Abstract
Colorectal cancer (CRC) is the third most common malignant tumor globally, and its development is closely related to interactions between the host and microbes. Recent studies have shown that the diversity of intratumoral microbiota significantly influences CRC progression and responses to immune therapy. This influence occurs through mechanisms such as immune microenvironment regulation, metabolic reprogramming, and epigenetic modifications. However, there is still a lack of systematic analysis regarding the diversity of intratumoral microbiota in CRC and its immune regulatory mechanisms, particularly in the metabolic and immune regulation. This article presents a systematic review of the compositional characteristics of intratumoral microbiota in CRC, the associated immune regulatory mechanisms, and their roles in chemotherapy and immunotherapy. It also discusses challenges like standardizing microbiome detection methods and the ethics of clinical translation, while proposing a strategy for integrating multi-omics using artificial intelligence. This article provides a theoretical basis for developing personalized treatment regimens that target the microbiota.
1 Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-related deaths worldwide. Epidemiological data indicate that CRC has become the third most common cancer worldwide, influenced by complex interactions among diet, lifestyle, and genetic factors (Sung et al., 2021). As traditional treatments face resistance challenges, tumor immunotherapy has brought new hope to advanced CRC patients by activating host anti-tumor immune responses. However, clinical data reveal that only 12%–15% of CRC patients have sustained responses to PD-1/PD-L1 inhibitors. This significant variability among patients is closely related to the heterogeneous immune environment within tumors (Cai et al., 2024).
Recent studies have identified distinct microbial communities in CRC tumor tissues that differ significantly in composition from the gut’s symbiotic microbiota (Nejman et al., 2020; Zhou P. et al., 2022). Intratumoral microbiota primarily colonize through three pathways: direct migration by disrupting the intestinal barrie, hematogenous invasion via the circulatory system, and migration from adjacent tissues (Cao et al., 2024). These colonizing microbiota significantly affect the immunogenicity of tumor cells through mechanisms including epigenetic regulation, secretion of metabolic products, and activation of pattern recognition receptors (Li et al., 2023). This finding offers a new perspective on how microbiota contribute to resistance against immunotherapy. Nevertheless, research on the microbiota-immune interaction network in CRC remains limited, particularly regarding metabolic-immune cross-regulation.
In assessing microbial community complexity, alpha and beta diversity serve as foundational metrics (Wei et al., 2025). Alpha diversity quantifies species richness and evenness within a single sample, commonly measured by: Shannon Index (reflecting species abundance and uniformity), Simpson Index (emphasizing dominance patterns), and Chao1 Index (estimating total species richness from rare taxa counts) (Maley et al., 2024). Conversely, beta diversity evaluates compositional differences across samples using distance-based metrics: Bray-Curtis Dissimilarity (comparing abundance profiles) and Weighted UniFrac Distance (incorporating phylogenetic relationships) (Kim et al., 2023; Lozupone and Knight, 2005). In CRC tumors, these indices map spatial heterogeneity—alpha diversity revealing niche-specific microbial load, while beta diversity distinguishes intratumoral sub-regions and correlates with molecular subtypes (Byrd et al., 2025; Zhao et al., 2021). This analytical framework is critical for decoding microbial-driven immunotherapy resistance.
Current research has two main limitations: on one hand, most studies focus on the association between gut microbiota and CRC immunotherapy, leaving the independent mechanisms of intratumoral microbiota in CRC unclear. On the other hand, research on the cross-regulation of the CRC intratumoral microbiota-metabolism-immune axis is still in early stages, especially concerning how bacterial metabolites influence immune cells and cytokines in the tumor microenvironment (Gutiérrez-Salmerón et al., 2021; Wang J. et al., 2021). Furthermore, the diversity of intratumoral microbiota, including its spatial distribution, microbial interactions, and links to CRC molecular subtypes, needs further clarification (Gao et al., 2022).
An in-depth exploration of the relationship between intratumoral microbiota and tumor immunotherapy in colorectal cancer will help us understand how tumors evade the immune system and may also reveal new strategies to optimize immunotherapy. This review aims to discuss the dual roles of intratumoral microbiota in CRC immunotherapy and the challenges in clinical translation. By integrating cutting-edge evidence, this article aims to provide a theoretical basis for optimizing targeted strategies for CRC immunotherapy and to promote the clinical translation of microbiota-immune interaction research.
2 Colorectal cancer and its intratumoral microbiota
2.1 Characteristics of the intratumoral microbiota in colorectal cancer
2.1.1 Detection technologies and methods for intratumoral microbiota in colorectal cancer
The primary detection technologies for intratumoral microbiota in CRC are 16S rRNA gene sequencing, metagenomic sequencing, and transcriptomic analysis. These technologies work together to reveal the composition, function, and interactions of microbial communities within the tumor microenvironment. 16S rRNA gene sequencing, the most widely used method for detecting intratumoral microbiota. It efficiently identifies microbes at the genus or species level by amplifying conserved 16S rRNA gene fragments, making it suitable for analyzing the diversity of large-scale samples (Chao 1 and Shannon index were used to estimate alpha diversity, beta diversity was used in QIIME) (Zwinsová et al., 2021). For example, this technology has been used to identify the microbial composition of 1,526 tumors and adjacent tissues from seven types of cancer (Nejman et al., 2020). Metagenomic sequencing offers detailed insights into microbial community structure and functional gene analysis, which are essential for understanding the role of microbes in tumor progression (Hardwick et al., 2018). Additionally, transcriptomic analysis can reveal interactions between microbes and host gene expression, further exploring the impact of microbes on the tumor microenvironment. Recently developed methods that combine single-cell metagenomics with spatial metabolomics enable the localization of microbial communities at the subcellar level (Xu and Zhao, 2018; Zhang et al., 2023). For instance, spatial transcriptomics and single-cell RNA sequencing have confirmed that Fusobacterium nucleatum in CRC is primarily enriched in CD45+ immune cells and CD66b+ myeloid cells (Galeano Niño et al., 2022). The combined application of these technologies provides technical support for comprehensively analyzing the composition, function, and spatial heterogeneity of intratumoral microbiota in CRC.
2.1.2 Composition and characteristics of intratumoral microbiota in colorectal cancer
The intratumoral microbiota in CRC exhibits specific compositional characteristics closely related to tumor stage, molecular subtype, and prognosis. For example, Fusobacterium nucleatum is widely recognized as a marker for advanced CRC. Its abundance in CRC tissues is positively correlated with TNM staging (Ma et al., 2024; Naghavian et al., 2023). Furthermore, other bacterial phyla, such as Bacteroides, Firmicutes, and Proteobacteria, are also enriched in CRC (Gao et al., 2022; Xu et al., 2023; Zhou X. et al., 2022). Studies have shown that the composition of intratumoral microbiota is closely associated with the host’s immune status, tumor stage, and prognosis. This suggests that microbes may influence tumor occurrence and development by regulating the tumor microenvironment (Zhang H. et al., 2024).
2.1.3 Clinical significance of heterogeneity in intratumoral microbiota in colorectal cancer
The diversity, abundance, and distribution of intratumoral microbiota in CRC are important for evaluating prognosis and treatment strategies. In CRC patients, the tumor microbiota usually exhibits higher diversity compared to that of healthy individuals (Zhang et al., 2025). For instance, the microbial alpha diversity index is lower in CRC tumor tissues than in adjacent tissues, and the differences in beta diversity are significantly linked to TNM staging (Long et al., 2024; Zhang et al., 2025). Studies indicate that intratumoral microbiota diversity is closely related to tumor prognosis, with lower microbial diversity usually associated with poorer prognosis (Jin et al., 2024; Zhang et al., 2025). A positive correlation exists between microbial diversity in CRC tumors and tumor diameter, following a power-law model. This suggests that larger tumors tend to have greater internal microbial diversity, indicating that intratumoral microbiota likely play a significant role in the tumor microenvironment and cancer progression (Dovrolis et al., 2024).
Bullman et al. used 16S rRNA gene sequencing combined with spatial transcriptomics to identify varied spatial distribution heterogeneity of bacterial communities such as Fusobacterium nucleatum, Bacteroides, and Leptotrichia in CRC tumor tissues (Galeano Niño et al., 2022). The abundance of Prevotella, E. coli, and Phascolarctobacterium significantly increased in CRC liver metastatic tissues (Gu et al., 2024). In CRC patients, the relative abundance of Alistipes and Blautia was positively correlated with survival probability. In contrast, in rectal cancer patients, the abundance of Porphyromonas was positively correlated with tumor stage (Wang L. et al., 2021).
Furthermore, studies have shown a significant relationship between the diversity of microbes in tumor tissues and the presence of tumor-infiltrating lymphocytes. This suggests that the composition of microbial communities may somewhat reflect the immune microenvironment of tumors (Bhatnagar et al., 2024). Longitudinal cohort studies indicate that antibiotic exposure can increase Fusobacterium levels in tumors while reducing CD8+ T cell infiltration. This implies that the stability of microbial community impacts immune surveillance (Popkes and Valenzano, 2020). Additionally, the stability of intratumoral microbiota is influenced by various factors, including diet, antibiotic use, and the host’s immune status (Bhatnagar et al., 2024). For example, certain probiotics, such as Lactobacillus rhamnosus GG, can enhance intestinal immune responses and promote the stability of microbial communities, which may potentially improving treatment outcomes for CRC patients (Owens et al., 2021). These findings provide valuable insights for improving early diagnosis and developing targeted therapies for CRC (Lee et al., 2021; Okuda et al., 2021) (Table 1).
TABLE 1
| Time | Source | High expression | Low expression | Methods | Results | References |
|---|---|---|---|---|---|---|
| 2022 | CRC OSCC |
Fusobacterium
Bacteroides Leptotrichia Parvimonas Peptoniphilus |
---- | spatial-profiling technologies single-cell RNA sequencing INVADEseq |
The intratumoral microbiota drives tumor heterogeneity by creating an immunosuppressive microenvironment and regulating the transcriptional programs of cancer cells | Farhadi Rad et al. (2024) |
| 2024 | Colorectal cancer liver metastasis | Prevotella Escherichia coli,E. coli,E.coli Phascolarctobacterium |
---- | 16S rDNA | E. coli promotes the progression of colorectal cancer liver metastasis by enhancing lactic acid production and facilitating macrophage M2 polarization | Gubernatorova et al. (2023) |
| 2024 | CRC | Right hemicolon Fusobacteria 、 Alistipes Left hemicolon Firmicutes 、 Bacteroidetes |
Bacteroidetes Firmicutes Verrucomicrobia Actinobacteria Euarchaeota |
The microbial community in CRC exhibits site and molecular subtype-specific distribution, regulating tumor progression through multiple pathways | Gopalakrishnan et al. (2018) | |
| 2024 | CRC treatment | ---- | ---- | Bird shotgun metagenomic sequencing | Orally administered hydrogel (Oxa@HMI) treats CRC by significantly increasing beneficial bacteria in the intestines and tumors while reducing pathogenic bacteria | Lozupone and Knight (2005) |
| 2023 | Locally advanced rectal cancer (LARC) | Metagenomic sequencing | Streptococcus equinus, Scardovia odontolytica and Clostridium hylemonae re significantly associated with the resistance of LARC patients to neoadjuvant chemoradiotherapy (nCRT) | Midha et al. (2023) | ||
| 2021 | CRC |
Fusobacterium
Peptostreptococcus Campylobacter Streptococcus Treponema Selenomonas Enterococcus Veillonella Leptotrichia Parvimonas Gemella Porphyromonas |
Bacteroides
Clostridium Prevotella Faecalibacterium |
16S rRNA | The development of CRC may be partially driven by the symbiotic microbiota through DNA damage or abnormal signaling pathways | Huang et al. (2023) |
| 2021 | Gastrointestinal cancer | Capnocytophaga and Helicobacter (Upper gastrointestinal tumors) Porphyromonas (Rectum Adenocarcinoma) |
---- | PLS-DA | The microbial characteristics of gastrointestinal cancer exhibit significant organ specificity, with distinct differences in the microbial communities of upper and lower digestive tract tumors | Gutiérrez-Salmerón et al. (2021) |
| 2021 | microsatellite instability-high CRC | Fusobacterium nucleatum | quantitative PCR | Fusobacterium nucleatum may influence the progression of MSI-high CRC by promoting tumor-associated immune responses | Jiang et al. (2023) | |
| 2018 | CRC | Bifidobacterium Fusobacterium nucleatum |
quantitative PCR | The content of bifidobacteria is significantly correlated with the proportion of signet-ring cells in CRC. | Scott et al. (2020) | |
| 2024 | Metastatic colorectal cancer (mCRC) |
Helicobacter cinaedi Sphingobium herbicidovorans |
--- | RNA-SEQ 16S rRNA |
The high expression of Helicobacter cinaedi and Sphingobium herbicidovorans is associated with poor prognosis in mCRC. | Sikavi et al. (2021) |
Comprehensive profiling of intratumoral microbiota in colorectal cancer: origins, methodologies, and clinical associations.
2.2 Impact of intratumoral microbiota on the tumor immune microenvironment in colorectal cancer
2.2.1 Recruitment and activation of immune cells
In CRC, the microbiota influences the immune environment of tumors by regulating immune cell recruitment, function, and the expression of immune checkpoint. For instance, as a typical pro-cancer bacterium in CRC, its abundance is significantly correlated with the polarization of tumor-associated macrophages (TAMs) and the methylation of the CDKN2A gene (Park et al., 2017). The FadA protein from Fusobacterium nucleatum activates the E-cadherin/β-catenin signaling pathway. This promotes IL-8 secretion, which leads to the formation of neutrophil extracellular traps (NETs) and inhibits the infiltration of CD8+ T cell (Rubinstein et al., 2013). Additionally, Fusobacterium nucleatum is closely related to immune evasion of tumor cells. By regulating immune cells function, these bacteria suppress anti-tumor immune responses, which in turn promotes tumor growth and metastasis (Hong et al., 2024; Jin et al., 2024). For example, Fusobacterium nucleatum can upregulate PD-L1 expression, directly inhibiting T cell activity, leading to immune evasion and affecting CRC efficacy (Gao et al., 2021; Gao et al., 2023). Furthermore, succinate produced by Prevotella spp. in CRC can weaken anti-tumor immune responses by binding to the SUCNR1 receptor. This binding inhibits mitochondrial oxidative phosphorylation (OXPHOS) and reduces IFN-γ secretion (Franke and Deppenmeier, 2018). Furthermore, the diversity of intratumoral microbiota may also influence the expression of immune checkpoints, impacting the effectiveness of immunotherapy. For instance, the enrichment of Proteobacteria in CRC can activate the TLR4 signaling pathway, inducing PD-L1 expression and leading to resistance to immunotherapy (Gopalakrishnan et al., 2018). Therefore, intratumoral microbiota not only affects the recruitment of immune cells but may also influence the immune response in CRC by regulating the activity of immune cells.
2.2.2 Production and regulation of cytokines
Intratumoral microbiota can regulate the secretion of important cytokines in TIME through metabolic products, pathogen-associated molecular patterns (PAMPs), and epigenetic modifications, thereby altering immune responses (Kyriazi et al., 2024). For example, intratumoral microbiota can stimulate intestinal epithelial cells and immune cells to produce various cytokines such as IL-6, TNF-α, and IFN-γ, driving chronic inflammatory diseases and accelerating tumor progression (Li et al., 2022). An imbalance in intratumoral microbiota can increase immunosuppressive factors, which help tumors evade the immune system (Azevedo et al., 2020; Pratap Singh et al., 2023). Certain microbial metabolites can influence the polarization state of tumor-associated macrophages, which in turn affect the cytokines they secrete and alter the immune status of the tumor microenvironment. For instance, Prevotella has been identified in CRC liver metastatic tissues (Gu et al., 2024). Therefore, intratumoral microbiota influences the immune microenvironment of tumors by recruiting immune cells. Additionally, it shapes the immune characteristics of the tumor microenvironment by regulating the production and release of cytokines.
2.3 Metabolic pathways and immune regulation mediated by intratumoral microbiota in colorectal cancer
2.3.1 Impact of intratumoral microbiota on colorectal cancer cell metabolism
Intratumoral microbiota in CRC not only affects immune evasion but can also alter the metabolic pathways of tumor cells by producing specific metabolites. For example, lactic acid, a common metabolic product of various bacteria, is often found in many tumors, including CRC. The use of lactate dehydrogenase inhibitors can significantly reduce tumor volume (Xue et al., 2023). Fusobacterium nucleatum can promote tumor cell glycolysis by activating HIF-1α and inhibiting OXPHOS, thereby providing energy and biosynthetic precursors for rapid tumor cell proliferation (Dipalma and Blattman, 2023; Midha et al., 2023). Additionally, Bullman et al. confirmed that Fusobacterium nucleatum promotes lipid synthesis in CRC by upregulating FASN (Mager et al., 2020). Furthermore, changes in the composition of intratumoral microbiota may lead to the reprogramming of host metabolic pathways, thereby affecting tumor growth and development. For instance, Bacteroides can promote CRC cell proliferation through the bile acid-FXR axis (Seki et al., 2021). The absence of beneficial bacteria, such as Bifidobacterium, is associated with tumor progression, while the enrichment of pathogenic bacteria, such as Fusobacterium, is closely related to malignant tumor progression (Long et al., 2024). Moreover, intratumoral microbiota can significantly impact targeted therapies for colorectal cancer by influencing host metabolic processes. For example, Bifidobacterium can enhance the efficacy of anti-PD-1 therapy through the inosine-ADORA2A pathway (Mager et al., 2020). Therefore, intervention strategies targeting intratumoral microbiota, such as using probiotics or altering dietary structures, may become new methods to improve CRC treatment outcomes (Sepich-Poore et al., 2021).
2.3.2 Effects of metabolites from intratumoral microbiota on immune cells
Intratumoral microbiota in CRC significantly affects the function of immune cells and the tumor microenvironment through the production of various metabolites. Bacterial metabolites primarily consist of short-chain fatty acids (SCFAs) like acetate, propionate, and butyrate, along with amino acids and vitamins. These compounds can regulate the proliferation, differentiation, and function of immune cells, thus impacting tumor immunity (Scott et al., 2020). For instance, a novel oral hydrogel Oxa@HMI can generate SCFAs, regulate the intratumoral microbiota, stimulate various immune cells to initiate an immune response, and improve the efficacy of CRC treatment (Li et al., 2024). Additionally, butyrate enhances Treg function through HDAC inhibition, which promotes immune tolerance. Meanwhile, propionate activates IL-12 secretion in dendritic cell via GPR43 (Liu C. et al., 2024). Furthermore, SCFAs can regulate the activity of immune cells by activating G protein-coupled receptors (such as GPR41 and GPR43), promoting anti-tumor immune responses (Shi et al., 2023). In CRC patients, the reduction of SCFAs is closely related to tumor progression and immunosuppressive states, indicating that SCFAs may serve as potential biomarkers or therapeutic targets (Jin et al., 2024). Therefore, regulating gut microbiota to increase SCFA production holds promise for improving immune responses and treatment outcomes in CRC patients.
2.3.3 Emerging evidence: Akkermansia muciniphila modulates antitumor immunity via metabolite-driven pathways
Recent advances highlight Akkermansia muciniphila (A. muciniphila) as a dual-functional modulator of colorectal cancer (CRC) through metabolite-immune crosstalk (Kropp et al., 2024; Zhu et al., 2024). As a mucin-degrading bacterium, A. muciniphila produces immunoregulatory short-chain fatty acids (SCFAs; e.g., propionate and acetate) that enhance intestinal barrier integrity and directly activate antitumor immunity (Lin et al., 2025). Propionate engages GPR43 on dendritic cells to amplify antigen presentation, driving CD8+T cell infiltration and IFN-γ production—critical for immune checkpoint inhibitor (ICI) efficacy (Gubernatorova et al., 2023; Li et al., 2025; Wang et al., 2023; Zhu et al., 2024). Concurrently, its surface protein Amuc_1100 activates TLR2/4 pathways, synergizing with SCFAs to suppress PD-L1 expression and bolster T cell-mediated tumor clearance (Wu et al., 2025). Paradoxically, A. muciniphila’s mucolytic activity may exacerbate inflammation in colitis-associated CRC contexts, underscoring context-dependent roles (Zhu et al., 2024). Crucially, elevated A. muciniphila abundance correlates with improved ICI responsiveness and survival in CRC patients, positioning it as both a predictive biomarker and a next-generation probiotic candidate for microbiota-directed therapies (Ni et al., 2022).
2.4 Intratumoral microbiota promotes colorectal cancer metastasis: mechanisms and pathways
Tumor-resident microbiota critically drive colorectal cancer metastasis through coordinated mechanisms targeting cellular plasticity and microenvironment remodeling. For example, Fusobacterium nucleatum induces epithelial-mesenchymal transition (EMT) by activating the PI3K/AKT signaling pathway, thereby enhancing the migration and invasion capabilities of tumor cells (Fan et al., 2024). Concurrently, microbial metabolites such as bile acids engage FXR/TGR5 receptors to induce IL-1β secretion by hepatic macrophages, disrupting endothelial barriers and priming pre-metastatic niches in distant organs (Liu L. et al., 2024). This spatial reprogramming is further amplified by LPS-stimulated neutrophil extracellular traps (NETs), which entrap circulating tumor cells to facilitate vascular adhesion and extravasation (Kyriazi et al., 2024). Notably, microbial communities exhibit context-dependent influences: For example, butyrate has been shown to inhibit the proliferation of tumor cells and induce apoptosis by regulating gene expression through the inhibition of histone deacetylases (HDAC), affecting the growth and migration of tumor cells (Kim et al., 2020). However, in certain cases, butyrate can promote the activation of signaling pathways related to metastasis in the tumor microenvironment (Kong et al., 2022; Zou et al., 2024). These findings establish intratumoral microbiota as master regulators of the metastatic cascade, offering mechanistic links to clinical treatment failures.
3 Role of intratumoral microbiota in colorectal cancer treatment
3.1 Impact of intratumoral microbiota on sensitivity to chemotherapy and radiotherapy in CRC
The composition of intratumoral microbiota can significantly influence the response of CRC to chemotherapy and radiotherapy. For example, Fusobacterium nucleatum is associated with chemotherapy resistance in CRC, possibly due to its promotion of immunosuppressive effects in the tumor microenvironment (Bullman, 2023). Additionally, the diversity of intratumoral microbiota in CRC is closely related to patient prognosis, with low-diversity microbiota typically associated with poorer treatment responses and survival rates (Gopalakrishnan et al., 2018). RNA sequencing of tissues from 105 rectal cancer patients identified 12 microbes linked to treatment response. This finding confirms that the bacterial composition in these tumors significantly differs based on treatment outcomes and serves as an independent biomarker for predicting responses to neoadjuvant chemotherapy and radiotherapy (nCRT) (Sun et al., 2023). Furthermore, a metagenomic sequencing study involving tissues from 73 patients with advanced rectal cancer found that key bacteria, such as Streptococcus, Fusobacterium, and Clostridium, are strongly linked to resistance against nCRT. This study also revealed that butyrate derived from the microbiome may play a role in reducing nCRT efficacy (Huang et al., 2023). In terms of radiotherapy, significant changes in the composition of intratumoral microbiota occur in patients undergoing radiotherapy, which may be related to the efficacy of radiotherapy (Zhang J. et al., 2024). For instance, some studies have found that Fusobacterium nucleatum and Fusobacterium canifelinum are enriched in hypoxic CRC and are closely related to patient prognosis, suggesting potential interactions between hypoxia, microbiota, and radiotherapy responses (Benej et al., 2024). Therefore, interventions targeting intratumoral microbiota may become a potential strategy to enhance CRC patients’ sensitivity to chemotherapy and radiotherapy (Cogdill et al., 2018).
3.2 Role of intratumoral microbiota in colorectal cancer immunotherapy
Intratumoral microbiota critically regulates immunotherapy responses by altering the TIME and immune checkpoint pathways. For instance, the purple photosynthetic bacterium Rhodopseudomonas palustris (RP) has a symbiotic relationship with the anaerobic bacteria Proteus mirabilis (PM) in tumors. This microbial symbiont, known as A-gyo, not only eliminates various tumors, including colorectal cancer (CRC), but also induces durable anti-tumor immunity, offering long-term specific protection (Goto et al., 2023). Moreover, Lactobacillus rhamnosus GG enhances CD8+ T cell responses in the gut, which is essential for strengthening anti-tumor immune responses (Owens et al., 2021). Furthermore, the diversity of intratumoral microbiota and the abundance of specific microbes significantly impact the efficacy of immune checkpoint inhibitors (ICIs). Low-diversity microbiota may contribute to immune suppression and diminish the effectiveness of immunotherapy (Farhadi Rad et al., 2024; Jiang et al., 2023). For instance, the abundance of Fusobacterium nucleatum is associated with tumor immune evasion mechanisms, potentially leading to resistance to ICIs (He et al., 2022). For patients with CRC liver metastasis, RIG-I lactylation inhibitors target E. coli in tumor tissues. This action suppresses M2 macrophage polarization and increases patient sensitivity to 5-fluorouracil (5-FU) (Gu et al., 2024). A study found that certain combinations of intratumoral microbiota can predict how patient respond to ICIs. This indicats that the composition of microbiota may serve as a potential biomarker for assessing treatment responses and prognosis PMID: 39439901. Modulating the composition of intratumoral microbiota could improve the efficacy of immunotherapy and potentially lead to better prognoses for patients (Shen et al., 2021a). Therefore, understanding the interactions between intratumoral microbiota and immunotherapy can help develop personalized treatment strategies to enhance outcomes for CRC patients.
4 Potential of intratumoral microbiota as diagnostic and prognostic biomarkers in colorectal cancer
4.1 Role of intratumoral microbiota in early diagnosis of colorectal cancer
Changes in intratumoral microbiota are strongly linked to the development of CRC, especially in its early stages. Studies comparing the microbiomes of tumor tissues to those of nearby normal tissues have found that some microbes are significantly more abundant in tumor tissues. For example, Fusobacterium nucleatum and Bacteroides fragilis have been shown to be associated with the malignancy of CRC and patient survival rates (Kunzmann et al., 2019; Mondal et al., 2025; Shen X. et al., 2021). Furthermore, microbiome detection in the blood of patients with stage II/III-IV CRC has shown that the detection rates of Escherichia coli, B. fragilis, and Candida albicans are significantly related to CRC liver metastasis and overall survival rates, emphasizing the prognostic value of these microbial biomarkers (Zhang Q. et al., 2024). Another study conducted quantitative PCR testing on tumor tissue samples from 1,313 CRC patients. It found that Bifidobacterium DNA was present in 30% of these samples, and its content was significantly related to the proportion of signet-ring cells in tumors. This suggests that Bifidobacterium could be an indicator of specific colorectal cancer characteristics (Kosumi et al., 2018). Additionally, liquid biopsy techniques based on intratumoral microbiota are becoming a promising non-invasive detection method that can assist in the early diagnosis of colorectal cancer (Ramos et al., 2023; Raza et al., 2022).
4.2 Role of intratumoral microbiota in predicting treatment responses and prognosis in colorectal cancer
Intratumoral microbiota shows significant potential in predicting treatment responses and prognosis in CRC. For example, the levels of intratumoral microbiota are associated with the number of tumor-infiltrating T cells, indicating that these microbes might influence treatment outcomes by altering immune responses (Luu et al., 2023; Pratap Singh et al., 2023). Additionally, the diversity of intratumoral microbiota is positively correlated with patient survival rates, indicating that changes in microbial abundance may serve as a new tool for predicting prognosis in CRC patients. Prospective cohort studies suggest that a dietary pattern known as the ‘sulfur microbiome diet’ may be associated with specific CRC molecular subtypes or the presence of intratumoral microbiota. Long-term adherence to the sulfur microbiome diet may be related to high expression of prostaglandin synthase 2 (PTGS2) and increased risk of distal CRC with Bifidobacterium negativity (Sikavi et al., 2021). These findings provide new insights into the relationship between diet, intratumoral microbiota, and CRC, suggesting that dietary modulation may serve as a preventive strategy for high-risk populations.
5 Challenges and future research directions
5.1 Technical challenges in intratumoral microbiome research
Research on intratumoral microbiomes encounters several technical challenges, mainly due to low microbial biomass and the inability to certain microbial species. These challenges complicate and hinder in-depth studies of tumor microbiota. Current research methods often have difficulty accurately capturing the diversity and changes in tumor microbiota, especially in different tumor microenvironments. To overcome these technical barriers, researchers are developing new methods with high spatial and temporal resolution, such as the combined application of multi-omics technologies (including genomics, transcriptomics, proteomics, and metabolomics). For example, comprehensive profiling of microbial communities in tumor samples through next-generation sequencing (NGS) can obtain complete lineages of microbes without the need for culturing and determine the composition and abundance of microbes in different digestive system tumors (Xuan et al., 2024). Other studies have confirmed strong correlations between Helicobacter cinaedi and Sphingobium herbicidovorans in metastatic colorectal cancer tissues with specific genes (such as SELENBP1 and SNORA38), providing new ideas for personalized treatment (Feng et al., 2024). These technological advancements will help to better understand the interactions between tumor microbiota and the tumor microenvironment, thereby providing new ideas and strategies for early diagnosis and treatment of cancer.
5.2 Ethical and safety issues in clinical applications
When translating the research findings of intratumoral microbiomes into clinical applications, ethical and safety issues cannot be overlooked. First, interventions in the microbiome could have unexpected effects on patient health, necessitating in-depth discussions of related ethical issues. For example, we need to address how to ensure patients receive adequate informed consent for microbiome interventions and how to assess and monitor the safety of these interventions. Additionally, individual differences in microbiomes make it challenging to create universal treatment plans (Liu et al., 2021). Therefore, future research should focus on developing personalized microbiome modulation treatment plans while ensuring ethical considerations to maintain patient safety and treatment effectiveness.
5.3 Heterogeneity in sample collection of intratumoral microbiota
There is significant heterogeneity in the collection of intratumoral microbiota samples, posing challenges to the reproducibility and reliability of research results. Various factors, including patients’ tumor microenvironments, treatment histories, and lifestyles, can influence the composition and function of microbiota (Zhang et al., 2020). For instance, CRC-related pathogenic bacteria (such as Clostridium, Bacteroides, Eubacterium, and Prevotella) exhibit high variability across different samples (Barc et al., 2008; Benjamin et al., 2012). Therefore, standardized protocols for collecting and processing samples must be established to reduce variability in microbiome studies. Additionally, specific sample collection strategies may need to be developed for different types of tumors to ensure that the true state of tumor microbiota can be captured. Future research should focus on the heterogeneity of samples and validate the relationships between microbiomes and tumor characteristics. This can be achieved through large-scale, multi-center clinical trials, which will enhance the application of microbiome research in cancer treatment.
6 Conclusion
This review reveals the mechanisms by which the intratumoral microbiome affects treatment response through a metabolic-immune regulatory network in CRC (Figure 1). This not only provides new biomarkers for predicting CRC progression but also offers potential new targets for cancer therapy. However, current research on intratumoral microbiota still has some controversies and gaps, and how to effectively integrate these discoveries into clinical applications is a pressing issue. Future studies should systematically explore the characteristics of microbial communities in different patient populations, tumor types, and treatment regimens, using big data and AI to pull out valuable insights and promote the progress of clinical translational research. In clinical practice, focusing on the impact of intratumoral microbiota could lead to new treatment strategies. By adjusting microbial communities, we hope to improve patients’ response rates to chemotherapy or immunotherapy, but we need to rigorously validate the effectiveness and safety. In summary, intratumoral microbiota have great potential in the research and development of CRC. Encouraging collaboration across disciplines can help us better understand their role in tumor biology, bringing new hope and treatment options to patients.
FIGURE 1
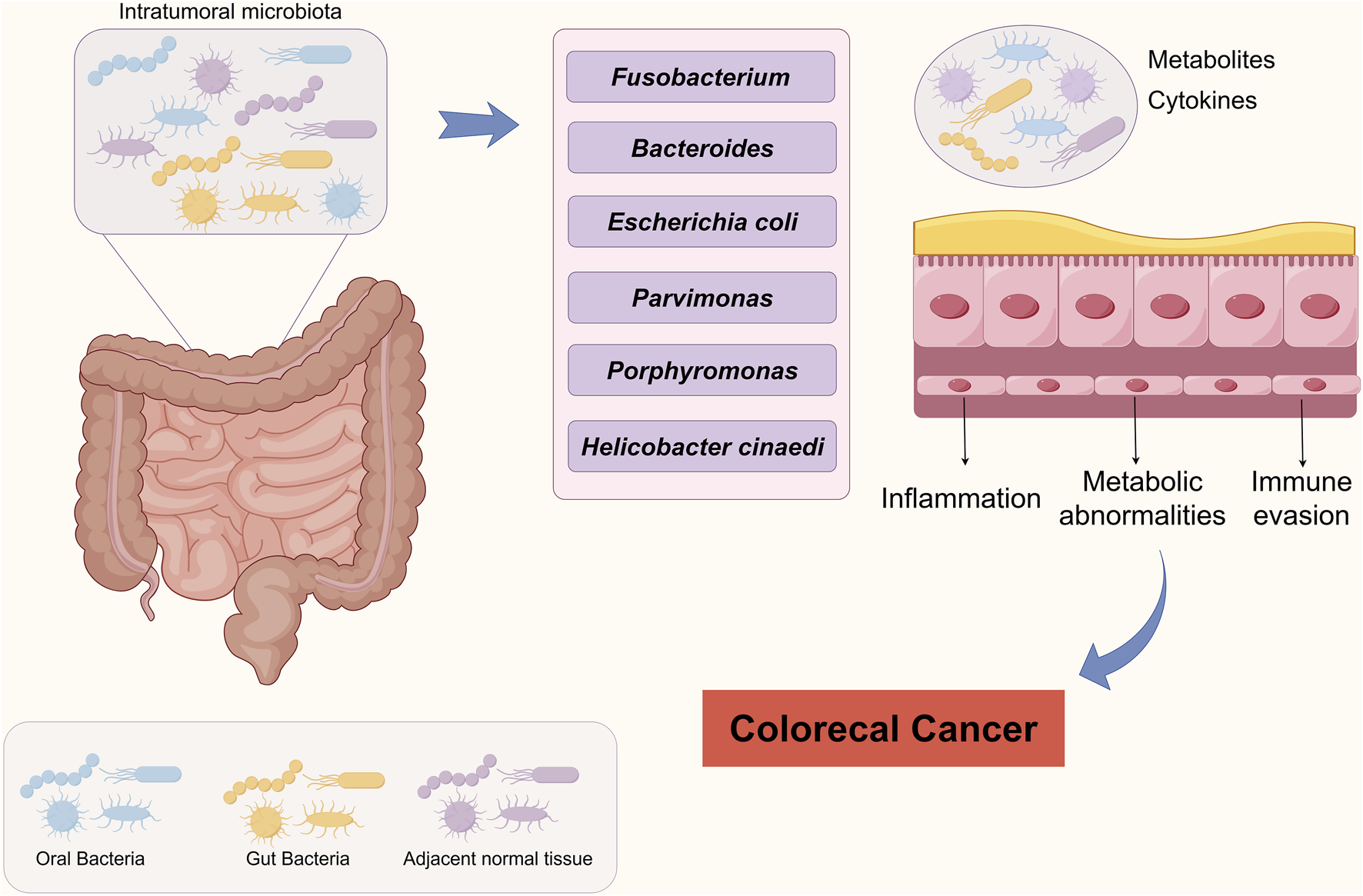
Schematic representation of intratumoral microbiota driving colorectal carcinogenesis.
Statements
Author contributions
MH: Funding acquisition, Writing – review and editing, Writing – original draft, Methodology. HX: Writing – review and editing, Data curation, Methodology. ML: Writing – original draft, Visualization, Data curation, Investigation. DJ: Project administration, Writing – review and editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by Department of Science and Technology of Jilin Province (No. 20230402003GH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Azevedo M. M. Pina-Vaz C. Baltazar F. (2020). Microbes and cancer: friends or faux?Int. J. Mol. Sci.21 (9), 3115. 10.3390/ijms21093115
2
Barc M. C. Charrin-Sarnel C. Rochet V. Bourlioux F. Sandré C. Boureau H. et al (2008). Molecular analysis of the digestive microbiota in a gnotobiotic mouse model during antibiotic treatment: influence of saccharomyces boulardii. Anaerobe14 (4), 229–233. 10.1016/j.anaerobe.2008.04.003
3
Benej M. Hoyd R. Kreamer M. Wheeler C. E. Grencewicz D. J. Choueiry F. et al (2024). The tumor microbiome reacts to hypoxia and can influence response to radiation treatment in colorectal cancer. Cancer Res. Commun.4 (7), 1690–1701. 10.1158/2767-9764.CRC-23-0367
4
Benjamin J. L. R. H. C. Koutsoumpas A. Ng S. C. McCarthy N. E. Prescott N. J. Whelan K. et al (2012). Smokers with active crohn's disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm. Bowel Dis.18 (6), 1092–1100. 10.1002/ibd.21864
5
Bhatnagar K. Jha K. Dalal N. Patki N. Gupta G. Kumar A. et al (2024). Exploring micronutrients and microbiome synergy: pioneering new paths in cancer therapy. Front. Immunol.15, 1442788. 10.3389/fimmu.2024.1442788
6
Bullman S. (2023). The intratumoral microbiota: from microniches to single cells. Cell.186 (8), 1532–1534. 10.1016/j.cell.2023.03.012
7
Byrd D. A. Damerell V. Morales M. Hogue S. R. Lin T. Ose J. et al (2025). The gut microbiome is associated with disease-free survival in stage I-III colorectal cancer patients. Int. J. Cancer157 (1), 64–73. 10.1002/ijc.35342
8
Cai L. Chen A. Tang D. (2024). A new strategy for immunotherapy of microsatellite-stable (MSS)-Type advanced colorectal cancer: multi-Pathway combination therapy with PD-1/PD-L1 inhibitors. Immunology173 (2), 209–226. 10.1111/imm.13785
9
Cao Y. Xia H. Tan X. Shi C. Ma Y. Meng D. et al (2024). Intratumoural microbiota: a new frontier in cancer development and therapy. Signal Transduct. Target Ther.9 (1), 15. 10.1038/s41392-023-01693-0
10
Cogdill A. P. Gaudreau P. O. Arora R. Gopalakrishnan V. Wargo J. A. (2018). The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol.39 (11), 900–920. 10.1016/j.it.2018.09.007
11
DiPalma M. P. Blattman J. N. (2023). The impact of microbiome dysbiosis on T cell function within the tumor microenvironment (TME). Front. Cell. Dev. Biol.11, 1141215. 10.3389/fcell.2023.1141215
12
Dovrolis N. Gazouli M. Rigal F. Whittaker R. J. Matthews T. J. Georgiou K. et al (2024). Power-law scaling in intratumoral microbiota of colorectal cancer. Gut Pathog.16 (1), 34. 10.1186/s13099-024-00631-x
13
Fan S. Zhou L. Zhang W. Wang D. Tang D. (2024). Role of imbalanced gut microbiota in promoting CRC metastasis: from theory to clinical application. Cell. Commun. Signal22 (1), 232. 10.1186/s12964-024-01615-9
14
Farhadi Rad H. Tahmasebi H. Javani S. Hemati M. Zakerhamidi D. Hosseini M. et al (2024). Microbiota and cytokine modulation: innovations in enhancing anticancer immunity and personalized cancer therapies. Biomedicines12 (12), 2776. 10.3390/biomedicines12122776
15
Feng L. Wang R. Zhao Q. Wang J. Luo G. Xu C. (2024). Racial disparities in metastatic colorectal cancer outcomes revealed by tumor microbiome and transcriptome analysis with bevacizumab treatment. Front. Pharmacol.14, 1320028. 10.3389/fphar.2023.1320028
16
Franke T. Deppenmeier U. (2018). Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol. Microbiol.109 (4), 528–540. 10.1111/mmi.14058
17
Galeano Niño J. L. Wu H. LaCourse K. D. Kempchinsky A. G. B. A. Barber B. Futran N. et al (2022). Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature611 (7937), 810–817. 10.1038/s41586-022-05435-0
18
Gao Y. Bi D. Xie R. Li M. Guo J. Liu H. et al (2021). Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct. Target Ther.6 (1), 398. 10.1038/s41392-021-00795-x
19
Gao F. Yu B. Rao B. Sun Y. Yu J. Wang D. et al (2022). The effect of the intratumoral microbiome on tumor occurrence, progression, prognosis and treatment. Front. Immunol.13, 1051987. 10.3389/fimmu.2022.1051987
20
Gao Y. Zou T. Xu P. Wang Y. Jiang Y. Chen Y. X. et al (2023). Fusobacterium nucleatum stimulates cell proliferation and promotes PD-L1 expression via IFIT1-related signal in colorectal cancer. Neoplasia35, 100850. 10.1016/j.neo.2022.100850
21
Gopalakrishnan V. Spencer C. N. Nezi L. Reuben A. Andrews M. C. Karpinets T. V. et al (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science359 (6371), 97–103. 10.1126/science.aan4236
22
Goto Y. Iwata S. Miyahara M. Miyako E. (2023). Discovery of intratumoral oncolytic bacteria toward targeted anticancer theranostics. Adv. Sci.10 (20), e2301679. 10.1002/advs.202301679
23
Gu J. Xu X. Li X. Yue L. Zhu X. Chen Q. et al (2024). Tumor-resident microbiota contributes to colorectal cancer liver metastasis by lactylation and immune modulation. Oncogene43 (31), 2389–2404. 10.1038/s41388-024-03080-7
24
Gubernatorova E. O. Gorshkova E. A. Bondareva M. A. Podosokorskaya O. A. Sheynova A. D. Yakovleva A. S. et al (2023). Akkermansia muciniphila - friend or foe in colorectal cancer?Front. Immunol.14, 1303795. 10.3389/fimmu.2023.1303795
25
Gutiérrez-Salmerón M. Lucena S. R. Chocarro-Calvo A. García-Martínez J. M. Martín Orozco R. M. García-Jiménez C. (2021). Remodelling of colorectal cancer cell signalling by microbiota and immunity in diabetes. Endocr. Relat. Cancer28 (6), R173–R190. 10.1530/ERC-20-0315
26
Hardwick S. A. Chen W. Y. Wong T. Kanakamedala B. S. Deveson I. W. Ongley S. E. et al (2018). Synthetic microbe communities provide internal reference standards for metagenome sequencing and analysis. Nat. Commun.9 (1), 3096. 10.1038/s41467-018-05555-0
27
He Z. Tian W. Wei Q. Xu J. (2022). Involvement of Fusobacterium nucleatum in malignancies except for colorectal cancer: a literature review. Front. Immunol.13, 968649. 10.3389/fimmu.2022.968649
28
Hong B. Y. Chhaya A. Robles A. Cervantes J. Tiwari S. (2024). The role of Fusobacterium nucleatum in the pathogenesis of Colon cancer. J. Investig. Med.72 (8), 819–827. 10.1177/10815589241277829
29
Huang X. Chen C. Xie W. Zhou C. Tian X. Zhang Z. et al (2023). Metagenomic analysis of intratumoral microbiome linking to response to neoadjuvant chemoradiotherapy in rectal cancer. Int. J. Radiat. Oncol. Biol. Phys.117 (5), 1255–1269. 10.1016/j.ijrobp.2023.06.2515
30
Jiang S. S. Xie Y. L. Xiao X. Y. Kang Z. R. Lin X. L. Zhang L. et al (2023). Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell. Host Microbe31 (5), 781–797.e9. 10.1016/j.chom.2023.04.010
31
Jin S. Zhong W. Li B. Wang K. Lai D. (2024). Multidimensional analysis of the impact of gemmatimonas, rhodothermus, and sutterella on drug and treatment response in colorectal cancer. Front. Cell. Infect. Microbiol.14, 1457461. 10.3389/fcimb.2024.1457461
32
Kim D. J. Yang J. Seo H. Lee W. H. Ho Lee D. Kym S. et al (2020). Colorectal cancer diagnostic model utilizing metagenomic and metabolomic data of stool microbial extracellular vesicles. Sci. Rep.10 (1), 2860. 10.1038/s41598-020-59529-8
33
Kim Y. Kim G. T. Kang J. (2023). Microbial composition and stool short chain fatty acid levels in fibromyalgia. Int. J. Environ. Res. Public Health20 (4), 3183. 10.3390/ijerph20043183
34
Kong F. Fang C. Zhang Y. Duan L. Du D. Xu G. et al (2022). Abundance and metabolism disruptions of intratumoral microbiota by chemical and physical actions unfreeze tumor treatment resistance. Adv. Sci.9 (7), e2105523. 10.1002/advs.202105523
35
Kosumi K. Hamada T. Koh H. Borowsky J. Bullman S. Twombly T. S. et al (2018). The amount of bifidobacterium genus in colorectal carcinoma tissue in relation to tumor characteristics and clinical outcome. Am. J. Pathol.188 (12), 2839–2852. 10.1016/j.ajpath.2018.08.015
36
Kropp C. Tambosco K. Chadi S. Langella P. Claus S. P. Martin R. (2024). Christensenella minuta protects and restores intestinal barrier in a colitis mouse model by regulating inflammation. NPJ Biofilms Microbiomes10 (1), 88. 10.1038/s41522-024-00540-6
37
Kunzmann A. T. Proença M. A. Jordao H. W. Jiraskova K. Schneiderova M. Levy M. et al (2019). Fusobacterium nucleatum tumor DNA levels are associated with survival in colorectal cancer patients. Eur. J. Clin. Microbiol. Infect. Dis.38 (10), 1891–1899. 10.1007/s10096-019-03649-1
38
Kyriazi A. A. Karaglani M. Agelaki S. Baritaki S. (2024). Intratumoral microbiome: Foe or friend in reshaping the tumor microenvironment landscape?Cells13 (15), 1279. 10.3390/cells13151279
39
Lee J. A. Yoo S. Y. Oh H. J. Jeong S. Cho N. Y. Kang G. H. et al (2021). Differential immune microenvironmental features of microsatellite-unstable colorectal cancers according to Fusobacterium nucleatum status. Cancer Immunol. Immunother.70 (1), 47–59. 10.1007/s00262-020-02657-x
40
Li X. Qi M. He K. Liu H. Yan W. Zhao L. et al (2022). Neospora caninum inhibits tumor development by activating the immune response and destroying tumor cells in a B16F10 melanoma model. Parasit. Vectors15 (1), 332. 10.1186/s13071-022-05456-8
41
Li X. Yan X. Wang Y. Kaur B. Han H. Yu J. (2023). The notch signaling pathway: a potential target for cancer immunotherapy. J. Hematol. Oncol.16 (1), 45. 10.1186/s13045-023-01439-z
42
Li L. He S. Liao B. Wang M. Lin H. Hu B. et al (2024). Orally administrated hydrogel harnessing intratumoral microbiome and microbiota-related immune responses for potentiated colorectal cancer treatment. Research7, 0364. 10.34133/research.0364
43
Li X. Rui W. Shu P. Sun Y. Yang J. (2025). Efficacy evaluation of selenium-enriched Akkermansia muciniphila in the treatment of Colon tumor mice. Probiotics Antimicrob. Proteins. 10.1007/s12602-025-10500-x
44
Lin J. Wang T. Zhou Y. Sha J. Chen X. Wang W. et al (2025). Molecular characterization of extended-spectrum β-lactamases from the akkermansia genus. Int. J. Food Microbiol.428, 110998. 10.1016/j.ijfoodmicro.2024.110998
45
Liu W. Zhang X. Xu H. Li S. Lau H. C. Chen Q. et al (2021). Microbial community heterogeneity within colorectal neoplasia and its correlation with colorectal carcinogenesis. Gastroenterology160 (7), 2395–2408. 10.1053/j.gastro.2021.02.020
46
Liu C. Yang L. Gao T. Yuan X. Bajinka O. Wang K. (2024a). A mini-review-cancer energy reprogramming on drug resistance and immune response. Transl. Oncol.49, 102099. 10.1016/j.tranon.2024.102099
47
Liu L. Li Y. Zheng X. Huang R. Huang X. Zhao Y. et al (2024b). Natural polysaccharides regulate intestinal microbiota for inhibiting colorectal cancer. Heliyon10 (10), e31514. 10.1016/j.heliyon.2024.e31514
48
Long J. Wang J. Xiao C. You F. Jiang Y. Li X. (2024). Intratumoral microbiota in colorectal cancer: focus on specific distribution and potential mechanisms. Cell. Commun. Signal22 (1), 455. 10.1186/s12964-024-01831-3
49
Lozupone C. Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol.71 (12), 8228–8235. 10.1128/AEM.71.12.8228-8235.2005
50
Luu K. Ye J. Y. Lagishetty V. Liang F. Hauer M. Sedighian F. et al (2023). Fecal and tissue microbiota are associated with tumor T-Cell infiltration and mesenteric lymph node involvement in colorectal cancer. Nutrients15 (2), 316. 10.3390/nu15020316
51
Ma S. Yao H. Si X. Huang Z. Wang R. Wan R. et al (2024). Orally available dextran-aspirin nanomedicine modulates gut inflammation and microbiota homeostasis for primary colorectal cancer therapy. J. Control Release370, 528–542. 10.1016/j.jconrel.2024.05.002
52
Mager L. F. Burkhard R. Pett N. Cooke N. C. A. Brown K. Ramay H. et al (2020). Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science369 (6510), 1481–1489. 10.1126/science.abc3421
53
Maley S. J. Yue Y. Burns K. F. Hovey K. M. Wactawski-Wende J. Freudenheim J. L. et al (2024). Alcohol consumption and the diversity of the oral microbiome in postmenopausal women. J. Nutr.154 (1), 202–212. 10.1016/j.tjnut.2023.10.025
54
Midha A. D. Zhou Y. Queliconi B. B. Barrios A. M. Haribowo A. G. Chew B. T. L. et al (2023). Organ-specific fuel rewiring in acute and chronic hypoxia redistributes glucose and fatty acid metabolism. Cell. Metab.35 (3), 504–516.e5. 10.1016/j.cmet.2023.02.007
55
Mondal T. Chattopadhyay D. Saha Mondal P. Das S. Mondal A. Das A. et al (2025). Fusobacterium nucleatum modulates the Wnt/β-catenin pathway in colorectal cancer development. Int. J. Biol. Macromol.299, 140196. 10.1016/j.ijbiomac.2025.140196
56
Naghavian R. Faigle W. Oldrati P. Wang J. Toussaint N. C. Qiu Y. et al (2023). Microbial peptides activate tumour-infiltrating lymphocytes in glioblastoma. Nature617 (7962), 807–817. 10.1038/s41586-023-06081-w
57
Nejman D. Livyatan I. Fuks G. Gavert N. Zwang Y. Geller L. T. et al (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science368 (6494), 973–980. 10.1126/science.aay9189
58
Ni J. J. Li X. S. Zhang H. Xu Q. Wei X. T. Feng G. J. et al (2022). Mendelian randomization study of causal link from gut microbiota to colorectal cancer. BMC Cancer22 (1), 1371. 10.1186/s12885-022-10483-w
59
Okuda S. Shimada Y. Tajima Y. Yuza K. Hirose Y. Ichikawa H. et al (2021). Profiling of host genetic alterations and intra-tumor microbiomes in colorectal cancer. Comput. Struct. Biotechnol. J.19, 3330–3338. 10.1016/j.csbj.2021.05.049
60
Owens J. A. Saeedi B. J. Naudin C. R. Hunter-Chang S. Barbian M. E. Eboka R. U. et al (2021). Lactobacillus rhamnosus GG orchestrates an antitumor immune response. Cell. Mol. Gastroenterol. Hepatol.12 (4), 1311–1327. 10.1016/j.jcmgh.2021.06.001
61
Park H. E. Kim J. H. Cho N. Y. Lee H. S. Kang G. H. (2017). Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch.471 (3), 329–336. 10.1007/s00428-017-2171-6
62
Popkes M. Valenzano D. R. (2020). Microbiota-host interactions shape ageing dynamics. Philos. Trans. R. Soc. Lond B Biol. Sci.375 (1808), 20190596. 10.1098/rstb.2019.0596
63
Pratap Singh R. Kumari N. Gupta S. Jaiswal R. Mehrotra D. Singh S. et al (2023). Intratumoral microbiota changes with tumor stage and influences the immune signature of oral squamous cell carcinoma. Microbiol. Spectr.11 (4), e0459622. 10.1128/spectrum.04596-22
64
Ramos P. Carvalho M. R. Chen W. Yan L. P. Zhang C. H. He Y. L. et al (2023). Microphysiological systems to study colorectal cancer: State-Of-The-Art. Biofabrication15 (3), 032001. 10.1088/1758-5090/acc279
65
Raza A. Khan A. Q. Inchakalody V. P. Mestiri S. Yoosuf Z. Bedhiafi T. et al (2022). Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J. Exp. Clin. Cancer Res.41 (1), 99. 10.1186/s13046-022-02318-0
66
Rubinstein M. R. Wang X. Liu W. Hao Y. Cai G. Han Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell. Host Microbe14 (2), 195–206. 10.1016/j.chom.2013.07.012
67
Scott S. A. Fu J. Chang P. V. (2020). Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U. S. A.117 (32), 19376–19387. 10.1073/pnas.2000047117
68
Seki D. Mayer M. Hausmann B. Pjevac P. Giordano V. Goeral K. et al (2021). Aberrant gut-microbiota-immune-brain axis development in premature neonates with brain damage. Cell. Host Microbe29 (10), 1558–1572.e6. 10.1016/j.chom.2021.08.004
69
Sepich-Poore G. Zitvogel L. Straussman R. Hasty J. Wargo J. A. Knight R. (2021). The microbiome and human cancer. Science371 (6536), eabc4552. 10.1126/science.abc4552
70
Shen L. Ye Y. Sun H. Su B. (2021a). ILC3 plasticity in microbiome-mediated tumor progression and immunotherapy. Cancer Cell.39 (10), 1308–1310. 10.1016/j.ccell.2021.08.002
71
Shen X. Li J. Li J. Zhang Y. Li X. Cui Y. et al (2021b). Fecal enterotoxigenic bacteroides fragilis-Peptostreptococcus stomatis-Parvimonas micra biomarker for noninvasive diagnosis and prognosis of colorectal laterally spreading tumor. Front. Oncol.11, 661048. 10.3389/fonc.2021.661048
72
Shi L. Xu Y. Feng M. (2023). Role of gut microbiome in immune regulation and immune checkpoint therapy of colorectal cancer. Dig. Dis. Sci.68 (2), 370–379. 10.1007/s10620-022-07689-0
73
Sikavi D. R. Nguyen L. H. Haruki K. Ugai T. Ma W. Wang D. D. et al (2021). The sulfur microbial diet and risk of colorectal cancer by molecular subtypes and intratumoral microbial species in adult men. Clin. Transl. Gastroenterol.12 (8), e00338. 10.14309/ctg.0000000000000338
74
Sun L. Qu J. Ke X. Zhang Y. Xu H. Lv N. et al (2023). Interaction between intratumoral microbiota and tumor mediates the response of neoadjuvant therapy for rectal cancer. Front. Microbiol.14, 1229888. 10.3389/fmicb.2023.1229888
75
Sung H. Ferlay J. Siegel R. Laversanne M. Soerjomataram I. Jemal A. et al (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71 (3), 209–249. 10.3322/caac.21660
76
Wang J. Wang Y. Li Z. Gao X. Huang D. (2021a). Global analysis of microbiota signatures in four major types of gastrointestinal cancer. Front. Oncol.11, 685641. 10.3389/fonc.2021.685641
77
Wang L. Peng F. Peng C. Du J. R. (2021b). Gut microbiota in tumor microenvironment: a critical regulator in cancer initiation and development as potential targets for Chinese medicine. Am. J. Chin. Med.49 (3), 609–626. 10.1142/S0192415X21500270
78
Wang T. Wu L. Wang S. Shi X. Liu H. Deng W. (2023). Chang wei qing decoction enhances the anti-tumor effect of PD-1 inhibitor therapy by regulating the immune microenvironment and gut microbiota in colorectal cancer. Chin. J. Nat. Med.21 (5), 333–345. 10.1016/S1875-5364(23)60451-0
79
Wei Y. Wu Z. Lan G. (2025). Distinct spatiotemporal patterns between fungal alpha and beta diversity of soil-plant continuum in rubber tree. Microbiol. Spectr.13 (2), e0209724. 10.1128/spectrum.02097-24
80
Wu X. Yu D. Ma Y. Fang X. Sun P. (2025). Function and therapeutic potential of Amuc_1100, an outer membrane protein of akkermansia muciniphila: a review. Int. J. Biol. Macromol.308 (Pt 4), 142442. 10.1016/j.ijbiomac.2025.142442
81
Xu Y. Zhao F. (2018). Single-cell metagenomics: challenges and applications. Protein Cell.9 (5), 501–510. 10.1007/s13238-018-0544-5
82
Xu Y. Zhao J. Ma Y. Liu J. Cui Y. Yuan Y. et al (2023). The microbiome types of colorectal tissue are potentially associated with the prognosis of patients with colorectal cancer. Front. Microbiol.14, 1100873. 10.3389/fmicb.2023.1100873
83
Xuan M. Gu X. Liu Y. Yang L. Li Y. Huang D. et al (2024). Intratumoral microorganisms in tumors of the digestive system. Cell. Commun. Signal22 (1), 69. 10.1186/s12964-023-01425-5
84
Xue C. Li G. Zheng Q. Gu X. Shi Q. Su Y. et al (2023). Tryptophan metabolism in health and disease. Cell. Metab.35 (8), 1304–1326. 10.1016/j.cmet.2023.06.004
85
Zhang X. Liu Q. Liao Q. Zhao Y. (2020). Pancreatic cancer, gut microbiota, and therapeutic efficacy. J. Cancer11 (10), 2749–2758. 10.7150/jca.37445
86
Zhang Z. Bao C. Jiang L. Wang S. Wang K. Lu C. et al (2023). When cancer drug resistance meets metabolomics (bulk, single-cell And/Or spatial): progress, potential, and perspective. Front. Oncol.12, 1054233. 10.3389/fonc.2022.1054233
87
Zhang H. Fu L. Leiliang X. Qu C. Wu W. Wen R. et al (2024a). Beyond the gut: the intratumoral microbiome's influence on tumorigenesis and treatment response. Cancer Commun.44 (10), 1130–1167. 10.1002/cac2.12597
88
Zhang J. Wang P. Wang J. Wei X. Wang M. (2024b). Unveiling intratumoral microbiota: an emerging force for colorectal cancer diagnosis and therapy. Pharmacol. Res.203, 107185. 10.1016/j.phrs.2024.107185
89
Zhang Q. Song J. Wu H. Wang L. Zhuo G. Li H. et al (2024c). Intratumoral microbiota associates with systemic immune inflammation state in nasopharyngeal carcinoma. Int. Immunopharmacol.141, 112984. 10.1016/j.intimp.2024.112984
90
Zhang X. Kong C. Chen K. Liu J. (2025). Colonization of stenotrophomonas and its associated microbiome between paired primary colorectal cancers and their lung metastatic tumors (experimental studies). Int. J. Surg.111 (2), 2291–2295. 10.1097/JS9.0000000000002121
91
Zhao L. Cho W. C. Nicolls M. R. (2021). Colorectal cancer-associated microbiome patterns and signatures. Front. Genet.12, 787176. 10.3389/fgene.2021.787176
92
Zhou P. Yang D. Sun D. Zhou Y. (2022a). Gut microbiome: new biomarkers in early screening of colorectal cancer. J. Clin. Lab. Anal.36 (5), e24359. 10.1002/jcla.24359
93
Zhou X. Kandalai S. Hossain F. Zheng Q. (2022b). Tumor microbiome metabolism: a game changer in cancer development and therapy. Front. Oncol.12, 933407. 10.3389/fonc.2022.933407
94
Zhu Z. Huang J. Zhang Y. Hou W. Chen F. Mo Y. Y. et al (2024). Landscape of tumoral ecosystem for enhanced anti-PD-1 immunotherapy by gut Akkermansia muciniphila. Cell. Rep.43 (6), 114306. 10.1016/j.celrep.2024.114306
95
Zou Q. Dong H. Cronan J. E. (2024). The enteric bacterium Enterococcus faecalis elongates and incorporates exogenous short and medium chain fatty acids into membrane lipids. Mol. Microbiol.122 (5), 757–771. 10.1111/mmi.15322
96
Zwinsová B. Petrov V. A. Hrivňáková M. Smatana S. Micenková L. Kazdová N. et al (2021). Colorectal tumour mucosa microbiome is enriched in oral pathogens and defines three subtypes that correlate with markers of tumour progression. Cancers (Basel)13 (19), 4799. 10.3390/cancers13194799
Summary
Keywords
colorectal cancer, intratumoral microbiota, tumor microenvironment, tumor therapy, chemotherapy, immunotherapy
Citation
Hao M, Xu H, Li M and Jiao D (2025) The role and challenges of intratumoral microbiota in colorectal cancer immunotherapy. Front. Pharmacol. 16:1634703. doi: 10.3389/fphar.2025.1634703
Received
25 May 2025
Accepted
28 July 2025
Published
07 August 2025
Volume
16 - 2025
Edited by
Baoming Wang, University of Technology Sydney, Australia
Reviewed by
Fan Ying, University Health Network (UHN), Canada
Lukasz Laczmanski, Polish Academy of Sciences, Poland
Updates
Copyright
© 2025 Hao, Xu, Li and Jiao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Jiao, jiaodan@jlu.edu.cn; Min Li, liming198@jlu.edu.cn
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.