- 1Department of Mathematics, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia
- 2Department of General Studies, Technical College, Technical and Vocational Training Corporation, Jeddah, Saudi Arabia
Human immunodeficiency virus (HIV) and hepatitis B virus (HBV) co-infection is common due to their shared transmission routes. Understanding their interaction within host cells is key to improving treatment strategies. Mathematical models are crucial tools for analyzing within-host viral dynamics and informing therapeutic interventions. This study presents a mathematical framework designed to investigate the interactions and progression of HIV-HBV co-infection within a host. The model captures the distinct biological characteristics of the two viruses: HBV primarily infects liver cells (hepatocytes), while HIV targets CD4+ T cells and can also infect hepatocytes. A system of seven non-linear delay differential equations (DDEs) is formulated to represent the dynamic interactions among uninfected and virus-infected hepatocytes, uninfected and HIV-infected CD4+T cells, as well as circulating HIV and HBV particles. The model incorporates two biologically significant time delays: the first represents the latency between the initial infection and the onset of productive infection in host cells, while the second accounts for the maturation duration of newly produced virions before they become infectious. The model's mathematical consistency is verified by showing that its solutions remain bounded and non-negative throughout the system's dynamics. Equilibrium points and their associated threshold parameters are identified, with conditions for existence and stability rigorously derived. Global stability of the equilibria is established through the application of carefully designed Lyapunov functionals in conjunction with Lyapunov-LaSalle asymptotic stability theorem, ensuring a rigorous and comprehensive analysis of the system's long-term behavior. The theoretical findings are corroborated by numerical simulations. We conducted a sensitivity analysis of the basic reproduction numbers, R0 for HIV and R1 for HBV. The effects of antiviral treatment and time delays on the HIV-HBV co-dynamics are discussed. Minimum efficacy thresholds for anti-HIV and anti-HBV therapies are Determined, and when drug effectiveness surpasses these levels, the model predicts the full elimination of both viruses from the host. Additionally, the length of the time delay interval plays a role similar to that of antiviral treatment, suggesting a potential strategy for developing drug therapies aimed at extending the time delay period. The results of this study highlight the importance of incorporating time delays in models of dual viral infection and support the development of treatment strategies that enhance therapeutic outcomes by extending these delays.
1 Introduction
Two deadly viruses for the human body are the human immunodeficiency virus (HIV) and hepatitis B virus (HBV). In several patients, co-infection with these two viruses occurs because they share common transmission routes, such as blood transfusions, use of contaminated syringes, unprotected sexual intercourse, and others [1]. It is estimated that 8% to 10% of people diagnosed with HIV are also infected with HBV worldwide, making it important to study the co-infection of these two viruses. The co-infection with both viruses may increase the symptoms of each disease. This is due to the weakening and deterioration of the patient's immune system and the difficulty of determining effective treatments for both viruses [2, 3]. HIV is a single-stranded RNA virus that primarily infects CD4+ T cells, which are key components of the adaptive immune system. Over time, if left untreated, the number of CD4+ T cells declines, eventually leading to a disease called acquired immunodeficiency syndrome (AIDS) [4]. AIDS infection exposes the patient to opportunistic diseases and cancers. Besides infecting CD4+ T cells, HIV is also capable of invading other immune cells like dendritic cells and macrophages, resulting in their reduction and impaired function. The HIV also has the ability to infect different liver cells, including hepatocytes, Kupffer cells, and infiltrating T cells [5, 6]. HBV is a double-stranded DNA virus that primarily targets liver cells (hepatocytes) [7]. Over time, if left untreated, this infection can lead to chronic hepatitis, which may progress to cirrhosis and hepatocellular carcinoma.
Studying the interactions between viruses and target cells within the host, as well as immune responses, through laboratory experiments is not only difficult but also expensive. Therefore, mathematical modeling and simulation have become essential and important tools for understanding the dynamics of viruses within the host, as well as the role of the immune system in combating them. Furthermore, mathematical models may help design appropriate therapeutic strategies. In [8, 9], the basic models of HIV and hepatitis B virus (HBV) infection, respectively, have been formulated. These models consist of three main compartments: uninfected target cells (CD4+ T cells for HIV and hepatocytes for HBV), infected cells, and free viral particles. To take into account different biological phenomena not covered by these basic models, various extensions and generalizations have been made. One of the most important of these phenomena is the time delay during biological reactions. Examples of time delays include the time delay until a cell becomes infected, the time required for activation of the virus released from the cells, and the delay in the immune response. Time delays contribute to the development of accurate models that help understand the evolution of the virus within the host. Numerous studies have explored the incorporation of time delays into HIV single-infection models (see, e.g., [10–16]). On the other hand, HBV single-infection models with delay have also been formulated in some studies (e.g., [17–20]).
Mathematical modeling of HIV and hepatitis B virus co-infection at the population level has received considerable attention from researchers. These models play an important role in understanding patterns of infectious disease transmission within populations. These models may also play a significant role in formulating epidemic control strategies. Therefore, several mathematical models have been developed for the coinfection of HBV and HIV (see, e.g., [3, 21–26]). In contrast, within-host models that examine the biological interactions and co-dynamics of HIV and HBV at the cellular level are relatively scarce. To our knowledge, only a limited number of studies, notably [27, 28], have addressed the within-host interplay between these two viruses. This highlights a significant gap in the literature, underscoring the need for more comprehensive investigations into their intracellular co-infection dynamics.
The HIV-HBV co-infection model proposed by [28] is formulated as follows:
At time t, the variables x(t), y1(t), y2(t), v1(t), and v2(t) denotes the concentrations of uninfected hepatocytes, hepatocytes infected with HIV, hepatocytes infected with HBV, free HIV virions, and free HBV virions, respectively. τ1 and τ2 denote the respective time delays associated with the development of HIV-infected and HBV-infected hepatocytes. The factor , i = 1, 2 represents the probability of survival of infected hepatocytes throughout the delay time [t − τi, t], and ni > 0.
Notably, the model presented in [28] has certain limitations:
(i) it overlooks the dynamic evolution of both uninfected and HIV-infected CD4+ T cell populations, opting instead to assume a fixed rate of HIV production from infected CD4+ T cells, thereby ignoring their temporal variation;
(ii) it does not account for the maturation delay of newly produced virions;
(iii) Global stability was established exclusively for the infection-free equilibrium.
Limitation (i) was partially resolved in the model proposed by [27], which incorporated the dynamics of CD4+ T lymphocyte populations. However, the mathematical analysis in that study was largely confined to establishing the boundedness and positivity of the solutions. In addition, only the infection-free equilibrium was identified and analyzed for local stability, while a comprehensive investigation of other possible equilibria was not pursued.
This study builds upon and extends the models presented in [27, 28] by addressing key limitations. We explicitly model the dynamics of both uninfected and HIV-infected CD4+ T lymphocytes, replace the assumption of a constant HIV production rate with a more realistic approach, and include a biologically relevant time delay to reflect delays in viral production and virion maturation. A rigorous mathematical analysis, including equilibrium identification and global stability examination, provides deeper insights into the system's long-term dynamics. These findings are supported by numerical simulations and sensitivity analyses that highlight the influence of critical parameters.
The main contributions of this study are:
• Model enhancement: Addressing limitations of previous models to better capture HIV-HBV co-infection dynamics.
• Explicit immune cell dynamics: Including both uninfected and infected CD4+ T cells, with state-dependent HIV production.
• Intracellular delay: Introducing a time delay to represent the lag in viral production and the maturation process.
• Mathematical analysis: Identifying equilibria and establishing global stability for a comprehensive understanding.
• Numerical simulations: Illustrating biologically relevant scenarios that validate theoretical results.
• Sensitivity analysis: Exploring parameter impacts to inform potential control strategies.
2 Model formulation
In this section formulate our proposed model on based on the next Hypotheses:
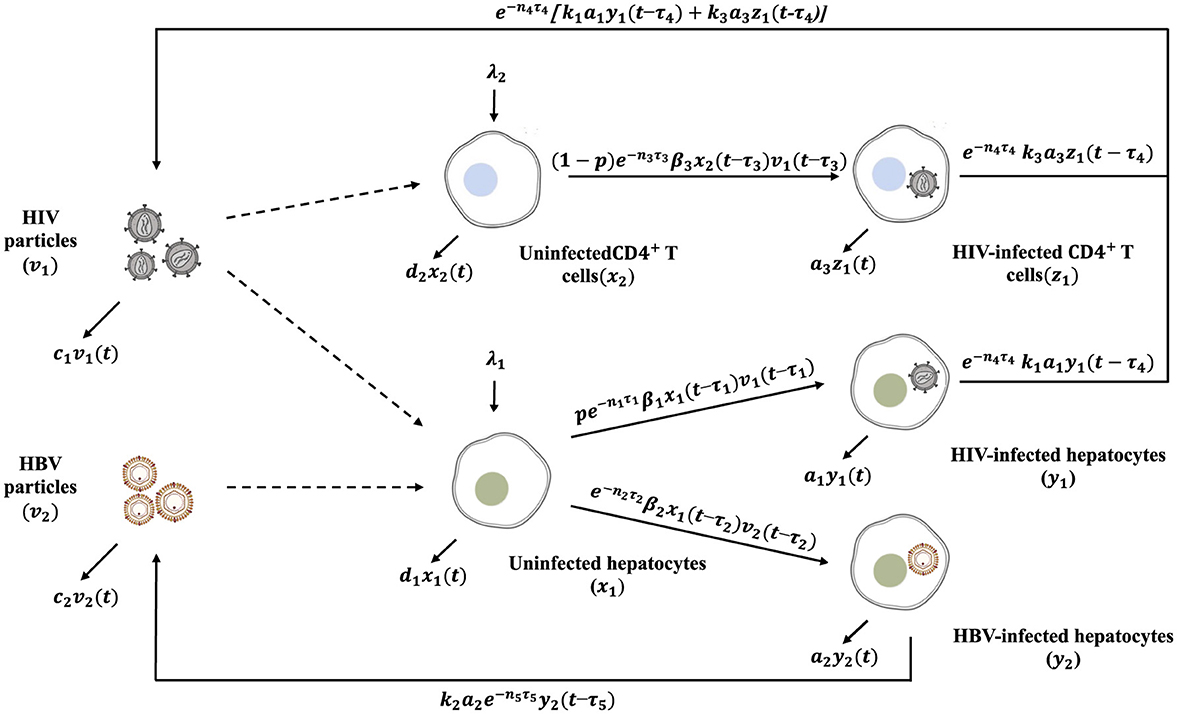
(H1): Seven populations are depicted in the model: Uninfected hepatocytes, x1(t), HIV-infected hepatocytes, y1(t), HBV-infected hepatocytes, y2(t), uninfected CD4+ T cells, x2(t), HIV-infected CD4+ T cells, z(t), free HIV, v1(t), free HBV, v2(t), where t is the time. Compartments (x1, y1, y2, x2, z1, v1, v2) die (or clear) at rates (d1x1, a1y1, a2y2, d2x2, a3z1, c1v1, c2v2), respectively. The co-dynamics of HIV and HBV are illustrated in the schematic diagram shown in Figure 1.
(H2): HBV targets uninfected hepatocytes of the liver [9], whereas HIV targets two cell types: uninfected hepatocytes [5, 6] and uninfected CD4+ T cells [8]. When HIV enters the liver, it has a probability (p) of infecting a hepatocyte, and a probability of 1 − p of infecting a CD4+ T cell [27] .
(H3): Uninfected hepatocytes are generated at rate λ1 and are infected via two competing viruses, HIV and HBV, with rates pβ1x1v1 and β2x1v2, respectively [27] (see Equation 1).
(H4): HIV-infected and HBV-infected hepatocytes are generated at rates of pβ1x1v1 and β2x1v2, respectively (see Equations 2, 3).
(H5): Uninfected CD4+ T cells are generated at rate λ2 and can be infected by HIV at rate of (1 − p)β1x1v1 [8, 27] (see Equation 4).
(H6): HIV-infected CD4+ T cells are formed at rate of (1 − p)β1x1v1 [27] (see Equation 5).
(H7): HIV particles are produced from two sources, HIV-infected hepatocytes and HIV-infected CD4+ T cells at rates of k1a1y1 and k3a3z1, respectively [8, 27] (see Equation 6).
(H8): HBV-infected hepatocytes produce HBV particles at a rate k2a2y2 [9, 27] (see Equation 7).
Based on Hypotheses (H1)-(H8), we formulate an HIV-HBV co-dynamics model with delays as follows:
The model incorporates five distinct time delays:
• The parameters τ1, τ2, and τ3 correspond to the delays associated with the development of HIV-infected hepatocytes, HBV-infected hepatocytes and HIV-infected CD4+ T cells, respectively [28, 29]. The expression for i = 1, 2, 3, denotes the likelihood that hepatocytes or CD4+ T cells survive during the interval [t − τi, t], where ni > 0.
• The delays τ4, and τ5 represent the time required for newly released HIV and HBV to become mature, respectively. Similarly, the term for i = 4, 5 reflects the survival probability of viral particles throughout the corresponding maturation period [t − τi, t], assuming ni > 0.
The initial conditions for model (Equations 1–7) are given as follows:
where , and is the Banach space of continuous functions. The norm is defined as for ϕi ∈ C, and i = 1, 2, ⋯ , 7. The standard theory of functional differential equations shows that, model (Equations 1–7) with initial conditions (Equation 2) has a unique solution [30, 31].
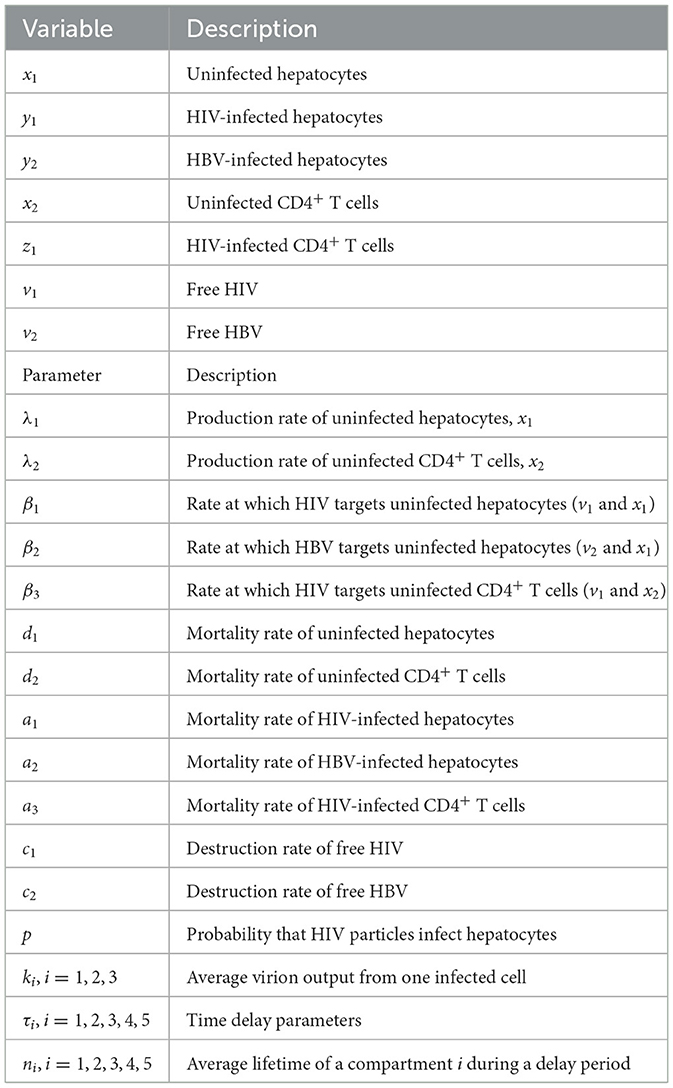
The model's variables and parameters are defined in Table 1.
3 Biological feasible domain
Let for i = 1, 2, ⋯ , 5, σ1 = min{d1, a1, a2} and σ2 = min{d2, a3}.
Lemma 1. The dynamics (Equations 1–7) with the initial states (Equation 2) admits nonnegative and ultimately bounded solutions.
Proof. Clearly, Equations 1, 4 of model (Equations 1–7) give ẋi∣xi = 0 = λi > 0, which implies that xi(t) > 0 for i = 1, 2 and all t ≥ 0. Moreover, for t ∈ [0, τm] we have
which gives (x1, y1, y2, x2, z1, v1, v2)(t) ≥ 0. Hence, by recursive argumentation, we obtain that (x1, y1, y2, x2, z1, v1, v2)(t) ≥ 0 for any t ≥ 0.
Next we prove the ultimately boundedness of the model's solution (x1, y1, y2, x2, z1, v1, v2). From Equation 1, we have lim supt → ∞ x1(t) ≤ Γ1, where . Concerning, y1(t) and y2(t), we define
Then, we get
Thus lim supt → ∞ ψ1(t) ≤ Γ2, and hence lim supt → ∞ y1(t) ≤ Γ2 and lim supt → ∞ y2(t) ≤ Γ2, where . From Equation 4, we have lim supt → ∞ x2(t) ≤ Γ3, where . Define
Then, we get
This gives lim supt → ∞ ψ2(t) ≤ Γ4, and hence lim supt → ∞ z1(t) ≤ Γ4 where .
Note that
Consequently, lim supt → ∞ v1(t) ≤ Γ5, where .
Similarly, we have
This yields lim supt → ∞ v2(t) ≤ Γ6, where .
Based on Lemma 1, one can show
is positively invariant w.r.t. system (Equation 1–7).
3.1 Equilibria and thresholds
This section analyzes the existence of equilibria for the proposed model. To compute the basic reproduction number, , associated with the HIV-HBV co-infection dynamics, we utilize the next-generation matrix approach as described by [32] (refer to the Appendix for details). The expression obtained for :
where and
Here, represents the basic reproduction number for an HIV infection in isolation, while corresponds to that of HBV infection alone (see Appendix for derivation). Let us define and and let the following two indices be denoted by i = 2, 3 and given by
Therefore, we obtain the following main result regarding the existence of the equilibria of the system (Equations 1–7).
Lemma 2.
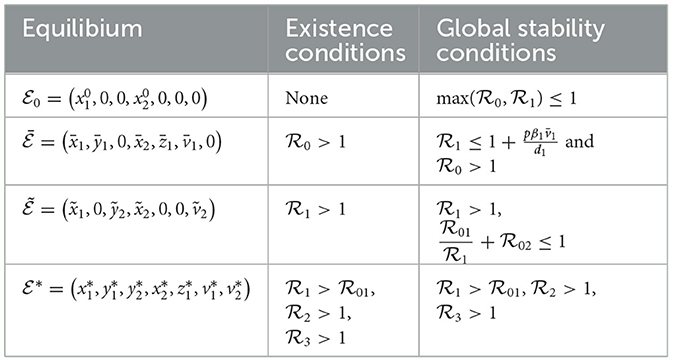
• The system (Equation 1–7) admits an infection-free equilibrium denoted by .
• If , then the system admits an HIV-only infection equilibrium denoted by
• When , the system admits an HBV-only infection equilibrium denoted by
• If , , and , the system possesses an HIV and HBV co-infection equilibrium denoted by
Proof. The equilibrium points are normally given when the time-derivatives are all set to zero, so that here we have the following:
We have the following cases:
1. If v1 = v2 = 0, the model has infection-free equilibrium .
2. If v1 ≠ 0 and v2 = 0, we obtain
and v1 satisfies the following equation
Define a function f by
Then we have
Thus, f(0) > 0 when . In fact, and denote, respectively, the total number of newly HIV-infected hepatocytes and HIV-infected CD4+ T cells generated from a single HIV-infected hepatocyte and CD4+ T cell at the onset of HIV infection. The parameter represents the basic reproductive number for a single HIV infection. We have f(v1) → −c1 < 0 whenever v1 → ∞. Moreover,
Thus, f is strictly decreasing, and hence, if , there exists a unique satisfying . Hence, , , , and , where satisfies the following quadratic equation:
which takes the form
where
Clearly, c < 0 if . Equation 8 has a positive solution
We obtain an HIV-only infection equilibrium
This implies that exists when .
3. If v1 = 0 and v2 ≠ 0, we obtain an HBV-only infection equilibrium
Clearly, exists when .
4. If v1 ≠ 0 and v2 ≠ 0, we obtain an HIV and HBV co-infection equilibrium
where
It is evident that exists if , , and . This equilibrium represents the coexistence of HIV and HBV infections.
4 Global stability
The global asymptotic stability of all equilibrium points in the system (Equations 1–7) is investigated using Lyapunov's direct method in this section. The development of Lyapunov functions is based on the approach outlined in [33]. Let represent the candidate Lyapunov function, and let Ω′ denote the largest invariant subset of . Consider the function Φ(X) = X − ln X − 1. Let us denote by (x1, y1, y2, x2, z1, v1, v2) = (x1, y1, y2, x2, z1, v1, v2)(t), and .
Theorem 1. The infection-free equilibrium is globally asymptotically stable (GAS) when .
Proof. Consider
Calculating along the solution of system (Equations 1–7) as
From Equations 1–7 we obtain
Collecting terms as
Substituting and , we obtain
Therefore, for all x1, y1, y2, x2, z1, v1, v2 > 0 we have when . Moreover, when , , , and . According to [30], solutions of system (Equation 1–7) asymptotically approach the set , which consists of elements satisfying , and
Let us consider four cases:
• If then and and from Equations 9, 10, we obtain v1 = v2 = 0. Since is invariant, we obtain . From Equations 6, 7, we have
and
Hence,
• If , we have , then ẋ1(t) = ẋ2(t) = 0. From Equations 1, 4, we have
Equations 11, 12 imply y1(t) = z1(t) = y2(t) = 0, ∀t ≥ 0. Hence, .
• If and then v1 = 0 and thus Equation 14 gives v2(t) = 0, ∀t ≥ 0 and from Equations 11, 12, we obtain y1 = z1 = y2 = 0. Hence, .
• Similarly, if and then v2 = 0 and thus Equation 13 provides v1(t) = 0, ∀t ≥ 0. From Equations 11, 12, we obtain y1 = z1 = y2 = 0. Hence, .
Lyapunov-LaSalle asymptotic stability theorem [34–36] reveals that is GAS when .
Theorem 2. If the HIV-only infection equilibrium exists () then it is GAS under the condition .
Proof. Define a function
We calculate as follows:
Collecting terms, we get
By using the equilibrium conditions
we obtain
Then
Using the following equalities:
we obtain
We have
Therefore,
It follows that when . Moreover, if and
We have two cases:
• . Then v2 = 0, and hence and from Equation 7 we have and thus y2(t) = 0, ∀t ≥ 0. Hence, .
• . From Equation 1 we have , hence v2(t) = 0, and from Equation 7 we obtain y2(t) = 0 for any t. Hence, .
Lyapunov-LaSalle asymptotic stability theorem indicates that is GAS when and .
Theorem 3. If the HBV-only infection equilibrium exists (if ) then it is GAS under the condition .
Proof. Consider
We calculate as:
Collecting terms as:
By using the conditions of
we obtain
Applying the following equality:
we obtain
Furthermore, we have
Then, we obtain the following:
If , then we obtain , where occurs at , and
We have two cases:
• , From Equation 4 we have
and from Equation 6 we have then
Hence,
• , then v1 = 0 and from Equation 16 we obtain y1 = z1 = 0. Hence,
Lyapunov-LaSalle asymptotic stability theorem indicates that is GAS when .
Theorem 4. If the HIV and HBV co-infection equilibrium exists (if , and ) then it is GAS.
Proof. Define be defined as:
Calculating as:
Collecting terms we obtain the following:
By using the equilibrium conditions
we obtain
Applying the following equalities:
we obtain
Therefore, we obtain for all x1, y1, y2, x2, z1, v1, v2 > 0. Moreover, when . Therefore, . Applying Lyapunov-LaSalle asymptotic stability theorem we obtain that is GAS.
Table 2 provides a concise overview of the conditions for existence and global stability of the four equilibria.
Remark 1. An important and promising direction for future investigation involves incorporating memory effects into our model by employing fractional differential equations (FDEs) [37, 38]. Unlike classical differential equations, FDEs are particularly effective in modeling systems where the current state depends not only on present conditions but also on the history of the system–an attribute often observed in complex biological processes, including viral infections and immune responses. The intrinsic non-locality and memory-retaining nature of FDEs make them especially suitable for capturing the nuanced dynamics of HIV-HBV co-infection. In recent studies, there has been growing interest in using the Lyapunov stability theory to establish the global behavior of fractional-order systems, as highlighted in the works of [39, 40]. The Lyapunov functions introduced in this section are not only essential for the integer-order formulation of our model but also form a foundational framework that can be extended to analyze the global stability of its fractional counterpart. Such an extension would offer deeper insights into the long-term progression and control of co-infection under memory-influenced dynamics.
5 Numerical simulations
5.1 Stability of equilibria
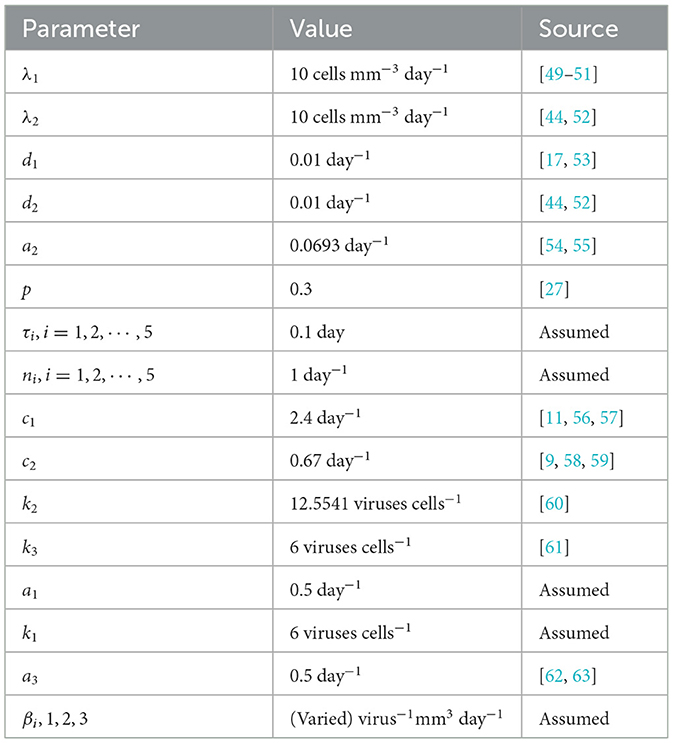
We conduct numerical simulations for the system defined by Equations 1–7, using parameter values listed in Table 3. Some of the parameter values, as presented in Table 3, were sourced from existing literature. The rest were estimated based on plausible assumptions intended purely for illustrative and computational exploration. This was necessitated by the lack of precise clinical data, especially concerning individuals co-infected with HIV and HBV. Challenges such as ethical considerations, logistical hurdles, and small patient cohorts contribute to this data gap, making accurate parameter estimation difficult. Nevertheless, the chosen values offer a meaningful foundation for analyzing the model's qualitative dynamics and serve as a stepping stone for future research as more empirical data become available.
The system of delay differential equations is solved using the MATLAB solver dde23. To validate the theoretical findings discussed in earlier sections, we vary selected parameters that have a notable impact on the threshold values and, in turn, on the system's stability behavior.
To validate the results of our global stability analysis, we demonstrate that the trajectories of the system, regardless of their starting points within the biologically feasible domain, consistently evolve toward the equilibrium point that satisfies the established stability criteria. This behavior confirms that the equilibrium state dictates the system's long-term dynamics for all permissible initial conditions. Drawing methodological inspiration from the study of [41], we proceed by examining the system's response to the following representative set of initial values:
To investigate the dynamical behavior of the system under varying infection pressures, we select different values for the transmission rates β1, β2, and β3. These values are chosen to illustrate distinct virological scenarios, ranging from disease eradication to full co-infection. The remaining parameters are fixed as presented in Table 3. Additionally, we set τi = 0.1 for i = 1, 2, ⋯ , 5. We analyze four representative cases:
Case 1: Complete clearance of both infections
When the infection rates are set to β1 = 0.0001, β2 = 0.00002 and β3 = 0.00003, the calculated basic reproduction numbers are and . Under these conditions, the system stabilizes at the infection-free equilibrium point , which is GAS. As illustrated in Figure 2, all trajectories converge to , indicating that both HIV and HBV are eradicated and the uninfected hepatocytes and CD4+ T cells return to their normal physiological levels. This outcome confirms the theoretical results stated in Theorem 1.
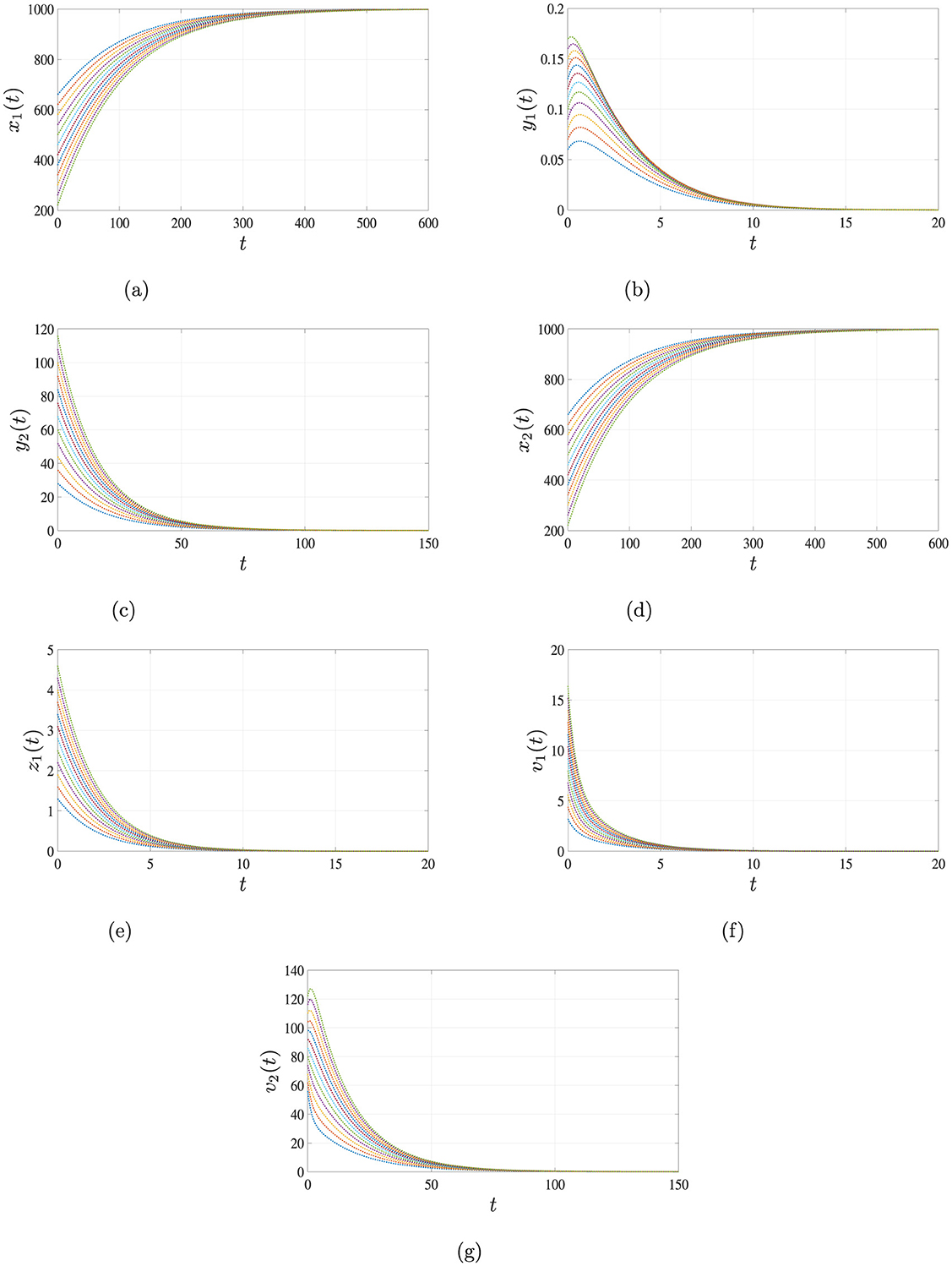
Figure 2. Simulation results for Case 1 demonstrating the global asymptotic stability of the infection-free equilibrium point . When the basic reproduction number satisfies , all trajectories of system Equations 1–7 converge to over time, regardless of the initial conditions. This indicates that the infection cannot persist below this threshold. (a) Uninfected hepatocytes. (b) HIV-infected hepatocytes. (c) HBV-infected hepatocytes. (d) Uninfected CD4+ T cells. (e) HIV-infected CD4+ T cells. (f) HIV. (g) HBV.
Case 2: HIV persistence with HBV elimination
For the parameter set β1 = 0.003, β2 = 0.00002 and β3 = 0.001, we obtain and . In this scenario, only the HIV infection remains active while HBV is cleared. The solution of the system converges to the equilibrium point , where HBV-related compartments approach zero. Figure 3 supports this outcome, which aligns with the theoretical conclusions presented in Theorem 2.
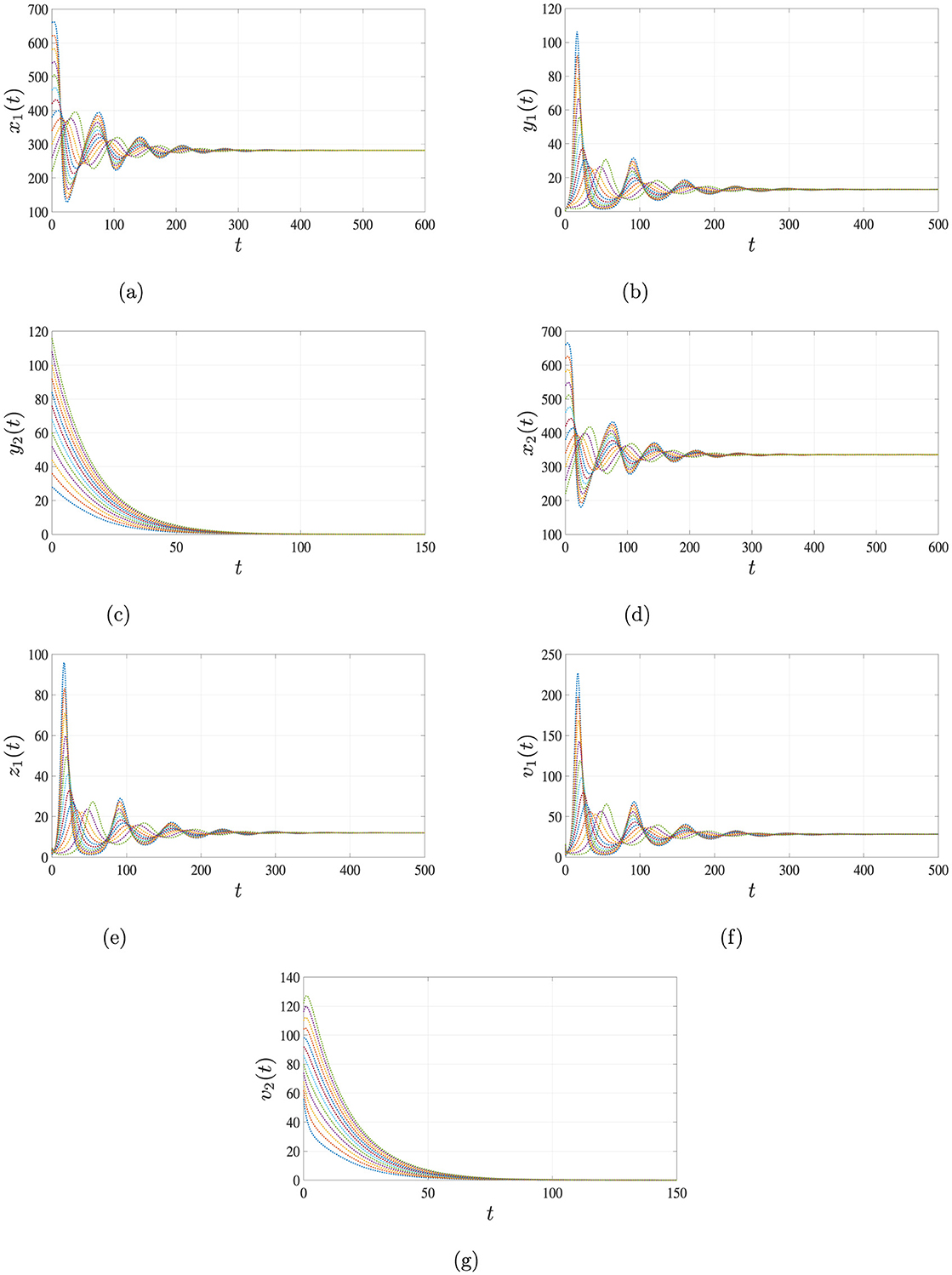
Figure 3. Simulation results for case 2 illustrating the persistence of HIV single-infection. When and the solutions of system Equations 1–7, starting from different initial conditions, converge to the equilibrium point . This indicates that HIV persists in the body, while HBV is cleared over time. (a) Uninfected hepatocytes. (b) HIV-infected hepatocytes. (c) HBV-infected hepatocytes. (d) Uninfected CD4+ T cells. (e) HIV-infected CD4+ T cells. (f) HIV. (g) HBV.
Case 3: HBV persistence with HIV clearance
Choosing β1 = 0.0017, β2 = 0.0002, and β3 = 0.00001 leads to , and . The system evolves toward the equilibrium , indicating persistence of HBV infection and clearance of HIV. The simulation results depicted in Figure 4 validate the analytical findings of Theorem 3.
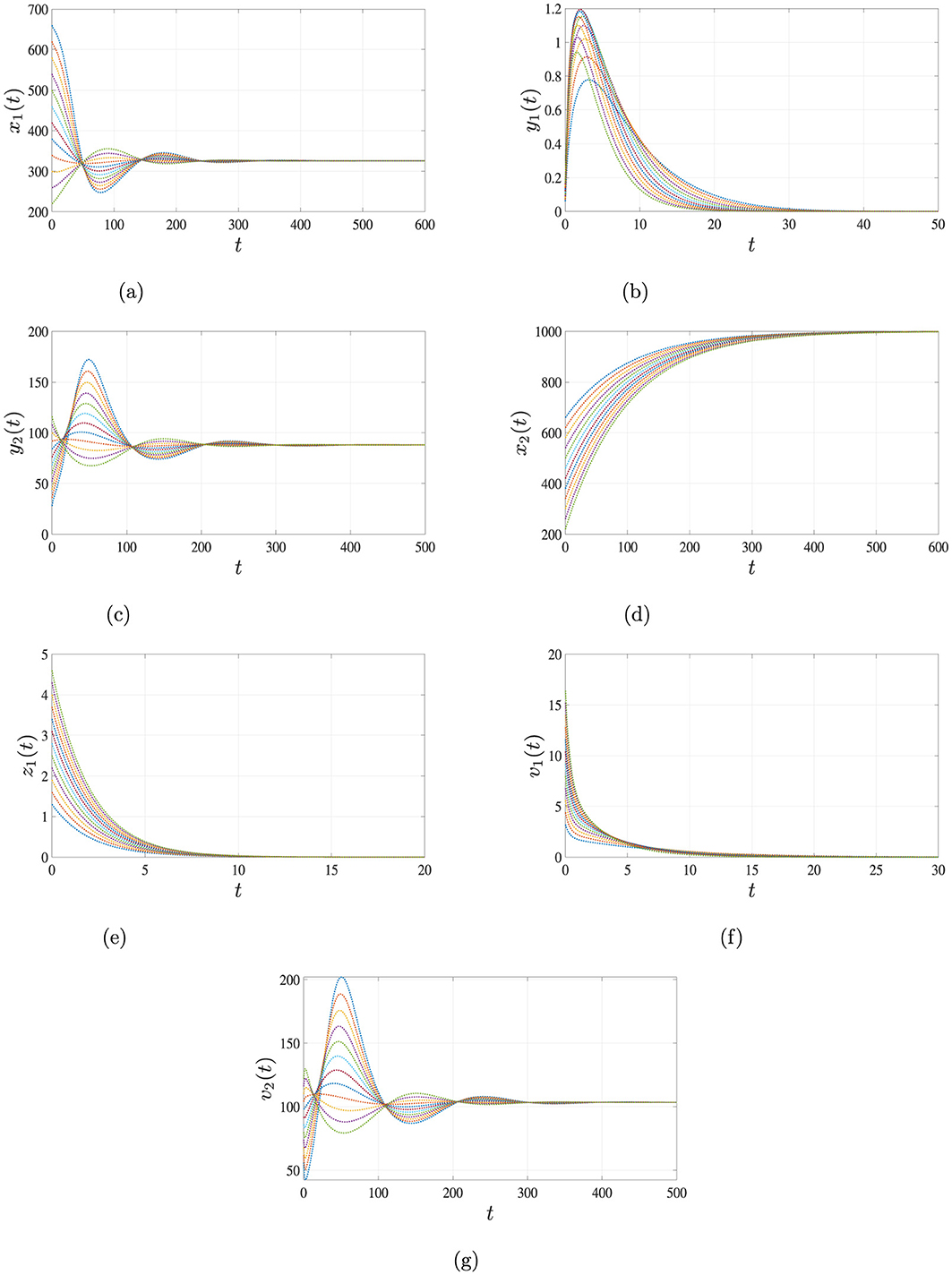
Figure 4. Simulation results for Case 3 illustrating the persistence of HBV single-infection. When and , the solutions of system Equations 1–7, starting from different initial conditions, converge to the equilibrium point . This indicates that HBV persists in the body, while HIV is cleared over time. (a) Uninfected hepatocytes. (b) HIV-infected hepatocytes. (c) HBV-infected hepatocytes. (d) Uninfected CD4+ T cells. (e) HIV-infected CD4+ T cells. (f) HIV. (g) HBV.
Case 4: Coexistence of HIV and HBV
Finally, for β1 = 0.00015, β2 = 0.0002 and β3 = 0.001, the system exhibits a more complex dynamic with , , . In this case, both HIV and HBV persist, and the system converges to a co-infected equilibrium , which is GAS. The long-term dynamics illustrated in Figure 5 confirm the results of Theorem 4, demonstrating stable co-infection of both viruses.
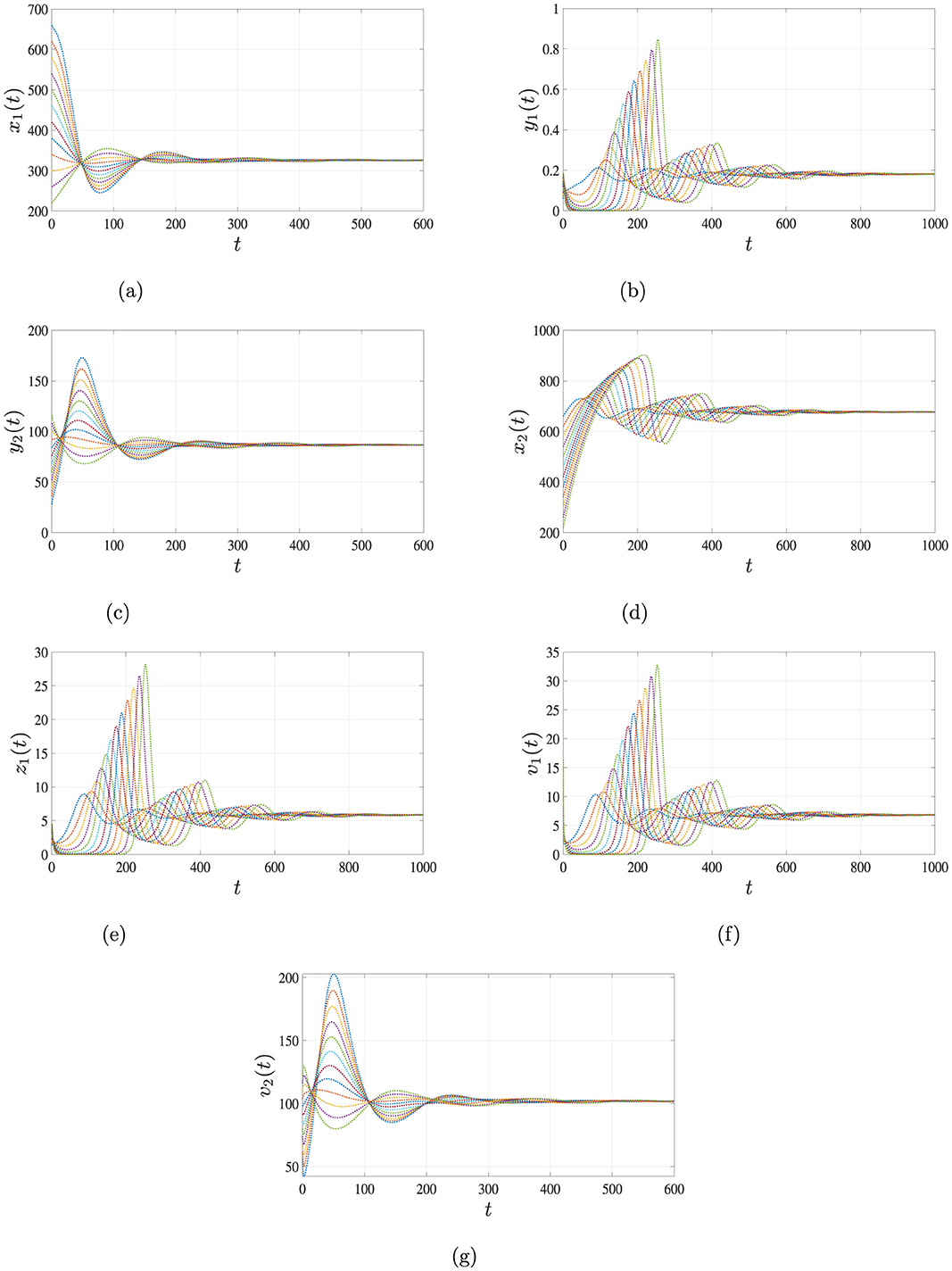
Figure 5. Simulation results for Case 4 illustrating the persistence of both HIV and HBV co-infection. When , , and , the solutions of system Equations 1–7, starting from three distinct initial conditions, converge to the endemic equilibrium point . This indicates that both HIV and HBV persist in the body under this scenario. (a) Uninfected hepatocytes. (b) HIV-infected hepatocytes. (c) HBV-infected hepatocytes. (d) Uninfected CD4+ T cells. (e) HIV-infected CD4+ T cells. (f) HIV. (g) HBV.
The outcomes of these four scenarios clearly illustrate the significant impact of infection rate parameters on the progression and stability of the system. Varying these rates leads to distinct biological outcomes, including total clearance of both viruses, dominance of one infection, or stable coexistence of HIV and HBV. The close agreement between simulation results and analytical predictions reinforces the model's reliability in representing the intricate dynamics of HIV-HBV co-infection.
5.2 Sensitivity analysis
Sensitivity analysis aims to assess how variations in model parameters influence the system's output dynamics [42]. A common method involves calculating sensitivity indices, which quantify the proportional change in a model outcome resulting from a small change in a specific parameter. This technique is particularly useful for identifying parameters that most significantly affect the basic reproduction numbers , where i = 0, 1. Given that and are differentiable functions of key parameters, their sensitivity indices can be derived using partial derivatives [43]. In this study, we focus on computing the sensitivity indices for both and , with the goal of understanding how different parameters contribute to the stability of the infection-free equilibrium . The normalized sensitivity index of w.r.t. a given parameter η is given as follows [42]:
5.2.1 Sensitivity analysis of
To investigate how variations in model parameters affect the basic reproduction number HIV-only infection , we compute normalized sensitivity indices. These indices reflect the proportional change in resulting from a small perturbation in a given parameter. The general formula used for this computation is based on partial derivatives of with respect to each parameter.
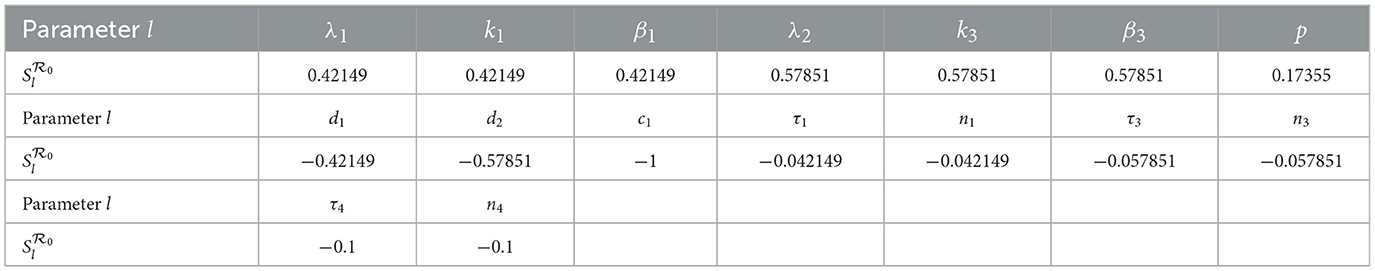
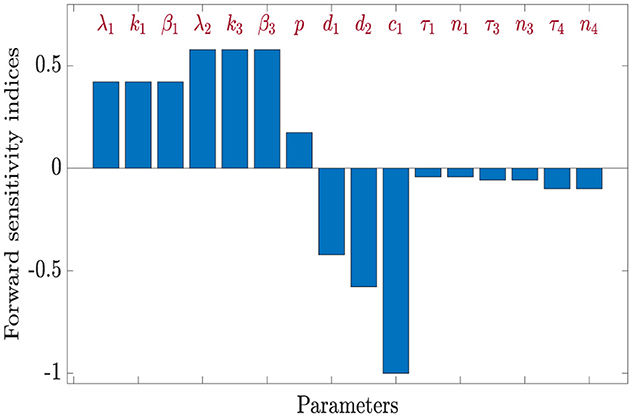
Using parameter values β1 = 0.0017, β2 = 0.0002, β3 = 0.001, and τi = 0.1 for i = 1, 2, ⋯ , 5 along with the values listed in Table 3, we calculate the sensitivity indices of as shown in Table 4.
From the computed indices, we observe that parameters λ1, k1, β1, λ2, k3, β3, and p positively influence ; increases in any of these raise the value of . On the other hand, parameters such as d1, d2, c1, τ1, n1, τ3, n3, τ4, and n4 have a negative effect on , meaning that increasing them leads to a reduction in viral transmission potential.
The impact of selected parameters can be summarized as follows:
• A 10% increase in λ1, k1, or β1 leads to a 4.2149% increase in , while the same percentage increase in λ2, k3, or β3 yields a 5.7851% rise in .
• Conversely, increasing d1 or d2 by 10% results in 4.2149% and 5.7851% reductions in , respectively.
• A 10% rise (or drop) in the parameter c1 results in a corresponding 10% reduction (or increase) in the value of .
• Parameter p has a more modest yet significant influence; a 10% increase in p increases by approximately 1.7355%.
• Time-delay-related parameters such as τ1 or n1 impact by about 4.215%, while τ3, n3 contribute about 5.785% each, and τ4, n4 account for a 10% effect.
These results indicate that is especially sensitive to λ2, k3, β3, p, d2 and c1, in agreement with biological expectations, since HIV predominantly targets CD4+ T cells.
5.2.2 Sensitivity analysis of
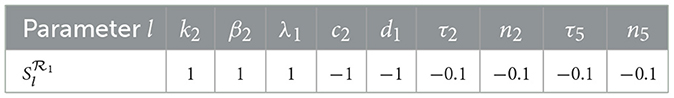
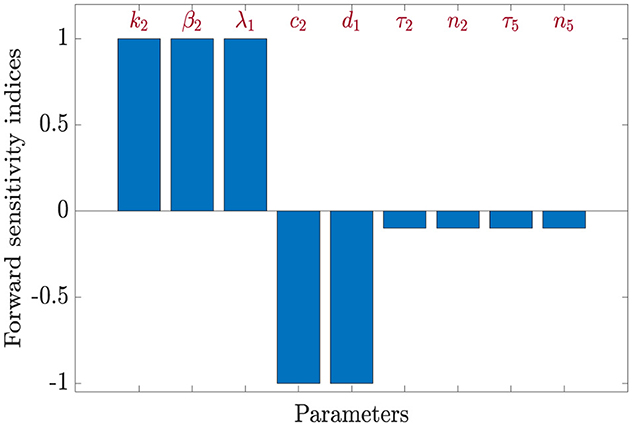
We now turn our attention to the basic reproduction number of HBV-only infection . The corresponding sensitivity indices are derived as follows: , and . Using the same parameter values as before, the calculated indices for are summarized in Table 5.
The analysis reveals that is positively affected by increases in k2, β2, and λ1, while it decreases with higher values of c2 and d1. Specifically:
• A 10% increase in k2, β2, or λ1 results in a 10% increase in .
• A 10% increase in c2 or d1 leads to a 10% decrease in .
• Likewise, increasing τ2 or n2 by 100% reduces by 10%, and the same applies to τ5 and n5
The reproduction number shows no sensitivity to the remaining parameters in system (Equation 1–7), indicating that the progression of HBV infection in this model is primarily governed by a smaller subset of variables.
5.3 Co-infection dynamics of HIV and HBV under antiviral therapies
To address the impact of antiviral therapies on the progression of HIV and HBV co-infection, we modify the original system of Equations 1–7 by incorporating two pharmacological interventions:
(i) an antiretroviral drug with efficacy parameter ϵ1∈[0, 1] targeting HIV [44], and
(ii) an anti-HBV agent with efficacy ϵ2∈[0, 1] aimed at suppressing hepatitis B viral replication [45].
The resulting treatment-augmented model is expressed as follows:
The therapeutic impact can be understood by analyzing the basic reproduction numbers for HIV and HBV in the presence of treatment. Referring to the basic reproduction numbers (for HIV) and (for HBV) as previously defined in Section 3.1, the modified reproduction numbers under drug action are computed as folllows:
To ensure long-term viral clearance, the aim is to suppress both reproduction numbers below unity:
which would guarantee global asymptotic stability of the disease-free equilibrium .
Solving the above inequalities yields the minimum efficacies–also termed critical drug efficacies–required for viral eradication:
• ensuring for all
• ensuring for all
Using model parameters including β1 = 0.00015, β2 = 0.0002, β3 = 0.001, τi = 0.1 for i = 1, 2, ⋯ , 5, along with values from Table 3, we compute the following thresholds:
•
•
The results provided in Figures 6, 7 can be interpreted as follows.
(i) If ϵ1 ≥ 0.34421 ≤ ≤ 1 and ϵ2 ≥ 0.67407, both HIV and HBV infections are effectively controlled, and the system stabilizes at the disease-free state (GAS).
(ii) If ϵ1 < 0.34421 and/or ϵ2 < 0.67407, then at least one reproduction number exceeds 1, causing the infection-free equilibrium to lose stability. In such cases, a chronic infection equilibrium may emerge and become globally attractive.
To further analyze therapeutic outcomes, we simulate the model system (Equations 17–23) under varying drug efficacies, employing the following set of initial functions for θ ∈ [−0.1, 0]:
We examine a range of ϵ1 and ϵ2 values as detailed in Table 6.
Simulation outcomes, summarized in Table 6 and illustrated in Figure 8, reveal that increasing antiviral efficacies leads to a marked decline in both and . This pharmacodynamic effect contributes to the restoration of healthy hepatocyte and CD4+ T cell populations, while infected compartments (e.g., y1, y2, z1) and viral loads (v1, v2) decrease accordingly.
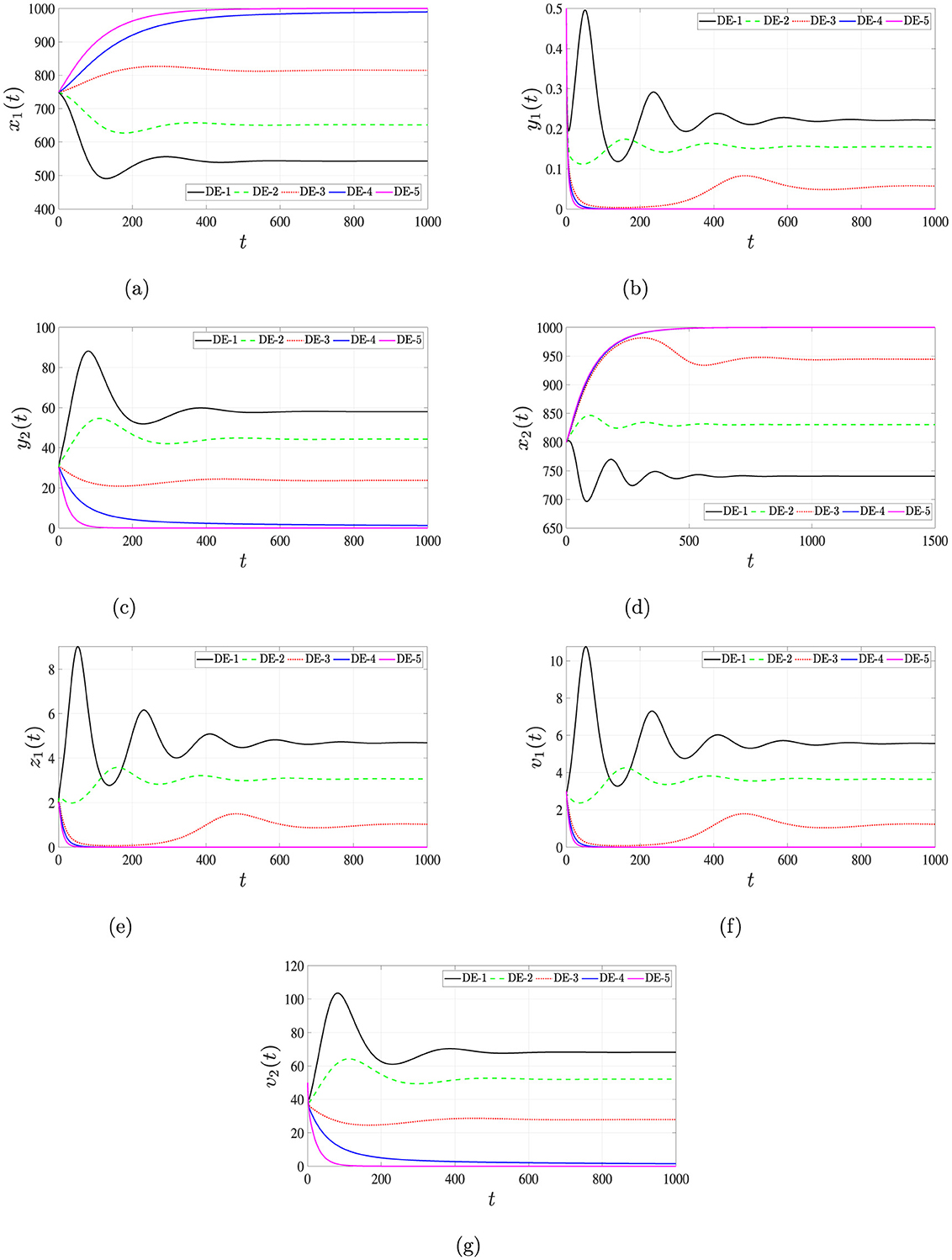
Figure 8. Solutions of system (Equations 16–22) for different antiviral effectiveness rates, ϵ1 and ϵ2. (a) Uninfected hepatocytes. (b) HIV-infected hepatocytes. (c) HBV-infected hepatocytes. (d) Uninfected CD4+ T cells. (e) HIV-infected CD4+ T cells. (f) HIV. (g) HBV.
These findings highlight the critical role of treatment efficacy in shifting the system dynamics toward recovery, emphasizing the need for optimal dosing strategies to achieve viral suppression and potential eradication. The analysis demonstrates that when antiviral drug efficacies exceed specific threshold values, the basic reproduction numbers fall below one, leading the system toward a globally stable, infection-free equilibrium. This underscores the importance of well-calibrated therapeutic interventions in effectively controlling and potentially eliminating HIV and HBV co-infections.
The set of equations defined by Equations 17–23 can be viewed through the framework of a nonlinear control system, in which the parameters ϵ1 and ϵ2 represent controllable inputs corresponding to the efficacies of anti-HIV and anti-HBV therapies, respectively. By treating these drug efficacies as control variables, we can employ tools from optimal control theory to determine treatment strategies that minimize viral loads and optimize patient outcomes [46–48]. This approach enables the systematic design of therapeutic protocols for individuals co-infected with HIV and HBV, ensuring both effectiveness and efficiency in long-term disease management.
5.3.1 Influence of time delays on the dynamics of HIV-HBV co-infection
Among the various time delay parameters influencing the system dynamics, it has been observed that the basic reproduction number is most sensitive to variations in the delay parameter τ4 (maturation delay of newly released HIV), while is similarly influenced by the delay τ5 (maturation delay of newly released HBV). Given this sensitivity, our analysis will focus on assessing how delays τ4 and τ5 affect the co-dynamics of HIV and HBV.
Using the transmission rate parameters β1 = 0.00015, β2 = 0.0002 and β3 = 0.001, along with other parameter values specified in Table 3, we fix τ1 = τ2 = τ3 = 0.01 and vary τ4 and τ5 to explore their impact on the stability of the disease-free equilibrium point . Since the expressions for and explicitly incorporate τ4 and τ5, respectively, any modification in these delays will directly affect the stability characteristics of . Notably, increasing either τ4 or τ5 tends to reduce the corresponding reproduction number, thus potentially promoting system stability.
The initial conditions for the delay differential system (Equation 1–7) are defined over the interval θ ∈ [−4, 0] as:
To determine the threshold values of τ4 and τ5 that ensure global asymptotic stability of the infection-free state, we express the reproduction numbers as functions of these delays:
To guarantee that and , we compute the critical values and as:
Substituting the parameter values from Table 3, we obtain the numerical approximations: and . Therefore:
• If and , then both and , ensuring that the infection-free equilibrium is GAS.
• Conversely, if or , the corresponding reproduction number(s) exceed 1, destabilizing and possibly rendering another equilibrium globally attractive.
As observed in Table 7, increasing the delays τ4 and τ5 leads to a reduction in the reproductive numbers and . Figure 9 presents the simulation outcomes for the full system, demonstrating the profound influence of time delays on the system dynamics. Increasing the delay durations τ4 and τ5 enhances the populations of uninfected hepatocytes and CD4+ T cells, while simultaneously decreasing the concentrations of infected cells and viral loads. This dynamic underscores the biological relevance of incorporating intracellular delays in modeling, as they exhibit effects analogous to therapeutic interventions.
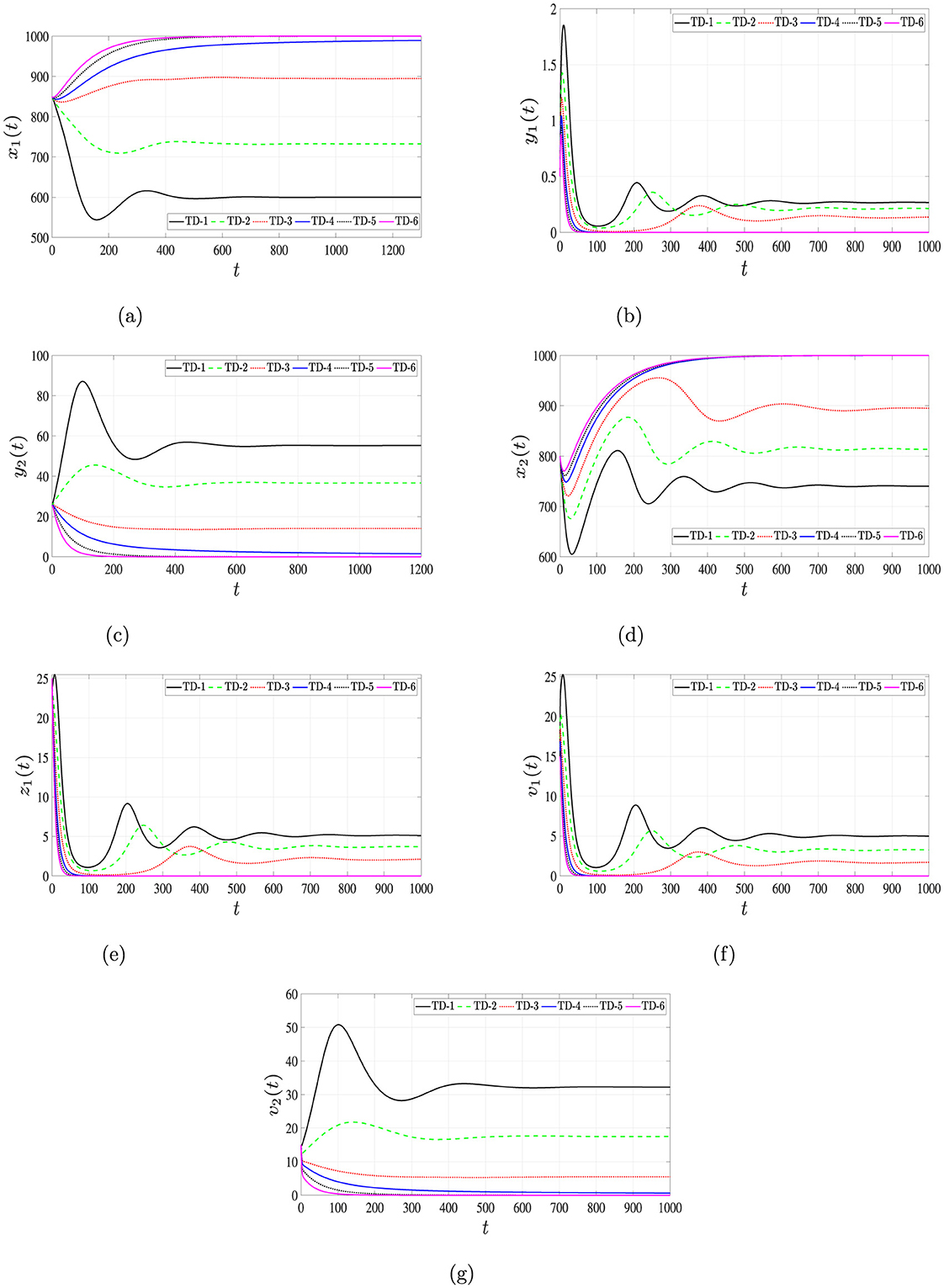
Figure 9. Solutions of system (Equation 1–7) for different time delays, τ4 and τ5. (a) Uninfected hepatocytes. (b) HIV-infected hepatocytes. (c) HBV-infected hepatocytes. (d) Uninfected CD4+ T cells. (e) HIV-infected CD4+ T cells. (f) HIV. (g) HBV.
In summary, incorporating biologically realistic time delays into the model not only changes the system's qualitative behavior but also offers insights into the development of novel therapies aimed at extending these delays. Such treatments could help slow down virus-cell interactions and prolong the maturation period of viruses released from infected cells. This, in turn, could significantly contribute to controlling the progression of viral activity within the host and enhancing the elimination of viruses from the body.
6 Conclusions
This study develops a mathematical model describing the within-host co-infection dynamics of HIV and HBV. In this framework, HBV primarily infects hepatocytes, while HIV targets both CD4+ T lymphocytes and hepatocytes. The system is represented by seven nonlinear delay differential equations (DDEs), which account for the interactions among uninfected hepatocytes, hepatocytes infected with either HIV or HBV, uninfected CD4+ T lymphocytes, HIV-infected CD4+ T lymphocytes, and free viral particles of both HIV and HBV. Two distinct delays are incorporated into the model: one representing the latency in cell infection and the other reflecting the maturation period of newly produced virions. The model ensures that all solutions are biologically feasible, remaining non-negative and bounded over time. Four equilibrium states are identified, and corresponding threshold quantities (for i = 0, 1, 2, 3) are derived to characterize their behavior. Conditions guaranteeing the global asymptotic stability of each equilibrium point are established using Lyapunov function techniques along with LaSalle's invariance principle. We performed numerical simulations and sensitivity analysis to validate the theoretical findings and to determine the most effective parameters on the basic reproduction numbers for HIV-only () and HBV-only () infections. We also studied the effects of two types of antiviral treatments: one for HIV and the other for HBV. We calculated the minimum treatment efficacy needed for clearing the virus from the body. Furthermore, we studied the effect of time delays on the dynamics of viral co-infection and found that these delays reduce the basic reproduction numbers. This highlights the role and importance of considering time delays in modeling viral co-infection, as neglecting these delays leads to an overestimation of the drug's efficacy required to clear the virus. It is also noted that longer delays show effects similar to antiviral therapy, which gives some insight into the idea that finding a treatment that extends the time delay period indicates a potential therapeutic benefit. These insights underscore the importance of incorporating time delays into treatment models and may contribute to the development of more effective strategies for managing coinfection.
The proposed model is capable of capturing a range of clinically observed scenarios such as
• Individuals who successfully avoid infection.
• Cases involving HIV infection alone.
• Cases involving HBV infection alone.
• Chronic co-infection with both HIV and HBV.
This framework also highlights the significant role of antiviral treatments in restoring immune competence and preserving liver function. Mathematical modeling of HIV-HBV co-infection at the within-host level has enhanced our understanding of how these viruses interact and influence disease progression. It also supports the optimization of therapeutic approaches tailored to individual patient needs.
The findings of this study contribute meaningfully to the clinical management of HIV-HBV co-infections. By identifying thresholds for treatment effectiveness and outlining conditions that lead to viral clearance or persistence, this study aids in refining treatment strategies, guiding personalized care and improving patient outcomes. Additionally, these insights can support broader public health efforts aimed at mitigating the long-term burden of co-infections.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
AE: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing. AA: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. AH: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fams.2025.1633039/full#supplementary-material
References
1. Corcorran MA, Kim HN. Chronic hepatitis B and HIV coinfection. Topics Antiviral Med. (2023) 31:14.
2. Weldemhret L. Epidemiology and challenges of HBV/HIV co-infection amongst HIV-infected patients in endemic areas. HIV/AIDS-Res Palliat Care. (2021) 13:485–490. doi: 10.2147/HIV.S273649
3. Ullah MA, Raza N, Omame A, Alqarni MS. A new co-infection model for HBV and HIV with vaccination and asymptomatic transmission using actual data from Taiwan. Physica Scripta. (2024) 99:065254. doi: 10.1088/1402-4896/ad4b6c
4. Wodarz D, Levy DN. Human immunodeficiency virus evolution towards reduced replicative fitness in vivo and the development of AIDS. Proc Biol Sci. (2007) 274:2481–91. doi: 10.1098/rspb.2007.0413
5. Zerbato JM, Avihingsanon A, Singh KP, Zhao W, Deleage C, Rosen E., et al. HIV DNA persists in hepatocytes in people with HIV-hepatitis B co-infection on antiretroviral therapy. Bullet Mathem Biol. (2023) 87:104391. doi: 10.1016/j.ebiom.2022.104391
6. Blackard JT, Sherman KE. HCV/HIV co-infection: time to re-evaluate the role of HIV in the liver? J Viral Hepatitis. (2008) 15:323–30. doi: 10.1111/j.1365-2893.2008.00970.x
7. Seeger C, Mason WS. Hepatitis B virus biology, microbiology and molecular. Biol Rev. (2000) 64:51–68. doi: 10.1128/MMBR.64.1.51-68.2000
8. Nowak MA, Bangham CRM. Population dynamics of immune responses to persistent viruses. Science. (1996) 272:74–9. doi: 10.1126/science.272.5258.74
9. Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci U S A. (1996) 93:4398–402. doi: 10.1073/pnas.93.9.4398
10. Lv L, Yang J, Hu Z, Fan D. Dynamics analysis of a delayed HIV model with latent reservoir and both viral and cellular infections. Mathem Methods Appl Sci. (2025) 48:6063–80. doi: 10.1002/mma.10655
11. Culshaw RV, Ruan S. A delay-differential equation model of HIV infection of CD4+ T-cells. Mathem Biosci. (2000) 165:27–39. doi: 10.1016/S0025-5564(00)00006-7
12. Prakash S, Umrao AK, Srivastava PK. Bifurcation and stability analysis of within host HIV dynamics with multiple infections and intracellular delay. Chaos. (2025) 35:013128. doi: 10.1063/5.0232978
13. Hmarrass H, Qesmi R. Global stability and Hopf bifurcation of a delayed HIV model with macrophages, CD4+ T cells with latent reservoirs and immune response. Eur Phys J Plus. (2025) 140:335. doi: 10.1140/epjp/s13360-025-06001-z
14. Rathnayaka NS, Wijerathna JK, Pradeep GSA. Stability properties and hopf bifurcation of a delayed HIV dynamics model with saturation functional response, absorption effect and cure rate. Int J Analy Appl. (2025) 23:92–92. doi: 10.28924/2291-8639-23-2025-92
15. Chen C, Ye Z, Zhou Y. Stability and Hopf bifurcation of a cytokine-enhanced HIV infection model with saturation incidence and three delays. Int J Bifurcat Chaos. (2024) 34:2450133. doi: 10.1142/S0218127424501335
16. Hu Z, Yang J, Li Q, Liang S, Fan D. Mathematical analysis of stability and Hopf bifurcation in a delayed HIV infection model with saturated immune response. Mathem Methods Appl Sci. (2024) 47:9834–57. doi: 10.1002/mma.10097
17. Gourley SA, Kuang Y, Nagy JD. Dynamics of a delay differential equation model of hepatitis B virus infection. J Biol Dynam. (2008) 2:140–53. doi: 10.1080/17513750701769873
18. Foko S Dynamical analysis of a general delayed HBV infection model with capsids and adaptive immune response in presence of exposed infected hepatocytes. J Mathem Biol. (2024) 88:75. doi: 10.1007/s00285-024-02096-7
19. Prakash M, Rakkiyappan R, Manivannan A, Zhu H, Cao J. Stability and bifurcation analysis of hepatitis B-type virus infection model. Mathem Methods Appl Sci. (2021) 44:6462–81. doi: 10.1002/mma.7198
20. Chen C, Zhou Y, Ye Z, Gu M. Stability and Hopf bifurcation of a HBV infection model with capsids and CTL immune response delay. Eur Phys J Plus. (2024) 139:1–22. doi: 10.1140/epjp/s13360-024-05764-1
21. Bowong S, Kamganga J, Tewa J, Tsanou B. Modelling and analysis of hepatitis B and HIV co-infections. In: Proceedings of the 10th African Conference on Research in Computer Science and Applied Mathematics. At Yamoussoukro, Ivory Cost (2010). p. 109–16.
22. Endashaw EE, Mekonnen TT. Modeling the effect of vaccination and treatment on the transmission dynamics of hepatitis B virus and HIV/aids coinfection. J Appl Mathemat. (2022) 2022:5246762. doi: 10.1155/2022/5246762
23. Endashaw EE, Gebru DM, Alemneh HT. Coinfection dynamics of HBV-HIV/AIDS with mother-to-child transmission and medical interventions. Comput Mathem Methods Med. (2022) 2022:4563577. doi: 10.1155/2022/4563577
24. Rasheed A, Raza A, Omame A. Analysis of a novel mathematical model for HBV and HIV incorporating vertical transmission and intervention measures. Model Earth Syst Environm. (2025) 11:222. doi: 10.1007/s40808-025-02408-w
25. Teklu SW. Workie AH. HIV/AIDS and HBV co-infection with optimal control strategies and cost-effectiveness analyses using integer order model. Sci Rep. (2025) 15:4004. doi: 10.1038/s41598-024-83442-z
26. Abebaw YF, Teklu SW. Dynamical analysis of HIV/AIDS and HBV co-infection model with drug-related kidney disease using optimal control theory. Model Earth Syst Environm. (2025) 11:1–34. doi: 10.1007/s40808-024-02203-z
27. Nampala H, Luboobi LS, Mugisha JY, Obua C, Jablonska-Sabuka M. Modelling hepatotoxicity and antiretroviral therapeutic effect in HIV/HBV co-infection. Mathem Biosci. (2018) 302:67–79. doi: 10.1016/j.mbs.2018.05.012
28. Nampala H, Jablonska-Sabuka M, Singull M. Mathematical analysis of the role of HIV/HBV latency in hepatocytes. J Appl Mathem. (2021) 2021:5525857. doi: 10.1155/2021/5525857
29. Herz AV, Bonhoeffer S, Anderson RM, May RM, Nowak MA. Viral dynamics in vivo: limitations on estimates of intracellular delay and virus decay. Proc Natl Acad Sci. (1996) 93:7247–51. doi: 10.1073/pnas.93.14.7247
30. Hale JK, Verduyn M, Lunel S. Introduction to Functional Differential Equations. New York: Springer-Verlag. (1993).
31. Kuang Y. Delay Differential Equations: With Applications in Population Dynamics. Cambridge, MA: Academic Press (1993).
32. Van den Driessche P, Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Mathem Biosci. (2002) 180:29–48. doi: 10.1016/S0025-5564(02)00108-6
33. Korobeinikov A Global properties of basic virus dynamics models. Bullet Mathem Biol. (2004) 66:879–83. doi: 10.1016/j.bulm.2004.02.001
36. Lyapunov AM. The General Problem of the Stability of Motion. London: Taylor & Francis, Ltd. (1992). doi: 10.1080/00207179208934253
37. Yaseen RM, Ali NF, Mohsen AA, Khan A, Abdeljawad T. The modeling and mathematical analysis of the fractional-order of Cholera disease: Dynamical and simulation. Partial Differ Equ Appl Math. (2024) 12:100978. doi: 10.1016/j.padiff.2024.100978
38. Hattaf K, A new mixed fractional derivative with applications in computational biology. Computation. (2024) 12:7. doi: 10.3390/computation12010007
39. Yaagoub Z, Sadki M, Allali K. A generalized fractional hepatitis B virus infection modelwith both cell-to-cell and virus-to-cell transmissions. Nonlinear Dynam. (2024) 112:16559–85. doi: 10.1007/s11071-024-09867-3
40. Naim M, Zeb A, Mohsen AA, Sabbar Y, Yıldız M. Local and global stability of a fractional viral infection model with two routes of propagation, cure rate and non-lytic humoral immunity. Mathem Model Numer Simul Appl. (2024) 4:94–115. doi: 10.53391/mmnsa.1517325
41. Song B, Zhang Y, Sang Y. Zhang L. Stability and Hopf bifurcation on an immunity delayed HBV/HCV model with intra-and extra-hepatic coinfection and saturation incidence. Nonlinear Dynam. (2023) 111:14485–511. doi: 10.1007/s11071-023-08580-x
42. Chitnis N, Hyman JM, Cushing JM. Determining important parameters in the spread of malaria through the sensitivity analysis of a mathematical model. Bullet Mathem Biol. (2008) 70:1272–96. doi: 10.1007/s11538-008-9299-0
43. Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty, sensitivity analysis in systems biology. J Theor Biol. (2008) 254:178–96. doi: 10.1016/j.jtbi.2008.04.011
44. Callaway DS, Perelson AS. HIV-1 infection and low steady state viral loads. Bullet Mathem Biol. (2002) 64:29–64. doi: 10.1006/bulm.2001.0266
45. Lewin SR, Ribeiro RM, Walters T, Lau GK, Bowden S, Locarnini S, Perelson AS. Analysis of hepatitis B viral load decline under potent therapy: complex decay profiles observed. Hepatology. (2001) 34:1012–20. doi: 10.1053/jhep.2001.28509
46. Joshi HR Optimal control of an HIV immunology model. Optim Control Appl Methods. (2002) 23:199–213. doi: 10.1002/oca.710
47. Mohsen AA, Naji RK. Dynamical analysis within-host and between-host for HIV/AIDS with the application of optimal control strategy. Iraqi J Sci. (2020) 61:1173–89. doi: 10.24996/ijs.2020.61.5.25
48. Nampala H, Jabłońska-Sabuka M, Singull M. Analysis of HIV therapy in the liver using optimal control and pharmacokinetics. J Mathem Indust. (2025) 15:2. doi: 10.1186/s13362-025-00167-y
49. Wodarz D. Hepatitis C. virus dynamics and pathology: the role of CTL and antibody responses. J General Virol. (2003) 84:1743–50. doi: 10.1099/vir.0.19118-0
50. Wodarz D. Mathematical models of immune effector responses to viral infections: Virus control versus the development of pathology. J Comput Appl Mathem. (2005) 184:301–19. doi: 10.1016/j.cam.2004.08.016
51. Webster G, Reignat S, Maini M, Whalley S, Ogg G, King A, et al. Incubation phase of acute hepatitis B in man: Dynamic of cellular immune mechanisms. Hepatology. (2000) 32:1117–24. doi: 10.1053/jhep.2000.19324
52. Mohri H, Bonhoeffer S, Monard S, Perelson AS, Ho DD. Rapid turnover of T lymphocytes in SIV-infected rhesus macaques. Science. (1998) 279:1223–7. doi: 10.1126/science.279.5354.1223
53. Dahari H, Lo A, Ribeiro R, Perelson A. Modeling hepatitis C virus dynamics: liver regeneration and critical drug efficacy. J Theoret Biol. (2007) 247:371–81. doi: 10.1016/j.jtbi.2007.03.006
54. Eikenberry S, Hews S, Nagy JD, Kuang Y. The dynamics of a delay model of hepatitis B virus infection with logistic hepatocyte growth. Mathem Biosci Eng. (2009) 6:283–99. doi: 10.3934/mbe.2009.6.283
55. Yousfi N, Hattaf K, Tridane A. Modeling the adaptive immune response in HBV infection. J Mathem Biol. (2011) 63:933–57. doi: 10.1007/s00285-010-0397-x
56. Perelson AS, Kirschner DE, De Boer R. Dynamics of HIV infection of CD4+ T cells. Mathem Biosci. (1993) 114:81–125. doi: 10.1016/0025-5564(93)90043-A
57. Wang Y, Zhou Y, Wu J, Heffernan J. Oscillatory viral dynamics in a delayed HIV pathogenesis model. Mathem Biosci. (2009) 219:104–12. doi: 10.1016/j.mbs.2009.03.003
58. Tsiang M, Rooney J, Toole J, Gibbs C. Biphasic clearance kinetics of hepatitis B virus from patients during adefovir dipivoxil therapy. Hepatology. (1999) 29:1863–9. doi: 10.1002/hep.510290626
59. Ciupe SM, Ribeiro RM, Nelson PW, Perelson AS. Modeling the mechanisms of acute hepatitis B virus infection. J Theor Biol. (2007) 247:23–35. doi: 10.1016/j.jtbi.2007.02.017
60. Harroudi S, Meskaf A, Allali K. Modelling the adaptive immune response in HBV infection model with HBV DNA-containing capsids. Differ Equ Dyn Syst. (2023) 31:371–93. doi: 10.1007/s12591-020-00549-1
61. Manna K Global properties of a HBV infection model with HBV DNA-containing capsids and CTL immune response. Int J Appl Comput Mathem. (2017) 3:2323–38. doi: 10.1007/s40819-016-0205-4
62. Shih Y, Liu C. Hepatitis C virus and hepatitis B virus co-infection. Viruses. (2020) 12:1–11. doi: 10.3390/v12070741
Keywords: HIV, HBV, co-infection, time delay, global dynamics, Lyapunov stability
Citation: Elaiw AM, Alhmadi AS and Hobiny AD (2025) Dynamics and stability of a within-host HIV-HBV co-infection model with time delays. Front. Appl. Math. Stat. 11:1633039. doi: 10.3389/fams.2025.1633039
Received: 22 May 2025; Accepted: 09 June 2025;
Published: 16 July 2025.
Edited by:
Khalid Hattaf, Centre Régional des Métiers de l'Education et de la Formation (CRMEF), MoroccoReviewed by:
Anthony O'Hare, University of Stirling, United KingdomAhmed Mohsen, University of Baghdad, Iraq
Copyright © 2025 Elaiw, Alhmadi and Hobiny. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed M. Elaiw, YWVsYWl3a3N1LmVkdS5zYUBrYXUuZWR1LnNh
 Ahmed M. Elaiw
Ahmed M. Elaiw Abdulaziz S. Alhmadi
Abdulaziz S. Alhmadi Aatef D. Hobiny
Aatef D. Hobiny