Abstract
Proper regulation of gene expression is required by bacterial pathogens to respond to continually changing environmental conditions and the host response during the infectious process. While transcriptional regulation is perhaps the most well understood form of controlling gene expression, recent studies have demonstrated the importance of post-transcriptional mechanisms of gene regulation that allow for more refined management of the bacterial response to host conditions. Yersinia species of bacteria are known to use various forms of post-transcriptional regulation for control of many virulence-associated genes. These include regulation by cis- and trans-acting small non-coding RNAs, RNA-binding proteins, RNases, and thermoswitches. The effects of these and other regulatory mechanisms on Yersinia physiology can be profound and have been shown to influence type III secretion, motility, biofilm formation, host cell invasion, intracellular survival and replication, and more. In this review, we discuss these and other post-transcriptional mechanisms and their influence on virulence gene regulation, with a particular emphasis on how these processes influence the virulence of Yersinia in the host.
Introduction
The three species of Yersinia pathogenic to humans encounter a variety of challenges throughout the course of their life cycles, including from the host immune system as well as from various environmental sources. Two of these species, Yersinia enterocolitica and Y. pseudotuberculosis, are gastrointestinal pathogens that are transmitted to mammals through the fecal-oral route from water and other natural reservoirs (Vantrappen et al., 1982; Rich et al., 1990). In these environmental niches they encounter a range of temperatures and pH as well as other potentially hostile microbes, to which Y. pseudotuberculosis and Y. enterocolitica must respond appropriately in order to survive and maintain homeostasis (Calvo et al., 1986; Pepe et al., 1994; Harrison et al., 2000; Palonen et al., 2010). Following ingestion by a mammalian host, the enteric Yersiniae must adapt to higher temperatures and pass through the acidic environment of the stomach before reaching the small intestine where invasion of the deeper tissue occurs (Miller and Falkow, 1988; Marra and Isberg, 1997; Nagel et al., 2001; Abdela et al., 2011). Upon invasion Y. enterocolitica and Y. pseudotuberculosis are confronted with host immune cells such as dendritic cells, macrophages, and neutrophils that the Yersiniae must resist in order to disseminate and propagate the infection (Simonet et al., 1985; Monack et al., 1998; Laws et al., 2011).
Similarly to its enteric ancestors, Y. pestis, the causative agent of plague, has two distinct phases of its life cycle (Perry and Fetherston, 1997; Gage and Kosoy, 2005). Y. pestis does not live freely in the soil or water, instead it typically colonizes fleas in order to be transmitted to mammals (Lorange et al., 2005), and maintains considerable genetic material dedicated to this part of its life cycle (Hinnebusch et al., 2002; Darby et al., 2005; Vadyvaloo et al., 2007; Sebbane et al., 2009; Chouikha and Hinnebusch, 2012). In addition, Y. pestis has been shown to colonize multiple mammalian organs, including the lymph nodes, spleen, lungs, and blood, and the infection of these distinct sites in the body results in the expression of different subsets of genes (Tieh and Landauer, 1948; Lathem et al., 2005; Sebbane et al., 2005; Lawson et al., 2006; Chauvaux et al., 2007).
The diversity and flux of conditions that Yersiniae must adapt to throughout their life cycles result in significant changes in metabolic, cell surface, and virulence factor gene expression, which are modulated through complex regulatory networks that allow the bacteria to respond appropriately and rapidly. These pathogenic Yersiniae, and bacteria in general, regulate gene transcription as a major means of controlling the cohort of factors expressed under any given condition. For instance, sigma factors, promoter sequences, and transcriptional activators and repressors all directly impact the frequency with which any particular gene is transcribed, and the contribution of transcriptional regulators to gene expression is directly linked to mechanisms that sense the environment and needs of the bacterium at any given moment (Cornelis et al., 1989, 1991; Iriarte et al., 1995a,b; Trulzsch et al., 2001; Nagel et al., 2003; Green and Darwin, 2004; Brzostek et al., 2007; Gao et al., 2008; Raczkowska et al., 2011). In addition, the production and use of bacterial virulence factors are energy intensive processes, and the Yersiniae must tightly control their synthesis in order to maximize survival, replication, and spread. For example, the Yop-Ysc type III secretion system (T3SS) of Yersinia, which is essential for mammalian virulence, requires a large energetic commitment from the bacteria for production and effector protein translocation, and inappropriate expression or mis-targeting of the system can have significant impact on the success of the bacteria in the host (Woestyn et al., 1994; Cornelis et al., 1998; Aepfelbacher and Heesemann, 2001; Blaylock et al., 2006; Shao, 2008). However, when the T3SS is deployed upon contact with host immune cells, the system prevents phagocytosis and activation of an effective immune response against the bacteria (Cornelis and Wolf-Watz, 1997; Shao, 2008). It is no surprise, then, that dozens of studies have revealed an intricate and multi-layered regulatory network that governs the expression and synthesis of the T3SS. Many of these studies have focused on the role of transcriptional regulation of this system; in recent years, however, there have been a number of reports that have elucidated post-transcriptional and post-translational mechanisms of regulation as well.
Post-transcriptional regulation is a key step in the control of bacterial gene expression, and our understanding of the mechanisms involved therein is rapidly growing. The regulatory elements involved in post-transcriptional regulation encompass any interaction of molecules with mRNA transcripts that affect translation of the message into protein products. This review focuses on the mechanisms that are specific to this step (as distinct from post-translational regulation, which occurs once protein has been synthesized). The post-transcriptional regulatory mechanisms of the Yersiniae include the use of RNA-binding proteins, small regulatory RNAs, other non-coding RNAs, thermosensors, RNases, and others. Post-transcriptional regulation provides a powerful way for the bacteria to more rapidly adjust to the changing environment during the Yersinia life cycle and to fine tune gene expression to the needs of the cell. Indeed, this is because translation can occur more quickly from existing transcripts rather than requiring de novo transcription. In this review we discuss specific examples of post-transcriptional regulation in Yersinia that may be involved in pathogenesis or other aspects of Yersinia physiology (Table 1) and we provide a comparative context for similar and/or divergent mechanisms in other pathogenic bacteria.
Table 1
| Name | Target | Function/mechanism | Reference |
|---|---|---|---|
| RNA-BINDING PROTEINS | |||
| YopD | 5′ UTR of yopQ in Y. enterocolitica (yopK); maybe others | Repression of transcription via ribosome competition or transcript degradation | Williams and Straley (1998), Chen and Anderson (2011) |
| LcrH | Same as YopD | Functions in complex with YopD | Anderson et al. (2002) |
| YscM1/YscM2 (LcrQ) | Not determined; predicted to be 5′ UTR of yop mRNAs | Negative regulation of T3SS; possibly same as YopD and/or with YopD | Cambronne and Schneewind (2002) |
| CsrA | GGA-motifs in the 5′ UTR; pleiotropic | Global carbon storage regulation; represses by ribosome competition or transcript degradation | Dubey et al. (2003), Baker et al. (2007), Heroven et al. (2008), Heroven et al. (2012) |
| SmpB | SsrA and A site of stalled ribosome | Ribosome rescue; molecular mimicry; enters into empty A site of a ribosome 1:1 ratio w/SsrA | Karzai et al. (1999), Okan et al. (2006), Okan et al. (2010), Neubauer et al. (2012) |
| Hfq | AU-rich regions of RNA; pleiotropic | sRNA chaperone; stabilizes interaction of sRNA w/mRNA | Nakao et al. (1995), Moller et al. (2002), Geng et al. (2009), Schiano et al. (2010) |
| NON-CODING RNAs | |||
| SsrA | Stalled ribosomes | Ribosome rescue; tRNA and mRNA Replaces incomplete transcript in ribosome; allows termination; tags for degradation | Karzai et al. (2000), Okan et al. (2006) |
| CsrB/CsrC | CsrA | Highly structured RNAs sequester CsrA w/multiple GGA-motifs | Liu et al. (1997), Romeo (1998), Heroven et al. (2012) |
| SgrS/Ysr150 | 5′ UTR of ptsG | Inhibits ptsG translation to stop the synthesis of new glucose transporters | Wadler and Vanderpool (2007), Horler and Vanderpool (2009), Wadler and Vanderpool (2009), Rice and Vanderpool (2011) |
| RybB/Ysr48 | 5′ UTR of many omp transcripts, incl. ompA, ompC, ompD, ompF, ompW | Regulates outer membrane protein composition; promotes accelerated mRNA degradation | Vogel and Papenfort (2006) |
| MicF | 5′ UTR of ompF | Same as RybB | Andersen et al. (1989), Schmidt et al. (1995), Delihas and Forst (2001), Delihas (2003), Vogel and Papenfort (2006) |
| MicA/Ysr7 | 5′ UTR of ompA | Same as RybB | Udekwu et al. (2005), Vogel and Papenfort (2006) |
| OmrA/Ysr149 | 5′ UTR of ompT | Same as RybB | Guillier and Gottesman (2006), Vogel and Papenfort (2006) |
| GlmY/Ysr147 | glmS transcript | Stabilizes glmS mRNA; positive regulation of cell wall synthesis | Kalamorz et al. (2007), Urban et al. (2007), Reichenbach et al. (2008), Gopel et al. (2011) |
| GlmZ/Ysr148 | GlmY – unknown if direct or indirect | Regulates amount of GlmY | Kalamorz et al. (2007), Urban et al. (2007), Reichenbach et al. (2008), Gopel et al. (2011) |
| GcvB/Ysr45 | dppA transcript | Repression of periplasmic-binding protein component of the dipeptide transport system | McArthur et al. (2006), Pulvermacher et al. (2008, 2009) |
| YenS | yenI transcript; potential secondary targets | Positive regulation of motility; inhibits translation and promotes degradation of yenI mRNA | Tsai and Winans (2011) |
| RyhB/Ysr48 | sdhCDAB operon transcript, others | Fur-repressed sRNAs; negative post-transcriptional regulation of targets | Masse and Gottesman (2002), Vecerek et al. (2003), Deng et al. (2012) |
| SraG | YPK_1206-05 operon transcript | Direct regulation; unknown function | Lu et al. (2012) |
| Ysr29 | ureC, ahpC, gst, frr, rpsA, ompA, groEL, dnaK | Hfq-dependent; negative regulation and positive regulation of different targets | Koo et al. (2011) |
| Ysr35 | Unknown | Role in virulence of Y. pestis and Y. pseudotuberculosis | Koo et al. (2011) |
| Antisense to pla | pla | Unknown; predicted to repress translation of Pla | Sodeinde and Goguen (1989) |
| Yp-sR7 | 50 s Ribosomal protein (rplK) | Expressed in exponential phase; cis-encoded; unknown function | Qu et al. (2012) |
| Yp-sR3 | YP_1329 and YP_1330 | Expressed in stationary phase; cis-encoded across operon; unknown function. May regulate these putative membrane proteins | Qu et al. (2012) |
| Yp-sR8 | Unknown | Expressed in stationary phase; unknown function | Qu et al. (2012) |
| RNases | |||
| RNase E | Many; T3SS | Inhibits export/secretion of T3SS effector proteins | Yang et al. (2008) |
| PNPase | Many; T3SS | Same as RNase E | Rosenzweig et al. (2005), Rosenzweig et al. (2007), Rosenzweig and Schesser (2007) |
| THERMOSENSORS | |||
| yscW-lcrF intergenic region | lcrF (virF) transcript; cis-acting RNA | Two-stem loop structure restricts access of ribosome to SD sequence at 25°C; but not at 37°C. Proper function required for virulence | Hoe and Goguen (1993), Bohme et al. (2012) |
| RIBOSWITCHES | |||
| mgtA | 5′ UTR of mgtA | High Mg2+ concentration leads to early Rho-dependent termination of mgtA transcription through conformational change in the RNA; regulates magnesium transporter production | Korth and Sigel (2012) |
Post-transcriptional regulators, targets, and functions in Yersinia spp.
Regulatory RNA-Binding Proteins
The ribosome is the primary RNA-binding protein in the cell and is required for the translation of mRNA into protein. However, there are other proteins that are known to bind to mRNA, and by doing so regulate translation initiation, mRNA stability, and half-life of the message (Anderson et al., 2002; Brennan and Link, 2007). These proteins often compete with the ribosome for binding to exert their regulatory effects (Anderson et al., 2002; Wang et al., 2005).
T3SS regulators
In Yersinia there have been several RNA-binding proteins implicated in the regulation of the T3SS. One of these proteins is YopD, which works in conjunction with YopB to form a pore in the eukaryotic target cell membrane that permits the translocation of T3S effectors into the host cell (Hakansson et al., 1993; Tardy et al., 1999). In addition to its role in translocation, YopD has also been implicated in the negative regulation of the so-called low calcium response (LCR; Williams and Straley, 1998). In Y. enterocolitica YopD, together with secretion chaperone LcrH, has been shown to bind yop mRNA in the 5′ untranslated region (5′ UTR) of yopQ transcripts (known as yopK in Y. pestis; Anderson et al., 2002). It has been proposed that this binding represses translation from yopQ mRNA, perhaps by promoting the degradation of the transcript or by competing with the ribosome for binding (Chen and Anderson, 2011). Furthermore, the half-life of yopH, yopE, and yscB transcripts are longer in a yopD deletion mutant of Y. pestis compared to wild-type bacteria when grown at 37°C in the presence of calcium, suggesting that the post-transcriptional control of secreted effectors by YopD is not limited to yopQ/K. The binding of the YopD-LcrH complex to target transcripts requires two specific AU-rich regions of mRNA that is common to many, but not all yop transcripts. The distance of the AU-rich regions from the Shine–Dalgarno site may affect the affinity of YopD for the transcript, suggesting a mechanism for a hierarchy of translation (Figure 1; Chen and Anderson, 2011). Interestingly, these AU-rich regions are not sufficient to confer YopD-LcrH binding to pla (plasminogen activator protease) mRNA in vivo, nor does mutation of these regions to a more GC-rich content abolish YopD-LcrH binding to yopH transcripts in vitro. This suggests that the post-transcriptional repression of yops is more complex than the effects of these two proteins and the AU-rich regions alone (Chen and Anderson, 2011).
Figure 1
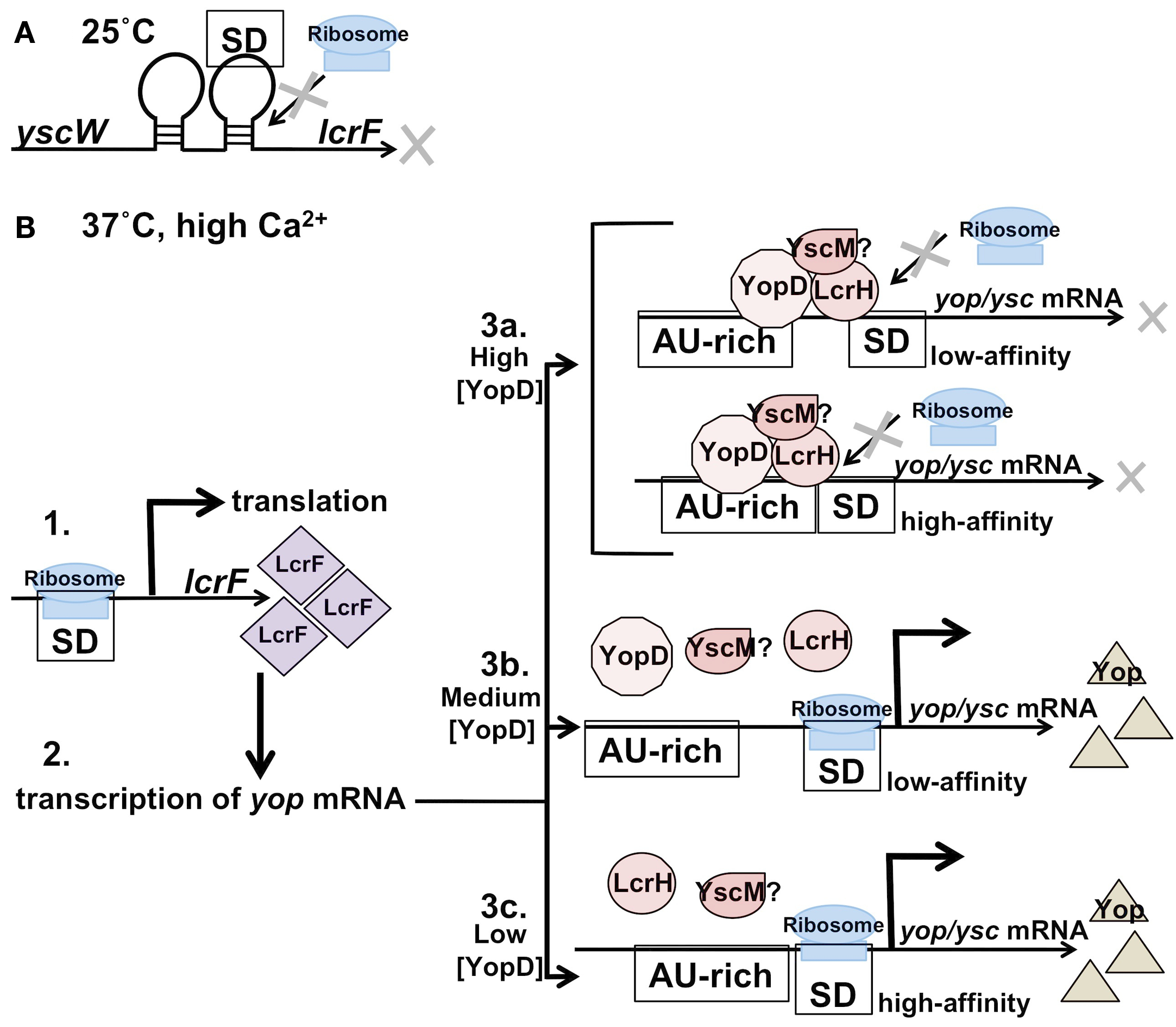
Post-transcriptional regulation of the Yersiniayop-ysc T3SS: LcrF thermosensor and the YopD-LcrH complex. (A) At 25°C the T3SS is repressed at the transcriptional level. The thermosensor in the UTR between the yscW and lcrF transcriptis in the closed conformation at 25°C, which prevents ribosome binding and translation initiation of lcrF. (B) 1. At 37°C the thermosensor of lcrF is in the open conformation, allowing translation, and production of LcrF to proceed. 2. LcrF activates transcription of yop/ysc genes. 3. Translation of a cohort of yop/ysc mRNAs is repressed by a YopD-LcrH (YscM) complex binding in the 5′ UTR until favorable translocation conditions exist. 3a. Under high YopD concentrations (little-to-no secretion exists) the YopD complex binds to both “high-affinity” and “low-affinity” AU-rich regions of specific yop transcripts. Intracellular YopD levels decrease as secretion is initiated. 3b. Under medium YopD concentrations the “low-affinity” sites are released and translation proceeds. 3c. Under low YopD concentrations the “high-affinity” sites are also released and translation of these proteins proceeds. This model suggests a hierarchy for yop/ysc translation that relies on the concentration of YopD in the cell, which in turn is dependent on extracellular conditions.
In addition, YscM1 and YscM2 (called LcrQ in Y. pestis and Y. pseudotuberculosis), which have previously been shown to be negative regulators of the T3SS via a mechanism of feedback inhibition, have also been implicated in the post-transcriptional regulation of yop genes in Y. enterocolitica (Allaoui et al., 1995; Cambronne and Schneewind, 2002). It has been proposed that the post-transcriptional regulation of the T3SS by the YscM proteins may occur through a mechanism similar to that of YopD-LcrH, and that they may function together with YopD-LcrH to prevent Yop translation (Cambronne and Schneewind, 2002).
Repression of effector Yop translation by YopD-LcrH/YscM1/YscM2 may play a significant role in allowing Yersinia to prepare for the translocation of effectors almost immediately upon host cell contact by transcribing low levels of required mRNA, while preventing translation of these effectors before the appropriate time. Although the transcription of yops is not strongly activated under non-secretion-inducing conditions, low-level transcription of these genes does occur at 37°C (Cornelis and Wolf-Watz, 1997), which leaves a gap in regulation that may be filled by YopD. During the course of infection Yersinia species must carefully balance energy use for growth versus energy use for production of virulence determinants in order to maximize survival and spread, which may explain why this system is so tightly regulated at the transcriptional, post-transcriptional, and post-translational levels. YopD has homologs in nearly every other bacterial pathogen that harbors a T3SS, including Aeromonas, Pseudomonas, and Vibrio species, although no RNA-binding post-transcriptional role for the YopD orthologs has yet been described.
Csr system
The RNA-binding protein CsrA is the central component of the global carbon storage regulatory system and has been characterized in a number of bacterial pathogens (Altier et al., 2000; Lenz et al., 2005; Heroven et al., 2008; Brencic and Lory, 2009). The Csr system is involved in the regulation of many factors involved in both general metabolism as well as virulence and the response to stress (Timmermans and Van Melderen, 2010). In Y. pseudotuberculosis, transcription of csrA itself is induced in stationary phase (Heroven et al., 2008). Post-transcriptional regulation by CsrA occurs through the binding of CsrA to nucleotides near the Shine–Dalgarno region of target mRNAs (Liu and Romeo, 1997; Baker et al., 2007). Generally, CsrA binds to GGA-motifs in the 5′ UTR and represses translation by competing with the 30 s ribosomal subunit (Dubey et al., 2003, 2005; Schubert et al., 2007). This also results in accelerated mRNA degradation (Liu et al., 1995). Two highly structured small non-coding regulatory RNA (sRNA) molecules, CsrB and CsrC, control the levels of free CsrA in the bacterial cell, and only CsrA not complexed with CsrB/C can bind to and regulate target mRNA (Liu et al., 1997; Romeo, 1998). In Yersinia, csrB is transcriptionally regulated by the two-component system BarA/UvrY in response to unknown extracellular signals that likely include nutrient availability, while the mechanism of csrC transcriptional regulation in Yersinia is not yet known (Heroven et al., 2008). Both CsrB and CsrC possess multiple GGA-motifs that are able to bind CsrA, which then derepresses the post-transcriptional targets of CsrA (Heroven et al., 2008). The sequences of csrB and csrC in Y. pseudotuberculosis are not well conserved with respect to E. coli and Salmonella, and seem to have a more complex transcriptional regulation than in these species (Heroven et al., 2012).
Microarray analysis of a csrA deletion mutant of Y. pseudotuberculosis compared with wild-type bacteria showed that CsrA influences, either directly or indirectly, the transcript levels of approximately 500 open reading frames, 3% of which are virulence- or stress-associated (Heroven et al., 2012). Phenotypically, CsrA mutants of Y. pseudotuberculosis are aflagellate and non-motile (Heroven et al., 2012). While it is not yet known if a csrA mutant in Yersinia is attenuated in animal models of infection, the microarray study suggests that csrA may regulate host cell invasion, which is a key step in virulence (Heroven et al., 2012). Finally, CsrA regulates the global transcriptional regulator RovA of Y. pseudotuberculosis, thereby indirectly controlling the expression of a number of genes, some of which are involved in virulence (Cathelyn et al., 2006; Heroven et al., 2008). With respect to other pathogens, CsrA has been shown to play a role in the regulation of Legionella pneumophila type IV secretion (Rasis and Segal, 2009), a key virulence determinant, and likely contributes to the virulence of other bacterial species as well.
Trans-translation
The small protein SmpB is a unique RNA-binding protein that functions in tandem with the small RNA SsrA to rescue stalled ribosomes from incomplete mRNA transcripts (Komine et al., 1994; Keiler et al., 1996; Karzai et al., 2000). Known as trans-translation, this is a vital process for bacteria as stalled ribosomes will stay on transcripts indefinitely and cannot be recycled for new rounds of protein synthesis, which would thereby result in growth arrest and the eventual death of the cell. The RNA molecule SsrA serves as a substitute message for the ribosome, however without SmpB, SsrA is unable to enter the ribosome to participate in trans-translation (Karzai et al., 1999). SmpB binds to aminoacylated-SsrA in a 1:1 ratio that structurally mimics a tRNA where SmpB acts as the anticodon loop, and this faux tRNA enters into the empty A site of a stalled ribosome (Neubauer et al., 2012). This is followed by a transpeptidation reaction linking the unfinished polypeptide chain to SsrA (Karzai et al., 2000). The errant mRNA is subsequently replaced by SsrA, which allows translation to continue until the ribosome reaches the built-in stop codon (Karzai et al., 2000). This results in the addition of an 11-amino acid residue tag to the end of the incomplete protein that marks it for degradation by intracellular proteases (Karzai et al., 2000). In E. coli RNase R is the RNase responsible for degradation of these errant mRNA molecules in an SmpB-SsrA-dependent manner (Richards et al., 2006). Normal (unstalled) ribosomes cannot accommodate SmpB due to the structure of SmpB, thereby ensuring only stalled ribosomes are affected (Neubauer et al., 2012). The C-terminal tail of SmpB is required for binding to the SsrA RNA (Miller et al., 2011). While SmpB plays a key role in ribosome rescue, more recently SmpB has been postulated to contribute to other forms of post-transcriptional regulation, although the mechanism remains elusive (Ansong et al., 2009). In addition to general ribosome recycling, SmpB-SsrA also may play an active role in post-transcriptional gene regulation (Abo et al., 2000; Withey and Friedman, 2002). One such example is seen in E. coli in which the binding of LacI to the lac operator results in incomplete lacI transcripts, which are then acted upon by SmpB-SsrA to ultimately result in the degradation of the transcript, thereby reducing the production of LacI. This is hypothesized to support rapid induction of lac operon expression in response to lactose availability (Abo et al., 2000).
Disruption of ssrA or smpB results in growth defects in many bacterial species under various conditions, including E. coli, Salmonella, Neisseria gonorrhoeae, Mycobacterium genitalium, and M. pneumoniae (Karzai et al., 2000). In Y. pseudotuberculosis, SsrA has been shown to contribute to the pathogenesis in a mouse model of infection (Okan et al., 2006), and similarly, SsrA mutants of Francisella tularensis and Salmonella are also defective for virulence (Ansong et al., 2009; Svetlanov et al., 2012). In vitro, Y. pseudotuberculosis lacking SmpB and SsrA is more sensitive to sublethal concentrations of translation-specific antibiotics such as streptomycin and chloramphenicol, and in tissue culture models of infection, this mutant shows a twofold decrease in intracellular survival in macrophages and significantly delayed cytotoxicity toward HeLa cells (Okan et al., 2006). Furthermore, transcription of the T3SS master regulator gene lcrF/virF is reduced by 50% in the absence of SmpB-SsrA and is not significantly up-regulated under secretion-inducing conditions (Okan et al., 2006). The exact mechanism by which SsrA regulates LcrF/VirF is unknown, although it has been proposed that SsrA may exert its effects by tagging for degradation the transcript of an unknown transcriptional repressor; hence in an ssrA-deficient strain the repressor would accumulate and prevent transcriptional activation of T3SS genes by LcrF. SsrA is also critical for the virulence of Y. pestis by both the intranasal and intravenous routes (Okan et al., 2010). Mice inoculated intranasally with Y. pestis lacking ssrA survived a secondary challenge with fully virulent Y. pestis, suggesting that this mutant could serve as a potential vaccine candidate to prevent pneumonic plague (Okan et al., 2010).
Hfq
The small RNA chaperone protein Hfq is a pleiotropic RNA-binding protein that plays a central role in the post-transcriptional regulation of large numbers of genes in bacteria. This protein is small (101 amino acids in Yersinia), and forms a homohexameric ring complex that allows Hfq to bind more than one RNA molecule simultaneously (Kajitani and Ishihama, 1991; Vytvytska et al., 1998; Sauter et al., 2003). Although not ubiquitous, Hfq has been identified in many bacterial species and is known to bind AU-rich regions of RNA (Sun et al., 2002; Zhang et al., 2002; Sobrero and Valverde, 2012). Binding of RNA to Hfq can either stabilize or promote the degradation of mRNA transcripts, depending on the specific interaction (Vytvytska et al., 1998; Masse et al., 2003; Meibom et al., 2009). Furthermore, Hfq also binds to sRNA molecules and enhances the RNA–RNA interaction between the sRNA and its target mRNA (Moller et al., 2002; Zhang et al., 2002; Vecerek et al., 2003). Hfq has been shown to be essential for the virulence of both Y. pestis and Y. pseudotuberculosis in mouse models of bubonic plague and Yersiniosis, respectively (Geng et al., 2009; Schiano et al., 2010). Although no study has yet been performed to evaluate the effects of an hfq deletion in Y. enterocolitica, Hfq has been implicated in the negative regulation of the heat stable enterotoxin gene yst and thus it is quite likely that Hfq plays a role in the virulence of this pathogen as well (Nakao et al., 1995).
The pleiotropic nature of Hfq, in that it facilitates many different sRNA-mRNA interactions, has made it difficult to elucidate all of the sRNAs and targets responsible for the Hfq-dependent phenotypes that have been observed so far. The loss of Hfq from Y. pestis leads to a significant growth defect when cultured in vitro at 37°C, but surprisingly the same growth defect is not observed for Y. pseudotuberculosis (Bai et al., 2010; Schiano et al., 2010; Bellows et al., 2012). Growth defects of hfq mutants in other bacterial species (Brucella abortus, Vibrio cholerae, and Salmonella) are more similar to the minor defects observed in vitro for Y. pseudotuberculosis, while more pronounced defects are seen in vivo (Robertson and Roop, 1999; Ding et al., 2004; Sittka et al., 2007). Consistent with these data, both Y. pestis and Y. pseudotuberculosis lacking hfq are impaired for intracellular survival in tissue culture cells compared to wild-type, suggesting that one or more Hfq-dependent sRNAs may directly or indirectly regulate critical factors required for intracellular survival (Geng et al., 2009; Schiano et al., 2010). Additionally, there is a significant reduction in T3SS effector protein production in the absence of Hfq in Y. pseudotuberculosis, which suggests that one or more sRNAs positively regulate the Yersinia T3SS (Figure 2; Schiano et al., 2010). Taken together, these data suggest that Yersinia require Hfq to with-stand the stressful conditions found within the mammalian host, although the Hfq-dependent virulence defects in the mammal may be due to general growth deficiencies in addition to the loss of properly functioning virulence determinants. Similarly, Hfq has been implicated in the virulence of other pathogens (Christiansen et al., 2004; Sittka et al., 2007; Kulesus et al., 2008; Fantappie et al., 2009; Meibom et al., 2009; Kendall et al., 2011). For example in Salmonella, HilD is positively regulated post-transcriptionally in an Hfq-dependent manner, but it is not yet known if the regulation is indirect through an RNA-binding protein or direct through association with an sRNA (Ellermeier and Slauch, 2007; Sittka et al., 2008). Hfq has been implicated in the regulation of T3SS in other bacteria as well, which suggests that the involvement of Hfq-dependent sRNAs here is a conserved theme in bacterial pathogens (Pfeiffer et al., 2007; Mitobe et al., 2009; Shakhnovich et al., 2009; Kendall et al., 2011).
Figure 2
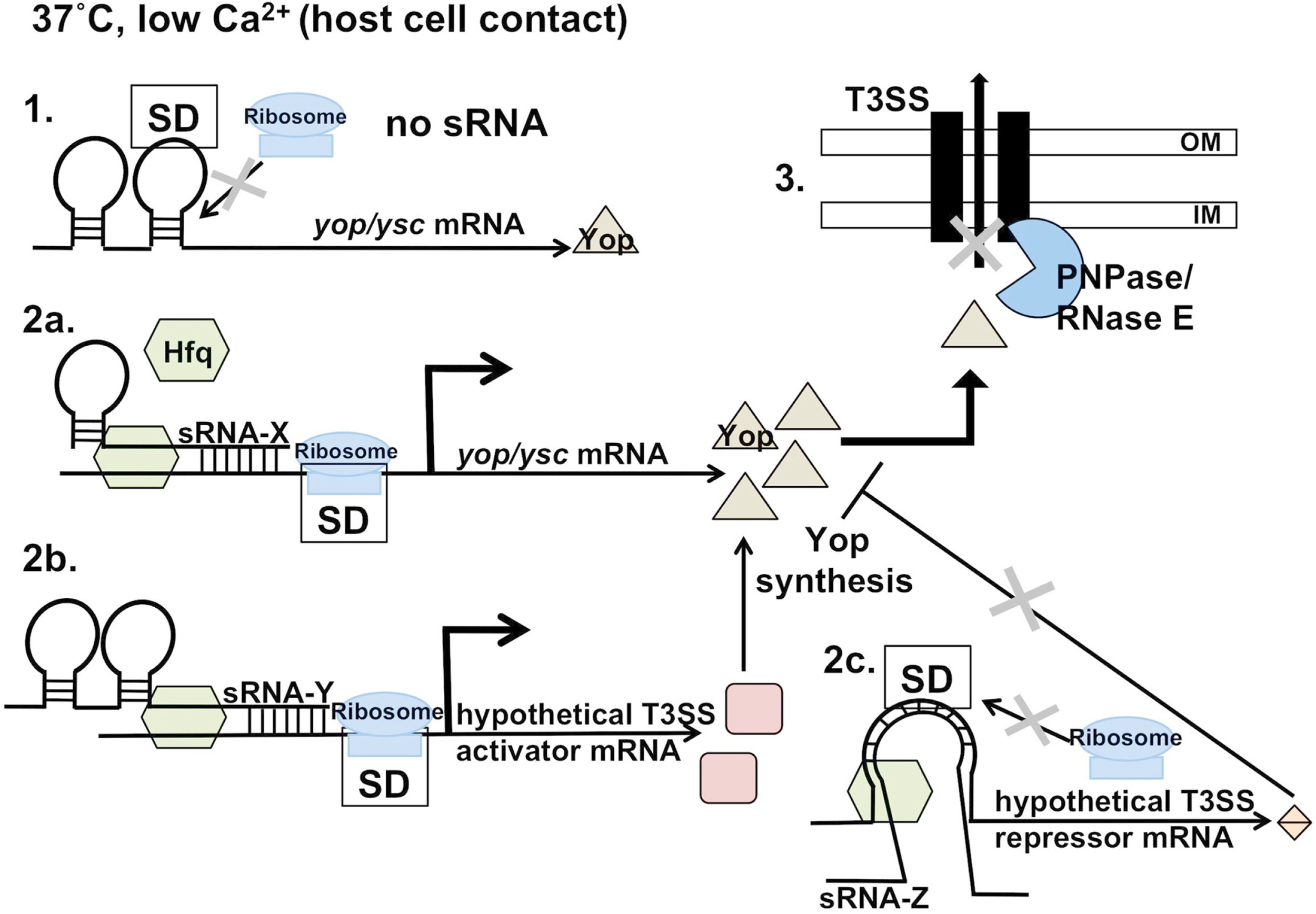
Hfq-mediated regulation of Yop/Ysc production and ribonuclease-mediated secretion. Upon host cell contact (or in vitro in low calcium concentrations), Yop production, and secretion/translocation occurs. 1. In the absence of one or more hypothetical, Hfq-dependent sRNAs, Yop production occurs at a lower level. 2a. Hfq-dependent sRNA stimulation of translation or stabilization of the transcript may be direct, through binding of the sRNA to the 5′ UTR of the mRNA, leading to increased Yop production. 2b. Alternatively, regulation may be indirect through Hfq-dependent stimulation/stabilization of a secondary post-transcriptional activator of the T3SS, or 2c. Hfq-dependent repression/degradation of a post-transcriptional repressor of T3SS. Both of these scenarios would result in increased Yop production in an sRNA-dependent, post-transcriptional manner. Upon Yop production, PNPase and RNase E may block secretion/translocation of the Yop at a terminal step if conditions are non-ideal.
As with other pathogens, the number of genes regulated through Hfq in Yersinia species is likely to be large. Microarray studies on the Hfq-affected transcriptome in Y. pestis revealed 243 genes with twofold or greater difference in transcript levels in the absence of hfq compared to wild-type bacteria (Geng et al., 2009). Of these, 139 were down-regulated and 104 were up-regulated, and genes belonging to metabolism functional classes are overrepresented in the transcriptome when compared to the genome at large (Geng et al., 2009). This study also found that 23% of these transcripts are related to pathogenicity or stress response processes, including the plasminogen activator protease gene pla, the F1 antigen gene cafI, the diguanylate cyclase gene hmsT, and half of the genes of the T3SS (Geng et al., 2009). Interestingly, the abundance of these T3SS-associated transcripts is elevated in the absence of hfq, suggesting that Hfq may be a negative regulator of these genes in Y. pestis; this, however, is in contrast to the positive regulation of T3S effector protein levels by Hfq in Y. pseudotuberculosis (Geng et al., 2009; Schiano et al., 2010). Hfq-dependent regulation of the T3SS may be divergent between the two species, but a more likely explanation is that in the absence of Hfq, decreased Yop protein production leads to increased transcription of yop mRNA through a feedback loop (Allaoui et al., 1995). In addition, there may be other targets of Hfq not identified by this study, as Hfq functions at the post-transcriptional level and thus its absence may not alter the transcript abundance of every Hfq-regulated gene (Koo et al., 2011).
Hfq also contributes to other virulence-associated phenotypes in the Yersiniae. For instance, Hfq represses the non-flagellar-dependent, swarming motility of Y. pseudotuberculosis, as a deletion mutant of hfq is hyper-motile on low percentage agar plates (although interestingly this phenomenon is not observed in Y. pestis; Schiano et al., 2010). The regulation of this form of motility via Hfq may play a role for Y. pseudotuberculosis and perhaps Y. enterocolitica during their free-living phase in environment, or may contribute to the cessation of swarming upon entering the mammalian host. Indeed, Hfq has also been implicated in the non-mammalian phase of the Y. pestis life cycle. Recently it was shown that Hfq is required for biofilm production in the proventriculus of the flea (Rempe et al., 2012), which suggests that Hfq enables the efficient transmission of Y. pestis from the flea vector to the mammalian host (Jarrett et al., 2004). In vitro, Hfq represses biofilm formation through the reduction of cyclic diguanylate (c-di-GMP) levels by reciprocally regulating the abundance of both HmsP and HmsT, which are the c-di-GMP phosphodiesterase and major diguanylate cyclase, respectively (Bellows et al., 2012). Hfq contributes to the regulation of HmsP at the transcriptional level, while HmsT is directly regulated via Hfq at the post-transcriptional level (Figure 3; Bellows et al., 2012). These coordinate but distinct regulatory processes may allow Y. pestis to link the post-transcriptional control of biofilm formation to environmental sensing, although it is not yet known whether this occurs in the flea, the mammal, or both. Hfq has also been implicated in the regulation of biofilms produced by uropathogenic E. coli and V. cholerae, suggesting that the contribution of Hfq to biofilm formation, though perhaps not the mechanism, is a conserved one in the bacterial world (Kulesus et al., 2008; Bardill et al., 2011).
Figure 3
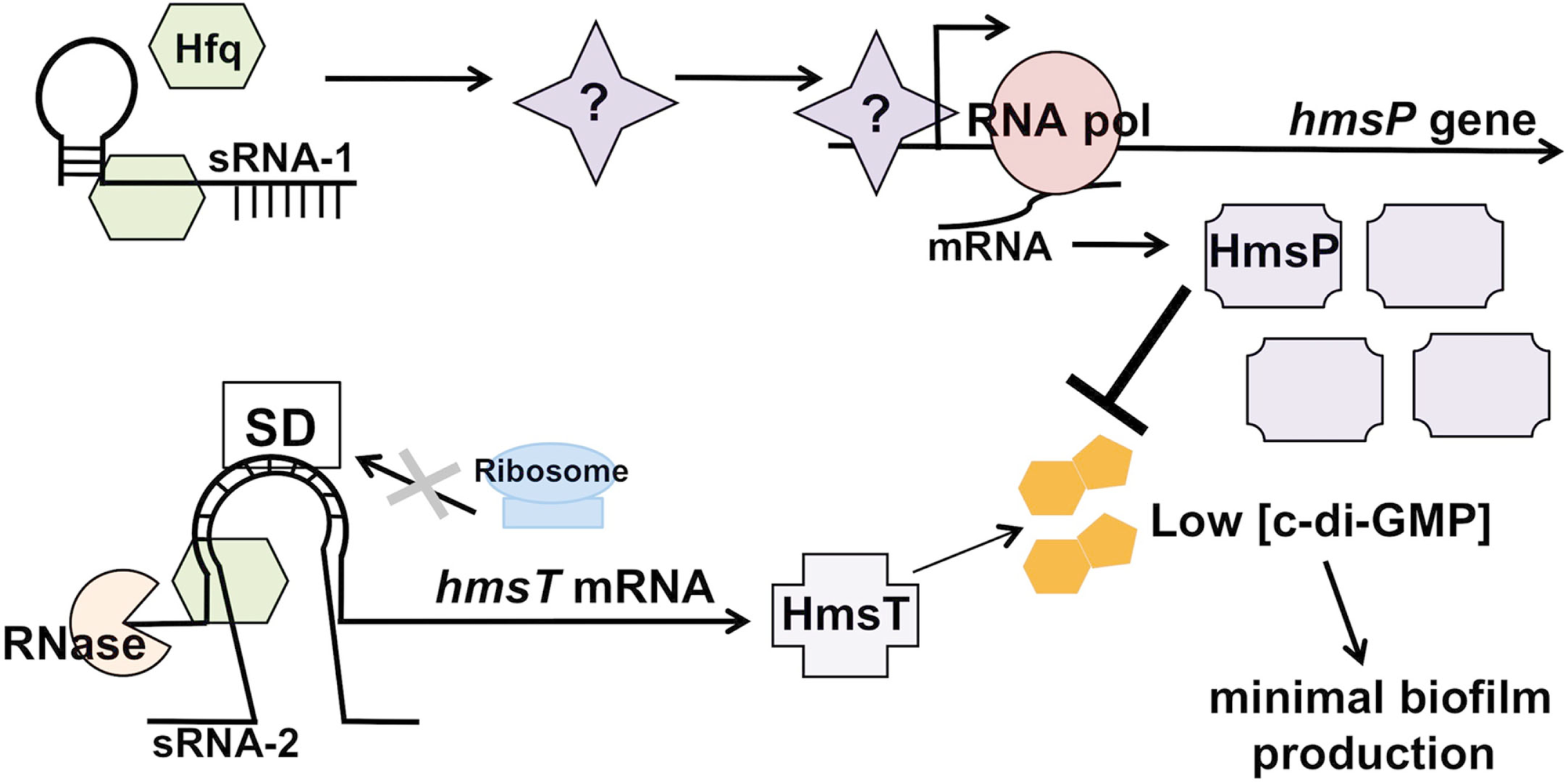
Hfq-dependent regulation of biofilm formation in Yersinia pestis. Hfq, in conjunction with one or more hypothetical sRNAs, is predicted to indirectly affect transcript levels of hmsP by either increasing the levels of a transcriptional activator or by decreasing levels of a repressor, thereby stimulating the synthesis of the c-di-GMP phosphodiesterase HmsP. Simultaneously, Hfq (and presumably a cognate hmsT-binding sRNA) represses translation of the transcript encoding the diguanylate cyclase HmsT through increasing transcript degradation and possibly blocking translation initiation. These coordinately regulated activities result in low c-di-GMP levels, leading to minimal biofilm production by Y. pestis.
Non-Coding RNAs
Non-coding RNAs encompass a large and diverse group of RNA molecules that do not result in the translation of a protein product. Instead, many are used in regulatory roles or other functional capacities upon transcription.
Cis- and trans-acting sRNAs
A major class of non-coding RNAs includes the trans-acting sRNAs, which by definition are encoded at genomic loci distal to those of their targets. These small RNAs have recently become appreciated for their roles in the post-transcriptional regulation of many bacterial genes, including virulence-associated genes in both Yersinia and other bacterial pathogens. In general, trans-acting sRNAs range in size from 50 to 500 nt in length, are highly structured, and usually base pair with target mRNA within the 5′ UTR to either repress or stimulate translation (Gottesman, 2005). The ability of the sRNA to base pair with its target is the basis for this type of regulation, although this base pairing is usually imperfect over the span of 7–10 nt (the “seed” sequence; Gottesman, 2005). Additionally, trans-acting sRNAs often require the chaperone protein Hfq for their expression, stability, and/or function. Hfq is thought to simultaneously bind to both the sRNA and mRNA, and in so doing facilitate the interaction of sRNAs with their targets. Alternately, cis-acting sRNAs generally do not require Hfq for base pairing as their genomic locations, usually antisense to their target mRNAs, allow for extensive Watson–Crick interactions between the sRNA and target (Gottesman and Storz, 2011). Several publications identifying various cohorts of non-coding sRNAs expressed by Yersiniae have recently been published. Using a deep sequencing-based approach, Koo et al. (2011) identified 150 previously unannotated sRNAs in Y. pseudotuberculosis (termed Ysrs for Yersinia small RNAs), and Qu et al. (2012) used cDNA cloning for the identification of 43 sRNAs in Y. pestis, 25 of which are predicted to be cis-acting. While the functions of most of these sRNAs have not yet been elucidated, those that have and other specific examples are discussed here.
sRNAs/Ysrs that target the outer membrane
The synthesis of outer membrane proteins is often highly regulated at the post-transcriptional level. RybB is a well conserved Hfq-binding sRNA in the enterobacteriaceae that has been shown in both E. coli and Salmonella to down-regulate the production of multiple outer membrane proteins including OmpA, OmpC, OmpD, OmpF, OmpW, and others by promoting accelerated mRNA degradation (Johansen et al., 2006; Papenfort et al., 2006). This sRNA (also known as Ysr48 in Yersinia) is present in all three pathogenic Yersinia species, but does not seem to play a major role in the pathogenesis of either Y. pestis or Y. pseudotuberculosis in mouse models of infection (although a more subtle role for RybB/Ysr48 in Yersinia virulence cannot not be ruled out; Koo et al., 2011). While the targets of RybB/Ysr48 in Yersiniae have not yet been identified, as many of the RybB-regulated genes in other species are conserved in Yersinia there is likely to be some overlap.
MicF is another highly conserved sRNA of the enterobacteriaceae that regulates the outer membrane protein OmpF in Yersinia species. The mechanism of MicF regulation is well understood in E. coli, with an approximately 20 base pair imperfect RNA duplex that forms within the Shine–Dalgarno region of the ompF mRNA (Schmidt et al., 1995). This base pairing prevents translation of OmpF and may lead to the degradation of the message (Andersen et al., 1989; Delihas and Forst, 2001). The binding of MicF to ompF in Yersinia is predicted to be very similar, with the same expected outcome of repression by translation inhibition and degradation of the ompF transcript (Delihas, 2003). Two other conserved sRNAs, MicA/Ysr7, and OmrA/Ysr149 that (based on their homologies with other species) are also predicted to target mRNAs encoding conserved outer membrane proteins in Yersinia (Udekwu et al., 2005; Guillier and Gottesman, 2006; Vogel and Papenfort, 2006). These sRNAs are the most abundantly expressed Ysrs during the invitro growth of Y. pseudotuberculosis at 37°C (Koo et al., 2011), although their contributions to the biology of the Yersiniae have not yet been established.
GcvB/Ysr45
The conserved sRNA gene gcvB/Ysr45 encodes not one but two sRNAs in Yersinia, and these sRNAs have been shown to be repressors of dppA, which encodes the periplasmic-binding protein component of the dipeptide transport system in Y. pestis (McArthur et al., 2006). The two GcvB molecules have different termination sites and are 130 and 206 nt in length, and in E. coli the 206-nt form is required for repression of dppA; whether this is conserved in Yersinia remains to be determined (Urbanowski et al., 2000; McArthur et al., 2006). As the regulation of dppA by GcvB occurs post-transcriptionally in E. coli (Pulvermacher et al., 2008, 2009), it is likely that the regulation of dppA in Yersinia is similar. Regulation of the dipeptide transport system in this way could play a role in the Yersinia adaptation to stress, as in E. coli the modulation of dppA occurs in response to environmental signals (Olson et al., 1991). A gcvB/Ysr45 deletion in Y. pestis results in changes both in colony morphology and generation time, suggesting that GcvB/Ysr45 may regulate multiple targets directly or indirectly (McArthur et al., 2006). As opposed to the high levels of the MicA/Ysr7 and OmrA/Ysr149 sRNAs at 37°C, when Y. pseudotuberculosis is cultured in vitro at lower temperatures (26°C), GcvB/Ysr45 is the most highly up-regulated Yersinia Ysr, although the implications of this are as yet unknown (Koo et al., 2011).
GlmY/Ysr147 and GlmZ/Ysr148
In Y. pseudotuberculosis there exists an interesting sRNA-target relationship between glmS, which encodes glucosamine-6-phosphate, a key enzyme in the cell wall synthesis pathway, and the sRNAs GlmY/Ysr147 and GlmZ/Ysr148 (Gopel et al., 2011). GlmZ/Ysr148 stabilizes the glmS transcript and therefore acts as a positive post-transcriptional regulator of cell wall synthesis, while GlmY/Ysr147 regulates the levels of GlmZ/Ysr148 through an unknown mechanism (Kalamorz et al., 2007; Reichenbach et al., 2008). This method of glmS regulation by GlmZ is highly conserved among the enterobacteriaceae (Urban et al., 2007). Based on the sheer number of targets and the evolutionary conservation of sRNAs that are involved in the regulation of outer membrane proteins, it is clear that the rapid adjustment of the outer membrane is a priority for bacterial species, and that post-transcriptional regulation mediated by these sRNAs is a major mechanism by which to accomplish such regulation.
YenS
In Y. enterocolitica, the sRNA gene yenS encodes two small, non-translated RNAs of 165 and 105 nt that have different termination sites but share the same 5′ end (Tsai and Winans, 2011). These two sRNAs were found to inhibit translation and promote degradation of the yenI mRNA. YenI is homologous to the autoinducer synthetase LuxI of Vibrio fischeri and produces the quorum sensing molecule 3-oxohexanoylhomoserine lactone (OHHL), which at high levels inhibits swarming motility (Shadel et al., 1990; Tsai and Winans, 2011). Thus, the YenS sRNA is an indirect positive regulator of motility in Y. enterocolitica that acts through the modulation of YenI production. Transcription of yenS is activated in response to low levels of OHHL through YenR, which is homologous to the response regulator LuxR in V. fischeri (Dunlap and Ray, 1989). Interestingly, in a yenI null strain that is hyper-motile, a second deletion in yenS suppresses the hyper-motility phenotype, suggesting a potential secondary role for YenS in the regulation of motility in Y. enterocolitica (Tsai and Winans, 2011). A similar non-coding RNA was found in the plant pathogen Pantoea stewartii subsp. stewartii that is regulated by the LuxR homolog in an OHHL-dependent way, suggesting a conservation of sRNA regulation by YenS in quorum sensing (Schu et al., 2009). While the genomes of Y. pestis and Y. pseudotuberculosis also encode homologs of the yenS gene, this sRNA was not identified during a global screen for sRNAs in Y. pseudotuberculosis; it is possible that YenS may not be expressed under the conditions examined in the study or did not meet the filtering/threshold criteria to be identified in the survey (Koo et al., 2011; Tsai and Winans, 2011). Post-transcriptional regulation of quorum sensing may be a common theme among bacterial pathogens and symbionts, as small non-coding RNAs have also been implicated in the regulation of quorum sensing in V. harveyi and V. cholera (albeit by a seemingly different mechanism; Lenz et al., 2004; Svenningsen et al., 2009; Tu et al., 2010).
RhyB/Ysr48
Iron acquisition is a key concern for bacterial pathogens; therefore it is not surprising that the enterobacteriaceae possess sRNAs that participate in the regulation of genes involved in such systems (Masse and Gottesman, 2002; Nairz et al., 2010). One such sRNA is RhyB, an Hfq-dependent, Fur-repressed sRNA responsible for the post-transcriptional regulation of the sdhCDAB operon and five other genes in E. coli and V. cholerae (Masse and Gottesman, 2002; Davis et al., 2005). The products of these genes are all iron-storing or iron-using and under low iron conditions, RyhB acts as a negative regulator to repress their translation. Two copies of this sRNA gene, RhyB1/Ysr48.1 and RhyB2/Ysr48.2, are found in Yersinia species, and both are thought to function similarly to the canonical RhyB of E. coli. Deng et al. (2012) recently showed that although RhyB1/Ysr48.1 requires Hfq for its stability, RhyB2/Ysr48.2 does not, and that the expression of both sRNAs is highly up-regulated in the lungs of mice infected with Y. pestis, known to be an iron-limiting environment. However, deletions of these sRNAs do not result in decreased bacterial burden in the lungs of infected mice, suggesting that they may not play a vital role in iron acquisition by Y. pestis during pneumonic plague. It remains unclear as to why Y. pestis carries two copies of RhyB/Ysr48 and if there is a biological consequence for this gene duplication during infection of either the mammal or the flea.
SgrS/Ysr150
An unusual sRNA conserved among enterobacteriaceae is SgrS (Horler and Vanderpool, 2009). This sRNA not only functions in the traditional sense by base-paring with a target mRNA to regulate translation, but in many bacteria is itself also translated into a small 43 amino acid protein known as SgrT (Wadler and Vanderpool, 2007). SgrS is induced under glucose-phosphate stress conditions and utilizes traditional Hfq-dependent base pairing within the 5′ UTR to negatively regulate ptsG [EIICBGlc of the phosphoenolpyruvate phosphotransferase system (PTS)] post-transcriptionally, which in E. coli prevents the synthesis of new glucose transporters. SgrT helps to rescue the bacterial cell from glucose-phosphate stress by inhibiting glucose transporter activity at the post-translational level (Wadler and Vanderpool, 2007). Recent evidence suggests that SgrS may also regulate other PTS genes (Rice and Vanderpool, 2011). The base-paring function of SgrS/Ysr150 with ptsG is conserved in Y. pestis, however the sRNA does not have the same 5′ end as the E. colisgrS, and therefore does not produce SgrT (Wadler and Vanderpool, 2009). The lack of SgrT in Y. pestis may alter the bacterial response to glucose-phosphate stress compared to other organisms, however there could also be an unknown redundant system that Y. pestis utilizes instead.
SraG
One conserved sRNA present in Yersinia with no assigned function in other enterobacteriaceae to date is SraG (Argaman et al., 2001; Sridhar et al., 2009). An attempt to determine targets of SraG in Y. pseudotuberculosis using a proteomic screen followed by genetic analyses identified the operon YPK_1206-05 as a potential target. The authors of this study conclude that SraG negatively regulates this operon directly at the post-transcriptional level (Lu et al., 2012). This operon has not been annotated, but secondary structure prediction suggests that it YPK_1206 is similar to an IHF-like DNA bending protein.
Ysr29 and Ysr35
Koo et al. (2011) detected almost all of the conserved sRNAs discussed above in a global analysis of non-coding RNAs expressed by Y. pseudotuberculosis, as well as an additional 118 previously unidentified, putative sRNAs that are specific for Y. pseudotuberculosis and/or Y. pestis, but absent from other enterobacterial species. In an effort to provide a consistent naming convention for these sRNAs across all Yersinia species, going forward the authors propose a standard nomenclature for newly identified non-coding RNAs in Yersiniae that adopts the “Ysr” naming convention. Twenty-nine of the Ysrs identified by Koo et al. (2011) were detected by northern blot analysis, and one Y. pseudotuberculosis-specific sRNA, Ysr29, was found to contribute significantly to mortality in a mouse model of Yersiniosis. By using 2D differential gel electrophoresis (DIGE) combined with mass spectrometry, eight genes were identified as potential regulated targets of Ysr29: ureC, ahpC, gst, frr, rpsA, ompA, groEL, and dnaK (Koo et al., 2011). None of these targets showed a significant change in transcript level in the absence of ysr29, suggesting that Ysr29 affects protein levels through post-transcriptional mechanisms of regulation. Each of these targets may contribute to the bacterial response to environmental or host stresses, demonstrating that Ysr29 is a major post-transcriptional regulator of the Y. pseudotuberculosis adaptation to stress (Koo et al., 2011). A second Yersinia-specific sRNA, Ysr35, was also examined for its contribution to virulence. Deletion of this conserved sRNA from the genomes of both Y. pseudotuberculosis and Y. pestis resulted in decreased virulence in mouse models of Yersiniosis and pneumonic plague, respectively, although it is not yet known if the Ysr35-regulated target(s) are also conserved between the species or if Ysr35 has adapted to specific and/or unique targets between the two (Koo et al., 2011). It is expected that future sRNA identification studies will reveal additional Ysrs not found here, and it will be of particular interest to determine the sRNA-omes of the Yersiniae during mammalian infection, flea colonization, and in environmental reservoirs.
Transcript antisense to pla
While the majority of non-coding sRNAs identified in Yersinia so far appear to be trans-encoded, a number of putative cis-acting sRNAs have also been identified. One of these transcripts is encoded on pPCP1, a plasmid specific to Y. pestis that carries the genes for pesticin, the pesticin immunity protein, and the plasminogen activator protease Pla (Sodeinde and Goguen, 1989). Pla is an essential virulence factor required by Y. pestis during both bubonic and pneumonic plague (Sodeinde et al., 1992; Lathem et al., 2007). Sodeinde and Goguen (1989) identified a transcript complementary to the coding sequence and upstream region of pla that may be expressed from a promoter with high similarity to the E. coli sigma70 -10 and -35 sites. While a potential 48-amino acid polypeptide may be encoded within this sequence, the authors were unable to detect a protein produced from the transcript. Instead, this antisense molecule may participate in the regulation of Pla synthesis, although no physiological role for the anti-pla transcript has been determined to date.
Yp-sR3, Yp-sR7, and Yp-sR8
Among the sRNAs identified by Qu et al. (2012) 25 are predicted to be cis-acting. Two of these cis-encoded sRNAs in Y. pestis strain 201 are Yp-sR3 and Yp-sR8, both of which are maximally expressed during stationary phase in vitro (Qu et al., 2012). Yp-sR3 spans a potential operon formed by YP_1329 and YP_1330 that encodes two putative membrane proteins (Qu et al., 2012). This suggests that Yp-sR3 could also participate in the regulation of the Yersinia membrane through an antisense mechanism. A third cis-encoded RNA found is Yp-sR7, which is transcribed from the strand opposite the gene rplK that encodes the 50S ribosomal protein (Qu et al., 2012), but its function is as yet unknown. Indeed, much work remains to be done to determine the contributions of these and other cis-acting sRNAs to the biology of the Yersiniae.
RNases
RNases cleave RNA transcripts internally (endonucleases) or from the 5′ or 3′ end (exonucleases). The average half-life of mRNA transcripts in E. coli ranges from 1 to 10 min, which allows the bacterial cell to respond quickly to various surroundings, and RNA degradation is mediated by a degradasome that includes the endonuclease RNase E (Feng and Niu, 2007). The breakdown of RNA transcripts by the degradasome is essential to normal bacterial cell metabolism, but targeted degradation of mRNA also plays an important role in the post-transcriptional regulation of gene expression. For example, binding of the sRNA RyhB to mRNA targets in the presence of Hfq results in RNase E-dependent degradation of the sRNA::transcript complex (Masse et al., 2003). Hfq associates with RNase E through the C-terminal scaffold of RNase E, which is thought to be a general mechanism for the repression of protein production through sRNAs (Aiba, 2007).
As in other Gram-negative bacteria, RNase E is conserved in Yersinia species. While it has not yet been possible to generate an rne mutant of Yersinia, Y. pseudotuberculosis carrying a dominant negative version of the gene for RNase E that lacks the C-terminal scaffolding domain is defective for survival within macrophage-like cells (Yang et al., 2008). RNase E has also been implicated in the regulation of the T3SS, as the dominant negative-containing mutant also showed decreased secretion of YopE into culture supernatant. Interestingly, the amount of YopE isolated from cell lysates is unchanged, suggesting that RNase E regulates the T3SS at a post-translational step and not by the degradation of T3SS effector mRNA transcripts at the post-transcriptional level (Yang et al., 2008). A second exonuclease known as polynucleotide phosphorylase (PNPase) also contributes to the regulation of the T3SS in Yersinia species in a manner that is predicted to function at the same level as RNase E (Figure 2; Rosenzweig et al., 2005, 2007). The S1 RNA-binding domain of PNPase is required for the optimal export of effectors by the T3SS, but is independent of the ribonuclease activity of PNPase, as the catalytic activity of PNPase is required for growth of Yersinia at low temperature (Goverde et al., 1998; Rosenzweig et al., 2005; Rosenzweig and Schesser, 2007). PNPase is also associated with the regulation of the T3SS of S. enterica, however in this case the regulation is indirect and occurs at the transcriptional level (Clements et al., 2002). There are numerous other RNases that play critical roles in the post-transcriptional regulation of virulence genes, and although many of these RNases are conserved in Yersinia, the mechanisms of action and the targets involved have not been elucidated (Arraiano et al., 2010).
Thermoswitches/Thermosensors
The regulation of virulence in Yersinia is largely dependent on the ability of the bacteria to sense the temperature of their surroundings, and as such a significant number of genes are thermally regulated. Whole genome DNA microarray analysis of Y. pestis, for example, revealed that over 400 genes are differentially regulated at 26 and 37°C, with 39% induced and 61% repressed between the two temperatures (Han et al., 2004). A number of these genes include those encoding the T3SS. The T3SS is activated upon transition from the environmental niche to the mammalian host by the de-repression of transcription of lcrF (also known as virF), a gene encoding an AraC-like transcriptional activator of the T3SS. LcrF globally up-regulates the transcription of the genes for both the secretion apparatus (yscs)and effectors (yops). It has long been known that the transcription of lcrF is thermoregulated, but new data demonstrate that, as predicted by Hoe and Goguen (1993), the translation of LcrF is also controlled in a temperature-dependent manner (Hoe and Goguen, 1993; Bohme et al., 2012). Translation of LcrF is modulated through a secondary structure within the intergenic region of the yscW-lcrF transcript that traps the Shine–Dalgarno sequence of lcrF at low temperatures and thus prevents translation (Figure 1). This secondary structure forms a motif that is also observed for heat-shock genes in many eubacteria (Kortmann and Narberhaus, 2012; Schumann, 2012), and in Yersinia the lcrF thermosensor consists of a two-stem loop structure that forms at 25°C (Bohme et al., 2012). At 37°C, however, the stem-loops denature, the Shine–Dalgarno site becomes accessible to the ribosome, and translation is initiated (Bohme et al., 2012). The authors of this study found that if the lcrF thermosensor is mutated to create a permanently open or permanently closed conformation, the virulence of Y. pseudotuberculosis is reduced in a mouse model of Yersiniosis, which suggests that the ability to fine tune the translation of LcrF – either up or down – is critical to the virulence of this pathogen. So far only one other virulence-associated thermosensor has been identified in a bacterial pathogen: the prfA gene (a positive regulator of listeriolysin) of Listeria monocytogenes (Johansson et al., 2002; Loh et al., 2012). Given the major contribution of temperature to the biology of Yersinia, it is possible that there may be other thermosensors that remain unidentified, including those that regulate virulence genes.
Riboswitches
One mechanism of post-transcriptional regulation that has not been well studied in Yersinia is the use of riboswitches to control gene expression. A riboswitch is similar to the thermosensor/thermoswitch of LcrF discussed above in that it is usually (but not always) cis-encoded within the 5′ UTR of the regulated mRNA and forms a secondary structure that prevents translation under certain conditions (Mandal and Breaker, 2004). Where a riboswitch differs, however, is that the relief of this secondary structure depends on the binding of a ligand rather than changes in temperature, and in response to the presence or absence of the ligand a riboswitch may influence mRNA processing or transcript termination in addition to translation initiation. This ligand is often a metabolite that serves as a sensor of available nutrients (Mandal and Breaker, 2004). For example, in V. cholerae a glycine-binding riboswitch upstream of the gcvT operon acts as an on-switch in the presence of glycine to increase production of proteins that form the glycine cleavage system (Mandal et al., 2004). While many riboswitches contribute to the regulation of general metabolism or cellular homeostasis, some have been shown to participate in virulence factor expression. For instance, in L. monocytogenes the trans-acting S-adenosylmethionine (SAM)-riboswitch SreA is activated in the presence of SAM, which is then able to bind to and repress PrfA translation at 37°C, thereby linking a metabolite-sensing riboswitch directly to virulence (Loh et al., 2009; Xayarath and Freitag, 2009). As translation of PrfA is also regulated by a thermosensor, the dual post-transcriptional regulation of pfrA by secondary RNA structures demonstrates the need for tight regulation of the pfrA regulon in L. monocytogenes.
Conserved riboswitches in Yersinia species have been found through bioinformatic analyses. The molybdenum cofactor (Moco)-sensing riboswitch, which controls expression of adjacent genes in response to the presence of Moco, is conserved in Y. pseudotuberculosis (Regulski et al., 2008). Additionally, the co-enzyme B12 riboswitch, which is responsible for the translational repression of cobalamin-transport protein production at high concentrations of the co-enzyme, is also conserved in Y. pestis (Nahvi et al., 2004). Neither of these conserved riboswitches have been experimentally validated in Yersinia. One riboswitch that has been recently experimentally tested in Y. enterocolitica, however, is found within the 5′ UTR of the gene mgtA, which responds to Mg2+ concentrations to regulate expression of the magnesium transporter protein (Korth and Sigel, 2012). In S. enterica high Mg2+ concentrations lead to the early Rho-dependent termination of mgtA transcription through a conformational change in the RNA, thereby regulating magnesium transporter production (Hollands et al., 2012). Given the widespread and integral nature of many riboswitches (Winkler and Breaker, 2005)it is likely that Yersinia may encode additional riboswitches.
Conclusion
Post-transcriptional regulation in Yersinia occurs through multiple different mechanisms, many of which have been shown enhance the survival and virulence of these species. The studies performed so far on post-transcriptional regulation in Yersinia have focused heavily on the T3SS, which is reasonable given the central role of the T3SS to pathogenesis. While the complexity of the T3SS in Yersinia is great, further investigation into the post-transcriptional mechanisms that govern the optimal expression of the T3SS, including the identification of additional sRNAs, thermosensors, and the like, may fill in some of the gaps that remain after analysis of transcriptional regulation of the T3SS is exhausted.
The number of putative sRNAs identified by both deep sequencing and cDNA cloning of the Yersinia transcriptome have provided many avenues for the study of post-transcriptional mechanisms of gene regulation in these species (Koo et al., 2011; Qu et al., 2012). Although many sRNAs are conserved between related bacterial species, of the Ysrs identified in Y. pseudotuberculosis by Koo et al. (2011) 79% were found to be Yersinia-specific and not present in other genera such as Escherichia or Salmonella. Additionally, 56% of the Yersinia-specific sRNAs have nucleotide mismatches or other sequence differences between Y. pseudotuberculosis and Y. pestis (Koo and Lathem, 2012). These disparities, though small, may be significant as a single mismatch in the RNA–RNA-binding site of an sRNA-mRNA pair can abolish function of the sRNA (Hao et al., 2011). Furthermore, Koo et al. (2011) found that many sRNAs with identical sequences were differentially expressed between the two species with respect to growth phase, temperature and requirement for Hfq. These differences should not be overlooked when examining the mechanisms of gene regulation between the species and may inform further studies on the evolution of Yersinia as a human pathogen (Lathem, 2012). The variation between the sRNA-omes of Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, both in vitro and in vivo, may hold clues that help explain the stark differences between the disease etiologies of the enteric Yersinia and the plague bacillus that are not made clear by analyses of simple virulence gene acquisitions or losses.
The mechanisms detailed in this review emphasize the importance of post-transcriptional regulation in Yersinia species. Rapid adaptation to changing conditions through the action of RNA-binding proteins and several classes of non-coding RNAs are key for survival and propagation of these bacteria. In particular, post-transcriptional regulation plays an integral role in virulence, and not just in the control of metabolic pathways. The variety of post-transcriptional mechanisms in Yersinia makes it an excellent model organism for studying this growing field of regulation.
Statements
Acknowledgments
The authors would like to thank members of the Lathem lab and Dr. Chris Waters for helpful discussions. This work was supported by NIH grant R01 AI093727 to Wyndham W. Lathem.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AbdelaW.GrahamM.TsegayeH.TemesgenS.YehualaeshetT. (2011). Effects of orange juice pH on survival, urease activity and DNA profiles of Yersinia enterocolitica and Yersinia pseudotuberculosis stored at 4 degree C. J. Food Saf.31, 487–496.10.1111/j.1745-4565.2011.00325.x
2
AboT.InadaT.OgawaK.AibaH. (2000). SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J.19, 3762–3769.10.1093/emboj/19.14.3762
3
AepfelbacherM.HeesemannJ. (2001). Modulation of Rho GTPases and the actin cytoskeleton by Yersinia outer proteins (Yops). Int. J. Med. Microbiol.291, 269–276.10.1078/1438-4221-00130
4
AibaH. (2007). Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol.10, 134–139.10.1016/j.mib.2007.03.010
5
AllaouiA.SchulteR.CornelisG. R. (1995). Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol. Microbiol.18, 343–355.10.1111/j.1365-2958.1995.mmi_18020343.x
6
AltierC.SuyemotoM.LawhonS. D. (2000). Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA. Infect. Immun.68, 6790–6797.10.1128/IAI.68.12.6790-6797.2000
7
AndersenJ.ForstS. A.ZhaoK.InouyeM.DelihasN. (1989). The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem.264, 17961–17970.
8
AndersonD. M.RamamurthiK. S.TamC.SchneewindO. (2002). YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J. Bacteriol.184, 1287–1295.10.1128/JB.184.5.1287-1295.2002
9
AnsongC.YoonH.PorwollikS.Mottaz-BrewerH.PetritisB. O.JaitlyN.et al (2009). Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS ONE4, e4809.10.1371/journal.pone.0004809
10
ArgamanL.HershbergR.VogelJ.BejeranoG.WagnerE. G.MargalitH.et al (2001). Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol.11, 941–950.10.1016/S0960-9822(01)00270-6
11
ArraianoC. M.AndradeJ. M.DominguesS.GuinoteI. B.MaleckiM.MatosR. G.et al (2010). The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev.34, 883–923.
12
BaiG.GolubovA.SmithE. A.McDonoughK. A. (2010). The importance of the small RNA chaperone Hfq for growth of epidemic Yersinia pestis, but not Yersinia pseudotuberculosis, with implications for plague biology. J. Bacteriol.192, 4239–4245.10.1128/JB.00504-10
13
BakerC. S.EoryL. A.YakhninH.MercanteJ.RomeoT.BabitzkeP. (2007). CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol.189, 5472–5481.10.1128/JB.00529-07
14
BardillJ. P.ZhaoX.HammerB. K. (2011). The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol. Microbiol.80, 1381–1394.10.1111/j.1365-2958.2011.07655.x
15
BellowsL. E.KoestlerB. J.KarabaS. M.WatersC. M.LathemW. W. (2012). Hfq-dependent, coordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol.86, 661–674.10.1111/mmi.12011
16
BlaylockB.RiordanK. E.MissiakasD. M.SchneewindO. (2006). Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol.188, 3525–3534.10.1128/JB.188.10.3525-3534.2006
17
BohmeK.SteinmannR.KortmannJ.SeekircherS.HerovenA. K.BergerE.et al (2012). Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog.8, e1002518.10.1371/journal.ppat.1002518
18
BrencicA.LoryS. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol.72, 612–632.10.1111/j.1365-2958.2009.06670.x
19
BrennanR. G.LinkT. M. (2007). Hfq structure, function and ligand binding. Curr. Opin. Microbiol.10, 125–133.10.1016/j.mib.2007.03.015
20
BrzostekK.BrzostkowskaM.BukowskaI.KarwickaE.RaczkowskaA. (2007). OmpR negatively regulates expression of invasin in Yersinia enterocolitica. Microbiology153, 2416–2425.10.1099/mic.0.2006/003202-0
21
CalvoC.MelisR.BraultJ.AlonsoJ. M.Ramos-CormenzanaA.MollaretH. H. (1986). Antagonism between Yersinia intermedia and Yersinia enterocolitica in water. Folia Microbiol. (Praha)31, 167–173.10.1007/BF02926836
22
CambronneE. D.SchneewindO. (2002). Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5′ untranslated region of yop mRNA. J. Bacteriol.184, 5880–5893.10.1128/JB.184.21.5880-5893.2002
23
CathelynJ. S.CrosbyS. D.LathemW. W.GoldmanW. E.MillerV. L. (2006). RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. U.S.A.103, 13514–13519.10.1073/pnas.0603456103
24
ChauvauxS.RossoM. L.FrangeulL.LacroixC.LabarreL.SchiavoA.et al (2007). Transcriptome analysis of Yersinia pestis in human plasma: an approach for discovering bacterial genes involved in septicaemic plague. Microbiology153, 3112–3124.10.1099/mic.0.2007/006213-0
25
ChenY.AndersonD. M. (2011). Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol. Microbiol.80, 966–980.10.1111/j.1365-2958.2011.07623.x
26
ChouikhaI.HinnebuschB. J. (2012). Yersinia-flea interactions and the evolution of the arthropod-borne transmission route of plague. Curr. Opin. Microbiol.15, 239–246.10.1016/j.mib.2012.02.003
27
ChristiansenJ. K.LarsenM. H.IngmerH.Sogaard-AndersenL.KallipolitisB. H. (2004). The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol.186, 3355–3362.10.1128/JB.186.11.3355-3362.2004
28
ClementsM. O.ErikssonS.ThompsonA.LucchiniS.HintonJ. C.NormarkS.et al (2002). Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A.99, 8784–8789.10.1073/pnas.132047099
29
CornelisG.SluitersC.De RouvroitC. L.MichielsT. (1989). Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol.171, 254–262.
30
CornelisG. R.BolandA.BoydA. P.GeuijenC.IriarteM.NeytC.et al (1998). The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev.62, 1315–1352.
31
CornelisG. R.SluitersC.DelorI.GeibD.KanigaK.Lambert De RouvroitC.et al (1991). ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol.5, 1023–1034.10.1111/j.1365-2958.1991.tb01875.x
32
CornelisG. R.Wolf-WatzH. (1997). The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol.23, 861–867.10.1046/j.1365-2958.1997.2731623.x
33
DarbyC.AnanthS. L.TanL.HinnebuschB. J. (2005). Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect. Immun.73, 7236–7242.10.1128/IAI.73.11.7236-7242.2005
34
DavisB. M.QuinonesM.PrattJ.DingY.WaldorM. K. (2005). Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol.187, 4005–4014.10.1128/JB.187.12.4005-4014.2005
35
DelihasN. (2003). Annotation and evolutionary relationships of a small regulatory RNA gene micF and its target ompF in Yersinia species. BMC Microbiol.3, 13.10.1186/1471-2180-3-13
36
DelihasN.ForstS. (2001). MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol.313, 1–12.10.1006/jmbi.2001.5029
37
DengZ.MengX.SuS.LiuZ.JiX.ZhangY.et al (2012). Two sRNA RyhB homologs from Yersinia pestis biovar microtus expressed in vivo have differential Hfq-dependent stability. Res. Microbiol.163, 413–418.10.1016/j.resmic.2012.05.006
38
DingY.DavisB. M.WaldorM. K. (2004). Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol.53, 345–354.10.1111/j.1365-2958.2004.04142.x
39
DubeyA. K.BakerC. S.RomeoT.BabitzkeP. (2005). RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA11, 1579–1587.10.1261/rna.2990205
40
DubeyA. K.BakerC. S.SuzukiK.JonesA. D.PanditP.RomeoT.et al (2003). CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol.185, 4450–4460.10.1128/JB.185.15.4450-4460.2003
41
DunlapP. V.RayJ. M. (1989). Requirement for autoinducer in transcriptional negative autoregulation of the Vibrio fischeri luxR gene in Escherichia coli. J. Bacteriol.171, 3549–3552.
42
EllermeierJ. R.SlauchJ. M. (2007). Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10, 24–29.10.1016/j.mib.2006.12.002
43
FantappieL.MetruccioM. M.SeibK. L.OrienteF.CartocciE.FerliccaF.et al (2009). The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect. Immun.77, 1842–1853.10.1128/IAI.01216-08
44
FengL.NiuD. K. (2007). Relationship between mRNA stability and length: an old question with a new twist. Biochem. Genet.45, 131–137.10.1007/s10528-006-9059-5
45
GageK. L.KosoyM. Y. (2005). Natural history of plague: perspectives from more than a century of research. Annu. Rev. Entomol.50, 505–528.10.1146/annurev.ento.50.071803.130337
46
GaoH.ZhouD.LiY.GuoZ.HanY.SongY.et al (2008). The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol.190, 3063–3075.10.1128/JB.01910-07
47
GengJ.SongY.YangL.FengY.QiuY.LiG.et al (2009). Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS ONE4, e6213.10.1371/journal.pone.0006213
48
GopelY.LuttmannD.HerovenA. K.ReichenbachB.DerschP.GorkeB. (2011). Common and divergent features in transcriptional control of the homologous small RNAs GlmY and GlmZ in enterobacteriaceae. Nucleic Acids Res.39, 1294–1309.10.1093/nar/gkq986
49
GottesmanS. (2005). Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet.21, 399–404.10.1016/j.tig.2005.05.008
50
GottesmanS.StorzG. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol.3, a003798.10.1101/cshperspect.a003798
51
GoverdeR. L.Huis In’t VeldJ. H.KustersJ. G.MooiF. R. (1998). The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5 degrees C). Mol. Microbiol.28, 555–569.10.1046/j.1365-2958.1998.00816.x
52
GreenR. C.DarwinA. J. (2004). PspG, a new member of the Yersinia enterocolitica phage shock protein regulon. J. Bacteriol.186, 4910–4920.10.1128/JB.186.15.4910-4920.2004
53
GuillierM.GottesmanS. (2006). Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol.59, 231–247.10.1111/j.1365-2958.2005.04929.x
54
HakanssonS.BergmanT.VanooteghemJ. C.CornelisG.Wolf-WatzH. (1993). YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect. Immun.61, 71–80.
55
HanY.ZhouD.PangX.SongY.ZhangL.BaoJ.et al (2004). Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol. Immunol.48, 791–805.
56
HaoY.ZhangZ. J.EricksonD. W.HuangM.HuangY.LiJ.et al (2011). Quantifying the sequence-function relation in gene silencing by bacterial small RNAs. Proc. Natl. Acad. Sci. U.S.A.108, 12473–12478.10.1073/pnas.1100432108
57
HarrisonW. A.PetersA. C.FieldingL. M. (2000). Growth of Listeria monocytogenes and Yersinia enterocolitica colonies under modified atmospheres at 4 and 8 degrees C using a model food system. J. Appl. Microbiol.88, 38–43.10.1046/j.1365-2672.2000.00848.x
58
HerovenA. K.BohmeK.DerschP. (2012). The Csr/Rsm system of Yersinia and related pathogens: A post-transcriptional strategy for managing virulence. RNA Biol.9, 379–391.10.4161/rna.19333
59
HerovenA. K.BohmeK.RohdeM.DerschP. (2008). A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol.68, 1179–1195.10.1111/j.1365-2958.2008.06218.x
60
HinnebuschB. J.RudolphA. E.CherepanovP.DixonJ. E.SchwanT. G.ForsbergA. (2002). Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science296, 733–735.10.1126/science.1069972
61
HoeN. P.GoguenJ. D. (1993). Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol.175, 7901–7909.
62
HollandsK.ProshkinS.SklyarovaS.EpshteinV.MironovA.NudlerE.et al (2012). Riboswitch control of rho-dependent transcription termination. Proc. Natl. Acad. Sci. U.S.A.109, 5376–5381.10.1073/pnas.1112211109
63
HorlerR. S.VanderpoolC. K. (2009). Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res.37, 5465–5476.10.1093/nar/gkp501
64
IriarteM.StainierI.CornelisG. R. (1995a). The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun.63, 1840–1847.
65
IriarteM.StainierI.MikulskisA. V.CornelisG. R. (1995b). The fliA gene encoding sigma 28 in Yersinia enterocolitica. J. Bacteriol.177, 2299–2304.
66
JarrettC. O.DeakE.IsherwoodK. E.OystonP. C.FischerE. R.WhitneyA. R.et al (2004). Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis.190, 783–792.10.1086/422695
67
JohansenJ.RasmussenA. A.OvergaardM.Valentin-HansenP. (2006). Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol.364, 1–8.10.1016/j.jmb.2006.09.004
68
JohanssonJ.MandinP.RenzoniA.ChiaruttiniC.CossartP. (2002). An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell110, 551–561.10.1016/S0092-8674(02)00905-4
69
KajitaniM.IshihamaA. (1991). Identification and sequence determination of the host factor gene for bacteriophage Q beta. Nucleic Acids Res.19, 1063–1066.10.1093/nar/19.5.1063
70
KalamorzF.ReichenbachB.MarzW.RakB.GorkeB. (2007). Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol.65, 1518–1533.10.1111/j.1365-2958.2007.05888.x
71
KarzaiA. W.RocheE. D.SauerR. T. (2000). The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol.7, 449–455.10.1038/75843
72
KarzaiA. W.SusskindM. M.SauerR. T. (1999). SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J.18, 3793–3799.10.1093/emboj/18.13.3793
73
KeilerK. C.WallerP. R.SauerR. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science271, 990–993.10.1126/science.271.5251.990
74
KendallM. M.GruberC. C.RaskoD. A.HughesD. T.SperandioV. (2011). Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86-24. J. Bacteriol.193, 6843–6851.10.1128/JB.06141-11
75
KomineY.KitabatakeM.YokogawaT.NishikawaK.InokuchiH. (1994). A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A.91, 9223–9227.10.1073/pnas.91.20.9223
76
KooJ. T.AlleyneT. M.SchianoC. A.JafariN.LathemW. W. (2011). Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence. Proc. Natl. Acad. Sci. U.S.A.108, E709–E717.10.1073/pnas.1101655108
77
KooJ. T.LathemW. W. (2012). Global discovery of small noncoding RNAs in pathogenic Yersinia species. Adv. Exp. Med. Biol.954, 305–314.10.1007/978-1-4614-3561-7_38
78
KorthM. M.SigelR. K. (2012). Unusually high-affinity Mg(2+) binding at the AU-rich sequence within the antiterminator hairpin of a Mg(2+) riboswitch. Chem. Biodivers.9, 2035–2049.10.1002/cbdv.201200031
79
KortmannJ.NarberhausF. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol.10, 255–265.10.1038/nrmicro2730
80
KulesusR. R.Diaz-PerezK.SlechtaE. S.EtoD. S.MulveyM. A. (2008). Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun.76, 3019–3026.10.1128/IAI.00022-08
81
LathemW. W. (2012). Identification of small, noncoding RNAs in pathogenic Yersinia species: implications for evolution and virulence. Virulence3, 154–156.10.4161/viru.19155
82
LathemW. W.CrosbyS. D.MillerV. L.GoldmanW. E. (2005). Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. U.S.A.102, 17786–17791.10.1073/pnas.0506840102
83
LathemW. W.PriceP. A.MillerV. L.GoldmanW. E. (2007). A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science315, 509–513.10.1126/science.1137195
84
LawsT. R.DaveyM. S.GreenC.CooperI. A.TitballR. W.LukaszewskiR. A. (2011). Yersinia pseudotuberculosis is resistant to killing by human neutrophils. Microbes Infect.13, 607–611.10.1016/j.micinf.2011.02.004
85
LawsonJ. N.LyonsC. R.JohnstonS. A. (2006). Expression profiling of Yersinia pestis during mouse pulmonary infection. DNA Cell Biol.25, 608–616.10.1089/dna.2006.25.608
86
LenzD. H.MillerM. B.ZhuJ.KulkarniR. V.BasslerB. L. (2005). CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol.58, 1186–1202.10.1111/j.1365-2958.2005.04902.x
87
LenzD. H.MokK. C.LilleyB. N.KulkarniR. V.WingreenN. S.BasslerB. L. (2004). The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell118, 69–82.10.1016/j.cell.2004.06.009
88
LiuM. Y.GuiG.WeiB.PrestonJ. F.IIIOakfordL.YukselU.et al (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem.272, 17502–17510.10.1074/jbc.272.6.3628
89
LiuM. Y.RomeoT. (1997). The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol.179, 4639–4642.
90
LiuM. Y.YangH.RomeoT. (1995). The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol.177, 2663–2672.
91
LohE.DussurgetO.GripenlandJ.VaitkeviciusK.TiensuuT.MandinP.et al (2009). A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell139, 770–779.10.1016/j.cell.2009.08.046
92
LohE.MemarpourF.VaitkeviciusK.KallipolitisB. H.JohanssonJ.SondenB. (2012). An unstructured 5′-coding region of the prfA mRNA is required for efficient translation. Nucleic Acids Res.40, 1818–1827.10.1093/nar/gkr850
93
LorangeE. A.RaceB. L.SebbaneF.Joseph HinnebuschB. (2005). Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J. Infect. Dis.191, 1907–1912.10.1086/429931
94
LuP.ZhangY.LiL.HuY.HuangL.LiY.et al (2012). Small non-coding RNA SraG regulates the operon YPK_1206-1205 in Yersinia pseudotuberculosis. FEMS Microbiol. Lett.331, 37–43.10.1111/j.1574-6968.2012.02548.x
95
MandalM.BreakerR. R. (2004). Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol.5, 451–463.10.1038/nrm1403
96
MandalM.LeeM.BarrickJ. E.WeinbergZ.EmilssonG. M.RuzzoW. L.et al (2004). A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science306, 275–279.10.1126/science.1100829
97
MarraA.IsbergR. R. (1997). Invasin-dependent and invasin-independent pathways for translocation of Yersinia pseudotuberculosis across the Peyer’s patch intestinal epithelium. Infect. Immun.65, 3412–3421.
98
MasseE.EscorciaF. E.GottesmanS. (2003). Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev.17, 2374–2383.10.1101/gad.1127103
99
MasseE.GottesmanS. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A.99, 4620–4625.10.1073/pnas.032066599
100
McArthurS. D.PulvermacherS. C.StaufferG. V. (2006). The Yersinia pestis gcvB gene encodes two small regulatory RNA molecules. BMC Microbiol.6, 52.10.1186/1471-2180-6-52
101
MeibomK. L.ForslundA. L.KuoppaK.AlkhuderK.DubailI.DupuisM.et al (2009). Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect. Immun.77, 1866–1880.10.1128/IAI.01496-08
102
MillerM. R.LiuZ.CazierD. J.GebhardG. M.HerronS. R.ZaherH. S.et al (2011). The role of SmpB and the ribosomal decoding center in licensing tmRNA entry into stalled ribosomes. RNA17, 1727–1736.10.1261/rna.2462011
103
MillerV. L.FalkowS. (1988). Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun.56, 1242–1248.
104
MitobeJ.Morita-IshiharaT.IshihamaA.WatanabeH. (2009). Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol.9, 110.10.1186/1471-2180-9-110
105
MollerT.FranchT.HojrupP.KeeneD. R.BachingerH. P.BrennanR. G.et al (2002). Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell9, 23–30.10.1016/S1097-2765(01)00436-1
106
MonackD. M.MecsasJ.BouleyD.FalkowS. (1998). Yersinia-induced apoptosis in vivo aids in the establishment of a systemic infection of mice. J. Exp. Med.188, 2127–2137.10.1084/jem.188.11.2127
107
NagelG.HerovenA. K.EitelJ.DerschP. (2003). Function and regulation of the transcriptional activator RovA of Yersinia pseudotuberculosis. Adv. Exp. Med. Biol.529, 285–287.10.1007/0-306-48416-1_54
108
NagelG.LahrzA.DerschP. (2001). Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol.41, 1249–1269.10.1046/j.1365-2958.2001.02522.x
109
NairzM.SchrollA.SonnweberT.WeissG. (2010). The struggle for iron – a metal at the host-pathogen interface. Cell. Microbiol.12, 1691–1702.10.1111/j.1462-5822.2010.01529.x
110
NakaoH.WatanabeH.NakayamaS.TakedaT. (1995). yst gene expression in Yersinia enterocolitica is positively regulated by a chromosomal region that is highly homologous to Escherichia coli host factor 1 gene (hfq). Mol. Microbiol.18, 859–865.10.1111/j.1365-2958.1995.18050859.x
111
NahviA.BarrickJ. E.BreakerR. R. (2004). Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res.32, 143–150.10.1093/nar/gkh167
112
NeubauerC.GilletR.KelleyA. C.RamakrishnanV. (2012). Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science335, 1366–1369.10.1126/science.1217039
113
OkanN. A.BliskaJ. B.KarzaiA. W. (2006). A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog.2, e6.10.1371/journal.ppat.0020006
114
OkanN. A.MenaP.BenachJ. L.BliskaJ. B.KarzaiA. W. (2010). The smpB-ssrA mutant of Yersinia pestis functions as a live attenuated vaccine to protect mice against pulmonary plague infection. Infect. Immun.78, 1284–1293.10.1128/IAI.00976-09
115
OlsonE. R.DunyakD. S.JurssL. M.PoormanR. A. (1991). Identification and characterization of dppA, an Escherichia coli gene encoding a periplasmic dipeptide transport protein. J. Bacteriol.173, 234–244.
116
PalonenE.LindstromM.KorkealaH. (2010). Adaptation of enteropathogenic Yersinia to low growth temperature. Crit. Rev. Microbiol.36, 54–67.10.3109/10408410903382581
117
PapenfortK.PfeifferV.MikaF.LucchiniS.HintonJ. C.VogelJ. (2006). SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol.62, 1674–1688.10.1111/j.1365-2958.2006.05524.x
118
PepeJ. C.BadgerJ. L.MillerV. L. (1994). Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol.11, 123–135.10.1111/j.1365-2958.1994.tb00295.x
119
PerryR. D.FetherstonJ. D. (1997). Yersinia pestis – etiologic agent of plague. Clin. Microbiol. Rev.10, 35–66.
120
PfeifferV.SittkaA.TomerR.TedinK.BrinkmannV.VogelJ. (2007). A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol.66, 1174–1191.10.1111/j.1365-2958.2007.05991.x
121
PulvermacherS. C.StaufferL. T.StaufferG. V. (2008). The role of the small regulatory RNA GcvB in GcvB/mRNA posttranscriptional regulation of oppA and dppA in Escherichia coli. FEMS Microbiol. Lett.281, 42–50.10.1111/j.1574-6968.2008.01068.x
122
PulvermacherS. C.StaufferL. T.StaufferG. V. (2009). Role of the Escherichia coli Hfq protein in GcvB regulation of oppA and dppA mRNAs. Microbiology155, 115–123.10.1099/mic.0.023432-0
123
QuY.BiL.JiX.DengZ.ZhangH.YanY.et al (2012). Identification by cDNA cloning of abundant sRNAs in a human-avirulent Yersinia pestis strain grown under five different growth conditions. Future Microbiol.7, 535–547.10.2217/fmb.12.13
124
RaczkowskaA.BrzostkowskaM.KwiatekA.BieleckiJ.BrzostekK. (2011). Modulation of inv gene expression by the OmpR two-component response regulator protein of Yersinia enterocolitica. Folia Microbiol. (Praha)56, 313–319.10.1007/s12223-011-0054-9
125
RasisM.SegalG. (2009). The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol. Microbiol.72, 995–1010.10.1111/j.1365-2958.2009.06705.x
126
RegulskiE. E.MoyR. H.WeinbergZ.BarrickJ. E.YaoZ.RuzzoW. L.et al (2008). A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol. Microbiol.68, 918–932.10.1111/j.1365-2958.2008.06208.x
127
ReichenbachB.MaesA.KalamorzF.HajnsdorfE.GorkeB. (2008). The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res.36, 2570–2580.10.1093/nar/gkn091
128
RempeK. A.HinzA. K.VadyvalooV. (2012). Hfq regulates biofilm gut blockage that facilitates flea-borne transmission of Yersinia pestis. J. Bacteriol.194, 2036–2040.10.1128/JB.06568-11
129
RiceJ. B.VanderpoolC. K. (2011). The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res.39, 3806–3819.10.1093/nar/gkq1219
130
RichE. J.McDonaldR. A.ChristieD. L. (1990). Yersinia pseudotuberculosis: report of a case with endoscopic findings. J. Pediatr. Gastroenterol. Nutr.10, 413–415.10.1097/00005176-199002000-00005
131
RichardsJ.MehtaP.KarzaiA. W. (2006). RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol.62, 1700–1712.10.1111/j.1365-2958.2006.05472.x
132
RobertsonG. T.RoopR. M.Jr. (1999). The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol.34, 690–700.10.1046/j.1365-2958.1999.01629.x
133
RomeoT. (1998). Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol.29, 1321–1330.10.1046/j.1365-2958.1998.01021.x
134
RosenzweigJ. A.ChromyB.EcheverryA.YangJ.AdkinsB.PlanoG. V.et al (2007). Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol. Lett.270, 255–264.10.1111/j.1574-6968.2007.00689.x
135
RosenzweigJ. A.SchesserK. (2007). Polynucleotide phosphorylase and the T3SS. Adv. Exp. Med. Biol.603, 217–224.10.1007/978-0-387-72124-8_19
136
RosenzweigJ. A.WeltmanG.PlanoG. V.SchesserK. (2005). Modulation of Yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem.280, 156–163.
137
SauterC.BasquinJ.SuckD. (2003). Sm-like proteins in eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res.31, 4091–4098.10.1093/nar/gkg480
138
SchianoC. A.BellowsL. E.LathemW. W. (2010). The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect. Immun.78, 2034–2044.10.1128/IAI.01046-09
139
SchmidtM.ZhengP.DelihasN. (1995). Secondary structures of Escherichia coli antisense micF RNA, the 5′-end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry34, 3621–3631.10.1021/bi00030a006
140
SchuD. J.CarlierA. L.JamisonK. P.Von BodmanS.StevensA. M. (2009). Structure/function analysis of the Pantoea stewartii quorum-sensing regulator EsaR as an activator of transcription. J. Bacteriol.191, 7402–7409.10.1128/JB.00994-09
141
SchubertM.LapougeK.DussO.OberstrassF. C.JelesarovI.HaasD.et al (2007). Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol.14, 807–813.10.1038/nsmb1285
142
SchumannW. (2012). Thermosensor systems in eubacteria. Adv. Exp. Med. Biol.739, 1–16.10.1007/978-1-4614-1704-0_1
143
SebbaneF.GardnerD.LongD.GowenB. B.HinnebuschB. J. (2005). Kinetics of disease progression and host response in a rat model of bubonic plague. Am. J. Pathol.166, 1427–1439.10.1016/S0002-9440(10)62360-7
144
SebbaneF.JarrettC.GardnerD.LongD.HinnebuschB. J. (2009). The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect. Immun.77, 1222–1229.10.1128/IAI.00950-08
145
ShadelG. S.DevineJ. H.BaldwinT. O. (1990). Control of the lux regulon of Vibrio fischeri. J. Biolumin. Chemilumin.5, 99–106.10.1002/bio.1170050205
146
ShakhnovichE. A.DavisB. M.WaldorM. K. (2009). Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol. Microbiol.74, 347–363.10.1111/j.1365-2958.2009.06856.x
147
ShaoF. (2008). Biochemical functions of Yersinia type III effectors. Curr. Opin. Microbiol.11, 21–29.10.1016/j.mib.2008.01.005
148
SimonetM.FauchereJ. L.BercheP. (1985). Role of virulence-associated plasmid in the uptake and killing of Yersinia pseudotuberculosis by resident macrophages. Ann. Inst. Pasteur Microbiol.136B, 283–294.10.1016/S0769-2609(85)80074-0
149
SittkaA.LucchiniS.PapenfortK.SharmaC. M.RolleK.BinnewiesT. T.et al (2008). Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet.4, e1000163.10.1371/journal.pgen.1000163
150
SittkaA.PfeifferV.TedinK.VogelJ. (2007). The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol.63, 193–217.10.1111/j.1365-2958.2006.05489.x
151
SobreroP.ValverdeC. (2012). The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol.38, 276–299.10.3109/1040841X.2012.664540
152
SodeindeO. A.GoguenJ. D. (1989). Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonellatyphimurium. Infect. Immun.57, 1517–1523.
153
SodeindeO. A.SubrahmanyamY. V.StarkK.QuanT.BaoY.GoguenJ. D. (1992). A surface protease and the invasive character of plague. Science258, 1004–1007.10.1126/science.1439793
154
SridharJ.KumarS. S.RafiZ. A. (2009). Small RNA identification in enterobacteriaceae using synteny and genomic backbone retention II. OMICS13, 261–284.10.1089/omi.2008.0067
155
SunX.ZhulinI.WartellR. M. (2002). Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res.30, 3662–3671.10.1093/nar/gkf508
156
SvenningsenS. L.TuK. C.BasslerB. L. (2009). Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J.28, 429–439.10.1038/emboj.2008.300
157
SvetlanovA.PuriN.MenaP.KollerA.KarzaiA. W. (2012). Francisella tularensis tmRNA system mutants are vulnerable to stress, avirulent in mice, and provide effective immune protection. Mol. Microbiol.85, 122–141.10.1111/j.1365-2958.2012.08093.x
158
TardyF.HombleF.NeytC.WattiezR.CornelisG. R.RuysschaertJ. M.et al (1999). Yersinia enterocolitica type III secretion-translocation system: channel formation by secreted Yops. EMBO J.18, 6793–6799.10.1093/emboj/18.23.6793
159
TiehT. H.LandauerE. (1948). Primary pneumonic plague in Mukden, 1946, and report of 39 cases with three recoveries. J. Infect. Dis.82, 52–58.10.1093/infdis/82.1.52
160
TimmermansJ.Van MelderenL. (2010). Post-transcriptional global regulation by CsrA in bacteria. Cell. Mol. Life Sci.67, 2897–2908.10.1007/s00018-010-0381-z
161
TrulzschK.RoggenkampA.PelludatC.RakinA.JacobiC.HeesemannJ. (2001). Cloning and characterization of the gene encoding periplasmic 2′,3′-cyclic phosphodiesterase of Yersinia enterocolitica O:8. Microbiology147, 203–213.
162
TsaiC. S.WinansS. C. (2011). The quorum-hindered transcription factor YenR of Yersinia enterocolitica inhibits pheromone production and promotes motility via a small non-coding RNA. Mol. Microbiol.80, 556–571.10.1111/j.1365-2958.2011.07595.x
163
TuK. C.LongT.SvenningsenS. L.WingreenN. S.BasslerB. L. (2010). Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol. Cell37, 567–579.10.1016/j.molcel.2010.01.022
164
UdekwuK. I.DarfeuilleF.VogelJ.ReimegardJ.HolmqvistE.WagnerE. G. (2005). Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev.19, 2355–2366.10.1101/gad.354405
165
UrbanJ. H.PapenfortK.ThomsenJ.SchmitzR. A.VogelJ. (2007). A conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J. Mol. Biol.373, 521–528.10.1016/j.jmb.2007.07.035
166
UrbanowskiM. L.StaufferL. T.StaufferG. V. (2000). The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol.37, 856–868.10.1046/j.1365-2958.2000.02051.x
167
VadyvalooV.JarrettC.SturdevantD.SebbaneF.HinnebuschB. J. (2007). Analysis of Yersinia pestis gene expression in the flea vector. Adv. Exp. Med. Biol.603, 192–200.10.1007/978-0-387-72124-8_16
168
VantrappenG.GeboesK.PonetteE. (1982). Yersinia enteritis. Med. Clin. North Am.66, 639–653.
169
VecerekB.MollI.AfonyushkinT.KaberdinV.BlasiU. (2003). Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol.50, 897–909.10.1046/j.1365-2958.2003.03727.x
170
VogelJ.PapenfortK. (2006). Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol.9, 605–611.10.1016/j.mib.2006.10.006
171
VytvytskaO.JakobsenJ. S.BalcunaiteG.AndersenJ. S.BaccariniM.Von GabainA. (1998). Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl. Acad. Sci. U.S.A.95, 14118–14123.10.1073/pnas.95.24.14118
172
WadlerC. S.VanderpoolC. K. (2007). A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. U.S.A.104, 20454–20459.10.1073/pnas.0708102104
173
WadlerC. S.VanderpoolC. K. (2009). Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res.37, 5477–5485.10.1093/nar/gkp591
174
WangX.DubeyA. K.SuzukiK.BakerC. S.BabitzkeP.RomeoT. (2005). CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol.56, 1648–1663.10.1111/j.1365-2958.2005.04648.x
175
WilliamsA. W.StraleyS. C. (1998). YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J. Bacteriol.180, 350–358.
176
WinklerW. C.BreakerR. R. (2005). Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol.59, 487–517.10.1146/annurev.micro.59.030804.121336
177
WitheyJ. H.FriedmanD. I. (2002). The biological roles of trans-translation. Curr. Opin. Microbiol.5, 154–159.10.1016/S1369-5274(02)00299-0
178
WoestynS.AllaouiA.WattiauP.CornelisG. R. (1994). YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol.176, 1561–1569.
179
XayarathB.FreitagN. E. (2009). A bacterial pathogen flips the riboswitch. Cell Host Microbe6, 400–402.10.1016/j.chom.2009.10.009
180
YangJ.JainC.SchesserK. (2008). RNase E regulates the Yersinia type 3 secretion system. J. Bacteriol.190, 3774–3778.10.1128/JB.01577-07
181
ZhangA.WassarmanK. M.OrtegaJ.StevenA. C.StorzG. (2002). The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell9, 11–22.10.1016/S1097-2765(02)00468-9
Summary
Keywords
Ysr, sRNA, T3SS, Hfq, plague, yersiniosis, thermosensor, riboswitch
Citation
Schiano CA and Lathem WW (2012) Post-Transcriptional Regulation of Gene Expression in Yersinia Species. Front. Cell. Inf. Microbio. 2:129. doi: 10.3389/fcimb.2012.00129
Received
06 September 2012
Accepted
03 October 2012
Published
09 November 2012
Volume
2 - 2012
Edited by
Matthew Francis, Umeå University, Sweden
Reviewed by
Eric D. Cambronne, Oregon Health and Science University, USA; Franz Narberhaus, Ruhr University Bochum, Germany
Copyright
© 2012 Schiano and Lathem.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Wyndham W. Lathem, Department of Microbiology-Immunology, Northwestern University Feinberg School of Medicine, 303 East Chicago Avenue, Chicago, IL 60611, USA. e-mail: lathem@northwestern.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.