- 1Department of Microbiology, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, India
- 2Department of Microbiology, Apollomedics Super Speciality Hospital, Lucknow, India
Background: Increasing use of colistin has led to the world-wide emergence of mobile colistin resistant gene (mcr). The present study aimed to identify and characterise mcr and other drug-resistant genes in colistin resistant Klebsiella pneumoniae clinical isolates.
Methods: Twenty-two colistin resistant K. pneumoniae were analysed for mcr and other drug-resistant genes, efflux pumps, and virulence genes, and for their biofilm forming ability. Pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST) were performed for all mcr-1 positive isolates. S1-PFGE and Southern hybridisation were performed for localisation of mcr-1 and blaNDM.
Results: Nineteen colistin resistant K. pneumoniae harboured mcr-1 and 3 had mgrB disruption. All isolates harboured blaOXA-48-type and ESBL genes; eight strains (five with mcr-1 and three with mgrB disruption) co-harboured blaNDM. Efflux pumps genes AcrAB and mdtK were detected in all 22 and tol-C in 21 isolates. Virulence-related genes entB and irp-1 were detected in all 22, mrkD in 20, and fimH-1 in 18 isolates; 11 isolates were strong biofilm producers. PFGE clustered mcr-1 positive isolates into eight groups based on ≥90% similarity; MLST revealed diverse sequence types, predominant being ST-15 (n = 4) and ST-16 (n = 4). Both mcr-1 and blaNDM were localised on plasmid and chromosome; mcr-1 was present on IncFII type and blaNDM on IncFIB and IncA/C type plasmids.
Conclusions: Colistin resistance in K. pneumoniae was predominantly mediated by mcr-1. Co-existence of colistin, carbapenem, and other drug-resistant genes along with efflux pumps indicates towards enormous genomic plasticity in K. pneumoniae with ability to emerge as super-spreader of drug-resistance.
Introduction
Increasing prevalence of multi-drug resistant (MDR) Gram-negative bacteria (GNB) is a serious public health concern since they are susceptible only to few antibiotics (Laws et al., 2019). The World Health Organization (WHO) has listed carbapenem-resistant Klebsiella pneumoniae among the priority pathogen group as it poses great threat to human health (WHO, 2017). K. pneumoniae belongs to the Enterobacteriaceae family and is a common nosocomial pathogen responsible for significant morbidity and mortality. Virulence factors such as capsular polysaccharides, lipopolysaccharide (LPS), siderophores and adherence factors help K. pneumoniae to circumvent host immune response and increase its pathogenicity. Biofilm formation also plays a significant role in drug resistance and inflammation resulting in persistent infections (Navon-Venezia et al., 2017).
Colistin is the last resort drug of choice for treatment of lethal infections caused by carbapenem resistant GNB. Colistin is a cationic polypeptide antibiotic that binds to the negatively charged phosphate group of LPS of GNB, which results in disarrangement of cell membrane. Ultimately, there is a loss of cell membrane integrity resulting in increased permeability of the cell, leakage of cell contents, and finally cell lysis (Baron et al., 2016). The re-introduction of colistin in clinical practice has resulted in its increased reports of resistance in GNB. Resistance to colistin is either chromosomal or plasmid mediated. Mobile colistin resistant gene (mcr-1) located either on chromosome or on plasmid encodes phosphoethanolamine transferase. Since the first report of mcr-1 in late 2015, ten different mcr variants (mcr-1 to mcr-10) have been reported (Wang et al., 2020).
In this study, we investigated the presence of mcr in colistin resistant K. pneumoniae strains. Such strains were also examined for the presence of other drug-resistant genes and also for virulence and efflux pumps genes and for their ability to form biofilm. Analyses of clonal relatedness and strain typing were performed in mcr-1 positive isolates. Further, characterisation of plasmids harbouring both mcr-1 and blaNDM was also performed.
Materials and Methods
Bacterial Strains
The study was conducted at Sanjay Gandhi Postgraduate Institute of Medical Sciences (Lucknow, India), a 900 bed tertiary care referral hospital in North India. Twenty-two colistin resistant K. pneumoniae isolates recovered from various clinical samples like pus, blood, endotracheal aspirate, tissue, and sputum between October 2016 and March 2017 were included in the study. All the isolates were identified using biochemical tests and MALDI-TOF MS (BioMérieux, Marcy l’Étoile, France). Prior to testing, all the isolates were stored in brain heart infusion broth (Becton, Dickinson and Company, Sparks, USA) supplemented with 20% glycerol (Sigma-Aldrich, MO, USA) at −80°C.
Demographic and Clinical Data
Demographic and clinical data of patients were obtained from the hospital information system available in the hospital intranet.
Antimicrobial Susceptibility Testing
Minimum inhibitory concentrations (MICs) were determined by broth microdilution method (BMD) in cation adjusted Mueller–Hinton broth following Clinical and Laboratory Standards Institute (CLSI) guidelines except colistin for which European Committee on Antimicrobial Susceptibility Testing breakpoints were followed (CLSI, 2017; EUCAST, 2017). Isolates were considered MDR if they were resistant to at least one antibiotic of three different classes among those tested (cephalosporins, carbapenems, fluoroquinolones, aminoglycosides, and polymyxins) according to Magiorakos et al. (2012).
DNA Isolation, Detection of Antibiotic Resistance, Efflux Pump and Virulence Genes
DNA was extracted from overnight grown culture using Wizard Genomic DNA Purification Kit (Promega, WI, USA). Genomic DNA quality was measured by NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, DE, USA). The integrity of genomic DNA was analysed by agarose gel electrophoresis. The extracted DNA was stored at −20°C.
The presence of mcr genes (mcr-1 to mcr-8) was analysed by conventional PCR, and the amplified products were confirmed by sequencing. The mcr positive isolates were also examined for the presence of carbapenemases (blaIMP, blaKPC, blaNDM, blaVIM, and blaOXA-48 type), extended spectrum β-lactamases (ESBLs; blaTEM, blaSHV, and blaCTX-M), 16S rRNA methyltransferases (armA and rmtA-F). List of primers is given in Table S1.
Chromosomal mutations were analysed in isolates negative for mcr. Conventional PCR was performed using specific primers (Table S1) to detect mutations in mgrB, phoP/phoQ, pmrA, and pmrB. The PCR products were purified, Sanger sequenced, and analysed to determine the mutations responsible for colistin resistance.
K. pneumoniae strains positive for mcr were also screened by conventional PCR for the presence of genes encoding for multidrug efflux pump systems like ArcAB, TolC, and MdtK, and virulence determinants such as regulator of mucoid phenotype (rmpA), type 1 and type 3 adhesins (fimH-1 and mrkD), iron siderophores (aerobactin synthase, lucC), bacteriocin biosynthesis [enterobactin (entB), and yersiniabactin (irP-1)], and serum resistance-associated outer membrane lipoprotein (traT).
Capsular Typing
Capsular typing based on wzi gene sequence was done as reported previously (Brisse et al., 2013). The PCR products were Sanger sequenced, and wzi alleles were identified, and corresponding capsular polysaccharide types (KL-types) were determined by comparing our wzi sequences with those available on the Klebsiella PasteurMLST sequence definition database (https://bigsdb.pasteur.fr/).
Biofilm Assay
Biofilm assay was performed by O’Toole and Kolter’s protocol with little modification (O’toole and Kolter, 1998). Briefly, 1 µl of overnight grown culture was inoculated into 100 µl of fresh tryptone soya broth (TSB) in 96 well sterile flat bottom polystyrene plates. After overnight incubation at 37°C, the cultures in wells were discarded. The wells were washed gently with water followed by air drying for 15 min. Biomass was stained with 125 µl of 0.1% (w/v) crystal violet for 20 min. Plates were rinsed off, air dried, and the dye bound to adherent biomass was eluted with 30% acetic acid. Absorbance was measured using automated microplate reader (MultiskanGO, Thermo Scientific, MA, USA) at 570 nm. Tests were performed in triplicate, and results were averaged. The results were interpreted according to Stepanovic et al. (2000). K. pneumoniae ATCC strain, ATCC 700603 was used as positive control whereas E. coli K-12 was used as negative control.
Clonal Diversity and Strain Typing
Clonal diversity among 19 mcr-1 positive K. pneumoniae isolates was examined by pulsed field gel electrophoresis (PFGE) according to previously reported protocol (Qin et al., 2014). Banding patterns were analysed using BioNumerics software version 7.6 (Applied-Maths, Sint-Martens-Latem, Belgium). Salmonella serotype Branderup strain (H9812) digested with XbaI (Promega, WI, USA) was used as reference strain.
Multi-locus sequence type (MLST) of 19 mcr-1 positive K. pneumoniae isolates was analysed as described previously (Diancourt et al., 2005). The seven housekeeping genes were amplified and sequenced. The sequence type (ST) was assigned by determining the allele number for each of the housekeeping genes using the database maintained by Pasteur Institute at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html/.
Conjugation Experiment and Plasmid Replicon Typing
Horizontal gene transfer ability of blaNDM and mcr-1 was determined using liquid mating assay for five K. pneumoniae isolates that harboured both mcr-1 and blaNDM. E. coli J53 was used as recipient strain, and transconjugants selection was performed on MacConkey agar plates containing meropenem (2 µg/ml) or colistin (1.0 µg/ml) as applicable and sodium azide (100 µg/ml). Transconjugants were tested for mcr-1 or blaNDM by PCR and antimicrobial susceptibility. Plasmid DNA was isolated from transconjugants following Kado and Liu method (Kado and Liu, 1981). PCR-based replicon typing (PBRT) was done to determine the plasmid incompatibility group (Carattoli et al., 2005).
S1-PFGE and Hybridisation
S1 PFGE and Southern hybridisation were performed for five strains that harboured both blaNDM and mcr-1. Bacterial DNA was prepared in agarose plugs, digested with S1 nuclease (Promega, WI, USA), and the linearised plasmid was then separated through PFGE. The gel was stained with ethidium bromide and transferred to nylon membrane (Hybond N, Amersham, UK) followed by hybridisation with digoxigenin labelled probes specific to mcr-1 or blaNDM. Probe labelling and signal detection were done by DIG DNA Labeling and Detection Kit (Roche Diagnostics, GmbH, Germany).
Results
Bacterial Isolates and Patient Details
Twenty-two colistin resistant K. pneumoniae isolates recovered from 22 (male 18) patients were analysed; 12 patients were from post-operative intensive care unit (ICU) and four from critical care medicine, three from nephrology, two from paediatric gastroenterology, and one from haematology wards. Most of the isolates were recovered from endotracheal aspirate (45.4%, 10/22), followed by blood (27.3%, 6/22) and sputum (9.1%, 2/22). All isolates except one were recovered after 48 h of admission. Among the 12 post-operative ICU patients, 66.7% (8/12) succumbed to their infection. Co-morbidities were present in 86.4% (19/22) of patients. Hypertension was present in 36.4% (8/22), followed by acute kidney injury (13.6%, 3/22), type-2 diabetes, and chronic liver disease in 9.1% (2/22) each. The clinical details of all patients are given in Table 1.
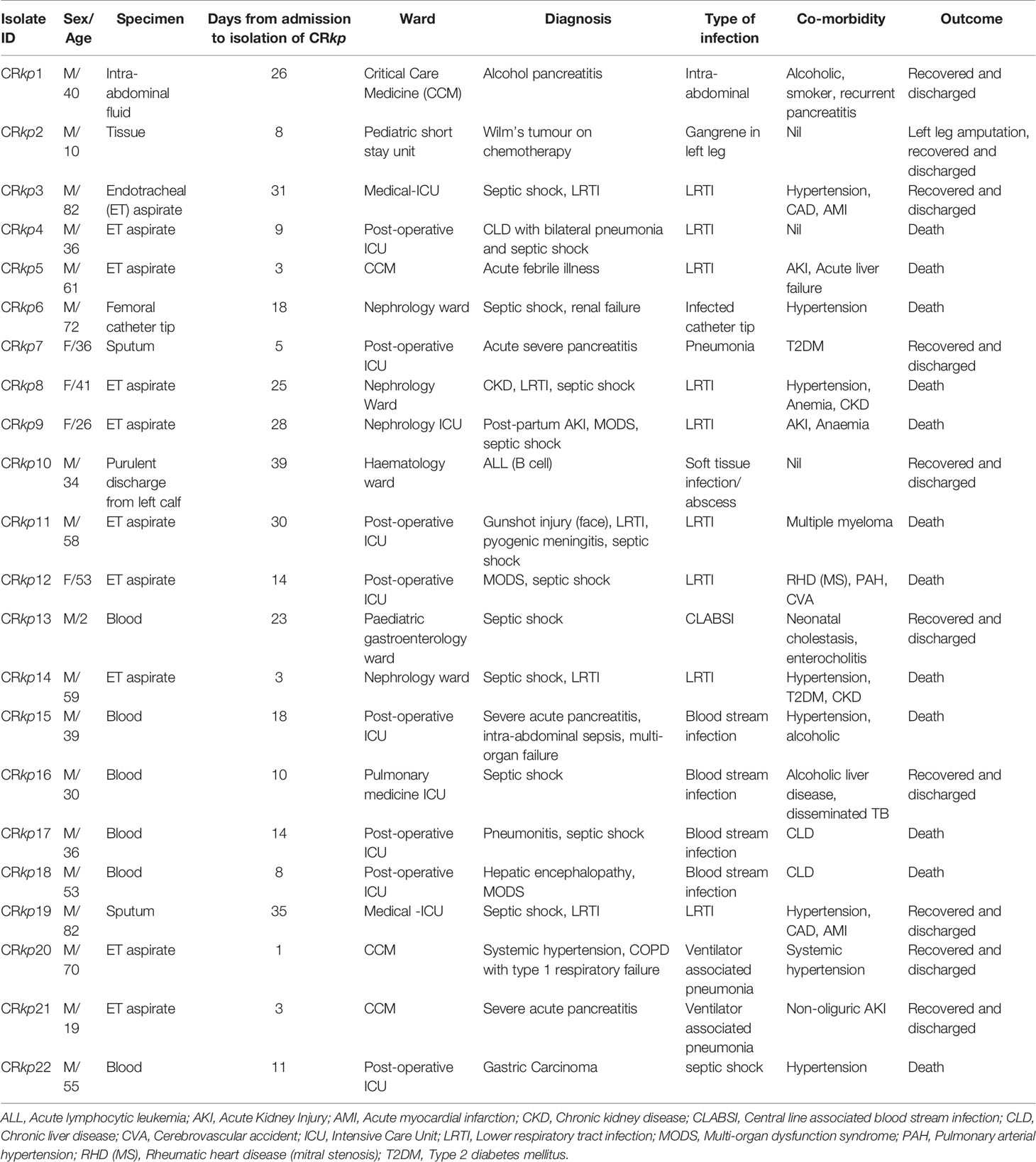
Table 1 Demographic and clinical features of patients infected with colistin resistant Klebsiella pneumoniae.
Antimicrobial Susceptibility Testing
The antimicrobial susceptibility profile showed that all the isolates were MDR as they were non-susceptible to at least one antibiotic from three or more antibiotics classes. All 22 isolates were resistant to carbapenems (imipenem and meropenem), 3rd generation cephalosporins (ceftazidime and ceftriaxone), monobactam (aztreonam), aminoglycoside (gentamicin), and fluoroquinolones (ciprofloxacin). The MIC values for colistin ranged from 8 to ≥512 mg/L. The antibiotic susceptibility results of 22 isolates are summarised in Table 2.
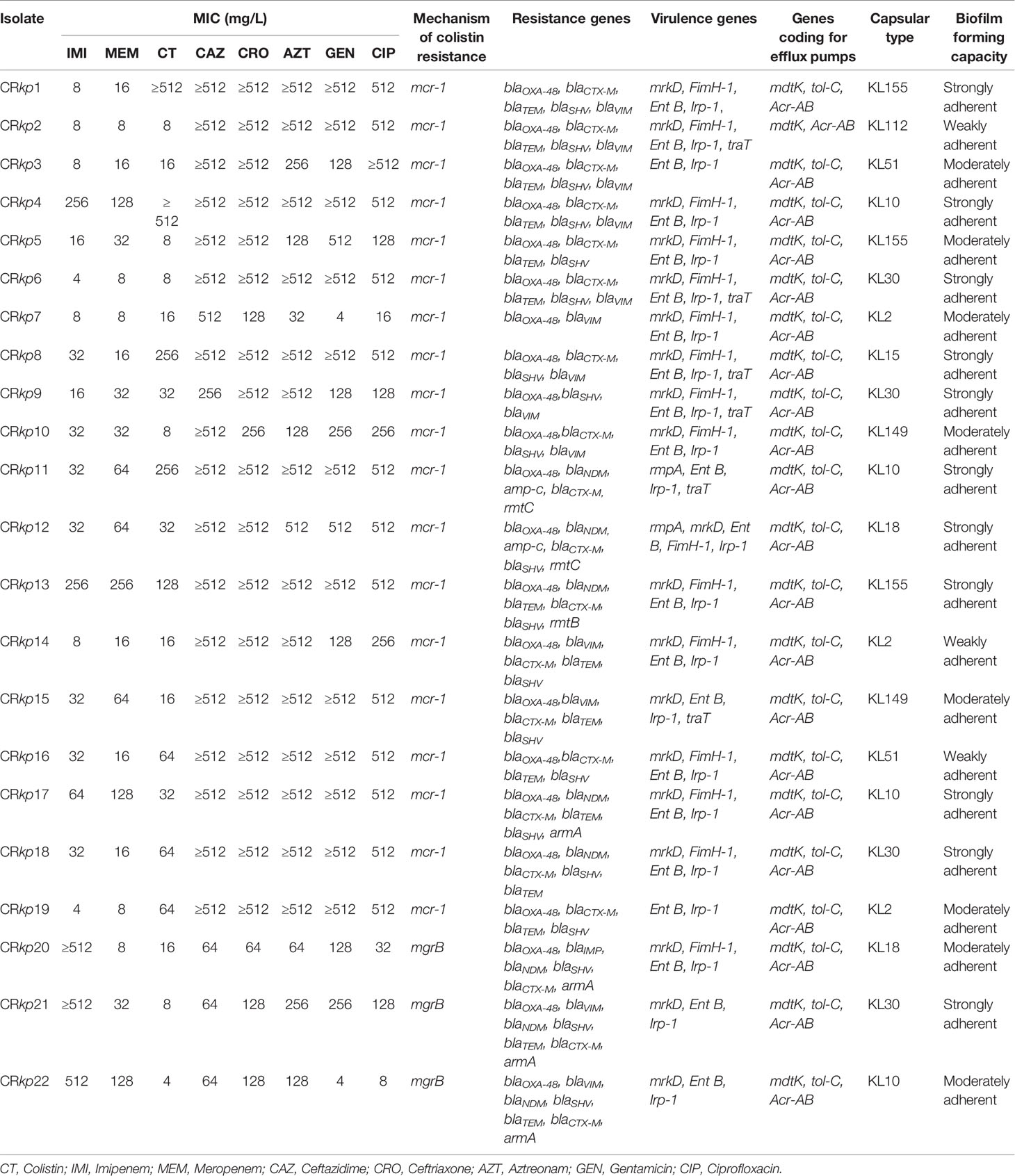
Table 2 Antimicrobial susceptibility profile and molecular characterisation of 22 colistin resistant Klebsiella pneumonia.
PCR Based Detection of Resistant Genes
Nineteen (86.4%) of 22 colistin resistant isolates harboured mcr-1, and the remaining three (13.6%) had mgrB disruption. Eight (36.4%) strains harboured blaNDM; five and three of them were positive for mcr-1 and mgrB disruption respectively. All 22 isolates carried blaOXA-48-type gene; blaVIM was detected in 13 (59.0%) isolates and blaIMP in one (4.5%) isolate. Twenty (90.1%) isolates harboured both blaCTX-M and blaSHV, whereas blaTEM was detected in 15 (68.2%) isolates. 16S r-RNA methyl transferase was detected in seven (31.8%) isolates (armA in four, rmtB in one, and rmtC in two isolates). Distributions of resistance genes in different combinations are given in Table 2.
PCR amplification of mgrB in three K. pneumoniae isolates (CRkp20, CRkp21, and CRkp22) revealed a larger (~1000 bp) amplicon. Sequencing of the amplicons showed IS elements mediated mgrB disruption. The IS elements involved in mgrB disruption belonged to IS1-like (777 bp) in CRkp20 and IS5-like families, 1,066 bp and 1,196 bp in CRkp21 and CRkp22 respectively. None of the isolates had mutation in phoP/phoQ, pmrA, and pmrBgene.
Detection of Efflux Pump and Virulence Genes
All 22 colistin resistant strains harboured AcrAB, mdtK, and tol-C efflux pumps except one isolate that lacked tol-C (Table 2). Mucoid phenotype regulator, rmpA, was identified in two isolates. The siderophore associated genes, entB and irp-1, were present in all the isolates. Other virulence genes fimH-1, mrkD, and traT were detected in 16 (72.7%), 19 (86.4%), and six (27.3%) isolates respectively. The distribution of virulence genes is shown in Table 2.
Biofilm Forming Capacity
In-vitro biofilm forming ability assay indicated that all 22 isolates were biofilm producers; 11 (50%) were strong, eight (36.4%) were moderate, and three (13.6%) were weak biofilm producers (Table 2).
Capsular Typing
Wzi based capsular typing of colistin resistant K. pneumoniae indicated a high diversity as it predicted 10 different capsular polysaccharide serotypes (KL155 (n = 3), KL112 (n = 1), KL51 (n = 2), KL10 (n = 4), KL30 (n = 2), KL2 (n = 3), KL15 (n = 1), KL30 (n = 2), KL149 (n = 2), KL18 (n = 2).
Clonal Diversity and Molecular Typing
All 19 mcr-1 positive K. pneumoniae isolates were typeable by PFGE. The maximum and minimum genetic similarity observed between the isolates was 99 and 86.5% respectively (Figure 1). Based on ≥90% similarity they were clustered into eight groups.

Figure 1 A dendrogram of the pulsed-field gel electrophoresis (PFGE) fingerprinting results and sequence types of 19 mcr-1 positive Klebsiella pneumoniae.
MLST analysis of 19 mcr-1 positive K. pneumoniae revealed eight different STs and their distributions were as follows: ST-15 (n = 4), ST-16 (n = 4), ST-231 (n = 3), and ST-147 (n = 3), ST-43 (n = 2) and one isolate each for ST-14, ST-11, and ST-23. The source of strains and their STs are shown in Figure 1.
Conjugation Experiment and Plasmid Replicon Typing
Conjugation experiments were performed for five mcr-1 positive K. pneumoniae, which also co-harboured blaNDM. PCR assay showed that mcr-1 was successfully transferred from four isolates (CRkp11, CRkp12, CRkp13, and CRkp17) by conjugation and failed to transfer in one isolate (CRkp18). All the transconjugants were phenotypically resistant to colistin but sensitive to imipenem and meropenem. PBRT showed transconjugants of CRkp11, CRkp12, and CRkp13 carried IncFII type plasmid, whilst transconjugants of CRkp17 carried an untypeable plasmid. Similarly, PCR assay showed that blaNDM was transferred successfully from all five isolates by conjugation, and PBRT results showed that transconjugants of CRkp11 and CRkp12 carried IncA/C type plasmid whilst transconjugants of CRkp13, CRkp17 and CRkp18 carried IncFIB type plasmid. Phenotypically, all blaNDM transconjugants were resistant to imipenem and meropenem but susceptible to colistin.
S1 PFGE and Southern Hybridisation
S1 PFGE followed by Southern hybridisation showed that mcr-1 was present both on plasmid and chromosome in three isolates (CRkp11, CRkp12, andCRkp13), whilst one each only on plasmid (CRkp17) and chromosome (CRkp18). The plasmid size in CRkp11, CRkp12, and CRkp17 was between ~138 and ~210 kb, whilst in CRkp13, mcr-1 was present on a small plasmid between ~33 and ~78 kb (Figure S1). The S1 nuclease digested genomic DNA from five K. pneumoniae was also probed with digoxigenin labelled blaNDM, and the results showed that blaNDM gene was present both on plasmid (between ~45 and ~400 kb) and chromosome in all five isolates (Figure S2).
GenBank Accession Numbers
The GenBank accession numbers assigned to nucleotide sequences of mcr-1 were MN652072-MN652090 and for nucleotide sequence of mgrB were MW389562–MW389564.
Discussion
The extensive use of antibiotics for treating infectious diseases has led to the emergence of bacterial antimicrobial resistance. The microbes have benefitted enormously from overuse of antibiotics in clinical practice, also in agricultural and livestock. Emergence and dissemination of transmissible colistin resistance have severely compromised the use of colistin for treatment of infections caused by carbapenem resistant Enterobacteriaceae. In studies reported across the world, mcr-1 has been predominantly reported in E. coli whereas K. pneumoniae accounts for less than 5% of the total mcr positive isolates to date (Sun et al., 2018; Nang et al., 2019). In contrast to global data, studies from India indicate that colistin resistance is more common in K. pneumoniae than in any other bacterial species (Pragasam et al., 2016; Singh et al., 2018; Sodhi et al., 2020). Also, very few studies are available on genomic characterisation of colistin resistant isolates. Hence, we investigated the mechanism of colistin resistance in clinical K. pneumoniae isolates and performed genetic characterisation of these isolates to expand our knowledge on colistin resistant K. pneumoniae.
mcr mediated colistin resistance has been reported across the world, but only few such reports are available from India (Singh et al., 2018; Gogry et al., 2019). We found mcr-1 mediated colistin resistance in 19 K. pneumoniae isolates, whilst insertional inactivation of mgrB gene by IS elements in three isolates. Insertional inactivation of mgrB activates the PhoP/Q two component signalling system that upregulates the arnBCADTEF operon which adds 4-amino-4-deoxy-L-arabinose to lipid A resulting in colistin resistance (Cannatelli et al., 2014). Insertional sequences of IS1and IS5 family are most common IS elements responsible for inactivation of mgrB gene (Azam et al., 2021). It is noteworthy to mention that we found coexistence of mcr-1 and blaNDM in five K. pneumoniae isolates; however studies suggest they are more commonly found in E. coli as compared to K. pneumoniae (Delgado-Blas et al., 2016; Zheng et al., 2017). Among carbapenemases, blaOXA-48 was found to be present in all the isolates. In recent years, blaOXA-48 has increasingly been reported from India; a multi-centric study from India reported the presence of blaOXA-48 in 80% of the carbapenem resistant isolates (Shankar et al., 2019a). We also observed 39% of our carbapenem resistant isolates were blaOXA-48 producers (unpublished data). Among MBLs, blaVIM was present in 59.1% (13/22) isolates. The unusually high prevalence of blaVIM (50% of blaNDM positive isolates) was also reported previously from our centre (Rahman et al., 2018). Another study from North India reported blaVIM in 18.4% (52/282) of carbapenem resistant isolates (Garg et al., 2019). Aminoglycosides in combination with other antibiotics such as tigecycline are often used for treating infections caused by carbapenem and colistin resistant K. pneumoniae (Petrosillo et al., 2019). In the current study, 16S RNA methyltransferase genes were found to be present in seven isolates that also harboured mcr-1 and blaNDM, which indicates towards a grim situation. Among twenty-two patients, twelve (54.5%) succumbed to their disease. We found that the patient death as outcome was attributed to lower respiratory tract infection, blood stream infections, and septic shock caused by MDR K. pneumoniae.
The resistance nodulation division acrAB-tolC efflux pumps are reported in diverse members of the Enterobacteriaceae family including K. pneumoniae. In K. pneumoniae acrAB-tolC efflux pumps have been associated with resistance to β-lactams, fluoroquinolones, and tetracycline (Li et al., 2015). Similarly, Multi-Antimicrobial Extrusion mdtK efflux pumps have been also been reported in K. pneumonia (Li et al., 2015). In the present study, we detected acrAB, tolc, and mdtK in MDR K. pneumoniae. Our results are in concordance with previous studies where authors had shown the presence of drug-resistant genes and efflux pumps in MDR K. pneumoniae (Maurya et al., 2019; Ni et al., 2020).
The role of virulence factors in colonisation, invasion, and pathogenicity of K. pneumoniae is well known (Paczosa and Mecsas, 2016). Mucoid regulator gene, rmpA, is involved in capsule biosynthesis and often associated with hypervirulence was detected in two K. pneumoniae isolates (Cheng et al., 2010). The other important virulence factors are siderophores; they are low molecular weight iron scavenging molecules secreted by many GNB that affect the iron homeostasis in host (Page, 2019). In this study, all the colistin resistant K. pneumoniae harboured ent B and irp-1 siderophores, which are also known to contribute towards inflammation and bacterial spread during infection (Holden et al., 2016). Adhesin associated genes fimH, a type 1 fimbria adhesive subunit and mrkD, a type 3 adhesive subunit have been detected in 72.7 and 86.4 isolates respectively. mrkD is known to facilitate binding to extracellular matrix which is responsible for bacterial adherence to tissue and indwelling devices such as endotracheal tubes (Paczosa and Mecsas, 2016). Serum resistant outer membrane lipoprotein (traT) was detected in 27.3% isolates and reported to play a crucial role in bacterial pathogenesis by blocking the action of membrane attack complex (Miajlovic and Smith, 2014). K. pneumoniae is known to produce biofilm which provides a layer of protection by preventing antibiotic penetration and reducing their efficacy. In our study all the isolates were biofilm producer with 50% of them producing strong biofilm, which suggests that MDR K. pneumoniae strains are associated with biofilm production.
PFGE is considered gold standard for molecular epidemiology of bacterial strains. PFGE data indicated that the clonal spread of K. pneumoniae was not responsible for colistin resistance. The isolates having more than 90% similarity most often were of same ST except in few cases where isolates of same STs clustered separately. Further, the MLST data showed that ST-15 and ST-16 were the most dominant clones followed by ST-231 and ST-147 amongst the mcr-1 positive K. pneumoniae. We found that ST15 K. pneumoniae isolate was associated with the presence of rmpA gene. Out of four ST-15 isolates, three harboured blaNDM and 16S rRNA methyltransferase [rmtB (n = 1) and rmtC (n = 2)]. All ST15 K. pneumoniae were associated with strong biofilm production, whilst the other dominant clone ST16 K. pneumoniae was moderate biofilm producers. Previous study from India also supports our data where authors had detected ST-231, ST-14, ST-147, ST-15, ST-16, ST-11, ST-23, and ST-43 in colistin resistant K. pneumoniae (Shankar et al., 2019b), whereas global data suggests the presence of heterogeneous STs in mcr-1 producing K. pneumoniae. The diversity in PFGE and ST was also supported by capsular serotyping which predicted eight serotypes based on wzi allele sequence. KL10 was the most common capsular serotype detected; in mcr-1 producing K. pneumoniae, KL10 capsulate serotype was associated with entB and irp-1 siderophores along with strong biofilm forming ability.
Conjugation experiments revealed that in four out of five K. pneumoniae isolates, mcr-1 was present on conjugative plasmid. Conjugative plasmids are self-transmissible and are often responsible for rapid spread of resistant traits. Three of the four mcr-1 transconjugants had IncFII type plasmid, which are conjugative plasmid with low copy number and size ranging between 45 and 200 kb (Rozwandowicz et al., 2018). The role of IncFII type plasmid in dissemination of mcr-1 is well known (Xavier et al., 2016; Wang et al., 2018). The mcr-1 harbouring IncFII plasmids were associated with ST15 K. pneumoniae. Conjugation experiments in the above five K. pneumoniae showed successful transfer of blaNDM to recipient E. coli J53 that suggests their location on conjugative plasmid. In two transconjugants blaNDM was present in IncA/C type plasmid whereas in three transconjugants blaNDM was present in IncFIB type plasmid. IncA/C type plasmids are broad host range, low copy number, and frequently found to be responsible for dissemination of blaNDM. Similarly, previous studies had shown that dissemination of blaNDM was linked to transferable IncA/C and IncFIB plasmids (Khan et al., 2017; Sugawara et al., 2019). IncF are considered as epidemic plasmids and linked with the global spread of K. pneumoniae ST258 (Rozwandowicz et al., 2018). The presence of multiple plasmids in MDR strains imparts fitness cost; however, it provides bacteria specific traits which help them to survive in stress conditions. S1-PFGE showed that majority of K. pneumoniae isolates harboured multiple plasmids. mcr-1 was present on plasmid of different sizes in these isolates. In three isolates, mcr-1 was present both on plasmid and chromosome. The chromosomal integration stabilises mcr-1 and enables it to be vertically transferred without the risk of plasmid loss. Co-existence of transferable blaNDM along with mcr-1 is a major threat to human health by compromising the available treatment options. Previous studies from USA, China, and Vietnam also reported the coexistence of mcr-1 and blaNDM in various members of Enterobacteriaceae and their potential to spread as extensively drug-resistant strains (Mediavilla et al., 2016; Feng et al., 2018; Jin et al., 2018).
In conclusion, K. pneumoniae has emerged as the most notorious pathogen among the members of Enterobacteriaceae. They are the reservoirs of diverse resistant traits and virulence genes. Moreover, their biofilm forming ability provides them survival and colonisation advantages. Co-existence of mcr-1 and blaNDM on the transmissible plasmids is a matter of concern as such plasmids possess significant risk of inter- and intra-species dissemination in the environmental and livestock pathogens. Therefore, strict epidemiological surveillance, infection control measures, and antibiotic stewardship are required to curb this menace of colistin resistance from dissemination.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MN652072-MN652090 https://www.ncbi.nlm.nih.gov/genbank/, MW389562-MW389564.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional ethics committee of Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow, India [2017-191-PhD-99(B)]. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
KP conceptualized and supervised the study. SS collected the sample, performed experiments, and drafted the manuscript. AP, MR, and AS performed the experiments and edited the manuscript. SN and CS collected the patient information and provided the demographic data. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Science and Engineering Research Board (SERB) (EMR/2015/001804), Government of India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SS acknowledges the Department of Science and Technology (DST/INSPIRE Fellowship/2015/IF150708), Government of India, for fellowship support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.666030/full#supplementary-material
Supplementary Figure 1 | S1-PFGE and Southern blot hybridization of mcr-1 and blaNDM producing Klebsiella pneumoniae; (A, C) S1 digested DNA analysed by PFGE; (B, D) hybridisation of S1-PFGE gel with digoxygenin labelled mcr-1gene probe; (A, C) Lane M- Salmonella Braenderup H9812; (A–D) lanes 1—CRkp11, 2—CRkp12, 3—CRkp13, 4—CRkp17 and 5—CRkp18.
Supplementary Figure 2 | S1-PFGE and Southern blot hybridisation of mcr-1 and blaNDM producing Klebsiella pneumoniae; (A) S1 digested DNA analysed by PFGE, (B) hybridisation of S1-PFGE gel with digoxygenin labelled blaNDM gene probe; (A) lane M—Salmonella Braenderup H9812, (A, B) lanes 1—CRkp11, 2—CRkp12, 3—CRkp13, 4—CRkp17, and 5—CRkp18.
References
Azam, M., Gaind, R., Yadav, G., Sharma, A., Upmanyu, K., Jain, M., et al. (2021). Colistin Resistance Among Multiple Sequence Types of Klebsiella Pneumoniae Is Associated With Diverse Resistance Mechanisms: A Report From India. Front. Microbiol. 12, 609840. doi: 10.3389/fmicb.2021.609840
Baron, S., Hadjadj, L., Rolain, J. M., Olaitan, A. O. (2016). Molecular Mechanisms of Polymyxin Resistance: Knowns and Unknowns. Int. J. Antimicrob. Agents 48, 583–591. doi: 10.1016/j.ijantimicag.2016.06.023
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Struve, C., et al. (2013). Wzi Gene Sequencing, a Rapid Method for Determination of Capsular Type for Klebsiella Strains. J. Clin. Microbiol. 51, 4073–4078. doi: 10.1128/JCM.01924-13
Cannatelli, A., Giani, T., D’andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014). Mgrb Inactivation Is a Common Mechanism of Colistin Resistance in KPC-Producing Klebsiella Pneumoniae of Clinical Origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/aac.03110-14
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., Threlfall, E. J. (2005). Identification of Plasmids by PCR-Based Replicon Typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., Peng, H. L. (2010). Rmpa Regulation of Capsular Polysaccharide Biosynthesis in Klebsiella Pneumoniae CG43. J. Bacteriol 192, 3144–3158. doi: 10.1128/jb.00031-10
CLSI (2017). “CLSI Supplement M100,” in Performance Standards for Antimicrobial Susceptibility Testing, 27th. Ed. Wayne, P. A. (Clinical and Laboratory Standards Institute). 2017.
Delgado-Blas, J. F., Ovejero, C. M., Abadia-Patiño, L., Gonzalez-Zorn, B. (2016). Coexistence of Mcr-1 and blaNDM-1 in Escherichia Coli From Venezuela. Antimicrob. Agents Chemother. 60, 6356–6358. doi: 10.1128/aac.01319-16
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., Brisse, S. (2005). Multilocus Sequence Typing of Klebsiella Pneumoniae Nosocomial Isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/jcm.43.8.4178-4182.2005
The European Committee on Antimicrobial Susceptibility Testing (EUCAST). (2017). Breakpoint tables for Interpretation of MICs and Zone Diameters. Version 7.1. Available at: http://www.eucast.org.
Feng, S., Shen, C., Chen, H., Zheng, X., Xia, Y., Zhong, L. L., et al. (2018). Co-production of MCR-1 and NDM-5 in Escherichia Coli Isolated From a Colonization Case of Inpatient. Infect. Drug Resist. 11, 1157–1161. doi: 10.2147/idr.s171164
Garg, A., Garg, J., Kumar, S., Bhattacharya, A., Agarwal, S., Upadhyay, G. C. (2019). Molecular Epidemiology & Therapeutic Options of Carbapenem-Resistant Gram-Negative Bacteria. Indian J. Med. Res. 149, 285–289. doi: 10.4103/ijmr.IJMR_36_18
Gogry, F. A., Siddiqui, M. T., Haq, Q. M. R. (2019). Emergence of Mcr-1 Conferred Colistin Resistance Among Bacterial Isolates From Urban Sewage Water in India. Environ. Sci. Pollut. Res. Int. 26, 33715–33717. doi: 10.1007/s11356-019-06561-5
Holden, V. I., Breen, P., Houle, S., Dozois, C. M., Bachman, M. A. (2016). Klebsiella Pneumoniae Siderophores Induce Inflammation, Bacterial Dissemination, and HIF-1α Stabilization During Pneumonia. mBio 7. doi: 10.1128/mBio.01397-16
Jin, L., Wang, R., Wang, X., Wang, Q., Zhang, Y., Yin, Y., et al. (2018). Emergence of Mcr-1 and Carbapenemase Genes in Hospital Sewage Water in Beijing, China. J. Antimicrob. Chemother. 73, 84–87. doi: 10.1093/jac/dkx355
Kado, C. I., Liu, S. T. (1981). Rapid Procedure for Detection and Isolation of Large and Small Plasmids. J. Bacteriol 145, 1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981
Khan, A. U., Maryam, L., Zarrilli, R. (2017). Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-Lactamase (NDM): A Threat to Public Health. BMC Microbiol. 17, 101. doi: 10.1186/s12866-017-1012-8
Laws, M., Shaaban, A., Rahman, K. M. (2019). Antibiotic Resistance Breakers: Current Approaches and Future Directions. FEMS Microbiol. Rev. 43, 490–516. doi: 10.1093/femsre/fuz014
Li, X. Z., Plésiat, P., Nikaido, H. (2015). The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/cmr.00117-14
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Maurya, N., Jangra, M., Tambat, R., Nandanwar, H. (2019). Alliance of Efflux Pumps With β-Lactamases in Multidrug-Resistant Klebsiella Pneumoniae Isolates. Microb. Drug Resist. 25, 1155–1163. doi: 10.1089/mdr.2018.0414
Mediavilla, J. R., Patrawalla, A., Chen, L., Chavda, K. D., Mathema, B., Vinnard, C., et al. (2016). Colistin- and Carbapenem-Resistant Escherichia Coli Harboring Mcr-1 and Blandm-5, Causing a Complicated Urinary Tract Infection in a Patient From the United States. mBio 7. doi: 10.1128/mBio.01191-16
Miajlovic, H., Smith, S. G. (2014). Bacterial Self-Defence: How Escherichia Coli Evades Serum Killing. FEMS Microbiol. Lett. 354, 1–9. doi: 10.1111/1574-6968.12419
Nang, S. C., Li, J., Velkov, T. (2019). The Rise and Spread of Mcr Plasmid-Mediated Polymyxin Resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841x.2018.1492902
Navon-Venezia, S., Kondratyeva, K., Carattoli, A. (2017). Klebsiella Pneumoniae: A Major Worldwide Source and Shuttle for Antibiotic Resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Ni, R. T., Onishi, M., Mizusawa, M., Kitagawa, R., Kishino, T., Matsubara, F., et al. (2020). The Role of RND-Type Efflux Pumps in Multidrug-Resistant Mutants of Klebsiella Pneumoniae. Sci. Rep. 10, 10876. doi: 10.1038/s41598-020-67820-x
O’toole, G. A., Kolter, R. (1998). Initiation of Biofilm Formation in Pseudomonas Fluorescens WCS365 Proceeds Via Multiple, Convergent Signalling Pathways: A Genetic Analysis. Mol. Microbiol. 28, 449–461. doi: 10.1046/j.1365-2958.1998.00797.x
Paczosa, M. K., Mecsas, J. (2016). Klebsiella Pneumoniae: Going on the Offense With a Strong Defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/mmbr.00078-15
Page, M. G. P. (2019). The Role of Iron and Siderophores in Infection, and the Development of Siderophore Antibiotics. Clin. Infect. Dis. 69, S529–s537. doi: 10.1093/cid/ciz825
Petrosillo, N., Taglietti, F., Granata, G. (2019). Treatment Options for Colistin Resistant Klebsiella Pneumoniae: Present and Future. J. Clin. Med. 8. doi: 10.3390/jcm8070934
Pragasam, A. K., Shankar, C., Veeraraghavan, B., Biswas, I., Nabarro, L. E., Inbanathan, F. Y., et al. (2016). Molecular Mechanisms of Colistin Resistance in Klebsiella Pneumoniae Causing Bacteremia From India-A First Report. Front. Microbiol. 7, 2135. doi: 10.3389/fmicb.2016.02135
Qin, S., Fu, Y., Zhang, Q., Qi, H., Wen, J. G., Xu, H., et al. (2014). High Incidence and Endemic Spread of NDM-1-Positive Enterobacteriaceae in Henan Province, China. Antimicrob. Agents Chemother. 58, 4275–4282. doi: 10.1128/aac.02813-13
Rahman, M., Prasad, K. N., Gupta, S., Singh, S., Singh, A., Pathak, A., et al. (2018). Prevalence and Molecular Characterization of New Delhi Metallo-Beta-Lactamases in Multidrug-Resistant Pseudomonas Aeruginosa and Acinetobacter Baumannii From India. Microb. Drug Resist. 24, 792–798. doi: 10.1089/mdr.2017.0078
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids Carrying Antimicrobial Resistance Genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Shankar, C., Mathur, P., Venkatesan, M., Pragasam, A. K., Anandan, S., Khurana, S., et al. (2019a). Rapidly Disseminating Bla(OXA-232) Carrying Klebsiella Pneumoniae Belonging to ST231 in India: Multiple and Varied Mobile Genetic Elements. BMC Microbiol. 19, 137. doi: 10.1186/s12866-019-1513-8
Shankar, C., Venkatesan, M., Rajan, R., Mani, D., Lal, B., Anandan, S., et al. (2019b). Molecular Characterization of Colistin-Resistant Klebsiella Pneumoniae & Its Clonal Relationship Among Indian Isolates. Indian J. Med. Res. 149, 199–207. doi: 10.4103/ijmr.IJMR_2087_17
Singh, S., Pathak, A., Kumar, A., Rahman, M., Singh, A., Gonzalez-Zorn, B., et al. (2018). Emergence of Chromosome-Borne Colistin Resistance Gene Mcr-1 in Clinical Isolates of Klebsiella Pneumoniae From India. Antimicrob. Agents Chemother. 62. doi: 10.1128/aac.01885-17
Sodhi, K., Mittal, V., Arya, M., Kumar, M., Phillips, A., Kajla, B. (2020). Pattern of Colistin Resistance in Klebsiella Isolates in an Intensive Care Unit of a Tertiary Care Hospital in India. J. Infect. Public Health 13, 1018–1021. doi: 10.1016/j.jiph.2019.10.013
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B., Svabic-Vlahovic, M. (2000). A Modified Microtiter-Plate Test for Quantification of Staphylococcal Biofilm Formation. J. Microbiol. Methods 40, 175–179. doi: 10.1016/s0167-7012(00)00122-6
Sugawara, Y., Akeda, Y., Hagiya, H., Sakamoto, N., Takeuchi, D., Shanmugakani, R. K., et al. (2019). Spreading Patterns of NDM-Producing Enterobacteriaceae in Clinical and Environmental Settings in Yangon, Myanmar. Antimicrob. Agents Chemother. 63 (3), e01924–18. doi: 10.1128/aac.01924-18
Sun, J., Zhang, H., Liu, Y. H., Feng, Y. (2018). Towards Understanding MCR-Like Colistin Resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., Zong, Z. (2020). Identification of Novel Mobile Colistin Resistance Gene Mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, R., Van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The Global Distribution and Spread of the Mobilized Colistin Resistance Gene Mcr-1. Nat. Commun. 9, 1179. doi: 10.1038/s41467-018-03205-z
WHO (2017). WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed (Geneva, Switzerland: World Health Organization).
Xavier, B. B., Lammens, C., Butaye, P., Goossens, H., Malhotra-Kumar, S. (2016). Complete Sequence of an IncFII Plasmid Harbouring the Colistin Resistance Gene Mcr-1 Isolated From Belgian Pig Farms. J. Antimicrob. Chemother. 71, 2342–2344. doi: 10.1093/jac/dkw191
Keywords: blaNDM, colistin resistance, Klebsiella pneumoniae, mcr-1, mgrB, sequence type
Citation: Singh S, Pathak A, Rahman M, Singh A, Nag S, Sahu C and Prasad KN (2021) Genetic Characterisation of Colistin Resistant Klebsiella pneumoniae Clinical Isolates From North India. Front. Cell. Infect. Microbiol. 11:666030. doi: 10.3389/fcimb.2021.666030
Received: 09 February 2021; Accepted: 13 May 2021;
Published: 21 June 2021.
Edited by:
Krisztina M. Papp-Wallace, Louis Stokes Cleveland VA Medical Center, United StatesReviewed by:
Laura Rojas, Case Western Reserve University, United StatesRafael Franco-Cendejas, National Institute of Rehabilitation Luis Guillermo Ibarra Ibarra, Mexico
Copyright © 2021 Singh, Pathak, Rahman, Singh, Nag, Sahu and Prasad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kashi Nath Prasad, a2FzaGlucHJhc2FkQGdtYWlsLmNvbQ==; ZHJrYXNoaW5hdGhfcEBhcG9sbG9ob3NwaXRhbHMuY29t
 Sanjay Singh
Sanjay Singh Ashutosh Pathak
Ashutosh Pathak Mohibur Rahman1
Mohibur Rahman1 Soumyabrata Nag
Soumyabrata Nag Kashi Nath Prasad
Kashi Nath Prasad