- 1Department of Laboratory Medicine, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
- 2Graduate School, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
- 3Department of Laboratory Medicine, Beijing Key Laboratory for Mechanisms Research and Precision Diagnosis of Invasive Fungal Diseases (BZ0447), Beijing, China
- 4Clinical Microbiology Laboratory, The First Affiliated Hospital of Hebei North University, Zhangjiakou, China
- 5Department of Infectious Diseases and Clinical Microbiology, Beijing Chaoyang Hospital, Beijing, China
- 6Department of Medical Research Center, Peking Union Medical College Hospital, Chinese Academy of Medical Science & Peking Union Medical College, Beijing, China
- 7Department of Laboratory Medicine, Daqing Oilfield General Hospital, Daqing, China
- 8Department of Laboratory Medicine, The First Affiliated Hospital of Harbin Medical University, Harbin, China
- 9Department of Laboratory Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 10Department of Laboratory Medicine, The Fourth Affiliated Hospital of Harbin Medical University, Harbin, China
- 11Department of Clinical Laboratory, Fujian Medical University Union Hospital, Fuzhou, China
- 12Department of Clinical Laboratory, The Affiliated Hospital of Qingdao University, Qingdao, China
Diutina catenulata (Candida catenulata) is an ascomycete yeast species widely used in environmental and industrial research and capable of causing infections in humans and animals. At present, there are only a few studies on D. catenulata, and further research is required for its more in-depth characterization and analysis. Eleven strains of D. catenulata collected from China Hospital Invasive Fungal Surveillance Net (CHIF-NET) and the CHIF-NET North China Program were identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry and internal transcribed spacer sequencing. The antifungal susceptibility of the Diutina catenulata strains was tested using the Clinical and Laboratory Standards Institute broth microdilution method and Sensititre YeastOne™. Furthermore, ERG11 and FKS1 were sequenced to determine any mutations related to azole and echinocandin resistance in D. catenulata. All isolates exhibited low minimum inhibitory concentration (MIC) values for itraconazole (0.06–0.12 μg/ml), posaconazole (0.06–0.12 μg/ml), amphotericin B (0.25–1 μg/ml), and 5-flucytosine (range, <0.06–0.12 μg/ml), whereas four isolates showed high MICs (≥4 μg/ml) for echinocandins. Strains with high MIC values for azoles showed common ERG11 mutations, namely, F126L/K143R. In addition, L139R mutations may be linked to high MICs of fluconazole. Two amino acid alterations reported to correspond to high MIC values of echinocandin, namely, F621I (F641) and S625L (S645), were found in the hot spot 1 region of FKS1. In addition, one new amino acid alteration, I1348S (I1368), was found outside of the FKS1 hot spot 2 region, and its contribution to echinocandin resistance requires future investigation. Diutina catenulata mainly infects patients with a weak immune system, and the high MIC values for various antifungals exhibited by these isolates may represent a challenge to clinical treatment.
Introduction
In recent years, Candida infections have been on the rise worldwide (Sanguinetti et al., 2015; Pristov and Ghannoum, 2019). Although Candida albicans remains the main causative agent of these infections, the rate of non–C. albicans candidial infections is increasing (Sanguinetti et al., 2015). Candida infections are not usually multidrug-resistant, but there are notable cases of drug resistance among non-albicans Candida (NAC) species. Candida auris shows in vitro multidrug resistance and is associated with outbreaks. Candida glabrata is the most common cause of candidemia and is resistant to azoles and echinocandins. Candida parapsilosis is another notorious pathogen isolated from patients, which causes outbreaks and multidrug resistance (Arastehfar et al., 2021). These Candida species, though rare, are clinically common and necessitate a better understanding of their pathogenic mechanisms. Diutina catenulata (C. catenulata), an ascomycete, can colonize the digestive tract of animals and humans and cause superficial or deep infections (Crozier, 1977; Radosavljevic et al., 1999; Ha et al., 2018; Cafarchia et al., 2019). Diutina catenulata is usually utilized in the production of industrial products (Joo et al., 2008; Subramanya et al., 2017; Babaei and Habibi, 2018; Cafarchia et al., 2019). Diutina catenulata belongs to the CTG-Ser clade and is closely related to Saccharomyces cerevisiae (O'Brien et al., 2018). The first time describing D. catenulata associated with disease in humans in 1977, and D. catenulata was recovered from the nail of a 50-year-old male Australian patient (Crozier, 1977). Subsequently, the first case of candidemia was diagnosed in a patient with cancer (Radosavljevic et al., 1999).
There are few reports of human infection with D. catenulata, and only a small number of studies are available to guide clinical treatment. Diutina catenulata generally exhibits antifungal sensitivity (Radosavljevic et al., 1999; Ha et al., 2018); however, some strains showing resistance to azoles and echinadines have been isolated from eggs and feces (Glushakova et al., 2021). Thus, it is important to consider the potential antifungal resistance of D. catenulata. However, the resistance mechanism of D. catenulata is poorly understood. Amino acid alternations in Erg11p and Fks1p are responsible for causing azole and echinocandin resistance in pathogenic Candida spp. (Berkow and Lockhart, 2017; Toutounji et al., 2019). However, the complete open reading frames of both ERG11 and FKS1 genes have not been revealed, and the gene polymorphisms of ERG11 and FKS1 in clinical D. catenulata isolates remain unknown.
Despite the three cases reported worldwide, the occurrence of D. catenulata in clinical specimens of the Chinese population remains unelucidated. Thus, we studied the antifungal susceptibility and potential drug resistance mechanisms of the clinical isolates of D. catenulata obtained in China over a period of 9 years.
Material and Methods
Ethics Statement
This study was approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (No. S-263). A written informed consent was obtained from all study participants to examine the isolates cultured from them for scientific research.
Isolates
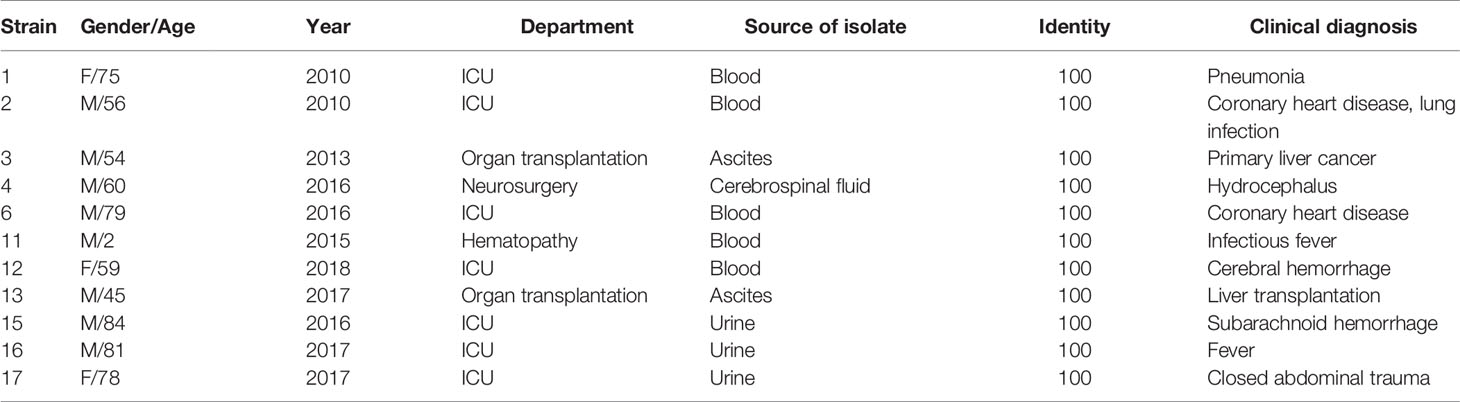
During the period from 2010 to 2018 (Table 1), 11 D. catenulata isolates were collected from seven different hospitals in five provinces from the China Hospital Invasive Fungal Surveillance Net (CHIF-NET) and the CHIF-NET North China Program. These isolates were mainly from invasive fungal infection specimens. Most specimens were obtained from patients with bloodstream infections, which indicates the seriousness of the situation for these patients.
Identification
All isolates were identified at the species level using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) conducted with Autof MS 1000 (Autobio, Zhengzhou, China) and Vitek MS (Bio Merieux, Marcy-l’Étoile, France). The species identification was confirmed via the sequencing of the rDNA internal transcribed spacer (ITS) region (ABI 3730XL, Thermo Fisher Scientific, Cleveland, OH, USA). PCR and sequencing of the amplicons were performed using the forward primers, V9G and ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′), and the reverse primers, LS266 and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Zhang et al., 2014; Hou et al., 2016). The phylogenetic tree of D. catenulata was constructed by alignment with the ITS gene sequences of the common Candida species in the NCBI gene library. Maximum-likelihood phylogenetic trees were constructed with IQ-TREE using an ultrafast bootstrap approximation approach with 1,000 replicates (Trifinopoulos et al., 2016).
Antifungal Susceptibility Testing
Minimum inhibitory concentrations (MICs, μg/ml) of nine antifungal agents, namely, anidulafungin, micafungin, caspofungin, fluconazole, posaconazole, voriconazole, itraconazole, amphotericin B, fluorocytosine, were determined for all isolates using the Sensititre YeastOne™ system (SYO, Trek Diagnostic Systems, Thermo Fisher Scientific, Cleveland, OH, USA) according to the manufacturer’s instructions. MICs of micafungin (Astellas, Japan), caspofungin (CAS; Merck, USA), fluconazole [(National Institute for Food and Drug Control (NIFDC), China], voriconazole (NIFDC, China), amphotericin B (NIFDC, China), and flucytosine (NIFDC, China) were determined using the CLSI broth microdilution method. MICs were determined after 36 h of culture. Candida krusei ATCC 6258 and C. parapsilosis ATCC 22019 were used as quality control strains. When the MIC obtained by the two methods fell within a twofold dilution gradient, the essential agreement (EA) between Sensititre™ YeastOne™ and CLSI was considered for D. catenulata. The clinical breakpoints of antifungals against D. catenulata in vitro have not yet been established by the Clinical and Laboratory Standards Institute (CLSI) or the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Consequently, the interpretative criteria were followed to test the in vitro susceptibility of Candida spp. according to the CLSI M59 guidelines (CLSI, 2017).
DNA Extraction and Sequencing of ERG11 and FKS1
On comparing published amino acid sequences of ERG11 (3641571, 1466526) and FKS1 (3639844, 856398) with D. catenulata CBS 565 whole-genome sequences (PJEZ00000000.1) and conducting gene annotation, we identified the ERG11 and FKS1 homologous proteins in C. catenulata. Amino acid alignment of Erg11p and Fks1p from published data and whole-genome sequencing of analyzed data is shown in Supplementary Figures 1, 2.
ERG11 and FKS1 Sequencing
Genomic ERG11 and FKS1 were amplified by PCR using specific primers and sequenced as previously described. The forward (5′- ACATTATTTATTGCCCCATG-3′) and reverse primers (5′-GCAAGTATCCCGCTTTTCCC-3′) for ERG11 and those for FKS1 are shown in Supplementary Table 1.
Nucleotide Sequence Accession Numbers
The ITS region sequences of strains found in this study were deposited in GenBank with accession numbers MW624477 to MW624482.
Literature Review
This literature review considered the available data regarding the susceptibility of the D. catenulata species to antifungals. The literature search was performed on September 3, 2021, using the following three databases: PubMed (https://pubmed.ncbi.nlm.nih.gov), Web of Science (https://webofknowledge.com), and Embase (https://www.embase.com). The terms “Diutina catenulates” or “Candida catenulate” or “Candida ravautii” were entered in the category of “Title/Abstract” in the PubMed Advanced Search Builder, and “TS=(Diutina catenulata)” or “TS=(Candida catenulata)” or “TS=(Candida ravautii)” was entered in the Web of Science databases. The search in Embase was conducted in the advanced search area, including the terms “‘diutina catenulate’:ti,ab,kw” or “‘candida catenulata’:ti,ab,kw” or “‘candida ravautii’:ti,ab,kw.” Studies describing the patients infected with D. catenulata were selected and summarized.
Results
Species Identification of D. catenulata Using MALDI-TOF and DNA Sequencing
All 11 clinical isolates were identified as D. catenulata by the Autof MS 1000 and Vitek MS. The ITS sequences of the study isolates exhibited 100% sequence identity to the corresponding ITS sequences from the reference D. catenulata isolates in GenBank (C. catenulata CBS 565). We performed a phylogenetic analysis using ITS sequences (Figure 1).
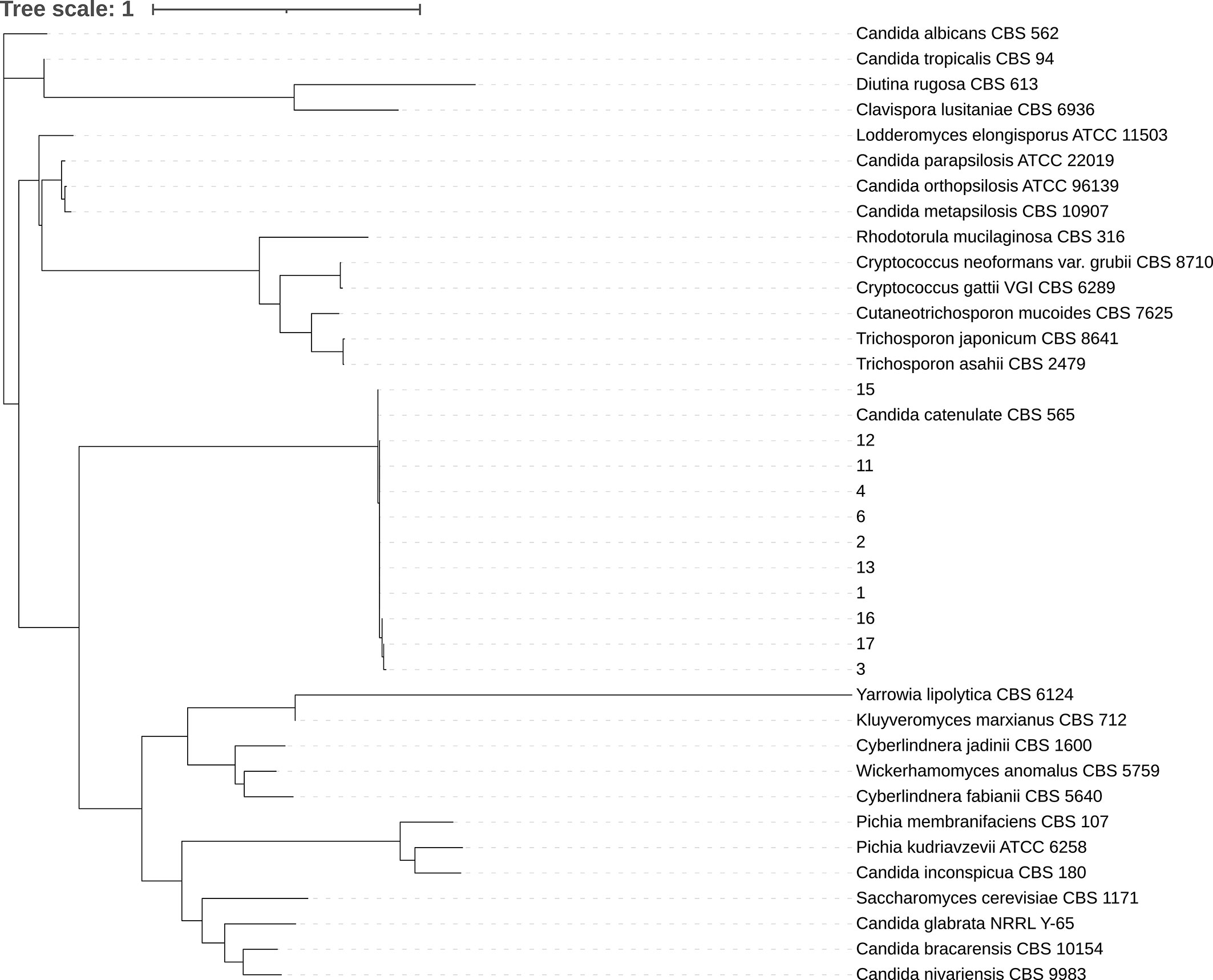
Figure 1 Phylogenetic tree. Internal transcribed spacer (ITS) sequences of nuclear rDNA of Candida spp. were used. The ITS sequence of the model strain was obtained from NCBI gene bank. CLC sequence viewer software was used for sequence alignment and IQ-Tree ultrafast bootstrap with 1,000 replicates was used for tree construction.
This study comprised 72.7% (8/11) males and 27.3% (3/11) females, with an average age of 58 years. Among the specimens, blood accounted for 45.5% (5/11), urine for 27.3% (3/11), ascites for 18.2% (2/11), and cerebrospinal fluid for 9.1% (1/11). The patients belonged to the following departments: intensive care unit (ICU) 45.5% (5/11), organ transplant department 18.2% (2/11), neurosurgery department 18.2% (2/11), hematology department 9.1% (1/11), and healthcare department 9.1% (1/11).
Antifungal Susceptibility
The quality control strains (C. krusei ATCC 6258 and C. parapsilosis ATCC 22019) showed MICs within the expected ranges. Aggregated MIC distributions of the nine antifungal agents tested against D. catenulata isolates by YeastOne™ are shown in Table 2. For all 11 D. catenulata isolates, the MICs of itraconazole and posaconazole were in the range of 0.06 to 0.12 μg/ml. In addition, 63.6% (7/11) of D. catenulata isolates exhibited high MIC values for fluconazole (MIC >8 μg/ml) and 81.8% (9/11) showed high MIC values (MIC ≥4 μg/ml) for caspofungin. A total of 54.5% (6/11) of D. catenulata isolates showed MICs of ≥1 μg/ml for anidulafungin. A total of 36.4% (4/11) of D. catenulata isolates showed high MICs for micafungin (MIC ≥1 μg/ml). The MICs of amphotericin B and 5-flucytosine against all D. catenulata isolates ranged from 0.25 to 1 and <0.06 to 0.12 μg/ml, respectively. Four D. catenulata isolates exhibiting increased MICs of ≥1 μg/ml for all three echinocandins were obtained in our study. Furthermore, the MIC values of echinocandins and fluconazole were high for five isolates of D. catenulata.
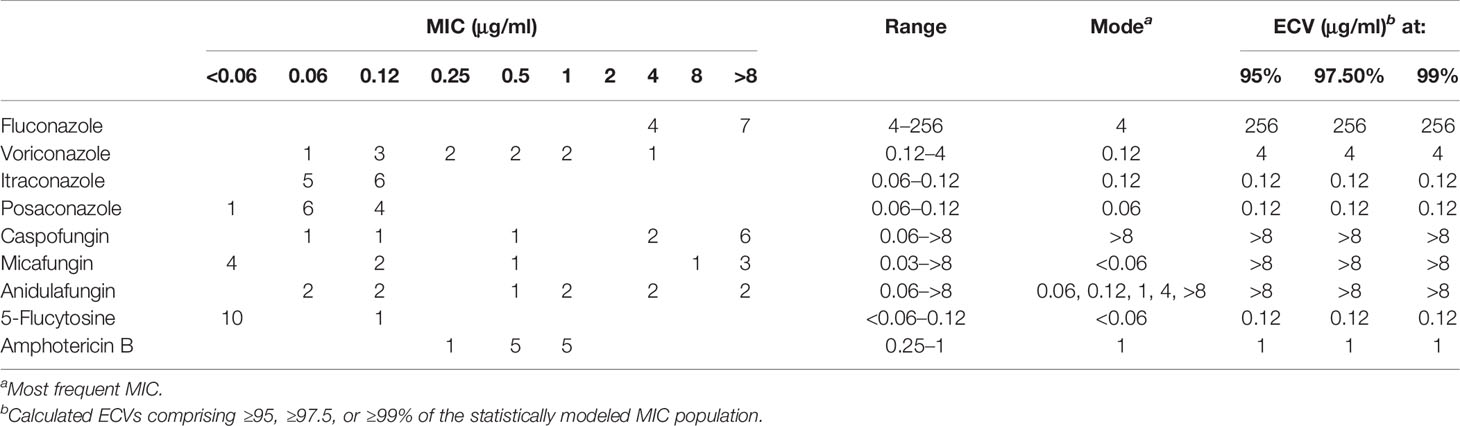
Table 2 Epidemiological cutoff values (ECV) of nine antifungal agents based on aggregated minimum inhibitory concentration (MIC) distributions of Diutina catenulata.
Agreement Between the CLSI Method and Sensititre YeastOne™
The EA values of the MICs between the CLSI method and YeastOne™ for a majority of the antifungal drugs tested were >90%. In triazoles, 100% EA values were obtained for amphotericin B and 5-flucytosine. EA values for caspofungin and micafungin were 81.9 (9/11) and 91% (10/11), respectively (Supplementary Table 2).
Sequence Analysis of the ERG11 Gene in the Clinical Isolates
The complete open reading frame of D. catenulata ERG11 gene, predicted using bioinformatics analysis, showed that ERG11 comprises 1,563 base pairs (Table 3). The ERG11 gene was sequenced for all 11 D. catenulata isolates. These isolates displayed amino acid substitutions in Erg11p compared with the sequences in GenBank. The SNP F126L (C378G) and synonymous mutations C681T, C888A, C999T, and C1164T were found in two isolates with high fluconazole MICs (>32 μg/ml). Amino acid substitutions, F126L (C378G) and G417K (L139F), and synonymous mutations, C681T, C888A, C999T, and C1164T, were found in three isolates with fluconazole MICs >64 μg/ml. Additionally, one isolate with K143R (A428G) and synonymous mutations, C681T, C888A, C999T, and C1164T, showed high resistance to fluconazole (MIC >256 μg/ml). Only one resistant strain had the K143R (A428G) mutation. In addition, synonymous mutations G306A and C852T were found in three isolates (Table 4). By aligning the amino acid sequences of Erg11P from C. albicans and S. cerevisiae, we found that Erg11p of D. catenulata shared 69.75 and 65.25% identity with that of C. albicans and S. cerevisiae, respectively. Notably, the C. catenulata Erg11p contained residues at three locations (F126, L139, K143) that corresponded with three residues (F126, L139, K143, respectively) in C. albicans ERG11 (Supplementary Figure 1) whose alterations (F126L, K143R) have been reported to be associated with azole resistance (Berkow and Lockhart, 2017).
Sequence Analysis of FKS1 Gene in Clinical Isolates
The complete open reading frame of D. catenulata FKS1, determined by bioinformatics analysis, showed that FKS1 comprises 5,652 base pairs (Table 3). The FKS1 gene was sequenced for all 11 D. catenulata clinical isolates. We found 27 SNPs (A591G, G732C, T1119C, C1194T, G1203A, C1299T, G1365T, A1554G, T1605C, T1861A, C1874T, T2079C, C2100T, C2682T, C2781G, G2871A, G3036A, C3273T, G3367A, T4043G, T4062G, G4230C, C4462T, C4704T, T4977C, C5006T, and 5088C) (Supplementary Table 3). The SNP T4062G was found in an isolate with high aggregated MIC distributions for echinocandin (≥8 μg/ml). By aligning the amino acid sequence of Fks1p of D. catenulata with those of the corresponding proteins in C. albicans and S. cerevisiae (which had 83.51 and 71.29% identities with D. catenulata, respectively), we found seven amino acid substitutions in the Fks1p of D. catenulata (Supplementary Table 3). Amino acid substitution F621I was found in one isolate (Isolate 2) with capofungin MIC >8 µg/ml. Substitution I1348S was found in three isolates with echinocandin MIC ≥4 µg/ml (isolates 15, 16, and 17). Additionally, amino acid substitutions S625L and F1354L were found in one isolate with an MIC ≥8 μg/ml for three types of echinocandins (Isolate 13) (Table 5).
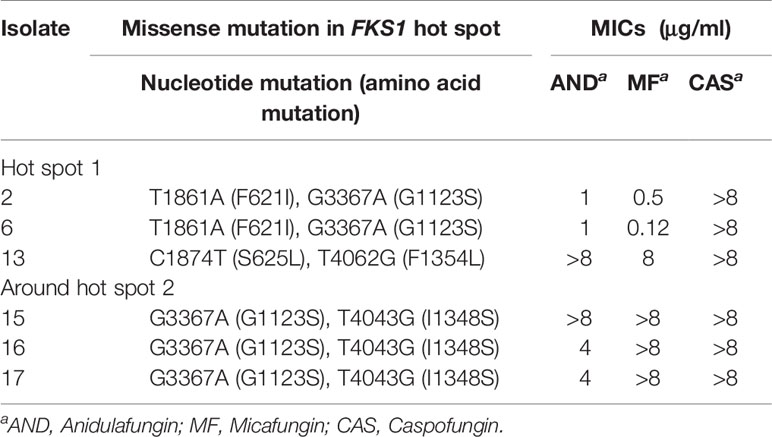
Table 5 The variability of FKS1 gene in the hot spot 1 and near hot spot 2 regions in clinical D. catenulata isolates.
Literature Review
Only three of the sourced articles had reported clinical cases of infection with D. catenulata, including one case, in which the D. catenulata was isolated from the nails and two cases of D. catenulata isolated from blood culture (Crozier, 1977; Radosavljevic et al., 1999; Ha et al., 2018). One of the patients was diagnosed with gastric cancer and the other with endocarditis. One of the isolates showed low MICs for amphotericin B, miconazole, econazole, tioconazole, ketoconazole (1 μg/ml), traconazole (0.25 μg/ml), and fluconazole (16 μg/ml). Other isolates showed low MICs for fluconazole and micafungin (2 and 0.06 μg/ml, respectively). In addition, based on the patients’ clinical information, we can conclude that broad-spectrum antibiotic therapy may be a risk factor for infection with D. catenulata (Supplementary Table 4).
Discussion
With the development of identification technologies, such as MALDI-TOF MS and high-throughput sequencing, many hitherto unknown species of microorganisms have been precisely identified (Qiu et al., 2019). Diutina catenulata is a rare yeast that causes infections in humans (Radosavljevic et al., 1999; Ha et al., 2018; Çakır et al., 2019). Therefore, we determined the proportion of D. catenulata isolates among clinical strains recovered from patients using CHIF-NET and the CHIF-NET North China Program. DNA sequencing and MALDI-TOF MS can be used to identify D. catenulata at the species level, and the proportions of the different species in the sample can be compared. ITS sequencing identified all 11 isolates, and the consistency of these clinical isolates with CBS 565 was 100%. Currently, ITS sequencing is considered a reliable marker for fungal identification (Wang et al., 2012).
Most Candida species are pathogens that cause infections in immunocompromised patients (Cernakova et al., 2019). In this study, D. catenulata infection mainly occurred in elderly male admitted ICU patients, and nearly half of the infections were bloodstream infections. There are differences in drug resistance rates among Candida species (Sanguinetti et al., 2015). Candida auris has attracted much attention worldwide due to its high drug resistance and high fatality rates (Lone and Ahmad, 2019). Despite the low frequency of the species, the high MIC values observed for D. catenulata potentially could be important. In vitro antifungal susceptibility results showed that all D. catenulata showed low MICs for itraconazole, posaconazole, amphotericin B, and 5-flucytosine, but four isolates showed high MICs (≥4 μg/ml) for the echinocandins. More than 36.4% of the isolates showed high MIC values for two echinocandins. High MICs of caspofungin may predict high MICs of micafungin and anidulafungin. More than 72.7% of the isolates showed high MICs for fluconazole. Thus, we suggest that D. catenulata with high MICs for fluconazole should be evaluated for its susceptibility to other antifungals. Micafungin has excellent effects in vivo against D. catenulata in infants (Çakır et al., 2019). However, in the present study, our in vitro antifungal susceptibility test showed that 36.4% of isolates showed high MICs (≥4 μg/ml) for echinocandins.
To analyze the molecular mechanisms of the antifungal resistance of D. catenulata, we sequenced the ERG11 and FKS1 genes. In agreement with the azole drug susceptibility profiles, we found three mutations in ERG11 of D. catenulata. Interestingly, F126L and K143R are the most prevalent residue substitutions in ERG11 (Berkow et al., 2017), but the L139F substitution is an unreported new observation. The mutations in FKS1 showed polymorphism. We identified two mutations in FKS1 hot spot 1 [F621I (F641) and S625L (S645)] and one near FKS1 hot spot 2 [I1348S (I1368)] associated with high MICs of caspofungin (Toutounji et al., 2019). S625L (S645) has been reported to cause failure of micafungin treatment against C. albicans (Slater et al., 2011). G3367A was found in isolates showing high and low MIC values against caspofungin and therefore may not be a potential of high MIC values against this antifungal. Of note, caspofungin was found to have a high interlaboratory variation (Espinel-Ingroff et al., 2013) and therefore the high MIC observed for this antifungal may not necessarily be a cause of concern as long as the MIC of the micafungin and anidulafungin are included and remain low. Polymorphisms of D. catenulata may also be used as a basis for typing.
Compared with the three articles have been described, we have noticed that the MICs of fluconazole and echinocandin of some of our strains were higher than 64 and 4 µg/mL, respectively, which are significantly higher than previous studies. This obvious difference also shows that there may be some evolutionary differences between Chinese and foreign isolates. In addition, our ICU patients also are treated with antibiotics. Therefore, patients maybe at high risk of D. catenulata infection. Similar to the case with most studies, lack of detailed clinical data on antifungal treatment is a major limitation of our study, which precludes us from reaching a conclusive statement about the choice of antifungal treatment and their potential outcome.
In conclusion, because of the low isolation rates, epidemiological knowledge of rare species is critical for clinical treatment. To the best of our knowledge, this study is the first to report the occurrence and distribution of D. catenulata in China, and its findings are expected to guide clinical treatment of D. catenulata infection in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
X-FC and WZ conceived and designed the experiment. XF, WK, GZ, HZ, W-HY, J-WW, D-WG, Z-YS, Z-JC, L-GZ, X-FD, Y-HP, BL, and HH contributed reagents/materials/analysis tools. X-FC, X-YL, J-JH, Y-XL, and WZ performed the experiments. X-FC and WZ analyzed the data and wrote the manuscript. Y-CX, XF, XH, and Y-XL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Beijing, China (Grant No. 7204288), National Natural Science Foundation of China (82002178 and 81971979), Special Foundation for National Science and Technology Basic Research Program of China (2019FY101200), Beijing Municipal Science and Technology Project (Z181100001618015), Beijing Key Clinical Specialty for Laboratory Medicine-Excellent Project (No. ZK201000), and Scientific Research Foundation Project of Hebei Provincial Health Commission (Grant No. 20210702, 20190904, 20180843).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank all the hospitals involved in the CHIF-NET study and Ms. Yan-Ting Cai (Autobio Labtec Instruments Co., Ltd.) and Ms. Li-Li Wang (Zhuhai Meihua Medical Technology Co., Ltd.) for technical support with MALDI-TOF MS identification of the Candida isolates. Dr. Bin Cheng (Beijing Miyun Hospital, Capital Medical University), Ms. Li-Li Wang, Ms. Yan-Ting Cai, Mr. Cheng-Hai Liu (Zhuhai DL Biotech Co., Ltd.), Mr. Wen-Pan Guo (Zhuhai DL Biotech Co., Ltd.), and Ms. Qi Shu (Bioyong Technologies Inc.) took macro and micro photos of the strains. Finally, we would like to thank Shu-Ying Yu, Pei-Yao Jia, Jing-Jia Zhang, and Ya-Ting Ning (Peking Union Medical College Hospital) for their valuable suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.739496/full#supplementary-material
References
Arastehfar, A., Daneshnia, F., Hilmioglu-Polat, S., Ilkit, M., Yasar, M., Polat, F., et al. (2021). Genetically Related Micafungin-Resistant Candida parapsilosis Blood Isolates Harbouring Novel Mutation R658G in Hotspot 1 of Fks1p: A New Challenge? J. Antimicrob. Chemother. 76, 418–422.
Babaei, F., Habibi, A. (2018). Fast Biodegradation of Diesel Hydrocarbons at High Concentration by the Sophorolipid-Producing Yeast Candida Catenulata KP324968. J. Mol. Microb. Biotech. 28, 240–253. doi: 10.1159/000496797
Berkow, E. L., Lockhart, S. R. (2017). Fluconazole Resistance in Candida species: A Current Perspective. Infect. Drug Resist. 10, 237–245. doi: 10.2147/IDR.S118892
Cafarchia, C., Iatta, R., Danesi, P., Camarda, A., Capelli, G., Otranto, D. (2019). Yeasts Isolated From Cloacal Swabs, Feces, and Eggs of Laying Hens. Med. Mycol. 57, 340–345. doi: 10.1093/mmy/myy026
Çakır, S., Çelebi, S., Özkan, H., Köksal, N., Dorum, B. A., Yeşil, E., et al. (2019). Results of the Use of Micafungin in Newborns. Mikrobiyol. Bul. 53, 70–80. doi: 10.5578/mb.67599
Cernakova, L., Light, C., Salehi, B., Rogel-Castillo, C., Victoriano, M., Martorell, M., et al. (2019). Novel Therapies for Biofilm-Based Candida Spp. Infections Adv. Exp. Med. Biol. 1214, 93–123. doi: 10.1007/5584_2019_400
Clinical and Laboratory Standards Institute [CLSI] (2020). Epidemiological Cutoff Values for Antifungal Susceptibility Testing. 3nd ed. CLSI supplement M59. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2017). Performance Standards for Antifungal Susceptibility Testing of Yeasts, CLSI Supplement M60, 1st Edn. Wayne, PA: CLSI.
Crozier, W. J. (1977). A Case of Onychomycosis Due to Candida Ravautii. Aust. J. Dermatol. 18, 139–140. doi: 10.1111/j.1440-0960.1977.tb00027.x
Espinel-Ingroff, A., Arendrup, M. C., Pfaller, M. A., Bonfietti, L. X., Bustamante, B., Canton, E., et al. (2013). Interlaboratory variability of Caspofungin MICs for Candida spp. Using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob. Agents Chemother. 57, 5836–5842.
Glushakova, A. M., Rodionova, E. N., Kachalkin, A. V. (2021). Yeasts in Feces of Pigeons (Columba Livia) in the City of Moscow. Curr. Microbiol. 78, 238–243. doi: 10.1007/s00284-020-02251-5
Ha, M. V., Choy, M. S., McCoy, D., Fernandez, N., Suh, J. S. (2018). Candida Catenulata Candidaemia and Possible Endocarditis in a Cirrhotic Patient Successfully De-Escalated to Oral Fluconazole. J. Clin. Pharm. Ther. 43, 910–913. doi: 10.1111/jcpt.12728
Hou, X., Xiao, M., Chen, S. C., Wang, H., Zhang, L., Fan, X., et al. (2016). Sequencer-Based Capillary Gel Electrophoresis (SCGE) Targeting the rDNA Internal Transcribed Spacer (ITS) Regions for Accurate Identification of Clinically Important Yeast Species. PloS One 11, e0154385. doi: 10.1371/journal.pone.0154385
Joo, H. S., Ndegwa, P. M., Shoda, M., Phae, C. G. (2008). Bioremediation of Oil-Contaminated Soil Using Candida Catenulata and Food Waste. Environ. Pollut. 156, 891–896. doi: 10.1016/j.envpol.2008.05.026
Lone, S. A., Ahmad, A. (2019). Candida Auris-the Growing Menace to Global Health. Mycoses 62, 620–637. doi: 10.1111/myc.12904
O'Brien, C. E., McCarthy, C. G. P., Walshe, A. E., Shaw, D. R., Sumski, D. A., Krassowski, T., et al. (2018). Genome Analysis of the Yeast Diutina Catenulata, a Member of the Debaryomycetaceae/Metschnikowiaceae (CTG-Ser) Clade. PloS One 13, e0198957. doi: 10.1371/journal.pone.0198957
Pristov, K. E., Ghannoum, M. A. (2019). Resistance of Candida to Azoles and Echinocandins Worldwide. Clin. Microbiol. Infect. 25, 792–798. doi: 10.1016/j.cmi.2019.03.028
Qiu, W., Huang, Y., Zhao, C., Lin, Z., Lin, W., Wang, Z. (2019). Microflora of Fresh White Button Mushrooms (Agaricus Bisporus) During Cold Storage Revealed by High-Throughput Sequencing and MALDI-TOF Mass Spectrometry Fingerprinting. J. Sci. Food Agric. 99, 4498–4503. doi: 10.1002/jsfa.9695
Radosavljevic, M., Koenig, H., Letscher-Bru, V., Waller, J., Maloisel, F., Lioure, B., et al. (1999). Candida Catenulata Fungemia in a Cancer Patient. J. Clin. Microbiol. 37, 475–477. doi: 10.1128/JCM.37.2.475-477.1999
Sanguinetti, M., Posteraro, B., Lass-Florl, C. (2015). Antifungal Drug Resistance Among Candida Species: Mechanisms and Clinical Impact. Mycoses 58 (Suppl 2), 2–13. doi: 10.1111/myc.12330
Slater, J. L., Howard, S. J., Sharp, A., Goodwin, J., Gregson, L. M., Alastruey-Izquierdo, A., et al. (2011). Disseminated Candidiasis Caused by Candida Albicans With Amino Acid Substitutions in Fks1 at Position Ser645 Cannot be Successfully Treated With Micafungin. Antimicrob. Agents Chemother. 55, 3075–3083. doi: 10.1128/AAC.01686-10
Subramanya, S. H., Sharan, N. K., Baral, B. P., Hamal, D., Nayak, N., Prakash, P. Y., et al. (2017). Diversity, in-Vitro Virulence Traits and Antifungal Susceptibility Pattern of Gastrointestinal Yeast Flora of Healthy Poultry, Gallus Gallus Domesticus. BMC Microbiol. 17, 113. doi: 10.1186/s12866-017-1024-4
Toutounji, M., Tokajian, S., Khalaf, R. A. (2019). Genotypic and Phenotypic Characterization of Candida Albicans Lebanese Hospital Isolates Resistant and Sensitive to Caspofungin. Fungal Genet. Biol. 127, 12–22. doi: 10.1016/j.fgb.2019.02.008
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., Minh, B. Q. (2016). W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Wang, H., Xiao, M., Chen, S. C., Kong, F., Sun, Z. Y., Liao, K., et al. (2012). In Vitro Susceptibilities of Yeast Species to Fluconazole and Voriconazole as Determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) Study. J. Clin. Microbiol. 50, 3952–3959. doi: 10.1128/JCM.01130-12
Zhang, L., Xiao, M., Wang, H., Gao, R., Fan, X., Brown, M., et al. (2014). Yeast Identification Algorithm Based on Use of the Vitek MS System Selectively Supplemented With Ribosomal DNA Sequencing: Proposal of a Reference Assay for Invasive Fungal Surveillance Programs in China. J. Clin. Microbiol. 52, 572–577. doi: 10.1128/JCM.02543-13
Keywords: Diutina catenulata (Candida catenulata), antifungal susceptibility, ERG11, FKS1, gene mutation, drug resistance mechanisms
Citation: Chen X-F, Zhang W, Fan X, Hou X, Liu X-Y, Huang J-J, Kang W, Zhang G, Zhang H, Yang W-H, Li Y-X, Wang J-W, Guo D-W, Sun Z-Y, Chen Z-J, Zou L-G, Du X-F, Pan Y-H, Li B, He H and Xu Y-C (2021) Antifungal Susceptibility Profiles and Resistance Mechanisms of Clinical Diutina catenulata Isolates With High MIC Values. Front. Cell. Infect. Microbiol. 11:739496. doi: 10.3389/fcimb.2021.739496
Received: 11 July 2021; Accepted: 28 September 2021;
Published: 29 October 2021.
Edited by:
Min Chen, Shanghai Changzheng Hospital, ChinaReviewed by:
Shuwen Deng, Suzhou High-tech Zone People’s Hospital, ChinaAylin Döğen, Mersin University, Turkey
Amir Arastehfar, Westerdijk Fungal Biodiversity Institute, Netherlands
Copyright © 2021 Chen, Zhang, Fan, Hou, Liu, Huang, Kang, Zhang, Zhang, Yang, Li, Wang, Guo, Sun, Chen, Zou, Du, Pan, Li, He and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-Chun Xu, eHljcHVtY2hAMTM5LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Xin-Fei Chen
Xin-Fei Chen Wei Zhang
Wei Zhang Xin Fan
Xin Fan Xin Hou
Xin Hou Xiao-Yu Liu1,3
Xiao-Yu Liu1,3 Jing-Jing Huang
Jing-Jing Huang Ge Zhang
Ge Zhang Han Zhang
Han Zhang Jin-Wen Wang
Jin-Wen Wang Da-Wen Guo
Da-Wen Guo Zi-Yong Sun
Zi-Yong Sun Bin Li
Bin Li Ying-Chun Xu
Ying-Chun Xu