- 1State Key Laboratory for Infectious Disease Prevention and Control, Chinese Centre for Disease Control and Prevention, National Institute for Communicable Disease Control and Prevention, Beijing, China
- 2Department of Respiratory Medicine, Zhejiang Provincial People's Hospital, People's Hospital of Hangzhou Medical College, Hangzhou, China
- 3Department of Bacteriology I, National Institute of Infectious Diseases, Tokyo, Japan
- 4Department of Health Sciences, Saitama Prefectural University, Saitama, Japan
- 5Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Hangzhou, China
Approximately 85% of cases of Legionnaires' disease are caused by Legionella pneumophila serogroup 1. In this study, we analyzed the distribution of lag-1 alleles, ORF 7 and ORF 8 genes of lipopolysaccharide (LPS) and sequence-based types of 616 L. pneumophila serogroup 1 strains isolated in Japan (206 clinical, 225 environmental) and China (13 clinical and 172 environmental). The lag-1 gene was harbored by significantly more of the clinical isolates compared with the environmental isolates (90.3 vs. 19.1% and 61.6 vs. 3.0%, respectively; both P < 0.001). ORF 7 genes were detected in 51.0% of Japanese clinical and 36.0% of Japanese environmental (P = 0.001) isolates, as well as 15.3% of Chinese clinical and 9.9% of Chinese environmental isolates (P = 0.544). ORF 8 genes were detected in 12.1% of Japanese clinical and 5.8% of Japanese environmental (P = 0.017) isolates, as well as 7.7% of Chinese clinical and 3.4% of Chinese environmental isolates (P = 0.388). The Japanese and Chinese isolates were assigned to 203 and 36 different sequence-types (ST), respectively. ST1 was predominant. Most isolates with the same ST also had the same lag-1, ORF 7, and ORF 8 gene subgroups. In conclusion, the lag-1 was present in most of the clinical isolates, but was absent from most of the environmental isolates from both China and Japan, regardless of the water source and SBT type. PCR-based serotyping and subgrouping methods can be used to define a hierarchy of virulence genotypes that require stringent surveillance to prevent human disease.
Introduction
Legionella, the causative agent of Legionnaires' disease, is a facultative intracellular Gram-negative bacteria that is ubiquitous in natural and man-made water systems (Rowbotham, 1980). These bacteria live parasitically in protozoa and can be found within biofilms (Rowbotham, 1980; Fields et al., 2002; Declerck, 2010; Stewart et al., 2012). Legionella is transmitted through inhaled aerosols and subsequently, enters and grows within human monocytes and alveolar macrophages, which can lead to a fatal form of pneumonia known as Legionnaires' disease (Isberg et al., 2009). To date, more than 60 species of Legionella and 70 serogroups (sg) have been identified (Viasus et al., 2019). Among them, Legionella pneumophila is the most common, causing ~90% of all cases of Legionnaires' disease (Viasus et al., 2019). At least 16 serogroups of L. pneumophila have now been identified, with serogroup 1 (sg1) predominating and causing approximately 85% of all cases of Legionnaires' disease (Fields et al., 2002; Yu et al., 2002; Carratala and Garcia-Vidal, 2010).
Lipopolysaccharide (LPS) is one of the most important outer membrane components and the major immune-dominant antigen for all the Legionella species (Ciesielski et al., 1986; Petzold et al., 2013). LPS consists of three parts: lipid A, core oligosaccharides, and polysaccharide (o) side-chains (Ciesielski et al., 1986). Furthermore, a genetic locus composed of at least 28 open reading frames (ORF) is essential in the biosynthesis of LPS core oligosaccharide and LPS polysaccharide O-chains (Petzold et al., 2013). The high diversity of LPS molecules provides the basis for classifying L. pneumophila into different serogroups and subgroups by detection using specific monoclonal antibodies (mAb) (Joly et al., 1983; Helbig et al., 1997, 2007). The Dresden Panel of mAbs, including mAb 3/1, was developed in 1997 to distinguish among heterogeneous bacterial groups belonging to L. pneumophila sg1 (Helbig et al., 2002). Moreover, the monoclonal types of sg1 were subdivided according to the presence or absence of the virulence-related epitope recognized by mAb 3/1. This epitope is not expressed by strains belonging to any of the other serogroups (Helbig et al., 2002). Surveys indicate that 65–100% of L. pneumophila sg1 clinical isolates react with mAb 3/1. In contrast, only 14.9–35% of L. pneumophila sg1 environmental isolates reacted with mAb 3/1 (Dournon et al., 1988; Harrison et al., 2007, 2009; Watkins et al., 2009). Mab 3/1 recognizes an epitope within the 8-O-acetylation group of the legionaminic acid of L. pneumophila sg1 LPS (Helbig et al., 1995; Zähringer et al., 1995; Zou et al., 1999). The lag-1 gene encodes an O-acetyltransferase responsible for the 8-O-acetylation of legionaminic acid (Zou et al., 1999). Studies have shown that all mAb 3/1-postitive isolates harbor the lag-1 gene, whereas mAb 3/1-negative isolates are either devoid of lag-1 or contain missense mutations or insertions within this gene (Bernander et al., 2003).
The distribution of lag-1 alleles and LPS genes in L. pneumophila sg1 isolates can be used to discriminate serogroups and monoclonal subgroups of the most common L. pneumophila serogroup 1. A previous study has established simple and rapid genotyping methods for culture-independent discrimination between serogroups of L. pneumophila and monoclonal subgroups of serogroup 1 (Thurmer et al., 2009). They identified a serogroup 1-specific genomic region, and developed two independent and suitable PCR assays for detecting serogroup 1 strains of L. pneumophila (Thurmer et al., 2009). Based on their research, we used conventional PCR methods to identify L. pneumophila sg1 subgroups in clinical and environmental isolates from Japan and China. We further characterized these isolates using the sequence-based typing (SBT) method, which is the most widely used molecular tool for epidemiological typing of L. pneumophila (Gaia et al., 2005; Ratzow et al., 2007).
Materials and Methods
Ethics
Ethical approval for this study was obtained from the meeting of ethics committee of National Institute for Communicable Disease Control and Prevention, China CDC.
L. pneumophila sg1 Isolates
Two hundred and six clinical isolates were collected from the National Institute of Infectious Diseases, Japan (NIID, Japan). All the isolates were gathered from different patients from 30 prefectures between 2000 and 2015. There was no known epidemiologic linkage among these isolates. One hundred and ninety-eight of the isolates were from sporadic cases, and eight were from separate outbreaks occurring in seven prefectures. Two hundred and twenty-five environmental isolates were also collected from the NIID, Japan. The sources of these isolates were: bath water (92 isolates), cooling tower water (49 isolates), soil (36 isolates), shower water (30 isolates), and fountain (18 isolates).
Thirteen clinical isolates were collected from the Chinese Center for Disease Control and Prevention (China CDC). All the isolates were gathered from different patients, and they were obtained from six provinces between 1988 and 2016. All the isolates were sporadic cases. One hundred and seventy-two environment isolates were also collected from the China CDC. They were obtained from seven provinces between 2005 and 2016. The sources of these isolates were: air conditioner water (40 isolates), hot spring water (25 isolates), cooling tower water (65 isolates), and pipe water (42 isolates).
All these isolates were cultured on buffered charcoal yeast extract (BCYE) agar plates (Oxoid, Germany) for 48 h at 37°C under a 5% CO2 atmosphere.
DNA Extraction and PCR Amplification
The target DNA of all the isolates were extracted using a QIAamp DNA Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. The sequences of all the serogroup 1-specific and subgroup-specific primers used are listed in Supplementary Table S1. The “lag-1” primers were used for amplification of the internal lag-1 fragment in all the stains. The “lag-1” primer pairs were used as “consensus” primers for all the strains. According to a previous study, lag-1 can be subtyped into “lag-1 Philadelphia,” “lag-1 Knoxville,” and “lag-1 Allentown” (Thurmer et al., 2009). The “intergenic region ORF 6–8,” “intergenic region ORF 7–9,” primers were used for amplification of “subgroup Benidorm/Bellingham” and “subgroup Knoxville” fragments, respectively, in the previous study (Thurmer et al., 2009). And in this study we named “intergenic region ORF 6–8,” “intergenic region ORF 7–9,” primers as “ORF 7,” “ORF 8,” respectively (listed in Supplementary Table S1). And the loci of “ORF 7,” “ORF 8” were shown in a schematic representation (Supplementary Figure S1). All PCRs were performed in thermal cyclers (SensoQuest LabCycler, Senso, Germany or LifeECO Thermal Cycler, BIOER, China) using Quick Taq HS DyeMix (Toyobo, Japan) by the following cycling parameters: denaturation at 94°C for 2 min; followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 1 min; and a final extension at 68°C for 7 min. The PCR products were resolved by 2.0% agarose gel electrophoresis and stained with GoldView or ethidium bromide, visualized under UV light, and analyzed using a Gel Doc system (BioRad, CA, USA).
Reactivity With lag-1, ORF 7, and ORF 8
For lag-1-positive strains, the lag-1 gene fragments were amplified by using the “lag-1” PCR primers shown in Supplementary Table S1. The lag-1 gene fragments cannot be detected in lag-1 (–) strains. The lag-1-positive strains were subtyped as “lag-1 Philadelphia,” “lag-1 Knoxville,” and “lag-1 Allentown” using the primers shown in Supplementary Table S1 and the positive strains were designated “lag-1P,” “lag-1K,” and “lag-1A,” respectively, whereas the negative strains were designated “lag-1O.” That is the “lag-1 Philadelphia,” “lag-1 Knoxville,” and “lag-1 Allentown” gene fragments cannot be detected in “lag-1O” strains (Tables 1–4).
All the strains were also subtyped by the “ORF 7” and “ORF 8” primers shown in Supplementary Table S1. The ORF 7-positive strains were defined as “variation of ORF 7” and the ORF 8-positive strains were defined as “variation of ORF 8,” respectively. Strains that were not detected by either “ORF 7” or “ORF 8” primers were defined as “normal ORF 7 and ORF 8” (Tables 1–4).
All these PCR products were verified by sequencing.
SBT
Genotyping was performed via the SBT protocol defined by the European Working Group for Legionella Infections (EWGLI) (www.ewgli.org). Genomic DNA was extracted and then amplified using primers targeting seven specific gene loci (i.e., flaA, pilE, asd, mip, mompS, proA, and neuA). Amplicons were sequenced with specific primers, and the resulting consensus sequences trimmed and compared to previously assigned allele numbers available on the website (http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/sbt_homepage.php). According to a pre-determined order (i.e., flaA, pilE, asd, mip, mompS, proA, and neuA), the combination of alleles was defined as seven-digit allelic profile (e.g., 1, 1, 1, 1, 1, 1, and 1) and a sequence Type (ST) represented by a number (e.g., ST1). Newly identified STs were submitted to the EWGLI SBT database (http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/sbt_homepage.php). Minimum spanning trees were constructed based on SBT profiles using BioNumerics 7.1 software (Applied Maths, Kortrijk, Belgium). A part of SBT profiles of some strains have been reported (Amemura-Maekawa et al., 2010, 2012, 2018).
Statistical Analysis
Data were analyzed with the Pearson χ2 test using SPSS version 17.0 software (IBM SPSS, United States). When any expected number in the 2 × 2 contingency table was <5 and ≥1, the P-value was calculated using a continuity correction test. P < 0.05 was considered to indicate statistical significance.
ATCC 33152
The DNA of legionella pneumophila type strain (pneumophila Philadelphia-1 ATCC 33152) was also extracted and amplified using primers listed in Supplementary Table S1.
Results
Subgroups of sg1 Isolates
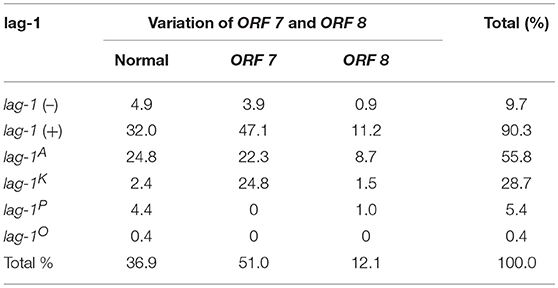
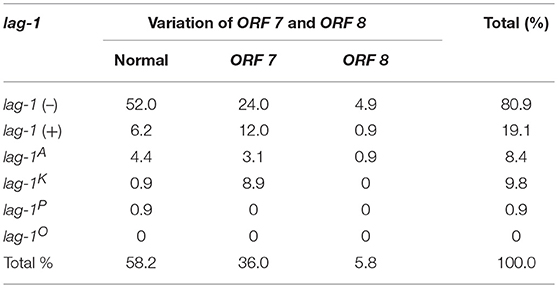
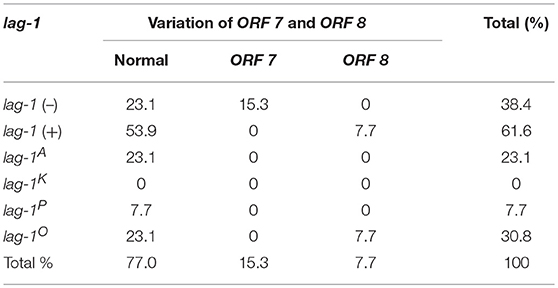
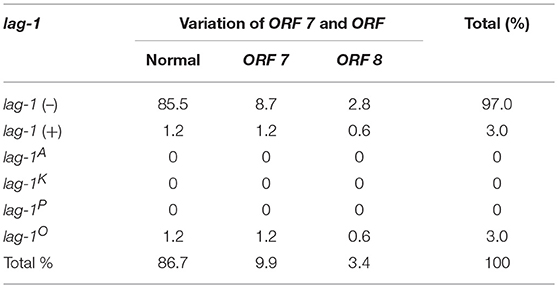
According to the reactivity with lag-1, ORF 7, and ORF 8, all the isolates were divided into 15 subgroups (Tables 1–4). Because the number of isolates for “lag-1, variation of ORF 8” is zero, there were 14 subgroups listed in Figure 1, finally.
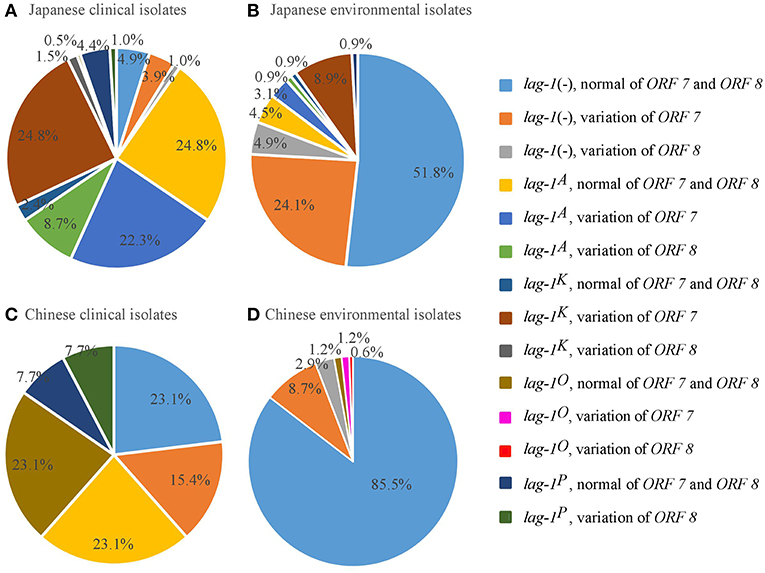
Figure 1. Distribution of lag-1 alleles, and ORF 7 and ORF 8 genes of LPS among L. pneumophila serogroup 1 isolates from Japan and China. (A) Distribution of lag-1 alleles, ORF 7 and ORF 8 genes of LPS among Japanese clinical isolates (A), Japanese environmental isolates (B), Chinese clinical isolates (C), and Chinese environmental isolates (D).
Among the Japanese isolates, 90.3% of clinical isolates and 19.1% of the environmental isolates harbored the lag-1 gene (P < 0.001 based on χ2 test on proportions) (Tables 1, 2, Figures 1A,B). Among the Chinese isolates, 61.6% of the clinical isolates and 3.0% of the environmental isolates harbored the lag-1 gene (P < 0.001 based on χ2 test on proportions) (Tables 3, 4, Figures 1C,D). These results demonstrated that the lag-1 was present in most of the clinical isolates but absent from most of the environmental isolates for both the Chinese and Japanese isolates.
Among the Japanese isolates, 51.0% of the clinical isolates and 36.0% of the environmental isolates harbored the ORF 7 gene (P < 0.001 based on χ2 test on proportions) (Tables 1, 2, Figures 1A,B). Among the Chinese isolates, 15.3% of the clinical isolates and 9.9% of the environmental isolates harbored the ORF 7 gene (P = 0.544 based on χ2 test on proportions) (Tables 3, 4, Figures 1C,D).
Among the Japanese isolates, 12.1% of clinical isolates and 5.8% of the environmental isolates harbored the ORF 8 gene (P = 0.017 based on χ2 test on proportions) (Tables 1, 2, Figures 1A,B). Among the Chinese isolates, 7.7% of the clinical isolates and 3.4% of the environmental isolates harbored the ORF 8 gene (P = 0.388 based on χ2 test on proportions) (Tables 3, 4, Figures 1C,D).
Environmental Strains Classified According to Different Sources
According to the presence of the specific alleles of lag-1, ORF 7, ORF 8, and the different sources, all the environmental strains were classified and are listed in Supplementary Tables S2, S3. Only 2% (1/49) of Japanese cooling tower water isolates and 6% (4/65) of Chinese cooling tower water isolates harbored the lag-1 gene. So as other environmental water sources. These data showed that, for both the Chinese and Japanese isolates, most of the environmental water isolates did not harbor the lag-1 gene, regardless of the source.
SBT Analysis
Based on SBT analysis, all the Japanese L. pneumophila sg1 isolates were assigned to 159 different STs (Supplementary Tables S4, S5) and all the Chinese L. pneumophila sg1 isolates were assigned to 36 different STs (Supplementary Tables S6, S7). Among these STs, ST1 was the most common for both the Japanese and Chinese isolates. For the clinical isolates, 2.4% (5/206) of the Japanese isolates and 0% of the Chinese isolates were classified as ST1. In contrast, for the environmental isolates, 35.6% (80/225) of the Japanese isolates and 57.6 % (99/172) of the Chinese isolates were classified as ST1.
Minimum spanning trees were constructed based on the SBT profile grouped according to countries and sources (Figure 2A), lag-1 subgroups (Figure 2B) and variations of ORF 7 and ORF 8 (Figure 2C). By comparing the three minimum spanning trees, we identified a correlation between ST and subgroups of sg1 (Figure 2). Most isolates with same ST also had the same lag-1, ORF 7 and ORF 8 gene subgroups. For example, all ST48 isolates had ORF 7 variation but no lag-1 gene; all ST42 isolates harbored the lag-1K gene and ORF 7 variation; all ST120 isolates harbored the lag-1A gene and ORF 7 variation; all ST23 isolates harbored the lag-1A gene and normal ORF 7 and ORF 8 genes; most ST1 isolates were environmental isolates devoid of the lag-1 gene and with normal ORF 7 and ORF 8 genes. This correlation between ST and subgroups was also presented by an evolutionary tree based on seven targeting specific gene loci (Figure 3).
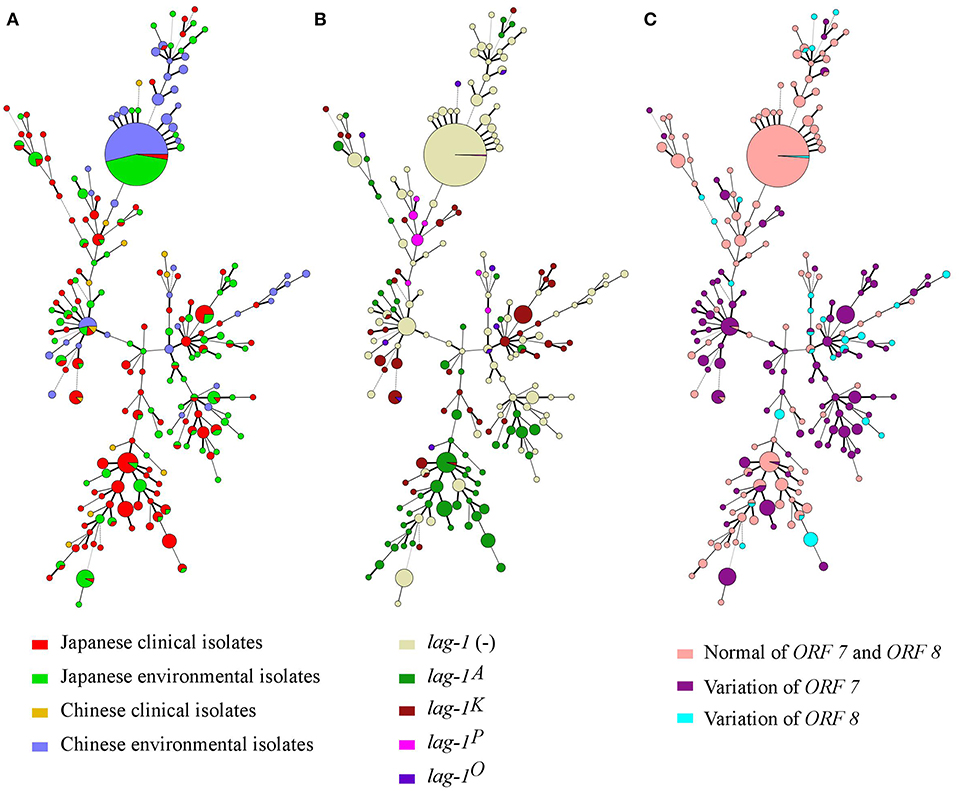
Figure 2. Minimum spanning tree analysis of 616 L. pneumophila serogroup 1 strains isolated in Japan and China. STs are shown as circles. The size of each circle indicates the number of isolates within this particular type. The 616 L. pneumophila serogroup 1 strains were divided into groups by countries and sources (A), lag-1 alleles (B), and Distribution of ORF 7 and ORF 8 genes of LPS (C).
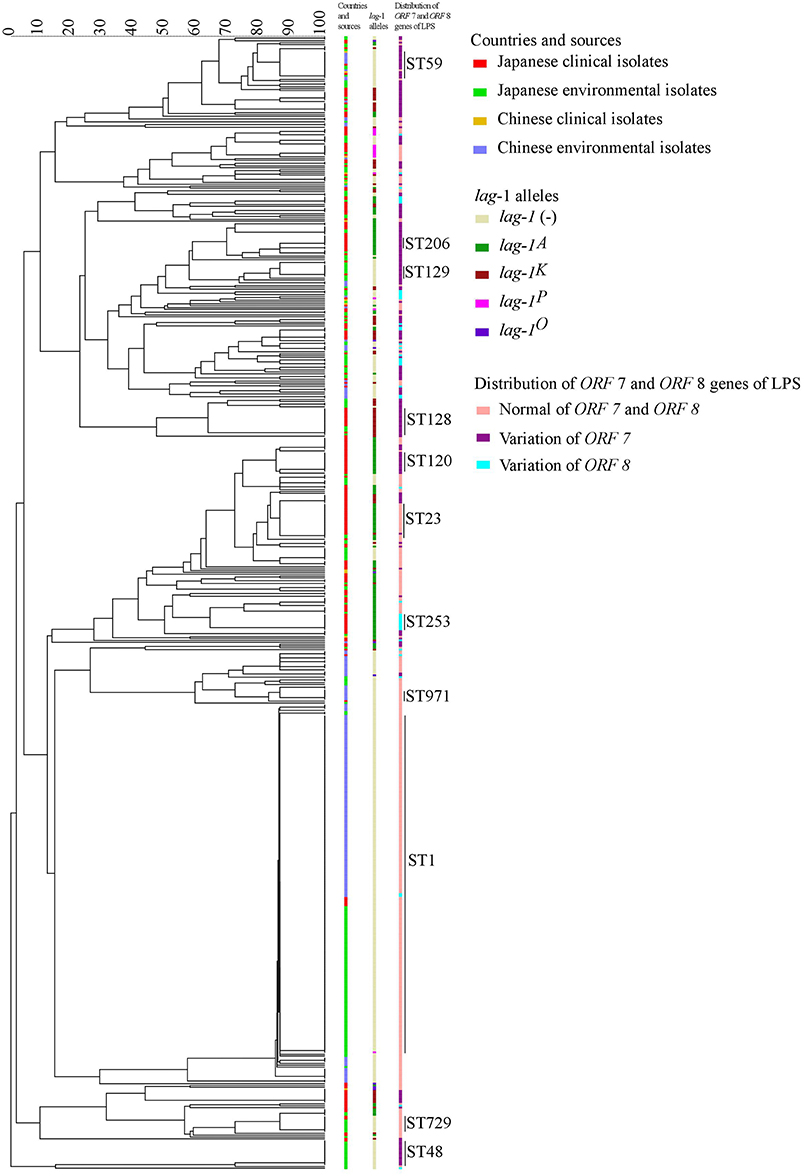
Figure 3. Evolutionary tree based on seven targeting specific gene loci. In the tree, the groups by countries and sources, lag-1 alleles and distribution of ORF 7 and ORF 8 genes of LPS were showed in different colors.
Subgroups of ATCC 33152
According to the reactivity with “lag-1,” “lag-1P,” “lag-1K,” “lag-1A,” “ORF 7,” and “ORF 8,” legionella pneumophila type strain ATCC 33152 harbored the lag-1 gene, subtyped as “lag-1O,” and didn't harbor either ORF 7 or ORF8 gene.
Discussion
Previous studies suggest that LPS is the major immunodominant antigen for all the Legionella species (Ciesielski et al., 1986; Petzold et al., 2013). Furthermore, the lag-1 gene encoding an O-acetyltransferase, which is responsible for the presence of the LPS epitope recognized by mAb 3/1, is exclusively found in sg1 strains (Petzold et al., 2013). Compared with other studies which used Dresden panel mentioned above, the other researchers usually used this method to differentiate the clinical sg1 strains into different subgroups (Helbig et al., 2002; Chasqueira et al., 2014). While, we used this method not only in clinical isolates but also in environmental strains, we also carried out comparative analysis. In our study, primers specific for the lag-1 gene were tested for their ability to differentiate Chinese and Japanese isolates into lag-1-positive and lag-1-negative strains. In the present study, the lag 1 gene was harbored by significantly more of the L. pneumophila sg1 clinical isolates than the environmental isolates, regardless of the country of origin. These findings are in accordance with a report of a study in the USA, in which 75% of the L. pneumophila sg1 clinical isolates but only 8% of environmental isolates harbored the lag-1 gene (P = 0.0001 based on χ2 test on proportions) (Kozak et al., 2009). Thus, it can be speculated that the lag-1 gene is present in the majority of clinical isolates, but is absent from most environmental isolates regardless of the country of origin, although the proportion of lag-1-positive clinical isolates varies in different countries. Furthermore, these results indicate that L. pneumophila sg1 infection exhibits unique virulence markers in every country. Due to the different national conditions in China and Japan, Legionellosis is not a legal infectious disease in China, and hospitals usually do not carry out clinical testing. Clinical strains are usually obtained through our active surveillance. Therefore, the number of Chinese clinical strains is lower.
In addition to lag-1, two intergenic regions expected to be specific for monoclonal subgroup Knoxville (ORF 7) and closely related subgroups Benidorm/Bellingham (ORF 8) were also used for selective genotyping. In a previous study in Germany (Thurmer et al., 2009), 31.7% (13/41) of the L. pneumophila sg1 clinical isolates and 50% (1/2) of the L. pneumophila sg1 environmental isolates harbored the ORF 7 gene, with no significant difference between the two groups of isolates (P = 0.085 based on χ2 test on proportions). In accordance with this, there was no significant difference in the proportions of clinical and environmental Chinese L. pneumophila sg1 isolates harboring the ORF 7 gene (15.3% of the clinical isolates and 9.9% of the environmental isolates; P = 0.544 based on χ2 test on proportions). In contrast, in the present study, a significantly higher proportion of the Japanese L. pneumophila sg1 clinical isolates harbored the ORF 7 gene compared with the environmental isolates (51.4% of the clinical isolates and 36.0% of the environmental isolates; P = 0.001 based on χ2 test on proportions). These results show marked differences in the ORF 7 characteristics of L. pneumophila sg1 isolates from these three countries, with a significant difference in the proportion of the clinical and environmental isolates harboring the ORF 7 gene observed only in the Japanese isolates. In the German study mentioned previously, 26.8% (11/41) of the L. pneumophila sg1 clinical isolates and none (0/2) of the L. pneumophila sg1 environmental isolates harbored the ORF 8 gene (P = 0.396 based on χ2 test on proportions). In our study, of the Japanese isolates, 12.1% of clinical isolates and 5.8% of environmental isolates harbored the ORF 8 gene (P = 0.017 based on χ2 test on proportions). Of the Chinese isolates, 7.7% of clinical isolates and 3.4% of environmental isolates harbored the ORF 8 gene (P = 0.388 based on χ2 test on proportions). Thus, similar to the observations for ORF 7, these results show marked differences in the ORF 8 characteristics of L. pneumophila sg1 isolates from these three countries, with a significant difference in the proportion of the clinical and environmental isolates harboring the ORF 8 gene observed only in the Japanese isolates. These results indicate that L. pneumophila sg1 infection exhibits unique virulence markers in Japan.
In addition, we carried out SBT analysis of the genetic relationship between L. pneumophila sg1 isolates from Japan and China. High SBT polymorphism was found in the analyzed strains. In accordance with previous reports on environmental L. pneumophila sg1 isolates (Amemura-Maekawa et al., 2012; Qin et al., 2014), ST1 was the dominant ST of environmental isolates in this study. Interestingly, we identified a correlation between STs and subgroups of sg1, as most isolates with same STs also had same lag-1, ORF 7 and ORF 8 gene subgroups. In addition, we observed that STs tend to associate with a single lag-1 allele type. This suggests the existence of clonal groups of L. pneumophila sg1 that rarely recombine. Further studies are required to evaluate the possibility that particular combination of SBT and lag-1 subgroups define a hierarchy of virulence genotypes that necessitates more stringent surveillance to prevent human diseases. Moreover, genome-wide sequencing techniques will facilitate the identification of lag-1, ORF 7, and ORF 8 gene subgroups, so the highly virulent strains of L. pneumophila can be monitored and identified not only at the species and serogroup levels, but also at the genetic level.
Data Availability
All datasets generated for this study are included in the manuscript/Supplementary Files.
Author Contributions
LJ and JA-M carried out the experiments of this study, except those documented in Table 4 which were performed by HR and YL, and took part in writing the manuscript. MS and HZ provided materials and participated in data analysis and took part in writing the manuscript. MM, BC, and MO participated in data analysis and wrote the manuscript. TQ participated in conceiving and coordinating experimental work, and took part in writing the manuscript. And she also provided materials for this study. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the National Science Foundation of China (81671985), the Science Foundation for the State Key Laboratory for Infectious Disease Prevention and Control of China (2015SKLID508), the Priority Project on Infectious Disease Control and Prevention (2018ZX10714002) of the Ministry of Science and Technology of the People's Republic of China, the Sanming Project of Medicine in Shenzhen (No. SZSM201811071), the Japan Agency for Medical Research and Development (AMED; JP18fk0108049), and the Health and Labor Sciences Research Grants (H28-Kenki-Ippan-006, H28-Shinko-Ippan-006).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank for T. Egawa (Bunkyo Public Health Center), H. Fujimoto (Setagaya Public Health Center), K. Furuhata (Azabu University), Y. Hanahara (Tottori Prefectural Institute of Public Health and Environmental Science), H. Inoue (Aquas Corporation), K. Iwabuchi (Institute for Environmental Sciences and Public Health of Iwate Prefecture), Y. Kanazawa (Wakayama City Institute of Public Health), S. Kawaguchi (Itabashi City Public Health Center), T. Koyama (Nagano Environmental Conservation Research Institute), H. Kurosawa (Gunma Prefectural Institute of Public Health and Environmental Sciences), H. Nakajima (Okayama Prefectural Institute for Environmental Science and Public Health), M. Nukina (Kobe Institute of Health), R. Okuno (Tokyo Metropolitan Institute of Public Health), T. Taguri (Nagasaki Prefectural Institute for Environmental Research and Public Health), Y. Takase (Arakawa City Public Health Center), A. Taketani and C. Hayashi (Amagasaki City Institute of Public Health), J. Uchida (Kagawa Prefectural Research Institute for Environmental Sciences and Public Health), Y. Watanabe (Kanagawa Prefectural Institute of Public Health), H. Yoshida (Fukuoka City Institute for Hygiene and the Environment), and S. Yoshino (Miyazaki Prefectural Institute for Public Health and Environment) for providing L. pneumophila strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00274/full#supplementary-material
References
Amemura-Maekawa, J., Kikukawa, K., Helbig, J. H., Kaneko, S., Suzuki-Hashimoto, A., Furuhata, K., et al. (2012). Distribution of monoclonal antibody subgroups and sequence-based types among Legionella pneumophila serogroup 1 isolates derived from cooling tower water, bathwater, and soil in Japan. Appl. Environ. Microbiol. 78, 4263–4270. doi: 10.1128/AEM.06869-11
Amemura-Maekawa, J., Kura, F., Chida, K., Ohya, H., Kanatani, J. I., Isobe, J., et al. (2018). Legionella pneumophila and other legionella species isolated from Legionellosis patients in Japan between 2008 and 2016. Appl. Environ. Microbiol. 84:e00721-18. doi: 10.1128/AEM.00721-18
Amemura-Maekawa, J., Kura, F., Helbig, J. H., Chang, B., Kaneko, A., Watanabe, Y., et al. (2010). Characterization of Legionella pneumophila isolates from patients in Japan according to serogroups, monoclonal antibody subgroups and sequence types. J. Med. Microbiol. 59(Pt 6), 653–659. doi: 10.1099/jmm.0.017509-0
Bernander, S., Jacobson, K., Helbig, J. H., Luck, P. C., and Lundholm, M. (2003). A hospital-associated outbreak of Legionnaires' disease caused by Legionella pneumophila serogroup 1 is characterized by stable genetic fingerprinting but variable monoclonal antibody patterns. J. Clin. Microbiol. 41, 2503–2508. doi: 10.1128/JCM.41.6.2503-2508.2003
Carratala, J., and Garcia-Vidal, C. (2010). An update on Legionella. Curr. Opin. Infect. Dis. 23, 152–157. doi: 10.1097/QCO.0b013e328336835b
Chasqueira, M. J., Rodrigues, L., Nascimento, M., Ramos, M., and Marques, T. (2014). Genetic diversity and evolutionary relationships among Legionella pneumophila clinical isolates, Portugal, 1987 to 2012. Euro Surveill. 19:20965. doi: 10.2807/1560-7917.ES2014.19.46.20965
Ciesielski, C. A., Blaser, M. J., and Wang, W. L. (1986). Serogroup specificity of Legionella pneumophila is related to lipopolysaccharide characteristics. Infect. Immun. 51, 397–404.
Declerck, P. (2010). Biofilms: the environmental playground of Legionella pneumophila. Environ. Microbiol. 12, 557–566. doi: 10.1111/j.1462-2920.2009.02025.x
Dournon, E., Bibb, W. F., Rajagopalan, P., Desplaces, N., and McKinney, R. M. (1988). Monoclonal antibody reactivity as a virulence marker for Legionella pneumophila serogroup 1 strains. J. Infect. Dis. 157, 496–501. doi: 10.1093/infdis/157.3.496
Fields, B. S., Benson, R. F., and Besser, R. E. (2002). Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526. doi: 10.1128/CMR.15.3.506-526.2002
Gaia, V., Fry, N. K., Afshar, B., Luck, P. C., Meugnier, H., Etienne, J., et al. (2005). Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 43, 2047–2052. doi: 10.1128/JCM.43.5.2047-2052.2005
Harrison, T. G., Afshar, B., Doshi, N., Fry, N. K., and Lee, J. V. (2009). Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000-2008). Eur. J. Clin. Microbiol. Infect. Dis. 28, 781–791. doi: 10.1007/s10096-009-0705-9
Harrison, T. G., Doshi, N., Fry, N. K., and Joseph, C. A. (2007). Comparison of clinical and environmental isolates of Legionella pneumophila obtained in the UK over 19 years. Clin. Microbiol. Infect. 13, 78–85. doi: 10.1111/j.1469-0691.2006.01558.x
Helbig, J. H., Benson, R. F., Pelaz, C., Jacobs, E., and Luck, P. C. (2007). Identification and serotyping of atypical Legionella pneumophila strains isolated from human and environmental sources. J. Appl. Microbiol. 102, 100–105. doi: 10.1111/j.1365-2672.2006.03057.x
Helbig, J. H., Bernander, S., Castellani Pastoris, M., Etienne, J., Gaia, V., Lauwers, S., et al. (2002). Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21, 710–716. doi: 10.1007/s10096-002-0820-3
Helbig, J. H., Kurtz, J. B., Pastoris, M. C., Pelaz, C., and Lück, P. C. (1997). Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species into serogroups. J. Clin. Microbiol. 35, 2841–2845.
Helbig, J. H., Luck, P. C., Knirel, Y. A., Witzleb, W., and Zahringer, U. (1995). Molecular characterization of a virulence-associated epitope on the lipopolysaccharide of Legionella pneumophila serogroup 1. Epidemiol. Infect. 115, 71–78. doi: 10.1017/S0950268800058131
Isberg, R. R., O'Connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24. doi: 10.1038/nrmicro1967
Joly, J. R., Chen, Y. Y., and Ramsay, D. (1983). Serogrouping and subtyping of Legionella pneumophila with monoclonal antibodies. J. Clin. Microbiol. 18, 1040–1046.
Kozak, N. A., Benson, R. F., Brown, E., Alexander, N. T., Taylor, T. H. Jr., Shelton, B. G., et al. (2009). Distribution of lag-1 alleles and sequence-based types among Legionella pneumophila serogroup 1 clinical and environmental isolates in the United States. J. Clin. Microbiol. 47, 2525–2535. doi: 10.1128/JCM.02410-08
Petzold, M., Thurmer, A., Menzel, S., Mouton, J. W., Heuner, K., and Luck, C. (2013). A structural comparison of lipopolysaccharide biosynthesis loci of Legionella pneumophila serogroup 1 strains. BMC Microbiol. 13:198. doi: 10.1186/1471-2180-13-198
Qin, T., Zhou, H., Ren, H., Guan, H., Li, M., Zhu, B., et al. (2014). Distribution of sequence-based types of Legionella pneumophila serogroup 1 strains isolated from cooling towers, hot springs, and potable water systems in China. Appl. Environ. Microbiol. 80, 2150–2157. doi: 10.1128/AEM.03844-13
Ratzow, S., Gaia, V., Helbig, J. H., Fry, N. K., and Luck, P. C. (2007). Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 45, 1965–1968. doi: 10.1128/JCM.00261-07
Rowbotham, T. J. (1980). Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33, 1179–1183. doi: 10.1136/jcp.33.12.1179
Stewart, C. R., Muthye, V., and Cianciotto, N. P. (2012). Legionella pneumophila persists within biofilms formed by Klebsiella pneumoniae, Flavobacterium sp., and Pseudomonas fluorescens under dynamic flow conditions. PLoS ONE 7:e50560. doi: 10.1371/journal.pone.0050560
Thurmer, A., Helbig, J. H., Jacobs, E., and Luck, P. C. (2009). PCR-based 'serotyping' of Legionella pneumophila. J. Med. Microbiol. 58(Pt 5), 588–595. doi: 10.1099/jmm.0.008508-0
Viasus, D., Calatayud, L., McBrown, M. V., Ardanuy, C., and Carratala, J. (2019). Urinary antigen testing in community-acquired pneumonia in adults: an update. Expert Rev. Anti Infect. Ther. 17, 107–115. doi: 10.1080/14787210.2019.1565994
Watkins, I. D., Tobin, J. O. H., Dennis, P. J., Brown, W., Newnham, R., and Kurtz, J. B. (2009). Legionella pneumophila serogroup 1 subgrouping by monoclonal antibodies – an epidemiological tool. J. Hygiene 95, 211–216. doi: 10.1017/S0022172400062641
Yu, V. L., Plouffe, J. F., Pastoris, M. C., Stout, J. E., Schousboe, M., Widmer, A., et al. (2002). Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186, 127–128. doi: 10.1086/341087
Zähringer, U., Knirel, Y. A., Lindner, B., Helbig, J. H., Sonesson, A., Marre, R., et al. (1995). The lipopolysaccharide of Legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog. Clin. Biol. Res. 392, 113–139.
Keywords: Legionella pneumophila, serogroup 1, lag-1 alleles, ORF 7 and ORF 8 genes, sequence-based typing
Citation: Jiang L, Amemura-Maekawa J, Ren H, Li Y, Sakata M, Zhou H, Murai M, Chang B, Ohnishi M and Qin T (2019) Distribution of lag-1 Alleles, ORF7, and ORF8 Genes of Lipopolysaccharide and Sequence-Based Types Among Legionella pneumophila Serogroup 1 Isolates in Japan and China. Front. Cell. Infect. Microbiol. 9:274. doi: 10.3389/fcimb.2019.00274
Received: 24 May 2019; Accepted: 17 July 2019;
Published: 05 August 2019.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Shaofu Qiu, Academy of Military Medical Sciences, ChinaChristophe Ginevra, Hospices Civils de Lyon, France
Changle Wang, Hebei Medical University, China
Copyright © 2019 Jiang, Amemura-Maekawa, Ren, Li, Sakata, Zhou, Murai, Chang, Ohnishi and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian Qin, cWludGlhbkBpY2RjLmNu
†These authors have contributed equally to this work
 Luxi Jiang
Luxi Jiang Junko Amemura-Maekawa3†
Junko Amemura-Maekawa3† Miho Sakata
Miho Sakata Haijian Zhou
Haijian Zhou Tian Qin
Tian Qin