- 1Department of Pharmacy, Faculty of Science, University of Rajshahi, Rajshahi, Bangladesh
- 2Division of Pharmacogenomics and Personalized Medicine, Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
- 3Laboratory for Pharmacogenomics, Somdech Phra Debaratana Medical Center (SDMC), Ramathibodi Hospital, Bangkok, Thailand
- 4Pharmacogenomics and Precision Medicine, The Preventive Genomics and Family Check-Up Services Center, Bumrungrad International Hospital, Bangkok, Thailand
- 5Faculty of Pharmaceutical Sciences, Burapha University, Saensuk, Chonburi, Thailand
Adverse drug reactions (ADRs) are gradually becoming a concerning health threat worldwide in patients undergoing acute or chronic therapy. Antibiotics are the main drugs that cause immune-mediated ADRs, such as severe cutaneous adverse reactions (SCARs), allergic reactions, and organ-specific diseases, representing a significant threat to patient safety. In this review, we present the current genetic evidence available for antibiotic-related toxicities from a pharmacogenomics (PGx) perspective. We also explore the current state of PGx-based dosing recommendations and the factors limiting their widespread application in routine clinical practice. Through a systematic literature review, this study identified at least 12 antibiotic–gene pairs (amikacin–MT-RNR1, gentamicin–MT-RNR1, kanamycin–MT-RNR1, streptomycin–MT-RNR1, neomycin–MT-RNR1, tobramycin–MT-RNR1, isoniazid–NAT2, dapsone–HLA-B, co-trimoxazole–HLA-B, HLA-C, flucloxacillin–HLA-B, daunorubicin–SLC28A3, and doxorubicin–SLC28A3) with moderate to high Pharmacogenomics Knowledgebase (PharmGKB) evidence levels for toxicity. However, PGx-based dosing guidelines, as recommended by the Clinical Pharmacogenetics Implementation Consortium (CPIC), the Dutch Pharmacogenetics Working Group (DPWG), and the Canadian Pharmacogenomics Network for Drug Safety (CPNDS), are currently available only for the following antibiotic–gene pairs: amikacin, gentamicin, kanamycin, streptomycin, neomycin, and tobramycin–MT-RNR1; flucloxacillin–HLA-B; dapsone–G6PD; nitrofurantoin–G6PD; and daunorubicin and doxorubicin–RARG, SLC28A3, and UGT1A6. Despite the established and growing genetic evidence for toxicity, particularly for Co-trimoxazole-induced SCARs by HLA-B and HLA-C, dapsone-induced SCARs by the HLA-B, and isoniazid-induced liver injury by the NAT2, insufficient approaches are being undertaken to translate these findings into routine clinical practice. The lack of validation of preliminary genetic associations, due to the scarcity of proper follow-up and large-scale replication, remains a key setback for PGx-based implementation of antibiotic therapy in clinical settings. More focused clinical studies, cost-effectiveness analyses, and polygenic risk score development are required to enable the PGx-based clinical use of antibiotics and optimize both safety and effectiveness in achieving precision medicine.
1 Introduction
Adverse drug reactions (ADRs) are gradually becoming a concerning health threat worldwide in patients undergoing acute or chronic therapy (Osanlou et al., 2018). Rawlins and Thompson grouped ADRs into two types: dose-dependent and predictable reactions (type A) and unpredictable dose-independent reactions (type B) (Dekker et al., 1997). Hypersensitivity reaction, a type-B ADR, is produced by cellular mediators released through both immunological and non-immune mechanisms (Doña et al., 2012). Allergic reactions are hypersensitivity reactions involving either an immunoglobulin E (IgE)-mediated or non-IgE (e.g., T cell)-mediated mechanism (Johansson et al., 2004). Severe cutaneous adverse reactions (SCARs) are potentially fatal T-cell-mediated delayed allergic reactions (Peter et al., 2017). The most prevalent SCARs, contributing to over 85% of the SCARs occurring in adults, are drug reaction with eosinophilia and systemic symptoms (DRESS), Stevens–Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), and acute generalized exanthematous pustulosis (AGEP) (Duong et al., 2017; Sassolas et al., 2010).
A high estimated mortality ranging from 10% to 40% for SJS/TEN, <5% for AGEP and 2%–10% for DRESS was reported (Chen et al., 2010; Duong et al., 2017; Firoz et al., 2012; Husain et al., 2013; Kloypan et al., 2021; Owen and Jones, 2021; Schneck et al., 2008). Globally, the prevalence of SCARs was said to be 0.4–1.2 per million/years (Verma et al., 2013). Nevertheless, a racial discrepancy in the prevalence of SCARs has also been recorded. For example, the incidence was reported to be as high as 1.53–1.89 per million/year in the German population, whereas among the Filipino population, the rate of SCARs was reported to be 6.25/10,000 people from 2011 to 2015 (Guzman and Paliza, 2018; Mockenhaupt, 2012; Tempark et al., 2022). Additionally, the prevalence of TEN and SJS was estimated to be 0.4–1.2 and 1-6 per million/year, respectively, among the European population, while the rate was 0.94–1.45 and 3.96–5.03 per million/year, respectively, for Koreans (Yang et al., 2016; Kang et al., 2021; Duong et al., 2017).
Antibiotics are the main drugs that cause immune-mediated ADRs, such as SCARs, allergic reactions, and organ-specific diseases, representing an indisputable threat to patient safety (Blumenthal et al., 2019). Several antibiotics (e.g., beta-lactams, co-trimoxazole, vancomycin, and dapsone) have been associated with drug-induced hypersensitivity reactions (DIHRs) and have been associated with different genetic variants (Konvinse et al., 2019; Sukasem et al., 2020; Tempark et al., 2017; Wang et al., 2024a). Apart from DIHRs, other ADRs are also attributable to antibiotics. For example, anti-tuberculosis drug-induced hepatotoxicity (ATDH) represents an important clinical challenge as it is associated with treatment failure and increased mortality. The risk of developing hepatotoxicity ranges from 2% to 18% (Devarbhavi et al., 2010; Ramappa and Aithal, 2013). Cardiotoxicity is another important ADR related to anthracycline antibiotics and is deemed the most critical ADR in childhood cancer therapy, contributing to substantial mortality and morbidity (Lipshultz et al., 2008). In addition to nephrotoxicity, cochleotoxicity (sensorineural hearing loss) and vestibulotoxicity are the well-established side effects of aminoglycosides, which are typically dose-dependent and occur in the long-term use of high-dose drugs. However, certain individuals have been reported to be sensitive to aminoglycoside-induced hearing loss, even with single doses, resulting in profound bilateral sensorineural hearing loss (Mcdermott et al., 2022; Dean and Kane, 2018).
Recently developed cutting-edge technologies have identified the molecular mechanisms of underlying DIHRs and other ADRs. Therefore, in this article, we present the current genetic evidence from a pharmacogenomics (PGx) perspective. We also explore how these PGx–antibiotic associations can be more effectively translated into clinical practice to optimize antibiotic safety or efficacy, thereby serving as a cornerstone of antibiotic precision medicine.
2 Methods
2.1 Literature searching
Following the PRISMA guidelines, an extensive literature search was undertaken on PubMed on 25/5/2025 with the following keywords: pharmacogenomics, hypersensitivity, antibiotics, beta-lactam, sulfonamide, co-trimoxazole, dapsone, vancomycin, fluoroquinolone, anticancer antibiotics, macrolide, aminoglycoside, cephalosporins, tetracyclines, and anti-tubercular drugs to identify relevant articles (Page et al., 2021). Articles were included if 1 the study was performed on human subjects, 2 the study assessed the pharmacogenomic association of an antibiotic drug, and 3 the study evaluated the association of any gene or variant with antibiotic-induced hypersensitivity or adverse reactions. Studies were excluded if 1 the genetic assessment was conducted only computationally, 2 the study reported the genetic frequency without associating the findings with any drug, 3 the analysis was in vitro or the studies was conducted in an animal model, and 4 the publication was something other than a research article (e.g., review articles, meta-analysis, book chapter, editorial, case report, letter, and conference paper),
We utilized Rayyan QCRI, a web-based tool for systematic reviews, to select the primary studies (Ouzzani et al., 2016). We obtained the full texts of initially selected studies and reviewed them carefully to determine the final set of studies for inclusion. Two researchers independently performed the study selection using Rayyan QCRI software, and any disagreements during data extraction were resolved through mutual discussion.
2.2 Identification of the PGx-based evidence level, drug label, and therapeutic and testing guidelines for antibiotics
To assess the current state of PGx-based evidence for gene variants involved in the toxicity, metabolism/pharmacokinetics (PK), and efficacy of antibiotics, we utilized clinical annotations provided by the Pharmacogenomics Knowledgebase (PharmGKB), which is a comprehensive PGx resource managed by Stanford University to support, expand, and promote the implementation and education of PGx knowledge. PGx-based drug label information for the antibiotics was sourced from various internationally acknowledged pharmacogenetics working bodies, namely, the Health Canada Santé Canada (HCSC)-approved drug label, the US Food and Drug Administration (FDA)-approved drug label, the Swissmedic (Swiss Agency of Therapeutic Products)-approved drug label, the Pharmaceuticals and Medical Devices Agency (Japan) (PMDA)-approved drug label, and the European Medicines Agency (EMA)-approved drug label. We accessed all the information from the PharmGKB website (Barbarino et al., 2018). To obtain current information on therapeutic and testing guidelines for antibiotics, we searched different guideline-providing PGx working groups and included recommendations from the Clinical Pharmacogenetics Implementation Consortium (CPIC), the Dutch Pharmacogenetics Working Group (DPWG), and the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) (CPIC, 2025; CPNDS, 2025; DPWG, 2025a).
3 Results
3.1 Literature search results
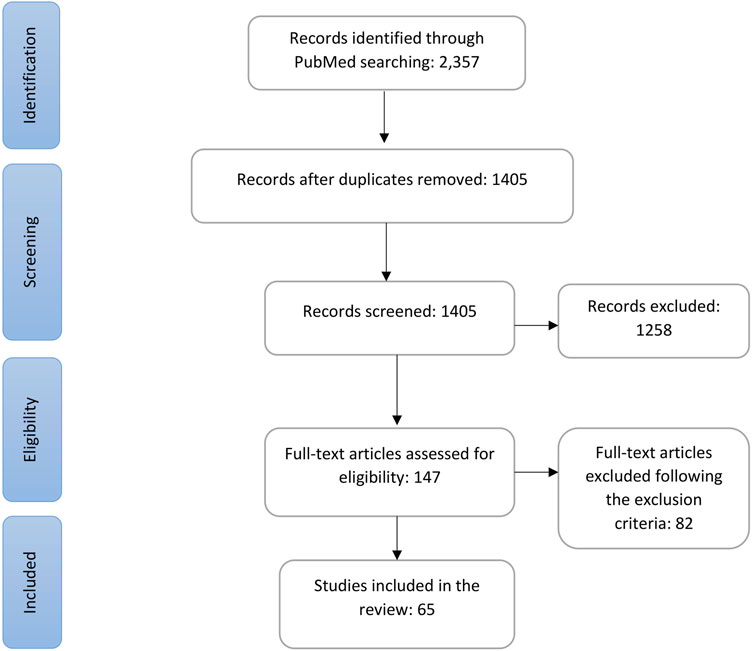
The strategic search using the aforementioned keywords generated 2,357 records, and after removal of duplicates, 1,405 remained for screening. Through initial screening with title and abstract, we excluded 1,258, and after another round of screening, we identified 147 articles for full-text eligibility assessment. Following the predefined inclusion and exclusion criteria (detailed in the Section 2), we identified 65 articles that examined the PGx associations of genes with the DIHRs and other adverse effects of antibiotics for inclusion in this review. The whole selection process is shown in a PRISMA flowchart in Figure 1.
Of the identified 65 articles, PGx assessments are presented for beta-lactams in 8 studies, anti-tuberculosis drugs in 25 studies, anticancer antibiotics in 13 studies, sulfonamides in 6 studies, aminoglycosides in 4 studies, and other antibiotics in the remaining 9 studies. Table 1 summarizes the key PGx associations for antibiotics from the included studies.
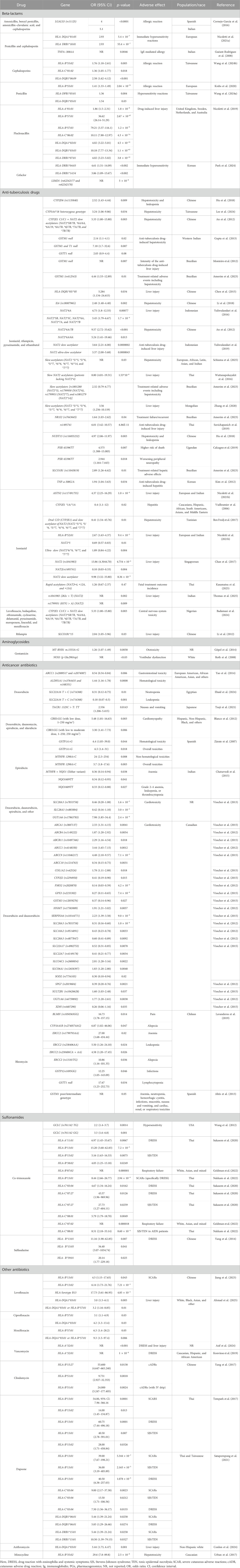
Table 1. Overview of the included studies that reported significant PGx associations of different genes/variants for antibiotic drugs.
3.2 Current evidence of PGx for antibiotic-induced hypersensitivity and adverse drug reactions
3.2.1 Beta-lactam antibiotics
We identified eight studies assessing the PGx associations of genes with beta-lactam antibiotics for DIHRs and other adverse effects. These studies primarily investigated the genetic associations with the DIHRs, with only one study examining the genetic link to flucloxacillin-induced liver injury (Wang et al., 2024b; Wang et al., 2024a; Park et al., 2024; Nicoletti et al., 2021a; Nicoletti et al., 2019; Krebs et al., 2020; Guéant-Rodriguez et al., 2008; Cornejo-García et al., 2016). Cornejo-García et al. (2016) proposed that LGALS3 could be a potential genetic predictor of immediate drug reactions and reported that rs11125 of LGALS3 (odds ratio, OR = 5.1 in the Italian population (p < 0.0001)) was strongly associated with beta-lactam (BL)-induced allergy. Mast cells release tumor necrosis factor-α (TNF-α) via an immunoglobulin E (IgE)-dependent mechanism. TNFA–308G>A is part of the extended haplotype HLA-A1-B8-DR3-DQ2 and influences the expression of the gene. Guéant-Rodriguez et al. (2008) evaluated this variant in relation to IgE-mediated reactions to BLs and reported its association with the BL-induced immediate allergic reactions. They observed that individuals carrying the –308AA genotype exhibited significantly higher specific IgE serum levels compared to those with the –308GA/GG genotype (p = 0.0046) (Guéant-Rodriguez et al., 2008).
Other studies aimed to evaluate the association between different HLA genes and DIHRs. Nicoletti et al. (2021a) identified HLA-DRB1*10:01 (OR = 2.93; p = 5.4 × 10−7) as a risk factor for immediate reaction with BLs even without the HLA-DQA1*01:05 allele (OR = 2.93, p = 5.4 × 10−7). Park et al. (2024) identified LIMD1 (rs62242177 and rs62242178) (significance level 5 × 10−8), HLA-DRB1*04:03 (OR = 4.61, 95% confidence interval (CI): 1.51–14.09, p < 0.002), and HLA-DRB1*14:54 (OR = 3.86, 95% CI: 1.09–13.67, p < 0.002) as potential factors influencing susceptibility to cefaclor-induced type I hypersensitivity. Krebs et al. (2020) provided robust evidence of HLA-B *55:01 (OR = 1.41; 95% CI: 1.33–1.49, p = 2.04 × 10−31) being associated with the occurrence of penicillin allergy through a genome-wide study. Wang et al. (2024a) reported HLA-DPB1*05:01 (OR = 1.36, p = 0.004) and HLA-DQB1*05:01 (OR = 1.54, p = 0.03) to be significantly linked with penicillin allergy among Taiwanese. For cephalosporin, on the other hand, Wang et al. (2024b) identified HLA-DQB1*06:09 (OR = 2.58, 95% CI: 1.62–4.12, p < 0.001), HLA-C*01:02 (OR = 1.36, 95% CI: 1.05–1.77, p = 0.018), and HLA-B*55:02 (OR = 1.76, 95% CI: 1.18–2.61, p = 0.005) alleles to be linked with cephalosporin-induced allergy. Nicoletti et al. (2019) performed a genome-wide association study and reported the following associations with flucloxacillin-induced liver injury: HLA-B *57:01 (allelic OR = 36.62, 95% CI: 26.14–51.29, p = 2.67 × 10−97), HLA-A *01:01(OR = 1.86, 95% CI: 1.5–2.31, p = 1.8 × 10−8), HLA-C*06:02 (OR = 10.11, 95% CI: 7.88–12.97, p = 4.3 × 10−74), HLA-B *57:03 (OR = 79.21, 95% CI: 3.37–116.1, p = 1.2 × 10−6), HLA-DQB1*03:03 (OR = 10.18, 95% CI: 7.77–13.34, p = 1.1 × 10−63), HLA-DRB1*07:01 (OR = 4.02, 95% CI: 3.23–5.02, p = 3.8 × 10−35), HLA-DQA1*02:01 (OR = 4.02, 95% CI: 3.22–5.01, p = 4.5 × 10−35). They also stated no association of HLA-B*57 with drug-induced liver injury (DILI) for other isoxazolyl penicillin or amoxicillin (Nicoletti et al., 2019).
These studies are population-based and involve varying sample sizes. Consequently, studies with smaller case numbers may either underestimate or overestimate the findings. Therefore, further evaluation with a larger sample size was encouraged for better understanding, rationalization, and integration of that information in clinical practice.
3.2.2 Aminoglycosides
We identified at least four studies that associated aminoglycoside-induced ototoxicity with MT-RNR1 mutations (Roth et al., 2008; Fischel-Ghodsian et al., 1997; Lu et al., 2010; Göpel et al., 2014). Göpel et al. (2014), using a multivariable logistic regression, demonstrated treatment with aminoglycosides in m.1555A>G-carriers was associated with the failed hearing screening (OR = 1.26; 95% CI: 1.07–1.49; p = 0.0058). They also observed the m.1555A>G mutation in all the mothers of the children carrying the m.1555A>G mutation, which was absent in the mothers of the non-carrier children of the m.1555A>G mutation. They suggested antenatal screening of the m.1555A>G mutation through maternal genotyping of pregnant women with preterm labor may potentially be a rational approach to identifying infants with an increased risk of permanent hearing loss (Göpel et al., 2014). Lu et al. (2010) observed 745A>G, 792C>T, 801A>G, 839A>G, 856A>G, 1027A>G, 1192C>T, 1192C>A, 1310C>T, 1331A>G, 1374A>G, and 1452T>C variants to confer increased sensitivity to nonsyndromic deafness or ototoxic drugs. Bilateral and sensorineural hearing loss was exhibited in 65 Chinese individuals who carried the 1555A>G mutation (Lu et al., 2010). Fischel-Ghodsian et al. (1997) explored the irreversible sensorineural hearing loss (SNHL) with the use of aminoglycosides (streptomycin, gentamicin, kanamycin, amikacin, and neomycin) due to m.1555A > G variants in mitochondrial 12S RNA and observed the presence of polymorphism in 17% of the total population having SNHL after aminoglycoside exposure, and among them, more than half had a family history of SNHL with aminoglycosides. Therefore, they recommended clinical screening and appropriate familial evaluation to avoid associated ototoxicity (Fischel-Ghodsian et al., 1997). Roth et al. (2008) stated that carriers of risk alleles of NOS3 (p.Glu298Asp), GSTZ1 (p.Lys32Glu), and GSTP1 (p.Ile105Val) are relevant for the elevated risk of vestibular dysfunction with gentamicin (p < 0.03).
3.2.3 Sulfonamides
We identified at least five studies that correlated co-trimoxazole/sulfamethoxazole/trimethoprim with genetic association (Nakkam et al., 2022; Goldman et al., 2022; Alfirevic et al., 2009; Wang et al., 2012; Sukasem et al., 2020). Similarly, one such study explored the genetic association with sulfasalazine-induced ADRs (Yang et al., 2014). Nakkam et al. (2022) reported that the HLA-B*13:01 allele was significantly associated with co-trimoxazole-induced SCARs, particularly DRESS (OR = 8.44, 95% CI: 2.66–26.77, p = 2.94 × 10−4). Additionally, the HLA-C*08:01 allele was observed to have a significant association with SJS/TEN induced by co-trimoxazole in HIV/AIDS patients [OR of 8.51, 95% CI: 2.18–33.14, p = 8.60 × 10−4] (Nakkam et al., 2022). Goldman et al. (2022) evaluated respiratory failure with trimethoprim/sulfamethoxazole and HLA and identified HLA-B *07:02 (p = 0.000001) and HLA-C *07:02 (p = 0.000018) to be significantly associated with the increased risk of respiratory failure. However, Alfirevic et al. (2009) stated that MHC polymorphisms were not a major predisposing factor for co-trimoxazole hypersensitivity, although a minor contribution cannot be ruled out. For sulfamethoxazole (SMX)-induced hypersensitivity in HIV/AIDS patients, Wang et al. (2012) reported that GCLC (rs761142 T>G) was significantly associated with hypersensitivity induced by SMX (adjusted p-value = 0.045). In a replicated cohort with 249 patients, the result was replicated (p = 0.025). For the combined cohort, homozygous and heterozygous carriers of the minor G allele were recorded for an increased risk of hypersensitivity (GT vs TT, OR = 2.2, 95% CI: 1.4–3.7, p = 0.0014; GG vs. TT, OR = 3.3, 95% CI: 1.6–6.8, p = 0.0010). Each minor allele copy increased the risk of developing hypersensitivity 1.9-fold (95% CI: 1.4–2.6, p = 0.00012) (Wang et al., 2012). Sukasem et al. (2020) identified HLA-C*08:01 (OR = 5.79, 95% CI: 1.79–18.70, p = 0.0049) and HLA-B*15:02 (OR = 5.16, 95% CI: 1.63–16.33, p = 0.0075) alleles as significantly associated with SJS/TEN induced by co-trimoxazole, and the HLA-B*13:01 allele was significantly linked to co-trimoxazole-induced DRESS (OR = 15.20, 95% CI: 3.68–62.83, p = 7.2 × 10−5). Additionally, significantly high frequency of HLA-B*13:01-C*03:04 (OR = 14.53, 95% CI: 3.74–56.47, p = 1.8 × 10−4) and HLA-A*11:01-B*15:02 (OR = 6.00, 95% CI: 1.72–20.88, p = 0.0074) haplotypes were observed in the group of co-trimoxazole-induced DRESS and SJS/TEN, respectively (Sukasem et al., 2020).
In the Chinese Han population, Yang et al. (2014) explored sulfasalazine-induced DRESS and identified HLA-B*13:01 as a potential biomarker for increasing the risk of DRESS since the distribution of the HLA-B*13:01 allele was significantly higher in sulfasalazine-induced DRESS patients than in sulfasalazine-tolerant patients (OR = 13.00, 95% CI: 1.76–95.80, p = 0.004) (Yang et al., 2014).
3.2.4 Anti-tuberculous drugs
We identified at least 25 studies evaluating the PGx associations of different genes with anti-tuberculous drug (ATD)-induced adverse effects (Amorim et al., 2023; An et al., 2012; Badamasi et al., 2024; Ben Fredj et al., 2017; Calcagno et al., 2019; Chan et al., 2017; Chen et al., 2015; Gupta et al., 2013; Hu et al., 2018; Kasamatsu et al., 2025; Kim et al., 2012; Lee et al., 2024; Li et al., 2012; Li et al., 2018; Monteiro et al., 2012; Nicoletti et al., 2021b; Schiuma et al., 2025; Suvichapanich et al., 2019; Thomas et al., 2025; Vuilleumier et al., 2006; Wattanapokayakit et al., 2016; Yamada et al., 2010; Yuliwulandari et al., 2019; Yuliwulandari et al., 2016; Zhang et al., 2020). Of these, the study by Li et al. evaluated the association of ATDs in pediatric patients and reported a striking difference in the allele distribution of rs1800796 in the IL6 gene between the control and case groups, and the G allele of rs1800796 was linked with an elevated risk for anti-tuberculosis drug-induced hepatotoxicity (OR = 2.48, 95% CI: 1.40–4.40, p = 0.002). After Bonferroni correction, no significant difference was observed in the allele and genotype distributions of the other SNPs in the IL6, XO, and NOS2 genes between the control and case groups (Li et al., 2018). Three studies evaluated the association of GSTM1 and GSTT1 with ATDs. They reported that the homozygous null mutation of the GSTM1 gene, either alone or in combination with T1, was significantly associated with anti-tuberculosis drug-induced hepatotoxicity (p < 0.02 and p < 0.007, respectively); one study further reported that the GSTM1 polymorphism (rs412543) (p = 0.01) was linked to an elevated risk of treatment-related adverse events, including hepatotoxicity. Conversely, another study found no significant role of the GSTM1 and GSTT1 null genotypes in anti-tuberculosis drug-induced liver injury, although there was evidence that GSTM1 polymorphisms may be related to the intensity of toxicity (p = 0.007) (Amorim et al., 2023; Gupta et al., 2013; Monteiro et al., 2012).
Yuliwulandari et al. (2019) found that the NAT2 slow-acetylator phenotype was significantly associated with the risk of AT-DILI (p = 2.7 × 10−7, OR = 3.64, 95% CI: 2.21–6.00). The NAT2 ultra-slow acetylator showed an even stronger association with AT-DILI risk in the subgroup analysis (p = 4.3 × 10−6, OR = 3.37, 95% CI: 2.00–5.68). In the Thai population, Suvichapanich et al. (2019) reported that the A allele of rs1495741, the top SNP in the intergenic region of NAT2 and PSD3, was significantly associated with anti-tuberculosis drug-induced liver injury (ATDILI) (OR = 6.01, 95% CI: 3.42–10.57, p = 6.86E-11), identifying that NAT2 ultra-slow acetylator as the most important risk factor for ATDILI. In the Indian population, Thomas et al. (2025) observed that allele T (rs1041983) (p = 0.002) and allele A (rs1799931) (p = 0.009) were associated with an elevated risk of drug-induced liver injury in patients receiving anti-tubercular drugs, compared to allele C and allele G, respectively. Schiuma et al. (2025) reported that NAT2*5/*5, *5/*6, *5/*7, *6/*6, *6/*7, *6/*14, and *7/*7 (grouped as the slow-acetylator phenotype) were linked to an increased likelihood of toxic liver disease during treatment with ethambutol and isoniazid/pyrazinamide/rifampin in individuals with tuberculosis (p = 0.03), compared to NAT2*1/*5, *1/*6, and *1/*7 (grouped as intermediate acetylator and rapid acetylator phenotypes). Three additional studies confirmed that slow NAT2 acetylators are a risk factor for ATDILI. Specifically, NAT2*6 was associated with an increased risk (OR = 4.75, 95% CI: 1.80–12.55, p = 0.00077), while no significant association was observed for NAT2*5 or *7. On the contrary, NAT2*4 was associated with a decreased risk of drug-induced liver injury (p = 1.8 × 10−6, OR = 0.2, 95% CI: 0.1–0.39); compared to intermediate or rapid acetylators (NAT2*4, NAT2*12A, and NAT2*13), slow acetylators due to NAT2 genotypes (NAT2*5B, NAT2*5C, NAT2*6A, NAT2*7A, and NAT2*7B) exhibited a higher risk of liver injury (p = 1.7 × 10−4, OR = 3.45, 95% CI: 1.79–6.67). Overall, the slow-acetylator type due to the polymorphism of NAT2 was considered a risk factor for ATDILI (OR = 3.56, 95% CI: 1.256–10.119), and slow NAT2 acetylators (patients lacking NAT2*4) showed a significant association with ATDILI risk (OR = 8.80; 95% CI = 4.01–19.31, p = 1.53 × 10−8) (Wattanapokayakit et al., 2016; Yuliwulandari et al., 2016; Zhang et al., 2020). In patients with tuberculosis, Kasamatsu et al. (2025) observed that rapid acetylators due to NAT2 polymorphism had a 1.26-fold higher incidence of fatal treatment outcomes (95% CI: 0.67–2.37) compared to intermediate acetylators.
Hu et al. (2018) reported an increased risk of leukopenia and hepatotoxicity associated with CYP2D6 rs1135840 and NUDT15 rs116855232, with ORs of 2.52 (95% CI: 1.43–4.44, p = 0.009) and 4.97 (95% CI: 2.06–11.97, p = 0.003), respectively. For multidrug-resistant tuberculosis treatment, Badamasi et al. (2024) reported a significant association between CNS toxicity and the dominant model of inheritance for the crude model (p = 0.024; OR = 3.57; 95% CI: 1.18–10.76) and the adjusted model (p = 0.031, OR = 3.92, 95% CI: 1.13–13.58). They reported that the AT + TT genotype of IL8 (rs4073) is associated with a 3.92-fold increased risk of CNS toxicity compared to the AA genotype (Badamasi et al., 2024).
Apart from the GSTM1 association as mentioned earlier, Amorim et al. (2023) also explored other genetic associations and stated that NAT2 slow acetylator status was linked with an increased risk of treatment-related adverse events, including hepatotoxicity, compared with rapid acetylator (OR = 2.32, 95% CI: 0.79–6.77). Treatment failure or recurrence was more likely among NAT2 rapid acetylators. Similarly, SLCO1B1 (p = 0.01) was linked with an elevated risk of treatment-related adverse events, including hepatotoxicity. Polymorphisms in NR1I2 were associated with decreased risk of adverse effects but increased risk of failure/recurrence (p = 0.04). Although in whole exome sequencing, hepatotoxicity was associated with a polymorphism in VTI1A, and the genes METTL17 and PRSS57, but none achieved genome-wide significance (Amorim et al., 2023). Calcagno et al. (2019) reported that NAT2 (rs1799930), SLCO1B1 (rs4149032), and PXR (rs2472677) variants affected isoniazid exposure. Genotype TT (rs2472677) was linked with an elevated peripheral nervous system disease (p = 0.018) and elevated death risk (p = 0.007) with treatment with ethambutol, isoniazid, efavirenz, and rifampin in people with HIV and tuberculosis compared with genotypes CC and CT.
Although univariate analyses by Chen et al. (2015) and Chan et al. (2017) found no statistically significant association between ATDILI and the frequency of HLA-DQB1 genotypes, multivariate analysis revealed that individuals carrying two DQB1*05 alleles had a higher risk of ATDILI compared to the control group (OR = 5.28 adjusted for use of liver-protective drugs and weight 10/88 VS 2/88, 95% CI: 1.134–24.615, p = 0.034). Regardless of the presence of pre-existing liver disease, the heterozygous CYP3A4*18 genotype was associated with anti-tuberculosis drug-induced hepatotoxicity (ATDH) in a study by Lee et al. (2024) (OR: 3.24, 95% CI: 1.06–9.86). Although among the subjects without having liver disease, CYP3A4*18 heterozygotes were observed to have a significantly higher risk of ATDH (OR: 9.10, 95% CI: 1.56–53.16), in subjects with previous liver disease, CYP3A4*18 heterozygotes had a lower risk of ATDH (OR: 0.21, 95% CI: 0.05–0.98) (Lee et al., 2024). The frequency of -308AG/AA carriers was found to be significantly higher in ATD-induced hepatitis patients than the ATD-tolerant patients (p = 0.034, OR = 1.94; 95% CI = 1.04–3.64) and the frequency of the A allele significantly differed between the two groups (p = 0.018, OR 1.95, 95% CI = 1.11–3.44). These results indicated that the TNFA-308G/A polymorphism was significantly associated with ATDH (Kim et al., 2012). An et al. (2012) deemed slow acetylators due to NAT2 genotypes (particularly, NAT2*6A/7B and NAT2*6A/6A) risk factors for drug-induced hepatotoxicity (DIH) (OR = 9.57; p < 0.001) for NAT2*6A/7B; OR 5.24 (p = 0.02) for NAT2*6A/6A). Although the CYP2E1 genotype was not significantly linked with the development of anti-tuberculosis DIH, the combination of the CYP2E1 C1/C1 genotype and the NAT2 genotype of slow acetylator was observed to increase the risk of anti-tuberculosis (OR = 5.33; p = 0.003) compared to the combination of the NAT2 rapid acetylator genotype paired with either a C1/C2 or C2/C2 genotype (An et al., 2012).
Six of the studies evaluated PGx’s association with the adverse effects of isoniazid alone. Chan et al. (2017), on the Singaporean population, performed a study and identified a significant association of two SNPs of NAT2 (rs1041983 and rs1495741) and NAT2 slow acetylators with isoniazid-induced liver injury (OR = 13.86, 95% CI: 4.30–44.70; OR = 0.10, 95% CI = 0.03–0.33 and OR = 9.98, 95% CI = 3.32–33.80, respectively). They also stated a model based on clinical and NAT2 acetylator status resulted in much better prediction for isoniazid-induced liver injury compared to a clinical model alone (area under the receiver operating characteristic curve = 0.863 vs. 0.766, respectively, p = 0.027) (Chan et al., 2017). A genome-wide association study by Nicoletti et al. identified rs117491755 in ASTN2 as being significantly associated with DILI in European patients only. HLA-B*52:01 was also found to be significant (OR = 2.67, 95% CI = 1.63–4.37, p = 9.4 × 10−5). The frequency of NAT2*5 was lower for cases (OR = 0.69, 95% CI = 0.57–0.83, p = 0.01). NAT2*6 and NAT2*7 were relatively common, homozygotes for NAT2*6 and/or NAT2*7 being enriched in cases (OR = 1.89, 95% CI = 0.84–4.22, p = 0.004). They reported that HLA genotypes made a minimal contribution to ATDILI and that the contribution of NAT2 was complex. However, their findings were consistent with previous studies when considering differences in metabolic effects between NAT2*5, NAT2*6, and NAT2*7 alleles (Nicoletti et al., 2021b). Two separate studies reported that NAT2 and CYP2E1 variants were not associated an increased risk of isoniazid-induced hepatotoxicity when analyzed independently; however, Vuilleumier et al. found that compared with other CYP2E1 genotypes, a significant association between the CYP2E1 *1A/*1A genotype and isoniazid-induced elevated liver enzymes, including hepatitis (OR: 3.4; 95% CI:1.1–12; p = 0.02), and a non-significant trend for isoniazid induced hepatotoxicity was also recorded (OR: 5.9; 95% CI: 0.69–270; p = 0.13). Similarly, Ben Fredj et al. stated that a combined analysis of the polymorphism in the NAT2/CYP2E1 gene revealed that individuals with both DraI C/D (CYP2E1) and slow acetylator (NAT2) genotypes have an elevated risk of isoniazid-induced hepatotoxicity as compared to other combined NAT2/CYP2E1 genotype profiles (OR: 8.41, p = 0.01, 95% CI: 1.54–45.76) (Ben Fredj et al., 2017; Vuilleumier et al., 2006). Yamada et al. (2010) found no association between isoniazid-induced hepatotoxicity SNPs and haplotypes at CES2 and CES1/CES4.
Li et al. (2012) evaluated the PGx association of rifampin and identified an association between SLCO1B1*15 and the increased risk of drug-induced liver injury (p = 0.03, OR = 2.04, 95% CI: 1.05–3.96). No such association was found for SLCO1B1*5 and *1.
3.2.5 Anticancer antibiotics
We identified at least 11 studies assessing the association of genes with the adverse effects of anthracyclines (Chaturvedi et al., 2015; Yao et al., 2014; Visscher et al., 2013; Visscher et al., 2015; Nyangwara et al., 2024; Ebaid et al., 2024; Visscher et al., 2012; Robinson et al., 2019; Zárate et al., 2007; Blanco et al., 2012; Tsuji et al., 2021). Five of them were on pediatric patients. Among those, Robinson et al. (2019) reported that G6PD deficiency did not have any effect on the hemolytic toxicities with daunorubicin during the induction treatment for acute lymphoblastic leukemia (p = 0.73). Blanco et al. (2012) observed the exposure of low-to-moderate doses of anthracyclines in individuals carrying the variant A allele (CBR1:GA/AA and/or CBR3:GA/AA) did not raise the risk of cardiomyopathy, but with similar doses, an increased risk of cardiomyopathy was observed in individuals with the CBR3 V244M homozygous G genotypes (CBR3:GG) compared to the individuals with the CBR3:GA/AA genotypes unexposed to anthracyclines (OR = 5.48; p = 0.003) and exposed to low-to-moderate doses of anthracyclines (OR = 3.30; p = 0.006). High doses of anthracyclines, irrespective of CBR genotype status, were associated with increased cardiomyopathy risk (Blanco et al., 2012). Visscher et al. identified a highly significant association with a synonymous coding variant, rs7853758 (L461L), in the SLC28A3 gene with anthracycline-induced cardiotoxicity in children (OR = 0.35; p = 1.8 × 10−5, single marker test). Additionally, other significant associations with protective and risk variants in other genes, including SLC28A1, ABCB1, ABCB4, and ABCC1, were present. For safer treatment options, combining genetic risk profiles may be considered (Visscher et al., 2012). In this replication cohort, Visscher et al. confirmed the association of rs17863783 (UGT1A6) and anthracycline-induced cardiotoxicity (p = 0.0062, OR = 7.98). Additionally, evidence for the association of rs885004 (p = 0.058, OR 0.42) and rs7853758 (p = 0.058, OR 0.46) in SLC28A3 was reported (combined p = 3.0 × 10−5 and p = 1.6 × 10−5, respectively). Unlike a previously constructed model for prediction, the improved prediction model constructed utilizing the replicated genetic variants alongside the clinical factors discriminated significantly better among cases and controls against only clinical factors, both in the original (AUC 0.77 vs. 0.68, p = 0.0031) and replication cohort (AUC 0.77 vs. 0.69, p = 0.060) (Visscher et al., 2013). In this study, Visscher et al. identified significant associations of SLC22A7 (rs4149178, p = 0.0034) and SLC22A17 (rs4982753, p = 0.0078) with anthracycline-induced cardiotoxicity in both discovery and replication cohort. Additionally, evidence was found for SULT2B1 and several other genes related to oxidative stress (Visscher et al., 2015).
Yao et al. (2014) observed in breast cancer patients that rs3764435 and rs168351 (ALDH1A1) were significantly associated with hematological toxicity (OR = 1.44, 95% CI: 1.16–1.78, p = 0.0008), and rs2889517 and rs2074087 (ABCC1) were significantly associated with gastrointestinal toxicity (OR = 0.54, 95% CI: 0.34–0.84, p = 0.006). Nyangwara et al. (2024), in a study on Zimbabwean breast cancer patients, found no significant association between doxorubicin-induced cardiotoxicity and SLC28A3 (rs7853758, p = 0.408), UGT1A6*4 (rs17863783, p = 0.354), or RARG (rs2229774, p = 0.471). Ebaid et al. (2024), in Egyptian breast cancer patients, reported that carriers of CBR1 C>T (rs20572) had significantly higher doxorubicin concentrations, but no significant association with hematological toxicity was observed. On the contrary, although no significant effect of SLC22A16 T>C (rs714368) on the plasma concentration was observed, it was significantly correlated with a lower risk of neutropenia (OR 0.31, 95% CI = 0.12–0.75, p = 0.01) and leucopenia (OR 0.18, 95% CI = 0.07–0.5, p = 0.001). Doxorubicin-related cardiotoxicity was associated with the cumulative doxorubicin dose (OR = 0.238, p = 0.017), but not with any of the two SNPs examined (Ebaid et al., 2024). Tsuji et al. (2021) reported that in breast cancer patients receiving triplet antiemetic combination regimens, ABCB1 2677G>T/A was not predictive of the antiemetic response. However, an association was observed between the TACR1 1323C>T polymorphism and complete response in the acute phase.
Among Indian breast cancer patients treated with 5-fluorouracil, epirubicin/methotrexate/adriamycin, and cyclophosphamide regimens, Chaturvedi et al. (2015) observed that grade 2–4 toxicity (anemia, leucopenia, or thrombocytopenia) was significantly associated with NQO1609TT (OR = 0.33, 95% CI: 0.12–0.88, p = 0.027). Further analysis for anemia found a significant association with NQO1609TT (OR = 0.34; 95% CI: 0.12–0.95; p = 0.041) and the combination of MTHFR + NQO1 (either variant) (OR = 0.36; 95% CI = 0.14–0.94; p = 0.038) (Chaturvedi et al., 2015). For breast cancer adjuvant therapy with anthracycline (epirubicin), Zárate et al. (2007) found that hematological GIII-IV toxicity was associated with GSTP1 polymorphism (p = 0.044, hazard ratio, HR = 6.4, 95% CI: 1.05–39). Evaluation of non-hematological toxicities revealed increased and significant HR for GIII-IV toxicities in the MTHFR-1298 AC + CC group (HR = 24, 95% CI = 2.3 to 254, p = 0.008). They identified GSTP1 and MTHFR-1298A>C polymorphisms as independent risk factors regarding overall toxicities (Zárate et al., 2007). Two studies establishing a genetic association with bleomycin-induced ADRs were selected for the study. The first one, by Altés et al. (2013), explored the use of bleomycin in Hodgkin lymphoma and found that the carrier of GSTM1 extensive or ultrahigh activity was linked to a decreased risk of grade III/IV toxicity development (p = 0.05), but with efficacy analysis, they concluded that compared to PGx determinants, clinical determinants could be more relevant for the Hodgkin lymphoma treatment. The other study explored the genetic association of toxicities with the bleomycin-containing regimen in Chilean testicular cancer patients and emphasized the need of PGx implementations for severe ADR prediction based on some robust genetic associations, including ERCC2 (rs1799793AA) and anemia (OR = 27.00, 95% CI = 1.68–434.44, p = 0.020), ERCC2 (rs238406AA) and leukopenia (OR = 5.50, 95% CI = 1.26–24.10, p = 0.024), GSTT1 null and lymphocytopenia (OR = 17.67, 95% CI = 1.23–252.73, p = 0.034), CYP3A41B (rs2740574GG) and alopecia (OR = 6.87, 95% CI = 1.02–46.06, p = 0.047), BLMH (rs1050565) and pain (OR = 16.73, 95% CI = 1.78–157.15, p = 0.014) and GSTP1 (rs1695GG) and infections (OR = 12.25, 95% CI = 1.05–143.09, p = 0.046) (Lavanderos et al., 2019).
3.2.6 Other antibiotics
A study of genetic association of levofloxacin-induced SCARs in the Chinese population by Jiang et al. revealed that compared to levofloxacin-tolerant patients, significantly higher frequencies of HLA-B*13:01 (OR: 4.50, 95% CI: 1.15–17.65, p = 0.043), HLA-B*13:02 (OR: 6.14, 95% CI: 1.73–21.76, p = 0.0072), and serotype B13 (OR: 17.73, 95% CI: 3.61–86.95, p = 4.85 × 10−5) were observed in patients with levofloxacin-induced SCARs. They proposed prospective screening or alternative therapy that may benefit the patient in concern (Jiang et al., 2023). Ahmad et al. (2025) found a significant association with HLA-DQA1*03:01 and HLA -B*57:01 for DILI induced by fluoroquinolones (ciprofloxacin, levofloxacin, and moxifloxacin). Details of the specific ORs are presented in Table 1.
Of the included studies, we identified two studies that evaluated the association of HLA with vancomycin-induced adverse effects, such as liver injury and DRESS. Asif et al. (2024) reported that HLA-A*32:01 was associated with vancomycin-induced liver injury and DRESS (p < 0.001). Konvinse et al. (2019) noted that the carriage of the HLA-A*32:01 allele is significantly associated (p = 1 × 10−8) with the development of DRESS induced by vancomycin.
Yang et al. (2017) evaluated the genetic association with clindamycin-induced cADRs in the Chinese population and observed that compared to the control and clindamycin-tolerant groups, the frequency of HLA-B*51:01 was significantly higher in the case group. They identified HLA-B*51:01 as a risk allele for clindamycin-related cADRs in the Han Chinese population, particularly with clindamycin administration via an intravenous drip (OR = 24.00, 95% CI: 3.25–177.41, p = 0.0024). HLA-B*15:27 was also found to have a link with clindamycin-induced cADRs (OR = 55.60, 95% CI: 4.647–665.24, p = 0.0046, pc = 0.0184) (Yang et al., 2017).
Urban et al. (2017) explored the genetic link with minocycline hepatotoxicity and noted HLA-B*35:02 to have a significant association with the risk for minocycline-induced liver injury (OR: 29.6, 95% CI: 7.8–89.8, p = 2.5 × 10−8). Sequence-based HLA typing verified this association (Urban et al., 2017).
Two of the included studies explored the PGx association of dapsone-induced SCARs. Tempark et al. (2017) reported that the HLA-B*13:01 allele had a significant association with SCARs induced by dapsone compared to the dapsone-tolerant controls (OR: 54.00, 95% CI: 7.96–366.16, p = 0.0001) and the general population (OR: 26.11, 95% CI: 7.27–93.75, p = 0.0001). Additionally, HLA-B*13:01 was found to be associated with dapsone-induced DRESS (OR: 60.75, 95% CI: 7.44–496.18, p = 0.0001) and SJS-TEN (OR: 40.50, 95% CI: 2.78–591.01, p = 0.0070) in non-leprosy Thai patients (Tempark et al., 2017). Of all HLA alleles, Satapornpong et al. (2021) reported that only the HLA-B*13:01 allele was significantly associated with dapsone-induced SCARs (OR = 39.00, 95% CI: 7.67–198.21, p = 5.3447 × 10−7), DRESS (OR = 40.50, 95% CI: 6.38–257.03, p = 1.0784 × 10−5), and SJS-TEN (OR = 36.00, 95% CI: 3.19–405.89, p = 2.1657 × 10−3) compared with dapsone-tolerant controls. The HLA-B*13:01 allele was also strongly associated with dapsone-induced SCARs among the Taiwanese population (OR = 31.50, 95% CI: 4.80–206.56, p = 2.5519 × 10−3) and Asians (OR = 36.00, 95% CI = 8.67–149.52, p = 2.8068 × 10−7) (Satapornpong et al., 2021). Compared to the control population, Conlon et al. (2024) observed a significant association with HLA-DQA1*03:01 for azithromycin-induced liver injury (OR = 3.44, 95% CI: 1.73, 6.47, p = 0.001) and recommend further exploration for a comprehensive understanding of the mechanism involved and clinical role (Conlon et al., 2024).
3.3 Current state of PGx-based clinical annotations and drug labels for antibiotics
We used the PharmGKB clinical annotations to determine the current PGx evidence level for the variants and genes involved in the safety and effectiveness of the antibiotics. Based on variant annotations and incorporating available variant-specific prescribing guidelines and FDA-approved drug labels, these annotations provide information on the drug–variant pairs. Following a scoring system, these annotations are then assigned a level of evidence ranging from level-4 (unsupported) to level-1A (high) (PharmGKB, 2025a; Whirl-Carrillo et al., 2021). Our search across PharmGKB revealed clinical annotations for at least 36 antibiotic drugs, each with various variants of at least 85 genes. These annotations are presented in Tables 2, 3.
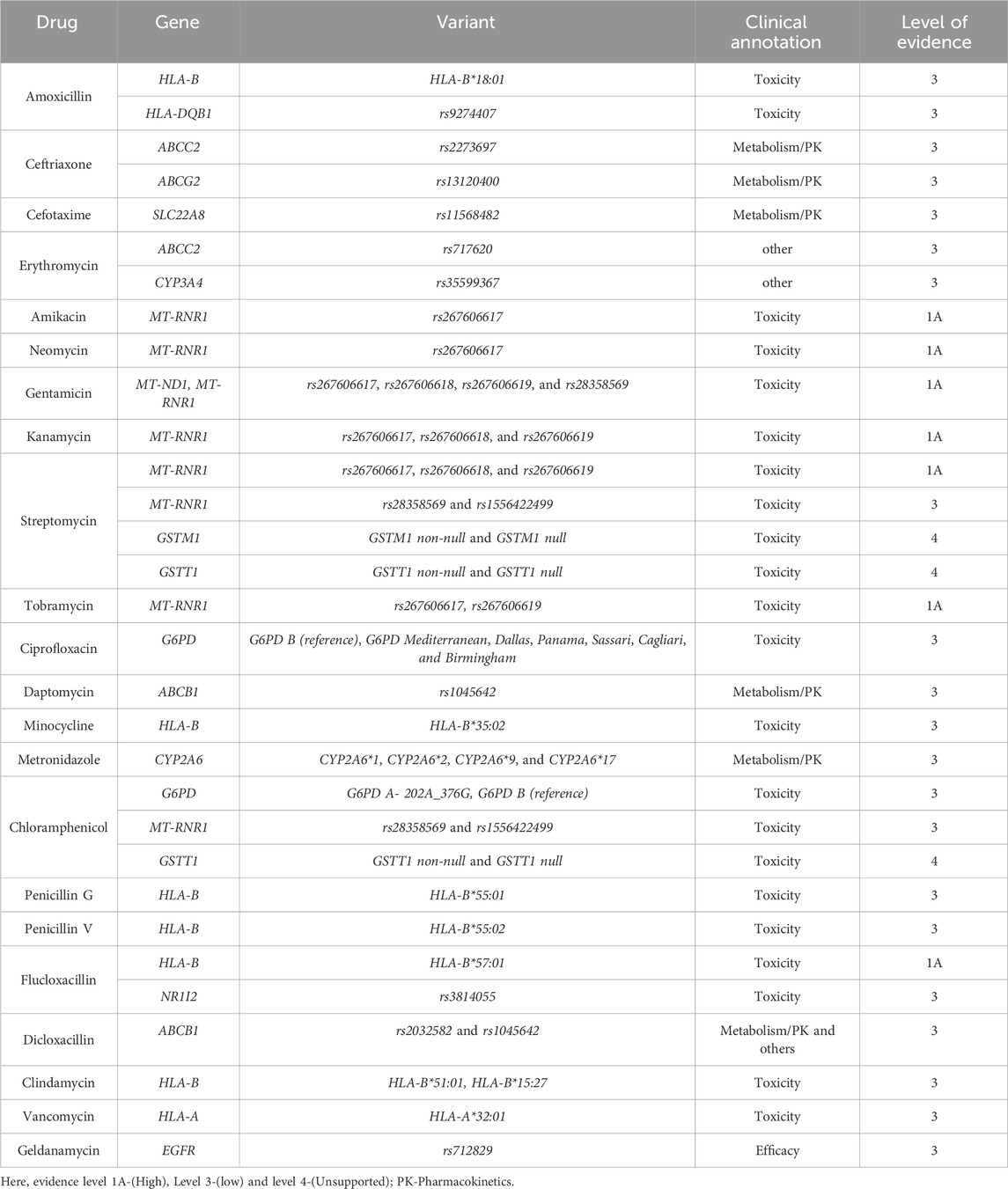
Table 2. Current PGx-based clinical annotations of various antibiotic–gene pairs with the PharmGKB level of evidence.
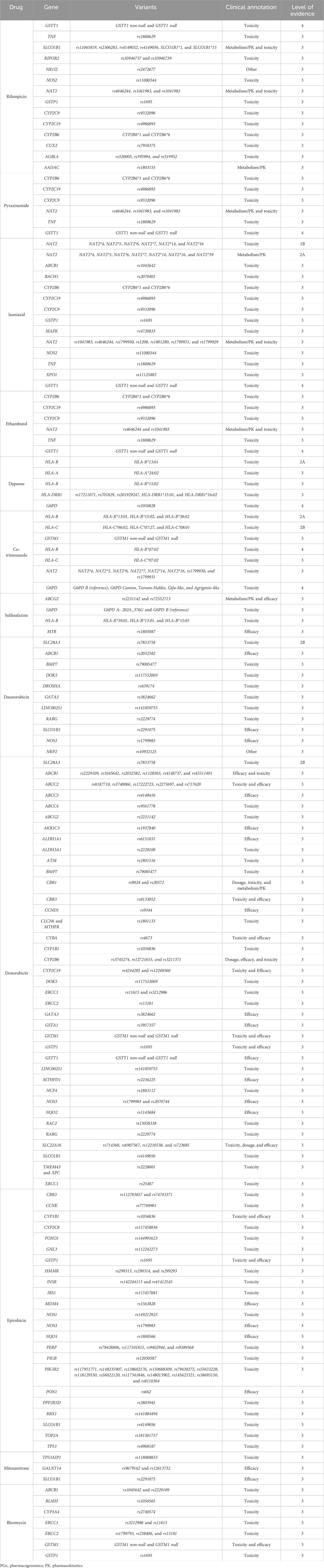
Table 3. Current PGx-based clinical annotations of various antibiotic–gene pairs with the PharmGKB level of evidence.
Although most of the annotations were assigned evidence level-3 (low), for a few antibiotics, we also identified some moderate (2A and 2B) and high (1A and 1B) levels of evidence. Aminoglycosides (amikacin, neomycin, gentamicin, kanamycin, streptomycin, and tobramycin) had a level-1A association for toxicity (ototoxicity) with different variants of MT-RNR1—rs267606617 being the variant common to all of them. Other variants are outlined in Tables 2, 3. For flucloxacillin, we observed another level-1A association with HLA-B*57:01 for drug-induced liver injury. For isoniazid induced toxicity, level-1B evidence was assigned with the NAT2 for the variants NAT2*1, NAT2*4, NAT2*5, NAT2*6, NAT2*7, NAT2*14, and NAT2*16.
Similarly, level-2A evidence was assigned with isoniazid for metabolism/PK for various variants of the NAT2 gene (i.e., NAT2*1, NAT2*4, NAT2*5, NAT2*6, NAT2*7, NAT2*14, NAT2*16, and NAT2*39). For drug-induced toxicity, an evidence level of 2A was assigned with various variants of HLA-B for co-trimoxazole (HLA-B*13:01, HLA-B*15:02, and HLA-B*38:02) and dapsone (HLA-B*13:01). Co-trimoxazole also had a level-2B association for toxicity with HLA-C*06:02, HLA-C*07:27, and HLA-C*08:01. Anthracycline antibiotics (doxorubicin and daunorubicin) had a level-2A association for drug-induced toxicity with SLC28A3 (rs7853758).
Considering the overall clinical annotations for antibiotics, we identified HLA-B (one level-1A, two level-2A, and eight level-3 associations), MT-RNR1 (six level-1A and two level-3 associations), and NAT2 (one level-1B, one level-2B, and five level-3 associations) as concerning genes for the safety and effectiveness of the antibiotic drug. The clinical annotations of level-1 and level-2 for antibiotics are outlined in Figure 2.
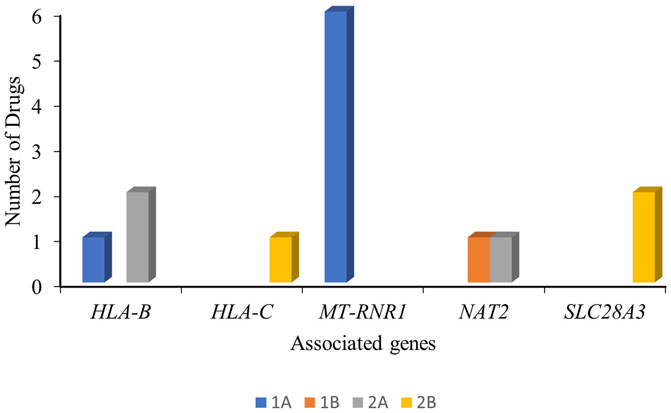
Figure 2. Clinical annotations (level-1 and level-2) of the antibiotic drugs and the associated genes with their PharmGKB evidence level.
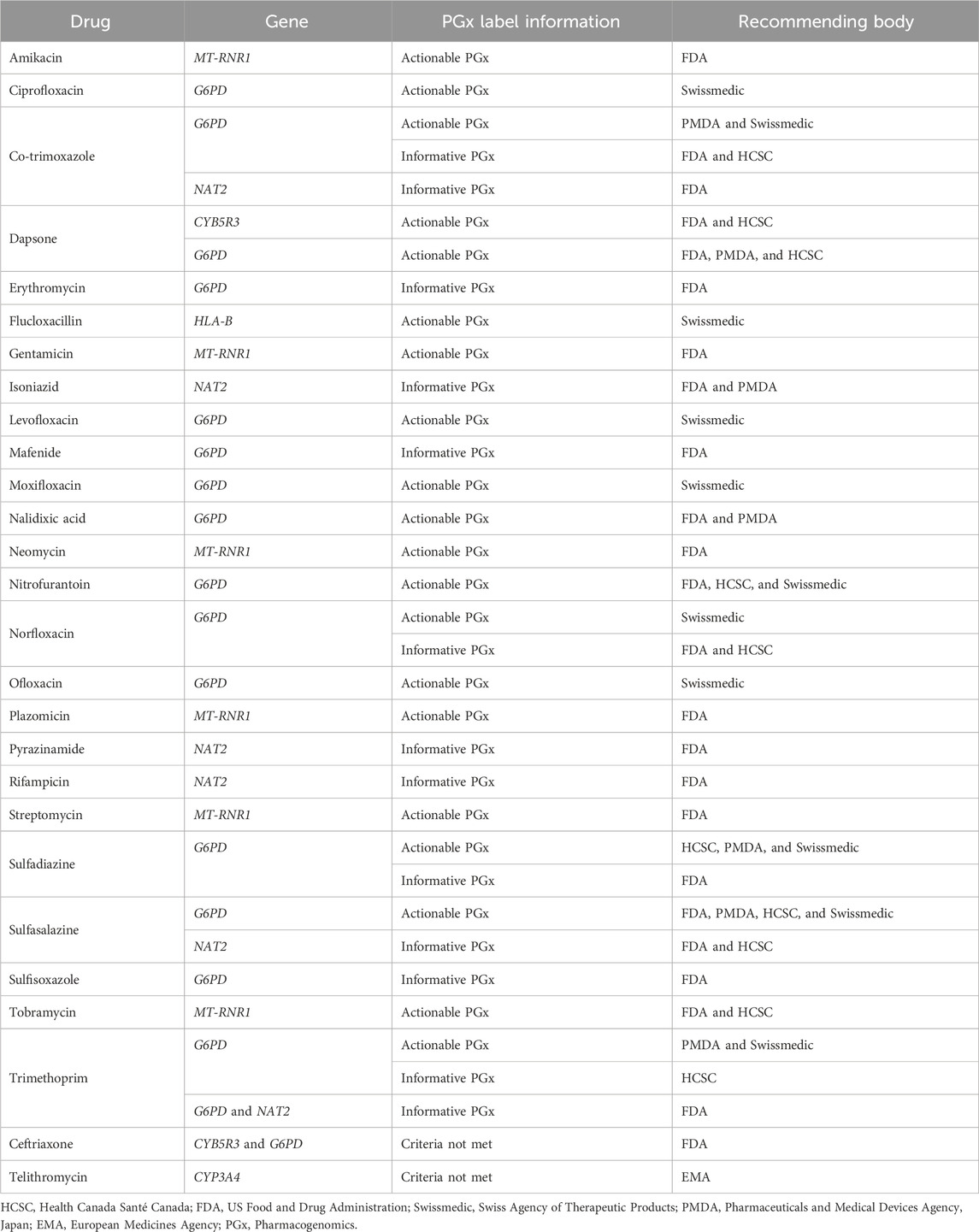
The PharmGKB curates and presents the PGx-based drug labels on its site. These labels are sourced from the FDA, EMA, PMDA, HCSC, and Swissmedic and are presented as testing required, testing recommended, actionable PGx, informative PGx, no clinical PGx, and criteria not met (PharmGKB, 2025b). Our search across the PharmGKB website revealed PGx label information for at least 27 antibiotic drugs, considering the polymorphisms of at least 6 genes (MT-RNR1, G6PD, NAT2, CYB5R3, CYP3A4, and HLA-B) involved. These labels are presented in Table 4. Although the majority of the drugs were labeled as actionable PGx, none were labeled as no clinical PGx, testing required, or testing recommended. Actionable PGx entails contraindication, dose alteration, alternative therapy, or other management for individuals with a specific metabolizer phenotype or genotype (if known). This label, however, does not recommend phenotype or genotype testing prior to the use of the drug. The informative PGx label provides information on a particular variant/gene/phenotype/protein that can potentially affect the metabolism, concentration, and frequency of side effects or impose a general risk for the patients. However, this label provides no further guidance for the actions to be undertaken in such situations (PharmGKB). The overall statistics of the PGx label of antibiotics are shown in Figure 3. The majority of these labels are sourced from the FDA-approved drug label with at least 11 actionable PGx and 12 informative PGx for antibiotic drugs. Swissmedic, with at least 11 actionable PGx, is another important source for PGx-based drug labels for antibiotics.
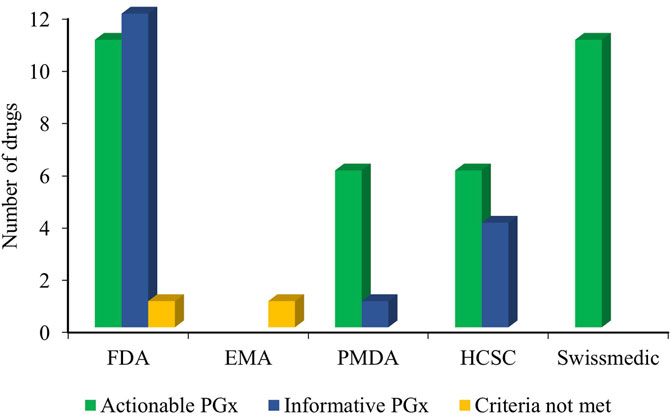
Figure 3. Overall PGx-based drug label of the antibiotics from the FDA, EMA, PMDA, HCSC, and Swissmedic (HCSC, Health Canada Santé Canada; FDA, US Food and Drug Administration; Swissmedic, Swiss Agency of Therapeutic Products; PMDA, Pharmaceuticals and Medical Devices Agency, Japan; EMA, European Medicines Agency; PGx, Pharmacogenomics).
3.4 Current state of PGx-based therapeutic and testing guidelines for antibiotics
The search for PGx-based guidelines across CPIC, DPWG, and CPNDS revealed at least six genes i.e., HLA-B, MT-RNR1, G6PD, RARG, SLC28A3, and UGT1A6. These PGx working bodies recommend therapy or testing for optimizing the effectiveness of several antibiotics based on the genetic variants of these six genes (Aminkeng et al., 2016; Gammal et al., 2023; Mcdermott et al., 2022, Dpwg, 2025b). For flucloxacillin-induced liver injury, DPWG deemed genotyping for HLA-B*57:01 to be beneficial and recommended alternative medicine for HLA-B*57:01-positive patients when bilirubin and/or liver enzyme levels are found elevated (Dpwg, 2025b). For aminoglycoside-induced hearing loss, CPIC provided a guideline considering the genotype of MT-RNR1, where they classified people into the categories normal, increased, and uncertain risk of aminoglycoside-induced hearing loss based on their genotype. In patients at increased risk, aminoglycoside use is strongly discouraged unless both the lack of safer alternatives and the severity of the infection outweigh the risk of ototoxicity (Mcdermott et al., 2022).
Based on the polymorphism in G6PD, the CPIC provided therapeutic guidelines for dapsone and nitrofurantoin. They classified individuals into normal, deficient, and deficient in chronic non-spherocytic hemolytic anemia (CNSHA) groups and variable and indeterminate groups based on the genotypes of G6PD. Avoidance of dapsone use is strongly recommended in deficient and deficient in CNSHA groups. On the contrary, for those deficient in the CNSHA group, avoidance of nitrofurantoin use is moderately recommended. They also suggested that in the deficient group, nitrofurantoin can be used in a standard dose, optionally with close monitoring for anemia (Gammal et al., 2023).
CPNDS, on the other hand, provided a guideline for anthracycline (doxorubicin, daunorubicin, and others)-induced cardiotoxicity based on the polymorphism of RARG, SLC28A3, and UGT1A6. They classified individuals according to their genotype into low, moderate, and high-risk groups. For the high-risk group, comprising individuals carrying RARG rs2229774A or UGT1A6*4, the CPNDS strongly recommended increased monitoring frequency and appropriate management of associated cardiovascular risk factors. They moderately encouraged the use of dexrazoxane and liposome-enclosed anthracycline preparations. As optional recommendations, they suggest slower infusion rates or continuous infusion, use of cardioprotective agents, or choosing alternative therapy with comparable efficacy (if available). For children receiving doxorubicin or daunorubicin therapy, CPNDS moderately recommended genetic testing for RARG rs2229774A, SLC28A3 rs7853758, and UGT1A6*4 rs17863783 variants. They, however, did not recommend genetic testing for children and adults receiving other types of anthracyclines (Aminkeng et al., 2016).
More details on these guidelines provided by DPWG, CPIC, and CPNDS are presented in Table 5.
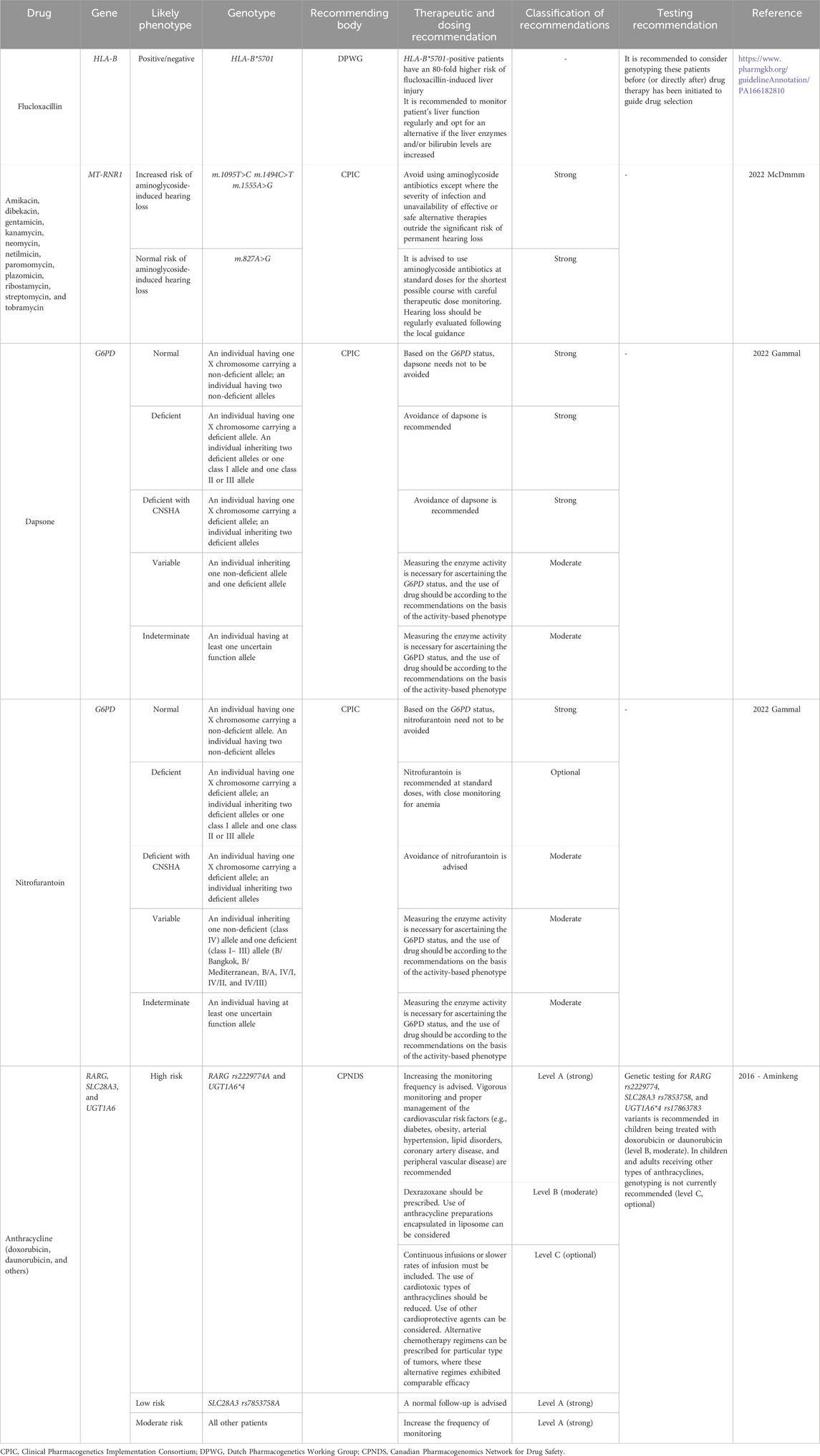
Table 5. Current PGx-based therapeutic and testing guidelines for antibiotics provided by the CPIC, CPNDS, and DPWG.
4 Discussion
This study identified a total of 65 clinical studies evaluating the genetic impact in producing different drug-induced adverse effects associated with antibiotic drugs. These studies provide a wide range of evidence reinforcing the need for PGx-based antibiotic therapy in clinical practice to achieve precision medicine. This evidence base explored a variety of gene variants associated with the ADRs—for example, beta-lactam-induced hypersensitivity reaction (with a varying OR of 1.36–5.1), flucloxacillin-induced DILI (associated with several HLA genes with ORs ranging from 1.86 to 79.21), anti-tuberculosis drug-induced hepatotoxicity (OR range 0.10–9.57), anthracycline-induced cardiotoxicity (reporting a varied ORs from 0.14 to 7.98), co-trimoxazole-induced SCARs (for a limited number of HLA genes with an OR range of 4.05–43.57), etc. A few of the protective biomarkers were identified during the literature search, such as NAT2*5 and NAT2 (rs1495741) (for isoniazid-induced liver injury, OR = 0.69 and 0.10, respectively), SLC22A16 T>C (rs714368) for doxorubicin-induced neutropenic and leukopenia (OR = 0.31 and 0.18, respectively), NQO1609TT (for epirubicin-induced anemia OR = 0.34 and grade 2–4 toxicity OR = 0.33), SLC28A3 (rs7853758), SLC28A3 (rs885004), ABCC10 (rs1214763), CYP2J2 (rs2294950), FMO2 (rs2020870), GPX3 (rs2233302), GSTM3 (rs12059276), SLC28A3 (rs7853758), SLC10A2 (rs9514091), SLC28A3 (rs4877847), SLC22A17 (rs4982753), SLC22A7 (rs4149178), SOD2 (rs7754103), SPG7 (rs2019604), and XDH (rs4407290) (for anthracycline-induced cardiotoxicity, OR = 0.46, 0.42, 0.34, 0.41, 0.14, 0.27, 0.37, 0.31, 0.43, 0.60, 0.52, 0.41, 0.30, 0.39, and 0.26, respectively) (Chan et al., 2017; Chaturvedi et al., 2015; Ebaid et al., 2024; Nicoletti et al., 2021b; Visscher et al., 2015; Visscher et al., 2012). We also explored the PharmGKB evidence level and PGx label information, which provided similar information on the genetic associations for the antibiotic drug-induced ADRs. However, to date, the clinical and dosing guidelines have been suggested for only a limited number of antibiotic drugs, with the aim of optimizing safety and effectiveness while reducing the incidence of ADRs through prediction. The findings of the current study, therefore, encourage policymakers to consider the growing evidence and take the necessary measures for its clinical adoption.
Although some robust literature-based associations were identified in the included studies, most of them provided preliminary associations of the genetic variants and adverse effects and recommended further exploration with a large number of subjects across the population for a comprehensive understanding, validation, and translation into implementable clinical guidelines (Amorim et al., 2023; An et al., 2012; Calcagno et al., 2019; Goldman et al., 2022; Guéant-Rodriguez et al., 2008; Gupta et al., 2013; Krebs et al., 2020; Nicoletti et al., 2021a; Nyangwara et al., 2024; Park et al., 2024; Sukasem et al., 2020; Suvichapanich et al., 2019; Tempark et al., 2017; Thomas et al., 2025; Vuilleumier et al., 2006; Wang et al., 2024b; Yang et al., 2017; Yuliwulandari et al., 2016). However, such proper large-scale follow-up studies were scarce, keeping these reported preliminary associations largely unexplored, which may contribute to the limited number of clinical guidelines available. Nevertheless, there are several antibiotic candidates with various genetic associations replicated in multiple studies and have moderate to high (level-1 and level-2) PharmGKB evidence level and PGx drug label information. For example, the association between isoniazid and the NAT2 genetic polymorphism has been well studied for toxicity, carries a high PharmGKB evidence level-1B, and has been labeled with informative PGx by the FDA and PMDA (Ben Fredj et al., 2017; Chan et al., 2017; Kasamatsu et al., 2025; Nicoletti et al., 2021b; Thomas et al., 2025). Similarly, the association between co-trimoxazole and HLA genes for SCARs has been reported in multiple clinical studies and has a moderate PharmGKB evidence level of 2A (for HLA-B) and 2B (for HLA-C) for drug-induced toxicity. However, this genetic association with HLA has no PGx label information (Goldman et al., 2022; Sukasem et al., 2020). It is evident that even after having some considerable and growing evidence for certain genetic associations for antibiotics and toxicity, sufficient measures are not being undertaken to translate them into clinical use. It is about time for the international PGx working bodies to develop PGx-dosing guidelines so that clinicians can easily incorporate recommendations into routine clinical practice.
As of now, no antibiotic drug has a testing-required or recommended label by the FDA, EMA, PMDA, HCSC, or Swissmedic. Nevertheless, several studies reported the importance of genetic testing in the prediction and management of adverse effects associated with antibiotics. For example, Gupta et al. (2013) informed that the early detection of GSTM1 and T1 null may help lower ATD-induced hepatotoxicity. To reduce the risk of AT-DILI, Yuliwulandari et al. (2016) recommended the NAT2 genotype and corresponding phenotype determination. For customizing the anthracycline therapy in cancer, Ebaid et al. (2024) emphasized the importance of genetic testing for SLC22A16 and CBR1. A prediction model based on both genetic and clinical risk factors was deemed beneficial by Visscher et al. (2013) in anthracycline therapy for identifying risk profiles for cardiotoxicity. For vancomycin-induced DRESS, Konvinse et al. (2019) stated that HLA-A*32:01 testing may improve safety and efficacy. For levofloxacin-induced SCARs, Jiang et al. (2023) informed prospective screening of serotype B13, and prescribing alternative drug therapy for the carriers significantly reduces the incidence of adverse effects. Satapornpong et al. (2021) supported the genotyping of the HLA-B*13:01 allele to avoid SCARs with dapsone therapy in the Asian population. Asif et al. (2024) recommended considering the screening of HLA-A*32:01 for risk stratification in long-term therapy with vancomycin (Asif et al., 2024; Blanco et al., 2012; Ebaid et al., 2024; Göpel et al., 2014; Gupta et al., 2013; Jiang et al., 2023; Konvinse et al., 2019; Satapornpong et al., 2021; Schiuma et al., 2025; Visscher et al., 2013; Wang et al., 2024b; Yuliwulandari et al., 2016).
Another limiting factor for the adoption of PGx in clinical practice for antibiotic therapy is the paucity of cost-effectiveness studies. Health economics plays a vital role in supporting policymakers in allocating limited resources, and therefore, cost-effectiveness studies are essential for evidence-based decision-making (Kategeaw et al., 2023; Leelahavarong et al., 2019). One such cost-effectiveness analysis conducted by Kategeaw et al. (2023), for preventing SCARs with co-trimoxazole therapy in HIV-infected Thai patients, revealed that the screening of HLA-B*13:01 before initiating the therapy was not likely to be cost-effective. Similar cost-effectiveness studies for the important antibiotic-genetic variant pairs in diverse populations are warranted to provide a comprehensive overview of the effects of PGx in antibiotic therapy and subsequent adoption in clinical practice.
Several complex traits, such as the sensitivity to adverse reactions and efficacy of the drug, are sometimes attributable to several different genetic variants. Owing to the remarkable progress in genome sequencing and genome-wide association studies, several polygenic risk scores, including some related to PGx, have been developed (Cross et al., 2022; Evans et al., 2009). For antibiotics, such multigene effects have also been recorded. For example, GSTM1 and T1 null genotypes had a significant association with ATD-induced hepatotoxicity (OR = 7.18, 95% CI: 1.7–32.6, p = 0.007), and for isoniazid-induced hepatotoxicity, individuals with both NAT2 slow acetylator and CYP2E1 DraI C/D had an elevated risk (Ben Fredj et al., 2017; Gupta et al., 2013). Exploring these and other genetic associations for different antibiotic drugs and further developing polygenic risk scores for them can be a rational approach for adopting PGx-based antibiotic use in clinical practice.
To the best of our knowledge, this is the first comprehensive review showing the current evidence of antibiotic-induced hypersensitivity reactions involving PGx. Furthermore, this review summarized the current state of PGx-based therapeutic and testing guidelines for antibiotics in clinical practice, taking into account PGx-based clinical annotations and drug label information.
Although this comprehensive review has insightful information regarding PGx associations of antibiotic-induced hypersensitivity reactions, there is a limitation of this review. The search for relevant literature was carried out in PubMed only, which may limit the possibility of obtaining all potential evidence.
5 Conclusion
In conclusion, this study identified at least 12 antibiotic–gene pairs (amikacin–MT-RNR1, gentamicin–MT-RNR1, kanamycin–MT-RNR1, streptomycin–MT-RNR1, neomycin–MT-RNR1, tobramycin–MT-RNR1, isoniazid–NAT2, dapsone–HLA-B, co-trimoxazole–HLA-B and HLA-C, flucloxacillin–HLA-B, daunorubicin–SLC28A3, and doxorubicin–SLC28A3) with moderate-to-high PharmGKB evidence level for toxicity. However, PGx-based dosing guidelines, as recommended by the CPIC, DPWG and CPNDS, are available for the following antibiotic–gene pairs: amikacin, gentamicin, kanamycin, streptomycin, neomycin, and tobramycin–MT-RNR1; flucloxacillin–HLA-B; dapsone–G6PD; nitrofurantoin–G6PD; and daunorubicin and doxorubicin–RARG, SLC28A3, and UGT1A6. Despite the established and growing genetic evidence for the toxicity, particularly co-trimoxazole-induced SCARs associated with HLA-B and HLA-C, dapsone-induced SCARs associated with HLA-B, and isoniazid-induced liver injury associated with NAT2, sufficient efforts have not been undertaken to translate findings into routine clinical practice. The lack of validation of preliminary genetic associations, due to the scarcity of proper follow-up and large-scale replication, represents a key setback for the PGx-based implementation of antibiotic therapy in clinical practice. More focused clinical studies, cost-effectiveness analyses, and polygenic risk score development are required for the PGx-based clinical use of antibiotics to optimize the safety and effectiveness.
Author contributions
MB: Conceptualization, Data curation, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing. MM: Data curation, Formal analysis, Writing – original draft. MA: Data curation, Formal analysis, Writing – review and editing. ME: Data curation, Visualization, Writing – review and editing. CS: Conceptualization, Supervision, Validation, Visualization, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, J., Dellinger, A., Nicoletti, P., Barnhart, H. X., Ghabril, M., Fontana, R. J., et al. (2025). Clinical and HLA associations of fluoroquinolone-induced liver injury: results from the drug-induced liver injury network. Am. J. Gastroenterol. doi:10.14309/ajg.0000000000003457
Alfirevic, A., Vilar, F. J., Alsbou, M., Jawaid, A., Thomson, W., Ollier, W. E., et al. (2009). TNF, LTA, HSPA1L and HLA-DR gene polymorphisms in HIV-positive patients with hypersensitivity to cotrimoxazole. Pharmacogenomics 10, 531–540. doi:10.2217/pgs.09.6
Altés, A., Paré, L., Esquirol, A., Xicoy, B., Rámila, E., Vicente, L., et al. (2013). Pharmacogenetic analysis in the treatment of hodgkin lymphoma. Leuk. Lymphoma 54, 1706–1712. doi:10.3109/10428194.2012.752080
Aminkeng, F., Ross, C. J., Rassekh, S. R., Hwang, S., Rieder, M. J., Bhavsar, A. P., et al. (2016). Recommendations for genetic testing to reduce the incidence of anthracycline-induced cardiotoxicity. Br. J. Clin. Pharmacol. 82, 683–695. doi:10.1111/bcp.13008
Amorim, G., Jaworski, J., Cordeiro-Santos, M., Kritski, A. L., Figueiredo, M. C., Turner, M., et al. (2023). Pharmacogenetics of tuberculosis treatment toxicity and effectiveness in a large Brazilian cohort. medRxiv, 23294860. doi:10.1101/2023.08.30.23294860
An, H. R., Wu, X. Q., Wang, Z. Y., Zhang, J. X., and Liang, Y. (2012). NAT2 and CYP2E1 polymorphisms associated with antituberculosis drug-induced hepatotoxicity in Chinese patients. Clin. Exp. Pharmacol. Physiol. 39, 535–543. doi:10.1111/j.1440-1681.2012.05713.x
Asif, B. A., Koh, C., Phillips, E. J., Gu, J., Li, Y. J., Barnhart, H., et al. (2024). Vancomycin-induced liver injury, DRESS, and HLA-A∗32:01. J. Allergy Clin. Immunol. Pract. 12, 168–174.e2. doi:10.1016/j.jaip.2023.09.011
Badamasi, I. M., Muhammad, M., Umar, A. A., Madugu, U. M., Gadanya, M. A., Aliyu, I. A., et al. (2024). Role of the IL8 rs4073 polymorphism in central nervous system toxicity in patients receiving multidrug-resistant tuberculosis treatment. J. Bras. Pneumol. 50, e20230338. doi:10.36416/1806-3756/e20230338
Barbarino, J. M., Whirl-Carrillo, M., Altman, R. B., and Klein, T. E. (2018). PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 10, e1417. doi:10.1002/wsbm.1417
Ben Fredj, N., Gam, R., Kerkni, E., Chaabane, A., Chadly, Z., Boughattas, N., et al. (2017). Risk factors of isoniazid-induced hepatotoxicity in Tunisian tuberculosis patients. Pharmacogenomics J. 17, 372–377. doi:10.1038/tpj.2016.26
Blanco, J. G., Sun, C. L., Landier, W., Chen, L., Esparza-Duran, D., Leisenring, W., et al. (2012). Anthracycline-related cardiomyopathy after childhood cancer: role of polymorphisms in carbonyl reductase genes--a report from the children's oncology group. J. Clin. Oncol. 30, 1415–1421. doi:10.1200/jco.2011.34.8987
Blumenthal, K. G., Peter, J. G., Trubiano, J. A., and Phillips, E. J. (2019). Antibiotic allergy. Lancet 393, 183–198. doi:10.1016/s0140-6736(18)32218-9
Calcagno, A., Cusato, J., Sekaggya-Wiltshire, C., Von Braun, A., Motta, I., Turyasingura, G., et al. (2019). The influence of pharmacogenetic variants in HIV/tuberculosis coinfected patients in Uganda in the SOUTH study. Clin. Pharmacol. Ther. 106, 450–457. doi:10.1002/cpt.1403
Chan, S. L., Chua, A. P. G., Aminkeng, F., Chee, C. B. E., Jin, S., Loh, M., et al. (2017). Association and clinical utility of NAT2 in the prediction of isoniazid-induced liver injury in Singaporean patients. PLoS One 12, e0186200. doi:10.1371/journal.pone.0186200
Chaturvedi, P., Tulsyan, S., Agarwal, G., Lal, P., Agrawal, S., Mittal, R. D., et al. (2015). Relationship of MTHFR and NQO1 pharmacogenetics and chemotherapy clinical outcomes in breast cancer patients. Biochem. Genet. 53, 211–222. doi:10.1007/s10528-015-9683-z
Chen, Y. C., Chiu, H. C., and Chu, C. Y. (2010). Drug reaction with eosinophilia and systemic symptoms: a retrospective study of 60 cases. Arch. Dermatol 146, 1373–1379. doi:10.1001/archdermatol.2010.198
Chen, R., Zhang, Y., Tang, S., Lv, X., Wu, S., Sun, F., et al. (2015). The association between HLA-DQB1 polymorphism and antituberculosis drug-induced liver injury: a case-control study. J. Clin. Pharm. Ther. 40, 110–115. doi:10.1111/jcpt.12211
Conlon, C., Li, Y. J., Ahmad, J., Barnhart, H., Fontana, R. J., Ghabril, M., et al. (2024). Clinical characteristics and HLA associations of azithromycin-induced liver injury. Aliment. Pharmacol. Ther. 60, 787–795. doi:10.1111/apt.18160
Cornejo-García, J. A., Romano, A., Guéant-Rodríguez, R. M., Oussalah, A., Blanca-López, N., Gaeta, F., et al. (2016). A non-synonymous polymorphism in galectin-3 lectin domain is associated with allergic reactions to beta-lactam antibiotics. Pharmacogenomics J. 16, 79–82. doi:10.1038/tpj.2015.24
Cpic (2025). Clinical pharmacogenetics implementation consortium. Available online at: https://cpicpgx.org/ (Accessed June 1, 2025).
Cpnds (2025). Canadian pharmacogenomics network for drug safety. Available online at: https://cpnds.ubc.ca/ (Accessed June 1, 2025).
Cross, B., Turner, R., and Pirmohamed, M. (2022). Polygenic risk scores: an overview from bench to bedside for personalised medicine. Front. Genet. 13, 1000667. doi:10.3389/fgene.2022.1000667
Dekker, J. W., Nizankowska, E., Schmitz-Schumann, M., Pile, K., Bochenek, G., Dyczek, A., et al. (1997). Aspirin-induced asthma and HLA-DRB1 and HLA-DPB1 genotypes. Clin. Exp. Allergy 27, 574–577. doi:10.1046/j.1365-2222.1997.540848.x
Devarbhavi, H., Dierkhising, R., and Kremers, W. K. (2010). Antituberculosis therapy drug-induced liver injury and acute liver failure. Hepatology 52, 798–799. doi:10.1002/hep.23805
Doña, I., Blanca-López, N., Torres, M. J., García-Campos, J., García-Núñez, I., Gómez, F., et al. (2012). Drug hypersensitivity reactions: response patterns, drug involved, and temporal variations in a large series of patients. J. Investig. Allergol. Clin. Immunol. 22, 363–371.
Dpwg (2025a). The Dutch guidelines November 2018 update. Available online at: https://api.pharmgkb.org/v1/download/file/attachment/DPWG_November_2018.pdf (Accessed June 1, 2025).
Dpwg (2025b). Dutch pharmacogenetics working group. Available online at: https://www.knmp.nl/dossiers/farmacogenetica (Accessed June 1, 2025).
Duong, T. A., Valeyrie-Allanore, L., Wolkenstein, P., and Chosidow, O. (2017). Severe cutaneous adverse reactions to drugs. Lancet 390, 1996–2011. doi:10.1016/s0140-6736(16)30378-6
Ebaid, N. F., Abdelkawy, K. S., Shehata, M. A., Salem, H. F., Magdy, G., Hussein, R. R. S., et al. (2024). Effects of pharmacogenetics on pharmacokinetics and toxicity of doxorubicin in Egyptian breast cancer patients. Xenobiotica 54, 160–170. doi:10.1080/00498254.2024.2330493
Evans, D. M., Visscher, P. M., and Wray, N. R. (2009). Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum. Mol. Genet. 18, 3525–3531. doi:10.1093/hmg/ddp295
Firoz, B. F., Henning, J. S., Zarzabal, L. A., and Pollock, B. H. (2012). Toxic epidermal necrolysis: five years of treatment experience from a burn unit. J. Am. Acad. Dermatol. 67, 630–635. doi:10.1016/j.jaad.2011.12.014
Fischel-Ghodsian, N., Prezant, T. R., Chaltraw, W. E., Wendt, K. A., Nelson, R. A., Arnos, K. S., et al. (1997). Mitochondrial gene mutation is a significant predisposing factor in aminoglycoside ototoxicity. Am. J. Otolaryngol. 18, 173–178. doi:10.1016/s0196-0709(97)90078-8
Gammal, R. S., Pirmohamed, M., Somogyi, A. A., Morris, S. A., Formea, C. M., Elchynski, A. L., et al. (2023). Expanded clinical pharmacogenetics implementation consortium guideline for medication use in the context of G6PD genotype. Clin. Pharmacol. Ther. 113, 973–985. doi:10.1002/cpt.2735
Goldman, J. L., Miller, J. O., Miller, N., Eveleigh, R., Gibson, A., Phillips, E. J., et al. (2022). HLA-B*07:02 and HLA-C*07:02 are associated with trimethoprim-sulfamethoxazole respiratory failure. Pharmacogenomics J. 22, 124–129. doi:10.1038/s41397-022-00266-8
Göpel, W., Berkowski, S., Preuss, M., Ziegler, A., Küster, H., Felderhoff-Müser, U., et al. (2014). Mitochondrial mutation m.1555A>G as a risk factor for failed newborn hearing screening in a large cohort of preterm infants. BMC Pediatr. 14, 210. doi:10.1186/1471-2431-14-210
Guéant-Rodriguez, R. M., Guéant, J. L., Viola, M., Tramoy, D., Gaeta, F., and Romano, A. (2008). Association of tumor necrosis factor-alpha -308G>A polymorphism with IgE-mediated allergy to betalactams in an Italian population. Pharmacogenomics J. 8, 162–168. doi:10.1038/sj.tpj.6500456
Gupta, V. H., Singh, M., Amarapurkar, D. N., Sasi, P., Joshi, J. M., Baijal, R., et al. (2013). Association of GST null genotypes with anti-tuberculosis drug induced hepatotoxicity in Western Indian population. Ann. Hepatol. 12, 959–965. doi:10.1016/s1665-2681(19)31302-x
Guzman, A. I., and Paliza, A. C. (2018). Epidemiology of severe cutaneous adverse drug reactions in a university hospital: a five-year review. J. Med. 2, 171–184. doi:10.35460/2546-1621.2017-0031
Hu, X., Zhang, M., Bai, H., Wu, L., Chen, Y., Ding, L., et al. (2018). Antituberculosis drug-induced adverse events in the liver, kidneys, and blood: clinical profiles and pharmacogenetic predictors. Clin. Pharmacol. Ther. 104, 326–334. doi:10.1002/cpt.924
Husain, Z., Reddy, B. Y., and Schwartz, R. A. (2013). DRESS syndrome: part I. Clinical perspectives. J. Am. Acad. Dermatol. 68, 693.e1–14. doi:10.1016/j.jaad.2013.01.033
Jiang, M., Yang, J., Yang, L., Wang, L., Wang, T., Han, S., et al. (2023). An association study of HLA with levofloxacin-induced severe cutaneous adverse drug reactions in han Chinese. iScience 26, 107391. doi:10.1016/j.isci.2023.107391
Johansson, S. G., Bieber, T., Dahl, R., Friedmann, P. S., Lanier, B. Q., Lockey, R. F., et al. (2004). Revised nomenclature for allergy for global use: report of the nomenclature review committee of the world allergy organization, October 2003. J. Allergy Clin. Immunol. 113, 832–836. doi:10.1016/j.jaci.2003.12.591
Kang, D. Y., Yun, J., Lee, S. Y., Koh, Y. I., Sim, D. W., Kim, S., et al. (2021). A nationwide study of severe cutaneous adverse reactions based on the multicenter registry in Korea. J. Allergy Clin. Immunol. Pract. 9, 929–936.e7. doi:10.1016/j.jaip.2020.09.011
Kasamatsu, A., Miyahara, R., Yoneoka, D., Toyo-Oka, L., Chiyasirinroje, B., Imsanguan, W., et al. (2025). One-year mortality of tuberculosis patients on isoniazid-based treatment and its association with rapid acetylator NAT2 genotypes. Int. J. Infect. Dis. 155, 107895. doi:10.1016/j.ijid.2025.107895
Kategeaw, W., Nakkam, N., Kiertiburanakul, S., Sukasem, C., Tassaneeyakul, W., and Chaiyakunapruk, N. (2023). Cost-effectiveness analysis of HLA-B*13:01 screening for the prevention of co-trimoxazole-induced severe cutaneous adverse reactions among HIV-Infected patients in Thailand. J. Med. Econ. 26, 1330–1341. doi:10.1080/13696998.2023.2270868
Kim, S. H., Kim, S. H., Yoon, H. J., Shin, D. H., Park, S. S., Kim, Y. S., et al. (2012). TNF-α genetic polymorphism− 308G/A and antituberculosis drug-induced hepatitis. Liver Int. 32, 809–814. doi:10.1111/j.1478-3231.2011.02697.x
Kloypan, C., Koomdee, N., Satapornpong, P., Tempark, T., Biswas, M., and Sukasem, C. (2021). A comprehensive review of HLA and severe cutaneous adverse drug reactions: implication for clinical pharmacogenomics and precision medicine. Pharm. (Basel) 14, 1077. doi:10.3390/ph14111077
Konvinse, K. C., Trubiano, J. A., Pavlos, R., James, I., Shaffer, C. M., Bejan, C. A., et al. (2019). HLA-A*32:01 is strongly associated with vancomycin-induced drug reaction with eosinophilia and systemic symptoms. J. Allergy Clin. Immunol. 144, 183–192. doi:10.1016/j.jaci.2019.01.045
Krebs, K., Bovijn, J., Zheng, N., Lepamets, M., Censin, J. C., Jürgenson, T., et al. (2020). Genome-wide study identifies association between HLA-B(∗)55:01 and self-reported penicillin allergy. Am. J. Hum. Genet. 107, 612–621. doi:10.1016/j.ajhg.2020.08.008
Lavanderos, M. A., Cayún, J. P., Roco, Á., Sandoval, C., Cerpa, L., Rubilar, J. C., et al. (2019). Association study among candidate genetic polymorphisms and chemotherapy-related severe toxicity in testicular cancer patients. Front. Pharmacol. 10, 206. doi:10.3389/fphar.2019.00206
Lee, S. W., Chen, P. T., Liu, C. W., Li, Y. H., and Wu, L. S. (2024). Polymorphism of CYP3A4*18 is associated with anti-tuberculosis drug-induced hepatotoxicity. Pharmacogenomics 25, 241–247. doi:10.1080/14622416.2024.2346069
Leelahavarong, P., Doungthipsirikul, S., Kumluang, S., Poonchai, A., Kittiratchakool, N., Chinnacom, D., et al. (2019). Health technology assessment in Thailand: institutionalization and contribution to healthcare decision making: review of literature. Int. J. Technol. Assess. Health Care 35, 467–473. doi:10.1017/s0266462319000321
Li, L. M., Chen, L., Deng, G. H., Tan, W. T., Dan, Y. J., Wang, R. Q., et al. (2012). SLCO1B1 *15 haplotype is associated with rifampin-induced liver injury. Mol. Med. Rep. 6, 75–82. doi:10.3892/mmr.2012.900
Li, Y., Tang, H., Qi, H., Shen, C., Sun, L., Li, J., et al. (2018). rs1800796 of the IL6 gene is associated with increased risk for anti-tuberculosis drug-induced hepatotoxicity in Chinese Han children. Tuberc. (Edinb) 111, 71–77. doi:10.1016/j.tube.2018.05.011
Lipshultz, S. E., Alvarez, J. A., and Scully, R. E. (2008). Anthracycline associated cardiotoxicity in survivors of childhood cancer. Heart 94, 525–533. doi:10.1136/hrt.2007.136093
Lu, J., Li, Z., Zhu, Y., Yang, A., Li, R., Zheng, J., et al. (2010). Mitochondrial 12S rRNA variants in 1642 han Chinese pediatric subjects with aminoglycoside-induced and nonsyndromic hearing loss. Mitochondrion 10, 380–390. doi:10.1016/j.mito.2010.01.007
Mcdermott, J. H., Wolf, J., Hoshitsuki, K., Huddart, R., Caudle, K. E., Whirl-Carrillo, M., et al. (2022). Clinical pharmacogenetics implementation consortium guideline for the use of aminoglycosides based on MT-RNR1 genotype. Clin. Pharmacol. Ther. 111, 366–372. doi:10.1002/cpt.2309
Mockenhaupt, M. (2012). Epidemiology of cutaneous adverse drug reactions. Chem. Immunol. Allergy 97, 1–17. doi:10.1159/000335612
Monteiro, T. P., El-Jaick, K. B., Jeovanio-Silva, A. L., Brasil, P. E., Costa, M. J., Rolla, V. C., et al. (2012). The roles of GSTM1 and GSTT1 null genotypes and other predictors in anti-tuberculosis drug-induced liver injury. J. Clin. Pharm. Ther. 37, 712–718. doi:10.1111/j.1365-2710.2012.01368.x
Nakkam, N., Saksit, N., Konyoung, P., Amornpinyo, W., Khunarkornsiri, U., Purimart, D., et al. (2022). Associations of HLA and drug-metabolizing enzyme genes in co-trimoxazole-induced severe cutaneous adverse reactions. Drug Metab. Pharmacokinet. 47, 100480. doi:10.1016/j.dmpk.2022.100480
Nicoletti, P., Aithal, G. P., Chamberlain, T. C., Coulthard, S., Alshabeeb, M., Grove, J. I., et al. (2019). Drug-induced liver injury due to flucloxacillin: relevance of multiple human leukocyte antigen alleles. Clin. Pharmacol. Ther. 106, 245–253. doi:10.1002/cpt.1375
Nicoletti, P., Carr, D. F., Barrett, S., Mcevoy, L., Friedmann, P. S., Shear, N. H., et al. (2021a). Beta-lactam-induced immediate hypersensitivity reactions: a genome-wide association study of a deeply phenotyped cohort. J. Allergy Clin. Immunol. 147, 1830–1837.e15. doi:10.1016/j.jaci.2020.10.004
Nicoletti, P., Devarbhavi, H., Goel, A., Venkatesan, R., Eapen, C. E., Grove, J. I., et al. (2021b). Genetic risk factors in drug-induced liver injury due to isoniazid-containing antituberculosis drug regimens. Clin. Pharmacol. Ther. 109, 1125–1135. doi:10.1002/cpt.2100
Nyangwara, V. A., Mazhindu, T., Chikwambi, Z., Masimirembwa, C., Campbell, T. B., Borok, M., et al. (2024). Cardiotoxicity and pharmacogenetics of doxorubicin in black Zimbabwean breast cancer patients. Br. J. Clin. Pharmacol. 90, 1782–1789. doi:10.1111/bcp.15659
Osanlou, O., Pirmohamed, M., and Daly, A. K. (2018). Pharmacogenetics of adverse drug reactions. Adv. Pharmacol. 83, 155–190. doi:10.1016/bs.apha.2018.03.002
Ouzzani, M., Hammady, H., Fedorowicz, Z., and Elmagarmid, A. (2016). Rayyan-a web and mobile app for systematic reviews. Syst. Rev. 5, 210. doi:10.1186/s13643-016-0384-4
Owen, C. E., and Jones, J. M. (2021). Recognition and management of severe cutaneous adverse drug reactions (including drug reaction with eosinophilia and systemic symptoms, Stevens-Johnson syndrome, and toxic epidermal necrolysis). Med. Clin. North Am. 105, 577–597. doi:10.1016/j.mcna.2021.04.001
Page, M. J., Mckenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Bmj 372, n71. doi:10.1136/bmj.n71
Park, S. Y., Park, S. Y., Seo, S., Kwon, H. S., Kim, S. H., Kim, S. H., et al. (2024). HLA-DRB1 is associated with cefaclor-induced immediate hypersensitivity. World Allergy Organ J. 17, 100901. doi:10.1016/j.waojou.2024.100901
Peter, J. G., Lehloenya, R., Dlamini, S., Risma, K., White, K. D., Konvinse, K. C., et al. (2017). Severe delayed cutaneous and systemic reactions to drugs: a global perspective on the science and art of current practice. J. Allergy Clin. Immunol. Pract. 5, 547–563. doi:10.1016/j.jaip.2017.01.025
Pharmgkb (2025a). Clinical annotation levels of evidence. Available online at: https://www.pharmgkb.org/page/clinAnnLevels (Accessed June 2, 2025).
Pharmgkb (2025b). Drug label information and legend. Available online at: https://www.pharmgkb.org/page/drugLabelLegend (Accessed June 2, 2025).
Ramappa, V., and Aithal, G. P. (2013). Hepatotoxicity related to anti-tuberculosis drugs: mechanisms and management. J. Clin. Exp. Hepatol. 3, 37–49. doi:10.1016/j.jceh.2012.12.001
Robinson, K. M., Yang, W., Karol, S. E., Kornegay, N., Jay, D., Cheng, C., et al. (2019). No evidence that G6PD deficiency affects the efficacy or safety of daunorubicin in acute lymphoblastic leukemia induction therapy. Pediatr. Blood Cancer 66, e27681. doi:10.1002/pbc.27681
Roth, S. M., Williams, S. M., Jiang, L., Menon, K. S., and Jeka, J. J. (2008). Susceptibility genes for gentamicin-induced vestibular dysfunction. J. Vestib. Res. 18, 59–68. doi:10.3233/ves-2008-18106
Sassolas, B., Haddad, C., Mockenhaupt, M., Dunant, A., Liss, Y., Bork, K., et al. (2010). ALDEN, an algorithm for assessment of drug causality in Stevens-Johnson syndrome and toxic epidermal necrolysis: comparison with case-control analysis. Clin. Pharmacol. Ther. 88, 60–68. doi:10.1038/clpt.2009.252
Satapornpong, P., Pratoomwun, J., Rerknimitr, P., Klaewsongkram, J., Nakkam, N., Rungrotmongkol, T., et al. (2021). HLA-B*13:01 is a predictive marker of dapsone-induced severe cutaneous adverse reactions in Thai patients. Front. Immunol. 12, 661135. doi:10.3389/fimmu.2021.661135
Schiuma, M., Dinegro, S., Battini, V., Torre, A., Covizzi, A., Civati, A., et al. (2025). NAT2 acetylation status predicts hepatotoxicity during antituberculosis therapy: cumulative risk analysis of a multiethnic cohort. Int. J. Mol. Sci. 26, 3881. doi:10.3390/ijms26083881
Schneck, J., Fagot, J. P., Sekula, P., Sassolas, B., Roujeau, J. C., and Mockenhaupt, M. (2008). Effects of treatments on the mortality of Stevens-Johnson syndrome and toxic epidermal necrolysis: a retrospective study on patients included in the prospective EuroSCAR study. J. Am. Acad. Dermatol. 58, 33–40. doi:10.1016/j.jaad.2007.08.039
Sukasem, C., Pratoomwun, J., Satapornpong, P., Klaewsongkram, J., Rerkpattanapipat, T., Rerknimitr, P., et al. (2020). Genetic association of co-trimoxazole-induced severe cutaneous adverse reactions is phenotype-specific: HLA class I genotypes and haplotypes. Clin. Pharmacol. Ther. 108, 1078–1089. doi:10.1002/cpt.1915
Suvichapanich, S., Wattanapokayakit, S., Mushiroda, T., Yanai, H., Chuchottawon, C., Kantima, T., et al. (2019). Genomewide association study confirming the association of NAT2 with susceptibility to antituberculosis drug-induced liver injury in Thai patients. Antimicrob. Agents Chemother. 63, e02692-18. doi:10.1128/aac.02692-18
Tempark, T., Satapornpong, P., Rerknimitr, P., Nakkam, N., Saksit, N., Wattanakrai, P., et al. (2017). Dapsone-induced severe cutaneous adverse drug reactions are strongly linked with HLA-B*13: 01 allele in the Thai population. Pharmacogenet Genomics 27, 429–437. doi:10.1097/fpc.0000000000000306
Tempark, T., John, S., Rerknimitr, P., Satapornpong, P., and Sukasem, C. (2022). Drug-induced severe cutaneous adverse reactions: insights into clinical presentation, immunopathogenesis, diagnostic methods, treatment, and pharmacogenomics. Front. Pharmacol. 13, 832048. doi:10.3389/fphar.2022.832048
Thomas, L., Batra, Y., Mathur, M., Kulavalli, S., Sv, C., Chaithra, S., et al. (2025). Pharmacogenomic heterogeneity of N-acetyltransferase 2: a comprehensive analysis of real world data in Indian tuberculosis patients and from literature and database review. Ann. Med. 57, 2478316. doi:10.1080/07853890.2025.2478316
Tsuji, D., Matsumoto, M., Kawasaki, Y., Kim, Y. L., Yamamoto, K., Nakamichi, H., et al. (2021). Analysis of pharmacogenomic factors for chemotherapy-induced nausea and vomiting in patients with breast cancer receiving doxorubicin and cyclophosphamide chemotherapy. Cancer Chemother. Pharmacol. 87, 73–83. doi:10.1007/s00280-020-04177-y
Urban, T. J., Nicoletti, P., Chalasani, N., Serrano, J., Stolz, A., Daly, A. K., et al. (2017). Minocycline hepatotoxicity: clinical characterization and identification of HLA-B∗35:02 as a risk factor. J. Hepatol. 67, 137–144. doi:10.1016/j.jhep.2017.03.010
Verma, R., Vasudevan, B., and Pragasam, V. (2013). Severe cutaneous adverse drug reactions. Med. J. Armed Forces India 69, 375–383. doi:10.1016/j.mjafi.2013.01.007
Visscher, H., Ross, C. J., Rassekh, S. R., Barhdadi, A., Dubé, M. P., Al-Saloos, H., et al. (2012). Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J. Clin. Oncol. 30, 1422–1428. doi:10.1200/jco.2010.34.3467
Visscher, H., Ross, C. J., Rassekh, S. R., Sandor, G. S., Caron, H. N., Van Dalen, E. C., et al. (2013). Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr. Blood Cancer 60, 1375–1381. doi:10.1002/pbc.24505
Visscher, H., Rassekh, S. R., Sandor, G. S., Caron, H. N., Van Dalen, E. C., Kremer, L. C., et al. (2015). Genetic variants in SLC22A17 and SLC22A7 are associated with anthracycline-induced cardiotoxicity in children. Pharmacogenomics 16, 1065–1076. doi:10.2217/pgs.15.61
Vuilleumier, N., Rossier, M. F., Chiappe, A., Degoumois, F., Dayer, P., Mermillod, B., et al. (2006). CYP2E1 genotype and isoniazid-induced hepatotoxicity in patients treated for latent tuberculosis. Eur. J. Clin. Pharmacol. 62, 423–429. doi:10.1007/s00228-006-0111-5
Wang, D., Curtis, A., Papp, A. C., Koletar, S. L., and Para, M. F. (2012). Polymorphism in glutamate cysteine ligase catalytic subunit (GCLC) is associated with sulfamethoxazole-induced hypersensitivity in HIV/AIDS patients. BMC Med. Genomics 5, 32. doi:10.1186/1755-8794-5-32
Wang, C. C., Chen, I. C., Lin, G. C., Chen, Y. M., and Shen, C. H. (2024a). Polymorphisms of HLA genes and hypersensitivity to penicillin among patients in a Taiwanese population. Int. J. Immunogenet. 51, 291–299. doi:10.1111/iji.12678
Wang, C. C., Shen, C. H., Lin, G. C., Chen, Y. M., and Chen, I. C. (2024b). Association of HLA alleles with cephalosporin allergy in the Taiwanese population. Sci. Rep. 14, 17167. doi:10.1038/s41598-024-68185-1
Wattanapokayakit, S., Mushiroda, T., Yanai, H., Wichukchinda, N., Chuchottawon, C., Nedsuwan, S., et al. (2016). NAT2 slow acetylator associated with anti-tuberculosis drug-induced liver injury in Thai patients. Int. J. Tuberc. Lung Dis. 20, 1364–1369. doi:10.5588/ijtld.15.0310
Whirl-Carrillo, M., Huddart, R., Gong, L., Sangkuhl, K., Thorn, C. F., Whaley, R., et al. (2021). An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 110, 563–572. doi:10.1002/cpt.2350
Yamada, S., Richardson, K., Tang, M., Halaschek-Wiener, J., Cook, V. J., Fitzgerald, J. M., et al. (2010). Genetic variation in carboxylesterase genes and susceptibility to isoniazid-induced hepatotoxicity. Pharmacogenomics J. 10, 524–536. doi:10.1038/tpj.2010.5
Yang, F., Gu, B., Zhang, L., Xuan, J., Luo, H., Zhou, P., et al. (2014). HLA-B*13:01 is associated with salazosulfapyridine-induced drug rash with eosinophilia and systemic symptoms in Chinese Han population. Pharmacogenomics 15, 1461–1469. doi:10.2217/pgs.14.69
Yang, M. S., Lee, J. Y., Kim, J., Kim, G. W., Kim, B. K., Kim, J. Y., et al. (2016). Incidence of Stevens-Johnson syndrome and toxic epidermal necrolysis: a nationwide population-based study using national health insurance database in Korea. PLoS One 11, e0165933. doi:10.1371/journal.pone.0165933
Yang, Y., Chen, S., Yang, F., Zhang, L., Alterovitz, G., Zhu, H., et al. (2017). HLA-B*51:01 is strongly associated with clindamycin-related cutaneous adverse drug reactions. Pharmacogenomics J. 17, 501–505. doi:10.1038/tpj.2016.61
Yao, S., Sucheston, L. E., Zhao, H., Barlow, W. E., Zirpoli, G., Liu, S., et al. (2014). Germline genetic variants in ABCB1, ABCC1 and ALDH1A1, and risk of hematological and gastrointestinal toxicities in a SWOG phase III trial S0221 for breast cancer. Pharmacogenomics J. 14, 241–247. doi:10.1038/tpj.2013.32
Yuliwulandari, R., Susilowati, R. W., Wicaksono, B. D., Viyati, K., Prayuni, K., Razari, I., et al. (2016). NAT2 variants are associated with drug-induced liver injury caused by anti-tuberculosis drugs in Indonesian patients with tuberculosis. J. Hum. Genet. 61, 533–537. doi:10.1038/jhg.2016.10
Yuliwulandari, R., Prayuni, K., Susilowati, R. W., As, M. S., Tokunaga, K., and Shin, J. G. (2019). NAT2 slow acetylator is associated with anti-tuberculosis drug-induced liver injury severity in indonesian population. Pharmacogenomics 20, 1303–1311. doi:10.2217/pgs-2019-0131
Zárate, R., González-Santigo, S., De La Haba, J., Bandres, E., Morales, R., Salgado, J., et al. (2007). GSTP1 and MTHFR polymorphisms are related with toxicity in breast cancer adjuvant anthracycline-based treatment. Curr. Drug Metab. 8, 481–486. doi:10.2174/138920007780866780
Keywords: antibiotics, hypersensitivity, severe cutaneous adverse drug reactions, liver injury, pharmacogenomics, precision medicine
Citation: Biswas M, Murad MA, Ashik MIH, Ershadian M and Sukasem C (2025) Pharmacogenomics of antibiotic-induced hypersensitivity reactions: current evidence and implications in clinical practice. Front. Pharmacol. 16:1651909. doi: 10.3389/fphar.2025.1651909
Received: 22 June 2025; Accepted: 08 September 2025;
Published: 01 October 2025.
Edited by:
Yaya Kassogue, Université des Sciences, des Techniques et des Technologies de Bamako, MaliReviewed by:
Nancy Hakooz, The University of Jordan, JordanMara Morelo Rocha Felix, Rio de Janeiro State Federal University, Brazil
Santenna Chenchula, All India Institute of Medical Sciences, Bhopal, India
Copyright © 2025 Biswas, Murad, Ashik, Ershadian and Sukasem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chonlaphat Sukasem, Y2hvbmxhcGhhdC5zdWtAbWFoaWRvbC5hYy50aA==
†ORCID: Chonlaphat Sukasem, orcid.org/0000-0003-0033-5321; Mohitosh Biswas, orcid.org/0000-0003-1432-7701; Murshadul Alam Murad, orcid.org/0009-0006-6113-0687
‡These authors have contributed equally to this work
 Mohitosh Biswas
Mohitosh Biswas Murshadul Alam Murad
Murshadul Alam Murad Md. Ismail Hossain Ashik
Md. Ismail Hossain Ashik Maliheh Ershadian
Maliheh Ershadian Chonlaphat Sukasem
Chonlaphat Sukasem