- 1Department of Genetics and Genomics, College of Medicine and Health Sciences, United Arab Emirates University, Al-Ain, United Arab Emirates
- 2Department of Biomedical Sciences, College of Health Sciences, Abu Dhabi University, Al-Ain, United Arab Emirates
Background and objectives: Angiotensin-converting enzyme inhibitors (ACEIs) are widely used to manage hypertension and cardiovascular diseases. However, dry cough is a common side effect, affecting 5%–35% of patients and often leading to discontinuation. This study aimed to investigate genetic variants involved in ACEI-induced cough and ACE plasma levels in UAE multi-ethnic hypertensive patients.
Method: The study cohort was pragmatically selected from the larger EmHeart Study (n = 900), a UAE-based pharmacogenomic initiative. Patients prescribed ACEIs were screened for inclusion. This multi-center, retrospective exploratory study involved genotyping 107 patients treated with ACEIs, including n = 35 in the cough group and n = 72 in the non-cough group. Variants of ACE; rs1799752 I/D, BDKRB2; rs1799722 (C>T), and KCNIP4; rs7675300 (C>A), rs1495509 (T>C), rs7661530 (T>C), and rs16870989 (T>A) were genotyped using standard technologies. A sandwich ELISA was done to investigate the ACE plasma levels in our cohort.
Results: We found that the ACE rs1799752 I/D genotype in the over-dominant model, was statistically significantly associated with ACEI-induced cough (p = 0.046) after adjusting for gender. Similarly, the T/T genotype of the KCNIP4 rs7661530 (T>C) variant was associated with significantly higher risk of cough compared to the combined C/C and T/C genotypes (p = 0.035). In contrast, the variants BDKRB2 rs1799722 (C>T), KCNIP4 rs7675300 (C>A), rs1495509 (T>C), and rs16870989 (T>A) were not significantly associated with ACEI-induced cough in our study. Moreover, ACE plasma levels were significantly lower in the cough group compared to the non-cough group (p = 0.0014). Stratified analysis by rs1799752 I/D genotypes revealed a significant difference within the I/D genotype (p = 0.0061), with higher levels in the non-cough group. No significant differences were found for the D/D or I/I genotypes.
Conclusion and limitations: Our data showed a significant association between ACEI-induced cough and the ACE rs1799752 I/D genotype, as well as lower ACE plasma levels in the cough group. This is the first study in the UAE and Middle East to report such findings and include all these variants in a single analysis. Although the sample size is small, our results contribute cumulative evidence on the genetic predisposition to ACEI-induced cough among hypertensive patients.
Introduction
Angiotensin-converting enzyme inhibitors (ACEIs) are considered the standard treatment for hypertension and have been widely used for many years. They exert their effects by inhibiting the angiotensin-converting enzyme (ACE), which prevents the conversion of angiotensin I to the vasoconstrictor angiotensin II. Moreover, ACEIs inhibit the release of the aldosterone hormone, leading to increased sodium and water excretion. These actions lead to a reduction in blood pressure (Altoum et al., 2023; Herman et al., 2025).
Despite their effectiveness, ACEIs are linked to side effects that may limit their use (Manoharan, 2023). The most frequently observed side effect is a dry cough, affecting between 5% and 35% of treated patients (Hallberg et al., 2017). The likelihood of experiencing an ACEI-induced cough is higher among females, individuals with respiratory conditions such as asthma, and the elderly (Ding et al., 2020; Hallberg et al., 2017). This cough is characterized by dryness and a tickling or scratching sensation in the throat. It may occur as early as after the first dose or gradually develop over weeks to months. Although the cough usually presents with mild to moderate severity, there are instances when it becomes severe enough to necessitate the discontinuation of the medication. While the cough may linger for up to 3 months after ceasing ACEIs, it typically resolves within one to 4 weeks (Dicpinigaitis, 2006).
The exact mechanism behind the cough induced by ACEIs remains elusive. However, it is suspected that several mediators, including the vasodilator bradykinin (BK), play a role in triggering this type of cough. BK is a vasodilator that reduces blood pressure by binding to the Bradykinin Receptor B2 (BDKRB2). Moreover, BK can cause bronchoconstriction by prompting the production of other mediators, such as prostaglandins, histamine, and leukotrienes. Subsequently, BK is rapidly degraded into its inactive metabolites, primarily by ACE. Accordingly, when ACEIs are administered, they inhibit BK’s metabolism by blocking the ACE’s action. This inhibition leads to an accumulation of BK in respiratory tracts and binds to the BDKRB2 resulting in bronchospasm and cough (Altoum et al., 2023; Pinto et al., 2020).
Given that the occurrence of cough due to ACEIs is unpredictable, it is reasonable to suggest that ACEI-induced cough is influenced by one or more genetic factors (Borghi et al., 2023). The ACE;rs1799752 (Insertion/Deletion), BDKRB2;rs1799722 (C>T), and variants located on Potassium Voltage-Gated Channel Interacting Protein 4 (KCNIP4); rs145489027 (G>A), rs7675300 (C>A), rs1495509 (T>C), rs7661530 (T>C), rs16870989 (T>A), and rs6838116 (A>C) variants have been identified as key genetic contributors involved in the intricate genetic basis of ACEI-induced cough in some populations (Mosley et al., 2016; Mu et al., 2019; Mukae et al., 2000). The ACE; rs1799752 variant located within intron 16 of the ACE is characterized by the Insertion (I) and/or Deletion (D) of 287 base pairs “Alu repeat” and is known to affect the ACE serum levels. The (D/D) carriers have higher ACE serum levels in contrast to those with heterozygous (I/D) and homozygous insertion (I/I) carriers, with intermediate and low ACE serum levels, respectively. The latter two groups are known to accumulate BK, which ACE normally degrades. Consequently, patients in these groups are at a higher risk of experiencing cough from ACEIs compared to those with the (D/D) genotype (Abbas et al., 2012; Mu et al., 2019).
Moreover, a study has shown a significant correlation between the rs1799722 (C>T) variant located in the BDKRB2 promoter region and ACEI-induced cough. BDKRB2: rs1799722 T/T genotype carriers were found to be more prone to developing cough than C/C genotype (Mukae et al., 2000). Finally, a genome-wide association study (GWAS) identified KCNIP4 single nucleotide polymorphisms (SNPs) within intron 4 to be associated with ACEI-induced cough (Mosley et al., 2016). Consequently, studies have documented the correlation between these genetic variants and ACEI-induced cough in various populations.
Despite the fact that ACEI-induced cough is not considered a severe side effect, in some cases, it results in the discontinuation of ACEIs (Borghi et al., 2023). It is well-known that the leading risk factor for heart failure, coronary heart disease, stroke, and end-stage renal disease is hypertension. Consequently, discontinuing hypertension treatment might seriously affect the patients (Rohman et al., 2018). Accordingly, we sought to investigate the genetic factors that might predict the occurrence of ACEI-induced cough. Although pharmacogenomics in cardiovascular disease research has been conducted in the United Arab Emirates (UAE) (Al-Mahayri et al., 2022; Alqasrawi et al., 2024; 2025; Khasawneh et al., 2024), studies on the pharmacogenomics of ACEI-induced cough have never been reported in the UAE or any other Middle Eastern populations.
Our current study aimed to assess the association between various genetic variants suggested by previous research in other populations with ACEI-induced cough among adult hypertensive patients in the UAE. The selected variants included ACE; rs1799752 I/D, BDKRB2; rs1799722 (C>T), and KCNIP4; rs7675300 (C>A), rs1495509 (T>C), rs7661530 (T>C), and rs16870989 (T>A). Additionally, we explored the relationship between ACE plasma levels and cough induction, correlating these findings with the genotypes present in the ACE I/D variant.
In addition, we aimed to examine the distribution of these variants across different ethnic groups, assessed linkage disequilibrium (LD) patterns, constructed haplotypes to explore combined genetic effects, and performed variant interaction analysis to evaluate potential gene-gene interactions. To our knowledge, this is the first study in the UAE and the Middle East to report such findings and the first to include all these variants in a single analysis.
Methods
Study design and patients
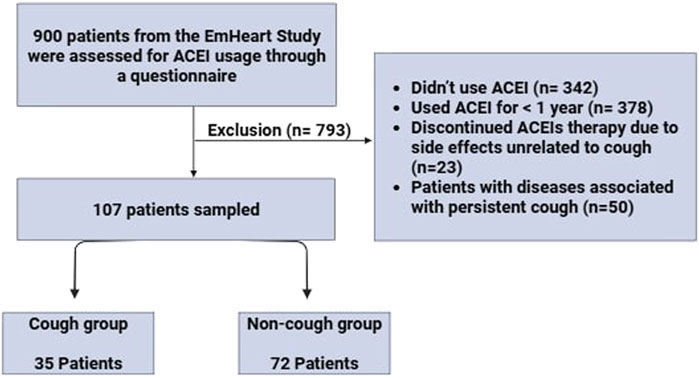
This is a retrospective multicenter study conducted at four recruitment sites, Tawam Hospital, The Heart Medical Center, and Mediclinic Al-Ain Hospital in Al-Ain, UAE, and Burjeel Day Surgery Center in Abu Dhabi, UAE, from October 2022 to October 2023. The study was conducted in accordance with the Declaration of Helsinki and approved by the Department of Health-Abu Dhabi for the Data from the UAEU (DOH/CVDC/2020/1187), (DOH/CVDC/2021/1519), (DOH/CVDC/2022/1458), (DOH/CVDC/2023/1952) (MCME.CR.213.MAIN.2021), and (SNA/FA/2020-14). Our sample was drawn from the EmHeart Study, the first pharmacogenomic implementation initiative conducted in the UAE, which focuses on integrating pharmacogenomic data into cardiovascular drug prescribing to improve safety and efficacy. The original cohort included 900 patients. We screened the entire cohort to identify those who had been prescribed ACEIs. The inclusion criteria included adults aged 18 years and above with hypertension who are currently taking ACEIs for at least 1 year or had used ACEIs before and discontinued due to ACEI-induced cough. The Exclusion criteria included (1) Those who didn’t receive ACEIs, (2) Those who had discontinued ACEI therapy due to side effects unrelated to cough, such as headache, dizziness, or angioedema, (3) Patients who were currently using ACEIs but for less than 1-year, (4) Patients with diseases associated with chronic cough including severe asthma, COPD, and GERD, (5) patients with any secondary cause of hypertension, and (6) patients who disagree to give blood for the study. A group of 107 hypertensive adult patients signed an informed consent form to participate in this study. Two distinct groups of patients were recruited: a non-cough group comprised of 72 patients who received any type of ACEIs for at least 1 year without experiencing ACEI-induced cough. The second group, the cough group, comprised 35 patients who had used ACEIs and developed ACEI-induced cough.
Data collection
Data were collected from the patient’s medical records, including demographic (age, gender, ethnicity), health behaviours (smoking status), and concomitant medications. To gather detailed information on ACEI usage and associated cough, we first conducted a thorough review of each patient’s electronic medical records (EMR) for physician documented evidence of ACEI-induced cough, including its onset timing and severity. Following this, we directly contacted patients to confirm ACEI use and further clarify the occurrence and characteristics of the cough. A structured questionnaire was developed to support this process. It initially confirmed whether the patient had received any ACEIs and then assessed the presence of ACEI-induced cough. The questionnaire included targeted questions on the reason for ACEI discontinuation, time of cough onset relative to ACEI initiation, frequency and severity of coughing episodes, including daytime vs nighttime patterns, as well as the specific ACEI type used. Figure 1 represents the flow of participant recruitment and the selection process within the study.
Blood sampling and DNA extraction
Blood samples were obtained from the peripheral vein and were collected in 3 mL vacuum tubes containing EDTA (BD Inc.). Genomic DNA was extracted from whole blood using QIAamp® DNA kit (Qiagen, Germany) according to the manufacturer’s protocol. The quality and quantity of DNA were determined using a Nanodrop One Spectrophotometer (Thermo Fisher Scientific, United States). Plasma samples were prepared by centrifuging 500 mL of whole blood at 2,500 rpm for 10 min at room temperature, and the resultant plasma was then stored at −80 °C for future use.
Amplification of the ACE, BDKRB2, and KCNIP4 variants
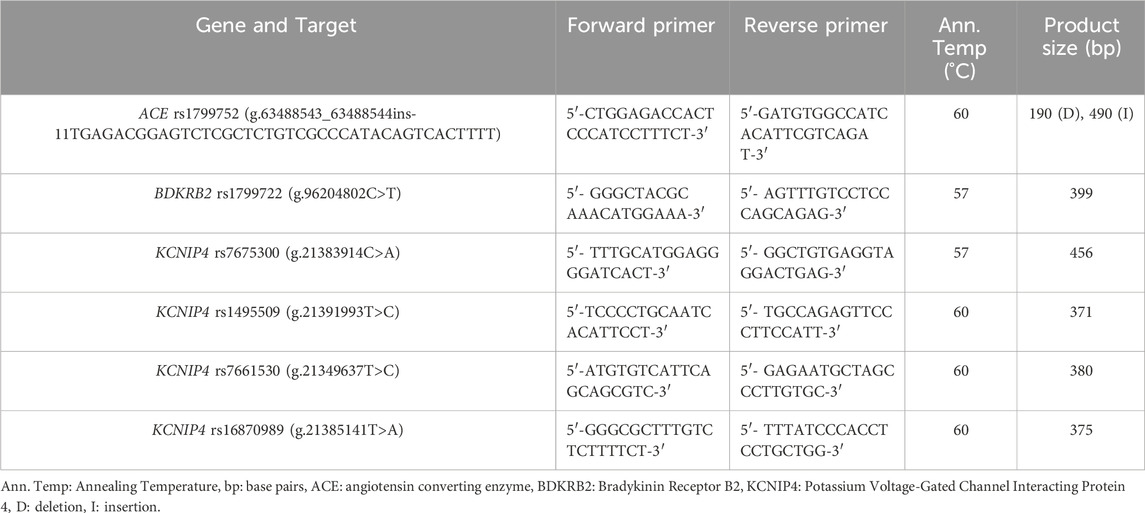
Primers for amplifying specific gene regions were designed using the Primer3 website (https://primer3.ut.ee/), and the primer sequences are provided in Table 1. The PCR reaction was carried out in a total volume of 25 μL, consisting of combining 13.5 µL of DNAse free water, 1x of 10x PCR Buffer, 1x of 5x Q-Solution, 200 µM of dNTPs, 1.5 units of Taq DNA Polymerase, 100–200 pmol of the forward primer, 100–200 pmol of the reverse primer and 296–350 ng of DNA. Amplification was performed using a SimpliAmp thermal cycler (ThermoFisher Scientific, Waltham, Massachusetts, U.S.A.) over 32 cycles, each cycle included pre-denaturation at 95 °C for 5 minutes, denaturation at 95 °C for 45 s, annealing at either 57 °C or 60 °C for 45 s, extension at 72 °C for 45 s, and a final extension at 72 °C for 7 min. PCR products were separated using electrophoresis on a 1.5% agarose gel stained with ethidium bromide and visualized under a UV illuminator.
Identification of genetic variants
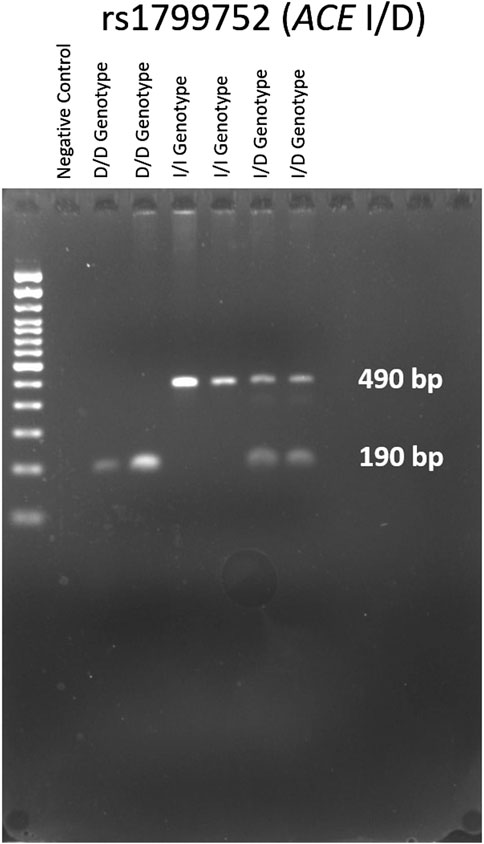
The genotypes of the ACE; rs1799752 I/D variant were determined through gel electrophoresis of the PCR products, as illustrated in Figure 2. The electrophoresis resulted in a 190 bp product corresponding to the D allele and a 490 bp product corresponding to the I allele. In heterozygote samples, both 490 and 190 bp bands were observed. Genotypes for BDKRB2; rs1799722 (C>T), and KCNIP4; rs7675300 (C>A), rs1495509 (T>C), rs7661530 (T>C), and rs16870989 (T>A) were identified by sanger sequencing using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Waltham, Massachusetts, United States). Sequencing analysis was conducted with the 3130xl Genetic Analyzer (Applied Biosystems, Waltham, Massachusetts, United States). The results were processed using Chromas software (Technelysium, Australia).
Measurement of ACE plasma levels
Sandwich Enzyme-Linked Immunosorbent Assay (ELISA) was employed to measure ACE protein levels in plasma samples from various patients using the Human ACE ELISA Kit (ab263889) (Abcam, Cambridge, United Kingdom) strictly following the manufacturer’s provided instructions. To ensure the accuracy of the ELISA results and minimize potential bias, the assay was performed on plasma samples collected exclusively from patients who were not currently taking ACEIs or had not taken the medication within 5 days prior to blood sample collection. This precaution was taken to avoid any possible interference from the medication on the assay outcomes. Additionally, to enhance the reliability and precision of the results, each plasma sample was tested in quadruplicate, with four separate wells assigned to each sample.
Variants distribution within ethnicities
To explore potential ethnicity-specific patterns, we performed a stratified analysis of genotype frequencies across the main ethnic groups represented in our cohort. Ethnicity information was initially collected from medical records and then confirmed directly with patients to ensure accuracy.
Linkage disequilibrium (LD)
To investigate the non-random association between alleles at different loci, we conducted LD analysis among the KCNIP4 rs7675300 (C>A), rs1495509 (T>C), rs7661530 (T>C), and rs16870989 (T>A) variants.
Haplotype analysis
Haplotype analysis was performed to investigate the combined effect of the KCNIP4 variants (rs7675300, rs1495509, rs7661530, and rs16870989) on ACEI-induced cough.
Variant interaction analysis
To evaluate potential synergistic effects among genetic variants we conducted variant interaction analysis involving ACE rs1799752 (I/D), BDKRB2 rs1799722 (C>T), and KCNIP4 rs7675300 (C>A).
Statistical analysis
Statistical analyses were conducted using SNPStats and SPSS version 29.0.2.0 (SPSS Inc., US) (Solé et al., 2006). ELISA results were processed with GraphPad Prism version 10.2.2 (GraphPad Software, Boston, Massachusetts, United States). Categorical variables and Hardy-Weinberg equilibrium (HWE) were analyzed using the Chi-square (χ2) test. Numerical data were presented as means with standard deviations. Odds ratios with 95% confidence intervals [OR; 95% CI] were computed to determine associations, considering p < 0.05 statistically significant. For ELISA results, the Mann-Whitney test was used to compare total ACE plasma levels between the two study groups and within the study groups for each ACE-rs1799752 I/D genotype, and the Kruskal–Wallis test was applied to analyze differences in ACE plasma levels across the three ACE I/D genotypes, with p < 0.05 considered statistically significant.
Chi-square tests were performed to assess the significance of variants distribution across different ethnic groups, with p < 0.05 considered statistically significant. Further, adjusted residuals greater than +2 or less than −2 was used to identify specific genotype-ethnicity associations. LD among KCNIP4 variants was measured by calculating D′ values using the SHEsisPlus platform (Shen et al., 2016). Haplotype construction was carried out using the SNPStats software. Logistic regression analysis was performed to calculate the ORs and 95% CIs for each haplotype compared to the reference. Finally, variant interaction analysis was conducted to examine the collective impact of all variants on the risk of ACEI-induced cough.
Results
Patient’s baseline demographics and clinical characteristics
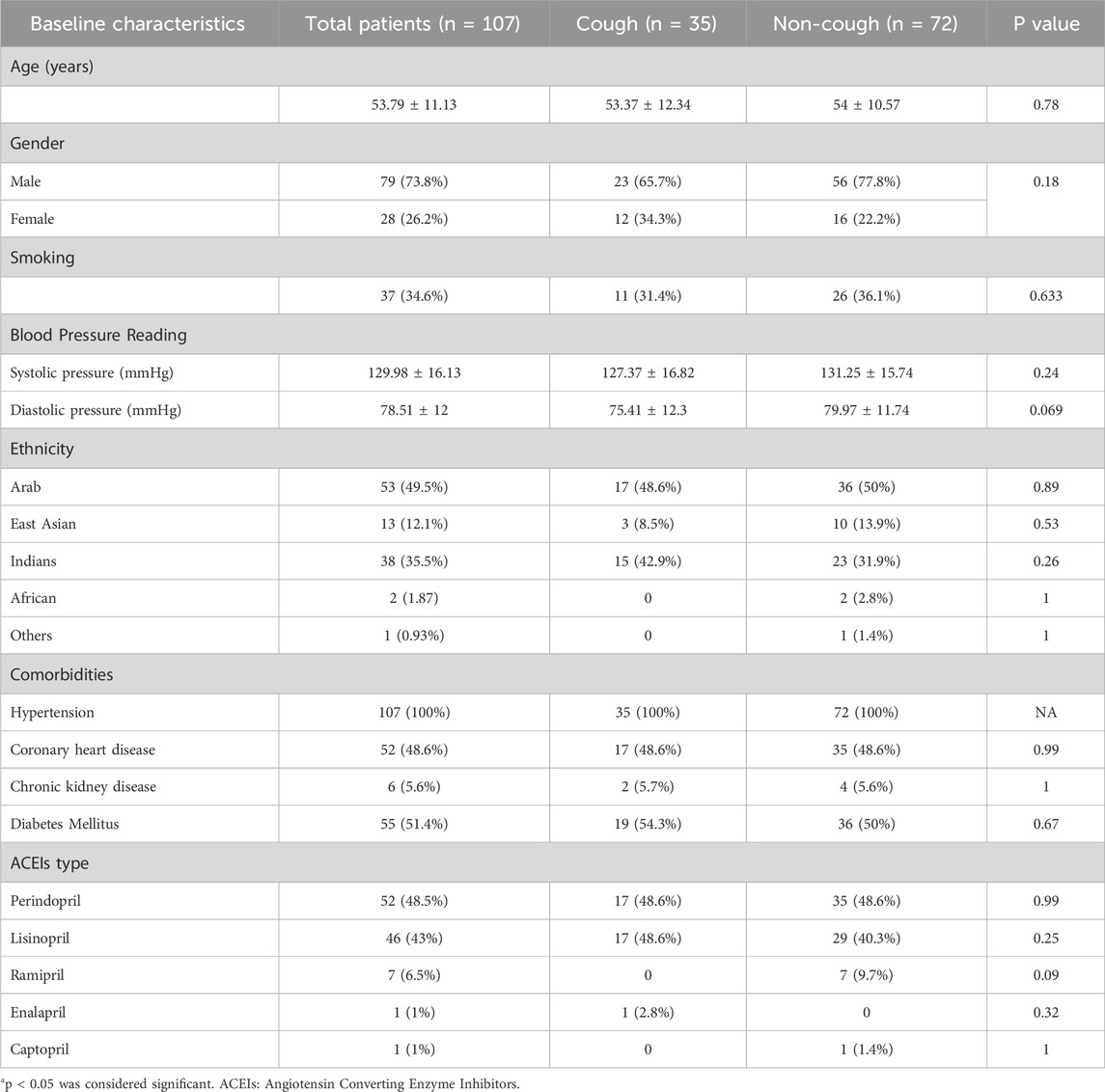
107 adult hypertensive patients were identified from the EmHeart study as ACEIs users and recruited for this study. The incidence of cough in our study was 32.7% (35 patients). The baseline characteristics and collected data, including demographics, comorbidities, and ACEIs type, are presented in Table 2. 73.8% of the participants were males, with no statistically significant difference in gender distribution between the cough and no-cough groups (P = 0.18). The average age across all patients was 53.79 (±11.13) years. Moreover, the participants represented a diverse ethnic background categorized into five distinct groups (Arabs, East Asians, Indians, Africans, and others). Coronary heart disease affected 48.6% of patients equally in both groups. Diabetes mellitus was slightly more common in the cough group (54.3%) than in the non-cough group (50%). Chronic kidney disease was the least prevalent, occurring in 5.6% of patients, with similar distribution in both groups.
Regarding ACEIs usage among patients, perindopril was the most commonly prescribed ACEIs to 48.5% of the total population, with almost identical usage in both the cough (48.6%) and non-cough groups (48.6%). Lisinopril was the second most frequently used ACEIs, accounting for 43% of total usage, with a slightly higher proportion in the cough group (48.6%) than the non-cough group (40.3%). Ramipril was used by 6.5% of patients, all of whom were in the non-cough group. The least commonly prescribed ACEIs were Enalapril and Captopril, each used by only 1% of the participants, with Enalapril used exclusively in the cough group and Captopril in the non-cough group. Overall, there was no statistically significant difference in the distribution of ACEIs type among the two groups.
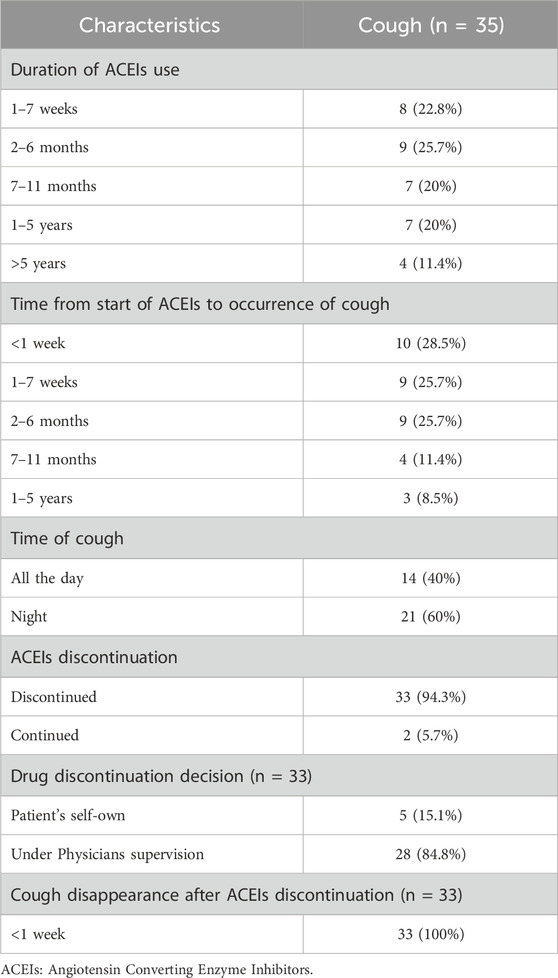
For patients in the cough group, information regarding the duration of ACEIs usage, onset of cough, and drug discontinuation is provided in Table 3. The duration of ACEIs usage varied, with the most common being 2–6 months and 1–7 weeks, each by 25.7% and 22.8% of the patients, respectively. A smaller percentage used ACEIs for longer durations. The onset of cough after starting ACEIs varied, with the highest percentages reported within the first week, 28.5%, and from 1–7 weeks to 2–6 months, each 25.7%. The cough occurred mostly at night in 60% of the patients. A total of 94.3% discontinued ACEIs due to the tolerable nature of the cough, and in all cases (100%), the cough resolved within less than 1 week after discontinuation.
Association of ACE, BDKRB2, and KCNIP4 alleles and genotypes with ACEI-induced cough
The genotype frequencies of ACE-rs1799752 I/D, BDKRB2-rs1799722 (C>T), and KCNIP4- rs7675300 (C>A), rs1495509 (T>C), rs7661530(T>C), and rs16870989 (C>A) variants were consistent with HWE in both cough and non-cough groups (p > 0.05). Detailed results are presented in Supplementary Table S1. Tables 4–9 present the crude analysis, as well as the analysis after adjusting for the confounding factor (gender) for the variants. The analysis was conducted using various genetic models, including codominant, dominant, recessive, and over-dominant models.
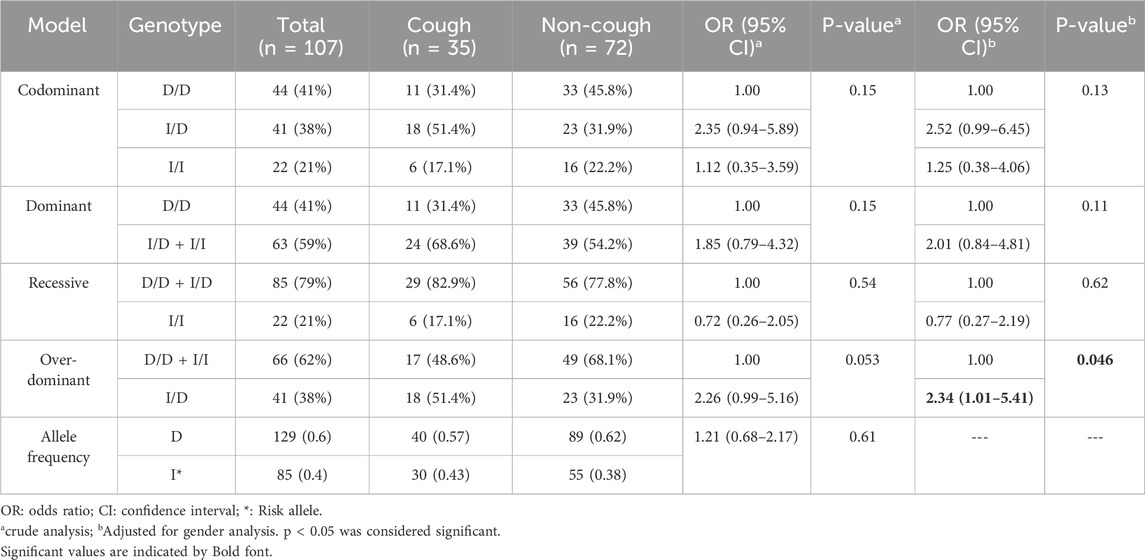
ACE rs1799752 (I / D) variant
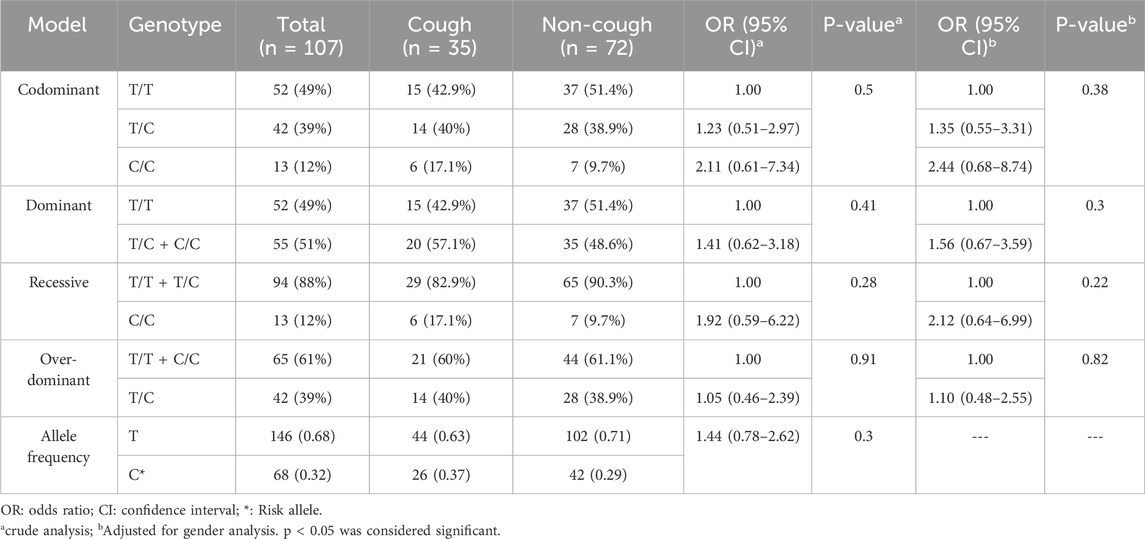
The genotypic and allelic frequencies of the ACE I/D variant are presented in Table 4. Among this variant, 31.4% were D/D, 51.4% were I/D, and 17.2% were I/I in the cough group. Among the non-cough, 46% were D/D, 32% were I/D, and 22% were I/I. The frequency of the I allele was 0.43 in the cough group and 0.38 in the non-cough group; however, there was no statistically significant association between the I/I genotype (p = 0.15) or the I allele (p = 0.61) and ACEI-induced cough. Nevertheless, after adjusting for gender, the over-dominant model showed the most statistically significant association with ACEI-induced cough, where the heterozygous I/D genotype is linked to a higher risk of ACEI-induced cough compared to the combined D/D and I/I genotypes, with an OR of 2.34 and p = 0.046.
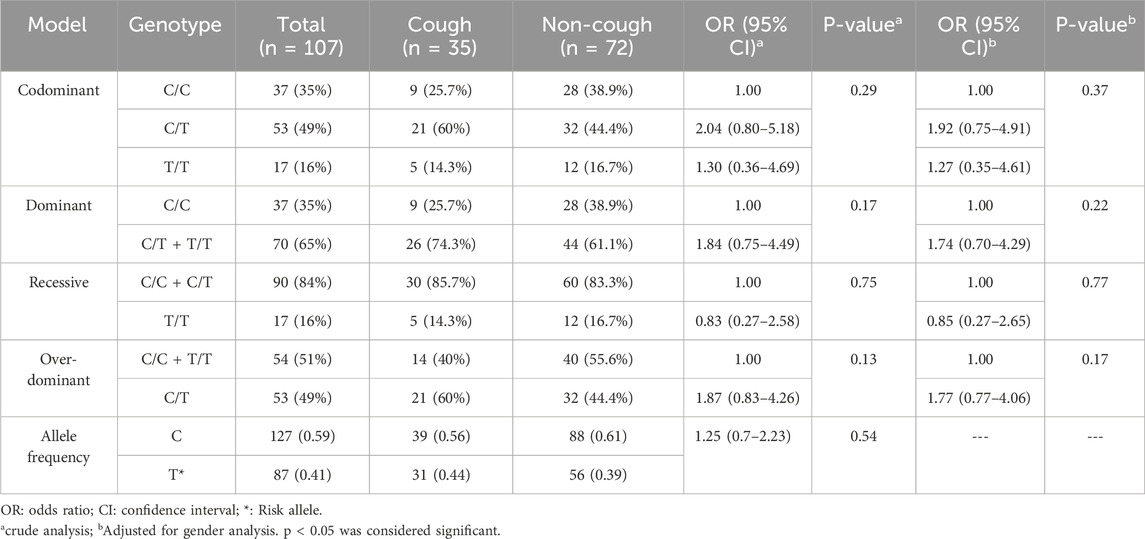
BDKRB2 rs1799752 (C>T) variant
The genotypic and allelic frequencies of the BDKRB2 rs1799752 (C>T) are presented in Table 5. Among this variant, 26% were C/C, 60% were C/T, and 14% were T/T in the cough group. Among the non-coughers, 39% were C/C, 44% were C/T, and 17% were T/T. The frequency of the T allele was 0.44 in the cough group and 0.39 in the non-cough group; however, there was no statistically significant association between the T allele (p = 0.54) or T/T genotype and ACEI-induced cough either in the crude analysis (p = 0.29) or after adjusting for gender (p = 0.37).
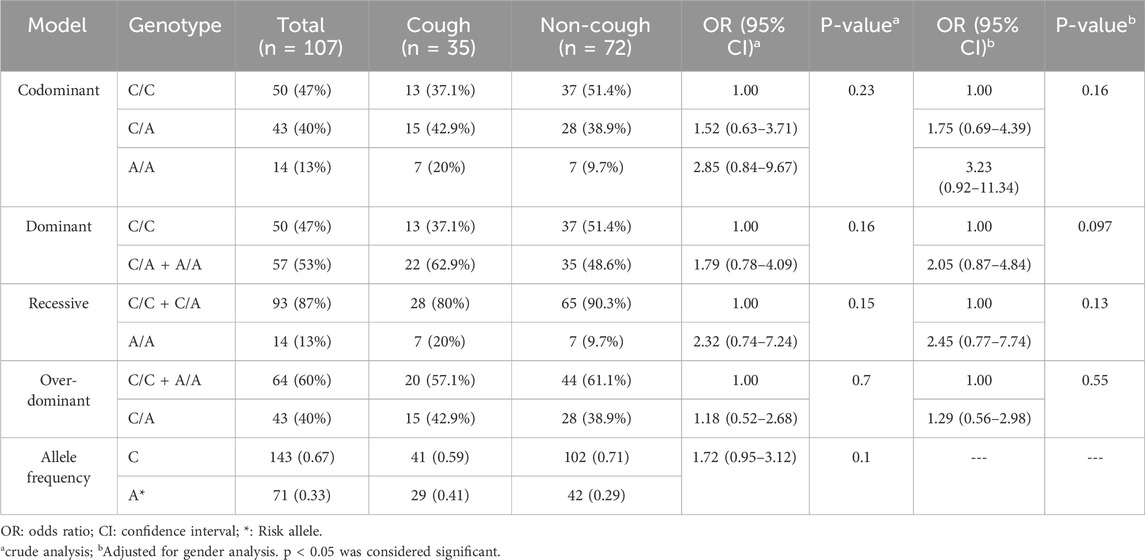
KCNIP4 rs7675300 (C>A) variant
The genotypic and allelic frequencies of the KCNIP4 rs7675300 (C>A) are presented in Table 6. Among this variant, 37% were C/C, 43% were C/A, and 20% were A/A in the cough group. Among the non-coughers, 51% were C/C, 39% were C/A, and 10% were A/A. The frequency of the A allele was 0.41 in the cough group and 0.29 in the non-cough group; however, there was no statistically significant association between the A allele (p = 0.1) or A/A genotype and ACEI-induced cough either in the crude analysis (p = 0.23) or after adjusting for gender (p = 0.16).
KCNIP4 rs1495509 (T>C) variant
The genotypic and allelic frequencies of the KCNIP4 rs1495509 (T>C) are presented in Table 7. Among this variant, 43% were T/T, 40% were T/C, and 17% were C/C in the cough group. Among the non-cough, 51% were T/T, 39% were T/C, and 10% were C/C. The frequency of the C allele was 0.37 in the cough group and 0.29 in the non-cough group; however, there was no statistically significant association between the C allele (p = 0.3) or C/C genotype and ACEI-induced cough either in the crude analysis (p = 0.5) or after adjusting for gender (p = 0.38).
KCNIP4 rs7661530 (T>C) variant
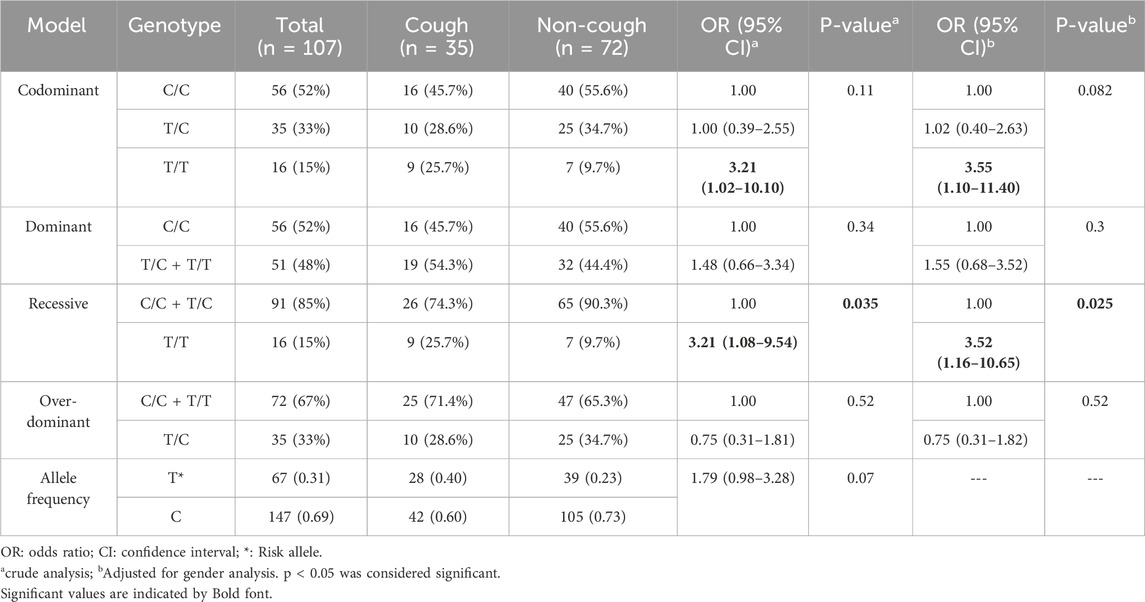
The genotypic and allelic frequencies of the KCNIP4 rs7661530 (T>C) are presented in Table 8. Among this variant, 26% were T/T, 29% were T/C, and 45% were C/C in the cough group. Among the non-cough, 10% were T/T, 35% were T/C, and 55% were C/C. The frequency of the T allele was 0.40 in the cough group and 0.23 in the non-cough group. Although a higher frequency of the T allele was observed in the cough group compared to the non-cough group, indicating an increased risk of ACEI-induced cough for the T/T genotype (OR = 3.21), this result did not reach statistical significance in the codominant model (p = 0.11). However, the recessive model shows a statistically significant association between the T/T genotype and ACEI-induced cough in both the crude and adjusted analyses. In the crude analysis, patients with the T/T genotype have a significantly increased risk of ACEI-induced cough compared to those with the combined C/C and T/C genotypes, with an OR of 3.21 and a p = 0.035. Similarly, in the adjusted-to-gender analysis, the T/T genotype is associated with a significantly increased risk of cough, with an OR of 3.52 and a p = 0.025.
KCNIP4 rs16870989 (T>A) variant
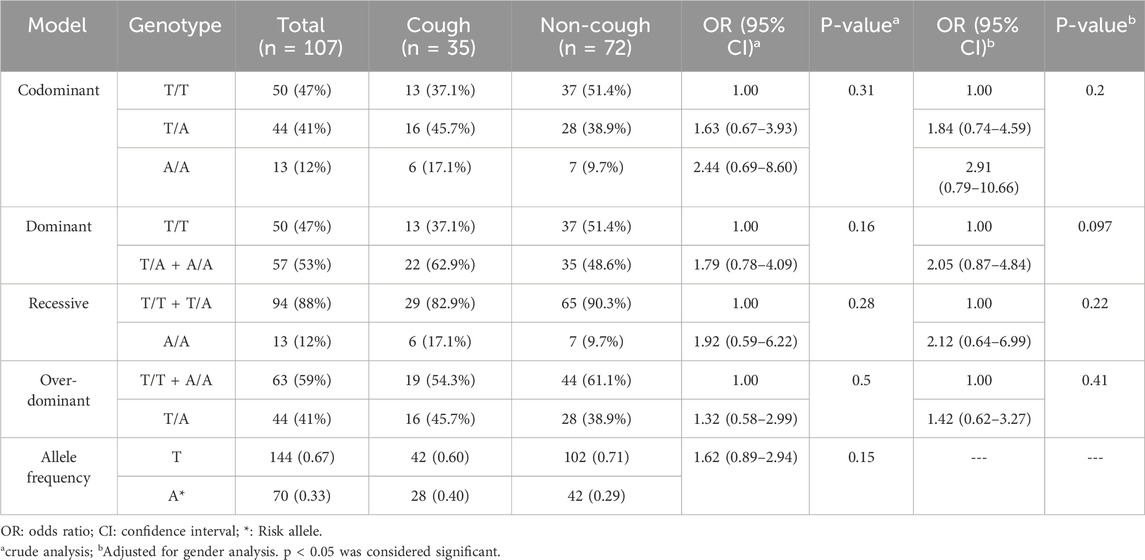
The genotypic and allelic frequencies of the KCNIP4 rs1495509 (T>C) are presented in Table 9. Among this variant, 37% were T/T, 46% were T/A and 17% were A/A in the cough group. Among the non-cough, 51% were T/T, 39% were T/A and 10% were A/A. The frequency of the A allele was 0.40 in the cough group and 0.29 in the non-cough group; however, there was no statistically significant association between the A allele (p = 0.15) or the A/A genotype and ACEI-induced cough either in the crude analysis (p = 0.31) or after adjusting for gender (p = 0.20).
ACE plasma levels
A total of 28 plasma samples were analysed using an ELISA kit, with 14 samples from each group (cough and non-cough), focusing on the distribution across the three rs1799752 ACE I/D genotypes. The median ACE plasma levels were significantly lower in the cough group compared to the non-cough group (423 ng/mL and 595.8 ng/mL, respectively; p = 0.0014, Figure 3A), suggesting a potential association between decreased ACE levels and the occurrence of ACEI-induced cough in the cough group. Furthermore, when stratified by the rs1799752 I/D genotypes, the median plasma levels for the D/D, I/D, and I/I genotypes were 499.9 ng/mL, 510.4 ng/mL, and 574.8 ng/mL, respectively. While there was a trend of higher levels in individuals with the I/I genotype compared to the D/D and I/D genotypes, the difference between the three genotypes was not statistically significant (p = 0.43) as shown in Figure 3B. Finally, comparing the plasma levels for each rs1799752 I/D genotype between the cough and non-cough groups, a statistically significant difference was found in the I/D genotype (p = 0.0061) where the cough group exhibited lower plasma levels of 437.8 ng/mL compared to the non-cough group 593.6 ng/ml as shown in Figure 4B. In contrast, no significant differences were observed for the D/D and I/I genotypes. For the D/D genotype in Figure 4A, the plasma levels were 408.1 ng/mL in the cough group and 569.8 ng/mL in the non-cough group, with p = 0.54. Similarly, for the I/I genotype in Figure 4C, the plasma levels were 455.4 ng/mL in the cough group and 616.8 ng/mL in the non-cough group, with a p = 0.19.
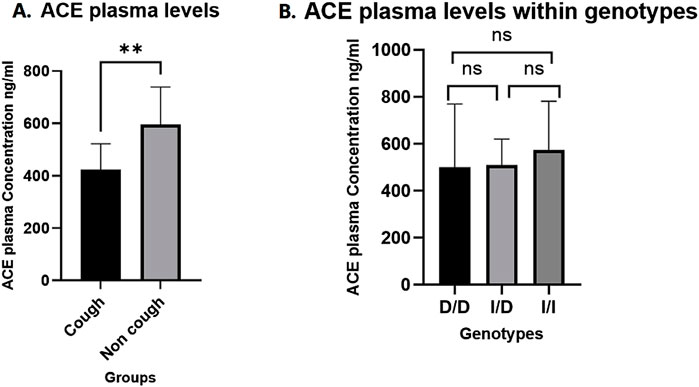
Figure 3. (A) Comparison of ACE plasma levels Between cough and non-cough groups. (B) Comparison ACE plasma levels across ACE I/D genotypes (D/D, I/D, and I/I genotypes). ** Indicates statistically significant difference (p < 0.05), ns: not significant.
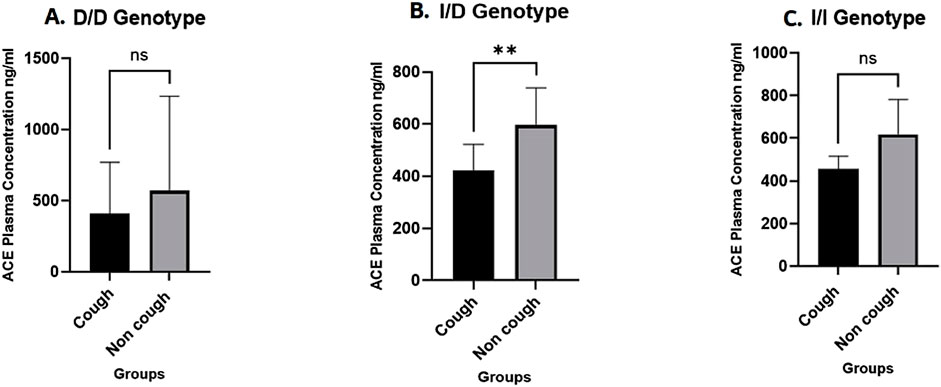
Figure 4. Comparison of ACE plasma levels by ACE I/D genotypes between the Cough and Non-cough Groups. (A) Comparison of ACE plasma levels within D/D genotype. (B) Comparison of ACE plasma levels within I/D genotype. (C) Comparison of ACE plasma levels within I/I genotype. ** Indicates statistically significant difference (p < 0.05), ns: not significant.
Variants distribution within ethnicities
A total of 107 patients were included in the analysis representing diverse ethnic backgrounds. The majority were Arab (n = 53), followed by Indian (n = 38), East Asian (n = 13), African (n = 2), and other ethnicities (n = 1). The stratified analysis of genotype distributions across these groups was conducted, and the detailed results are presented in Supplementary Tables S2–S7.
For the ACE rs1799752 (I/D) variant (Supplementary Table S2), a p < 0.01 indicated a statistically significant difference in genotype distribution among ethnic groups, with the Arab group showing a notably higher-than-expected frequency of the D/D genotype (adjusted residual = + 4.4) while the Indian group showed a significantly lower-than-expected frequency of the same genotype (adjusted residual = - 4.4). In contrast, for the BDKRB2 rs1799722 (C>T) variant (Supplementary Table S3), the p = 0.15 indicated no statistically significant difference in distribution across ethnicities.
For the KCNIP4 rs7675300 (C>A) variant (Supplementary Table S4), a p = 0.04 indicated a statistically significant difference in genotype distribution across ethnic groups. The Arab group showed a higher-than-expected frequency of the C/C genotype (adjusted residual = + 3.2), while the Indian group showed a lower-than-expected frequency of the same genotype (adjusted residual = - 2.7). In contrast, the East Asian group had a higher-than-expected frequency of the A/A genotype (adjusted residual = + 3). Similarly, for KCNIP4 rs1495509 (T>C) variant (Supplementary Table S5), a p = 0.04 indicated a statistically significant difference in genotype distribution across ethnic groups. The Arab group had a higher-than-expected frequency of the T/T genotype (adjusted residual = + 2.8), while the Indian group showed a higher-than-expected frequency of the T/C genotype (adjusted residual = + 2.5). For KCNIP4 rs7661530 (T>C) variant (Supplementary Table S6), the p = 0.23 indicated no statistically significant difference in genotype distribution across ethnicities. Finally, for the KCNIP4 rs16870989 (T>A) variant (Supplementary Table S7), a p = 0.04 indicated a statistically significant difference, with the Arab group showing a higher-than-expected frequency of the T/T genotype (adjusted residual = + 3.2), and the Indian group displaying a higher-than-expected frequency of the T/A genotype (adjusted residual = + 2.2).
Linkage disequilibrium analysis
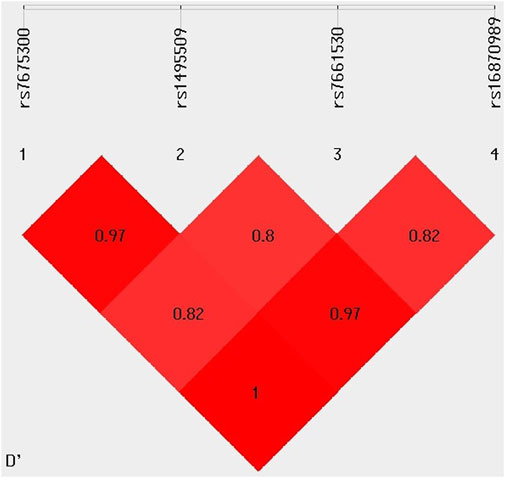
The LD analysis of four KCNIP4 variants (rs7675300, rs1495509, rs7661530, and rs16870989) revealed strong non-random associations between them. The D′ values indicated a high degree of co-inheritance, with rs7675300 and rs16870989 showing complete LD (D' = 1), and rs7675300 and rs1495509 demonstrating very strong LD (D' = 0.97). Similar strong linkage was observed between rs1495509 and rs16870989, suggesting that these variants are frequently inherited together within the study population. These findings are presented in Figure 5.
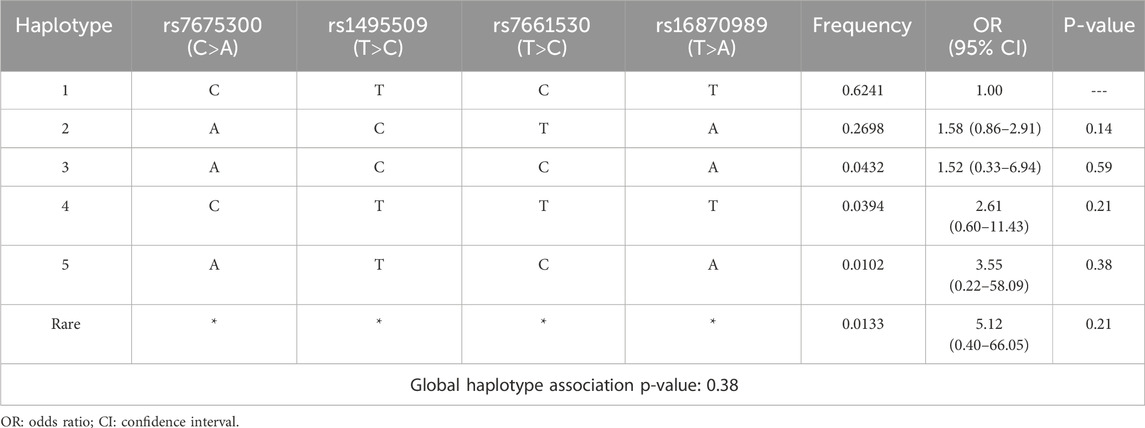
Haplotype association with ACEI-induced cough
Haplotypes were constructed based on the LD analysis. Haplotype 1 (CTCT), with a frequency of 0.6241, was the most prevalent in the study population and served as the reference. Although some haplotypes exhibited high ORs indicative of potential increased risk, none reached statistical significance. Additionally, the global haplotype association test yielded a p = 0.38, indicating no significant overall association between the haplotypes and ACEI-induced cough. The detailed results of this analysis are presented in Table 10.
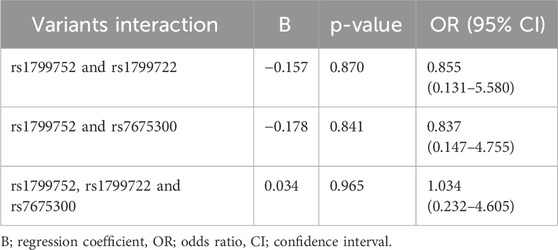
Variant interaction analysis
Due to high LD among the four KCNIP4 variants, rs7675300 (C>A) was selected as a representative. The results, summarized in Table 11, show that the interaction between rs1799752 (I/D) and rs1799722 (C>T) had a p = 0.870, the interaction between rs1799752 (I/D) and rs7675300 (C>A) had a p = 0.841, and the three-way interaction among rs1799752 (I/D), rs1799722 (C>T), and rs7675300 (C>A) had a p = 0.965. These findings indicate no statistically significant synergistic effect of these variants on the risk of ACEI-induced cough.
Discussion
The current study provides valuable insights into the genetic determinants influencing ACEI-induced cough within a multi-ethnic population. We validated the suggested variants and examined their effect on patients complaining of this side effect. We further evaluated the effect of one common variant in the ACE gene on ACE plasma levels.
ACEIs are commonly prescribed for treating hypertension and other cardiovascular diseases, but a persistent dry cough is a clinically important side effect affecting up to 35% of patients. This side effect is dose-independent, unpredictable, and represents a unique reaction in patients who are predisposed, and it can occur at any time from 1 week up to a year after starting the medication. Although the cough is typically mild to moderate in severity, there are cases where it becomes severe enough to require discontinuation of the treatment and switching to alternatives, such as angiotensin receptor blockers (ARBs) (Borghi et al., 2023; Vegter and De Jong-van den Berg, 2010; Zamora and Parodi, 2011).
Diagnosing ACEI-induced cough is difficult due to the absence of physical biomarkers, and its exact mechanism remains unclear. However, it is thought that BK and substance P play vital roles. ACE normally metabolizes BK, and ACEIs lead to BK accumulation in the respiratory tract, which binds to BDKRB2 receptors, causing histamine release, resulting in bronchospasm and cough. The reason why only some ACEIs users experience cough remains to be determined. However, research suggests that genetic predispositions, including ACE I/D, BDKRB2, and KCNIP4 variants, may contribute. This suggests that multiple mechanisms may be involved rather than a single factor (Ancion et al., 2019; Mosley et al., 2016; Yılmaz, 2019; Zamora and Parodi, 2011). The relationship between these gene variants and susceptibility to ACEI-induced cough has been studied in limited studies.
A meta-analysis of the ACE rs1799752 I/D variant found that individuals with the I/I genotype had a significant association with ACE inhibitor-induced cough, particularly in East Asian populations (Nishio et al., 2011). In contrast, other studies reported no link between rs1799752 and ACEI-induced cough in African and Caucasian populations (Moholisa et al., 2013; Rohman et al., 2018). Additionally, regarding the BDKRB2 rs1799722 (C>T) SNP, Mukae and coworkers investigated the relationship between the T/T genotype and ACEI-induced cough, particularly in Japanese females (Mukae et al., 2000). Their later study further supported a strong association between the T/T genotype and the T allele with ACEI-induced cough, suggesting a genetic predisposition to this side effect in the Japanese population (Mukae et al., 2002). However, there was no association between rs1799722 (C>T) and ACEI-induced cough in Korean and South African populations (Moholisa et al., 2013; Woo et al., 2009). Furthermore, a Genome-Wide Association Study (GWAS) identified a connection between the rs7675300 (C>A), rs1495509 (T>C), rs7661530 (T>C), and rs16870989 (T>A) SNPs on the KCNIP4 gene and ACEI-induced cough (Mosley et al., 2016). In contrast, a separate GWAS on a Swedish population found no association between these SNPs and ACEI-induced cough (Hallberg et al., 2017).
Considering the contradictory findings across various studies regarding the association between these variants and ACEI-induced cough, and recognizing the lack of research on this relationship in Middle Eastern countries and populations, we aimed to investigate the relationship between these variants and ACEI-induced cough within the UAE population. We also investigated the difference in ACE plasma levels between the two study groups and the effect of ACE rs1799752 I/D genotypes on ACE plasma levels in this population. Notably, this study is the first to include all these variants in a single analysis. In the present study, we identified that four variants, rs1799722 (C>T) on BDKRB2, rs7675300 (C>A), rs1495509 (T>C), and rs16870989 (T>A) on KCNIP4, showed no statistically significant association with ACEI-induced cough. However, noteworthy exceptions emerged: the I/D genotype of rs1799752 (I/D) on ACE was significantly associated with an increased risk of ACEI-induced cough following gender adjustment in the over-dominant model. Furthermore, we observed a significant association between the T/T genotype of rs7661530 (T>C) in KCNIP4 and ACEI-induced cough in both codominant and recessive models, both before and after adjusting for gender.
Moreover, the current study’s findings reveal a significant difference in ACE plasma levels between the cough and non-cough groups, with the non-cough group having elevated ACE levels. This result suggests a potential association between reduced ACE plasma levels and the occurrence of cough (p < 0.01). However, a unique pattern emerged when we examined ACE plasma levels in relation to the ACE rs1799752 (I/D) genotype (D/D, I/D, I/I) and cough status. In particular, within the I/D genotype, the non-cough group exhibited significantly higher ACE levels than the cough group (p < 0.01). In contrast, no significant differences were observed between the cough and non-cough groups for the D/D and I/I genotypes. This suggests that the I/D genotype may be a significant factor in explaining the variations in ACE plasma levels associated with cough status (cough and non-cough). This noteworthy discovery in the I/D group may explain why this genotype was the only one previously linked to cough in our study, possibly suggesting a molecular relationship between the incidence of cough in this genotype and ACE plasma levels within the study population.
Furthermore, in our stratified analysis by ethnicity, we observed significant differences in the distribution of genotypes among ethnic groups, particularly for the ACE rs1799752 (I/D) and KCNIP4 variants (rs7675300 (C>A), rs1495509 (T>C), rs16870989 (T>A)). The Arab population showed a consistently higher frequency of the D/D genotype in ACE rs1799752 and the C/C and T/T genotypes in KCNIP4 rs7675300 and rs1495509 variants respectively, whereas the East Asian and Indian groups displayed higher frequencies of alternative genotypes. These differences highlight the genetic diversity across ethnicities and may partially explain variations in ACEI-induced cough among populations. The findings support the importance of considering population background in pharmacogenomic research and future clinical implementation. Further large-scale, prospective studies with balanced representation of ethnic groups are needed to validate these associations and explore their clinical significance.
Additionally, results showed that variants of the KCNIP4; rs7675300 (C>A), rs1495509 (T>C), rs7661530 (T>C), and rs1687089 (T>A) are in high LD with each other, indicating they are often inherited together within the population studied. This strong LD suggests the presence of haplotype blocks, and based on this association, we have constructed haplotypes further to investigate the potential implications for ACEI-induced cough. However, the global association p = 0.38 indicates that the haplotypes, when considered collectively, did not exhibit a statistically significant correlation with ACEI-induced cough. Furthermore, we assessed the combined effect of ACE; rs1799752 (I/D), BDKRB2; rs1799722 (C>T), and KCNIP4; rs7675300 (C>A) on ACEI-induced cough. The variants interaction analysis did not reveal any statistically significant associations indicating a lack of strong evidence for any meaningful effect of these variant’s interactions on ACEI-induced cough.
However, adjustments for additional covariates such as age, smoking status, comorbidities, and ACEIs type using multivariable models were not feasible due to the small sample size. Additionally, no multiple testing correction was applied, therefore, these results should be interpreted with appropriate caution. These limitations are acknowledged to provide a balanced context for our findings.
Finally, this study represents the first investigation of the association between these six genetic variants and ACEI-induced cough in an Arab population and in the Middle East. However, the findings require replication in independent cohorts to confirm their reproducibility and generalizability. Future studies involving larger and ethnically diverse Middle Eastern populations are essential to validate these pharmacogenomic associations. Such replication will help to establish robust genetic markers for predicting ACEI-induced cough and guide personalized treatment strategies in different populations. In addition, further functional assay studies are needed to explore the potential impact of the ACE rs1799752 (I/D) variant on ACE expression and activity to clarify the mechanistic role of this variant and enhance the clinical interpretation of our findings.
Conclusion
This study provides valuable insights into the genetic susceptibility to ACEI-induced cough by examining variants in ACE, BDKRB2, and KCNIP4. The significant associations identified between the I/D genotype of ACE: rs1799752 I/D, and the T/T genotype of KCNIP4, rs7661530 (T>C) with ACEI-induced cough emphasize the role of genetic factors in this side effect. Furthermore, the observation that total ACE plasma levels were significantly higher in the non-cough group, particularly among individuals with the I/D genotype, underscores the potential effect of ACE levels on cough development. These findings underscore the significance of genetic factors and ACE plasma levels in determining the risk of ACEI-induced cough, which may inform personalized treatment strategies for patients at higher risk.
Limitations
The current study has limitations that should be acknowledged. First, the relatively small sample size may limit the generalizability of the findings, as a larger cohort would likely yield more robust and statistically reliable results. Second, the study focused primarily on perindopril and lisinopril, as these were the most commonly prescribed ACE inhibitors at the recruitment sites. This focus may restrict the applicability of the findings to other ACEIs, such as enalapril or ramipril, which were underrepresented in the cohort.
Data availability statement
The data presented in this study are not publicly available due to participant privacy concerns and data sharing restrictions set by the institutional ethics board. De-identified data may be made available upon reasonable request to the corresponding author, subject to ethical approval.
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Department of Health-Abu Dhabi for the Data from the UAEU (DOH/CVDC/2020/1187), (DOH/CVDC/2021/1519), (DOH/CVDC/2022/1458), (DOH/CVDC/2023/1952) (MCME.CR.213.MAIN.2021), and (SNA/FA/2020-14). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
SA: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft. ZA-M: Conceptualization, Formal Analysis, Writing – review and editing. LK: Data curation, Investigation, Writing – review and editing. MA: Data curation, Investigation, Writing – review and editing. LD: Data curation, Investigation, Writing – review and editing. DH: Data curation, Investigation, Writing – review and editing. BA: Resources, Supervision, Writing – review and editing, Funding acquisition, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We acknowledge the UAE Ministry of Education’s financial support through grant number 1570604941 (UAEU code 21M139). LK and MA are supported by UAEU Tuition waiver fellowships.
Acknowledgments
We would like to thank the patients for taking part in this project as well as nurses and other staff at the recruitment hospitals for their help and support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1655617/full#supplementary-material
References
Abbas, D., Ezzat, Y., Hamdy, E., and Gamil, M. (2012). Angiotensin-converting enzyme (ACE) serum levels and gene polymorphism in Egyptian patients with systemic lupus erythematosus. Lupus 21 (1), 103–110. doi:10.1177/0961203311418268
Al-Mahayri, Z. N., Khasawneh, L. Q., Alqasrawi, M. N., Altoum, S. M., Jamil, G., Badawi, S., et al. (2022). Pharmacogenomics implementation in cardiovascular disease in a highly diverse population: initial findings and lessons learned from a pilot study in United Arab Emirates. Hum. Genomics 16, 42. doi:10.1186/s40246-022-00417-9
Alqasrawi, M. N., Al-Mahayri, Z. N., Alblooshi, H., Alsafar, H., and Ali, B. R. (2024). Utilizing pharmacogenomic data for a safer use of statins among the Emirati population. Curr. Vasc. Pharmacol. 22 (3), 218–229. doi:10.2174/0115701611283841231227064343
Alqasrawi, M. N., Al-Mahayri, Z. N., AlBawa’neh, A. S., Khasawneh, L. Q., Dabaghie, L., Altoum, S. M., et al. (2025). Pharmacogenomic insights into atorvastatin and rosuvastatin adverse effects: a prospective observational study in the UAE’s multiethnic population. Hum. Genomics 19 (1), 44. doi:10.1186/s40246-025-00753-6
Altoum, S. M., Al-Mahayri, Z. N., and Ali, B. R. (2023). Antihypertensives associated adverse events: a review of mechanisms and pharmacogenomic biomarkers available evidence in multi-ethnic populations. Front. Pharmacol. 14, 1286494. doi:10.3389/fphar.2023.1286494
Ancion, A., Tridetti, J., Nguyen Trung, M.-L., Oury, C., and Lancellotti, P. (2019). A review of the role of bradykinin and nitric oxide in the cardioprotective action of angiotensin-converting enzyme inhibitors: focus on perindopril. Cardiol. Ther. 8 (2), 179–191. doi:10.1007/s40119-019-00150-w
Borghi, C., Cicero, A. F., Agnoletti, D., and Fiorini, G. (2023). Pathophysiology of cough with angiotensin-converting enzyme inhibitors: how to explain within-class differences? Eur. J. Intern. Med. 110, 10–15. doi:10.1016/j.ejim.2023.01.005
Dicpinigaitis, P. V. (2006). Angiotensin-converting enzyme inhibitor-induced cough: ACCP evidence-based clinical practice guidelines. CHEST 129 (1), 169S-173S–173S. doi:10.1378/chest.129.1_suppl.169S
Ding, H., Shi, C., Xu, X., and Yu, L. (2020). Drug-induced chronic cough and the possible mechanism of action. Ann. Palliat. Med. 9 (5), 3562–3570. doi:10.21037/apm-20-819
Hallberg, P., Persson, M., Axelsson, T., Cavalli, M., Norling, P., Johansson, H.-E., et al. (2017). Genetic variants associated with angiotensin-converting enzyme inhibitor-induced cough: a genome-wide association study in a Swedish population. Pharmacogenomics 18 (3), 201–213. doi:10.2217/pgs-2016-0184
Herman, L. L., Padala, S. A., Ahmed, I., and Bashir, K. (2025). “Angiotensin converting enzyme inhibitors (ACEI),” in StatPearls (Treasure Island (FL): StatPearls Publishing). Available online at: http://www.ncbi.nlm.nih.gov/books/NBK431051/.
Khasawneh, L. Q., Alsafar, H., Alblooshi, H., Allam, M., Patrinos, G. P., and Ali, B. R. (2024). The diversity and clinical implications of genetic variants influencing clopidogrel bioactivation and response in the Emirati population. Hum. Genomics 18 (1), 2. doi:10.1186/s40246-023-00568-3
Manoharan, S. (2023). Is it still relevant to discover new ACE inhibitors from natural products? YES, but only with comprehensive approaches to address the patients’ real problems: chronic dry cough and angioedema. Molecules 28 (11), 4532. doi:10.3390/molecules28114532
Moholisa, R. R., Rayner, B. R., Patricia Owen, E., Schwager, S. L. U., Stark, J. S., Badri, M., et al. (2013). Association of B 2 receptor polymorphisms and ACE activity with ACE inhibitor–induced angioedema in black and mixed-race South Africans. J. Clin. Hypertens. 15 (6), 413–419. doi:10.1111/jch.12104
Mosley, J. D., Shaffer, C. M., Van Driest, S. L., Weeke, P. E., Wells, Q. S., Karnes, J. H., et al. (2016). A genome-wide association study identifies variants in KCNIP4 associated with ACE inhibitor-induced cough. Pharmacogenomics J. 16 (3), 231–237. doi:10.1038/tpj.2015.51
Mu, G., Xiang, Q., Zhou, S., Xie, Q., Liu, Z., Zhang, Z., et al. (2019). Association between genetic polymorphisms and angiotensin-converting enzyme inhibitor-induced cough: a systematic review and meta-analysis. Pharmacogenomics 20 (3), 189–212. doi:10.2217/pgs-2018-0157
Mukae, S., Aoki, S., Itoh, S., Iwata, T., Ueda, H., and Katagiri, T. (2000). Bradykinin B2 receptor gene polymorphism is associated with angiotensin-converting enzyme inhibitor–related cough. Hypertension 36 (1), 127–131. doi:10.1161/01.HYP.36.1.127
Mukae, S., Itoh, S., Aoki, S., Iwata, T., Nishio, K., Sato, R., et al. (2002). Association of polymorphisms of the renin-angiotensin system and bradykinin B2 receptor with ACE-inhibitor-related cough. J. Hum. Hypertens. 16 (12), 857–863. doi:10.1038/sj.jhh.1001486
Nishio, K., Kashiki, S., Tachibana, H., and Kobayashi, Y. (2011). Angiotensin-converting enzyme and bradykinin gene polymorphisms and cough: a meta-analysis. World J. Cardiol. 3 (10), 329–336. doi:10.4330/wjc.v3.i10.329
Pinto, B., Jadhav, U., Singhai, P., Sadhanandham, S., and Shah, N. (2020). ACEI-Induced cough: a review of current evidence and its practical implications for optimal CV risk reduction. Indian Heart J. 72 (5), 345–350. doi:10.1016/j.ihj.2020.08.007
Rohman, M. S., Fajar, J. K., Kuncahyo, B. H., Yunita, L., Sidarta, E. P., Saka, P. N. B., et al. (2018). Angiotensin-converting enzyme (ACE) I/D and bradykinin B2 receptor T/C genes polymorphism in patients with ACE inhibitors-related cough. Egypt. J. Med. Hum. Genet. 19 (4), 307–313. doi:10.1016/j.ejmhg.2018.05.006
Shen, J., Li, Z., Chen, J., Song, Z., Zhou, Z., and Shi, Y. (2016). SHEsisPlus, a toolset for genetic studies on polyploid species. Sci. Rep. 6 (1), 24095. doi:10.1038/srep24095
Solé, X., Guinó, E., Valls, J., Iniesta, R., and Moreno, V. (2006). SNPStats: a web tool for the analysis of association studies. Bioinforma. Oxf. Engl. 22, 1928–1929. doi:10.1093/bioinformatics/btl268
Vegter, S., and De Jong-van den Berg, L. T. W. (2010). Misdiagnosis and mistreatment of a common side-effect – angiotensin-converting enzyme inhibitor-induced cough. Br. J. Clin. Pharmacol. 69 (2), 200–203. doi:10.1111/j.1365-2125.2009.03571.x
Woo, S. W., Bang, S., Chung, M. W., Jin, S. K., Kim, Y. S., and Lee, S. H. (2009). Lack of association between ACE and bradykinin B2 receptor gene polymorphisms and ACE inhibitor-induced coughing in hypertensive koreans. J. Clin. Pharm. Ther. 34 (5), 561–567. doi:10.1111/j.1365-2710.2009.01028.x
Yılmaz, İ. (2019). Angiotensin-converting enzyme inhibitors induce cough. Turkish Thorac. J. 20 (1), 36–42. doi:10.5152/TurkThoracJ.2018.18014
Keywords: hypertension, ACE inhibitors, cough, UAE, pharmacogenomics and personalized medicine, ACE I/D polymorphism, BDKRB2, KCNIP4
Citation: Altoum SM, Al-Mahayri ZN, Khasawneh LQ, Alqasrawi MN, Dabaghie L, Hamza D and Ali BR (2025) Pharmacogenomic biomarkers of ACE inhibitor–induced cough in a multi-ethnic UAE cohort. Front. Pharmacol. 16:1655617. doi: 10.3389/fphar.2025.1655617
Received: 28 June 2025; Accepted: 19 August 2025;
Published: 25 September 2025.
Edited by:
Michal Korostynski, Institute of Pharmacology PAS, PolandCopyright © 2025 Altoum, Al-Mahayri, Khasawneh, Alqasrawi, Dabaghie, Hamza and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bassam R. Ali, YmFzc2FtLmFsaUB1YWV1LmFjLmFl
†ORCID: Bassam R. Ali, orcid.org/0000-0003-1306-6618
 Sahar M. Altoum
Sahar M. Altoum Zeina N. Al-Mahayri2
Zeina N. Al-Mahayri2 Mais N. Alqasrawi
Mais N. Alqasrawi Bassam R. Ali
Bassam R. Ali