- 1College of Pharmacy, Dali University, Dali, China
- 2Yunnan Key Laboratory of Southern Medicine Utilization, Yunnan Branch of Institute of Medicinal Plant Development Chinese Academy of Medical Sciences, Peking Union Medical College, Jinghong, China
Background: Tetrastigma plants are widely utilized in traditional medicine (such as Tetrastigma. obtectum and Tetrastigma. serrulatum, two important commonly medicinal plants), primarily for their properties in promoting blood circulation, strengthening bones and tendons, and so on. However, the high diversity of species differentiation poses a challenge in accurately identifying the various Tetrastigma species without specialized taxonomic knowledge.
Materials and methods: To screen the candidate barcode sequences of Tetrastigma species, we first report the complete chloroplasts (CP) genomes of T. obtectum and T. serrulatum obtained via high throughput Illumina sequencing and compare them with fourteen previously sequenced species. Furthermore, we collected fresh leaf samples from T. obtectum and T. serrulatum (totally 37 samples) and evaluated the discriminatory efficacy of the nuclear DNA Internal Transcribed Spacer 2 (ITS2) fragment through comparative analysis of sequence variations and secondary structures. Finally, to analyze the phylogenetic position of Tetrastigma species, we constructed a Maximum Likelihood (ML) phylogenetic tree using CP genome sequences of 46 species from seven genera within the Vitaceae family.
Results and discussion: The CP genomes of Tetrastigma exhibited a typical circular tetramerous structure, including a large single-copy region (LSC) (87,381–88,979 bp), a small single-copy region (SSC) (18,649–19,339 bp), and a pair of inverted repeats (IRa and IRb) (26,288–26,934 bp). The guanine-cytosine content of the CP genomes is 37.35%–37.62%. The codon usage shows a significant preference for end with A/T. Then, the results of nucleotide diversity analysis showed that ten polymorphic hotspots (psbM-trnD-GUC, ndhF-rpl32, trnS-GCU-trnG-UCC, ycf1, rpl32-trnL-UAG, trnS-UGA-psbZ, psbE-petL, matK-rps16, rpl16, and rpl22) could be the candidate DNA marker suitable for Tetrastigma species. Furthermore, our results demonstrate that the ITS2 sequence could effectively discriminate T. obtectum and T. serrulatum, whereas the secondary structure cannot, proving that ITS2 can be used as an efficient barcode fragment to accurately identify the two species. The aim of this study was not only to determine the identification efficiency of the CP genome and ITS2 for T. obtectum and T. serrulatum but also to clarify the phylogenetic relationship and screen the candidate DNA marker suitable for Tetrastigma species, provide valuable data support for further accurate identification of the Tetrastigma genus.
1 Introduction
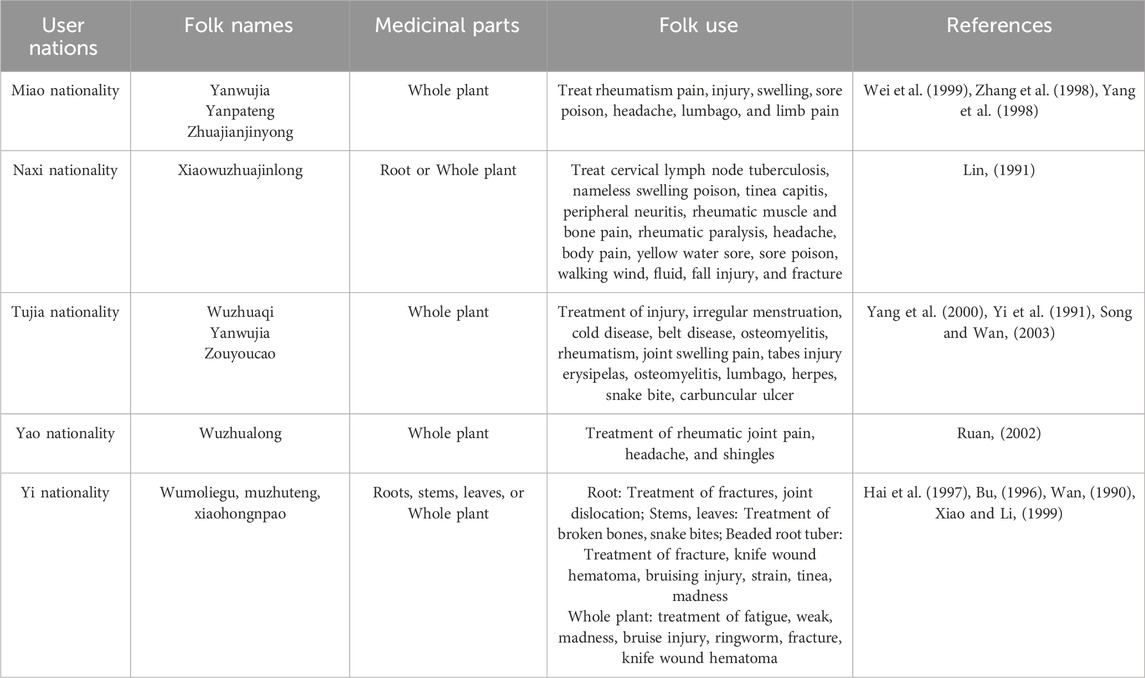
Tetrastigma obtectum (Wall.) Planch. Belongs to the genus Tetrastigma (Miq.) Planch. (Vitaceae), is an evergreen liana plant found mainly in China, Bhutan, Nepal, and Vietnam. It is a crucial commonly medicinal plant used in folk, which primarily had the functions of promoting blood circulation and clearing collaterals, reinforcing bones and tendons, clearing heat and cooling blood, etc., and had different applications in different nationalities (Table 1), such as Miao nationality, Naxi nationality, Tujia nationality, Yao nationality, and Yi nationality. However, due to the high diversity of species differentiation and the relatively concentrated distribution area of the species, it is difficult to accurately identify the genus without all the species knowledge in a relatively large area, and the phenomenon of invalid naming is very likely to occur (Li and Wu, 1995). The same is true for T. obtectum, especially for medicinal materials often found on the market. During our field investigation, we found that T. obtectum is usually confused with similar plant species of the same genus in the medicinal market, such as Tetrastigma serrulatum (Roxb.) Planch, residents often confuse the two species because of their similar appearance, similar distribution areas, similar habitat environment, and primary distribution in multi-ethnic regions. T. serrulatum could also be used as a medicinal material, and it appears to have a similar medicinal value to T. obtectum. Previous studies of our research group showed significant differences between T. serrulatum and T. obtectum in terms of compound composition, active ingredients, and pharmacological activities (The data has not yet been published). If the two are confused, it will inevitably affect the clinical application effect and have a non-negligible adverse impact on the patient’s health recovery and the further development of the medicinal material industry of the two species.
As one of the important organelles in green plants, chloroplasts (CP) responsible for metabolic and photosynthetic processes, provide essential energy for plant growth and development (Daniell et al., 2016; Dobrogojski et al., 2020). Because of the relatively conserved genome structure characteristics that with a closed circular DNA molecule composed of a typical quadripartite structure: a large single-copy region (LSC), a small single-copy region (SSC), and a pair of mirrored inverted repeat sequences (IRa and IRb) (Sato et al., 1999; Ferrarini et al., 2013; Ahmad et al., 2022), CP genomes are often used for revealing evolutionary patterns and phylogenetic relationships of interspecies and intraspecies (Fan and Ma, 2022; Wicke et al., 2011; Jin and Daniell, 2015; Daniell et al., 2021). Researchers have demonstrated that the evolutionary rate of synonymous substitution in the CP genome is half that of the nuclear genome (Wang et al., 2024). Furthermore, the whole CP genome as a super-barcode has been widely used in species identification (Zhang et al., 2024; Chen et al., 2010), and the sequences selected from the highly-variation regions of the whole CP genome have been used for distinguishing closely related species and counterfeit medicinal materials, especially in tightly related taxa (Li et al., 2015; Yang et al., 2021; Zhang et al., 2024). In addition, the cognomen, characterized by maternal inheritance, is an ideal molecular data resource widely employed in research fields such as genetic engineering, breeding, and evolutionary biology (Wei et al., 2021; Dong et al., 2012). Therefore, investigating the CP genome is important for revealing evolutionary patterns, phylogenetic relationships, species identification, molecular breeding, and so on (Jin and Daniell, 2015; Daniell et al., 2021).
In addition, DNA barcoding has been applied more and more widely because of the overcoming of the limitations of traditional identification methods and allowed accurate identification of materials with similar morphologies and chemical structures (Gregory, 2005). In various DNA barcodes, the ribosomal DNA Internal Transcribed Spacer 2 (ITS2) sequence has been widely used to identify plant species because of its universality and specificity (Chen et al., 2023). Chen et al. (2010) first proposed the ITS2 region as an efficient barcoding tool for medicinal plants and their closely related species due to its relatively short length, consistent performance in distinguishing closely related species, and ease of amplification with a single set of universal primers, they also proposed that ITS2 can serve as a novel universal barcode for the identification of a broader range of plant taxa, they tested the discrimination ability of ITS2 in more than 6,600 plant samples belonging to 4,800 species from 753 distinct genera. They found that the rate of successful identification with the ITS2 was 92.7% at the species level. Yao et al. (2010) subsequently proposed ITS2 as a DNA barcode for plant and animal medicine based on an investigation of delimitation ability in 50,790 plants and 12,221 animals. The research results show that the success rates of the ITS2 region in identifying different taxonomic groups were 67.1%–91.7% at the species level (Yao et al., 2010). In sum, ITS2 remains the best and most widely used single-locus barcode for the delimitation and identification of medicinal plants, as it is easy to amplify and has enough variability to distinguish even closely related species (Han et al., 2016; Hu et al., 2022; Yao et al., 2022; Ma et al., 2017), despite the peculiarity of multiple-copy sequence and the identification ability differs among species and taxonomic levels (Sokołowska et al., 2022). Moreover, research on molecular markers aids in identifying the sources of adulterated medicinal materials, thus addressing market chaos and the challenges in tracing the origins of these materials. This has a significant positive impact on the safety of clinical medications and the recovery of patients’ health.
Here, we first sequenced the complete CP genomes of T. obtectum and its adulterant species T. serrulatum, using the Illumina HiSeq4000 sequencing platform. We assembled and annotated the CP genomes of the 2 Tetrastigma species. We compared them with 14 previously sequenced Tetrastigma species, examining their structural differences and identifying polymorphic hotspots suitable for candidate DNA marker development. Furthermore, we analyzed the published CP genomes of the Vitaceae family, constructing phylogenetic trees based on methods such as Maximum Likelihood (ML), and examined their phylogenetic relationships. Finally, we evaluated the identification efficiency of ITS2 fragments for 37 collected T. obtectum and T. serrulatum samples (17 and 20 samples, respectively). This study aimed to determine the identification efficiency of CP genome and DNA barcode fragment ITS2 for T. obtectum and T. serrulatum, and to clarify the relationship between the phylogenetic analysis.
2 Materials and methods
2.1 Plant material, DNA extraction and sequencing
T. obtectum and T. serrulatum collected in Yunnan. Fresh green leaves were sampled, washed, dried, and stored at −80°C till DNA extraction. The voucher specimens were deposited in the herbarium, Yunnan branch of the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences herbarium (voucher numbers: IMDY2022091005, IMDY2022100011) and identified by Zhonglian Zhang. Total genomic DNA was extracted from fresh leaves using the TIANGEN plant Genomic DNA kit (Tianjin Biotech, Beijing, Co., Ltd.) The purity of total DNA was evaluated using electrophoresis on 1.0% agarose gels. And the concentration was measured using a Nanodrop spectrophotometer 2000 (Thermo Fisher Scientific Inc., Waltham, MA, United States). The OD260/280 value ranges from 1.8 to 2.2, and ≥2 µg was equally pooled from individuals that could be used to construct the library. The DNA was sheared into fragments at approximately 500 bp long using the Covaris M220 focused ultrasonicator (Covaris, Woburn, MA, United States) for paired-end library construction. The DNA library was constructed using the Illumina TruSeq™ Nano DNA Sample Prep Kit (Illumina, San Diego, CA, United States). The library was sequenced using the Illumina HiSeq4000 sequencing platform at Biozeron (Shanghai, China).
2.2 Genome assembly and annotations
The number of raw reads obtained for T. obtectum and T. serrulatum were checked (Q ≥ 25) using the Fast QC Toolkit (Brown et al., 2017). Low quality reads were filtered out from the raw data to obtain high-quality data (clean reads) for subsequent analysis, with the published CP genome of Tetrastigma species T. hemsleyanum and T. planicaule as the reference sequence. The CP genome was De novo assembled using Get Organelle (Jin et al., 2020), perform local assembly and optimization of the assembly results, as well as internal hole repairs. Producing circular assemblies for T. obtectum and T. serrulatum that conform to the quadripartite structure of CP genomes. The filtered files were visualized in Bandage v.0.8.1 (Wick et al., 2015). Bowite 2 in Geneious v.9.0.2 (Kearse et al., 2012) was used to align the raw sequence to the assembled CP genome to verify the assembly results. Then, the reference genome was used to correct the starting position of the CP assembly sequence and determine the position and direction of 4 CP regions (LSC, IRa, SSC, and IRb). The assembly results were imported into Geneiousv.9.0.2 (Kearse et al., 2012) for annotation, and annotations were manually refined using the Geneiousv.9.0.2 (Kearse et al., 2012). Then, the CP genome map was drawn using the online website (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Lohse et al., 2013). At the default Settings, tRNAscanSE (Lowe and Chan, 2016) and DOGMA (Wyman et al., 2004) were used to further validate tRNA genes. Finally, we obtained the gb. File and submit our report to the NCBI. Whole CP genome sequences of T. obtectum and T. serrulatum were deposited into GenBank with entry numbers PV454981, PV454982, respectively.
2.3 Sequence analysis and repeat sequence analysis
Use CodonW software (University of Texas, Houston, TX, United States) to analyze codon usage and relative synonymous codon usage (RSCU), calculate RSCU values, and visualize (Sharp and Li, 1987). Use MEGA X to analyze guanine-cytosine (GC) content (Kumar et al., 2018). The MISA-web (http://pgrc.ipk-gatersleben.de/misa/) was used to identify Simple Sequence Repeats (SSRs) in the T. obtectum and T. serrulatum CP genomes, with the parameters set to encompass mononucleotide SSRs with 10 repeat units, di- and tri-nucleotide SSRs with 5 and 4 repeat units, respectively, and tetra-, penta-, and hexa-nucleotide SSRs with 3 repeat units. The Long repeat sequences of T. obtectum and T. serrulatum were detected by REPuter, including forward, palindromic, reverse, and complementary repeats (Kurtz et al., 2001).
2.4 Genome comparison and sequence divergence analyses
The mVISTA (v2.0) program in Shuffle-LAGAN mode was used for Tetrastigma species CP genome comparison analysis (Frazer et al., 2004). The whole CP genomes were initially aligned using the MAFFT software (Katoh et al., 2019). Then, we used DnaSP 6 (Rozas et al., 2017) software to calculate the nucleotide variability (Pi) with a sliding window of 600 bp and a step size of 200 bp.200 bp step size and a 600 bp window length and the results were visualized using Excel. IRScope software was used to detect the contraction and expansion of the IR/SC region boundaries (Amiryousefi et al., 2018).
2.5 ITS2 barcode and secondary structure identification
We collected fresh leaf samples from T. obtectum (17 samples) and T. serrulatum (20 samples) in Yunnan and identified the two species using conventional barcode ITS2 amplification sequencing. The sample number and location information are listed in Supplementary Table S1. Construct the secondary structures of the ITS2 base sequences of T. obtectum and T. serrulatum obtained from sequencing using the online tool available at https://www.vectorbuilder.cn/tool/dna-secondary-structure.html. Analyze the differences in their secondary structures by comparing the remaining parameters using an energy minimization-based dynamic programming algorithm.
2.6 Phylogenetic analysis
To investigate the phylogenetic relationships within the genus Tetrastigma and its placement within the family Vitaceae, we constructed a Maximum Likelihood (ML) phylogenetic tree using CP genome sequences of 46 species from seven genera within the Vitaceae family (Tetrastigma, Parthenocissus, Vitis, Ampelopsis, Ampelocissus, Cissus, and Leea), with Celastrus vaniotii and Tripterygium wilfordii serving as outgroups. The whole CP genomes were initially aligned using the MAFFT software (Katoh et al., 2019). The aligned sequences were then imported into MEGA11 software (Tamura et al., 2021) to determine the best DNA model maximum likelihood (ML). And the best-fit substitution models were selected by ModelTest-NG (Darriba et al., 2020) for ML trees.
3 Results and discussion
3.1 Chloroplast genome features of Tetrastigma species
We organized and analyzed the CP genome characteristics of T. obtectum and T. serrulatum. The whole genome sequence lengths of T. obtectum and T. serrulatum CP are 159,478 and 160,515 bp, respectively, both of which have a typical quadripartite structure (Figure 1). The complete CP genomes of T. obtectum and T. serrulatum consist of a pair of inverted repeats (IRa and IRb) measuring 26,526 bp and 26,480 bp, respectively, an SSC region of 19,045 bp and 19,137 bp, and an LSC region of 87,381 bp and 88,418 bp. The CP genome of T. serrulatum is 1,037 bp longer than that of T. obtectum. In the whole CP genomes of two Tetrastigma species, a total of 131 genes were detected, including 86 different protein-coding genes, 8 different rRNA genes, and 37 different tRNA genes. The gene distribution of the two CP genomes is identical: the LSC region has a total of 81 genes, including 60 protein-coding genes and 21 tRNA genes, while the SSC region contains 11 protein-coding genes and 1 tRNA gene; the IR region replicates a total of 20 genes, including 8 protein-coding genes, 8 tRNA genes, and 4 rRNA genes (Figure 1; Supplementary Table S2).
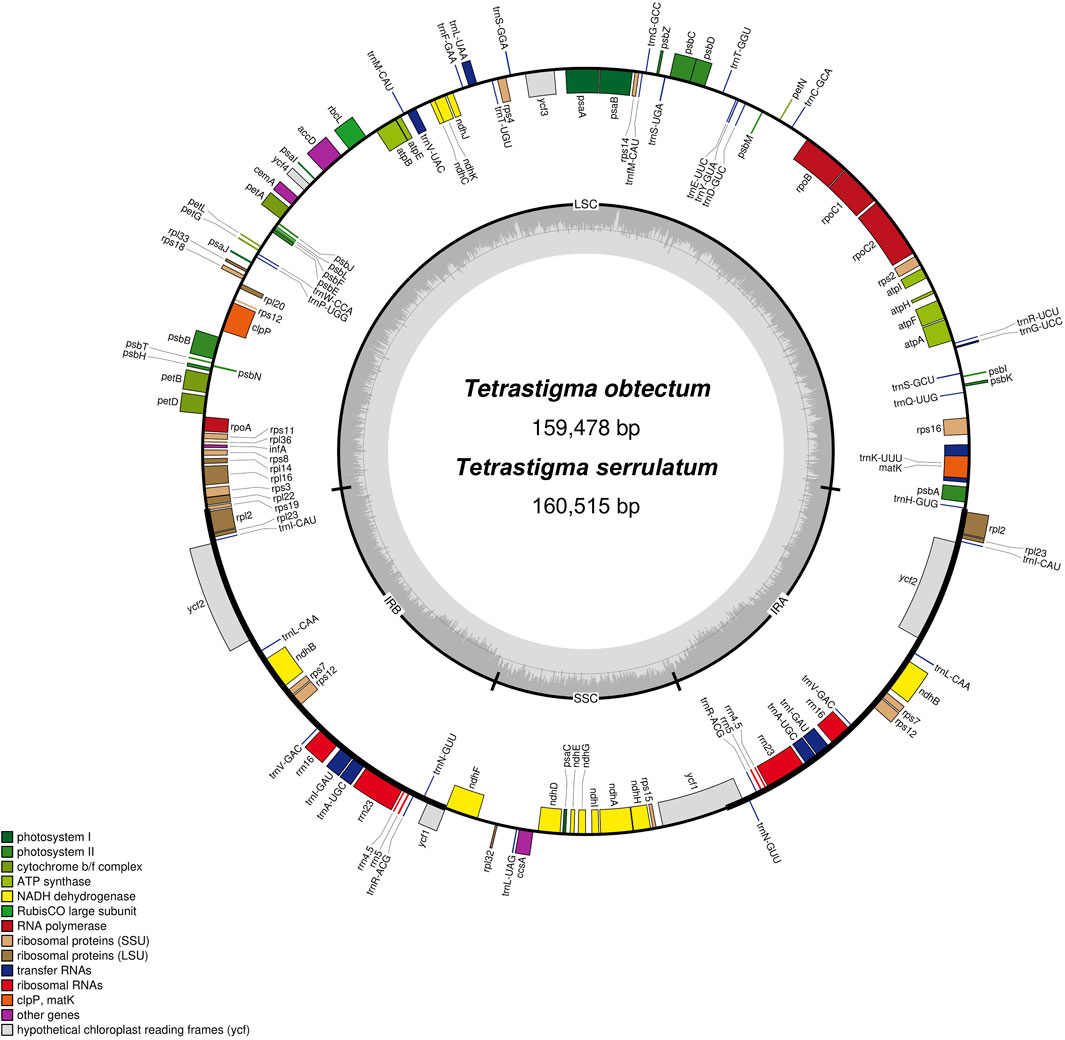
Figure 1. Gene map of two Tetrastigma complete chloroplast genomes. Genes on the inside of the outer circle are transcribed in a clockwise direction, while genes on the outside of the outer circle are transcribed in a counterclockwise direction. Genes belonging to different functional categories are different color-coded. The inner circle indicates the range of the LSC, SSC, and IRs. Also, the darker gray area in the inner circle corresponds to the GC content, whereas the lighter gray area corresponds to the AT content.
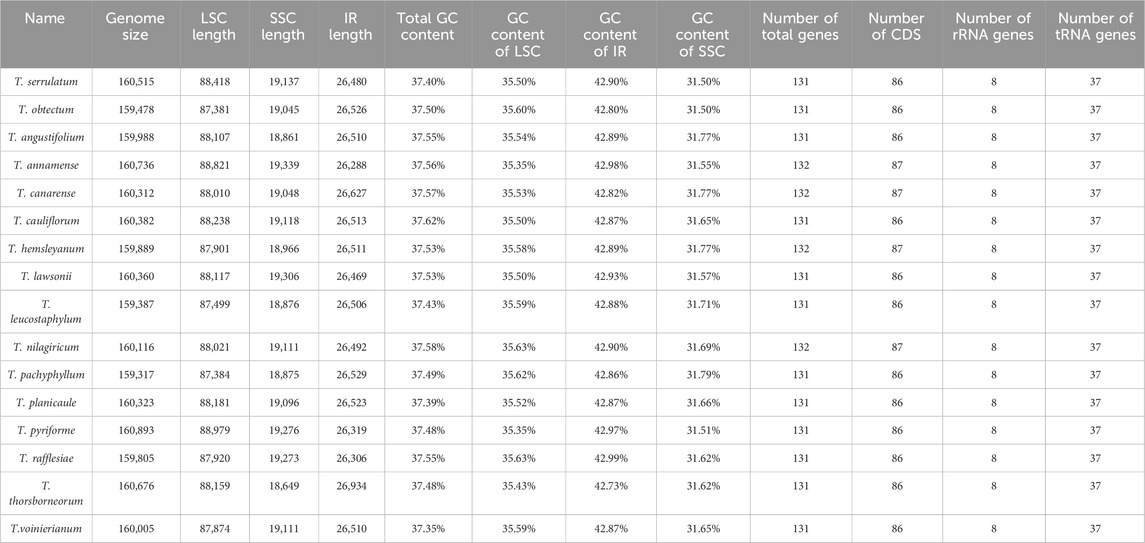
Additionally, the CP genomes of 14 other Tetrastigma species were downloaded from NCBI, allowing for the analysis of 16 species. All CP genomes feature a characteristic quadripartite structure comprising a large single-copy region (LSC), two inverted repeats (IRs), and a small single-copy region (SSC). The chloroplast (CP) genome sizes of the 16 Tetrastigma species (Table 2) range from 159,317 base pairs (bp) in T. pachyphyllum to 160,893 bp in T. pyriforme. The total GC content varies from 37.35% in T. voinierianum to 37.62% in T. cauliflorum. The length of the LSC region ranges from 87,381 bp in T. obtectum to 88,979 bp in T. pyriforme. The IR region varies from 26,288 bp in T. annamense to 26,934 bp in T. thorsborneorum, while the SSC region ranges from 18,649 bp in T. thorsborneorum to 19,339 bp in T. annamense. The corresponding GC content is 35.34%–35.63%, 42.73%–42.99%, and 31.41%–31.79%, respectively.
3.2 Codon usage
RSCU ratio is an indicator used to measure whether synonymous and non-synonymous codons are used uniformly in a coding sequence. When the RSCU ratio is < 1.00, the codon usage frequency is lower than expected; when the RSCU ratio is > 1.00, the codon usage frequency is higher than expected (Sharp and Li, 1987; Liu et al., 2018). The codon usage levels of the CP genomes of T. obtectum and T. serrulatum are shown in (Figure 2; Supplementary Table S3). All protein-coding genes comprise 26,909 and 26,857 codons from the two CP genomes. Among them, leucine (Leu) is the most frequent, with an occurrence frequency of 10.39% and 10.41%, followed by isoleucine (Ile), with an occurrence frequency of 8.56% and 8.53%, and cysteine (Cys) is the least frequent, with an occurrence frequency of 1.19% and 1.20%. In the CP genomes of two Tetrastigma species, usage of the codons AUG and UGG-encoding methionine and tryptophan, respectively is not biased (RSCU ratio = 1.00). Most protein-coding genes in the CP genome of land plants use the standard AUG initiator, and the AUG codon shows no bias in the CP genomes of T. obtectum and T. serrulatum (RSCU = 1). Codons ending in A and/or U account for 69.90% of all protein-coding genes in the CP genomes of T. obtectum and T. serrulatum. In both CP genomes, codons ending in A and/or T (U) generally have higher RSCU ratios, such as GCU (1.82) encoding alanine and UUA (1.81) encoding leucine. The codon usage pattern can be determined by whether there is a high proportion of A/T component bias. In the CP genomes of higher terrestrial plants, preferred codons are often very high, and the preference for A/T is widespread (Kim and Lee, 2004; Zhang et al., 2021a). Our results also showed that except for Leu-UUG, all types of preferred synonymous codons (RSCU ratio >1.00) in the two Tetrastigma species end with A or U. A high RSCU ratio may be attributed to the function of amino acids or the peptide structure needed to avoid transcription errors (Gao et al., 2019; Zhang et al., 2021b). This phenomenon suggests that stable CP genome evolution helps protect CP genes from harmful mutations while improving adaptability to selective pressure (Wang et al., 2016; Ivanova et al., 2017).
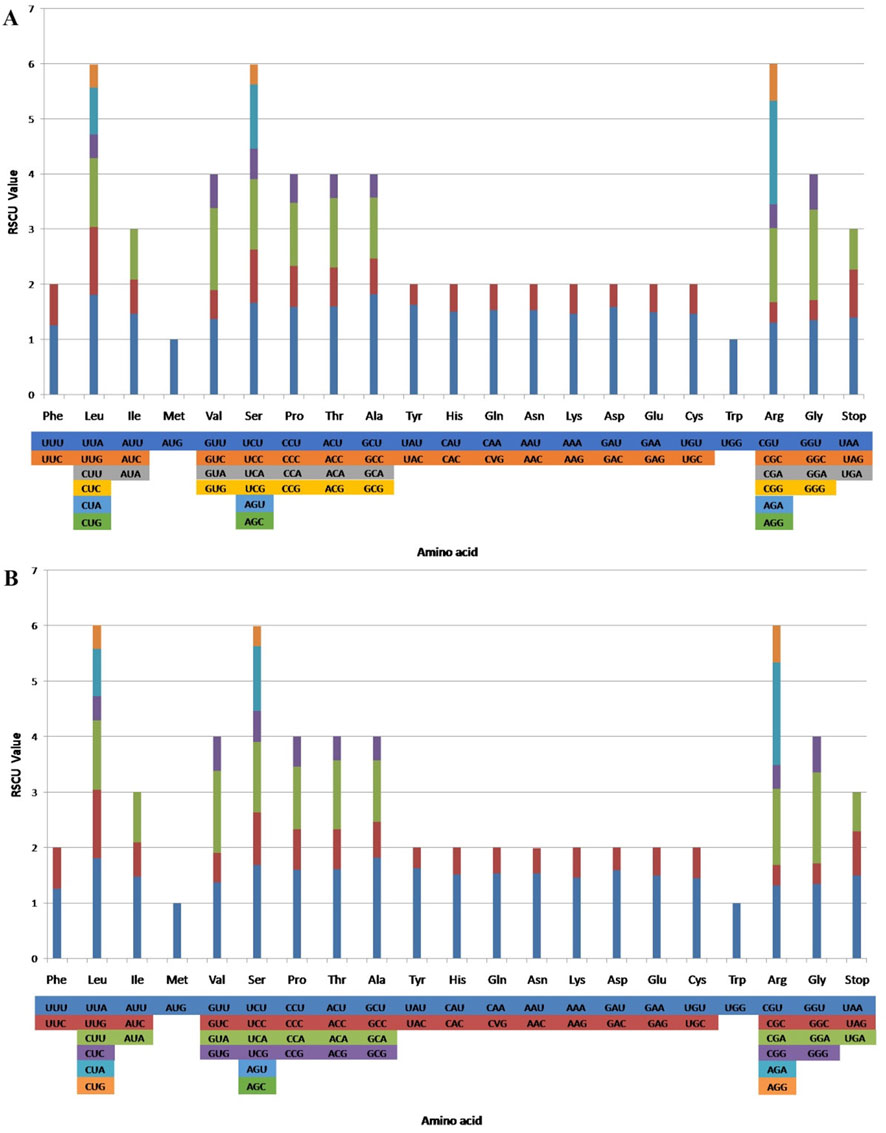
Figure 2. Codon contents in all protein-coding genes in the T. obtectum (A) and T. serrulatum (B) complete chloroplast (CP) genome. RSCU, relative synonymous codon usage.
3.3 Simple sequence repeats analyses
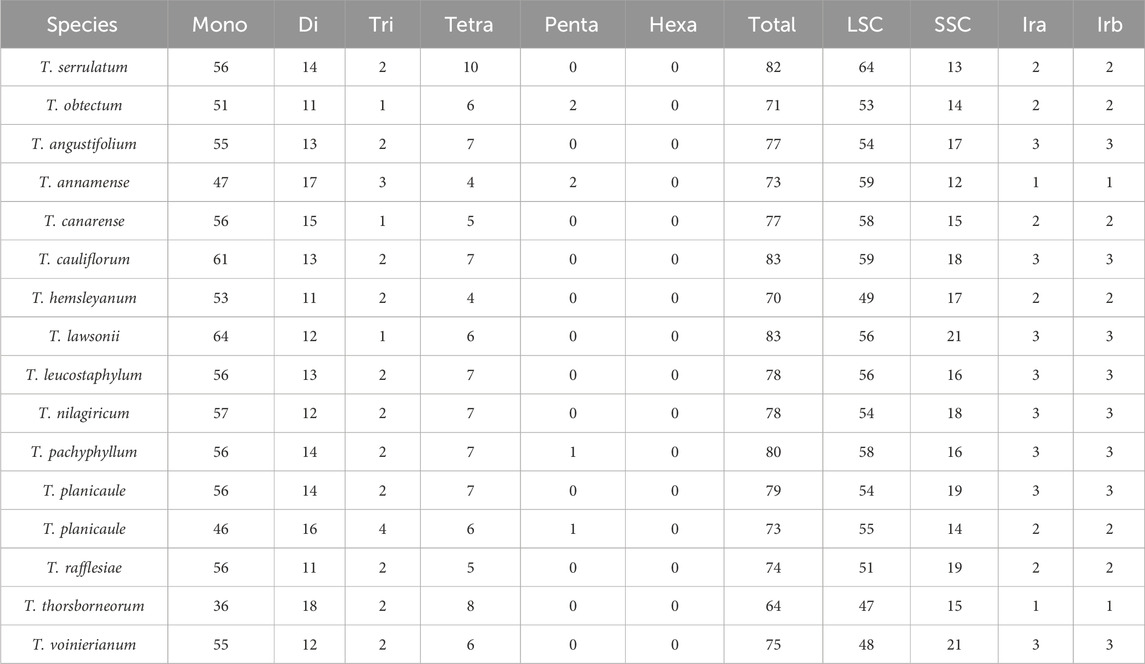
Simple sequence repeats are sequences composed of 1-6 nucleotide repeat units, which are widely distributed in CP genomes and are commonly used in population genetics and molecular phylogenetic studies (George et al., 2015; Zhou J. et al., 2018). We analyzed the distribution and types of SSRs in the CP genome by combining 14 species of the genus Tetrastigma published by NCBI. A total of 1,217 SSRs were detected. Among them, T. lawsonii and T. cauliflorum have the highest number of SSRs, 83, while T. thorsborneorum has the lowest number of SSRs, with only 64. The SSRs in the sixteen species are primarily distributed in the LSC region of the CP genome, followed by the SSC region and the IR region (Table 3). Mononucleotide repeats (56.25∼77.11%) are the most abundant type, followed by dinucleotides (14.46%–28.13%); tetranucleotide (5.48%–12.5%), trinucleotide (1.2%–5.48%), and pentanucleotide repeat sequences (0%–2.82%). Regarding nucleotide composition types, a total of twenty-one repeat types were detected, mainly A/T and AT/AT (Figure 3). A/T repeat sequences (46.36∼57.63%) are the most abundant, followed by AT/TA dinucleotide repeat sequences (20.34∼28.68%) and AAAT/ATTT tetranucleotide repeat sequences (5.93∼8.18%). A detailed analysis of different nucleotide composition types reveals that A and T are the most commonly used bases, while repeat sequences containing C and G bases are infrequent. Our results are consistent with previous studies reporting that CP SSRs usually consist of short poly-A or poly-T repeats (Kuang et al., 2011; Zhang et al., 2021a). Interestingly, among the 16 species of the genus Tetrastigma, only T. obtectum, T. annamense, T. pachyphyllum, and T. pyriforme have five nucleotide SSRs. In contrast, no species of the genus Tetrastigma has six-nucleotide SSRs. Currently, SSR markers are widely used in fields such as genetic diversity and population structure assessment, comparative genomics, genetic map development, and marker-assisted selection breeding (Flannery et al., 2006; Cui et al., 2019). The repeat sequences identified in this study are a valuable resource for species identification and research on the genetic diversity and population structure of Tetrastigma plants.
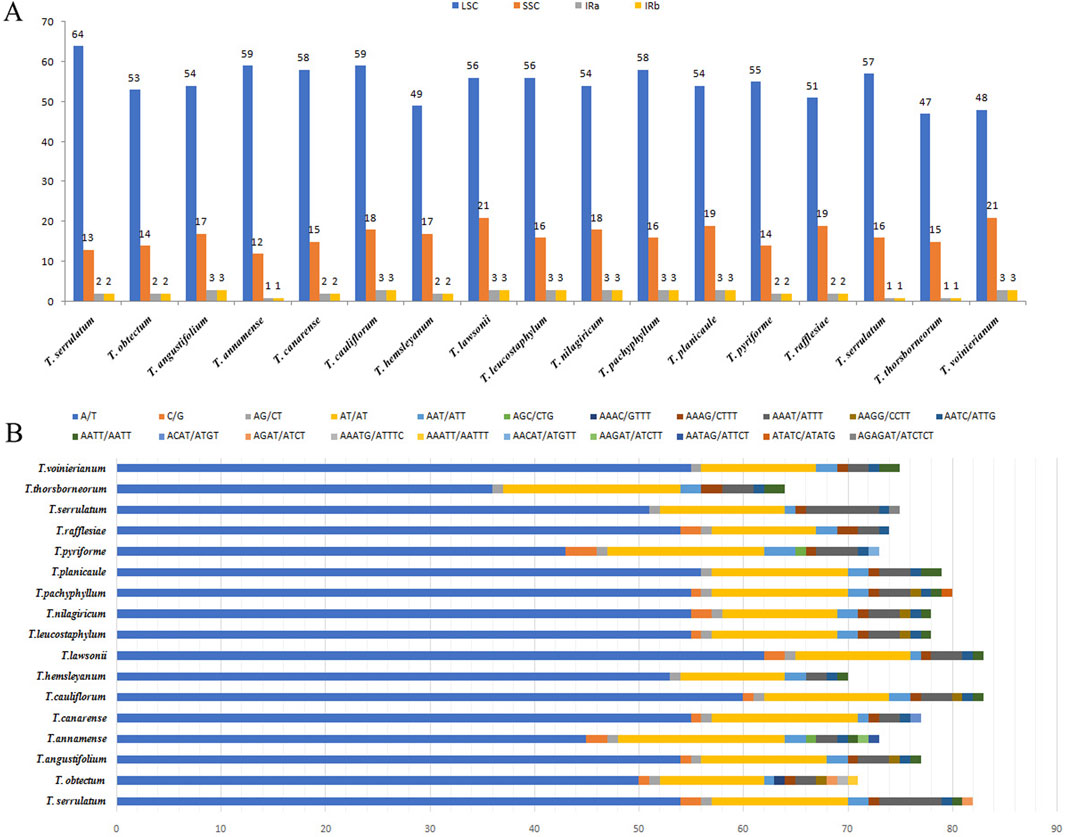
Figure 3. Analysis of chloroplast genome SSRs in 16 Tetrastigma species. (A) Frequencies of identified SSRs in the large single copy (LSC), SSC and inverted repeats (IRs) regions; (B) Frequency of identified SSRs in different repeat class types. The repetitive types of T. obtectum and T. serrulatum include: mononucleotides, dinucleotides, trinucleotides, tetranucleotides, and pentanucleotides.
3.4 Contraction and expansion of IRs
The CP genome of higher plants is highly conserved. Many researchers believe that the main reason for the variation in CP genome size is the contraction and expansion of the IR regions and the boundaries of the LSC/SSC regions (Wang et al., 2022; Zhang et al., 2024). In this study, we compared the IR/LSC and IR/SSC boundary structures of 16 species of Tetrastigma. The expansion and contraction of the IRs are shown in (Figure 4). The results indicate that the length of the IR region in the 16 species of ivy ranges from 26,306 to 26,934 bp, with no significant differences. The psbA gene positions of all species in the genus Tetrastigma are similar and are completely located in the LSC region. The trnH gene is also located within the LSC region, with a distance to the boundary ranging from 9 to 90 bp. The rps19 gene in T. rafflesiae is located in the LSC region, while in the other 15 species of the genus Tetrastigma, it spans the LSC/IRb boundary, with the portion within IRb ranging from 26 to 107 bp. The ndhF genes of 16 species of the genus Tetrastigma are all located in the SSC region, with distances from the IRb/SSC boundary ranging from 5 to 110 bp. The ycf1 gene (5,576–5,645 bp) spans the SSC/IRa boundary, and a portion of the ycf1 gene (1,034–1,265) spans the IRb/SSC region were annotated as pseudogenes. This also leads to the ycf1 pseudogene and the gene ndhF having partial overlap in the SSC region in the three species T. thorsborneorum, T. leucostaphylum, and T. obtectum. The pseudogenization and copy positions of ycf1 are also frequently observed in other higher plants (Lu et al., 2022; Zhu et al., 2024). In summary, while the CP genomes of the 16 Tetrastigma species exhibit high conservation in gene number and genomic structure, there are notable differences in the lengths of specific genes and the IR, LSC, and SSC regions. This phenomenon suggests that the expansion and contraction of the IR region may primarily drive changes in CP genome length and serve as a catalyst for variations in plant CP genomes (Kim and Lee, 2004; Pei et al., 2022; Zhang et al., 2024).
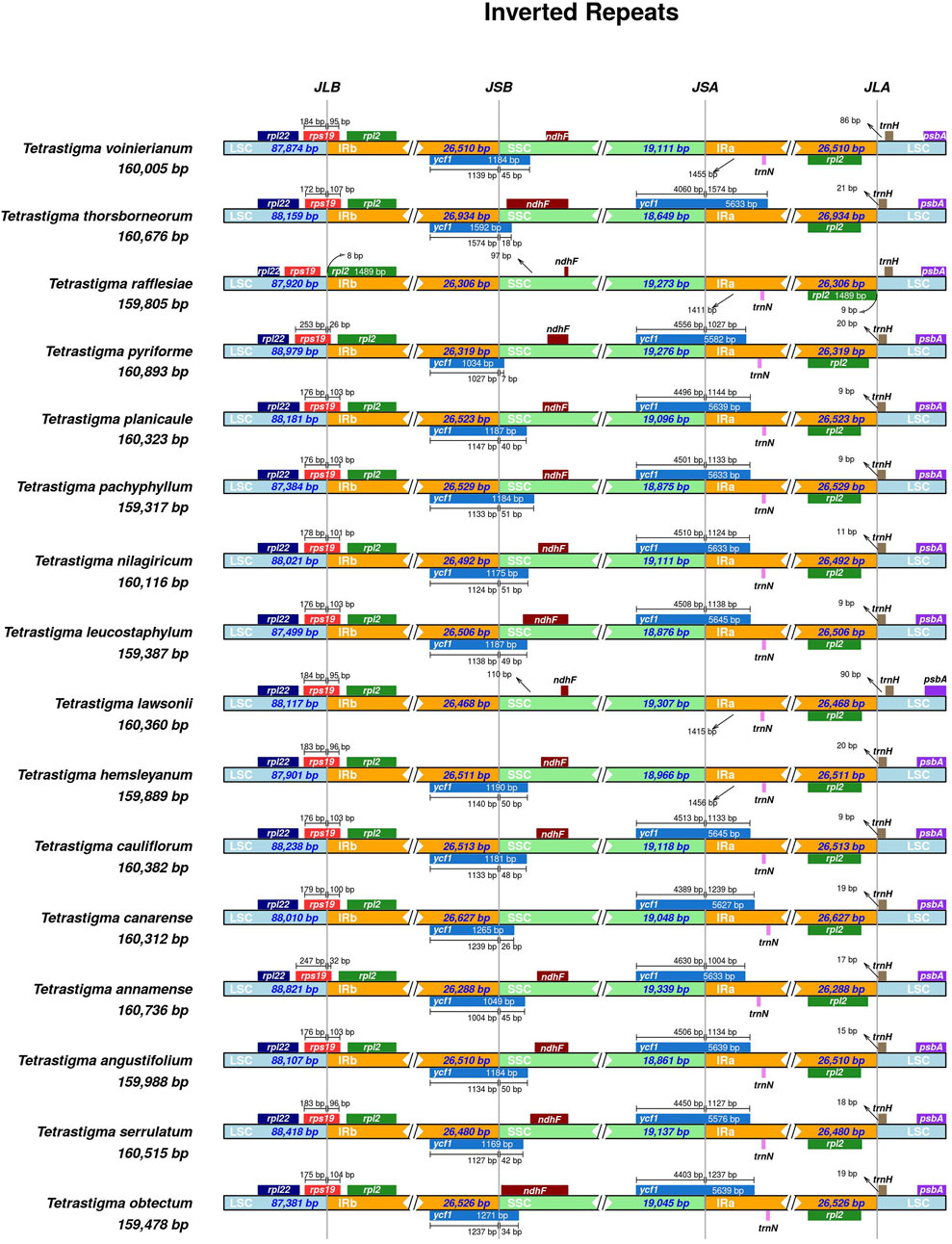
Figure 4. Comparison of the borders of the LSC, SSC, IRs regions among four CP genomes of Tetrastigma. JLB: boundary of the LSC and the IRb. JSB: boundary of the SSC and the IRb. JSA: boundary of the SSC and the IRa. JLA: boundary of the LSC and the IRa.
3.5 Comparative chloroplast genome analysis
The genomic structure of plant CP is highly conserved, and through comparative analysis, high mutation regions can be easily identified. These variations contribute to elucidating the genetic structure and evolutionary relationships of this genus of plants (Daniell et al., 2016; Hong et al., 2020). This study conducted a comparative analysis of a total of 16 Tetrastigma CP genomes, and the results showed the overall differences among the 16 CP genomes are minimal, with the sequence differences in the two IR regions being smaller than those in the LSC and SSC regions (Figure 5). Additionally, we found that the sequence differences are more prominent in non-coding regions, whereas the differences in exons and untranslated regions (UTRs) are more minor. The most differentiated non-coding regions include rps16-trnG-UUC,psbM-trnD-GUC, psbE-petL, ndhF-rpl32-trnL-UAG, and the most differentiated coding regions include rpl16, rpl22,ycf1. This result further supports the view held by many scholars that the coding regions of higher plants are more conserved than non-coding regions, and the IR regions are more conserved than the LSC and SSC regions (Nazareno et al., 2015; Fan and Ma, 2022; Zhang et al., 2024). In the study Elimination of deleterious mutations in plastid genomes by gene conversion, Khakhlova believes that due to gene conversion correcting mutations in the IR sequence (Khakhlova and Bock, 2006).

Figure 5. The alignment and comparative analysis of the whole CP genome for sixteen Tetrastigma species using mVISTA, and T. obtectum as a reference. Gray arrows and thick black lines above the alignments indicate gene orientations. White peaks represent differences among CP genomes. Exons, introns, and conserved noncoding sequences (CNSs) were displayed in different colors. A similarity cut-off value of 70% was used for the plots, and the Y-axis represents the percentage similarity (50%–100%).
3.6 Discovery of candidate markers based on nucleotide diversity analyses
Nucleotide polymorphism (Pi) analysis of CP genomes from 16 Tetrastigma species was conducted using DnaSP software, identifying highly variable regions (Figure 6). The IR region exhibits lower variability than the LSC and SSC regions, similar to previous studies. The results showed that the average Pi value of 16 species of the genus Tetrastigma was 0.0065 (Supplementary Table S4). A total of 6 nucleotide polymorphism hotspots with Pi > 0.02 were detected, with 3 found in the LSC region: trnS-GCU-trnG-UCC, psbM-trnD-GUC, trnS-UGA-psbZ, and 3 found in the SSC region: ndhF-rpl32, rpl32-trnL-UAG, ycf1. These results suggest that the LSC and SSC regions of Tetrastigma species may have experienced rapid nucleotide substitution, which is crucial for species identification and phylogenetic analysis. This study performed a nucleotide polymorphism (Pi) analysis on the CP genomes of 16 Tetrastigma species. The ten regions with the highest variability detected are psbM-trnD-GUC, ndhF-rpl32, trnS-GCU-trnG-UCC, ycf1, rpl32-trnL-UAG, trnS-UGA-psbZ, psbE-petL, matK-rps16, rpl16, and rpl22. These highly variable regions can be candidate markers for species identification within the genus Tetrastigma. Previous molecular identification studies on Panax, Dracaena, and Gentiana have demonstrated that CP genetic markers possess high identification capability (Nguyen et al., 2017; Zhou T. et al., 2018).
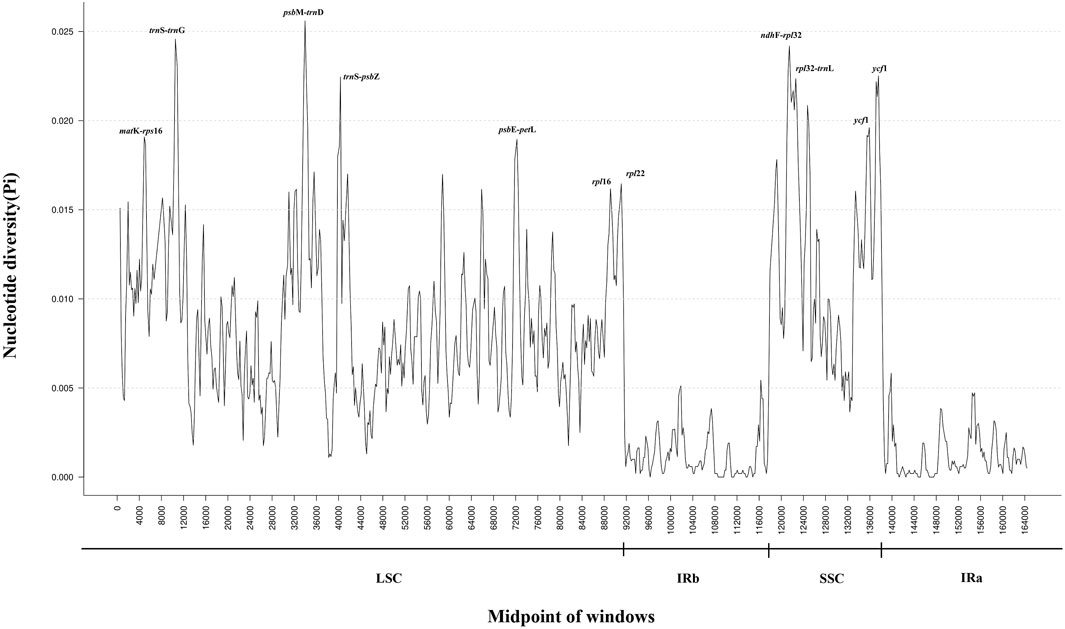
Figure 6. Sliding window analysis based on the complete chloroplast (CP) genomes of sixteen Tetrastigma species. Window length: 600 bp; step size: 200 bp. X-axis: position of the midpoint of a window. Y-axis: nucleotide diversity of each window.
3.7 ITS2 barcode and secondary structure identification
To identify the most effective barcode markers for species within the genus Tetrastigma, we utilized the conventional DNA barcode ITS2 to amplify two Tetrastigma species and evaluated their identification efficiency. We constructed an NJ phylogenetic tree using conventional barcodes (Figure 7). The results show that variable sites in the ITS2 base sequence can effectively distinguish T. obtectum from T. serrulatum. Additionally, we constructed the secondary structures of the ITS2 sequences for T. obtectum and T. serrulatum using an online tool, as depicted in Figure 8. According to the secondary structure diagrams of the ITS2 base sequences, T. serrulatum exhibits a unique secondary structure (Figure 8A), whereas T. obtectum displays two distinct secondary structures (Figures 8A,B). Notably, the structure in Figure 8A shares the same stem and loop configuration as that of T. serrulatum. Consequently, it is not feasible to differentiate these two species based on the shape of the ITS2 secondary structures alone. In this study, the traditional barcode ITS2 base sequence successfully distinguished T. obtectum from T. serrulatum. However, the limited number of sampled species in this study necessitates an expansion of sampling in future research. It is recommended that future studies increase the sampling scope within the genus and incorporate CP barcode markers for more comprehensive species identification.
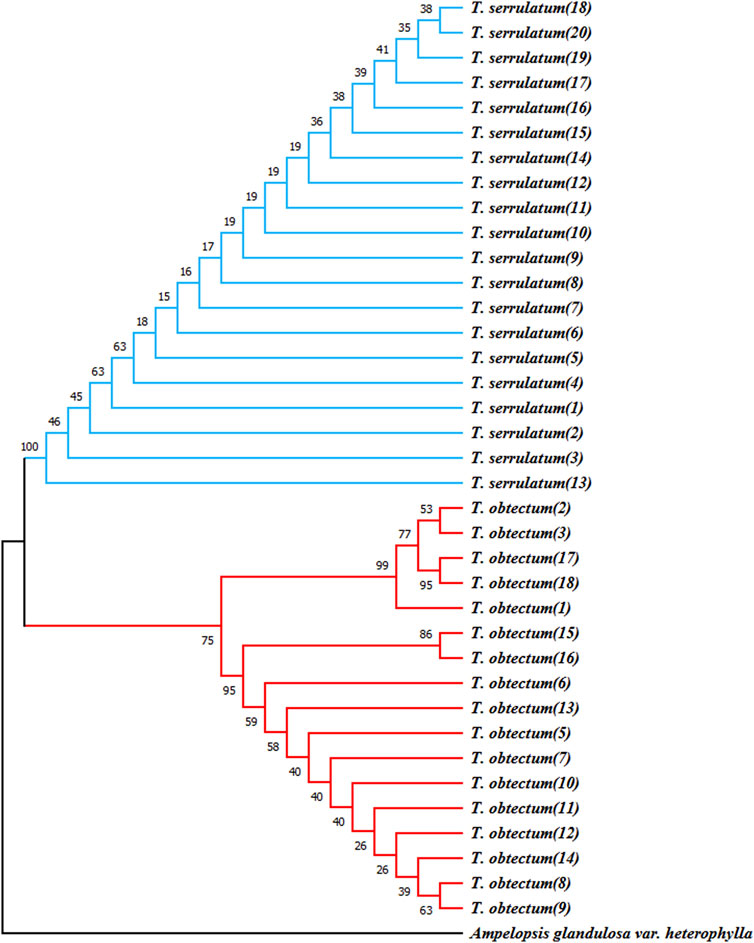
Figure 7. Neighbor-joining bootstrap trees (based on Kimura-2-Parameter) illustrating the resolution of T. obtectum and T. serrulatum for the “ITS2” barcoding.
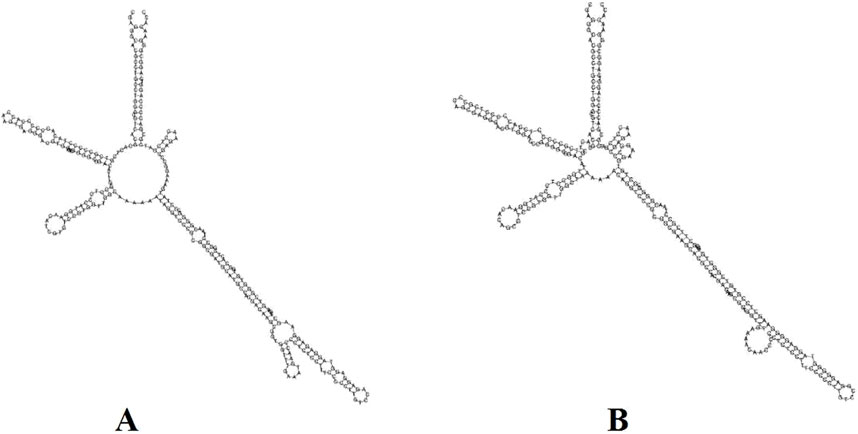
Figure 8. Constructing secondary structures T. serrulatum and T. obtectum based on ITS2 base sequence, (A) is the secondary structure of T. serrulatum, while the secondary structure of T. obtectum includes both (A, B).
3.8 Phylogenetic relationships of Vitaceae species
The CP genome sequence is crucial for investigating the phylogenetic relationships, genetic structure, and taxonomic classification of higher plants. To investigate the phylogenetic relationships within the genus Tetrastigma and its placement within the family Vitaceae, we constructed a Maximum Likelihood (ML) phylogenetic tree using CP genome sequences of 46 species from Vitaceae family (Figure 9). And all nodes received strong support. The phylogenetic tree indicates that the L. asiatica and L. indica from the genus Leea are the first to split into a separate branch. At the same time, the remaining species are grouped into another significant branch. Then, the six species of the Cissus divided into one branch, while the species of the remaining five genera split into a large branch. Next, the species of the genus Tetrastigma gathered on one branch, and each species was separated. In contrast, the species of Parthenocissus, Vitis, Ampelopsis, and Ampelocissus gathered on another branch, with a support rate of 100%. In Tetrastigma, T. obtectum, T. serrulatum, T. annamense, T. thorsborneorum, and T. pyriforme form one clade, while the remaining species form another clade, both supported by 100% bootstrap values. This result aligns with the conclusion of Zhu et al. (2024) that species of the genus Tetrastigma constitute a monophyletic group. In the classification study by Zhu et al. (2024), the close phylogenetic relationship between T. thorsborneorum and T. pyriforme has been confirmed. Additionally, the phylogenetic analysis of the species from the Vitaceae family included in this study is more comprehensive, resulting in a more stable and accurate phylogenetic relationship. Therefore, we believe the CP genome can be a reliable tool for species identification within the genus Tetrastigma. As a highly effective super barcode, the CP genome sequence offers a robust method for identifying higher plant species. Furthermore, we believe that as more species are included in the analysis, the species relationships within this genus will change and become clearer. The genus Tetrastigma includes numerous species, many of which have uncertain classification statuses and Tetrastigma relationships. Future phylogenetic analyses should incorporate more CP genome samples. Our results provide valuable references and a foundation for species identification using the CP genome to reveal the evolutionary relationships within this genus and help to improve the understanding of the phylogeny of the Tetrastigma genus.
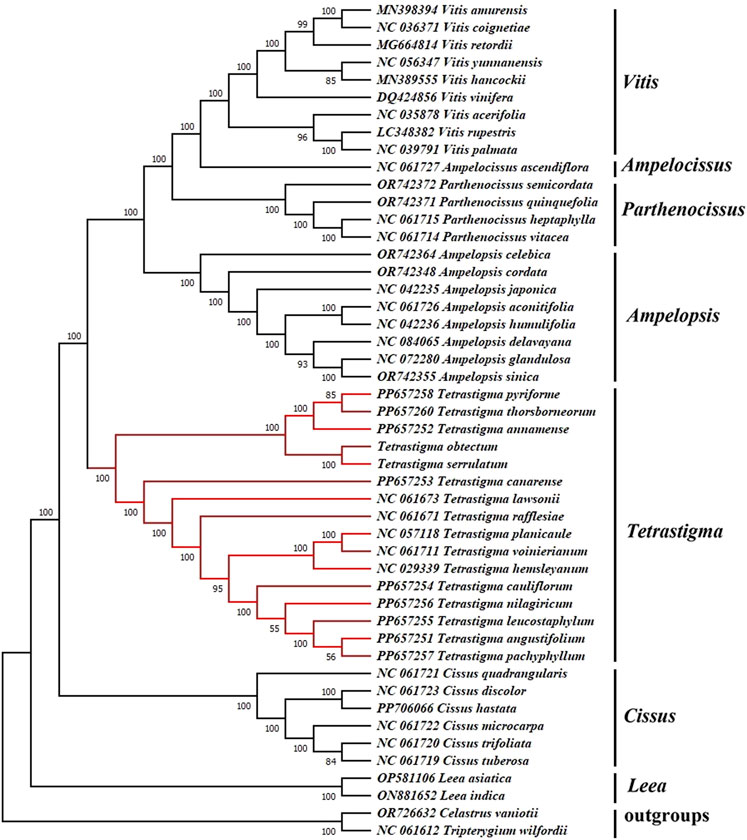
Figure 9. Maximum Likelihood (ML) phylogenetic tree based on complete chloroplast genomes, with Celastrus vaniotii and Tripterygium wilfordii as outgroups. Bootstrap values are displayed on the branch phylogenetic tree, the number above each node referred to the bootstrap value from 1,000 replicates and the optimal model selected by ModelTest-NG is “GTR + I + G”.
4 Conclusion
This study assembled and annotated the complete CP genomes of T. obtectum (159,478 bp) and T. serrulatum (160,515 bp) and analyzed them in combination with the existing CP genomes of 14 Tetrastigma species from NCBI. The CP genomes of Tetrastigma exhibited a typical circular tetramerous structure, including a large single-copy region (87,381–88,979 bp), a small single-copy region (18,649–19,339 bp), and a pair of inverted repeats (26,288–26,934 bp). The guanine-cytosine content of the CP genomes is 37.35%–37.62%. The codon usage of Tetrastigma plants shows a significant preference for ending with A/T. A total of 1,217 SSRs were detected in 16 species of Tetrastigma. We then compared the analysis of the CP genome sequences of Tetrastigma species and analyzed their sequence characteristics. Nucleotide diversity was analyzed based on the alignment of complete CP genome sequences, identifying ten polymorphic hotspots suitable for DNA marker development. Using the ITS2 sequence and its secondary structure for species identification of T. obtectum and T. serrulatum, our results demonstrate that the ITS2 sequence can effectively discriminate between the two species, whereas the secondary structure cannot. Finally, to investigate the phylogenetic relationships within the genus Tetrastigma and its placement within the family Vitaceae, we constructed a Maximum Likelihood (ML) phylogenetic tree using CP genome sequences of 46 species from seven genera within the Vitaceae family (Tetrastigma, Parthenocissus, Vitis, Ampelopsis, Ampelocissus, Cissus, and Leea).
Data availability statement
The GenBank data presented in the study are deposited in the NCBI (Nucleotide) repository, available at: https://www.ncbi.nlm.nih.gov/nuccore/2987893153 and https://www.ncbi.nlm.nih.gov/nuccore/PV454981. Further inquiries can be directed to the corresponding authors.
Author contributions
XL: Conceptualization, Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing. YZ: Conceptualization, Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing. MS: Data curation, Validation, Writing – original draft, Writing – review and editing. NX: Data curation, Validation, Writing – original draft, Writing – review and editing. LQ: Investigation, Resources, Writing – original draft, Writing – review and editing. HL: Investigation, Resources, Writing – original draft, Writing – review and editing. YW: Writing – original draft, Writing – review and editing, Investigation, Resources. BD: Writing – original draft, Writing – review and editing, Funding acquisition, Project administration, Resources. ZZ: Writing – original draft, Writing – review and editing, Funding acquisition, Project administration, Resources, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the program of Yunnan “Xingdian Talent Support Program” young talents unique project, CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-032), and Selection Special Programme of Yunnan Province High-level Technological Talents and Innovative Teams (202405AS350020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1607947/full#supplementary-material
References
Ahmad, W., Asaf, S., Khan, A., Al-Harrasi, A., Al-Okaishi, A., and Khan, A. L. (2022). Complete chloroplast genome sequencing and comparative analysis of threatened dragon trees Dracaena serrulata and Dracaena cinnabari. Sci. Rep. 12 (1), 16787. doi:10.1038/s41598-022-20304-6
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34 (17), 3030–3031. doi:10.1093/bioinformatics/bty220
Brown, J., Pirrung, M., and Mccue, L. A. (2017). FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33 (19), 3137–3139. doi:10.1093/bioinformatics/btx373
Bu, R. E. (1996). Pharmacognostical study on Mongolian folk medicine--Erdos Biriyanggu. Chin. J. Ethnomedicine Ethnopharmacy 20, 39.
Chen, S., Yao, H., Han, J., Liu, C., Song, J., Shi, L., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5 (1), e8613. doi:10.1371/journal.pone.0008613
Chen, S., Yin, X., Han, J., Sun, W., Yao, H., Song, J., et al. (2023). DNA barcoding in herbal medicine: retrospective and prospective. J. Pharm. Anal. 13 (5), 431–441. doi:10.1016/j.jpha.2023.03.008
Cui, Y., Nie, L., Sun, W., Xu, Z., Wang, Y., Yu, J., et al. (2019). Comparative and phylogenetic analyses of ginger (Zingiber officinale) in the family zingiberaceae based on the complete chloroplast genome. Plants 8, 283. doi:10.3390/plants8080283
Daniell, H., Jin, S., Zhu, X. G., Gitzendanner, M. A., Soltis, D. E., and Soltis, P. S. (2021). Green giant—a tiny chloroplast genome with mighty power to produce high-value proteins: history and phylogeny. Plant Biotechnol. J. 19 (3), 430–447. doi:10.1111/pbi.13556
Daniell, H., Lin, C., Yu, M., and Chang, W. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi:10.1186/s13059-016-1004-2
Darriba, D., Posada, D., Kozlov, A. M., Stamatakis, A., Morel, B., and Flouri, T. (2020). ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37 (1), 291–294. doi:10.1093/molbev/msz189
Dobrogojski, J., Adamiec, M., and Luciński, R. (2020). The chloroplast genome: a review. Plant. 42, 98. doi:10.1007/s11738-020-03089-x
Dong, W., Liu, J., Yu, J., Wang, L., and Zhou, S. (2012). Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One 7 (4), e35071. doi:10.1371/journal.pone.0035071
Fan, Z., and Ma, C. (2022). Comparative chloroplast genome and phylogenetic analyses of Chinese Polyspora. Sci. Rep. 12 (1), 15984. doi:10.1038/s41598-022-16290-4
Ferrarini, M., Moretto, M., Ward, J. A., Surbanovski, N., Stevanovic, V., Giongo, L., et al. (2013). An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genomics 14, 670. doi:10.1186/1471-2164-14-670
Flannery, M. L., Mitchell, F. J. G., Coyne, S., Kavanagh, T. A., Burke, J. I., Salamin, N., et al. (2006). Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor. Appl. Genet. 113 (7), 1221–1231. doi:10.1007/s00122-006-0377-0
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic. acids. Res. 32 (Web Server), W273–W279. doi:10.1093/nar/gkh458
Gao, B., Yuan, L., Tang, T., Hou, J., Pan, K., and Wei, N. (2019). The complete chloroplast genome sequence of Alpinia oxyphylla Miq. and comparison analysis within the Zingiberaceae family. PLoS One 14 (6), e0218817. doi:10.1371/journal.pone.0218817
George, B., Bhatt, B. S., Awasthi, M., George, B., and Singh, A. K. (2015). Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. Curr. Genet. 61 (4), 665–677. doi:10.1007/s00294-015-0495-9
Gregory, T. R. (2005). DNA barcoding does not compete with taxonomy. Nature 434, 1067. doi:10.1038/4341067a
Hai, P., Zhou, S. X., Zhao, G. L., and Shang, H. (1997). Pharmacological study on Tibetan medicine Tanggute Qinglan against cerebral hypoxia. J. Med. and Pharm. Chin. Minorities. 3, 42–43. doi:10.16041/j.cnki.cn15-1175.1997.3.036
Han, J., Pang, X., Liao, B., Yao, H., Song, J., and Chen, S. (2016). An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci. Rep. 18723, 18723. doi:10.1038/srep18723
Hong, Z., Wu, Z., Zhao, K., Yang, Z., Zhang, N., Guo, J., et al. (2020). Comparative analyses of five complete chloroplast genomes from the genus pterocarpus (fabacaeae). Int. J. Mol. Sci. 21, 3758. doi:10.3390/ijms21113758
Hu, J. L., Ci, X. Q., Liu, Z. F., Dormontt, E. E., Conran, J. G., Lowe, A. J., et al. (2022). Assessing candidate DNA barcodes for Chinese and internationally traded timber species. Mol. Ecol. Resour. 22, 1478–1492. doi:10.1111/1755-0998.13546
Ivanova, Z., Sablok, G., Daskalova, E., Zahmanova, G., Apostolova, E., Yahubyan, G., et al. (2017). Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 8, 204. doi:10.3389/fpls.2017.00204
Jin, J., Yu, W., Yang, J., Song, Y., Depamphilis, C. W., Yi, T., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi:10.1186/s13059-020-02154-5
Jin, S., and Daniell, H. (2015). The engineered chloroplast genome just got smarter. Trends Plant Sci. 20 (10), 622–640. doi:10.1016/j.tplants.2015.07.004
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20 (4), 1160–1166. doi:10.1093/bib/bbx108
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 (12), 1647–1649. doi:10.1093/bioinformatics/bts199
Khakhlova, O., and Bock, R. (2006). Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 46, 85–94. doi:10.1111/j.1365-313X.2006.02673.x
Kim, K., and Lee, H. (2004). Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11, 247–261. doi:10.1093/dnares/11.4.247
Kuang, D., Wu, H., Wang, Y., Gao, L., Zhang, S., and Lu, L. (2011). Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome 54 (8), 663–673. doi:10.1139/g11-026
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic. acids. Res. 29, 4633–4642. doi:10.1093/nar/29.22.4633
Li, C. L., and Wu, Z. Y. (1995). A taxonomical study on tetrastigma(mio.)planch. China. Chin. J. Appl. Environ. Biol. 4, 307–333.
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y., and Chen, S. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. Camb. Philos. Soc. 90 (1), 157–166. doi:10.1111/brv.12104
Lin, Y. (1991). Pharmacognostic identification of Mongolian medicine Scutellaria mucilaginosa. Chin. Traditional Herb. Drugs 22 (4), 179–180.
Liu, X., Li, Y., Yang, H., and Zhou, B. (2018). Chloroplast genome of the folk medicine and vegetable plant Talinum paniculatum (Jacq.) gaertn.: gene organization, comparative and phylogenetic analysis. Molecules 23, 857. doi:10.3390/molecules23040857
Lohse, M., Drechsel, O., Kahlau, S., and Bock, R. (2013). OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic. acids. Res. 41, W575–W581. doi:10.1093/nar/gkt289
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic. acids. Res. 44 (W1), W54–W57. doi:10.1093/nar/gkw413
Lu, Q., Chang, X., Gao, J., Wu, X., Wu, J., Qi, Z., et al. (2022). Evolutionary comparison of the complete chloroplast genomes in convallaria species and phylogenetic study of asparagaceae. Genes 13 (10), 1724. doi:10.3390/genes13101724
Ma, S., Lv, Q., Zhou, H., Fang, J., Cheng, W., Jiang, C., et al. (2017). Identification of traditional she medicine shi-liang tea species and closely related species using the ITS2 barcode. Appl. Sci. 7 (3), 195. doi:10.3390/app7030195
Nazareno, A. G., Carlsen, M., and Lohmann, L. G. (2015). Complete chloroplast genome of Tanaecium tetragonolobum: the first bignoniaceae plastome. PLoS One 10 (6), e0129930. doi:10.1371/journal.pone.0129930
Nguyen, V. B., Park, H., Lee, S., Lee, J., Park, J. Y., and Yang, T. (2017). Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences. J. Agric. Food. Chem. 65 (30), 6298–6306. doi:10.1021/acs.jafc.7b00925
Pei, J., Wang, Y., Zhuo, J., Gao, H., Vasupalli, N., Hou, D., et al. (2022). Complete chloroplast genome features of Dendrocalamus farinosus and its comparison and evolutionary analysis with other bambusoideae species. Genes 13 (9), 1519. doi:10.3390/genes13091519
Rozas, J., Ferrer-Mata, A., Sánchez-Delbarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34 (12), 3299–3302. doi:10.1093/molbev/msx248
Ruan, X. Z. (2002). Folk prescription of honeysuckle used by She nationality. China's Naturop. 11, 58–59. doi:10.19621/j.cnki.11-3555/r.2002.11.088
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., and Tabata, S. (1999). Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 6 (5), 283–290. doi:10.1093/dnares/6.5.283
Sharp, P. M., and Li, W. H. (1987). The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic. acids. Res. 15, 1281–1295. doi:10.1093/nar/15.3.1281
Sokołowska, J., Fuchs, H., and Celiński, K. (2022). Assessment of ITS2 region relevance for taxa discrimination and phylogenetic inference among pinaceae. Plants 11, 1078. doi:10.3390/plants11081078
Song, B. Z., and Wan, D. G. (2003). Investigation on commonly used botanical drugs (vitis) of Tujia nationality in hubei Province. Chin. J. Ethnomedicine Ethnopharmacy 13 (2), 117.
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi:10.1093/molbev/msab120
Wan, D. R. (1990). Commonly used botanical drugs of Tujia nationality in hubei Province (ranunculaceae). J. Chin. Med. Mater. 3, 13–15. doi:10.13863/j.issn1001-4454.1990.03.006
Wang, J., Kan, S., Liao, X., Zhou, J., Tembrock, L. R., Daniell, H., et al. (2024). Plant organellar genomes: much done, much more to do. Trends Plant Sci. 29, 754–769. doi:10.1016/j.tplants.2023.12.014
Wang, Y., Wen, F., Hong, X., Li, Z., Mi, Y., and Zhao, B. (2022). Comparative chloroplast genome analyses of Paraboea (Gesneriaceae): insights into adaptive evolution and phylogenetic analysis. Front. Plant Sci. 13, 1019831. doi:10.3389/fpls.2022.1019831
Wang, Y., Zhan, D., Jia, X., Mei, W., Dai, H., Chen, X., et al. (2016). Complete chloroplast genome sequence of Aquilaria sinensis (lour.) gilg and evolution analysis within the malvales order. Front. Plant Sci. 7, 280. doi:10.3389/fpls.2016.00280
Wei, Q. H., Qian, Z. G., and Chen, Y. X. (1999). Pharmacognostical study on the national medicine jinglingdou. Chin. J. Ethnomedicine Ethnopharmacy 39, 224.
Wei, X., Shen, F., Zhang, Q., Liu, N., Zhang, Y., Xu, M., et al. (2021). Genetic diversity analysis of Chinese plum (Prunus salicina L.) based on whole-genome resequencing. Tree Genet. Genomes. 17, 26. doi:10.1007/s11295-021-01506-x
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization ofde novo genome assemblies. Bioinformatics 31 (20), 3350–3352. doi:10.1093/bioinformatics/btv383
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Müller, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi:10.1007/s11103-011-9762-4
Wyman, S. K., Jansen, R. K., and Boore, J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20 (17), 3252–3255. doi:10.1093/bioinformatics/bth352
Xiao, C. W., and Li, Y. Y. (1999). Folk folk collection of Dong nationality. Chin. J. Ethnomedicine Ethnopharmacy 36, 60–61.
Yang, H., Wang, L., Chen, H., Jiang, M., Wu, W., Liu, S., et al. (2021). Phylogenetic analysis and development of molecular markers for five medicinal Alpinia species based on complete plastome sequences. BMC Plant Biol. 21, 431. doi:10.1186/s12870-021-03204-1
Yang, L. Y., Hu, C. G., Zhao, J. H., Pan, L. T., and Li, Y. (2000). Animal medicine commonly used by Gelao people. Chin. J. Ethnomedicine Ethnopharmacy 55, 103–105.
Yang, Y. H., Bai, W., Li, M. L., and Feng, D. Q. (1998). Lahu medicine geese mother frame that this. Chin. J. Ethnomedicine Ethnopharmacy 5, 38–39.
Yao, H., Song, J., Liu, C., Luo, K., Han, J., Li, Y., et al. (2010). Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One 5, e13102. doi:10.1371/journal.pone.0013102
Yao, Q., Zhu, X., Han, M., Chen, C., Li, W., Bai, H., et al. (2022). Decoding herbal materials of TCM preparations with the multi-barcode sequencing approach. Sci. Rep. 12, 5988. doi:10.1038/s41598-022-09979-z
Yi, J. H., Yan, X. Z., Luo, Z. Y., and Zhong, C. C. (1991). Studies on the chemical constituents of the roots of lamiophlomis rotata. Acta Pharm. Sin. 1, 37–41. doi:10.16438/j.0513-4870.1991.01.008
Zhang, R. P., Tang, J. C., Chen, S. X., Zeng, Y. Q., and Zeng, Y. L. (1998). Pharmacognostic study on the national medicine Nitraria tangutorum. J. Yunnan Coll. Traditional Chin. Med. 1, 24–26. doi:10.19288/j.cnki.issn.1000-2723
Zhang, Y., Song, M., Li, Y., Sun, H., Tang, D., Xu, A., et al. (2021a). Complete chloroplast genome analysis of two important medicinal alpinia species: Alpinia galanga and Alpinia kwangsiensis. Front. Plant Sci. 12, 705892. doi:10.3389/fpls.2021.705892
Zhang, Y., Song, M., Tang, D., Li, X., Xu, N., Li, H., et al. (2024). Comprehensive comparative analysis and development of molecular markers for Lasianthus species based on complete chloroplast genome sequences. BMC Plant Biol. 24, 867. doi:10.1186/s12870-024-05383-z
Zhang, Y., Song, M. F., Li, H. T., Sun, H. F., and Zhang, Z. L. (2021b). DNA barcoding identification of original plants of a rare medicinal material Resina Draconis and related Dracaena species. China J. Chin. Mater. Med. 46, 2173–2181. doi:10.19540/j.cnki.cjcmm.20210124.104
Zhou, J., Cui, Y., Chen, X., Li, Y., Xu, Z., Duan, B., et al. (2018a). Complete chloroplast genomes of Papaver rhoeas and Papaver orientale: molecular structures, comparative analysis, and phylogenetic analysis. Molecules 23, 437. doi:10.3390/molecules23020437
Zhou, T., Wang, J., Jia, Y., Li, W., Xu, F., and Wang, X. (2018b). Comparative chloroplast genome analyses of species in Gentiana section cruciata (Gentianaceae) and the development of authentication markers. Int. J. Mol. Sci. 19, 1962. doi:10.3390/ijms19071962
Keywords: Tetrastigma, Tetrastigma obtectum, Tetrastigma serrulatum, chloroplast genome, species identification, phylogenetic relationship
Citation: Li X, Zhang Y, Song M, Xu N, Qu L, Li H, Wang Y, Duan B and Zhang Z (2025) Molecular identification and phylogenetic analysis of confusing Tetrastigma species based on DNA barcoding and chloroplast genome. Front. Pharmacol. 16:1607947. doi: 10.3389/fphar.2025.1607947
Received: 08 April 2025; Accepted: 11 June 2025;
Published: 11 July 2025.
Edited by:
Ruyu Yao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Fardous Mohammad Safiul Azam, Neijiang Normal University, ChinaDa-Cheng Hao, Dalian Jiaotong University, China
Copyright © 2025 Li, Zhang, Song, Xu, Qu, Li, Wang, Duan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baozhong Duan, YnpkdWFuQDEyNi5jb20=; Zhonglian Zhang, enpsMDYwNUAxNjMuY29t
 Xianjing Li
Xianjing Li Yue Zhang
Yue Zhang Meifang Song
Meifang Song Niaojiao Xu1
Niaojiao Xu1 Baozhong Duan
Baozhong Duan Zhonglian Zhang
Zhonglian Zhang