Abstract
Acinetobacter baumannii is undoubtedly one of the most successful pathogens responsible for hospital-acquired nosocomial infections in the modern healthcare system. Due to the prevalence of infections and outbreaks caused by multi-drug resistant A. baumannii, few antibiotics are effective for treating infections caused by this pathogen. To overcome this problem, knowledge of the pathogenesis and antibiotic resistance mechanisms of A. baumannii is important. In this review, we summarize current studies on the virulence factors that contribute to A. baumannii pathogenesis, including porins, capsular polysaccharides, lipopolysaccharides, phospholipases, outer membrane vesicles, metal acquisition systems, and protein secretion systems. Mechanisms of antibiotic resistance of this organism, including acquirement of β-lactamases, up-regulation of multidrug efflux pumps, modification of aminoglycosides, permeability defects, and alteration of target sites, are also discussed. Lastly, novel prospective treatment options for infections caused by multi-drug resistant A. baumannii are summarized.
Introduction
Acinetobacter spp. are glucose-non-fermentative, non-motile, non-fastidious, catalase-positive, oxidative-negative, aerobic Gram-negative coccobacilli (Lin and Lan, 2014). Due to clusters of closely related species, it is difficult to distinguish Acinetobacter taxonomy using phenotypic traits and chemotaxonomic methods. Because antibiotic susceptibility and clinical relevance are significantly different between different genomic species, exact identification of Acinetobacter species are required (Bergogne-Berezin and Towner, 1996; Dijkshoorn et al., 1996; Houang et al., 2003; Lee et al., 2007). Many genomic fingerprinting methods have been developed, including repetitive extragenic palindromic sequence-based polymerase chain reaction (rep-PCR), pulsed-field gel electrophoresis (PFGE), matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry, ribotyping, amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, multilocus sequence typing (MLST), RNA spacer fingerprinting, amplified fragment length polymorphism analysis, and sequence analysis of 16S-23S rRNA intergene spacer regions or the rpoB and gyrB genes (Koeleman et al., 1998; Chang et al., 2005; La Scola et al., 2006; Croxatto et al., 2012; Higgins et al., 2012; Lee C. R. et al., 2015; Li X. M. et al., 2016).
Among Acinetobacter species, Acinetobacter baumannii is the most important member associated with hospital-acquired infections worldwide (Lin and Lan, 2014). This aerobic Gram-negative coccobacillus had been regarded as a low-grade pathogen, but it is a successful pathogen responsible for opportunistic infections of the skin, bloodstream, urinary tract, and other soft tissues (Peleg et al., 2008). Because many A. baumannii infections have suddenly been reported among veterans and soldiers who served in Iraq and Afghanistan (Centers for Disease and Prevention, 2004), A. baumannii is referred to as “Iraqibacter.” Multidrug-resistant (MDR) A. baumannii has spread to civilian hospitals in part by cross-infection of injured military patients repatriated from war zones (Peleg et al., 2008). Most A. baumannii infections occur in critically ill patients in the intensive care unit (ICU) setting (Fournier and Richet, 2006) and account for up to 20% of infections in ICUs worldwide (Vincent et al., 2009). Furthermore, the frequency of community-acquired A. baumannii infections has been increasing gradually (Lin and Lan, 2014). Several virulence factors have been identified by genomic and phenotypic analyses, including outer membrane porins, phospholipases, proteases, lipopolysaccharides (LPS), capsular polysaccharides, protein secretion systems, and iron-chelating systems (Antunes et al., 2011; McConnell et al., 2013; Lin and Lan, 2014).
Many reports have shown that A. baumannii rapidly develops resistance to antimicrobials, and multidrug-resistant strains have been isolated (McConnell et al., 2013). The WHO declared that A. baumannii is one of the most serious ESKAPE organisms (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter species) that effectively escape the effects of antibacterial drugs (Boucher et al., 2009). A number of A. baumannii resistance mechanisms are known, including enzymatic degradation of drugs, target modifications, multidrug efflux pumps, and permeability defects (Gordon and Wareham, 2010; Kim et al., 2012; Lin and Lan, 2014). In this review, we summarize the virulence factors of A. baumannii, antibiotic resistance mechanisms, and the therapeutic options available for treating A. baumannii infections. Figure 1 depicts all the features described in this review.
Figure 1
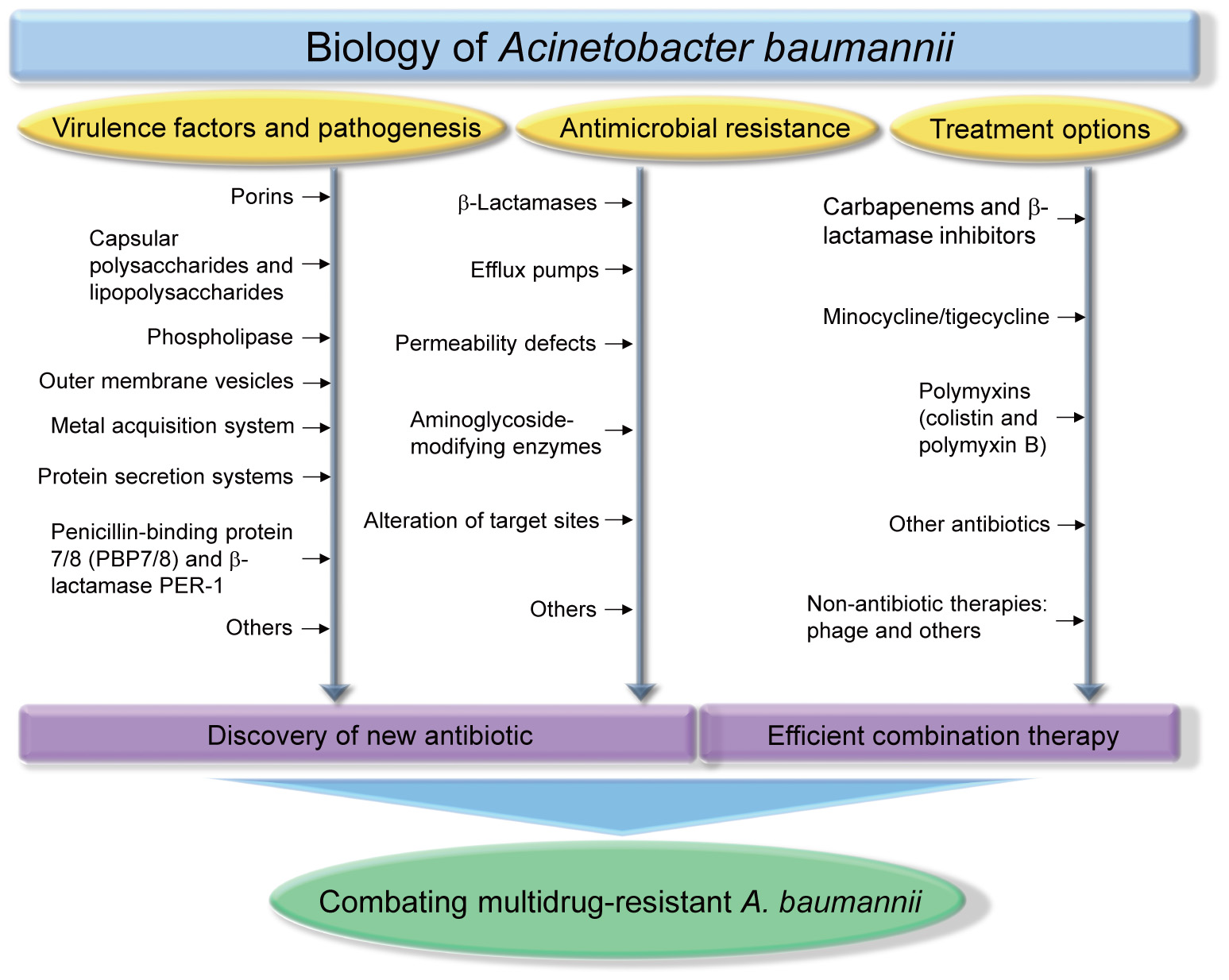
Biology of Acinetobacter baumannii. Studies of virulence factors, pathogenesis, antimicrobial resistance, treatment options of A. baumannii will provide an important aid for discovering new antibiotics and determining efficient combination therapy, which are essential strategies for combating multidrug-resistant A. baumannii infections.
Acinetobacter baumannii virulence factors and pathogenesis
Although recent genomic and phenotypic analyses of A. baumannii have identified several virulence factors responsible for its pathogenicity, relatively few virulence factors have been identified in A. baumannii, compared to those in other Gram-negative pathogens (McConnell et al., 2013). The proposed A. baumannii virulence factors are summarized in Table 1.
Table 1
| Virulence factor | Proposed role in pathogenesis | References |
|---|---|---|
| Porin (OmpA, Omp33-36, Omp22, CarO, OprD-like) | Adherence and invasion, induction of apoptosis, serum resistance, biofilm formation, persistence | Choi et al., 2005, 2008b; Gaddy et al., 2009; Kim et al., 2009; Lee et al., 2010; Fernandez-Cuenca et al., 2011; Smani et al., 2012, 2013; Rumbo et al., 2014; Wang et al., 2014; Huang et al., 2016 |
| Capsular polysaccharide | Growth in serum, survival in tissue infection, biofilm formation | Russo et al., 2010; Iwashkiw et al., 2012; Lees-Miller et al., 2013 |
| Lipopolysaccharide (LPS) | Serum resistance, survival in tissue infection, evasion of the host immune response | Luke et al., 2010; Lin et al., 2012; McQueary et al., 2012; McConnell et al., 2013 |
| Phospholipase (PLC and PLD) | Serum resistance, invasion, in vivo survival | Camarena et al., 2010; Jacobs et al., 2010; Stahl et al., 2015; Fiester et al., 2016 |
| Outer membrane vesicle (OMV) | Delivery of virulence factors, horizontal transfer of antibiotic resistance gene | Kwon et al., 2009; Jin et al., 2011; Rumbo et al., 2011; Moon et al., 2012; Jun et al., 2013; Li Z. T. et al., 2015 |
| Iron acquisition system (acinetobactin and NfuA) | In vivo survival, persistence, killing of host cells | Gaddy et al., 2012; Penwell et al., 2012; Zimbler et al., 2012; Fiester et al., 2016; Megeed et al., 2016 |
| Zinc acquisition system (ZnuABC and ZigA) | In vivo survival | Hood et al., 2012; Nairn et al., 2016 |
| Manganese acquisition system (MumC and MumT) | In vivo survival | Juttukonda et al., 2016 |
| Type II protein secretion system | In vivo survival | Johnson et al., 2015; Elhosseiny et al., 2016; Harding et al., 2016 |
| Type VI protein secretion system | Killing of competing bacteria, host colonization | Carruthers et al., 2013; Wright et al., 2014; Jones et al., 2015; Repizo et al., 2015; Ruiz et al., 2015 |
| Type V protein secretion system | Biofilm formation, adherence | Bentancor et al., 2012b |
| Penicillin-binding protein 7/8 and β-lactamase PER-1 | Serum resistance, in vivo survival, adherence | Sechi et al., 2004; Russo et al., 2009 |
| CipA | Serum resistance, invasion | Koenigs et al., 2016 |
| Tuf | Serum resistance | Koenigs et al., 2015 |
| RecA | In vivo survival | Aranda et al., 2011 |
| SurA1 | Serum resistance, in vivo survival | Liu D. et al., 2016 |
| GigABCD | In vivo survival, killing of host cells | Gebhardt et al., 2015 |
| UspA | In vivo survival, killing of host cells | Elhosseiny et al., 2015; Gebhardt et al., 2015 |
| GacS and PaaE | Neutrophil influx | Cerqueira et al., 2014; Gebhardt et al., 2015; Bhuiyan et al., 2016 |
| Pili | Adherence, biofilm formation | Tomaras et al., 2003, 2008 |
| OmpR/EnvZ | Killing of host cells | Tipton and Rather, 2016 |
| FhaBC | Adherence, killing of host cells | Perez et al., 2016 |
| AbeD | Killing of host cells | Srinivasan et al., 2015 |
Identified virulence factors of Acinetobacter baumannii..
Porins
Porins are outer membrane proteins associated with modulating cellular permeability. OmpA is a β-barrel porin and one of the most abundant porins in the outer membrane. In A. baumannii, OmpA is the very well-characterized virulence factor with a variety of interesting biological properties identified in in vitro model systems (Smith et al., 2007; McConnell et al., 2013). A random mutagenesis screen showed that the A. baumannii ompA mutant is defective in inducing apoptosis in human epithelial cells (Choi et al., 2005). Purified OmpA binds host epithelial cells, targets mitochondria, and induces apoptosis by releasing proapoptotic molecules, such as cytochrome c and apoptosis-inducing factor (Choi et al., 2005; Lee et al., 2010). Another study showed that OmpA translocates to the nucleus by a novel monopartite nuclear localization signal and induces cell death (Choi et al., 2008a). OmpA also plays a major role in adherence and invasion of epithelial cells by interacting with fibronectin (Choi et al., 2008b; Gaddy et al., 2009; Smani et al., 2012), and binds to factor H in human serum (Kim et al., 2009), which may allow A. baumannii to avoid complement-mediated killing. The ompA gene is necessary for persistence of A. baumannii in the mouse lung (Wang et al., 2014).
Furthermore, OmpA is also involved in antimicrobial resistance of A. baumannii (Sugawara and Nikaido, 2012; Smani et al., 2014). The major A. baumannii porin is OmpA, which has 70-fold lower pore-forming activity than that of OmpF (Sugawara and Nikaido, 2012). Furthermore, disrupting the ompA gene significantly decreases the minimal inhibitory concentrations (MICs) of several antibiotics (chloramphenicol, aztreonam, and nalidixic acid), suggesting that OmpA participates in the extrusion of antibiotics from the periplasmic space through the outer membrane and couples with inner membrane efflux systems (Smani et al., 2014). OmpA enhances survival and persistence of A. baumannii by facilitating surface motility and biofilm formation (Gaddy et al., 2009; Clemmer et al., 2011; McConnell et al., 2013). OmpA also regulates biogenesis of outer membrane vesicles (Moon et al., 2012). These results suggest that the OmpA protein is an attractive target for developing novel antibiotics and prevention strategies. Two recent reports based on immuno-proteomics and reverse vaccinology suggested that OmpA is a potential vaccine candidate against A. baumannii (Fajardo Bonin et al., 2014; Hassan et al., 2016). Actually, the OmpA protein is immunogenic in healthy individuals and patients with A. baumannii invasive infections (Zhang et al., 2016). In a mouse model of A. baumannii infection, mice immunized with OmpA had a significantly higher survival rate than that of control mice (Luo et al., 2012; Lin L. et al., 2013; Zhang et al., 2016).
The 33- to 36-kDa Omp protein (Omp33-36), which acts as a water passage channel, is another outer membrane porin associated with A. baumannii cytotoxicity (Smani et al., 2013; Rumbo et al., 2014). The omp33-36 deletion strain significantly reduces adherence and invasion of human lung epithelial cells and cytotoxicity to these cells (Smani et al., 2013). Deletion of the omp33-36 gene in a murine sepsis model attenuates lethality and reduces bacterial concentrations in the spleen and lungs (Smani et al., 2013). One study showed that purified Omp33-36 induces apoptosis in several different cell types, including immune and connective tissue cells, by activating caspases and modulating autophagy (Rumbo et al., 2014). Omp33-36 is also involved in antibiotic resistance. A. baumannii strain JC10/01 resistant to carbapenem antibiotics (imipenem and meropenem) exhibits loss of Omp33-36 and episomal expression of Omp33-36 in this strain clearly reduces the MICs of imipenem and meropenem (del Mar Tomas et al., 2005).
Omp22 has also been identified as a novel, conserved, and safe antigen for developing effective vaccines to control A. baumannii infections (Huang et al., 2016), although the contribution of Omp22 to A. baumannii pathogenicity has not been determined. Both active and passive immunizations with Omp22 increase the survival rates of mice, suppress bacterial burdens in the organs and peripheral blood, and reduce serum levels of inflammatory cytokines and chemokines (Huang et al., 2016). Other porins, such as carbapenem-associated outer membrane protein (CarO) and OprD-like, are also virulence-related factors associated with attenuated virulence in a mouse model (Fernandez-Cuenca et al., 2011).
Capsular polysaccharides and lipopolysaccharides (LPS)
Beyond OmpA, the A. baumannii envelope is associated with many factors that contribute to pathogenicity. Among these, capsular exopolysaccharides and LPS are A. baumannii pathogenicity factors. Notably, many isolates from patients with A. baumannii infections express surface capsular polysaccharides and contain a conserved gene cluster, called the K locus, which may determine production of capsular polysaccharides (Koeleman et al., 2001; Hu et al., 2013; Kenyon and Hall, 2013; Geisinger and Isberg, 2015). A random transposon screening to identify genes essential for growth in an inflammatory exudative fluid lead to the identification of the ptk and epsA genes, which are predicted to be required for capsule polymerization and assembly (Russo et al., 2010). The ptk and epsA mutants are deficient in capsule production and have a growth defect in human serum, resulting in a highly significant decrease in survival in soft tissue infection sites (Russo et al., 2010). Mutation in the pglC or pglL gene, which is responsible for synthesis of the O-pentasaccharide found on glycoproteins and capsular polysaccharides, also attenuate lethality in a mouse septicemia model and form abnormal biofilm structures (Iwashkiw et al., 2012; Lees-Miller et al., 2013). Therefore, capsular polysaccharides have been proposed to be a target for protective antibody-based interventions (passive immunization; Russo et al., 2013).
One study showed that capsular polysaccharides are involved in antimicrobial resistance of A. baumannii (Geisinger and Isberg, 2015). Mutants deficient in capsular polysaccharides have lower intrinsic resistance to peptide antibiotics. In addition, the presence of antibiotics induces hyperproduction of capsular polysaccharides (Geisinger and Isberg, 2015). Antibiotic-induced production of capsular polysaccharides increases resistance to killing by host complement and increases virulence in a mouse model of systemic infection (Geisinger and Isberg, 2015). That study also demonstrated that increased capsule production after exposure to an antibiotic depends on transcriptional increases in K locus gene expression, and that expression of K locus genes is regulated by the bfmRS two-component regulatory system (Geisinger and Isberg, 2015). bfmR is a gene essential for growth in human ascites, which is an ex vivo medium that reflects the infection environment (Umland et al., 2012), and is important for persistence in the lung in a murine pneumonia model (Wang et al., 2014). BfmS is also a virulence factor that plays an important role in biofilm formation, adherence to eukaryotic cells, and resistance to human serum (Liou et al., 2014). On report showed BfmR-mediated resistance to complement-mediated bactericidal activity and resistance to the clinically important antimicrobials (meropenem and colistin; Russo et al., 2016). However, that study suggested that BfmR effects are independent of capsular polysaccharide production. Therefore, the relationship between BfmRS and capsular polysaccharides must be described in more detail.
LPS is the major component of the outer leaflet of the outer membrane in most Gram-negative bacteria and is an immunoreactive molecule that induces release of tumor necrosis factor and interleukin 8 from macrophages in a Toll-like receptor 4 (TLR4)-dependent manner (Erridge et al., 2007). LPS is composed of an endotoxic lipid A moiety, an oligosaccharide core, and a repetitive O-antigen (Lee et al., 2013b). In A. baumannii, LPS plays a major role in virulence and survival of A. baumannii (Luke et al., 2010; Lin et al., 2012; McQueary et al., 2012). Mutant cell lacking LpsB glycotransferase have a highly truncated LPS glycoform containing only two carbohydrate residues bound to lipid A, resulting in decreased resistance to human serum and decreased survival in a rat model of soft tissue infection (Luke et al., 2010; McConnell et al., 2013). Inhibiting LpxC, an enzyme involved in the lipid A biosynthesis, dose not inhibit growth of the bacterium, but suppresses A. baumannii LPS-mediated activation of TLR4 (Lin et al., 2012). Inhibition of LpxC in mouse model enhances clearance of A. baumannii by enhancing opsonophagocytic killing and reduces serum LPS concentration and inflammation, which completely protects mice from lethal infection (Lin et al., 2012; Lee et al., 2013b). These results indicate that blocking LPS synthesis is a powerful strategy for discovering novel antibiotics. Modification of LPS contributes to resistance to antimicrobials. Many studies have shown that modifications in LPS decrease the susceptibility of A. baumannii to many clinical important antibiotics, such as colistin (Moffatt et al., 2010; Arroyo et al., 2011; Beceiro et al., 2011; Pelletier et al., 2013; Boll et al., 2015; Chin et al., 2015).
Phospholipase
Phospholipase is a lipolytic enzyme essential for phospholipid metabolism and is a virulence factor in many bacteria, such as P. aeruginosa, Legionella monocytogenes, and Clostridium perfringens (Camarena et al., 2010; Flores-Diaz et al., 2016). Three classes of phospholipases, such as phospholipase A (PLA), phospholipase C (PLC), and phospholipase D (PLD) have been defined based on the cleavage site. PLA hydrolyzes fatty acids from the glycerol backbone, whereas PLC cleaves the phosphorylated head group from the phospholipid. PLD is a transphosphatidylase that only cleaves off the head group. Degradation of phospholipids affects the stability of host cell membranes, and the cleaved head group can interfere with cellular signaling, resulting in changes in the host immune response (Songer, 1997; Flores-Diaz et al., 2016). PLC and PLD have been identified as virulence factors in A. baumannii (Camarena et al., 2010; Jacobs et al., 2010; Stahl et al., 2015). Acinetobacter baumannii ATCC17978 has two PLCs (A1S_0043 and A1S_2055) and inactivation of the A1S_0043 gene leads to a modest reduction in the cytotoxic effect of A. baumannii on epithelial cells compared to that of the parental strain (Camarena et al., 2010; Fiester et al., 2016). Disrupting one (A1S_2989) of the two PLD genes present in A. baumannii strain 98-37-09 results in reduced resistance to human serum, decreased capacity for invading epithelial cells, and decreased virulence in a murine model of pneumonia (Jacobs et al., 2010). Another report showed that A. baumannii ATCC 19606 has three PLD genes and all three play important roles in virulence and host cell invasion in a concerted manner (Stahl et al., 2015). These results suggest that phospholipase enzymes are important virulence factors in A. baumannii pathogenesis.
Outer membrane vesicles (OMVs)
OMVs are spherical, 20–200 nm diameter vesicles secreted by the outer membranes of various Gram-negative pathogenic bacteria (Kulp and Kuehn, 2010). They are composed of LPS, outer membrane and periplasmic proteins, phospholipids, and DNA or RNA, and are recognized as delivery vehicles for bacterial effectors to host cells (Ellis and Kuehn, 2010). OMVs deliver diverse virulence factors to the interior of host cells simultaneously and allow the pathogens to interact with the host without close contact between bacteria and host cells (Jun et al., 2013). Many A. baumannii strains secrete OMVs containing various virulence factors, including OmpA (Kwon et al., 2009; Jin et al., 2011; Moon et al., 2012), proteases (Kwon et al., 2009), and phospholipases (Kwon et al., 2009). OMVs derived from A. baumannii interact with host cells and deliver bacterial effectors to host cells via lipid rafts, resulting in cytotoxicity (Jin et al., 2011). Purified OMVs of A. baumannii ATCC 19606 induce expression of pro-inflammatory cytokine genes in epithelial cells in a dose-dependent manner (Jun et al., 2013). Notably, OMVs treated with proteinase do not induce a significant increase in the expression of pro-inflammatory cytokine genes, suggesting that the membrane proteins in OMVs are responsible for eliciting a potent innate immune response (Jun et al., 2013). One study supports the role of OMVs in A. baumannii pathogenesis. An A. baumannii strain that produces abundant OMVs with more virulence factors induces a stronger innate immune response and is more cytotoxic compared with those of a strain producing fewer OMVs (Li Z. T. et al., 2015).
Due to the importance of OMVs in A. baumannii virulence, several reports have shown that A. baumannii OMVs could be used as an acellular vaccine to elevate protective immunity (McConnell et al., 2011; Huang et al., 2014). In a mouse model of disseminated sepsis, vaccination with A. baumannii ATCC 19606 strain OMVs protects mice from challenge with homologous bacteria and provides protection against other clinical isolates (McConnell et al., 2011). Similar results were obtained in a pneumonia mouse model. Bacterial burden, inflammatory cell infiltration, and inflammatory cytokine accumulation in the pneumonia model were significantly suppressed by both active and passive immunization with OMVs (Huang et al., 2014). These results indicate that A. baumannii OMVs can be used as an acellular vaccine to effectively control A. baumannii infections. Interestingly, A. baumannii OMVs are also related with the spread of antibiotic resistance and induce the horizontal transfer of the OXA-24 carbapenemase gene (Rumbo et al., 2011).
Metal acquisition system
Although iron is one of the most abundant elements in environmental and biological systems, ferric iron is relatively unavailable to bacteria in the preferred state, because of its poor solubility (10−17 M solubility limit for ferric iron) under aerobic and neutral pH conditions as well as due to chelation by low-molecular-weight compounds, such as heme, or high-affinity iron-binding compounds, such as lactoferrin and transferrin (Rakin et al., 2012; Saha et al., 2013). To overcome this iron limitation, most aerobic bacteria produce a high-affinity iron chelator known as a siderophore (Saha et al., 2013). Siderophores are low molecular weight compounds (400–1,000 kDa) with high affinity for iron. The range of Fe3+-siderophore association constants is 1012–1052 (Saha et al., 2013). Siderophores have been classified into catecholates, hydroxymates, and a mixed type based on the moiety that donates oxygen ligands to coordinate Fe3+ (Saha et al., 2013). Acinetobacter baumannii also has iron siderophores and acinetobactin, the best-characterized A. baumannii siderophore, is a mixed type siderophore with an oxazoline ring derived from threonine (McConnell et al., 2013). Acinetobactin is an A. baumannii virulence factor (Gaddy et al., 2012; Penwell et al., 2012; Megeed et al., 2016). Impaired acinetobactin biosynthesis and transport functions significantly reduce the ability of A. baumannii ATCC 19606 cells to persist within epithelial cells and cause cell damage and animal death (Gaddy et al., 2012). Mutation in the entA gene, which is essential for biosynthesis of the acinetobactin precursor 2,3-dihydroxybenzoic acid, also significantly reduces the capacity of A. baumannii ATCC 19606 cells to persist within human alveolar epithelial cells and diminishes the ability to infect and kill Galleria mellonella larvae (Penwell et al., 2012). One study showed that acinetobactin production occurs significantly more frequently in MDR A. baumannii isolates than that in avirulent isolates (Megeed et al., 2016).
The A. baumannii NfuA Fe-S scaffold protein, that participates in the formation of Fe-S clusters and plays a role in cell responses to iron chelation and oxidative stress, has also been identified as a virulence factor (Zimbler et al., 2012). The nfuA mutant is more sensitive to reactive oxygen species (ROS), such as hydrogen peroxide and cumene hydroperoxide, and shows significantly reduced growth in human epithelial cells. In addition, a G. mellonella infection model showed that more than 50% of injected G. mellonella larvae die 6 days after infection with the parental strain, whereas less than 30% of the larvae die when infected with the nfuA mutant (Zimbler et al., 2012). One report showed that iron starvation increases production of PLCs, which increase hemolytic activity of A. baumannii (Fiester et al., 2016). These reports indicate that iron acquisition functions play a critical role in A. baumannii virulence.
The innate immune metal-chelating protein calprotectin inhibits bacterial growth by host-mediated chelation of metals, such as zinc (Zn2+ and Zn) and manganese (Mn2+ and Mn) (Corbin et al., 2008). However, A. baumannii can cause disease in the presence of this nutritional immune protein in vivo (Juttukonda et al., 2016). To combat the zinc limitation, A. baumannii uses a zinc acquisition system (ZnuABC), which is up-regulated under Zn-limiting conditions, and the znuB mutant strain experiences Zn starvation at higher Zn concentrations than that of the wild-type (Hood et al., 2012). ZnuB contributes to the pathogenesis of A. baumannii pulmonary infections. Notably, a zinc limitation reduces the imipenem MIC of MDR A. baumannii to below the clinical breakpoint for imipenem resistance in A. baumannii (Hood et al., 2012), possibly because many carbapenemases are metalloenzymes that require Zn for their hydrolyzing activity. Besides the ZnuABC system, the novel Zn metallochaperone ZigA has been characterized in A. baumannii (Nairn et al., 2016). ZigA tightly interacts with Zn, which is required for bacterial growth under Zn starvation conditions and for disseminated infection in mice (Nairn et al., 2016).
The mechanism employed by A. baumannii to overcome a Mn limitation has been identified. Calprotectin induces Mn starvation in A. baumannii, which increases transcription of an NRAMP (Natural Resistance-Associated Macrophage Proteins) family Mn transporter and a urea carboxylase to resist the antimicrobial activities of calprotectin (Juttukonda et al., 2016). A urea carboxylase enzyme (MumC) is important for growth of A. baumannii in the presence of calprotectin and an NRAMP family transporter (MumT) contributes to the fitness of A. baumannii in a murine pneumonia model (Juttukonda et al., 2016), suggesting that the two proteins are virulence factors. Acinetobacter baumannii can utilize urea as a sole nitrogen source, and this urea utilization is required for MumC (Juttukonda et al., 2016). Based on the contribution of MumC to A. baumannii resistance to calprotectin, the authors suggest a connection between metal starvation and metabolic stress, such as nitrogen starvation.
Protein secretion systems
Several protein secretion systems have been identified in A. baumannii (Weber et al., 2015a). The most recently described A. baumannii secretion system is a type II secretion system (T2SS) (Johnson et al., 2015). The T2SS is a multi-protein complex that is structurally very similar to type IV pili systems, which is an appendage that is commonly found in Gram-negative bacteria (Korotkov et al., 2012). T2SS translocates a wide range of proteins from the periplasmic space to the extracellular milieu out of the cell or the outer membrane surface. The T2SS is composed of 12–15 proteins comprised of four sub-assemblies: a pseudopilus, a cytoplasmic secretion ATPase, an inner-membrane platform assembly, and a dodecameric outer-membrane complex (Korotkov et al., 2012; Harding et al., 2016). Secretion by T2SS is a two-step process. The target proteins are first translocated to the periplasm by the general secretory (Sec) system or the twin arginine transport (Tat) system, where the target proteins are then secreted out of the cell through the T2SS (Korotkov et al., 2012). Deleting A. baumannii genes for the T2SS components, gspD or gspE, results in loss of LipA secretion, indicating that LipA is a T2SS substrate (Johnson et al., 2015). Because LipA is a lipase that breaks down long-chain fatty acids, lipA, gspD, and gspE mutant strains are incapable of growing on long-chain fatty acids as a sole carbon source and are defective in in vivo growth in a neutropenic murine model of bacteremia (Johnson et al., 2015). The role of a functional T2SS for full virulence of A. baumannii has been shown in G. mellonella and murine pulmonary infection models (Harding et al., 2016). Lipases (LipA, LipH, and LipAN) and the metallopeptidase CpaA have been identified as T2SS substrates (Elhosseiny et al., 2016; Harding et al., 2016). Notably, two proteins (LipA and CpaA) among these secreted proteins require specific chaperones for secretion. These chaperones are encoded adjacently to their cognate effector, and their inactivation abolishes secretion of LipA and CpaA (Harding et al., 2016).
Acinetobacter baumannii also has a type VI secretion system (T6SS). The T6SS was first identified in Vibrio cholera and P. aeruginosa (Mougous et al., 2006; Pukatzki et al., 2006). Many bacteria use the T6SS to inject effector proteins, providing a colonization advantage during infection of eukaryotic hosts (Mougous et al., 2006) or to kill competing bacteria (Basler et al., 2013). The T6SS leads to DNA release and horizontal gene transfer in V. cholera, which may contribute to spread of antibiotic resistance (Borgeaud et al., 2015). The T6SS is composed of many conserved structural proteins and accessory factors, and bears a contractile bacteriophage sheath-like structure forming a needle or spike structure used to penetrate the target cell (Shneider et al., 2013). Hcp is a structural protein forming a polymerized tubular structure that is secreted out of the cell, and VgrGs are involved in attaching effector domains to the spike, and a proline-alanine-alanine-arginine (PAAR) repeat protein forms the sharp tip of the distinctive needle-like structure (Shneider et al., 2013; Zoued et al., 2014).
The presence of T6SS in A. baumannii was initially predicted by bioinformatic analysis (Weber et al., 2013). Although the role of T6SS in A. baumannii ATCC 17978 has not been determined (Weber et al., 2013), research on A. baumannii strain M2 showed that this strain produces a functional T6SS and that the T6SS mediates killing of competing bacteria (Carruthers et al., 2013). Another study showed that the T6SS is active in six pathogenic strains of A. baumannii (Ruiz et al., 2015). However, the T6SS seems to play an important role in A. baumannii virulence in a strain-specific manner (Repizo et al., 2015). They compared T6SS functionality of several A. baumannii strains, including ATCC17978 (a type strain), various MDR strains implicated in hospital outbreaks (Ab242, Ab244, and Ab825), and DSM30011 (a non-clinical isolate). Although the T6SS genomic locus is present in all of these strains, only DSM30011 has a fully active T6SS that mediates E. coli killing (Repizo et al., 2015). In addition, the T6SS of DSM30011 is required for host colonization of the G. mellonella model organism (Repizo et al., 2015). Similar results were obtained from a comparative analysis of the genomes of MDR A. baumannii clinical strains (Wright et al., 2014; Jones et al., 2015). A. baumannii isolates of a particular clade exhibit complete loss of the T6SS genomic locus. Therefore, these results suggest that more extensive investigations are required to analyze the role of T6SS in A. baumannii virulence, even though this system seems to play an important role in A. baumannii virulence in some strains. Notably, one study showed that several MDR A. baumannii strains have a large, self-transmissible plasmid that carries negative regulators for T6SS (Weber et al., 2015b). The T6SS is silenced in plasmid-containing, antibiotic-resistant cells, whereas plasmid-losing cells have an active T6SS. Although plasmid-losing cells are capable of T6SS-mediated killing of competing bacteria, they become susceptible to antibiotics (Weber et al., 2015b). This result suggests a molecular switch between T6SS and antibiotic resistance.
The type V system autotransporter Ata has also been characterized in A. baumannii (Bentancor et al., 2012a). This is a trimeric membrane protein that mediates biofilm formation, adherence to extracellular matrix components such as collagen I, III, and IV, and virulence in a murine systemic model of Acinetobacter infection (Bentancor et al., 2012a). Another experiment using a pneumonia model of infection in immunocompetent and immunocompromised mice showed that Ata is a vaccine candidate against A. baumannii infections (Bentancor et al., 2012b). A type IV secretion system present in the plasmid was bioinformatically identified in A. baumannii (Liu C. C. et al., 2014), but no experimental evidence describing its function has been presented.
Penicillin-binding protein 7/8 (PBP7/8) and β-lactamase PER-1
Although PBPs are commonly involved in resistance to β-lactam antibiotics, PBP7/8 encoded by the pbpG gene is a virulence factor in A. baumannii. The pbpG mutant strain grows similar to its wild-type strain in Luria-Bertani medium, but the mutant shows reduced growth in human serum and its survival significantly decreases in rat soft-tissue infection and pneumonia models (Russo et al., 2009). An investigation of bacterial morphology using electron microscopy suggested that loss of PBP7/8 may have affected peptidoglycan structure, which may affect susceptibility to host defense factors (Russo et al., 2009).
Interestingly, β-lactamase PER-1 has been suggested to be an A. baumannii virulence factor. PER-1 is an extended-spectrum-β-lactamase (ESBL), but this gene is associated with cell adhesion (Sechi et al., 2004). Nine PER-1-producing strains adhere to the Caco2 cell lines, whereas all PER-1-negative strains are negative for cell adhesion (Sechi et al., 2004). Notably, many β-lactamases are associated with virulence in various pathogenic bacteria, such as E. coli (Dubois et al., 2009), P. aeruginosa (Moya et al., 2008), and K. pneumoniae (Sahly et al., 2008). However, no general mechanisms have been proposed (Beceiro et al., 2013).
Others
Acinetobacter baumannii CipA is a novel plasminogen binding and complement inhibitory protein that mediates serum resistance (Koenigs et al., 2016). CipA-binding plasminogen is converted to active plasmin that degrades fibrinogen and complement C3b, which contributes to serum resistance of A. baumannii. Therefore, the cipA mutant strain is efficiently killed by human serum and also shows a defect in the penetration of endothelial monolayers (Koenigs et al., 2016). Similar to CipA, the A. baumannii translation elongation factor Tuf is also a plasminogen-binding protein. Tuf-binding plasminogen can be converted to active plasmin, which proteolytically degrades fibrinogen as well as component C3b (Koenigs et al., 2015). RecA, which is involved in homologous recombination and the SOS response, has been identified as an A. baumannii virulence factor. The recA mutant shows significantly reduced survival within macrophages and decreases lethality in a mouse model of systemic infection (Aranda et al., 2011). The surface antigen protein 1 (SurA1) plays an important role in fitness and virulence of A. baumannii (Liu D. et al., 2016). Serum resistance of the surA1 mutant significantly decreases compared with that of the wild-type strain CCGGD201101. In the G. mellonella insect model, a surA1 mutant strain exhibits a lower survival rate and decreased dissemination (Liu D. et al., 2016).
A growth analysis of 250,000 A. baumannii transposon mutants within G. mellonella larvae identified 300 genes required for survival or growth of A. baumannii inside G. mellonella larvae (Gebhardt et al., 2015). The 300 genes were classified into six categories of micronutrient acquisition, cysteine metabolism/sulfur assimilation, aromatic hydrocarbon metabolism, cell envelope/membrane/wall, stress response genes, antibiotic resistance, and transcriptional regulation. Among them, four transcriptional regulators required for growth in G. mellonella larvae were called the gig (growth in Galleria) genes. Loss of these genes (gigA-D) led to a significant defect in both growth within and killing of G. mellonella larvae (Gebhardt et al., 2015). This study identified stress proteins, such as UspA, as factors required for growth in G. mellonella. Another study showed that UspA is essential for pneumonia and sepsis pathogenesis of A. baumannii (Elhosseiny et al., 2015). Among the 300 genes, several genes are involved in aromatic hydrocarbon metabolism (Gebhardt et al., 2015). Another study showed that GacS, which is a transcriptional factor that regulates expression of genes, such as paaE, and is responsible for the phenylacetic acid catabolic pathway, affects A. baumannii virulence (Cerqueira et al., 2014). Experiments using a paaE deletion mutant confirmed the role of aromatic hydrocarbon metabolism in A. baumannii virulence (Cerqueira et al., 2014), but its molecular mechanism remains unknown. Interestingly, a recent report showed that accumulation of phenylacetate in A. baumannii induces rapid neutrophil influx to a localized site of infection and increases bacterial clearance (Bhuiyan et al., 2016). They suggested that phenylacetate is a neutrophil chemoattractant inducing bacterial-guided neutrophil chemotaxis. This report may reveal a novel molecular mechanism about the role of the phenylacetic acid catabolic pathway in A. baumannii virulence.
Biofilm formation plays an important role in immune evasion by A. baumannii (de Breij et al., 2010), and pili are essential for A. baumannii adherence to and biofilm formation on abiotic surfaces as well as virulence (Tomaras et al., 2003, 2008). Notably, imipenem treatment of the imipenem-resistant A. baumannii isolate induces expression of important genes responsible for synthesis of type IV pili (Dhabaan et al., 2015), suggesting that the ability to overproduce pili confers a biological advantage to A. baumannii.
Other virulence-related proteins have been identified, including OmpR/EnvZ (Tipton and Rather, 2016), FhaBC (Perez et al., 2016), and the resistance-nodulation-division-type membrane transporter AbeD (Srinivasan et al., 2015), but their molecular mechanisms remain unknown.
Antimicrobial resistance of A. baumannii
Acinetobacter baumannii has become one of the most successful pathogens in modern healthcare because of its amazing ability to acquire antimicrobial resistance. Several strains of A. baumannii are highly resistant to most clinically available antibiotics (Lin and Lan, 2014). A. baumannii has a number of resistance mechanisms, including β-lactamases, aminoglycoside-modifying enzymes, efflux pumps, permeability defects, and modifications of target sites. The accumulation of several resistance mechanisms in A. baumannii has gradually decreased the number of antibiotic classes available to treat A. baumannii infections in clinical practice. Table 2 shows the antibiotic resistance mechanisms found in A. baumannii. We will discuss the details below.
Table 2
| Resistance mechanism | Class/subgroup | Protein | References |
|---|---|---|---|
| β-Lactamases | Class A | TEM-1 | Chen et al., 2006; Adams et al., 2008; Krizova et al., 2013 |
| TEM-92 | Endimiani et al., 2007 | ||
| GES-1 | Al-Agamy et al., 2016 | ||
| GES-5 | Al-Agamy et al., 2016 | ||
| GES-11 | Moubareck et al., 2009; Bogaerts et al., 2010; Chihi et al., 2016 | ||
| GES-12 | Bogaerts et al., 2010 | ||
| GES-14 | Bogaerts et al., 2010 | ||
| PER-1 | Jeong et al., 2005; Poirel et al., 2005a; Aly et al., 2016 | ||
| PER-2 | Pasteran et al., 2006 | ||
| PER-7 | Bonnin et al., 2011b | ||
| CTX-M-2 | Nagano et al., 2004 | ||
| CTX-M-15 | Potron et al., 2011 | ||
| SCO-1 | Poirel et al., 2007 | ||
| VEB-1 | Fournier et al., 2006; Naas et al., 2006; Pasteran et al., 2006; Adams et al., 2008; Poirel et al., 2009 | ||
| KPC-2 | Martinez et al., 2016 | ||
| KPC-10 | Robledo et al., 2010 | ||
| CARB-4 | Ramirez et al., 2010b | ||
| CARB-10 | Potron et al., 2009 | ||
| Class B | IMP-1 | Tognim et al., 2006 | |
| IMP-2 | Riccio et al., 2000 | ||
| IMP-4 | Chu et al., 2001 | ||
| IMP-5 | Koh et al., 2007 | ||
| IMP-6 | Gales et al., 2003 | ||
| IMP-8 | Lee M. F. et al., 2008 | ||
| IMP-11 | Yamamoto et al., 2011 | ||
| IMP-19 | Yamamoto et al., 2011 | ||
| IMP-24 | Lee M. F. et al., 2008 | ||
| VIM-1 | Tsakris et al., 2006, 2008; Papa et al., 2009 | ||
| VIM-2 | Yum et al., 2002; Lee M. F. et al., 2008 | ||
| VIM-3 | Lee M. F. et al., 2008 | ||
| VIM-4 | Tsakris et al., 2008; Papa et al., 2009 | ||
| VIM-11 | Lee M. F. et al., 2008 | ||
| NDM-1 | Chen et al., 2011; Pfeifer et al., 2011; Bonnin et al., 2012; Voulgari et al., 2016 | ||
| NDM-2 | Espinal et al., 2011 | ||
| NDM-3 | Kumar, 2016 | ||
| SIM-1 | Lee et al., 2005 | ||
| Class C | AmpC | Bou and Martinez-Beltran, 2000; Corvec et al., 2003; Segal et al., 2004; Hujer et al., 2005; Heritier et al., 2006; Liu and Liu, 2015 | |
| Class D | |||
| OXA-2 subgroup | OXA-21 | Vila et al., 1997 | |
| OXA-10 subgroup | OXA-128 | Giannouli et al., 2009 | |
| OXA-20 subgroup | OXA-37 | Navia et al., 2002 | |
| OXA-23 subgroup | OXA-23 | Heritier et al., 2005b; Naas et al., 2005; Corvec et al., 2007; Koh et al., 2007; Perez et al., 2007; Valenzuela et al., 2007; Wang et al., 2007; Adams et al., 2008; Stoeva et al., 2008; Kohlenberg et al., 2009; Kuo et al., 2010; Mugnier et al., 2010; Bonnin et al., 2011a; Lee et al., 2011; Lin et al., 2011b; Koh et al., 2012; Mosqueda et al., 2013; Chagas et al., 2014; Principe et al., 2014; Li Y. et al., 2015 | |
| OXA-133 | Mendes et al., 2009 | ||
| OXA-239 | Gonzalez-Villoria et al., 2016 | ||
| OXA-24 subgroup | OXA-24 | Bou et al., 2000b; Merino et al., 2010; Acosta et al., 2011; Pailhories et al., 2016 | |
| OXA-25, OXA-26, OXA-27 | Afzal-Shah et al., 2001 | ||
| OXA-40 | Heritier et al., 2003; Lolans et al., 2006; Quinteira et al., 2007; Ruiz et al., 2007 | ||
| OXA-72 | Wang et al., 2007; Lu et al., 2009; Goic-Barisic et al., 2011; Dortet et al., 2016; Kuo et al., 2016 | ||
| OXA-143 | Higgins et al., 2009 | ||
| OXA-182 | Kim et al., 2010 | ||
| OXA-51 subgroup | OXA-51 | Brown et al., 2005; Hu et al., 2007; Ruiz et al., 2007; Adams et al., 2008; Chen et al., 2010; Fang et al., 2016 | |
| OXA-64, OXA-65, OXA-66, OXA-68, OXA-70, OXA-71 | Hamouda et al., 2010; Biglari et al., 2016 | ||
| OXA-69, OXA-75, OXA-76, OXA-77 | Heritier et al., 2005a | ||
| OXA-79, OXA-80, OXA-104, OXA-106~ OXA-112 | Evans et al., 2007 | ||
| OXA-82, OXA-83, OXA-83, OXA-84 | Turton et al., 2006b; Evans et al., 2007 | ||
| OXA-86, OXA-87 | Vahaboglu et al., 2006 | ||
| OXA-88, OXA-91, OXA-93, OXA-94, OXA-95, OXA-96 | Koh et al., 2007 | ||
| OXA-92 | Tsakris et al., 2007 | ||
| OXA-113 | Naas et al., 2007 | ||
| OXA-58 subgroup | OXA-58 | Dijkshoorn et al., 1996; Poirel et al., 2005b; Pournaras et al., 2006; Chen et al., 2008; Qi et al., 2008; Donnarumma et al., 2010; Gogou et al., 2011; Ravasi et al., 2011; Hou and Yang, 2015 | |
| OXA-96 | Koh et al., 2007 | ||
| OXA-97 | Poirel et al., 2008 | ||
| OXA-143 subgroup | OXA-253 | de Sa Cavalcanti et al., 2016 | |
| OXA-235 subgroup | OXA-235 | Higgins et al., 2013 | |
| Efflux pumps | Resistance-nodulation-division superfamily | AdeABC | Magnet et al., 2001; Marchand et al., 2004; Peleg et al., 2007; Ruzin et al., 2007; Lin et al., 2015; Sun et al., 2016 |
| AdeFGH | Coyne et al., 2010; He X. et al., 2015 | ||
| AdeIJK | Damier-Piolle et al., 2008 | ||
| Major facilitator superfamily | TetA | Ribera et al., 2003a | |
| TetB | Vilacoba et al., 2013 | ||
| CmlA | Coyne et al., 2011 | ||
| CraA | Roca et al., 2009 | ||
| AmvA | Rajamohan et al., 2010 | ||
| AbaF | Sharma et al., 2016 | ||
| Multidrug and toxic compound extrusion family | AbeM | Su et al., 2005 | |
| Small multidrug resistance family | AbeS | Srinivasan et al., 2009 | |
| Other efflux pumps | EmrAB-TolC | Nowak-Zaleska et al., 2016 | |
| A1S_1535, A1S_2795, and ABAYE_0913 | Li L. et al., 2016 | ||
| Permeability defects | Porin | OmpA | Smani et al., 2014; Wu et al., 2016 |
| CarO | Mussi et al., 2005, 2007; Siroy et al., 2005; Catel-Ferreira et al., 2011; Jin et al., 2011 | ||
| Omp22-33 | Bou et al., 2000a | ||
| Omp33-36 | del Mar Tomas et al., 2005 | ||
| Omp37 | Quale et al., 2003 | ||
| Omp43 | Dupont et al., 2005 | ||
| Omp44 | Quale et al., 2003 | ||
| Omp47 | Quale et al., 2003 | ||
| Aminoglycoside-modifying enzymes | Aminoglycoside acetyltransferases | AAC3 (aacC1, aacC2) | Nemec et al., 2004 |
| AAC(6′) (aacA4) | Doi et al., 2004; Cho et al., 2009; Zhu et al., 2009; Lin et al., 2010; Lin M. F. et al., 2013; Bakour et al., 2014 | ||
| Aminoglycoside adenyltransferases | ANT(2″) (aadB) | Nemec et al., 2004 | |
| ANT(3″) (aadA1) | Cho et al., 2009; Lin et al., 2010; Lin M. F. et al., 2013 | ||
| Aminoglycoside phosphotransferases | APH(3′) (aphA1) | Gallego and Towner, 2001 | |
| APH(3″) | Cho et al., 2009 | ||
| Alteration of target sites | Change of penicillin binding protein(PBP) | PBP2 | Gehrlein et al., 1991 |
| 16S rRNA methylation | ArmA | Yu et al., 2007; Cho et al., 2009; Karthikeyan et al., 2010; Brigante et al., 2012; Hong et al., 2013; Bakour et al., 2014; Tada et al., 2014 | |
| Ribosomal protection | TetM | Ribera et al., 2003b | |
| DNA gyrase | GyrA/ParC | Higgins et al., 2004 | |
| Dihydrofolate reductase | DHFR | Mak et al., 2009; Lin M. F. et al., 2013 | |
| FolA | Mak et al., 2009 | ||
| Lipopolysaccharide | PmrC, LpxA, LpxC, LpxD | Adams et al., 2009; Moffatt et al., 2010; Arroyo et al., 2011 | |
| Other mechanisms | S-adenosyl-L-methionine-dependent methyltransferase | Trm | Chen et al., 2014; Trebosc et al., 2016 |
| 1-Acyl-sn-3-phosphate acyltransferase | PlsC | Li X. et al., 2015 | |
| Peptidase C13 family | Abrp | Li X. et al., 2016 | |
| Cell division proteins | BlhA, ZipA, ZapA, and FtsK | Knight et al., 2016 | |
| SOS response | RecA | Aranda et al., 2011, 2014; Norton et al., 2013 |
Resistance mechanisms in Acinetobacter baumannii.
β-lactamases
Inactivation of β-lactams by β-lactamases is a major antibiotic resistance mechanism in A. baumannii. Based on sequence homology, β-lactamases are grouped into molecular classes, A, B, C, and D (Jeon et al., 2015). All four classes of β-lactamases were identified in A. baumannii. Recent studies have shown that A. baumannii has natural competence to incorporate exogenous DNA and its genome has foreign DNA at high frequencies, implying frequent horizontal gene transfer in this pathogen (Ramirez et al., 2010a; Touchon et al., 2014; Traglia et al., 2014). Additionally, albumin, a main protein in blood, enhances natural competence of A. baumannii (Traglia et al., 2016). Therefore, natural competence of A. baumannii may contribute to identification of a large number of β-lactamases in this threatening human pathogen.
Class A β-lactamases inhibited by clavulanate hydrolyze penicillins and cephalosporins more efficiently than carbapenems, except for some KPC type enzymes (Jeon et al., 2015). A number of class A β-lactamases, including TEM, SHV, GES, CTX-M, SCO, PER, VEB, KPC, and CARB, have been identified in A. baumannii (Table 2). Some of these enzymes, such as TEM-1, CARB-4, and SCO-1, are narrow-spectrum β-lactamases, whereas other enzymes (e.g., PER-1, TEM-92, CARB-10, SHV-5, PER-2, CTX-M-2, CTX-M-15, VEB-1, GES-14, and PER-7) are ESBLs. Some carbapenemases, such as GES-14 and KPC-2, have been detected in A. baumannii (Moubareck et al., 2009; Bogaerts et al., 2010).
Unlike the serine-dependent β-lactamases (classes A, C, and D), class B β-lactamases are metallo-β-lactamases (MBLs) that require zinc or another heavy metal for catalysis (Jeon et al., 2015). Due to a broad substrate spectrum, MBLs catalyze the hydrolysis of virtually all β-lactam antibiotics including carbapenems, but not monobactams (Jeon et al., 2015). A variety of class B β-lactamases have been identified in A. baumannii (Table 2).
Class C β-lactamases pose therapeutic problems because they can confer resistance to cephamycins (cefoxitin and cefotetan), penicillins, cephalosporins, and β-lactamase inhibitor combinations, but are not significantly inhibited by clinically used β-lactamase inhibitors, such as clavulanic acid (Jeon et al., 2015). Acinetobacter baumannii has an intrinsic AmpC cephalosporinase (Gordon and Wareham, 2010). An analysis of 23 MDR A. baumannii clinical isolates in Taiwan showed that all isolates had AmpC-type β-lactamases (Lin et al., 2011a). Several clinical isolates of A. baumannii have the ampC gene transcribed from a strong promoter contained within a putative insertion sequence element (ISAba1-like sequence), which results in high resistance to ceftazidime (Corvec et al., 2003; Segal et al., 2004). This sequence has been identified in ceftazidime-resistant A. baumannii isolates, but is absent in ceftazidime-susceptible A. baumannii isolates (Heritier et al., 2006).
Class D β-lactamases are called OXAs (oxacillinases), because they commonly hydrolyze isoxazolylpenicillin oxacillin much faster than benzylpenicillin (Jeon et al., 2015). More than 400 OXA-type enzymes have been identified and many variants actually possess carbapenemase activity. The presence of carbapenem-hydrolyzing class D β-lactamases or MBLs is one of the major carbapenem resistance mechanisms in A. baumannii (Lin and Lan, 2014). The subgroups of carbapenem-hydrolyzing OXAs, such as the OXA-23, OXA-24, OXA-51, and OXA-58 subgroups, are prevalent in A. baumannii (Table 2). The OXA-23 enzyme was first identified in an A. baumannii isolate in the United Kingdom in 1985 (Perez et al., 2007). The blaOXA-23 gene has been disseminated worldwide, and the frequency of OXA-23-producing A. baumannii strains is significantly high (Mugnier et al., 2010; Al-Agamy et al., 2016). One recent report from Lebanon showed 76.5% of 119 A. baumannii isolates are resistant to carbapenems, and OXA-23 β-lactamases have been found in 82 isolates (Al Atrouni et al., 2016). Insertion of ISAba1 in the blaOXA-23 promoter sequence has been reported to be associated with overexpression of blaOXA-23, blaOXA-51, or blaOXA-58 in A. baumannii (Turton et al., 2006a). One report from India showed that blaOXA-51 and blaOXA-23 were present in all 103 carbapenem-resistant A. baumannii isolates and almost 80% of the isolates had ISAba1 upstream of the blaOXA-23 gene, indicating the prevalence of the ISAba1 insertion (Vijayakumar et al., 2016).
Efflux pumps
Efflux pumps are associated with resistance against many different classes of antibiotics, such as imipenem (Hu et al., 2007) and tigecycline (Peleg et al., 2007; Ruzin et al., 2007), in A. baumannii. Reversal of antibiotic resistance by efflux pump inhibitors, such as 1-(1-naphthylmethyl)-piperazine and carbonyl cyanide 3-chlorophenyl-hydrazone, supports the importance of efflux pumps in A. baumannii antibiotic resistance (Pannek et al., 2006; Deng et al., 2014). Four categories of efflux pumps, such as the resistance-nodulation-division superfamily, the multidrug and toxic compound extrusion family, the major facilitator superfamily, and the small multidrug resistance family transporters, are related to antimicrobial resistance in A. baumannii (Table 2; Lin and Lan, 2014).
AdeABC in the resistance-nodulation-division superfamily is associated with aminoglycoside resistance (Magnet et al., 2001) and with decreasing susceptibility to tigecycline (Ruzin et al., 2007) and non-fluoroquinolone antibiotics (Higgins et al., 2004). AdeABC seems to be cryptic in wild-type A. baumannii because of stringent control by the AdeRS two-component system (Marchand et al., 2004), but point mutations or insertion of the ISAba1 sequence in the adeS gene leads to overexpression of AdeABC (Marchand et al., 2004; Sun et al., 2012, 2016; Hammerstrom et al., 2015). Cell density (Fernando and Kumar, 2012) and the BaeSR two-component system (Lin et al., 2014, 2015), which is involved in an envelope stress response, also seem to regulate transcription of the adeA gene and thus affect tigecycline susceptibility. Other resistance-nodulation-division type efflux pumps, such as AdeFGH and AdeIJK, are synergistically associated with tigecycline resistance (Damier-Piolle et al., 2008). AdeFGH and AdeIJK expression is regulated by the LysR-type transcriptional regulator AdeL and the TetR-type transcriptional regulator AdeN (Coyne et al., 2010; Rosenfeld et al., 2012).
Acinetobacter baumannii clinical isolates possess a strong ability to form biofilms (Rodriguez-Bano et al., 2008). Notably, the subinhibitory concentrations of antibiotics encountered by low-dose therapy seem to strongly induce biofilm formation (Kaplan, 2011). A recent result revealed the mechanism. Overexpression of the AdeFGH efflux pump by low-dose antimicrobial therapy increases the synthesis and transport of autoinducer molecules, which induce biofilm formation (He X. et al., 2015). These results suggest a link between low-dose antimicrobial therapy and a high risk for biofilm infections caused by A. baumannii.
CmlA and CraA are major facilitator superfamily efflux pumps related with chloramphenicol (Fournier et al., 2006; Roca et al., 2009), and TetA is associated with tetracycline resistance (Ribera et al., 2003a). The novel efflux pump AmvA mediates resistance to different classes of antibiotics, disinfectants, detergents, and dyes, such as erythromycin, acriflavine, benzalkonium chloride, and methyl viologen (Rajamohan et al., 2010). AbaF was recently identified as a novel efflux pump associated with fosfomycin resistance (Sharma et al., 2016).
AbeM is in the multidrug and toxic compound extrusion family and confers resistance to imipenem and fluoroquinolones (Su et al., 2005). AbeS is the small multidrug resistance family transporter and affects resistance to various antimicrobial compounds. Deletion of the abeS gene results in increased susceptibility to various antimicrobial compounds, such as chloramphenicol, nalidixic acid, and erythromycin (Srinivasan et al., 2009).
Some other efflux pumps, such as MacAB-TolC (Kobayashi et al., 2001) and EmrAB-TolC (Lomovskaya and Lewis, 1992), have been well described in E. coli, but their role in A. baumannii has been recently explored. The EmrAB-TolC efflux pump is also present in A. baumannii where it conferred resistance to netilmicin, tobramycin, and imipenem (Nowak-Zaleska et al., 2016). Another report identified three novel efflux pumps (A1S_1535, A1S_2795, and ABAYE_0913) in A. baumannii using multiplexed phenotypic screening (Li L. et al., 2016). A1S_1535 confers resistance to various antibiotics, including gentamicin, kanamycin, chloroxylenol, oxytetracycline, 1,10-phenanthroline, and chloramphenicol (Li L. et al., 2016). A1S_2795 is the first major facilitator superfamily efflux pump found to confer resistance to the sulphonamide sulfathiazole, and ABAYE_0913 is associated with resistance to chloramphenicol and fusidic acid (Li L. et al., 2016).
Permeability defects
A change in envelope permeability can influence antibiotic resistance. For example, porins form channels that allow transport of molecules across the outer membrane and play a significant role in A. baumannii virulence (Table 1). Because porins affect membrane permeability, they also play a significant role in the mechanism of resistance. Reduced expression of some porins, including CarO (Mussi et al., 2005, 2007; Siroy et al., 2005; Catel-Ferreira et al., 2011; Jin et al., 2011), Omp22-33 (Bou et al., 2000a), Omp33-36 (del Mar Tomas et al., 2005; Hood et al., 2010), Omp37 (Quale et al., 2003), Omp43 (Dupont et al., 2005), Omp44 (Quale et al., 2003), and Omp47 (Quale et al., 2003), is associated with carbapenem resistance in A. baumannii. Loss of Omp29 in A. baumannii producing OXA-51-like or OXA-23-like carbapenemases results in increased imipenem resistance (Jeong et al., 2009; Fonseca et al., 2013). OmpA is also related with resistance to aztreonam, chloramphenicol, and nalidixic acid (Smani et al., 2014). One study showed that OmpA and CarO physically interact with OXA-23 carbapenemase, and these interactions are associated with antibiotic resistance (Wu et al., 2016). These results provide a novel view to increase understanding of bacterial antibiotic resistance mechanisms.
Besides outer membrane proteins, envelope components, such as LPS and peptidoglycans, also affects antibiotic resistance of A. baumannii. Loss or modification of LPS decreases membrane integrity and increases colistin resistance in A. baumannii (Adams et al., 2009; Moffatt et al., 2010).
Aminoglycoside-modifying enzymes
Aminoglycoside-modifying enzymes are the major mechanism by which A. baumannii confers resistance to aminoglycosides. Aminoglycoside-modifying enzymes can be classified into acetyltransferases, adenyltransferases, and phosphotransferases. These enzymes are typically present on transposable elements and are transferred among pathogenic bacteria (Lin and Lan, 2014). Several reports show that many MDR A. baumannii isolates produce a combination of aminoglycoside-modifying enzymes (Gallego and Towner, 2001; Nemec et al., 2004). A study from China identified a MDR A. baumannii strain carrying four aminoglycoside-modifying enzymes (Zhu et al., 2009). Another study from Greece reported that all A. baumannii strains contain aminoglycoside-modifying enzymes (Ploy et al., 1994), indicating the high prevalence of these enzymes in A. baumannii.
Alteration of target sites
Modifications in antibiotic target sites for antibiotics can induce antibiotic resistance in A. baumannii. In the absence of other known resistance mechanisms, only overexpression of altered PBPs with a low affinity for imipenem induce imipenem resistance (Gehrlein et al., 1991). Quinolone resistance is associated with modifications in GyrA (one subunit of DNA gyrase) and ParC (one subunit of topoisomerase IV) in epidemiologically unrelated A. baumannii isolates (Vila et al., 1995). Acinetobacter baumannii TetM, which has 100% homology with S. aureus TetM, has been proposed to be associated with tetracycline resistance through ribosomal protection (Ribera et al., 2003b). Similar to other pathogenic bacteria, dihydrofolate reductases (DHFR and FolA) responsible for trimethoprim resistance have been found in nosocomial MDR A. baumannii isolates (Mak et al., 2009; Lin M. F. et al., 2013; Taitt et al., 2014). The 16S rRNA methylase ArmA responsible for aminoglycoside resistance is also found in many A. baumannii strains and always coexists with OXA type carbapenemases such as OXA-23 (Yu et al., 2007; Cho et al., 2009; Karthikeyan et al., 2010; Brigante et al., 2012; Hong et al., 2013; Bakour et al., 2014; Tada et al., 2014; Hasani et al., 2016). As described above, many studies have shown that modifications or/and loss of LPS decrease the susceptibility of A. baumannii to many clinical important antibiotics, such as colistin.
Others
AdeABC is associated with decreased susceptibility to tigecycline (Ruzin et al., 2007). However, some clinical isolates without overexpressed AdeABC, AdeFGH, and AdeIJK have decreased susceptibility to tigecycline. Several reports have suggested the mechanism. One study analyzed eight A. baumannii clinical isolates and revealed that the deletion mutation in the trm gene, which encodes S-adenosyl-L-methionine-dependent methyltransferase, decreases susceptibility to tigecycline (Chen et al., 2014). The same result was reported using a highly efficient and versatile genome-editing platform enabling markerless modification of the A. baumannii genome. Deletion of AdeR, a transcription factor that regulates AdeABC efflux pump expression in tigecycline-resistant A. baumannii, reduces the MIC of tigecycline. However, 60% of the clinical isolates remained nonsusceptible to tigecycline after the adeR deletion according to a highly efficient and versatile genome-editing platform (Trebosc et al., 2016). Whole-genome sequencing in two tigecycline-resistant adeR deletion strains revealed that a mutation in the trm gene makes the adeR mutant resistant to tigecycline. In addition, a trm disruption was identified in most tigecycline-resistant clinical isolates (Trebosc et al., 2016). However, its exact mechanism was not determined. Another study revealed that a frameshift mutation in plsC, encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase, is associated with decreased susceptibility to tigecycline (Li X. et al., 2015).
The abrp gene, which encodes the peptidase C13 family, is associated with decreased susceptibility to tetracycline, minocycline, doxycycline, tigecycline, chloramphenicol, and fosfomycin (Li X. et al., 2016). Deletion of abrp increases cell membrane permeability, displays slower cell growth rate, and confers reduced susceptibility to these antibiotics (Liu X. et al., 2016). However, its exact mechanism was not determined. Some genes involved in cell division, including blhA, zipA, zapA, and ftsK, are associated with intrinsic β-lactam resistance in A. baumannii (Knight et al., 2016).
Increased expression of mutagenesis-related genes, such as the SOS response genes, is a well-understood mechanism of E. coli and other bacteria to obtain antibiotic resistance (Cirz and Romesberg, 2007). Acinetobacter baumannii also seems to have an inducible DNA damage response in which RecA plays a major regulatory role and seems to acquire antibiotic resistances under clinically relevant DNA-damaging conditions (Aranda et al., 2011, 2014; Norton et al., 2013). Furthermore, RecA is involved in the A. baumannii pathogenicity (Aranda et al., 2011).
Prospective treatment options
Although carbapenems are effective antibiotics to treat A. baumannii infections (Cisneros and Rodriguez-Bano, 2002; Turner et al., 2003), the rate of carbapenem-resistant A. baumannii isolates has been increasing gradually (Mendes et al., 2010; Kuo et al., 2012; Su et al., 2012). Only a few effective antibiotic options are available to treat MDR A. baumannii infections (Gordon and Wareham, 2009; Lee J. H. et al., 2015, 2016). To combat MDR or pandrug-resistant (PDR) A. baumannii, which are resistant to all available antibiotics, combination therapies, including colistin/imipenem, colistin/meropenem, colistin/rifampicin, colistin/tigecycline, colistin/sulbactam, colistin/teicoplanin, and imipenem/sulbactam, have been extensively studied. Prospective treatment options of Acinetobacter baumannii infections are summarized in Table 3. We will discuss the most recent published reports.
Table 3
| Drugs | Type of research | Type of A. baumannii | Findings | References |
|---|---|---|---|---|
| Carbapenem +ampicillin+sulbactam+ | In vivo | Carbapenem-resistant | Combination therapy with ampicillin-sulbactam and meropenem is effective against skin and soft tissue infection | Hiraki et al., 2013 |
| In vivo | Multidrug-resistant | The combination of a carbapenem and ampicillin/sulbactam was associated with a better outcome than the combination of a carbapenem and amikacin, or a carbapenem alone | Kuo et al., 2007 | |
| Carbapenem +minocycline | In vitro | Multidrug-resistant | Minocycline in combination with rifampicin, imipenem, and colistin showed bactericidal synergy in most of the isolates which did not harbor the tetB gene, but the combinations were not synergistic in tetB-positive isolates | Rodriguez et al., 2015 |
| Carbapenem +tigecycline+colistin | Case report | Multidrug-resistant, colistin-susceptible | A patient with bacteremia had a favorable clinical outcome by a meropenem/colistin/tigecycline combination therapy | Candel et al., 2010 |
| Carbapenem +colistin | In vitro/case report | Extensively drug-resistant, colistin-susceptible | Effective; 80% of patients were treated successfully | Ozbek and Senturk, 2010 |
| In vitro | Multidrug-resistant, colistin-susceptible | Imipenem/colistin showed best synergy effects | Pongpech et al., 2010 | |
| In vitro/case report | Multidrug-resistant, colistin-susceptible | Meropenem/colistin can inhibit bacterial regrowth at 24 h | Lee C. H. et al., 2008 | |
| In vitro | Colistin-susceptible and colistin-resistant | Subinhibitory meropenem/colistin showed synergy against 49 of 52 strains at 24 h | Pankuch et al., 2008 | |
| In vitro | Extensively drug-resistant, colistin-susceptible | Combinations of colistin/rifampicin, colistin/meropenem, colistin/minocycline and minocycline/meropenem are synergistic | Liang et al., 2011 | |
| A retrospective study | Extensively drug-resistant, colistin-susceptible | Colistin/carbapenem and colistin/sulbactam resulted in significantly higher microbiological eradication rates, relatively higher cure and 14-day survival rates, and lower in-hospital mortality compared to colistin monotherapy in patients with bloodstream infections | Batirel et al., 2014 | |
| In vitro | Carbapenem-resistant, colistin-susceptible | Synergistic effects against all 12 isolates | Liu X. et al., 2016 | |
| In vivo | Extensively drug-resistant, colistin-susceptible and colistin-resistant | Colistin/fusidic acid and colistin/rifampicin were synergistic in a murine thigh-infection model; The colistin-meropenem combination was also effective when the colistin MIC is ≤32 mg/L. | Fan et al., 2016 | |
| In vitro | Extensively drug-resistant | The daptomycin-colistin combination was the most effective; the colistin/imipenem combination was also effective | Cordoba et al., 2015 | |
| Carbapenem +colistin+rifampicin | Case report | Multidrug-resistant, colistin-susceptible | Successful treatment by a meropenem/colistin/rifampicin combination therapy in a case of multifocal infection | Biancofiore et al., 2007 |
| Carbapenem+plazomicin | In vitro | Carbapenem-resistant | Synergistic activity | Garcia-Salguero et al., 2015 |
| Imipenem+polymyxin B | In vitro | Carbapenem-resistant | Doripenem, meropenem, or imipenem displayed similar pharmacodynamics in combination with polymyxin B | Lenhard et al., 2016b |
| Meropenem+ polymyxin B | In vitro | Multidrug-resistant | Combinations of polymyxin B/meropenem and polymyxin B/meropenem/fosfomycin showed high synergistic activity | Menegucci et al., 2016 |
| In vitro/in vivo | Carbapenem-resistant | Intensified meropenem dosing in combination with polymyxin B synergistically killed carbapenem-resistant strains, irrespective of the meropenem MIC | Lenhard et al., 2016a | |
| In vitro | Carbapenem-resistant | Doripenem, meropenem, or imipenem displayed similar pharmacodynamics in combination with polymyxin B | Lenhard et al., 2016b | |
| Doripenem+tigecycline | In vitro | Multidrug-resistant, doripenem-resistant | Synergistic activity | Principe et al., 2013 |
| Doripenem+colistin | In vitro | Multidrug-resistant, doripenem-resistant | Synergistic activity | Principe et al., 2013 |
| Doripenem+polymyxin B | In vitro | Carbapenem-resistant | Doripenem, meropenem, or imipenem displayed similar pharmacodynamics in combination with polymyxin B | Lenhard et al., 2016b |
| In vitro | Polymyxin-heteroresistant | The polymyxin B/doripenem combination resulted in rapid and extensive initial killing within 24 h, which was sustained over 10 days | Rao et al., 2016a | |
| Doripenem+amikacin | In vitro | Multidrug-resistant, doripenem-resistant | Synergistic activity | Principe et al., 2013 |
| Ampicillin+sulbactam | In vitro/in vivo | Multi-drug resistant | Ampicillin/sulbactam therapy significantly decreased the risk of death in patients with bloodstream infections | Smolyakov et al., 2003 |
| Sulbactam+colistin | A retrospective study | Extensively drug-resistant, colistin-susceptible | Colistin/carbapenem and colistin/sulbactam resulted in significantly higher microbiological eradication rates, relatively higher cure and 14-day survival rates, and lower in-hospital mortality compared to colistin monotherapy in patients with bloodstream infections | Batirel et al., 2014 |
| A retrospective study | Multidrug-resistant | The colistin/sulbactam combination therapy is promising in patients with ventilator-associated pneumonia | Kalin et al., 2014 | |
| Tazobactam+colistin | In vivo | Colistin-susceptible | Tazobactam plus colistin showed synergy | Sakoulas et al., 2016 |
| Minocycline+colistin | In vitro | Extensively drug-resistant | Combinations of colistin/rifampicin, colistin/meropenem, colistin/minocycline and minocycline/meropenem are synergistic | Liang et al., 2011 |
| In vitro/in vivo | Minocycline-resistant | Minocycline/colistin synergistically killed minocycline-resistant isolates; minocycline/colistin also significantly improved the survival of mice and reduced the number of bacteria present in the lungs of mice | Yang et al., 2016 | |
| In vitro | Multidrug-resistant | Minocycline in combination with rifampicin, imipenem, and colistin showed bactericidal synergy in most of the isolates which did not harbor the tetB gene, but the combinations were not synergistic in tetB-positive isolates | Rodriguez et al., 2015 | |
| Minocycline+rifampicin | In vivo | Multidrug-resistant | Synergistic effect of minocycline/rifampicin and minocycline/amikacin combinations in a mouse lung infection model | He S. et al., 2015 |
| In vitro | Multidrug-resistant | Minocycline in combination with rifampicin, imipenem, and colistin showed bactericidal synergy in most of the isolates which did not harbor the tetB gene, but the combinations were not synergistic in tetB-positive isolates | Rodriguez et al., 2015 | |
| Minocycline+amikacin | In vivo | Multidrug-resistant | Synergistic effect of minocycline/rifampicin and minocycline/amikacin combinations in a mouse lung infection model | He S. et al., 2015 |
| Tigecycline+colistin | In vitro | Carbapenem-resistant, colistin-susceptible | Good synergy | Ozbek and Senturk, 2010; Sheng et al., 2011 |
| In vitro | Extensively drug-resistant, colistin-susceptible | Good synergy | Dizbay et al., 2010 | |
| In vitro | Tigecycline-non-susceptible | Good synergy | Principe et al., 2009 | |
| In vitro | Carbapenem-resistant, colistin-susceptible and colistin-resistant | Good synergy | Peck et al., 2012 | |
| In vitro/in vivo | Extensively drug-resistant | In vitro synergistic activity; no statistically significant differences were found between colistin, tigecycline, and combination treatments in terms of efficacy on bacterial counts in lung tissue of a rat pneumonia model | Mutlu Yilmaz et al., 2012 | |
| Tigecycline+polymyxin B | In vitro | Carbapenem-resistant, polymyxin-heteroresistant | Combination of polymyxin B-with higher tigecycline concentrations result in sustained bactericidal activity | Rao et al., 2016b |
| In vitro | Carbapenem-resistant | Synergistic effects in combination therapy with simulated exposures of polymyxin B and tigecycline at an aggressive dose | Hagihara et al., 2014 | |
| Tigecycline+amikacin | In vitro | Multidrug-resistant | Synergistic bactericidal activities | Moland et al., 2008 |
| Colistin+rifampicin | In vitro/in vivo | Multidrug-resistant, colistin-susceptible | Efficacy in vitro and in experimental models of pneumonia and meningitis | Pachon-Ibanez et al., 2010 |
| Case report | Carbapenem-resistant, colistin-susceptible | Efficacy in 7 of 10 patients with ventilator-associated pneumonia | Song et al., 2008 | |
| Case report | Multidrug-resistant, colistin-susceptible | Efficacy in 22 of 29 critically ill patients with pneumonia and bacteremia | Bassetti et al., 2008 | |
| In vivo | Multidrug-resistant, colistin-susceptible | Synergistic effect in prolonging survival | Pantopoulou et al., 2007 | |
| Clinical trial | Multidrug-resistant, colistin-susceptible | Favorable for all 26 nosocomial infection patients | Motaouakkil et al., 2006 | |
| In vitro | Carbapenem-resistant, colistin-susceptible | Effective for strains highly resistant to imipenem and moderately resistant to rifampicin | Montero et al., 2004 | |
| In vitro | Multidrug-resistant, colistin-susceptible | Synergistic effect against 11 of 13 isolates | Hogg et al., 1998 | |
| In vitro | Extensively drug-resistant | Combinations of colistin/rifampicin, colistin/meropenem, colistin/minocycline and minocycline/meropenem are synergistic | Liang et al., 2011 | |
| In vitro | Multidrug-resistant, colistin-susceptible | Colistin/rifampicin was fully synergistic against 4 of 5 isolates; colistin/meropenem and colistin/azithromycin were synergistic against 3 of 5 isolates; colistin/doxycycline was partially synergistic or additive against 5 isolates | Timurkaynak et al., 2006 | |
| Case report | Carbapenem-resistant, colistin-susceptible | Rifampicin/colistin and ampicillin/sulbactam resulted in microbiological clearance in 9 of 14 critically ill patients | Petrosillo et al., 2005 | |
| In vitro | Carbapenem-resistant, colistin-heteroresistant | Rifampicin/colistin and imipenem/colistin were synergistic against heteroresistant isolates and prevented the development of colistin-resistant strains | Rodriguez et al., 2010 | |
| Case report | Carbapenem-resistant, colistin-susceptible | Synergistic effect in patients with ventilator-associated pneumonia | Aydemir et al., 2013 | |
| In vivo | Extensively drug-resistant, colistin-susceptible and colistin-resistant | Colistin/fusidic acid and colistin/rifampicin were synergistic in a murine thigh-infection model; The colistin-meropenem combination was also effective when the colistin MIC is ≤32 mg/L. | Fan et al., 2016 | |
| In vitro | Colistin-resistant | The most effective combinations were colistin-rifampin and colistin-teicoplanin; both combinations showed synergistic effect against 8 of 9 colistin-resistant strains | Bae et al., 2016 | |
| Colistin+teicoplanin | In vitro/in vivo | Multidrug-resistant, colistin-susceptible | Synergistic effect of colistin/daptomycin and colistin/teicoplanin in a mouse model | Cirioni et al., 2016 |
| In vitro | Multidrug-resistant, colistin-susceptible | Significant synergy | Wareham et al., 2011 | |
| In vitro | Colistin-resistant | The most effective combinations were colistin-rifampin and colistin-teicoplanin; both combinations showed synergistic effect against 8 of 9 colistin-resistant strains | Bae et al., 2016 | |
| Colistin+daptomycin | In vitro/in vivo | Multidrug-resistant, colistin-susceptible | Synergistic effect of colistin/daptomycin and colistin/teicoplanin in a mouse model | Cirioni et al., 2016 |
| In vitro | Extensively drug-resistant | The daptomycin-colistin combination was the most effective; the colistin/imipenem combination was also effective | Cordoba et al., 2015 | |
| Colistin+vancomycin | In vitro/in vivo | Multidrug-resistant, colistin-susceptible | Highly active both in vitro and in an animal model of Galleria mellonella | Hornsey and Wareham, 2011 |
| Colistin+fosfomycin | In vitro | Carbapenem-resistant, colistin-susceptible | Good synergy; no synergy between colistin and sulbactam, colistin and imipenem | Santimaleeworagun et al., 2011 |
| Colistin+fusidic acid | In vitro | Carbapenem-resistant, colistin-susceptible and colistin-resistant | In vitro synergy between colistin and fusidic acid that is comparable to the synergy between colistin and vancomycin; the synergy with fusidic acid is strain-dependent and applicable to strains for which the colistin MICs are relatively low | Bowler et al., 2016 |
| In vitro | Carbapenem-resistant, colistin-susceptible and colistin-resistant | Robust synergy between fusidic acid and colistin against multidrug-resistant clinical strains, including some colistin-resistant strains | Phee et al., 2015 | |
| In vivo | Extensively drug-resistant, colistin-susceptible and colistin-resistant | Colistin/fusidic acid and colistin/rifampicin were synergistic in a murine thigh-infection model; The colistin-meropenem combination was also effective when the colistin MIC is ≤32 mg/L. | Fan et al., 2016 | |
| Colistin+amikacin | Case report | Multidrug-resistant, colistin-susceptible | Successful clinical and microbiological outcomes | Fulnecky et al., 2005 |
| Colistin+trimethoprim-sulfamethoxazole | In vitro | Carbapenem-resistant | Colistin/trimethoprim-sulfamethoxazole killed effectively all carbapenem-resistant strains | Nepka et al., 2016 |
| Polymyxin B+netropsin | In vitro/in vivo | Colistin-resistant | The survival of infected Galleria mellonella was significantly higher when treated with polymyxin B and netropsin in combination than when treated with polymyxin B or netropsin alone | Chung et al., 2016 |
| Trimethoprim-sulfamethoxazole | In vitro | Carbapenem-resistant | Trimethoprim-sulfamethoxazole killed effectively all carbapenem-resistant strains | Nepka et al., 2016 |
| Novobiocin | In vitro | Carbapenem-susceptible | Inhibition of frequency of the occurrence of rifampin resistance mutants | Jara et al., 2015 |
| Bacteriophages | In vitro/in vivo | Carbapenem-resistant, carbapenem-susceptible | Strong lytic activities and the improvement of survival rates | Jeon et al., 2016; Kusradze et al., 2016 |
| Endolysin (LysABP-01)+colistin | In vitro | Multidrug-resistant | Synergistic activity | Thummeepak et al., 2016 |
| Artilysins | In vitro | Carbapenem-resistant, carbapenem-susceptible | Artilysins are effective in vitro and in vivo | Briers et al., 2014; Yang et al., 2015; Defraine et al., 2016; Thandar et al., 2016 |
| Antimicrobial peptides | In vitro | Multidrug-resistant | Good antimicrobial activities | Pires et al., 2015; Barksdale et al., 2016 |
| Rose bengal+ carbapenem | In vitro | Carbapenem-resistant | Imipenem or meropenem with rose bengal showed synergistic effects | Chiu et al., 2016 |
| β-Aminoketone (MD3)+colistin | In vitro | Colistin-susceptible, colistin-resistant | Synergistic effect targeting to strains with specific colistin resistance mechanisms; synergy against both colistin-susceptible strains and colistin-resistant strains with mutations in pmrB and phosphoethanolamine modification of lipid A, but not against colistin-resistant strains with loss of lipopolysaccharide | Martinez-Guitian et al., 2016 |
| Bulgecin A+ carbapenem | In vitro | Carbapenem-resistant | Synergistic activity | Skalweit and Li, 2016 |
| Farnesol+colistin | In vitro | Colistin-resistant | Farnesol increased sensitivity to colistin for colistin-resistant strains | Kostoulias et al., 2015 |
| Oleanolic acid+gentamicin or kanamycin | In vitro | Carbapenem-susceptible | Synergistic activity | Shin and Park, 2015 |
| Cyanide 3-chlorophenylhydrazone (CCCP)+colistin | In vitro | Colistin-resistant | CCCP reversed colistin resistance and inhibited the regrowth of the resistant subpopulation | Ni et al., 2016 |
| In vitro | Colistin-resistant | Synergistic activity | Park and Ko, 2015 | |
| ABEPI1 or ABEPI2+minocycline | In vitro | Carbapenem-susceptible | Synergistic activity | Blanchard et al., 2014 |
| Gallium nitrate | In vitro/in vivo | Multidrug-resistant | Good antimicrobial activities; protection of Galleria mellonella larvae from lethal A. baumannii infection; synergistic activity with colistin | Antunes et al., 2012 |
| Gallium protoporphyrin IX | In vitro/in vivo | Multidrug-resistant | Good antimicrobial activities | Arivett et al., 2015 |
| Gallium nitrate+colistin | In vitro/in vivo | Multidrug-resistant | Good antimicrobial activities; protection of Galleria mellonella larvae from lethal A. baumannii infection; synergistic activity with colistin | Antunes et al., 2012 |
| D-amino acids | In vitro/in vivo | Carbapenem-susceptible | Some D-amino acids (D-histidine and D-cysteine) can inhibit bacterial growth, biofilm formation and adherence to eukaryotic cells | Rumbo et al., 2016 |
| Bifidobacterium breve strain Yakult | In vivo | Multidrug-resistant | Protection against fatal intestinal infection in a murine infection model | Asahara et al., 2016 |
| Clarithromycin | In vivo | Multidrug-resistant | Inhibition of bacterial growth and biofilm formation; immunomodulator | Konstantinidis et al., 2016 |
| Lysophosphatidylcholine+carbapenem | In vivo | Multidrug-resistant strain | Lysophosphatidylcholine in combination with colistin, tigecycline, or imipenem markedly enhanced the bacterial clearance from the spleen and lungs and reduced bacteremia and mouse mortality rates | Parra Millan et al., 2016 |
| Lysophosphatidylcholine+tigecycline | In vivo | Multidrug-resistant strain | Lysophosphatidylcholine in combination with colistin, tigecycline, or imipenem markedly enhanced the bacterial clearance from the spleen and lungs and reduced bacteremia and mouse mortality rates | Parra Millan et al., 2016 |
| Lysophosphatidylcholine+colistin | In vivo | Multidrug-resistant strain | Lysophosphatidylcholine in combination with colistin, tigecycline, or imipenem markedly enhanced the bacterial clearance from the spleen and lungs and reduced bacteremia and mouse mortality rates | Parra Millan et al., 2016 |
Prospective treatment options of Acinetobacter baumannii infections.
Carbapenems and β-lactamase inhibitors
Carbapenems, including imipenem, meropenem, and doripenem, have generally been considered the agents to treat A. baumannii infections, due to their effective activity against this organism and their favorable safety (Doi et al., 2015). However, the decreased susceptibility of A. baumannii to carbapenems has forced clinicians and researchers to explore alternative therapeutic approaches (Doi et al., 2015). Because carbapenem-resistant A. baumannii strains are often resistant to all other commonly used antibiotics as well, these strains remain susceptible to only limited antibiotics, such as minocycline/tigecycline and polymyxins (colistin and polymyxin B; Lin and Lan, 2014; Doi et al., 2015). Carbapenem therapies combined with a few effective antibiotics was extensively tested and many cases showed a synergistic effect against A. baumannii infections (Table 3). However, recent increase of tigecycline- or colistin-resistant A. baumannii increasingly poses a serious threat to public health worldwide (Peleg et al., 2007; Hornsey et al., 2010; Cai et al., 2012).
Sulbactam is a β-lactamase inhibitor and also has affinity for penicillin-binding proteins of A. baumannii (Rafailidis et al., 2007; Doi et al., 2015). Combined therapy of ampicillin with sulbactam is effective for treating bloodstream infections due to MDR A. baumannii (Smolyakov et al., 2003). Ampicillin/sulbactam/carbapenem combination therapy is also effective for treating MDR A. baumannii bacteremia (Kuo et al., 2007) and skin and soft tissue infection of carbapenem-resistant A. baumannii (Hiraki et al., 2013), but not in ventilator-associated pneumonia (Kalin et al., 2014). The population pharmacokinetics and pharmacodynamics of sulbactam were determined in critically ill patients with severe sepsis caused by A. baumannii (Jaruratanasirikul et al., 2016) and in patients with impaired renal function (Yokoyama et al., 2015). Another β-lactamase inhibitor tazobactam increases the activity of peptide antibiotics, such as colistin and daptomycin, in a murine model of A. baumannii pneumonia (Sakoulas et al., 2016). The authors suggested that β-lactamase inhibitors may exert similar effects in potentiating peptide antibiotics, because of structural similarities between β-lactamase inhibitors and peptide antibiotics (Sakoulas et al., 2016).
Minocycline/tigecycline
Minocycline, is a broad-spectrum tetracycline antibiotic that has been proposed for treating drug-resistant A. baumannii based on its high degree of susceptibility to this drug and its favorable pharmacokinetic profile (Ritchie and Garavaglia-Wilson, 2014). The mean susceptibility rate of A. baumannii to minocycline is approximately 80% worldwide (Castanheira et al., 2014). Therefore, minocycline therapy has high treatment success rates and good tolerability (Ritchie and Garavaglia-Wilson, 2014). However, since the introduction of minocycline, approximately 20% of A. baumannii isolates are not susceptible to minocycline. The TetB efflux pump is the main determinants of minocycline resistance (Vilacoba et al., 2013). Minocycline therapy combined with colistin is effective for treating minocycline-resistant A. baumannii infections (Yang et al., 2016), and minocycline therapy combined with rifampicin, colistin, or imipenem has a synergistic effect in most of isolates without the tetB gene, but combined therapies are not synergistic in isolates with the tetB gene (Rodriguez et al., 2015).
Tigecycline is the first glycylcycline class antibiotic that exhibits bacteriostatic activity by binding to the 30S ribosomal subunit, and is active against A. baumannii infections (Pachon-Ibanez et al., 2004; Anthony et al., 2008; Koomanachai et al., 2009). Tigecycline shows a synergistic effect with some classes of antibiotics, such as amikacin (Moland et al., 2008) and colistin (Mutlu Yilmaz et al., 2012). However, limitations of tigecycline use have emerged with its increasing use. Tigecycline is less effective than imipenem to treat pneumonia in a murine pneumonia model (Pichardo et al., 2010). β-Lactam or carbapenem instead of tigecycline was recommended for A. baumannii infections with tigecycline MIC of more than 2 mg/L, due to high mortality from the tigecycline treatment (Curcio and Fernandez, 2008). In a study of 266 patients with MDR A. baumannii infections, tigecycline-based therapy was not more effective than non-tigecycline-based therapies (Lee Y. T. et al., 2013). Tigecycline resistance associated with overexpression of efflux pumps, such as AdeABC, has been reported in clinical isolates of A. baumannii (Peleg et al., 2007; Ruzin et al., 2007; Hornsey et al., 2010, 2011). Multiple MDR A baumannii clones resistant to tigecycline have been reported in many medical centers (Navon-Venezia et al., 2007). Therefore, tigecycline can only be used in limited cases for treating A. baumannii infections.
Polymyxins (colistin and polymyxin B)
Polymyxins are a group of polycationic peptide antibiotics that were discovered more than 60 years ago and exhibit potent efficacy against most Gram-negative bacteria (Liu Q. et al., 2014; Lee C. R. et al., 2016). Among all five polymyxins (A–E), only polymyxin B and E (colistin) with a one amino acid difference are used clinically. Colistin is a key component of combination therapies used to treat MDR A. baumannii infections (Cai et al., 2012). The rate of colistin resistance (10.4%) in MDR A. baumannii isolates is lower than that of rifampicin (47.8%) or tigecycline (45.5%) resistance (Chang et al., 2012). Similar results were reported in another study (Muthusamy et al., 2016). Therefore, colistin seems to be the only effective antimicrobial agent against MDR A. baumannii infections. Many colistin-based combined therapies, including colistin/rifampicin (Liang et al., 2011; Aydemir et al., 2013), colistin/minocycline (Liang et al., 2011), colistin/carbapenem (Liang et al., 2011; Batirel et al., 2014; Liu X. et al., 2016), colistin/sulbactam (Batirel et al., 2014), colistin/tigecycline (Principe et al., 2009; Ozbek and Senturk, 2010; Sheng et al., 2011; Peck et al., 2012), colistin/daptomycin (Cirioni et al., 2016), colistin/fusidic acid (Bowler et al., 2016; Fan et al., 2016), and colistin/teicoplanin (Wareham et al., 2011; Cirioni et al., 2016), are synergistic in vivo or in vitro against A. baumannii infections. Colistin therapy combined with rifampin or fusidic acid seems to be the most effective for treating a MDR A. baumannii in a murine thigh-infection model (Fan et al., 2016). Another report comparing colistin/daptomycin, colistin/imipenem, and imipenem/ertapenem showed that the daptomycin-colistin combination was the most effective (Cordoba et al., 2015).
Unfortunately, the emergence of colistin-resistant A. baumannii strains has increased worldwide (Cai et al., 2012). The mechanisms of colistin resistance include loss of LPS (Moffatt et al., 2010) and the addition of phosphoethanolamine to LPS by the PmrAB two-component system (Adams et al., 2009). Mutations in pmrA and pmrB activate pmrC, which adds phosphoethanolamine to the hepta-acylated form of lipid A (Beceiro et al., 2011). Interestingly, an investigation of the in vitro activities of various antimicrobial combinations against colistin-resistant A. baumannii showed that the most effective combinations against colistin-resistant A. baumannii are colistin-rifampin and colistin-teicoplanin, indicating that colistin is the most common constituent of antimicrobial combinations even against colistin-resistant A. baumannii (Bae et al., 2016). Similarly, minocycline therapy in combination with colistin is effective to treat infections caused by minocycline-resistant A. baumannii. Minocycline/colistin therapy significantly improves survival of mice infected with minocycline-resistant A. baumannii and reduces the number of bacteria present in the lungs of mice (Yang et al., 2016).
A urinary tract Enterococcus faecalis isolate that apparently requires vancomycin to grow was reported in 1994, and this phenomenon is called “antimicrobial agent dependence.” Colistin dependence was reported in an A. baumannii–A. calcoaceticus complex (Hawley et al., 2007). Partial colistin dependence has been detected in several LPS-deficient strains with mutations in lpxA, lpxC, and lpxD (Garcia-Quintanilla et al., 2015). Many colistin-susceptible A. baumannii isolates develop colistin dependence in vitro after exposure to colistin (Hong et al., 2016). Although the clinical implication of colistin dependence and its molecular mechanism remain unclear, it is interesting that patients with colistin-dependent A. baumannii isolates show a high rate of treatment failure (Hong et al., 2016).
Unlike colistin, polymyxin B is not converted from a prodrug form into an active form; thus, plasma concentrations of polymyxin B more quickly reach target levels (Sandri et al., 2013). In addition, polymyxin B is available for direct parenteral administration (Zavascki et al., 2007). Despite the favorable pharmacokinetics of polymyxin B, dose-related nephrotoxicity limits the concentration of polymyxin B used in combination therapy (Dubrovskaya et al., 2015). Therefore, almost all studies on polymyxins are carried out for colistin. However, because some carbapenems have comparatively safer dose modulation to optimize killing during combination therapy (Cannon et al., 2014), several studies have analyzing the pharmacodynamics of carbapenems in combination with polymyxin B (Lenhard et al., 2016a,b; Rao et al., 2016a). One study showed that intensified meropenem dosing combined with polymyxin B is a good strategy to treat carbapenem-resistant A. baumannii, regardless of the meropenem MIC (Lenhard et al., 2016a). Combination therapy with doripenem and polymyxin B also showed similar results. Early aggressive dosing of doripenem combined with polymyxin B is effective for treating heteroresistant A. baumannii infections (Rao et al., 2016a). A combined pharmacodynamics analysis of four different carbapenems with polymyxin B showed that doripenem, meropenem, or imipenem display similar pharmacodynamics in combination, and the decision to use carbapenem in combination with polymyxin B is usually based on toxicodynamic profiles (Lenhard et al., 2016b). Polymyxin B also shows good bactericidal activity in combination with high tigecycline concentrations (Hagihara et al., 2014; Rao et al., 2016b). Therefore, polymyxin B combination therapies seem to be one of the most promising options for minimizing the emergence of polymyxin resistance. Increasing the dose intensity of polymyxin B amplifies polymyxin B resistance in A. baumannii (Cheah et al., 2016; Tsuji et al., 2016). In conclusion, although polymyxin B displays dose-related nephrotoxicity, it is a potential therapeutic alternative to colistin when use together with intensified doses of other antibiotics. Large-scale screening of Streptomyces secondary metabolites was performed to develop a novel combination therapy using minimal concentrations of polymyxin B, and the reliable polymyxin synergist netropsin was identified (Chung et al., 2016). Survival of G. mellonella infected with colistin-resistant clinical A. baumannii isolates is significantly higher when treated with polymyxin B combined with netropsin than when treated with polymyxin B or netropsin alone (Chung et al., 2016).
Other antibiotics
Trimethoprim-sulfamethoxazole is a two antibiotics combination that exerts a synergistic effect by inhibiting successive steps in the folate synthesis pathway against a number of bacteria (Wormser et al., 1982). The in vitro killing activity of trimethoprim-sulfamethoxazole against carbapenem-resistant A. baumannii was recently studied. Trimethoprim-sulfamethoxazole alone effectively kills all carbapenem-resistant A. baumannii strains and trimethoprim-sulfamethoxazole combined with colistin also rapidly kills all strains for up to 24 h (Nepka et al., 2016). These results suggest that trimethoprim-sulfamethoxazole might be an effective therapy for severe carbapenem-resistant A. baumannii infections. Plazomicin is a next-generation aminoglycoside synthetically derived from sisomicin that enhances activity against many MDR Gram-negative bacteria (Garcia-Salguero et al., 2015). A synergistic effect was observed with carbapenems along with plazomicin during treatment of A. baumannii infections (Garcia-Salguero et al., 2015), indicating the potential utility of plazomicin combined with carbapenems.
The inducible DNA damage response in A. baumannii plays an important role in acquiring antibiotic resistance under clinically relevant DNA-damaging conditions (Aranda et al., 2011, 2014; Norton et al., 2013). The aminocoumarin novobiocin is a well-established antimicrobial agent that inhibits the DNA damage response in Gram-positive bacteria by interfering with ATPase activity of DNA gyrase (Schroder et al., 2013). One study showed that novobiocin also inhibits acquisition of antimicrobial resistance in MDR A. baumannii through DNA damage-induced mutagenesis (Jara et al., 2015).
Non-antibiotic therapies: phage and others
The worldwide spread of MDR pathogens has renewed interest in the therapy using bacteriophage, which is a virus that infects and lyses bacteria. Various lytic A. baumannii bacteriophages, such as vB_Ab-M-G7 (Kusradze et al., 2016) and Bϕ-C62 (Jeon et al., 2016), have been used to treat infections caused by MDR A. baumannii. Bacteriophage-encoded endolysin has also received attention. Endolysin is a lytic enzyme that degrades the cell wall of bacterial hosts and shows promise as a novel class of antibacterials with a unique mode of action (Defraine et al., 2016). For example, endolysin from A. baumannii bacteriophage ØABP-01 degrades the crude cell wall of A. baumannii strains and elevates antibacterial activity when combined with colistin (Thummeepak et al., 2016). However, most Gram-negative pathogens are generally not susceptible to endolysins, due to their protective outer membrane (Lee et al., 2013a). To overcome this problem, endolysins have recently been engineered with specific outer membrane-destabilizing peptides to obtain the ability to penetrate outer membrane and these engineered endolysins are called “artilysins” (Rodriguez-Rubio et al., 2016). Several engineered artilysins have been developed to combat MDR A. baumannii and show highly effective antimicrobial activity against A. baumannii (Briers et al., 2014; Yang et al., 2015; Defraine et al., 2016; Thandar et al., 2016). These results suggest that artilysins can be a treatment option for MDR A. baumannii. The diversity of the phage population was determined by analysis of viromes, endolysins, and CRISPR spacers (Davison et al., 2016). These results can be used to assist in finding an effective endolysin for combating MDR A. baumannii. Various peptides, such as American alligator plasma peptide (Barksdale et al., 2016) and antimicrobial peptide dendrimer G3KL (Pires et al., 2015), have in vitro antimicrobial activity against MDR A. baumannii. However, the use of antimicrobial enzymes or peptides also has some important problems, such as their short half-life in serum and high production costs compared with those of smaller molecules.
An in silico analysis predicted that OXA-58, OXA-23, and OXA-83 are translocated to the periplasm via the Sec system (Liao et al., 2015; Chiu et al., 2016). A SecA inhibitor (rose bengal) inhibits periplasmic translocation of these carbapenem-hydrolyzing class D β-lactamases, indicating that these β-lactamases are selectively released via a Sec-dependent system (Liao et al., 2015; Chiu et al., 2016). Imipenem or meropenem combined with rose bengal shows synergistic effects for carbapenem-resistant A. baumannii clinical isolates (Chiu et al., 2016). Similarly, β-aminoketone (MD3), an inhibitor of bacterial type I signal peptidases that cleaves the amino-terminal signal peptides of translocated proteins, shows a synergistic effect when combined with colistin against colistin-resistant A. baumannii strains (Martinez-Guitian et al., 2016).
Bulgecin A is a natural product of P. mesoacidophila and a lytic transglycosylase inhibitor that works synergistically with β-lactams (Skalweit and Li, 2016). Bulgecin A restores the efficacy of meropenem in suppressing growth of carbapenem-resistant A. baumannii strains, suggesting that Bulgecin A may be an adjunctive compound to extend the life of carbapenems against A. baumannii infections (Skalweit and Li, 2016). Similarly, farnesol, a natural product of Candida albicans for quorum-sensing, disrupts A. baumannii cell membrane integrity, alters cell morphology, and increases sensitivity of MDR A. baumannii strains to colistin (Kostoulias et al., 2015). Many herbal active compounds have potent antibacterial activities against many bacteria including carbapenem-resistant A. baumannii (Lin et al., 2015). For example, oleanolic acid is a triterpenoid compound that widely exists in food, medicinal herbs, and many plants and can potently inhibit various pathogenic bacteria. One study showed that oleanolic acid increases aminoglycoside uptake by changing membrane permeability and energy metabolism in A. baumannii (Shin and Park, 2015).
Cyanide 3-chlorophenylhydrazone (CCCP) is an efflux pump inhibitor that decreases the MIC of colistin in colistin-susceptible and colistin-resistant A. baumannii strains (Park and Ko, 2015; Ni et al., 2016). Other efflux pump inhibitors, such as ABEPI1 and ABEPI2, inhibit efflux-mediated minocycline tolerance of A. baumannii. Adding these compounds during growth in human serum leads to the accumulation of minocycline within A. baumannii and inhibits efflux potential of the bacterium (Blanchard et al., 2014).
Gallium is a semi-metallic element in group 13 of the periodic table that binds to biological complexes containing Fe3+ and disrupts essential redox-driven biological processes (Bernstein, 1998). Gallium has been used as a simple inorganic or organic salt or complexed with organic compounds. Several studies have shown that gallium nitrate or gallium protoporphyrin IX could be a viable therapeutic option for treating MDR A. baumannii (Antunes et al., 2012; Arivett et al., 2015). Some d-amino acids, such as d-His and d-Cys, inhibit bacterial growth, biofilm formation, and adherence to eukaryotic cells in A. baumannii (Rumbo et al., 2016).
Probiotics are “live microorganisms that confer a health benefit on the host when administered in adequate amounts” (Reid et al., 2005) and assist in protecting against MDR A. baumannii infections. For example, the ability of the probiotic Bifidobacterium breve to protect against MDR A. baumannii infections has been investigated (Asahara et al., 2016). This probiotic markedly potentiates protection against fatal intestinal infections caused by MDR A. baumannii (Asahara et al., 2016). With probiotics, immunomodulators, such as lysophosphatidylcholine (Parra Millan et al., 2016) and macrolide antibiotics such as clarithromycin (Konstantinidis et al., 2016), can reduce A. baumannii infection severity by stimulating the immune response, when combined with antibiotics such as colistin, tigecycline, or imipenem.
Conclusion
The number of studies about A. baumannii is increasing dramatically because of its increasing clinical importance. Use of animal models has produced important data regarding virulence factors that contribute to A. baumannii pathogenesis. Notably, some studies on metal acquisition and protein secretion systems are interesting. Besides iron acquisition systems such as acinetobactin, the discovery of zinc and manganese acquisition systems in A. baumannii broadens our understanding of A. baumannii pathogenesis. More extensive studies on various protein secretion systems present in A. baumannii are required. About 300 genes required for in vivo survival of A. baumannii were identified using transposon screening in G. mellonella larvae (Gebhardt et al., 2015). Because many of these genes were not known to be associated with A. baumannii pathogenesis, more detailed studies are required to determine whether these genes are related to the pathogenesis of A. baumannii. In addition, transposon screening in other model animals will provide novel insight into A. baumannii pathogenesis. Knowledge of virulence factors responsible for A. baumannii pathogenicity will be the cornerstone for developing novel antibiotics. For example, LPS is an important virulence factor and LpxC inhibitor, which inhibits LPS synthesis, completely protects mice from lethal infection (Lin et al., 2012). These results indicate that blocking LPS synthesis is a powerful strategy for discovering novel antibiotics. However, despite recent extensive studies about A. baumannii pathogenesis, the toxicity and pathogenicity of A. baumannii remain unclear.
Recent interest about A. baumannii is mostly due to its seemingly endless capacity to acquire antibiotic resistance. A. baumannii has almost all bacterial resistance mechanisms. All class β-lactamases have been detected in A. baumannii and the frequency of carbapenem-resistant A. baumannii isolates is very high. Furthermore, almost all A. baumannii contain aminoglycoside-modifying enzymes and many efflux pumps responsible for resistance to various clinically important antibiotics have been identified in A. baumannii. Due to these abilities, available antibiotics to treat A. baumannii infections are significantly limited. Colistin is used as the antibiotic treatment of last resort, due to its relatively low resistance rate. However, emergence of colistin-resistant A. baumannii strains has increased worldwide with increasing use of colistin. Notably, some more recent studies have proposed that another polymyxin antibiotic, polymyxin B, is a potential therapeutic alternative to colistin (Lenhard et al., 2016a,b; Rao et al., 2016a; Repizo et al., 2015). Polymyxin B has not been a good antibiotic owing to dose-dependent nephrotoxicity, but recent reports show that a novel combination therapy with carbapenems or tigecycline using minimal concentrations of polymyxin B can be a good strategy to treat carbapenem-resistant A. baumannii infections. These results indicate the requirement for extensive studies that analyze the pharmacodynamics of polymyxin B in combination therapy.
Various trials to identify a novel alternative to carbapenem or colistin have been performed. Among them, engineered endolysins (artilysins) are particularly interesting, despite evident defects. A lytic enzyme degrading peptidoglycan of bacteria is a promising novel class of antimicrobial agents due to its unique mode of action. Similar to β-lactam antibiotics that are one of the most successful antibiotics, inhibition of peptidoglycan synthesis is a promising target of antimicrobial agents. Because lytic enzymes directly degrade peptidoglycans, but not proteins, the possibility of the emergence of a resistance mechanism is relatively low. In addition, enzymes with relatively high molecular weight are not inhibited by efflux pumps. If the short stability of artilysin in serum and high cost in its production compared with small molecules can be resolved, the improved artilysin can be a good treatment option for carbapenem- or colistin-resistant A. baumannii infections. In conclusion, novel, rationally designed strategies and screening-based approaches are required to discover new classes of antibiotics. If we continue to take all efforts at maintaining the effectiveness of antibiotics and developing novel antibiotics, effective control of A. baumannii infections can be successful.
Funding
This review was supported by the Cooperative Research Program for Agriculture Science and Technology Development (No. PJ01103103) of Rural Development Administration in Republic of Korea; the Environmental Health Action Program (No. 2016001350004) funded by the Ministry of Environment (MOE) in Republic of Korea; and the National Research Foundation of the Ministry of Education, Republic of Korea (NRF-2015R1C1A1A02037470).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Statements
Author contributions
CL, JL, MP, and SL contributed to the conception and the design of the review and CL, JL, MP, KP, IB, YK, CC, BJ, and SL researched and wrote the review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AcostaJ.MerinoM.ViedmaE.PozaM.SanzF.OteroJ. R.et al. (2011). Multidrug-resistant Acinetobacter baumannii Harboring OXA-24 carbapenemase, Spain. Emerg. Infect Dis.17, 1064–1067. 10.3201/eid/1706.091866.
2
AdamsM. D.GoglinK.MolyneauxN.HujerK. M.LavenderH.JamisonJ. J.et al. (2008). Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol.190, 8053–8064. 10.1128/JB.00834-08
3
AdamsM. D.NickelG. C.BajaksouzianS.LavenderH.MurthyA. R.JacobsM. R.et al. (2009). Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother.53, 3628–3634. 10.1128/AAC.00284-09
4
Afzal-ShahM.WoodfordN.LivermoreD. M. (2001). Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother.45, 583–588. [Epub ahead of print]. 10.1128/AAC.45.2.583-588.2001
5
Al-AgamyM. H.JeannotK.El-MahdyT. S.ShiblA. M.KattanW.PlesiatP.et al. (2016). First Detection of GES-5 Carbapenemase-Producing Acinetobacter baumannii Isolate. Microb. Drug Resist. [Epub ahead of print]. 10.1089/mdr.2016.0152
6
Al AtrouniA.HamzeM.JisrT.LemarieC.EveillardM.Joly-GuillouM. L.et al. (2016). Wide spread of OXA-23-producing carbapenem-resistant Acinetobacter baumannii belonging to clonal complex II in different hospitals in Lebanon. Int. J. Infect. Dis.52, 29–36. 10.1016/j.ijid.2016.09.017
7
AlyM. M.Abu AlsoudN. M.ElrobhM. S.Al JohaniS. M.BalkhyH. H. (2016). High prevalence of the PER-1 gene among carbapenem-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur. J. Clin. Microbiol. Infect Dis.35, 1759–1766. 10.1007/s10096-016-2723-8
8
AnthonyK. B.FishmanN. O.LinkinD. R.GasinkL. B.EdelsteinP. H.LautenbachE. (2008). Clinical and microbiological outcomes of serious infections with multidrug-resistant gram-negative organisms treated with tigecycline. Clin. Infect. Dis.46, 567–570. 10.1086/526775
9
AntunesL. C.ImperiF.CarattoliA.ViscaP. (2011). Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS ONE6:e22674. 10.1371/journal.pone.0022674
10
AntunesL. C.ImperiF.MinandriF.ViscaP. (2012). In vitro and in vivo antimicrobial activities of gallium nitrate against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother.56, 5961–5970. 10.1128/AAC.01519-12
11
ArandaJ.BardinaC.BeceiroA.RumboS.CabralM. P.BarbeJ.et al. (2011). Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J. Bacteriol.193, 3740–3747. 10.1128/JB.00389-11
12
ArandaJ.LopezM.LeivaE.MaganA.AdlerB.BouG.et al. (2014). Role of Acinetobacter baumannii UmuD homologs in antibiotic resistance acquired through DNA damage-induced mutagenesis. Antimicrob. Agents Chemother.58, 1771–1773. 10.1128/AAC.02346-13
13
ArivettB. A.FiesterS. E.OhneckE. J.PenwellW. F.KaufmanC. M.RelichR. F.et al. (2015). Antimicrobial activity of Gallium Protoporphyrin IX against Acinetobacter baumannii strains displaying different antibiotic resistance phenotypes. Antimicrob. Agents Chemother.59, 7657–7665. 10.1128/AAC.01472-15
14
ArroyoL. A.HerreraC. M.FernandezL.HankinsJ. V.TrentM. S.HancockR. E. (2011). The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother.55, 3743–3751. 10.1128/AAC.00256-11
15
AsaharaT.TakahashiA.YukiN.KajiR.TakahashiT.NomotoK. (2016). Protective effect of a synbiotic against multidrug-resistant Acinetobacter baumannii in a Murine Infection Model. Antimicrob. Agents Chemother.60, 3041–3050. 10.1128/AAC.02928-15
16
AydemirH.AkdumanD.PiskinN.ComertF.HoruzE.TerziA.et al. (2013). Colistin vs. the combination of colistin and rifampicin for the treatment of carbapenem-resistant Acinetobacter baumannii ventilator-associated pneumonia. Epidemiol. Infect.141, 1214–1222. 10.1017/S095026881200194X
17
BaeS.KimM. C.ParkS. J.KimH. S.SungH.KimM. N.et al. (2016). In vitro synergistic activity of antimicrobial agents in combination against clinical isolates of colistin-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 6774–6779. 10.1128/AAC.00839-16
18
BakourS.AlsharapyS. A.TouatiA.RolainJ. M. (2014). Characterization of Acinetobacter baumannii clinical isolates carrying blaOXA-23carbapenemase and 16S rRNA methylase armA genes in Yemen. Microb. Drug Resist.20, 604–609. 10.1089/mdr.2014.0018
19
BarksdaleS. M.HrifkoE. J.ChungE. M.van HoekM. L. (2016). Peptides from American alligator plasma are antimicrobial against multi-drug resistant bacterial pathogens including Acinetobacter baumannii. BMC Microbiol.16:189. 10.1186/s12866-016-0799-z
20
BaslerM.HoB. T.MekalanosJ. J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell152, 884–894. 10.1016/j.cell.2013.01.042
21
BassettiM.RepettoE.RighiE.BoniS.DiverioM.MolinariM. P.et al. (2008). Colistin and rifampicin in the treatment of multidrug-resistant Acinetobacter baumannii infections. J. Antimicrob. Chemother.61, 417–420. 10.1093/jac/dkm509
22
BatirelA.BalkanI. I.KarabayO.AgalarC.AkalinS.AliciO.et al. (2014). Comparison of colistin-carbapenem, colistin-sulbactam, and colistin plus other antibacterial agents for the treatment of extremely drug-resistant Acinetobacter baumannii bloodstream infections. Eur. J. Clin. Microbiol. Infect. Dis.33, 1311–1322. 10.1007/s10096-014-2070-6
23
BeceiroA.LlobetE.ArandaJ.BengoecheaJ. A.DoumithM.HornseyM.et al. (2011). Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother.55, 3370–3379. 10.1128/AAC.00079-11
24
BeceiroA.TomasM.BouG. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world?Clin. Microbiol. Rev.26, 185–230. 10.1128/CMR.00059-12
25
BentancorL. V.Camacho-PeiroA.Bozkurt-GuzelC.PierG. B.Maira-LitranT. (2012a). Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J. Bacteriol.194, 3950–3960. 10.1128/JB.06769-11
26
BentancorL. V.RoutrayA.Bozkurt-GuzelC.Camacho-PeiroA.PierG. B.Maira-LitranT. (2012b). Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect. Immun.80, 3381–3388. 10.1128/IAI.06096-11
27
Bergogne-BerezinE.TownerK. J. (1996). Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol Rev. 9, 148–165.
28
BernsteinL. R. (1998). Mechanisms of therapeutic activity for gallium. Pharmacol. Rev. 50, 665–682.
29
BhuiyanM. S.EllettF.MurrayG. L.KostouliasX.CerqueiraG. M.SchulzeK. E.et al. (2016). Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc. Natl. Acad. Sci. U.S.A.113, 9599–9604. 10.1073/pnas.1523116113
30
BiancofioreG.TasciniC.BisaM.GemignaniG.BindiM. L.LeonildiA.et al. (2007). Colistin, meropenem and rifampin in a combination therapy for multi-drug-resistant Acinetobacter baumannii multifocal infection. A case report. Minerva. Anestesiol. 73, 181–185.
31
BiglariS.HanafiahA.Mohd PuziS.RamliR.RahmanM.LopesB. S. (2016). Antimicrobial resistance mechanisms and genetic diversity of multidrug-resistant Acinetobacter baumannii isolated from a teaching hospital in Malaysia. Microb. Drug Resist. [Epub ahead of print]. 10.1089/mdr.2016.0130
32
BlanchardC.BarnettP.PerlmutterJ.DunmanP. M. (2014). Identification of Acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob. Agents Chemother.58, 6360–6370. 10.1128/AAC.03535-14
33
BogaertsP.NaasT.El GarchF.CuzonG.DeplanoA.DelaireT.et al. (2010). GES extended-spectrum β-lactamases in Acinetobacter baumannii isolates in Belgium. Antimicrob. Agents Chemother.54, 4872–4878. 10.1128/AAC.00871-10
34
BollJ. M.TuckerA. T.KleinD. R.BeltranA. M.BrodbeltJ. S.DaviesB. W.et al. (2015). Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. MBio6, e00478–e00415. 10.1128/mBio.00478-15
35
BonninR. A.PoirelL.LickerM.NordmannP. (2011a). Genetic diversity of carbapenem-hydrolysing β-lactamases in Acinetobacter baumannii from Romanian hospitals. Clin. Microbiol. Infect.17, 1524–1528. 10.1111/j.1469-0691.2011.03622.x
36
BonninR. A.PoirelL.NaasT.PirsM.SemeK.SchrenzelJ.et al. (2012). Dissemination of New Delhi metallo-β-lactamases-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect.18, E362–E365. 10.1111/j.1469-0691.2012.03928.x
37
BonninR. A.PotronA.PoirelL.LecuyerH.NeriR.NordmannP. (2011b). PER-7, an extended-spectrum β-lactamases with increased activity toward broad-spectrum cephalosporins in Acinetobacter baumannii. Antimicrob. Agents Chemother.55, 2424–2427. 10.1128/AAC.01795-10
38
BorgeaudS.MetzgerL. C.ScrignariT.BlokeschM. (2015). The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science.347, 63–67. 10.1126/science.1260064
39
BouG.CerveroG.DominguezM. A.QueredaC.Martinez-BeltranJ. (2000a). Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38, 3299–3305.
40
BouG.Martinez-BeltranJ. (2000). Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamases in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44, 428–432. 10.1128/AAC.44.2.428-432.2000
41
BouG.OliverA.Martinez-BeltranJ. (2000b). OXA-24, a novel class D β-lactamases with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44, 1556–1561. 10.1128/AAC.44.6.1556-1561.2000
42
BoucherH. W.TalbotG. H.BradleyJ. S.EdwardsJ. E.GilbertD.RiceL. B.et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis.48, 1–12. 10.1086/595011
43
BowlerS. L.SpychalaC. N.McElhenyC. L.MettusR. T.DoiY. (2016). In vitro activity of fusidic acid-containing combinations against carbapenem-resistant Acinetobacter baumannii clinical strains. Antimicrob. Agents Chemother.60, 5101. 10.1128/AAC.01124-16
44
BriersY.WalmaghM.Van PuyenbroeckV.CornelissenA.CenensW.AertsenA.et al. (2014). Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. MBio5, e01379–14. 10.1128/mBio.01379-14
45
BriganteG.MigliavaccaR.BramatiS.MottaE.NucleoE.ManentiM.et al. (2012). Emergence and spread of a multidrug-resistant Acinetobacter baumannii clone producing both the carbapenemase OXA-23 and the 16S rRNA methylase ArmA. J. Med. Microbiol.61, 653–661. 10.1099/jmm.0.040980-0
46
BrownS.YoungH. K.AmyesS. G. (2005). Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect.11, 15–23. 10.1111/j.1469-0691.2004.01016.x
47
CaiY.ChaiD.WangR.LiangB.BaiN. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother.67, 1607–1615. 10.1093/jac/dks084
48
CamarenaL.BrunoV.EuskirchenG.PoggioS.SnyderM. (2010). Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog.6:e1000834. 10.1371/journal.ppat.1000834
49
CandelF. J.CalvoN.HeadJ.SanchezA.MatesanzM.CulebrasE.et al. (2010). A combination of tigecycline, colistin, and meropenem against multidrug-resistant Acinetobacter baumannii bacteremia in a renal transplant recipient: pharmacodynamic and microbiological aspects. Rev. Esp. Quimioter. 23, 103–108.
50
CannonJ. P.LeeT. A.ClarkN. M.SetlakP.GrimS. A. (2014). The risk of seizures among the carbapenems: a meta-analysis. J. Antimicrob. Chemother.69, 2043–2055. 10.1093/jac/dku111
51
CarruthersM. D.NicholsonP. A.TracyE. N.MunsonR. S.Jr. (2013). Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS ONE8:e59388. 10.1371/journal.pone.0059388
52
CastanheiraM.MendesR. E.JonesR. N. (2014). Update on Acinetobacter species: mechanisms of antimicrobial resistance and contemporary in vitro activity of minocycline and other treatment options. Clin. Infect. Dis. 59 (Suppl. 6), S367–S373. 10.1093/cid/ciu706
53
Catel-FerreiraM.CoadouG.MolleV.MugnierP.NordmannP.SiroyA.et al. (2011). Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J. Antimicrob. Chemother.66, 2053–2056. 10.1093/jac/dkr267
54
Centers for Disease Prevention (2004). Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002–2004. MMWR Morb. Mortal. Wkly. Rep. 53, 1063–1066.
55
CerqueiraG. M.KostouliasX.KhooC.AibinuI.QuY.TravenA.et al. (2014). A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis.210, 46–55. 10.1093/infdis/jiu024
56
ChagasT. P.CarvalhoK. R.de Oliveira SantosI. C.Carvalho-AssefA. P.AsensiM. D. (2014). Characterization of carbapenem-resistant Acinetobacter baumannii in Brazil (2008-2011): countrywide spread of OXA-23-producing clones (CC15 and CC79). Diagn. Microbiol. Infect. Dis.79, 468–472. 10.1016/j.diagmicrobio.2014.03.006
57
ChangH. C.WeiY. F.DijkshoornL.VaneechoutteM.TangC. T.ChangT. C. (2005). Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol.43, 1632–1639. 10.1128/JCM.43.4.1632-1639.2005
58
ChangK. C.LinM. F.LinN. T.WuW. J.KuoH. Y.LinT. Y.et al. (2012). Clonal spread of multidrug-resistant Acinetobacter baumannii in eastern Taiwan. J. Microbiol. Immunol. Infect.45, 37–42. 10.1016/j.jmii.2011.09.019
59
CheahS. E.LiJ.TsujiB. T.ForrestA.BulittaJ. B.NationR. L. (2016). Colistin and polymyxin B dosage regimens against Acinetobacter baumannii: differences in activity and the emergence of resistance. Antimicrob. Agents Chemother.60, 3921–3933. 10.1128/AAC.02927-15
60
ChenC. H.YoungT. G.HuangC. C. (2006). Predictive biomarkers for drug-resistant Acinetobacter baumannii isolates with blaTEM-1, AmpC-type bla and integrase 1 genotypes. J. Microbiol. Immunol. Infect. 39, 372–379.
61
ChenQ.LiX.ZhouH.JiangY.ChenY.HuaX.et al. (2014). Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J. Antimicrob. Chemother.69, 72–76. 10.1093/jac/dkt319
62
ChenT. L.LeeY. T.KuoS. C.HsuehP. R.ChangF. Y.SiuL. K.et al. (2010). Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents Chemother.54, 4575–4581. 10.1128/AAC.00764-10
63
ChenT. L.WuR. C.ShaioM. F.FungC. P.ChoW. L. (2008). Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother.52, 2573–2580. 10.1128/AAC.00393-08
64
ChenY.ZhouZ.JiangY.YuY. (2011). Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother.66, 1255–1259. 10.1093/jac/dkr082
65
ChihiH.BonninR. A.BourouisA.MahroukiS.BesbesS.MoussaM. B.et al. (2016). GES-11-producing Acinetobacter baumannii clinical isolates from Tunisian hospitals: long-term dissemination of GES-type carbapenemases in North Africa. J. Glob. Antimicrob. Resist.5, 47–50. 10.1016/j.jgar.2016.03.005
66
ChinC. Y.GreggK. A.NapierB. A.ErnstR. K.WeissD. S. (2015). A PmrB-regulated deacetylase required for lipid A modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother.59, 7911–7914. 10.1128/AAC.00515-15
67
ChiuC. H.LiuY. H.WangY. C.LeeY. T.KuoS. C.ChenT. L.et al. (2016). In vitro activity of SecA inhibitors in combination with carbapenems against carbapenem-hydrolysing class D β-lactamase-producing Acinetobacter baumannii. J. Antimicrob. Chemother.71, 3441–3448. 10.1093/jac/dkw331
68
ChoY. J.MoonD. C.JinJ. S.ChoiC. H.LeeY. C.LeeJ. C. (2009). Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn. Microbiol. Infect. Dis. 64, 185–190. 10.1016/j.diagmicrobio.2009.02.010
69
ChoiC. H.HyunS. H.LeeJ. Y.LeeJ. S.LeeY. S.KimS. A.et al. (2008a). Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell. Microbiol.10, 309–319. 10.1111/j.1462-5822.2007.01041.x
70
ChoiC. H.LeeE. Y.LeeY. C.ParkT. I.KimH. J.HyunS. H.et al. (2005). Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol.7, 1127–1138. 10.1111/j.1462-5822.2005.00538.x
71
ChoiC. H.LeeJ. S.LeeY. C.ParkT. I.LeeJ. C. (2008b). Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol.8:216. 10.1186/1471-2180-8-216
72
ChuY. W.Afzal-ShahM.HouangE. T.PalepouM. I.LyonD. J.WoodfordN.et al. (2001). IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45, 710–714. 10.1128/AAC.45.3.710-714.2001
73
ChungJ. H.BhatA.KimC. J.YongD.RyuC. M. (2016). Combination therapy with polymyxin B and netropsin against clinical isolates of multidrug-resistant Acinetobacter baumannii. Sci. Rep.6:28168. 10.1038/srep28168
74
CirioniO.SimonettiO.PierpaoliE.BaruccaA.GhiselliR.OrlandoF.et al. (2016). Colistin enhances therapeutic efficacy of daptomycin or teicoplanin in a murine model of multiresistant Acinetobacter baumannii sepsis. Diagn. Microbiol. Infect. Dis.86, 392–398. 10.1016/j.diagmicrobio.2016.09.010
75
CirzR. T.RomesbergF. E. (2007). Controlling mutation: intervening in evolution as a therapeutic strategy. Crit. Rev. Biochem. Mol. Biol.42, 341–354. 10.1080/10409230701597741
76
CisnerosJ. M.Rodríguez-BañoJ. (2002). Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin. Microbiol. Infect. 8, 687–693. 10.1046/j.1469-0691.2002.00487.x
77
ClemmerK. M.BonomoR. A.RatherP. N. (2011). Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology157, 2534–2544. 10.1099/mic.0.049791-0
78
CorbinB. D.SeeleyE. H.RaabA.FeldmannJ.MillerM. R.TorresV. J.et al. (2008). Metal chelation and inhibition of bacterial growth in tissue abscesses. Science319, 962–965. 10.1126/science.1152449
79
CordobaJ.Coronado-AlvarezN. M.ParraD.Parra-RuizJ. (2015). In vitro activities of novel antimicrobial combinations against extensively drug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother.59, 7316–7319. 10.1128/AAC.00493-15
80
CorvecS.CaroffN.EspazeE.GiraudeauC.DrugeonH.ReynaudA. (2003). AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother.52, 629–635. 10.1093/jac/dkg407
81
CorvecS.PoirelL.NaasT.DrugeonH.NordmannP. (2007). Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA−23 in Acinetobacter baumannii. Antimicrob. Agents Chemother.51, 1530–1533. 10.1128/AAC.01132-06
82
CoyneS.CourvalinP.PerichonB. (2011). Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother.55, 947–953. 10.1128/AAC.01388-10
83
CoyneS.RosenfeldN.LambertT.CourvalinP.PerichonB. (2010). Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother.54, 4389–4393. 10.1128/AAC.00155-10
84
CroxattoA.Prod'homG.GreubG. (2012). Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev.36, 380–407. 10.1111/j.1574-6976.2011.00298.x
85
CurcioD.FernandezF. (2008). Tigecycline for Acinetobacter baumannii infection: other considerations. Clin. Infect. Dis. 46, 1797–1798; author reply 1798–1799. 10.1086/588051
86
Damier-PiolleL.MagnetS.BremontS.LambertT.CourvalinP. (2008). AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother.52, 557–562. 10.1128/AAC.00732-07
87
DavisonM.TreangenT. J.KorenS.PopM.BhayaD. (2016). Diversity in a polymicrobial community revealed by analysis of viromes, endolysins and CRISPR spacers. PLoS ONE11:e0160574. 10.1371/journal.pone.0160574
88
de BreijA.DijkshoornL.LagendijkE.van der MeerJ.KosterA.BloembergG.et al. (2010). Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii?PLoS ONE5:e10732. 10.1371/journal.pone.0010732
89
DefraineV.SchuermansJ.GrymonprezB.GoversS. K.AertsenA.FauvartM.et al. (2016). Efficacy of artilysin Art-175 against resistant and persistent Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 3480–3488. 10.1128/AAC.00285-16
90
del Mar TomasM.BeceiroA.PerezA.VelascoD.MoureR.VillanuevaR.et al. (2005). Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother.49, 5172–5175. 10.1128/AAC.49.12.5172-5175.2005
91
DengM.ZhuM. H.LiJ. J.BiS.ShengZ. K.HuF. S.et al. (2014). Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob. Agents Chemother.58, 297–303. 10.1128/AAC.01727-13
92
de Sa CavalcantiF. L.Mendes-MarquesC. L.VasconcelosC. R.de Lima CamposT.RezendeA. M.XavierD. E.et al. (2016). High frequency of OXA-253-producing Acinetobacter baumannii in different hospitals in Recife, Brazil: a new threat?Antimicrob. Agents Chemother.61:e01309–16. 10.1128/AAC.01309-16
93
DhabaanG. N.AbuBakarS.CerqueiraG. M.Al-HaroniM.PangS. P.HassanH. (2015). Imipenem treatment induces expression of important genes and phenotypes in a resistant Acinetobacter baumannii isolate. Antimicrob. Agents Chemother.60, 1370–1376. 10.1128/AAC.01696-15
94
DijkshoornL.AuckenH.Gerner-SmidtP.JanssenP.KaufmannM. E.GaraizarJ.et al. (1996). Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34, 1519–1525.
95
DizbayM.TozluD. K.CirakM. Y.IsikY.OzdemirK.ArmanD. (2010). In vitro synergistic activity of tigecycline and colistin against XDR-Acinetobacter baumannii. J. Antibiot. (Tokyo)63, 51–53. 10.1038/ja.2009.117
96
DoiY.MurrayG. L.PelegA. Y. (2015). Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med.36, 85–98. 10.1055/s-0034-1398388
97
DoiY.WachinoJ.YamaneK.ShibataN.YagiT.ShibayamaK.et al. (2004). Spread of novel aminoglycoside resistance gene aac(6′)-Iad among Acinetobacter clinical isolates in Japan. Antimicrob. Agents Chemother.48, 2075–2080. 10.1128/AAC.48.6.2075-2080.2004
98
DonnarummaF.SergiS.IndoratoC.MastromeiG.MonnanniR.NicolettiP.et al. (2010). Molecular characterization of acinetobacter isolates collected in intensive care units of six hospitals in Florence, Italy, during a 3-year surveillance program: a population structure analysis. J. Clin. Microbiol.48, 1297–1304. 10.1128/JCM.01916-09
99
DortetL.BonninR. A.BernabeuS.EscautL.VittecoqD.GirlichD.et al. (2016). First occurrence of OXA-72-producing Acinetobacter baumannii in Serbia. Antimicrob. Agents Chemother.60, 5724–5730. 10.1128/AAC.01016-16
100
DuboisD.PrasadaraoN. V.MittalR.BretL.Roujou-GrisM.BonnetR. (2009). CTX-M β-lactamase production and virulence of Escherichia coli K1. Emerg. Infect. Dis.15, 1988–1990. 10.3201/eid1512.090928
101
DubrovskayaY.PrasadN.LeeY.EsaianD.FigueroaD. A.TamV. H. (2015). Risk factors for nephrotoxicity onset associated with polymyxin B therapy. J. Antimicrob. Chemother.70, 1903–1907. 10.1093/jac/dkv014
102
DupontM.PagesJ. M.LafitteD.SiroyA.BolletC. (2005). Identification of an OprD homologue in Acinetobacter baumannii. J. Proteome. Res.4, 2386–2390. 10.1021/pr050143q
103
ElhosseinyN. M.AminM. A.YassinA. S.AttiaA. S. (2015). Acinetobacter baumannii universal stress protein A plays a pivotal role in stress response and is essential for pneumonia and sepsis pathogenesis. Int. J. Med. Microbiol.305, 114–123. 10.1016/j.ijmm.2014.11.008
104
ElhosseinyN. M.El-TayebO. M.YassinA. S.LoryS.AttiaA. S. (2016). The secretome of Acinetobacter baumannii ATCC 17978 type II secretion system reveals a novel plasmid encoded phospholipase that could be implicated in lung colonization. Int. J. Med. Microbiol.306, 633–641. 10.1016/j.ijmm.2016.09.006
105
EllisT. N.KuehnM. J. (2010). Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev.74, 81–94. 10.1128/MMBR.00031-09
106
EndimianiA.LuzzaroF.MigliavaccaR.MantengoliE.HujerA. M.HujerK. M.et al. (2007). Spread in an Italian hospital of a clonal Acinetobacter baumannii strain producing the TEM-92 extended-spectrum β-lactamase. Antimicrob. Agents Chemother.51, 2211–2214. 10.1128/AAC.01139-06
107
ErridgeC.Moncayo-NietoO. L.MorganR.YoungM.PoxtonI. R. (2007). Acinetobacter baumannii lipopolysaccharides are potent stimulators of human monocyte activation via Toll-like receptor 4 signalling. J. Med. Microbiol.56, 165–171. 10.1099/jmm.0.46823-0
108
EspinalP.FugazzaG.LopezY.KasmaM.LermanY.Malhotra-KumarS.et al. (2011). Dissemination of an NDM-2-producing Acinetobacter baumannii clone in an Israeli rehabilitation center. Antimicrob. Agents Chemother.55, 5396–5398. 10.1128/AAC.00679-11
109
EvansB. A.BrownS.HamoudaA.FindlayJ.AmyesS. G. (2007). Eleven novel OXA-51-like enzymes from clinical isolates of Acinetobacter baumannii. Clin. Microbiol. Infect.13, 1137–1138. 10.1111/j.1469-0691.2007.01828.x
110
Fajardo BoninR.ChapeaurougeA.PeralesJ.da SilvaJ. G.Jr.do NascimentoH. J.D'Alincourt Carvalho AssefA. P.et al. (2014). Identification of immunogenic proteins of the bacterium Acinetobacter baumannii using a proteomic approach. Proteomics Clin. Appl.8, 916–923. 10.1002/prca.201300133
111
FanB.GuanJ.WangX.CongY. (2016). Activity of colistin in combination with meropenem, tigecycline, fosfomycin, fusidic acid, rifampin or sulbactam against extensively drug-resistant Acinetobacter baumannii in a murine thigh-infection model. PLoS ONE11:e0157757. 10.1371/journal.pone.0157757
112
FangF.WangS.DangY. X.WangX.YuG. Q. (2016). Molecular characterization of carbapenemase genes in Acinetobacter baumannii in China. Genet. Mol. Res.15:gmr7432. 10.4238/gmr.15017432
113
Fernandez-CuencaF.SmaniY.Gomez-SanchezM. C.Docobo-PerezF.Caballero-MoyanoF. J.Dominguez-HerreraJ.et al. (2011). Attenuated virulence of a slow-growing pandrug-resistant Acinetobacter baumannii is associated with decreased expression of genes encoding the porins CarO and OprD-like. Int. J. Antimicrob. Agents38, 548–549. 10.1016/j.ijantimicag.2011.08.002
114
FernandoD.KumarA. (2012). Growth phase-dependent expression of RND efflux pump- and outer membrane porin-encoding genes in Acinetobacter baumannii ATCC 19606. J. Antimicrob. Chemother.67, 569–572. 10.1093/jac/dkr519
115
FiesterS. E.ArivettB. A.SchmidtR. E.BeckettA. C.TicakT.CarrierM. V.et al. (2016). Iron-regulated phospholipase C activity contributes to the cytolytic activity and virulence of Acinetobacter baumannii. PLoS ONE11:e0167068. 10.1371/journal.pone.0167068
116
Flores-DiazM.Monturiol-GrossL.NaylorC.Alape-GironA.FliegerA. (2016). Bacterial sphingomyelinases and phospholipases as virulence factors. Microbiol. Mol. Biol. Rev.80, 597–628. 10.1128/MMBR.00082-15
117
FonsecaE. L.ScheideggerE.FreitasF. S.CiprianoR.VicenteA. C. (2013). Carbapenem-resistant Acinetobacter baumannii from Brazil: role of carO alleles expression and blaOXA-23 gene. BMC Microbiol.13:245. 10.1186/1471-2180-13-245
118
FournierP. E.RichetH. (2006). The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis.42, 692–699. 10.1086/500202
119
FournierP. E.VallenetD.BarbeV.AudicS.OgataH.PoirelL.et al. (2006). Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet.2:e7. 10.1371/journal.pgen.0020007
120
FulneckyE. J.WrightD.ScheldW. M.KanawatiL.ShohamS. (2005). Amikacin and colistin for treatment of Acinetobacter baumannii meningitis. J. Infect.51, e249–e251. 10.1016/j.jinf.2005.04.003
121
GaddyJ. A.ArivettB. A.McConnellM. J.Lopez-RojasR.PachonJ.ActisL. A. (2012). Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun.80, 1015–1024. 10.1128/IAI.06279-11
122
GaddyJ. A.TomarasA. P.ActisL. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun.77, 3150–3160. 10.1128/IAI.00096-09
123
GalesA. C.TognimM. C.ReisA. O.JonesR. N.SaderH. S. (2003). Emergence of an IMP-like metallo-enzyme in an Acinetobacter baumannii clinical strain from a Brazilian teaching hospital. Diagn. Microbiol. Infect. Dis. 45, 77–79. 10.1016/S0732-8893(02)00500-X
124
GallegoL.TownerK. J. (2001). Carriage of class 1 integrons and antibiotic resistance in clinical isolates of Acinetobacter baumannii from northern Spain. J. Med. Microbiol.50, 71–77. 10.1099/0022-1317-50-1-71
125
Garcia-QuintanillaM.Carretero-LedesmaM.Moreno-MartinezP.Martin-PenaR.PachonJ.McConnellM. J. (2015). Lipopolysaccharide loss produces partial colistin dependence and collateral sensitivity to azithromycin, rifampicin and vancomycin in Acinetobacter baumannii. Int. J. Antimicrob. Agents46, 696–702. 10.1016/j.ijantimicag.2015.07.017
126
Garcia-SalgueroC.Rodriguez-AvialI.PicazoJ. J.CulebrasE. (2015). Can plazomicin alone or in combination be a therapeutic pption against carbapenem-resistant Acinetobacter baumannii?Antimicrob. Agents Chemother.59, 5959–5966. 10.1128/AAC.00873-15
127
GebhardtM. J.GallagherL. A.JacobsonR. K.UsachevaE. A.PetersonL. R.ZurawskiD. V.et al. (2015). Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. MBio6, e01660–15. 10.1128/mBio.01660-15
128
GehrleinM.LeyingH.CullmannW.WendtS.OpferkuchW. (1991). Imipenem resistance in Acinetobacter baumanii is due to altered penicillin-binding proteins. Chemotherapy37, 405–412.
129
GeisingerE.IsbergR. R. (2015). Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog.11:e1004691. 10.1371/journal.ppat.1004691
130
GiannouliM.TomasoneF.AgodiA.VahabogluH.DaoudZ.TriassiM.et al. (2009). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii strains in intensive care units of multiple Mediterranean hospitals. J. Antimicrob. Chemother.63, 828–830. 10.1093/jac/dkp032
131
GogouV.PournarasS.GiannouliM.VoulgariE.PiperakiE. T.ZarrilliR.et al. (2011). Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 year study in Greece (2000–09). J. Antimicrob. Chemother.66, 2767–2772. 10.1093/jac/dkr390
132
Goic-BarisicI.TownerK. J.KovacicA.Sisko-KraljevicK.TonkicM.NovakA.et al. (2011). Outbreak in Croatia caused by a new carbapenem-resistant clone of Acinetobacter baumannii producing OXA-72 carbapenemase. J. Hosp. Infect.77, 368–369. 10.1016/j.jhin.2010.12.003
133
Gonzalez-VilloriaA. M.Tamayo-LegorretaE.Garza-RamosU.BarriosH.Sanchez-PerezA.Rodriguez-MedinaN.et al. (2016). A multicenter study in Mexico finds Acinetobacter baumannii clinical isolates belonging to clonal complexes 636B (113B) and 92B harboring OXA-72, OXA-239, and OXA-469. Antimicrob. Agents Chemother.60, 2587–2588. 10.1128/AAC.02042-15
134
GordonN. C.WarehamD. W. (2009). A review of clinical and microbiological outcomes following treatment of infections involving multidrug-resistant Acinetobacter baumannii with tigecycline. J. Antimicrob. Chemother.63, 775–780. 10.1093/jac/dkn555
135
GordonN. C.WarehamD. W. (2010). Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents35, 219–226. 10.1016/j.ijantimicag.2009.10.024
136
HagiharaM.HousmanS. T.NicolauD. P.KutiJ. L. (2014). In vitro pharmacodynamics of polymyxin B and tigecycline alone and in combination against carbapenem-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother.58, 874–879. 10.1128/AAC.01624-13
137
HammerstromT. G.BeaboutK.ClementsT. P.SaxerG.ShamooY. (2015). Acinetobacter baumannii repeatedly evolves a hypermutator phenotype in response to tigecycline that effectively surveys evolutionary trajectories to resistance. PLoS ONE10:e0140489. 10.1371/journal.pone.0140489
138
HamoudaA.EvansB. A.TownerK. J.AmyesS. G. (2010). Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of blaOXA-51-like genes. J. Clin. Microbiol.48, 2476–2483. 10.1128/JCM.02431-09
139
HardingC. M.KinsellaR. L.PalmerL. D.SkaarE. P.FeldmanM. F. (2016). Medically relevant Acinetobacter species require a type II secretion system and specific membrane-associated chaperones for the export of multiple substrates and full virulence. PLoS Pathog.12:e1005391. 10.1371/journal.ppat.1005391
140
HasaniA.SheikhalizadehV.Ahangarzadeh RezaeeM.Rahmati-YamchiM.HasaniA.GhotaslouR.et al. (2016). Frequency of aminoglycoside-modifying enzymes and ArmA among different sequence groups of Acinetobacter baumannii in Iran. Microb. Drug Resist.22, 347–353. 10.1089/mdr.2015.0254
141
HassanA.NazA.ObaidA.ParachaR. Z.NazK.AwanF. M.et al. (2016). Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genomics.17:732. 10.1186/s12864-016-2951-4
142
HawleyJ. S.MurrayC. K.JorgensenJ. H. (2007). Development of colistin-dependent Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Antimicrob. Agents Chemother.51, 4529–4530. 10.1128/AAC.01115-07
143
HeS.HeH.ChenY.ChenY.WangW.YuD. (2015). In vitro and in vivo analysis of antimicrobial agents alone and in combination against multi-drug resistant Acinetobacter baumannii. Front. Microbiol.6, 507. 10.3389/fmicb.2015.00507
144
HeX.LuF.YuanF.JiangD.ZhaoP.ZhuJ.et al. (2015). Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob. Agents Chemother.59, 4817–4825. 10.1128/AAC.00877-15
145
HeritierC.PoirelL.AubertD.NordmannP. (2003). Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47, 268–273. 10.1128/AAC.47.1.268-273.2003
146
HeritierC.PoirelL.FournierP. E.ClaverieJ. M.RaoultD.NordmannP. (2005a). Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother.49, 4174–4179. 10.1128/AAC.49.10.4174-4179.2005
147
HeritierC.PoirelL.LambertT.NordmannP. (2005b). Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother.49, 3198–3202. 10.1128/AAC.49.8.3198-3202.2005
148
HeritierC.PoirelL.NordmannP. (2006). Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect.12, 123–130. 10.1111/j.1469-0691.2005.01320.x
149
HigginsP. G.JanssenK.FresenM. M.WisplinghoffH.SeifertH. (2012). Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J. Clin. Microbiol.50, 3493–3500. 10.1128/JCM.01759-12
150
HigginsP. G.Perez-LlarenaF. J.ZanderE.FernandezA.BouG.SeifertH. (2013). OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother.57, 2121–2126. 10.1128/AAC.02413-12
151
HigginsP. G.PoirelL.LehmannM.NordmannP.SeifertH. (2009). OXA-143, a novel carbapenem-hydrolyzing class D β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother.53, 5035–5038. 10.1128/AAC.00856-09
152
HigginsP. G.WisplinghoffH.StefanikD.SeifertH. (2004). Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J. Antimicrob. Chemother.54, 821–823. 10.1093/jac/dkh427
153
HirakiY.YoshidaM.MasudaY.InoueD.TsujiY.KamimuraH.et al. (2013). Successful treatment of skin and soft tissue infection due to carbapenem-resistant Acinetobacter baumannii by ampicillin-sulbactam and meropenem combination therapy. Int. J. Infect. Dis.17, e1234–e1236. 10.1016/j.ijid.2013.05.002
154
HoggG. M.BarrJ. G.WebbC. H. (1998). In-vitro activity of the combination of colistin and rifampicin against multidrug-resistant strains of Acinetobacter baumannii. J Antimicrob Chemother. 41, 494–495.
155
HongS. B.ShinK. S.HaJ.HanK. (2013). Co-existence of blaOXA-23 and armA in multidrug-resistant Acinetobacter baumannii isolated from a hospital in South Korea. J. Med. Microbiol.62, 836–844. 10.1099/jmm.0.055384-0
156
HongY. K.LeeJ. Y.WiY. M.KoK. S. (2016). High rate of colistin dependence in Acinetobacter baumannii. J. Antimicrob. Chemother.71, 2346–2348. 10.1093/jac/dkw121
157
HoodM. I.JacobsA. C.SayoodK.DunmanP. M.SkaarE. P. (2010). Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob. Agents Chemother.54, 1029–1041. 10.1128/AAC.00963-09
158
HoodM. I.MortensenB. L.MooreJ. L.ZhangY.Kehl-FieT. E.SugitaniN.et al. (2012). Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog.8:e1003068. 10.1371/journal.ppat.1003068
159
HornseyM.EllingtonM. J.DoumithM.ThomasC. P.GordonN. C.WarehamD. W.et al. (2010). AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J. Antimicrob. Chemother.65, 1589–1593. 10.1093/jac/dkq218
160
HornseyM.LomanN.WarehamD. W.EllingtonM. J.PallenM. J.TurtonJ. F.et al. (2011). Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J. Antimicrob. Chemother.66, 1499–1503. 10.1093/jac/dkr168
161
HornseyM.WarehamD. W. (2011). In vivo efficacy of glycopeptide-colistin combination therapies in a Galleria mellonella model of Acinetobacter baumannii infection. Antimicrob. Agents Chemother.55, 3534–3537. 10.1128/AAC.00230-11
162
HouC.YangF. (2015). Drug-resistant gene of blaOXA-23, blaOXA-24, blaOXA-51and blaOXA-58 in Acinetobacter baumannii. Int. J. Clin. Exp. Med. 8, 13859–13863.
163
HouangE. T.ChuY. W.ChuK. Y.NgK. C.LeungC. M.ChengA. F. (2003). Significance of genomic DNA group delineation in comparative studies of antimicrobial susceptibility of Acinetobacter spp. Antimicrob. Agents Chemother. 47, 1472–1475. 10.1128/AAC.47.4.1472-1475.2003
164
HuD.LiuB.DijkshoornL.WangL.ReevesP. R. (2013). Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS ONE8:e70329. 10.1371/journal.pone.0070329
165
HuW. S.YaoS. M.FungC. P.HsiehY. P.LiuC. P.LinJ. F. (2007). An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother.51, 3844–3852. 10.1128/AAC.01512-06
166
HuangW.YaoY.LongQ.YangX.SunW.LiuC.et al. (2014). Immunization against multidrug-resistant Acinetobacter baumannii effectively protects mice in both pneumonia and sepsis models. PLoS ONE9:e100727. 10.1371/journal.pone.0100727
167
HuangW.YaoY.WangS.XiaY.YangX.LongQ.et al. (2016). Immunization with a 22-kDa outer membrane protein elicits protective immunity to multidrug-resistant Acinetobacter baumannii. Sci. Rep.6:20724. 10.1038/srep20724
168
HujerK. M.HamzaN. S.HujerA. M.PerezF.HelfandM. S.BethelC. R.et al. (2005). Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 β-lactamase: defining a unique family of class C enzymes. Antimicrob. Agents Chemother.49, 2941–2948. 10.1128/AAC.49.7.2941-2948.2005
169
IwashkiwJ. A.SeperA.WeberB. S.ScottN. E.VinogradovE.StratiloC.et al. (2012). Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog.8:e1002758. 10.1371/journal.ppat.1002758
170
JacobsA. C.HoodI.BoydK. L.OlsonP. D.MorrisonJ. M.CarsonS.et al. (2010). Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun.78, 1952–1962. 10.1128/IAI.00889-09
171
JaraL. M.Perez-VarelaM.CorralJ.ArchM.CortesP.BouG.et al. (2015). Novobiocin inhibits the antimicrobial resistance acquired through DNA damage-induced mutagenesis in Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 637–639. 10.1128/AAC.01810-15
172
JaruratanasirikulS.WongpoowarakW.WattanavijitkulT.SukarnjanasetW.SamaengM.NawakitrangsanM.et al. (2016). Population pharmacokinetics and pharmacodynamics modeling to optimize dosage regimens of sulbactam in critically ill patients with severe sepsis caused by Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 7236–7244. 10.1128/AAC.01669-16
173
JeonJ. H.LeeJ. H.LeeJ. J.ParkK. S.KarimA. M.LeeC. R.et al. (2015). Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int. J. Mol. Sci.16, 9654–9692. 10.3390/ijms16059654
174
JeonJ.RyuC. M.LeeJ. Y.ParkJ. H.YongD.LeeK. (2016). In vivo application of bacteriophage as a potential therapeutic agent to control OXA-66-like carbapenemase-producing Acinetobacter baumannii strains belonging to sequence type 357. Appl. Environ. Microbiol.82, 4200–4208. 10.1128/AEM.00526-16
175
JeongH. W.CheongH. J.KimW. J.KimM. J.SongK. J.SongJ. W.et al. (2009). Loss of the 29-kilodalton outer membrane protein in the presence of OXA-51-like enzymes in Acinetobacter baumannii is associated with decreased imipenem susceptibility. Microb. Drug Resist.15, 151–158. 10.1089/mdr.2009.0828
176
JeongS. H.BaeI. K.KwonS. B.LeeK.YongD.WooG. J.et al. (2005). Investigation of a nosocomial outbreak of Acinetobacter baumannii producing PER-1 extended-spectrum β-lactamase in an intensive care unit. J. Hosp. Infect.59, 242–248. 10.1016/j.jhin.2004.09.025
177
JinJ. S.KwonS. O.MoonD. C.GurungM.LeeJ. H.KimS. I.et al. (2011). Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS ONE6:e17027. 10.1371/journal.pone.0017027
178
JohnsonT. L.WaackU.SmithS.MobleyH.SandkvistM. (2015). Acinetobacter baumannii is dependent on the type II secretion system and its substrate LipA for lipid utilization and in vivo fitness. J. Bacteriol.198, 711–719. 10.1128/JB.00622-15
179
JonesC. L.ClancyM.HonnoldC.SinghS.SnesrudE.Onmus-LeoneF.et al. (2015). Fatal outbreak of an emerging clone of extensively drug-resistant Acinetobacter baumannii with enhanced virulence. Clin. Infect. Dis.61, 145–154. 10.1093/cid/civ225
180
JunS. H.LeeJ. H.KimB. R.KimS. I.ParkT. I.LeeJ. C.et al. (2013). Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins. PLoS ONE8:e71751. 10.1371/journal.pone.0071751
181
JuttukondaL. J.ChazinW. J.SkaarE. P. (2016). Acinetobacter baumannii coordinates urea metabolism with metal import to resist host-mediated metal limitation. MBio7:e01475–16. 10.1128/mBio.01475-16
182
KalinG.AlpE.AkinA.CoskunR.DoganayM. (2014). Comparison of colistin and colistin/sulbactam for the treatment of multidrug resistant Acinetobacter baumannii ventilator-associated pneumonia. Infection.42, 37–42. 10.1007/s15010-013-0495-y
183
KaplanJ. B. (2011). Antibiotic-induced biofilm formation. Int. J. Artif. Organs.34, 737–751. 10.5301/ijao.5000027
184
KarthikeyanK.ThirunarayanM. A.KrishnanP. (2010). Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother.65, 2253–2254. 10.1093/jac/dkq273
185
KenyonJ. J.HallR. M. (2013). Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS ONE8:e62160. 10.1371/journal.pone.0062160
186
KimC. K.LeeY.LeeH.WooG. J.SongW.KimM. N.et al. (2010). Prevalence and diversity of carbapenemases among imipenem-nonsusceptible Acinetobacter isolates in Korea: emergence of a novel OXA-182. Diagn. Microbiol. Infect. Dis.68, 432–438. 10.1016/j.diagmicrobio.2010.07.014
187
KimS. W.ChoiC. H.MoonD. C.JinJ. S.LeeJ. H.ShinJ. H.et al. (2009). Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol. Lett.301, 224–231. 10.1111/j.1574-6968.2009.01820.x
188
KimY. J.KimS. I.KimY. R.HongK. W.WieS. H.ParkY. J.et al. (2012). Carbapenem-resistant Acinetobacter baumannii: diversity of resistant mechanisms and risk factors for infection. Epidemiol. Infect.140, 137–145. 10.1017/S0950268811000744
189
KnightD.DimitrovaD. D.RudinS. D.BonomoR. A.RatherP. N. (2016). Mutations decreasing intrinsic β-lactam resistance are linked to cell division in the nosocomial pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 3751–3758. 10.1128/AAC.00361-16
190
KobayashiN.NishinoK.YamaguchiA. (2001). Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol.183, 5639–5644. 10.1128/JB.183.19.5639-5644.2001
191
KoelemanJ. G.StoofJ.BiesmansD. J.SavelkoulP. H.Vandenbroucke-GraulsC. M. (1998). Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J. Clin. Microbiol. 36, 2522–2529.
192
KoelemanJ. G.van der BijlM. W.StoofJ.Vandenbroucke-GraulsC. M.SavelkoulP. H. (2001). Antibiotic resistance is a major risk factor for epidemic behavior of Acinetobacter baumannii. Infect. Control Hosp Epidemiol.22, 284–288. 10.1086/501901
193
KoenigsA.StahlJ.AverhoffB.GottigS.WichelhausT. A.WallichR.et al. (2016). CipA of Acinetobacter baumannii is a novel plasminogen binding and complement inhibitory protein. J. Infect. Dis.213, 1388–1399. 10.1093/infdis/jiv601
194
KoenigsA.ZipfelP. F.KraiczyP. (2015). Translation elongation factor Tuf of Acinetobacter baumannii is a plasminogen-binding protein. PLoS ONE10:e0134418. 10.1371/journal.pone.0134418
195
KohT. H.SngL. H.WangG. C.HsuL. Y.ZhaoY. (2007). IMP-4 and OXA beta-lactamases in Acinetobacter baumannii from Singapore. J. Antimicrob. Chemother.59, 627–632. 10.1093/jac/dkl544
196
KohT. H.TanT. T.KhooC. T.NgS. Y.TanT. Y.HsuL. Y.et al. (2012). Acinetobacter calcoaceticus-Acinetobacter baumannii complex species in clinical specimens in Singapore. Epidemiol. Infect.140, 535–538. 10.1017/S0950268811001129
197
KohlenbergA.BrummerS.HigginsP. G.SohrD.PieningB. C.de GrahlC.et al. (2009). Outbreak of carbapenem-resistant Acinetobacter baumannii carrying the carbapenemase OXA-23 in a German university medical centre. J. Med. Microbiol.58, 1499–1507. 10.1099/jmm.0.012302-0
198
KonstantinidisT.KambasK.MitsiosA.PanopoulouM.TsironidouV.DellaportaE.et al. (2016). Immunomodulatory role of clarithromycin in Acinetobacter baumannii infection via formation of neutrophil extracellular traps. Antimicrob. Agents Chemother.60, 1040–1048. 10.1128/AAC.02063-15
199
KoomanachaiP.KimA.NicolauD. P. (2009). Pharmacodynamic evaluation of tigecycline against Acinetobacter baumannii in a murine pneumonia model. J. Antimicrob. Chemother.63, 982–987. 10.1093/jac/dkp056
200
KorotkovK. V.SandkvistM.HolW. G. (2012). The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol.10, 336–351. 10.1038/nrmicro2762
201
KostouliasX.MurrayG. L.CerqueiraG. M.KongJ. B.BantunF.MylonakisE.et al. (2015). Impact of a cross-kingdom signaling molecule of Candida albicans on Acinetobacter baumannii physiology. Antimicrob. Agents Chemother.60, 161–167. 10.1128/AAC.01540-15
202
KrizovaL.PoirelL.NordmannP.NemecA. (2013). TEM-1 β-lactamase as a source of resistance to sulbactam in clinical strains of Acinetobacter baumannii. J. Antimicrob. Chemother.68, 2786–2791. 10.1093/jac/dkt275
203
KulpA.KuehnM. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol.64, 163–184. 10.1146/annurev.micro.091208.073413
204
KumarM. (2016). Identification of a novel NDM variant, blaNDM−3, from a multidrug-resistant Acinetobacter baumannii. Infect. Control. Hosp. Epidemiol.37, 747–748. 10.1017/ice.2016.66
205
KuoH. Y.HsuP. J.ChenJ. Y.LiaoP. C.LuC. W.ChenC. H.et al. (2016). Clonal spread of blaOXA-72-carrying Acinetobacter baumannii sequence type 512 in Taiwan. Int. J. Antimicrob. Agents48, 111–113. 10.1016/j.ijantimicag.2016.04.020
206
KuoH. Y.YangC. M.LinM. F.ChengW. L.TienN.LiouM. L. (2010). Distribution of blaOXA-carrying imipenem-resistant Acinetobacter spp. in 3 hospitals in Taiwan. Diagn. Microbiol. Infect. Dis. 66, 195–199. 10.1016/j.diagmicrobio.2009.09.013
207
KuoL. C.LaiC. C.LiaoC. H.HsuC. K.ChangY. L.ChangC. Y.et al. (2007). Multidrug-resistant Acinetobacter baumannii bacteraemia: clinical features, antimicrobial therapy and outcome. Clin. Microbiol. Infect.13, 196–198. 10.1111/j.1469-0691.2006.01601.x
208
KuoS. C.ChangS. C.WangH. Y.LaiJ. F.ChenP. C.ShiauY. R.et al. (2012). Emergence of extensively drug-resistant Acinetobacter baumannii complex over 10 years: nationwide data from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program. BMC Infect Dis.12:200. 10.1186/1471-2334-12-200
209
KusradzeI.KarumidzeN.RigvavaS.DvalidzeT.KatsitadzeM.AmiranashviliI.et al. (2016). Characterization and testing the efficiency of Acinetobacter baumannii phage vB-GEC_Ab-M-G7 as an antibacterial agent. Front. Microbiol.7:1590. 10.3389/fmicb.2016.01590
210
KwonS. O.GhoY. S.LeeJ. C.KimS. I. (2009). Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol. Lett.297, 150–156. 10.1111/j.1574-6968.2009.01669.x
211
La ScolaB.GundiV. A.KhamisA.RaoultD. (2006). Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol.44, 827–832. 10.1128/JCM.44.3.827-832.2006
212
LeeC. H.TangY. F.SuL. H.ChienC. C.LiuJ. W. (2008). Antimicrobial effects of varied combinations of meropenem, sulbactam, and colistin on a multidrug-resistant Acinetobacter baumannii isolate that caused meningitis and bacteremia. Microb. Drug Resist.14, 233–237. 10.1089/mdr.2008.0840
213
LeeC. R.ChoI. H.JeongB. C.LeeS. H. (2013a). Strategies to minimize antibiotic resistance. Int. J. Environ. Res. Public Health.10, 4274–4305. 10.3390/ijerph10094274
214
LeeC. R.LeeJ. H.JeongB. C.LeeS. H. (2013b). Lipid a biosynthesis of multidrug-resistant pathogens - a novel drug target. Curr. Pharm. Des. 19, 6534–6550. 10.2174/13816128113199990494
215
LeeC. R.LeeJ. H.ParkK. S.JeongB. C.LeeS. H. (2015). Quantitative proteomic view associated with resistance to clinically important antibiotics in Gram-positive bacteria: a systematic review. Front. Microbiol.6:828. 10.3389/fmicb.2015.00828
216
LeeC. R.LeeJ. H.ParkK. S.KimY. B.JeongB. C.LeeS. H. (2016). Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol.7:895. 10.3389/fmicb.2016.00895
217
LeeJ. H.ChoiC. H.KangH. Y.LeeJ. Y.KimJ.LeeY. C.et al. (2007). Differences in phenotypic and genotypic traits against antimicrobial agents between Acinetobacter baumannii and Acinetobacter genomic species 13TU. J. Antimicrob. Chemother.59, 633–639. 10.1093/jac/dkm007
218
LeeJ. H.LeeJ. J.ParkK. S.LeeS. H. (2015). Urgent need for β-lactam-β-lactamase inhibitors. Lancet Infect. Dis.15, 876–877. 10.1016/S1473-3099(15)00143-7
219
LeeJ. H.ParkK. S.KarimA. M.LeeC. R.LeeS. H. (2016). How to minimise antibiotic resistance. Lancet Infect. Dis.16, 17–18. 10.1016/S1473-3099(15)00467-3
220
LeeJ. S.ChoiC. H.KimJ. W.LeeJ. C. (2010). Acinetobacter baumannii outer membrane protein A induces dendritic cell death through mitochondrial targeting. J. Microbiol.48, 387–392. 10.1007/s12275-010-0155-1
221
LeeK.YumJ. H.YongD.LeeH. M.KimH. D.DocquierJ. D.et al. (2005). Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother.49, 4485–4491. 10.1128/AAC.49.11.4485-4491.2005
222
LeeM. F.PengC. F.HsuH. J.ChenY. H. (2008). Molecular characterisation of the metallo-β-lactamase genes in imipenem-resistant Gram-negative bacteria from a university hospital in southern Taiwan. Int. J. Antimicrob. Agents32, 475–480. 10.1016/j.ijantimicag.2008.07.009
223
LeeY.KimC. K.LeeH.JeongS. H.YongD.LeeK. (2011). A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the blaOXA-23 gene and disrupting the outer membrane protein gene carO in Acinetobacter baumannii. Antimicrob. Agents Chemother.55, 361–363. 10.1128/AAC.01672-09
224
LeeY. T.TsaoS. M.HsuehP. R. (2013). Clinical outcomes of tigecycline alone or in combination with other antimicrobial agents for the treatment of patients with healthcare-associated multidrug-resistant Acinetobacter baumannii infections. Eur. J. Clin. Microbiol Infect Dis.32, 1211–1220. 10.1007/s10096-013-1870-4
225
Lees-MillerR. G.IwashkiwJ. A.ScottN. E.SeperA.VinogradovE.SchildS.et al. (2013). A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol.89, 816–830. 10.1111/mmi.12300
226
LenhardJ. R.BulittaJ. B.ConnellT. D.King-LyonsN.LandersdorferC. B.CheahS. E.et al. (2016a). High-intensity meropenem combinations with polymyxin B: new strategies to overcome carbapenem resistance in Acinetobacter baumannii. J. Antimicrob. Chemother.72, 153–165. 10.1093/jac/dkw355
227
LenhardJ. R.GallJ. S.BulittaJ. B.ThamlikitkulV.LandersdorferC. B.ForrestA.et al. (2016b). Comparative pharmacodynamics of four different carbapenems in combination with polymyxin B against carbapenem-resistant Acinetobacter baumannii. Int. J. Antimicrob. Agents48, 719–724. 10.1016/j.ijantimicag.2016.07.024
228
LiL.HassanK. A.BrownM. H.PaulsenI. T. (2016). Rapid multiplexed phenotypic screening identifies drug resistance functions for three novel efflux pumps in Acinetobacter baumannii. J. Antimicrob. Chemother.71, 1223–1232. 10.1093/jac/dkv460
229
LiX.LiuL.JiJ.ChenQ.HuaX.JiangY.et al. (2015). Tigecycline resistance in Acinetobacter baumannii mediated by frameshift mutation in plsC, encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase. Eur. J. Clin. Microbiol. Infect. Dis.34, 625–631. 10.1007/s10096-014-2272-y
230
LiX. M.ChoiJ. A.ChoiI. S.KookJ. K.ChangY. H.ParkG.et al. (2016). Development and evaluation of species-specific PCR for detection of nine Acinetobacter species. Ann. Clin. Lab. Sci. 46, 270–278.
231
LiX.QuanJ.YangY.JiJ.LiuL.FuY.et al. (2016). Abrp, a new gene, confers reduced susceptibility to tetracycline, glycylcine, chloramphenicol and fosfomycin classes in Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis.35, 1371–1375. 10.1007/s10096-016-2674-0
232
LiY.GuoQ.WangP.ZhuD.YeX.WuS.et al. (2015). Clonal dissemination of extensively drug-resistant Acinetobacter baumannii producing an OXA-23 β-lactamase at a teaching hospital in Shanghai, China. J. Microbiol. Immunol. Infect.48, 101–108. 10.1016/j.jmii.2014.04.005
233
LiZ. T.ZhangR. L.BiX. G.XuL.FanM.XieD.et al. (2015). Outer membrane vesicles isolated from two clinical Acinetobacter baumannii strains exhibit different toxicity and proteome characteristics. Microb. Pathog.81, 46–52. 10.1016/j.micpath.2015.03.009
234
LiangW.LiuX. F.HuangJ.ZhuD. M.LiJ.ZhangJ. (2011). Activities of colistin- and minocycline-based combinations against extensive drug resistant Acinetobacter baumannii isolates from intensive care unit patients. BMC Infect. Dis.11:109. 10.1186/1471-2334-11-109
235
LiaoY. T.KuoS. C.ChiangM. H.LeeY. T.SungW. C.ChenY. H.et al. (2015). Acinetobacter baumannii extracellular OXA-58 is primarily and selectively released via outer membrane vesicles after Sec-dependent periplasmic translocation. Antimicrob. Agents Chemother.59, 7346–7354. 10.1128/AAC.01343-15
236
LinL.TanB.PantapalangkoorP.HoT.BaquirB.TomarasA.et al. (2012). Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. MBio3:e00312–12. 10.1128/mBio.00312-12
237
LinL.TanB.PantapalangkoorP.HoT.HujerA. M.TaracilaM. A.et al. (2013). Acinetobacter baumannii rOmpA vaccine dose alters immune polarization and immunodominant epitopes. Vaccine31, 313–318. 10.1016/j.vaccine.2012.11.008
238
LinM. F.ChangK. C.LanC. Y.ChouJ.KuoJ. W.ChangC. K.et al. (2011a). Molecular epidemiology and antimicrobial resistance determinants of multidrug-resistant Acinetobacter baumannii in five proximal hospitals in Taiwan. Jpn. J. Infect. Dis. 64, 222–227.
239
LinM. F.ChangK. C.YangC. Y.YangC. M.XiaoC. C.KuoH. Y.et al. (2010). Role of integrons in antimicrobial susceptibility patterns of Acinetobacter baumannii. Jpn. J. Infect. Dis. 63, 440–443.
240
LinM. F.KuoH. Y.YehH. W.YangC. M.SungC. H.TuC. C.et al. (2011b). Emergence and dissemination of blaOXA-23-carrying imipenem-resistant Acinetobacter sp. in a regional hospital in Taiwan. J. Microbiol. Immunol. Infect.44, 39–44. 10.1016/j.jmii.2011.01.008
241
LinM. F.LanC. Y. (2014). Antimicrobial resistance in Acinetobacter baumannii: from bench to bedside. World J. Clin. Cases.2, 787–814. 10.12998/wjcc.v2.i12.787
242
LinM. F.LinY. Y.LanC. Y. (2015). The role of the two-component system BaeSR in disposing chemicals through regulating transporter systems in Acinetobacter baumannii. PLoS ONE10:e0132843. 10.1371/journal.pone.0132843
243
LinM. F.LinY. Y.YehH. W.LanC. Y. (2014). Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol.14:119. 10.1186/1471-2180-14-119
244
LinM. F.LiouM. L.TuC. C.YehH. W.LanC. Y. (2013). Molecular epidemiology of integron-associated antimicrobial gene cassettes in the clinical isolates of Acinetobacter baumannii from northern Taiwan. Ann. Lab. Med.33, 242–247. 10.3343/alm.2013.33.4.242
245
LiouM. L.SooP. C.LingS. R.KuoH. Y.TangC. Y.ChangK. C. (2014). The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J. Microbiol. Immunol. Infect.47, 275–281. 10.1016/j.jmii.2012.12.004
246
LiuC. C.KuoH. Y.TangC. Y.ChangK. C.LiouM. L. (2014). Prevalence and mapping of a plasmid encoding a type IV secretion system in Acinetobacter baumannii. Genomics.104, 215–223. 10.1016/j.ygeno.2014.07.011
247
LiuD.LiuZ. S.HuP.CaiL.FuB. Q.LiY. S.et al. (2016). Characterization of surface antigen protein 1 (SurA1) from Acinetobacter baumannii and its role in virulence and fitness. Vet. Microbiol.186, 126–138. 10.1016/j.vetmic.2016.02.018
248
LiuQ.LiW.FengY.TaoC. (2014). Efficacy and safety of polymyxins for the treatment of Acinectobacter baumannii infection: a systematic review and meta-analysis. PLoS ONE9:e98091. 10.1371/journal.pone.0098091
249
LiuX.ZhaoM.ChenY.BianX.LiY.ShiJ.et al. (2016). Synergistic killing by meropenem and colistin combination of carbapenem-resistant Acinetobacter baumannii isolates from Chinese patients in an in vitro pharmacokinetic/pharmacodynamic model. Int. J. Antimicrob. Agents48, 559–563. 10.1016/j.ijantimicag.2016.07.018
250
LiuY.LiuX. (2015). Detection of AmpC β-lactamases in Acinetobacter baumannii in the Xuzhou region and analysis of drug resistance. Exp. Ther. Med.10, 933–936. 10.3892/etm.2015.2612
251
LolansK.RiceT. W.Munoz-PriceL. S.QuinnJ. P. (2006). Multicity outbreak of carbapenem-resistant Acinetobacter baumannii isolates producing the carbapenemase OXA-40. Antimicrob. Agents Chemother.50, 2941–2945. 10.1128/AAC.00116-06
252
LomovskayaO.LewisK. (1992). Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 89, 8938–8942.
253
LukeN. R.SauberanS. L.RussoT. A.BeananJ. M.OlsonR.LoehfelmT. W.et al. (2010). Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect. Immun.78, 2017–2023. 10.1128/IAI.00016-10
254
LuoG.LinL.IbrahimA. S.BaquirB.PantapalangkoorP.BonomoR. A.et al. (2012). Active and passive immunization protects against lethal, extreme drug resistant-Acinetobacter baumannii infection. PLoS ONE7:e29446. 10.1371/journal.pone.0029446
255
LuP. L.DoumithM.LivermoreD. M.ChenT. P.WoodfordN. (2009). Diversity of carbapenem resistance mechanisms in Acinetobacter baumannii from a Taiwan hospital: spread of plasmid-borne OXA-72 carbapenemase. J. Antimicrob. Chemother.63, 641–647. 10.1093/jac/dkn553
256
MagnetS.CourvalinP.LambertT. (2001). Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother.45, 3375–3380. 10.1128/AAC.45.12.3375-3380.2001
257
MakJ. K.KimM. J.PhamJ.TapsallJ.WhiteP. A. (2009). Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemother.63, 47–54. 10.1093/jac/dkn454
258
MarchandI.Damier-PiolleL.CourvalinP.LambertT. (2004). Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother.48, 3298–3304. 10.1128/AAC.48.9.3298-3304.2004
259
Martinez-GuitianM.Vazquez-UchaJ. C.OdingoJ.ParishT.PozaM.WaiteR. D.et al. (2016). Synergy between colistin and the signal peptidase inhibitor MD3 is dependent on the mechanism of colistin resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 4375–4379. 10.1128/AAC.00510-16
260
MartinezT.MartinezI.VazquezG. J.AquinoE. E.RobledoI. E. (2016). Genetic environment of the KPC gene in Acinetobacter baumannii ST2 clone from Puerto Rico and genomic insights into its drug resistance. J. Med. Microbiol.65, 784–792. 10.1099/jmm.0.000289
261
McConnellM. J.ActisL.PachonJ. (2013). Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev.37, 130–155. 10.1111/j.1574-6976.2012.00344.x
262
McConnellM. J.RumboC.BouG.PachonJ. (2011). Outer membrane vesicles as an acellular vaccine against Acinetobacter baumannii. Vaccine29, 5705–5710. 10.1016/j.vaccine.2011.06.001
263
McQuearyC. N.KirkupB. C.SiY.BarlowM.ActisL. A.CraftD. W.et al. (2012). Extracellular stress and lipopolysaccharide modulate Acinetobacter baumannii surface-associated motility. J. Microbiol.50, 434–443. 10.1007/s12275-012-1555-1
264
MegeedA. A.HayssamM. A.SalemM. Z.El-ShikhM. S.TaleaI. A.AlogaibiY. A. (2016). Investigation of the virulence factors and molecular characterization of the clonal relations of multidrug-resistant Acinetobacter baumannii isolates. J. AOAC Int. [Epub ahead of print]. 10.5740/jaoacint.16-0139
265
MendesR. E.BellJ. M.TurnidgeJ. D.CastanheiraM.JonesR. N. (2009). Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J. Antimicrob. Chemother.63, 55–59. 10.1093/jac/dkn434
266
MendesR. E.FarrellD. J.SaderH. S.JonesR. N. (2010). Comprehensive assessment of tigecycline activity tested against a worldwide collection of Acinetobacter spp. (2005–2009). Diagn. Microbiol. Infect. Dis. 68, 307–311. 10.1016/j.diagmicrobio.2010.07.003
267
MenegucciT. C.AlbieroJ.MiglioriniL. B.AlvesJ. L.VianaG. F.MazucheliJ.et al. (2016). Strategies for the treatment of polymyxin B-resistant Acinetobacter baumannii infections. Int. J. Antimicrob. Agents47, 380–385. 10.1016/j.ijantimicag.2016.02.007
268
MerinoM.AcostaJ.PozaM.SanzF.BeceiroA.ChavesF.et al. (2010). OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother.54, 2724–2727. 10.1128/AAC.01674-09
269
MoffattJ. H.HarperM.HarrisonP.HaleJ. D.VinogradovE.SeemannT.et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother.54, 4971–4977. 10.1128/AAC.00834-10
270
MolandE. S.CraftD. W.HongS. G.KimS. Y.HachmeisterL.SayedS. D.et al. (2008). In vitro activity of tigecycline against multidrug-resistant Acinetobacter baumannii and selection of tigecycline-amikacin synergy. Antimicrob. Agents Chemother.52, 2940–2942. 10.1128/AAC.01581-07
271
MonteroA.ArizaJ.CorbellaX.DomenechA.CabellosC.AyatsJ.et al. (2004). Antibiotic combinations for serious infections caused by carbapenem-resistant Acinetobacter baumannii in a mouse pneumonia model. J. Antimicrob. Chemother.54, 1085–1091. 10.1093/jac/dkh485
272
MoonD. C.ChoiC. H.LeeJ. H.ChoiC. W.KimH. Y.ParkJ. S.et al. (2012). Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J. Microbiol.50, 155–160. 10.1007/s12275-012-1589-4
273
MosquedaN.EspinalP.CosgayaC.ViotaS.PlasensiaV.Alvarez-LermaF.et al. (2013). Globally expanding carbapenemase finally appears in Spain: nosocomial outbreak of Acinetobacter baumannii producing plasmid-encoded OXA-23 in Barcelona, Spain. Antimicrob. Agents Chemother.57, 5155–5157. 10.1128/AAC.01486-13
274
MotaouakkilS.CharraB.HachimiA.NejmiH.BenslamaA.ElmdaghriN.et al. (2006). Colistin and rifampicin in the treatment of nosocomial infections from multiresistant Acinetobacter baumannii. J. Infect.53, 274–278. 10.1016/j.jinf.2005.11.019
275
MoubareckC.BremontS.ConroyM. C.CourvalinP.LambertT. (2009). GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother.53, 3579–3581. 10.1128/AAC.00072-09
276
MougousJ. D.CuffM. E.RaunserS.ShenA.ZhouM.GiffordC. A.et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science312, 1526–1530. 10.1126/science.1128393
277
MoyaB.JuanC.AlbertiS.PerezJ. L.OliverA. (2008). Benefit of having multiple ampD genes for acquiring β-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob. Agents Chemother.52, 3694–3700. 10.1128/AAC.00172-08
278
MugnierP. D.PoirelL.NaasT.NordmannP. (2010). Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis.16, 35–40. 10.3201/eid1601.090852
279
MussiM. A.LimanskyA. S.VialeA. M. (2005). Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of b-barrel outer membrane proteins. Antimicrob. Agents Chemother.49, 1432–1440. 10.1128/AAC.49.4.1432-1440.2005
280
MussiM. A.RellingV. M.LimanskyA. S.VialeA. M. (2007). CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Lett.581, 5573–5578. 10.1016/j.febslet.2007.10.063
281
MuthusamyD.SudhishnaaS.BoppeA. (2016). In vitro activities of polymyxins and rifampicin against carbapenem resistant Acinetobacter baumannii at a tertiary care hospital from south India. J. Clin. Diagn. Res. 10, DC15–DC18. 10.7860/JCDR/2016/19968.8535
282
Mutlu YilmazE.SunbulM.AksoyA.YilmazH.GuneyA. K.GuvencT. (2012). Efficacy of tigecycline/colistin combination in a pneumonia model caused by extensively drug-resistant Acinetobacter baumannii. Int. J. Antimicrob. Agents40, 332–336. 10.1016/j.ijantimicag.2012.06.003
283
NaasT.CoignardB.CarbonneA.BlanckaertK.BajoletO.BernetC.et al. (2006). VEB-1 Extended-spectrum β-lactamase-producing Acinetobacter baumannii, France. Emerg. Infect. Dis.12, 1214–1222. 10.3201/eid1208.051547
284
NaasT.LevyM.HirschauerC.MarchandinH.NordmannP. (2005). Outbreak of carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-23 in a tertiary care hospital of Papeete, French Polynesia. J. Clin. Microbiol.43, 4826–4829. 10.1128/JCM.43.9.4826-4829.2005
285
NaasT.NamdariF.Reglier-PoupetH.PoyartC.NordmannP. (2007). Panresistant extended-spectrum β-lactamase SHV-5-producing Acinetobacter baumannii from New York City. J. Antimicrob. Chemother.60, 1174–1176. 10.1093/jac/dkm366
286
NaganoN.NaganoY.CordevantC.ShibataN.ArakawaY. (2004). Nosocomial transmission of CTX-M-2 β-lactamase-producing Acinetobacter baumannii in a neurosurgery ward. J. Clin. Microbiol.42, 3978–3984. 10.1128/JCM.42.9.3978-3984.2004
287
NairnB. L.LonerganZ. R.WangJ.BraymerJ. J.ZhangY.CalcuttM. W.et al. (2016). The Response of Acinetobacter baumannii to Zinc Starvation. Cell Host Microbe.19, 826–836. 10.1016/j.chom.2016.05.007
288
NaviaM. M.RuizJ.VilaJ. (2002). Characterization of an integron carrying a new class D β-lactamase (OXA-37) in Acinetobacter baumannii. Microb. Drug Resist.8, 261–265. 10.1089/10766290260469516
289
Navon-VeneziaS.LeavittA.CarmeliY. (2007). High tigecycline resistance in multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemother.59, 772–774. 10.1093/jac/dkm018
290
NemecA.DolzaniL.BrisseS.van den BroekP.DijkshoornL. (2004). Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol.53, 1233–1240. 10.1099/jmm.0.45716-0
291
NepkaM.PerivoliotiE.KraniotakiE.PolitiL.TsakrisA.PournarasS. (2016). In vitro bactericidal activity of trimethoprim-sulfamethoxazole alone and in combination with colistin, against carbapenem-resistant Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 10.1128/AAC.01082-16
292
NiW.LiY.GuanJ.ZhaoJ.CuiJ.WangR.et al. (2016). Effects of efflux pump inhibitors on colistin resistance in multidrug-resistant Gram-negative gacteria. Antimicrob. Agents Chemother.60, 3215–3218. 10.1128/AAC.00248-16
293
NortonM. D.SpilkiaA. J.GodoyV. G. (2013). Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J. Bacteriol.195, 1335–1345. 10.1128/JB.02176-12
294
Nowak-ZaleskaA.WieczorM.CzubJ.NierzwickiL.KotlowskiR.MikuckaA.et al. (2016). Correlation between the number of Pro-Ala repeats in the EmrA homologue of Acinetobacter baumannii and resistance to netilmicin, tobramycin, imipenem and ceftazidime. J. Glob. Antimicrob. Resist.7, 145–149. 10.1016/j.jgar.2016.09.004
295
OzbekB.SenturkA. (2010). Postantibiotic effects of tigecycline, colistin sulfate, and levofloxacin alone or tigecycline-colistin sulfate and tigecycline-levofloxacin combinations against Acinetobacter baumannii. Chemotherapy56, 466–471. 10.1159/000321015
296
Pachon-IbanezM. E.Docobo-PerezF.Lopez-RojasR.Dominguez-HerreraJ.Jimenez-MejiasM. E.Garcia-CurielA.et al. (2010). Efficacy of rifampin and its combinations with imipenem, sulbactam, and colistin in experimental models of infection caused by imipenem-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother.54, 1165–1172. 10.1128/AAC.00367-09
297
Pachon-IbanezM. E.Jimenez-MejiasM. E.PichardoC.LlanosA. C.PachonJ. (2004). Activity of tigecycline (GAR-936) against Acinetobacter baumannii strains, including those resistant to imipenem. Antimicrob. Agents Chemother.48, 4479–4481. 10.1128/AAC.48.11.4479-4481.2004
298
PailhoriesH.KempfM.BelmonteO.Joly-GuillouM. L.EveillardM. (2016). First case of OXA-24-producing Acinetobacter baumannii in cattle from Reunion Island, France. Int. J. Antimicrob. Agents48, 763–764. 10.1016/j.ijantimicag.2016.09.005
299
PankuchG. A.LinG.SeifertH.AppelbaumP. C. (2008). Activity of meropenem with and without ciprofloxacin and colistin against Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother.52, 333–336. 10.1128/AAC.00689-07
300
PannekS.HigginsP. G.SteinkeP.JonasD.AkovaM.BohnertJ. A.et al. (2006). Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-b-naphthylamide. J. Antimicrob. Chemother.57, 970–974. 10.1093/jac/dkl081
301
PantopoulouA.Giamarellos-BourboulisE. J.RaftogannisM.TsaganosT.DontasI.KoutoukasP.et al. (2007). Colistin offers prolonged survival in experimental infection by multidrug-resistant Acinetobacter baumannii: the significance of co-administration of rifampicin. Int. J. Antimicrob. Agents29, 51–55. 10.1016/j.ijantimicag.2006.09.009
302
PapaA.KoulouridaV.SouliouE. (2009). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in a newly established Greek hospital. Microb. Drug Resist.15, 257–260. 10.1089/mdr.2009.0060
303
ParkY. K.KoK. S. (2015). Effect of carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on killing Acinetobacter baumannii by colistin. J. Microbiol.53, 53–59. 10.1007/s12275-015-4498-5
304
Parra MillanR.Jimenez MejiasM. E.Sanchez EncinalesV.Ayerbe AlgabaR.Gutierrez ValenciaA.Pachon IbanezM. E.et al. (2016). Efficacy of lysophosphatidylcholine in combination with antimicrobial agents against Acinetobacter baumannii in experimental murine peritoneal sepsis and pneumonia models. Antimicrob. Agents Chemother.60, 4464–4470. 10.1128/AAC.02708-15
305
PasteranF.RapoportM.PetroniA.FacconeD.CorsoA.GalasM.et al. (2006). Emergence of PER-2 and VEB-1a in Acinetobacter baumannii strains in the Americas. Antimicrob. Agents Chemother.50, 3222–3224. 10.1128/AAC.00284-06
306
PeckK. R.KimM. J.ChoiJ. Y.KimH. S.KangC. I.ChoY. K.et al. (2012). In vitro time-kill studies of antimicrobial agents against blood isolates of imipenem-resistant Acinetobacter baumannii, including colistin- or tigecycline-resistant isolates. J. Med. Microbiol.61, 353–360. 10.1099/jmm.0.036939-0
307
PelegA. Y.AdamsJ.PatersonD. L. (2007). Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob. Agents Chemother.51, 2065–2069. 10.1128/AAC.01198-06
308
PelegA. Y.SeifertH.PatersonD. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev.21, 538–582. 10.1128/CMR.00058-07
309
PelletierM. R.CasellaL. G.JonesJ. W.AdamsM. D.ZurawskiD. V.HazlettK. R.et al. (2013). Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob. Agents Chemother.57, 4831–4840. 10.1128/AAC.00865-13
310
PenwellW. F.ArivettB. A.ActisL. A. (2012). The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS ONE7:e36493. 10.1371/journal.pone.0036493
311
PerezA.MerinoM.Rumbo-FealS.Alvarez-FragaL.VallejoJ. A.BeceiroA.et al. (2016). The FhaB/FhaC two-partner secretion system is involved in adhesion of Acinetobacter baumannii AbH12O-A2 strain. Virulence. [Epub ahead of print]. 10.1080/21505594.2016.1262313
312
PerezF.HujerA. M.HujerK. M.DeckerB. K.RatherP. N.BonomoR. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother.51, 3471–3484. 10.1128/AAC.01464-06
313
PetrosilloN.ChinelloP.ProiettiM. F.CecchiniL.MasalaM.FranchiC.et al. (2005). Combined colistin and rifampicin therapy for carbapenem-resistant Acinetobacter baumannii infections: clinical outcome and adverse events. Clin. Microbiol. Infect.11, 682–683. 10.1111/j.1469-0691.2005.01198.x
314
PfeiferY.WilharmG.ZanderE.WichelhausT. A.GottigS.HunfeldK. P.et al. (2011). Molecular characterization of blaNDM−1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother.66, 1998–2001. 10.1093/jac/dkr256
315
PheeL. M.BettsJ. W.BharathanB.WarehamD. W. (2015). Colistin and fusidic acid, a novel potent synergistic combination for treatment of multidrug-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother.59, 4544–4550. 10.1128/AAC.00753-15
316
PichardoC.Pachon-IbanezM. E.Docobo-PerezF.Lopez-RojasR.Jimenez-MejiasM. E.Garcia-CurielA.et al. (2010). Efficacy of tigecycline vs. imipenem in the treatment of experimental Acinetobacter baumannii murine pneumonia. Eur. J. Clin. Microbiol. Infect. Dis.29, 527–531. 10.1007/s10096-010-0890-6
317
PiresJ.SiriwardenaT. N.StachM.TinguelyR.KasraianS.LuzzaroF.et al. (2015). In vitro activity of the novel antimicrobial peptide dendrimer G3KL against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Antimicrob. Agents Chemother.59, 7915–7918. 10.1128/AAC.01853-15
318
PloyM. C.GiamarellouH.BourliouxP.CourvalinP.LambertT. (1994). Detection of aac(6′)-I genes in amikacin-resistant Acinetobacter spp. by PCR. Antimicrob. Agents Chemother. 38, 2925–2928.
319
PoirelL.CabanneL.VahabogluH.NordmannP. (2005a). Genetic environment and expression of the extended-spectrum β-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob. Agents Chemother.49, 1708–1713. 10.1128/AAC.49.5.1708-1713.2005
320
PoirelL.CorvecS.RapoportM.MugnierP.PetroniA.PasteranF.et al. (2007). Identification of the novel narrow-spectrum β-lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob. Agents Chemother.51, 2179–2184. 10.1128/AAC.01600-06
321
PoirelL.MansourW.BouallegueO.NordmannP. (2008). Carbapenem-resistant Acinetobacter baumannii isolates from Tunisia producing the OXA-58-like carbapenem-hydrolyzing oxacillinase OXA-97. Antimicrob. Agents Chemother.52, 1613–1617. 10.1128/AAC.00978-07
322
PoirelL.MarqueS.HeritierC.SegondsC.ChabanonG.NordmannP. (2005b). OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother.49, 202–208. 10.1128/AAC.49.1.202-208.2005
323
PoirelL.MugnierP. D.TolemanM. A.WalshT. R.RapoportM. J.PetroniA.et al. (2009). ISCR2, another vehicle for blaVEB gene acquisition. Antimicrob. Agents Chemother.53, 4940–4943. 10.1128/AAC.00414-09
324
PongpechP.AmornnopparattanakulS.PanapakdeeS.FungwithayaS.NannhaP.DhiraputraC.et al. (2010). Antibacterial activity of carbapenem-based combinations againts multidrug-resistant Acinetobacter baumannii. J. Med. Assoc. Thai. 93, 161–171.
325
PotronA.Munoz-PriceL. S.NordmannP.ClearyT.PoirelL. (2011). Genetic features of CTX-M-15-producing Acinetobacter baumannii from Haiti. Antimicrob. Agents Chemother.55, 5946–5948. 10.1128/AAC.05124-11
326
PotronA.PoirelL.CroizeJ.ChanteperdrixV.NordmannP. (2009). Genetic and biochemical characterization of the first extended-spectrum CARB-type b-lactamase, RTG-4, from Acinetobacter baumannii. Antimicrob. Agents Chemother.53, 3010–3016. 10.1128/AAC.01164-08
327
PournarasS.MarkogiannakisA.IkonomidisA.KondyliL.BethimoutiK.ManiatisA. N.et al. (2006). Outbreak of multiple clones of imipenem-resistant Acinetobacter baumannii isolates expressing OXA-58 carbapenemase in an intensive care unit. J. Antimicrob. Chemother.57, 557–561. 10.1093/jac/dkl004
328
PrincipeL.CaponeA.MazzarelliA.D'ArezzoS.BordiE.Di CaroA.et al. (2013). In vitro activity of doripenem in combination with various antimicrobials against multidrug-resistant Acinetobacter baumannii: possible options for the treatment of complicated infection. Microb. Drug Resist.19, 407–414. 10.1089/mdr.2012.0250
329
PrincipeL.D'ArezzoS.CaponeA.PetrosilloN.ViscaP. (2009). In vitro activity of tigecycline in combination with various antimicrobials against multidrug resistant Acinetobacter baumannii. Ann. Clin. Microbiol. Antimicrob.8:18. 10.1186/1476-0711-8-18
330
PrincipeL.PiazzaA.GianiT.BraccoS.CaltagironeM. S.ArenaF.et al. (2014). Epidemic diffusion of OXA-23-producing Acinetobacter baumannii isolates in Italy: results of the first cross-sectional countrywide survey. J. Clin. Microbiol.52, 3004–3010. 10.1128/JCM.00291-14
331
PukatzkiS.MaA. T.SturtevantD.KrastinsB.SarracinoD.NelsonW. C.et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A.103, 1528–1533. 10.1073/pnas.0510322103
332
QiC.MalczynskiM.ParkerM.ScheetzM. H. (2008). Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J. Clin. Microbiol.46, 1106–1109. 10.1128/JCM.01877-07
333
QualeJ.BratuS.LandmanD.HeddurshettiR. (2003). Molecular epidemiology and mechanisms of carbapenem resistance in Acinetobacter baumannii endemic in New York City. Clin. Infect. Dis.37, 214–220. 10.1086/375821
334
QuinteiraS.GrossoF.RamosH.PeixeL. (2007). Molecular epidemiology of imipenem-resistant Acinetobacter haemolyticus and Acinetobacter baumannii isolates carrying plasmid-mediated OXA-40 from a Portuguese hospital. Antimicrob. Agents Chemother.51, 3465–3466. 10.1128/AAC.00267-07
335
RafailidisP. I.IoannidouE. N.FalagasM. E. (2007). Ampicillin/sulbactam: current status in severe bacterial infections. Drugs67, 1829–1849. 10.2165/00003495-200767130-00003
336
RajamohanG.SrinivasanV. B.GebreyesW. A. (2010). Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J. Antimicrob. Chemother.65, 1919–1925. 10.1093/jac/dkq195
337
RakinA.SchneiderL.PodladchikovaO. (2012). Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia. Front. Cell. Infect. Microbiol.2:151. 10.3389/fcimb.2012.00151
338
RamirezM. S.DonM.MerkierA. K.BistueA. J.ZorreguietaA.CentronD.et al. (2010a). Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J. Clin. Microbiol.48, 1488–1490. 10.1128/JCM.01264-09
339
RamirezM. S.PineiroS.Argentinian Integron Study GroupCentronD. (2010b). Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob. Agents Chemother.54, 699–706. 10.1128/AAC.01392-08
340
RaoG. G.LyN. S.BulittaJ. B.SoonR. L.San RomanM. D.HoldenP. N.et al. (2016a). Polymyxin B in combination with doripenem against heteroresistant Acinetobacter baumannii: pharmacodynamics of new dosing strategies. J. Antimicrob. Chemother.71, 3148–3156. 10.1093/jac/dkw293
341
RaoG. G.LyN. S.DiepJ.ForrestA.BulittaJ. B.HoldenP. N.et al. (2016b). Combinatorial pharmacodynamics of polymyxin B and tigecycline against heteroresistant Acinetobacter baumannii. Int. J. Antimicrob. Agents48, 331–336. 10.1016/j.ijantimicag.2016.06.006
342
RavasiP.LimanskyA. S.RodriguezR. E.VialeA. M.MussiM. A. (2011). ISAba825, a functional insertion sequence modulating genomic plasticity and blaOXA-58 expression in Acinetobacter baumannii. Antimicrob. Agents Chemother.55, 917–920. 10.1128/AAC.00491-10
343
ReidG.Food Agricultural Organization of the United Nation the WHO (2005). The importance of guidelines in the development and application of probiotics. Curr. Pharm. Des. 11, 11–16. 10.2174/1381612053382395
344
RepizoG. D.GagneS.Foucault-GrunenwaldM. L.BorgesV.CharpentierX.LimanskyA. S.et al. (2015). Differential Role of the T6SS in Acinetobacter baumannii Virulence. PLoS ONE10:e0138265. 10.1371/journal.pone.0138265
345
RiberaA.RocaI.RuizJ.GibertI.VilaJ. (2003a). Partial characterization of a transposon containing the tet(A) determinant in a clinical isolate of Acinetobacter baumannii. J. Antimicrob. Chemother.52, 477–480. 10.1093/jac/dkg344
346
RiberaA.RuizJ.VilaJ. (2003b). Presence of the Tet M determinant in a clinical isolate of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47, 2310–2312. 10.1128/AAC.47.7.2310-2312.2003
347
RiccioM. L.FranceschiniN.BoschiL.CaravelliB.CornagliaG.FontanaR.et al. (2000). Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44, 1229–1235. 10.1128/AAC.44.5.1229-1235.2000
348
RitchieD. J.Garavaglia-WilsonA. (2014). A review of intravenous minocycline for treatment of multidrug-resistant Acinetobacter infections. Clin. Infect. Dis. 59 (Suppl. 6), S374–S380. 10.1093/cid/ciu613
349
RobledoI. E.AquinoE. E.SanteM. I.SantanaJ. L.OteroD. M.LeonC. F.et al. (2010). Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother.54, 1354–1357. 10.1128/AAC.00899-09
350
RocaI.MartiS.EspinalP.MartinezP.GibertI.VilaJ. (2009). CraA, a major facilitator superfamily efflux pump associated with chloramphenicol resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother.53, 4013–4014. 10.1128/AAC.00584-09
351
Rodriguez-BanoJ.MartiS.SotoS.Fernandez-CuencaF.CisnerosJ. M.PachonJ.et al. (2008). Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin. Microbiol. Infect.14, 276–278. 10.1111/j.1469-0691.2007.01916.x
352
RodriguezC. H.De AmbrosioA.BajukM.SpinozziM.NastroM.BombicinoK.et al. (2010). In vitro antimicrobials activity against endemic Acinetobacter baumannii multiresistant clones. J. Infect. Dev. Ctries. 4, 164–167. 10.3855/jidc.604
353
RodriguezC. H.NastroM.VayC.FamigliettiA. (2015). In vitro activity of minocycline alone or in combination in multidrug-resistant Acinetobacter baumannii isolates. J. Med. Microbiol.64, 1196–1200. 10.1099/jmm.0.000147
354
Rodriguez-RubioL.ChangW. L.GutierrezD.LavigneR.MartinezB.RodriguezA.et al. (2016). ‘Artilysation’ of endolysin lambdaSa2lys strongly improves its enzymatic and antibacterial activity against streptococci. Sci. Rep.6:35382. 10.1038/srep35382
355
RosenfeldN.BouchierC.CourvalinP.PerichonB. (2012). Expression of the resistance-nodulation-cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob. Agents Chemother.56, 2504–2510. 10.1128/AAC.06422-11
356
RuizF. M.SantillanaE.Spinola-AmilibiaM.TorreiraE.CulebrasE.RomeroA. (2015). Crystal Structure of Hcp from Acinetobacter baumannii: a Component of the Type VI Secretion System. PLoS ONE10:e0129691. 10.1371/journal.pone.0129691
357
RuizM.MartiS.Fernandez-CuencaF.PascualA.VilaJ. (2007). High prevalence of carbapenem-hydrolysing oxacillinases in epidemiologically related and unrelated Acinetobacter baumannii clinical isolates in Spain. Clin. Microbiol. Infect.13, 1192–1198. 10.1111/j.1469-0691.2007.01825.x
358
RumboC.Fernandez-MoreiraE.MerinoM.PozaM.MendezJ. A.SoaresN. C.et al. (2011). Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother.55, 3084–3090. 10.1128/AAC.00929-10
359
RumboC.TomasM.Fernandez MoreiraE.SoaresN. C.CarvajalM.SantillanaE.et al. (2014). The Acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infect. Immun.82, 4666–4680. 10.1128/IAI.02034-14
360
RumboC.VallejoJ. A.CabralM. P.Martinez-GuitianM.PerezA.BeceiroA.et al. (2016). Assessment of antivirulence activity of several D-amino acids against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Antimicrob. Chemother.71, 3473–3481. 10.1093/jac/dkw342
361
RussoT. A.BeananJ. M.OlsonR.MacDonaldU.CoxA. D.St. MichaelF.et al. (2013). The K1 capsular polysaccharide from Acinetobacter baumannii is a potential therapeutic target via passive immunization. Infect. Immun.81, 915–922. 10.1128/IAI.01184-12
362
RussoT. A.LukeN. R.BeananJ. M.OlsonR.SauberanS. L.MacDonaldU.et al. (2010). The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun.78, 3993–4000. 10.1128/IAI.00366-10
363
RussoT. A.MacDonaldU.BeananJ. M.OlsonR.MacDonaldI. J.SauberanS. L.et al. (2009). Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J. Infect. Dis.199, 513–521. 10.1086/596317
364
RussoT. A.ManoharA.BeananJ. M.OlsonR.MacDonaldU.GrahamJ.et al. (2016). The response regulator BfmR is a potential drug target for Acinetobacter baumannii. mSphere1:e00082–16. 10.1128/mSphere.00082-16
365
RuzinA.KeeneyD.BradfordP. A. (2007). AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Antimicrob. Chemother.59, 1001–1004. 10.1093/jac/dkm058
366
SahaR.SahaN.DonofrioR. S.BesterveltL. L. (2013). Microbial siderophores: a mini review. J. Basic Microbiol.53, 303–317. 10.1002/jobm.201100552
367
SahlyH.Navon-VeneziaS.RoeslerL.HayA.CarmeliY.PodschunR.et al. (2008). Extended-spectrum β-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesins in Klebsiella pneumoniae. Antimicrob. Agents Chemother.52, 3029–3034. 10.1128/AAC.00010-08
368
SakoulasG.RoseW.BertiA.OlsonJ.MungiaJ.NonejuieP.et al. (2016). Classical β-lactamase inhibitors potentiate the activity of daptomycin against methicillin-resistant Staphylococcus aureus and colistin against Acinetobacter baumannii. Antimicrob. Agents Chemother.61:e01745–16. 10.1128/AAC.01745-16
369
SandriA. M.LandersdorferC. B.JacobJ.BoniattiM. M.DalarosaM. G.FalciD. R.et al. (2013). Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin. Infect. Dis.57, 524–531. 10.1093/cid/cit334
370
SantimaleeworagunW.WongpoowarakP.ChayakulP.PattharachayakulS.TansakulP.GareyK. W. (2011). In vitro activity of colistin or sulbactam in combination with fosfomycin or imipenem against clinical isolates of carbapenem-resistant Acinetobacter baumannii producing OXA-23 carbapenemases. Southeast Asian J. Trop. Med. Public Health. 42, 890–900.
371
SchroderW.GoerkeC.WolzC. (2013). Opposing effects of aminocoumarins and fluoroquinolones on the SOS response and adaptability in Staphylococcus aureus. J. Antimicrob. Chemother. 68, 529–538. 10.1093/jac/dks456
372
SechiL. A.KaradenizliA.DeriuA.ZanettiS.KolayliF.BalikciE.et al. (2004). PER-1 type β-lactamase production in Acinetobacter baumannii is related to cell adhesion. Med. Sci. Monit. 10, BR180–BR184.
373
SegalH.NelsonE. C.ElishaB. G. (2004). Genetic environment and transcription of ampC in an Acinetobacter baumannii clinical isolate. Antimicrob. Agents Chemother. 48, 612–614. 10.1128/AAC.48.2.612-614.2004
374
SharmaA.SharmaR.BhattacharyyaT.BhandoT.PathaniaR. (2016). Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter-AbaF. J. Antimicrob. Chemother.72, 68–74. 10.1093/jac/dkw382
375
ShengW. H.WangJ. T.LiS. Y.LinY. C.ChengA.ChenY. C.et al. (2011). Comparative in vitro antimicrobial susceptibilities and synergistic activities of antimicrobial combinations against carbapenem-resistant Acinetobacter species: Acinetobacter baumannii versus Acinetobacter genospecies 3 and 13TU. Diagn. Microbiol. Infect. Dis.70, 380–386. 10.1016/j.diagmicrobio.2011.03.003
376
ShinB.ParkW. (2015). Synergistic effect of oleanolic acid on aminoglycoside antibiotics against Acinetobacter baumannii. PLoS ONE10:e0137751. 10.1371/journal.pone.0137751
377
ShneiderM. M.ButhS. A.HoB. T.BaslerM.MekalanosJ. J.LeimanP. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature500, 350–353. 10.1038/nature12453
378
SiroyA.MolleV.Lemaitre-GuillierC.VallenetD.Pestel-CaronM.CozzoneA. J.et al. (2005). Channel formation by CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. Antimicrob. Agents Chemother.49, 4876–4883. 10.1128/AAC.49.12.4876-4883.2005
379
SkalweitM. J.LiM. (2016). Bulgecin A as a β-lactam enhancer for carbapenem-resistant Pseudomonas aeruginosa and carbapenem-resistant Acinetobacter baumannii clinical isolates containing various resistance mechanisms. Drug Des. Devel. Ther.10, 3013–3020. 10.2147/DDDT.S110193
380
SmaniY.Dominguez-HerreraJ.PachonJ. (2013). Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii. J. Infect. Dis.208, 1561–1570. 10.1093/infdis/jit386
381
SmaniY.FabregaA.RocaI.Sanchez-EncinalesV.VilaJ.PachonJ. (2014). Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob. Agents Chemother.58, 1806–1808. 10.1128/AAC.02101-13
382
SmaniY.McConnellM. J.PachonJ. (2012). Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PLoS ONE7:e33073. 10.1371/journal.pone.0033073
383
SmithS. G.MahonV.LambertM. A.FaganR. P. (2007). A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett.273, 1–11. 10.1111/j.1574-6968.2007.00778.x
384
SmolyakovR.BorerA.RiesenbergK.SchlaefferF.AlkanM.PorathA.et al. (2003). Nosocomial multi-drug resistant Acinetobacter baumannii bloodstream infection: risk factors and outcome with ampicillin-sulbactam treatment. J. Hosp. Infect. 54, 32–38. 10.1016/S0195-6701(03)00046-X
385
SongerJ. G. (1997). Bacterial phospholipases and their role in virulence. Trends Microbiol.5, 156–161. 10.1016/S0966-842X(97)01005-6
386
SongJ. Y.LeeJ.HeoJ. Y.NohJ. Y.KimW. J.CheongH. J.et al. (2008). Colistin and rifampicin combination in the treatment of ventilator-associated pneumonia caused by carbapenem-resistant Acinetobacter baumannii. Int. J. Antimicrob. Agents32, 281–284. 10.1016/j.ijantimicag.2008.04.013
387
SrinivasanV. B.RajamohanG.GebreyesW. A. (2009). Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob. Agents Chemother.53, 5312–5316. 10.1128/AAC.00748-09
388
SrinivasanV. B.VenkataramaiahM.MondalA.RajamohanG. (2015). Functional characterization of AbeD, an RND-type membrane transporter in antimicrobial resistance in Acinetobacter baumannii. PLoS ONE10:e0141314. 10.1371/journal.pone.0141314
389
StahlJ.BergmannH.GottigS.EbersbergerI.AverhoffB. (2015). Acinetobacter baumannii virulence is mediated by the concerted action of three phospholipases D. PLoS ONE10:e0138360. 10.1371/journal.pone.0138360
390
StoevaT.HigginsP. G.BojkovaK.SeifertH. (2008). Clonal spread of carbapenem-resistant OXA-23-positive Acinetobacter baumannii in a Bulgarian university hospital. Clin. Microbiol. Infect.14, 723–727. 10.1111/j.1469-0691.2008.02018.x
391
SuC. H.WangJ. T.HsiungC. A.ChienL. J.ChiC. L.YuH. T.et al. (2012). Increase of carbapenem-resistant Acinetobacter baumannii infection in acute care hospitals in Taiwan: association with hospital antimicrobial usage. PLoS ONE7:e37788. 10.1371/journal.pone.0037788
392
SugawaraE.NikaidoH. (2012). OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J. Bacteriol.194, 4089–4096. 10.1128/JB.00435-12
393
SunJ. R.JengW. Y.PerngC. L.YangY. S.SooP. C.ChiangY. S.et al. (2016). Single amino acid substitution Gly186Val in AdeS restores tigecycline susceptibility of Acinetobacter baumannii. J. Antimicrob. Chemother.71, 1488–1492. 10.1093/jac/dkw002
394
SunJ. R.PerngC. L.ChanM. C.MoritaY.LinJ. C.SuC. M.et al. (2012). A truncated AdeS kinase protein generated by ISAba1 insertion correlates with tigecycline resistance in Acinetobacter baumannii. PLoS ONE7:e49534. 10.1371/journal.pone.0049534
395
SuX. Z.ChenJ.MizushimaT.KurodaT.TsuchiyaT. (2005). AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother.49, 4362–4364. 10.1128/AAC.49.10.4362-4364.2005
396
TadaT.Miyoshi-AkiyamaT.ShimadaK.ShimojimaM.KirikaeT. (2014). Dissemination of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and emergence of OXA-72 carbapenemase coproducers in Japan. Antimicrob. Agents Chemother.58, 2916–2920. 10.1128/AAC.01212-13
397
TaittC. R.LeskiT. A.StockelmanM. G.CraftD. W.ZurawskiD. V.KirkupB. C.et al. (2014). Antimicrobial resistance determinants in Acinetobacter baumannii isolates taken from military treatment facilities. Antimicrob. Agents Chemother.58, 767–781. 10.1128/AAC.01897-13
398
ThandarM.LoodR.WinerB. Y.DeutschD. R.EulerC. W.FischettiV. A. (2016). Novel engineered peptides of a phage lysin as effective antimicrobials against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 2671–2679. 10.1128/AAC.02972-15
399
ThummeepakR.KittiT.KunthalertD.SitthisakS. (2016). Enhanced antibacterial activity of Acinetobacter baumannii bacteriophage OABP-01 endolysin (LysABP-01) in combination with colistin. Front. Microbiol.7:1402. 10.3389/fmicb.2016.01402
400
TimurkaynakF.CanF.AzapO. K.DemirbilekM.ArslanH.KaramanS. O. (2006). In vitro activities of non-traditional antimicrobials alone or in combination against multidrug-resistant strains of Pseudomonas aeruginosa and Acinetobacter baumannii isolated from intensive care units. Int. J. Antimicrob. Agents27, 224–228. 10.1016/j.ijantimicag.2005.10.012
401
TiptonK. A.RatherP. N. (2016). An ompR/envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J. Bacteriol. [Epub ahead of print]. 10.1128/JB.00705-16
402
TognimM. C.GalesA. C.PenteadoA. P.SilbertS.SaderH. S. (2006). Dissemination of IMP-1 metallo- beta -lactamase-producing Acinetobacter species in a Brazilian teaching hospital. Infect. Control Hosp. Epidemiol.27, 742–747. 10.1086/504356
403
TomarasA. P.DorseyC. W.EdelmannR. E.ActisL. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology149, 3473–3484. 10.1099/mic.0.26541-0
404
TomarasA. P.FlaglerM. J.DorseyC. W.GaddyJ. A.ActisL. A. (2008). Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology154, 3398–3409. 10.1099/mic.0.2008/019471-0
405
TouchonM.CuryJ.YoonE. J.KrizovaL.CerqueiraG. C.MurphyC.et al. (2014). The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol. Evol.6, 2866–2882. 10.1093/gbe/evu225
406
TragliaG. M.ChuaK.CentronD.TolmaskyM. E.RamirezM. S. (2014). Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol. Evol.6, 2235–2239. 10.1093/gbe/evu176
407
TragliaG. M.QuinnB.SchrammS. T.Soler-BistueA.RamirezM. S. (2016). Serum Albumin and Ca2+ Are Natural Competence Inducers in the Human Pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 4920–4929. 10.1128/AAC.00529-16
408
TreboscV.GartenmannS.RoyetK.ManfrediP.TotzlM.SchellhornB.et al. (2016). A novel genome-editing platform for drug-resistant Acinetobacter baumannii reveals an AdeR-unrelated tigecycline resistance mechanism. Antimicrob. Agents Chemother.60, 7263–7271. 10.1128/AAC.01275-16
409
TsakrisA.IkonomidisA.PoulouA.SpanakisN.VrizasD.DiomidousM.et al. (2008). Clusters of imipenem-resistant Acinetobacter baumannii clones producing different carbapenemases in an intensive care unit. Clin. Microbiol. Infect.14, 588–594. 10.1111/j.1469-0691.2008.01996.x
410
TsakrisA.IkonomidisA.PournarasS.TzouvelekisL. S.SofianouD.LegakisN. J.et al. (2006). VIM-1 metallo-β-lactamase in Acinetobacter baumannii. Emerg. Infect. Dis. 12, 981–983. 10.3201/eid1206.051097
411
TsakrisA.IkonomidisA.SpanakisN.PournarasS.BethimoutiK. (2007). Identification of a novel blaOXA-51 variant, blaOXA-92, from a clinical isolate of Acinetobacter baumannii. Clin. Microbiol. Infect.13, 348–349. 10.1111/j.1469-0691.2006.01598.x
412
TsujiB. T.LandersdorferC. B.LenhardJ. R.CheahS. E.ThamlikitkulV.RaoG. G.et al. (2016). Paradoxical effect of polymyxin B: high drug exposure amplifies resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 3913–3920. 10.1128/AAC.02831-15
413
TurnerP. J.GreenhalghJ. M.MYSTIC Study Group (Europe) (2003). The activity of meropenem and comparators against Acinetobacter strains isolated from European hospitals, 1997–2000. Clin. Microbiol. Infect. 9, 563–567. 10.1046/j.1469-0691.2003.00591.x
414
TurtonJ. F.WardM. E.WoodfordN.KaufmannM. E.PikeR.LivermoreD. M.et al. (2006a). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett.258, 72–77. 10.1111/j.1574-6968.2006.00195.x
415
TurtonJ. F.WoodfordN.GloverJ.YardeS.KaufmannM. E.PittT. L. (2006b). Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin Microbiol.44, 2974–2976. 10.1128/JCM.01021-06
416
UmlandT. C.SchultzL. W.MacDonaldU.BeananJ. M.OlsonR.RussoT. A. (2012). In vivo-validated essential genes identified in Acinetobacter baumannii by using human ascites overlap poorly with essential genes detected on laboratory media. MBio. 3:e00113–12. 10.1128/mBio.00113-12
417
VahabogluH.BudakF.KasapM.GacarG.TorolS.KaradenizliA.et al. (2006). High prevalence of OXA-51-type class D β-lactamases among ceftazidime-resistant clinical isolates of Acinetobacter spp.: co-existence with OXA-58 in multiple centres. J. Antimicrob. Chemother.58, 537–542. 10.1093/jac/dkl273
418
ValenzuelaJ. K.ThomasL.PartridgeS. R.van der ReijdenT.DijkshoornL.IredellJ. (2007). Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J. Clin. Microbiol.45, 453–460. 10.1128/JCM.01971-06
419
VijayakumarS.GopiR.GunasekaranP.BharathyM.WaliaK.AnandanS.et al. (2016). Molecular characterization of invasive carbapenem-resistant Acinetobacter baumannii from a tertiary care hospital in south India. Infect. Dis. Ther.5, 379–387. 10.1007/s40121-016-0125-y
420
VilacobaE.AlmuzaraM.GuloneL.TragliaG. M.FigueroaS. A.SlyG.et al. (2013). Emergence and spread of plasmid-borne tet(B)::ISCR2 in minocycline-resistant Acinetobacter baumannii isolates. Antimicrob. Agents Chemother.57, 651–654. 10.1128/AAC.01751-12
421
VilaJ.NaviaM.RuizJ.CasalsC. (1997). Cloning and nucleotide sequence analysis of a gene encoding an OXA-derived β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 41, 2757–2759.
422
VilaJ.RuizJ.GoniP.MarcosA.Jimenez de AntaT. (1995). Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 39, 1201–1203.
423
VincentJ. L.RelloJ.MarshallJ.SilvaE.AnzuetoA.MartinC. D.et al. (2009). International study of the prevalence and outcomes of infection in intensive care units. JAMA302, 2323–2329. 10.1001/jama.2009.1754
424
VoulgariE.PolitiL.PitirigaV.DendrinosJ.PoulouA.GeorgiadisG.et al. (2016). First report of an NDM-1 metallo-β-lactamase-producing Acinetobacter baumannii clinical isolate in Greece. Int. J. Antimicrob. Agents48, 761–762. 10.1016/j.ijantimicag.2016.09.006
425
WangH.GuoP.SunH.WangH.YangQ.ChenM.et al. (2007). Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from Chinese hospitals. Antimicrob. Agents Chemother.51, 4022–4028. 10.1128/AAC.01259-06
426
WangN.OzerE. A.MandelM. J.HauserA. R. (2014). Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio5, e01163–14. 10.1128/mBio.01163-14
427
WarehamD. W.GordonN. C.HornseyM. (2011). In vitro activity of teicoplanin combined with colistin versus multidrug-resistant strains of Acinetobacter baumannii. J. Antimicrob. Chemother.66, 1047–1051. 10.1093/jac/dkr069
428
WeberB. S.HardingC. M.FeldmanM. F. (2015a). Pathogenic Acinetobacter: from the Cell Surface to Infinity and Beyond. J. Bacteriol.198, 880–887. 10.1128/JB.00906-15
429
WeberB. S.LyP. M.IrwinJ. N.PukatzkiS.FeldmanM. F. (2015b). A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A.112, 9442–9447. 10.1073/pnas.1502966112
430
WeberB. S.MiyataS. T.IwashkiwJ. A.MortensenB. L.SkaarE. P.PukatzkiS.et al. (2013). Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS ONE8:e55142. 10.1371/journal.pone.0055142
431
WormserG. P.KeuschG. T.HeelR. C. (1982). Co-trimoxazole (trimethoprim-sulfamethoxazole): an updated review of its antibacterial activity and clinical efficacy. Drugs24, 459–518.
432
WrightM. S.HaftD. H.HarkinsD. M.PerezF.HujerK. M.BajaksouzianS.et al. (2014). New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. MBio5, e00963–e00913. 10.1128/mBio.00963-13
433
WuX.ChavezJ. D.SchweppeD. K.ZhengC.WeisbrodC. R.EngJ. K.et al. (2016). In vivo protein interaction network analysis reveals porin-localized antibiotic inactivation in Acinetobacter baumannii strain AB5075. Nat. Commun.7:13414. 10.1038/ncomms13414
434
YamamotoM.NagaoM.MatsumuraY.MatsushimaA.ItoY.TakakuraS.et al. (2011). Interspecies dissemination of a novel class 1 integron carrying blaIMP−19 among Acinetobacter species in Japan. J. Antimicrob. Chemother.66, 2480–2483. 10.1093/jac/dkr336
435
YangH.WangM.YuJ.WeiH. (2015). Antibacterial activity of a novel peptide-modified lysin against Acinetobacter baumannii and Pseudomonas aeruginosa. Front. Microbiol.6:1471. 10.3389/fmicb.2015.01471
436
YangY. S.LeeY.TsengK. C.HuangW. C.ChuangM. F.KuoS. C.et al. (2016). In vivo and in vitro efficacy of minocycline-based combination therapy for minocycline-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother.60, 4047–4054. 10.1128/AAC.02994-15
437
YokoyamaY.MatsumotoK.IkawaK.WatanabeE.MorikawaN.TakedaY. (2015). Population pharmacokinetic-pharmacodynamic target attainment analysis of sulbactam in patients with impaired renal function: dosing considerations for Acinetobacter baumannii infections. J. Infect. Chemother.21, 284–289. 10.1016/j.jiac.2014.12.005
438
YumJ. H.YiK.LeeH.YongD.LeeK.KimJ. M.et al. (2002). Molecular characterization of metallo-β-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the blaVIM-2 gene cassettes. J. Antimicrob. Chemother. 49, 837–840. 10.1093/jac/dkf043
439
YuY. S.ZhouH.YangQ.ChenY. G.LiL. J. (2007). Widespread occurrence of aminoglycoside resistance due to ArmA methylase in imipenem-resistant Acinetobacter baumannii isolates in China. J. Antimicrob. Chemother.60, 454–455. 10.1093/jac/dkm208
440
ZavasckiA. P.GoldaniL. Z.LiJ.NationR. L. (2007). Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother.60, 1206–1215. 10.1093/jac/dkm357
441
ZhangX.YangT.CaoJ.SunJ.DaiW.ZhangL. (2016). Mucosal immunization with purified OmpA elicited protective immunity against infections caused by multidrug-resistant Acinetobacter baumannii. Microb. Pathog.96, 20–25. 10.1016/j.micpath.2016.04.019
442
ZhuJ.WangC.WuJ.JiangR.MiZ.HuangZ. (2009). A novel aminoglycoside-modifying enzyme gene aac(6′)-Ib in a pandrug-resistant Acinetobacter baumannii strain. J. Hosp. Infect.73, 184–185. 10.1016/j.jhin.2009.05.012
443
ZimblerD. L.ParkT. M.ArivettB. A.PenwellW. F.GreerS. M.WoodruffT. M.et al. (2012). Stress response and virulence functions of the Acinetobacter baumannii NfuA Fe-S scaffold protein. J. Bacteriol.194, 2884–2893. 10.1128/JB.00213-12
444
ZouedA.BrunetY. R.DurandE.AschtgenM. S.LoggerL.DouziB.et al. (2014). Architecture and assembly of the Type VI secretion system. Biochim. Biophys. Acta1843, 1664–1673. 10.1016/j.bbamcr.2014.03.018
Summary
Keywords
antimicrobial resistance, Acinetobacter baumannii, treatment option, resistance mechanism, virulence factor
Citation
Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha C-J, Jeong BC and Lee SH (2017) Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 7:55. doi: 10.3389/fcimb.2017.00055
Received
19 December 2016
Accepted
13 February 2017
Published
13 March 2017
Volume
7 - 2017
Edited by
Ghassan M. Matar, American University of Beirut, Lebanon
Reviewed by
Sang Sun Yoon, Yonsei University, South Korea; Ravi Jhaveri, University of North Carolina Hospitals, USA
Updates
Copyright
© 2017 Lee, Lee, Park, Park, Bae, Kim, Cha, Jeong and Lee.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang Hee Lee sangheelee@mju.ac.kr
†These authors have contributed equally to this work.
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.