- 1Aquatic Ecosystem Research, University of Duisburg-Essen, Essen, Germany
- 2Department of Integrative Biology, University of Guelph, Guelph, ON, Canada
- 3Centre for Water and Environmental Research, University of Duisburg-Essen, Essen, Germany
- 4Emschergenossenschaft-Lippeverband, Essen, Germany
Freshwaters face some of the highest rates of species loss, caused by strong human impact. To decrease or even revert this strong impact, ecological restorations are increasingly applied to restore and maintain the natural ecological status of freshwaters. Their ecological status can be determined by assessing the presence of indicator species (e.g., certain fish species), which is called biomonitoring. However, traditional biomonitoring of fish, such as electrofishing, is often challenging and invasive. To augment traditional biomonitoring of fish, the analysis of environmental DNA (eDNA) has recently been proposed as an alternative, sensitive approach. The present study employed this modern approach to monitor the Rhine sculpin (Cottus rhenanus), a fish species that has been reintroduced into a recently restored stream within the Emscher catchment in Germany, in order to validate the success of the applied restorations and to monitor the species’ dispersal. We monitored the dispersal of the Rhine sculpin using replicated 12S end-point nested PCR eDNA surveillance at a fine spatial and temporal scale. In that way, we investigated if eDNA analysis can be applied for freshwater assessments. We also performed traditional electrofishing in one instance to validate our eDNA-based approach. We could track the dispersal of the Rhine sculpin and showed a higher dispersal potential of the species than we assumed. eDNA detection indicated the species’ dispersal across a potential dispersal barrier and showed a steep increase of positive detections once the reintroduced population had established. In contrast to that, false negative eDNA results occurred at early reintroduction stages. Our results show that eDNA detection can be used to confirm and monitor reintroductions and to contribute to the assessment and modeling of the ecological status of streams.
Introduction
Freshwaters are heavily impacted due to habitat fragmentation caused by wetland drainage, river straightening, and dam building, in combination with poor water quality caused by agricultural and industrial pollution (Jensen et al., 2006). This strong human impact can cause a decrease in the ecological status of freshwaters. Monitoring their ecological status is consequently an important task to assess if actions are needed for restoring and maintaining these ecosystems. Their ecological status can be determined by assessing indicator species, which include algae, benthic macroinvertebrates, and fish (Resh and Unzicker, 1975; Karr, 1981; Bellinger and Sigee, 2015).
The Rhine sculpin (Cottus rhenanus Freyhof et al., 2005) and the European bullhead (Cottus gobio Linnaeus, 1758) are two closely related freshwater sculpin species (previously treated as one species) that prefer similar habitats. They require well-oxygenated streams (Colleye et al., 2013) with gravelly to stony stream beds (Wittkugel, 2005), show a stationary behavior with limited home ranges (Ovidio et al., 2009), and are presumed to be incapable of crossing barriers higher than 18–20 cm (based on a study on the European bullhead; Utzinger et al., 1998). These requirements make them good indicators for the structure and passability of flowing waters due to their limited movement behavior. The Rhine sculpin used to be resident in the Emscher catchment (North Rhine-Westphalia, Germany) but became locally extinct during the 19th century when the catchment was used as an open sewer system for wastewater disposal (Brink-Kloke et al., 2006). The species survived in only one tributary stream that was less anthropogenically impacted during that period (Donoso-Büchner, 2009). Today, one of the largest European infrastructure projects supports comprehensive river restructuring of the Emscher catchment, with 4.5 billion Euro invested in the project as of 2015 (Böhmer, 2015). The project aims to restore the river and all its tributaries to a near-natural ecological status (Schnelle and Wilts, 2016). These efforts make it possible to reintroduce individuals from the isolated Rhine sculpin population into the restored streams, which can additionally confirm the successful restoration of suitable ecological conditions.
Monitoring the reintroduction success of the Rhine sculpin is, however, logistically challenging. Electrofishing, where fish are temporarily stunned using an electronic device, is the established method for collecting individuals. This traditional approach is invasive for ecosystems and not always feasible (Platts et al., 1983; Bohlin et al., 1989). The application of environmental DNA (eDNA) analysis is a promising and non-invasive alternative for the detection and biomonitoring of species in aquatic environments (Thomsen and Willerslev, 2015). While eDNA studies face several limitations because of eDNA degradation, PCR inhibition, and uncertainties about eDNA production and transportation (Goldberg et al., 2015), it was nevertheless shown that eDNA analysis has potential to effectively infer the richness of fish in streams, lakes, and the ocean (Jerde et al., 2011; Dejean et al., 2012; Takahara et al., 2013; Miya et al., 2015; Hänfling et al., 2016; Yamamoto et al., 2016). A recent large-scale comparison has shown reliability and higher success rates when inferring the presence of fish species in rivers via eDNA analysis as compared to electrofishing (Pont et al., 2018).
Environmental DNA analysis is established for monitoring the dispersal of invasive fish species (Takahara et al., 2013; Laramie et al., 2015; Adrian-Kalchhauser and Burkhardt-Holm, 2016) and has recently been used in a study to monitor and validate the reintroduction success of a locally extinct fish (Riaz et al., 2019) and amphibian species (Rojahn et al., 2018). The present study uses eDNA analysis for the same application but at a much finer temporal and spatial scale to investigate upstream dispersal. The aim of the present study was to use eDNA analysis for monitoring the reintroduction success and dispersal of the Rhine sculpin in a restored stream of the Emscher catchment. In the context of the present study, the recently restored stream Borbecker Mühlenbach within the Emscher catchment was considered suitable for re-establishing the Rhine sculpin post-restoration. Successful re-establishment of the target species in restored streams indicates that a good ecological status has been achieved, supporting the successful restoration. We monitored the fish’s dispersal at a fine spatial and temporal scale to test two hypotheses: (1) the reintroduced Rhine sculpin individuals will disperse faster upstream than expected from the species’ typical stationary behavior (maximum moving distance of 149 m in 27 days in an established population = 5.51 m/day; Ovidio et al., 2009) because of the high density of individuals at the reintroduction sites reinforcing post-release exploratory behavior; and (2) the reintroduced individuals are not able to cross a potential dispersal barrier and will not be detected upstream of that barrier. To validate our eDNA results with traditional methods, we carried out electrofishing in one instance toward the end of our eDNA sampling period.
Materials and Methods
Field Site
The Borbecker Mühlenbach is a small urban stream categorized as German stream type 6 (fine-grained, carbonic mountain streams; Sommerhäuser and Pottgiesser, 2005; Figure 1A) that has its source in Essen, North Rhine-Westphalia, Germany. Like most streams within the Emscher catchment, the Borbecker Mühlenbach was used as an open sewer system for wastewater disposal from the 19th century on. Recently, the stream was ecologically restored to a large extent (mainly carried out in 2011, finished in 2014) and today consists of both piped underground sections and restored above-ground sections. This study focuses on a 1050 m long above-ground section of the Borbecker Mühlenbach that represents a young freshwater ecosystem due to its recent restoration. A loose stone dam is located within the studied stream section, which was deemed to represent a potential dispersal barrier for the Rhine sculpin as it is approximately 40 cm high and blocks the entire width of the stream (Figure 1B). Further upstream of the stone dam, the stream Kesselbach enters the Borbecker Mühlenbach. After the studied stream section had been deemed suitable for reintroducing the Rhine sculpin post restoration, 118 Rhine sculpin individuals taken from the river Boye (Bottrop, Germany) were released into the stream at sampling sites 3–5 (Figure 1C) on August 23, 2017. To prevent individuals from drifting downstream into underground pipes, a net was installed in front of the pipes.
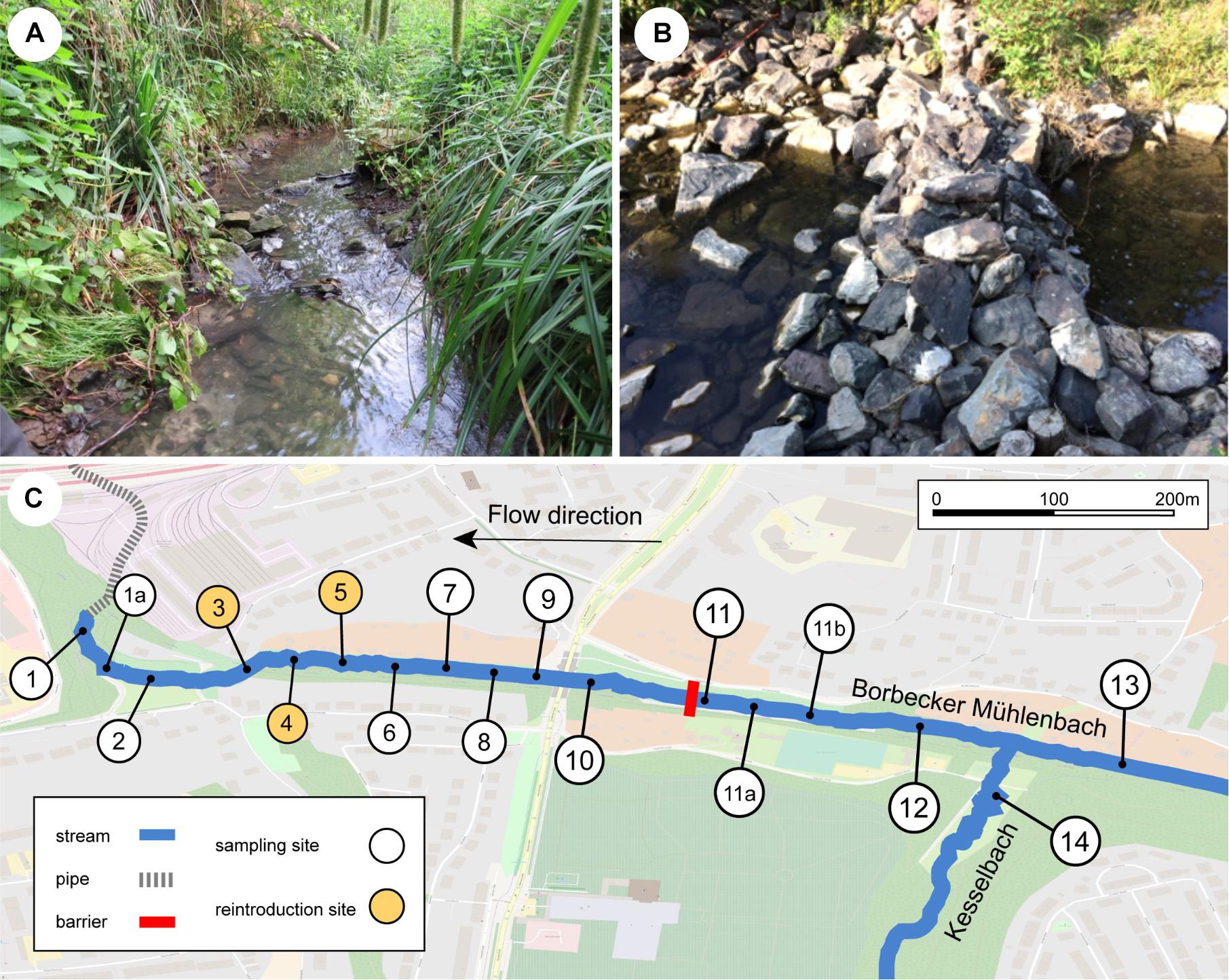
Figure 1. (A) Borbecker Mühlenbach, (B) barrier downstream of site 11, which was deemed to represent a dispersal barrier for the Rhine sculpin, and (C) detailed map of the sampling sites at the stream. For coordinates of and distances between sampling sites, see Supplementary Appendix B.
Sampling
eDNA Sampling
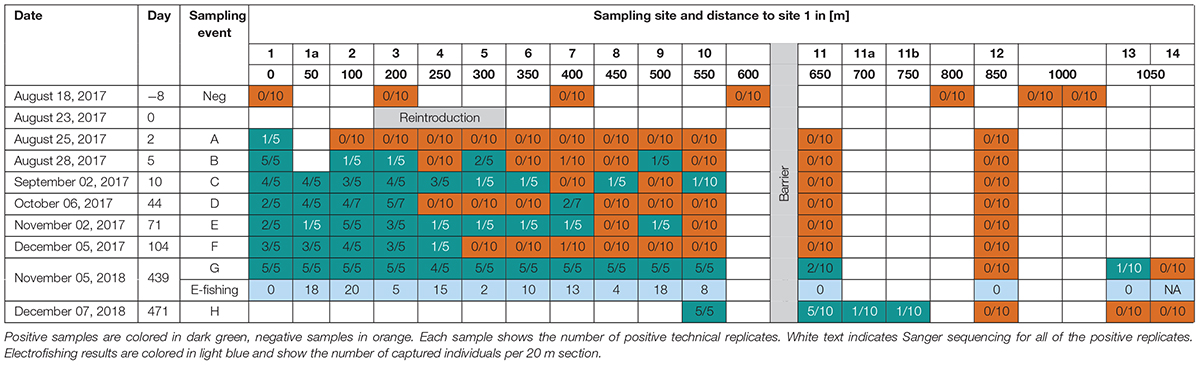
Environmental DNA samples were taken 8 days before the reintroduction of the Rhine sculpin (negative control samples), and 2, 5, 10, 44, 71, 104, 439, and 471 days after the reintroduction. Seven sites were sampled for the negative control samples, 14 sites on the first two sampling days, and 15 sites on the following sampling days (Figure 1C; sampling site 1a was added 50 m upstream of site 1 after two sampling days). The second last sampling day included both eDNA collection and electrofishing. Due to unexpected findings, we took samples at the last sampling day from site 10 on (sampling sites 11a and 11b were added 50 and 100 m upstream of site 11). Sampling dates and distances between sampling sites are shown in Table 1; capital letters A–H indicate the respective sampling days.
For every eDNA sample, 1 L of water was collected in bottles (sterilized in 4% chlorine bleach overnight after each use) and filtered through sterile 0.45 μm cellulose nitrate filters (Thermo Fisher ScientificTM NalgeneTM filtration units, Thermo Fisher Scientific, Waltham, MA, United States) using a vacuum pump. The filtering took place in the field to prevent cross-contamination by other laboratory samples. A separate filter was used for every 500 mL (two filters per sample). Additionally, an extra filter was exposed to air and then included to the other samples at each sampling day. This was done to check for cross-contamination by air on-site (field blank control). The filters were preserved in 96% ethanol and stored at −20°C until extraction.
Electrofishing
On November 5, 2018, electrofishing was carried out after taking water samples for eDNA analyses. One person experienced in electrofishing was using a portable backpack device (EFGI 650, Bretschneider, Chemnitz, Germany) and a round anode net with a diameter of 50 cm and a mesh size of 6 mm. The voltage was set to 115 V, the current to 3 A and the pulse frequency to 60 Hz. Another person was walking behind the first, additionally looking for stunned fish. Every sampling site was electrofished along a stretch of 20 m in the upstream direction from the point of eDNA sampling, for approximately 15 min. Sites 11a and 11b were not electrofished because they were added after the electrofishing survey, and site 14 was not accessible for electrofishing due to the presence of dense vegetation. We are aware that a stretch of 20 m length does not meet requirements of official electrofishing standards (required is a stretch of 40 times the stream width at minimum; Barbour et al., 1999; Dußling, 2014). However, our goal was not to conduct a traditional survey on the entire fish inventory but to verify the presence of the Rhine sculpin at the respective sites. To not overfish the small stream for that purpose, we decided to deviate from the standard approach and to reduce the electrofished stream stretches to 20 m.
Laboratory Work
All lab work was carried out in an eDNA laboratory under sterile PCR hoods, and the whole lab, as well as the individual PCR hoods, were UV-light sterilized for 45 min between separate work cycles. DNA was extracted from filters using a modified salt extraction protocol [original protocol by Sunnucks and Hales (1996); for modified protocol see Weiss and Leese (2016)]. For the purification, either the NucleoSpin Gel and PCR Clean-up kit (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany) or the MinElute Reaction Cleanup Kit (QIAGEN Inc., Germantown, MD, United States) were used, following the manufacturer’s instructions. During each extraction and purification step, it was ensured that only one sample was opened at a time to avoid cross-contamination. The final DNA extracts were eluted into 20 μL of water. The success of all extractions and purifications was verified using agarose gel electrophoresis.
Primers targeting the hypervariable mitochondrial 12S rRNA gene of the Rhine sculpin were designed using a reference database containing 272 sequences of the 12S rRNA gene from 57 fish species (Hänfling et al., 2016). These included all native fish species resident in the Borbecker Mühlenbach and Kesselbach. The complete mitochondrial genome of the Rhine sculpin was downloaded from NCBI GenBank (MF326941/NC_036147, both identical), which was representative for the study population in the 12S rRNA gene sequence, and added to the downloaded 12S rRNA dataset [for final Mafft (Katoh and Standley, 2013) alignment of the 12S rRNA gene sequences, see Supplementary Appendix A]. The newly designed primers C_12S_377F (5′–AGGCCCAAGTTGACAAACAC–3′) and C_12S_731R (5′ – GGCGGGTAAAACAAGGAAGG–3′) amplify a 344 bp long region of the Rhine sculpin’s 12S rRNA, which is located within the 12S rRNA fragment targeted by the universal fish 12S rRNA primers 12S_30F and 12S_1380R (Hänfling et al., 2016). The application of a nested PCR assay design (nPCR) was chosen as this method is known to significantly improve detectability compared to conventional PCR (Davison et al., 2019). The designed primers are universal for 20 out of 24 species of the Cottus complex with available 12S sequences on GenBank including Baikal sculpins, according to PrimerBlast (NCBI)1. No other Cottus species has been reported from the Emscher catchment (Gunnar Jacobs, personal communication), therefore, it is extremely unlikely that other Cottus species rather than the re-stocked C. rhenanus would be the source of captured eDNA.
The presence of Rhine sculpin DNA within the eDNA extracts was tested with end-point nPCRs using the QIAGEN Multiplex PCR Plus Kit (QIAGEN Inc., Hilden, Germany). We included negative PCR controls as well as field blank controls in the PCR runs to check for a clean PCR setup and cross-contamination. During each PCR step, it was ensured that only one 8-sample PCR microtube strip was opened at a time to avoid cross-contamination. For the first PCR step, 2 μL of the extracted eDNA, 0.25 μL of 100 μM universal fish 12S rRNA gene primers 12S_30F and 12S_1380R (Hänfling et al., 2016), 25 μL of the Multiplex MasterMix, and 22.5 μL water were used per reaction. PCR conditions consisted of an initial incubation at 95°C for 15 min followed by 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 50 s, and final elongation at 72°C for 10 min. For the second PCR step, 1 μL template of each PCR product from step 1, 0.25 μL of 100 μM newly designed primers C_12S_377F and C_12S_731R, 12.5 μL of the Multiplex MasterMix, and 11.25 μL water were used per reaction. PCR conditions consisted of an initial incubation at 95°C for 15 min followed by 30 cycles of 95°C for 20 s, 63°C for 30 s, and 72°C for 1 min, and final incubation at 72°C for 5 min.
Half of the extracted DNA of every sample (10 μL) was used as a template for the first PCR step to minimize stochasticity effects, leading to five PCR replicates per sample containing 2 μL of the extracted DNA each. The presence of the Rhine sculpin was confirmed if a band was present at the expected amplicon length using agarose gel electrophoresis (with 1% agarose gels run for 15 min at 80 V). In case the Rhine sculpin was not detected in the first half of the eluate, the remaining eluate was used as an input for further PCRs. If this approach also did not show any signal, then the Rhine sculpin was considered to be absent at the sampling site.
Samples from each sampling day were tested up to site 12. If the species was detected behind the barrier (sites 11 and 12), then sites 13 and 14 were also tested.
To validate that the newly designed primer pair amplified our target species C. rhenanus in field samples, PCR amplicons of 22 positive replicates were Sanger sequenced (Eurofins Genomics, Ebersberg, Germany). These included replicates from all positive samples upstream of the barrier, as well as all from samples that resulted in just one positive replicate (excluding sample 9 B, for which Sanger sequencing failed). All Sanger sequenced replicates confirmed the detection of the Rhine sculpin.
Results
No DNA was found in the field blank controls, excluding cross-contamination during filtering on-site and confirming clean DNA extraction and purification. Moreover, no DNA was found in any negative PCR control of the end-point nPCRs, confirming a clean PCR setup.
The Rhine sculpin was not detected in any negative control sample, confirming its absence in the stream section prior to the reintroduction (Table 1). Within the first 3 months, the Rhine sculpin was detected at least once at every sampling site downstream of the barrier (site 1–10) and not detected upstream of the barrier (site 11 and 12). Samples 9 B and 10 C were tested positive, confirming the fish’s dispersal of 200 m upstream within the first 5 days and 250 m within the first 10 days, respectively. However, the species was not detected in several samples despite the presence of the species further upstream on the same day. In general, positive detections strongly varied among technical replicates, from five detections in five replicates to one detection in 10 replicates. Positive detections in all technical replicates of one sample were rare within the first 3 months, indicating low DNA template concentration in the stream.
After 1 year, the species was detected at every sampling site downstream of the barrier, with positive detections in all technical replicates of each sample (apart from sample 4 G, of which only four of five technical replicates were positive). In addition, the Rhine sculpin was detected at sites 11 G and 13 G upstream of the barrier.
In contrast to that, we detected Rhine sculpin individuals at every site downstream of the barrier (apart from site 1) by conducting electrofishing on the same day, but no single individual upstream of the barrier. We detected a total number of 113 individuals, including juveniles, with 2–20 individuals found per site (Table 1).
After one more month, positive eDNA detections of the species again confirmed the species’ presence at sites 11 H, 11a H, and 11b H (Table 1).
Discussion
Dispersal of the Rhine Sculpin
We assumed the detection of Rhine sculpin eDNA at a given site if at least one replicate of our end-point nPCR approach confirmed amplification. We did not have the option for more sensitive qPCR or ddPCR, yet our applied end-point nPCR approach is known to lead to reliable detections (Nathan et al., 2014; Davison et al., 2019). Furthermore, end-point PCRs can have low detection limits (e.g., Qu and Stewart, 2017), which is why we considered that our results were more impacted by low detection rates than cross-contamination.
Based on end-point nPCR eDNA detections, we showed that individuals dispersed at least 200 m within the first 5 days (positive sample 9 B), on average at least 40 m/day, which greatly exceeds daily moving distances of the Rhine sculpin in an established population observed by Ovidio et al. (2009; maximum of 5.51 m/day). This verifies our first hypothesis that the reintroduced individuals disperse faster from their reintroduction sites than expected from their known stationary behavior. An explanation for this pattern is that the initially high density of individuals at each reintroduction site led to high intraspecific competition for habitats. A density-dependent, exploratory behavioral mechanism to compensate for this high density is the dispersal of smaller individuals to sub-optimal habitats (Davey et al., 2005). Note that maximum moving distances of up to 395 m for the Rhine sculpin during 1 year and over 250 m for the closely related European bullhead during several months have been observed (Knaepkens et al., 2005, 2006; Ovidio et al., 2009). Furthermore, seasonal migration has been shown for the European bullhead (Crisp et al., 1984). However, none of these studies investigated the fish’s movement at a temporal scale comparable to the present study.
The Rhine sculpin was not detected in eDNA samples upstream of site 10 within the first 3 months and furthermore not detected with electrofishing after 1 year at these sites, indicating a dispersal limitation by the barrier before site 11. However, eDNA detections of the species upstream of the barrier after 1 year (detections at sites 11 G and 13 G) and moreover after one additional month (detections at sites 11 H, 11a H, and 11b H) indicate that at minimum one Rhine sculpin individual was present upstream of the barrier. This result was unexpected (see the second hypothesis) and not supported by our electrofishing survey. We based our second hypothesis on the finding that the European bullhead (which used to be a cryptic species including the Rhine sculpin) is unable to cross solid stream barriers of 18–20 cm height (Utzinger et al., 1998). However, the barrier in the present study consists of loose stones and is occasionally flooded during heavy rainfall events. It is possible that juveniles were able to swim through gaps in the loose stone dam or that individuals were able to cross the barrier after heavy rainfalls. Another explanation for positive eDNA detection upstream of the barrier is that predators (for example, gray heron) that preyed upon Rhine sculpin individuals defecated fish DNA into the upstream section. Human-induced translocation of individuals across the barrier (for example, by hand-netting) can also not be excluded, as the stream is urban and frequently visited by pedestrians. Although we consider predator- or human-induced translocation of Rhine sculpin individuals or DNA as unlikely, we cannot exclude it. Our results indicate a higher dispersal potential of the species than previously assumed because the loose stone dam was deemed to represent a substantial dispersal barrier for small, benthic fish, but this indication cannot truly be evaluated, due to the impossibility to rule out other dispersal mechanisms within the scope of this study.
The inconsistency between eDNA-based and traditional monitoring results for sites upstream of the barrier was unexpected. An explanation for the failure in detecting the species using electrofishing is that individuals were located upstream of the electrofishing sites and hence could not be detected via electrofishing. In that case, they were still releasing eDNA that was transported downstream and was collected, making the detection of the individuals possible. However, a more comprehensive electrofishing survey would be needed to confirm this explanation. In contrast to that, the positive detection of Rhine sculpin DNA could also be predator-induced, as earlier discussed. In that case, no actual individuals of the species would have been present upstream of the barrier, which would explain the failed electrofishing detection.
After 1 year, the Rhine sculpin was detected at every sampling site downstream of the barrier using eDNA and at every sampling site downstream of the barrier except for site 1 using electrofishing. This verified previous expectations and motivations for the reintroduction of the Rhine sculpin into the stream section, justified by the presence of suitable habitats for the species. Additional sampling following the Rhine sculpins’ next reproduction period is needed to investigate the fish’s further dispersal and mid-term reintroduction success.
The finding of juveniles during the electrofishing survey confirmed the reproduction of the species. The confirmed reproduction, together with the fact that we captured 113 individuals with electrofishing in a fraction of the stream habitat while 118 individuals were released in total, implies a substantial increase in population size. This increase is a likely source for the highly increased number of positive eDNA-based detections after 1 year, although the adsorption-release dynamics of eDNA might also have facilitated the detection, i.e., more eDNA being released in contrast to the initial phase (Spear et al., 2015; Shogren et al., 2017).
Our results confirm successful reintroduction, dispersal, and establishment of the species within the stream Borbecker Mühlenbach. Additionally, the species’ establishment indicates good water quality, habitat structure and passability of the stream and consequently indicates a successful stream restoration.
Applicability of eDNA for Biomonitoring of Fish
Our electrofishing observations suggest that the Rhine sculpin was not able to cross a minor barrier in the stream, which is congruent with previous observations for the closely related European bullhead (Utzinger et al., 1998). However, the results of our eDNA analysis indicate that the species was present upstream of the barrier, which would mean that the mobility of the Rhine sculpin and its capacity to cross instream obstacles is better than previously assumed. Greater electrofishing efforts, which are typically performed in standardized surveys, might have led to certainty about the presence or absence of the Rhine sculpin upstream of the barrier. Nevertheless, our observed trend is consistent with previous studies showing that overlooking individuals is the rule rather than the exception with electrofishing (Jerde et al., 2011; Pont et al., 2018). Thus, although analyzing eDNA cannot be used for estimating individual numbers or age structure, its application in biomonitoring presents a promising, non-invasive, and sensitive tool for species detection.
Nevertheless, the applicability of eDNA analysis as a monitoring tool is still limited, as many characteristics of eDNA are poorly understood yet. It is known that eDNA release can be affected by various conditions of an individual, for example, size, health, sex, fecundity, or diet (Goldberg et al., 2015; Klymus et al., 2015); the extent and variation between species and even individuals remain, however, unclear. Moreover, eDNA durability in aquatic environments is strongly influenced by degradation through temperature, pH value, conductivity, UV light, and microbes (Barnes et al., 2014), reducing eDNA concentration and hence causing additional bias. Furthermore, eDNA movement processes such as downstream transport, soil retention, and soil resuspension constantly affect eDNA concentration (Shogren et al., 2017). Apart from these ecological issues, methodological difficulties can lead to failure in detecting eDNA, caused by stochastic effects and PCR inhibition (Taberlet et al., 1996; Thomsen and Willerslev, 2015). Together, these limitations may lead to inexplicable patterns of eDNA detection in some studies [observed by Jane et al. (2015) and Laramie et al. (2015)].
eDNA can be transported downstream over long distances in flowing waters (Deiner and Altermatt, 2014; Jane et al., 2015; Deutschmann et al., 2019), which implies that Rhine sculpin eDNA was transported downstream from actual residence sites in our study. This effect was observed for sample 1 G, where the species was detected with eDNA even though electrofishing failed to detect individuals up to 20 m upstream of the site. Consequently, eDNA detections of the Rhine sculpin downstream of the uppermost detection site do not equal true local detection. Visual observation of individuals by traditional surveys is still necessary for the validation of local presence at sites downstream of actual residence sites. However, when validating the general presence of a fish species in flowing waters, this effect might be beneficial, as there is no need to sample the entire stream or river.
In this study, the Rhine sculpin was not detected at several sites despite the presence of the species further upstream on the same day. These occurrences represent false negatives, whose presence is consistent with comparable studies (Foote et al., 2012; Jane et al., 2015; Laramie et al., 2015) and can be explained by dilution of DNA by high flows, increasing distance from the DNA source, or PCR inhibition (Jane et al., 2015); eDNA dynamics in the system (e.g., retention to stream bottom; Shogren et al., 2017); and the less sensitive end-point nPCR approach we used (Turner et al., 2014). Inhibition is especially problematic in eDNA studies, as it can mask even high eDNA copy numbers (Jane et al., 2015) and hence can lead to false negatives although the target organism is present (Goldberg et al., 2015; Thomsen and Willerslev, 2015).
Our results show that false negatives can be reduced by both high numbers of technical replicates and periodical sampling. An increase from 5 to 10 technical replicates led to positive detection of the Rhine sculpin in several cases (Table 1, samples 7 B, 10 C, 7 F, 13 G, 11a H, 11b H), and periodical sampling enabled the detection of the species at sites where detection failed at previous sampling events (Table 1, samples 4 C, 6 C, 8 C, 4 E, 5 E, 6 E). However, addressing the previously mentioned challenges in eDNA studies, especially PCR inhibition, will further decrease the number of false negatives in future eDNA studies for target species detection.
Conclusion
Our study shows that eDNA surveys even via conventional end-point nested PCR can provide detailed insights into reintroduction success and fish dispersal. eDNA analysis allowed for monitoring the dispersal of the Rhine sculpin in a small German stream at a fine temporal and spatial scale. The method revealed a substantially higher realized dispersal potential than previously assumed and verified a successful stream restoration. We conclude that eDNA analysis is thus applicable to investigate the ecological status and fragmentation of streams by proving the presence of an indicator species, making it a useful tool for biomonitoring. Consequently, eDNA analysis holds the potential for freshwater assessments and is an effective, non-invasive approach that can be used to augment traditional methods, although several aspects need to be further understood to correctly interpret false negative results.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The animal study was reviewed and approved by the Fisheries Institution Untere Fischereibehörde of the city Essen.
Author Contributions
CH, AB, BP, FL, GJ, J-NM, and VE designed the study. CH, BP, GJ, J-NM, T-HM, and VE acquired the data. CH wrote the manuscript with the support of AB and FL. All authors analyzed and interpreted the data, and commented on and approved the final manuscript.
Funding
This project was in part supported by an award of the Ruediger Kurt Bode Foundation (Deutscher Stifterverband).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The electrofishing survey in the Borbecker Mühlenbach was approved by the Fisheries Institution Untere Fischereibehörde of the city of Essen. We thank Lars Hasselsweiler and Saskia Rohrer for their support in the electrofishing survey and Alexander M. Weigand for helpful discussions. FL, VE, J-NM, and T-HM are members of and supported by EU COST Action DNAqua-Net CA15219. This manuscript has been released as a Pre-Print at PeerJ Preprints.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00081/full#supplementary-material
Footnotes
References
Adrian-Kalchhauser, I., and Burkhardt-Holm, P. (2016). An eDNA assay to monitor a globally invasive fish species from flowing freshwater. PLoS One 11:e0147558. doi: 10.1371/journal.pone.0147558
Barbour, M. T., Gerritsen, J., Snyder, B. D., and Stribling, J. B. (1999). Rapid Bioassessment Protocols For Use in Streams and Wadeable Rivers: Periphyton, Benthic Macroinvertebrates, and Fish, 2ND Edn. Washington, DC: EPA. doi: 10.1093/gerona/60.10.1351
Barnes, M. A., Turner, C. R., Jerde, C. L., Renshaw, M. A., Chadderton, W. L., and Lodge, D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48, 1819–1827. doi: 10.1021/es404734p
Bellinger, E. G., and Sigee, D. C. (2015). Freshwater Algae - Identification, Enumeration and Use as Bioindicators, 2nd Edn. Chichester: John Wiley & Sons Ltd.
Bohlin, T., Hamrin, S., Heggberget, T. G., Rasmussen, G., Saltveit, S. J., and Nielsen, J. (1989). Electrofishing - Theory and practice with special emphasis on salmonids. Hydrobiologia 173, 9–43. doi: 10.1007/bf00008596
Brink-Kloke, H., Rückheim, S., Derks, P., Molkenthin, R., Winter, K., von Stillfried, I., et al. (2006). Alles fließt – das Wasser der Emscher. Heim. Dortmund 2, 1–46.
Colleye, O., Ovidio, M., Salmon, A., and Parmentier, E. (2013). Contribution to the study of acoustic communication in two Belgian river bullheads (Cottus rhenanus and C. perifretum) with further insight into the sound-producing mechanism. Front. Zool. 10:71. doi: 10.1186/1742-9994-10-71
Crisp, D. T., Mann, R. H. K., and Cubby, P. R. (1984). Effects of impoundment upon fish populations in afferent streams at cow green reservoir. J. Appl. Ecol. 21, 739–756.
Davey, A. J. H., Hawkins, S. J., Turner, G. F., and Doncaster, C. P. (2005). Size-dependent microhabitat use and intraspecific competition in Cottus gobio. J. Fish Biol. 67, 428–443. doi: 10.1111/j.0022-1112.2005.00736.x
Davison, P. I., Falcou-Préfol, M., Copp, G. H., Davies, G. D., Vilizzi, L., and Créach, V. (2019). Is it absent or is it present? Detection of a non-native fish to inform management decisions using a new highly-sensitive eDNA protocol. Biol. Invasions 21, 2549–2560. doi: 10.1007/s10530-019-01993-z
Deiner, K., and Altermatt, F. (2014). Transport distance of invertebrate environmental DNA in a natural river. PLoS One 9:e0088786. doi: 10.1371/journal.pone.0088786
Dejean, T., Valentini, A., Miquel, C., Taberlet, P., Bellemain, E., and Miaud, C. (2012). Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. J. Appl. Ecol. 49, 953–959. doi: 10.1111/j.1365-2664.2012.02171.x
Deutschmann, B., Müller, A. K., Hollert, H., and Brinkmann, M. (2019). Assessing the fate of brown trout (Salmo trutta) environmental DNA in a natural stream using a sensitive and specific dual-labelled probe. Sci. Total Environ. 655, 321–327. doi: 10.1016/j.scitotenv.2018.11.247
Donoso-Büchner, R. (2009). Zur Bestandssituation der Emschergroppe, Cottus cf. rhenanus, aus dem Einzugsbereich der Boye im Emschersystem. Solingen: Verlag Natur & Wissenschaft.
Dußling, U. (2014). Kurzbeschreibung des fischbasierten Bewertungssystems für Fließgewässer – fiBS, Version 8.1.1. Langenargen: Fischereiforschungsstelle Baden-Württemberg
Foote, A. D., Thomsen, P. F., Sveegaard, S., Wahlberg, M., Kielgast, J., Kyhn, L. A., et al. (2012). Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS One 7:e0041781. doi: 10.1371/journal.pone.0041781
Freyhof, J., Kottelat, M., and Nolte, A. (2005). Taxonomic diversity of European Cottus with descritpion of eight new species. Ichthyol. Explor. Freshwaters 16, 107–172.
Goldberg, C. S., Strickler, K. M., and Pilliod, D. S. (2015). Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms. Biol. Conserv. 183, 1–3. doi: 10.1016/j.biocon.2014.11.040
Hänfling, B., Lawson Handley, L., Read, D. S., Hahn, C., Li, J., Nichols, P., et al. (2016). Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 25, 3101–3119. doi: 10.1111/mec.13660
Jane, S. F., Wilcox, T. M., Mckelvey, K. S., Young, M. K., Schwartz, M. K., Lowe, W. H., et al. (2015). Distance, flow and PCR inhibition: EDNA dynamics in two headwater streams. Mol. Ecol. Resour. 15, 216–227. doi: 10.1111/1755-0998.12285
Jensen, K., Trepel, M., Merritt, D., and Rosenthal, G. (2006). Restoration ecology of river valleys. Basic Appl. Ecol. 7, 383–387. doi: 10.1016/j.baae.2006.05.008
Jerde, C. L., Mahon, A. R., Chadderton, W. L., and Lodge, D. M. (2011). “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 4, 150–157. doi: 10.1111/j.1755-263X.2010.00158.x
Karr, J. R. (1981). Assessment of biotic integrity using fish communities. Fisheries 6, 21–27. doi: 10.1577/1548-84461981006
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Klymus, K. E., Richter, C. A., Chapman, D. C., and Paukert, C. (2015). Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 183, 77–84. doi: 10.1016/j.biocon.2014.11.020
Knaepkens, G., Baekelandt, K., and Eens, M. (2005). Assessment of the movement behaviour of the bullhead (Cottus gobio), an endangered European freshwater fish. Anim. Biol. 55, 219–226. doi: 10.1163/1570756054472845
Knaepkens, G., Baekelandt, K., and Eens, M. (2006). Fish pass effectiveness for bullhead (Cottus gobio), perch (Perca fluviatilis) and roach (Rutilus rutilus) in a regulated lowland river. Ecol. Freshw. Fish 15, 20–29. doi: 10.1111/j.1600-0633.2005.00117.x
Laramie, M. B., Pilliod, D. S., and Goldberg, C. S. (2015). Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biol. Conserv. 183, 29–37. doi: 10.1016/j.biocon.2014.11.025
Miya, M., Sato, Y., Fukunaga, T., Sado, T., Poulsen, J. Y., Sato, K., et al. (2015). MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2:150088. doi: 10.1098/rsos.150088
Nathan, L. M., Simmons, M., Wegleitner, B. J., Jerde, C. L., and Mahon, A. R. (2014). Quantifying environmental DNA signals for aquatic invasive species across multiple detection platforms. Environ. Sci. Technol. 48, 12800–12806. doi: 10.1021/es5034052
Ovidio, M., Detaille, A., Bontinck, C., and Philippart, J. C. (2009). Movement behaviour of the small benthic Rhine sculpin Cottus rhenanus (Freyhof, Kottelat & Nolte, 2005) as revealed by radio-telemetry and pit-tagging. Hydrobiologia 636, 119–128. doi: 10.1007/s10750-009-9941-9943
Platts, W. S., Megahan, W. F., and Minshall, G. W. (1983). Methods for Evaluating Stream, Riparian, and Biotic Conditions. Odgen: Intermountain Forest Experiment Station. doi: 10.1007/978-3-662-02568-0_33
Pont, D., Rocle, M., Valentini, A., Civade, R., Jean, P., Maire, A., et al. (2018). Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci. Rep. 8, 1–13. doi: 10.1038/s41598-018-28424-28428
Qu, C., and Stewart, K. A. (2017). Evaluating monitoring options for conservation: comparing traditional and environmental DNA tools for a critically endangered mammal. PeerJ Prepr. 106:9. doi: 10.1007/s00114-019-1605-1601
Resh, V. H., and Unzicker, J. D. (1975). Water quality monitoring and aquatic organisms?: the importance of species identification. Water Pollut. Control Fed. 47, 9–19.
Riaz, M., Kuemmerlen, M., Wittwer, C., Cocchiararo, B., Khaliq, I., Pfenninger, M., et al. (2019). Combining environmental DNA and species distribution modeling to evaluate reintroduction success of a freshwater fish. Ecol. Appl. 0, ea2034. doi: 10.1002/eap.2034
Rojahn, J., Gleeson, D., and Furlan, E. M. (2018). Monitoring post-release survival of the northern corroboree frog, Pseudophryne pengilleyi, using environmental DNA. Wildl. Res. 45, 620–626. doi: 10.1071/WR17179
Schnelle, K., and Wilts, S. (2016). Kläranlagen - So wird Abwasser wieder flusstauglich. Essen: EGLV.
Shogren, A. J., Tank, J. L., Andruszkiewicz, E., Olds, B., Andrew, R., Jerde, C. L., et al. (2017). Controls on eDNA movement in streams: transport. retention, and resuspension. Sci. Rep. 7:5065. doi: 10.1038/s41598-017-05223-5221
Sommerhäuser, M., and Pottgiesser, T. (2005). Die Fließgewässertypen Deutschlands als Beitrag zur Umsetzung der EG-Wasserrahmenrichtlinie. Limnol. Aktuell 11, 13–27.
Spear, S. F., Groves, J. D., Williams, L. A., and Waits, L. P. (2015). Using environmental DNA methods to improve detectability in a hellbender (Cryptobranchus alleganiensis) monitoring program. Biol. Conserv. 183, 38–45. doi: 10.1016/j.biocon.2014.11.016
Sunnucks, P., and Hales, D. F. (1996). Numerous transposed sequences mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobian (Hemiptera: Aphididae). Mol. Biol. Evol. 13, 510–524. doi: 10.1093/oxfordjournals.molbev.a025612
Taberlet, P., Griffin, S., Goossens, B., Questiau, S., Manceau, V., Escaravage, N., et al. (1996). Reliable genotyping of samll samples with very low DNA quantities using PCR. Nucleic Acid Res. 24, 3189–3194. doi: 10.1093/nar/24.16.3189
Takahara, T., Minamoto, T., and Doi, H. (2013). Using environmental DNA to estimate the distribution of an invasive fish species in ponds. PLoS One 8:e0056584. doi: 10.1371/journal.pone.0056584
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA - An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Turner, C. R., Barnes, M. A., Xu, C. C. Y., Jones, S. E., Jerde, C. L., and Lodge, D. M. (2014). Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol. Evol. 5, 676–684. doi: 10.1111/2041-210X.12206
Utzinger, J., Roth, C., and Peter, A. (1998). Effects of environmental parameters on the distribution of bullhead Cottus gobio with particular consideration of the effects of obstructions. J. Appl. Ecol. 35, 882–892. doi: 10.1111/j.1365-2664.1998.tb00006.x
Weiss, M., and Leese, F. (2016). Widely distributed and regionally isolated! Drivers of genetic structure in Gammarus fossarum in a human-impacted landscape. BMC Evol. Biol. 16:723. doi: 10.1186/s12862-016-0723-z
Wittkugel, V. C. (2005). Untersuchungen zum Vorkommen der Groppe (Cottus gobio) im Bereich der Rheinvorstreckung am Bodensee. Vorarlberg: Zobodat, 93–104.
Keywords: biomonitoring, ecological restoration, eDNA, reintroduction, fish, freshwaters
Citation: Hempel CA, Peinert B, Beermann AJ, Elbrecht V, Macher J-N, Macher T-H, Jacobs G and Leese F (2020) Using Environmental DNA to Monitor the Reintroduction Success of the Rhine Sculpin (Cottus rhenanus) in a Restored Stream. Front. Ecol. Evol. 8:81. doi: 10.3389/fevo.2020.00081
Received: 10 October 2019; Accepted: 12 March 2020;
Published: 21 April 2020.
Edited by:
Frederic J. J. Chain, University of Massachusetts Lowell, United StatesReviewed by:
Chris Wilson, Ontario Ministry of Natural Resources, CanadaAndres Aguilar, California State University, Los Angeles, United States
Copyright © 2020 Hempel, Peinert, Beermann, Elbrecht, Macher, Macher, Jacobs and Leese. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Florian Leese, Zmxvcmlhbi5sZWVzZUB1bmktZHVlLmRl
†Present address: Bianca Peinert, Ennepe-Ruhr-Kreis, Wasserwirtschaft, Schwelm, Germany; Vasco Elbrecht, Centre for Molecular Biodiversity Research, Zoological Research Museum Alexander Koenig, Bonn, Germany; Jan-Niklas Macher, Marine Biodiversity Group, Naturalis Biodiversity Center, Leiden, Netherlands
 Christopher A. Hempel
Christopher A. Hempel Bianca Peinert1†
Bianca Peinert1† Jan-Niklas Macher
Jan-Niklas Macher Florian Leese
Florian Leese