- 1Department of Veterinary Pathology and Public Health, Agronomic and Veterinary Institute Hassan II, Rabat, Morocco
- 2African Genome Center, Mohammed VI Polytechnic University, Ben Guerir, Morocco
- 3Milken Institute School of Public Health, George Washington University, Washington, DC, United States
- 4Medical Biology Department, Molecular Biology Laboratory, Pasteur Institute of Morocco, Casablanca, Morocco
Fowl adenoviruses (FAdV) are prevalent in chickens worldwide, responsible for several poultry diseases, including inclusion body hepatitis (IBH), hepatitis-hydropericardium syndrome (HHS), and gizzard erosion (GE), which result in significant economic losses in the poultry industry. Consequently, detection and efficient identification of FAdV serotypes are becoming extremely urgent to monitor outbreaks and develop vaccination strategies. Conventional PCR (cPCR) tests, combined with Restriction Fragment Length Polymorphism (RFLP) or sequencing, were developed for FAdV diagnosis. Although these molecular tests have considerably improved the accuracy of FAdV diagnosis compared with conventional methods, certain drawbacks remain unresolved, including lack of sensitivity and post-PCR analysis. Subsequently, advanced molecular technologies such as real-time PCR (qPCR), Loop Isothermal Amplification (LAMP), Cross-Priming Amplification (CPA), Recombinase Polymerase Amplification (RPA), Digital Droplet Polymerase Chain Reaction (ddPCR), Dot Blot Assay Combined with cPCR, Nanoparticle-Assisted PCR (nano-PCR), PCR-Refractory Quantitative Amplification (ARMS-qPCR), CRISPR/Cas13a Technology, and High-Resolution Melting Curve (HRM), have been developed to improve FAdV diagnosis.
1 Introduction
Fowl adenoviruses (FAdVs) belong to Adenoviridae family, and the aviadenovirus genus, which comprises avian adenoviruses that share a common antigen (1). Based on whole genome analysis and viral neutralization (VN) test, FAdVs are divided into 5 species (A to E) and 12 serotypes (1-8a, 8b-11), respectively (2, 3). Countries such as the United States of America (USA), the European Union (EU), Australia, and Japan have established their classification systems based on local strains. However, this lack of standardization in serotype numbering can lead to confusion and misinterpretation when comparing articles and research results (4). Therefore, the international committee on taxonomy of viruses (ICTV) has published an international classification system that researchers are required to use in their publications (5).
Due to their vertical and horizontal transmission (6–10), FAdVs are widespread throughout the world (11, 12) and are associated with significant economic losses in the poultry industry. Although most infections are subclinical, some FAdV serotypes are associated with impactful poultry diseases such as inclusion body hepatitis (IBH) (13, 14), adenoviral gizzard erosion (AGE) (15, 16), pancreatic necrosis (17), and hepatitis-hydropericardium syndrome (HHS) (18). The latter is particularly worrying due to its association with high mortality rates from 30 to 80% (18–20). Consequently, efficient diagnosis of FAdV has become highly urgent, as well as developing effective vaccination strategies (21).
FAdV diagnosis is initially based on macroscopic features, followed by histological examination (22). Furthermore, electron microscopy enables a direct visualization of icosahedral viral particles in infected tissues, confirming the FAdV diagnosis (3, 23, 24). On the other hand, various serological tests have been widely employed for FAdV detection, including agar gel immunodiffusion (25), double immunodiffusion (26, 27), immunofluorescence (28), counter-immunoelectrophoresis (29), and agar gel precipitation test (30). However, the VN test represents the gold standard for its ability to differentiate between FAdV serotypes (31, 32). Despite this accuracy, the VN test cannot be used for mass detection due to their significant compromises in terms of cost, time, cell culture, and reference strains (32). Various versions of enzyme-linked immunosorbent assay (ELISA) have been developed to overcome these limitations imposed by conventional serological tests (33–36). Although these tests are economical, rapid, and suitable for mass detection, they present many limitations, notably in terms of cross-reactivity and low sensitivity (37).
Compared with other techniques, molecular techniques offer significant advantages in terms of sensitivity, specificity, rapidity, and safety (38). Several reports have employed in situ hybridization (ISH) to detect FAdV DNA using specific probes (39–41). Unlike other molecular techniques, these probes can be directly applied to suspected lesions, enabling the confirmation of FAdV involvement. However, this method is no longer widely used today due to the complexity of its application and the availability of more convenient and reliable diagnostic methods. Polymerase chain reaction (PCR) is a widely used technique for the diagnosis of various infections (42). On the other hand, novel versions of PCR have recently been applied for FAdVs diagnosis, including loop-mediated isothermal amplification (LAMP), cross-primed amplification (CPA), recombinase polymerase amplification (RPA), droplet digital polymerase chain reaction (ddPCR), dot blot assay combined with cPCR, nanoparticle-assisted PCR (nano-PCR), PCR-refractory quantitative amplification (ARMS-qPCR), and CRISPR technology. These tests aim to improve the detection, quantification, and genotyping of FAdVs involved in avian diseases. This review covers the molecular methods used for FAdV detection and genotyping, highlighting their role in overcoming the limitations of traditional diagnostic approaches. It also discusses the strengths and drawbacks of these molecular techniques, offering a detailed analysis of their effectiveness and potential challenges.
2 Conventional PCR
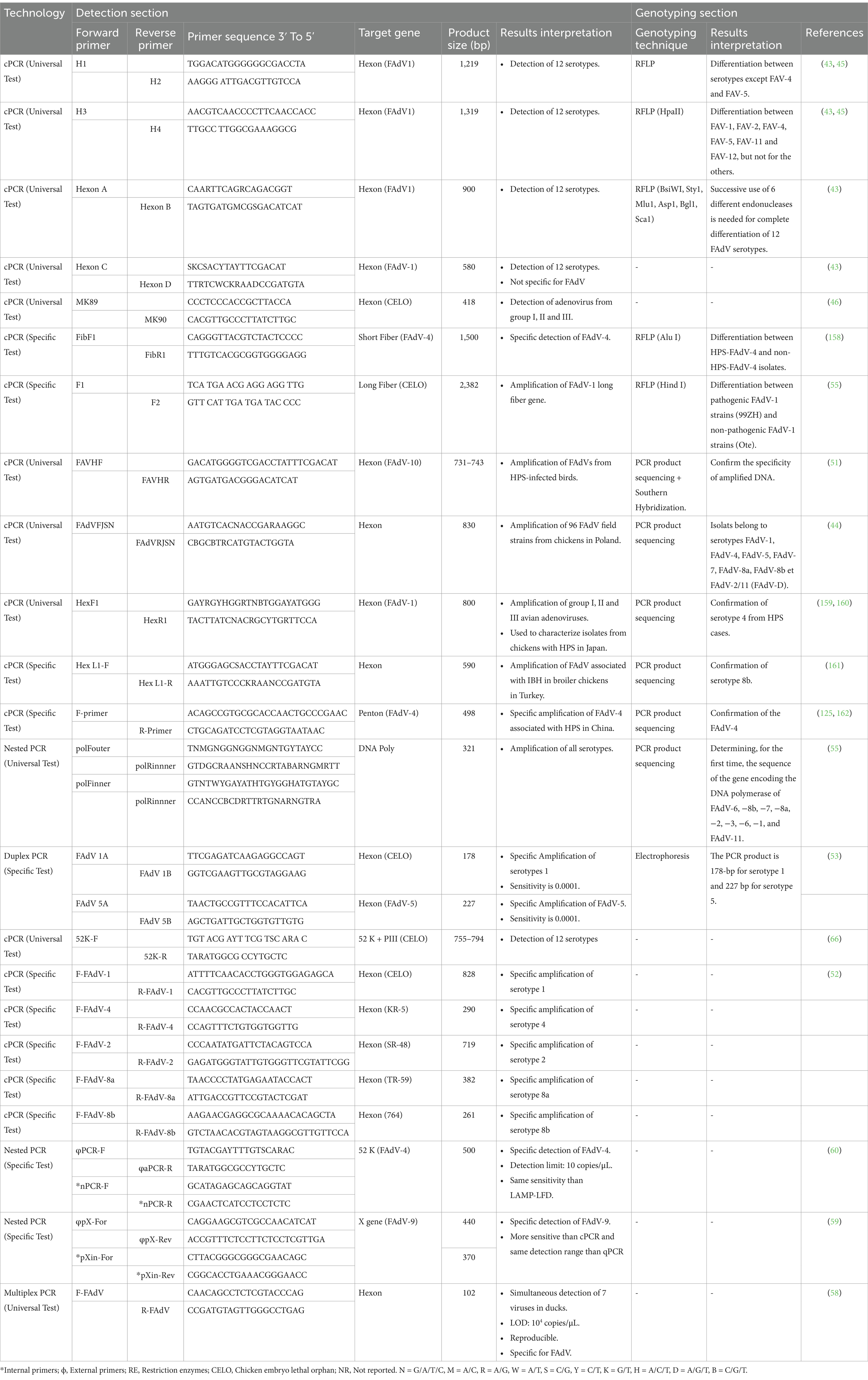
Initially, cPCR was used for FAdV diagnosis due to its sensitivity and simplicity. Several PCR assays specifically targeting the hexon gene were initially developed for FAdV diagnosis (43–46) (Table 1). The hexon gene is the longest gene in the FAdV genome and encodes a capsid structural protein, specifically the antigenic determinants of either type, group, and subgroup (47). It has 2 functional components: Pedestals regions P1 and P2, conserved between FAdV serotypes (4, 48), and L1-L4 loops that form hypervariable regions (HVR1-4) (49). These HVRs have been identified exclusively in the L1 and L2 regions (43). Except for the L3 region, these loops are surface exposed and interact with the host immune response, making them targeted in taxonomy and FAdV genotyping (4). It has been reported that analysis of the HVR1 region distinguishes between strains of the same serotype from different geographical regions (44).
Primers for cPCR were designed in both conserved and variable regions on the FAdV hexon gene. The use of universal primers H1/H2, H3/H4, HexonA/HexonB, HexF/HexR, FAdVF JSN/FAdVR JSN, and HexL1-F/HexL1-R, which hybridize to conserved regions on the hexon gene enabling the amplification of L1 hypervariable regions was used for detection of FAdVs. However, To ensure a universal detection, most of these primers are degenerated, including various alternative sequences to cover all minor variations between the 12 serotypes (43). Nevertheless, it was reported that H1/H2 primers failed to amplify FAdV-3 from the supernatant of infected cell cultures (43). Comparison of the sense primer (H1) with the FAdV-3 hexon gene sequence revealed the existence of 3 mismatches located in the last 9 nucleotides on the 3′ end of H1 (nucleotides at positions 11, 14, and 17) (43). The same study showed that the MK89/MK90 primers amplified only FAdV-1 due to the lack of identity of these primers with the other 11 serotypes. This contrasts with the findings of Xie et al. (46), who have reported that MK89/MK90 is a universal primer. Furthermore, the hexon C/hexon D primers are less specific, as they enabled the amplification of the EDS virus, which belongs to group III of avian adenoviruses. This lower specificity is probably associated with the higher degraded level of the Hexon C/Hexon D primers compared to others (43). Consequently, the use of H1/H2, HexonC/HexonD, and MK89/MK90 primers is not suitable for universal detection as they will inevitably lead to false-negative PCR results. These tests are often associated with enzymatic digestion or sequencing for serotype identification. The use of universal hexon A/Hexon B primers followed by sequencing of the product remains the reference technique used for FAdV serotype identification (12). However, a study revealed that Hexon A/Hexon B primers did not amplify FAdV-5 (50), raising questions about their effectiveness for detecting this serotype.
On the other hand, using specific primers targeting hypervariable regions of the hexon gene, in particular, the L1 loop, which shows a higher degree of variability than the L2 loop, FAdV-4 was successfully detected from HHS cases in India by PCR coupled with Southern hybridization (51). Moreover, FAdV-8a, −8b, −1, −2, −4 were detected from IBH, HHS, and AGE cases in Japan using serotype-specific primers targeting specific regions within the hexon gene (52). In addition, primers specific to FAdV-1 and FAdV-5 have been used in duplex PCR for the simultaneous detection of both serotypes in a single reaction. The size of the PCR product differentiates between these 2 serotypes in the case of co-infections. This technique has proved to be a fast, efficient, specific, and highly effective tool for FAdV-5/1 detection (53).
Besides, fiber genes are also used for specific detection of certain FAdV serotypes as they encode type-specific neutralizing epitopes, non-type-specific neutralizing epitopes, and type-specific neutralizing epitopes for the subgenus (54). Primers targeting fiber genes 1 and 2 have been designed to detect FAdV-4 involved in HHS. Moreover, PCR combined with Restriction Fragment Length Polymorphism (RFLP) of the fiber gene, has allowed distinguishing between pathogenic FAdV-1 isolates involved in GE and non-pathogenic isolates from healthy chickens (55). The technique has also been successfully used to differentiate FAdV-4 pathogenic strains from non-pathogenic strains isolated from healthy chickens in Japan, India, and Pakistan.
Compared to Uniplex PCR, Multiplex PCR (m-PCR) offers significant advantages, such as time savings and the ability to diagnose multiple viruses in a single reaction, making it an effective method for rapid diagnosis of mixed infections (56, 57). In this context, an m-PCR test was developed for the simultaneous detection of 7 viruses causing significant economic losses in the poultry industry, including FAdVs (58). Specific primers were designed for each virus, and the tests for specificity, sensitivity, reproducibility, and repeatability were conducted. The m-PCR test showed no cross-reactivity between these 7 viruses or with other common duck pathogens. In addition, the test was able to detect co-infection with several viruses in clinical samples. However, the assay is not highly sensitive, with a detection limit of 104 copies/μL, raising questions about their effectiveness for FAdV detection.
A nested PCR targeting X gene and 52 k gene with 2 successive cycles using PCR-F/PCR-R as external primers in the first amplification cycle and nPCR primers (nPCR-F/nPCR-R) in the second cycle has been developed for specific detection of FAdV-9, and FAdV-4, receptively (59, 60). This method enhances both specificity and sensitivity, with sensitivity being 100 to 1,000 times greater than conventional PCR. Other cPCR tests targeting the penton gene and the gene encoding for DNA polymerase have also been developed (61).
Although conventional PCR targeting specific genes, such as hexon gene and fiber gene, is considered as an efficient, specific, and reliable tool for FAdVs diagnosis, its considered a not very-sensitive tool. Moreover, these tests require post-PCR steps, including RFLP, Electrophoresis, PCR product sequencing, and interpretation of sequencing results, which increases the cost, time, complexity of the analysis, and the risk of contamination. Additionally, conventional PCR cannot quantify the virus, which is necessary for evaluating the effectiveness of infection control and surveillance measures. Therefore, real-time PCR tests have been developed later to overcome these limitations.
3 Real-time PCR
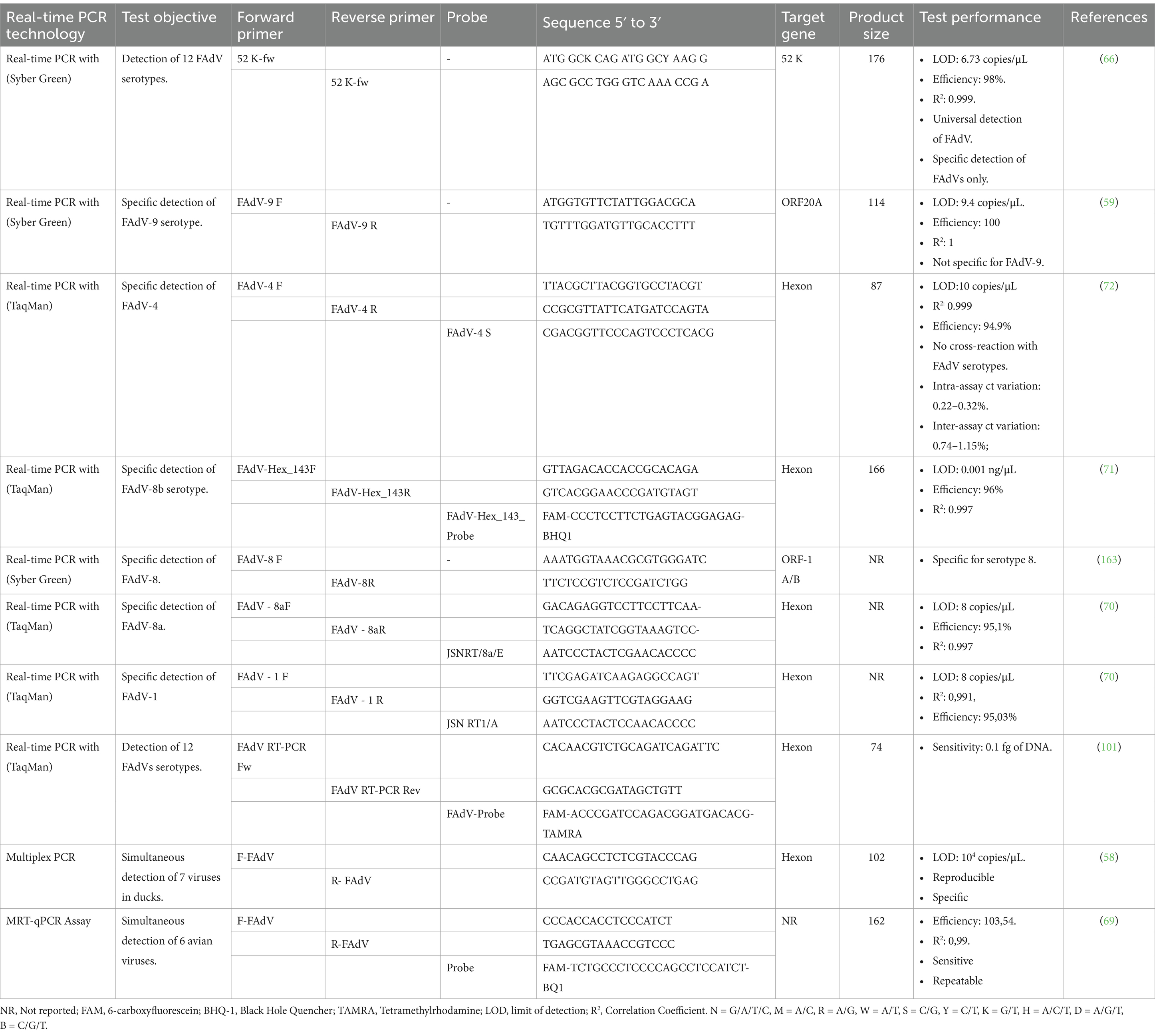
Real-time PCR, also known as quantitative polymerase chain reaction (qPCR), represents a significant advance in molecular biology, offering a powerful molecular diagnostic tool for the detection of various human and animal pathogens (62–64). The method is widely adopted due to its high sensitivity, simplicity, reproducibility, and specificity (65). Studies have suggested that it is ten times more sensitive than cPCR, making it extremely valuable in molecular diagnosis (66). qPCR also offers the possibility of performing real-time quantification, making it an invaluable tool in biomedical research, genetic studies, and diagnostic applications where high accuracy and sensitivity are crucial (67, 68).
Real-time PCR method using SYBR Green, a fluorescent dye that binds to double-stranded DNA emitting detectable fluorescence, was initially developed to detect and quantify FAdV-9 genome in various tissues (59). In this assay, a region located at the right end of the FAdV-9 genome, corresponding to ORF 20A, was used as a target (Table 2). However, the qPCR assay was not specific to FAdV-9 since it also detected other serotypes, such as FAdV-1, FAdV-2, FAdV-8, and FAdV-10. This suggests that the selected region is not specific for FAdV-9, and other regions should be examined. In terms of sensitivity, the test showed a sensitivity of 9.4 copies/μL, which is comparable to nested PCR and 100 times more sensitive than conventional PCR (59).
Subsequently, a universal SYBR Green-based qPCR test was developed by targeting a conserved region of the 52 K gene. This assay demonstrated high sensitivity and specificity, enabling precise detection and quantification of 5 FAdV species (FAdV-A to FAdV-E) with a detection limit of 6.73 copies/μL of FAdV DNA using standards and control vectors. To establish the standard curve, different regions of FAdV genomes are isolated, meticulously prepared, and cloned into plasmid vectors. The concentration of the plasmid DNA is determined using spectrophotometry, and the number of DNA copies is calculated according to the following formula: [(g/μL of DNA)/(length of the plasmid in base pairs × 660) × 6.022 × 1023]. The plasmid DNA is then diluted to several concentrations and used to establish the standard curve, which is included in each qPCR reaction (69). qPCR using SYBR Green represents a simple, sensitive, and cost-effective quantitative approach. However, SYBER Green can bind to non-specific products or contaminants, leading to false-positive results.
Consequently, analysis of melting curves and negative controls is required to interpret qPCR results correctly. Other real-time PCR methods, using fluorogenic probes complementary to the target sequences and doubly labeled with a fluorophore and a quencher, have been developed for specific detection of certain FAdV serotypes, such as FAdV-4, FAdV-8b, FAdV-8a, and FAdV-1 (70–72). The use of specific primers that hybridize to type-specific regions allows qPCR to simultaneously perform real-time molecular detection, quantification, and typing in a single reaction. Notably, TaqMan-based qPCR demonstrated superior efficiency compared to SYBR Green-based qPCR, while both methods exhibited similar sensitivity (72).
Recently, a multiplex real-time PCR has been developed, called multiplex reverse transcription real-time quantitative PCR (MRT-qPCR) (69). This technique was designed to detect co-infection of 6 vertically transmitted or immunosuppressive avian viruses, including Marek’s Disease Virus (MDV), Reticuloendotheliosis Virus (REV), Avian Reovirus (ARV), Chicken Infectious Anemia Virus (CIAV), Infection Bursal Disease (IBD), and FAdVs. Six specific probes were designed, each complementary to one virus, and labeled with a unique fluorophore, allowing differentiation of the signal emitted by the 6 probes during the qPCR reaction. A series of validation and optimization tests were carried out, confirming the high specificity, sensitivity, and repeatability of the MRT-qPCR assay. These characteristics ensure the reliability and relevance of this method in diagnosing viral co-infections in poultry, making it an excellent first-line screening tool for a wide range of viruses before moving on to more genus-specific tests.
4 Recent breakthroughs in molecular diagnostics of FAdV infections
Although molecular diagnostic tests, such as conventional PCR have considerably improved diagnostic accuracy compared with traditional methods, certain drawbacks linked to operational complexity remain unsolved. Thus, to improve FAdVs detection in terms of sensitivity, cost, and process time, other advanced molecular tests have been developed.
4.1 Loop-mediated isothermal amplification
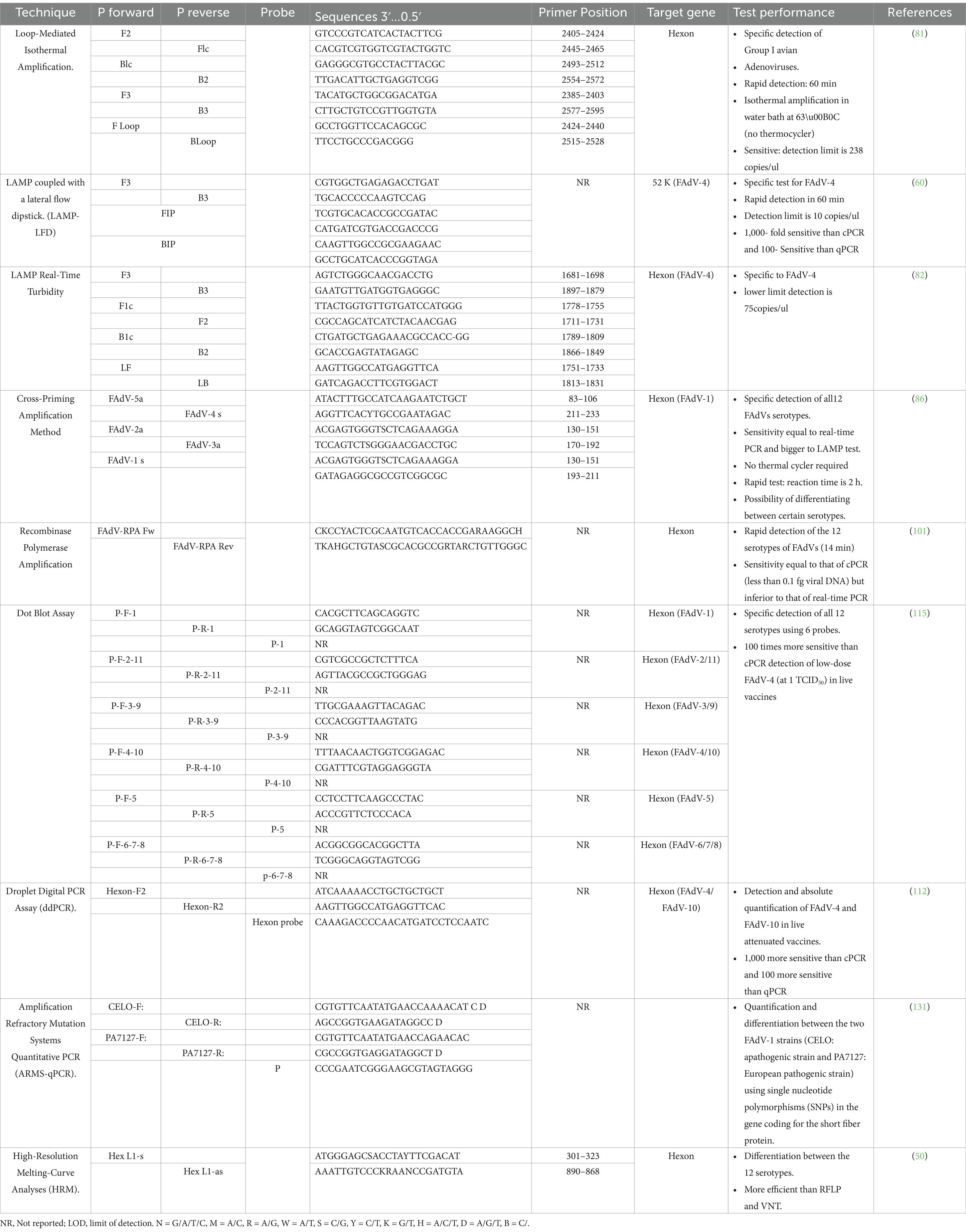
The LAMP technique was first described by Notomi et al. (73). Since then, it has attracted significant interest due to its simplicity and rapidity compared to other amplification methods. Unlike PCR, which requires a series of temperature cycles, LAMP is an isothermal amplification method that proceeds at a constant temperature between 60°C and 65°C, eliminating the need for a thermocycler (73). The LAMP reaction involves 2 external primers (F3 and B3) and 2 pairs of internal primers (FIB and BIP). One or 2 additional primer pairs, known as ‘loop primers’, can be incorporated to accelerate the reaction and improve its sensitivity (74). The LAMP technique consists of 2 significant steps: synthesis of the initiator serving as a template, followed by cyclic amplification, which produces a DNA mixture of stem ring DNA and cauliflower DNA of different sizes. This method can also be applied to RNA by Reverse Transcription-LAMP (RT-LAMP) (75). Amplification products can be detected by several techniques, including agarose gel electrophoresis, which produces multiple ladder bands, or by real-Time turbidity measurement due to the formation of manganese pyrophosphate precipitates. The addition of hydroxy naphthol blue, calcein, or SYBR Green to the reaction system also offers colorimetric detection of the product (74).
LAMP assay has been successfully employed to detect many avian viruses, including Infectious Bronchitis Virus (IBV) (75), Chicken Anemia Virus (76), Avian Influenza Virus (AIV) (77), Newcastle Disease Virus (NDV) (78), IBDV (79), and Marek’s Disease Virus (MDV) (80). Recently, a LAMP assay was specifically developed and optimized for FAdVs detection (81). Based on an analysis of their Hexon genes, 4 primer pairs were designed for a conserved region (Figure 1). The LAMP reaction was performed in a water bath at 63°C for 60 min. The addition of SYBR Green fluorescent dye to the reaction gave positive samples a greenish color under ultraviolet (UV) light. Additionally, the formation of white sediment due to precipitated pyrophosphate was a distinctive feature of positive samples. The test shows a detection limit of 238 copies/μL.
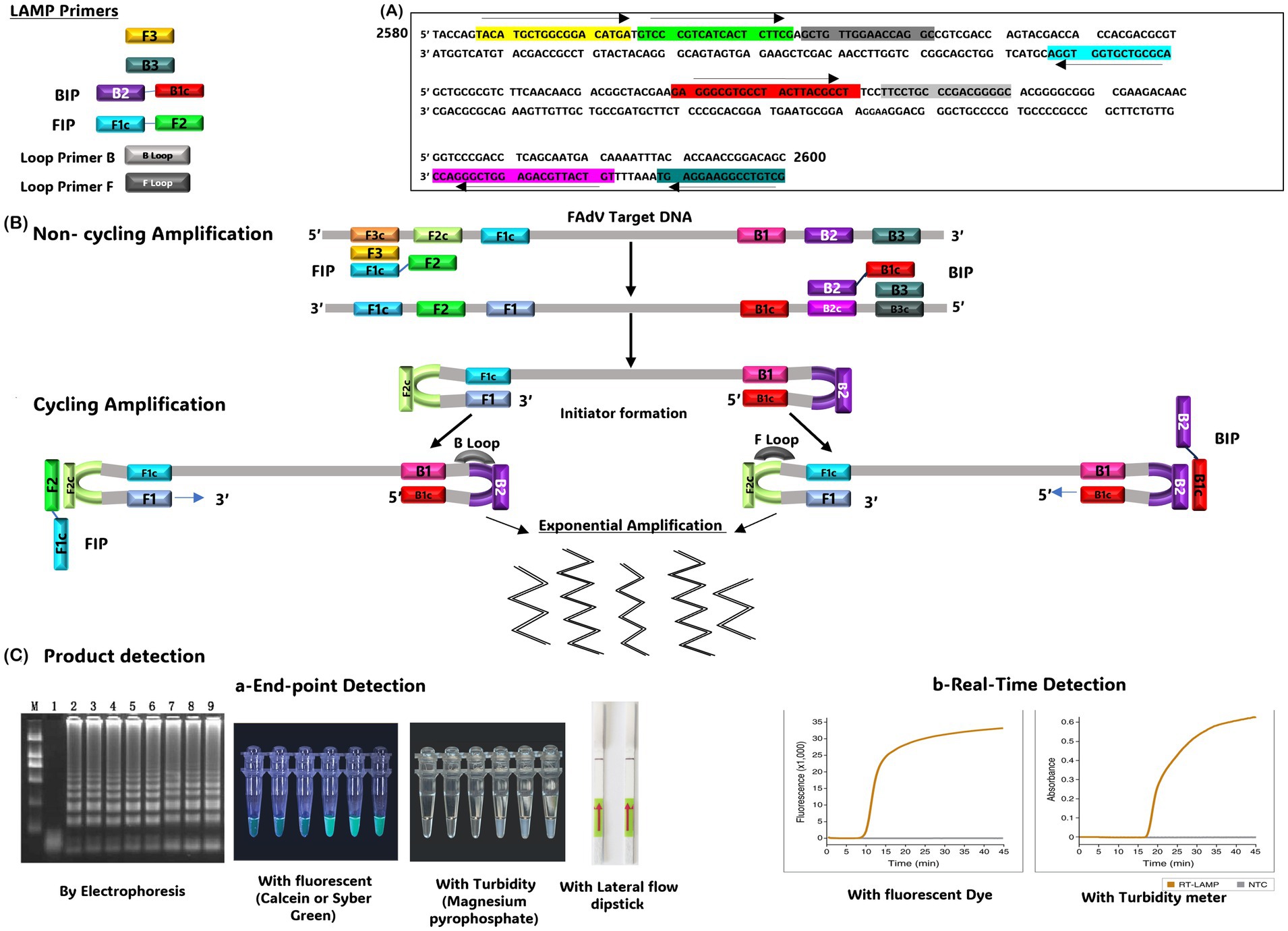
Figure 1. (A) Position of the LAMP primers on the FAdV target gene. (B) Schematic representation of the Loop-Mediated Isothermal Amplification (LAMP) technique. (C) Methods used to detect the LAMP product, including End-point detection (using electrophoresis, addition of fluorescent substrates, turbidity measurement, or lateral flow dipstick or (real-time detection)). The gel image presented in this figure are adapted from previous work (81) and are included to ensure the figure is comprehensive and meaningful.
To improve the LAMP sensitivity, a real-time LAMP assay has been developed for specific FAdV-4 detection (82). The Mg2P2O7 precipitate produced during the reaction is detected by measuring reaction turbidity every 6 s using a real-time turbidimeter. The assay shows a detection limit of 75 copies/μL of FAdV-4 DNA. Subsequently, a new version of LAMP coupled with lateral flow dipstick (LAMP-LFD) was developed for rapid and specific detection of fowl Adenovirus serotype 4 (60). This test can be completed in 60 min at 65°C, with a detection limit of 10 copies/μL of FAdV-4 DNA, making it more sensitive than real-time LAMP (75 copies/μL) (82), and 1,000 times more sensitive than conventional PCR. However, it has the same detection limit as Nested PCR (60), and specific qPCR (72). Although it remains less sensitive than universal qPCR assay (6.9 copies/μL) (66), it can potentially require less than half time and reagents. These advantages enable LAMP-LFD to be applied in resource-limited areas, such as small farms and basic veterinary laboratories (83).
4.2 Cross-priming amplification method
CPA is a technique primarily developed by Ustar Biotechnologies (Hangzhou, China) and initially described for the detection of various pathogens such as Mycobacterium, and Penaeid shrimp white spot syndrome virus. This technique can be divided into single-crossing CPA and double-crossing CPA (84), requiring specific polymerases such as Bst, Bsm, or Gsp SSD (85). Recently, a double-crossing CPA assay has been optimized explicitly for FAdVs detection (86). The assay uses 5 specific primers corresponding to a conserved region of the hexon gene (151-bp) and induces cross-priming amplification with the formation of a hairpin intermediate product (Figure 2). The optimal temperature and incubation time were determined at 68°C for 2 h, respectively. The amplification product was visualized by adding SYBR Green I to the reaction. The test is specific for all FAdV serotypes, and no cross-reactivity was observed with other avian viruses. Its sensitivity was equivalent to that of real-time PCR, reaching 10−2 TCID50 (TCID50: Tissue Culture Infectious Dose). However, the CPA method is faster and cheaper compared to real-time PCR. Consequently, the CPA-FAdV assay has been effectively used to detect 30 field adenovirus strains, representing 7 distinct serotypes (FAdV-1, FAdV-2/11, FAdV-4, FAdV-5, FAdV-7, FAdV-8a, and FAdV-8b) (86).
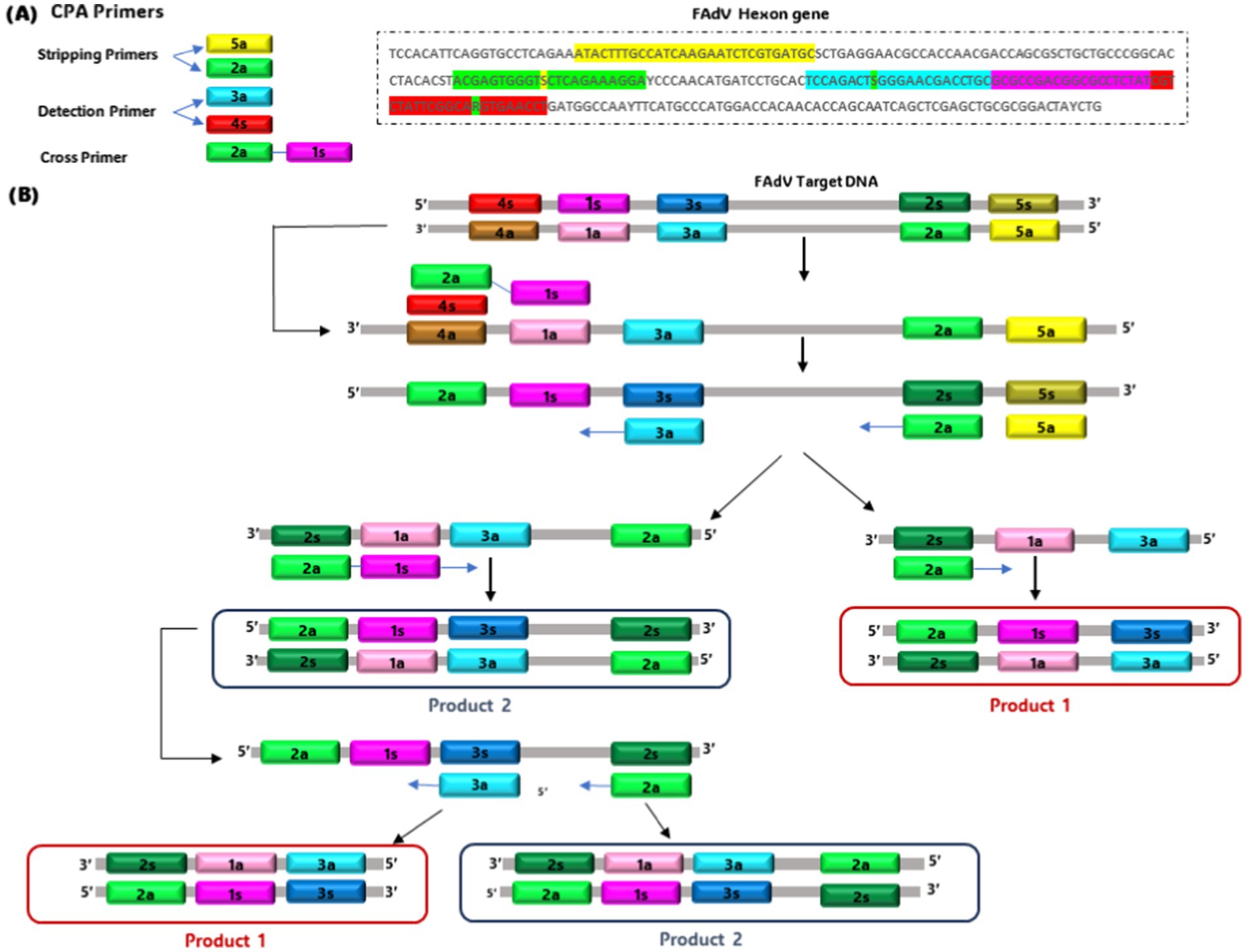
Figure 2. Schematic of single cross-priming amplification technology. (A) Primer design site. (B) Generation of template with cross-primer sites.
4.3 Recombinase polymerase amplification
RPA is an advanced isothermal technique that was discovered in 2006 by Piepenburg et al. (87). Since the availability of the TwistAmp Basic commercial RPA kit in 2014, the RPA technique has been widely used in the diagnosis of numerous pathogens, such as Human Immunodeficiency Virus 1 (HIV) (88), Ebola Virus (89), Dengue Virus (90), Porcine Circovirus Type 2 (91), Pseudorabies Virus (92), and Foot-And-Mouth Disease Virus (93). The technique relies on the use of T4 phage UvsX recombinase and its cofactor as an essential component that binds to forward and reverse primers (94). The strands are then exchanged after the Single-Stranded Binding Protein (SSB) combines with the parental strand, allowing amplification to continue with the template strand.
DNA polymerase initiates the synthesis of the template strand from the 3′ end of the primers, forming a new duplex DNA. In this way, a specific fragment is amplified exponentially (95). The amplification occurs between 37 and 42°C (96). However, the amplified signal can be detected by electrophoresis, lateral flow dipstick (LFD), or in real-time using a fluorogenic probe. Real-time RPA and RPA-LFD assays have been developed as attractive and promising tools for rapid, convenient, and reliable detection of M. ovipneumoniae in sheep (97), African Fever Virus (AFV) (98), Actinobacillus Pleuropneumonia (99) in swine and Peste Petits Ruminants Virus (PPRV) in small ruminants (100). Recently, an RPA assay has been developed for FAdV detection (101). Primers targeting a conserved region between the 12 serotypes were selected for this assay (Figure 3). Amplification was performed under isothermal conditions (from 26 to 42°C) without using sophisticated thermocyclers in just 14 min. This time is considerably shorter than that of conventional PCR (98 min), while offering similar sensitivity (as low as 0.1 fg viral DNA). However, its sensitivity remains lower than that of real-time PCR (66). The RPA test has revolutionary potential for rapid diagnosis of FAdV. Its rapidity, specificity, simplicity, and adaptability to moderate temperatures make it an ideal technology for large-scale screening of samples, particularly where laboratory resources are limited.
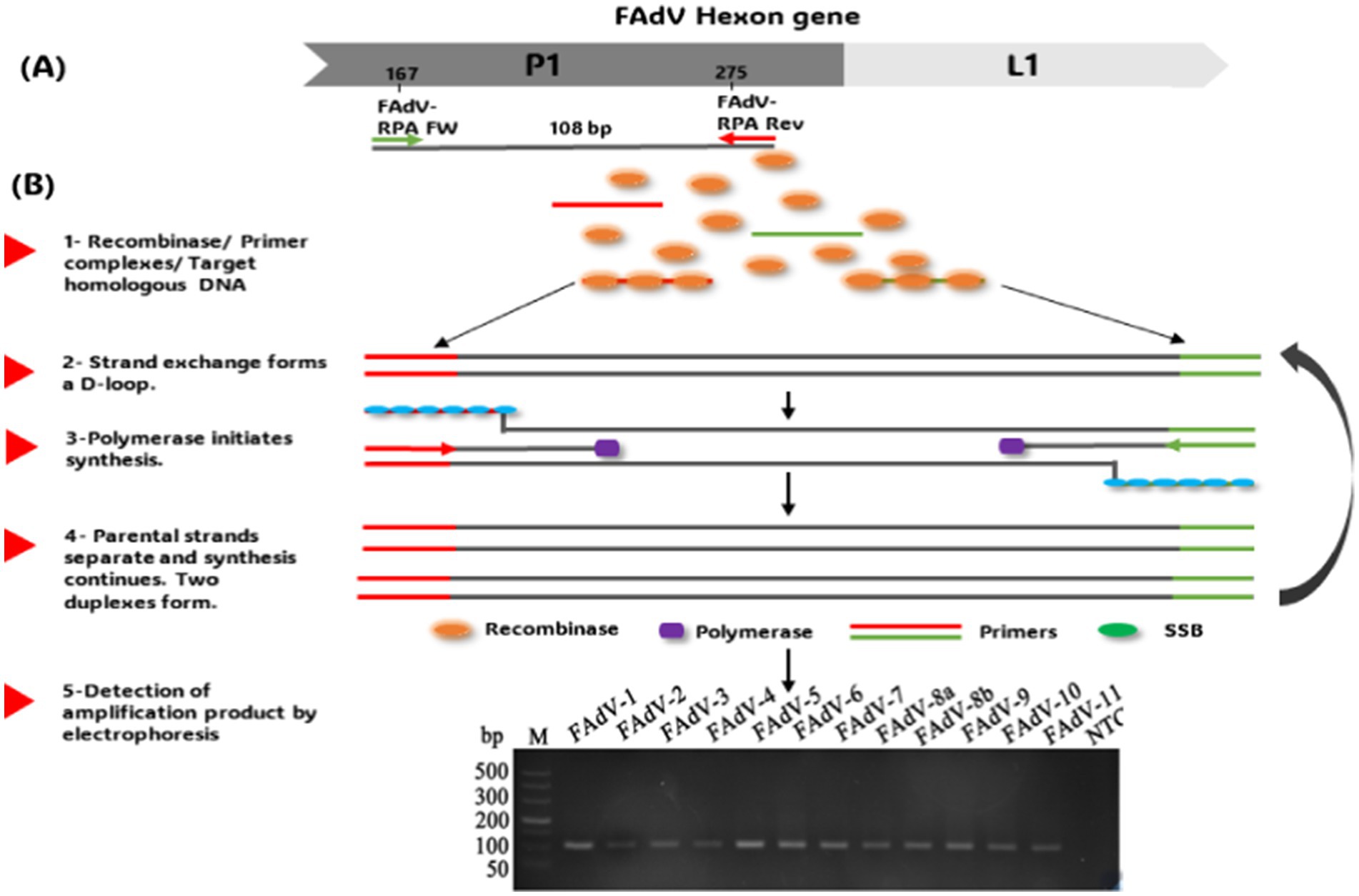
Figure 3. (A) RPA primer design. (B) Schematic of the recombinase polymerase amplification technique workflow and product detection by Electrophoresis. The gel images presented in this figure are adapted from previous work (101) and are included to ensure the figure is comprehensive and meaningful.
4.4 Digital droplet polymerase chain reaction
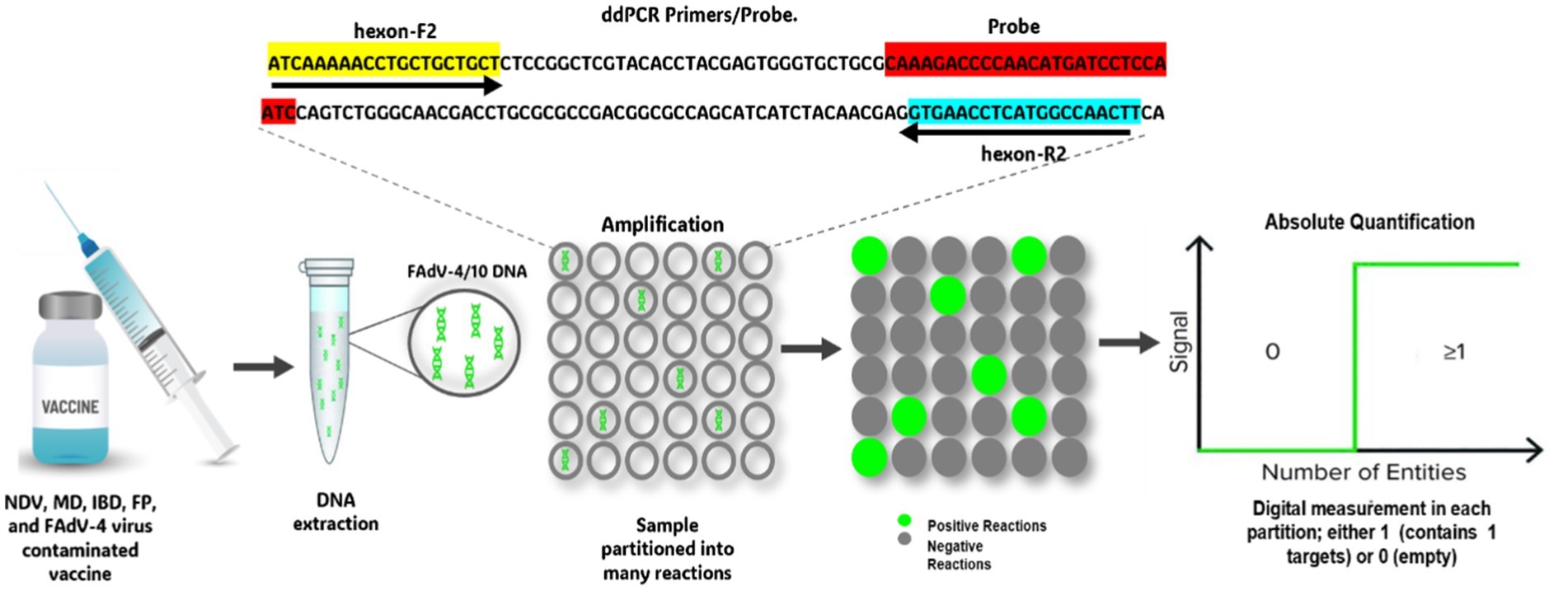
ddPCR represents a significant advance in the precise quantification of nucleic acids, particularly useful in cases where the quantity of DNA or RNA is very low (102). Unlike quantitative real-time PCR, which uses standard curves to estimate the target quantity, ddPCR enables absolute quantification by counting the number of copies present in each sample (103). This method relies on the partition of the PCR reaction into numerous tiny droplets of water in oil. Each droplet contains either zero or a single copy of the target sequence (Figure 3) (104). After PCR amplification, the positive droplets emit a detectable fluorescent signal, while the negative droplets do not. Using statistical calculations, the absolute number of target copies can be determined with great precision (105), highlighting that ddPCR offers exceptional sensitivity and reliability, making it an ideal method for robust quantitative analysis, even in samples with low viral loads or compromised quality (102). This technique has been successfully employed for the precise quantification of defective genomic segments in influenza A virus, providing a highly sensitive approach for detecting these particles in viral stocks (106, 107). Additionally, ddPCR has been applied to detect Chicken Anemia Virus (CIAV) in vaccines, demonstrating its sensitivity in identifying viral contamination (108). Moreover, despite stringent biosafety measures, contamination of live attenuated vaccines with Fowl Adenovirus serotype 4 (FAdV-4) remains a concern, as documented by Yang (109). The use of such contaminated vaccines has been implicated in large-scale outbreaks of Hepatitis-Hepatitis Syndrome (HHS) and Infectious Bronchitis-like Hepatitis (IBH) in poultry populations (110). This is due to the use of vaccines manufactured from chicken embryos with Specific Pathogens Free (SPF), but susceptible to infection by exogenous viruses such as FAdV-4, Avian Leukosis Virus (ALV), and Reticuloendotheliosis Virus (REV) (54, 109, 111). These contaminating viruses can escape detection by most molecular tools, necessitating a highly sensitive detection technique. Consequently, a ddPCR assay has been developed for sensitive detection of FAdV-4 and FAdV-10 in attenuated vaccines (112) (Figure 4). The efficacy of this ddPCR test in detecting FAdV-4 contamination in attenuated vaccines was evaluated in comparison with qPCR and cPCR. Results showed that ddPCR assay could detect FAdV-4 contamination at a concentration of 0.1 EID50/1,000 feathers (EID50 for Median Infectious Dose), while cPCR and qPCR could detect FAdV-4 contamination at concentrations of 102 EID50/1,000 feathers and 1 EID50/1,000 feather, respectively. Thus, the ddPCR assay looks 1,000 times more sensitive than conventional PCR detection and ten times more sensitive than real-Time PCR. In addition, the ddPCR assay showed high specificity for FAdV-4/10, generating no positive signals for other FAdVs (112). This makes ddPCR an effective diagnostic technology, particularly for detecting FAdV-4 contamination in live attenuated vaccines. Despite its high cost, the high sensitivity and specificity may contribute to the use of this technique for virus control.
Dot Blot assay is a widely used technique in molecular biology to identify target DNA or RNA.
4.5 Dot blot assay combined with cPCR
The Dot Blot assay is a widely used technique in molecular biology to identify a target DNA or ARN fragments with high sensitivity (113). The combination of dot blot with PCR significantly increases test sensitivity (114). Recently, a Dot Blot test has been developed for FAdVs detection (115). The 12 FAdV serotypes have been grouped into 6 categories based on their hexon gene sequence. Subsequently, a conserved region for each category was selected as a probe. Results showed that these probes can efficiently identify the corresponding serotypes, with a detection limit of 10 copies/μL.
Furthermore, the use of a hybrid probe combining all 6 probes at an optimal concentration considerably improved test sensitivity, enabling the detection of one copy of DNA for certain serotypes, which is more sensitive than conventional PCR. The test’s sensitivity was also determined on live attenuated vaccines artificially contaminated with FAdV-4. The results showed that the Dot Blot test can effectively identify exogenous FAdV-4 with an extremely low concentration (1 TCID50), whereas conventional PCR can only detect a contaminated vaccine with a viral concentration over 100 TCID50 per bottle, demonstrating that the Dot Blot test is 100 times higher sensitivity than cPCR. The same analysis was repeated using vaccines contaminated with mixed serotypes of FAdV, and the same conclusion was reached. In addition, the Dot Blot test was successfully used to diagnose co-infection of FAdV and vertically-transmitted immunosuppressive viruses (CIAV, REV, ALV) in parental flocks with IBH (116). In conclusion, the Dot Blot test, designed based on traditional PCR, is a simple, sensitive, reliable, efficient, and cost-effective tool for the universal detection of all 12 FAdV serotypes.
4.6 Nanoparticle-assisted PCR
Nano-PCR is an advanced form of PCR in which solid gold nanoparticles (1 to 100 nm) from colloidal nanofluids are used to improve reaction efficiency, sensitivity, and time (117–119). Compared to other PCR techniques, nano-PCR using nanofluids reaches the target temperature more rapidly, reducing analysis time and nonspecific amplification (120). This technique has been successfully employed in the detection of various viruses such as Pseudorabies virus (121), Porcine Bocavirus (122), Epidemic Diarrhea Virus (123), and Porcine Transmissible Gastroenteritis Virus (124).
Recently, a nano-PCR test has been developed to detect FAdV-4, using primers specific for the FAdV-4 penton gene (125). Test results indicated that nano-PCR has a reasonable specificity, repeatability, and high sensitivity (54 copies/μL), which is ten times higher than that of conventional PCR (cPCR), making it suitable for clinical diagnosis and field surveillance of FAdV-4 infections. Subsequently, a Triplex Nanoparticle-Assisted PCR test has been developed, enabling simultaneous detection of FADV, CAV, and IBDV in one reaction (126). This innovative assay utilizes PCR primers designed to target specific genes of each virus. The test was specific to FAdV, CAV, and IBDV, with a detection limit of 27.2 femtograms (fg) for all 3 viruses’ DNA. This makes it 1,000 times more sensitive than multiplex PCR using identical primers, which provides a simple method for detecting FAdV, CAV, and IBDV infections.
4.7 Quantitative PCR refractory amplification (ARMS-qPCR)
Quantitative PCR with Refractory Amplification (ARMS-qPCR) also is an innovative molecular tool specially designed to detect and quantify target DNA with high specificity. Unlike other real-time PCR techniques, ARMS-qPCR incorporates refractory amplification, making it highly sensitive and specific for the detection of genetic variations such as Single Nucleotide Polymorphisms (SNPs). This technique is particularly valuable for diagnosing genetic diseases (127, 128) and monitoring the evolution of viruses, bacterial resistance to antibiotics (129), or discrimination between strains of the same genotype based on changes in a few nucleotides (130). In a recent study, ARMS-qPCR was used to quantify and distinguish the European pathogenic strain (FAdV-1/PA7127) from the apathogenic strain (CELO). This distinction is based on SNPs identified in the gene coding for the short-fibre protein (Fiber-2) (131) (Figure 5). Fecal, liver, and gizzard samples from chickens vaccinated with the apathogenic strain (CELO) and challenged with the pathogenic strain (FAdV-1/PA7127) were analyzed by ARMS-qPCR to quantify consensus FAdV-1 DNA as well as FAdV-1 DNA variants (CELO or PA7127). Two pairs of primers, each specific to an FAdV-1 strain, with a hydrolysis probe, were used in this assay. Specificity of discrimination between FAdV-1 strains was ensured using primers targeting SNPs on the 3′ side of each primer. The results confirmed the effectiveness of this test in discriminating between the vaccine and pathogen strains. Furthermore, it was observed that even though chickens were fully protected, they continued to excrete the challenge strain. This observation was achieved for the first-time using ARMS-qPCR. By combining the benefits of refractory amplification with the precision and quantification capabilities of qPCR, ARMS-qPCR represents a powerful method for monitoring vaccines in chicken flocks.
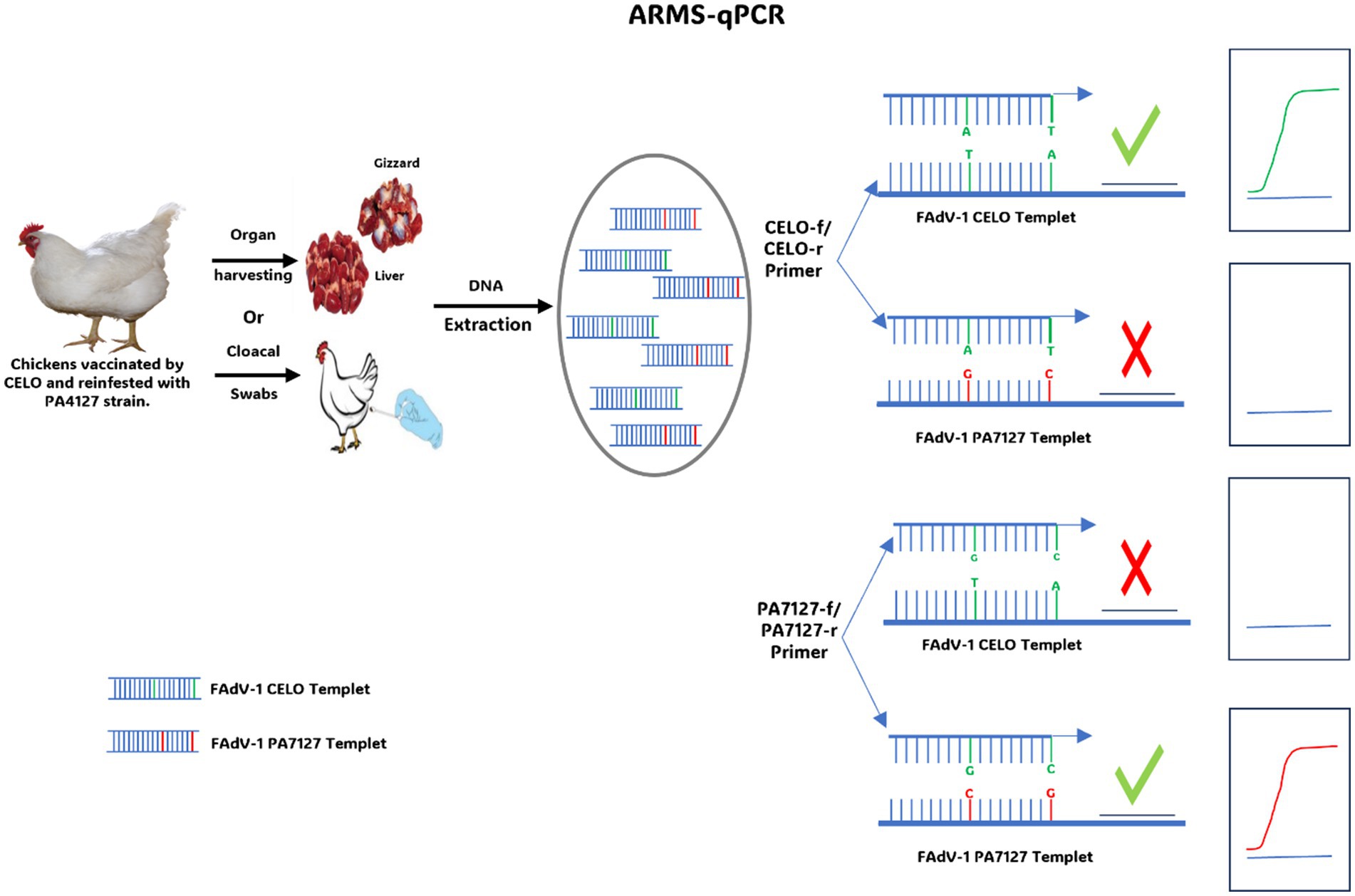
Figure 5. Diagram illustrating the use of ARMS-qPCR to discriminate between the CELO strain and the PA7127 challenge strain.
4.8 CRISPR/Cas13a-based lateral flow assay
Over the past decade, Clustered Regularly Interspersed Short Palindromic Repeats (CRISPR) associated with Cas (CRISPR-associated) proteins have attracted considerable interest due to their exceptional characteristics, notably their ability to cut DNA with outstanding sensitivity and specificity (132). This has enabled researchers to use it as a molecular scissor for genome engineering (133). CRISPR system is an immune system acquired by bacteria that protects them against viral invasions (134). It works by scanning the DNA of the viral aggressor, degrading it with Cas enzymes, and incorporating segments of the foreign viral genes into regions called “CRISPR Arrays” in the bacterial genome (135, 136). These regions are then transcribed into specific non-coding RNAs (lncRNAs), which direct Cas proteins with endonuclease activity to identify and degrade viral DNA sequences during subsequent reinfections by the same virus (117).
Over the last few years, researchers have discovered that specific Cas proteins, such as Cas13a and cas12a, possess additional cleavage activity, laying the groundwork for a revolutionary new molecular diagnostic method hailed as one of the most impactful “disruptive” innovations of our time (137, 138). By combining CRISPR/Cas13a technology with RPA (CRISPR/Cas13a or cas12a/RPA) (139), this technique has been effectively used to detect AIV (140–142) and other human viruses (143–146).
Similarly, CRISPR/Cas13a technology, combined with recombinase polymerase amplification (RPA) and lateral flow test strips, has been developed for the rapid, sensitive, efficient, and simple detection of FAdV-4 (147) (Figure 6). This method operates under isothermal conditions at 37°C and enables visual detection through lateral flow strips, with a detection limit of 10 copies/μL. Additionally, a CRISPR/Cas12a assay integrated with LAMP has been developed as a fast, convenient, and cost-effective platform for detecting FAdV-4, offering a detection limit as low as one copy (148) This makes it particularly useful for early viral diagnosis and point-of-care testing.
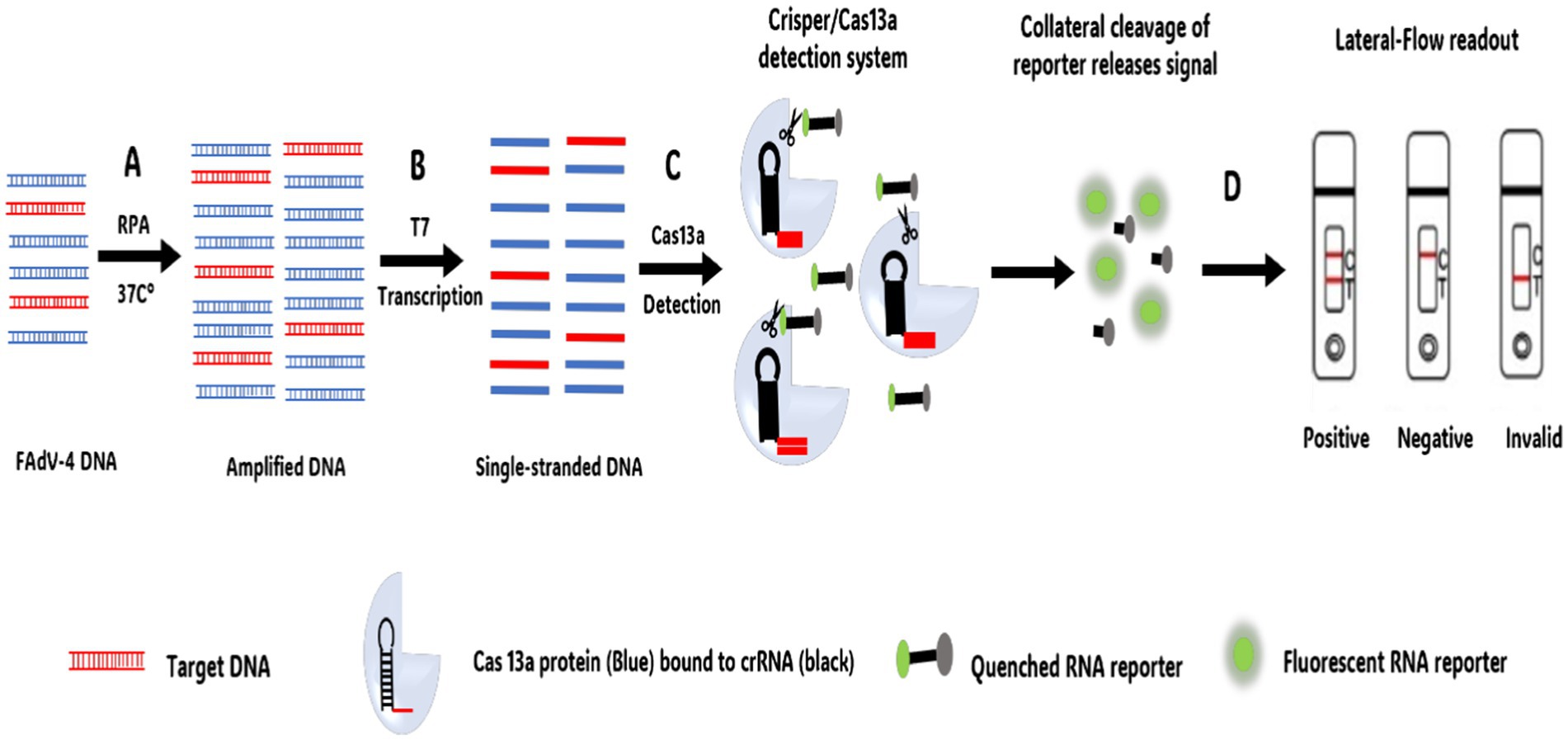
Figure 6. CRISPR-Cas13a DNA combined with lateral flow strips for FAdV-4 detection workflow. (A) Recombinase Polymerase Amplification (RPA) performed at a single temperature of 37°C. (B) Generation of single-stranded DNA by T7 Polymerase. (C) Target sequences complementary to cRNA bind to the CRISPR/Cas13a system. Once the target sequence is present within the system, the non-specific RNA cleavage activity of Cas13a is activated and the reporter RNA is cleaved, resulting in the activation of the fluorescence signal. (D) Lateral flow-based detection can be read from strips with a colored positive/negative band using a FAM/Biotin.
5 Genotyping techniques
Identifying the serotype involved in each case of FAdV infection is essential for understanding pathogenicity, monitoring circulating strains within each country, and developing effective vaccine strategies to control these economically significant diseases (28, 149, 150). Typically, virus typing requires isolation in cell culture or embryonated eggs, followed by a VN test, which was considered as the gold standard protocol for FAdV typing (2, 151). However, this method remains laborious, time-consuming, and requires reference strains and materials. Furthermore, Cross-reaction between serotypes is very pronounced between specific serotypes, which makes results inconclusive (152).
Initially, REA was used to classify FAdVs based on restriction profiles of the whole genome. These restriction profiles, generated by BamHI or HindIII enzymes, classified 17 FAdV strains representing 12 serotypes into 5 species (Avian Adenovirus A to Avian Adenovirus E) (153). The question of whether FAdV serotypes 4 and 10 should be considered as distinct serotypes or reclassified as a single serotype has been raised (152). This debate stems from the observation of cross-neutralization between serotypes 4 and 10 based on the use of antisera produced in rabbits. Additionally, cross-protection in vivo has been detected in chickens vaccinated with the CFA15 strain (serotype 4) and subsequently exposed to CFA20 (serotype 10) through natural infection, demonstrating a strong serological relationship between these 2 serotypes (152). However, whole-genome analysis of both serotypes using restriction enzyme analysis with E.R. BamHI, Dra I, Sma I, and Bgl II has revealed considerable variations (Figure 7).
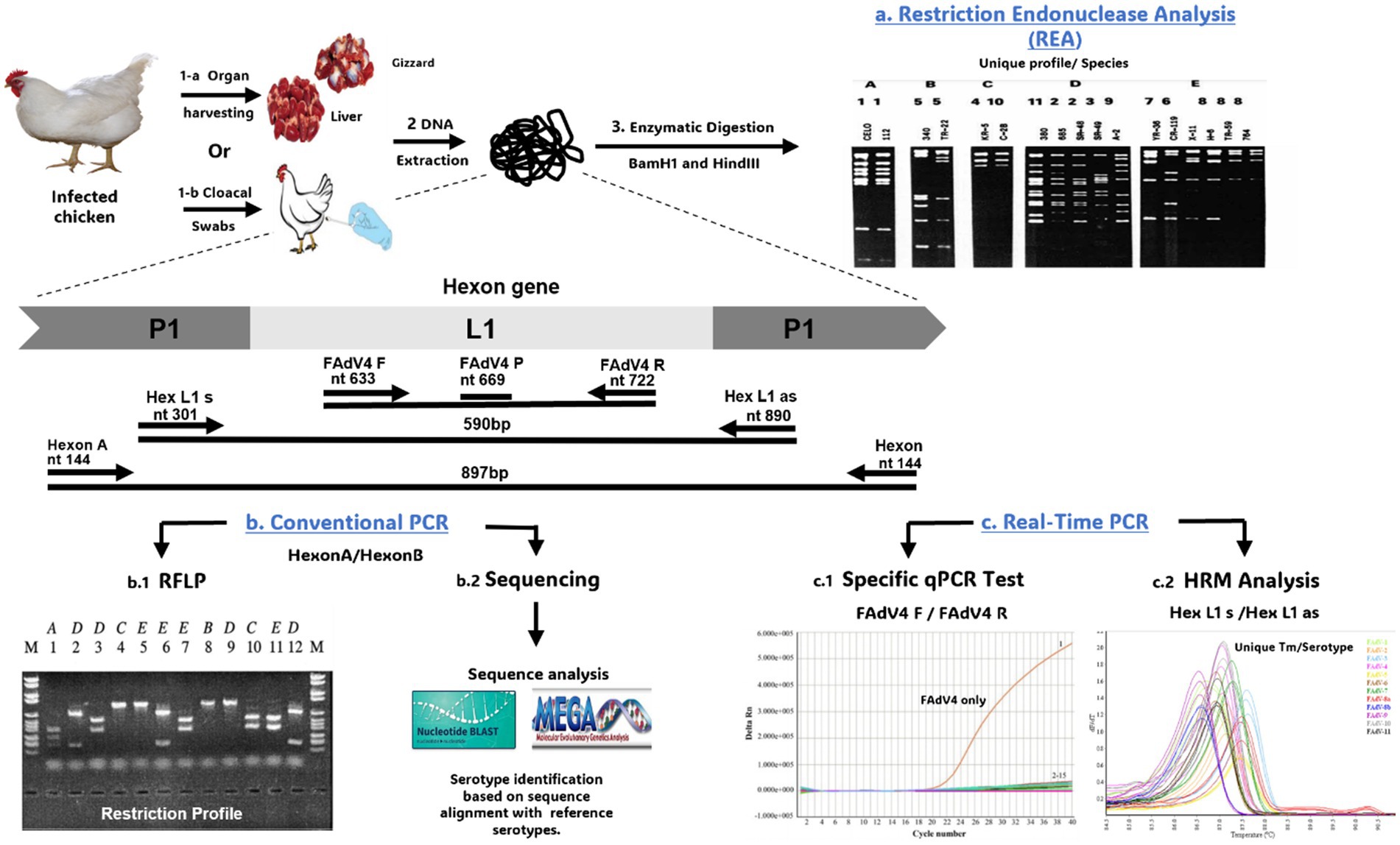
Figure 7. Overview of techniques used for FAdVs genotyping. (a) REA digesting the whole genome using BamH1 and HindIII enzymes. (b.1) PCR/RFLP digesting the PCR product derived from HexonA/HexonB primers with restriction enzymes. (b.2) Sequencing of PCR product followed by sequence analysis using BLAST and alignment with other FAdVs sequences on GenBank. (c.1) Specific qPCR test using a specific primer of serotype (e.g., FAdV4 F/FAdV4 R specific for FAdV-4). (c.2) Analysis of High-Resolution Melting (HRM) curve derived from the Hex L1 s/Hex L1 as primer. The gel images presented in this figure are adapted from previous work (43) and are included to ensure the figure is comprehensive and meaningful.
Since adenovirus neutralization relies on the hexon and fiber proteins, more accurate evolutionary profiles can be expected from the L1 loop rather than the entire DNA restriction profile. It has been reported that the L1 region shows more significant variability than the L2 loop (45). In this context, several conventional PCR assays have targeted the HVR regions of the L1 loop, while restriction enzyme analysis of these PCR products generates serotype-specific restriction profiles (43, 45). Hexon A/Hexon B primers is one of the most widely used primers for FAdV genotyping (43). Subsequent digestion of the PCR product with BsiWI, Sty1, and Mlu1 generates specific restriction profiles for 6 FAdVs serotypes. However, other restriction enzymes must be used to differentiate the remaining serotypes: FAdV-2 and FAdV-11 (Asp1), FAdV-4 and FAdV-9 (Bgl1), or FAdV-7 and FAdV-11 (Sca1).
Nevertheless, one study reported that RFLP using Hexon A/Hexon B failed to classify FAdV-2 and FAdV-11 with the previously mentioned enzymes, and other enzymes must be used (43). Similarly, digestion of the PCR product generated by H1/H2 primer with HaeII differentiated some serotypes, but identical restriction profiles were found for other serotypes. Therefore, we can conclude that multiple digestions using various restriction enzymes might be able to distinguish between 12 FAdV serotypes or that other regions showing high variability between FAdV serotypes should be chosen for RFLP typing.
Besides, PCR product sequencing, generated by Hexon A/Hexon B, followed by analysis using the BLAST bioinformatics tool, is widely used as a reliable tool for genotyping FAdV isolates (see Table 3) (44). However, this technique, like REA and PCR/RFLP, is considered as a multi-step process, making it expensive, time-consuming, and resource-intensive.
Recently, a number of studies have shown that real-time PCR combined with High-Resolution Melting Curve (HRM) analysis is a valuable and cost-effective alternative for rapid and efficient genotyping, especially for mass detection, facilitating epidemiological investigations (50, 154). This technique involves the integration of a DNA intercalating fluorochrome into the amplified DNA. The gradual increase in temperature at the end of PCR cycles causes denaturation of the DNA, leading to the elimination of the fluorochrome and resulting in a decrease in fluorescence. These data are recorded by a fluorescence detection system and used to generate a melting curve representing fluorescence changes based on temperature. Slight variations in DNA levels result in variations in the melting temperature (Tm), thus altering the shape of the curve. By comparing the melting curve of a unknown sample with reference curves, it is possible to detect genetic variations, DNA mutations, or even molecular typing (155, 156). HRM technique is widely used in genetic research and molecular diagnostics for its rapidity, sensitivity, and affordability. In 2009, Penelope A. Steer established for the first time a robust closed-tube PCR-HRM genotyping technique for FAdV classification (50). Three primers pairs amplifying 3 different regions within FAdVs hexon gene (Hexon A/B, HexL1-s/Hex L1-as, and HEX-S F/HEX-S R), and generating 3 products of various sizes (897, 590, and 191 bp) were tested.
HRM curve analysis of the PCR product generated by the HexL1-s/Hex L1-as primer proved to be highly sensitive and specific for FAdV genotyping. All serotypes generated one or more significant peaks and were visually distinct from each other in their melting curve profiles, with a confidence level greater than 99%. The applicability of the HRM/PCR Hex L1 assay was also tested on a collection of fields strains from 6 European countries: Pakistan, India, Kuwait, Mexico, Peru, Ecuador, an Australian vaccine, as well as reference strains representing the 12 serotypes, demonstrating that HRM/PCR Hex L1 test is a successful genotyping tool capable of accurately differentiating field isolates from geographically distant regions (50, 154). Subsequently, the PCR/HRM Hex L1 technique was employed to genotype FAdVs from 26 IBH cases in Australian broiler flocks, while cross-neutralization was observed between FAdV-11 and FAdV-2 reference sera using the VN test (157). These findings confirmed that the PCR/HRM Hex L1 assay is a rapid, cost-effective, and more reliable alternative for FAdV genotyping, offering greater accuracy than the VN test, PCR/RFLP, or sequencing for large-scale detection.
6 Conclusion
In general, molecular tools play a crucial role in diagnosing and managing FAdV infections in poultry. Unlike conventional diagnostic techniques, real-time PCR has revolutionized FAdV diagnosis, offering highye sensitivity, specificity, suitability for mass detection. In certain cases, more sensitive techniques such as ddPCR are recommended, particularly for detecting vaccine contaminants. For FAdV genotyping, conventional PCR followed by sequencing remains the most reliable method. Additionally, a specific real-time PCR test serves as a valuable tool, enabling detection, quantification, and genotyping in a single reaction, which makes the serotype identification process flexible. However, HRM analysis is an emerging technique that allows the detection, quantification, and identification of FAdV serotypes in a single step, streamlining the diagnostic process and reducing the impact of these infections on the poultry industry. Prioritizing multiplex and highly specific tests for agents involved in conditions like IBH, HHP, and AGE will further streamline the diagnostic process and help mitigate the impact of these infections on the poultry industry.
Author contributions
AK: Methodology, Writing – original draft. FS: Writing – review & editing. AA: Writing – review & editing. KF: Project administration, Supervision, Writing – review & editing. OI: Formal analysis, Writing – review & editing. JT: Formal analysis, Visualization, Writing – review & editing. BA: Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Benkő, M, Aoki, K, Arnberg, N, Davison, A, Echavarria, M, Hess, M, et al. ICTV virus taxonomy profile: Adenoviridae 2022. J Gen Virol. (2022) 103:001721. doi: 10.1099/jgv.0.001721
2. Hess, M. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. (2000) 29:195–206. doi: 10.1080/03079450050045440
3. Williams, SM. A laboratory manual for the isolation, identification and characterization of avian pathogens. Jacksonville, FL: American Association of Avian Pathologists (2016).
4. Meulemans, G, Couvreur, B, Decaesstecker, M, Boschmans, M, and van den Berg, TP. Phylogenetic analysis of fowl adenoviruses. Avian Pathol. (2004) 33:164–70. doi: 10.1080/03079450310001652086
5. International Committee on Taxonomy of VirusesVan Regenmortel, MHVInternational Union of Microbiological Societies. Virus taxonomy: Classification and nomenclature of viruses: Seventh report of the international committee on taxonomy of viruses: Seventh report of the international committee on taxonomy of viruses. San Diego: Academic Press (2000).
6. Gomis, S, Goodhope, R, Ojkic, D, and Willson, P. Inclusion body hepatitis as a primary disease in broilers in Saskatchewan, Canada. Avian Dis. (2006) 50:550–5. doi: 10.1637/7577-040106R.1
7. Grafl, B, Aigner, F, Liebhart, D, Marek, A, Prokofieva, I, Bachmeier, J, et al. Vertical transmission and clinical signs in broiler breeders and broilers experiencing adenoviral gizzard Erosion. Avian Pathol. (2012) 41:599–604. doi: 10.1080/03079457.2012.740614
8. Grgić, H, Philippe, C, Ojkić, D, and Nagy, E. Study of vertical transmission of fowl adenoviruses. Canadian J Vet Res. (2006) 70:230–3.
9. Mazaheri, A, Prusas, C, Voss, M, and Hess, M. Vertical transmission of fowl adenovirus serotype 4 investigated in specified pathogen-free birds after experimental infection. Archiv fur Geflugelkunde. (2003) 67:6–10. doi: 10.1016/S0003-9098(25)00062-1
10. Ono, M, Okuda, Y, Shibata, I, Sato, S, and Okad, K. Reproduction of adenoviral gizzard Erosion by the horizontal transmission of fowl adenovirus serotype 1. J Vet Med Sci. (2007) 69:1005–8. doi: 10.1292/jvms.69.1005
11. Jakab, S, Bali, K, Homonnay, Z, Kaszab, E, Ihász, K, Fehér, E, et al. Genomic epidemiology and evolution of fowl adenovirus 1. Animals. (2023) 13:2819. doi: 10.3390/ani13182819
12. Kiss, I, Homonnay, ZG, Mató, T, Bányai, K, and Palya, V. Research note: An overview on distribution of fowl adenoviruses. Poult Sci. (2021) 100:101052. doi: 10.1016/j.psj.2021.101052
13. Alvarado, IR, Villegas, P, El-Attrache, J, Jensen, E, Rosales, G, Perozo, F, et al. Genetic characterization, pathogenicity, and protection studies with an avian adenovirus isolate associated with inclusion body hepatitis. Avian Dis. (2007) 51:27–32. doi: 10.1637/0005-2086(2007)051[0027:GCPAPS]2.0.CO;2
14. McFerran, JB, McCracken, RM, Connor, TJ, and Evans, RT. Isolation of viruses from clinical outbreaks of inclusion body hepatitis. Avian Pathol. (1976) 5:315–24. doi: 10.1080/03079457608418201
15. Grafl, B, Garcia-Rueda, C, Cargill, P, Wood, A, Schock, A, Liebhart, D, et al. Fowl aviadenovirus serotype 1 confirmed as the aetiological agent of gizzard erosions in replacement pullets and layer flocks in Great Britain by laboratory and in vivo studies. Avian Pathol. (2018) 47:63–72. doi: 10.1080/03079457.2017.1367364
16. Ono, M, Okuda, Y, Yazawa, S, Shibata, I, Tanimura, N, Kimura, K, et al. Epizootic outbreaks of gizzard Erosion associated with adenovirus infection in chickens. Avian Dis. (2001) 45:268–75. doi: 10.2307/1593040
17. Nakamura, K, Tanaka, H, Mase, M, Imada, T, and Yamada, M. Pancreatic necrosis and ventricular Erosion in adenovirus-associated Hydropericardium syndrome of broilers. Vet Pathol. (2002) 39:403–6. doi: 10.1354/vp.39-3-403
18. Ye, J, Liang, G, Zhang, J, Wang, W, Song, N, Wang, P, et al. Outbreaks of serotype 4 fowl adenovirus with novel genotype, China. Emerg Microbes Infect. (2016) 5:1–12. doi: 10.1038/emi.2016.50
19. Chen, H, Dou, Y, Zheng, X, Tang, Y, Zhang, M, Zhang, Y, et al. Hydropericardium hepatitis syndrome emerged in Cherry Valley ducks in China. Transbound Emerg Dis. (2017) 64:1262–7. doi: 10.1111/tbed.12500
20. Wei, Z, Liu, H, Diao, Y, Li, X, Zhang, S, Gao, B, et al. Pathogenicity of fowl adenovirus (FAdV) serotype 4 strain SDJN in Taizhou geese. Avian Pathol. (2019) 48:477–85. doi: 10.1080/03079457.2019.1625305
21. Schachner, A, Matos, M, Grafl, B, and Hess, M. Fowl adenovirus-induced diseases and strategies for their control – a review on the current global situation. Avian Pathol. (2018) 47:111–26. doi: 10.1080/03079457.2017.1385724
22. Gallina, AM, Winterfield, RW, and Fadly, AM. Adenovirus infection and disease. II. Histopathology of natural and experimental disease. Avian Dis. (1973) 17:343–53. doi: 10.2307/1589218
23. Ganesh, K, Raghavan, R, Gowda, RNS, Satyanarayana, ML, and Suryanarayana, VVS. Purification and characterization of the Aetiological agent of Hydropericardium hepatitis syndrome from infected liver tissues of broiler chickens. Trop Anim Health Prod. (2002) 34:7–17. doi: 10.1023/A:1013777509538
24. McFerran, JB, and Adair, BM. Avian adenoviruses--a review. Avian Pathol. (1977) 6:189–217. doi: 10.1080/03079457708418228
25. Verma, KC, Malik, BS, and Kumar, S. Preliminary report on the isolation and characterization of chicken embryo-lethal-orphan (CELO) virus from poultry. Indian J Anim Sci. (1971)
26. Adair, BM, McFerran, JB, and Calvert, VM. Development of a microtitre fluorescent antibody test for serological detection of adenovirus infection in birds. Avian Pathol. (1980) 9:291–300. doi: 10.1080/03079458008418414
27. McFerran, JB, Adair, B, and Connor, TJ. Adenoviral Antigens (CELO, QBV, GAL). Am J Vet Res. (1975) 36:527–9. doi: 10.2460/ajvr.1975.36.04.527
28. Balamurugan, V, and Kataria, JM. The Hydropericardium syndrome in poultry--a current scenario. Vet Res Commun. (2004) 28:127–48. doi: 10.1023/b:verc.0000012115.86894.1e
29. Berg, NW. Rapid detection of infectious bursal disease antibodies by Counterimmunoelectrophoresis. Avian Pathol. (1982) 11:611–4. doi: 10.1080/03079458208436136
30. Kumar, R, Chandra, R, Shukla, SK, Agrawal, DK, and Kumar, M. Hydropericardium syndrome (HPS) in India: a preliminary study on the causative agent and control of the disease by inactivated autogenous vaccine. Trop Anim Health Prod. (1997) 29:158–64. doi: 10.1007/bf02633014
31. Mirzazadeh, A, Grafl, B, Berger, E, Schachner, A, and Hess, M. Longitudinal serological monitoring of commercial broiler breeders for fowl adenoviruses (FAdVs)—presence of antibodies is linked with virus excretion. Avian Dis. (2020) 65:177–87. doi: 10.1637/aviandiseases-D-20-00107
32. American Association of Avian Pathologists. A laboratory manual for the isolation and identification of avian pathogens. Jacksonville, FL: American Association of Avian Pathologists (1998).
33. Junnu, S, Lertwatcharasarakul, P, Jala, S, Phattanakunanan, S, Moonjit, P, and Songserm, T. Developing an indirect ELISA based on recombinant Hexon protein for serological detection of inclusion body hepatitis in chickens. J Vet Med Sci. (2014) 76:289–93. doi: 10.1292/jvms.13-0196
34. Mockett, AP, and Cook, JK. The use of an enzyme-linked immunosorbent assay to detect IgG antibodies to serotype-specific and group-specific antigens of fowl adenovirus serotypes 2, 3 and 4. J Virol Methods. (1983) 7:327–35. doi: 10.1016/0166-0934(83)90086-1
35. Pan, Q, Wang, J, Gao, Y, Cui, H, Liu, C, Qi, X, et al. Development and application of a novel ELISA for detecting antibodies against group I fowl adenoviruses. Appl Microbiol Biotechnol. (2020) 104:853–9. doi: 10.1007/s00253-019-10208-3
36. Rajasekhar, R, and Roy, P. Recombinant Hexon antigen based single serum dilution ELISA for rapid serological profiling against fowl Adenovirus-4 causing Hydropericardium syndrome in chickens. J Virol Methods. (2014) 207:121–7. doi: 10.1016/j.jviromet.2014.06.017
37. Feichtner, F, Schachner, A, Berger, E, and Hess, M. Fiber-based fluorescent microsphere immunoassay (FMIA) as a novel multiplex serodiagnostic tool for simultaneous detection and differentiation of all clinically relevant fowl adenovirus (FAdV) serotypes. J Immunol Methods. (2018) 458:33–43. doi: 10.1016/j.jim.2018.03.002
38. Mirmajlessi, SM, Destefanis, M, Gottsberger, RA, Mänd, M, and Loit, E. PCR-based specific techniques used for detecting the Most important pathogens on strawberry: a systematic review. Syst Rev. (2015) 4:9. doi: 10.1186/2046-4053-4-9
39. Goodwin, MA, Latimer, KS, Resurreccion, RS, Miller, PG, and Campagnoli, RP. DNA in situ hybridization for the rapid diagnosis of massive necrotizing avian adenovirus hepatitis and pancreatitis in chicks. Avian Dis. (1996) 40:828–31. doi: 10.2307/1592305
40. Latimer, KS, Niagro, FD, Williams, OC, Ramis, A, Goodwin, MA, Ritchie, BW, et al. Diagnosis of avian adenovirus infections using DNA in situ hybridization. Avian Dis. (1997) 41:773–82. doi: 10.2307/1592329
41. Ramis, A, Latimer, KS, Niagro, FD, Campagnoli, RP, Ritchie, BW, and Pesti, D. Diagnosis of psittacine beak and feather disease (PBFD) viral infection, avian polyomavirus infection, adenovirus infection and herpesvirus infection in psittacine tissues using DNA in situ hybridization. Avian Pathol. (1994) 23:643–57. doi: 10.1080/03079459408419034
42. Yang, S, and Rothman, RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. (2004) 4:337–48. doi: 10.1016/S1473-3099(04)01044-8
43. Meulemans, G, Boschmans, M, Van Den Berg, TP, and Decaesstecker, M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. (2001) 30:655–60. doi: 10.1080/03079450120092143
44. Niczyporuk, JS. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Arch Virol. (2016) 161:33–42. doi: 10.1007/s00705-015-2635-4
45. Raue, R, and Hess, M. Hexon based PCRs combined with restriction enzyme analysis for rapid detection and differentiation of fowl adenoviruses and egg drop syndrome virus. J Virol Methods. (1998) 73:211–7. doi: 10.1016/S0166-0934(98)00065-2
46. Xie, Z, Fadl, AA, Girshick, T, and Khan, MI. Detection of avian adenovirus by polymerase chain reaction. Avian Dis. (1999) 43:98–105. doi: 10.2307/1592767
47. Norrby, E. The relationship between the soluble antigens and the Virion of adenovirus type 3: IV. Immunological complexity of soluble components. Virology. (1969) 37:565–76. doi: 10.1016/0042-6822(69)90274-8
48. Ganesh, K, Suryanarayana, V, Raghavan, R, and Gowda, S. Nucleotide sequence of L1 and part of P1 of Hexon gene of fowl adenovirus associated with Hydropericardium hepatitis syndrome differs with the corresponding region of other fowl adenoviruses. Vet Microbiol. (2001) 78:1–11. doi: 10.1016/s0378-1135(00)00288-1
49. Athappily, FK, Murali, R, Rux, JJ, Cai, Z, and Burnett, RM. The refined crystal structure of Hexon, the major coat protein of adenovirus type 2, at 2·9 Å resolution. J Mol Biol. (1994) 242:430–55. doi: 10.1006/jmbi.1994.1593
50. Steer, PA, Kirkpatrick, NC, O’Rourke, D, and Noormohammadi, AH. Classification of fowl adenovirus serotypes by use of high-resolution melting-curve analysis of the Hexon gene region. J Clin Microbiol. (2009) 47:311–21. doi: 10.1128/jcm.01567-08
51. Kondabattula, G, Suryanarayana, V, and Raghavan, R. Detection of fowl adenovirus associated with Hydropericardium hepatitis syndrome by a polymerase chain reaction. Vet Res Commun. (2002) 26:73–80. doi: 10.1023/A:1013361906791
52. Mase, M, Hiramatsu, K, Nishijima, N, Iseki, H, and Watanabe, S. Identification of specific serotypes of fowl adenoviruses isolated from diseased chickens by PCR. J Vet Med Sci. (2021) 83:130–3. doi: 10.1292/jvms.20-0400
53. Niczyporuk, JS, Samorek-Salamonowicz, E, and Czekaj, H. Incidence and detection of Aviadenoviruses of serotypes 1 and 5 in poultry by PCR and duplex PCR. Bulletin Vet Institute Puławy. (2010) 54:451–5.
54. Zhao, P, Dong, X, and Cui, Z. Isolation, identification, and Gp85 characterization of a subgroup a avian Leukosis virus from a contaminated live Newcastle disease virus vaccine, first report in China. Poult Sci. (2014) 93:2168–74. doi: 10.3382/ps.2014-03963
55. Okuda, Y, Ono, M, Shibata, I, Sato, S, and Akashi, H. Comparison of the polymerase chain reaction-restriction fragment length polymorphism pattern of the Fiber gene and Pathogenicity of Serotype-1 fowl adenovirus isolates from gizzard erosions and from feces of clinically healthy chickens in Japan. J Vet Diagn Invest. (2006) 18:162–7. doi: 10.1177/104063870601800204
56. Almeida, S, Dorneles, EMS, Diniz, C, Abreu, V, Sousa, C, Alves, J, et al. Quadruplex PCR assay for identification of Corynebacterium Pseudotuberculosis differentiating Biovar Ovis and Equi. BMC Vet Res. (2017) 13:290. doi: 10.1186/s12917-017-1210-5
57. Hao, L, Xie, J, Chen, S, Wang, S, Gong, Z, Ling, K-S, et al. A multiple RT-PCR assay for simultaneous detection and differentiation of latent viruses and Apscarviroids in apple trees. J Virol Methods. (2016) 234:16–21. doi: 10.1016/j.jviromet.2016.04.003
58. Yao, M, Zhang, X, Gao, Y, Song, S, Danning, X, and Yan, L. Development and application of multiplex PCR method for simultaneous detection of seven viruses in ducks. BMC Vet Res. (2019) 15:103. doi: 10.1186/s12917-019-1820-1
59. Romanova, N, Corredor, JC, and Nagy, É. Detection and quantitation of fowl adenovirus genome by a real-time PCR assay. J Virol Methods. (2009) 159:58–63. doi: 10.1016/j.jviromet.2009.02.026
60. Zhai, X, Mei, X, Xuan, W, Zuo, L, Zhou, L, Tian, Y, et al. A loop-mediated isothermal amplification coupling with a lateral flow dipstick for rapid and specific detection of fowl adenovirus Serotype-4. J Virol Methods. (2019) 270:79–86. doi: 10.1016/j.jviromet.2019.04.026
61. Kaján, G, Sameti, S, and Benkő, M. Partial sequence of the DNA-dependent DNA polymerase gene of fowl adenoviruses: a reference panel for a general diagnostic PCR in poultry. Acta Vet Hung. (2011) 59:279–85. doi: 10.1556/avet.2011.006
62. Abdul-Careem, MF, Hunter, BD, Nagy, É, Read, LR, Babak Sanei, J, Spencer, L, et al. Development of a real-time PCR assay using SYBR green chemistry for monitoring Marek’s disease virus genome load in feather tips. J Virol Methods. (2006) 133:34–40. doi: 10.1016/j.jviromet.2005.10.018
63. Watanabe, M, Kohdera, U, Kino, M, Haruta, T, Nukuzuma, S, Suga, T, et al. Detection of adenovirus DNA in clinical samples by SYBR green real-time polymerase chain reaction assay. Pediatr Int. (2005) 47:286–91. doi: 10.1111/j.1442-200x.2005.02057.x
64. Willoughby, K, Valdazo-González, B, Maley, M, Gilray, J, and Nettleton, PF. Development of a real time RT-PCR to detect and type ovine Pestiviruses. J Virol Methods. (2006) 132:187–94. doi: 10.1016/j.jviromet.2005.10.007
65. Mackay, I, Arden, K, and Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. (2002) 30:1292–305. doi: 10.1093/nar/30.6.1292
66. Günes, A, Marek, A, Grafl, B, Berger, E, and Hess, M. Real-time PCR assay for universal detection and quantitation of all five species of fowl adenoviruses (FAdV-A to FAdV-E). J Virol Methods. (2012) 183:147–53. doi: 10.1016/j.jviromet.2012.04.005
67. Bustin, SA, Benes, V, Garson, JA, Hellemans, J, Huggett, J, Kubista, M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. (2009) 55:611–22. doi: 10.1373/clinchem.2008.112797
68. Slana, I, Kralik, P, Kralova, A, and Pavlik, I. On-farm spread of Mycobacterium avium subsp. paratuberculosis in raw milk studied by IS900 and F57 competitive real time quantitative PCR and culture examination. Int J Food Microbiol. (2008) 128:250–7. doi: 10.1016/j.ijfoodmicro.2008.08.013
69. Li, X, Keran Zhang, Y, Pei, JX, Ruan, S, and Zhang, G. Development and application of an MRT-qPCR assay for detecting coinfection of six vertically transmitted or immunosuppressive avian viruses. Front Microbiol. (2020) 11:1581. doi: 10.3389/fmicb.2020.01581
70. Niczyporuk, JS, and Czekaj, H. A comparative pathogenicity analysis of two adenovirus strains, 1/a and 8a/E, isolated from poultry in Poland. Arch Virol. (2018) 163:3005–13. doi: 10.1007/s00705-018-3965-9
71. Ugwu, C, Bejo, M, Omar, A, Isa, N, and Ideris, A. TaqMan probe-based qPCR method for specific detection and quantification of fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. Open Vet J. (2023) 13:171–8. doi: 10.5455/OVJ.2023.v13.i2.4
72. Wang, J, Wang, J, Chen, P, Liu, L, and Yuan, W. Development of a TaqMan-based real-time PCR assay for rapid and specific detection of fowl aviadenovirus serotype 4. Avian Pathol. (2017) 46:338–43. doi: 10.1080/03079457.2016.1278428
73. Notomi, T, Okayama, H, Masubuchi, H, Yonekawa, T, Watanabe, K, Amino, N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. (2000) 28:63e–663e. doi: 10.1093/nar/28.12.e63
74. Tomita, N, Mori, Y, Kanda, H, and Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. (2008) 3:877–82. doi: 10.1038/nprot.2008.57
75. Chen, H-t, Zhang, J, Ma, Y-p, Ma, L-n, Ding, Y-z, Liu, X-t, et al. Reverse transcription loop-mediated isothermal amplification for the rapid detection of infectious bronchitis virus in infected chicken tissues. Mol Cell Probes. (2010) 24:104–6. doi: 10.1016/j.mcp.2009.10.001
76. Huang, C-H, Lai, G-H, Lee, M-S, Lin, W-H, Lien, Y-Y, Hsueh, S-C, et al. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of chicken Anaemia virus. J Appl Microbiol. (2010) 108:917–24. doi: 10.1111/j.1365-2672.2009.04481.x
77. Postel, A, Letzel, T, Frischmann, S, Grund, C, Beer, M, and Harder, T. Evaluation of two commercial loop-mediated isothermal amplification assays for detection of avian influenza H5 and H7 hemagglutinin genes. J Vet Diagn Invest. (2010) 22:61–6. doi: 10.1177/104063871002200110
78. Pham, HM, Nakajima, C, Ohashi, K, and Onuma, M. Loop-mediated isothermal amplification for rapid detection of Newcastle disease virus. J Clin Microbiol. (2005) 43:1646–50. doi: 10.1128/JCM.43.4.1646-1650.2005
79. Xu, J, Zhang, Z, Yin, Y, Cui, S, Shouzhen, X, Guo, Y, et al. Development of reverse-transcription loop-mediated isothermal amplification for the detection of infectious bursal disease virus. J Virol Methods. (2009) 162:267–71. doi: 10.1016/j.jviromet.2009.07.010
80. Woźniakowski, G, Samorek-Salamonowicz, E, and Kozdruń, W. Rapid detection of Marek’s disease virus in feather follicles by loop-mediated amplification. Avian Dis. (2011) 55:462–7. doi: 10.1637/9668-012711-ResNote.1
81. Xie, Z, Tang, Y, Fan, Q, Liu, J, Pang, Y, Deng, X, et al. Rapid detection of group I avian adenoviruses by a loop-mediated isothermal amplification. Avian Dis. (2011) 55:575–9. doi: 10.1637/9719-031611-Reg.1
82. Yuan, X-Y, Wang, Y-L, Meng, K, Zhang, Y-X, Huai-Ying, X, and Ai, W. LAMP real-time turbidity detection for fowl adenovirus. BMC Vet Res. (2019) 15:256. doi: 10.1186/s12917-019-2015-5
83. Zhang, X, Lowe, SB, and Gooding, JJ. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens Bioelectron. (2014) 61:491–9. doi: 10.1016/j.bios.2014.05.039
84. Yulong, Z, Xia, Z, Hongwei, Z, Wei, L, Wenjie, Z, and Xitai, H. Rapid and sensitive detection of Enterobacter sakazakii by cross-priming amplification combined with immuno-blotting analysis. Mol Cell Probes. (2010) 24:396–400. doi: 10.1016/j.mcp.2010.09.001
85. Zheng, F, Li, S, Wang, S, Feng, T, Jiang, Z, and Pan, J. Cross-priming isothermal amplification combined with nucleic acid test strips for detection of meat species. Anal Biochem. (2020) 597:113672. doi: 10.1016/j.ab.2020.113672
86. Niczyporuk, JS, Woźniakowski, G, and Samorek-Salamonowicz, E. Application of cross-priming amplification (CPA) for detection of fowl adenovirus (FAdV) strains. Arch Virol. (2015) 160:1005–13. doi: 10.1007/s00705-015-2355-9
87. Piepenburg, O, Williams, CH, Stemple, DL, and Armes, NA. DNA detection using recombination proteins. PLoS Biol. (2006) 4:e204. doi: 10.1371/journal.pbio.0040204
88. Boyle, DS, Lehman, DA, Lillis, L, Peterson, D, Singhal, M, Armes, N, et al. Rapid detection of HIV-1 Proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio. (2013) 4:e00135–13. doi: 10.1128/mBio.00135-13
89. James, AS, Todd, S, Pollak, NM, Marsh, GA, and Macdonald, J. Ebolavirus diagnosis made simple, comparable and faster than molecular detection methods: preparing for the future. Virol J. (2018) 15:75. doi: 10.1186/s12985-018-0985-8
90. Tan, K-K, Azizan, NS, Yaacob, CN, Seri, NAACM, Samsudin, NI, Teoh, B-T, et al. Operational utility of the reverse-transcription recombinase polymerase amplification for detection of dengue virus. BMC Infect Dis. (2018) 18:169. doi: 10.1186/s12879-018-3065-1
91. Yang, Y, Qin, X, Zhang, W, Li, Z, Zhang, S, Li, Y, et al. Development of an isothermal recombinase polymerase amplification assay for rapid detection of pseudorabies virus. Mol Cell Probes. (2017) 33:32–5. doi: 10.1016/j.mcp.2017.03.005
92. Yang, Y, Qin, X, Sun, Y, Cong, G, Li, Y, and Zhang, Z. Development of isothermal recombinase polymerase amplification assay for rapid detection of porcine circovirus type 2. BioMed Res Int. (2017) 2017:e8403642. doi: 10.1155/2017/8403642
93. Liu, L, Wang, J, Zhang, R, Lin, M, Shi, R, Han, Q, et al. Visual and equipment-free reverse transcription recombinase polymerase amplification method for rapid detection of foot-and-mouth disease virus. BMC Vet Res. (2018) 14:263. doi: 10.1186/s12917-018-1594-x
94. Wu, S, Ping, X, Xiangbin, X, and Liu, S-B. Research Progress and prospects of nucleic acid isothermal amplification technology. Biocell. (2023) 47:2385–95. doi: 10.32604/biocell.2023.029687
95. Del Río, JS, Lobato, IM, Mayboroda, O, Katakis, I, and O’Sullivan, CK. Enhanced solid-phase recombinase polymerase amplification and electrochemical detection. Anal Bioanal Chem. (2017) 409:3261–9. doi: 10.1007/s00216-017-0269-y
96. Li, J, Macdonald, J, and von Stetten, F. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst. (2019) 144:31–67. doi: 10.1039/C8AN01621F
97. Wang, X, Xie, S, Chen, X, Peng, C, Xiaoli, X, Wei, W, et al. A rapid and convenient method for on-site detection of MON863 maize through real-time fluorescence recombinase polymerase amplification. Food Chem. (2020) 324:126821. doi: 10.1016/j.foodchem.2020.126821
98. Miao, F, Zhang, J, Li, N, Chen, T, Wang, L, Zhang, F, et al. Rapid and sensitive recombinase polymerase amplification combined with lateral flow strip for detecting African swine fever virus. Front Microbiol. (2019) 10:1004. doi: 10.3389/fmicb.2019.01004
99. Li, R, Wang, J, Liu, L, Zhang, R, Hao, X, Han, Q, et al. Direct detection of Actinobacillus pleuropneumoniae in swine lungs and tonsils by real-time recombinase polymerase amplification assay. Mol Cell Probes. (2019) 45:14–8. doi: 10.1016/j.mcp.2019.03.007
100. Li, Y, Li, L, Fan, X, Zou, Y, Zhang, Y, Wang, Q, et al. Development of real-time reverse transcription recombinase polymerase amplification (RPA) for rapid detection of Peste des Petits ruminants virus in clinical samples and its comparison with real-time PCR test. Sci Rep. (2018) 8:17760. doi: 10.1038/s41598-018-35636-5
101. Zhang, J, Liu, J, An, D, Fan, Y, Cheng, Z, Tang, Y, et al. A novel recombinase polymerase amplification assay for rapid detection of epidemic fowl adenovirus. Poult Sci. (2020) 99:6446–53. doi: 10.1016/j.psj.2020.08.021
102. Vogelstein, B, and Kinzler, KW. Digital PCR. Proc Natl Acad Sci. (1999) 96:9236–41. doi: 10.1073/pnas.96.16.9236
103. Hindson, BJ, Ness, KD, Masquelier, DA, Belgrader, P, Heredia, NJ, Makarewicz, AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. (2011) 83:8604–10. doi: 10.1021/ac202028g
104. Kopylova, KV, Ed, W, Kasparov, IVM, and Smolnikova, MV. Digital PCR as a highly sensitive diagnostic tool: a review. Mol Biol. (2023) 57:793–801. doi: 10.1134/S0026893323050059
105. Morisset, D, Štebih, D, Milavec, M, Gruden, K, and Žel, J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS One. (2013) 8:e62583. doi: 10.1371/journal.pone.0062583
106. Schwartz, SL, and Lowen, AC. Droplet digital PCR: a novel method for detection of influenza virus defective interfering particles. J Virol Methods. (2016) 237:159–65. doi: 10.1016/j.jviromet.2016.08.023
107. Yan, Y, Jia, X-j, Wang, H-h, Xiao-fei, F, Ji, J-m, He, P-y, et al. Dynamic quantification of avian influenza H7N9(a) virus in a human infection during clinical treatment using droplet digital PCR. J Virol Methods. (2016) 234:22–7. doi: 10.1016/j.jviromet.2016.04.001
108. Li, Q, Zhang, Y, Meng, F, Jiang, H, Guanlong, X, Ding, J, et al. A new strategy for the detection of chicken infectious Anemia virus contamination in attenuated live vaccine by droplet digital PCR. Biomed Res Int. (2019) 2019:2750472–9. doi: 10.1155/2019/2750472
109. Li, Y, Jiayuan, F, Chang, S, Fang, L, Cui, S, Wang, Y, et al. Isolation, identification, and Hexon gene characterization of fowl adenoviruses from a contaminated live Newcastle disease virus vaccine. Poult Sci. (2017) 96:1094–9. doi: 10.3382/ps/pew405
110. Su, Q, Li, Y, Meng, F, Cui, Z, Chang, S, and Zhao, P. Newcastle disease virus-attenuated vaccine co-contaminated with fowl adenovirus and chicken infectious Anemia virus results in inclusion body hepatitis-Hydropericardium syndrome in poultry. Vet Microbiol. (2018) 218:52–9. doi: 10.1016/j.vetmic.2018.03.019
111. Li, Y, Cui, S, Cui, Z, Chang, S, and Zhao, P. Genome analysis and pathogenicity of reticuloendotheliosis virus isolated from a contaminated vaccine seed against infectious bursal disease virus: first report in China. J Gen Virol. (2016) 97:2809–15. doi: 10.1099/jgv.0.000588
112. Dong, G, Meng, F, Zhang, Y, Cui, Z, Lidan, H, Chang, S, et al. Development and evaluation of a droplet digital PCR assay for the detection of fowl adenovirus serotypes 4 and 10 in attenuated vaccines. J Virol Methods. (2019) 265:59–65. doi: 10.1016/j.jviromet.2018.09.005
113. Kafatos, FC, Weldon Jones, C, and Efstratiadis, A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. (1979) 7:1541–52. doi: 10.1093/nar/7.6.1541
114. Cheng, L-T, Zeng, Y-J, Chu, C-Y, and Wang, H-Y. Development of a quick dot blot assay for the titering of bovine ephemeral fever virus. BMC Vet Res. (2019) 15:313. doi: 10.1186/s12917-019-2059-6
115. Hou, L, Qi, S, Zhang, Y, Liu, D, Mao, Y, and Zhao, P. Development of a PCR-based dot blot assay for the detection of fowl adenovirus. Poult Sci. (2022) 101:101540. doi: 10.1016/j.psj.2021.101540
116. Meng, F, Dong, G, Zhang, Y, Tian, S, Cui, Z, Chang, S, et al. Co-infection of fowl adenovirus with different immunosuppressive viruses in a chicken flock. Poult Sci. (2018) 97:1699–705. doi: 10.3382/ps/pex414
117. Ali, Z, Jin, G, Zhili, H, Wang, Z, Khan, MA, Dai, J, et al. A review on NanoPCR: history, mechanism and applications. J Nanosci Nanotechnol. (2018) 18:8029–46. doi: 10.1166/jnn.2018.16390
118. Khaliq, RA, Kafafy, R, Salleh, HM, and Faris, WF. Enhancing the efficiency of polymerase chain reaction using graphene Nanoflakes. Nanotechnology. (2012) 23:455106. doi: 10.1088/0957-4484/23/45/455106
119. Sang, F-M, Li, X, and Liu, J. Development of Nano-polymerase chain reaction and its application. Chin J Anal Chem. (2017) 45:1745–53. doi: 10.1016/S1872-2040(17)61051-X
120. Li, H, and Rothberg, L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Natl Acad Sci USA. (2004) 101:14036–9. doi: 10.1073/pnas.0406115101
121. Ma, X, Cui, Y, Qiu, Z, Zhang, B, and Cui, S. A nanoparticle-assisted PCR assay to improve the sensitivity for rapid detection and differentiation of wild-type pseudorabies virus and gene-deleted vaccine strains. J Virol Methods. (2013) 193:374–8. doi: 10.1016/j.jviromet.2013.07.018
122. Wang, X, Bai, A, Zhang, J, Kong, M, Cui, Y, Ma, X, et al. A new nanoPCR molecular assay for detection of porcine Bocavirus. J Virol Methods. (2014) 202:106–11. doi: 10.1016/j.jviromet.2014.02.029
123. Yuan, W, Li, Y, Li, P, Song, Q, Li, L, and Sun, J. Development of a nanoparticle-assisted PCR assay for detection of porcine epidemic diarrhea virus. J Virol Methods. (2015) 220:18–20. doi: 10.1016/j.jviromet.2015.04.008
124. Zhu, Y, Liang, L, Luo, Y, Wang, G, Wang, C, Cui, Y, et al. A sensitive duplex nanoparticle-assisted PCR assay for identifying porcine epidemic diarrhea virus and porcine transmissible gastroenteritis virus from clinical specimens. Virus Genes. (2017) 53:71–6. doi: 10.1007/s11262-016-1405-z
125. Wang, L, Zheng, L, FuQiang, L, XiuLi, L, WeiKe, R, Chao, L, et al. “Establishment and application of Nano PCR assay for detection on Fowl aviadenovirus serotype 4,” Zhongguo Yufang Shouyi Xuebao (Chinese J. Prev. Vet. Med.). Chinese Association of Preventive Veterinary Medicine. (2019) 41:701–4.
126. Luan, Q, Jiang, Z, Wang, D, Wang, S, Yin, Y, and Wang, J. A sensitive triple nanoparticle-assisted PCR assay for detection of fowl adenovirus, infectious bursal disease virus and chicken anemia virus. J Virol Methods. (2022) 303:114499. doi: 10.1016/j.jviromet.2022.114499
127. Tan, Y, Jian, H, Zhang, R, Wang, J, Zhou, C, Xiao, Y, et al. Applying amplification refractory mutation system technique to detecting cell-free fetal DNA for single-gene disorders purpose. Front Genet. (2023) 14:1071406. doi: 10.3389/fgene.2023.1071406
128. Waqar, A, Haq, I., and Chaudhry, B.. (2013). Tetra-primer amplification refractory mutation system for screening T2D, TCF7L2 variant. Available online at: https://www.semanticscholar.org/paper/Tetra-primer-Amplification-Refractory-Mutation-for-Waqar-Haq/a2a88f992c4fc9ff71b9589a7f4e793af58aefb0
129. Furtado, LF, Viana, C, Zuccherato, LW, Alves, WP, de Oliveira, VNGM, da Silva, VJ, et al. First identification of the benzimidazole resistance-associated F200Y SNP in the beta-tubulin gene in Ascaris lumbricoides. PLoS One. (2019) 14:e0224108. doi: 10.1371/journal.pone.0224108
130. Chen, J, Xinyun, X, Dalhaimer, P, and Zhao, L. Tetra-primer amplification-refractory mutation system (ARMS)—PCR for genotyping mouse leptin gene mutation. Animals. (2022) 12:2680. doi: 10.3390/ani12192680
131. Grafl, B, Prokofieva, I, Wernsdorf, P, Steinborn, R, and Hess, M. Infection with an Apathogenic fowl adenovirus Serotype-1 strain (CELO) prevents adenoviral gizzard Erosion in broilers. Vet Microbiol. (2014) 172:177–85. doi: 10.1016/j.vetmic.2014.05.020
132. Doudna, JA, and Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science. (2014) 346:1258096. doi: 10.1126/science.1258096
133. Hsu, PD, Lander, ES, and Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. (2014) 157:1262–78. doi: 10.1016/j.cell.2014.05.010
134. Horvath, P, and Barrangou, R. CRISPR/Cas, the immune system of Bacteria and Archaea. Science. (2010) 327:167–70. doi: 10.1126/science.1179555
135. Pausch, P, Al-Shayeb, B, Bisom-Rapp, E, Tsuchida, CA, Li, Z, Cress, BF, et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. (2020) 369:333–7. doi: 10.1126/science.abb1400
136. Newsom, S, Parameshwaran, HP, Martin, L, and Rajan, R. The CRISPR-Cas mechanism for adaptive immunity and alternate bacterial functions fuels diverse biotechnologies. Front Cell Infect Microbiol. (2021) 10:619763. doi: 10.3389/fcimb.2020.619763
137. Kaminski, MM, Abudayyeh, OO, Gootenberg, JS, Zhang, F, and Collins, JJ. CRISPR-based diagnostics. Nature Biomed Eng. (2021) 5:643–56. doi: 10.1038/s41551-021-00760-7
138. Maxmen, A. Faster, better, cheaper: the rise of CRISPR in disease detection. Nature. (2019) 566:437. doi: 10.1038/d41586-019-00601-3
139. Gootenberg, JS, Abudayyeh, OO, Lee, JW, Essletzbichler, P, Dy, AJ, Joung, J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. (2017) 356:438–42. doi: 10.1126/science.aam9321
140. Zhou, X, Wang, S, Ma, Y, Li, Y, Deng, G, Shi, J, et al. Rapid detection of avian influenza virus based on CRISPR-Cas12a. Virol J. (2023) 20:1–10. doi: 10.1186/s12985-023-02232-7
141. Wu, Y, Zhan, J, Shan, Z, Li, Y, Liu, Y, Li, Y, et al. CRISPR-Cas13a-based detection method for avian influenza virus. Front Microbiol. (2023) 14:1288951. doi: 10.3389/fmicb.2023.1288951
142. Huang, Z, Fang, J, Zhou, M, Gong, Z, and Xiang, T. CRISPR-Cas13: a new technology for the rapid detection of pathogenic microorganisms. Front Microbiol. (2022) 13:1011399. doi: 10.3389/fmicb.2022.1011399
143. Gootenberg, JS, Abudayyeh, OO, Kellner, MJ, Joung, J, Collins, JJ, and Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. (2018) 360:439–44. doi: 10.1126/science.aaq0179
144. Myhrvold, C, Freije, CA, Gootenberg, JS, Abudayyeh, OO, Metsky, HC, Durbin, AF, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. (2018) 360:444–8. doi: 10.1126/science.aas8836
145. Ning, B, Tao, Y, Zhang, S, Huang, Z, Tian, D, Lin, Z, et al. A smartphone-Read ultrasensitive and quantitative saliva test for COVID-19. Sci Adv. (2021) 7:eabe3703. doi: 10.1126/sciadv.abe3703
146. Tian, Y, Fan, Z, Ling, X, Cao, Y, Chen, S, Pan, Z, et al. CRISPR/Cas13a-assisted rapid and portable HBV DNA detection for low-level viremia patients. Emerg Microbes Infect. (2023) 12:e2177088. doi: 10.1080/22221751.2023.2177088
147. Yin, D, Yin, L, Wang, J, Dai, Y, Shen, X, Zhao, R, et al. Visual detection of fowl adenovirus serotype 4 via a portable CRISPR/Cas13a-based lateral flow assay. Avian Pathol. (2023) 52:438–45. doi: 10.1080/03079457.2023.2254253
148. Yu, Z, Shao, Y, Shi, D, Yanli Dong, Y, Zhang, FC, Wang, Z, et al. A rapid, ultrasensitive, and highly specific method for detecting fowl adenovirus serotype 4 based on the LAMP-CRISPR/Cas12a system. Poult Sci. (2024) 103:104048. doi: 10.1016/j.psj.2024.104048
149. Jadhao, SJ, Deepak, JN, Kataria, JM, Kataria, RS, Tiwari, AK, Somvanshi, R, et al. Characterisation of fowl adenoviruses from chickens affected with infectious Hydropericardium during 1994-1998 in India. Indian J Exp Biol. (2003) 41:321–7.
150. Li, S, Zhao, R, Yang, Q, Meihua, W, Ma, J, Wei, Y, et al. Phylogenetic and pathogenic characterization of current fowl adenoviruses in China. Infect Genet Evol. (2022) 105:105366. doi: 10.1016/j.meegid.2022.105366
151. Grimes, TM, Culver, DH, and King, DJ. Virus-neutralizing antibody titers against 8 avian adenovirus serotypes in breeder hens in Georgia by a microneutralization procedure. Avian Dis. (1977) 21:220–9. doi: 10.2307/1589342
152. Erny, K, Pallister, J, and Sheppard, M. Immunological and molecular comparison of fowl adenovirus serotypes 4 and 10. Arch Virol. (1995) 140:491–501. doi: 10.1007/BF01718426
153. Zsák, L, and Kisary, J. Grouping of fowl adenoviruses based upon the restriction patterns of DNA generated by bam HI and Hin dIII: restriction patterns of DNA generated by bam HI and hind III. Intervirology. (2008) 22:110–4. doi: 10.1159/000149541
154. Marek, A, Günes, A, Schulz, E, and Hess, M. Classification of fowl adenoviruses by use of phylogenetic analysis and high-resolution melting-curve analysis of the Hexon L1 gene region. J Virol Methods. (2010) 170:147–54. doi: 10.1016/j.jviromet.2010.09.019
155. Herrmann, MG, Durtschi, JD, Kathryn Bromley, L, Wittwer, CT, and Voelkerding, KV. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem. (2006) 52:494–503. doi: 10.1373/clinchem.2005.063438
156. Herrmann, MG, Durtschi, JD, Wittwer, CT, and Voelkerding, KV. Expanded instrument comparison of amplicon DNA melting analysis for mutation scanning and genotyping. Clin Chem. (2007) 53:1544–8. doi: 10.1373/clinchem.2007.088120
157. Steer, P, O’Rourke, D, Ghorashi, S, and Noormohammadi, A. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust Vet J. (2011) 89:184–92. doi: 10.1111/j.1751-0813.2011.00695.x
158. Mase, M, Nakamura, K, and Imada, T. Characterization of fowl adenovirus serotype 4 isolated from chickens with Hydropericardium syndrome based on analysis of the short Fiber protein gene. J Vet Diagn Invest. (2010) 22:218–23. doi: 10.1177/104063871002200207
159. Mase, M, Chuujou, M, Inoue, T, Nakamura, K, Yamaguchi, S, and Imada, T. Genetic characterization of fowl adenoviruses isolated from chickens with Hydropericardium syndrome in Japan. J Vet Med Sci. (2009) 71:1455–8. doi: 10.1292/jvms.001455
160. Mase, M, Mitake, H, Inoue, T, and Imada, T. Identification of group I-III avian adenovirus by PCR coupled with direct sequencing of the Hexon gene. J Vet Med Sci. (2009) 71:1239–42. doi: 10.1292/jvms.71.1239
161. Cizmecigil, UY, Umar, S, Yilmaz, A, Bayraktar, E, Turan, N, Tali, B, et al. Characterisation of fowl adenovirus (FAdV-8b) strain concerning the geographic analysis and pathological lesions associated with inclusion body hepatitis in broiler flocks in Turkey. J. Vet Res. (2020) 64:231–7. doi: 10.2478/jvetres-2020-0026
162. Wang, L, Zheng, L, Jiang, S, Li, X, Chao, L, Zhang, L, et al. Isolation, identification and genetic characterization analysis of a fowl Aviadenovirus serotype 4 strain from Tianjin, China. Infect Genet Evol. (2021) 96:105078. doi: 10.1016/j.meegid.2021.105078
Keywords: fowl adenovirus, molecular diagnosis, polymerase chain reaction, real-time PCR, isothermal amplification, CRISPR/Cas13, genotyping, HRM
Citation: Kardoudi A, Siham F, Abdelmounaaim A, Faouzi K, Ikram O, Thomas J and Abdelouaheb B (2025) A snapshot on molecular technologies for diagnosing FAdV infections. Front. Vet. Sci. 12:1558257. doi: 10.3389/fvets.2025.1558257
Edited by:
Francisco Javier Salguero, UK Health Security Agency (UKHSA), United KingdomReviewed by:
Soyoun Hwang, Animal and Plant Health Inspection Service (USDA), United StatesAhmed R. Elbestawy, Menoufia University, Egypt
Copyright © 2025 Kardoudi, Siham, Abdelmounaaim, Faouzi, Ikram, Thomas and Abdelouaheb. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amina Kardoudi, YW1pbmEua2FyZG91ZGkyQGdtYWlsLmNvbQ==
 Amina Kardoudi
Amina Kardoudi Fellahi Siham
Fellahi Siham Allaoui Abdelmounaaim2
Allaoui Abdelmounaaim2 Kichou Faouzi
Kichou Faouzi