- 1Department of Ecological and Biological Science, University of Tuscia, Viterbo, Italy
- 2Department of Biology and Biotechnologies “Charles Darwin”, University of Rome “La Sapienza”, Rome, Italy
- 3Natural History Museum and Botanical Garden, University of Calabria, Rende, Italy
- 4Aspromonte National Park, Santo Stefano in Aspromonte, Italy
Unprecedented rates of biodiversity loss raise the urgency for preserving species ability to cope with ongoing global changes. An approach in this direction is to target intra-specific hotspots of genetic diversity as conservation priorities. However, these hotspots are often identified by sampling at a spatial resolution too coarse to be useful in practical management of threatened species, hindering the long-appealed dialog between conservation stakeholders and conservation genetic researchers. Here, we investigated the spatial and temporal variation in species presence, genetic diversity, as well as potential risk factors, within a previously identified hotspot of genetic diversity for the endangered Apennine yellow-bellied toad Bombina pachypus. Our results show that this hotspot is neither a geographically homogeneous nor a temporally stable unit. Over a time-window spanning 10–40 years since previous assessments, B. pachypus populations declined in large portions of their hotspot, and their genetic diversity levels decreased. Considering the demographic trend, genetic and epidemiological data, and models of current and future climatic suitability, populations at the extreme south of the hotspot area still qualify for urgent in-situ conservation actions, whereas northern populations would be better managed through a mix of in-situ and ex-situ actions. Our results emphasize that identifying hotspots of genetic diversity, albeit an essential step, does not suffice to warrant on-ground conservation of threatened species. Hotspots should be analyzed at finer geographic and temporal scales, to provide conservation stakeholders with key knowledge to best define conservation priorities, and to optimize resource allocation to alternative management practices.
Introduction
Unprecedented rates of biodiversity loss raise the urgency for preserving species ability to cope with ongoing global changes (Crandall et al., 2000; Moritz, 2002; Hendry et al., 2010; Eizaguirre and Baltazar-Soares, 2014 and references therein). However, conservation policies have traditionally been oriented toward the protection of species diversity and habitats, rarely considering the evolutionary potential of individual populations within species (Mace and Purvis, 2008; Laikre, 2010; Mimura et al., 2017). A direct approach in this direction, which has been largely debated for decades, is to directly target populations with high levels of genetic diversity (Frankham, 2010). In fact, owing to the intimate links between genetic diversity and effective population size, these populations are, at the same time, less likely to be affected by the detrimental consequences of inbreeding depression and genetic drift, and more likely to warrant evolvability and adaptive potential (Allendorf and Luikart, 2009; Frankham, 2010; Allendorf et al., 2012; Lanfear et al., 2014). When different populations, all with relatively high levels of genetic diversity, are concentrated in the same area, it is possible to define an intra-specific hotspot of genetic diversity (Hewitt, 2000, 2004; Petit et al., 2003; Hampe and Petit, 2005), clearly representing a conservation priority. Importantly, as pointed out by Pérez-Espona and ConGRESS Consortium (2017), genetic diversity data useful to detect hotspots are now available for a large number of taxa, at least within some intensively studied areas. However, in spite of both their extreme value and data availability, hotspots of genetic diversity have been largely ignored in the context of biodiversity management strategies, highlighting an existing gap between conservation geneticists' achievements and conservation stakeholders' priorities (Vernesi et al., 2008; Pérez-Espona and ConGRESS Consortium, 2017).
Traditionally, hotspots have been identified over large areas (e.g., considering the entire range of a species) using the analytical tools and the sampling scheme of phylogeography (Hewitt, 2011), providing therefore a spatial resolution that can hardly be considered in practical conservation exercises (Cañadas and Vázquez, 2014). Consequently, important questions for on-ground management practices still remain largely unexplored. Can a hotspot be considered as a homogeneous regional unit for management purposes? Are all populations “created” equal within hotspot areas? Or, in order to achieve optimal resource allocation and maximum chance of success, should populations be prioritized as well? Answering these open questions could have major practical implications for the apportionment of financial budgets among alternative management practices, as well as for the design of protected areas.
Here, we explore the importance of addressing these questions in a collaborative framework between academic researchers and conservation authorities (governmental, protected area managers). To this aim, we analyzed spatial and temporal patterns of variation in species presence, genetic diversity, as well as potential risk factors, within a previously identified hotspot of genetic diversity for the endangered Apennine yellow-bellied toad Bombina pachypus. Indeed, substantial previous knowledge available on this species makes it especially suitable to reach the study aim. The Apennine yellow-bellied toad is an amphibian endemic to the Italian peninsula, whose hotspot of genetic diversity has been identified at the southernmost portion of its range (Canestrelli et al., 2006). Extensive population declines in the last decades have raised severe concerns for the conservation status of this species (Barbieri et al., 2004), that is now listed as endangered on the Red List of IUCN (Andreone et al., 2009). Previous studies have suggested the potential conservation value of the hotspot area for this species, showing that: (i) B. pachypus populations are facing a dramatic demographic decline in the whole range, except for this area (Barbieri et al., 2004); (ii) genetic diversity levels in this area are by far higher than in the rest of the range (Canestrelli et al., 2006); and (iii) populations from this area are reported as viable despite the long-term co-occurrence of the “killer fungus” Batrachochytrium dendrobatidis (Canestrelli et al., 2013), considered one of the main drivers of the decline (Stagni et al., 2004).
In this paper, we investigated whether the hotspot area can be considered—and managed—as a single and temporally stable geographic unit, or if substantial variation exists within this area in parameters of key importance for the conservation of B. pachypus populations. To achieve this goal, we integrated previous knowledge with the results of three experimental steps. First, we surveyed the whole region to assess whether occurrence pattern of populations has remained stable over time, in the whole area or in parts of it. Second, we assessed the genetic structure of populations, quantified their levels of genetic diversity, and compared them with those of populations sampled in the same area in the past. Third, we performed an analysis of current potential risk factors by focusing on the present-day occurrence of the chytrid pathogen B. dendrobatidis, as well as by estimating variations in bioclimatic suitability of the hotspot area, from current to future climatic scenarios.
Materials and Methods
Species Presence Data
Barbieri et al. (2004) indicated southern Italy (Calabria administrative region) as the only area within the distribution range where the yellow-bellied toad was not facing demographic declines at the end of 1990s. In order to confirm this finding and/or to characterize spatial and temporal patterns of variation, we carried out extensive field surveys within this region. Field activities were carried out from April to October, corresponding with the reproductive season of the species, from 2013 to 2016. Field activities and sampling procedures were approved by the Italian Ministry of the Environment (protocol numbers: 0042634/PNM, and 0007727/PNM).
We screened peer-reviewed and gray literature and museum collections for records of the presence of the toad in Calabria. We retained only 56 records with detailed information about location and year of observation and we divided the region into 4 areas where most of the records are concentrated: Catena Costiera, Sila, Serre and Aspromonte (Figure 1). The boundaries of these 4 areas were defined arbitrarily, although roughly based on mountain ranges.
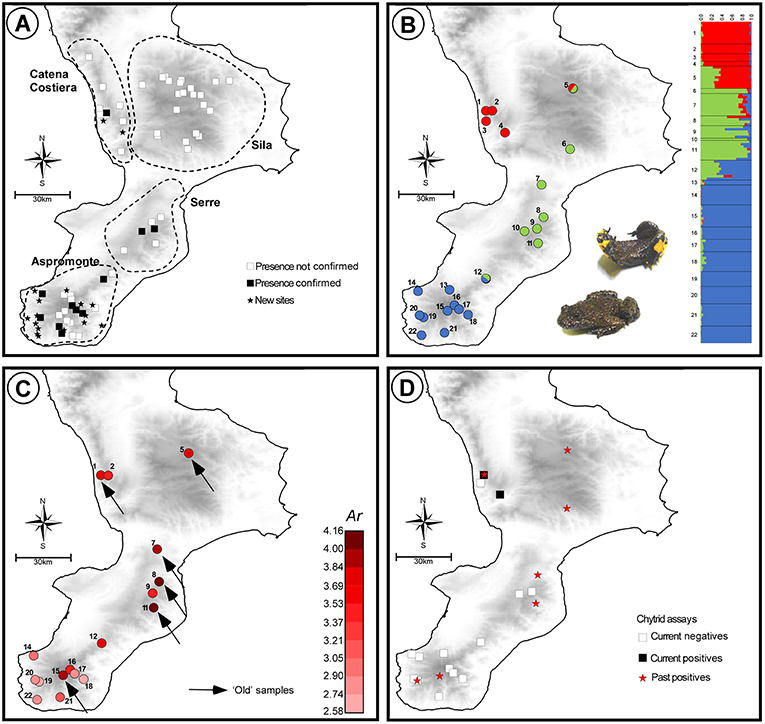
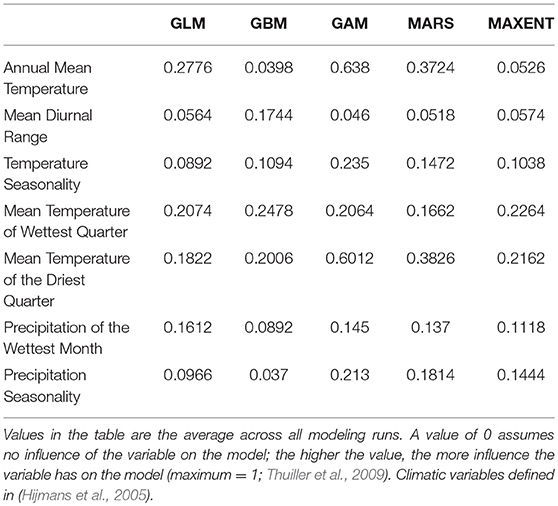
Figure 1. Spatial and temporal patterns of variation in species presence, genetic diversity, and chytrid occurrence, for the Apennine yellow-bellied toad in southern Italy (Calabria region). Sources of historical data and samples are reported within the main text. (A) Sites of past and current presence of Bombina pachypus in the area; dashed shapes show the four subregions considered in this study. (B) Results of the Bayesian clustering analysis used to assess the population genetic structure of B. pachypus in the area; admixture proportions of each sampled individual for the 3 genetic clusters identified as the best clustering option by the method implemented in TESS. (C) Geographic variation in estimates of genetic diversity (Ar: allelic richness), among populations of B. pachypus sampled either for the present study or in the past. (D) Past and current presence of the chytrid pathogen Batrachochytrium dendrobatidis among the sampled populations of B. pachypus from the study area.
Each site of historical presence was visited three times during the whole campaign. We considered the species as currently present in a site when at least one evidence of its presence (either adults, or tadpoles, or clutches) was observed in at least one visit. Field research was also opportunistically extended to other 80 non-reported but potentially suitable sites in the surroundings, in order to search for new records.
Samples for subsequent laboratory procedures were collected from the adult individuals encountered during the last visit at each site. In order to limit the stress associated to handling procedures for the individual toad, each individual was subjected to a single sampling procedure (i.e., either toe-clipping or skin-swabbing; see below).
Genetic Diversity and Population Structure
Population genetic structure and diversity were assessed by genotyping 130 individuals from 22 sites, ranging over the whole region (Table 1 and Figure 1). Individuals from 15 of these sites (“new” samples in Table 1) were sampled by toe-clipping after anaesthetization in a 0.1% solution of MS222 (3-aminobenzoic acid ethyl ester). Toe-clipping is a permanent marking procedure, allowing to avoid re-sampling DNA from the same individual in case of multiple sampling session, as is the case in the present study. In principle, this procedure could also be used to estimate population size, using mark-recapture experimental designs. However, we did not use it for this additional purpose, as multiple captures of the same individual were never observed during this study. All individuals were immediately released at the collection sites, while tissue samples were stored in 95% alcohol. Tissue samples from the remaining 7 sites (“old” samples in Table 1) were collected during previous studies carried out from 1978 to 2006 (see Canestrelli et al., 2006, 2013 and references therein), and were stored in 95% alcohol.
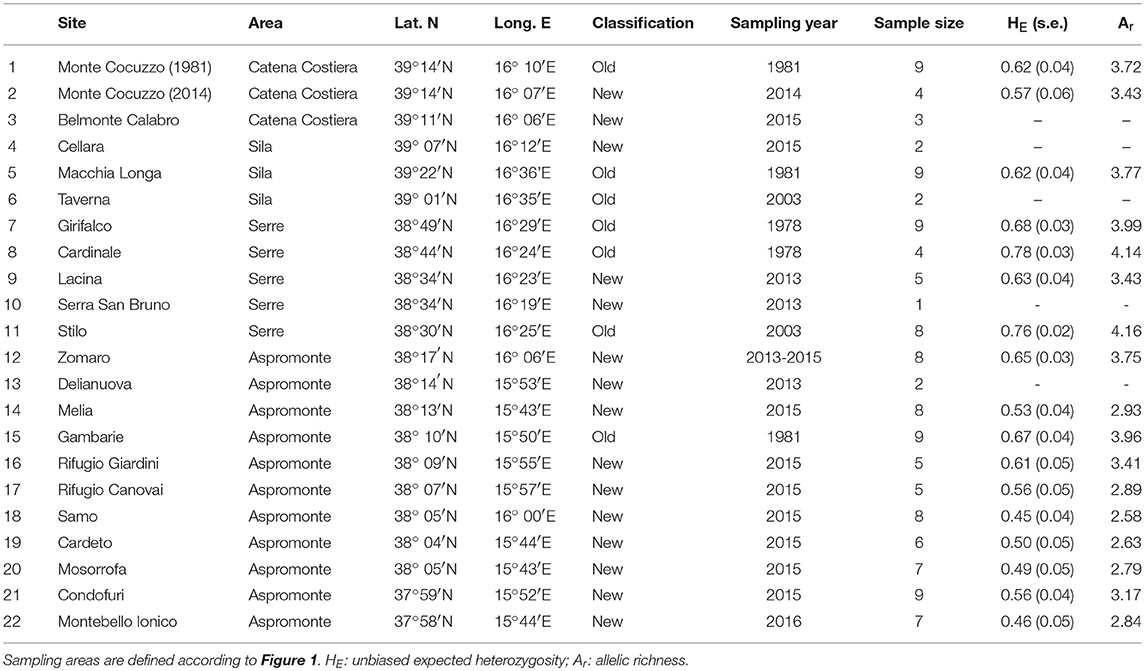
Table 1. Geographic location, sample size, sampling year, and estimates of genetic diversity, for the 22 populations of Bombina pachypus sampled for the analysis of genetic variation in southern Italy.
Genomic DNA was extracted using the ZR Genomic DNA™—Tissue MiniPrep kit (Zymoresearch) following the manufacturer's instructions. We initially tested 11 microsatellite loci that previously proved to amplify in the sister species B. variegata: Bv11.7, Bv32.7, B13, B14, F22, 1A, 5F, 8A, 9H, 10F, 12F (Nürnberger et al., 2003; Stuckas and Tiedemann, 2006; Hauswaldt et al., 2007). We excluded from the analyses the loci Bv32.7 (failed the amplification step for our samples), B13 (yielded inconsistent reactions), B14 (yielding missing data in 26% of the individuals), and 12F (monomorphic in preliminary trials based on 30 randomly selected individuals). PCR conditions for the 7 loci analyzed in this study, along with fluorescent dyes used to label forward primers and multiplex assembly are shown in Supplementary Material. Fragment analysis of PCR products was performed by Macrogen Inc. on an ABI 3730xl Genetic Analyser (Applied Biosystems) with a 400HD size standard.
Allele calling was performed with GENEMAPPER® 4.1 checking electropherograms by eye. Microsatellite dataset was then analyzed with MICRO-CHECKER 2.2.3 (Van Oosterhout et al., 2004) to test for the presence of null alleles, large alleles drop-out or scoring errors. We used GENEPOP 4.5.1 (Raymond and Rousset, 1995; Rousset, 2008) to test for departures from Hardy-Weinberg equilibrium and for genotypic linkage disequilibrium. Population genetic diversity was estimated as expected heterozigosity (He) and allelic richness (Ar) using GENETIX 4.05.2 (Belkhir et al., 1996) and FSTAT 2.9.3.2 (Goudet, 2001), respectively. These analyses were carried out excluding sites where we sampled <4 individuals.
Genetic population structure was inferred by means of the Bayesian clustering algorithm implemented in TESS 2.3.1 (Chen et al., 2007), with the admixture model, under the conditional autoregressive model (CAR), and the geographical coordinates of individuals as prior information. TESS was used as clustering method since it has been shown to perform better than similar methods when the number loci is limited and/or when genetic structure is shallow (Chen et al., 2007; François and Durand, 2010). We performed a set of preliminary analyses with 20,000 runs, discarding the first 5,000 as burn-in, and with 10 replicates for each value of K from 2 to 10 to test model performance. For the final analysis we ran 100 replicates for each value of K from 2 to 6, with 100,000 steps and discarding the first 50,000 as burn-in. The spatial interaction parameter was set to the default value (0.6) and the option of update this parameter was activated. The clustering model that best fitted the data was inferred by the deviance information criterion (DIC), averaging the DIC values over the 100 replicates for each K and selecting the K value at which the average DIC reached a plateau. The 10 runs with the lowest DIC values for the inferred K were finally selected, and the estimated admixture proportions were averaged over them using CLUMPP 1.1.2 (Jakobsson and Rosenberg, 2007).
The extent of genetic differentiation between the inferred clusters was analyzed by estimating pairwise Fst values (Weir and Cockerham, 1984), by means of FSTAT. Individuals of mixed ancestry were assigned to the cluster with the highest admixture proportion. The statistical significance of the estimated Fst values was assessed through 3000 permutations, and the nominal 5% level of significance was adjusted for multiple comparisons, using the Bonferroni correction.
Pathogen Assay
Skin swabs were collected from 105 individuals collected in 15 sites (Figure 1). Genomic DNA was extracted from swabs following the protocol of Boyle et al. (2004) as modified by Zampiglia et al. (2013). The molecular diagnostic assay was conducted in 25 μl reaction volumes using a nested PCR protocol as developed by Goka et al. (2009). The assay was performed once for each sample and it included a positive (DNA extracted from B. dendrobatidis zoospores JEL423, kindly provided by Prof. Joyce Longcore) and a negative (DNA-free distilled water) control. PCR products were loaded on a 1% agarose gel and checked for the diagnostic band at approximately 300 bp size to be considered as B. dendrobatidis positive. A geographic representative subset of samples (20%) was analyzed in duplicate.
Changes in Bioclimatic Suitability
We calibrated species distribution models (SDM) with a dataset of 361 occurrences covering the entire distribution range of the species and a set of bioclimatic variables. The occurrences were obtained by pooling the data collected during the field campaign (this study) and different data sources: (Stoch, 2000–2005), Global Biodiversity Information Facility (www.GBIF.org), Observado (www.observation.org) and Canestrelli et al. (2006). The dataset was filtered to remove duplicated records and data georeferenced with uncertainty. To limit spatial autocorrelation, we thinned the raw occurrences using the SPTHIN R package (Aiello-Lammens et al., 2015), obtaining five alternative calibration subsets with 182 occurrence data (see Supplementary Material for details). Considering the same data sources available for the presence, we collected all data available for the same study area for species of reptiles and amphibians. We collected a total of 7614 records that we used in model calibration as background points in order to limit the effects of the existing sampling bias (Ranc et al., 2017).
The bioclimatic variables for current climate were obtained from Hijmans et al. (2005). Following a variance inflation factor (VIF) analysis on the original 18 bioclimatic variables, we considered in the analyses only the following 7 variables with VIF <5: annual mean temperature, mean diurnal range, temperature seasonality, mean temperature of wettest quarter, mean temperature of driest quarter, precipitation of wettest month, and precipitation seasonality. For the future climate, we considered the same 7 variables calculated under three general circulation models (CCSM4, MIROC-ESM, MPI-ESM-P) and two emission scenarios (RCP2.6 and RCP8.5) adopted by the IPCC5 (Intergovernmental Panel on Climate Change).
Using an ensemble forecasting approach (Araújo and New, 2007), we calibrated the SDM considering the current climate, the five thinned occurrence sets, the set of background points, and the following five algorithms as implemented in the BIOMOD2 R packages (Thuiller et al., 2009): generalized linear model (GLM); generalized additive model (GAM); generalized boosted models (GBM); multivariate adaptive regression spline (MARS), and maximum entropy (MAXENT).
For model evaluation, we considered again five sets of points, each including the 179 occurrence points excluded through the thinning procedure from the calibration datasets and 7488 random background points (maintaining the same prevalence as in the calibration dataset). For each model, we measured the area under the curve (AUC) of the receiver operating characteristic (ROC) curve. Models with AUC value >0.7 (Swets, 1988) were projected under the current and future climate over the entire peninsular Italy. Then, for each projection we obtained a final consensus SDM, calculated as the weighted average of all available models (weights for each model based on the respective AUC score, as in Marmion et al., 2009).
Results
Species Presence in Space and Time
We found evidence of the presence of B. pachypus in 28 localities out of the original 136 (21%), including 11 sites of historical presence (out of the original 56) and 17 new records. The highest number of historical sites with confirmed presence was found in the Aspromonte (8 out of 19; 42%), followed by the Serre (2 out of 6; 33%) and the Catena Costiera (1 out of 8; 13%). No presence was confirmed for the Sila (0 presences out of the original 23). The 17 new records of presences were unevenly distributed among regions, with 15 new sites found in the Aspromonte, 2 sites found in the Catena Costiera, and no new site found in Sila and Serre regions. The Aspromonte region was also the area with the highest number of individuals observed per site/day, with an average of 21.6 individuals (s.d. 18.3) compared to 3.5 individuals (s.d. 1.9) observed on average in the other regions.
Genetic Diversity and Population Structure
The number of alleles observed at each microsatellite locus was as follows: 1A = 11, 8A = 13, 5F = 12, 10F = 12, 9H = 25, Bv11 = 3 and F22 = 9. All loci were used for downstream analyses, since none of them showed evidence of null alleles in more than one sampling site. Missing data accounted for 3.7% of the whole dataset. No evidence of genotypic linkage disequilibrium between loci was observed, and no departures from the Hardy-Weinberg equilibrium was detected after the Bonferroni correction was applied (Rice, 1989).
Expected heterozigosity ranged from 0.45 (site 18) to 0.78 (site 8), while allelic richness ranged from 2.58 (site 18) to 4.16 (site 11; Figure 1C). Estimates of both the expected heterozigosity and the allelic richness for each sampled site are shown in Table 1. Among the “old” samples, the highest values of both heterozigosity and allelic richness were observed within the Serre region, whereas among the “new” samples, samples with the highest values of genetic diversity were observed within the Aspromonte region. Although the location and size of our samples prevented us from carrying out quantitative temporal comparisons, it is worth noting that “old” samples systematically yielded higher values of genetic diversity when compared with the nearest “new” samples from the same area (see Table 1).
Bayesian clustering analysis of population structure revealed the presence of three main population clusters within the study area. Pie-charts displaying the contribution of the three clusters to the genetic pool of each site and bar-plots indicating individual admixture proportions are shown in Figure 1B. The three clusters showed a clear geographic structure along the north-south axis: one was restricted to the north, in Catena Costiera and northern Sila, one had a central distribution, ranging from southern Sila to Serre, and the third one was restricted to the south, in Aspromonte. Evidence of admixture between clusters was observed in some individuals, particularly from sites located in intermediate areas (sites 5 and 12). The northern cluster was the most genetically differentiated, with pairwise Fst values of 0.11 and 0.18 with the central and the southern clusters, respectively (both P < 0.01). Instead, the extent of differentiation was lower between the central and southern the clusters, with a Fst value of 0.05 (P < 0.01).
Occurrence of Batrachochytrium dendrobatidis
The diagnostic tests for B. dendrobatidis occurrence yielded positive results for the presence of the pathogen on the skin of 3 individuals from 2 sites in the Catena Costiera. Instead, and in contrast to previous assessments (Canestrelli et al., 2013), no positive tests were observed for individuals sampled in the Serre and Aspromonte (Figure 1D). All the analyses carried out in duplicate yielded fully consistent results.
Species Distribution Modeling
We calibrated 25 models (5 presence-background sets for 5 modeling techniques) with, on average, good model performance (mean AUC = 0.80; s.d. AUC = 0.04). GBM was the model with the highest AUC values (mean AUC = 0.86) and the GLM with the lowest AUC (mean AUC = 0.76). Temperature was by far the most important bioclimatic factor for explaining the distribution of the yellow-bellied toad, with mean temperature of the driest quarter, mean temperature of the wettest quarter, and annual mean temperature being the most important variables, followed by the two precipitation variables (Table 2).
Regardless of the climate scenario considered, the future climate suitability is predicted to decrease substantially (Figure 2), especially in the north and central Apennines, where bioclimatic suitability drops dramatically in some areas. A much higher climatic stability in time is predicted for the southern part of the distribution range, where however the models predict a future range shift toward higher elevations (Figure 2).
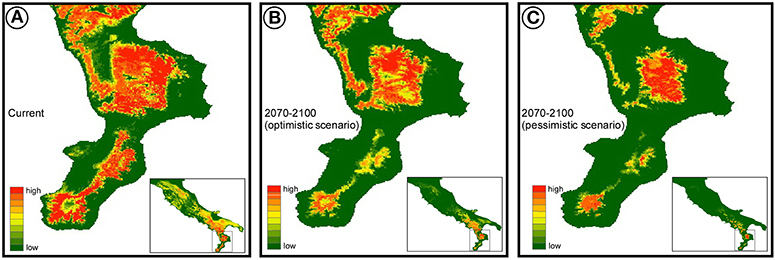
Figure 2. Species distribution model estimated for Bombina pachypus under current bioclimatic conditions (A), and its projection to the near future (2070–2100), under optimistic (B) and pessimistic (C) scenarios of greenhouse gas emission (RCP2.6 and RCP8.5, respectively).
Discussion
Identifying hotspots of intraspecific genetic diversity is now increasingly seen as a key step toward conservation practices effective in the long-term (Thomassen et al., 2011; Brooks et al., 2015). For the first time, however, our results show that this fundamental step does not suffice, and that finer spatial and temporal scales of analysis, which are characteristic of monitoring programs (Brodersen and Seehausen, 2014; Mimura et al., 2017), should be adopted in order to provide conservation stakeholders with important knowledge to best define conservation priorities and management practices.
The hotspot of intraspecific genetic diversity of B. pachypus is not a homogeneous geographic unit, either in terms of genetic diversity, or in terms of species presence and risk factors' distribution. Moreover, our results provide evidence of geographically structured temporal changes in the analyzed features. In the next sections we will discuss these spatio-temporal patterns of variation, and how they could help to identify priorities in the context of both short-term and long-term conservation programs.
Spatial and Temporal Patterns of Variation
Over <15 years since the last assessment of species presence (Barbieri et al., 2004), patterns of B. pachypus occurrence in southern Italy have dramatically changed, although with marked differences among sub-regions. At the extreme of these differences are the Sila and the Aspromonte massifs. Throughout the Sila region we completely failed to identify sites of current B. pachypus presence, despite the particular sampling effort we devoted to this area (41% of the inspected sites of historical presence were located within this area). While this lack of evidence is not a conclusive argument that the species completely disappeared from the area, it clearly testifies to a dramatic decline in the number of sites of occurrence. On the other hand, in the Aspromonte region several sites of historical presence were confirmed, and new ones identified. Together with the higher number of individuals per site observed in this area than elsewhere, the Aspromonte region has clearly appeared as the least affected by population declines. Further indication of diffused declines comes also from the observed temporal decreases in genetic diversity estimates, suggesting widespread bottlenecks in population size throughout the hotspot area in southern Italy (Figure 1C).
These findings rise worrisome questions for the conservation of this endangered toad. Why did B. pachypus start to decline even in southern Italy? And why is there a geographic structure in this trend? First of all, our results allow us to exclude some of the most commonly invoked answers to these and similar questions. Habitat degradation is probably not a factor in this trend. The Aspromonte and Sila regions are largely covered by national parks, while the Serre region falls within a regional park. Also, in most instances, no evidence of alteration of the physical habitat or of the ecological community was observed at the sites visited. Thus, while a role in species decline at single sites cannot be excluded (Barbieri et al., 2004), habitat degradation can hardly explain the general pattern of declines throughout the study area. Likewise, bioclimatic factors are unlikely to explain the pattern, at least as the single factor. Indeed, the Sila massif, where no site of presence was found, does not suffer of a limited bioclimatic suitability, as compared with neighboring areas, and most historical sites of presence are located exactly within the area of bioclimatic optimum. Finally, not even B. dendrobatidis can be reliably invoked as the “lone-killer” (Pounds and Coloma, 2008 and references therein). As a matter of fact, B. dendrobatidis was widespread in the area (including the Aspromonte) at least since the early ‘80s (Canestrelli et al., 2013), that is, well before the observed declines begun. Nonetheless, its role in the observed declines in conjunction with other factors cannot be excluded. Indeed, contrary to results of the previous assessment, B. dendrobatidis occurrence was confirmed in the northern but not in the southern portion of the study area.
These results lead to at least two non-mutually-excluding hypotheses, useful to attempt answering the above questions. First, the overall reduction of the chytrid geographic distribution [and its absence from sites that are very close to those where high chytrid prevalence was reported until 2003] might be the outcome of epidemic dynamics related to environmental changes. In fact, experimental evidence showed chytrid pathogenicity to decrease at increasing temperatures, with possible implications even at microgeographic scales (Greenspan et al., 2017a), as well as increased host vulnerability to climate change driven by infection (Greenspan et al., 2017b). Second, geographic differences in the mechanisms of resistance of the amphibian host might be implicated (Luquet et al., 2012). In this regard, it is worth recalling that geographic patterns of variation in B. pachypus presence and chytrid distribution, are congruent with the geographic structure of the genetic variation among B. pachypus populations. Indeed, all the most abundant populations, currently free from chytrid infection, belong to the southern cluster confined to the Aspromonte area. Although not conclusive in disentangling these two scenarios, our results set the stage for future experimental efforts along this road, based on testable research hypotheses.
An interesting corollary to the first scenario comes from bioclimatic suitability predicted for future scenarios in the Aspromonte region. Indeed, B. pachypus climate optimum in the Aspromonte massif was predicted to shift upward in elevation in the near future, possibly leading to a selective scenario with two contrasting forces acting on populations, B. pachypus being on the horns of a dilemma. Sites of current presence will likely lose their bioclimatic suitability in the near future, but in a direction predicted to favor the toad host over the chytrid pathogenicity (i.e., increasing environmental temperatures). On the other hand, colonizing sites at higher elevation might allow B. pachypus to continue matching its bioclimatic optimum, but at the expense of more favorable climatic conditions for its chytrid pathogen. Intriguingly, although speculative for the time being, this scenario might be experimentally evaluated through monitoring environmental variables, epidemiological parameters, and B. pachypus performance traits, among populations from the Aspromonte area. Since this is also the only area where the species appeared relatively abundant, such a monitoring and research effort might be one of the last options available to shed light on the mechanisms behind its decline.
Geographic Structure of Conservation Targets
A thorough understanding of mechanisms underlying species' decline is an essential step toward better informed conservation efforts, but the observed spatial patterns and temporal variations clearly call for timely actions, aimed at preventing further populations' disappearance in the short-term. We see four major implications of our results, useful to identify priorities and to optimize resource allocation for such a “short-term” program.
First, and contrarily to what was previously inferred, not all populations within the hotspot area should be considered as at high priority for species' conservation programs in situ. Indeed, in most of this area, declines appeared at an advanced stage (if not ultimate). Since an optimal target for in situ actions should be maximizing the chance of species' persistence, Aspromonte populations appear the ones whose demographic, genetic, and epidemiological features make them suitable for in situ actions, including stringent protection and monitoring of breeding sites.
Second, in those areas (Catena Costiera and Serre) where the species is still present, but where data suggest strong negative demographic trends, pilot experiments of captive-breeding with mixed stocks (individuals of both local and Aspromonte origin) should be implemented. Under the hypothesis of better resistance performance of Aspromonte populations to the chytrid pathogen, such experiments might favor assisted flows of beneficial alleles within local populations (Jones, 2013). If successful under controlled (semi-natural) experimental conditions, these experiments might provide material for subsequent reintroduction programs aimed at species' recovery.
Third, our results showed a drastic reduction of the geographic occurrence of the chytrid pathogen on its B. pachypus host, if compared to what was observed in the recent past (Canestrelli et al., 2013), but they do not imply pathogen decline in the area, since we did not analyze the whole range of its potential hosts in southern Italy. Thus, monitoring its occurrence in situ both in B. pachypus and other potential hosts (Simoncelli et al., 2005; Zampiglia et al., 2013), as well as within environmental matrices, will be a mandatory step of in situ actions, as will be also a proper prophylaxis in ex situ programs.
Fourth, but not least, our results showed a possible drastic reduction of habitat suitability expected by 2070–2100, along with an upward range shift. Conservation plans should consider this expected trend, and should plan actions accordingly. In particular, captive-breeding programs should be designed to assess species performance along elevation clines. Useful approaches in this regard might be the assessment of comparative performances (with and without the chytrid pathogen) in experimental climate chambers, or the use of replicated semi-natural settings located along environmental clines. Such approaches might also provide data of extreme value to help identify priority sites for future restocking or reintroduction programs of this species, both in southern Italy and elsewhere. Finally, such approaches might provide the necessary information to address the possible role of the Sila region in B. pachypus conservation strategies. That is, whether, under the changing bioclimatic scenarios, this area might become a bioclimatic refugium for B. pachypus, and so a major area for reintroduction programs, or if the apparent disappearance of the species from this area is just a “canary in mine” of what will soon happen throughout the region.
Ethics Statement
Field activities and sampling procedures were approved by the Italian Ministry of the Environment (protocol numbers: 0042634/PNM, and 0007727/PNM).
Author Contributions
DC and GN conceived the study. DC and MZ designed the experiments. DC, MZ, GA, FP, GM, CM, and AS performed fieldwork and sampling. MZ, RB, and AC performed labwork. AP and LM performed SDM analyses. MZ, DC, RB, and AC analyzed the data and interpreted results. MZ and DC drafted the manuscript, with inputs from RB, AC, and LM. All authors discussed and revised the manuscript, and gave final approval for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Marco Puleo, Antonio Mancuso, and Antonio Barca, for their help with sample collection. This study was funded by the Italian Ministry of the Environment and Protection of Land and Sea, The Aspromonte National Park, The Italian Ministry of Education, University and Research (PRIN project 2012FRHYRA to DC). This article is published as a preprint in the biorXiv server (Zampiglia et al., 2018).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00205/full#supplementary-material
References
Aiello-Lammens, M. E., Boria, R. A., Radosavljevic, A., Vilela, B., and Anderson, R. P. (2015). spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38, 541–545. doi: 10.1111/ecog.01132
Allendorf, F. W., and Luikart, G. (2009). Conservation and the Genetics of Populations. Malden, MA: John Wiley and Sons.
Allendorf, F. W., Luikart, G., and Aitken, S. N. (2012). Conservation and the Genetics of Populations. 2nd Edn. Oxford: JohnWiley and Sons.
Andreone, F., Corti, C., Sindaco, R., Romano, A., Giachi, F., Vanni, S., et al. (2009). Bombina Pachypus. (Errata Version Published in 2016) The IUCN Red List of Threatened Species 2009:e.T54450A86629977 (accessed 1 February 2018).
Araújo, M. B., and New, M. (2007). Ensemble forecasting of species distributions. Trends Ecol. Evol. 22, 42–47. doi: 10.1016/j.tree.2006.09.010
Barbieri, F., Bernini, F., Guarino, F., and Venchi, A. (2004). Distribution and conservation status of Bombina variegata in Italy (Amphibia, Bombinatoridae). Ital. J. Zool. 71, 83–90. doi: 10.1080/11250003.2004.9525541
Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N., and Bonhomme, F. (1996). GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire génome, populations, interactions, CNRS UMR, 5000, 1996–2004.
Boyle, D. G., Boyle, D. B., Olsen, V., Morgan, J. A. T., and Hyatt, A. D. (2004). Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Org. 60, 141–148. doi: 10.3354/dao060141
Brodersen, J., and Seehausen, O. (2014). Why evolutionary biologists should get seriously involved in ecological monitoring and applied biodiversity assessment programs. Evol. Appl. 7, 968–983. doi: 10.1111/eva.12215
Brooks, T. M., Cuttelod, A., Faith, D. P., Garcia-Moreno, J., Langhammer, P., and Pérez-Espona, S. (2015). Why and how might genetic and phylogenetic diversity be reflected in the identification of key biodiversity areas? Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:20140019. doi: 10.1098/rstb.2014.0019
Cañadas, A., and Vázquez, J. A. (2014). Conserving cuvier's beaked whales in the Alboran Sea (SW Mediterranean): Identification of high density areas to be avoided by intense man-made sound. Biol. Conserv. 178, 155–162. doi: 10.1016/j.biocon.2014.07.018
Canestrelli, D., Cimmaruta, R., Costantini, V., and Nascetti, G. (2006). Genetic diversity and phylogeography of the Apennine yellow-bellied toad Bombina pachypus, with implications for conservation. Mol. Ecol. 15, 3741–3754. doi: 10.1111/j.1365-294X.2006.03055.x
Canestrelli, D., Zampiglia, M., and Nascetti, G. (2013). Widespread occurrence of Batrachochytrium dendrobatidis in contemporary and historical samples of the endangered Bombina pachypus along the Italian Peninsula. PLoS ONE 8:e63349. doi: 10.1371/journal.pone.0063349
Chen, C., Durand, E., Forbes, F., and François, O. (2007). Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol. Ecol. Notes 7, 747–756. doi: 10.1111/j.1471-8286.2007.01769.x
Crandall, K. A., Bininda-Emonds, O. R., Mace, G. M., and Wayne, R. K. (2000). Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 15, 290–295. doi: 10.1016/S0169-5347(00)01876-0
Eizaguirre, C., and Baltazar-Soares, M. (2014). Evolutionary conservation-evaluating the adaptive potential of species. Evol. Appl. 7, 963–967. doi: 10.1111/eva.12227
François, O., and Durand, E. (2010). Spatially explicit bayesian clustering models in population genetics. Mol. Ecol. Resour. 10, 773–784. doi: 10.1111/j.1755-0998.2010.02868.x
Frankham, R. (2010). Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 143, 1919–1927. doi: 10.1016/j.biocon.2010.05.011
Goka, K., Yokoyama, J., Une, Y., Kuroki, T., Suzuki, K., Nakahara, M., et al. (2009). Amphibian chytridiomycosis in Japan: distribution, haplotypes and possible route of entry into Japan. Mol. Ecol. 18, 4757–4774. doi: 10.1111/j.1365-294X.2009.04384.x
Goudet, J. (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Lausanne: Institute of Ecology.
Greenspan, S. E., Bower, D. S., Roznik, E. A., Pike, D. A., Marantelli, G., Alford, R. A., et al. (2017b). Infection increases vulnerability to climate change via effects on host thermal tolerance. Sci. Rep. 7:9349. doi: 10.1038/s41598-017-09950-3
Greenspan, S. E., Bower, D. S., Webb, R. J., Roznik, E. A., Stevenson, L. A., Berger, L., et al. (2017a). Realistic heat pulses protect frogs from disease under simulated rainforest frog thermal regimes. Funct. Ecol. 31, 2274–2286. doi: 10.1111/1365-2435.12944
Hampe, A., and Petit, R. J. (2005). Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 8, 461–467. doi: 10.1111/j.1461-0248.2005.00739.x
Hauswaldt, J. S., Schröder, C., and Tiedemann, R. (2007). Nine new tetranucleotide microsatellite markers for the fire-bellied toad (Bombina bombina). Mol. Ecol. Notes 7, 49–52. doi: 10.1111/j.1471-8286.2006.01516.x
Hendry, A. P., Lohmann, L. G., Conti, E., Cracraft, J., Crandall, K. A., Faith, D. P., et al. (2010). Evolutionary biology in biodiversity science, conservation, and policy: a call to action. Evolution 64, 1517–1528. doi: 10.1111/j.1558-5646.2010.00947.x
Hewitt, G. M. (2000). The genetic legacy of the quaternary ice ages. Nature 405, 907–913. doi: 10.1038/35016000
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the quaternary. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 183–195. doi: 10.1098/rstb.2003.1388
Hewitt, G. M. (2011). Quaternary phylogeography: the roots of hybrid zones. Genetica 139, 617–638. doi: 10.1007/s10709-011-9547-3
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G., and Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. doi: 10.1002/joc.1276
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Laikre, L. (2010). Genetic diversity is overlooked in international conservation policy implementation. Conserv. Genet. 11, 349–354. doi: 10.1007/s10592-009-0037-4
Lanfear, R., Kokko, H., and Eyre-Walker, A. (2014). Population size and the rate of evolution. Trends Ecol. Evol. 29, 33–41. doi: 10.1016/j.tree.2013.09.009
Luquet, E., Garner, T. W., Léna, J. P., Bruel, C., Joly, P., Lengagne, T., et al. (2012). Genetic erosion in wild populations makes resistance to a pathogen more costly. Evolution 66, 1942–1952. doi: 10.1111/j.1558-5646.2011.01570.x
Mace, G. M., and Purvis, A. (2008). Evolutionary biology and practical conservation: bridging a widening gap. Mol. Ecol. 17, 9–19. doi: 10.1111/j.1365-294X.2007.03455.x
Marmion, M., Parviainen, M., Luoto, M., Heikkinen, R. K., and Thuiller, W. (2009). Evaluation of consensus methods in predictive species distribution modelling. Divers. Distributions 15, 59–69. doi: 10.1111/j.1472-4642.2008.00491.x
Mimura, M., Yahara, T., Faith, D. P., Vázquez-Domínguez, E., Colautti, R. I., Araki, H., et al. (2017). Understanding and monitoring the consequences of human impacts on intraspecific variation. Evol. Appl. 10, 121–139. doi: 10.1111/eva.12436
Moritz, C. (2002). Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 51, 238–254. doi: 10.1080/10635150252899752
Nürnberger, B., Hofman, S., Förg-Brey, B., Praetzel, G., Maclean, A, Szymura, J. M., et al. (2003). A linkage map for the hybridising toads Bombina bombina and B. variegata (Anura: Discoglossidae). Heredity 91, 136–142. doi: 10.1038/sj.hdy.6800291
Pérez-Espona, S. ConGRESS Consortium (2017). Conservation genetics in the European union – biases, gaps and future directions. Biol. Conserv. 209, 130–136. doi: 10.1016/j.biocon.2017.01.020
Petit, R. J., Aguinagalde, I., de Beaulieu, J. L., Bittkau, C., Brewer, S., Cheddadi, R., et al. (2003). Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300, 1563–1565. doi: 10.1126/science.1083264
Pounds, J. A., and Coloma, L. A. (2008). Beware the lone killer. Nat. Clim. Change 2, 57–59. doi: 10.1038/climate.2008.37
Ranc, N., Santini, L., Rondinini, C., Boitani, L., Poitevin, F., Angerbjörn, A., et al. (2017). Performance trade-offs in target-group bias correction for species distribution models. Ecography 40, 1076–1087. doi: 10.1111/ecog.02414
Raymond, M., and Rousset, F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249.
Rice, W. E. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x
Rousset, F. (2008). GENEPOP'007: a complete re-implementation of the GENEPOP software for windows and linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Simoncelli, F., Fagotti, A., Dall'Olio, R., Vagnetti, D., Pascolini, R., and Di Rosa, I. (2005). Evidence of Batrachochytrium dendrobatidis infection in water frogs of the Rana esculenta complex in central Italy. EcoHealth 2:307. doi: 10.1007/s10393-005-8337-8
Stagni, G., Dall'olio, R., Fusini, U., Mazzotti, S., Scoccianti, C., and Serra, A. (2004). Declining populations of Apennine yellow-bellied toad Bombina pachypus in the northern apennines (Italy): Is Batrachochytrium dendrobatidis the main cause? Ital. J. Zool. 71, 151–154. doi: 10.1080/11250000409356625
Stoch, F. (2000–2005). CKmap 5.3.8. Ministero dell'Ambiente e della Tutela del Territorio e del Mare. Direzione Generale per la Protezione della Natura e del Mare. Available online at: http://www.minambiente.it/index.php?id_sezione=1930.
Stuckas, H., and Tiedemann, R. (2006). Eight new microsatellite loci for the critically endangered fire-bellied toad Bombina bombina and their cross-species applicability among anurans. Mol. Ecol. Notes 6, 150–152. doi: 10.1111/j.1471-8286.2005.01171.x
Swets, J. A. (1988). Measuring the accuracy of diagnostic systems. Science 240, 1285–1293. doi: 10.1126/science.3287615
Thomassen, H. A., Fuller, T., Buermann, W., Mila, B., Kieswetter, C. M., Cameron, S. E., et al. (2011). Mapping evolutionary process: a multi-taxa approach to conservation prioritization. Evol. Appl. 4, 397–413. doi: 10.1111/j.1752-4571.2010.00172.x
Thuiller, W., Lafourcade, B., Engler, R., and Araújo, M. B. (2009). BIOMOD–a platform for ensemble forecasting of species distributions. Ecography 32, 369–373. doi: 10.1111/j.1600-0587.2008.05742.x
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Vernesi, C., Bruford, M. W., Bertorelle, G., Pecchioli, E., Rizzoli, A., and Hauffe, H. C. (2008). Where is the conservation in conservation genetics? Conserv. Biol. 22, 802–804. doi: 10.1111/j.1523-1739.2008.00911.x
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.2307/2408641
Zampiglia, M., Bisconti, R., Maiorano, L., Aloise, G., Siclari, A., Pellegrino, F., et al. (2018). Drilling down hotspots of intraspecific diversity to bring them into on-ground conservation of threatened species. Biorxiv:419788. doi: 10.1101/419788
Keywords: evolutionary potential, amphibian declines, genetic diversity hotspots, management strategies, Bombina pachypus, southern Italy, Batrachochytrium dendrobatidis, species distribution models
Citation: Zampiglia M, Bisconti R, Maiorano L, Aloise G, Siclari A, Pellegrino F, Martino G, Pezzarossa A, Chiocchio A, Martino C, Nascetti G and Canestrelli D (2019) Drilling Down Hotspots of Intraspecific Diversity to Bring Them Into On-Ground Conservation of Threatened Species. Front. Ecol. Evol. 7:205. doi: 10.3389/fevo.2019.00205
Received: 09 November 2018; Accepted: 20 May 2019;
Published: 14 June 2019.
Edited by:
Melanie April Murphy, University of Wyoming, United StatesReviewed by:
Joana Isabel Robalo, University Institute of Psychological, Social and Life Sciences, PortugalCatherine I. Cullingham, University of Alberta, Canada
Copyright © 2019 Zampiglia, Bisconti, Maiorano, Aloise, Siclari, Pellegrino, Martino, Pezzarossa, Chiocchio, Martino, Nascetti and Canestrelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberta Bisconti, YmlzY29udGlAdW5pdHVzLml0
 Mauro Zampiglia
Mauro Zampiglia Roberta Bisconti
Roberta Bisconti Luigi Maiorano
Luigi Maiorano Gaetano Aloise
Gaetano Aloise Antonino Siclari4
Antonino Siclari4 Alice Pezzarossa
Alice Pezzarossa Andrea Chiocchio
Andrea Chiocchio Daniele Canestrelli
Daniele Canestrelli