- Key Laboratory of Tropical Biological Resources of Ministry of Education, School of Life and Pharmaceutical Sciences, Hainan University, Haikou, China
The superbacteria Aeromonas veronii displays not only a strong pathogenicity but also the resistance to nine kinds of antibiotics, resulting in the economic losses and health hazards. Small Protein B (SmpB) plays an important role in protein quality control, virulence, and stress reactions. Transcriptomic data revealed that expressions of the type IV pilus assembly and type VI secretion system (T6SS) proteins were downregulated in SmpB deficiency, indicating that the virulence of A. veronii might be attenuated. Although SmpB deletion decreased colonization in the mouse spleen and liver, LD50 of the smpB mutant was not altered as expected, compared with the wild type. Further, the transcriptomic and quantitative RT-PCR analyses showed that the combination of the downregulated AvrA and the upregulated iron-sulfur protein activator IscR, mediated the oxidative tolerance in smpB deletion. Next a reporter plasmid was constructed in which the promoter of iscR was applied to control the expression of the enhanced green fluorescent protein (eGFP) gene. When the reporter plasmid was co-expressed with the AvrA expression into E. coli, the relative fluorescence intensity was decreased significantly, suggesting that AvrA bound to iscR mRNA by base pairing, which in turn relieved the inhibition of iscR and intensified the downstream iron-sulfur proteins. Collectively, the smpB mutant exhibited an attenuated virulence in mice and enhanced tolerances to oxidative stress. This study demonstrates the complexity of gene regulation networks mediated by sRNA in systems biology, and also reflects the strong adaptability of superbacteria A. veronii in the process of evolution.
Introduction
With the application of antimicrobial drugs, a wide range of mechanisms have evolved for bacteria to combat the huge selection pressures of antibacterial agents. Superbacteria have accumulated in a long evolutionary history, while the abuse of antibiotics has accelerated its formation. The superbacterium Aeromonas veronii (A. veronii) has been isolated, which has displayed not only a strong pathogenicity but also the resistance to nine kinds of antibiotics (Liu et al., 2016, 2018). As A. veronii infects fish, it causes fish canker and perforated disease, eventually resulting in economic losses. Furthermore, A. veronii increases human diseases such as neonatal sepsis, diarrhea and dysentery (Aguilera-Arreola et al., 2007). In view of the wide array of infections to the host and a strong tolerance to antibiotics, A. veronii is worth researching further, as a model strain.
Proteins are the material basis of life. If there is a mistake in the protein synthesis process, ribosome rescue should occur in order to maintain life. Trans-translation which is composed of small protein B(SmpB) and transfer-message RNA (tmRNA) is the predominant form of ribosome rescue (Himeno et al., 2014). The malfunction of trans-translation exhibits more sensitivity to virulence and tolerance in most pathogens such as Mycobacterium tuberculosis, Legionella pneumophila, and Neisseria gonorrhoeae (Keiler, 2008, 2015). However, the alternative pathways have evolved to compensate for the deficiency of trans-translation. For example, the alternative rescue factor A (ArfA) acts to release the stocked mRNA in Proteobacteria and mycobacteria, and the alternative rescue factor B (ArfB) appears in 34% of bacteria (Kurita et al., 2014; Huter et al., 2017; Fiedler et al., 2018). Therefore, superbacteria might have alternate ways for SmpB defects, while no reports indicate that other functions of SmpB are maintained by small RNA (sRNA).
In this study, the transcriptome analysis was compared between wild type and smpB knockout strains, showing that the expressions of the pilus and virulence protein were downregulated after smpB deletion. However, the capacities of oxidative stress and iron were increased by sRNA downregulation in smpB knockout. Non-coding sRNA modulates bacterial metabolism and stress by base pairing with target genes and reacts faster than transcription factor regulation (Durand et al., 2015; Oliva et al., 2015). Here a sRNA designated as AvrA was revealed to negatively regulate the expression of IscR (iron-sulfur cluster assembly transcriptional regulator) IscR governs iron hemostasis as a global regulator during growth and stress responses (Aguilera-Arreola et al., 2007), which exhibits a resistance to oxidants (Mandin et al., 2016), and represses iron-sulfur metabolism under iron starvation (Haines et al., 2015; Carrier et al., 2017). The overproduction of IscR favored the tolerance under the nutrient deficiency and oxidative, iron-limited and membrane stresses when smpB was knocked out and followed by AvrA downregulation.
Survival under detrimental stresses is preliminary when pathogens are infected to the host and escape from macrophage recognition in the immune system (Mandin et al., 2016). And then the bacteria are transported to tissues using blood and lymph as channels, eventually causing tissue damage. Iron metabolism is of vital importance to bacteria. The proteins composed of iron/sulfur clusters participate in multiple cellular processes, such as gene expression, DNA repair, RNA modification, central metabolism and respiration (Roche et al., 2013). Iron sulfur proteins also relate to oxidoreductase which is involved in the basic metabolism of bacteria (Xu and Moller, 2008; Ilbert and Bonnefoy, 2013). So, overexpression of IscR is vital for bacteria to survive in adverse conditions.
In summary, SmpB plays an important role in protein quality control, virulence, gene transcription, and antibiotic and stress reaction (Liang and Deutscher, 2010; Li et al., 2013; Mu et al., 2013; Mraheil et al., 2017). Unlike Salmonella Typhimurium, the virulence of the A. veronii ΔsmpB strain did not become avirulent but maintained a strong pathogenicity. Loss of function caused by smpB deletion can be compensated through different pathways. For instance, some bacteria employ ArfA and ArfB to rescue stalled ribosomes when smpB is deficient (Keiler, 2015; Liu et al., 2016; Huter et al., 2017). Additionally, sRNA AvrA releases the trapped IscR promotor to activate the expression of iron sulfur proteins, remedying the iron deficiency after smpB destruction. In the evolutionary history of bacteria, it offers additional evidence for the formation of superbacteria.
Materials and Methods
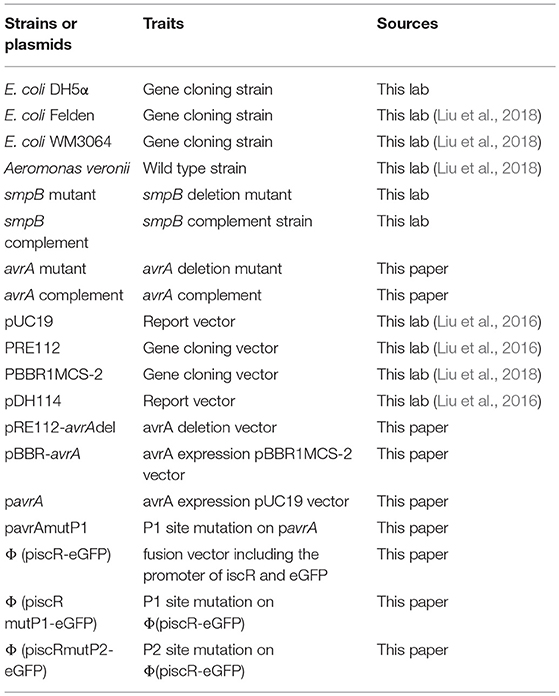
Plasmids
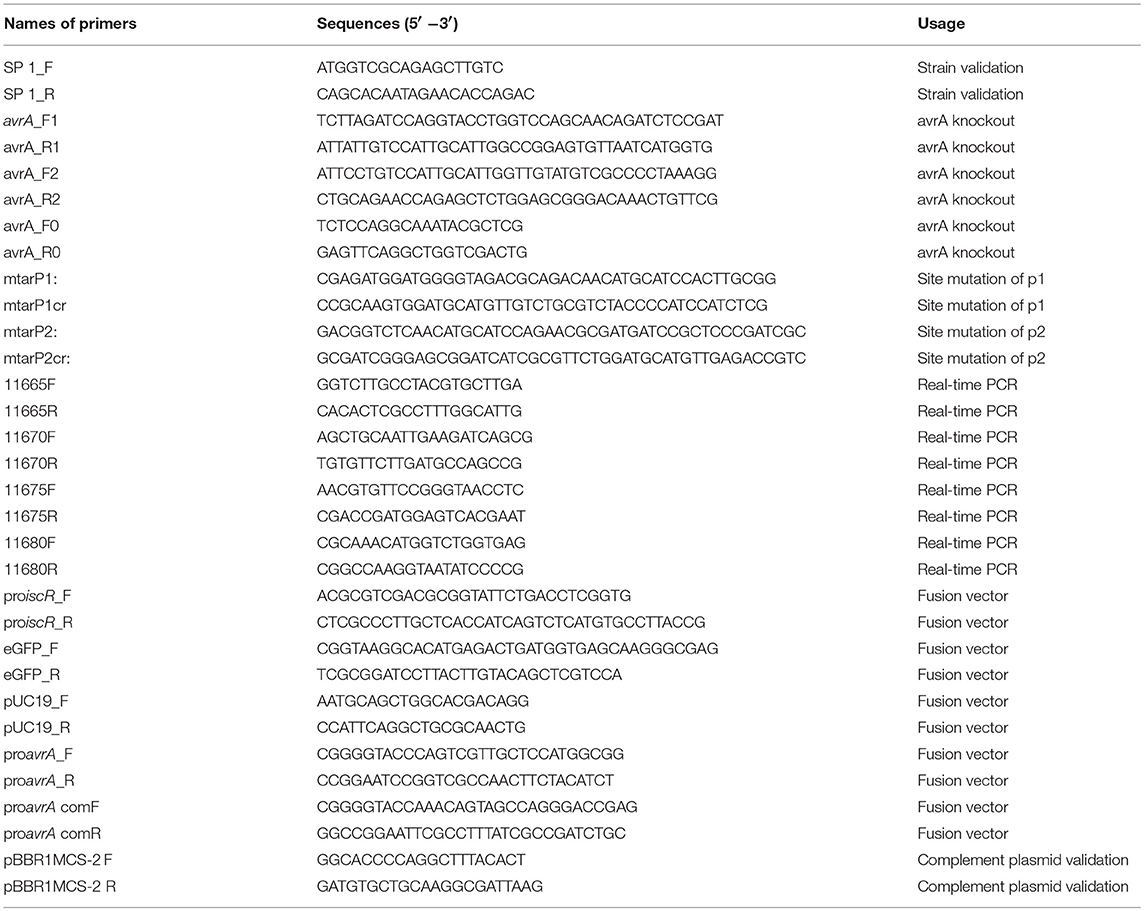
The plasmids and primers are listed in Tables 1, 2, respectively.
For the construction of plasmid piscR-eGFP, the 200 bp-DNA fragments containing the promoter of iscR was amplified from the genomic DNA of A. veronii with primers proiscR_F/proiscR_R. In the meantime, the 750 bp-encoding region of eGFP was amplified from a plasmid pDH114 with primer eGFP_F/eGFP_R. The fusion was produced using the above two fragments as the templates by overlapping the PCR with primers F_PiscR/eGFP_R and by insertion into the backbone of the plasmid pUC19 to construct the Φ(piscR-eGFP) vector. The plasmid pavrA was constructed in a similar manner using the primers proavrA_F and proavrA_R. To construct the gene knockout strain of ΔavrA, the flanking sequences of ΔavrA were amplified and inserted into the plasmid pRE112, resulting in the gene knockout vector pRE112-avrAdel. The complementary plasmid pBBR-avrA was applied to express the extra AvrA using plasmid pBBR1MCS-2 as the backbone.
Strains and Culture Conditions
The strains used are listed in Table 1. The derivative A. veronii strains were picked from frozen stocks and cultured at 30°C in Luria-Bertani (LB) broth or M9 medium with aeration. The Antibiotics included 50 μg/mL ampicillin, 25 μg/mL chloramphenicol, 50 μg/mL kanamycin. The E. Coli Felden strain was chosen for the expression of eGFP and sRNA (Liu et al., 2015). E. Coli WM3064 was selected for gene knockout by homologous recombination, which is capable of growth at 37°C in LB broth with 0.3 mM diaminopimelic acid (DAP).
The plasmid pRE112-avrAKO was transformed into the competent E. coli WM3064 and delivered into A. veronii by conjugation (Liu et al., 2016). The A. veronii ΔavrA was screened on LB agar supplemented with 6% sucrose and ampicillin and was further checked by sequencing.
The plasmid pBBR-avrA was transformed into the competent E. coli WM3064, and mobilized into the ΔavrA strain by conjugation, resulting in complementary A. veronii ΔavrA::avrA.
RNA Sequencing and Bioinformatics Analysis
Strains were grown in a 10 mL M9 medium containing 50 μg/mL ampicillin at 30°C, 150 rpm for 20 h. The cells were collected and lysed. RNA samples were extracted by phenol-chloroform. The concentration and quality of RNA was detected by the Agilent 2100 Bio analyzer. The samples were treated with DNase I to eliminate double-stranded and single-stranded DNA, followed by the depletion of rRNA with the Ribo-Zero Magnetic Kit. First-strand cDNA was generated using a random primer reverse transcription, followed by a second-strand cDNA synthesis. The end of the synthesized cDNA was subjected to repair and adenylate. The adapters were ligated to the ends of these 3′-adenylated cDNA fragments. The cDNA fragments were enriched by PCR amplification with a PCR Primer Cocktail, and the purified products were sequenced on the Hiseq Xten (Illumina, San Diego, CA, USA) using the gel-free protocol. Libraries were sequenced as 100 bp paired-end reads to a minimum target depth of 2G clean data per sample. The software HISAT (V2.0.1-beta) was used for the reference genome alignment, and CPC (coding potential calculator) was used to characterize the translation possibility of new transcripts. To identify the differential expression between wild type and ΔsmpB, Bowtie 2(v2.2.5) was used to analyze the mRNA expression. Fragments per kilobase of transcript per million fragments sequenced (FPKM) were used to normalize the gene expression levels. For each gene, the p-value was evaluated, and Benjamini-Hochberg false discovery rate (FDR) was applied for the correction. The differential expressions of the transcripts were estimated by the virtue of ≥1 absolute log2 -fold change and < 0.001 FDR adjusted p-value. These results were sorted by GO categories using Perl scripts. The GO and KEGG pathway enrichments of differential uni-genes were analyzed. The hypergeometric test was used to compare differential expression genes in the pathway with the entire genome background. Pathways with a p ≤ 0.05 were considered as significant. GEO accession number GSE120603.
LD50 Test in Mice
The derivative of A. veronii was streaked onto M9 medium and grown for 18–20 h at 30°C. The cells were re-suspended in sterile PBS. ICR mice (male, 6–8 weeks old) were infected by intraperitoneal injection with 103–107 CFU/g of each strain (n = 8) and monitored daily for weight loss and signs of illness. The animals were euthanized if they accorded with alternate endpoint criteria including the loss of mobility. The 50% lethal dose was calculated by linear regression (Torii et al., 2015).
Organ Coefficient
The coefficient of organs was calculated from the ratio of organ weight and body weight of the same mouse. The change of coefficient was used to characterize the degree of injury by contrasting the pathological with the normal tissue (Zhang et al., 2014).
All animal experiments were approved by the Committee of the Ethics on Animal Care and Experiments at Hainan University, and were carried out in accordance with the university guidelines.
Measurement of Growth Curve
The condition of iron deficiency was equipped with LB medium containing 200 mM dipyridyl. M9 medium was considered as the nutrient limited condition. The strains were cultured at logarithmic phase, and then inoculated into 50 ml media at an initial OD600 of 0.05. The ODs were monitored with a spectrophotometer (INESA 970CRT, Shanghai, China) at regular intervals.
Survival Under Oxidative Stress
The derivative of A. veronii was grown until to the OD600 value of 0.6 in the LB broth appended with 200 mM dipyridyl and subsequently challenged with 0–10 mM H2O2. The cultures were continuously incubated at 30°C with shaking for 10 min, and the survival colonies were counted on the LB agar plate with 50 μg/mL ampicillin.
Real-Time PCR Experiment
The wild type and mutant strains of A. veronii were grown in LB and M9 supplemented with 50 μg/mL ampicillin at 30°C till to stationary stage. The total amount of RNA was extracted for relative expression analysis of the genes, including smpB, the predicted targets of AvrA (t15190, t21425, t18380, t22295, t14250), Fe/S cluster assembly regulation factor (iscR) and its regulated targets iscA, iscU, iscS.
Florescent Measurement
The cells conferring eGFP expression were suspended in PBS buffer, and loaded to a 96 well microtiter plate (Greiner BIOONE, Nurnberg, Germany). The fluorescent intensity was measured at 488 nm excitation and 540 nm emission wavelengths with a fluorescence microplate reader (Infinite® 200 PRO, Tecan, Shanghai, China). The relative fluorescence was calculated as the total fluorescence divided by OD600 value.
Statistical Analysis
Statistical data were analyzed using the statistical Package for the Social Science (SPSS) version 20.0 (SPSS, Chicago, IL, United States) and GraphPad Prism version 6.0 (GraphPad, San Diego, CA, United States). The results were presented as mean values of three independent experiments with standard deviation (SD) using one-way analysis of variance (ANOVA). P < 0.05 or 0.01 were represented as significant or extremely significant.
Results
Transcriptomic Analysis
To determine the differences in pathways and gene expressions between wild type and smpB knockout (ΔsmpB) in the M9 medium, the transcriptome assembly of A. veronii was generated from 91.17 million paired-end RNA-seq reads using Illumina Xten technology and a total of 22,055 transcripts were mapped by genome NZ_CP012504.1 (Figure 1A). Sixteen pathways were significantly affected by SmpB knockout, including sulfur metabolism, biosynthesis of siderophore group non-ribosomal peptides, bacterial chemotaxis, arginine and peptidoglycan biosynthesis, and microbial metabolism in a diverse environment (Figure 1B). Among them, 21 and 20 genes were downregulated, respectively in the type IV pilus assembly and the type VI secretion system at the stationary phase (Figure 1C). The type IV pili are ubiquitously expressed on the surface of many Gram-negative bacteria and are important virulence factors that facilitate host-pathogen interactions and persistence of infection. T6SS is the molecular machine, which transports the virulence effectors from the interior cytoplasm or cytosol into an adjacent target cell across the cellular envelope. The downregulated pilus and secretion system proteins revealed that toxicity might be attenuated in ΔsmpB.
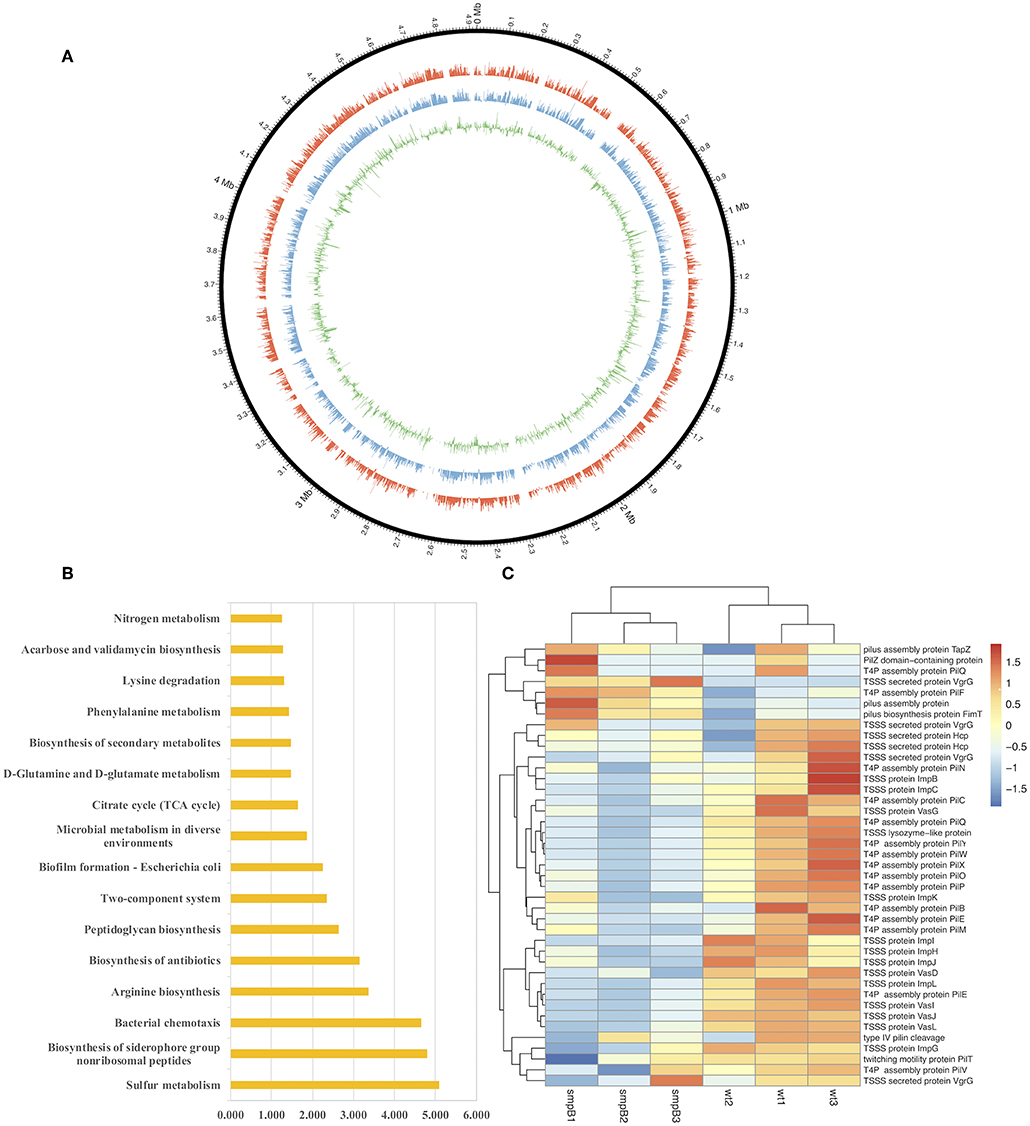
Figure 1. Transcriptomic analysis between wild type and smpB knockout. (A) Circos diagram of read distribution and gene expression. From inside, green (fold change of gene expression between ΔsmpB and wild type), blue (mRNA expression of wild type A. veronii by rpkm), red (mRNA expression of ΔsmpB by rpkm), black (reads distribution on the reference genome NZ_CP012504.1). (B) Pathways affected by smpB. The horizontal and vertical axes represented as –log(p) value and the pathway names (p < 0.05). (C) Heat map representing the expressions of type IV pilus assembly, type VI secretion system and outer membrane genes.
SmpB Deletion Decreases the Colonization in Host Tissue
To compare the virulence between wild type and ΔsmpB, Institute of Cancer Research (ICR) mice were divided into 10 groups with 104–108 CFU/g bacterial infection. The survival rate of ICR mice revealed that the virulence of the wild type was no different compared with the smpB deletion (Figures 2A,B).
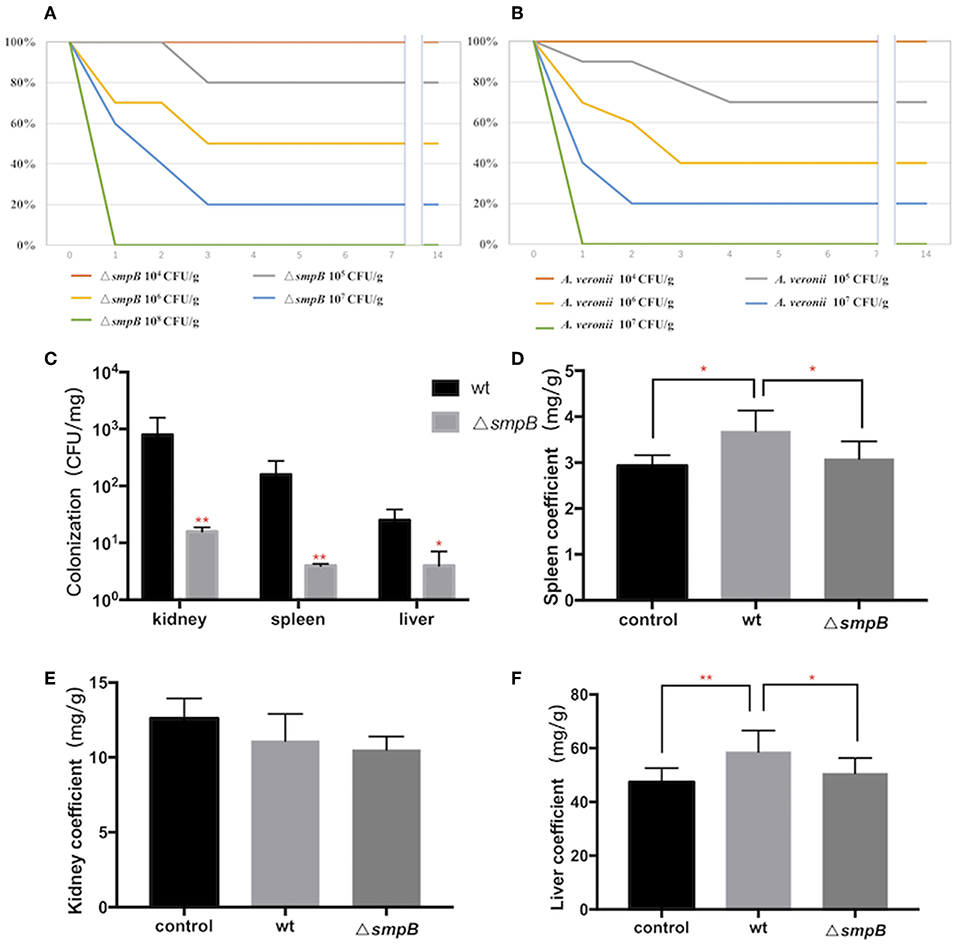
Figure 2. SmpB deletion decreases the colonization in host tissue. (A) Survival rate of mice treated by wild type A. veronii. (B) Survival rate of mice treated by ΔsmpB strain. (C) Colonization of wild type and ΔsmpB in spleen, kidney, and liver of ICR mice. (D) Spleen coefficient (mg/g). (E) Kidney coefficient (mg/g). (F) Liver coefficient (mg/g). Error bars were shown as SD. **p < 0.01, *p < 0.05, following by one-way ANOVA, independent t-tests.
After the mice were intraperitoneally injected with 5 × 105 CFU/g bacterial strains, the liver, spleen and kidneys were collected, and tissue suspension was cultured on LB plates supplemented with ampicillin. The wild type was colonized significantly more than ΔsmpB in the kidney, liver, and spleen (Figure 2C), and the results agreed with the downregulated expressions of the type IV pilus assembly proteins (Figure 1C). Organ coefficient is a common toxicology indicator of ratio of organ and body weight reflecting animal damage (Zhang et al., 2014). In our study, the coefficients of spleen and liver infected by the wild type were 3.68 ± 0.45 and 58.65 ± 7.94 mg/g, which were significantly higher than the control group (2.93 ± 0.23 and 47.38 ± 5.27 mg/g) and ΔsmpB group (3.08 ± 0.38 and 50.65 ± 5.62 mg/g) (Figures 2D,E), while little difference was observed in the kidney (Figure 2E). Next the organ coefficient was evaluated for the visceral injury between the treatment and the control group. The coefficients of the spleen and liver infected by the wild type were significantly higher than the control group and ΔsmpB group (Figures 2D–F), while little difference was observed in the kidney (Figure 2E). This also suggested that the virulence of wild type A. veronii was not affected by smpB deletion. Consistently, the acute LD50 for ΔsmpB strain in mice was calculated as 5 × 105 CFU/g, which was close to that of 106 CFU/g for the wild type. Because of the contradiction between the decreased pathogenicity and unvaried lethality, we speculated that the bacterial resistance had enhanced in the host.
The Combination of the Downregulated AvrA and Upregulated IscR Mediates the Oxidative Tolerance in smpB Deletion
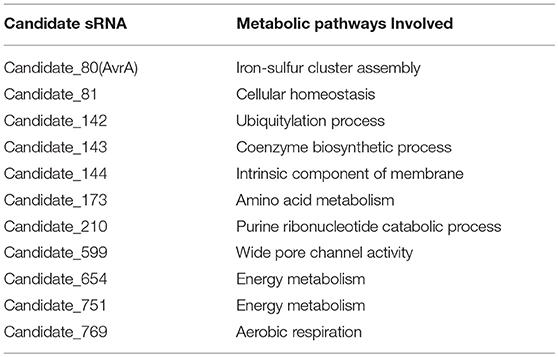
Expressions of transcripts were compared between the wild type and smpB mutant in the transcriptomic data. A total of 466 non-coding transcripts were defined as sRNA, including 50–500 nt. Among them, 11 sRNAs were analyzed to express specifically between the wild type and ΔsmpB at the stationary stage in the M9 medium (Figure 3A). There were 62 differentially expressed target genes of the 11 sRNAs involved in stress response pathways, pathogenesis, and also the regulations of metabolism, transport, quorum sensing (Supplementary Table 1). The pathways were associated with the synthesis of cell membrane, protein translocation, ATPase, Fe/S protein assembly and energy metabolism (Table 3). The candidate_80 sRNA, designated as AvrA (Aeromonas veronii non-coding RNA A) was potentially associated with the target gene iscR in Fe/S assembly. After smpB was knocked out, AvrA expression declined (Figure 3A) resulting in the increased expression of the predicted targets isc operon genes which encoded iron-sulfur proteins (Figure 3B). The similar tendency of AvrA and its target genes was also turned out by Real-time PCR (Figure 3C).
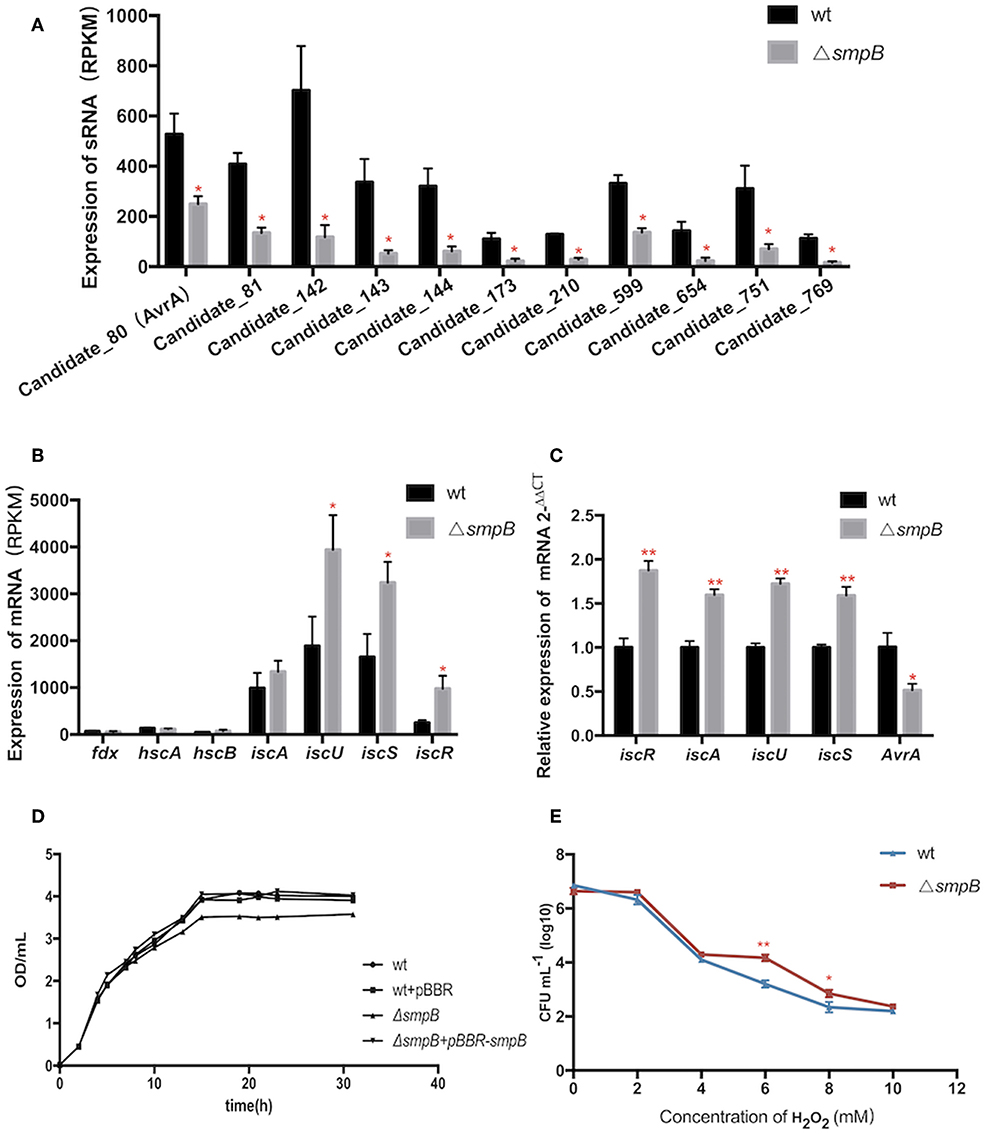
Figure 3. The AvrA downregulation and IscR upregulation mediate the oxidative tolerance of ΔsmpB. (A) Transcriptomic comparisons of 11 sRNA between wild type and ΔsmpB. (B) Transcriptomic comparisons of iscR operon genes between wild type and ΔsmpB. (C) Realtime qPCR detection of iscR operon and AvrA. (D) The growth curves of the derivative A. veronii. (E) The survivals of ΔsmpB and wild type in H2O2 concentrations ranging from 0 to 10 mM. **p < 0.01, *p < 0.05, following by one-way ANOVA, independent t-tests.
To evaluate whether smpB mutation lived better in stress conditions than the wild type, the iron deficiency condition was performed to mimic the host environment and the tolerance to hydrogen peroxide oxidation (H2O2) was tested. The smpB mutation exhibited a slow growth compared with the wild type and the complementary strain (Figure 3D). The ΔsmpB survived better than the wild type when the H2O2 concentration ranged from 6 to 10 mM, revealing that ΔsmpB was endowed with a stronger capability of growth under adverse conditions (Figure 3E).
AvrA Deletion Increases the Tolerances to Iron Deficiency and Oxidative Stress by Upregulating Iron-Sulfur Gene Expression
AvrA was a non-coding RNA of 253nt, and its secondary structure was predicted by the software RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) (Figure 4A). The growth results demonstrated that the AvrA deletion presented a better ability to grow under dipyridyl (DIP)-chelated iron deficiency state (Figure 4B). Also, the oxidation resistance of ΔavrA was evaluated under 0–10 mM H2O2 in comparison with the wild type and the complemented strains, showing that the antioxidation capacity was enhanced after AvrA knockout (Figure 4C). Realtime qPCR results showed that the transcriptional expressions of the iron-sulfur genes iscR, iscU, iscS, and iscA were significantly increased (Figure 4D).
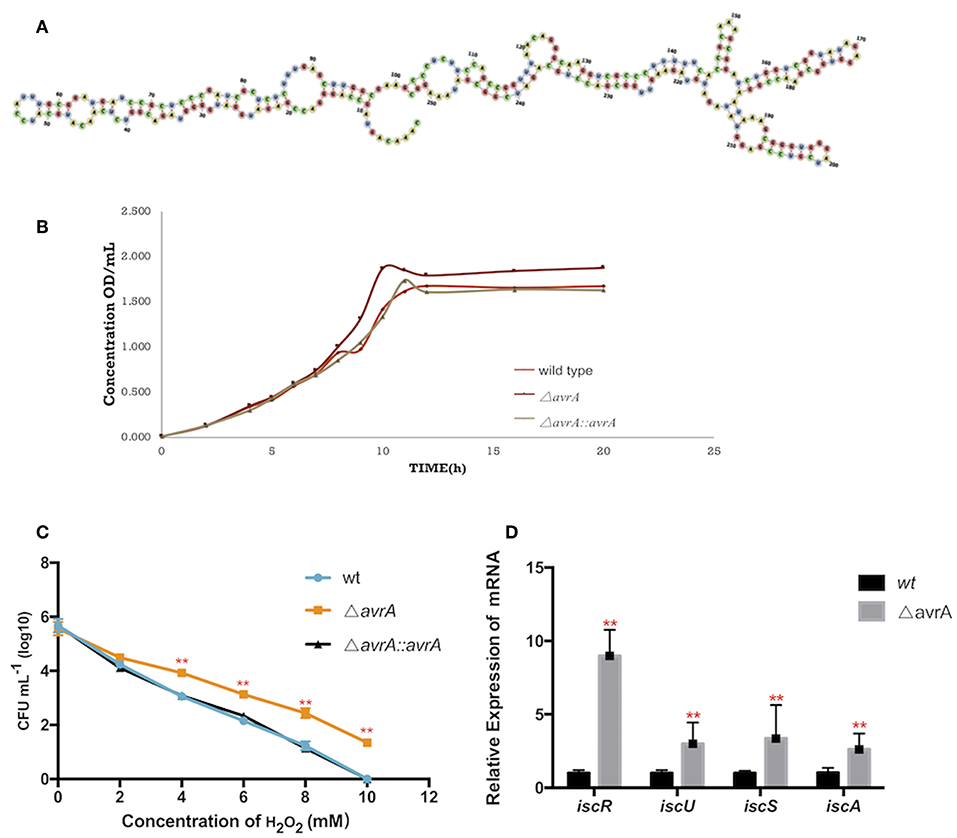
Figure 4. AvrA increases the tolerances to oxidative stress and iron deficiency. (A) The secondary structure prediction of AvrA by software RNAfold. (B) Growth curves of wild type, ΔavrA and the complemented strain in iron deficiency treatment. (C) The survivals of wild type, ΔavrA and the complemented strain under H2O2 stress in the concentrations ranging from 0 to 10 mM (D) Relative expression of iscR operon at stationary phage in iron deficiency treating with DIP. **p < 0.01, following by one-way ANOVA, independent t-tests.
AvrA Binds and Downregulates iscR mRNA by Base Pairing
The potential interaction site was predicted between AvrA and the iscR promotor using the RNAplex program. Interestingly, the base-pairing region encompassed the ribosome binding site and the start codon AUG of iscR, and the binding site spanned from C37 to A63 of 5′-end AvrA (Figure 5A). Two mutations, P1 (from A2 to C5, ACUC) and P2 (from C−18 to G−13, GCGUUC), were introduced in the iscR promoter (piscR) and fused with the enhanced green fluorescent protein (eGFP) to produce Φ(piscR mutP1-eGFP) and Φ(piscRmutP2-eGFP), respectively (Figures 5A,B). When AvrA was co-expressed with Φ(piscR-eGFP), the fluorescence was repressed, suggesting that AvrA could interact with the promotor of iscR by base pairing (Figure 5C). Furthermore, the fluorescence recovered when AvrA was co-transformed with Φ(piscRmutP1-eGFP), revealing that P1 was the exact site for interaction (Figure 5C). Next, a mutation named pavrAmutP1 was introduced in the AvrA expression plasmid pavrA to match Φ (piscR mutP1-eGFP), resulting in fluorescent repression. Conversely the mutated pavrAmutP1 did not reduce the fluorescence when co-expressed with the wild type Φ(iscR-eGFP) (Figure 5D). The results concluded that AvrA repressed the expression of iscR by base pairing with the iscR translation initiation region P1.
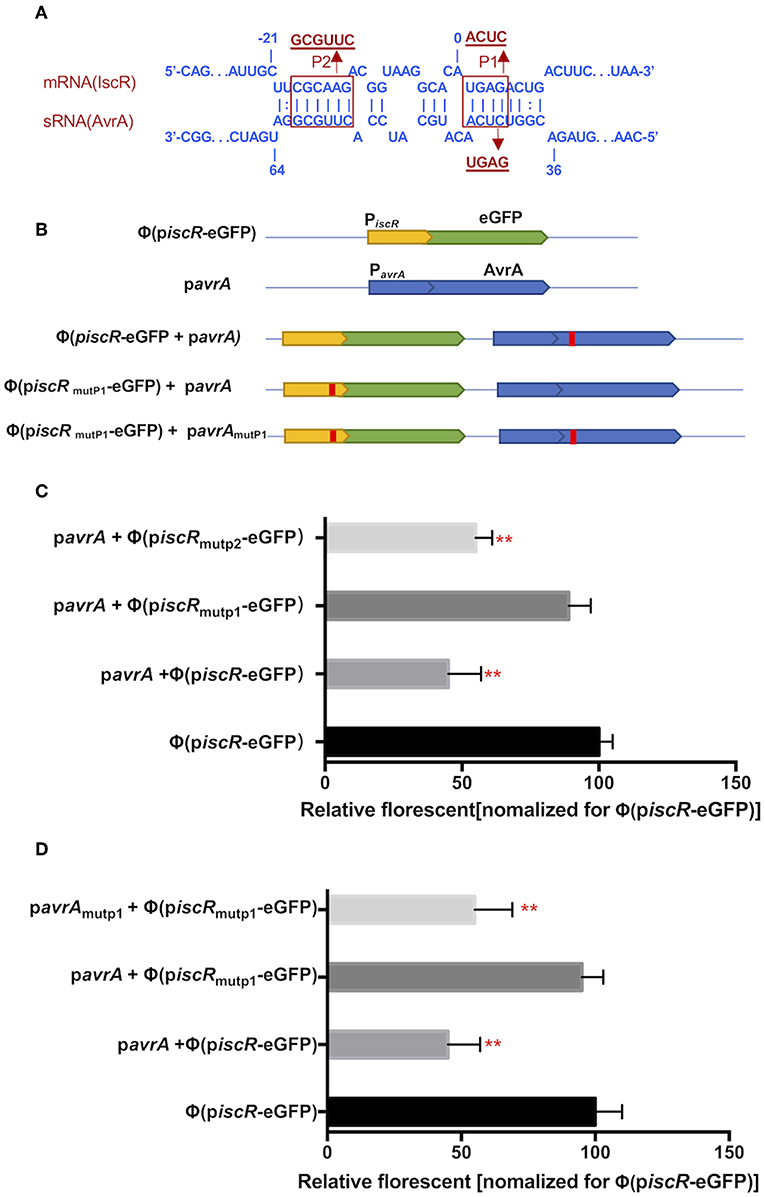
Figure 5. AvrA interacts with iscR mRNA by base pairing. (A) Predicted interaction between AvrA and iscR mRNA by the RNAplex program. (B) Basic architecture of the plasmid constructs. (C,D) The relative florescence was measured at 488 nm excitation and 520 nm emission lights after the corresponding constructs were co-transformed in E. coli strain. **p < 0.01, following by one-way ANOVA, independent t-tests.
Discussion
SmpB is not only the key component of the trans-translation system, but also plays an important role as a transcription regulator in the global regulation of bacteria (Liu et al., 2016). In our study, the smpB deletion affected the transcriptional expression of the genes related to the bacterial growth, metabolism and protein secretion in multiple metabolic pathways in A. veronii. Previously Type II, III, and VI secretion systems were reported in multiple strains of Aeromonas bacteria, which function by exporting bacterial virulence proteins and by infecting host cells (Suarez et al., 2009; Journet and Cascales, 2016). The gene expression of the Type III secretion system had extremely low abundance, while those of Type VI performed higher in the transcriptome. Most of all, virulence-related genes which mediated the pili assembly (type IV pilus) and protein secretion (type VI secretion system, TSSS) were totally downregulated (Figure 1). Previously, the type VI secretion system was identified to function as the virulence factor in Vibrio hollisae and Aeromonas hydrophila (Russell et al., 2014). The colonization of wild type A. veronii was significantly higher than that of ΔsmpB. However, the survival rates of the infected mice showed no significant differences between the wild type and ΔsmpB strain. Previously the smpB deletion mutant of Listeria monocytogenes and Salmonella Typhimurium were avirulent and deficient in the intramacrophage survival (Kivisaar, 2003; Mraheil et al., 2017) The results were inconsistent with past reports, indicating that there are other pathways maintaining the virulence in A. veronii smpB mutants. Subsequently, sRNA AvrA was revealed as an alternative for virulence compensation. The downregulation of AvrA affected the expression of IscR in ΔsmpB A. veronii. In case of iron deficiency, IscR enhanced iron-sulfur cluster assembly and iron sulfur protein synthesis (Santos et al., 2014), and amended the antioxidant ability of A. veronii.
When bacteria infect the host, they are subjected to stressful conditions such as strong oxidization, deficient nutrition and metal ion. The harsh conditions cause starvation, accumulation of secondary metabolites, decreased fidelity of DNA replication and the reduction in DNA repair activity which are similarly encountered during the stationary stage (De Biase et al., 1999; Kivisaar, 2003). Most sRNAs are expressed as important regulators in the process of gene regulation under adverse stress (Michaux et al., 2014; Amin et al., 2016; Holmqvist and Wagner, 2017). The sRNA AvrA was uncovered to function by base pairing with its target gene iscR. IscR regulated a set of genes involved in iron-sulfur cluster assembly in adversity like iron deficiency and oxidative stress (Figure 6). However, the transcription factors associated with oxidative stress, including Fur (ferric uptake regulation protein), OxyR (hydrogen peroxide-inducible genes activator), SoxR (hydrogen peroxide-inducible genes activator) were insignificantly expressed by comparing ΔsmpB strains the with wild type strains (Supplementary Table 2). The mutation of smpB resulted in the downregulation of sRNA AvrA, which leads to the upregulation of iscR expression and the enhancement of the adverse resistance. Combined with the attenuated virulence and the increased survival in adversity in ΔsmpB, the strains performed a similar lethality in mice compared with the wild type. Virulence is a trait that has been selected in evolutionary history (Rafaluk et al., 2015; Gerstein and Nielsen, 2017). Therefore, the survival viability in the macrophage and the tolerance to adverse environments caused by smpB mutation compensates for the loss of function in virulence, causing A. veronii to evolve as a superbug.
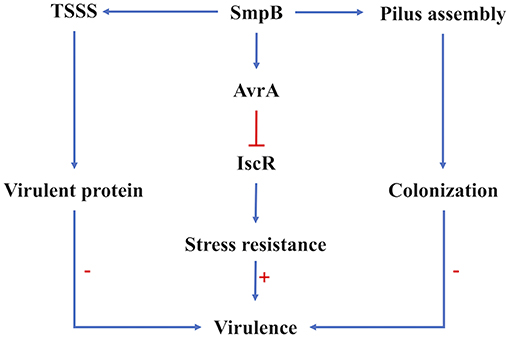
Figure 6. Regulation of SmpB in virulence regulation. The virulence of A. veronii was affected by the expression of virulent proteins the ability of host colonization and the resistance to adversity. Type VI secretion system and pilus assembly were affected on the transcription and translation level by SmpB regulation directly. The sRNA AvrA functioned the stress resistance directly, which was mediated by SmpB indirectly. The arrows in blue showed the positive regulation, and the line in red showed repression. The symbol “+”, “–” indicated upregulation and downregulation, respectively.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
ZL, XM, and DW contributed the conception and design of the study. DW, HL, YT, HT, and ZL performed the statistical analysis. DW, HL, XH, and ZL drafted the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This work was supported by grants from the Hainan Natural Science Foundation Nos. 317015 (to YT) and 319QN161 (to HL), and the Science and Technology Program of Hainan Province ZDYF2017020 (to ZL).This work was also partially supported by grants from the National Natural Science Foundation of China Nos. 31560021, 31772887 (to ZL), and 31860676 (to YT).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00142/full#supplementary-material
References
Aguilera-Arreola, M. G., Hernandez-Rodriguez, C., Zuniga, G., Figueras, M. J., Garduno, R. A., and Castro-Escarpulli, G. (2007). Virulence potential and genetic diversity of Aeromonas caviae, Aeromonas veronii, and Aeromonas hydrophila clinical isolates from Mexico and Spain: a comparative study. Can. J. Microbiol. 53, 877–887. doi: 10.1139/W07-051
Amin, S. V., Roberts, J. T., Patterson, D. G., Coley, A. B., Allred, J. A., and Denner, J. M., et al. (2016). Novel small RNA (sRNA) landscape of the starvation-stress response transcriptome of Salmonella enterica serovar typhimurium. RNA Biol. 13, 331–342. doi: 10.1080/15476286.2016.1144010
Carrier, M.-C., Bourassa, J.-S., and Massé, E. (2017). Cellular homeostasis: a small RNA at the crossroads of iron and photosynthesis. Curr. Biol. 27, R380–R383. doi: 10.1016/j.cub.2017.04.003
De Biase, D., Tramonti, A., Bossa, F., and Visca, P. (1999). The response to stationary-phase stress conditions in Escherichia coli : role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32, 1198–1211.
Durand, S., Tomasini, A., Braun, F., Condon, C., and Romby, P. (2015). sRNA and mRNA turnover in gram-positive bacteria. FEMS Microbiol. Rev. 39, 316–330. doi: 10.1093/femsre/fuv007
Fiedler, M. R. M., Cairns, T. C., Koch, O., Kubisch, C., and Meyer, V. (2018). Conditional expression of the small GTPase ArfA impacts secretion, morphology, growth, and actin ring position in Aspergillus niger. Front. Microbiol. 9:878. doi: 10.3389/fmicb.2018.00878
Gerstein, A. C., and Nielsen, K. (2017). It's not all about us: evolution and maintenance of Cryptococcus virulence requires selection outside the human host. Yeast 34, 143–154. doi: 10.1002/yea.3222
Haines, S., Arnaud-Barbe, N., Poncet, D., Reverchon, S., Wawrzyniak, J., and Nasser, W., et al. (2015). IscR regulates synthesis of colonization factor antigen I fimbriae in response to iron starvation in enterotoxigenic Escherichia coli. J. Bacteriol. 197, 2896–2907. doi: 10.1128/JB.00214-15
Himeno, H., Kurita, D., and Muto, A. (2014). tmRNA-mediated trans-translation as the major ribosome rescue system in a bacterial cell. Front. Genet. 5:66. doi: 10.3389/fgene.2014.00066
Holmqvist, E., and Wagner, E. G. H. (2017). Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 45, 1203–1212. doi: 10.1042/BST20160363
Huter, P., Muller, C., Arenz, S., Beckert, B., and Wilson, D. N. (2017). Structural basis for ribosome rescue in bacteria. Trends Biochem. Sci. 42, 669–680. doi: 10.1016/j.tibs.2017.05.009
Ilbert, M., and Bonnefoy, V. (2013). Insight into the evolution of the iron oxidation pathways. Biochim. Biophys. Acta 1827, 161–175. doi: 10.1016/j.bbabio.2012.10.001
Journet, L., and Cascales, E. (2016). The type VI secretion system in Escherichia coli and related species. EcoSal Plus 7:20. doi: 10.1128/ecosalplus.ESP-0009-2015
Keiler, K. C. (2008). Biology of trans-translation. Annu. Rev. Microbiol. 62, 133–151. doi: 10.1146/annurev.micro.62.081307.162948
Keiler, K. C. (2015). Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 13, 285–297. doi: 10.1038/nrmicro3438
Kivisaar, M. (2003). Stationary phase mutagenesis: mechanisms that accelerate adaptation of microbial populations under environmental stress. Environ. Microbiol. 5, 814–827. doi: 10.1046/j.1462-2920.2003.00488.x
Kurita, D., Miller, M. R., Muto, A., Buskirk, A. R., and Himeno, H. (2014). Rejection of tmRNA·SmpB after GTP hydrolysis by EF-Tu on ribosomes stalled on intact mRNA. RNA 20, 1706–1714. doi: 10.1261/rna.045773.114
Li, J., Ji, L., Shi, W., Xie, J., and Zhang, Y. (2013). Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrobial Chemother. 68, 2477–2481. doi: 10.1093/jac/dkt231
Liang, W., and Deutscher, M. P. (2010). A novel mechanism for ribonuclease regulation: transfer-messenger RNA (tmRNA) and its associated protein SmpB regulate the stability of RNase R. J. Biol. Chem. 285, 29054–29058. doi: 10.1074/jbc.C110.168641
Liu, P., Chen, Y., Wang, D., Tang, Y., Tang, H., and Song, H., et al. (2016). Genetic selection of peptide aptamers that interact and inhibit both small protein B and alternative ribosome-rescue factor A of Aeromonas veronii C4. Front. Microbiol. 7:1228. doi: 10.3389/fmicb.2016.01228
Liu, Z., Hu, K., Tang, Y., Li, H., Tang, H., and Hu, X., et al. (2018). SmpB down-regulates proton-motive force for the persister tolerance to aminoglycosides in Aeromonas veronii. Biochem. Biophys. Res. Commun. 507, 407–413. doi: 10.1016/j.bbrc.2018.11.052
Liu, Z., Liu, P., Liu, S., Song, H., Tang, H., and Hu, X. (2015). Small protein B upregulates sensor kinase bvgS expression in Aeromonas veronii. Front. Microbiol. 6:579. doi: 10.3389/fmicb.2015.00579
Mandin, P., Chareyre, S., and Barras, F. (2016). A regulatory circuit composed of a transcription factor, IscR, and a regulatory RNA, RyhB, controls Fe-S cluster delivery. mBio 7:e00966-16. doi: 10.1128/mBio.00966-16
Michaux, C., Verneuil, N., Hartke, A., and Giard, J.-C. (2014). Physiological roles of small RNA molecules. Microbiology 160(Pt. 6), 1007–1019. doi: 10.1099/mic.0.076208-0
Mraheil, M. A., Frantz, R., Teubner, L., Wendt, H., Linne, U., and Wingerath, J., et al. (2017). Requirement of the RNA-binding protein SmpB during intracellular growth of Listeria monocytogenes. Int. J. Med. Microbiol. 307, 166–173. doi: 10.1016/j.ijmm.2017.01.007
Mu, X., Huan, H., Xu, H., Gao, Q., Xiong, L., and Gao, R., et al. (2013). The transfer-messenger RNA-small protein B system plays a role in avian pathogenic Escherichia coli pathogenicity. J. Bacteriol. 195, 5064–5071. doi: 10.1128/JB.00628-13
Oliva, G., Sahr, T., and Buchrieser, C. (2015). Small RNAs, 5′ UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS Microbiol. Rev. 39, 331–349. doi: 10.1093/femsre/fuv022
Rafaluk, C., Gildenhard, M., Mitschke, A., Telschow, A., Schulenburg, H., and Joop, G. (2015). Rapid evolution of virulence leading to host extinction under host-parasite coevolution. BMC Evol. Biol. 15:112. doi: 10.1186/s12862-015-0407-0
Roche, B., Aussel, L., Ezraty, B., Mandin, P., Py, B., and Barras, F. (2013). Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta 1827, 455–469. doi: 10.1016/j.bbabio.2012.12.010
Russell, A. B., Wexler, A. G., Harding, B. N., Whitney, J. C., Bohn, A. J., and Goo, Y. A., et al. (2014). A type VI secretion-related pathway in bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236. doi: 10.1016/j.chom.2014.07.007
Santos, J. A., Alonso-Garcia, N., Macedo-Ribeiro, S., and Pereira, P. J. (2014). The unique regulation of iron-sulfur cluster biogenesis in a Gram-positive bacterium. Proc. Natl. Acad. Sci. U S A. 111, E2251–E2260. doi: 10.1073/pnas.1322728111
Suarez, G., Sierra, J. C., Erova, T. E., Sha, J., Horneman, A. J., and Chopra, A. K. (2009). A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168. doi: 10.1128/jb.01260-09
Torii, Y., Goto, Y., Nakahira, S., Kozaki, S., Kaji, R., and Ginnaga, A. (2015). Comparison of Systemic toxicity between botulinum toxin subtypes A1 and A2 in mice and rats. Basic Clin. Pharmacol. Toxicol. 116, 524–528. doi: 10.1111/bcpt.12351
Xu, X. M., and Moller, S. G. (2008). Iron-sulfur cluster biogenesis systems and their crosstalk. Chembiochem 9, 2355–2362. doi: 10.1002/cbic.200800384
Keywords: oxidative, stress, iron deficiency, iron/sulfur clusters, virulence
Citation: Wang D, Li H, Ma X, Tang Y, Tang H, Hu X and Liu Z (2019) Small RNA AvrA Regulates IscR to Increase the Stress Tolerances in SmpB Deficiency of Aeromonas veronii. Front. Cell. Infect. Microbiol. 9:142. doi: 10.3389/fcimb.2019.00142
Received: 14 March 2019; Accepted: 17 April 2019;
Published: 08 May 2019.
Edited by:
Xiangmin Lin, Fujian Agriculture and Forestry University, ChinaReviewed by:
Shengkang Li, Shantou University, ChinaZhihong Xie, Yantai Institute of Coastal Zone Research (CAS), China
Copyright © 2019 Wang, Li, Ma, Tang, Tang, Hu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinwen Hu, aHV4aW53ZW5AaGFpbnUuZWR1LmNu
Zhu Liu, emh1bGl1QGhhaW51LmVkdS5jbg==
 Dan Wang
Dan Wang Hong Li
Hong Li