- 1Division of Clinical Immunology and Allergy, Children's Hospital Los Angeles, Los Angeles, CA, United States
- 2Department of Pediatrics, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States
Summary
The current paper suggests that egg allergy may arise due to microbial proteins that are homologous to egg allergens. These microbial proteins elicit an allergic response and lead to the development of specific microbial IgE molecules. These molecules cross-react with egg allergens and result in egg allergy. Some examples of microbial proteins that share similar sequences as egg allergens are presented in this paper.
Food allergy has a negative impact on the quality of life and nutrition. In addition, it can lead to life-threatening reactions. The pathogenesis of food allergy is still not fully understood. Many young children develop food allergy with no known prior ingestion of the food allergen in question. The strong connection between food allergy and atopic dermatitis has been well-documented. While the prevalence of food allergy in the general pediatric population is 4–5%, the prevalence of food allergy in atopic dermatitis is at least 20% (1). This connection between food allergy and atopic dermatitis has led to the suggestion that the skin may be the site of food IgE sensitization, leading to food allergy. The dual-allergen-exposure hypothesis suggests that food allergens are sensitized via eczema, whereas early gastro-intestinal exposure leads to tolerance (2). This hypothesis is supported by multiple basic studies that provide evidence for IgE sensitization via the skin [reviewed in (3)]. In addition, it has been shown that environmental level of peanut allergens is increased in children who developed peanut allergy (4). The early introduction of peanut has led to the prevention of peanut allergy (5). Likewise, early introduction of egg has also met with some success, although the results were not as consistent (3). It has been suggested that improvement in eczema, in addition to early introduction of egg, is needed for successful prevention of egg allergy (6). This suggestion further highlights the importance of skin in the pathogenesis of food allergy. A hypothesis is proposed here that the interaction between the neonatal skin and microbial proteins is important for the development of IgE sensitization and egg allergy.
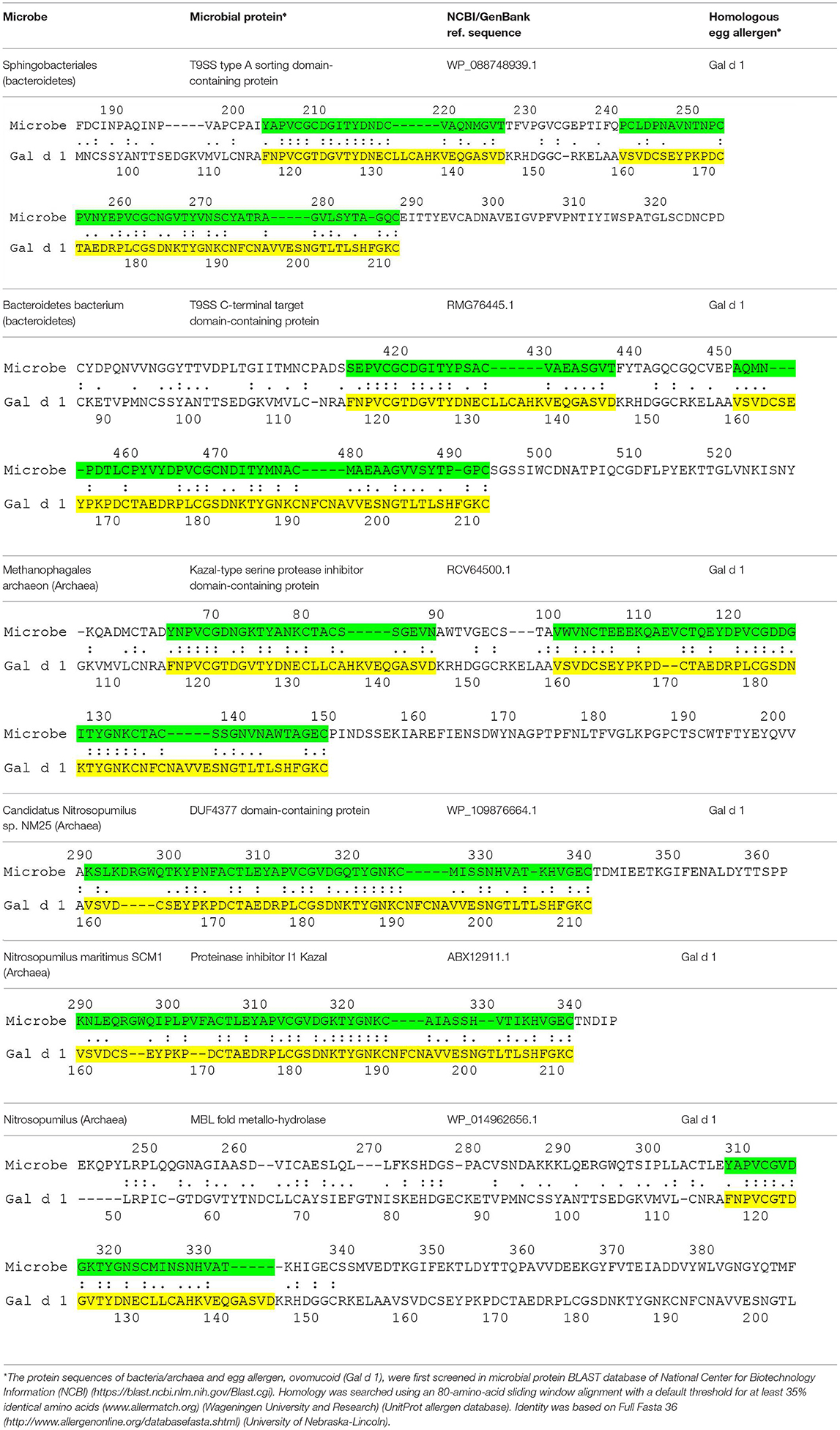
The current hypothesis predicts the presence of microbial proteins that are homologous to egg allergens. Table 1 shows the microbial proteins that share homology with the IgE-binding domains of Gal d 1 (ovomucoid) (7). Four out of the 6 microbial proteins share > 60 % identity with a clinically-relevant IgE-binding region of Gal d 1 (FNPVCGTDGVTYDN) (8). Significant homology was also found between microbial proteins and Gal d 2 (ovalbumin), but no homology was found in the IgE-binding domains of Gal d 2 (9) (data not shown). To further confirm the correlation between microbial proteins and Gal d 1, prospective studies may be carried out to look for these microbial pathogens in atopy-prone neonates and correlate with neonates who eventually develop allergy to Gal d 1. These microbial pathogens can also be inoculated in animal models to show the development of specific IgE that cross-react with Gal d 1. Atopy-prone neonates who are born to parent with atopic dermatitis, asthma or allergic rhinitis have inherent skin barrier defects that predispose them to develop atopic dermatitis. Microbial pathogens are capable of evading these barrier defects to interact with the cutaneous immune system in these children. The processing of microbial proteins by antigen-presenting cells and subsequent presentation of antigenic peptides to T helper type 2 cells leads to the production of IL-4 and IL-13, which induce B cells to express specific IgE molecules. Bacterial allergy has been described more than half a century ago (10). It is also known that specific IgE to staphylococcal toxins can develop in young children with atopic dermatitis (11). Bacteria such as bacteroides can be acquired during birth or they can be part of neonates' microbiome (12). The skin also contains a wide array of microbial pathogens that can participate in the development of allergy (13). More recent data suggests that many microbial organisms (e.g., proteobacteria and archaea) previously thought to exist only in the environment such as soil, fresh or marine waters are now found to be part of the human skin microbiome (14, 15). Proteobacteria and bacteroides are also known to be present in atopic dermatitis lesions (16). The current proposal is conceptual that microbial proteins can be a sensitizing source in the development of food allergy in predisposed children. Preliminary data also suggests the presence of homologous proteins between microbes and other food allergens including peanut and cow's milk (unpublished data). Whether these microbial pathogens have a direct interaction with neonates, leading to the development of specific IgE, remains to be proven. It is possible that the prevention of egg allergy requires a different approach by targeting microbial pathogens.
Author Contributions
PO conceived the idea, generated the data, and wrote the paper.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The current hypothesis is developed from the project Prediction of eczema, infections and allergy sponsored by The Albert and Bettie Sacchi Foundation.
References
1. Martin PE, Eckert JK, Koplin JJ, Lowe AJ, Gurrin LC, Dharmage SC, et al. Which infants with eczema are at risk of food allergy? Results from a population-based cohort? Clin Exp Allergy. (2015) 45:255–64. doi: 10.1111/cea.12406
2. Lack G. Epidemiologic risks for food allergy. J Allergy Clin Immunol. (2008) 121:1331–6. doi: 10.1016/j.jaci.2008.04.032
3. Tordesillas L, Berin MC, Sampson HA. Immunology of food allergy. Immunity. (2017) 121:1331–6. doi: 10.1016/j.immuni.2017.07.004
4. Bough HA, Liu AH, Sicherer S, Makinson K, Douiri A, Brown SJ, et al. Atopic dermatitis increases the effect of exposure to peanut protein antigen in dust on peanut sensitization and likely peanut allergy. J Allergy Clin Immunol. (2015) 135:164–70. doi: 10.1016/j.jaci.2014.10.007
5. Du Toit G, Roberts G, Sayre PH, Bahson HT, Santos AF, Brough HA, et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. (2015) 372:803–13. doi: 10.1056/NEJMoa1414850
6. Matsumoto K, Mori R, Miyazaki C, Ohya Y, Saito H. Are both early egg introduction and eczema treatment necessary for primary prevention of egg allergy? J Allergy Clin Immunol. (2018) 141:1997–2001. doi: 10.1016/j.jaci.2018.02.033
7. Mine Y, Bresler M. The major allergen from hen's egg white: ovomucoid (Gal d 1). Internet Sympos Food Allergy. (1999) 1:137–46.
8. Martinez-Botas J, Cerecedo I, Zamora J, Vlaicu C, Dieguez MC, Gomez-Coronado D, et al. Mapping of the IgE and IgG4 sequential epitopes of ovomucoid with a peptide microarray immunoassay. Int Arch Allergy Immunol. (2013) 161:11–20. doi: 10.1159/000343040
9. Mine Y, Rupa P. Fine Mapping and structural analysis of immunodominant IgE allergenic epitopes in chicken egg ovalbumin. Prot Eng. (2003) 16:747–52. doi: 10.1093/protein/gzg095
11. Ong PY, Patel M, Ferdman RM, Dunaway T, Church JA. Association of staphylococcal superantigen-specific immunoglobulin E with mild and moderate atopic dermatitis. J Pediatr. (2008) 153:803–6. doi: 10.1016/j.jpeds.2008.05.047
12. Wexler HM. Bacteroides: the good, the bad and the nitty-gritty. Clin Microbiol Rev. (2007) 20:593–621. doi: 10.1128/CMR.00008-07
13. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. (2011) 9:244–53. doi: 10.1038/nrmicro2537
14. Cosseau C, Romano Bertrand S, Duplan H, Lucas O, Ingrassia I, Pigasse C, et al. Proteobacteria from the human skinmicrobiota: Species level diversity and hypotheses. One Health. (2016) 2:33–41. doi: 10.1016/j.onehlt.2016.02.002
15. Probst AJ, Auerbach AK, Moissl-Eichinger C. Archaea on human skin. PLoS ONE. (2013) 8:e65388. doi: 10.1371/journal.pone.0065388
Keywords: archaea, bacteria, skin, atopic dermatitis, cross-reactivity, egg allergy, IgE sensitization
Citation: Ong PY (2019) The PATHOGENesis of Food Allergy. Front. Pediatr. 7:484. doi: 10.3389/fped.2019.00484
Received: 24 July 2019; Accepted: 04 November 2019;
Published: 21 November 2019.
Edited by:
George N. Konstantinou, 424 General Military Hospital, GreeceReviewed by:
Anastasios Panagiotis Konstantinopoulos, Independent Researcher, Athens, GreeceCopyright © 2019 Ong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peck Y. Ong, cHlvbmdAY2hsYS51c2MuZWR1
 Peck Y. Ong
Peck Y. Ong