces cardiac oxidative stress, decreases apoptosis, restores the autophagolysosomal degradation process that is inhibited by DOX, and increases autophagy (Li C. et al., 2023).
In addition, studies on PCSK6 expression in cell lines revealed a PCSK6 variant (rs7181043) that was significantly associated with the plaque fat content in atherosclerotic patients (Suur et al., 2021; Bordicchia et al., 2019). PCSK6 variants are predicted to decrease gene function, which may increase DOX-induced cardiotoxicity. Consistently, our study reported that rs8038528 in the PCSK6 gene, a low-response variant, was associated with a lower AFP% reduction after DOX therapy. Based on the available findings, the PCSK6 expression level appears to be related to the AFP% change in HCC patients and is potentially related to DOX treatment.
We also identified the rs12440990 and rs4774596 SNPs, which are located within the DMXL2 gene, as high-response variants. Carriage of these variants is likely linked to an AFP% reduction. DMXL2 is a transmembrane protein that is overexpressed in resistant breast cancer cell lines, is an activator of the epithelial-to-mesenchymal transition, and is a potential new biomarker for hormone-positive breast cancer (Valter et al., 2023; Faronato et al., 2015). Nevertheless, the exact mechanisms of DMXL2 involvement in HCC are still being investigated (Ikeda et al., 2014).
The marker rs1998853, located in NPAS3 on chromosome 14, also showed a greater response, with an AFP% reduction following DOX treatment. NPAS3 is a member of the neuronal PAS transcription factor gene family, which has a variety of functions, including cancer development and neurobehaviour. NPAS3, a transcription factor, has a tumour-suppressive role in manipulating the progression of certain types of cancer (Moreira et al., 2011; Yu et al., 2024). The absence of NPAS3 expression in glioblastomas (a type of brain cancer) was reported to be associated with shorter OS. Overexpression of NPAS3 in malignant glioma cell lines dramatically slowed transformation, whereas lower expression spurred more aggressive development (Moreira et al., 2011). Although NPAS3 was historically connected to neurogenesis, it has emerged as a predictive hallmark for triple-negative breast cancer as a tumour suppressor that drives the progression of breast cancer via the modulation of autophagy (Yu et al., 2024).
4.3 Death-related variants in HCC
The highest detected death signal (P = 1.78 × 10−09) was for the rs1582409 variant located in the DENND1B (DENN domain containing 1B) locus on chromosome 1. Previously, DENND1B was associated with autoimmune liver disease in genome-wide meta-analyses (Huang et al., 2023; Liu et al., 2010). It has also been implicated in certain types of cancer. For example, a circular RNA form of DENND1B (circ_DENND1B) was found to inhibit tumorigenicity in clear cell renal cell carcinoma (ccRCC) (Chen et al., 2022). These findings suggest that DENND1B may have tumour-suppressive properties in certain contexts. Furthermore, a few SNPs in DENND1B have been reported to increase the risks of pancreatic, renal and gastric cancers (Cotterchio et al., 2015; Nookala et al., 2012).
DOX can cause significant side effects, including cardiotoxicity (Nyangwara et al., 2024; Hertz et al., 2016; Wojnowski et al., 2005; Gándara-Mireles et al., 2021). The rs1582409 variant in the DENND1B gene might influence the likelihood or severity of cardiotoxicity leading to death.
The SNP rs6725318, which is located in the TMEM169 gene on chromosome 2, may play a role as a risk factor for death in HCC patients receiving DOX therapy. TMEM is a protein that functions as a channel for the transport of certain chemicals across biological membranes. TMEM9 was found to be abnormally expressed in liver cancers (Zhang et al., 2016), and it was described as an oncogene implicated in tumour development, invasion, and chemoresistance (Schmit and Michiels, 2018). A TMEM169 gene mutation (rs6725318) is predicted to contribute to DOX resistance by lowering transmembrane protein activity, possibly altering drug uptake, efflux, and accumulation in cells. These properties may partially explain the suppression of the apoptosis pathway, which is a mechanism of DOX resistance.
The present study also revealed a strong association between rs73004405 in RNF152 on chromosome 18 and the death incidence. RNF152, a membrane-associated RING finger ubiquitin ligase, is found in lysosomes and is thought to play a role in lysosome-related apoptosis, resulting in the inhibition of HCC cell growth (Ueda et al., 2024).
Furthermore, RING finger-related E3 ubiquitin ligases play a role in carcinogenesis and can act as oncogenes or tumour suppressors, depending on the target proteins. RNF152 was previously reported to be highly downregulated in HCC and associated with decreased OS and PFS (Wan et al., 2021). Similarly, the expression of RNF152 in colorectal cancer (CRC) tissues is considerably lower than its expression in adjacent noncancerous tissues. In patients with CRC, high expression levels of RNF152 are associated with a better prognosis, whereas low expression of RNF152 is associated with lymphatic metastases (Cui et al., 2018). RNF152 is known to play a role in regulating apoptosis and autophagy (Ueda et al., 2024).
Another RNF gene (RNF6) was also identified by Cai et al. (Cai et al., 2019) as an independent predictor of poor outcomes in patients with HCC. In a previous HCC study, RNF6 silencing inhibited radioresistance and the EMT both in vivo and in vitro. Forkhead box protein A1 (FoxA1), a crucial transcriptional repressor of the EMT, is directly bound and ubiquitylated by RNF6, suggesting that FoxA1 degradation is partially responsible for the carcinogenic effect of RNF6 on HCC (Liu et al., 2010). Overall, these results are consistent with our findings, which indicate an important association between the rs73004405 variant in RNF152 and the survival of HCC patients treated with DOX. Given the effects of DOX on apoptotic pathways, some interplay may exist between the drug’s action and RNF152’s function in regulating cell death.
4.4 Summary outcomes
The genes explored in our study are novel and different from those previously reported. As illustrated in Supplementary Table S1, most of the previously reported genes are known to influence DOX PKs (e.g., distribution (transport) and metabolism) rather than DOX PDs (e.g., efficacy and toxicity) (Gonzalez-Covarrubias et al., 2009; Mordente et al., 2003; Nyangwara et al., 2024; Ruiz-Pinto et al., 2018) in heterogeneous types of cancer (Hertz et al., 2016; Lal et al., 2016; Faraji et al., 2016; Wojnowski et al., 2005; Windsor et al., 2011). These genes include SLC transporters, which mediate DOX uptake into cancer cells [e.g., SLC22A16 (organic cation transporter)]; ABC transporters, which are involved in the efflux of DOX and contribute to drug resistance [e.g., ABCB1 (MDR1), ABCC1 (MRP1), and ABCG2 (BCRP)]; RABLP, which may influence the intracellular trafficking of DOX; and CBR enzymes [e.g., CBR1 and CBR3] or AKR1A, which are involved in DOX metabolism and detoxification. In contrast, our GWAS explored various genes (AK3, TRPM3, CDH4, LINC00504, GRIN2D, HPGD, RC3H2, PCSK6, NPAS3, and DMXL2), which had significantly variable impacts on clinical outcomes of DOX after TACE in HCC patients. These genes are suggested to be involved in diverse DOX PD-related biological processes, such as nucleotide metabolism (AK3), calcium signalling (TRPM3 and GRIN2D), cell adhesion and invasion (CDH4 and DMXL2), posttranscriptional and posttranslational regulation (RC3H2 and PCSK6), transcription regulation (NPAS3) and prostaglandin metabolism (HPGD).
4.5 Limitations
First, the associations obtained in this study are limited to the tested Arabic cohort and may not be generalizable unless they are replicated in diverse populations. While evidence of a shared genetic architecture for many traits has been reported, significant differences exist in risk allele frequencies and effect sizes between populations of different ethnicities (Chen et al., 2022). Second, GWAS findings are highly replicable, with some studies showing replication rates of up to 94% for markers that meet the GWAS cut-off level (P ≤ 5.0 × 10−8) (Ntzani et al., 2011). The reported associations with the PFS and the AFP response of patients with HCC did not reach the genome-wide significance threshold; thus, replication in another independent study is recommended. Third, although the detected risk-of-death markers met the GWAS significance level, the analysis of a small number of death incidents (n = 8) may lack sufficient statistical power to detect true associations. Hence, further replication studies may be necessary to confirm our findings. Fourth, genotype-based association tests were not performed as substantially larger sample sizes would typically be required to achieve a satisfactory power. This can be more resource-intensive and challenging to implement in the current study. Finally, this study report focuses on genetic factors influencing specific disease outcomes, such as the DOX response and survival, to analyze their roles in inter-individual variability. It identifies key genetic variants and their functional implications, uncovering the potential biomarkers and pathways contributing to treatment outcomes. However, non-genetic factors such as environmental exposures, lifestyle, and comorbidities may also play a significant role in shaping disease outcomes. Future larger sample studies could allow us to integrate findings from both genetic and non-genetic analyses for more effective risk prediction, treatment optimization, and personalized patient care.
5 Conclusion
In summary, this study identified novel genetic variants that may play significant roles in mediating DOX resistance and variants linked with a good prognosis in a real-world cohort of HCC patients. However, further replication in larger multiethnic studies remains mandatory before their implementation in precision medicine practice. Additionally, the findings suggest that biochemical investigations targeting these variants could lead to the development of different treatment options to overcome the resistance and limited efficacy of DOX-based therapies for HCC patients. Further research is necessary to fully elucidate these mechanisms and explore therapeutic strategies that could mitigate the impacts of these markers on drug resistance, PFS, and/or OS of patients with HCC.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the Institutional Review Board (IRB) of Princess Nourah University (PNU), Riyadh, Saudi Arabia [IRB Log Number: 23-0177]. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
SS: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review and editing. NK: Conceptualization, Data curation, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing. MA: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. AAls: Data curation, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. AAla: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. OAE-B: Conceptualization, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing. RW: Conceptualization, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research project was funded by the Deanship of Scientific Research and Libraries, Princess Nourah bint Abdulrahman University, through the Program of Research Project Funding After Publication, grant No. (RPFAP-39-1445).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1604473/full#supplementary-material
References
Aacr Project GENIE Consortium (2017). AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 7, 818–831. doi:10.1158/2159-8290.CD-17-0151
Abudeif, A. (2019). Epidemiology and risk factors of hepatocellular carcinoma in Egypt. Sohag Med. J. 23, 8–12. doi:10.21608/smj.2019.13376.1019
Balinsky, D., Greengard, O., Cayanis, E., and Head, J. F. (1984). Enzyme activities and isozyme patterns in human lung tumors. Cancer Res. 44, 1058–1062.
Bessar, A. A., Farag, A., Monem, S. M. A., Wadea, F. M., Shaker, S. E., Ebada, M. A., et al. (2021). Transarterial chemoembolisation in patients with hepatocellular carcinoma: low-dose doxorubicin reduces post-embolisation syndrome without affecting survival-prospective interventional study. Eur. Radiol. Exp. 5, 10. doi:10.1186/s41747-021-00204-6
Bisht, A., Avinash, D., Sahu, K. K., Patel, P., Das Gupta, G., and Kurmi, B. D. (2024). A comprehensive review on doxorubicin: mechanisms, toxicity, clinical trials, combination therapies and nanoformulations in breast cancer. Drug Deliv. Transl. Res. 15, 102–133. doi:10.1007/s13346-024-01648-0
Bordicchia, M., Spannella, F., Ferretti, G., Bacchetti, T., Vignini, A., Di Pentima, C., et al. (2019). PCSK9 is expressed in human visceral adipose tissue and regulated by insulin and cardiac natriuretic peptides. Int. J. Mol. Sci. 20, 245. doi:10.3390/ijms20020245
Bruix, J. (2021). Endpoints in clinical trials for liver cancer and their value in evidence-based clinical decision making: an unresolved Gordian knot. J. Hepatol. 74 (6), 1483–1488. Epub 2021 Feb 5. PMID: 33556420. doi:10.1016/j.jhep.2021.01.033
Cai, J., Xiong, Q., Jiang, X., Zhou, S., and Liu, T. (2019). RNF6 facilitates metastasis and radioresistance in hepatocellular carcinoma through ubiquitination of FoxA1. Exp. Cell Res. 374, 152–161. doi:10.1016/j.yexcr.2018.11.019
Chao, S., Liu, S., Wang, K., Xie, M., and Liu, B. (2024). Identification of glycolysis related genes for prognosis prediction in patients with breast cancer. Front Oncol. 10. doi:10.21203/rs.3.rs-4113335/v1
Chen, P., Yuan, P., Chen, B., Sun, J., Shen, H., and Qian, Y. (2017). Evaluation of drug-eluting beads versus conventional transcatheter arterial chemoembolization in patients with unresectable hepatocellular carcinoma: a systematic review and meta-analysis. Clin. Res. Hepatol. Gastroenterol. 41, 75–85. doi:10.1016/j.clinre.2016.05.013
Chen, D., Zhang, Y., Meng, L., Lu, L., and Meng, G. (2022). circRNA DENND1B inhibits tumorigenicity of clear cell renal cell carcinoma via miR-122-5p/TIMP2 axis. Open Med. 17, 2085–2097. doi:10.1515/med-2022-0536
Clapham, D. E., Julius, D., Montell, C., and Schultz, G. (2005). International union of pharmacology. XLIX. Nomenclature and structure-function relationships of transient receptor potential channels. Pharmacol. Rev. 57, 427–450. doi:10.1124/pr.57.4.6
Cotterchio, M., Lowcock, E., Bider-Canfield, Z., Lemire, M., Greenwood, C., Gallinger, S., et al. (2015). Association between variants in atopy-related immunologic candidate genes and pancreatic cancer risk. PLoS One 10, e0125273. doi:10.1371/journal.pone.0125273
Criss, W. E., Sapico, V., and Litwack, G. (1970). Rat liver adenosine triphosphate:adenosine monophosphate phosphotransferase activity. J. Biol. Chem. 245, 6346–6351. doi:10.1016/s0021-9258(18)62616-x
Cui, X., Shen, W., Wang, G., Huang, Z., Wen, D., Yang, Y., et al. (2018). Ring finger protein 152 inhibits colorectal cancer cell growth and is a novel prognostic biomarker. Am. J. Transl. Res. 10, 3701–3712.
Duan, W., and Liu, Y. (2018). Targeted and synergistic therapy for hepatocellular carcinoma: monosaccharide modified lipid nanoparticles for the co-delivery of doxorubicin and sorafenib. Drug Des. Dev. Ther. 12, 2149–2161. doi:10.2147/DDDT.S166402
Eliades, N. G., and Eliades, D. G. (2009). Haplotype analysis: software for analysis of haplotype data; genetics and forest tree breeding. Germany: Georg-August University Goettingen. Available online at: https://www.uni-goettingen.de/en/134935.html.
El-Kassas, M., and Elbadry, M. (2022). Hepatocellular carcinoma in Africa: challenges and opportunities. Front. Med. 9, 899420. doi:10.3389/fmed.2022.899420
Fan, Y. P., Liao, J. Z., Lu, Y. Q., Tian, D. A., Ye, F., Zhao, P. X., et al. (2017). MiR-375 and doxorubicin co-delivered by liposomes for combination therapy of hepatocellular carcinoma. Mol. Ther. Nucleic acids 7, 181–189. doi:10.1016/j.omtn.2017.03.010
Faraji, A., Manshadi, H. R. D., Mobaraki, M., Zare, M., and Houshmand, M. (2016). Association of ABCB1 and SLC22A16 gene polymorphisms with incidence of doxorubicin-induced febrile neutropenia: a survey of Iranian breast cancer patients. PLoS One 11, e0168519. doi:10.1371/journal.pone.0168519
Faronato, M., Nguyen, V. T. M., Patten, D. K., Lombardo, Y., Steel, J. H., Patel, N., et al. (2015). DMXL2 drives epithelial to mesenchymal transition in hormonal therapy resistant breast cancer through Notch hyper-activation. Oncotarget 6, 22467–22479. doi:10.18632/oncotarget.4164
Feng, F., Jiang, Q., Jia, H., Sun, H., Chai, Y., Li, X., et al. (2018). Which is the best combination of TACE and Sorafenib for advanced hepatocellular carcinoma treatment? A systematic review and network meta-analysis. Pharmacol. Res. 135, 89–101. doi:10.1016/j.phrs.2018.06.021
Fujisawa, K., Wakazaki, M., Matsuzaki, A., Matsumoto, T., Yamamoto, N., Noma, T., et al. (2022). Adenylate kinase isozyme 3 regulates mitochondrial energy metabolism and knockout alters hela cell metabolism. Int. J. Mol. Sci. 23, 4316. doi:10.3390/ijms23084316
Gándara-Mireles, J. A., Lares-Asseff, I., Reyes Espinoza, E. A., Blanco, J. G., Font, A. E. G., Hurtado, L. P. C., et al. (2021). Association of genetic polymorphisms NCF4 rs1883112, CBR3 rs1056892, and ABCC1 rs3743527 with the cardiotoxic effects of doxorubicin in children with acute lymphoblastic leukemia. Pharmacogenet. Genom 31, 108–115. doi:10.1097/fpc.0000000000000428
Gelderblom, H., Blay, J. Y., Seddon, B. M., Leahy, M., Ray-Coquard, I., Sleijfer, S., et al. (2014). Brostallicin versus doxorubicin as first-line chemotherapy in patients with advanced or metastatic soft tissue sarcoma: an European organisation for research and treatment of cancer soft tissue and bone sarcoma group randomised phase II and pharmacogenetic study. Eur. J. Cancer 50, 388–396. doi:10.1016/j.ejca.2013.10.002
Gkika, D., and Prevarskaya, N. (2009). Molecular mechanisms of TRP regulation in tumor growth and metastasis. Biochim. Biophys. Acta 1793, 953–958. doi:10.1016/j.bbamcr.2008.11.010
Gonzalez-Covarrubias, V., Zhang, J., Kalabus, J. L., Relling, M. V., and Blanco, J. G. (2009). Pharmacogenetics of human carbonyl reductase 1 (CBR1) in livers from black and white donors. Drug Metab. Dispos. Biol. Fate Chem. 37, 400–407. doi:10.1124/dmd.108.024547
Han, T., Yang, X., Zhang, Y., Li, G., Liu, L., Chen, T., et al. (2019). The clinical safety and efficacy of conventional transcatheter arterial chemoembolization and drug-eluting beads-transcatheter arterial chemoembolization for unresectable hepatocellular carcinoma: a meta-analysis. Biosci. Trends 13, 374–381. doi:10.5582/bst.2019.01153
Harris, P. A., Taylor, R., Minor, B. L., Elliott, V., Fernandez, M., O'Neal, L., et al. (2019). The REDCap consortium: building an international community of software platform partners. J. Biomed. Inf. 95, 103208. doi:10.1016/j.jbi.2019.103208
Hashim, M. J., Rizvi, S. S., and Khan, G. (2021). “Hepatocellular carcinoma in the United Arab Emirates,” in Liver cancer in the Middle East. Editor B. I. Carr (Cham, Switzerland: Springer International Publishing), 101–108.
He, H., Zhang, S., Yang, H., Xu, P., Kutschick, I., Pfeffer, S., et al. (2022). Identification of genes associated with liver metastasis in pancreatic cancer reveals PCSK6 as a crucial mediator. Cancers 15, 241. doi:10.3390/cancers15010241
Hertz, D. L., Caram, M. V., Kidwell, K. M., Thibert, J. N., Gersch, C., Seewald, N. J., et al. (2016). Evidence for association of SNPs in ABCB1 and CBR3, but not RAC2, NCF4, SLC28A3 or TOP2B, with chronic cardiotoxicity in a cohort of breast cancer patients treated with anthracyclines. Pharmacogenomics 17, 231–240. doi:10.2217/pgs.15.162
Hou, X. R., Zhang, Z. D., Cao, X. L., and Wang, X. P. (2023). Long noncoding RNAs, glucose metabolism and cancer (Review). Oncol. Lett. 26, 340. doi:10.3892/ol.2023.13925
Hu, X., Yasuda, T., Yasuda-Yosihara, N., Yonemura, A., Umemoto, T., Nakachi, Y., et al. (2023). Downregulation of 15-PGDH enhances MASH-HCC development via fatty acid-induced T-cell exhaustion. JHEP Rep. 5, 100892. doi:10.1016/j.jhepr.2023.100892
Huang, W., Jiang, R., Zeng, R., Ma, Y., Zhang, L., Tong, S., et al. (2023). Rare comorbidity between inflammatory bowel disease and primary biliary cholangitis: evidence from causality, shared genetic architecture and transcriptomics. medRxiv 2023.03.01, 2023–2003. doi:10.1101/2023.03.01.23286611
Ikeda, A., Shimizu, T., Matsumoto, Y., Fujii, Y., Eso, Y., Inuzuka, T., et al. (2014). Leptin receptor somatic mutations are frequent in HCV-infected cirrhotic liver and associated with hepatocellular carcinoma. Gastroenterology 146, 222–232. doi:10.1053/j.gastro.2013.09.025
Karim, H., Hashemi, J., Larsson, C., Moshfegh, A., Fotoohi, A. K., and Albertioni, F. (2011). The pattern of gene expression and gene dose profiles of 6-Mercaptopurine- and 6-Thioguanine-resistant human leukemia cells. Biochem. Biophys. Res. Commun. 411, 156–161. doi:10.1016/j.bbrc.2011.06.120
Kassner, N., Huse, K., Martin, H. J., Gödtel-Armbrust, U., Metzger, A., Meineke, I., et al. (2008). Carbonyl reductase 1 is a predominant doxorubicin reductase in the human liver. Drug Metab. Dispos. 36, 2113–2120. doi:10.1124/dmd.108.022251
Kciuk, M., Gielecińska, A., Mujwar, S., Kołat, D., Kałuzińska-Kołat, Ż., Celik, I., et al. (2023). Doxorubicin-an agent with multiple mechanisms of anticancer activity. Cells 12, 659. doi:10.3390/cells12040659
Klepinin, A., Zhang, S., Klepinina, L., Rebane-Klemm, E., Terzic, A., Kaambre, T., et al. (2020). Adenylate kinase and metabolic signaling in cancer cells. Front. Oncol. 10, 660. doi:10.3389/fonc.2020.00660
Konyn, P., Ahmed, A., and Kim, D. (2021). Current epidemiology in hepatocellular carcinoma. Expert Rev. Gastroenterol. Hepatol. 15, 1295–1307. doi:10.1080/17474124.2021.1991792
Lal, S., Sutiman, N., Ooi, L. L., Wong, Z. W., Wong, N. S., Ang, P. C. S., et al. (2016). Pharmacogenetics of ABCB5, ABCC5 and RLIP76 and doxorubicin pharmacokinetics in Asian breast cancer patients. Pharmacogenomics J. 17, 337–343. doi:10.1038/tpj.2016.17
Lencioni, R., and Llovet, J. (2010). Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin. Liver Dis. 30, 52–60. doi:10.1055/s-0030-1247132
Li, L., Zhang, J. W., Jenkins, G., Xie, F., Carlson, E. E., Fridley, B. L., et al. (2016). Genetic variations associated with gemcitabine treatment outcome in pancreatic cancer. Pharmacogenet Genom 26, 527–537. doi:10.1097/FPC.0000000000000241
Li, Z., Su, D., Ying, L., Yu, G., and Mao, W. (2017). Study on expression of CDH4 in lung cancer. World J. Surg. Oncol. 15, 26. doi:10.1186/s12957-016-1083-2
Li, W. F., Liu, Y. W., Wang, C. C., Yong, C. C., Lin, C. C., and Yen, Y. H. (2023a). Radiographic tumor burden score is useful for stratifying the overall survival of hepatocellular carcinoma patients undergoing resection at different Barcelona clinic liver cancer stages. Langenbeck's Arch. Surg. 408, 169. doi:10.1007/s00423-023-02869-6
Li, C. J., Tsai, H. W., Chen, Y. L., Wang, C. I., Lin, Y. H., Chu, P. M., et al. (2023b). Cisplatin or doxorubicin reduces cell viability via the PTPIVA3-JAK2-STAT3 cascade in hepatocellular carcinoma. J. Hepatocell. Carcinoma 10, 123–138. doi:10.2147/JHC.S385238
Li, C., Guo, Z., Liu, F., An, P., Wang, M., Yang, D., et al. (2023c). PCSK6 attenuates cardiac dysfunction in doxorubicin-induced cardiotoxicity by regulating autophagy. Free Radic. Biol. Med. 203, 114–128. doi:10.1016/j.freeradbiomed.2023.04.005
Liang, K. H., Lin, C. L., Chen, S. F., Chiu, C. W., Yang, P. C., Chang, M. L., et al. (2016). GALNT14 genotype effectively predicts the therapeutic response in unresectable hepatocellular carcinoma treated with transcatheter arterial chemoembolization. Pharmacogenomics 17, 353–366. doi:10.2217/pgs.15.179
Liu, X., Invernizzi, P., Lu, Y., Kosoy, R., Lu, Y., Bianchi, I., et al. (2010). Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat. Genet. 42, 658–660. doi:10.1038/ng.627
Liu, B., Huang, J. W., Li, Y., Hu, B. S., He, X., Zhao, W., et al. (2015). Single-agent versus combination doxorubicin-based transarterial chemoembolization in the treatment of hepatocellular carcinoma: a single-blind, randomized, phase II trial. Oncology 89, 23–30. doi:10.1159/000371522
Liu, G., Ouyang, Q., Xia, F., Fan, G., Yu, J., Zhang, C., et al. (2019). Alpha-fetoprotein response following transarterial chemoembolization indicates improved survival for intermediate-stage hepatocellular carcinoma. HPB 21, 107–113. doi:10.1016/j.hpb.2018.06.1800
Ma, Z., Guo, D., Wang, Q., Liu, P., Xiao, Y., Wu, P., et al. (2019). Lgr5-mediated p53 Repression through PDCD5 leads to doxorubicin resistance in Hepatocellular Carcinoma. Theranostics 9, 2967–2983. doi:10.7150/thno.30562
Masior, Ł., Krasnodębski, M., Kuncewicz, M., Karaban, K., Jaszczyszyn, I., Kruk, E., et al. (2023). Alpha-fetoprotein response after first transarterial chemoembolization (TACE) and complete pathologic response in patients with hepatocellular cancer. Cancers 15, 3962. doi:10.3390/cancers15153962
Mathias, C., Groeneveld, C. S., Trefflich, S., Zambalde, E. P., Lima, R. S., Urban, C. A., et al. (2021). Novel lncRNAs co-expression networks identifies LINC00504 with oncogenic role in luminal A breast cancer cells. Int. J. Mol. Sci. 22, 2420. doi:10.3390/ijms22052420
McLaren, W., Gil, L., Hunt, S. E., Riat, H. S., Ritchie, G. R. S., Thormann, A., et al. (2016). The ensembl variant effect predictor. Genome Biol. 17, 122. doi:10.1186/s13059-016-0974-4
Micallef, I., and Baron, B. (2021). The mechanistic roles of ncRNAs in promoting and supporting chemoresistance of colorectal cancer. Non-Coding RNA 7, 24. doi:10.3390/ncrna7020024
Mordente, A., Minotti, G., Martorana, G. E., Silvestrini, A., Giardina, B., and Meucci, E. (2003). Anthracycline secondary alcohol metabolite formation in human or rabbit heart: biochemical aspects and pharmacologic implications. Biochem. Pharmacol. 66, 989–998. doi:10.1016/s0006-2952(03)00442-8
Moreira, F., Kiehl, T. R., So, K., Ajeawung, N. F., Honculada, C., Gould, P., et al. (2011). NPAS3 demonstrates features of a tumor suppressive role in driving the progression of Astrocytomas. Am. J. Pathol. 179, 462–476. doi:10.1016/j.ajpath.2011.03.044
Nagy, M., Attya, M., and Patrinos, G. P. (2020). Unraveling heterogeneity of the clinical pharmacogenomic guidelines in oncology practice among major regulatory bodies. Pharmacogenomics 21, 1247–1264. doi:10.2217/pgs-2020-0056
Nookala, R. K., Langemeyer, L., Pacitto, A., Ochoa-Montaño, B., Donaldson, J. C., Blaszczyk, B. K., et al. (2012). Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2, 120071. doi:10.1098/rsob.120071
Ntzani, E. E., Liberopoulos, G., Manolio, T. A., and Ioannidis, J. P. A. (2011). Consistency of genome-wide associations across major ancestral groups. Hum. Genet. 131, 1057–1071. doi:10.1007/s00439-011-1124-4
Nyangwara, V. A., Mazhindu, T., Chikwambi, Z., Masimirembwa, C., Campbell, T. B., Borok, M., et al. (2024). Cardiotoxicity and pharmacogenetics of doxorubicin in black Zimbabwean breast cancer patients. Br. J. Clin. Pharmacol. 90, 1782–1789. doi:10.1111/bcp.15659
Pugazhendhi, A., Edison, T. N. J. I., Velmurugan, B. K., Jacob, J. A., and Karuppusamy, I. (2018). Toxicity of doxorubicin (Dox) to different experimental organ systems. Life Sci. 200, 26–30. doi:10.1016/j.lfs.2018.03.023
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Qin, F., Lao, L., Huang, M., Tan, H., Jin, X., Ma, X., et al. (2020). Evaluation of the TRPM protein family as potential biomarkers for various types of human cancer using public database analyses. Exp. Ther. Med. 20, 770–785. doi:10.3892/etm.2020.8739
Ren, X., Su, D., Shi, D., and Xiang, X. (2023). The improving strategies and applications of nanotechnology-based drugs in hepatocellular carcinoma treatment. Front. Bioeng. Biotechnol. 11, 1272850. doi:10.3389/fbioe.2023.1272850
Ruiz-Pinto, S., Martin, M., Pita, G., Caronia, D., de la Torre-Montero, J. C., Moreno, L. T., et al. (2018). Pharmacogenetic variants and response to neoadjuvant single-agent doxorubicin or docetaxel: a study in locally advanced breast cancer patients participating in the NCT00123929 phase 2 randomized trial. Pharmacogenet. Genom 28, 245–250. doi:10.1097/fpc.0000000000000354
Rumgay, H., Ferlay, J., de Martel, C., Georges, D., Ibrahim, A. S., Zheng, R., et al. (2022). Global, regional and national burden of primary liver cancer by subtype. Eur. J. Cancer 161, 108–118. doi:10.1016/j.ejca.2021.11.023
Schmit, K., and Michiels, C. (2018). TMEM proteins in cancer: a review. Front. Pharmacol. 9, 1345. doi:10.3389/fphar.2018.01345
Shan, Y., and Li, P. (2019). Long intergenic non-protein coding RNA 665 regulates viability, apoptosis, and autophagy via the MiR-186-5p/MAP4K3 Axis in hepatocellular carcinoma. Yonsei Med. J. 60, 842–853. doi:10.3349/ymj.2019.60.9.842
Shi, J., Cheng, C., Ma, J., Liew, C. C., and Geng, X. (2018). Gene expression signature for detection of gastric cancer in peripheral blood. Oncol. Lett. 15, 9802–9810. doi:10.3892/ol.2018.8577
Singal, A. G., Llovet, J. M., Yarchoan, M., Mehta, N., Heimbach, J. K., Dawson, L. A., et al. (2023). AASLD practice guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatol. Baltim. Md 78, 1922–1965. doi:10.1097/HEP.0000000000000466
Suur, B. E., Chemaly, M., Jin, H., Kronqvist, M., Lengquist, M., Van Der Laan, S. W., et al. (2021). Proprotein convertase subtilisin/kexin 6 is involved in lipid metabolism in liver and adipose tissue. Atherosclerosis 331, e4. doi:10.1016/j.atherosclerosis.2021.06.016
Tam, K. (2013). The roles of doxorubicin in hepatocellular carcinoma. ADMET and DMPK 1, 29–44. doi:10.5599/admet.1.3.7
Thermo Fisher Scientific (2024). Axiom™ precision medicine diversity array plus. Available online at: https://www.thermofisher.com/order/catalog/product/951961 (accessed on January 1, 2024).
Thorn, C. F., Klein, T. E., and Altman, R. B. (2013). PharmGKB: the pharmacogenomics knowledge base. Pharmacogenomics Methods Protoc. 1015, 311–320. doi:10.1007/978-1-62703-435-7_20
Ueda, R., Hashimoto, R., Fujii, Y., Menezes, J. C. J. M. D. S., Takahashi, H., Takeda, H., et al. (2024). Membrane-associated ubiquitin ligase RING finger protein 152 orchestrates melanogenesis via tyrosinase ubiquitination. Membranes 14, 43. doi:10.3390/membranes14020043
Uhlen, M., Zhang, C., Lee, S., Sjöstedt, E., Fagerberg, L., Bidkhori, G., et al. (2017). A pathology atlas of the human cancer transcriptome. Science 357, eaan2507. doi:10.1126/science.aan2507
Valter, A., Luhari, L., Pisarev, H., Truumees, B., Planken, A., Smolander, O. P., et al. (2023). Genomic alterations as independent prognostic factors to predict the type of lung cancer recurrence. Gene 885, 147690. doi:10.1016/j.gene.2023.147690
van der Zanden, S. Y., Qiao, X., and Neefjes, J. (2021). New insights into the activities and toxicities of the old anticancer drug doxorubicin. FEBS J. 288, 6095–6111. doi:10.1111/febs.15583
Wan, J., Liu, S., Sun, W., Yu, H., Tang, W., Liu, W., et al. (2021). Ring finger protein 152-dependent degradation of TSPAN12 suppresses hepatocellular carcinoma progression. Front. Pharmacol. 21, 122. doi:10.1186/s12935-021-01806-1
Wang, J., Wong, C. H., Zhu, Y., Yao, X., Ng, K. K. C., Zhou, C., et al. (2023). Identification of GRIN2D as a novel therapeutic target in pancreatic ductal adenocarcinoma. Biomark. Res. 11, 74. doi:10.1186/s40364-023-00514-4
Whirl-Carrillo, M., McDonagh, E. M., Hebert, J. M., Gong, L., Sangkuhl, K., Thorn, C. F., et al. (2012). Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92, 414–417. doi:10.1038/clpt.2012.96
Whirl-Carrillo, M., Huddart, R., Gong, L., Sangkuhl, K., Thorn, C. F., Whaley, R., et al. (2021). An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 110, 563–572. doi:10.1002/cpt.2350
Windsor, R. E., Strauss, S. J., Kallis, C., Wood, N. E., and Whelan, J. S. (2011). Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: a pilot study. Cancer 118, 1856–1867. doi:10.1002/cncr.26472
Witte, J. S., Elston, R. C., and Schork, N. J. (1996). Genetic dissection of complex traits. Nat. Genet. 12 (4), 355–356. doi:10.1038/ng0496-355
Wojnowski, L., Kulle, B., Schirmer, M., Schlüter, G., Schmidt, A., Rosenberger, A., et al. (2005). NAD(P)H oxidase and multidrug resistance protein genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Circulation 112, 3754–3762. doi:10.1161/circulationaha.105.576850
Xie J., J., Lan, T., Zheng, D. L., Ding, L. C., and Lu, Y. G. (2023). CDH4 inhibits ferroptosis in oral squamous cell carcinoma cells. BMC Oral Health 23, 329. doi:10.1186/s12903-023-03046-3
Xie Z., Z., Huang, J., Li, Y., Zhu, Q., Huang, X., Chen, J., et al. (2023). Single-cell RNA sequencing revealed potential targets for immunotherapy studies in hepatocellular carcinoma. Sci. Rep. 13, 18799. doi:10.1038/s41598-023-46132-w
Yu, B., Xing, Z., Tian, X., and Feng, R. (2024). A prognostic risk signature of two autophagy-related genes for predicting triple-negative breast cancer outcomes. Breast Cancer (Dove Med. Press) 16, 529–544. doi:10.2147/BCTT.S475007
Zhang, Q., Fan, L., Hou, F., Dong, A., Wang, Y.-X., and Tong, Y. (2015). New insights into the RNA-binding and E3 ubiquitin ligase activities of roquins. Sci. Rep. 5, 15660. doi:10.1038/srep15660
Zhang, Y. I., Ran, Y. A. N., Xiong, Y. A. N., Zhong, Z. B., Wang, Z. H., Fan, X. L., et al. (2016). Effects of TMEM9 gene on cell progression in hepatocellular carcinoma by RNA interference. Oncol. Rep. 36, 299–305. doi:10.3892/or.2016.4821
Zhang, Y. G., Jin, M. Z., Zhu, X. R., and Jin, W. L. (2022). Reclassification of hepatocellular cancer with neural-related genes. Front. Oncol. 12, 877657. doi:10.3389/fonc.2022.877657
Zhang, Z., Chen, W., Luo, C., and Zhang, W. (2022). Exploring a four-gene risk model based on doxorubicin resistance-associated lncRNAs in hepatocellular carcinoma. Front. Pharmacol. 13, 1015842. doi:10.3389/fphar.2022.1015842
Zhang, Z. (2020). Silencing LINC00504 inhibits cell proliferation, invasion as well as migration and promotes cell apoptosis in lung cancer cells via upregulating miR-876-3p. Cytotechnology 72, 807–817. doi:10.1007/s10616-020-00424-5
Keywords: genome-wide association study, hepatocellular carcinoma, doxorubicin, transarterial chemoembolization, array genotyping
Citation: Shilbayeh SAR, Khedr NF, Alshabeeb MA, Alsaleh AA, Alanizi AH, Abd El-Baset OA and Werida RH (2025) Genetic polymorphisms as predictors of the response of hepatocellular carcinoma patients to doxorubicin chemotherapy: a genome-wide association study. Front. Pharmacol. 16:1604473. doi: 10.3389/fphar.2025.1604473
Received: 01 April 2025; Accepted: 14 May 2025;
Published: 04 June 2025.
Edited by:
Khaled Abdelkawy Ibrahim, Kafrelsheikh University, EgyptReviewed by:
Yazun Jarrar, Al-Balqa Applied University, JordanAhmed Amin, Kafrelsheikh University, Egypt
Copyright © 2025 Shilbayeh, Khedr, Alshabeeb, Alsaleh, Alanizi, Abd El-Baset and Werida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sireen Abdul Rahim Shilbayeh, c3NhYmR1bHJhaGltQHBudS5lZHUuc2E=
†ORCID: Sireen Abdul Rahim Shilbayeh, orcid.org/0000-0002-5020-6838; Naglaa F. Khedr, orcid.org/0000-0003-4539-8940; Mohammad A. Alshabeeb, orcid.org/0000-0002-8885-6254; Abdulmonem Ali Alsaleh, orcid.org/0000-0002-1188-1451; Abdalrhman Hamdan Alanizi, orcid.org/0000-0002-0754-7493; Rehab H. Werida, orcid.org/0000-0002-5983-3993
 Sireen Abdul Rahim Shilbayeh
Sireen Abdul Rahim Shilbayeh Naglaa F. Khedr
Naglaa F. Khedr Mohammad A. Alshabeeb
Mohammad A. Alshabeeb Abdulmonem Ali Alsaleh4†
Abdulmonem Ali Alsaleh4† Omnia A. Abd El-Baset
Omnia A. Abd El-Baset Rehab H. Werida
Rehab H. Werida