- 1College of Animal Science and Technology, Northeast Agricultural University, Harbin, China
- 2Lanxi Breeding Farm, Lanxi, China
- 3Zhejiang Mebolo Swine Breeding Co., Ltd., Jinhua, China
- 4Zhejiang Key Laboratory of Livestock and Poultry Biotech Breeding, Institute of Animal Husbandry and Veterinary Science, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
ANXA5 is a pleiotropic candidate gene, however its effect on piglet production trait remains unclear. In this study, a novel SNP: g.-519 A > G was identified in ANXA5 promoter. In Min pigs, association analysis showed the birth weight of AA animals at g.-674 C > A was higher than that of the AC or CC piglets (p < 0.05), while the heterozygous of g.-519 A > G had a higher weight than the homozygote at day 3, 7, 14, 21 after birth (p < 0.05). Porcine ANXA5 and NR4A1 existed in piglets’ gastrointestinal tract, and NR4A1 localized to the nucleus and cytoplasm which regulated the expression of ANXA5. Luciferase reporter analysis demonstrated that the deletion of predictive NR4A1 binding region decreased the luciferase activity of porcine ANXA5 promoter (p < 0.05), and the A allele of g.-519 A > G within this region had significantly higher luciferase activity than the G allele (p < 0.01). In conclusion, this research suggested that g.-519 A > G was a piglet weight variant that regulated the transcription of ANXA5 partially by NR4A1.
1 Introduction
The genetic improvement of reproduction trait is important for the porcine production. In the past decades, gene-based selection for female fertility has increased the ovulation rate and the number of piglets born (1). However, the proportion of low birth weight piglets increased accordingly (2–4). Additionally, selection for larger litter size produced sows with a repeatable low average litter birth weight phenotype (3). Piglets’ low birth weight is associated with increased mortality, high infection risk, poor growth and meat quality (5, 6). Hence, it is encouraged to identify pleiotropic genes or genetic variations, which contributes to understand the genetic mechanism of complex traits and benefits porcine production.
As an important member of Annexin protein family, AnnexinA5 (ANXA5) is linked to multiple functions, such as placental anticoagulant, cell apoptosis, immune response, and so on (7, 8). Häggman and Uimari (9) located a putative lethal haplotype on SSC8 in Yorkshire boars, confirmed the effect of this haplotype on the number of stillborn piglets, and suggested ANXA5 as the positional and functional candidate gene for swine reproduction and fertility traits. Intra-uterine growth restriction (IUGR) is a critical factor for piglets’ mortality, which impairs individual development and reduces the birth weight. Wang et al. (10) found ANXA5 expressed differentially in the jejunal mucosa of IUGR and normal piglets. In order to understand the genetic interplay between litter size and production traits in the Yorkshire, Wei et al. (11) reported a set of pleiotropic candidate genes by integrative genomic strategy. Among them, ANXA5 was associated with total number of piglets born (TNB), number of piglets born alive (NBA) and mean litter weight (MLW). In human, it is clear that the H2 haplotype on ANXA5 promoter increases the risks of placental thrombus and pregnancy loss through reducing the expression of ANXA5 (12–14). Consistently, our recent study confirmed the existence of functional genetic variation associated with semen traits in porcine ANXA5 promoter (15). However, the effect of these variations on the piglets’ production trait is still unknown.
The Min pig is the most famous indigenous breed in Northeast China. Compared to the global general breeds, Min pigs have superior meat quality, high reproductive capacity, and strong stress resistance1. However, the slow growth rate of Min pigs limits their utilization in commercial production. In terms of the multifunction of ANXA5, the purpose of this study was to evaluate the effect of ANXA5 on the growth of Min piglets, explore the biological function of the identified mutations, thereby providing new insights into the pleiotropic genetic architecture, the conservation and utilization of Min pigs.
2 Materials and methods
2.1 Animals
Tissue samples (stomach, duodenum, jejunum, ileum, cecum, colon, and rectum) from 4 piglets were described by Niu et al. (16). A total of 186 Min piglets were raised in the Lanxi Breeding Farm (Lanxi, Heilongjiang, China). All these piglets were weighed (at birth, day 3, 7, 14, and 21) and weaned at day 35. Average daily gain (ADG) was calculated using the formula: ADG = (21-day weight - birth weight) /21. 84 Jinhua pigs were raised in the Zhejiang Mebolo Swine Breeding farm, and 16 Durocs were provided by Haining Yangdu Science and Technology Ranch of Zhejiang Academy of Agricultural Sciences, China.
2.2 Genotyping SNPs within the promoter of porcine ANXA5
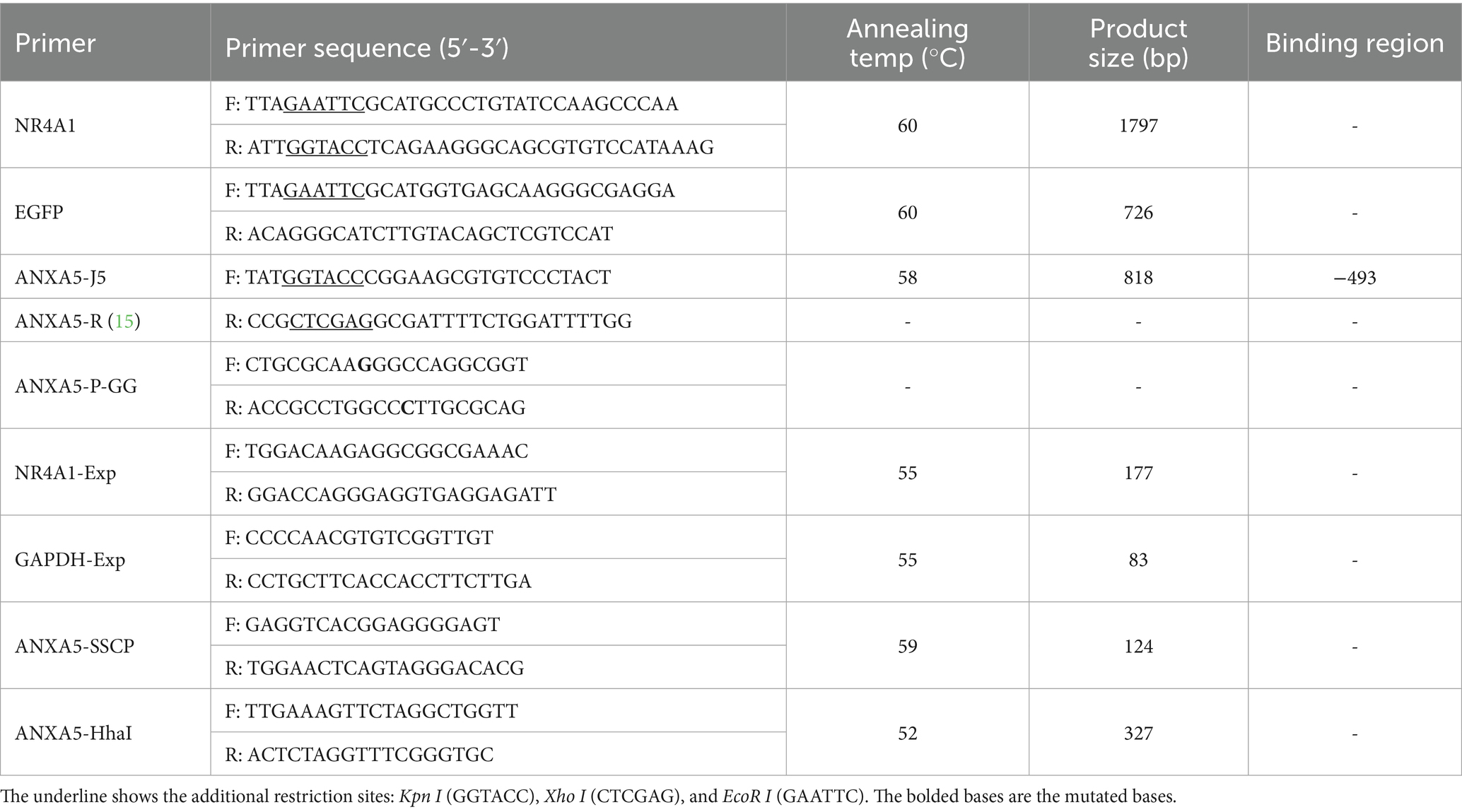
All the primers were designed based on porcine ANXA5 (GenBank accession: XM_003129218.5) and NR4A1 (Ensembl: ENSSSCT00000059252.3) by Primer 5 (Table 1). For the PCR, the 20 μL reactions contained 100–150 ng DNA, 10 μL 2 × Taq Master Mix (Takara, Dalian, China), and 0.5 μM primers (Table 1), performed under the following conditions: 94 °C for 4 min; 35 cycles of 94 °C/30 s, primer-specific annealing (Table 1)/30 s, 72 °C/1 min; final extension at 72 °C/10 min. For the Hha I PCR-RFLP, the DNA fragments containing SNP: g.-674 C > A were amplified using primer pair ANXA5-HhaI (Table 1) from 186 Min pigs. Then, the 8.5 μL of PCR-amplified products, 1 μL of 10 × M Buffer, and 0.5 μL of Hha I enzyme (Takara, Dalian, China) were mixed, incubated at 37 °C for 8 h and detected by 1.5% agarose gels electrophoresis.
The sequence of ANXA5 promoter from Min pigs and Landraces obtained in our previous study (15) was re-aligned by DNAMAN2. The putative functional transcription factor binding sites were analyzed by JASPAR3. For the SNP: g.-519 A > G, the DNA fragments containing this SNP were amplified using primer pair ANXA5-SSCP in 186 Min pigs (Table 1). Then, the 1 μL PCR product was mixed with 9 μL denaturing buffer and denatured at 98 °C for 10 min. After this, the mixture was immediately cooled in ice water for 5 min, separated by 14% polyacrylamide gel electrophoresis (PAGE), and finally analyzed by silver staining. Alternatively, the PCR products containing this SNP were amplified by primers: ANXA5-S-F/R (15), purified and sequenced commercially (Sangon, Shanghai, China).
2.3 Plasmids construction
The pGL3-ANXA5-J3 containing the putative NR4A1 and ESR1 binding sites and the pGL3-ANXA5-P containing A allele at g.-519 A > G of the ANXA5 promoter were constructed in our previous study (15). In this study, with the pGL3-ANXA5-J3 as template, the 5’ NR4A1 binding region deletion fragments were produced using primers: ANXA5-J5-F/R (Table 1) and named ANXA5-J5. With pGL3-ANXA5-P as template, a mutant fragment with G allele which caused the loss of the putative NR4A1 binding region was amplified using mutagenic primer: ANXA5-P-GG-F/R (Table 1) and termed ANXA5-GG. Then, the fragment of ANXA5-GG was digested by Kpn I and Xho I (Takara, Dalian, China), inserted into pGL3-Basic vector (Promega, Madison, WI, USA), confirmed by DNA sequence and double restriction endonuclease digestion, which was named pGL3-ANXA5-GG, respectively.
The complete coding sequence of porcine NR4A1 was amplified using primers NR4A1-CDS-F/R (Table 1), digested and cloned into the Kpn I/EcoR I restriction sites of the pCMV-HA expression vector, confirmed and named pCMV-HA-NR4A1.
The coding sequence of both EGFP and porcine NR4A1 were amplified using primer pairs EGFP-F/R and NR4A1-F/R, respectively (Table 1). With the mixture of these two fragments as template, overlap extension PCR was performed with primers EGFP-F and NR4A1-R to generate the NR4A1-EGFP fusion fragment. After purification and sequencing, the NR4A1-EGFP fragment was inserted into the EcoR I/ Kpn I sites of pCMV-HA to construct the pCMV-HA-EGFP-NR4A1.
2.4 Cell transfection and luciferase reporter assay
The ST and IPEC-J2 cells were cultured with DMEM supplemented with 10% FBS (Gibco, Carlsbad, CA, USA) and 1% penicillin–streptomycin solution (Beyotime, Shanghai, China), maintained at 37 °C in the atmosphere of 5% CO2. For the promoter activity assay, cells were seeded in 24-well plates. To explore the role of the putative NR4A1 binding site within the ANXA5 promoter, with 1.5 μL of LP2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA), the cells were transiently transfected with 0.5 μg of the corresponding luciferase reporter gene vectors (pGL3- ANXA5-J3, pGL3- ANXA5-J4 and pGL3- ANXA5-J5), or 0.005 μg of pGL3-basic as negative control and pRL-TK as internal control. To validate the role of NR4A1 on the transcription of ANXA5, the cells were co-transfected with 0.25 μg of the pCMV-HA-NR4A1 or pCMV-HA, 0.25 μg of the pGL3-ANXA5-P(AA) or pGL3- ANXA5-GG, and 0.005 μg of pRL-TK, with 1.5 μL of LP2000 (Invitrogen, Carlsbad, CA, USA). 24 h after transfection, in line with the Dual Luciferase Reporter Assay System (Beyotime, Shanghai, China), all the cells were harvested and lysed, and the enzymatic activity of firefly and Renilla luciferase were assessed using the Sirius L Luminometer (Berthold, Pforzheim, Germany). Relative luciferase activity was calculated as the ratio of firefly to Renilla luciferase activity (Fluc/Rluc). All transfections were performed in triplicate and repeated three times.
2.5 The subcellular distribution of NR4A1
The localization of porcine NR4A1 in the cells was firstly predicted by PSORT II4 and Uniprot5. Then, the pCMV-HA-EGFP-NR4A1 or pCMV-HA-EGFP were transfected into the cells, respectively. After 48 h, 1 mL of 1 × Hoechst 33342 was added into each well, incubated at room temperature in the dark for 10 min. Then, the staining solution were discarded, all the cells were washed with PBS for three times. The subcellular distribution of NR4A1-EGFP was observed using the Leica TCS SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany). The experiment was repeated three times with consistent results.
2.6 Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from specific tissues of four Min piglets, ST and IPEC-J2 cells (n = 3 biological replicates) using TRIzol (Takara, Dalian, China), reverse-transcribed into cDNA by PrimeScript RT Master Mix (Takara, Dalian, China). According to the manufacturer’s instructions of the TB Green Premix Ex Taq II (Tli RNaseH Plus) kit (Takara, Dalian, China), the qRT-PCR reactions (20 μL) contained 100 ng cDNA, 10 μL SYBR mix (Takara, Dalian, China), and 0.2 μM primers (Table 1), performed on the ABI QuantStudio 3 system (Applied Biosystems, Foster City, CA, USA) with: 95 °C/30s; 40 cycles of 95 °C/5 s and 60–62 °C/35 s; melt curve analysis (60–95 °C, 0.3 °C/s). The qRT-PCR for each sample was performed in technical triplicate. GAPDH-normalized mRNA levels were quantified by the 2−ΔΔCT method.
2.7 Statistical analysis
Statistical analyses were performed using SPSS (IBM, Armonk, NY, USA). The mRNA expression of ANXA5 or NR4A1 in different tissues were analyzed by ANOVA (Analysis of Variance). The differences of gene expression between the cells, or the transcriptional activity between groups were evaluated using the two-tailed Student’s t-test. All data are presented as the mean ± standard error of the mean.
The effect of SNP on growth traits of 186 piglets was evaluated by general linear model (GLM) of SAS version 9.2.21 (SAS, Cary, NC, USA) as follow:
Yij = μ + Gi + Sj + eij, where Yij is the observed traits under study, μ is the mean value of the trait, Gi is the genotype effects, Sj is the maternal effects and eij is the random residual. Significance was declared at p < 0.05 or p < 0.01.
3 Results
3.1 g.-674 C > A in the promoter of ANXA5 was associated with piglets’ weight in Min pigs
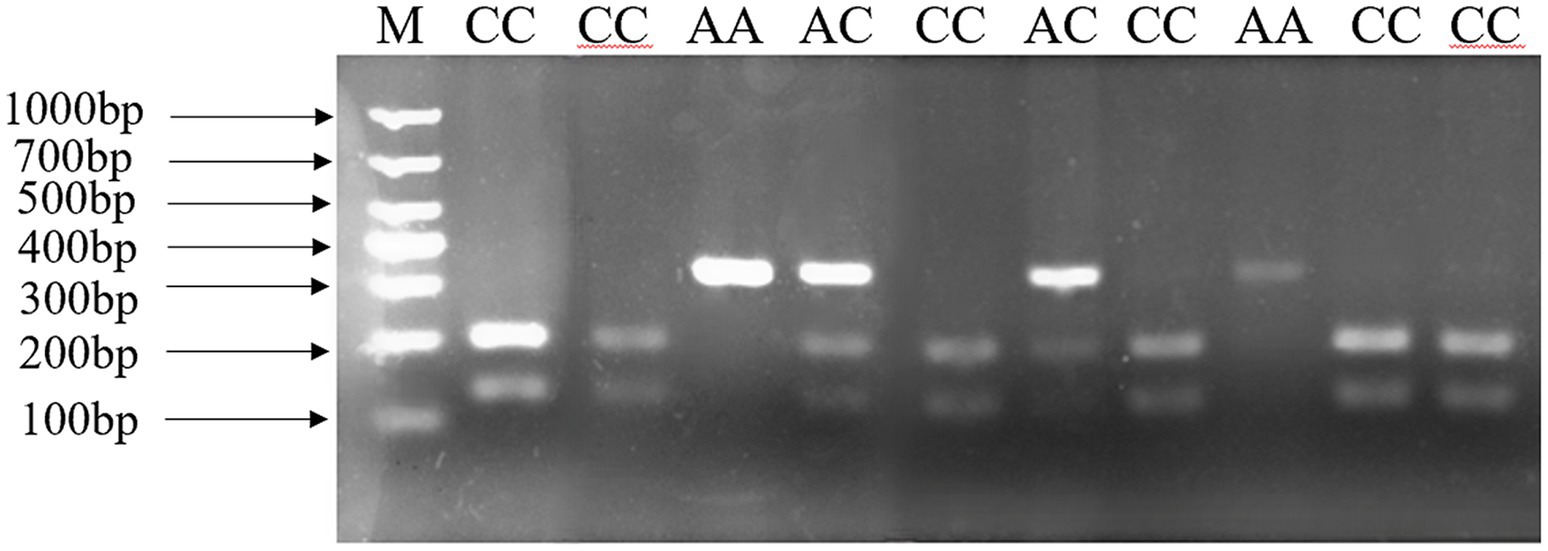
In our previous research (15), three tightly linked SNPs which constructed one haplotype were found in the ANXA5 promoter. In this study, a Hha I PCR-RFLP assay based on SNP: g.-674C > A was established to genotype this haplotype. As shown in Figure 1, a 327 bp fragment was produced by PCR and three genotypes including AA (327 bp), AC (327, 198, 129 bp) and CC (198, 129 bp) were distinguished after the Hha I digestion and the agarose gels electrophoresis. Association analysis in Min pigs showed that the birth weight of the AA animals was significantly higher than that of the AC and CC genotypes (p < 0.05) (Table 2).
3.2 g.-519 A > G in the promoter of ANXA5 was associated with piglets’ weight in Min pigs
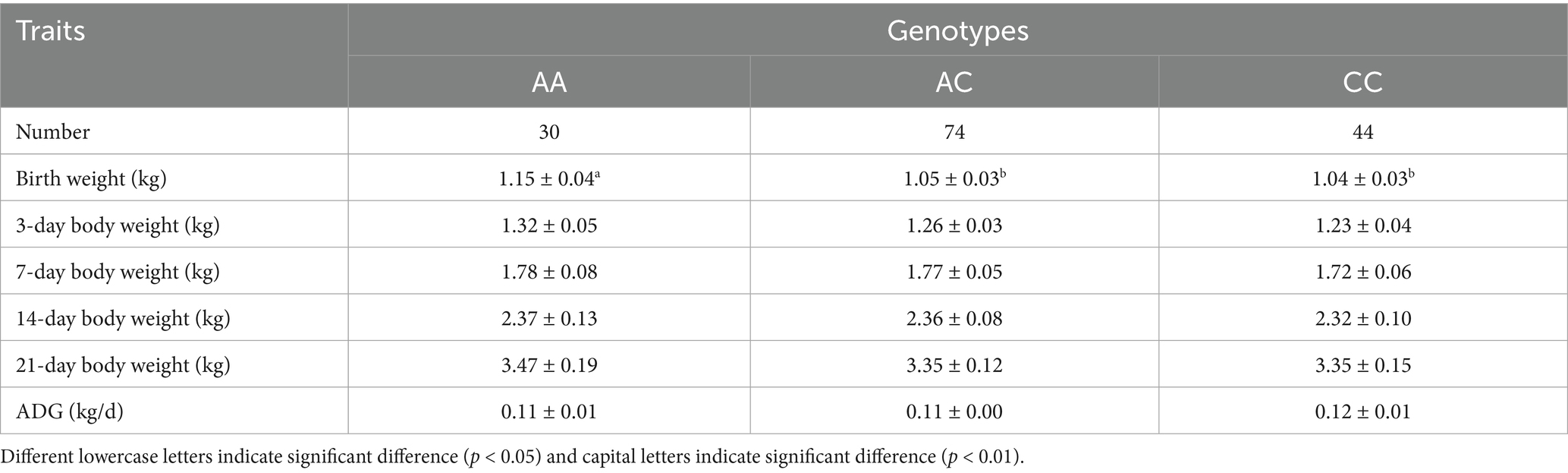
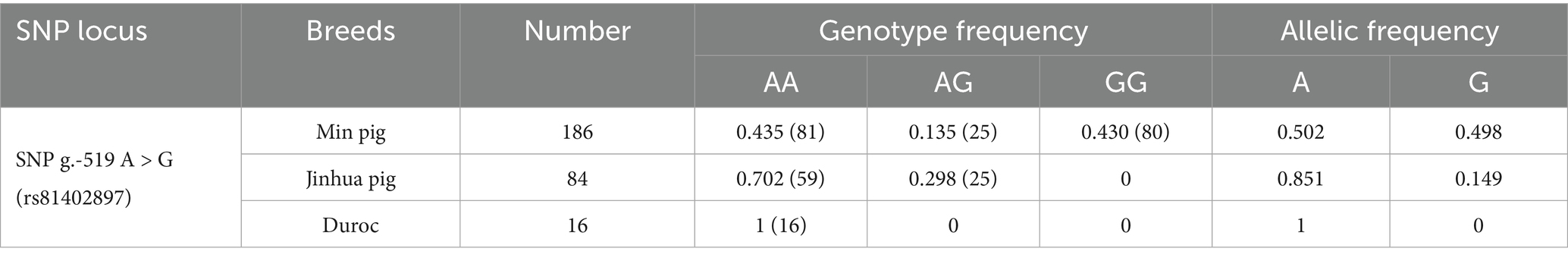
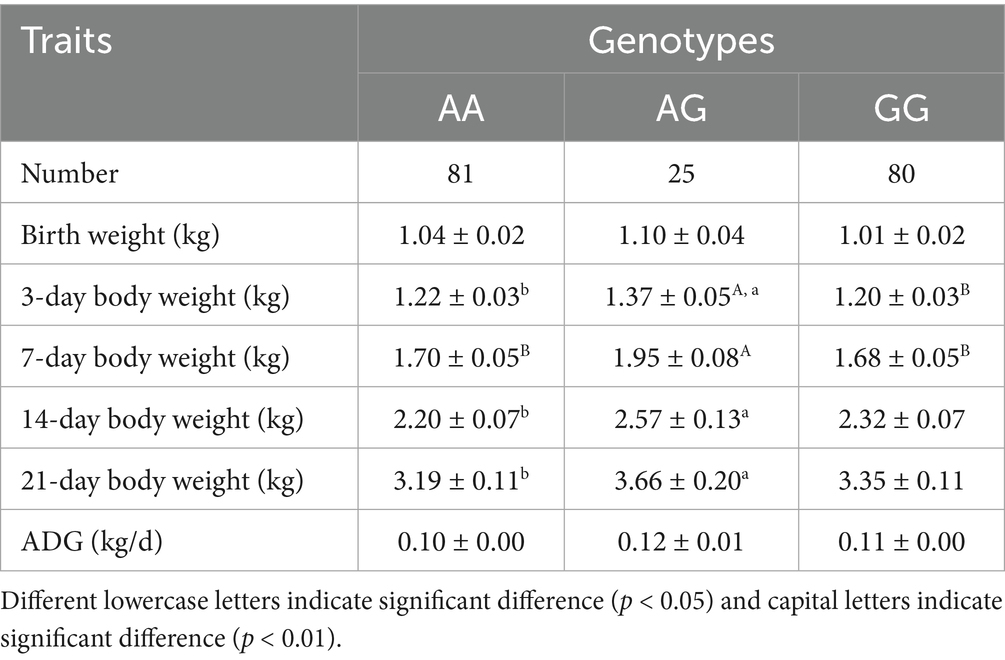
By sequence alignment, a novel g.-519 A > G located in the NR4A1 binding site (−523 bp to −512 bp before the ATG) of the ANXA5 promoter was identified in Min pigs instead of Landraces or Yorkshires (Figures 2A,B). As predicted by JASPAR, transcription factor NR4A1 would bind to the ANXA5 promoter with A allele but not the G allele (Figure 2C). Then, a PCR-SSCP assay was established to genotype this SNP in Min pigs, where the 124-bp fragment was obtained using primer pairs ANXA5-SSCP-F/R (Table 1) and three genotypes (AA, AG and GG) were observed through polyacrylamide gel electrophoresis (Figure 2D). This novel SNP was genotyped in Min pigs, Jinhua pigs and Durocs. As shown in Table 3, the A allele frequency was 0.50 in Min pigs, while it was 0.85 in Jinhua pigs and fixed in Durocs. Statistical analysis in Min pigs showed that AG animals had higher weight at day 3 and day 7 when compared with GG (p < 0.01) or AA piglets (p < 0.05 or p < 0.05), and the weight of AG individuals at day 14 or 21 were higher than that of AA piglets (p < 0.05) (Table 4).
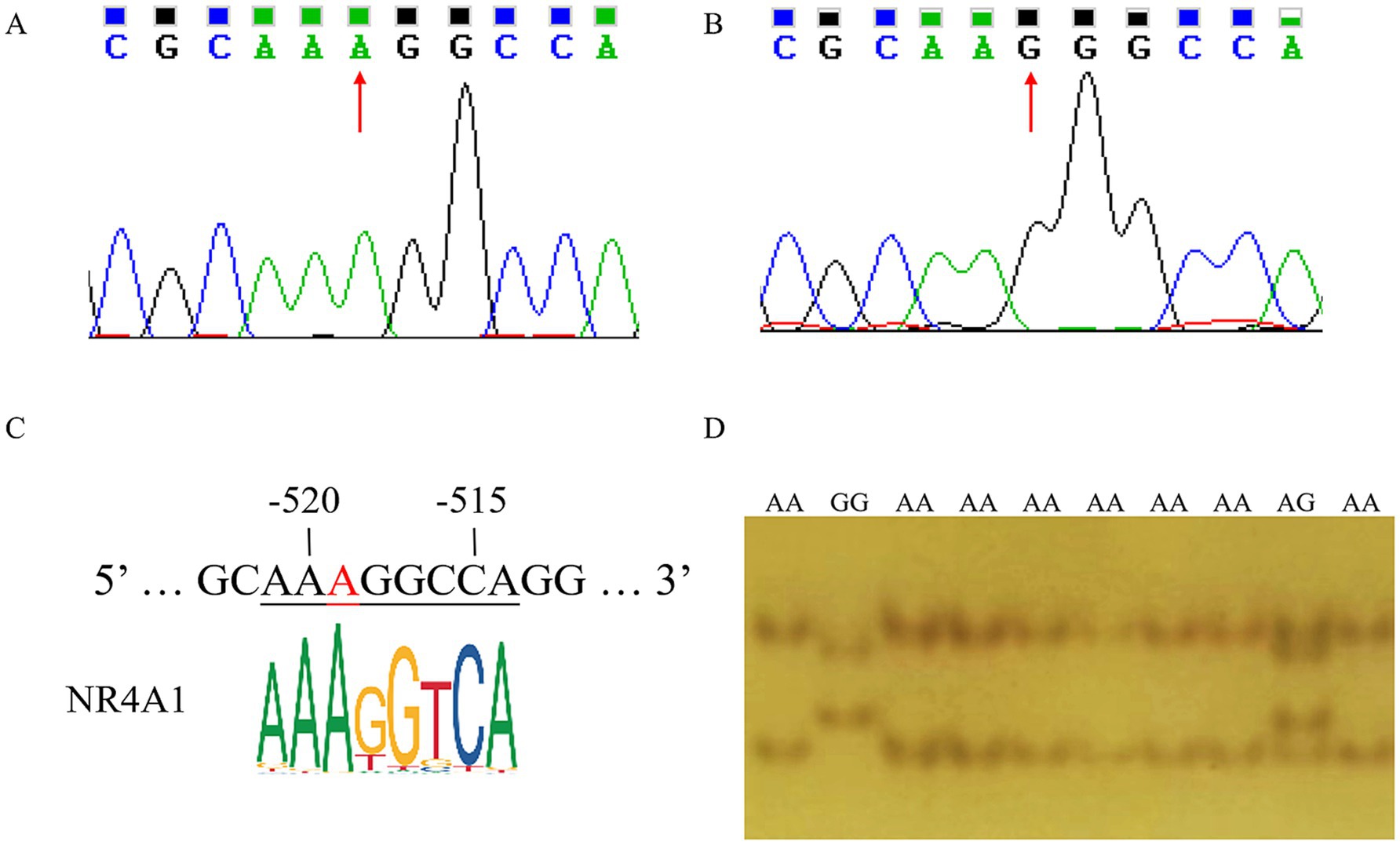
Figure 2. Identification of g.-519 A > G in the promoter of porcine ANXA5. (A) Sequencing result of A allele. (B) Sequencing result of G allele. (C) The transcription factor NR4A1 binding site. (D) PCR-SSCP assay for g.-519 A > G in the promoter of porcine ANXA5.
3.3 The subcellular distribution of NR4A1
Based on UniProt, Figure 3A presents a schematic of the subcellular localization of NR4A1. Using PSORT II, the subcellular localization of NR4A1 was predicted as follows: 30.4% nuclear, 21.7% cytoplasmic, 13.0% mitochondrial, 13.0% Golgi, 8.7% endoplasmic reticulum, 8.7% vesicles of the secretory system, and 4.3% peroxisomal (Figure 3A). To clarify the subcellular distribution of NR4A1 in swine cells, the 1797 bp coding sequence of NR4A1 was amplified, inserted into eukaryotic expression plasmid pCMV-HA-EGFP and verified using double-restriction enzyme digestion (Figure 3B). The pCMV-HA-EGFP-NR4A1 expression vector was transfected into IPEC-J2. Fluorescence and confocal analyses in IPEC-J2 showed that porcine NR4A1-EGFP fusion proteins located within the cell nucleus and cytoplasm (Figure 3C), which was consistent with the prediction of PSORT II.
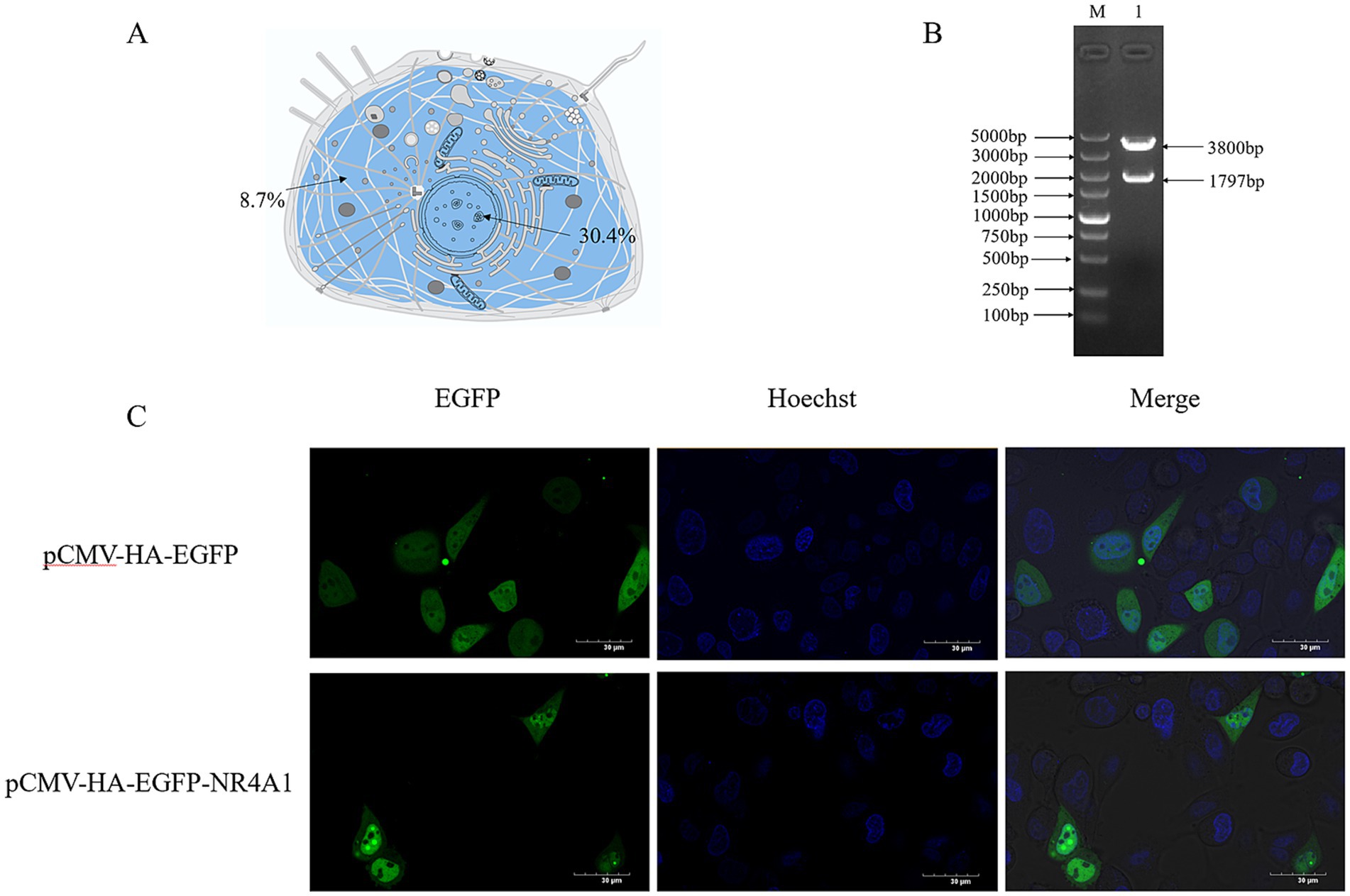
Figure 3. The subcellular distribution of NR4A1. (A) Prediction of cellular localization of NR4A1. (B) Construction of pCMV-HA-NR4A1. (C) The distribution of NR4A1-EGFP in the IPEC-J2 cell. Scale bar: 30 μm.
3.4 NR4A1 promotes the mRNA expression of ANXA5
The mRNA expression of NR4A1 and ANXA5 in the gastrointestinal tract of weaned piglets was quantification by qRT-PCR. As shown in Figure 4A, both NR4A1 and ANXA5 were expressed in all the tissues including stomach, duodenum, jejunum, ileum, colon, cecum and rectum, where the expression of ANXA5 were higher than that of the NR4A1 gene (p < 0.05 or p < 0.01). Then, the NR4A1 expression vector pCMV-HA-NR4A1 were constructed, verified and transfected into IPEC-J2 and ST. qRT-PCR showed that NR4A1 overexpression significantly increased the mRNA expression of ANXA5 in both IPEC-J2 (p < 0.01) and ST (p < 0.01) (Figures 4B,C).
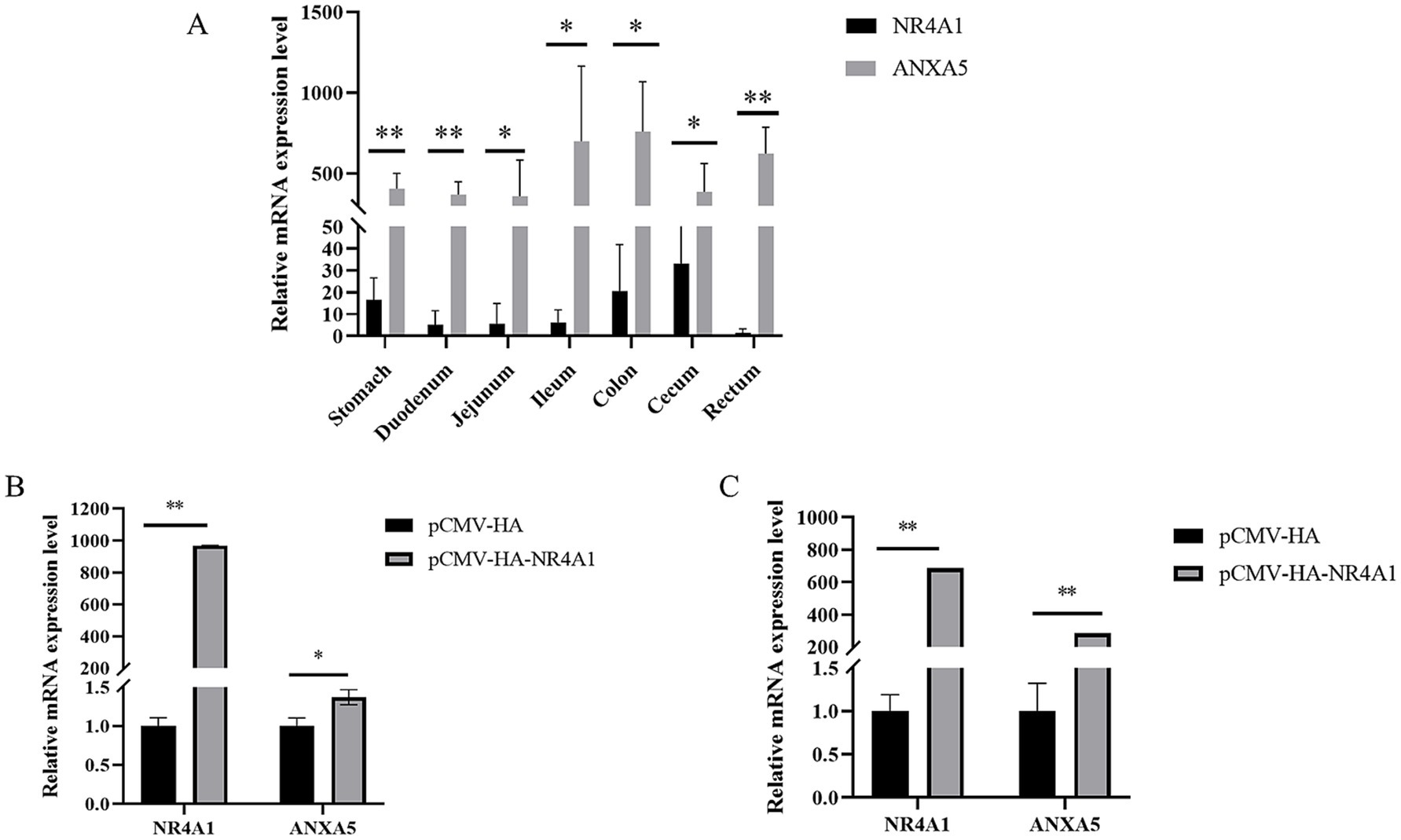
Figure 4. NR4A1 promotes the mRNA expression of ANXA5. (A) Tissue expression profiles of the porcine ANXA5 and NR4A1. (B) Overexpression of NR4A1 increases the mRNA expression of ANXA5 in the IPEC-J2 cell. (C) Overexpression of NR4A1 increases the mRNA level of ANXA5 in the ST cell. Values were shown as the mean ± SEM (n = 3). *p < 0.05, **p < 0.01.
3.5 Identification of the putative NR4A1 binding site of porcine ANXA5 promoter
To explore the function of this putative NR4A1 binding site, an 818-bp 5′-deletion fragment targeting the NR4A1 binding site was amplified to construct the corresponding pGL3-ANXA5-J5 (Figure 5A). Then, pGL3-ANXA5-J5, pGL3-ANXA5-J3 and pGL3-ANXA5-J4, were transfected into ST, respectively. As shown in Figure 5B, the luciferase activity of pGL3-ANXA5-J5 was weaker than that of pGL3-ANXA5-J3 (p < 0.01), but stronger when compared with pGL3-ANXA5-J4 (p < 0.01), indicating the promotive effect of the predictive NR4A1 binding region (−523 bp to -512 bp) on the transcription of porcine ANXA5.
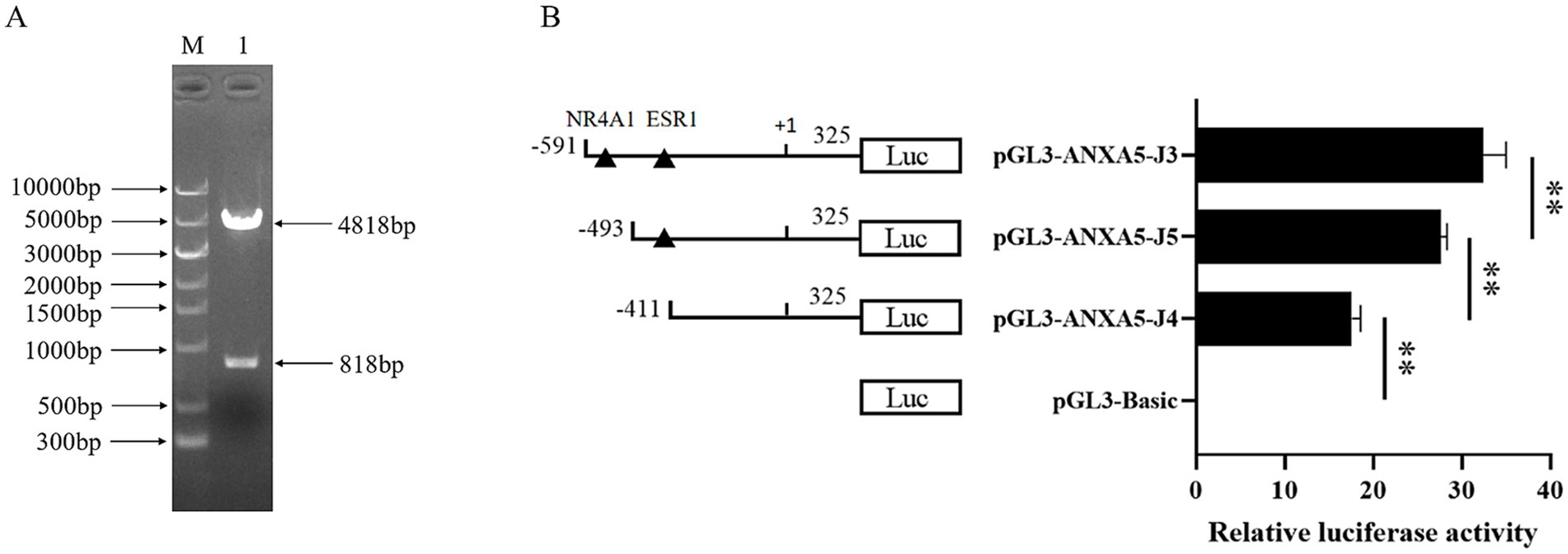
Figure 5. Identification of the putative NR4A1 binding site of porcine ANXA5 promoter. (A) Construction of pGL3-ANXA5-J5. (B) Dual luciferase activity analysis of pGL3-ANXA5-J3 ~ J5. Values were shown as the mean ± SEM (n = 3). *p < 0.05, **p < 0.01.
3.6 g.-519 A > G modulates the ANXA5 transcription partially through NR4A1
According to our previous study, the luciferase reporter pGL3-ANXA5 containing A allele of SNP: g.-519 A > G, which was renamed as pGL3-ANXA5-P(AA) in the current study. To verify the effects of this SNP on ANXA5 transcription, a fragment containing G allele was obtained based on pGL3-ANXA5-P(AA) and the mutation primer (Table 1) to construct pGL3-ANXA5-GG (Figure 6A). Both the pGL3-ANXA5-P(AA) and pGL3-ANXA5-GG were transfected into ST cells, respectively. Statistical analysis showed that the luciferase activity of pGL3-ANXA5-GG was lower than its wild type promoter (p < 0.01) (Figure 6B). Then, NR4A1 expression vector was co-transfected into IPEC-J2 or ST with pGL3-ANXA5-P(AA) or pGL3-ANXA5-GG. As shown in Figure 6C, overexpression of NR4A1 in IPEC-J2 significantly increased the luciferase activity of the pGL3-ANXA5-P(AA) (p < 0.01) or pGL3-ANXA5-GG (p < 0.01), and the luciferase activity of pGL3-ANXA5-P(AA) had higher luciferase activity when compared with pGL3-ANXA5-GG (p < 0.05). In the ST cells, NR4A1 overexpression enhanced the luciferase activity of the pGL3-ANXA5-P(AA) (p < 0.01), but not the pGL3-ANXA5-GG (Figure 6D). Irrespective of NR4A1 overexpression, the luciferase activity of pGL3-ANXA5-P(AA) remained higher than that of pGL3-ANXA5-GG (p < 0.01) in Figure 6D.
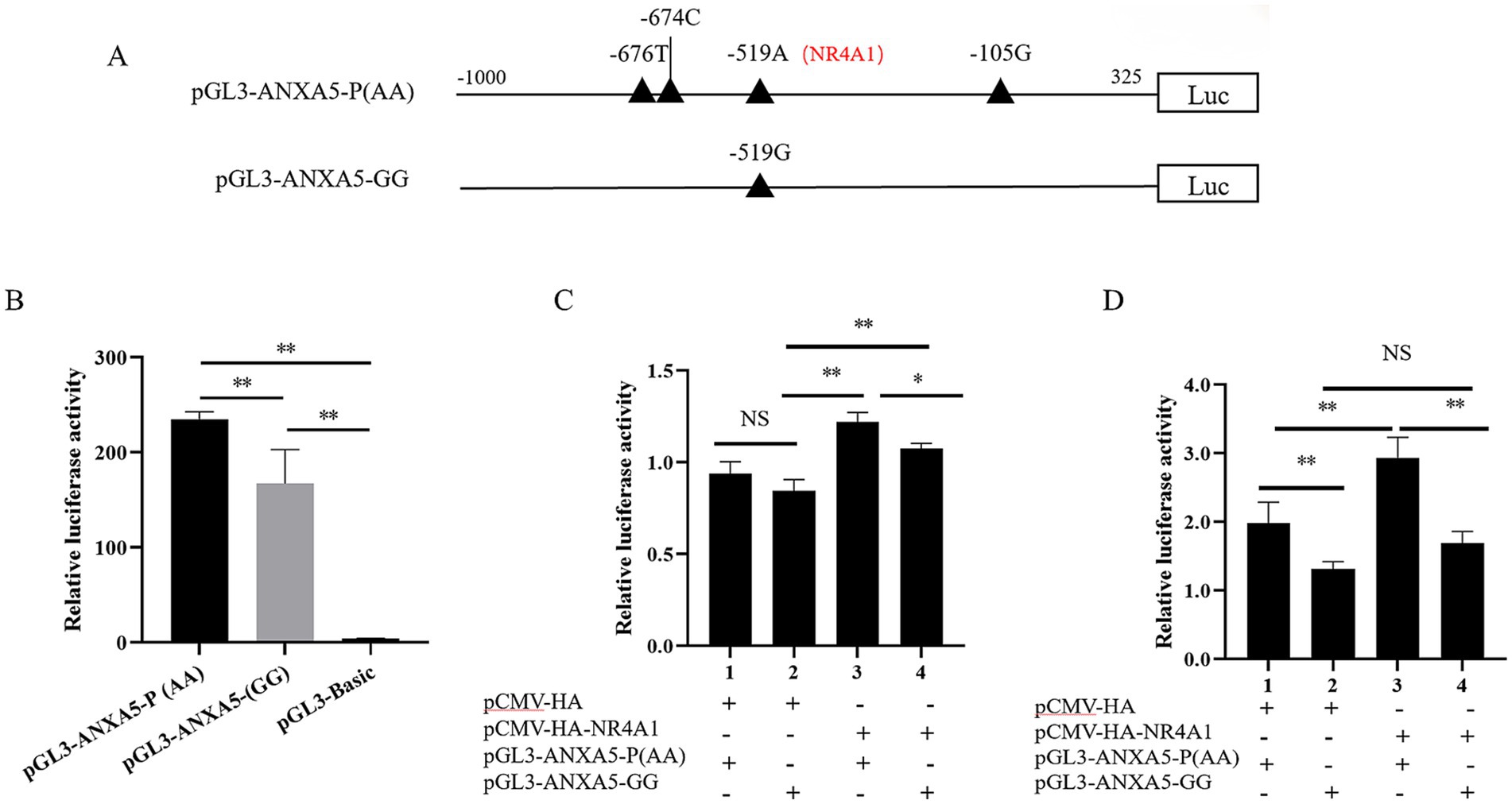
Figure 6. -519 A > G modulates the ANXA5 transcription partially through NR4A1. (A) A schematic diagram of SNP point mutations in the promoter region of the porcine ANXA5 gene. (B) Dual luciferase activity analysis of the SNP g.-519 A > G mutation plasmids. (C) Effect of NR4A1 overexpression on the promoter activity of different alleles of SNP g.-519 A > G in IPEC-J2 cells. (D) Effect of the transcription factor NR4A1 on the promoter activity of different alleles of SNP g.-519 A > G in ST cells. Values were shown as the mean ± SEM (n = 3). *p < 0.05, **p < 0.01.
4 Discussion
4.1 Effects of g.-674 C > A and g.-519 A > G on piglet growth
Previous studies have revealed the effects of ANXA5 polymorphism on reproduction traits in Yorkshires (9, 15), it is necessary to evaluate the genetic effect of ANXA5 on piglets’ performance trait before the genome selection. In this study, a Hha I PCR-RFLP assay was established to detect an identified haplotype (SNP: g.-676 T > C, g.-674 C > A and g.-105G > T) by genotyping g.-674 C > A. According to Han et al. (15), the semen traits of AA animals were lower than AG boars. In the current study, although AA piglets had higher birth weight than the AC and CC animals, the g.-674 C > A was not associated with the later weight (from 3 day to the 21 day) and ADG. It seems the genetic selection based on g.-674 C > A might improve boar reproduction traits without negative effect on piglet weaning weight and growth. Additionally, this PCR-RFLP approach is more accurate and cost-effective when it compares with DNA sequencing. Hence, the HhaI PCR-RFLP is suggested to distinguish the identified haplotype in the promoter of ANXA5.
The abundance of genetic variation in local breeds is higher than that in commercial breeds. In current study, a novel g.-519 A > G in the promoter of ANXA5 was identified in Min and Jinhua pigs, but fixed as A allele in Yorkshires and Durocs. It is known that the human-driven selection resulted in substantial phenotypic diversity among indigenous and commercial pigs (17–19). Hence, one possible explanation for the existence of g.-519 A > G in the Min pig and the Jinhua pig might be the weak artificial selection pressure. Chinese indigenous pigs are divided into North China, Central China, South Chinese, Lower Yangtze River Basin, Southwest, and Plateau, based on geographic distribution, historical origin and morphological characteristics (20, 21). The Min pig is well-adapted to the conditions of North China, while the Jinhua pig belongs to Central China. The geographic differences might be the second explanation for the allelic diversity among these breeds (17, 22, 23). Additionally, the heterozygous Min pig had better growth performance, which might be caused by the epistatic effect of non-alleles, or the interaction between the gene and environment. Min and Jinhua pigs’ high allelic diversity in ANXA5 gene suggested the merit role of indigenous animals in swine genetic improvement.
4.2 g.-519 A > G regulates ANXA5 transcription partially by NR4A1
Nuclear receptor subfamily 4 group A Member 1 (NR4A1) is an immediate-early response gene which regulates diverse biological processes, including cell proliferation, apoptosis, and inflammatory responses (24). The current study demonstrated the existence of both NR4A1 and ANXA5 in the piglet’s intestine, and NR4A1 promoted the transcription and expression of porcine ANXA5 in IPEC-J2 cells. It is well known that the spatiotemporal expression of genes is closely linked to their functions. Hence, we presume that NR4A1 might take important part in the intestine development or health partially by regulating the transcription of ANXA5, thereby affecting piglets’ growth. This hypothesis was supported by literature. For example, both NR4A1 and ANXA5 were associated with ulcerative colitis (UC) (8, 25); NR4A1 expressed in intestine tissues, inflammatory cells and epithelium (26, 27), regulated the differentiation and function of Paneth cell (28); ANXA5 had differential expression in the jejunal mucosa of IUGR and normal piglets (10).
The subcellular localization of one protein partially determines its function. NR4A1 generally resides in the nucleus, but can be translocated to the mitochondria in specific cells or upon stimulation. For example, H2O2 (29), Celastrol (24) or Gly-Pro-Ala (GPA) peptide (30) can promote the nucleocytoplasmic shuttling of NR4A1, induces autophagy, apoptosis, inflammation or other biological process. Here, the NR4A1-EGFP fusion proteins were observed in the nucleus and cytoplasm, indicating that NR4A1 might regulate ANXA5 as a transcription factor in IPEC-J2 cells.
Nowadays, it has been accepted that non-coding regions harbor abundant variations (31–33). For example, g.128G > A influenced granulosa cell apoptosis and sow fertility by altering the RBP-Binding sit of LncRNA NORSF (34); g.-283G > C and g.-271C > T in the miR-23a promoter affected Large White fertility traits partially by altering the transcription of miR-23a in porcine GCs (35). The roles of promoters and enhancers in complex traits have emphasized by accumulating swine genome-wide chromatin landscapes (36–38). The current study focused on the cis-elements of porcine ANXA5, the increased effect of NR4A1 on the luciferase activity of the A allele suggested that g.-519 A > G might alter ANXA5 expression dynamics partially through the modulation of NR4A1 binding. It should be noted that A allele has high transcription activity, but the heterozygote piglets exhibit higher weight. The explanation is as follows: Firstly, post-transcriptional regulation or post-translational modifications of ANXA5 potentially weaken the functional impact of g.-519 A > G; Secondly, allelic interactions (e.g., overdominance effect) strengthen heterozygote phenotypes. Additionally, although g.-519 A > G was identified as a functional variant, it is still unclear whether this SNP alters the transcription of ANXA5 by binding the cis-element or interacting with other transcriptional factors or small RNA. Further study will focus on clarifying the regulatory mechanism of ANXA5. Given the pleiotropic nature of ANXA5, future studies are warranted to evaluate the genetic effects or function of g.-519 A > G on Min pigs’ reproductive trait.
5 Conclusion
This study demonstrated that both g.-674 C > A and g.-519 A > G were associated with piglets’ weight in the Min pig, and it is efficient and convenient to genotype g.-674 C > A by Hha I PCR-RFLP. Porcine NR4A1 was located in the nucleus and cytoplasm, regulated the transcription and expression of ANXA5 in piglet intestinal epithelial cells. SNP: g.-519 A > G might have an allele-specific effect on ANXA5 transcription partially by varying affinity for NR4A1.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal study was approved by Experimental Animal Management Committee of Northeast Agricultural University (NEAUEC20200212). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MH: Writing – original draft, Methodology, Data curation. YiZ: Data curation, Methodology, Writing – original draft. YuZ: Data curation, Writing – original draft. DY: Writing – original draft, Data curation. SS: Data curation, Writing – original draft. XL: Writing – original draft, Data curation. XY: Writing – review & editing. SD: Writing – original draft, Resources. XW: Resources, Writing – original draft. JC: Writing – original draft, Resources. LC: Resources, Writing – original draft. LD: Writing – review & editing, Project administration, Funding acquisition, Methodology, Formal analysis, Writing – original draft, Resources. BN: Funding acquisition, Writing – review & editing, Project administration, Formal analysis, Writing – original draft, Methodology, Data curation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (2021C02068-4), Municipal Agricultural Research Institute Alliance Project of Zhejiang Province (2023SJLM16), Science and Technology Plan Project of Jinhua City (2022-2-09) and Student Innovation Practical Training of Northeast Agricultural University (X202410224005).
Conflict of interest
JC was employed by Lanxi Breeding Farm. LC was employed by Zhejiang Mebolo Swine Breeding Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Kemp, B, Da Silva, CLA, and Soede, NM. Recent advances in pig reproduction: focus on impact of genetic selection for female fertility. Reprod Domest Anim. (2018) 53:28–36. doi: 10.1111/rda.13264
2. Da Silva, CLA, van den Brand, H, Laurenssen, BF, Broekhuijse, ML, Knol, EF, Kemp, B, et al. Relationships between ovulation rate and embryonic and placental characteristics in multiparous sows at 35 days of pregnancy. Animal. (2016) 10:1192–9. doi: 10.1017/s175173111600015x
3. Patterson, J, Bernardi, ML, Allerson, M, Hanson, A, Holden, N, Bruner, L, et al. Associations among individual gilt birth weight, litter birth weight phenotype, and the efficiency of replacement gilt production. J Anim Sci. (2020) 98:skaa331. doi: 10.1093/jas/skaa331
4. Faccin, JEG, Tokach, MD, Goodband, RD, DeRouchey, JM, Woodworth, JC, and Gebhardt, JT. Gilt development to improve offspring performance and survivability. J Anim Sci. (2022) 100:skac128. doi: 10.1093/jas/skac128
5. Magnabosco, D, Bernardi, M, Wentz, I, Cunha, E, and Bortolozzo, FJLS. Low birth weight affects lifetime productive performance and longevity of female swine. Livest Sci. (2016) 184:119–25. doi: 10.1016/j.livsci.2015.12.008
6. Stange, K, and Röntgen, M. Myotube formation and cellular fusion are diminished due to low birth weight in piglets. Int J Mol Sci. (2025) 26:2847. doi: 10.3390/ijms26072847
7. Ueki, H, Mizushina, T, Laoharatchatathanin, T, Terashima, R, Nishimura, Y, Rieanrakwong, D, et al. Loss of maternal annexin A5 increases the likelihood of placental platelet thrombosis and foetal loss. Sci Rep. (2012) 2:827. doi: 10.1038/srep00827
8. Hua, R, Qiao, G, Chen, G, Sun, Z, Jia, H, Li, P, et al. Single-cell RNA-sequencing analysis of colonic Lamina propria immune cells reveals the key immune cell-related genes of ulcerative colitis. J Inflamm Res. (2023) 16:5171–88. doi: 10.2147/jir.S440076
9. Häggman, J, and Uimari, P. Novel harmful recessive haplotypes for reproductive traits in pigs. J Anim Breed Genet. (2017) 134:129–35. doi: 10.1111/jbg.12240
10. Wang, X, Zhu, Y, Feng, C, Lin, G, Wu, G, Li, D, et al. Innate differences and colostrum-induced alterations of jejunal mucosal proteins in piglets with intra-uterine growth restriction. Br J Nutr. (2018) 119:734–47. doi: 10.1017/s0007114518000375
11. Wei, R, Zhang, Z, Han, H, Miao, J, Yu, P, Cheng, H, et al. Integrative genomic analysis reveals shared loci for reproduction and production traits in Yorkshire pigs. BMC Genomics. (2025) 26:310. doi: 10.1186/s12864-025-11416-0
12. Markoff, A, Gerdes, S, Feldner, S, Bogdanova, N, Gerke, V, and Grandone, E. Reduced allele specific annexin A5 mRNA levels in placentas carrying the M2/ANXA5 allele. Placenta. (2010) 31:937–40. doi: 10.1016/j.placenta.2010.08.002
13. Tiscia, GL, Dørum, E, Myklebust, CF, Grandone, E, Sandset, PM, and Skretting, G. Functional characterization of annexin A5 gene promoter allelic variants. Thromb Res. (2016) 144:93–9. doi: 10.1016/j.thromres.2016.06.009
14. Cai, Z, Zheng, X, Chen, Y, Chen, F, Chen, L, and Deng, X. Genetic analysis of ANXA5 haplotype and its effect on recurrent pregnancy loss. Mol Med Rep. (2022) 25:43. doi: 10.3892/mmr.2021.12559
15. Han, M, Yao, D, Song, Y, Liu, Y, Chen, Z, Li, J, et al. Identification of functional SNP associated with sperm quality in porcine ANXA5 that contributes to the growth of immature Sertoli cell. Front Vet Sci. (2025) 12:1576566. doi: 10.3389/fvets.2025.1576566
16. Niu, B, Liu, L, Chen, Z, Kou, M, Yang, X, Sun, Y, et al. Characterization, mRNA expression profile, subcellular distribution and association analysis with piglet diarrhea of porcine matrix metallopeptidase 7 (pMMP7). Gene. (2022) 821:146319. doi: 10.1016/j.gene.2022.146319
17. Liu, Y, Fu, Y, Yang, Y, Yi, G, Lian, J, Xie, B, et al. Integration of multi-omics data reveals cis-regulatory variants that are associated with phenotypic differentiation of eastern from western pigs. Genet Sel Evol. (2022) 54:62. doi: 10.1186/s12711-022-00754-2
18. Fu, W, Xie, Q, Yu, P, Liu, S, Xu, L, Ye, X, et al. Pig jejunal single-cell RNA landscapes revealing breed-specific immunology differentiation at various domestication stages. Front Immunol. (2025) 16:1530214. doi: 10.3389/fimmu.2025.1530214
19. Cao, C, Miao, J, Xie, Q, Sun, J, Cheng, H, Zhang, Z, et al. A near telomere-to-telomere genome assembly of the Jinhua pig: enabling more accurate genetic research. Gigascience. (2025) 14:giaf048. doi: 10.1093/gigascience/giaf048
20. Zhang, ZG, Li, B, and Chen, X. Pig breeds in China. Shanghai: Shanghai Scientific and Technical Publisher (1986).
21. Tong, X, Hou, L, He, W, Mei, C, Huang, B, Zhang, C, et al. Whole genome sequence analysis reveals genetic structure and X-chromosome haplotype structure in indigenous Chinese pigs. Sci Rep. (2020) 10:9433. doi: 10.1038/s41598-020-66061-2
22. Zhu, Y, Li, W, Yang, B, Zhang, Z, Ai, H, Ren, J, et al. Signatures of selection and interspecies introgression in the genome of Chinese domestic pigs. Genome Biol Evol. (2017) 9:2592–603. doi: 10.1093/gbe/evx186
23. Gao, Y, Feng, X, Diao, S, Liu, Y, Zhong, Z, Cai, X, et al. Deciphering genetic characteristics of South China and North China indigenous pigs through selection signatures. BMC Genomics. (2024) 25:1191. doi: 10.1186/s12864-024-11119-y
24. Hu, M, Luo, Q, Alitongbieke, G, Chong, S, Xu, C, Xie, L, et al. Celastrol-induced Nur77 interaction with TRAF2 alleviates inflammation by promoting mitochondrial ubiquitination and autophagy. Mol Cell. (2017) 66:141–53.e6. doi: 10.1016/j.molcel.2017.03.008
25. Wu, H, Li, XM, Wang, JR, Gan, WJ, Jiang, FQ, Liu, Y, et al. NUR77 exerts a protective effect against inflammatory bowel disease by negatively regulating the TRAF6/TLR-IL-1R signalling axis. J Pathol. (2016) 238:457–69. doi: 10.1002/path.4670
26. Hamers, AA, van Dam, L, Teixeira Duarte, JM, Vos, M, Marinković, G, van Tiel, CM, et al. Deficiency of nuclear receptor Nur77 aggravates mouse experimental colitis by increased NFκB activity in macrophages. PLoS One. (2015) 10:e0133598. doi: 10.1371/journal.pone.0133598
27. Yu, Z, Yong, Y, Liu, X, Ma, X, Abd El-Aty, AM, Li, L, et al. Insights and implications for transcriptomic analysis of heat stress-induced intestinal inflammation in pigs. BMC Genomics. (2024) 25:1110. doi: 10.1186/s12864-024-10928-5
28. Cui, C, Wang, X, Zheng, Y, Wu, L, Li, L, Wei, H, et al. Nur77 as a novel regulator of Paneth cell differentiation and function. Mucosal Immunol. (2024) 17:752–67. doi: 10.1016/j.mucimm.2023.09.001
29. Xu, A, Liu, J, Liu, P, Jia, M, Wang, H, and Tao, L. Mitochondrial translocation of Nur77 induced by ROS contributed to cardiomyocyte apoptosis in metabolic syndrome. Biochem Biophys Res Commun. (2014) 446:1184–9. doi: 10.1016/j.bbrc.2014.03.089
30. Deng, Z, Liu, Q, Wang, M, Wei, HK, and Peng, J. GPA peptide-induced Nur77 localization at mitochondria inhibits inflammation and oxidative stress through activating autophagy in the intestine. Oxidative Med Cell Longev. (2020) 2020:1–18. doi: 10.1155/2020/4964202
31. Farh, KK, Marson, A, Zhu, J, Kleinewietfeld, M, Housley, WJ, Beik, S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. (2015) 518:337–43. doi: 10.1038/nature13835
32. Finucane, HK, Bulik-Sullivan, B, Gusev, A, Trynka, G, Reshef, Y, Loh, PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. (2015) 47:1228–35. doi: 10.1038/ng.3404
33. Xu, Z, Lin, Q, Cai, X, Zhong, Z, Teng, J, Li, B, et al. Integrating large-scale meta-GWAS and PigGTEx resources to decipher the genetic basis of 232 complex traits in pigs. Natl Sci Rev. (2025) 12:nwaf048. doi: 10.1093/nsr/nwaf048
34. Wang, M, Sheng, W, Zhang, J, Cao, Q, Du, X, and Li, Q. A mutation losing an RBP-binding site in the LncRNA NORSF transcript influences granulosa cell apoptosis and sow fertility. Adv Sci. (2024) 11:e2404747. doi: 10.1002/advs.202404747
35. Wang, S, Li, Y, Du, X, and Li, Q. Two single nucleotide variants in the miR-23a promoter affect granulosa cell apoptosis. Anim Genet. (2023) 54:207–10. doi: 10.1111/age.13284
36. Pan, Z, Yao, Y, Yin, H, Cai, Z, Wang, Y, Bai, L, et al. Pig genome functional annotation enhances the biological interpretation of complex traits and human disease. Nat Commun. (2021) 12:5848. doi: 10.1038/s41467-021-26153-7
37. Zhao, Y, Hou, Y, Xu, Y, Luan, Y, Zhou, H, Qi, X, et al. A compendium and comparative epigenomics analysis of cis-regulatory elements in the pig genome. Nat Commun. (2021) 12:2217. doi: 10.1038/s41467-021-22448-x
Keywords: ANXA5 , NR4A1 , functional SNP, production trait, Min pigs
Citation: Han M, Zhang Y, Zhang Y, Yao D, Sun S, Li X, Yang X, Di S, Wang X, Cai J, Chen L, Dai L and Niu B (2025) Single nucleotide variations in the ANXA5 promoter regulated piglet weight in the Min pig. Front. Vet. Sci. 12:1644944. doi: 10.3389/fvets.2025.1644944
Edited by:
Martijn Derks, Wageningen University and Research, NetherlandsReviewed by:
Jun He, Hunan Agricultural University, ChinaTânia Fernandes Martins, Universidade Federal de Viçosa, Brazil
Copyright © 2025 Han, Zhang, Zhang, Yao, Sun, Li, Yang, Di, Wang, Cai, Chen, Dai and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihe Dai, ZGFpbGhAemFhcy5hYy5jbg==; Buyue Niu, bml1YnV5dWVAbmVhdS5lZHUuY24=
†These authors have contributed equally to this work
 Mingyang Han1†
Mingyang Han1† Xiuqin Yang
Xiuqin Yang Buyue Niu
Buyue Niu