- 1National Engineering Laboratory for Digital and Material Technology of Stomatology, Beijing Key Laboratory of Digital Stomatology, Department of Periodontology, National Clinical Research Center for Oral Diseases, Peking University School and Hospital of Stomatology, Beijing, China
- 2Laboratory of Environmental Microbiology, Department of Energy and Resources Engineering, College of Engineering, Peking University, Beijing, China
Periodontal microorganisms not only colonize subgingival pockets, but also are detected on various mucous membranes in patients with periodontitis. The object of this pilot study was, using the next-generation sequencing of 16S RNA gene, to characterize the microbiota in two oral habitats (buccal mucosas and subgingival pockets) in patients with different forms of periodontitis. Thirty-two buccal swab samples and 113 subgingival samples were obtained from eleven subjects with chronic periodontitis (ChP), twelve subjects with aggressive periodontitis (AgP), and nine periodontally healthy individuals (HP). Using Miseq Sequencing of 16S rRNA gene, we found that the subgingival and buccal mucosa microbiome of ChP and AgP patients both differed from HP. Meanwhile, Veillonella, Treponema, Filifactor, Fretibacterium, Peptostreptococcaceae_[XI][G-6], Peptostreptococcaceae_[XI][G-5], Bacteroidetes_[G-5], Bacteroidetes_[G-3], Peptostreptococcaceae_[XI][G-4], Peptostreptococcaceae_[XI][G-2] significantly increased both in buccal and subgingival plaque samples in periodontitis subjects (ChP and AgP) compared with HP. Moreover, the results based on the Unweighted UniFrac distance showed that buccal and subgingival plaque samples from the same individuals show higher community divergence than same habitats from different subject samples. This study demonstrated that the microbiome of buccal mucosa can be influenced by periodontitis. However, subgingival and buccal mucosa microbiome seem to be characterized by species-specific colonization patterns. This pilot study provides a glimpse at the changes of subgingival and buccal mucosa associated with periodontitis from a holistic view. Further studies should be taken to illuminate the interplay between these detected changes and periodontitis development.
Introduction
The bacteria colonizing the hard and soft tissues of the oral cavity profoundly influence oral health and disease. Microbiologic studies have identified over 700 species of microorganisms in oral cavity (Aas et al., 2005), including a large number of uncultivable species (~50%), more than 400 of which have been detected in the subgingival crevice/pocket (Paster et al., 2006), the remaining species have been identified from other oral habitats such as the tongue, oral mucous membranes, and carious lesions.
Periodontitis is a chronic inflammatory disease initiated by colonization of subgingival periodontal pathogens. It has been shown that periodontal microorganisms are not only restricted to subgingival pockets, but are also found on various mucous membranes in patients with periodontitis (Van Winkelhoff et al., 1986; Eger et al., 1996; Gohler et al., 2018). Several studies have shown that buccal mucosa surfaces harbor periodontal pathogens (Danser et al., 1996; Socransky et al., 1999), and have suggested that it may serve as reservoirs for infection or reinfection of the periodontal tissues and deserve therapeutic attention (Zambon et al., 1981; Quirynen et al., 2001).
Aggressive periodontitis (AgP) is recognized as a group of periodontal diseases with faster destruction of connective tissue attachment that are different from chronic periodontitis (ChP) (Lang et al., 1999). Decades of studies have tried to distinguish AgP from ChP, however, the notion that AgP has a different microbiological pathogenesis from ChP has still not been confirmed. Notably, several studies have found that sampling oral soft tissues was a reliable method for monitoring the presence of A. actinomycetemcomitans (Müller et al., 1995; Eger et al., 1996), which has been considered to be closely related to the development of periodontitis, especially the development of AgP.
Microbiota on buccal mucosa can be collected easily, non-invasively, and repetitively. Therefore, sampling the microbiota on oral mucosa seems to be a promising way to diagnose periodontal diseases. Some investigations have demonstrated that the buccal mucosa microbiota in periodontally healthy and periodontitis subjects differ, but almost were based on traditional identification of bacteria in cultivations or culture-independent molecular studies. Recently, the next-generation sequencing (NGS) of bacterial 16S rRNA gene makes it possible to broaden our knowledge of the complexity of the microbiome, which has the advantage of detecting the composition of microbial communities in unprecedented depth. Several studies using NGS were to assess the composition of the polymicrobial periodontal community (Liu et al., 2012; Abusleme et al., 2013; Szafranski et al., 2015; Schulz et al., 2019). However, there is scarce evidence on microbiota that colonize the buccal mucosa of different forms of periodontitis.
Thus, the primary objective of this study was, using NGS of the16S RNA gene, to characterize the microbiota in two oral habitats (buccal mucosas and subgingival pockets) in patients with ChP or AgP, and to compare them with samples of subjects with periodontal health (HP).
Materials and Methods
Study Population and Clinical Examination
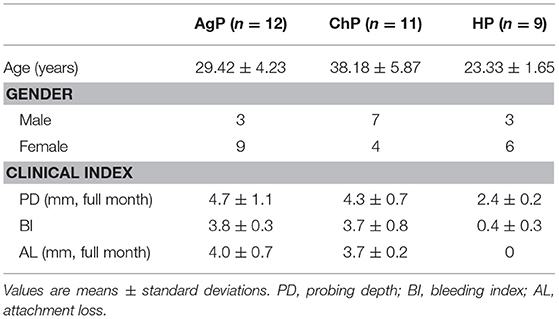
From January 2017 to July 2017, eleven patients with ChP and twelve patients with AgP were recruited from the Department of Periodontology, Peking University School and Hospital of Stomatology, and nine HP subjects were selected as controls. The clinical inclusion criteria for all participants were that: (1) they agreed to join the research and signed an informed consent, (2) they were free of systemic disease and not pregnant or lactating, (3) they had not received any kind of periodontal treatment in the past half year, nor had taken antibiotics within the past 3 months, (4) without removable or fixed denture in month, (5) without salivary glands disease, (6) without aggressive caries, untreated pulp disease or periapical problems, (7) they were non-smoker. The clinical inclusion criteria for periodontally healthy individuals and the diagnostic criteria for ChP and AgP have been previously described in detail (Shi et al., 2018).
Full-mouth clinical examinations (including PD, AL, and BOP at six sites per tooth) were carried out by one practitioner (WH). His reliability was calibrated as described by Shi (Shi et al., 2018). Full-mouth periapical radiographs were taken as diagnostic basis.
The study protocol was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the Peking University Health Science Center (approval number: PKUSSIRB-201631135). All enrolled individuals gave written informed consent.
Sample Collection
Buccal and subgingival plaque samples were collected 1 week after the full-mouth periodontal examination. All participants were requested to refrain from food for 8 h and oral hygiene (brushing or flossing the teeth) for 12 h before sampling. Samples from all subjects were collected and stored at −80°C for subsequent processing.
The buccal swab sample was collected before the subgingival plaque sample. Buccal swab sample was obtained from mucosa of both cheeks with a sterile cotton swab and placed in a separate sterile 1.5 ml Eppendorf microcentrifuge tube containing 1 ml TE buffer (50 mM Tris-HCl, 1 mM EDTA; pH 8.0). All subgingival plaque samples were collected from each participant from mesiobuccal sites of molars. For each patient with periodontitis, three to five samples were collected from the mesiobuccal site of molars with PD ≥ 4 mm. For each HP subject, two subgingival plaque samples were collected from the mesiobuccal site of molars with PD ≤ 3 mm without AL and BOP. The samples were obtained and processed was based on our previous research (Shi et al., 2018).
Extraction of Total Genomic DNA
Total DNA was extracted from the buccal and subgingival plaque using a QIAamp DNA Mini Kit (QIAGEN Sciences, USA) following the manufacturer's instructions, with an extra lysozyme treatment (3 mg/mL, 1.5 h) for bacterial cell lysis. The final concentration was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA) and monitored on 1% agarose gels. The results showed that the A260:A280 ratios were 1.8–2.1 and the DNA concentrations were all more than 20 ng/μL. Based on the quantity and the quality of the DNA extracted, samples were diluted to 1 ng/μL using sterile water and stored at −80°C until use.
PCR Amplification and Sequencing Analysis
PCR amplification of the 16S ribosomal RNA gene V4-V5 region was performed using primers 338F (5', ACTCCTACGGGAGGCAGCAG, 3') and 806R (5', GGACTACNNGGGTATCTAAT, 3'), where barcode is an eight-base sequence unique to each sample. The PCR cycling conditions were used: initial temperature of 95°C for 3 min, followed by 25 cycles consisting of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 45 s and a final extension step at 72°C for 10 min. PCR reactions were performed in triplicate 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA.PCR products were separated by 2% agarose gels electrophoresis, and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) according to the manufacturer's instructions and quantified with a QuantiFluor™ -ST Handheld Fluorometer with UV/Blue Channel (Promega, U.S.).
Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 250) on an Illumina MiSeq platform at Majorbio Bio-Pharm Technology (Shanghai, China). Raw sequencing data was filtered and trimmed by using QIIME (version 1.17), and classified into operational taxonomic units (OTUs) with 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/). The taxonomy of each 16S rRNA gene sequence was analyzed by Ribosomal Database Project Classifier tool (http://rdp.cme.msu.edu/) against the Human Oral Microbiome Database using a default confidence threshold of 0.7 (Dewhirst et al., 2010). The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP173111).
Bioinformatic Analysis, Statistical Analysis, and Visualization
Normality tests were conducted in each group of data. The mean clinical parameters were then compared via a paired Student's t-test (PD, AL) or Mann–Whitney U-test (BI). After rarefying the OTU table, two metrics were calculated for the evaluation of alpha diversity: abundance-based-coverage (ACE) estimates the species abundance; and the diversity of the sample microbiota was estimated by the Shannon index. The Mann–Whitney U test was used to compare significant differences of the alpha diversity indexes between the different groups (p < 0.05). For evaluating the similarity of microbial community structure among all samples, a principal coordinates analysis (PCoA) was performed on the OTU level. Analysis of similarities was calculated to compare the intra- and inter-group similarities based on the Unweighted UniFrac distance, a phylogenetic measures of beta diversity which was calculated by QIIME. The Mann–Whitney U-test was used to analyze the differences between the Unweighted UniFrac distances among groups and the results were visualized by constructing a scatter diagram and box plot. PCoA analysis, scatter diagram and box plot and ternary plots were performed using R 3.2.5. The taxonomy compositions and abundances of different samples were analyzed and visualized by GraphPad PRISM® software (version 4.0). To compare the taxa in buccal and subgingival plaque samples between HP, AgP, and ChP, the Mann–Whitney U-test was used. The Mann–Whitney test, Student's t-test were performed using SPSS 20.0.
Results
Increasing Diversity in Buccal and Subgingival Microbiota of Patients With Periodontitis
Thirty-two subjects were recruited in the study, including twenty-three patients with periodontitis: eleven of them were diagnosed as ChP, twelve of them were diagnosed as AgP, and other nine subjects were defined as HP. Consequently, 32 buccal swab samples (showed as HP/ChP/AgP_B), and 113 subgingival samples (showed as HP_Sub: 1-2 sample(s) per/subject, ChP/AgP_Sub: 3-5 samples per/patient, for a total of 16 samples belonged to HP_Sub, 46 samples belonged to ChP_Sub and 51 samples belonged to AgP_Sub) were collected. The demographic and clinical metrics of subjects on baseline are detailed in Table 1. As expected, BOP, PD, and AL values were greater in the group with periodontitis than for HP subjects. Sequences were clustered to 959 OTUs at a 97% similarity level. A total of 12 phyla, 31 classes, 56 orders, 103 families, and 195 genera were detected (Table 2).
In the HP_B, ChP_B, and AgP_B groups, the five most abundant of the 12 phyla were Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Fusobacteria, which represented more than 95% of the total sequences. In samples of subgingival plaque, HP_Sub, ChP_Sub, and AgP_Sub, the aforementioned five phyla also constituted the majority of the sequences, but the abundance of TM7, Spirochaetes, and Synergistetes increased, especially in ChP_Sub and AgP_Sub (Figure 1).
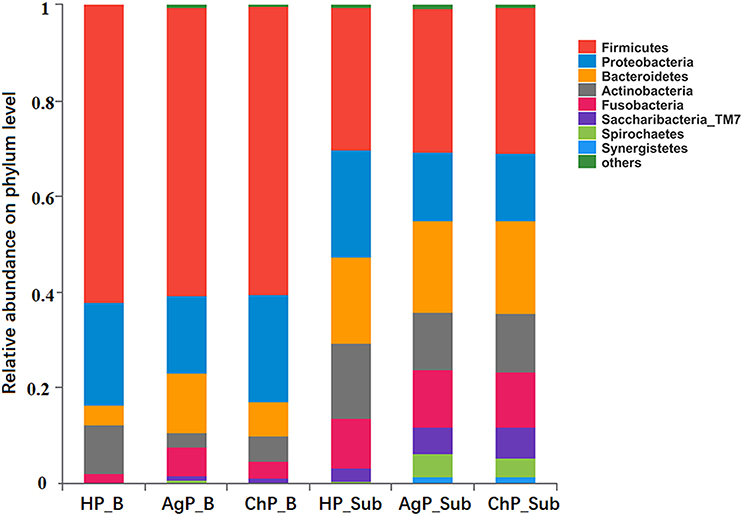
Figure 1. Relative abundance of bacterial composition on phylum level of buccal (shown as B) and subgingival plaque (shown as Sub) samples in health group (HP), chronic periodontitis group (ChP), and aggressive periodontitis group (AgP).
At the genus level, among the 195 genera detected, the five most abundant in buccal samples were Streptococcus, Haemophilus, Gemella, Neisseria, and Veillonella. These constituted more than 60% of the total sequences. In subgingival plaque samples, Streptococcus, Prevotella, Neisseria, Fusobacterium, Actinomyces, Capnocytophaga, Veillonella, and Leptotrichia represented more than 50% of the total sequences (Figure 2).
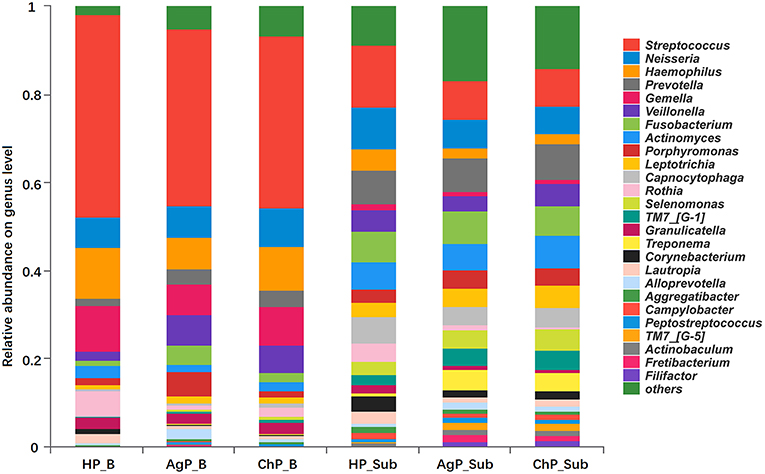
Figure 2. Relative abundance of bacterial composition on genus level of buccal (shown as B) and subgingival plaque (shown as Sub) samples in health group (HP), chronic periodontitis group (ChP), and aggressive periodontitis group (AgP).
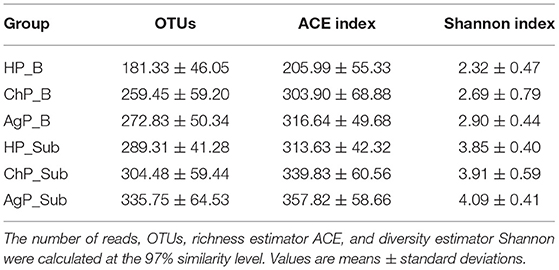
The richness of the total amount of bacteria was estimated by ACE index. The diversity of the microbiota was estimated by Shannon index (Table 3). The ACE index in buccal microbiota of periodontitis patients compared to that of HP_B samples was significantly increased. The Shannon index was also higher in ChP_B and AgP_B, although there were no statistically significant differences among HP_B and ChP_B (Figure 3). As for HP_Sub, ChP_Sub, and AgP_Sub, ACE index and Shannon index had identical trend with the buccal samples. The results showed that the alpha-diversity of subjects with aggressive periodontitis was greater than that in healthy individuals, both in buccal and subgingival plaque samples.
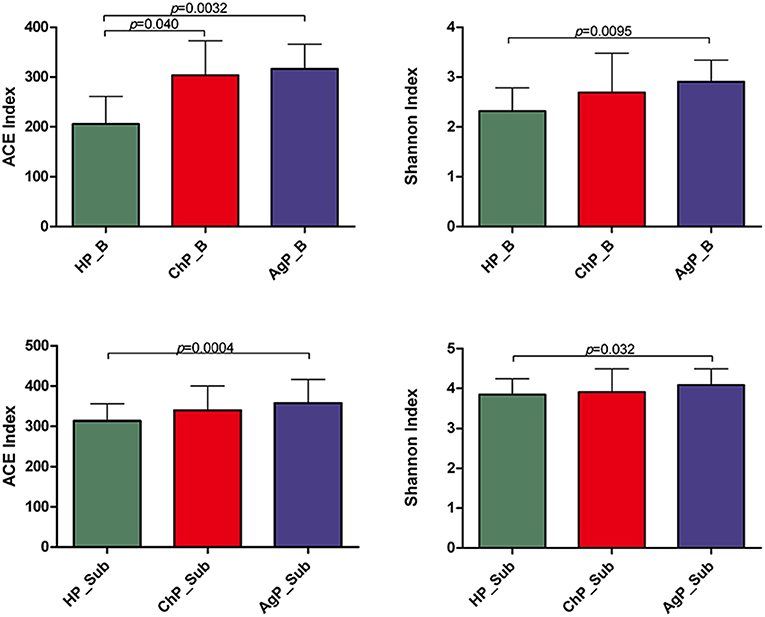
Figure 3. Bacterial community alpha-diversity as calculated by ACE and Shannon index of buccal (shown as B) and subgingival plaque (shown as Sub) samples in health group (HP), chronic periodontitis group (ChP), and aggressive periodontitis group (AgP). The error bars indicate mean with standard error.
Venn diagram was used to show the number of different OTUs common/unique to the buccal and subgingival plaque samples in three groups. In the HP_B, ChP_B, and AgP_B groups, 319, 345, 337, 467 OTUs shared by samples HP_B/ChP_B/AgP_B, HP_B/ChP_B, HP_B/AgP_B, ChP_B/AgP_B, respectively. Measurements of unique OTUs in the HP_B, ChP_B, and AgP_B groups were 18, 81, and 75, respectively. As in the HP_Sub, ChP_Sub, and AgP_Sub groups, 514, 517, 530, 708 OTUs shared by samples HP_Sub/ChP_Sub/AgP_Sub, HP_Sub/ChP_Sub, HP_Sub/AgP_Sub, ChP_Sub/AgP_Sub, respectively. Measurements of unique OTUs in the HP_Sub, ChP_Sub and AgP_Sub groups were 7, 30, and 111, respectively (Figure 4).
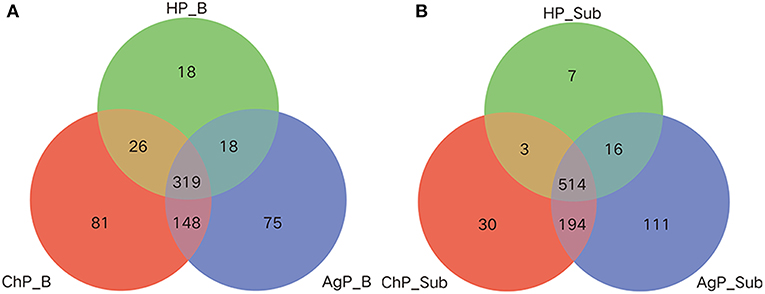
Figure 4. Venn diagram of the number of different OTUs common/unique to the buccal (shown as B) and subgingival plaque (shown as Sub) samples in health group (HP), chronic periodontitis group (ChP), and aggressive periodontitis group (AgP). The overlapping areas represent the number of OTUs shared by the counterpart samples. (A) A total of 685 kind of OTUs were detected in buccal samples of three groups. Three hundred nineteen kind of OTUs were shared by the HP_B, AgP_B, and ChP_B groups, while 18, 75, and 81 kind of OTUs were unique to the respective groups. (B) A total of 875 kind of OTUs were detected in the subgingival plaque samples of three groups. Five hundred fourteen kind of OTUs were shared by the HP_Sub, AgP_Sub, and ChP_Sub groups, and 7, 111, and 30 kind of OTUs were unique to the respective groups.
Comparison of Buccal and Subgingival Microbiota Between Health and Periodontitis
To analyze the distribution of microbiota among various groups, principal coordinates analysis (PCoA) were conducted based on the OTU abundances. The ChP_B and AgP_B samples clustered together and could not be distinguished from each other, while the HP_B samples appeared more dispersive (Figure 5). As for subgingival plaque samples, the trend was in agreement with buccal samples, however all subgingival plaque samples clustered tighter when compared with buccal samples.
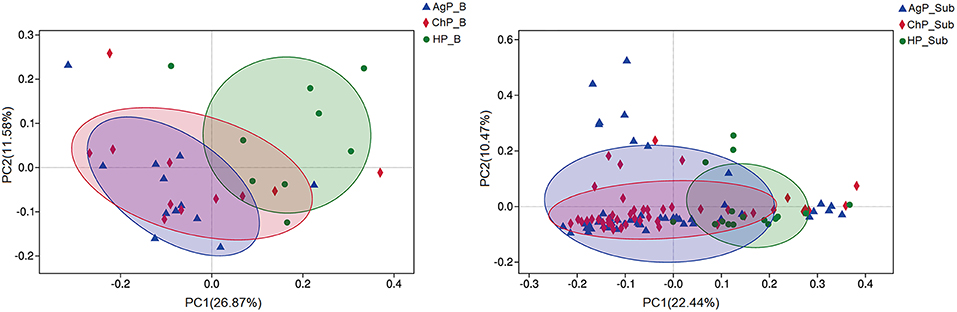
Figure 5. β-diversity of buccal (Left) and subgingival plaque (Right) samples in three groups. Principal coordinates analysis (PCoA) of unweighted UniFrac distance were performed based on the OTU abundances.
We analyzed our data in phyla level using ternary plots based on three main status: the intraoral environment of periodontal healthy status (HP), and the intraoral environment of periodontitis (ChP and AgP). According to the most abundant OTUs (Figure 6), HP_B harbored a high abundance of Actinobacteria, whereas ChP_B had a high abundance of Proteobacteria and AgP_B had a relatively high abundance of Firmicutes, Bacteroidetes and Synergistetes. Meanwhile, Fusobacteria and Saccharibacteria_TM7 were associated with both ChP_B and AgP_B bacterial communities. As for the subgingival plaque from periodontal pockets, the situation became different and complicated. The diversities increased and OTUs clustered more together between all groups than that in buccal samples, which was consistent with previous PCoA results.
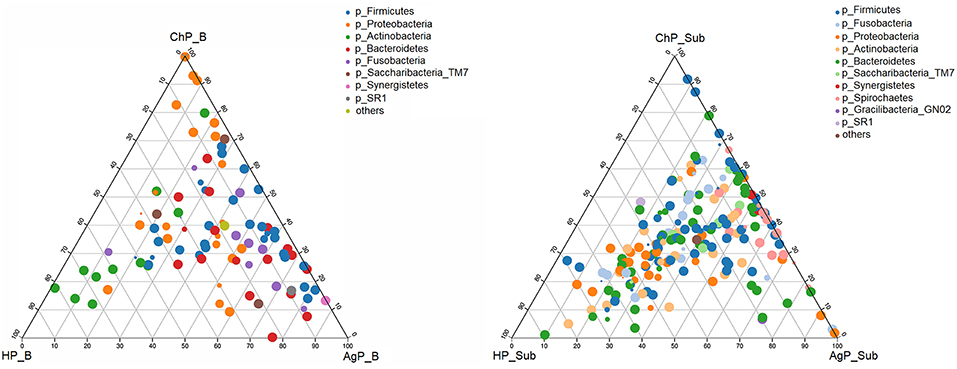
Figure 6. The ternary plots analysis of buccal (Left) and subgingival plaque (Right) samples on the OTU level which based on three main status: the intraoral environment of periodontal healthy status (HP), and the intraoral environment of periodontitis (ChP/AgP). The dots represented as OTU, the size of the dots represented the OTU abundances and the color represented as different phylum. The OTUs whose abundance <0.01 in all samples were classified into others.
The Unweighted UniFrac distances were calculated and compared between groups to investigate the similarity of all samples (Figure 7). The distances between buccal bacterial samples and subgingival plaque from the same individuals (Intra Sub-B) were maximal in HP, ChP, and AgP groups. As for subgingival plaque samples, the Unweighted UniFrac distances between samples from the same subjects (Intra Sub-Sub) were significantly lower than those between samples from different individuals in the same groups (Inter Sub-Sub).
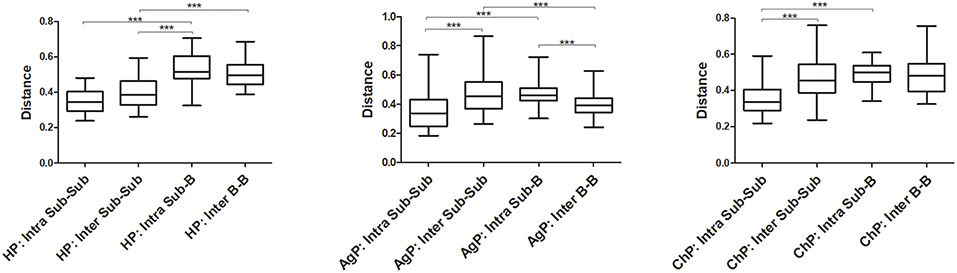
Figure 7. The Unweighted UniFrac distances were calculated and compared between groups to evaluate the similarity of all samples. The distances between subgingival plaque from the same (Intra Sub-Sub) or different individuals (Inter Sub-Sub), the distances between buccal bacterial samples and subgingival plaque from the same individuals (Intra Sub-B) and the distances between buccal bacterial samples from different individuals (Inter B-B) were presented in three groups. ***p < 0.001. The error bars indicate mean with standard error.
We compared microbiota between groups at various levels using the Mann-Whitney U-test. There were significant differences between all groups at several levels in both buccal and subgingival plaque sample (Supplementary Figures 1–3). The relative abundance of Veillonella, Treponema, Filifactor, Fretibacterium, Peptostreptococcaceae_[XI][G-6], Peptostreptococcaceae_[XI][G-5], Bacteroidetes_[G-5], Bacteroidetes_[G-3], Peptostreptococcaceae_[XI][G-4] and Peptostreptococcaceae_[XI][G-2] increased significantly in both buccal and subgingival plaque samples of periodontitis groups (ChP and AgP) compared to HP group (Figure 8). However, only Alloprevotella had significant difference between ChP and AgP both in two kind of samples.
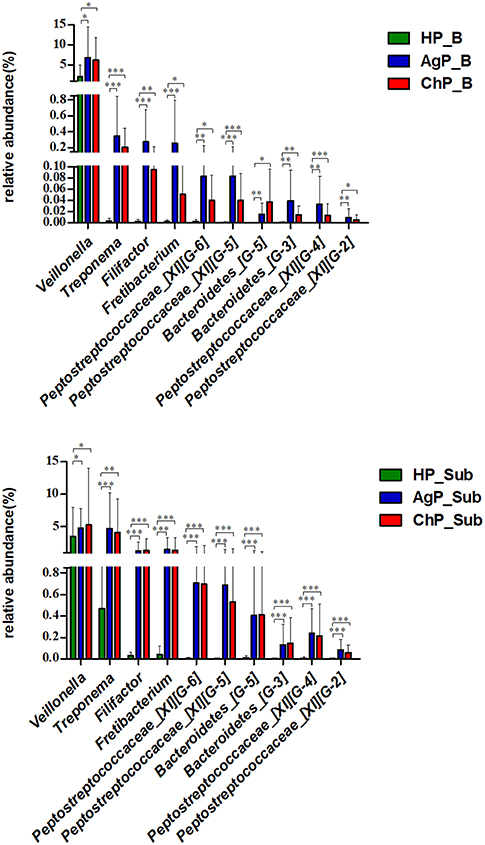
Figure 8. Comparisons of microbiota that presented significantly different contents in both buccal (up) and subgingival plaque (down) samples of periodontitis groups (AgP/ChP) shown the same trend of differences when compared with HP group at genus level. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Periodontitis are polymicrobial infections of the tooth supporting tissues. The primary etiology is plaque biofilm. Periodontitis-associated microorganisms colonize not only subgingival pockets but also other habitats like the buccal mucosa. The current study evaluated the composition of microbiomes on buccal mucosa and subgingival plaques in health and periodontitis using 16S rRNA gene next-generation sequencing analysis. In our current research, we found that higher levels of alpha diversity in buccal and subgingival microbiota of periodontitis patients. While the results also indicated that, along with the changes in periodontal tissue health, the relative abundance of several bacteria would change with them in both two groups of periodontitis and two types of samples. In addition, the results based on the Unweighted UniFrac distance analysis showed that buccal and subgingival plaque samples from the same individuals show higher community divergence than same habitats from different subject samples, which corroborated a notion that individual oral habitats are preferentially populated by different and somewhat unique sets of microbes.
In our research, we used 16S rRNA gene sequencing to evaluate and compare the different characteristics of subgingival plaque and buccal plaque between HP, ChP, and AgP patients. We found that periodontal destruction was associated with higher alpha diversity of microbiota from buccal mucosa samples and subgingival pockets. This may due to the formation and accumulation of dental plaque biofilm, which is the onset of periodontitis; meanwhile, along with the severity and the extent of periodontal tissue destruction, the deepening of the periodontal pockets can provide more spaces and more stable, ideal environment for dental plaque biofilm and bacteria might have the chance to transfer to other oral locations, in our case, buccal mucous.
Previous research reported bacterial community diversity was higher in chronic periodontitis patients than in healthy individuals (Griffen et al., 2012; Abusleme et al., 2013; Camelo-Castillo et al., 2015). It can be considered as the result of a nutritionally richer environment or a reduced immune competence. It is noteworthy that the trend of bacterial shifts accompanying periodontitis is on the opposite situation compared with other polymicrobial infection-related diseases, such as caries (Xiao et al., 2016) or children with black stain (Li et al., 2015) have been associated with a decrease in bacterial diversity. This kind of difference brings up another hint that the progress of periodontitis has relatively special aspects over others. Besides the microbiological reasons, the host defenses and the different environment where the disease occurs may also need to be considered.
Significant differences of oral microbiomes were detected between the health and periodontitis individuals at phyla and genus level in both buccal mucosa and subgingival plaque samples. Segata et al. (2012) reported that over 98% of the buccal mucosa bacteria consisted of 5 main phyla: Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Fusobacteriaex by exploring the microbiota of 18 body sites (including buccal mucosal surface) in over 200 individuals. In our current study, the bacterial composing of buccal samples from both health and periodontitis corresponded to the results of HMP. While HP_B harbored a high abundance of Actinobacteria, ChP_B had a high abundance of Proteobacteria and AgP_B had a relatively high abundance of Firmicutes, Bacteroidetes, and Synergistetes.
We found 10 genera in both buccal and subgingival plaque samples of periodontitis groups (ChP and AgP) significantly increased when compared with HP group. Among them, Peptostreptococcus, Treponema, and Filifactor had been reported in previous studies (Kumar et al., 2005; Li et al., 2014; Shi et al., 2015), which were relatively more abundant at the genus level in ChP, and Treponema were found at increased abundance in AgP compared with HP (Han et al., 2017). In these genera, some well-known destructive periodontal pathogens were included, such as Treponema denticola (Treponema) (Socransky et al., 1998). The cluster model of Socransky has been regarded as valid, and the main focus is on the “red” and “orange” complexes that are strongly associated with periodontitis (Socransky et al., 1998). Micromonas micro, a member of the genus Peptostreptococcus, is a member of the orange complex and is known as a potential pathogen associated with periodontitis. However, the role of some genera in the pathogenesis of periodontitis, such as Fretibacterium were not well-studied yet. Based on our current results, when periodontitis occurred in the cavity, the proportion of periodontal pathogens not only increased in subgingival plaques, but also in the samples of buccal plaques. The findings above gave us reason to speculate that some of the periodontal pathogens or opportunistic pathogen could co-aggregate on buccal surfaces or even had the chance to invade into the buccal tissue, as previously shown by other investigators that the A. actinomycetemcomitans and P. gingivalis both can invade buccal cells (Rudney et al., 2001; Leung et al., 2006).
The PCoA showed all subgingival plaque samples clustered tighter with each other compared with buccal samples. These results agree with those described by Gohler, each oral habitat appears to be preferentially populated by different and somewhat unique sets of microbes (Gohler et al., 2018).
Moreover, the results based on the Unweighted UniFrac distance showed that the distances between buccal swab samples and subgingival plaque from the same individuals (Intra Sub-B) were maximal in each group. The oral microbiota of hard and soft tissues appeared to be characterized by species-specific colonization patterns, which is in accordance with previous study by Mager et al. They reported differences in the bacterial profiles of 40 oral cultivable species on various oral habitats in healthy individuals and found that the profiles of soft tissues were more similar to each other when compared to those from subgingival plaques (Mager et al., 2003). In our study, we presented evidence that fine-scale biogeography variation within the human oral cavity is larger than inter-subject variability in the structure of either subgingival or mucosal communities.
To the best of our knowledge, this is the first study using NGS of the16S RNA gene to characterize the microbiota in two oral habitats (buccal mucosas and subgingival pockets) in patients with ChP or AgP, and to compare them with samples of subjects with periodontal health (HP). We found that buccal and subgingival microbial communities in healthy samples and periodontitis largely differed, with higher abundance in periodontitis. Some genera had significant difference between health and periodontitis both in buccal and subgingival plaque samples. While buccal mucosa and subgingival pockets to be characterized by species-specific colonization patterns.
Conclusions
In our study, we found that the subgingival and buccal mucosa microbiome in health and periodontitis largely differed, with higher bacterial abundance in periodontitis. In addition, we found 10 genera in both buccal and subgingival plaque samples of periodontitis groups (ChP and AgP) significantly increased when compared with HP group. Moreover, the results based on the Unweighted UniFrac distance showed that buccal and subgingival plaque samples from the same individuals show higher community divergence than same habitats from different subject samples, which corroborated a notion that individual oral habitats are characterized by species-specific colonization patterns. Within the limitation of the relatively small sample size, this pilot study provides a glimpse at the changes in the microbiota of subgingival and buccal mucosa associated with periodontitis from a holistic view.
Author Contributions
WH and XW: conception and design. YW, MS, MZ, and WH: recruitment of patients and collection of oral samples. MS, YW, CW, and YN: analysis and interpretation of data. YW and MS: manuscript preparation. YW, MS, MZ, WH, and XW: manuscript revisions.
Funding
This work was supported by National Natural Science Foundation of China (No.81672077) and Peking University Clinical Scientist Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00053/full#supplementary-material
References
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., and Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005
Abusleme, L., Dupuy, A. K., Dutzan, N., Silva, N., Burleson, J. A., Strausbaugh, L. D., et al. (2013).The subgingival microbiome in health and periodontitis and itsrelationship with community biomass and inflammation. ISME J. 7, 1016–1025. doi: 10.1038/ismej.2012.174
Camelo-Castillo, A. J., Mira, A., Pico, A., Nibali, L., Henderson, B., Donos, N., et al. (2015). Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front. Microbiol. 6:119. doi: 10.3389/fmicb.2015.00119
Danser, M. M., Timmerman, M. F., van Winkelhoff, A. J., and van der Velden, U. (1996). The effect of periodontal treatment on periodontal bacteria on the oral mucous membranes. J Periodontol. 67, 478–485. doi: 10.1902/jop.1996.67.5.478
Dewhirst, F. E., Chen, T., Izard, J., Pasteur, B. J., Tanner, A. R. C., Yu, W. H., et al. (2010). The human oral microbiome. J. Bacteriol. 192, 5002–5017. doi: 10.1128/JB.00542-10
Eger, T., Zoller, L., Muller, H. P., Hoffmann, S., and Lobinsky, D. (1996). Potential diagnostic value of sampling oral mucosal surfaces for Actinobacillus actinomycetemcomitans in young adults. Eur. J. Oral Sci. 104, 112–117. doi: 10.1111/j.1600-0722.1996.tb00054.x
Gohler, A., Samietz, S., Schmidt, C. O., Kocher, T., Steinmetz, I., and Holtfreter, B. (2018). Comparison of oral microbe quantities from tongue samples and subgingival pockets. Int. J. Dent. 26:2048390. doi: 10.1155/2018/2048390
Griffen, A. L., Beall, C. J., Campbell, J. H., Firestone, N. D., Kumar, P. S., Yang, Z. K., et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185. doi: 10.1038/ismej.2011.191
Han, J., Wang, P., and Ge, S. (2017). The microbial community shifts of subgingival plaque in patients with generalized aggressive periodontitis following non-surgical periodontal therapy: a pilot study. Oncotarget 8:10609. doi: 10.18632/oncotarget.12532
Kumar, P. S., Griffen, A. L., Moeschberger, M. L., and Leys, E. J. (2005). Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43, 3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005
Lang, N., Bartold, P. M., Cullinan, M., Jeffcoat, M., Mombelli, A., Murakami, S., et al. (1999). Consensus report: aggressive periodontitis. Ann. Periodontol. 4:53. doi: 10.1902/annals.1999.4.1.53
Leung, N. M., Chen, R., and Rudney, J. D. (2006). Oral bacteria in plaque and invading buccal cells of young orthodontic patients. Am. J. Orthod. Dentofacial Orthop. 130, 698.e11–18. doi: 10.1016/j.ajodo.2006.05.028
Li, Y., He, J., He, Z., Zhou, Y., Yuan, M., Xu, X., et al. (2014). Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 8, 1879–1891. doi: 10.1038/ismej.2014.28
Li, Y., Zhang, Q., Zhang, F., Liu, R., Liu, H., and Chen, F. (2015). Analysis of the Microbiota of Black Stain in the Primary Dentition. PLoS ONE.10:e0137030. doi: 10.1371/journal.pone.0137030
Liu, B., Faller, L. L., Klitgord, N., Mazumdar, V., Ghodsi, M., Sommer, D. D., et al. (2012). Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS ONE. 7:e37919. doi: 10.1371/journal.pone.0037919
Mager, D. L., Ximenez-Fyvie, L. A., Haffajee, A. D., and Socransky, S. S. (2003). Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30, 644–654. doi: 10.1034/j.1600-051X.2003.00376.x
Müller, H. -P., Eickholz, P., Heinecke, A., Pohl, S, Muller, R. F., and Lange, D. E. (1995). Simultaneous isolation of Actinobacillus actinomycetemcomitans from subgingival and extracrevicular locations of the mouth. J. Clin. Periodontol. 22, 413–419. doi: 10.1111/j.1600-051X.1995.tb00169.x
Paster, B. J., Olsen, I., Aas, J. A., and Dewhirst, F. E. (2006). The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 42, 80–87. doi: 10.1111/j.1600-0757.2006.00174.x
Quirynen, M., De Soete, M., Dierickx, K., and van Steenberghe, D. (2001). The intra-oral translocation of periodontopathogens jeopardises the outcome of periodontal therapy. A review of the literature. J. Clin. Periodontol. 28, 499–507. doi: 10.1034/j.1600-051x.2001.028006499.x
Rudney, J. D., Chen, R., and Sedgewick, G. J. (2001). Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69, 2700–2707. doi: 10.1128/IAI.69.4.2700-2707.2001
Schulz, S., Porsch, M., Grosse, I., Hoffmann, K., Schaller, H. G., and Reichert, S. (2019). Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch. Oral Biol. 99, 169–176. doi: 10.1016/j.archoralbio.2019.01.015
Segata, N., Haake, S. K., Mannon, P., Lemon, K. P., Waldron, L., Gevers, D., et al. (2012). Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13:R42. doi: 10.1186/gb-2012-13-6-r42
Shi, B., Chang, M., Martin, J., Mitreva, M., Lux, R., Klokkevold, P., et al. (2015). Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. MBio 6, e01926–e01914. doi: 10.1128/mBio.01926-14
Shi, M., Wei, Y., Hu, W., Nie, Y., Wu, X., and Lu, R. (2018). The subgingival microbiome of periodontal pockets with different probing depths in chronic and aggressive periodontitis: a pilot study. Front. Cell. Infect. Microbiol. 8:124. doi: 10.3389/fcimb.2018.00124
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., and Kent, R. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x
Socransky, S. S., Haffajee, A. D., Ximenez-Fyvie, L. A., Feres, M., and Mager, D. (1999). Ecological considerations in the treatment of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis periodontal infections. Periodontol. 2000 20, 341–362. doi: 10.1111/j.1600-0757.1999.tb00165.x
Szafranski, S. P., Wos-Oxley, M. L., Vilchez-Vargas, R., Jauregui, R., Plumeier, I., Klawonn, F., et al. (2015). High-resolution taxonomic profiling of the subgingival microbiome for biomarker discovery and periodontitis diagnosis. Appl. Environ. Microbiol. 81, 1047–1058. doi: 10.1128/AEM.03534-14
Van Winkelhoff, A. J., Van der Veiden, U., Winkel, E. G., and de Graaff, J. (1986). Black-pigmented bacteroides and motile organisms on oral mucosal surfaces in individuals with and without periodontal breakdown. J. Periodont. Res. 21, 434–439. doi: 10.1111/j.1600-0765.1986.tb01477.x
Xiao, C., Ran, S., Huang, Z., and Liang, J. (2016). Bacterial diversity and community structure of supragingival plaques in adults with dental health or caries revealed by 16S Pyrosequencing. Front. Microbiol. 7:1145. doi: 10.3389/fmicb.2016.01145
Keywords: 16S sequencing, subgingival microbiome, buccal mucosa microbiome, chronic periodontitis, aggressive periodontitis
Citation: Wei Y, Shi M, Zhen M, Wang C, Hu W, Nie Y and Wu X (2019) Comparison of Subgingival and Buccal Mucosa Microbiome in Chronic and Aggressive Periodontitis: A Pilot Study. Front. Cell. Infect. Microbiol. 9:53. doi: 10.3389/fcimb.2019.00053
Received: 15 January 2019; Accepted: 19 February 2019;
Published: 11 March 2019.
Edited by:
Georgios N. Belibasakis, Karolinska Institutet (KI), SwedenReviewed by:
Ashu Sharma, University at Buffalo, United StatesMaribasappa Karched, Kuwait University, Kuwait
Copyright © 2019 Wei, Shi, Zhen, Wang, Hu, Nie and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjie Hu, aHV3ZW5qaWVAcGt1c3MuYmptdS5lZHUuY24=
Yong Nie, bmlleW9uZ0Bwa3UuZWR1LmNu
†These authors have contributed equally to this work
 Yiping Wei
Yiping Wei Meng Shi
Meng Shi Min Zhen
Min Zhen Cui Wang1
Cui Wang1 Yong Nie
Yong Nie Xiaolei Wu
Xiaolei Wu