- 1Tropical Dermatology Infection Division, Department of Dermatology and Venereology, Faculty of Medicine, Universitas Indonesia, Central Jakarta, Indonesia
- 2Plastic and Reconstructive Surgery Division, Department of Ophthalmology, Faculty of Medicine, Universitas Indonesia, Central Jakarta, Indonesia
- 3Department of Medical Biology, Faculty of Medicine, Universitas Indonesia, Central, Jakarta, Indonesia
- 4Faculty of Public Health, Universitas Indonesia, Jakarta, Indonesia
- 5Department of Dermatology, Erasmus Medical Centre, Rotterdam, Netherlands
Background: Indonesia ranks third globally in leprosy, with the eastern part of the country, particularly Ambon, still reporting high prevalence rates. In Ambon, the capital city of Maluku Province, the prevalence is 8.27 per 10.000 persons, exceeding the national rate of 0.61. While multidrug therapy (MDT) including Dapsone remains the standard treatment for leprosy, it carries the risk of a severe adverse reaction known as Dapsone Hypersensitivity Syndrome (DHS). DHS has been reported in eastern Indonesia, sometimes with fatal outcomes, including in Ambon. Recent research suggests a link between the HLA-B 13*01allele and DHS. Therefore, this study aims to investigate the association between HLA-B*13:01 and DHS within Ambon and surrounding regions.
Methods: This case-control study included leprosy patients undergoing or having completed MDT therapy in Maluku. Cases were included if they met two or more Richardus and Smith’s criteria and were recruited from several hospital in Ambon and Tual Cities. Controls were leprosy patients with >8 weeks of Dapsone exposure without DHS symptoms. Peripheral blood samples were collected, and DNA was extracted from lymphocytes using the QIAGEN QIAmp kit with a modified 2-hour incubation. HLA-B*13:01 genotyping was performed using PCR with specific primers, followed by gel electrophoresis and Sanger sequencing. Statistical analysis was performed using SPSS. Associations between categorical variables were assessed using the Chi-square test, with Fisher’s exact test used when expected cell counts were <5. Significance was set at p<0.05.
Results: A total of 8 cases and 53 controls were included. The HLA-B*13:01 was present in 100% of DHS cases and 6% of controls. Statistical analysis using Fisher’s exact test revealed a significant association between HLA-B*13:01 and DHS (p < 0.001).
Conclusions: This study demonstrates a significant association between HLA-B 13*01 and DHS in Ambon’s leprosy patients. These findings support the potential use of HLA-B*13:01 as a screening tool to identify individuals at high DHS risk before dapsone initiation, potentially improve patient safety by preventing fatal adverse events, optimizing treatment outcomes for leprosy patients by avoiding delayed treatment, and ultimately reducing leprosy transmission rates in Ambon, its surrounding regions, and other high-prevalence areas.
1 Introduction
Leprosy remains a significant public health issue in Indonesia, which ranks third globally in the number of leprosy cases, following India and Brazil (1). According to data from the Ministry of Health, Republic of Indonesia, in 2023, Maluku was identified as one of 12 provinces where leprosy remains endemic, with a prevalence rate of 2.5 per 10.000 population, considerably higher than the national prevalence rate of 0.61 per 10.000 population in Indonesia (2). In contrast, Ambon and Tual cities continue to struggle with high prevalence rates of 8.27 and 4.14 per 10.000 population, respectively (2). The high prevalences in these areas highlights the importance of implementing safe treatment approaches, especially considering risks such as Dapsone Hypersensitivity Syndrome.
The national leprosy elimination strategy prioritizes early detection- before disability manifests, and complete treatment using globally standardized multidrug therapy (MDT), which is provided free of charge at public health centers (2). MDT, which includes rifampicin, clofazimine dan dapsone (3), has proven highly effective in controlling the disease. However, despite its efficacy, one of the key concerns is the potential for severe adverse reactions, particularly Dapsone Hypersensitivity Syndrome (DHS). Dapsone, first introduced as a leprosy treatment in 1945, functions by inhibiting bacterial folic acid synthesis (4). Nevertheless, it has been associated with DHS, a serious complication first reported in Nigeria in 1949 as “glandular fever”. DHS is characterized by at least two of following symptoms: fever, lymphadenopathy, generalized rash, and hepatitis, typically appearing 5–6 weeks after dapsone initiation (5). The global prevalence of DHS is approximately 1.4% with a case-fatality rate of 9.9% (6).
Previous study in China showed that alelle HLA-B*13:01 significantly found in leprosy patients with DHS (7). It was demonstrated that individuals carrying this allele have a 34-fold increased risk of developing DHS compared to those without it (7). A study on DHS in leprosy patients in Indonesia has already been conducted in another eastern region of the country, namely Papua. The study found that HLA-B*13:01 was the most significant allele with a notable prevalence. The HLA-B*13:01 gene is present in 50% of the cases and 1.9% of the controls, and the difference between the two populations is statistically significant. Additionally, this research has validated HLA-B*13:01 as a genetic biomarker for DHS in the local population (8). The genetic landscape across Indonesia is diverse, including eastern populations, making it essential to investigate whether findings from Papua can be replicated in Ambon to ensure the biomarker’s broader relevance.
Given Ambon’s status as a leprosy-endemic region and the risks associated with dapsone treatment, this study aims to investigate the relationship between the HLA-B*13:01 allele and the occurrence of DHS among leprosy patients in Ambon and Tual. While both cities were included as study sites due to their high disease burden, the majority of samples were collected in Ambon. Whereas in Tual, being a remote area with limited healthcare resources and no permanent dermatologist, could only be accessed by researchers once a week, resulting in fewer available samples. Furthermore, although the global DHS fatality rate is estimated at 9.9% (6), local epidemiological data in Indonesia are limited, with most existing reports coming from Ambon (9, 10). These factors highlight the relevance of focusing research efforts in this region, where findings may contribute to evidence-based policies for leprosy management in high-risk populations.
2 Materials and methods
2.1 Study design
This study is a case-control study aimed at examining the association between the HLA-B*13:01 allele and the occurrence of dapsone hypersensitivity syndrome (DHS) in Ambon. The case group consists of DHS patients who were treated by dermatology and venereology specialists in five hospitals spread across Ambon and Tual. Meanwhile, the control group consists of leprosy patients who are either undergoing multidrug therapy (MDT) or have completed MDT for six months without experiencing DHS symptoms at community health centers in Ambon. The study protocol was reviewed and approved by the Ethics Committee of the Faculty of Medicine, Universitas Indonesia – Dr. Cipto Mangunkusumo Hospital (Komite Etik Penelitian Kesehatan Fakultas Kedokteran Universitas Indonesia – RSUPN Dr. Cipto Mangunkusumo). The approval date was June 6, 2024, valid for one year from the date of approval. The protocol number assigned to this study is KET- KET-818/UN2.F1/ETIK/PPM.00.02/2024.
2.2 Participants
The control patients were recruited from two community health centers in Ambon, namely Benteng and Rumahtiga, while case patients were enrolled during follow-up visits after DHS hospitalization across five hospitals in Ambon and Tual. In this study, we addressed equity in access to genetic screening by requesting control participants to visit the same clinic, regardless of their geographic origin.
An informed consent form was explained and provided to patients, which included details on data confidentiality, research objectives, inclusion criteria, as well as the risks and benefits of the study.
All leprosy patients diagnosed with DHS during the study period and within the study locations (Ambon and Tual) were selected for the case group using a total sampling method, given the rarity of the condition and the limited number of eligible subjects. The control group consisted of leprosy patients without DHS, drawn from the same population and time frame. Due to the small number of cases available in the population, no formal sample size calculation was conducted. Instead, the analysis was based on the entire accessible population of cases to explore potential risk factors associated with DHS.
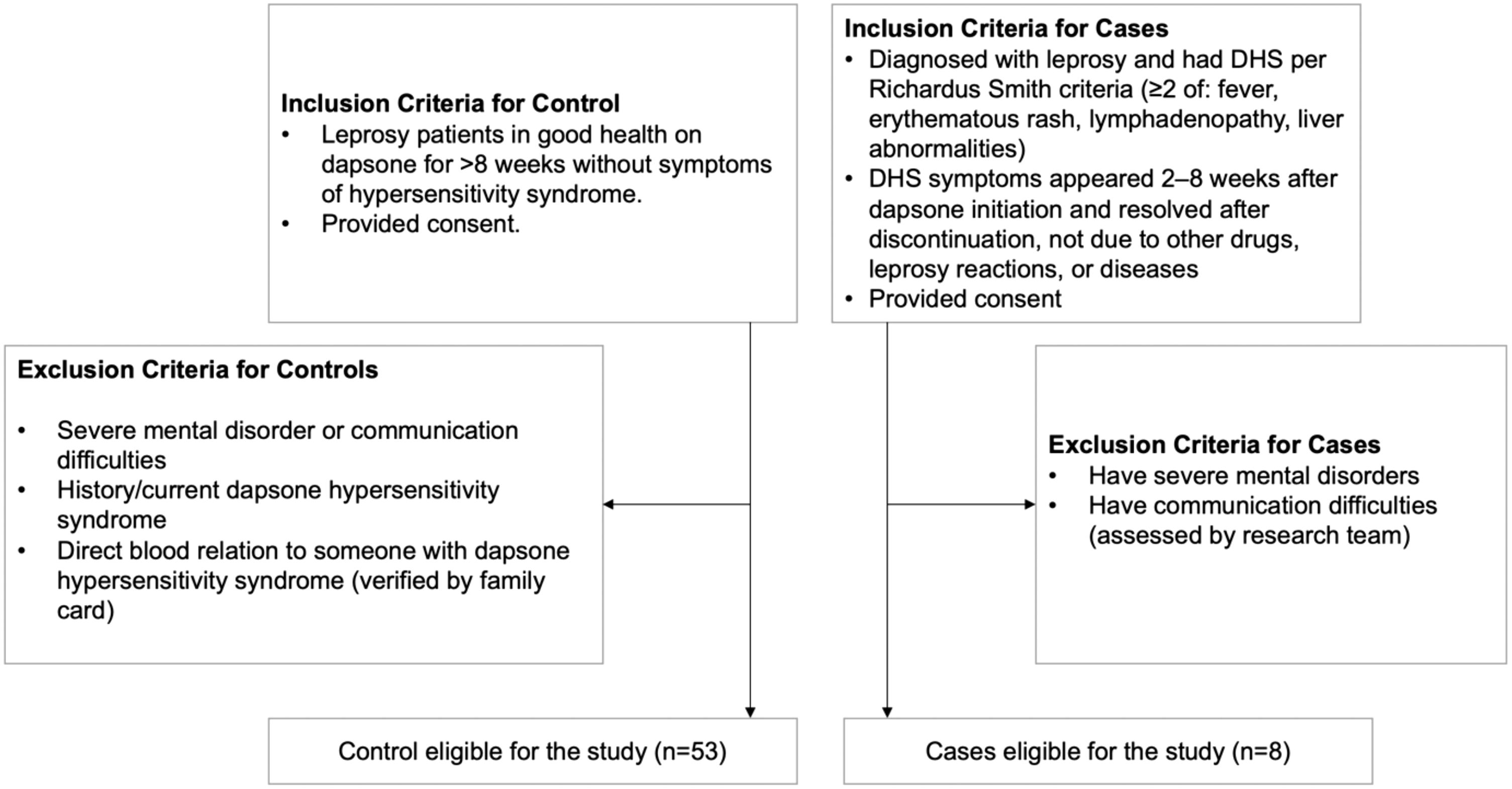
The inclusion and exclusion criteria are described in Figure 1. The inclusion criteria for case patients were: (1) diagnosed with leprosy and had a history of DHS according to the Richardus Smith criteria (11), which requires at least two of the following symptoms: fever, erythematous rash, lymphadenopathy, liver abnormalities (hepatomegaly, hepatitis, jaundice, or abnormal liver function test results). The DHS-related symptoms must have appeared 2–8 weeks after the first administration of dapsone and resolved after discontinuation of the drug. The symptoms must not be associated with other suspected drugs taken concurrently or be a manifestation of leprosy reaction. Additionally, the symptoms must not be linked to other diseases. (2) patients had provided consent to participate in the study.
The exclusion criteria for case patients were patients suffering from severe mental disorders or having communication difficulties, as assessed by the research team.
The inclusion criteria for control patients were: (1) patients in good health who had taken dapsone for more than 8 weeks without experiencing symptoms related to dapsone hypersensitivity syndrome, (2) patients had provided consent to participate in the study.
The exclusion criteria for control patients were: (1) patients suffering from severe mental disorders or having communication difficulties, as assessed by the research team, (2) patients who had taken dapsone and had experienced (or were currently experiencing) dapsone hypersensitivity syndrome, (3) patients who had a direct blood relationship with someone diagnosed with (or previously diagnosed with) dapsone hypersensitivity syndrome, as verified through the family registration card. The blood relationship were verified through family registration card and pedigree charts developed during the study.
The 8-week threshold was chosen based on the typical onset time of DHS, which most commonly develops within the first 2 to 8 weeks after the initiation of dapsone therapy (11). This timeframe allows for sufficient exposure to the drug while minimizing the inclusion of individuals with possible subclinical or evolving DHS. Therefore, controls were selected from patients who had received dapsone for at least 8 weeks without showing clinical features consistent with DHS, ensuring that they had passed the critical window of risk for the syndrome.
2.3 Data collection
A 10 ml blood sample was collected from peripheral blood vessels and stored in two ethylenediaminetetraacetic acid (EDTA) tubes for DNA extraction. The samples were homogenized and kept at room temperature, ensuring they were not exposed to direct sunlight. The specimens were then sent to the Department of Biology, Faculty of Medicine, Universitas Indonesia (FKUI) in Jakarta, within a maximum of five days after collection. The shipment of the samples was facilitated through Prodia laboratorium.
We extracted DNA from lymphocytes using the standard procedure of the QIAGEN QIAamp DNA Blood Maxi KIT (Catalog No. 51194). The incubation period was extended to two hours, following modifications from the study by Krismawati et al. (8) The extracted DNA was then quantified and subjected to PCR using specific primers to detect the presence of the HLA-B*13:01 allele. The PCR products were analyzed via gel electrophoresis, targeting a DNA band of 193 base pairs. Samples that passed quality control (QC) through electrophoresis proceeded to the next stage, Sanger sequencing. The sequencing output in FASTA format was trimmed using FincTV software. This trimming step was necessary because the initial base positions often appeared disordered or contained noise, as is common in Sanger sequencing results. Trimming was performed to remove these unreliable base calls, allowing for accurate alignment across all samples. Subsequently, nucleotide alignment was performed using BioEdit by comparing the sequences with the reference allele obtained from the IPD-IMGT database.
2.4 Data analysis
Data entry was conducted using Microsoft Excel Version 16.89.1, and statistical analysis was performed using SPSS Statistics 26 (IBM Corp, Armonk, NY, USA). Spatial data was visualized using Google My Maps (Google LLC, Mountain View, CA, USA). The Chi-Square test was used to evaluate the association between two categorical parameters, with a significance level of p < 0.05. If any dataset contained a nominal value of 0, Fisher’s Exact Test was applied for statistical analysis.
3 Result
3.1 Sociodemographic characteristic
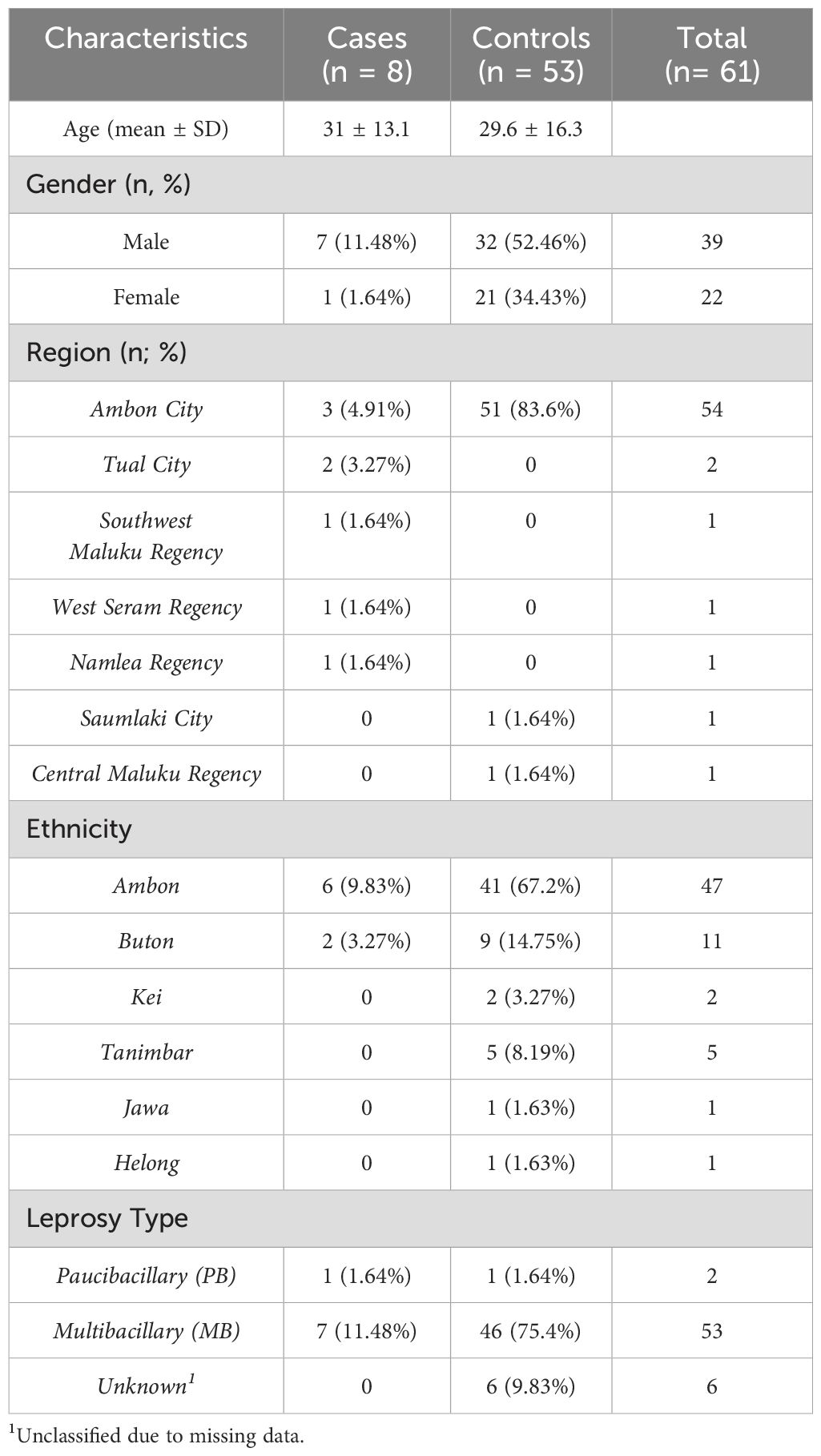
The study included 61 leprosy patients: 53 controls (asymptomatic after ≥8 weeks of MDT) and 8 cases (DHS based on Richardus Smith (11) criteria). Baseline characteristics are detailed in Table 1.
Comparison of baseline characteristics showed no significant differences in age (p = 0.681), gender distribution (p = 0.239), ethnicity (p = 0.585) and leprosy type (p = 0.193). But it did differ significantly in region (p <0.005). Due to perfect separation observed in the data, multivariable logistic regression adjustment was not feasible. Therefore, we performed Fisher’s exact test to evaluate the association between HLA-B*13:01 and DHS.
3.2 Clinical signs of dapsone hypersensitivity syndrome cases
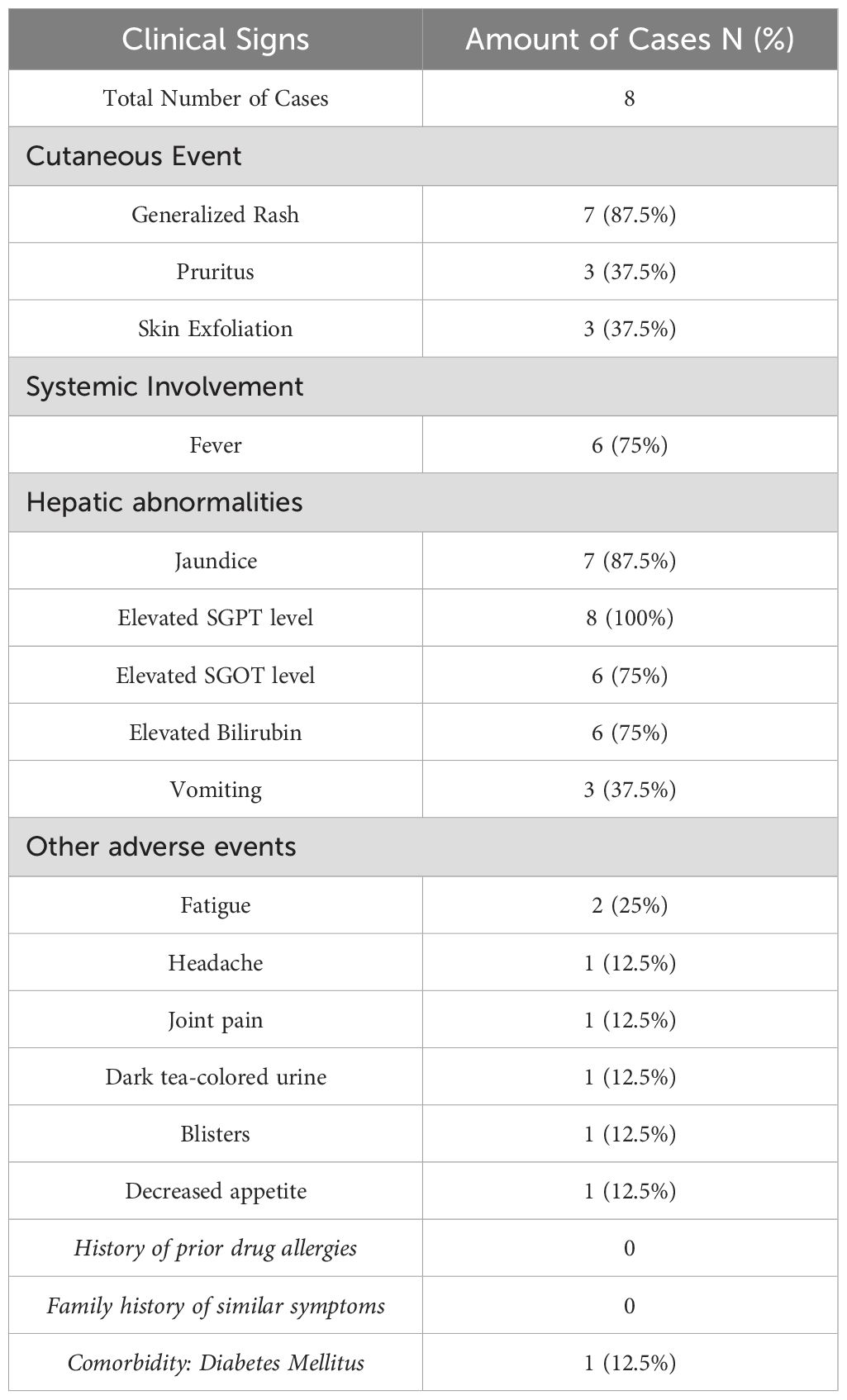
Clinical signs observed in dapsone hypersensitivity syndrome cases are summarized in Table 2. The most common cutaneous manifestation was generalized rash (87.5%), accompanied by pruritus and skin exfoliation (each 37.5%). Systemic symptoms such as fever (75%) and hepatic abnormalities, including elevated SGPT (100%) and jaundice (87.5%), were also frequently reported. Other adverse events included fatigue, headache, and gastrointestinal symptoms in a smaller proportion of cases.
3.3 HLA-B*13:01 is associated with DHS in Ambonese people
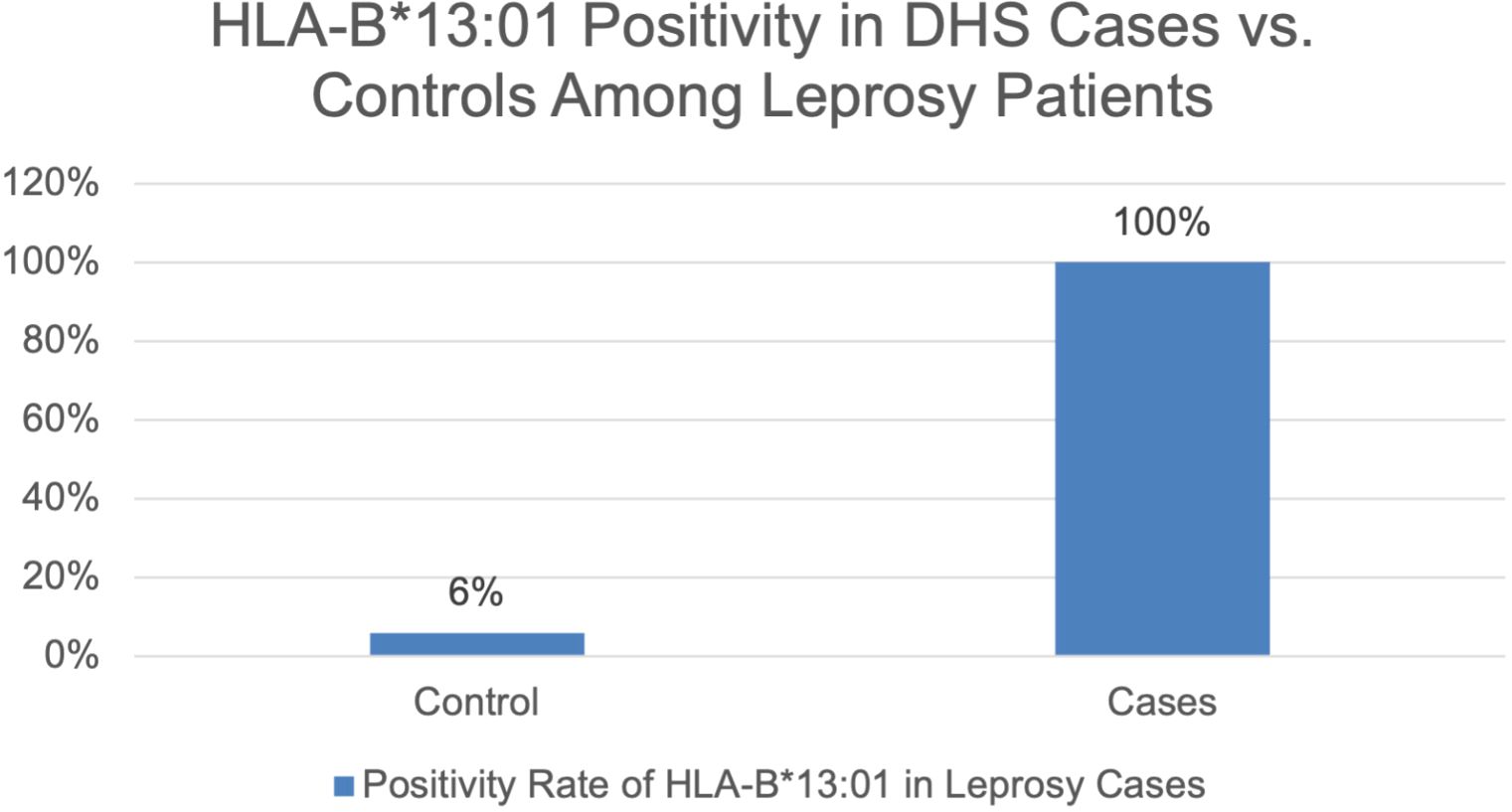
All cases (100%) carried the HLA-B*13:01 allele, whereas only 3 controls (6%) were positive (Figure 2). Both in cases and controls carried homozygous HLA-B*13:01 allele. Statistical analysis using Fisher’s Exact Test yielded a p-value of < 0.001, indicating a significant association between the HLA-B*13:01 allele and the occurrence of dapsone hypersensitivity syndrome in people in Ambon.
Three individuals in control group who carried the HLA_B*13:01 allele consisted of two females (66.7%) and one male (33.3%). All participants (100%) were multibacillary leprosy (MB) and originated from Ambon. Ages ranged from 31 to 36 years, with a median age of 35 years.
4 Discussion
Our study confirms a robust association between the HLA-B*13:01 allele and Dapsone Hypersensitivity Syndrome (DHS) in leprosy patients in Ambon, Indonesia (100% of cases vs. 6% of controls, p < 0.001). The complete penetrance (100% vs. 6%) suggests HLA-B13:01 may be a near-necessary factor for DHS in this population, though environmental cofactors likely contribute. Due to the absence of DHS cases among HLA-B*13:01 negative individuals, the odds ratio could not be calculated, and the association appears infinite. However, the Fisher’s exact test showed a highly significant association (p < 0.001), supporting a strong link between HLA-B13:01 positivity and DHS occurrence. This finding aligns with growing evidence of a strong genetic predisposition to DHS, particularly linked to the HLA-B*13:01 allele, which markedly increases susceptibility upon dapsone exposure (12).
DHS is associated with a significant risk of morbidity and mortality if not treated promptly (13). The role of HLA-B*13:01 in DHS pathogenesis is thought to involve altered antigen presentation, leading to aberrant immune responses against drug-modified self-peptides (14). While the association between HLA-B*13:01 and DHS has been explored, studies focusing on leprosy populations remain limited. Most existing research originated from China (7), while in Indonesia, investigations have only been conducted in Papua (8).
The highly significant association (p < 0.001) between HLA-B*13:01 and DHS in our Ambon population strongly supports previous findings linking this allele to an increased risk of DHS from China (7), Korea (15), Thailand (16), and Papua, Indonesia (8) (Figure 3). This clearly suggests that individuals carrying the HLA-B*13:01 allele have a substantially higher genetic predisposition to developing DHS when treated with dapsone. Our findings in Ambon, where 100% of DHS cases carried the HLA-B13:01 allele, suggest a potentially higher penetrance compared to other region in Eastern Indonesia (Papua, 50% in cases) (8). The higher prevalence of the allele in cases compared to controls reinforces the clinical implications of this finding, suggesting the potential for screening and approaches to minimize the risk of this severe adverse reaction.
The significant association detected in our study may be attributed to the unique peptide-binding residues of the HLA-B*13:01 allele, found predominantly in Asian populations, while being absent in Caucasian and African populations (17). This population specificity aligns with the distribution of DHS risk in Asian population, specifically Ambonese population. HLA-B*13:01, a member of the HLA class I heavy-chain paralogs, presents peptides from the endoplasmic reticulum lumen (7). Its distinct residues at amino acid positions 94, 95, and 145 within the binding groove are critical for peptide binding specificity, with polymorphisms at residue 95 potentially altering the recognition of dapsone-related or drug-modified-self peptides (7). Furthermore, computational docking study suggested that dapsone binds to a specific sub-pocket within the HLA-B*13:01 molecule, called “pocket F”, inducing structural changes in antigen-recognition site. This may trigger aberrant T-cell responses and clinically manifested as dapsone hypersensitivity syndrome (14). This mechanism resembling the ‘altered peptide theory’ described for abacavir-induced hypersensitivity reactions associated with HLA-B*57:01 (18, 19).
Our findings align with global reports on Dapsone Hypersensitivity Syndrome (DHS), as described in the literature. The high incidence of jaundice (87.5%), fever (75%), rash (50%), and elevated liver enzymes (100% for SGPT, 75% for SGOT) among our patients reflects the well-documented triad of dermatitis, hepatitis, and systemic symptoms commonly associated with DHS (20). Previous studies have highlighted similar patterns of hepatic dysfunction, characterized by elevated bilirubin and transaminases, as well as severe dermatological reactions, including erythema, exfoliative dermatitis, and pruritus (21, 22). These findings strengthen the need for early recognition and prompt discontinuation of dapsone in suspected DHS cases to prevent life-threatening complications.
While 87.5% of DHS patients in our study were male, statistical analysis revealed no significant association between gender and the occurrence of DHS (p = 0.239). Similar findings were reported in previous studies ([Papua, Eastern Indonesia]), Thailand, China] investigating HLA-B*13:01 and DHS, which also did not find gender to be a significant risk factor (2, 7, 8, 16). However, the predominance of male patients may reflect the generally higher prevalence of leprosy among men, as documented in numerous epidemiological studies regardless of DHS status (2, 8).
This study has several strengths, including the use of genetic sequencing for accurate HLA typing and the well-defined case and control groups, all contributing to the reliability of our findings. Furthermore, our study focuses on a population in the Ambon region, which is an area with a high prevalence of leprosy in Indonesia and where DHS has been reported.
Our findings have important implications for clinical practice and public health policy in leprosy-endemic regions. The strong association between HLA-B*13:01 and DHS suggests that screening for this allele before dapsone administration could be a valuable strategy to identify individuals at high risk and prevent severe adverse reactions. This is particularly relevant in settings like Ambon, where leprosy prevalence remains high. Implementing HLA-B*13:01 screening could lead to more personalized treatment approaches, potentially reducing morbidity and mortality associated with DHS.
4.1 Future directions
Given the high prevalence of leprosy in Ambon and other endemic regions, particularly among multibacillary (MB) cases with a high bacterial load, exploring expanded treatment options beyond the standard multi-drug therapy (MDT) is crucial. As an alternative therapy to traditional MDT regimen containing dapsone, researches have indicated that alternative regimens such as ROM (rifampicin, ofloxacin, and minocycline) showed high cure rates (23, 24). Among the three drugs in the ROM regimen, ofloxacin is the most accessible and available. Further research is needed to evaluate the effectiveness of a two-drug regimen (MDT without dapsone), particularly in MB patients from endemic areas. A study conducted in Jakarta, Indonesia found that adding ofloxacin into the MDT regimen was effective (25).
Future research should focus on validating our findings in larger, multi-center studies encompassing diverse populations. Investigating other genetic markers and non-genetic risk factors for DHS is also crucial to develop a comprehensive risk prediction model. Furthermore, cost-effectiveness analyses of HLA-B*13:01 screening programs are needed to guide policy decisions regarding implementation.
4.2 Limitation
This study has several limitations. First, the case and control participants were recruited primarily from hospitals and community health centers in Ambon and Tual, which may introduce selection bias. Second, the small number of DHS cases (n = 8) may lead to an overestimation of the association between HLA-B*13:01 and DHS. Third, while our study was conducted in Ambon, the study population included individuals from multiple ethnic backgrounds, including Butonese, Tanimbar, Kei, Javanese, and Helong origins. These groups may differ in allele frequency and genetic susceptibility to DHS, but our sample size was not powered to detect inter-ethnic differences, and ethnicity data were primarily self-reported. Fourth, a significant difference in geographical region between cases and controls was observed. Although we attempted to adjust for confounding factors, the presence of perfect separation in the data prevented reliable multivariable analysis, and residual confounding by geographical region cannot be excluded.
5 Conclusion
In conclusion, our study provides strong evidence for a significant association between the HLA-B*13:01 allele and Dapsone Hypersensitivity Syndrome in leprosy patients in Ambon, Indonesia (p < 0.001). This finding highlights that HLA-B*13:01 screening before dapsone therapy could avert life-threatening DHS in Ambon’s leprosy patients. Future work should validate these findings in larger cohorts, assess cost-effectiveness, and evaluate alternative regimens (e.g., ROM) for carriers.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by Komite Etik Penelitian Kesehatan FKUI-RSCM. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
SM: Writing – review & editing, Funding acquisition, Writing – original draft, Data curation, Visualization, Formal Analysis, Conceptualization, Methodology. MP: Visualization, Formal Analysis, Data curation, Writing – original draft, Writing – review & editing, Conceptualization, Methodology. SW: Writing – original draft, Writing – review & editing, Conceptualization. YI: Conceptualization, Writing – review & editing, Writing – original draft. YA: Conceptualization, Writing – review & editing, Writing – original draft. DS: Writing – review & editing, Writing – original draft, Conceptualization. MM: Writing – review & editing, Writing – original draft, Conceptualization. IA: Writing – review & editing, Writing – original draft, Conceptualization. EK: Writing – review & editing, Writing – original draft, Formal Analysis, Data curation. MY: Formal Analysis, Writing – review & editing, Writing – original draft, Data curation. HT: Writing – review & editing, Writing – original draft, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Internationally Indexed Publication Grant Program (PUTI) Q1 Universitas Indonesia for the Fiscal Year 2024-2025 under Grant Number NKB-307/UN2.RST/HKP.05.00/2024.
Acknowledgments
We sincerely thank KATAMATAKU Team for their dedication and contributions to the field of leprosy, Indra Muhiardi for his expertise in DNA sequencing, dr. Amanda Manuputty, Sp.DVE, dr. Hanny Tanasal, Sp.DVE, dr. Rita Sugiono Tanamal, Sp.DVE and dr. Fitri Kadarsih Bandjar, Sp.DVE for their support in case selection. We also appreciate dr. Illine Michaela and dr. Prilly Pricillya Theodorus for assisting in control recruitment, as well as the healthcare workers in Ambon for the help in sample and data collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Generative AI was used to assist in refining the text, ensuring clarity and coherence in between every paragraphs. However, all content has been reviewed and verified by the authors to maintain accuracy and originality.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2025.1597392/full#supplementary-material
References
1. de Moraes PC, Eidt LM, Koehler A, Ransan LG, and Scrofeneker ML. Epidemiological characteristics of leprosy from 2000 to 2019 in a state with low endemicity in southern Brazil. Bras Dermatol. (2023) 98:602–10. doi: 10.1016/j.abd.2022.08.009
2. Ministry of Health Republic of Indonesia and Directorate General of Disease Prevention and Control. Evaluation of the 2023 Leprosy Prevention and Control Program. Jakarta: Ministry of Health of the Republic of Indonesia (2024).
3. Celestino IC, Antunes DE, Santos DF, Gimenes VL, de Souza FM, and Goulart IMB. Adverse reactions induced by MDT/WHO (Rifampicin+Clofazimine+Dapsone) and ROM (Rifampicin+Ofloxacin+Minocycline) regimens used in the treatment of leprosy: a cohort study in a National Reference Center in Brazil. Front Pharmacol. (2024) 15. doi: 10.3389/fphar.2024.1346169
4. Kurien G, Jamil RT, and Preuss CV. Dapsone. StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing (2025). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK470552/ (Accessed February 13, 2025).
5. Zhao Q, Sun L, Sun Y, Naisbitt D, Liu H, Zhang F, et al. Dapsone hypersensitivity syndrome. Chin Med J (Engl). (2023) 136:1560–2. doi: 10.1097/CM9.0000000000002492
6. Lorenz M, Wozel G, and Schmit J. Hypersensitivity reactions to dapsone: A systematic review. Acta Dermato-Venereol. (2012) 92:194–9. doi: 10.2340/00015555-1268
7. Zhang FR, Liu H, Irwanto A, Fu XA, Li Y, Yu GQ, et al. HLA-B*13:01 and the dapsone hypersensitivity syndrome. N Engl J Med. (2013) 369:1620–8. doi: 10.1056/NEJMoa1213096
8. Krismawati H, Irwanto A, Pongtiku A, Irwan ID, Maladan Y, Sitanggang YA, et al. Validation study of hla-b* 13:01 as a biomarker of dapsone hypersensitivity syndrome in leprosy patients in Indonesia. PloS Negl Trop Dis. (2020) 14:1–11. doi: 10.1371/journal.pntd.0008746
9. Bandjar F. Sindrom hipersensitivitas dapson pada pasien morbus hansen multi-basiler: laporan kasus. Pattimura Medical Review (2024). Available online at: https://ojs3.unpatti.ac.id/index.php/pameri/article/view/12348 (Accessed April 23 2025).
10. Bandjar FK, Asmin E, Sulfiana S, Noya FC, Rahawarin H, De Lima FVI, et al. Pengetahuan dan sikap masyrakat terhadap penyakit kusta di daerah endemis Kusta, Kota Tual, Maluku: Kajian berbasis komunitas. Molucca Med [Internet]. (2024) 17:6–11. https://ojs3.unpatti.ac.id/index.php/moluccamedica/article/view/14233 (Accessed January 13, 2025).
11. Richardus JH and Smith TC. Increased incidence in leprosy of hypersensitivity reactions to dapsone after introduction of multidrug therapy. Lepr Rev. (1989) 60:267–73. doi: 10.5935/0305-7518.19890033
12. Tangamornsuksan W and Lohitnavy M. Association between HLA-B*1301 and dapsone-induced cutaneous adverse drug reactions a systematic review and meta-analysis. JAMA Dermatol Am Med Assoc. (2018) 154:441–6. doi: 10.1001/jamadermatol.2017.6484
13. Ansah R, Arkoh E, Quao B, and Groger M. Lack of suspicion of dapsone hypersensitivity syndrome in a leprosy patient: case report with fatal outcome. Res Rep Trop Med. (2023) 14:135–9. doi: 10.2147/RRTM.S434947
14. Watanabe H, Watanabe Y, Tashiro Y, Mushiroda T, Ozeki T, Hashizume H, et al. A docking model of dapsone bound to HLA-B*13:01 explains the risk of dapsone hypersensitivity syndrome. J Dermatol Sci. (2017) 88:320–9. doi: 10.1016/j.jdermsci.2017.08.007
15. Park HJ, Park JW, Kim SH, Choi SY, Kim HK, Jung CG, et al. The HLA-B*13:01 and the dapsone hypersensitivity syndrome in Korean and Asian populations: genotype- and meta-analyses. Expert Opin Drug Saf. (2020) 19(10):1349–56. doi: 10.1080/14740338.2020.1796965
16. Satapornpong P, Pratoomwun J, Rerknimitr P, Klaewsongkram J, Nakkam N, Rungrotmongkol T, et al. HLA-B*13 :01 is a predictive marker of dapsone-induced severe cutaneous adverse reactions in Thai patients. Front Immunol. (2021) 12. doi: 10.3389/fimmu.2021.661135
17. Li H, Dai Y, Huang H, Li L, Leng S, Cheng J, et al. HLA-B*13:01 as a biomarker for genetic susceptibility to hypersensitivity dermatitis induced by trichloroethylene among workers in China. Environ Health Perspect. (2007) 115:1553–6. doi: 10.1289/ehp.10325
18. Illing PT, Mifsud NA, and Purcell AW. Allotype specific interactions of drugs and HLA molecules in hypersensitivity reactions. Curr Opin Immunol. (2016) 42:31–40. doi: 10.1016/j.coi.2016.05.003
19. Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. (2012) 486:554–8. doi: 10.1038/nature11147
20. Kosseifi SG, Guha B, Nassour DN, Chi DS, and Krishnaswamy G. The Dapsone Hypersensitivity Syndrome revisited: A potentially fatal multisystem disorder with prominent hepatopulmonary manifestations. J Occup Med Toxicol. (2006) 1:1–9. doi: 10.1186/1745-6673-1-9
21. Sener O, Doganci L, Safali M, Besirbellioglu B, Bulucu F, and Pahsa A. Severe dapsone hypersensitivity syndrome. J Invest Allergol Clin Immunol. (2006) 16:268–70.
22. Vinod KV, Arun K, and Dutta TK. Dapsone hypersensitivity syndrome: A rare life threatening complication of dapsone therapy. J Pharmacol Pharmacother. (2013) 4:158–60. doi: 10.4103/0976-500X.110917
23. Girdhar A, Kumar A, and Girdhar B. A randomized controlled trial assessing the effect of adding clarithromycin to Rifampicin, ofloxacin and minocycline in the treatment of single lesion paucibacillary leprosy in Agra District, India. Lepr Rev. (2011) 82:46–54. doi: 10.47276/lr.82.1.46
24. Kumar A, Girdhar A, and Girdhar B. A randomized controlled trial to compare cure and relapse rate of paucibacillary multidrug therapy with monthly rifampicin, ofloxacin, and minocycline among paucibacillary leprosy patients in Agra District, India. Indian J Dermatol Venereol Leprol. (2015) 81:356–62. doi: 10.4103/0378-6323.159929
25. Priyanto MH, Yunifananda MS, Menaldi SLS, Nelwan EJ, and Marissa M. Optimizing treatment of lepromatous form of leprosy using ofloxacin on top of standard multi-drug therapy in National Referral Hospital, Jakarta, Indonesia. F1000Res. (2025) 14:252. https://f1000research.com/articles/14-252/v1 (Accessed March 10, 2025).
Keywords: HLA-B*13:01, leprosy, dapsone hypersensitivity syndrome, Ambon, Indonesia HLA-B13:01 DHS patients
Citation: Menaldi SLSW, Priyanto MH, Widaty S, Irawati Y, Aswin YA, Suryandari DA, Marissa M, Ariawan I, Kartika E, Yunifananda MS and Thio HB (2025) Identification of HLA-B*13:01 allele in leprosy patients with dapsone hypersensitivity syndrome in Ambon, Indonesia. Front. Trop. Dis. 6:1597392. doi: 10.3389/fitd.2025.1597392
Received: 21 March 2025; Accepted: 08 May 2025;
Published: 03 June 2025.
Edited by:
Natália Aparecida De Paula, University of Sao Paulo, BrazilReviewed by:
Leticia Gomes De Pontes, Departamento de Bioquímica e Imunologia da Universidade de Minas Gerais, BrazilEderson Ederson, University of Sao Paulo, Brazil
Copyright © 2025 Menaldi, Priyanto, Widaty, Irawati, Aswin, Suryandari, Marissa, Ariawan, Kartika, Yunifananda and Thio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mufqi Handaru Priyanto, bXVmcWkuaGFuZGFydTAxQHVpLmFjLmlk
 Sri Linuwih Susetyo Wardhani Menaldi
Sri Linuwih Susetyo Wardhani Menaldi Mufqi Handaru Priyanto
Mufqi Handaru Priyanto Sandra Widaty
Sandra Widaty Yunia Irawati
Yunia Irawati Yulia Ariani Aswin3
Yulia Ariani Aswin3 Malika Sabrina Yunifananda
Malika Sabrina Yunifananda Hok Bing Thio
Hok Bing Thio