- 1Department of Microbiology and Preclinical Research, National Medical Research Center for Phthisiopulmonology and Infection Disease, Yekaterinburg, Russia
- 2Department of Global Public Health and Primary Care, University of Bergen, Bergen, Norway
- 3LHL International, Oslo, Norway
- 4Department Phthisiopulmonology, Northern State Medical University, Arkhangelsk, Russia
- 5Research Department, Northern Arctic Federal University, Arkhangelsk, Russia
- 6Administration, National Medical Research Center of Phthisiopulmonology and Infection Disease, Moscow, Russia
Background: Nearly half of new tuberculosis patients in Sverdlovsk Oblast were diagnosed with multidrug-resistant tuberculosis (MDR-TB) and often exhibited fluoroquinolone resistance (FQ-R). This study aimed to (1) determine the number of MDR-TB patients who had Mycobacterium tuberculosis exhibiting the same genetic patterns (a unique combination of genotypes and mutations in genes associated with drug resistance) using the Russian microarray assay TB-TEST and (2) assess the correlation between these patterns and patient characteristics.
Materials and methods: We analyzed 345 MDR-TB DNA samples from patients with pulmonary TB in Sverdlovsk Oblast between 2017 and 2020 using the TB-TEST. We assumed that isolates with unique patterns, which were seen in only one patient, indicated minimal transmission of M. tuberculosis, while the presence of more isolates with the same pattern suggested a more recent transmission. All patients were categorized into three groups to ensure that each group was approximately of the same size: Group 1 consisted of unique patterns; Group 2 (the low-frequency patterns group) included patterns shared by 2–6 patients; and Group 3, (the high-frequency patterns group, “dominant”) included patterns shared by 7–18 patients.
Results: In total, 174 distinct genetic patterns were identified: Group 1 included unique patterns, accounting for 36.8%; Group 2 included low-frequency patterns, accounting for 31.0%; and Group 3 included high-frequency patterns, accounting for 32.2%. The Beijing B0/W148 genotype was found in 72.4% of cases, and it was significantly less frequent in patients with unique patterns (59.1% vs. 66.4% vs. 93.7%). Mutations in gyrA/B genes were found in 50.4% of all samples; however, these mutations were significantly more common in the group with unique patterns (73% vs. 43.9% and 30.6%). This suggests that the mutations in gyrA/B genes may have developed over the years because of inadequate chemotherapy regimens. Nevertheless, these mutations have not yet spread widely, possibly due to lower transmission potential or recent emergence. Patients with M. tuberculosis–positive sputum who had undergone treatment for more than 12 months demonstrated a significantly higher proportion of unique patterns and a higher rate of fluoroquinolone resistance.
Conclusion: Patients with unique patterns were found to have MDR-TB. However, a higher proportion of M. tuberculosis with mutations in gyrA/B genes in the group with unique patterns may indicate reduced transmissibility.
1 Introduction
In Russia, the incidence rate of tuberculosis (TB) decreased by 5.7% per year during 2010–2019, reaching 50 per 100,000 in 2019. However, the proportion of patients with multidrug-resistant (MDR) TB (rifampicin + isoniazid resistance) increased to an estimated 35% in new cases in 2019 (1). The prevalence of MDR-TB varies significantly within the country, from 7.8% in the Chechen Republic to 66.7% in the Nenets Autonomous Okrug in 2020 (2). Sverdlovsk Oblast also reported a high prevalence, with 48% of MDR-TB in new patients in 2020. Furthermore, in 2019, it was found that 23.5% of MDR-TB patients in Russia exhibited resistance to both fluoroquinolone (FQ) (i.e., pre-extensively drug-resistant [pre-XDR]) and second-line injectable drugs (3). The proportion of MDR-TB cases has increased since 1990, likely due to inadequate treatment regimens, non-adherence to treatment, limited financing for TB support services (4), and increasing levels of HIV infection. In 2019, there were 16,453 (23.3%) TB patients who were HIV positive in Russia (3). Drug resistance can be acquired during ineffective TB treatment or transmitted from individuals who already have MDR-TB (5). The success of bacterial strains depends on many factors, including increased transmission because of delay in diagnosis (of TB and resistance), crowded living conditions, resistant strains when adequate regimens are not followed, and the biological properties of Mycobacterium tuberculosis (6), one of which may belong to a certain genetic line.
Beijing B0/W148 is recognized as an evolutionarily successful genetic clone of M. tuberculosis; it has been detected in several regions of Russia (7–9), including Sverdlovsk Oblast (10, 11), and is often associated with MDR-TB (10, 12, 13). However, the question remains regarding the contribution of different genotypes of M. tuberculosis, each with different combinations of drug-resistant mutations, to the MDR-TB epidemic. It is known that a higher number of resistant mutations is associated with reduced transmission rates, suggesting a global decrease in fitness on gradual accumulation of mutations conferring resistance (14). Many studies describe “compensatory mutations” that enable bacteria to restore their viability and adapt to competing conditions (15–17). Compensatory evolution allows resistant bacteria to survive even in the absence of antibiotic selection pressure (18, 19). Therefore, not all resistant bacterium variants have the same epidemiological significance, but those with additional genetic advantages tend to transmit more successfully than bacteria without these advantages.
Genotyping serves as a powerful tool for unraveling the epidemiological significance of diverse microbial genetic variants. Since 2014, the microarray-based TB-TEST (BIOCHIP-IMB, Russia) has been included in the Russian clinical recommendations (20), allowing the study of specific genotypes (21) endemic to Russia and mutations associated with drug resistance to rifampicin, isoniazid, ethambutol, aminoglycosides, and fluoroquinolones.
The specific objectives of this study, using data collected between 2017 and 2020 from MDR-TB patients at the Ural Research Institute of Phthisiopulmonology (UNIIF), were (1) to determine the number of MDR-TB patients who had M. tuberculosis with the same pattern (a unique combination of genotype and drug-resistant mutations) using the Russian microarray assay TB-TEST and (2) to assess the correlation between groups of these patterns and patient characteristics (gender, age, HIV/hepatitis co-infection, duration of treatment, and type of clinical material). We assumed that isolates with unique patterns indicated minimal transmission of M. tuberculosis, while more samples of M. tuberculosis sharing the same pattern suggested a more recent transmission.
2 Materials and methods
2.1 Setting
The population of Sverdlovsk Oblast was 4,310,681 in 2020, including 1,526,000 in Yekaterinburg, the administrative center of the Ural Federal District and Sverdlovsk Oblast. The notification rate of new TB cases per 100,000 population in Sverdlovsk Oblast decreased from 101 in 2003 to 52 in 2020. The proportion of MDR-TB among new cases gradually increased from 4% in 2003 to a very high but stable level of 47% in 2017 and 48% in 2020. HIV infection rates among TB patients in Sverdlovsk Oblast increased from 5% in 2006 to 40% in 2020 (3, 23). In 2018, among newly diagnosed TB patients with resistance to at least rifampicin, the treatment outcomes were as follows: 62% achieved successful treatment, 12% experienced failure, 14% resulted in death, 7.4% had treatment interruptions, and 4.6% continued treatment (22).
2.2 Study population
The UNIIF, a branch of the National Medical Research Center of Phthisiopulmonology and Infection Disease (NMRC), is located in Yekaterinburg. The institute provides advanced medical care to TB patients from different regions of Russia, with 60%−70% of patients coming from Sverdlovsk Oblast. The main reasons for hospitalization include diagnosis (particularly for complex cases where M. tuberculosis is absent in sputum), surgical treatment of TB (pulmonary resections and thoracoplasties), and treatment for other lung pathologies and MDR-TB. The lack of clinical and radiological improvement, despite receiving appropriate chemotherapy for TB, indicates the need for hospitalization in our clinic for further chemotherapy or surgical intervention. HIV-positive TB patients are admitted to the UNIIF only if their CD4 count is above 400 cells/ml; patients with lower CD4 counts are usually referred to specialized AIDS-TB hospitals in the Sverdlovsk region.
In this study, we included only patients diagnosed with pulmonary TB who were residing in Sverdlovsk Oblast and receiving treatment at the UNIIF–NMRC. The samples were processed in the microbiology laboratory and underwent polymerase chain reaction (PCR) testing between January 2017 and December 2020, with corresponding TB-TEST results available. We included both previously untreated TB patients (new cases) and TB patients who had been previously treated with first- and second-line drugs.
2.3 Bacteriological and molecular-genetic methods
The biological materials included sputum, bronchoalveolar lavage fluid, and lung tissues. Each sample was examined using microscopy, culture techniques, and molecular genetic tests.
DNA extraction and real-time PCR for the genetic markers of M. tuberculosis complex were carried out with the Amplitub-RV kit (Syntol, Russia), following the manufacturer's instructions. If the DNA from M. tuberculosis was detected in the clinical sample in sufficient quantity to identify mutations associated with drug resistance, the sample was further examined using the TB-TEST (BIOCHIP-IMB, Russia), according to the manufacturer's instructions. The assay is based on the amplification of 17 fragments of the M. tuberculosis genome using the universal primer adapter technique and heat pulses at the elongation step, followed by hybridization on a microarray. The TB-TEST determines the genotypes of M. tuberculosis that are endemic to the Russian Federation. These genotypes include Beijing, Beijing B0/W148, LAM, Haarlem, and Ural, among others. This TB-TEST also detects 116 mutations carrying drug resistance in the following genes: rpoB, katG, inhA, ahpC, gyrA, gyrB, rrs, eis, and embB.
In 2015, the World Health Organization (WHO) recognized the UNIIF-NMRC as a National Centre of Excellence for the WHO TB Supranational Reference Laboratory Network. Since 2017, this laboratory has taken part in national and international quality assurance programs for testing TB drug susceptibility using the TB-TEST.
The diagnostic material was inoculated into Löwenstein–Jensen and Mycobacteria Growth Indicator Tube (MGIT) media following the Russian clinical recommendations (20). If drug-resistance mutations were not obtained directly from the biological materials, a culture was used.
We only analyzed the results of the phenotypic drug-susceptibility test (DST) in cases where an obtained M. tuberculosis culture had the mutation status of “Associated with R–Interim” or “Uncertain significance,” according to the Catalog of Mutations in Mycobacterium Tuberculosis Complex (24). During 2017–2020, our laboratory employed the absolute concentration method for drug susceptibility testing using the Löwenstein–Jensen medium, Sensititre Mycobacterium tuberculosis MYCOTBI AST Plate, and BACTEC MGIT system.
2.4 Variables and data collection
The database with drug-resistance mutations and genotypes also included patient information on gender, age, diagnosis, region of residence, comorbidities (HIV and hepatitis), vulnerable groups, treatment duration and treatment regimen, and sample types (respiratory or surgical). The data sources included clinical records that were extracted and entered into a spreadsheet. The database was accessed between June 2022 and March 2023.
2.5 Analysis
We defined genetic patterns based on a combination of M. tuberculosis genotypes and drug-resistance mutations and counted the number of patients who were infected by M. tuberculosis with the same combinations (patterns). We assumed that M. tuberculosis with unique patterns represented minimal transmission, and the more patients with the same M. tuberculosis pattern, the more that transmission had probably taken place, hence biologically successful variants. We analyzed the respiratory and surgical samples separately, considering the tuberculosis treatment duration. Cross-tabulation was done, and the data were analyzed using a chi-square test. Differences were considered significant at p < 0.01.
2.6 Ethical approval
This study was approved by the Local Ethics Committee of the National Medical Research Center for Phthisiopulmonology and Infection Disease, Ekaterinburg, Russia (approval protocol No 102, 24.11.2021).
3 Results
From January 2017 to December 2020, a total of 2,735 TB patients were examined in the UNIIF-NMRC clinic, and 748 had positive DNA results for M. tuberculosis from clinical samples and/or culture. Of these patients, we only included patients from Sverdlovsk Oblast who had bacterial mutations conferring resistance to rifampicin and isoniazid, which accounted in 345 patients; of these 345 patients, 193 (55.9%) had culture results available.
3.1 General characteristics of patients
Table 1 shows the clinical and demographic characteristics of the patients. Of all the patients, 67.5% were men, and the median age was 37 years. There were 62 patients (17.9%) with HIV/TB co-infection and 82 (23.8%) with hepatitis; 76 (22.0%) of them were former prisoners; 62 (17.9%) patients were Yekaterinburg citizens, while the others were from different places in Sverdlovsk Oblast. In total, 142 (41.2%) DNA samples were isolated from the sputum or bronchoalveolar lavage fluid samples collected from patients before TB treatment and during treatment with second-line TB drugs for varying durations; 125 (88.0%) respiratory samples were culture-positive. In total, 203 (58.8%) DNA samples were obtained from the biological material, of which 142 (69.9%) had M. tuberculosis–positive sputum before surgery and had different durations of TB treatment with second-line TB drugs; only 68 (33.5%) surgical samples were culture-positive. The remaining 61 (30.1%) samples were M. tuberculosis–negative before surgery, and these patients neither received treatment nor were treated with first- or second-line drugs before surgery.
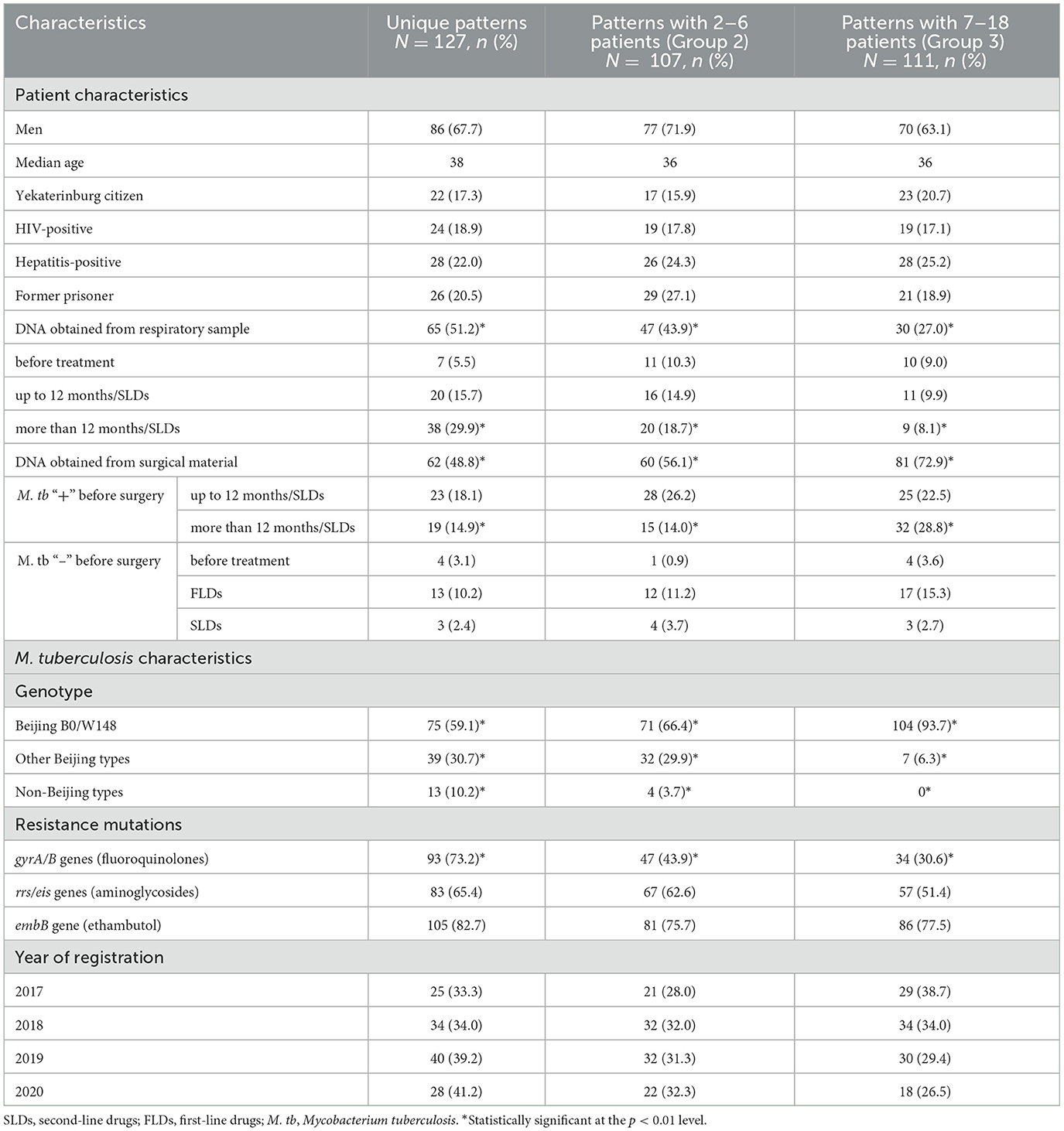
Table 1. Patterns of pulmonary MDR tuberculosis by patient and M. tuberculosis characteristics in Sverdlovsk oblast, Russia, 2017–2020.
3.2 Genotype and drug-resistant mutations
Table 1 also shows the number of patients infected by M. tuberculosis belonging to various genotypes and mutations in three pattern groups. Figure 1 illustrates the combination of mutations in the gyrA/B genes (FQs), rrs/eis genes (aminoglycosides), and embB gene (ethambutol) for all M. tuberculosis cases. The Beijing genotype was found in 328 (95.1%) cases and, of these, 250 (72.4%) belonged to the Beijing B0/W148 cluster. In total, 272 cases (78.8%) of M. tuberculosis had mutations in the embB gene, 207 cases (60.0%) had mutations in the rrs/eis genes, and 174 cases (50.4%) had mutations in the gyrA/B genes.
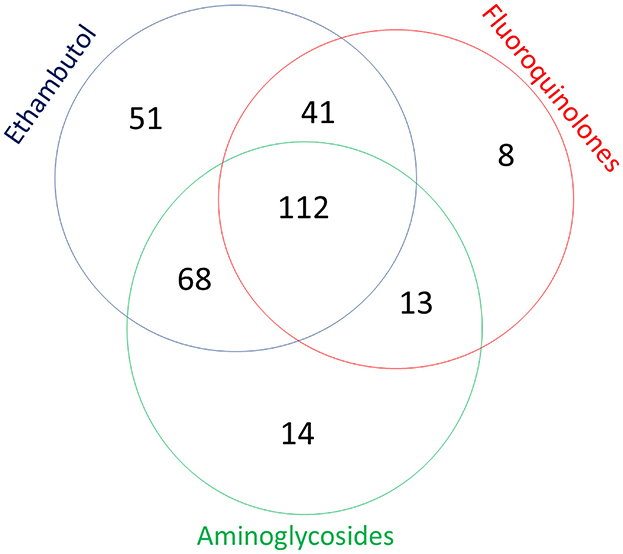
Figure 1. Combination of mutations in genes associated with resistance to fluoroquinolones, aminoglycosides, and ethambutol of 345 Mycobacterium tuberculosis cases detected in Sverdlovsk Oblast, 2017–2020.
Table 2 shows the types of drug-resistant mutations by genotype. The majority of identified mutations were within hotspot codons. The most frequent mutation to rifampicin was rpoBS531L, and it was present in 306 (88.7%) samples. For isoniazid resistance, the mutation katGS315T(1) was identified in 311 (90.1%) samples. In one case, mutation katG I335V with uncertain significance was detected, but a clinical isolate was not obtained. There were 29 different types of mutations and their combinations in gyrA and/or gyrB genes; the most common mutations were M. tuberculosis with gyrAD94G plus gyrAS95T mutations (61; 35.1%) and gyrAA90V (46; 26.4%). There were four samples with gyrAG88A mutation (uncertain significance), according to phenotypic DST. All isolates were found to be resistant to ofloxacin (2 mg/L) using the absolute concentration method. Additionally, two isolates were found to be resistant to moxifloxacin (0.25 mg/L) using the Bactec MGIT system, while resistance testing was not conducted for the other two isolates.
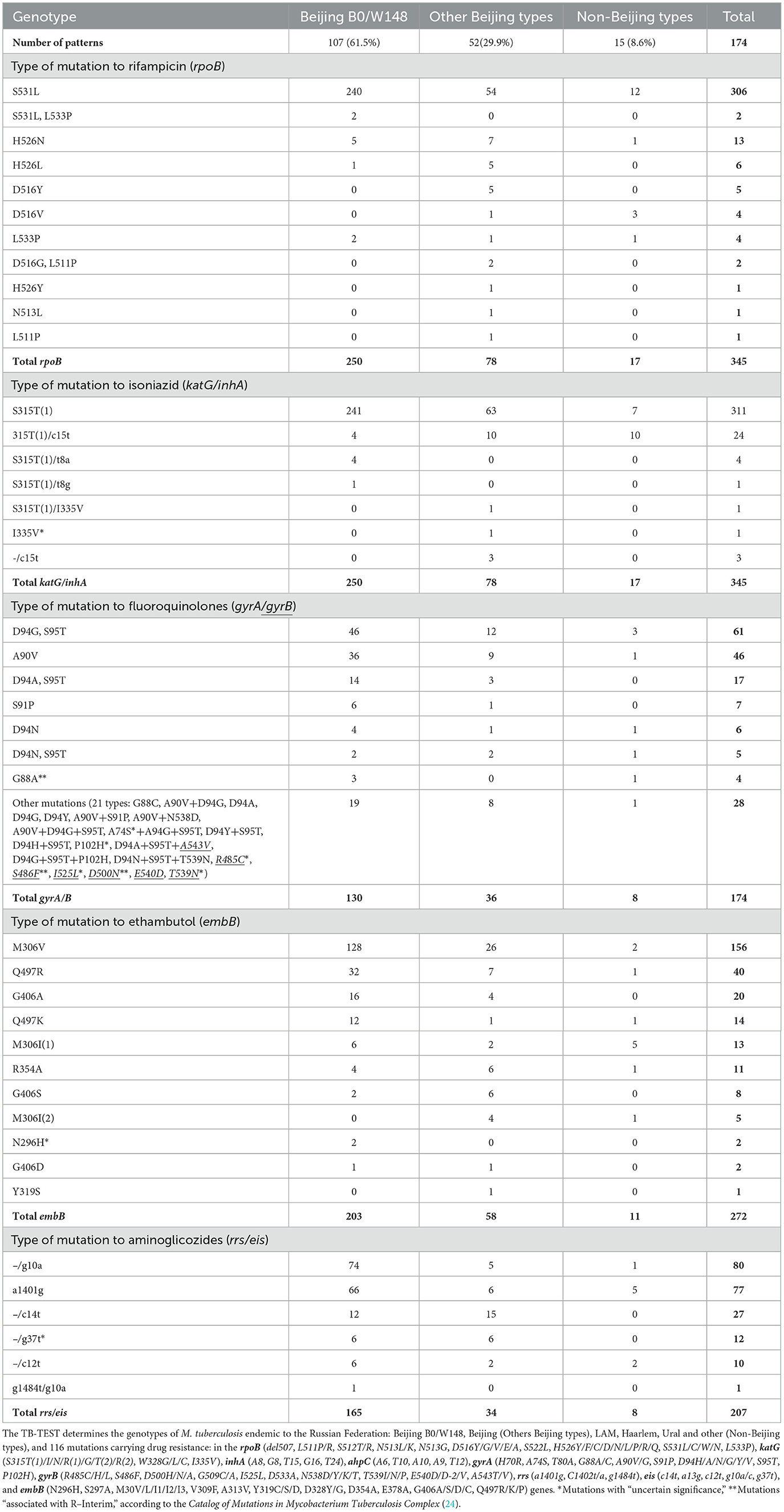
Table 2. Mutations in genes associated with drug resistance in M. tuberculosis by genotype, in Sverdlovsk Oblast, 2016–2020.
In six cases, mutations in the gyrB gene were detected. Clinical isolates were not obtained for two patients with M. tuberculosis containing gyrB mutations: R485C (uncertain significance) and S486F (associated with R–Interim). Clinical isolates with resistance to ofloxacin (2 mg/L) (absolute concentration method) and moxifloxacin (0.25 mg/L) using the BACTEC MGIT system were obtained for patients with M. tuberculosis gyrB mutations: T539N and I525L (uncertain significance) and D500N (associated with R–Interim). There were 11 different mutations in the embB gene associated with ethambutol resistance, and M306V was present in 156 (57.3%) samples. For two cases with M. tuberculosis embB mutation N296H, a clinical isolate was obtained from one case, and it was resistant to ethambutol (MIC 8 mg/L on Sensititre Mycobacterium tuberculosis MYCOTBI AST Plate). For resistance to aminoglycosides, two mutations were dominant: rrs a1401g, found in 77 (37.2%) samples, and eis g10a, found in 80 (38.6%) samples. Of the 12 M. tuberculosis samples with eis g37t mutation (uncertain significance), clinical isolates were obtained for 8 samples, and 7 of these isolates were resistant to kanamycin (30 mg/L, absolute concentration method).
3.3 Genetic patterns of MDR M. tuberculosis
In total, 174 different genetic patterns (a combination of M. tuberculosis genotypes and drug resistance mutations) were obtained from the 345 M. tuberculosis samples. We divided the patients into three groups: Group 1 included 127 patients with unique M. tuberculosis genetic patterns; Group 2 consisted of 107 patients with low-frequency patterns (37 patterns), where each pattern had 2–6 patients (21 sets of 2 patients, 7 sets of 3 patients, 4 sets of 4 patients, 2 sets of 5 patients, and 3 sets of 6 patients); and Group 3 comprised of 111 patients with high-frequency genetic patterns (10 patterns) that had more than 7 patients. The categorization into Group 2 and Group 3 was made conditionally so that the groups would be approximately the same size, allowing us to compare the impact of genetic diversity.
The proportions of patients belonging to the three groups of patterns are presented in Table 1. The proportion of Beijing B0/W148 in Group 3 (93.7%) was significantly higher than in Group 2 (66.7%) and Group 1 (59.1%). The proportion of M. tuberculosis with mutations in gyrA/B genes was significantly higher in Group 1 (73.2%) than in Group 2 (43.9%) and Group 3 (30.6%). The proportion of patients with M. tuberculosis obtained from respiratory material was significantly higher in Group 1 (51.2%) than in Group 2 (43.9%) and Group 3 (27.0%). For surgical samples, the proportions of patients were 48.8% in Group 1, 56.1% in Group 2, and 72.9% in Group 3.
An analysis of patterns across different categories of patients revealed that unique patterns were significantly more prevalent in patients with M. tuberculosis–positive sputum and a treatment duration exceeding 12 months (median: 44 months; 29.9%; Table 1). This patient group also exhibited a higher proportion of mutations in gyrA/B genes (91.0%) and included HIV-positive patients (43.3%), hepatitis B/C–positive patients (44.8%), and former prisoners (50.7%; Table 3).
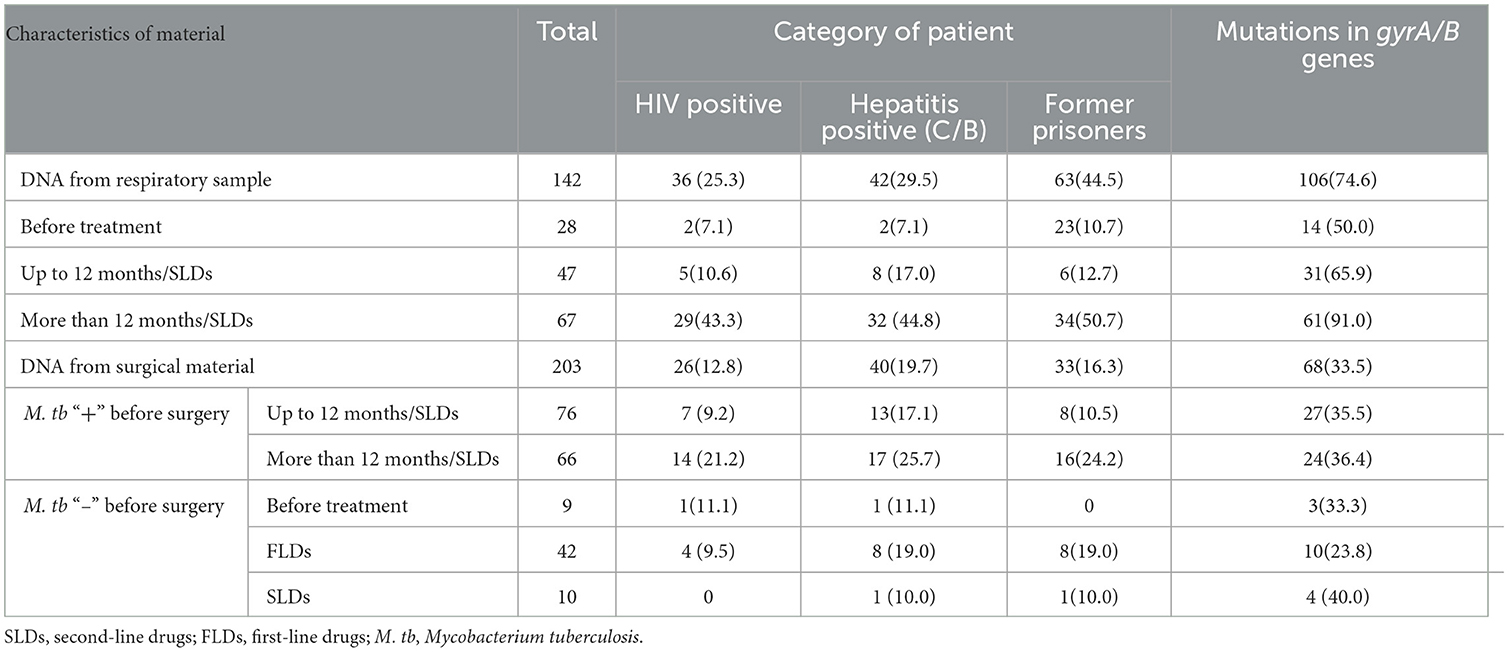
Table 3. Characteristics of patients with multidrug-resistant pulmonary tuberculosis and their examined surgical material in Sverdlovsk Oblast, 2017–2020.
In patients with M. tuberculosis–positive sputum, the proportion of mutations in gyrA/B genes depended on the duration of treatment: before treatment (50.0%), up to 12 months (65.9%), and more than 12 months (91.0%; Table 3). The three groups of patterns were not significantly different in the other variables.
In Table 4, the genetic patterns in Group 3 (patterns with more than seven patients) are listed. All patterns except one belonged to the Beijing B0/W148 genotype and had the same mutations katG S315T1 and rpoB S531L. The variability was due to the presence or absence of mutations in embB (M306V or Q497R), gyrA (D94G+S95T or A90V), rrs (a1401), and eis (g10a) genes and, for each drug, only two types of mutations were noted. The proportion of categories of patients (new cases, duration of treatment, HIV/hepatitis, and prisoners) varied among the patterns. In general, the duration of tuberculosis treatment before obtaining samples was longer in cases in which mutations in gyrA/B genes were present (Group 3 patterns). However, due to the small number of samples in each pattern, no statistically significant differences were identified.
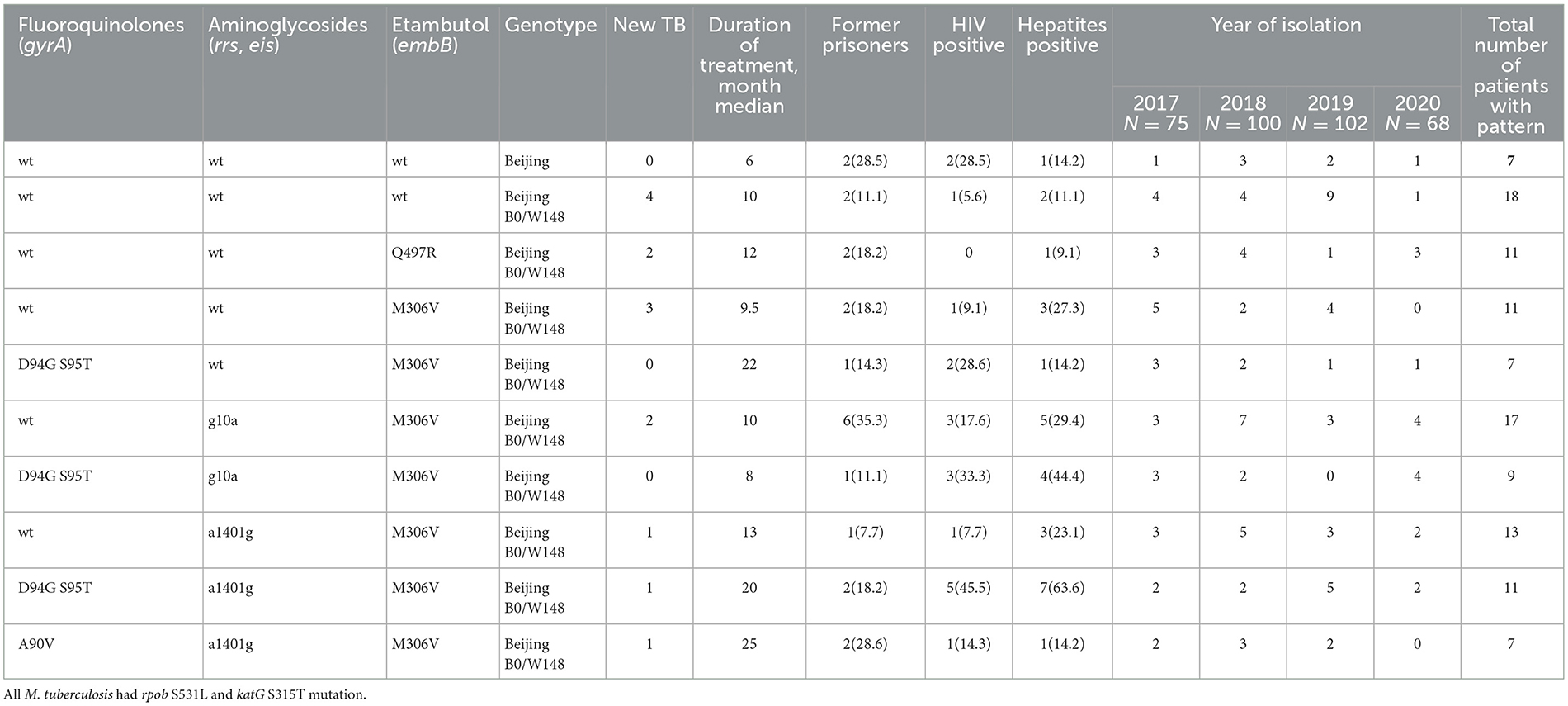
Table 4. Mutations and genotypes of the most common genetic patterns (Group 3) of Mycobacterium tuberculosis and clinical characteristics of patients, in Sverdlovsk Oblast, 2017–2020.
4 Discussion
In this study, we identified 174 distinct M. tuberculosis genetic patterns among 345 patients with MDR-TB. Of these, 36.8% were unique, 31.0% shared the same pattern with 2–6 others, and 32.2% shared the same pattern with 7–18 others. The Beijing B0/W148 genotype was more frequent in the group with 7–18 identical combinations; unique patterns were significantly more common in the patient group with M. tuberculosis–positive respiratory samples and a treatment duration exceeding 12 months. This group also exhibited a higher proportion of mutations in gyrA/B genes and included HIV-positive patients, hepatitis (B/C)–positive patients, and former prisoners.
These observations suggest that patients with “unique” M. tuberculosis patterns have poorer treatment adherence and may have been the primary source of the new pathogen variants, mainly due to the emergence of mutations in gyrA/B genes. The emergence of FQ resistance (often with a wide range of mutations and a larger number of unique profiles) may lead to less effective chemotherapy (or may result from inadequate chemotherapy), consequently resulting in ongoing bacterial excretion. The pathogen's biological traits (fitness) or the brief period after their emergence may have contributed to our observation that the variants were identified as unique.
The apparent increase in FQ resistance may also be related to the quality of laboratory diagnostics. Until 2014, in Sverdlovsk Oblast, identifying FQ susceptibility was based on ofloxacin using the absolute concentration method. Molecular genetic testing, since 2014, and the BACTEC MGIT, since 2018, determined mutations to FQ and susceptibility to levofloxacin and moxifloxacin, respectively. The WHO, in 2018, and Russian clinical recommendations, in 2022, lowered the critical concentration for BACTEC MGIT from 1.5 mg/L to 1 mg/L for levofloxacin and from 0.5 mg/L to 0.25 mg/L for moxifloxacin. This change suggests that, in previous years, we may have underdiagnosed real FQ resistance based on phenotypic tests, leading to inadequate TB treatment and a potential increase in TB drug resistance. These factors may account for the wide range of unique genetic patterns associated with different mutation types in gyrA/B genes, which we detected in our study.
Our data confirm that several factors are associated with the high levels of MDR/pre-XDR TB in Sverdlovsk Oblast. One significant factor is the dominance of the Beijing genotype (in particular, Beijing B0/W148, which was more frequent in the high-frequency pattern group). This strain has been circulating in Sverdlovsk Oblast (10, 11) and other regions inside and outside of Russia (7–9, 12, 14) for several decades. It is known that a higher mutation rate accounts for the higher adaptation ability and drug-resistance rates of the Beijing lineage (25, 26). The second factor is the high prevalence of HIV infection in the Sverdlovsk region, which may facilitate the circulation of drug-resistant mycobacteria with reduced fitness, allowing for further evolution. The relatively low treatment success rate of MDR-TB patients (62% in 2020; (22)) indicates that M. tuberculosis has the opportunity to evolve and acquire new mechanisms of drug resistance.
Upon analyzing the trend in the proportion of M. tuberculosis belonging to different pattern groups, we observed that over the 4-year period, the proportion of M. tuberculosis patterns gradually increased from 33.3% to 41.2%, while the proportion of M. tuberculosis with dominant patterns gradually decreased from 38.7% to 26.5%. This may be because new mutations accumulate faster than the spread of the pathogen's successful variants; however, a longer observation period is necessary to establish statistical significance.
The studies from different regions of Russia and beyond indicate that FQ resistance is significantly more likely to be acquired than other resistance genotypes, suggesting that gyrAB mutants may have impaired transmission fitness, which impedes the spread of MDR/FQ resistant clones (11, 27–29). An analysis of whole-genome sequencing data regarding Beijing B0/W148 isolates revealed that the gradual accumulation of resistance mutations is consistently negatively associated with epidemiological success; however, this association can be weakened by compensatory mutations in XDR isolates, which can neutralize the negative impact of resistance mutations on mycobacterial fitness (14). In this study, we identified several dominant patterns (combined 7–18 patients), including those involving mutations in gyrA/B genes (mainly gyrA D94G S95T). These patterns were observed across all patient groups, suggesting their potential for successful transmission. This finding warrants further investigation.
A limitation of the study was the insufficient discriminative power of the TB-TEST for genotyping, which only allowed categorization based on the detection of single nucleotide polymorphisms in some specific loci. As a result, we could not identify clusters that require MIRU-VNTR (mycobacterial interspersed repetitive units, variable number of tandem repeats) or whole-genome sequencing, and we lacked information about epidemiological links. However, the TB-TEST is useful for monitoring trends in genotype frequency and resistance mutation patterns, especially in cases when M. tuberculosis isolates are not obtained, and for identifying groups that need special follow-up.
Considering that the clinical isolates of M. tuberculosis were obtained from only 55.9% of patients, we performed phenotypic drug susceptibility tests only in cases with positive culture results and when mutations categorized as “uncertain significance” or “associated with R–Interim,” according to the Catalog of Mutations in Mycobacterium Tuberculosis Complex (24), were detected. We observed a strong correlation between the results of the TB-TEST and the phenotypic methods in these cases.
We included all patients hospitalized in the UNIIF-NMRC clinic with pulmonary MDR-TB during a 4-year study period (2017–2020), who had TB-TEST results; however, estimates suggest that this sample size represented only approximately 5% of all MDR-TB cases in Sverdlovsk Oblast, indicating that it was a highly selected group. Therefore, the proportion of patients with unique strains is likely higher than the rest of the MDR cases in the oblast. However, within our selected group of patients, we found different transmission potentials in our sample groups, which is of great interest from a public health perspective. However, the patients included in our study were a selected group of “problem cases,” and we cannot be certain that they represent the whole oblast or all MDR-TB cases, especially new TB cases. We likely overlooked TB patients with advanced HIV, as those with CD4 count of <400 cells/ml were not admitted to the UNIIF hospital.
Monitoring drug resistance in TB is an important aspect of evaluating a TB program. This study offers new insights into categorizing TB patients based on the patterns of M. tuberculosis genotypes and resistance mutations using a standard microarray assay TB-TEST as a screening method. Given the high prevalence of MDR-TB in Sverdlovsk Oblast, these data allow us to identify the most common patterns, suggesting the biological success of the TB pathogen and its transmission.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
TU: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Validation, Writing – original draft, Writing – review & editing. EM: Writing – review & editing. LL: Writing – review & editing. SH: Supervision, Writing – review & editing. EH: Conceptualization, Methodology, Supervision, Writing – review & editing. AM: Supervision, Writing – review & editing, Methodology. DV: Writing – review & editing. IV: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and publication of this article. LHL-International with support of the Norwegian Ministry of Health and Care Services funded the SORT IT Course, which helped prepare this study. The University of Bergen funded the publication fees.
Acknowledgments
We would like to thank Nina Nizovtseva, Aleksandra Staritsyna, and the Easy Breathing Charity Fund for their excellent organization of the SORT IT Course.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HW declared a shared affiliation with the author(s) SH to the handling editor at the time of review.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Global Tuberculosis Report 2020. Geneva: World Health Organization (2020).
2. Sterlikov SA, Nechaeva OB, Son IM, Ponomarev SB, Kucherryavaya DA, Obuhova OV, et al. Industry and Economic Indicators of TB Work in 2019–2020. Analytical Review of Key Indicators and - Statistic Materials. Moscow: RIO TsNIIOIZ (2021).
3. World Health Organization. Tuberculosis Surveillance and Monitoring in Europe 2021: 2019 daTa. Geneva: World Health Organization (2021).
4. Nechaeva OB, Skachkova EI, Fomina NI. Drug resistance of Mycobacterium tuberculosis in the Sverdlovsk Region. Probl Tuberk. (2002) 9:8–11. Available online at: https://pubmed.ncbi.nlm.nih.gov/12524978/
5. Kendall EA, Fofana MO, Dowdy DW. The burden of transmitted multi-drug resistance among epidemics epidemics of tuberculosis : a transmission model. Lancet Respir Med. (2015) 3:963–72. doi: 10.1016/S2213-2600(15)00458-0
6. Alame Emane AK, Guo X, Takiff HE, Liu S. Highly transmitted M. tuberculosis strains are more likely to evolve MDR/XDR and cause outbreaks, but what makes them highly transmitted? Tuberculosis. (2021) 129:102092. doi: 10.1016/j.tube.2021.102092
7. Vyazovaya A, Proshina E, Gerasimova A, Avadenii I, Solovieva N, Zhuravlev V, et al. Increased transmissibility of Russian successful strain Beijing B0/W148 of Mycobacterium tuberculosis: indirect clues from history and demographics: drug-resistant Tuberculosis in Russia. Tuberculosis. (2020) 122:101937. doi: 10.1016/j.tube.2020.101937
8. Vyazovaya AA, Lebedeva IA, Ushakova NB, Pavlov VV, Gerasimova AA, Solovieva NS, et al. Molecular and genetic analysis of Mycobacterium tuberculosis population in the Vologda Region with low tuberculosis incidence. Russ J Infect Immun. (2021) 11:497–505. doi: 10.15789/2220-7619-MAG-1545
9. Pasechnik OA, Vyazovaya AA, Bloch AI, Yarusova IV, Tatarintseva MP, Mokrousov IV. Assessment of the prevalence and epidemic spread of strains of ancient, and modern sublineages of the Mycobacterium tuberculosis beijing genotype in Omsk region. Epidemiol i Vaktsinoprofilaktika. (2020) 19:20–9. doi: 10.31631/2073-3046-2020-19-4-20-29
10. Umpeleva TV, Vyazovaya AA, Eremeeva NI, Kravchenko MA, Navarskaya OV, Skornyakov SN. Specific genetic features of tuberculous mycobacteria in Ural federal district of Russia. Tuberc lung Dis. (2016) 94:60–5. doi: 10.21292/2075-1230-2016-94-8-60-65
11. Umpeleva T, Belousova K, Golubeva L, Boteva T, Morozova I, Vyazovaya A, et al. Molecular characteristics of Mycobacterium tuberculosis in the “closed” Russian town with limited population migration. Infect Genet Evol. (2020) 79:104174. doi: 10.1016/j.meegid.2020.104174
12. Gupta A, Sinha P, Nema V, Gupta PK, Chakraborty P, Kulkarni S, et al. Detection of Beijing strains of MDR M. tuberculosis and their association with drug resistance mutations in katG, rpoB, and embB genes. BMC Infect Dis. (2020) 20:1–7. doi: 10.1186/s12879-020-05479-5
13. Afanas'ev MV, Ikryannikova LN, Il'Ina EN, Kuz'Min AV, Larionova EE, Smirnova TG, et al. Molecular typing of Mycobacterium tuberculosis circulated in Moscow, Russian Federation. Eur J Clin Microbiol Infect Dis. (2011) 30:181–91. doi: 10.1007/s10096-010-1067-z
14. Merker M, Rasigade JP, Barbier M, Cox H, Feuerriegel S, Kohl TA, et al. Transcontinental spread and evolution of Mycobacterium tuberculosis W148 European/Russian clade toward extensively drug resistant tuberculosis. Nat Commun. (2022) 13:5105. doi: 10.1038/s41467-022-32455-1
15. Li QJ, Jiao WW, Yin QQ, Xu F, Li JQ, Sun L, et al. Compensatory mutations of rifampin resistance are associated with transmission of multidrug-resistant Mycobacterium tuberculosis Beijing genotype strains in China. Antimicrob Agents Chemother. (2016) 60:2807–12. doi: 10.1128/AAC.02358-15
16. Zur Wiesch PS, Engelstädter J, Bonhoeffer S. Compensation of fitness costs and reversibility of antibiotic resistance mutations. Antimicrob Agents Chemother. (2010) 54:2085–95. doi: 10.1128/AAC.01460-09
17. Gagneux S. Fitness cost of drug resistance in Mycobacterium tuberculosis. Clin Microbiol Infect. (2009) 15:66–8. doi: 10.1111/j.1469-0691.2008.02685.x
18. Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. (2012) 44:106–10. doi: 10.1038/ng.1038
19. Ma P, Luo T, Ge L, Chen Z, Wang X, Zhao R, et al. Compensatory effects of M. tuberculosis rpoB mutations outside the rifampicin resistance-determining region. Emerg Microbes Infect. (2021) 10:743–52. doi: 10.1080/22221751.2021.1908096
20. Chernousova LN, Sevastyanova EV, Larionova EE, Smirnova TG, Andreevskaya SN, Popov SA et al. Federal clinical recommendation for microbiology and molecular-genetic diagnostic of tuberculosis. Moscow: Russian Society of Phthisiologists (2015) [In Russian].
21. Zimenkov DV, Kulagina EV, Antonova OV, Zhuravlev VY, Gryadunov DA. Simultaneous drug resistance detection and genotyping of Mycobacterium tuberculosis using a low-density hydrogel microarray. J Antimicrob Chemother. (2016) 71:1520–31. doi: 10.1093/jac/dkw015
22. Tsybikova T. Incidence of tuberculosis in subjects of the Russian federation in 2020. Soc Asp Popul Heal. (2022) 68:10 [In Russian]. doi: 10.21045/2071-5021-2021-68-2-10
23. Podgaeva VA. Epidemic Situation on Tuberculosos and TB Service in the Ural in 2011 (statistic materials). Yekaterinburg: UNIIF. (2012).
24. World Health Organization. Catalogue of Mutations in Mycobacterium Tuberculosis Complex and Their Association with Drug Resistance. Second edition. Geneva: World Health Organization (2023).
25. Hakamata M, Takihara H, Iwamoto T, Tamaru A, Hashimoto A, Tanaka T, et al. Higher genome mutation rates of Beijing lineage of Mycobacterium tuberculosis during human infection. Sci Rep. (2020) 10:1–11. doi: 10.1038/s41598-020-75028-2
26. Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. (2013) 45:784–90. doi: 10.1038/ng.2656
27. Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Kontsevaya I, Corander J, et al. Evolution and transmission of drug resistant tuberculosis in a Russian population. Nat Genet. (2014) 46:279–89. doi: 10.1038/ng.2878
28. Andreevskaya SN, Smirnova TG, Chernousova LN, Larionova EE, Kiseleva EA EA. The nature of genotypic resistance to fluoroquinolones in Mycobacterium tuberculosis circulating in Russian Federation. Bull RSMU. (2022) 5:15–22. doi: 10.24075/brsmu.2022.054
Keywords: tuberculosis, MDR, genotyping, mutation, TB-TEST
Citation: Umpeleva T, Mazurina E, Lavrenchuk L, Hinderaker SG, Heldal E, Mariandyshev A, Vakhrusheva D and Vasilieva I (2025) Patterns of Mycobacterium tuberculosis genotypes and resistance mutations in patients with pulmonary multidrug-resistant tuberculosis in Sverdlovsk Oblast, Russia. Front. Tuberc. 3:1406536. doi: 10.3389/ftubr.2025.1406536
Received: 22 April 2024; Accepted: 28 April 2025;
Published: 23 May 2025.
Edited by:
Seyed Ehtesham Hasnain, Indian Institute of Technology Delhi, IndiaReviewed by:
Djaltou Aboubaker Osman, Center of Study and Research of Djibouti (CERD), EthiopiaJavaid Ahmad Sheikh, Jamia Hamdard University, India
Harald Gotten Wiker, University of Bergen, Norway
Copyright © 2025 Umpeleva, Mazurina, Lavrenchuk, Hinderaker, Heldal, Mariandyshev, Vakhrusheva and Vasilieva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sven G. Hinderaker, c3Zlbi5oaW5kZXJha2VyQHVpYi5ubw==
 Tatiana Umpeleva
Tatiana Umpeleva Elena Mazurina1
Elena Mazurina1 Sven G. Hinderaker
Sven G. Hinderaker Einar Heldal
Einar Heldal Andrei Mariandyshev
Andrei Mariandyshev