- 1Oral Microbiology Research Laboratory, Faculty of Dentistry, Kuwait University, Kuwait City, Kuwait
- 2Functional Proteomics and Metabolomics Unit, Dasman Diabetes Institute, Kuwait City, Kuwait
Despite reports on the occurrence of Granulicatella adiacens in infective endocarditis, few mechanistic studies on its virulence characteristics or pathogenicity are available. Proteins secreted by this species may act as determinants of host-microbe interaction and play a role in virulence. Our aim in this study was to investigate and functionally characterize the secretome of G. adiacens. Proteins in the secretome preparation were digested by trypsin and applied to nanoLC-ESI-MS/MS. By using a combined mass spectrometry and bioinformatics approach, we identified 101 proteins. Bioinformatics tools predicting subcellular localization revealed that 18 of the secreted proteins possessed signal sequence. More than 20% of the secretome proteins were putative virulence proteins including serine protease, superoxide dismutase, aminopeptidase, molecular chaperone DnaK, and thioredoxin. Ribosomal proteins, molecular chaperones, and glycolytic enzymes, together known as “moonlighting proteins,” comprised fifth of the secretome proteins. By Gene Ontology analysis, more than 60 proteins of the secretome were grouped in biological processes or molecular functions. KEGG pathway analysis disclosed that the secretome consisted of enzymes involved in biosynthesis of antibiotics. Cytokine profiling revealed that secreted proteins stimulated key cytokines, such as IL-1β, MCP-1, TNF-α, and RANTES from human PBMCs. In summary, the results from the current investigation of the G. adiacens secretome provide a basis for understanding possible pathogenic mechanisms of G. adiacens.
Introduction
Granulicatella adiacens is part of the normal microbiota in the oral cavity, urogenital, and intestinal tract (Ruoff, 1991). It can occasionally cause serious infections such as infective endocarditis (Lin and Hsu, 2007), but may also participate in periodontitis (Belstrøm et al., 2014), caries (Kanasi et al., 2010), and endodontic infections (Siqueira and Rôças, 2006). It is a small Gram-positive, non-motile, non-spore-forming, oxidase-, and catalase-negative, facultatively anaerobic coccus. As previously members of Nutritionally Variant Streptococci and later of genus Abiotrophia, the current taxonomy separates three Abiotrophia species under a novel genus, Granulicatella (Collins and Lawson, 2000). The nutritional requirements of G. adiacens are complex and pyridoxal or L-cysteine in the growth medium is essential for normal growth. Absence of these supplements results in elongated cell morphology (Karched et al., 2015) and an altered protein expression (Karched et al. unpublished data).
Protein secretion helps bacteria in their normal growth and physiology, e.g., nutrient acquisition, but it can also function as a virulence mechanism in host colonization or by modulating host immune responses (Finlay and Falkow, 1997; Lee and Schneewind, 2001). Bacteria have devised dedicated secretory systems (Natale et al., 2008; Green and Mecsas, 2016) for protein secretion; Gram-positive species secrete mainly via general secretory system (sec-dependent) (Schneewind and Missiakas, 2012) or twin-arginine transport (Tat) pathway (Goosens et al., 2014) while Gram-negative bacteria use more complex secretory systems (Type I to Type VIII) (Costa et al., 2015). Little knowledge is available of the protein secretion of Granulicatella species. Recently, a close phylogenetic relative of G. adiacens, Granulicatella elegans, was shown to secrete arginine deiminase, which through citrullination inhibits proliferation of human peripheral blood mononuclear cells in vitro (Kanamoto et al., 2007), but also inhibits biofilm surface attachment of certain dental plaque bacteria (Abdullah et al., 2013) and may associate with the pathogenesis of periodontitis and certain systemic diseases (Olsen et al., 2018). In this study, we investigated the components of the secretome of G. adiacens. We also obtained preliminary information on the immunoinflammatory response induced by the secretome of G. adiacens.
Methods
Bacteria and Culture Conditions
Reference bacterial strain G. adiacens CCUG 27809 was cultured on chocolate blood agar (CBA) with 0.001% pyridoxal hydrochloride for 2 days at 37°C and in 5% CO2 in air as we previously reported (Christensen and Facklam, 2001; Karched et al., 2015).
Extracellular Protein Release
A loopful (1 μl) of bacterial colonies harvested from CBA plates were inoculated into 5 ml brucella broth (supplemented with 0.001% pyridoxal hydrochloride) and incubated in 5% CO2 in air at 37°C. No-bacteria control was incubated in parallel. After 24 h (in exponential growth phase) broth cultures were centrifuged at 5,000 × g for 5 min. Viability of bacteria was checked by culturing a 100-μl aliquot on CBA. Supernatants containing extracellularly released proteins were separated and filtered through 0.2 μm sterile filter to remove residual bacterial cells. All experiments were performed in duplicates and were repeated three times.
Preparation of Secretome
The replicates of extracellular proteins released in supernatant broth samples were extracted by tri-chloroacetic acid (TCA) precipitation method as described previously (Deatherage Kaiser et al., 2015) with modifications. One volume of TCA stock (100% w/v) was mixed with four volumes of supernatant culture broth and incubated for 30 min at −20°C. Extracted proteins in broth were recovered in pellet form by centrifugation at 14,000 × g for 20 min at 4°C on Beckman J2-M1 High-Speed centrifuge. The pellet was washed twice with 0.5 ml cold acetone to remove traces of acid followed by complete air-drying in a fume hood. Desalting of the samples was achieved by diluting protein samples to 0.5 ml volume (each time) in lysis buffer and washing three times by ultrafiltration through 3K Ultra-0.5 centrifugal filter devices (Amicon) at 14,000 × g for 15 min at 4°C. Flow through were discarded and concentrates in the columns were finally eluted from columns in upside down position on collection tubes by centrifugation at 1,000 × g for 2 min at 4°C. A “no-bacteria” broth control which was incubated in parallel was used as negative control.
Bacterial Cell Lysate/Whole Cell Protein Preparation
Harvested colonies from CBA plates were washed once in sterile PBS, followed by centrifugation at 5,000 × g for 5 min. The pellet recovered was resuspended in lysis buffer containing 1 mg/ml lysozyme and 1 mM phenyl methyl sulfonyl fluoride (PMSF) and incubated for 4 h at 4–8°C. The samples were then sonicated in Omni Ruptor at a pulse rate 40 for 8 times (1 min sonication with 1 min interval on ice). Cell lysates after sonication were centrifuged at 10,000 × g for 10 min at 4°C. Whole cell lysate of G. adiacens was prepared to use as a control in western blot analysis along with extracellular protein extract of the same.
Determination of Protein Concentration
Protein concentrations in extracellular protein extract and cell lysate were estimated by Quick StartTM Bradford protein microplate standard assay (Bio-Rad) as per manufacturer instructions.
SDS-PAGE
For SDS-PAGE analysis, protein samples were mixed with 5× Laemmli sample buffer (125 mM tris, pH 6.8; 6% glycerol, 2% SDS; 5% beta-mercapthoethanol; 0.025% bromophenol blue) followed by boiling at 95°C for 5 min. After cooling at room temperature, samples were loaded on a 15% SDS-PAGE gel [4% stacking gel (4% acrylamide; 68 mM tris, pH 6.8; 0.2% SDS), 15% separating gel; 375 mM tris, pH 8.8; 0.1% SDS]. Electrophoresis was run at 150 V for 75 min (Mini-protein II Dual Slab Cell, Bio Rad). After the run, protein bands were visualized using coommassie blue.
Western Blot Analysis
To rule out the possibility of cell lysis of G. adiacens cells and the release of cellular proteins in secretome preparations, western blot analysis of whole cell lysate and secretome preparation was performed. Proteins were transferred from the gel onto a PVDF membrane using Trans-Blot® Turbo™ transfer system (Bio-Rad). Membrane was blocked with 5% skimmed milk overnight at 4°C. An antibody against the cytoplasmic marker protein, Ftsz (Filamenting temperature sensitive mutant z) (Agrisera AB, Sweden) was used as a primary antibody at 1:1,000 dilution and incubated on a shaker for 1 h at room temperature. The membrane was then incubated as above with a peroxidase conjugated goat antirabbit IgG F (ab') 2 s Ab secondary antibody (1:5,000). The membrane was washed between each antibody treatment with tris-buffer saline containing Tween-20 (TBST). The membrane was finally treated with SuperSignalTM West Pico chemiluminescence substrate (Pierce) and images were acquired in G:Box Imaging System (Syngene).
nanoLC-ESI-MS/MS
Protein identification using nanoLC-ESI-MS/MS was performed by Proteome Factory (Proteome Factory AG, Berlin, Germany). The LC-MS system consisted of an Agilent 1100 nanoHPLC system (Agilent, Waldbronn, Germany), PicoTip electrospray emitter (New Objective, Woburn, MA) and an LTQ-FT Ultra mass spectrometer (ThermoFisher Scientific, Bremen, Germany). Replicate samples from secretome preparations were pooled and 400 ng protein were reduced, alkylated and digested by trypsin (Promega, Mannheim, Germany) and applied to nanoLC-ESI-MS/MS. Peptides were trapped and desalted on the enrichment column (Zorbax SB C18, 0.3 × 5 mm, Agilent) for 5 min using 1% acetonitrile/0.5% formic acid as eluent, then peptides were separated on a Zorbax 300 SB C18, 75 μm × 150 mm column (Agilent) using an acetonitrile/0.1% formic acid gradient from 5 to 40% acetonitrile within 120 min. MS spectra were automatically recorded by the mass spectrometer according to manufacturer's instrument settings for nanoLC-ESI-MSMS analyses. Proteins were identified by submitting all MS/MS spectra to the Mascot search engine (Matrix Science, London, England) and non-redundant protein database; NCBI-nr (National Center for Biotechnology Information, Bethesda, USA, version 20151202) and taxonomy Bacteria including 54,860,673 sequences. Ion charge in search parameters for ions from ESI-MS/MS data acquisition were set to “1+, 2+, or 3+.” Search parameters were as following: Fixed modifications: Carbamidomethyl (C); variable modifications: Deamidated (NQ), Oxidation (M); Peptide Mass Tolerance: ± 5 ppm; Fragment Mass Tolerance: ± 0.6 Da; Missed Cleavages: 2. Only peptides matched with a score of 20 or above were accepted and included in protein identification.
Bioinformatics Analyses of the Secreted Proteins
The signal peptides in the secreted proteins were determined by using SignalP (http://www.cbs.dtu.dk/services/SignalP/) (Bendtsen et al., 2004) Phobius (http://phobius.sbc.su.se/) (Kall et al., 2007), and PSORTb (http://www.psort.org/psortb/) (Yu et al., 2010) and a most-votes approach was used to interpret the results. To identify lipoproteins, LipoP (http://www.cbs.dtu.dk/services/LipoP/) and PRED-LIPO (http://bioinformatics.biol.uoa.gr/PRED-LIPO/input.jsp) (Bagos et al., 2008) were used to search for lipoboxes. TatP (http://www.cbs.dtu.dk/services/TatP/) (Bendtsen et al., 2005b) and TatFind (http://signalfind.org/tatfind.html) (Rose et al., 2002) were used to predict proteins secreted via Twin-arginine translocation (Tat) pathway. To identify proteins secreted by non-classical secretory system, SecretomeP2.0 was used (http://www.cbs.dtu.dk/services/SecretomeP/) (Bendtsen et al., 2005a). Proteins that were also positive for signal peptide were disregarded. Transmembrane alpha helices were predicted combining the tools TMHMM v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001), SVMtm (http://ccb.imb.uq.edu.au/svmtm/) (Yuan et al., 2004), and SOSUI (http://harrier.nagahama-i-bio.ac.jp/sosui/sosuiG/sosuigsubmit.html) (Hirokawa et al., 1998). A theoretical 2-dimensional electrophoresis (2-DE) image of the secreted proteins was acquired using the software JVirGel vs. 2.0 (Hiller et al., 2003).
For functional annotation of the secreted proteins, Gene Ontology (GO) IDs and terms were assigned using the tool “WEGO” (http://wego.genomics.org.cn/) (Ye et al., 2018) and the program Blast2GO. Proteins with enzymatic functions, possibly involved in metabolic pathways, were predicted by KEGG Pathway Database (http://www.genome.jp/kegg/pathway.html). Putative virulence proteins were predicted by the software “VirulentPred” (http://203.92.44.117/virulent/) (Garg and Gupta, 2008), and by using Virulence Factor DataBase (VFDB, http://www.mgc.ac.cn/VFs/) (Chen et al., 2016). The secreted proteins were grouped into functional protein association networks using STRING (https://string-db.org/) (von Mering et al., 2005). MultitaskProtDB (http://wallace.uab.es/multitask) was used for identifying “moonlighting proteins” in the G. adiacens secretome (Hernández et al., 2014).
Isolation of Human Peripheral Blood Mononuclear Cells (PBMCs)
Ethical approval for blood collection from a healthy human volunteer was obtained from Health Science Center Ethical Committee, Kuwait University. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood of a systemically healthy human volunteer as described earlier (Fuss et al., 2009; Bhardwaj et al., 2018). Blood was collected by venipuncture into tubes containing heparin vacutainer (4 ml /tube). PBMCs were fractionated by Ficoll-Paque density gradient centrifugation method. Under careful aseptic conditions, the blood was carefully layered over the Ficoll-PaqueTMPlus (GE Healthcare) solution in the test tube. The tube was centrifuged at 3,400 rpm at room temperature for 10 min and the resulting buffy coat layer containing PBMCs was transferred to another clean tube. After washing twice in 5 ml of RPMI medium the tube was centrifuged at 2,000 rpm for 5 min to recover the cell pellet. The supernatant was discarded and the cell pellet was finally resuspended in 1 ml of RPMI medium (supplemented with 10% heat-inactivated fetal bovine serum and 2% of GibcoTM 100× antibiotic-antimycotic solution). Cells were enumerated using 10 μl of homogenous cell suspension in hemocytometer under ×400 magnification of the microscope.
PBMCs Treatment With Secretome
PBMCs were stimulated with secretome preparations for 24 h. One hundred microliter of the secretome preparation was added into each well-containing 0.5 ml of PBMCs (106 cells per ml). The plate was incubated for 24 h in 5% CO2 in air at 37°C. PBS, which was the medium for secretome, was used as a negative control.
Cytokine Profiling Using the Membrane Arrays
Cytokines produced by PBMCs on stimulation with G. adiacens secretome were detected using a human cytokine array kit (Proteome ProfilerTM Antibody Arrays R&D SystemsTM). Nitrocellulose membrane with 36 selected capture antibodies spotted in duplicate was used to determine the relative levels of cytokines. The array membrane was blocked with assay buffer for 1 h at room temperature to prevent non-specific binding. The secretome-stimulated PBMC sample (1.5 ml) was diluted in assay buffer with 15 μl of reconstituted human cytokine array detection antibody cocktail and incubated at room temperature for 1 h. Following three washes in wash buffer, the array was treated with streptavidin HRP for 30 min at room temperature on a rocking platform shaker. Washed array was finally incubated with chemiluminescence reagent for 10 min and images were acquired in Syngene G:Box Imaging System. The positive signals seen on the array were identified by comparing it with the transparency overlay template with the pairs of reference spots in three corners of each array. Pixel densities (signals) in each spot on the array were collected, mean spot pixel density was created and analyzed image analysis software provided with G:Box Imaging System. The experiments were run in duplicates and repeated three times.
Results
Analysis of the Secretome of G. adiacens
Secretome preparation (Figure 1A) from G. adiacens was analyzed by LC-MS/MS. Database search (NCBI-nr) revealed 101 proteins (Table S1). As depicted in a theoretical 2DE map of the secretome, the MW of the secreted proteins ranged between 3.7 and 148 kDa (Figure 1B). The secretome proteins formed two clusters with respect to predicted isoelectric point (pI) values. Majority of the proteins belonged to the cluster with the pI range of 4.0–5.5, while the other cluster was of the proteins between pI values 9.5 and 11. To exclude the possibility that the secretome preparation contained proteins that originated from bacterial cell lysis, we used in western blot analysis a cytoplasmic cell lysis marker, Fts-Z protein, which was detected only in the total protein preparation from G. adiacens but not in the secretome preparation (Figure 1C). Further, plating of an aliquot of the 24-h broth culture confirmed the viability of bacteria during the experiment. To determine subcellular localization of the 101 secreted proteins detected with LC-MS/MS, PsortB tool was used. We found that 63 proteins were predicted to be cytoplasmic (60%), 10 cytoplasmic membrane (9.6%), 6 extracellular (5.7%), 2 cell wall anchored (1.9%), whereas the localization of 23 proteins (22%) could not be predicted.
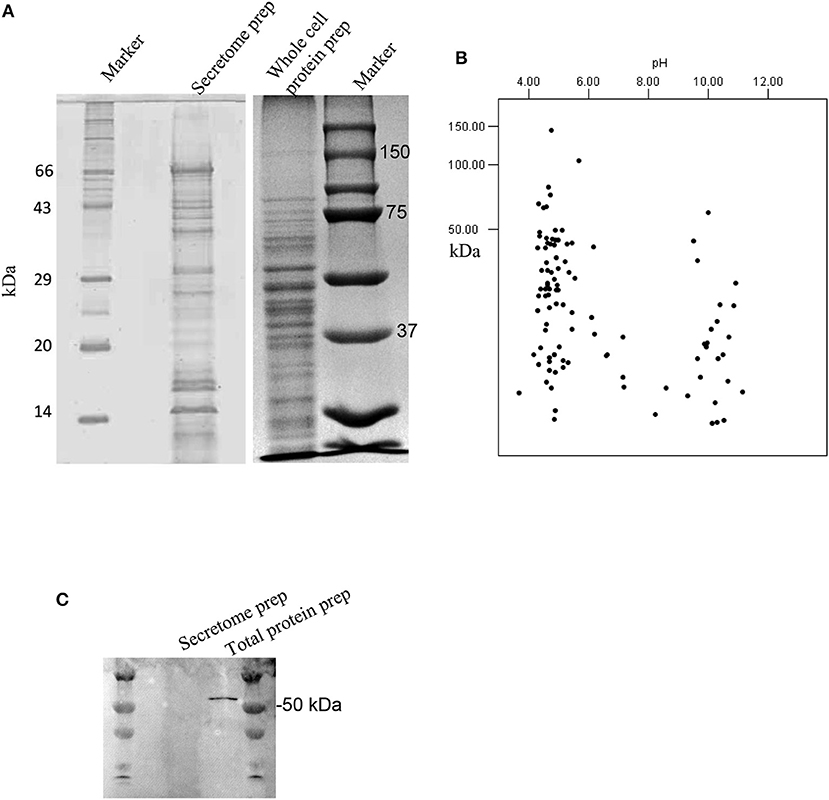
Figure 1. Analysis of the secretome of G. adiacens. (A) SDS-PAGE gel showing protein bands from secretome preparation. (B) Protein sequences from LC-MS analysis of the secretome were analyzed by an in silico 2DE tool. (C) Western blot analysis showing the absence of the cytoplasmic marker protein FtsZ in the secretome preparation.
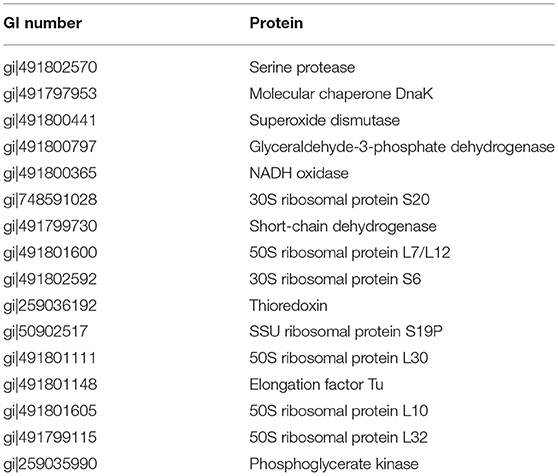
Protein sequences of the secretome of G. adiacens were analyzed for the route of their extracellular release by various bioinformatics tools as described in the methods section. As predicted by SignalP tool, 18 of the secreted proteins were predicted to possess signal sequence, suggesting the “Sec” pathway for their secretion. TatP prediction tool showed that 8 of the sequences contained TatP signal sequence. There were a total of 9 lipoproteins (Pred-Lipo, LipoP) in the secretome. Using the tool SecretomeP 2.0, 31 sequences were predicted to be secreted via non-classical secretion pathway. However, since 15 of them contained a signal sequence and hence concluded to be secreted via Sec pathway, those were discarded and only 16 proteins were finally considered to be secreted via non-classical pathway. Transmembrane alpha helices were found in 9 proteins. Two of the 9 protein sequences that were predicted to have at least 2 transmembrane domains were considered as putatively membrane attached and therefore not included in further analyses. Table 1 lists the 16 proteins from the secretome which were identified as “moonlighting proteins.”
Potential Virulence Proteins in G. adiacens Secretome
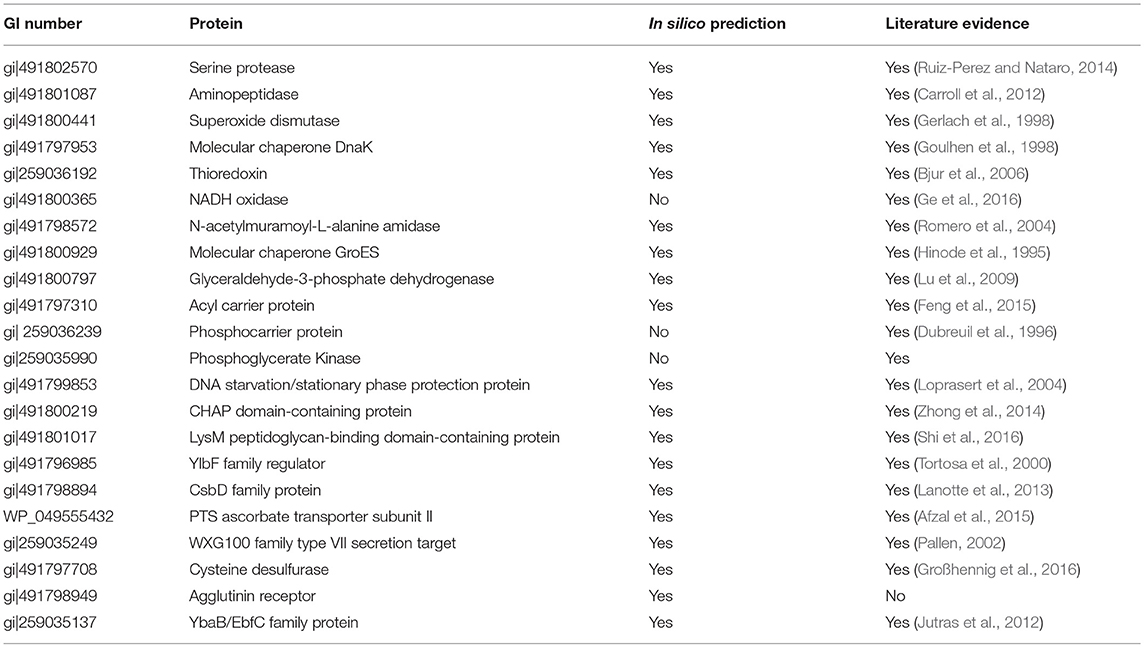
Virulence potential of the G. adiacens secretome was assessed by manually searching for their associations with virulence activities in other species, since little is known of the virulence factors of G. adiacens. Additionally, in silico prediction of virulence factors was performed using the online tools “VirulentPred” and “VFDB” (Virulence Factor DataBase). Table 2 shows the list of 22 proteins from the secretome that were deduced from in silico prediction and/or based on evidence from the literature. Thioredoxin, serine proteinase, aminopeptidase, molecular chaperones DnaK and GroES, Superoxide dismutase, N-acetylmuramoyl-L-alanine amidase, Glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, and acyl carrier protein are the major proteins with demonstrated virulence properties in other bacterial species.
Gene Ontology Analysis
Gene Ontology (GO) analysis of the amino acid FASTA sequences of the G. adiacens secretome was achieved by using the tools Blast2GO and WEGO. In the case of WEGO, the XML file from InterPro analysis was used (Figure 2). Overall, 73 of the 104 sequences were assigned with GO annotation. The secreted proteins were divided into 3 groups based on GO terms: 63 proteins in “biological process,” 31 proteins in the “cellular component” group, and 65 proteins in the group “molecular function.”
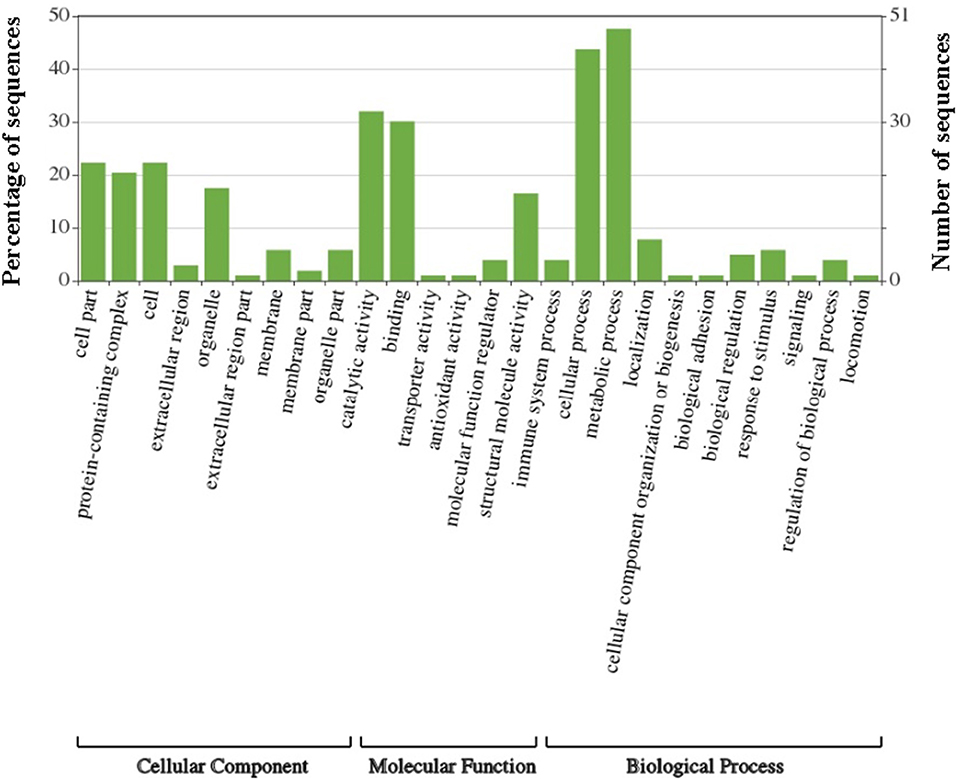
Figure 2. Gene Ontology analysis of G. adiacens secreted proteins. Gene ontology annotation was achieved using Blast2GO and an online software “WEGO.” Protein sequences were grouped into 3 categories based on their properties and functions.
KEGG Pathway Analysis
All protein sequences from the G. adiacens secretome were subjected to KEGG pathway annotation and analysis (Figure 3). Of all the pathways identified, 6 proteins were predicted to be involved in the biosynthesis of antibiotics, followed by 5 proteins that occurred in the glycolysis pathway. Fructose and mannose metabolism, and purine metabolism pathways contained 3 proteins each. One of the secreted proteins, transketolase, was predicted to be occurring in the biosynthesis of ansamycins, a family of bacterial secondary metabolites with antimicrobial activity.
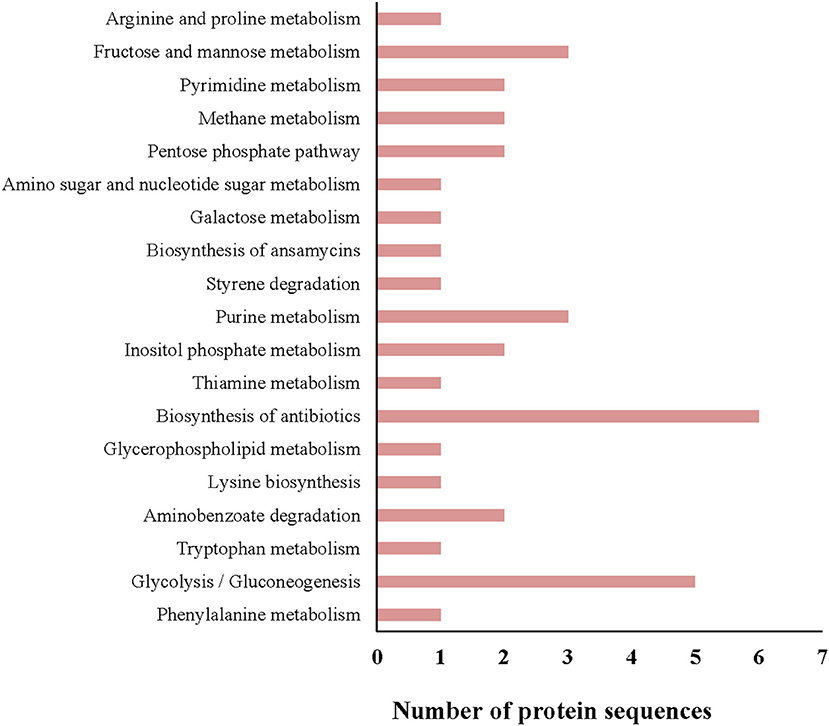
Figure 3. KEGG pathway analysis of the G. adiacens secretome. All protein sequences from the secretome were analyzed by KEGG pathway tool, which was included as a “plugin” tool within Blast2GO software.
Functional Protein Association Network Analysis
As seen in Figure 4, G. adiacens secretome proteins formed three major groups in the STRING network, i.e., sugar metabolism, ribosomal proteins and heat shock proteins/chaperones. Components of the sugar metabolism network were phosphoglycerokinase, enolase, triose phosphate isomerase, Fructose-1,6-bisphosphate aldolase, and phosphocarrier protein. Putative virulence-associated proteins super oxide dismutase, thioredoxin, molecular chaperones (DnaK, GroS, and GrpE) NADH oxidase and HtrA, a trypsin-like protease, formed another cluster. The ribosomal protein group consisted mainly of the secreted ribosomal proteins (Figure 4).
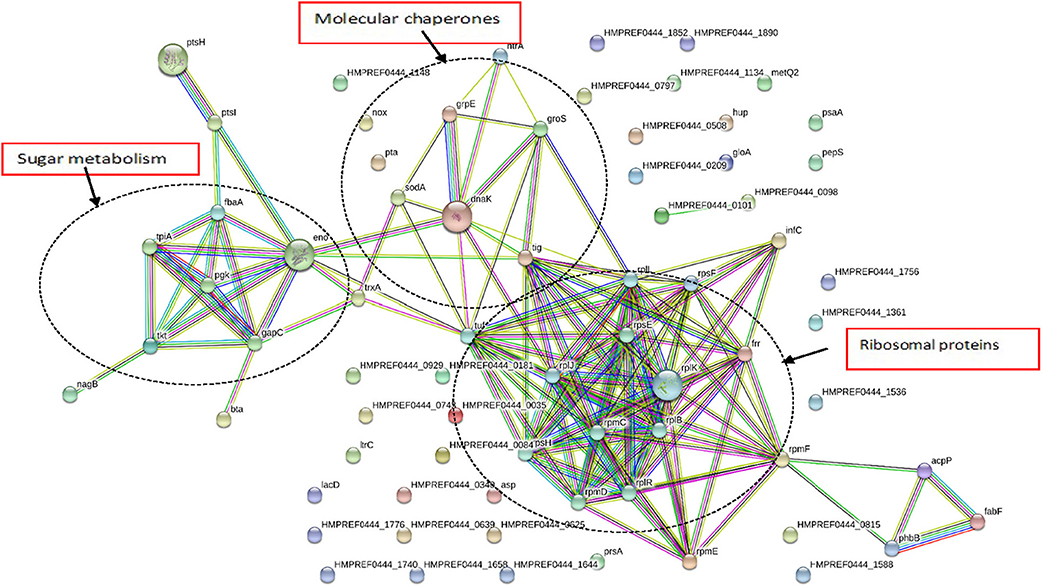
Figure 4. Functional protein association networks of G. adiacens secretome. The online tool STRING was used for grouping the secreted proteins on the basis of functional networks. Minimum interaction scores were set at a strong confidence level of 0.7. The three major network groups formed are shown in dotted circles. Seven different colored link a number of nodes and represent seven types of evidence used in predicting associations. A red line indicates the presence of fusion evidence; a green line represents neighborhood evidence, a blue line represents co-occurrence evidence; a purple line represents experimental evidence; a yellow line represents textmining evidence; a light blue line represents database evidence and a black line represents coexpression evidence.
Cytokine Stimulation by G. adiacens Secretome Preparation
When human PBMCs were stimulated with G. adiacens secretome preparation, semiquantitative analysis showed that IL-1β and MCP-1 were the cytokines found at highest relative quantities, followed by TNF-α and RANTES (Figure 5). Other important cytokines detected were IL-8, IL-6, G-CSF, GM-CSF, MIP1-α, and MIP1-β. When the PBMCs were stimulated with total proteins of G. adiacens, CCL-1, CCL2 (MCP-1), and G-CSF were not detected (data not shown).
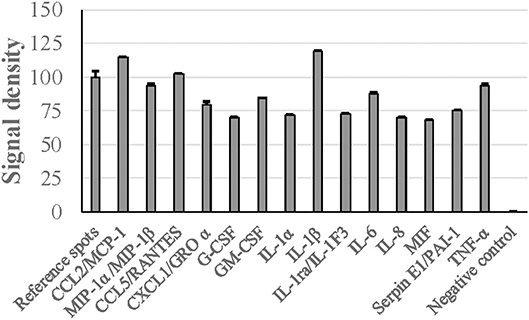
Figure 5. Cytokine induction from human PBMCs by G. adiacens secretome preparation. Fractionated human PBMCs were stimulated by G. adiacens secretome preparation for 24 h. The cytokines produced were detected by using Proteome Profiler™ membrane array. Means (SD) of signal densities of spots were determined using Gene Tools analysis software in Syngene Imaging System.
Discussion
While protein secretion is a well-established virulence mechanism in bacteria, little is known of the secretome of G. adiacens. Recently, G. elegans, a close phylogenetic relative of G. adiacens, was shown to secrete arginine deaminase (Kanamoto et al., 2007), a citrullinating enzyme that was shown to inhibit proliferation of human PBMCs in vitro and may associate with pathogenesis of periodontal and certain systemic diseases (Olsen et al., 2018). In this study, we took a qualitative proteomics approach to obtain a protein profile of the G. adiacens secretome. Since the objective of this study was to identify the proteins in the G. adiacens secretome by mass spectrometry, qualitative proteomics was adequate (Zijnge et al., 2012; Bao et al., 2015, 2017; Mohammed et al., 2017; Suriyanarayanan et al., 2018) without the necessity for validating the identified proteins as is the case for quantitative proteomics. We found that the secretome was enriched with a large number of putative virulence factors utilizing various bioinformatics analysis tools, we were able to characterize most of the secretome proteins in silico. The secretome proteins were predicted to be released via various secretion systems such as, sec-dependent, Tat pathway and via a non-classical secretory system.
To rule out the possibility of contamination of the secretome with subcellular proteins we used an established cytoplasmic marker protein Fts-Z (Terrasse et al., 2015) which remained absent in all G. adiacens secretome preparations.
By combining in silico analysis with experimental evidence and available bibliography, we were able to identify more than 20 putative virulence-associated proteins in G. adiacens secretome. This is in line with secretomes of other much-studied oral bacteria such as A. actinomycetemcomitans (Zijnge et al., 2012) and Porphyromonas gingivalis (Stobernack et al., 2016) Remarkably, several well-known virulence factors in other bacteria, such as serine protease, thioredoxin, superoxide dismutase, phosphocarrier and acyl carrier proteins were also detected in the G. adiacens secretome. Superoxide dismutase converts superoxide anions into oxygen and hydrogen peroxide. In streptococci, superoxide dismutase is displayed on the cell surface as well as released extracellularly (Gerlach et al., 1998; McMillan et al., 2004). Since G. adiacens is a catalase-negative organism, SOD might play an important role in the detoxification of oxidative burst against them by the host cells. G. adiacens apparently is equipped with more strategies to survive during oxidative stress. Indeed, gpoA gene, encoding for glutathione peroxidase was found in G. adiacens genome (Sequence ID WP_005604890.1). Superoxide dismutase is required not only for H2O2 resistance in S. mutans, but also is needed for coexistence with S. sanguinis (Fujishima et al., 2013). Whether SOD plays such a role in G. adiacens needs to be studied.
Virulence potential of several other proteins in G. adiacens secretome has been studied previously. For example, S. mutans, S. sanguinis and other species require NADH oxidase for biofilm formation (Ge et al., 2016). Streptococci secrete glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is known to help during bacterial invasion (Nelson et al., 2001). Serine proteases cause cytopathic effects and exhibit enterotoxin activity. They degrade mucins, including leukocyte surface O-glycoproteins with vital roles in numerous cellular functions, resulting in advantage for mucosal colonization and immune modulation (Dutta et al., 2002; Ruiz-Perez et al., 2011; Ruiz-Perez and Nataro, 2014). In Salmonella enterica, thioredoxin helps the bacterium in intracellular replication and virulence in a mouse model (Bjur et al., 2006). Serine protease of Fusobacterium nucleatum and S. mutans are shown to be critical in the survival and pathogenicity of these species (Diaz-Torres and Russell, 2001; Doron et al., 2014). Thus, it would be of great interest to study how G. adiacens utilizes this arsenal of putative virulence proteins for its own survival and to cause an infection.
Although G. adiacens secretome comprised of many proteins of cytoplasmic origin according to the prediction tools, interestingly, several of these belong to a so-called group “moonlighting proteins” (Jeffery, 1999; Henderson and Martin, 2014), which have a known function inside the bacterial cell but also participate in different biological processes in the extracellular medium after their secretion. That G. adiacens secretome consisted of several moonlighting proteins is of great significance since they are shown to play a role in bacterial virulence (Henderson and Martin, 2011; Wang et al., 2014). Major ribosomal proteins detected in G. adiacens secretome were 50S proteins L10, L11, L7/L12, L15, L32, and 30S proteins S5, S6, S8, and S20. Importantly, in other bacteria ribosomal protein L7/L12 is highly antigenic and immunogenic (Oliveira and Splitter, 1996; Ribeiro et al., 2002). Of the glycolytic enzymes, phosphoglycerate kinase, triose-phosphate isomerase, aldolase, and enolase possess moonlighting properties, e.g., they function as adhesins (Tunio et al., 2010), receptors for transferrin (Modun et al., 2000), neutrophil evasion proteins (Terao et al., 2006), immunomodulators (Madureira et al., 2007) and participate in extracellular polysaccharide synthesis (Lu et al., 2009) Oral bacteria express a number of molecular chaperones, including DnaK (hsp60) and GroES (GroEL) found in G. adiacens secretome. They express on the cell surface to use them as adhesins and can release them into the extracellular milieu to act as signaling virulence factors (Hinode et al., 1995; Goulhen et al., 1998; Henderson et al., 2006). The multifunctioning potential of moonlighting proteins may help G. adiacens propagate in its natural habitats as well as in sterile body areas.
More than 60 of G. adiacens secretome proteins were grouped into biological processes and molecular functions ontology groups. When we obtained GO annotations for the G. adiacens whole genome, we found that about 1,000 predicted proteins were grouped into “molecular functions,” followed by about 900 and 400 proteins in the groups “biological processes” and “cellular composition,” respectively (data not shown). The secretome proteins mapped to 19 different KEGG pathways, with antibiotic biosynthesis and glycolysis being most represented. The antibiotic biosynthesis pathway consisted of the enzymes phosphopyruvate hydratase, transketolase, glycolaldehyde transferase, triosephosphate isomerase, aldolase, and phosphoglycerate kinase, several of which have been experimentally shown to be essential for antibiotic biosynthesis (Barnard-Britson et al., 2012; Liu et al., 2016). Other prominent pathways were purine metabolism, fructose metabolism and aminobenzene degradation. These results suggest that G. adiacens secretome proteins with metabolic activities might help the bacterium in utilizing nutrients available in the extracellular milieu (Cezairliyan and Ausubel, 2017).
Functional associations among the secreted proteins using STRING network analysis showed three distinct network groups, i.e., sugar metabolism, ribosomal proteins, and putative virulence factors. Enzymes involved in sugar metabolism, i.e., phosphoglycerate kinase, triose-phosphate isomerase, 2-phosphoglycerate dehydratase, transketolase formed a cluster. Several of these enzymes seem to have overlapping functions, i.e., they were also involved in antibiotic biosynthesis as predicted by KEGG. In the cluster that consisted of putative virulence factors, molecular chaperone DnaK showed interactions with other chaperones GrpE, GroS, thioredoxin, and PPlase. This group also showed interactions with other virulence factors such as superoxide dismutase and trypsin-like protein HtrA. Molecular chaperones aid bacterial pathogenesis by helping bacteria in coping with stressful host environment, e.g., acidified phagosome, oxidative burst, and phagosome fusion with lysosomes (Hosogi and Duncan, 2005). Further, chaperones are potent immunogens and possess direct activating effect on different cell populations including lymphoid, myeloid, vascular endothelial and bone cells (Lewthwaite et al., 1998).
In several pathogenic bacteria, secreted proteins are known to modulate host immune responses (Finlay and Falkow, 1997; Lee and Schneewind, 2001). To get preliminary knowledge of the cytokine stimulatory potential of the G. adiacens secretome, we used human PBMCs as target host cells. Major cytokines such as IL-1β, TNF-α, MCP-1 were found at high levels as evidenced by higher signal densities than the reference spots. Although cytokine induction of PBMCs from total protein preparation from G. adiacens was similar, MCP-1 and G-CSF were absent. This was also observed in our previous study where biofilms failed to stimulate these cytokines but the biofilm supernatants did (Bhardwaj et al., 2018). While secreted components of different bacterial species have been previously shown to elicit inflammatory response from host cells (Oscarsson et al., 2008; Dapunt et al., 2016), a protein of Brucella suis was able to inhibit TNF-α production from macrophages when it was released extracellularly (Caron et al., 1996). Therefore, specific stimulation of certain cytokines by secreted proteins, but not the total bacterial proteins, is suggestive of possible mechanisms G. adiacens might use for systemic stimulation.
In conclusion, we unraveled the secretome of G. adiacens, an oral bacterium well-documented in infective endocarditis, but also recently shown to be involved in oral infections. Importantly, the secretome of G. adiacens comprised of a large number of putative virulence factors. Of particular importance is the finding that the G. adiacens secretome comprised of a number of “moonlighting” proteins, which in other species are shown to enhance bacterial colonization and virulence through their multifunctional roles (Pavkova et al., 2017; Graf et al., 2019). Thus, our results provide a basis for investigating the role of secreted proteins of G. adiacens in oral infections as well as in infective endocarditis.
Data Availability
Mass spectrometry data has been submitted to “PRIDE Archive” (https://www.ebi.ac.uk/pride/archive/) repository with the project accession number PXD013000. The data files can be accessed with the username:cmV2aWV3ZXI1Mzk2MUBlYmkuYWMudWs=and password: VmJUzTGl.
Ethics Statement
This study was carried out in accordance with the recommendations of the Ethical Committee of Health Sciences Center (HSC), Kuwait University. Written informed consent was obtained from the human volunteer and the consent was in accordance with the Declaration of Helsinki. The protocol was approved by the Ethical Committee of the HSC, Kuwait University.
Author Contributions
MK conceived and designed the study, bioinformatics analyses. RB and MK performed the laboratory experiments. MK, RB, AT, and SA wrote the manuscript.
Funding
This study was funded by Kuwait University, Grant Number SRUL 01/14.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Research Administration of Kuwait University for generous funding of the Oral Microbiology Research Laboratory, where this study was conducted. We also thank National Unit for Environmental Research and Services, (SRUL 01/13).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00104/full#supplementary-material
Table S1. List of proteins identified in G. adiacens secretome. Secretome preparation from G. adiacens was analyzed by LC-MS/MS and further revealed by database search (NCBI-nr).
References
Abdullah, S. N., Farmer, E. A., Spargo, L., Logan, R., and Gully, N. (2013). Porphyromonas gingivalis peptidylarginine deiminase substrate specificity. Anaerobe 23, 102–108. doi: 10.1016/j.anaerobe.2013.07.001
Afzal, M., Shafeeq, S., and Kuipers, O. P. (2015). Ascorbic acid-dependent gene expression in Streptococcus pneumoniae and the activator function of the transcriptional regulator UlaR2. Front. Microbiol. 6:72. doi: 10.3389/fmicb.2015.00072
Bagos, P. G., Tsirigos, K. D., Liakopoulos, T. D., and Hamodrakas, S. J. (2008). Prediction of lipoprotein signal peptides in gram-positive bacteria with a hidden markov model. J. Proteome Res. 7, 5082–5093. doi: 10.1021/pr800162c
Bao, K., Bostanci, N., Selevsek, N., Thurnheer, T., and Belibasakis, G. N. (2015). Quantitative proteomics reveal distinct protein regulations caused by Aggregatibacter actinomycetemcomitans within subgingival biofilms. PLoS ONE 10:e0119222. doi: 10.1371/journal.pone.0119222
Bao, K., Bostanci, N., Thurnheer, T., and Belibasakis, G. N. (2017). Proteomic shifts in multi-species oral biofilms caused by Anaeroglobus geminatus. Sci. Rep. 7:4409. doi: 10.1038/s41598-017-04594-9
Barnard-Britson, S., Chi, X., Nonaka, K., Spork, A. P., Tibrewal, N., Goswami, A., et al. (2012). Amalgamation of nucleosides and amino acids in antibiotic biosynthesis: discovery of an L-threonine:uridine-5'-aldehyde transaldolase. J. Am. Chem. Soc. 134, 18514–18517. doi: 10.1021/ja308185q
Belstrøm, D., Fiehn, N. E., Nielsen, C. H., Kirkby, N., Twetman, S., Klepac-Ceraj, V., et al. (2014). Differences in bacterial saliva profile between periodontitis patients and a control cohort. Journal of clinical periodontology 41, 104–112. doi: 10.1111/jcpe.12190
Bendtsen, J. D., Kiemer, L., Fausbøll, A., and Brunak, S. (2005a). Non-classical protein secretion in bacteria. BMC Microbiol. 5:58. doi: 10.1186/1471-2180-5-58
Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: signalp 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Bendtsen, J. D., Nielsen, H., Widdick, D., Palmer, T., and Brunak, S. (2005b). Prediction of twin-arginine signal peptides. BMC Bioinform. 6:167. doi: 10.1186/1471-2105-6-167
Bhardwaj, R. G., Al-Khabbaz, A., and Karched, M. (2018). Cytokine induction of peripheral blood mononuclear cells by biofilms and biofilm supernatants of Granulicatella and Abiotrophia spp. Microb. Pathog. 114, 90–94. doi: 10.1016/j.micpath.2017.11.037
Bjur, E., Eriksson-Ygberg, S., Aslund, F., and Rhen, M. (2006). Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 74, 5140–5151. doi: 10.1128/IAI.00449-06
Caron, E., Gross, A., Liautard, J. P., and Dornand, J. (1996). Brucella species release a specific, protease-sensitive, inhibitor of TNF-alpha expression, active on human macrophage-like cells. J. Immunol. 156, 2885–2893.
Carroll, R. K., Robison, T. M., Rivera, F. E., Davenport, J. E., Jonsson, I. M., Florczyk, D., et al. (2012). Identification of an intracellular M17 family leucine aminopeptidase that is required for virulence in Staphylococcus aureus. Microbes Infect. 14, 989–999. doi: 10.1016/j.micinf.2012.04.013
Cezairliyan, B., and Ausubel, F. M. (2017). Investment in secreted enzymes during nutrient-limited growth is utility dependent. Proc. Natl. Acad. Sci. U.S.A. 114, E7796–E7802. doi: 10.1073/pnas.1708580114
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis−10 years on. Nucl. Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Christensen, J. J., and Facklam, R. R. (2001). Granulicatella and Abiotrophia species from human clinical specimens. J. Clin. Microbiol. 39, 3520–3523. doi: 10.1128/JCM.39.10.3520-3523.2001
Collins, M. D., and Lawson, P. A. (2000). The genus Abiotrophia (Kawamura et al.) is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella elegans comb. nov. and Granulicatella balaenopterae comb. nov. Int. J. Syst. Evol. Microbiol. 50, 365–369. doi: 10.1099/00207713-50-1-365
Costa, T. R., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Dapunt, U., Giese, T., Stegmaier, S., Moghaddam, A., and Hänsch, G. M. (2016). The osteoblast as an inflammatory cell: production of cytokines in response to bacteria and components of bacterial biofilms. BMC Musculoske. Disord. 17:243. doi: 10.1186/s12891-016-1091-y
Deatherage Kaiser, B. L., Wunschel, D. S., Sydor, M. A., Warner, M. G., Wahl, K. L., and Hutchison, J. R. (2015). Improved proteomic analysis following trichloroacetic acid extraction of Bacillus anthracis spore proteins. J. Microbiol. Methods 118, 18–24. doi: 10.1016/j.mimet.2015.08.008
Diaz-Torres, M. L., and Russell, R. R. (2001). HtrA protease and processing of extracellular proteins of Streptococcus mutans. FEMS Microbiol. Lett. 204, 23–28. doi: 10.1111/j.1574-6968.2001.tb10856.x
Doron, L., Coppenhagen-Glazer, S., Ibrahim, Y., Eini, A., Naor, R., Rosen, G., et al. (2014). Identification and characterization of fusolisin, the Fusobacterium nucleatum autotransporter serine protease. PLoS ONE 9:e111329. doi: 10.1371/journal.pone.0111329
Dubreuil, J. D., Jacques, M., Brochu, D., Frenette, M., and Vadeboncoeur, C. (1996). Surface location of HPr, a phosphocarrier of the phosphoenolpyruvate: sugar phosphotransferase system in Streptococcus suis. Microbiology 142, 837–843. doi: 10.1099/00221287-142-4-837
Dutta, P. R., Cappello, R., Navarro-García , F., and Nataro, J. P. (2002). Functional comparison of serine protease autotransporters of enterobacteriaceae. Infect. Immun. 70, 7105–7113. doi: 10.1128/IAI.70.12.7105-7113.2002
Feng, S. X., Ma, J. C., Yang, J., Hu, Z., Zhu, L., Bi, H. K., et al. (2015). Ralstonia solanacearum fatty acid composition is determined by interaction of two 3-ketoacyl-acyl carrier protein reductases encoded on separate replicons. BMC Microbiol. 15:223. doi: 10.1186/s12866-015-0554-x
Finlay, B. B., and Falkow, S. (1997). Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61, 136–169.
Fujishima, K., Kawada-Matsuo, M., Oogai, Y., Tokuda, M., Torii, M., and Komatsuzawa, H. (2013). dpr and sod in Streptococcus mutans are involved in coexistence with S. sanguinis, and PerR is associated with resistance to H2O2. Appl. Environ. Microbiol. 79, 1436–1443. doi: 10.1128/AEM.03306-12
Fuss, I. J., Kanof, M. E., Smith, P. D., and Zola, H. (2009). Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protocol Immunol. Chap. 7:1. doi: 10.1002/0471142735.im0701s85
Garg, A., and Gupta, D. (2008). VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinform. 9:62. doi: 10.1186/1471-2105-9-62
Ge, X., Yu, Y., Zhang, M., Chen, L., Chen, W., Elrami, F., et al. (2016). Involvement of NADH oxidase in competition and endocarditis virulence in Streptococcus sanguinis. Infect. Immun. 84, 1470–1477. doi: 10.1128/IAI.01203-15
Gerlach, D., Reichardt, W., and Vettermann, S. (1998). Extracellular superoxide dismutase from Streptococcus pyogenes type 12 strain is manganese-dependent. FEMS Microbiol. Lett. 160, 217–224.
Goosens, V. J., Monteferrante, C. G., and van Dijl, J. M. (2014). The Tat system of Gram-positive bacteria. Biochim. Biophys. Acta 1843, 1698–1706. doi: 10.1016/j.bbamcr.2013.10.008
Goulhen, F., Hafezi, A., Uitto, V. J., Hinode, D., Nakamura, R., Grenier, D., et al. (1998). Subcellular localization and cytotoxic activity of the GroEL-like protein isolated from Actinobacillus actinomycetemcomitans. Infect. Immun. 66, 5307–5313.
Graf, A. C., Leonard, A., Schäuble, M., Rieckmann, L. M., Hoyer, J., Maaß, S., et al. (2019). Virulence factors produced by Staphylococcus aureus biofilms have a moonlighting function contributing to biofilm integrity. Mol. Cell Proteomics. 1–47. doi: 10.1074/mcp.RA118.001120
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4, 1–19. doi: 10.1128/microbiolspec.VMBF-0012-2015
Großhennig, S., Ischebeck, T., Gibhardt, J., Busse, J., Feussner, I., and Stülke, J. (2016). Hydrogen sulfide is a novel potential virulence factor of Mycoplasma pneumoniae: characterization of the unusual cysteine desulfurase/desulfhydrase HapE. Mol. Microbiol. 100, 42–54. doi: 10.1111/mmi.13300
Henderson, B., Allan, E., and Coates, A. R. (2006). Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect. Immun. 74, 3693–3706. doi: 10.1128/IAI.01882-05
Henderson, B., and Martin, A. (2011). Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491. doi: 10.1128/IAI.00179-11
Henderson, B., and Martin, A. C. (2014). Protein moonlighting: a new factor in biology and medicine. Biochem. Soc. Trans. 42, 1671–1678. doi: 10.1042/BST20140273
Hernández, S., Ferragut, G., Amela, I., Perez-Pons, J., Piñol, J., Mozo-Villarias, A., et al. (2014). MultitaskProtDB: a database of multitasking proteins. Nucl. Acids Res. 42, D517–520. doi: 10.1093/nar/gkt1153
Hiller, K., Schobert, M., Hundertmark, C., Jahn, D., and Münch, R. (2003). JVirGel: calculation of virtual two-dimensional protein gels. Nucl. Acids Res. 31, 3862–3865. doi: 10.1093/nar/gkg536
Hinode, D., Grenier, D., and Mayrand, D. (1995). Purification and characterization of a DnaK-like and a GroEL-like protein from Porphyromonas gingivalis. Anaerobe 1, 283–290.
Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379.
Hosogi, Y., and Duncan, M. J. (2005). Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect. Immun. 73, 2327–2335. doi: 10.1128/IAI.73.4.2327-2335.2005
Jutras, B. L., Verma, A., Adams, C. A., Brissette, C. A., Burns, L. H., Whetstine, C. R., et al. (2012). BpaB and EbfC DNA-binding proteins regulate production of the Lyme disease spirochete's infection-associated Erp surface proteins. J. Bacteriol. 194, 778–786. doi: 10.1128/JB.06394-11
Kall, L., Krogh, A., and Sonnhammer, E. L. (2007). Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35, W429–W432. doi: 10.1093/nar/gkm256
Kanamoto, T., Sato, S., Nakashima, H., and Inoue, M. (2007). Proliferation of mitogen-stimulated human peripheral blood mononuclear cells is inhibited by extracellular arginine deiminase of Granulicatella elegans isolated from the human mouth. J. Infect. Chemother. 13, 353–355. doi: 10.1007/s10156-007-0546-3
Kanasi, E., Dewhirst, F. E., Chalmers, N. I., Kent, R. Jr., Moore, A., Hughes, C. V., et al. (2010). Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 44, 485–497. doi: 10.1159/000320158
Karched, M., Bhardwaj, R. G., and Asikainen, S. E. (2015). Coaggregation and biofilm growth of Granulicatella spp. with Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans. BMC Microbiol. 15:114. doi: 10.1186/s12866-015-0439-z
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Lanotte, P., Perivier, M., Haguenoer, E., Mereghetti, L., Burucoa, C., Claverol, S., et al. (2013). Proteomic biomarkers associated with Streptococcus agalactiae invasive genogroups. PLoS ONE 8:e54393. doi: 10.1371/journal.pone.0054393
Lee, V. T., and Schneewind, O. (2001). Protein secretion and the pathogenesis of bacterial infections. Genes. Dev. 15, 1725–1752. doi: 10.1101/gad.896801
Lewthwaite, J., Skinner, A., and Henderson, B. (1998). Are molecular chaperones microbial virulence factors? Trends Microbiol. 6, 426–428.
Lin, C. H., and Hsu, R. B. (2007). Infective endocarditis caused by nutritionally variant streptococci. Am. J. Med. Sci. 334, 235–239. doi: 10.1097/MAJ.0b013e3180a6eeab
Liu, Y., Chen, S., Zhang, J., and Gao, B. (2016). Growth, microcystin-production and proteomic responses of Microcystis aeruginosa under long-term exposure to amoxicillin. Water Res. 93, 141–152. doi: 10.1016/j.watres.2016.01.060
Loprasert, S., Whangsuk, W., Sallabhan, R., and Mongkolsuk, S. (2004). DpsA protects the human pathogen Burkholderia pseudomallei against organic hydroperoxide. Arch. Microbiol. 182, 96–101. doi: 10.1007/s00203-004-0694-0
Lu, G. T., Xie, J. R., Chen, L., Hu, J. R., An, S. Q., Su, H. Z., et al. (2009). Glyceraldehyde-3-phosphate dehydrogenase of Xanthomonas campestris pv. campestris is required for extracellular polysaccharide production and full virulence. Microbiology 155, 1602–1612. doi: 10.1099/mic.0.023762-0
Madureira, P., Baptista, M., Vieira, M., Magalhães, V., Camelo, A., Oliveira, L., et al. (2007). Streptococcus agalactiae GAPDH is a virulence-associated immunomodulatory protein. J. Immunol. 178, 1379–1387. doi: 10.4049/jimmunol.178.3.1379
McMillan, D. J., Davies, M. R., Good, M. F., and Sriprakash, K. S. (2004). Immune response to superoxide dismutase in group A streptococcal infection. FEMS Immunol. Med. Microbiol. 40, 249–256. doi: 10.1016/S0928-8244(04)00003-3
Modun, B., Morrissey, J., and Williams, P. (2000). The staphylococcal transferrin receptor: a glycolytic enzyme with novel functions. Trends Microbiol. 8, 231–237. doi: 10.1016/S0966-842X(00)01728-5
Mohammed, M. M. A., Pettersen, V. K., Nerland, A. H., Wiker, H. G., and Bakken, V. (2017). Quantitative proteomic analysis of extracellular matrix extracted from mono- and dual-species biofilms of Fusobacterium nucleatum and Porphyromonas gingivalis. Anaerobe 44, 133–142. doi: 10.1016/j.anaerobe.2017.03.002
Natale, P., Brüser, T., and Driessen, A. J. (2008). Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane–distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756. doi: 10.1016/j.bbamem.2007.07.015
Nelson, D., Goldstein, J. M., Boatright, K., Harty, D. W., Cook, S. L., Hickman, P. J., et al. (2001). pH-regulated secretion of a glyceraldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: purification, characterization, and cloning of the gene encoding this enzyme. J. Dent Res. 80, 371–377. doi: 10.1177/00220345010800011301
Oliveira, S. C., and Splitter, G. A. (1996). Immunization of mice with recombinant L7/L12 ribosomal protein confers protection against Brucella abortus infection. Vaccine 14, 959–962.
Olsen, I., Singhrao, S. K., and Potempa, J. (2018). Citrullination as a plausible link to periodontitis, rheumatoid arthritis, atherosclerosis and Alzheimer's disease. J. Oral Microbiol. 10:1487742. doi: 10.1080/20002297.2018.1487742
Oscarsson, J., Karched, M., Thay, B., Chen, C., and Asikainen, S. (2008). Proinflammatory effect in whole blood by free soluble bacterial components released from planktonic and biofilm cells. BMC Microbiol. 8:206. doi: 10.1186/1471-2180-8-206
Pallen, M. J. (2002). The ESAT-6/WXG100 superfamily – and a new Gram-positive secretion system? Trends Microbiol. 10, 209–212. doi: 10.1016/S0966-842X(02)02345-4
Pavkova, I., Kopeckova, M., Klimentova, J., Schmidt, M., Sheshko, V., Sobol, M., et al. (2017). The Multiple Localized Glyceraldehyde-3-Phosphate Dehydrogenase contributes to the attenuation of the francisella tularensis dsba deletion mutant. Front. Cell Infect. Microbiol. 7:503. doi: 10.3389/fcimb.2017.00503
Ribeiro, L. A., Azevedo, V., Le Loir, Y., Oliveira, S. C., Dieye, Y., Piard, J. C., et al. (2002). Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis: a first step towards food-grade live vaccines against brucellosis. Appl. Environ. Microbiol. 68, 910–916. doi: 10.1128/AEM.68.2.910-916.2002
Romero, P., López, R., and Garcia, E. (2004). Characterization of LytA-like N-acetylmuramoyl-L-alanine amidases from two new Streptococcus mitis bacteriophages provides insights into the properties of the major pneumococcal autolysin. J. Bacteriol. 186, 8229–8239. doi: 10.1128/JB.186.24.8229-8239.2004
Rose, R. W., Brüser, T., Kissinger, J. C., and Pohlschröder, M. (2002). Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45, 943–950. doi: 10.1046/j.1365-2958.2002.03090.x
Ruiz-Perez, F., and Nataro, J. P. (2014). Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell Mol. Life Sci. 71, 745–770. doi: 10.1007/s00018-013-1355-8
Ruiz-Perez, F., Wahid, R., Faherty, C. S., Kolappaswamy, K., Rodriguez, L., Santiago, A., et al. (2011). Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc. Natl. Acad. Sci. U.S.A 108, 12881–12886. doi: 10.1073/pnas.1101006108
Schneewind, O., and Missiakas, D. M. (2012). Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R Soc. Lond. B Biol. Sci. 367, 1123–1139. doi: 10.1098/rstb.2011.0210
Shi, X. Z., Feng, X. W., Sun, J. J., Yang, M. C., Lan, J. F., Zhao, X. F., et al. (2016). Involvement of a LysM and putative peptidoglycan-binding domain-containing protein in the antibacterial immune response of kuruma shrimp Marsupenaeus japonicus. Fish Shellfish Immunol. 54, 489–498. doi: 10.1016/j.fsi.2016.04.134
Siqueira, J. F. Jr., and Rôças, I. N. (2006). Catonella morbi and Granulicatella adiacens: new species in endodontic infections. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 102, 259–264. doi: 10.1016/j.tripleo.2005.09.021
Stobernack, T., Glasner, C., Junker, S., Gabarrini, G., de Smit, M., de Jong, A., et al. (2016). Extracellular proteome and citrullinome of the oral Pathogen Porphyromonas gingivalis. J. Proteome Res. 15, 4532–4543. doi: 10.1021/acs.jproteome.6b00634
Suriyanarayanan, T., Qingsong, L., Kwang, L. T., Mun, L. Y., Truong, T., and Seneviratne, C. J. (2018). Quantitative proteomics of strong and weak biofilm formers of enterococcus faecalis reveals novel regulators of biofilm formation. Mol. Cell Proteomics 17, 643–654. doi: 10.1074/mcp.RA117.000461
Terao, Y., Yamaguchi, M., Hamada, S., and Kawabata, S. (2006). Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J. Biol. Chem. 281, 14215–14223. doi: 10.1074/jbc.M513408200
Terrasse, R., Amoroso, A., Vernet, T., and Di Guilmi, A. M. (2015). Streptococcus pneumoniae GAPDH Is Released by Cell Lysis and interacts with peptidoglycan. PLoS ONE 10:e0125377. doi: 10.1371/journal.pone.0125377
Tortosa, P., Albano, M., and Dubnau, D. (2000). Characterization of ylbF, a new gene involved in competence development and sporulation in Bacillus subtilis. Mol. Microbiol. 35, 1110–1119. doi: 10.1046/j.1365-2958.2000.01779.x
Tunio, S. A., Oldfield, N. J., Berry, A. D. A, Ala'Aldeen, Wooldridge, K. G., and Turner, D. P. (2010). The moonlighting protein fructose-1, 6-bisphosphate aldolase of Neisseria meningitidis: surface localization and role in host cell adhesion. Mol. Microbiol. 76, 605–615. doi: 10.1111/j.1365-2958.2010.07098.x
von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., et al. (2005). STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucl. Acids Res. 33, D433–D437. doi: 10.1093/nar/gki005
Wang, G., Xia, Y., Cui, J., Gu, Z., Song, Y., Chen, Y. Q., et al. (2014). The roles of moonlighting proteins in bacteria. Curr. Issues Mol. Biol. 16, 15–22. doi: 10.21775/cimb.016.015
Ye, J., Zhang, Y., Cui, H., Liu, J., Wu, Y., Cheng, Y., et al. (2018). WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 46, W71–W75. doi: 10.1093/nar/gky400
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Yuan, Z., Mattick, J. S., and Teasdale, R. D. (2004). SVMtm: support vector machines to predict transmembrane segments. J. Comput. Chem. 25, 632–636. doi: 10.1002/jcc.10411
Zhong, Q., Zhao, Y., Chen, T., Yin, S., Yao, X., Wang, J., et al. (2014). A functional peptidoglycan hydrolase characterized from T4SS in 89K pathogenicity island of epidemic Streptococcus suis serotype 2. BMC Microbiol. 14:73. doi: 10.1186/1471-2180-14-73
Keywords: Granulicatella, secretome, oral, infective endocarditis, virulence, periodontitis
Citation: Karched M, Bhardwaj RG, Tiss A and Asikainen S (2019) Proteomic Analysis and Virulence Assessment of Granulicatella adiacens Secretome. Front. Cell. Infect. Microbiol. 9:104. doi: 10.3389/fcimb.2019.00104
Received: 21 January 2019; Accepted: 26 March 2019;
Published: 24 April 2019.
Edited by:
Georgios N. Belibasakis, Karolinska Insitutet (KI), SwedenReviewed by:
Thuy Do, University of Leeds, United KingdomChaminda Jayampath Seneviratne, National Dental Centre of Singapore, Singapore
Copyright © 2019 Karched, Bhardwaj, Tiss and Asikainen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maribasappa Karched, bWthcmNoZWRAaHNjLmVkdS5rdw==
 Maribasappa Karched
Maribasappa Karched Radhika G. Bhardwaj
Radhika G. Bhardwaj Ali Tiss
Ali Tiss Sirkka Asikainen1
Sirkka Asikainen1