- Avicenna Biosciences Inc., Durham, NC, United States
Polypharmacology has long been an aspect of drug design for small molecules, and the multi-target pursuit has frequently behaved more akin to divine chance rather than controllable science. Targets unknown or once thought undesirable can often be revealed to be key points of intervention for the positive effects of a drug later in the development of a program or even after its approval. In this review, we look at historical examples of molecular pleiotropism and evaluate how new insights from computational systems biology and small molecule design can aid the rational design of Selective Targeters of Multiple Proteins (STaMPs).
1 Introduction
Medicinal chemistry arose out of the German dye industry at the turn of the 20th century, and the tuning of these chromophores into pharmacophores was guided at that time by phenotypic evaluation in animal models of disease (Manavi et al., 2024). The concept of DNA as the genetic code would be decades away (let alone the notion of proteins as targets) when the first synthetic drugs were discovered (Prescott, 2000). Paracetamol, still one of the most successful contemporary analgesics discovered in that early era, highlights how this early phenotypic process was able to produce extraordinary clinical utility without mechanistic insight into the mode of action (Graham and Scott, 2005). However, these early multi-target small molecules frequently had undesirable effects, and a movement emerged seeking to reduce the interactions of a ligand to a defined set of targets. The last two decades of the 20th century saw the emergence of “one-target-one disease” as the dominant philosophy of drug design with notable exceptions like antidepressant or antipsychotic drug design (Wermuth, 2004; Casas et al., 2019; Bartolomeis et al., 2022). A new appreciation of deliberate design of multi-target system modulation through a single molecule has appeared over the last 20 years of medicinal chemistry, though this multi-target approach is still not in the mainstream of drug design strategies (Proschak et al., 2019). The goals of this review are to define the properties of the modern, intentionally-designed pleiotropic drug, to highlight the potential therapeutic improvements of systems biology disruption, as well as highlight the new computational and data science tools like artificial intelligence/machine learning platforms which will make the design of such pleiotropic drugs facile.
2 Modern multi-target drugs
The shift from animal-based phenotypic screening to the “one-target, one-disease” approach to drug design has produced many effective drugs in the modern Pharmacopeia (examples of clinically transformative modalities include the BCR-Abl, Hepatitis C polymerase and TNFα inhibitors) (Martinelli et al., 2005; Sofia, 2016; Brekke and Sandlie, 2003). However, the focus on the design of highly specific agents which did not have the cross-reactivity of older medicines may have resulted in experimental therapeutics which were safe and yet not effective in clinical trials. As the single-target, hyperselective drugs designed in 80s, 90s and 00s moved into the clinic, the development success likelihood declined significantly (Smieta et al., 2016). Interestingly, there were far fewer failures in Phases I and II due to toxicological findings and far more Phase II failures due to lack of efficacy compared to earlier periods of development (Fogel, 2018). While there are some recent positive improvements in clinical trial success rates for Phase II and Phase III, it is still the case that experimental drugs have only a 10% success rate on average when entering clinical development which is much lower than 4–5 decades ago (Dowden and Munro, 2019). Economic factors related to the 2007 financial crisis could play a role in the decline in clinical success, although the sustained low success rate across therapeutic indications into the 2020s suggests that there are other factors causing the decline in translation success. One of these factors is that the deliberate design away from pleiotropism has reduced the ability of experimental drugs to meaningfully engage disease (Hopkins, 2007; Reddy and Zhang, 2013). As the clinical pharmacology and drug design communities bear down on diseases of unhealthy aging, neurodegeneration and inflammation, there has been renewed interest in systems biology and systems pharmacology. Designing agents which can engage multiple aspects of systems pathology in a deliberate and selective manner may offer a way to increase efficacy while not introducing limiting toxicological effects.
The pioneering work of Morphy and Rankovic published in 2004 first outlined the three approaches to polypharmacology (Morphy et al., 2004; Morphy and Rankovic, 2005). In this work, multitarget engagement could be achieved by (1) administering a drug cocktail, (2) a single drug comprised of multiple active pharmaceutical ingredients (e.g., Atripla®) or (3) a multiple ligand–a single small molecule drug that can modulate multiple targets concurrently. Since their original formulation, the idea of a multiple ligand has expanded beyond their original conception with emergence of targeting chimeras (TACs, e.g., PROTACs and AUTOTACs) and molecular glues: agents that indeed are multiple ligands but interact with their targets to generate synthetic biological complexes with the ability to induce degradation (vepdegestrant, a PROTAC) or stabilize macromolecular complexes (tacrolimus, a molecular glue) (Békés et al., 2022; Schreiber, 2024). Molecular glues and PROTACs are distinct from multitarget modulators in that multitarget modulators tend to be non-naturally occurring small molecules of a molecular weight <600 Da in a dissimilar chemical space from PROTACs and molecular glues (S et al., 2024; Apprato et al., 2024). Therefore, we wish to standardize a subtype of multiple ligand distinct from the PROTAC or molecular glue classes: the Selective Targeter of Multiple Proteins, or STaMP. The reason for this standardization is that molecular glues and PROTACs have distinct design criteria from STaMPs from both a chemistry perspective and a pharmacology perspective. Beyond the chemical space dissimilarities between PROTACs, molecular glues and STaMPs, a PROTAC or molecular glue may functionally target only a single point of intervention in a way that does not engage multiple points of a pathological system. This distinction in mechanism will impact the computational methods considered for STaMP design versus PROTAC or molecular glue design.
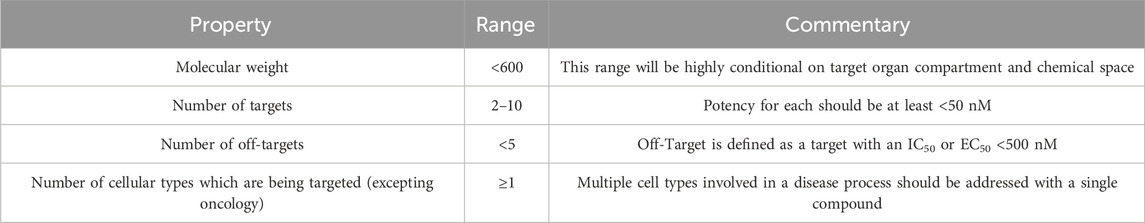
In order to highlight new techniques for the design of STaMPs, this manuscript defines a STaMP according to the framework outlined in Table 1. The goal of the range of Targets and Off-Targets is to include the atypical antipsychotics within the STaMP definition; (University of North Carolina at Chapel Hill and the United States National Institute of Mental Health) although, the number of targets modulated by the atypicals may be an outlier as this review anticipates that modern antidegeneration STaMPs will have between 2 and 7 low nanomolar target/off-target interactions in total. The proposed profile in Table 1 is for a compound with typical chemical properties for small molecule medicinal chemistry with respect to molecular weight, lipophilicity and other chemical constraints (Di and Kerns, 2016). Additionally, the number of targets/off-targets that are maximally permissible for a STaMP drug discovery campaign will depend on the targets’ physiology and any associated synergistic toxicology (Kabir and Muth, 2022; Rao et al., 2019). While there will likely be opportunities for STaMP design against single cellular types (e.g., survival pathway disruption in oncology) (Lazarte et al., 2025), this review proposes that a STaMP will find maximum therapeutic impact by disrupting pathological systems across cell lineages involved in disease (e.g., neuroinflammation, glial dysfunction and neural pathology in some types of neurodegeneration) (Castro-Gomez and Heneka, 2024; Stevenson et al., 2020).
3 Target combination identification for STaMPs
With this definition of a STaMP established, we turn our attention to highlighting key challenges in design as well as providing a survey of computational techniques which can aid in the rational design of these selective, multi-target ligands. Multifactor processes involved in the pathogenesis of metabolic diseases, neurodegeneration and inflammatory disorders are increasingly being understood in terms of systems biology and the interplay between several molecular partners (Proschak et al., 2019; Lillich et al., 2021; Artasensi et al., 2020; Ramsay et al.; Ma et al., 2020; Cavalli et al., 2008; Jana et al., 2022; Diaz-Beltran et al., 2013). The first key aspect to the development of a STaMP is the identification of biological target combinations which offer synergistic disease antagonism when those targets are modulated in the correct combination. This synergistic modulation may occur in a variety of cellular types in a tissue or an organ, and key considerations here will be considered in the review of technologies which can aid the design of STaMPs. The focus of this section will be on the developments for target selection that have been achieved across the systems biology, multi-omics and machine learning spaces, and how these new techniques offer promise for the design of STaMPs. Additionally, there are informatics approaches which can make use of clinical pharmacology databases or screen approved drug combinations in a high throughput format, however these methods have been extensively reviewed elsewhere (Proschak et al., 2019).
One of the emerging areas for the identification of targets for pleiotropic agents is the development of integrative -omics techniques (Noor et al., 2023). Transcriptomics, proteomics and metabolomics evaluate large scale biological experiments for changes in transcription, protein interaction and metabolism (Hasin et al., 2017). Although, the ability to identify key nodes of disease in patient samples and decipher the complexities of pathology in biological systems across biological layers (proteins, mRNA, etc.) remains a fundamental challenge for the -omics approach. That being said, significant progress in the integration of -omics data sets to provide novel biological insight to disease processes has been accomplished through the use of network analysis and machine learning (Borrego-Yaniz et al., 2024; Sanches et al., 2024). There are several excellent reviews for designing multi-omic studies, and these reviews offer mitigating strategies for dealing with potential pitfalls due to data heterogeneity, noise versus biological variation and poorly formulated questions for a given experimental technique (Borrego-Yaniz et al., 2024; Paananen and Fortino, 2019). Examples of correlation-based approaches for biological network analysis include Cytoscape (gene-metabolite method) (Cline et al., 2007) and Similarity Network Fusion methods (various -omics data networks created independently and then iteratively merged across biological layers) (Wang et al., 2014; Mu Yang et al., 2023). The work of Felsky and colleagues has demonstrated multi-omic integration by Similarity Network Fusion as a capable technique for analyzing human frontal cortex samples for -omics molecular subtypes associated with cognitive decline and neuropathology (Mu Yang et al., 2023). Additionally, Martin et al. explored the utility integrative-omics methods when applied to immune-mediated inflammatory diseases and reported that these approaches offer a comprehensive platform for understanding how multiple biological targets are involved in complex, multifactor disease processes across changes in lipids, proteins, transcription events and epigenetics (Borrego-Yaniz et al., 2024).
While network analysis methods can serve as an isolated method for multi-omics, the advent of machine learning applications in bioinformatics can aid in causal linkage discovery and/or integration between-omics and clinical parameters like demographics, presentation and outcome (Casas et al., 2019; Stafford et al., 2020; Jiang et al., 2017). This was the case with systemic autoimmune disease (SAD) patients where an unsupervised clustering method was used to integrate whole blood transcriptome and methylome data of SAD patients for seven types of systemic inflammatory disease (e.g., people with systemic lupus, rheumatoid arthritis, Sjögrens’s and systemic sclerosis) and healthy volunteers (Barturen et al., 2021). This revealed three distinct molecular clusters termed inflammatory, lymphoid and interferon which were stable across the seven clinical diagnoses. Not only were there discrete target sets enriched in each distinct cluster, but the three clusters were stable over time suggesting these clusters to be related to the fundamental disease biology for systemic inflammation. This categorization into potential multi-target phenotypes is therefore not only important for drug design from a preclinical perspective for STaMP design, but it is also important from a patient recruitment perspective for eventual clinical trials. These unsupervised clustering methods that can be used on both patient and animal samples can ensure that the STaMP target pleiotropic profile is linked with a disease model that shares the same molecular fingerprint as the eventual patient population cluster, given patient cluster subtypes will respond to therapies at differing rates (Khamashta et al., 2016).
Complementary to the multi-omics approaches reviewed above, there is also a large body of work evaluating protein-protein interactions (PPIs) as a key area to identify new targets involved in the systems biology of disease. The work of target identification through genome-wide association studies is important and impactful for finding drugs (Trajanoska et al., 2023; Nelson et al., 2015; Floris et al., 2018; Namba et al., 2022); however, work has demonstrated that PPI networks can reveal potentially druggable protein targets that, while not directly genetically associated themselves, are key points of interaction for targets which are genetically associated and yet less easily druggable (Fang et al., 2019). Fang et al. demonstrated that integration of functional genomic data and PPI networks led to a discovery of a set of targets that were key points of interaction with genetically linked proteins in a GWAS, and these identified nodes could be successfully modulated in cellular models of inflammatory disease (e.g., ICAM1 and its interaction with RhoA in ankylosing spondylitis, systemic lupus erythematosus and juvenile idiopathic arthritis, among others) (Fang et al., 2019). Interesting informatics methods for validating targets identified through genome-wide association studies and/or protein-protein interaction networks are being developed alongside the target identification methods discussed. A high throughput method for combining genetic evidence for potential targets and CRISPER/Cas9 modulation of these targets was developed by Yu et al. which allowed for the evaluation of candidate target according to their biologically importance for hepatic stellate cell activation in the presence of TGF-β stimulation (Yu et al., 2022). This work along with additional systems biology work could serve as an excellent starting point for exploring synergistic targets of interest for STaMP design to disrupt the multifactor fibrosis cascade driving liver fibrosis in advanced liver disease (Bashir et al., 2022).
As important as it is to select the targets one ought to modulate, the targets that a compound should avoid interacting with at physiologically relevant concentrations can be a difficult and yet equally important design consideration to address. While clear off-targets have emerged like hERG, it is often the case that desirable targets related to the pathogenesis of a disease of interest have dose-limiting on-target toxicological effects which preclude clinical development against that target (Lin et al., 2019; Bendels et al., 2019). Machine learning approaches are being tailored for predicting the toxicology of a target of interest, and Gao et al. have explored a deep learning approach using Genetic Profile-Activity Relationships (GPAR) to predict toxic phenotypes given some input chemical structure with reported examples of serotonin transporter inhibition, Na+/K+ ATPase inhibition and NF-κB inhibition being evaluated as use cases (Gao et al., 2021). While the method of Gao et al. requires correlating the changes in gene expression data when a cellular is exposed to a compound’s structure, there are emerging techniques employing the same gene expression data that evaluate targets for possible toxicology independently of chemical structure (Masarone et al., 2025). The use of networks across multi-omics information can also be useful for building methods like ComptoxAI which attempts to link chemical modulation with pathways and systems that account for an observed toxicologic effect (Romano et al., 2022). Although Comptox also requires input structures like the method of Gao et al., this review proposes that a candidate target of interest regularly implicated in similar toxicology across a variety of pharmacophores by ComptoxAI would allow the target to be deprioritized for STaMP design. Additionally, there have been advances in employing transcriptomics techniques which can evaluate a toxicogenomics phenotype consisting of more than 1,300 genes and involving 250 million datapoints to construct a predictive toxicogenomic space (PTGS) which could be employed to predict liver toxicity for STaMPs which are being designed at the discovery stage (Kohonen et al., 2017). While this work is an impressive initial inroad to anticipating liver toxicity, complex, organ-specific toxic events like drug-induced liver injury can be difficult to reduce to a single target (Yuan and Kaplowitz, 2013; Andrade et al., 2019). Unfortunately, a reductionistic method that can predict synergistic target combinations causing organ toxicity is not yet available. Several methods exist for predicting toxicology for chemical structures across a variety of organs without respect to the targets of said strucutres (Cavasotto and Scardino, 2022), but teasing out potential toxic combinations of drug targets remains a key area of interest for the development of computational methods to aid in the design of STaMPs.
4 Techniques for the design of STaMP ligands across biological targets
Ample chemical starting points exist for the design of a drug be the drug profile selective or pleiotropic. This is such the case that the true size of the space that contains all drugs be they natural product or synthetic is unintelligible (Lachance et al., 2012; Reymond, 2015). Given the synthetic inefficiency that currently limits natural product modification (despite the utility for new medicine discovery) (Truax and Romo, 2020), we accordingly constrain ourselves to synthetic molecules as opportunities for fine-tuned and rational STaMP design and will therefore limit this section to the review of methods which can be impactful in the synthetic small molecule chemical space.
Physics-based methods have made major improvements in accuracy over older methods used to predict potency against a biological target (e.g., GLIDE) (Jorgensen and Thomas, 2008; Kirkpatrick, 2004; Abel et al., 2017). These advances in methods like thermodynamic integration (TI) and free energy perturbation (FEP) have been driven by an increase in computing power serving both the refinement in methodology as well as improving the compound throughput markedly (Harder et al., 2016; Damm et al., 2025; Giese and York, 2018; Lee et al., 2017). Importantly for the design of STaMPs, these improvements allow physics-based methods to serve both a “dial-in” approach where additional target affinities are designed into a given scaffold and a “dial-out” approach where undesired target affinities are removed through the use of computational techniques. While limited examples exist of FEP being used to rationally design STaMPs, the work of Patel et al. highlights the utility of FEP in balancing affinity against two desired targets: human monophosphate kinase and hepatitis C viral RNA-dependent RNA polymerase (Patel et al., 2022). Antiviral nucleoside prodrugs must be converted to active triphosphates by a host nucleoside monophosphate kinase in order to be active against the viral polymerase, and the work of Patel et al. demonstrated that FEP could be employed to rationally design the multi-target constraints of a drug design program.
Machine learning has also been applied extensively for designing potency in concert with balancing other drug-like properties (Kosu et al., 2020; Antontsev et al., 2021; Kaiser et al., 2020; Han et al., 2023; Fowles et al., 2025). The ability to navigate large datasets and extract chemically meaningful information for the drug design problem of interest is a strength of machine learning, as long as the experimenter is careful with question phrasing. An example of machine learning in designing STaMP-like compounds was conducted by Bajorath et al. where they employed random forests to design dual monoamine oxidase B-acetylcholinesterase inhibitors (Feldmann et al., 2021). Here, the team focused on determining coherent substructures shared by ligands across both targets, and it was found that explainable ML methods were capable of building a bridge between chemical intuition and rational drug design. Although machine learning requires information on which to train, recent work has explored the utility of physics-based methods for augmenting machine learning datasets to facilitate machine learning algorithm construction (Burger et al., 2024; Ramaswamy et al., 2025; Thompson et al., 2022; Lonsdale et al., 2025). Such a hybrid approach will facilitate either “dial-in” or “dial-out” approaches for the design of STaMPs where there exists either a novel desirable target, or new insights reveal a target to be undesirable.
5 Conclusion
Given the stupefying progress achieved in the just the past decade with regard to informatics, -omics and computation, there is a great opportunity for the rational design of modern multi-target small molecules defined here as STaMPs. New insights in systems biology, mulit-omics, physics-based methods and machine learning can be employed to identify not only synergistic target combinations but also molecular targets with potentially toxic effects. Additionally, advances in computational methods can make it more efficient to optimize a chemical series for an experimentally determined target profile. While serendipity will always play a role in the discovery of new medicines, the technologies reviewed herein point to a new kind of medicinal chemistry: the rational design of STaMPs through advanced computation.
Author contributions
TK: Writing – review and editing, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
TK wishes to acknowledge helpful conversations with Pieter Burger, Sandro Belvedere and Christopher Meldrum.
Conflict of interest
Author TK was employed by Avicenna Biosciences Inc.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abel, R., Wang, L., Harder, E. D., Berne, B. J., and Friesner, R. A. (2017). Advancing drug discovery through enhanced free energy calculations. Accounts Chem. Res. 50, 1625–1632. doi:10.1021/acs.accounts.7b00083
Andrade, R. J., Chalasani, N., Björnsson, E. S., Suzuki, A., Kullak-Ublick, G. A., Watkins, P. B., et al. (2019). Drug-induced liver injury. Nat. Rev. Dis. Prim. 5, 58. doi:10.1038/s41572-019-0105-0
Antontsev, V., Jagarapu, A., Bundey, Y., Hou, H., Khotimchenko, M., Walsh, J., et al. (2021). A hybrid modeling approach for assessing mechanistic models of small molecule partitioning in vivo using a machine learning-integrated modeling platform. Sci. Rep. 11, 11143. doi:10.1038/s41598-021-90637-1
Apprato, G., Poongavanam, V., Jimenez, D. G., Atilaw, Y., Erdelyi, M., Ermondi, G., et al. (2024). Exploring the chemical space of orally bioavailable PROTACs. Drug Discov. Today 29, 103917. doi:10.1016/j.drudis.2024.103917
Artasensi, A., Pedretti, A., Vistoli, G., and Fumagalli, L. (2020). Type 2 diabetes mellitus: a review of multi-target drugs. Molecules 25, 1987. doi:10.3390/molecules25081987
Bartolomeis, A. d., Barone, A., Begni, V., and Riva, M. A. (2022). Present and future antipsychotic drugs: a systematic review of the putative mechanisms of action for efficacy and a critical appraisal under a translational perspective. Pharmacol. Res. 176, 106078. doi:10.1016/j.phrs.2022.106078
Barturen, G., Babaei, S., Català-Moll, F., Martínez-Bueno, M., Makowska, Z., Martorell-Marugán, J., et al. (2021). Integrative analysis reveals a molecular stratification of systemic autoimmune diseases. Arthritis Rheumatology 73, 1073–1085. doi:10.1002/art.41610
Bashir, A., Duseja, A., De, A., Mehta, M., and Tiwari, P. (2022). Non-alcoholic fatty liver disease development: a multifactorial pathogenic phenomena. Liver Res. 6, 72–83. doi:10.1016/j.livres.2022.05.002
Békés, M., Langley, D. R., and Crews, C. M. (2022). PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200. doi:10.1038/s41573-021-00371-6
Bendels, S., Bissantz, C., Fasching, B., Gerebtzoff, G., Guba, W., Kansy, M., et al. (2019). Safety screening in early drug discovery: an optimized assay panel. J. Pharmacol. Toxicol. Methods 99, 106609. doi:10.1016/j.vascn.2019.106609
Borrego-Yaniz, G., Terrón-Camero, L. C., Kerick, M., Andrés-León, E., and Marti, J. (2024). A holistic approach to understanding immune-mediated inflammatory diseases: bioinformatic tools to integrate omics data. Comput. Struct. Biotechnol. J. 23, 96–105. doi:10.1016/j.csbj.2023.11.045
Brekke, O. H., and Sandlie, I. (2003). Therapeutic antibodies for human diseases at the dawn of the twenty-first century. Nat. Rev. Drug Discov. 2, 52–62. doi:10.1038/nrd984
Burger, P. B., Hu, X., Balabin, I., Muller, M., Stanley, M., Joubert, F., et al. (2024). FEP augmentation as a means to solve data paucity problems for machine learning in chemical biology. J. Chem. Inf. Model. 64, 3812–3825. doi:10.1021/acs.jcim.4c00071
Casas, A. I., Hassan, A. A., Larsen, S. J., Schmidt, H. H. H. W., Elbatreek, M., Kleikers, P. W. M., et al. (2019). From single drug targets to synergistic network pharmacology in ischemic stroke. Proc. Natl. Acad. Sci. 116 (14), 7129–7136. doi:10.1073/pnas.1820799116
Castro-Gomez, S., and Heneka, M. T. (2024). Innate immune activation in neurodegenerative diseases. Immunity 57, 790–814. doi:10.1016/j.immuni.2024.03.010
Cavalli, A., Bolognesi, M. L., Minarini, A., Rosini, M., Tumiatti, V., Recanatini, M., et al. (2008). Multi-target-directed ligands to combat neurodegenerative diseases. J. Med. Chem. 51, 347–372. doi:10.1021/jm7009364
Cavasotto, C. N., and Scardino, V. (2022). Machine learning toxicity prediction: latest advances by toxicity end point. ACS Omega 7, 47536–47546. doi:10.1021/acsomega.2c05693
Cline, M. S., Smoot, M., Cerami, E., Kuchinsky, A., Landys, N., Workman, C., et al. (2007). Integration of biological networks and gene expression data using cytoscape. Nat. Protoc. 2, 2366–2382. doi:10.1038/nprot.2007.324
Damm, W., Dajnowicz, S., Ghoreishi, D., Yu, Y., Ganeshan, K., Madin, O., et al. (2025). OPLS5: addition of polarizability and improved treatment of metals. ChemRxiv.
Di, L., and Kerns, E. H. (2016). Drug-like properties: concepts, structure design and methods from ADME to toxicity optimization. Academic Press.
Diaz-Beltran, L., Cano, C., Wall, D. P., and Esteban, F. J. (2013). Systems biology as a comparative approach to understand complex gene expression in neurological diseases. Behav. Sci. 3, 253–272. doi:10.3390/bs3020253
Dowden, H., and Munro, J. (2019). Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 18, 495–496. doi:10.1038/d41573-019-00074-z
Fang, H., Consortium, T. U.-D., Wolf, H. D., Knezevic, B., Burnham, K. L., Osgood, J., et al. (2019). A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet. 51, 1082–1091. doi:10.1038/s41588-019-0456-1
Feldmann, C., Philipps, M., and Bajorath, J. (2021). Explainable machine learning predictions of dual-target compounds reveal characteristic structural features. Sci. Rep. 11, 21594. doi:10.1038/s41598-021-01099-4
Floris, M., Olla, S., Schlessinger, D., and Cucca, F. (2018). Genetic-driven druggable target identification and validation. Trends Genet. 34, 558–570. doi:10.1016/j.tig.2018.04.004
Fogel, D. B. (2018). Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: a review. Contemp. Clin. Trials Commun. 11, 156–164. doi:10.1016/j.conctc.2018.08.001
Fowles, D. J., Connaughton, B. J., Carter, J. W., Mitchell, J. B. O., and Palmer, D. S. (2025). Physics-based solubility prediction for organic molecules. Chem. Rev. 125, 7057–7098. doi:10.1021/acs.chemrev.4c00855
Gao, S., Han, L., Luo, D., Liu, G., Xiao, Z., Shan, G., et al. (2021). Modeling drug mechanism of action with large scale gene-expression profiles using GPAR, an artificial intelligence platform. BMC Bioinforma. 22, 17. doi:10.1186/s12859-020-03915-6
Giese, T. J., and York, D. M. (2018). A GPU-accelerated parameter interpolation thermodynamic integration free energy method. J. Chem. Theory Comput. 14, 1564–1582. doi:10.1021/acs.jctc.7b01175
Graham, G. G., and Scott, K. F. (2005). Mechanism of action of paracetamol. Am. J. Ther. 12 (1), 46–55. doi:10.1097/00045391-200501000-00008
Han, R., Yoon, H., Kim, G., Lee, H., and Lee, Y. (2023). Revolutionizing medicinal chemistry: the application of artificial intelligence (AI) in early drug discovery. Pharmaceuticals 16, 1259. doi:10.3390/ph16091259
Harder, E., Damm, W., Maple, J., Wu, C., Reboul, M., Xiang, J. Y., et al. (2016). OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 12, 281–296. doi:10.1021/acs.jctc.5b00864
Hasin, Y., Seldin, M., and Lusis, A. (2017). Multi-omics approaches to disease. Genome Biol. 18, 83. doi:10.1186/s13059-017-1215-1
Hopkins, A. L. (2007). Network pharmacology. Nat. Biotechnol. 25, 1110–1111. doi:10.1038/nbt1007-1110
Jana, A., Bhattacharjee, A., Das, S. S., Srivastava, A., Choudhury, A., Bhattacharjee, R., et al. (2022). Molecular insights into therapeutic potentials of hybrid compounds targeting alzheimer's disease. Mol. Neurobiol. 59, 3512–3528. doi:10.1007/s12035-022-02779-6
Jiang, F., Jiang, Y., Zhi, H., Dong, Y., Li, H., Ma, S., et al. (2017). Artificial intelligence in healthcare: past, present and future. Stroke Vasc. Neurology 2, 230–243. doi:10.1136/svn-2017-000101
Jorgensen, W. L., and Thomas, L. L. (2008). Perspective on free-energy perturbation calculations for chemical equilibria. J. Chem. Theory Comput. 4, 869–876. doi:10.1021/ct800011m
Kabir, A., and Muth, A. (2022). Polypharmacology: the science of multi-targeting molecules. Pharmacol. Res. 176, 106055. doi:10.1016/j.phrs.2021.106055
Kaiser, T. M., Dentmon, Z. W., Dalloul, C. E., Sharma, S. K., and Liotta, D. C. (2020). Accelerated discovery of novel ponatinib analogs with improved properties for the treatment of parkinson's disease. ACS Med. Chem. Lett. 11, 491–496. doi:10.1021/acsmedchemlett.9b00612
Khamashta, M., Merrill, J. T., Werth, V. P., Furie, R., Kalunian, K., Illei, G. G., et al. (2016). Sifalimumab, an anti-interferon-α monoclonal antibody, in moderate to severe systemic lupus erythematosus: a randomised, double-blind, placebo-controlled study. Ann. Rheumatic Dis. 75, 1909–1916. doi:10.1136/annrheumdis-2015-208562
Kohonen, P., Parkkinen, J. A., Willighagen, E. L., Ceder, R., Wennerberg, K., Kaski, S., et al. (2017). A transcriptomics data-driven gene space accurately predicts liver cytopathology and drug-induced liver injury. Nat. Commun. 8, 15932. doi:10.1038/ncomms15932
Kosugi, Y., and Hosea, N. (2020). Direct comparison of total clearance prediction: computational machine learning model versus Bottom-Up approach using in vitro assay. Mol. Pharm. 17, 2299–2309. doi:10.1021/acs.molpharmaceut.9b01294
Lachance, H., Wetzel, S., Kumar, K., and Waldmann, H. (2012). Charting, navigating, and populating natural product chemical space for drug discovery. J. Med. Chem. 55, 5989–6001. doi:10.1021/jm300288g
Lazarte, J. M. S., Ofosu-Asante, K., Tilghman, S. L., and Lamang, N. S. (2025). PCAIs stimulate MAPK, PI3K/AKT pathways and ROS-mediated apoptosis in aromatase inhibitor-resistant breast cancer cells while disrupting actin filaments and focal adhesion. Oncotarget 16, 621–641. doi:10.18632/oncotarget.28759
Lee, T.-S., Hu, Y., Sherborne, B., Guo, Z., and York, D. M. (2017). Toward fast and accurate binding affinity prediction with pmemdGTI: an efficient implementation of GPU-accelerated thermodynamic integration. J. Chem. Theory Comput. 13, 3077–3084. doi:10.1021/acs.jctc.7b00102
Lillich, F. F., Imig, J. D., and Proschak, E. (2021). Multi-target approaches in metabolic syndrome. Front. Pharmacol. 11, 554961. doi:10.3389/fphar.2020.554961
Lin, A., Giuliano, C. J., Palladino, A., John, K. M., Abramowicz, C., Yuan, M. L., et al. (2019). Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 11, eaaw8412. doi:10.1126/scitranslmed.aaw8412
Lonsdale, R., Glancy, J., Kalash, L., Marcus, D., and Wall, I. D. (2025). Active learning FEP: impact on performance of AL protocol and chemical diversity. J. Chem. Theory Comput. 21, 4867–4883. doi:10.1021/acs.jctc.5c00128
Ma, H., Huang, B., and Zhang, Y. (2020). Recent advances in multitarget-directed ligands targeting G-protein-coupled receptors. Drug Discov. Today 25, 1682–1692. doi:10.1016/j.drudis.2020.07.004
Manavi, M. A., Salehi, M., Jafari, R. M., and Dehpour, A. R. (2024). From dyes to drugs: the historical impact and future potential of dyes in drug discovery. Arch. Pharm. 357 (11), e2400532. doi:10.1002/ardp.202400532
Martinelli, G., Soverini, S., Rosti, G., Cilloni, D., and Baccarani, M. (2005). New tyrosine kinase inhibitors in chronic myeloid leukemia. Haematologica 90, 534–541.
Masarone, S., Beckwith, K. V., Wilkinson, M. R., Tuli, S., Lane, A., Windsor, S., et al. (2025). Advancing predictive toxicology: overcoming hurdles and shaping the future. Digit. Discov. 4, 303–315. doi:10.1039/d4dd00257a
Morphy, R., and Rankovic, Z. (2005). Designed multiple ligands. An emerging drug discovery paradigm. J. Med. Chem. 48, 6523–6543. doi:10.1021/jm058225d
Morphy, R., Kay, C., and Rankovic, Z. (2004). From magic bullets to designed multiple ligands. Drug Discov. Today 9, 641–651. doi:10.1016/S1359-6446(04)03163-0
Mu Yang, S. M.-L., Wang, Y., De Jager, P. L., Bennett, D. A., Felsky, D., and Felsky, D. (2023). Multi-omic integration via similarity network fusion to detect molecular subtypes of ageing. Brain Commun. 5, fcad110. doi:10.1093/braincomms/fcad110
Namba, S., Konuma, T., Wu, K.-H., Zhou, W., Initiative, G. B. M.-a., and Okada, Y. (2022). A practical guideline of genomics-driven drug discovery in the era of global biobank meta-analysis. Cell Genomics 2, 100190. doi:10.1016/j.xgen.2022.100190
Nelson, M. R., Tipney, H., Painter, J. L., Shen, J., Nicoletti, P., Shen, Y., et al. (2015). The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860. doi:10.1038/ng.3314
Noor, F., Asif, M., Ashfaq, U. A., Qasim, M., and Qamar, M. T. u. (2023). Machine learning for synergistic network pharmacology: a comprehensive overview. Briefings Bioinforma. 24, bbad120. doi:10.1093/bib/bbad120
Paananen, J., and Fortino, V. (2019). An omics perspective on drug target discovery platforms. Briefings Bioinforma. 21, 1937–1953. doi:10.1093/bib/bbz122
Patel, D., Cox, B. D., Kasthuri, M., Mengshetti, S., Bassit, L., Verma, K., et al. (2022). In silico design of a novel nucleotide antiviral agent by free energy perturbation. Chem. Biol. and Drug Des. 99, 801–815. doi:10.1111/cbdd.14042
Prescott, L. F. (2000). Paracetamol: past, present, and future. Am. J. Ther. 7 (2), 143–148. doi:10.1097/00045391-200007020-00011
Proschak, E., Stark, H., and Merk, D. (2019). Polypharmacology by design: a medicinal chemist’s perspective on multitargeting compounds. J. Med. Chem. 62, 420–444. doi:10.1021/acs.jmedchem.8b00760
Ramaswamy, V. K., Habgood, M., and Mackey, M. D. (2025). Active learning FEP using 3D-QSAR for prioritizing bioisosteres in medicinal chemistry. ACS Med. Chem. Lett. 16, 984–990. doi:10.1021/acsmedchemlett.4c00554
Ramsay, R. R., Majekova, M., Medina, M., and Valoti, M. (2016). Key targets for multi-target ligands designed to combat neurodegeneration. Front. Neurosci. 10, 375. doi:10.3389/fnins.2016.00375
Rao, M. S., Gupta, R., Liguori, M. J., Hu, M., Huang, X., Mantena, S. R., et al. (2019). Novel computational approach to predict off-target interactions for small molecules. Front. Big Data 2, 25. doi:10.3389/fdata.2019.00025
Reddy, A. S., and Zhang, S. (2013). Polypharmacology: drug discovery for the future. Expert Rev. Clin. Pharmacol. 6, 41–47. doi:10.1586/ecp.12.74
Reymond, J.-L. (2015). The chemical space project. Accounts Chem. Res. 48, 722–730. doi:10.1021/ar500432k
Romano, J. D., Hao, Y., Moore, J. H., and Penning, T. M. (2022). Automating predictive toxicology using ComptoxAI. Chem. Res. Toxicol. 35, 1370–1382. doi:10.1021/acs.chemrestox.2c00074
S, H. K., Venkatachalapathy, M., Sistla, R., and Poongavanam, V. (2024). Advances in molecular glues: exploring chemical space and design principles for targeted protein degradation. Drug Discov. Today 29, 104205. doi:10.1016/j.drudis.2024.104205
Sanches, P. H. G., deMelo, N. C., Porcari, A., and Carvalho, L. d. (2024). Integrating molecular perspectives: strategies for comprehensive multi-omics integrative data analysis and machine learning applications in transcriptomics, proteomics, and metabolomics. Biology 13, 848. doi:10.3390/biology13110848
Schreiber, S. L. (2024). Molecular glues and bifunctional compounds: therapeutic modalities based on induced proximity. Cell Chem. Biol. 31, p1050–p1063. doi:10.1016/j.chembiol.2024.05.004
Smietana, K., Siatkowski, M., and Møller, M. (2016). Trends in clinical success rates. Nat. Rev. Drug Discov. 15, 379–380. doi:10.1038/nrd.2016.85
Sofia, M. J. (2016). Enter sofosbuvir: the path to curing HCV. Cell 167, 25–29. doi:10.1016/j.cell.2016.08.044
Stafford, I. S., Kellermann, M., Mossotto, E., Beattie, R. M., MacArthur, B. D., and Ennis, S. (2020). A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. npj Digit. Med. 3, 30. doi:10.1038/s41746-020-0229-3
Stevenson, R., Samokhina, E., Rossetti, I., Morley, J. W., and Buskila, Y. (2020). Neuromodulation of glial function during neurodegeneration. Front. Cell. Neurosci. 14, 278. doi:10.3389/fncel.2020.00278
Thompson, J., Walters, W. P., Feng, J. A., Pabon, N. A., Xu, H., Maser, M., et al. (2022). Optimizing active learning for free energy calculations. Artif. Intell. Life Sci. 2, 100050. doi:10.1016/j.ailsci.2022.100050
Trajanoska, K., Bhérer, C., Taliun, D., Zhou, S., Richards, J. B., and Mooser, V. (2023). From target discovery to clinical drug development with human genetics. Nature 620, 737–745. doi:10.1038/s41586-023-06388-8
Truax, N. J., and Romo, D. (2020). Bridging the gap between natural product synthesis and drug discovery. Nat. Product. Rep. 37, 1436–1453. doi:10.1039/d0np00048e
University of North Carolina at Chapel Hill and the United States National Institute of Mental Health. Available online at: https://pdsp.unc.edu/databases/pdsp.php?knowID=0&kiKey=&receptorDD=&receptor=&speciesDD=&species=&sourcesDD=&source=&hotLigandDD=&hotLigand=&testLigandDD=&testFreeRadio=testFreeRadio&testLigand=olanzapine&referenceDD=&reference=&KiGreater=&KiLess=&kiAllRadio=all&doQuery=Submit+Query (Accessed August 17, 2025).
Wang, B., Mezlini, A. M., Demir, F., Fiume, M., Tu, Z., Brudno, M., et al. (2014). Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 11, 333–337. doi:10.1038/nmeth.2810
Wermuth, C. G. (2004). Multitargeted drugs: the end of the “one-target-one-disease” philosophy? Drug Discov. Today 9 (19), 826–827. doi:10.1016/S1359-6446(04)03213-1
Yu, S., Ericson, M., Fanjul, A., Erion, D. M., Paraskevopoulou, M., Smith, E. N., et al. (2022). Genome-wide CRISPR screening to identify drivers of TGF-β-Induced liver fibrosis in human hepatic stellate cells. ACS Chemcial Biol. 17, 918–929. doi:10.1021/acschembio.2c00006
Keywords: synthetic biology, drug design, polypharmacology, computational drug design, machine learning, systems biology, medicinal chemistry, pleiotropism
Citation: Kaiser TM (2025) The computationally guided design of selective targeters of multiple proteins (STaMPs) as a new opportunity for small molecule drug discovery. Front. Pharmacol. 16:1691119. doi: 10.3389/fphar.2025.1691119
Received: 22 August 2025; Accepted: 25 September 2025;
Published: 07 October 2025.
Edited by:
Hugo Gutiérrez De Teran, Uppsala University, SwedenReviewed by:
Rubén Prieto-Díaz, University of Santiago de Compostela, SpainCopyright © 2025 Kaiser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas M. Kaiser, dGthaXNlckBhdmljZW5uYS1iaW8uY29t
†ORCID: Thomas M. Kaiser, orcid.org/0000-0001-5174-9183
 Thomas M. Kaiser
Thomas M. Kaiser