Abstract
MHC class I antigen processing is an underappreciated area of nonviral host–pathogen interactions, bridging both immunology and cell biology, where the pathogen’s natural life cycle involves little presence in the cytoplasm. The effective response to MHC-I foreign antigen presentation is not only cell death but also phenotypic changes in other cells and stimulation of the memory cells ready for the next antigen reoccurrence. This review looks at the MHC-I antigen processing pathway and potential alternative sources of the antigens, focusing on Mycobacterium tuberculosis (Mtb) as an intracellular pathogen that co-evolved with humans and developed an array of decoy strategies to survive in a hostile environment by manipulating host immunity to its own advantage. As that happens via the selective antigen presentation process, reinforcement of the effective antigen recognition on MHC-I molecules may stimulate subsets of effector cells that act earlier and more locally. Vaccines against tuberculosis (TB) could potentially eliminate this disease, yet their development has been slow, and success is limited in the context of this global disease’s spread. This review’s conclusions set out potential directions for MHC-I-focused approaches for the next generation of vaccines.
Introduction
Mycobacterium tuberculosis (Mtb) is a causative agent of tuberculosis—an infectious disease responsible for ten million cases and over a million deaths every year (WHO, 2020). Despite the availability of antibiotics (Paulson, 2013) and vaccination with an attenuated form of Mycobacterium bovis, the Bacilli Calmette-Guérin (BCG), TB eventually kills 45% of HIV-negative people and nearly all HIV-positive individuals (WHO Key Facts, 2022).
Major histocompatibility complex receptor classes I and II (MHC-I and MHC-II) are two families of receptors involved in the recognition of “self” and surveillance of “foreign” antigens. While class II receptors evolved for the defense against pathogens and are present primarily in the immune cells, class I receptors are much more ubiquitous, also on MHC-II-negative cells. Ubiquitous presence may play a unique role in multiple-tissue surveillance, pathogen detection, and restriction of disease dissemination. MHC-I (or HLA class I) contributes both to innate immunity through the engagement of natural killer (NK) cells and to adaptive immunity through peptides presented to cytotoxic T cells (Jiang et al., 2019; Uzhachenko and Shanker, 2019). In contrast, MHC-II is solely dedicated to adaptive immunity (Leddon and Sant, 2010).
MHC-I molecules constitute a fundamental aspect of self in self-organizing networks of multicellular organisms starting from jawed vertebrates. Self is a process of continuous rebalancing of different biochemical and intercellular interactions rather than a constitutive feature of a single cell, as proposed initially by Turing (1990). MHC-I molecules present a snapshot of the intracellular proteome from all cell compartments by uploading self-peptides, byproducts of the reactions taking place in the cytosol. This proteome is recognized by T cells selected both in the thymus and periphery to have autoreactive cells eliminated or repressed (Perreault, 2010). MHC-I molecules can also present self-peptides that are mutated, as well as pathogen-derived mimicry peptides (Trost et al., 2010). The aim of the antigen presentation is the recurring recognition of cumulative signals up to the threshold where the “non-self” cell is destructed by adaptive immune cells, mainly CD8+ T cells, while protecting healthy cells from NK-mediated cytolysis. This requires coordinated action involving a series of brief encounters between antigen-presenting cells and their immune counterparts. As MHC-I molecules feature on every nucleated cell of the organism, they visualize pathological processes that occur in the organism at an early stage. Peptide presentation on MHC-I molecules, recognition by T-cell receptors (TCR), and immune cell activation are three stages in which cell interactions are inspected for affinity binding and initial signal strength, duration of stimulation, and decay (Segura et al., 2008; Pathni et al., 2022). Historically, the MHC-I system has been linked to transplant immunology and viral infections. MHC-I antigen presentation may contribute to the development of new vaccination strategies against chronic bacterial infections.
Structure and diversity of MHC-I molecules in macrophage
The major histocompatibility complex (MHC) locus, found on the short arm of chromosome 6 in humans, encodes genes from three classes of proteins (MHC-I, MHC-II, and MHC-III). There are three MHC-I groups, termed classical human leukocyte antigens A, B, and C (HLA), and several non-classical HLA groups, such as HLA-E, F, and G, a cluster of differentiation 1 (CD1) molecules, and MHC-I-related protein (MR1). Classical MHC-I molecules show a high degree of polymorphism, resulting in allele diversity predicted to encompass eight to nine million variants, although 80% of these occur only rarely and are represented by alleles differing by single base point mutations (Robinson et al., 2017). In humans, the most commonly found alleles are clustered into three major groups A–C, and 42 core classical types further grouped into 12 supertypes based on the similarities in peptide-binding motifs (Sette and Sidney, 1999). HLA class I supertypes have been linked to susceptibility and severity of tuberculosis (Balamurugan et al., 2004). Conversely, genes coding nonclassical HLA-E, HLA-F, and HLA-G are highly conserved as one or two alleles, often tissue-restricted, and involved in molecular mechanisms underpinning immune tolerance and fetus acceptance in pregnancy (Moscoso et al., 2006). Tissue expression of MHC-I molecules varies. In some cell subsets, such as postmitotic neurons, it is very limited (Neumann et al., 1997); in others, such as lymphocytes, MHC-I constitutive expression reaches up to 1% of the membrane protein content (Joly et al., 1991). It is also subject to transcriptional activation regulators TAF1, USF1/2, and CIITA (Howcroft et al., 2003).
Classical MHC-I molecules are made with a heavy glycoprotein α chain, composed of three immunoglobulin-like domains α-1, α-2, and α-3, a transmembrane segment with a cytosolic tail, and a smaller noncovalently attached light β chain (β2-microglobulin). Processed antigen fragments in the form of 8 to 10 amino acid peptide chains are affinity bound with each amino acid contributing to the overall MHC affinity score (Lundegaard et al., 2010). The binding pocket is formed by two conformational domains of the heavy chain, α1 and α2, supported by the β2 chain and its own residues to prevent binding of the longer peptides that would need to bulge outside the pocket. There are usually two anchor residues within the octo- or nonameric peptide, one of which is predominantly at the C-terminus (Falk and Rötzschke, 1993). It is different in MHC-II class molecules where two similarly sized α and β chains form an open-ended binding site so that typically 13–18 amino acid long peptides can extend out (Germain, 1994). In both MHC-I and MHC-II molecules, there are pockets present in the binding groove that accommodate side chains of the peptides; physicochemical features of these pockets underpin allele-specific consensus motifs (Falk et al., 1991). In MHC-I molecules, the conformational stability of the whole molecule is equally dependent on MHC and the bound peptide; it disassociates with any of the components’ removal, leading to MHC-I recycling from the cell surface. The peptide-MHC (pMHC) complex is recognized by the T-cell receptors and its CD8+ co-receptor, which binds to the nonpolymorphic α3 domain in the heavy chain (Huppa et al., 2010).
Nonclassical MHC-like, MHC-Ib, or MHC-related molecules resemble the classical receptors with their heavy chains but do not always associate with β2 microglobulin, forming homodimers instead. They include MR1, HLA-E, HLA-F, HLA-G, and CD1 molecules. MR1 is an MHC-Ib molecule that uniquely binds small molecules of the microbial metabolome. Several isoforms of MR1 have been detected, albeit only one, MR1A bears a close resemblance to the classical MHC-I molecules and is fully functional (Riegert et al., 1998). Other isoforms are either nonfunctional or with uncertain properties, like MR1B, which forms homodimers that do not associate with the β2 light chain. Although these isoforms may not be functional in the antigen-presenting process, they can still play role in intracellular trafficking. HLA-E or Qa-1 binds leader sequences of other MHC-I molecules and, as such, is a sensor for their expression and checkpoint in antigen presentation to NK cells. In homeostasis, HLA-E binds only a restricted set of nonamers (O’Callaghan and Bell, 1998).
The CD1 proteins’ binding groove is deeper and lined almost entirely with nonpolar or hydrophobic amino acid chains. This groove can accommodate a long hydrophobic lipid tail inside its pockets and expose the hydrophilic part on the surface, where it directly contacts the T-cell antigen receptor. In contrast to MHC-I molecules, where the peptides are trimmed to 9–11Aa-long peptides, lipids bound by CD1 proteins are not cleaved but adapted within the hydrophobic clefts, which can accommodate up to C70–C80-long chains. In humans, there are four members of antigen-presenting CD proteins: CD1a, CD1b, CD1c, and CD1d, and CD1e, which is a soluble carrier in the endolysosomal network for other CD1-lipid complexes (Moody et al., 1999; Ly and Moody, 2014; Moody and Suliman, 2017).
Finally, for completeness, in humans, there are MHC class I chain-related molecules that are not involved in antigen presentation but are still involved in immune responses. These are MICA and EPCR proteins, with MICA being highly polymorphic glycoproteins that are expressed ubiquitously and serve as a “danger signal” for NK cells, γδ T cells, and CD8+ T cells via NKG2D receptors (Schrambach et al., 2007). EPCR, a 46-kDa protein, shares ~20% homology with the CD1d molecule. They are expressed in the vascular system (endothelium) as well as on various innate immune cells; they are ligands for TCR of Vδ2 γδ T cells. MICA and EPCR proteins, respectively, regulate inflammation and coagulation (Willcox et al., 2012).
Mycobacterium tuberculosis as an intracellular microbe: Role of the phagosome
Mtb is an intracellular microbe that can infect any tissue, but the lung is the main niche for its transmission (Torrelles and Schlesinger, 2017). The presence of the asymptomatic latent phase in a significant proportion of immunocompetent infected individuals makes this pathogen life cycle akin to other human pathobionts that cause harm only under host–pathogen disequilibrium (Hakansson et al., 2018). The typical Mtb life cycle includes a phase of the primary disease followed by a stage of long-term occult and persistent intracellular infection. During this “latent” period, bacilli survive in a non-vegetative dormant form characterized by a thick lipid-rich bacterial cell wall and lipid accumulations (Vázquez et al., 2014) protecting from the degrading activity of host autophagolysosomal enzymes (Figure 1).
Figure 1
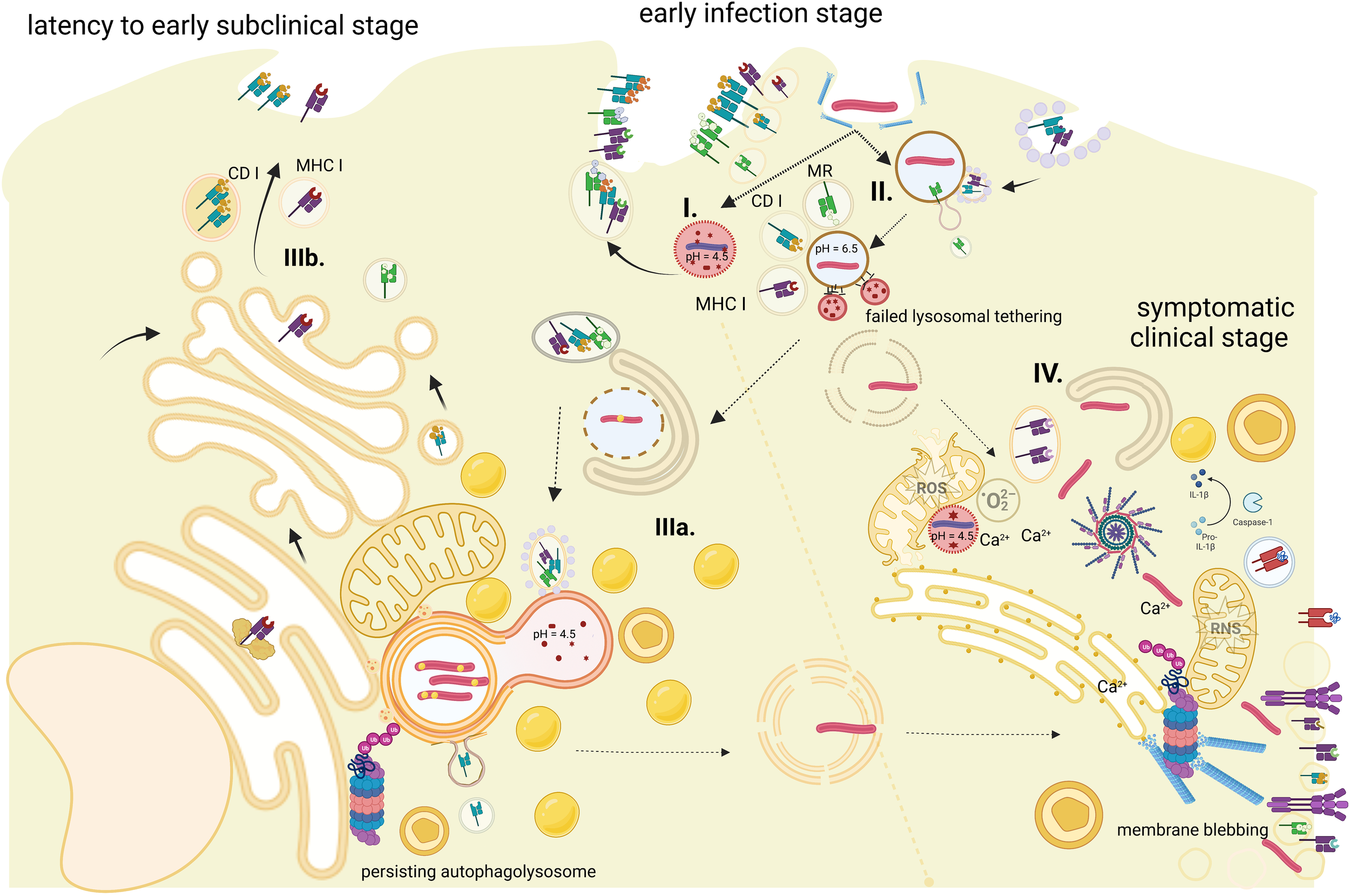
Antigen processing for MHC-I presentation during the course of Mtb infection. The figure represents stages of phagocytosed bacilli (pathways are time- and site-specific). (I) The phagolysosome, which disables pathogen—presented antigens are from fragmented dead bacilli. (II) Persistent phagosomes where fusion with lysosome was blocked successfully by the bacilli—presented antigens are virulent factors released by live Mtb. (III) Autophagolysosomes with persistent latent bacilli—presented antigens are scanty and a by-product of pathogen–host cellular organelles interactions: (IIIa) stage of autophagosome nucleation initiation and (IIIb) a stage of persisting autophagolysosome via contact sites with ER and classical MHC-I antigen processing route. (IV) Cytosol bacilli—stage of active infection with Mtb overtaking cell innate defense, generalized disruption of cell functions, and likely progress to cell death. Dashed arrows represent pathogen transition; solid arrows represent sources of the antigen. Created with BioRender.com.
Alveolar macrophages serve as the first port of entry and primary host for Mtb, killing it and presenting antigens while also sheltering persistent bacilli (van Crevel et al., 2003). The process of phagosomal uptake involves a range of cell receptors, including cell surface pattern recognition receptors (PRR), which bind pathogen-associated molecular patterns (PAMPs). Toll-like receptors (TLRs)on the cell surface (TLR1, TLR2, TLR4, TLR6) and endosomal membranes (TLR3, TLR7, TLR8, TLR9) are key PRRs that are responsible for the induction of the signaling pathways downstream and the production of cytokines (Underhill et al., 1999). Interestingly, mycobacteria-led use of TLR3 appears to enhance IL-10 production (Bai et al., 2014), suggesting that the binding of Mtb RNA may influence cross-talk with other intracellular signaling pathways. It has also been shown that other receptors, like C-type lectin receptors, scavenger receptors, nucleotide-binding-oligomerization domain (NOD)-like receptors (NLRs), opsonin receptors, ficolin, C1q, complement receptors CR1, CR3, and CR4, and Fc receptors for bacilli opsonized with immunoglobulins, promote phagocytosis but not necessarily pathogen eradication (Hossain and Norazmi, 2013).
The arrest of phagosome maturation is an important stage in the Mtb life cycle (Figure 1). Mycobacterium tuberculosis influences acidification of the phagosome in the early phase of infection by preventing tethering of V-ATPase to Mtb-containing vacuole and stabilizing its pH at 6.3–6.5 with tyrosine phosphatase PtpA (Wong et al., 2011). Early phagosomes undergo a series of encounters with other endocytic organelles to acquire various molecules, Rab5, EEA1, PI(3)P, and VPS 34 to name a few, before they progress to the late phagosome stage. Mtb prolongs that stage by hydrolyzing PI3P by the action of PI3P-specific acid phosphatase SapM (Vergne et al., 2005); mycobacterial ManLAM blocks the transport of acidic cargo from the trans-Golgi network via interference with early endosomal antigen EEA1-syntaxin 6 interactions (Fratti et al., 2003) and protein kinase G (pknG) induces continued accumulation of Rab5 and prevents Rab7 acquisition, further delaying phagosome maturation (Roberts et al., 2006). That temporary blocking effect is eventually overcome by the IFN-γ-activated macrophage. The immune response to mycobacterial early-conserved immunodominant epitopes of the Esx secretion system triggers pH to drop to 5.0 in the phagolysosome. However, the time gained allows Mtb to alter its ability to use carbon sources with the preference of glucose usage and storage of neutral lipids (triacylglycerols (TAG)) in droplets (Vázquez et al., 2014) followed by mycobacterial entry into the dormant phase.
Early phagosomal stage
During the early stage, phagosomes fuse with recycling endosomes containing classical MHC-I molecules endocytosed from the cell surface and traffic nonclassical MR1 molecules from the trans-Golgi network (Harriff et al., 2016). Mtb is thought to directly load its antigens on these MHC-I molecules and release them to the cell surface without going through the proteasomal and cytosolic processing route. Peptides in the phagosomes are derived from the action of phagosomal proteases, in particular, cathepsins belonging to aspartic (D, E), cysteine (B, C, F, H, K, L, O, S, V, X, and W), and serine (A and G) proteases (Pires et al., 2016). Mycobacterium tuberculosis was shown to downregulate not only cathepsins but also cathepsin inhibitors and cystatins. In effect, the antigen processing activity may actually be improved, as it was shown that the high proteolytic potential of these enzymes leads otherwise to epitope destruction via cleavage into extremely short peptide sequences no longer able to anchor into the binding groove of MHC-I molecules (Moss et al., 2005). The loading process is enhanced by endosomal TLR signaling and supported by a number of accessory molecules, some stationary for phagosome and others shuttled from the endoplasmic reticulum (ER). There is little knowledge about the “quality” of presented antigens in respect of infected cell recognition and destruction; however, the tight control over the timing of lysosome fusion by the pathogen may favor antigen processing of small, highly immunodominant proteins from the Esx family with, possibly, a role for some members of proline-glutamate (PE) motif-containing PE-PPE family of mycobacterial proteins (Vordermeier et al., 2012) in the absence of key lysosomal proteases—some of these virulence factors are listed in Table 1.
Table 1
| Protein | Size | Role | Putative location | Reference |
|---|---|---|---|---|
| Phagosome immunodominance | ||||
| SapM (Rv3310) | 299aa | Dephosphorylates phosphatidylinositol 3-phosphate (PI3P) catalyzing its hydrolysis; inhibition of phagosome maturation; binding to the small GTPase RAB7 delaying autophagy flux | Phagosome | Vergne et al. (2005) and Hu et al. (2015) |
| PPE57 (Rv3425) | 176aa | TLR2 stimulation/proinflammatory cytokines/macrophage maturation with upregulation of MHC-II | Phagosome | Xu et al. (2015) |
| PPE17 (Rv1168) | 346aa | TLR2 stimulation/proinflammatory cytokines; immunodominant epitopes | Phagosome | Udgata et al. (2016) |
| PPE39 (Rv2353) | 354aa | Enhances macrophage maturation and upregulation of MHC-I and MHC-II molecules; induces production of proinflammatory cytokines and Th1 responses | Phagosome | Choi et al. (2019) |
| PE-PGRS11 (Rv0754) | 584aa | TLR2, proinflammatory cytokines and Cox2 expression stimulation; induction of antiapoptotic Bcl2 | Phagosome, autophagosome | Bansal et al. (2010) and Chaturvedi et al. (2010) |
| Ppa (Rv3628) | 162aa | TLR2 stimulation/proinflammatory cytokines and Th1 immune responses | Phagosome | Kim et al. (2016) |
| Rv1507A (Rv1507) | 231aa | Induction of proinflammatory cytokines and upregulation of MHC-I and MHC-II molecules | Phagosome | Arora et al. (2020) |
| TB27.3/cfp32 (Rv0577) | 261aa | TLR2 stimulation/proinflammatory cytokines/upregulation of MHC-I and MHC-II on DCs | Phagosome, cell exit | Byun et al. (2012) |
| EsxL (Rv1198) | 94aa | TLR2 stimulation/TNF-α, IL-6 production | Phagosome | Pattanaik et al. (2021) |
| EspC (Rv3615) | 103aa | Stimulation of proinflammatory cytokines; contains broadly recognized CD4+ and CD8+T-cell epitopes | Phagosome, cell exit | Millington et al. (2011) |
| EsxV-EsxW (Rv3619/20) | 94aa-98aa | Elicits increased levels of IFN-gamma, IL-12, and IgG(2a) as a dimer | Phagosome, cell exit | Mahmood et al. (2011) |
| Persistence | ||||
| DnaK (Rv0350) | Bacterial chaperone protein stimulates macrophage for higher arginase activity, diverts it from the iNOS pathway, and switches on IL-10 production | Autophagosome | Lopes et al. (2016) | |
| PtpA (Rv2234) | 163aa | Dephosphorylates host VPS33B protein, which induces a block of the host phagosome maturation; antagonizes host protein TRIM27, which acts as E3 ubiquitin ligase that promotes innate immune responses and cell apoptosis | Phagosome, Autophagosome | Wang et al. (2016) |
| CpsY-cpsA (Rv0806/Rv3484) | 532aa | Glucose epimerases and stealth proteins conserved from bacteria to higher eukaryotes; diverting host glycosylation pathways; CpsA prevents recruitment of NADPH oxidase to the phagosome | Autophagosome | Sperisen et al. (2005) and Köster et al. (2017) |
| PPE34 (Rv1917) | 1459aa | Induced maturation of dendritic cells, via antigen presentation, induces Th2 responses with IL-10 production | Autophagosome | Bansal et al. (2010) |
| Erm37 (Rv1988) | 179aa | Localized to host chromatin serving as a functional methyltransferase that demethylates an arginine residue at H3R42 to repress a range of host genes involved in reactive oxygen species (ROS) | Autophagosome | Yaseen et al. (2015) |
| Eis (Rv4216) | 402aa | Secreted protein, which increases acetylation of host histone H3 to upregulate IL-10 and suppress autophagy and inhibits ERK1/2, JAK pathway, and subsequent production of tumor necrosis factor-alpha (TNF-alpha) and interleukin-4 (IL-4); inhibits ROS production via acetylation of host DUSP/MKP-7 phosphatase | Autophagosome | Lella and Sharma (2007); Samuel et al. (2007), and Kim et al. (2012) |
| LprG (Rv1411) | 236aa | Lipoprotein inhibits MHC-II antigen processing, enhances recognition of Mtb acetylated glycolipids by TLR2, induces mitochondrial fission, and lowers respiratory cell rate | Autophagosome | Gehring et al. (2004); Drage et al. (2010), and Aguilar-López et al. (2019) |
| PPE18 (Rv1196) | 391aa | Downregulation of Th1 responses, upregulation of Th2 responses | Autophagosome | Nair et al. (2009); Bhat et al. (2012), and Udgata et al. (2016) |
| PE-PGRS30 (Rv1651) | 1011aa | Downregulation of Th1 responses, phagolysosome block | Phagosome, Autophagosome | Chatrath et al. (2016) |
| PPE2 (Rv0256) | 556aa | Contains a eukaryotic-like nuclear signal, which allows it to be directly translocated into the host nucleus, where it binds to the NOS2 promoter and limits host ROS production | Autophagosome | Bhat et al. (2017) |
| TlyA (Rv1694) | 268aa | Downregulation of Th1 and Th17 responses | Autophagosome | Rahman et al. (2015) |
| LpqT (Rv1016) | 226aa | Mannosylated protein; downregulation of Th1 and Th17 responses; inhibits maturation of dendritic cells and decreases the production of proinflammatory cytokines | Autophagosome | Su et al. (2019) |
| LpnQ (Rv0583) | 228aa | Secreted, directly interacts with the human E3 ubiquitin ligase CBL | Autophagosome | Penn et al. (2018) |
| PE35/PPE68 (Rv3872/73) | 99aa/368aa | Located in RD1 region; stimulates the secretion of IL-10 and MCP-1 via TLR2 activation | Autophagosome | Tiwari et al. (2014) |
| DlaT (Rv2215) | 553aa | Function with Lpd as NADH-dependent peroxidase and peroxynitrite reductase that provides protection against nitrosative stress | Phagosome, autophagolysosome | Shi and Ehrt (2006) |
| Mce2D (Rv0592) | 508aa | Less TNF-α and IL-6; all mce1–4 take part in adaptation to adverse conditions of autophagosome | Autophagosome | Singh et al. (2016) |
| Cell exit | ||||
| PE-PGRS33 (Rv1818) | 236aa/498aa | Influence mitochondrial dynamics; precipitates macrophage apoptosis via mitochondrial CytC activation, leading to an increase in caspase-3 and caspase-9, induces TNF-α and TNF receptors 1A; highly immunodominant for both cellular and humoral responses | Cell exit | Basu et al. (2007); Cohen et al. (2014), and Aguilar-López et al. (2019) |
| LpqH (Rv3763) | 159aa | Induces interleukin 1-beta and IL-12 p40 (IL12B) and TNF-α; inhibits MHC-II expression and antigen processing in the host; traffics via MHC-I processing pathway and via bacilli extracytoplasmic vesicles; may reduce vacuolar MHC-I processing; induces macrophage apoptosis via loss of mitochondrial membrane potential, release cytochrome c, and release of mitochondrial apoptosis-inducing factor AIF followed by upregulation of death receptor signaling and caspase-8 and caspase-3 | Autophagosome, cell exit | Noss et al. (2001); Tobian et al. (2003); Stewart et al. (2005), and Sánchez et al. (2012) |
| PE-PGRS17 (Rv0978) | 331aa | TLR2 stimulation/proinflammatory cytokines | Phagosome, cell exit | Chen et al. (2013) and Moodley et al. (2022) |
| EsxT (Rv3444) | 100a | Induces macrophage apoptosis via activation of NF-kappa-B and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL); secreted via esx-4 T7S | Cell exit | Shi et al. (2014) |
| EspB (Rv3881) | 460aa | The secreted processed form of EspB binds to phosphatidic acid and phosphatidylserine-inducing host death; inhibits IFN-gamma-induced autophagy; binds to human serum amyloid A, acute phase protein; and facilitates cell entry of opsonized Mtb | Cell exit | Chen et al. (2013); Huang and Bao (2016); Kawka et al. (2021) |
| PE9-PE10 (Rv1088/89) | 144/120aa | Induces macrophage apoptosis, downregulation of IL-1B increases IFNB, has a number of highly immunodominant T-cell epitopes | Cell exit | Tiwari et al. (2015) and Sunita et al. (2020) |
| PE25/PPE41 (Rv2430/31) | 194aa/99aa | As dimer induces macrophage necrosis, induces maturation of mouse dendritic cells, and drives Th2-biased immune responses | Cell exit | Tundup et al. (2014) and Chen et al. (2016) |
Selected Mycobacterium tuberculosis virulence factors participating in host virulence resistance.
Late phagosomal stage
During the late phagolysosome stage, more lipids are present in the phagolysosome, which is reflected in antigen presentation shifting from proteins to lipids. It was shown that CD1d and CD1b molecules have an endosome-targeting motif regulated also by the interaction with MHC-II complexes and are able to withstand lower pH in the phagolysosome (Jayawardena-Wolf et al., 2001). Antigenic lipid-CD1 complexes traffic to the cell surface for iNKT cell activation (Sillé et al., 2011).
Cytosolic bacteria
Bacilli that escape from phagosomes or “leaky” phagosomes do so through the damaged primary phagosome membrane, using virulence factors and activating host cytoplasmic phospholipase A2 (cPLA2). Activation of cPLA2 has the additional effect of releasing arachidonic acid from plasma membranes. Arachidonic acid is the precursor of small lipid molecules and eicosanoid biosynthetic pathway derivates: lipoxins, prostaglandins, and leukotrienes. Their mechanisms of action create a network of regulatory counterbalances. Prostaglandins (PGE4) promote plasma membrane repair, whereas lipoxin A4 supports mitochondrial damage and macrophage necrosis via inhibition of PGE4-producing cyclooxygenase 2 (Chen et al., 2008). Virulent strains of Mtb have more propensity to inhibit macrophage apoptosis and antigen cross-presentation stimulating 5-lipoxygenase-dependent pathways (Divangahi et al., 2010; Jamwal et al., 2016). The burden of escaping cytosolic bacilli depends on their intra-phagosomal replication, which is higher for more virulent strains and lower for effective innate responses that either induce Mtb dormancy or lead to bacilli killing. Many cytosolic bacilli and partially damaged phagolysosomes are captured by a system of intracellular membranes which restructures into double-membrane autophagosomes. It is characterized by a series of sequential steps: nucleation, elongation, and completion. ER, mitochondria, and other organelles serve as sources of membranes and various other molecules facilitating the transfer of MHC-II proteins into the intravesical lumen (ten Broeke et al., 2013; Kimmey and Stallings, 2016).
Persistence of the autophagosome
The inhospitable environment of the autophagosome and autophagolysosome leads bacilli to re-enter their dormant form upon activation of the two-component transcriptional program dormancy-survival regulator, DosR-DosS, by hypoxic conditions. The acidic environment of autophagolysosomes contains enzymes that cause osmotic and redox damage. Bacilli become resistant, building up intracellular lipid content, thickening cell capsule, metabolic downshift, and glyoxylate shunt for efficient maintenance of tricarboxylic cycle (TCA) components and low levels of DNA synthesis (Lipworth et al., 2016; Murima et al., 2016). Eventually, persistent bacilli are thought to become nonreplicating, metabolically inert, and survive as hidden from host subpopulations. Suppressed metabolism and bacilli dormancy decrease overall antigen presentation to the host and impair host lytic attack to disassemble bacilli. While still confined, the secreted pathogen antigens that get through the autophagosome membrane are thought to have molecular signal sequences that direct them into different intracellular compartments where they influence host metabolism and respiration (Jamwal et al., 2013). These molecules, often PE/PPE proteins, are smaller in quantity but have structural modifications that disable efficient cytosol processing of MHC-I molecules (Koh et al., 2009; Saini et al., 2016). Some of these molecules are listed in Table 1. While it was reported that bacilli resuscitation may happen a number of years after the initial infection, the majority of current reports indicate a period of about 2 years (Behr et al., 2018). Beyond that, the control of dormant bacilli over the host macrophage metabolism, if not actively progressed, fades away, resulting in infection resolution (Stephenson and Byard, 2020).
Cell exit
Periodically, or upon a trigger such as a drop in immune surveillance, resuscitating and then actively replicating bacilli puncture the autophagosome membranes with its pore-forming virulence factors from the esx family. Bacilli release membrane vesicles (Prados-Rosales et al., 2011) that contain a high density of acetylated PIM, phospholipids, polyacetylated trehalose, and phenolic glycolipids (PGL), as well as Mtb virulence factors—Ag85 complex, CFP10, and lipoproteins (LPR family, PstS1). Microdisruptions of the cell membrane elicit immediate cell response to re-seal the lesion, maintain the continuity of the membrane, or form a new autophagosome to prevent the acidic content from leaking out, risking cell death (Zhen et al., 2021). Host proteins involved in either sensing membrane damage or its repair processes, like synaptotagmin Syt7, galectins Gal 3, Gal9, Annexin, SNARE proteins, and VPS4 are also known to be selectively disrupted by Mycobacterium tuberculosis (Gan et al., 2008; Divangahi et al., 2009; Chávez-Galán et al., 2017; López-Jiménez et al., 2018). Subsequently, infected macrophages may undergo a process of remodeling and fusion into Langhans multinucleate giant cells, with the possible contribution of phenolic glycolipids on the bacilli part (Cambier et al., 2017). The process of disruption of intracellular membrane continuity may also lead to cell death via either apoptosis, necrosis (Divangahi et al., 2009), pyroptosis (Behar et al., 2010), or ferroptosis (Amaral et al., 2019)—each endowed with separate characteristics that determine the degree of antigen cross-presentation uptake from host membrane exosomes (Giri et al., 2010) or engulfment by other phagocytes (Gan et al., 2008; Amaral et al., 2019). The possible routes of host–pathogen interactions are schematically represented in Figure 1. Bacilli persistence is a granuloma-specific phenomenon, as is bacilli resuscitation and return to active replication upon a change in the environmental conditions. Bacilli resuscitation in small quantities may be also a stochastic process (Buerger et al., 2012). While the former is a more generalized event and a result of weakening immune responses (Riaño et al., 2012), the latter might be more localized, confined to several bacilli that show differential patterns of resuscitation factors (Mukamolova et al., 2002; Tantivitayakul et al., 2020) and DosR-S regulon expression (Domenech et al., 2017). Successful “scouts” active at the border between the necrotic center and host cellular wall of defense (Davies et al., 2008) spread out to set up distant tubercle satellites in other lung lobes or cause multiple foci of inflammation in miliary tuberculosis (Liu et al., 2015).
Beyond phagosome: Sources of the antigen for MHC-I presentation in Mtb infection
1. Autophagosome-to-cytosol pathway: small molecules. Various molecules are actively secreted by live Mtb cells. Nearly 30% of the Mtb proteome is composed of small proteins defined as <200Aa. Development of the strategies that disable these molecules facilitates processing and antigen presentation. If the autophagosome establishes direct membrane contact with ER via fusion (Guermonprez et al., 2003), smaller size molecules (Table 1) can be transported out of the autophagosome either via ER-specific sec61 channel into ER, where post-translational modification are removed and then out to cytosol (Römisch, 2017). Retrograde transit facilitates entry of these molecules to the 19S unit of the proteasome and then back to the ER to form pMHC-I complexes via an antigen-processing transporter (TAP)-dependent pathway. Candidates could include Mtb 19-kDa glycosylated lipoprotein LpqH, fibronectin attachment protein Mpt32, and superoxide dismutase SodC (Mehaffy et al., 2019). Specific Mtb proteins lack fixed conformational structure, e.g., the PGRS part in PE-PGRS proteins or the PPE part in PE-PPE proteins. This results in their nonspecific interference with host signaling pathways by allosteric mimicry (Sharma et al., 2022). Cytosolic polyubiquitination of lysine residues in hydrophobic stretches of these proteins stabilizes their structure while directing them to the proteasome. Finally, the mycobacterial counterpart to the host sec61 channel is 8 membrane-bound Sec proteins that form translocation apparatus transporting many of Mtb-secreted proteins. In conjunctions with pore-forming type VII secretion system of esx1, esx3, and esx5-sec transport circumvent host ER sec-61 by directing small proteins out of Mtb and via punctured membrane into the cytosol (Divangahi et al., 2009).
2. Autophagosome-to-cytosol pathway: bigger molecules. Partially digested bacterial cells, e.g., BCG, molecular complexes, or even whole live bacilli, are transferred from the autophagosome to the cytosol in any conditions that significantly impair phagosome membrane stability, as described previously in Mycobacterium tuberculosis as an intracellular microbe: role of the phagosome. It was previously shown that in BCG-infected phagosomes, molecules as large as 70 kDa could access the cytosol (Teitelbaum et al., 1999), which may happen at a late stage when the phagolysosome membrane is partially degraded. Of note, both sec61 active transport and membrane disruption mode are not mutually exclusive. The repair patches of the plasma membrane are of ER origin and therefore sources of embedded molecules that may not be characteristic of the endosomal membrane itself.
3. Endosomal processing pathways. Mtb persistence in the phagosome (Mycobacterium tuberculosis as an intracellular microbe: role of the phagosome) is linked to the inhibition of its acidification. Less harsh conditions increase the likelihood of early antigens loading on classical MHC-I molecules. Conversely, loading on CD1 requires displacement of smaller self-lipids by bigger Mtb lipids in low pH, characteristic of late phagolysosomes (Ly and Moody, 2014), and is supported by CD1d and CD1b. Peptide-MHC-I loading is enhanced by endosomal TLR3, TLR7, an TLR9 signaling and any additional transport of surface recycled MHC-I vesicles or ER vesicles that may fuse with phagosomes/phagolysosomes and provide clusters of MHC-I pathway components. One of those is the TAP supplied in retrograde transport between endosome and ER (Harriff et al., 2013). In fact, in certain cases, ER vesicles with peptides processed in the cytosol can fuse to phagosomes for further antigen processing and MHC-I loading (Guermonprez et al., 2003; Gardiner et al., 2013). Late endosomes were also reported to be involved in MR1-specific molecule trafficking and release to the cell surface (Harriff et al., 2014).
Uptake of the antigenic molecules from apoptotic (apoptotic blebs, exosomes) or necrotic infected cells by dendritic cells, specifically CD8+, takes place via phagocytosis, receptor-mediated endocytosis, and micropinocytosis. Exosomes contain various cell wall and membrane byproducts, i.e., PIM, LAM, LM, lipoproteins (LpqH), trehalose dimycolate, and monoglycosylated PGL (Layre, 2020). Dendritic cells have an extensive network of specialized vesicular transport pathways (Montealegre and Van Endert, 2019). While this route is a critical source of Mtb pMHC-I complex presentation to CD8+ cells, it is less influenced by live intracellular bacilli, which disrupt phagosomal antigen processing and interfere with antigens present in the cytosol (Cruz et al., 2017).
Immune surveillance of mycobacterial antigen presentation on MHC-I molecules
Mtb survival strategy as an intracellular pathogen deploys various, often synergizing mechanisms to manipulate antigen presentation. Part of decoy strategies forms a tactic to “be recognized but not eradicated,” counteracting its host drive to “recognize and eliminate.” Cellular immunity is primarily involved in the local control of the infection via a cytokine-mediated feedback loop between antigen-presenting cells (APCs) and T cells. Sentinel lymph nodes, defined as first granuloma-draining lymph nodes (Nieweg et al., 2001), are the site for priming and expansion of the cognate T cells; they are also the main residence of various unconventional subsets of T cells that act as tissue-resident immunity. Lymph nodes can harbor pathogens themselves, transported here from lung parenchyma and directly infecting lymphatic endothelial cells (Lerner et al., 2016).
Cytotoxic T cells are the main responders to MHC-I presentation. The majority of them are classically (MHC-Ia) restricted; these are CD8+ T cells that recognize pMHC-I complex via TCR. Upon activation, they produce cytotoxic granules and discharge their content via direct contact with the target cell; they can also produce Th1-type cytokines IFN-γ, TNF-α, and IL-2. The functional profile of cytokine secretion differs between active and latent TB varying from poly- to monofunctional T cells (Rozot et al., 2013). CD8+ T cells preferentially recognize and destroy heavily infected macrophages and represent the sensor of the intracellular bacilli burden (Lewinsohn et al., 2003); or conversely, only heavily infected macrophages are able to stimulate cytotoxic T-cell granule exocytosis. Uniquely, CD8+ T cells recognize pMHC-I complexes on MHC-II-negative infected cells like some epithelia and can limit infection propagation in the local alveolar environment (Harriff et al., 2014), at least at an early stage. It is known that Mtb-infected cells, albeit recognized, do not elicit effective cytotoxic responses until late in infection, which hints at a possible decoy strategy of the bacilli, known to have many conserved T-cell epitopes (Comas et al., 2010). This appears to be the case for the TB10.4 (EsxH) antigen. It elicits a dominant CD8+ T-cell response which poorly recognizes Mtb-infected macrophages and is unable to lyse them (Yang et al., 2018). A substantial proportion of TB10.4-specific CD8+ T cells are directed to a single epitope, TB10.44-11, at the start of this 96aa small protein sequence, overshadowing other epitopes. This is an interesting example of a diversion strategy against CD8+ T cells. As intact TB10.4 or EsxH is reported to inhibit the endosomal sorting complex required for transport (ESCR) that processes antigens for MHC-II epitopes loading in phagosomes (Portal-Celhay et al., 2016), the gain is doubled.
A smaller population of unconventional cytotoxic T cells is MHC-Ib (HLA-E-H, CD1, MR1) restricted. HLA-E presents peptide sequences from other MHC-I molecules, i.e., VMAPRTLIL, VMAPRTLVL, VMAPRTLLL, VMAPRALLL, and VMAPRTLTL (O’Callaghan et al., 1998); it was reported that HLA-E may present a set of Mtb-derived peptides, i.e., VMATRRNVL, VLRPGGHFL, VMTTVLATL, and RLPAKAPLL (Caccamo et al., 2015), some with striking similarities to self-derived leader sequences. Among HLA-E-restricted nonclassical tolerogenic T cells, there has been also a smaller subset identified with Th2 cytokines (IL-4,5,13) secreting properties that activate B cells (Joosten et al., 2010). These cells, upon recognizing Mtb epitopes presented on HLA-E, are not only diverted from their cytolytic functions but induced secretion of cytokines is also acting to suppress other T cells in the vicinity (van Meijgaarden et al., 2015).
CD1 molecules are highly conserved and specialize in presenting to CD1-restricted T cells like double-negative (CD4−CD8−) and iNKT cells (Arora et al., 2013). CD1a receptors present Mtb mycoketides (Moody et al., 2004). CD1d is adapted for presenting phosphoglycolipids such as phosphatidylinositol mannosidase (PIM) (Fischer et al., 2004). Other CD molecules also participate in host cell responses to Mtb, with CD1c presenting lipids such as phosphodolichols, phosphomycoketides, and N-terminally acylated lipopeptides (Van Rhijn et al., 2009) and CD1b presenting mycobacterial mycolates and glycolipids, glucose-6-O-monomycolate, glycerol monomycolate, and sulfoglycolipids (Moody et al., 1997; Lopez et al., 2020). Invariant NKT cells’ recognition of mycobacterial fractions targets glycolipids via CD1d-restricted invariant TCRα chain paired with limited TCRβ chain; the percentage of these cells decreases in active TB as they express programmed death 1 (PD-1) molecule that marks their exhaustion. They are also present in local pleural effusion, where they can produce IL-21, taking part in the stimulation of humoral responses (Wu et al., 2015). Other subtypes of NKT cells produce cytokines like IFN-γ, IL-4, or IL-17a. iNKT cells can directly inhibit the intracellular growth of Mtb through the granulocyte-macrophage-colony stimulating factor (GM-CSF) (Rothchild et al., 2014; Di Carlo et al., 2022). Cytokines IL-12 and IL-18 stimulate GM-CSF production by iNKTs. GM-CSF is a potent cytokine involved in macrophage differentiation to M1 phenotype, upregulation of CD11c and MHC II markers and, intracellularly, shifting the balance from antioxidant responses to inflammasome processing and secretion IL-1β while protecting from DNA damages (Di Carlo et al., 2022; Vico et al., 2022). The role of IL-1β in responses to Mtb is complex: first in resistance to infection, then protection against cell death, and finally influencing the aforementioned death modality with the shift from necrosis to pyroptosis and the subsequent effect on antigen cross-presentation. Autophagosomal Mtb appears to actively counteract host cell inflammasomes via modulating its own antigen secretion and interference with host signaling pathways (Rastogi and Briken, 2022). Cytosolic virulent bacilli do the opposite (Beckwith et al., 2020).
Mucosa-associated invariant T cells (MAIT) are present in both the upper and lower respiratory tract and are likely to detect Mtb not only in macrophage host but also in infected epithelia while in transit (Harriff et al., 2014). MAIT cells express TCR receptors of restricted diversity and recognize small-molecule microbial metabolites (Chen et al., 2017) presented on MR1 receptors. MR1 ligands in Mtb infection are derivatives of riboflavin and folic acid synthesis pathway and possibly other transiently expressed molecules of mycobacterial metabolome like photolumazine I (Harriff et al., 2018). Mtb genome contains a family of RibA-H genes, indicating an essential requirement for riboflavin endogenous biosynthesis by the pathogen in the absence of a riboflavin transporter to acquire it or transport it back to the host (Long et al., 2010). The utilization of riboflavin derivatives intersects with iron and bacterial coenzyme F420 redox metabolism. Microbial colonization of mucosal surfaces drives the expansion of MAIT cells; there is a proportionally higher abundance of MAIT cells in the jejunum than in lung tissue (Provine and Klenerman, 2020), and overexpression of MR1 ligands has been shown to provide higher protection against TB disease in preclinical models (Dey et al., 2022). The phenotype of MAIT cells is defined as CD161hiVa7.2+ T cells, predominantly CD8+. These cells, abundant in the blood, periphery (Greene et al., 2017), and nonlymphoid organs, respond locally and at an earlier stage than the adaptive CD8+ ones (Godfrey et al., 2019; Provine and Klenerman, 2020). MAIT cells, similar to classical CD8+ cells, produce both cytotoxic granules and cytokines—TNF-α, IFN-γ, IL-17A, IL-2, IL-22, and IL-13, depending on the local microenvironment stimuli, but unlike MHC-Ia-restricted CD8+ cells, they are capable of effector functions immediately after leaving the thymus. They have a primarily effector memory phenotype, contribute directly to mycobacterial burden reduction (Chua et al., 2012), and are involved in inflammatory responses to infection.
γδ T cells are a subset of lymphocytes that enrich epithelial tissues. Their major blood subset of Vγ9Vδ2 cells can directly recognize microbial phosphoantigens in a non-MHC-dependent fashion; Vγ9Vδ2 T cells recognize not only molecules belonging to the family of butyrophilin but also mycobacterial 6-O-methyl-glucose containing lipopolysaccharides and phosphomonoester molecules. These are known as phosphoantigens, byproducts of the Mtb mevalonate metabolic pathway. The smaller periphery-bound subset of Vδ1 T cells bind to antigens displayed by several subtypes of MHC-I-like molecules, i.e., MHC-I chain-related gene A (MICA), via the NKG2D receptor shared also with CD8+ and NK cells, CD1, and endothelial protein C receptor (EPCR) (Witherden and Havran, 2012). Like other potentially highly cytotoxic cells, γδ T cells express CD94/NKG2A that inhibits MHC-I cells’ destruction by Vγ9Vδ2 ones. All γδ T cells are endowed with innate immune functions, allowing them to directly lyse infected cells as well as produce cytokines that stimulate αβ T cells.
Although not the subject of this review, it is worth remarking that CD4+ T cells can also display cytotoxic properties, and the plasticity of the immune response can include smaller populations of either double-negative T cells or MHC-II-restricted CD8+ T cells. Linked via their cytotoxicity properties are also NK cells, which contribute to overall immune responses to Mtb. Leader signal peptides presented via HLA-E downregulate NK cells via CD94/NKG2A receptors when processed via TAP antigen processing machinery. In cases of TAP inhibition, HLA-E is more likely to present exogenous antigens.
Therapeutic trio: MHC-I antigen processing for TB vaccine development
Despite considerable efforts in constructing a vaccine prototype that would effectively stimulate cytotoxic T-cell responses, the positive results have been moderate so far (Behar et al., 2007) and usually more pronounced in preclinical testing than as an outcome in immunogenicity testing of clinical trial samples (Rodo et al., 2019). Although it gets more accepted that protective immunity to TB may include other than CD4+ cell subsets, aggregated CD8+ responses constitute, so far, the main readout for current TB vaccine candidates, as summarized in Table 2.
Table 2
| Vaccine | Strategy tested in a clinical trial | Reference | |
|---|---|---|---|
| Viral vector | Trial population | Outcomes related to CTL detection | |
|
MVA85A
Modified Vaccinia Ankara virus (MVA): attenuated, replication-deficient poxvirus expressing Ag85A |
1. Phase I (NCT00460590): healthy adolescents/adults, any BCG status Phase I (NCT00480558): asymptomatic adults: any BCG status: LTBI, HIV+ only, LTBI+HIV+ 2. Phase I/II (NCT03681860): safety and immunogenicity of: MVA85A/ChAdOx1 85A/BCG re-vaccination in healthy, BCG-vaccinated adolescents and adults 3. Phase I trial (NCT01829490) in healthy BCG-vaccinated (up to 6 months before the study) adults; ChAdOx1 85A/ChAdOx1 85A+ MVA85A (boost) |
1. Low level of CD8+ IFN-γ+ and TNF-α+ 2. Induction of Ag85A-specific polyfunctional IFN-γ+, TNFα+ CD8+ T cells boosted by MVA85A 3. Ag85A-specific CD8+ polyfunctional T cells highest with regimen ChAdOx185A + MVA85A |
McShane et al. (2005) and Ndiaye et al. (2015) |
|
ChAdOx185A
Recombinant vector formed of simian adenovirus and MVA expressing Ag85A |
Dicks et al. (2012); Stylianou et al. (2015), and Wilkie et al. (2020) | ||
|
Ad5 Ag85A
Recombinant human type 5 adenovirus-expressing Ag85A |
Phase I trial (NCT00800670): healthy adults with any BCG status | CD8+ was detected in BCG+ individuals with a peak at 2 weeks and sustained TNF-α+ IL-2+ secretion | Smaill et al. (2013) |
|
TB-FLU-04L
A negative, single-stranded RNA virus attenuated and genetically manipulated to express TB genes |
Phase I study (NCT02501421): BCG vaccinated (up to 6 months before the study) healthy adults | Reported CD4+/CD8+ antigen-specific responses with a peak at 21 days | Walker et al. (2016) |
| Recombinant BCG | 1. Phase I trial (NCT00749034): healthy adults (Germany), any historic BCG but not in the last 10 years 2. Phase Ib trial (NCT01113281): healthy adults (South Africa), any historic BCG but not in the last 10 years 3. Phase II trial (NCT01479972): newborn infants, BCG naïve (South Africa) |
1. Reported increase in proliferative CD8+ responses at days 57 and 180 with an increase in multifunctional CD8+ at days 29 and 57 2. No reported significant CD8+ responses 3. Reported increase in CD8+IL-17+ at 16-week and 6-month timepoint but not CD8+ IFN-γ, TNF-α, or IL-2 either single or multifunctional |
Grode et al. (2005); Farinacci et al. (2012); Grode et al. (2013); Saiga et al. (2015); Hoft et al. (2016); Loxton et al. (2017), and Nieuwenhuizen et al. (2017) |
|
VPM1002
Recombinant BCG with urease C-deficient listeriolysin O |
|||
| Subunit antigen and adjuvant | 1. Phase IIa trial (NCT00600782) in healthy adults, any BCG status 2. Phase II trials (NCT00621322) in healthy adults, any BCG status 3. Phase IIb trial (NCT01755598; healthy adults, any BCG status with add-on sub-study for biomarkers NCT02097095 |
1. Reported monofunctional CD8+, IFN-γ+, TNF-α+, IL-2+, and IL-17+, 7 days after each vaccine dose with pattern indicated boosting of pre-existing responses rather than the generation of de novo ones; no effect on PD-1 upregulation; CD8+Ki67+ upregulation in TST > 10mm group 2. No reported significant CD8+ responses 3. No reported significant CD8+ responses |
Didierlaurent et al. (2017) |
|
M72-AS01:
Antigens PPE18 and PepA with the liposome-based adjuvant AS01: monophosphoryl lipid A and saponin QS21 |
|||
|
ID93+GLA-SE
GLA-SE: TLR4 agonist glucopyranosyl lipid/CpG ODN. ID93: PPE42, esxV, esxW, Rv1813 |
1. Phase Ib trial (NCT01927159) in healthy, BCG-vaccinated adults (not in the last 5 years). 2. Phase I trial (NCT01599897) in healthy adults; BCG naïve |
1. Reported very low and not statistically different IFN-γ, TNF-α, IL-2, and IL-17 CD8+ 2. Baseline to very low CD8+ responses detected |
Coler et al. (2018); Penn-Nicholson et al. (2018), and Kwon et al. (2019) |
Summary of selected clinical trials for new vaccines against TB that include outcomes related to the cytotoxic T cell.
Antigen
Mycobacterium tuberculosis consists of over 4,000 open reading frames (ORF). Currently, there are 11 antigens in the clinical testing phase. Broadly summarizing, the selection of new candidates includes MHC-I-directed methods and T-cell-specific methods. The former includes bioinformatics approaches of antigen selection based on the prediction databases as one category and immunopurification of naturally presented epitopes in infected cell lines as another. T-cell-specific methods use T cells from infected patients to evaluate their proliferation and polyfunctionality when exposed to peptide pools of selected antigens. The aim is to find antigens that sensitized immune cells respond to the most (Tang et al., 2011; Lewinsohn et al., 2013). In view of conserved immunodominant epitopes present in many of the Mtb virulent proteins that participate at early stages of the infection, isolation of antigen-specific T cells is likely to result in the identification of these highly virulent secreted proteins. Indeed, these are the antigens that have been so far tested in clinical trials, with TB10.4 being one of them. Although some of them are still in testing, antigens abundant in subdominant epitopes that elicit weaker natural immune responses are currently considered to be better vaccine candidates (Orr et al., 2014). That reverses the hierarchy of importance between results of in vitro T-cell stimulation assays and the validation of antigens as new vaccine candidates. More recently, CD8+T- cell-specific methods (Lewinsohn et al., 2017) identified members of the PE-PPE family currently tested as potential vaccine candidates (Stylianou et al., 2018). Improvements in analytical technology, mainly mass spectrometry (Purcell et al., 2019), led to reinvigorated research in immunoproteomics and related immunopeptidomics. In the tuberculosis field, the first demonstrations to identify Mtb-specific antigens showed not only many secreted Esx family of protein members but also membrane-associated proteins and some molecules involved in lipid biosynthesis and transport. That confirmed the potential usefulness of this method, promising new avenues for further research (Bettencourt et al., 2020).
Vaccine delivery system and adjuvants
The methods used to boost MHC-I antigen presentation and CTL responses include viral vectors for delivery of mycobacterial antigens (Stylianou et al., 2015; Humphreys and Sebastian, 2018) and adjuvants like the AS01 system deployed in the M72-AS01vaccine, as shown in Table 2. AS01 adjuvant consists of saponin extract from Quillaja saponaria, QS21, and was reported to strongly induce CD8+ responses via antigen cross-presentation (Garçon and Van Mechelen, 2011). On the other hand, the Listeria monocytogenes toxin, listeriolysin O, which acts as a membrane hole puncher to release live bacilli into the cytosol, was also deployed in combination with BCG to improve cytosolic processing and antigen presentation in the VPM1002 vaccine (Farinacci et al., 2012; Saiga et al., 2015; Nguyen et al., 2019).
Nonclassically restricted cytotoxic T cells in TB vaccine responses
Nonclassically restricted cytotoxic cells are potent effector cells endowed with variable levels of memory-like functions. Their residency in the periphery and in blood, cytotoxic properties, and lack of donor MHC-I allelic restriction make them potentially first responders to infection and therefore an attractive target for vaccine strategies. As endowed with highly cytotoxic properties and abundantly present at the periphery, they are also tightly controlled in the environment of multimicrobial presence and sensitive tissue to protect mucosal integrity as a mechanical barrier.
Early attempts at the deployment of MR1 ligands in TB vaccine research in the macaque model of TB led to MAIT cell dysfunction and indicated a narrow margin between lung MAIT cell stimulation and exhaustion in this model (Sakai et al., 2021). Lack of margin could be potentially disadvantageous from the therapeutical point of view, rendering MAIT cells easily overstimulated. MAIT cells were also functionally impaired and displayed exhaustion markers in HIV/SIV-MTB co-infection in the cynomolgus macaque model, although SIV did not prevent MAIT recruitment from blood to sites of infection in the lungs (Ellis et al., 2020).
In humans, proinflammatory, cytokine-secreting TRAV1-2+ CD8+ CD26+MAIT cells were identified in the lungs of individuals with active TB (Wong et al., 2019) while depleted in the blood. Transcriptomic analysis of blood samples from LTBI versus noninfected individuals showed a lower frequency of MR1tet+ CD8+ cells in LTBI (Pomaznoy et al., 2020), but in another study, the correlation between blood MAIT frequency and TB status was completely absent for both active and latent TB individuals (Suliman et al., 2020). The frequency of MAIT cells was unchanged in samples from the phase I study [NCT01119521] investigating the safety and reactogenicity of BCG revaccination with isoniazid pretreatment in LTBI adults, although changes were observed in the usage of TCR clonotypes (James et al., 2022), indicating the changes in MAIT cells are discrete and qualitative rather than quantitative.
The first identification of mycobacterial antigens stimulating γδ T cells showed the abundance of γδ T cells is proportional to mycobacterial pathogenicity (Constant et al., 1995). It was also observed that this subset of T cells is inducible by cross-reactive antigens from environmental mycobacteria, which at least partially explains the confounding results in the studies measuring their level as a correlate of vaccine efficacy (Hoft et al., 1998). γδ T cells from BCG-vaccinated responders show reactivity to whole-cell Mycobacterium tuberculosis lysates rather than secreted components of the culture filtrate or heat-inactivated whole bacilli. They display memory-like phenotype and support the expansion of CD4+ and CD8+ cells by secretion of IFN-γ. Partially due to the cross-reactivity in small metabolite molecules’ metabolism, BCG-specific γδ T cells are currently investigated as nonspecific immunomodulators to high-grade nonmuscle invasive bladder cancer and HIV-infected cells (Garrido et al., 2018).
NKT-like cells, defined as CD3+ and CD56+ expressing IFN-γ, TNF, and IL-2, were increased after vaccination with H4:IC31. H4 consists of Ag85B and TB10.4 and H4:IC31, a prototype of H56:IC31, was dropped off the WHO TB new vaccine pipeline in 2018 (WHO, 2017). The H4:IC31 phase I trial [NCT02075203] was composed of the interventional arm testing the H4:IC31 vaccine, while the comparator arms for this study were placebo and BCG revaccination (1:1:1). The immunogenicity outcomes included CD4+ and CD8+ as main outcome readouts, and a further in-depth flow cytometry strategy was designed to detail all antigen-specific responders. Indeed, the analysis showed that BCG revaccination stimulated donor-unrestricted responses at just slightly lower levels than conventional CD4+ T cells. It showed equal proportions of the presence of γδT cells and MAIT cells alongside the same level of innate NK cells. These cells were predominantly monofunctional IFN-γ producers (Rozot et al., 2020).
Finally, linked by their cytotoxic properties rather than lineage, NK cells appear overlooked yet potentially important strategic partners for new vaccine candidates. CD27+NK cells accumulated in the LTBI model of nonhuman primates (Esaulova et al., 2021), as well as being present in patients with active TB, where they appeared to enhance the cytotoxicity of CD8+ T cells with potential for innate-like memory (Choreño Parra et al., 2017). BCG revaccination increased the number of IFN-γ-producing NK cells and was linked to the nonspecific expansion of this subset in the H4:IC31 vaccine administration (Rozot et al., 2020).
Summary and conclusions
Mycobacterium tuberculosis’ adaptation to its host has been refined through thousands of years of coevolution. The renewed interest in intracellular antigen processing and presentation on MHC-I molecules has arisen in an attempt to better define immune correlates of protection against TB infection. Although classical CD8+ cells are still considered the main effector for peptides presented by MHC-I molecules and one of the main outcomes for immunogenicity assays, growing interest in donor unrestricted T cells (DURTs) may soon change that readout (Gela et al., 2022). In the majority of already completed trials, BCG vaccination is either a comparator or an inclusion criterion for patients’ eligibility. It is also a frequent Mtb surrogate for any in vitro and preclinical experiments. Although BCG-induced immune responses set a high testing threshold of efficacy for any new vaccine candidate entering clinical testing, our incomplete understanding of how these immune responses are induced and why they are insufficient in many TB-endemic countries outlines questions yet to be answered both scientifically and therapeutically. Together with new technological developments in cytometry, forthcoming clinical trials indicate a dynamic landscape in TB vaccinology with new, yet unexplored directions ahead.
Statements
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Acknowledgments
The author would like to acknowledge the advice she received from Assistant Professor Paulo JG Bettencourt from the Centre for Interdisciplinary Research in Health, Catholic University of Portugal, Lisbon, Portugal.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Aguilar-López B. A. Correa F. Moreno-Altamirano M. M.B. Espitia C. Hernández-Longoria R. Oliva-Ramírez J. et al . (2019). LprG and PE_PGRS33 mycobacterium tuberculosis virulence factors induce differential mitochondrial dynamics in macrophages. Scandinavian J. Immunol.89 (1), e12728. doi: 10.1111/sji.12728
2
Amaral E. P. Costa D. L. Namasivayam S. Riteau N. Kamenyeva O. Mittereder L. et al . (2019). A major role for ferroptosis in mycobacterium tuberculosis-induced cell death and tissue necrosis. J. Exp. Med.216 (3), 556–570. doi: 10.1084/jem.20181776
3
Arora P. Foster E. L. Porcelli S. A. (2013). CD1d and natural killer T cells in immunity to mycobacterium tuberculosis. Adv. Exp. Med. Biol.783, 199–223. doi: 10.1007/978-1-4614-6111-1_11
4
Arora S. K. Alam A. Naqvi N. Ahmad J. Sheikh J. A. Rahman S. A. et al . (2020). Immunodominant mycobacterium tuberculosis protein Rv1507A elicits Th1 response and modulates host macrophage effector functions. Front. Immunol.11. doi: 10.3389/fimmu.2020.01199
5
Bai W. Liu H. Ji Q. Zhou Y. Liang L. Zheng R. et al . (2014). TLR3 regulates mycobacterial RNA-induced IL-10 production through the PI3K/AKT signaling pathway. Cell. Signalling26 (5), 942–950. doi: 10.1016/j.cellsig.2014.01.015
6
Balamurugan A. Sharma S. K. Mehra N. K. (2004). Human leukocyte antigen class I supertypes influence susceptibility and severity of tuberculosis. J. Infect. Dis.189 (5), 805–811. doi: 10.1086/381689
7
Bansal K. Elluru S. R. Narayana Y. Chaturvedi R. Patil S. A. Kaveri S. V et al . (2010). PE_PGRS antigens of mycobacterium tuberculosis induce maturation and activation of human dendritic cells. J. Immunol.184 (7), 3495–3504. doi: 10.4049/jimmunol.0903299
8
Basu S. Pathak S. K. Banerjee A. Pathak S. Bhattacharyya A. Yang Z et al . (2007). Execution of macrophage apoptosis by PE_PGRS33 of mycobacterium tuberculosis is mediated by toll-like receptor 2-dependent release of tumor necrosis factor-alpha. J. Biol. Chem.282 (2), 1039–1050. doi: 10.1074/jbc.M604379200
9
Beckwith K. S. Beckwith M. S. Ullmann S. Sætra R. S. Kim H. Marstad A. et al . (2020). Plasma membrane damage causes NLRP3 activation and pyroptosis during mycobacterium tuberculosis infection. Nat. Commun.11 (1), 2270. doi: 10.1038/s41467-020-16143-6
10
Behar S. M. Divangahi M. Remold H. G. (2010). Evasion of innate immunity by mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol.8 (9), 668–674. doi: 10.1038/NRMICRO2387
11
Behar S. M. Woodworth J. S. M. Wu Y. (2007). Next generation: tuberculosis vaccines that elicit protective CD8+ T cells. Expert Rev. Vaccines6 (3), 441–456. doi: 10.1586/14760584.6.3.441
12
Behr M. A. Edelstein P. H. Ramakrishnan L. (2018). Revisiting the timetable of tuberculosis. BMJ (Clinical Res. ed.)362, 10. doi: 10.1136/BMJ.K2738
13
Bettencourt P. Muller J. Nicastri A. Cantillon D. Madhavan M. Charles P. D. et al . (2020). Identification of antigens presented by MHC for vaccines against tuberculosis. NPJ Vaccines5, 2. doi: 10.1038/s41541-019-0148-y
14
Bhat K. H. Ahmed A. Kumar S. Sharma P. Mukhopadhyay S. . (2012). Role of PPE18 protein in intracellular survival and pathogenicity of mycobacterium tuberculosis in mice. PloS One7 (12), e52601. doi: 10.1371/journal.pone.0052601
15
Bhat K. H. Srivastava S. Kotturu S. K. Ghosh S. Mukhopadhyay S. . (2017). The PPE2 protein of mycobacterium tuberculosis translocates to host nucleus and inhibits nitric oxide production. Sci. Rep.7, 13. doi: 10.1038/srep39706
16
Buerger S. Spoering A. Gavrish E. Leslin C. Ling L. Epstein S. S. et al . (2012). Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl. Environ. Microbiol.78 (9), 3221–3228. doi: 10.1128/AEM.07307-11
17
Byun E. Kim W. S. Kim J. Jung I. D. Park Y. Kim H. et al . (2012). Mycobacterium tuberculosis Rv0577, a novel TLR2 agonist, induces maturation of dendritic cells and drives Th1 immune response. FASEB J.26 (6), 2695–2711. doi: 10.1096/fj.11-199588
18
Caccamo N. Pietra G. Sullivan L. C. Brooks A. G. Prezzemolo T. La Manna M. P. et al . (2015). Human CD8 T lymphocytes recognize mycobacterium tuberculosis antigens presented by HLA-e during active tuberculosis and express type 2 cytokines. Eur. J. Immunol.45 (4), 1069–1081. doi: 10.1002/eji.201445193
19
Cambier C. J. O’Leary S. M. O’Sullivan M. P. Keane J. Ramakrishnan L. . (2017). Phenolic glycolipid facilitates mycobacterial escape from microbicidal tissue-resident macrophages. Immunity47 (3), 552–565.e4. doi: 10.1016/j.immuni.2017.08.003
20
Chávez-Galán L. Ramon-Luing L. Carranza C. Garcia I. Sada-Ovalle I. . (2017). Lipoarabinomannan decreases galectin-9 expression and tumor necrosis factor pathway in macrophages favoring mycobacterium tuberculosis intracellular growth. Front. Immunol.8 (NOV). doi: 10.3389/FIMMU.2017.01659
21
Chatrath S. Gupta V. K. Dixit A. Garg L. C. . (2016). PE_PGRS30 of mycobacterium tuberculosis mediates suppression of proinflammatory immune response in macrophages through its PGRS and PE domains. Microbes Infection18 (9), 536–542. doi: 10.1016/j.micinf.2016.04.004
22
Chaturvedi R. Bansal K. Narayana Y. Kapoor N. Sukumar N. Togarsimalemath S. K. et al . (2010). The multifunctional PE-PGRS11 protein from mycobacterium tuberculosis plays a role in regulating resistance to oxidative stress. J. Biol. Chem.285 (40), 30389–30403. doi: 10.1074/jbc.M110.135251
23
Chen J. M. Zhang M. Rybniker J. Boy-Röttger S. Dhar N. Pojer F. et al . (2013). Mycobacterium tuberculosisEspB binds phospholipids and mediates EsxA-independent virulence. Mol. Microbiol.89 (6), 1154–1166. doi: 10.1111/mmi.12336
24
Chen M. Divangahi M. Gan H. Shin D. S.J. Hong S. Lee D. M. et al . (2008). Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J. Exp. Med.205 (12), 2791–2801. doi: 10.1084/JEM.20080767
25
Chen W. Bao Y. Chen X. Burton J. Gong X. Gu D. et al . (2016). Mycobacterium tuberculosis PE25/PPE41 protein complex induces activation and maturation of dendritic cells and drives Th2-biased immune responses. Med. Microbiol. Immunol.205 (2), 119–131. doi: 10.1007/s00430-015-0434-x
26
Chen Z. Wang H. D’Souza C. Sun S. Kostenko L. Eckle S. B.G. et al . (2017). Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol.10 (1), 58–68. doi: 10.1038/mi.2016.39
27
Choi H. H. Kwon K. W. Han S. J. Kang S. M. Choi E. Kim A. et al . (2019). PPE39 of the mycobacterium tuberculosis strain Beijing/K induces Th1-cell polarization through dendritic cell maturation. J. Cell Sci.132 (17), 14. doi: 10.1242/JCS.228700
28
Choreño Parra J. A. Martínez Zúñiga N. Jiménez Zamudio L. A. Jiménez Álvarez L. A. Salinas Lara C. Zúñiga J. et al . (2017). Memory of natural killer cells: A new chance against mycobacterium tuberculosis? Front. Immunol.8. doi: 10.3389/fimmu.2017.00967
29
Chua W.-J. Truscott S. M. Eickhoff C. S. Blazevic A. Hoft D. F. Hansen T. H. et al . (2012). Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infection Immun.80 (9), 3256–3267. doi: 10.1128/IAI.00279-12
30
Cohen I. Parada C. Acosta-Gío E. Espitia C. . (2014). The PGRS domain from PE_PGRS33 of mycobacterium tuberculosis is target of humoral immune response in mice and humans. Front. Immunol.5 (MAY). doi: 10.3389/fimmu.2014.00236
31
Coler R. N. Day T. A. Ellis R. Piazza F. M. Beckmann A. M. Vergara J. et al . (2018). The TLR-4 agonist adjuvant, GLA-SE, improves magnitude and quality of immune responses elicited by the ID93 tuberculosis vaccine: first-in- human trial. NPJ Vaccines3, 34. doi: 10.1038/s41541-018-0057-5
32
Comas I. Chakravartti J. Small P. M. Galagan J. Niemann S. Kremer K. et al . (2010). Human T cell epitopes of mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet.42 (6), 498–503. doi: 10.1038/ng.590
33
Constant P. Poquet Y. Peyrat M. A. Davodeau F. Bonneville M. Fournie - J.J. et al . (1995). The antituberculous mycobacterium bovis BCG vaccine is an attenuated mycobacterial producer of phosphorylated nonpeptidic antigens for human γδ T cells. Infection Immun.63 (12), 4628–4633. doi: 10.1128/iai.63.12.4628-4633.1995
34
Cruz F. M. Colbert J. D. Merino E. Kriegsman B. A. Rock K. L. . (2017). The biology and underlying mechanisms of cross-presentation of exogenous antigens on MHC-I molecules. Annu. Rev. Immunol.35, 149–176. doi: 10.1146/ANNUREV-IMMUNOL-041015-055254
35
Davies A. P. Dhillon A. P. Young M. Henderson B. McHugh T. D. Gillespie S. H. et al . (2008). Resuscitation-promoting factors are expressed in mycobacterium tuberculosis-infected human tissue. Tuberculosis88 (5), 462–468. doi: 10.1016/j.tube.2008.01.007
36
Dey R. J. Dey B. Harriff M. Canfield E. T. Lewinsohn D. M. Bishai W. R. et al . (2022). Augmentation of the riboflavin-biosynthetic pathway enhances mucosa-associated invariant T (MAIT) cell activation and diminishes mycobacterium tuberculosis virulence. mBio13 (1), e0386521. doi: 10.1128/mbio.03865-21
37
Di Carlo S. Häcker G. Gentle I. E. (2022). GM-CSF suppresses antioxidant signaling and drives IL-1β secretion through NRF2 downregulation. EMBO Rep.23 (8), e54226. doi: 10.15252/embr.202154226
38
Dicks M. D. J. Spencer A. J. Edwards N. J. Wadell G. Bojang K. Gilbert S. C. et al . (2012). A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PloS One7 (7), e40385. doi: 10.1371/journal.pone.0040385
39
Didierlaurent A. M. Laupèze B. Di Pasquale A. Hergli N. Collignon C. Garçon N. (2017). Adjuvant system AS01: helping to overcome the challenges of modern vaccines. Expert Rev. Vaccines16 (1), 55–63. doi: 10.1080/14760584.2016.1213632
40
Divangahi M. Chen M. Gan H. Desjardins D. Hickman T. T. Lee D. M. et al . (2009). Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat. Immunol.10 (8), 899–906. doi: 10.1038/ni.1758
41
Divangahi M. Desjardins D. Nunes-Alves C. Remold H. G. Behar S. M. . (2010). Eicosanoid pathways regulate adaptive immunity to mycobacterium tuberculosis. Nat. Immunol.11 (8), 751–758. doi: 10.1038/ni.1904
42
Domenech P. Zou J. Averback A. Syed N. Curtis D. Donato S. et al . (2017). Unique regulation of the DosR regulon in the Beijing lineage of mycobacterium tuberculosis. J. Bacteriology199 (2), 696–716. doi: 10.1128/JB.00696-16
43
Drage M. G. Tsai H. C. Pecora N. D. Cheng T. Y. Arida A. R. Shukla S. et al . (2010). Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of toll-like receptor 2. Nat. Struct. Mol. Biol.17 (9), 1088–1095. doi: 10.1038/nsmb.1869
44
Ellis A. L. Balgeman A. J. Larson E. C. Rodgers M. A. Ameel C. Baranowski T. et al . (2020). MAIT cells are functionally impaired in a Mauritian cynomolgus macaque model of SIV and mtb co-infection. PloS Pathog.16 (5), e1008585. doi: 10.1371/journal.ppat.1008585
45
Esaulova E. Das S. Singh D. K. Choreño-Parra J. A. Swain A. Arthur L. et al . (2021). The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe29 (2), 165–178.e8. doi: 10.1016/j.chom.2020.11.013
46
Falk K. Rötzschke O. (1993). Consensus motifs and peptide ligands of mhc class i molecules. Semin. Immunol.5 (2), 81–94. doi: 10.1006/smim.1993.1012
47
Falk K. Rötzschke O. Stevanović S. Jung G. Rammensee H. G. (1991). Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature351 (6324), 290–296. doi: 10.1038/351290a0
48
Farinacci M. Weber S. Kaufmann S. H. E. (2012). The recombinant tuberculosis vaccine rBCG ΔureC::hly(+) induces apoptotic vesicles for improved priming of CD4(+) and CD8(+) T cells. Vaccine30 (52), 7608–7614. doi: 10.1016/j.vaccine.2012.10.031
49
Fischer K. Scotet E. Niemeyer M. Koebernick H. Zerrahn J. Maillet S. et al . (2004). Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc. Natl. Acad. Sci. United States America101 (29), 10685–10690. doi: 10.1073/pnas.0403787101
50
Fratti R. A. Chua J. Vergne I. Deretic V. et al . (2003). Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc. Natl. Acad. Sci. United States America100 (9), 5437–5442. doi: 10.1073/pnas.0737613100
51
Gan H. Lee J. Ren F. Chen M. Kornfeld H. Remold H. G. et al . (2008). Mycobacterium tuberculosis blocks crosslinking of annexin-1 and apoptotic envelope formation on infected macrophages to maintain virulence. Nat. Immunol.9 (10), 1189–1197. doi: 10.1038/ni.1654
52
Garçon N. Van Mechelen M. (2011). Recent clinical experience with vaccines using MPL- and QS-21-containing adjuvant systems. Expert Rev. Vaccines10 (4), 471–486. doi: 10.1586/ERV.11.29
53
Gardiner G. J. Deffit S. N. McLetchie S. Perez L. Walline C. C. Blum J. S. . (2013). A role for NADPH oxidase in antigen presentation. Front. Immunol.4. doi: 10.3389/fimmu.2013.00295
54
Garrido C. Clohosey M. L. Whitworth C. P. Hudgens M. Margolis D. M. Soriano- Sarabia N. et al . (2018). γδ T cells: an immunotherapeutic approach for HIV cure strategies. JCI Insight3 (12), e120121. doi: 10.1172/jci.insight.120121
55
Gehring A. J. Dobos K. M. Belisle J. T. Harding C. V Boom W. H. . (2004). Mycobacterium tuberculosis LprG ( Rv1411c ): A novel TLR-2 ligand that inhibits human macrophage class II MHC antigen processing. J. Immunol.173 (4), 2660–2668. doi: 10.4049/jimmunol.173.4.2660
56
Gela A. Murphy M. Rodo M. Hadley K. Hanekom W. A. Boom W. H. et al . (2022). Effects of BCG vaccination on donor unrestricted T cells in two prospective cohort studies. EBioMedicine76, 16. doi: 10.1016/J.EBIOM.2022.103839
57
Germain R. N. (1994). MHC-dependent antigen processing and peptide presentation: Providing ligands for T lymphocyte activation. Cell76 (2), 287–299. doi: 10.1016/0092-8674(94)90336-0
58
Giri P. K. Kruh N. A. Dobos K. M. Schorey J. S. . (2010). Proteomic analysis identifies highly antigenic proteins in exosomes from m. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics10 (17), 3190–3202. doi: 10.1002/PMIC.200900840
59
Godfrey D. I. Koay H. F. McCluskey J. Gherardin N. A. . (2019). The biology and functional importance of MAIT cells. Nat. Immunol.20 (9), 1110–1128. doi: 10.1038/s41590-019-0444-8
60
Greene J. M. Dash P. Roy S. McMurtrey C. Awad W. Reed J. S. et al . (2017). MR1-restricted mucosal-associated invariant T (MAIT) cells respond to mycobacterial vaccination and infection in nonhuman primates. Mucosal Immunol.10 (3), 802–813. doi: 10.1038/mi.2016.91
61
Grode L. Ganoza C. A. Brohm C. Weiner J. 3rd Eisele B. Kaufmann S. H.E. . (2013). Safety and immunogenicity of the recombinant BCG vaccine VPM1002 in a phase 1 open-label randomized clinical trial. Vaccine31 (9), 1340–1348. doi: 10.1016/j.vaccine.2012.12.053
62
Grode L. Seiler P. Baumann S. Hess J. Brinkmann V. Nasser Eddine A. et al . (2005). Increased vaccine efficacy against tuberculosis of recombinant mycobacterium bovis bacille calmette-guerin mutants that secrete listeriolysin. J. Clin. Invest.115 (9), 2472–2479. doi: 10.1172/JCI24617
63
Guermonprez P. Saveanu L. Kleijmeer M. Davoust J. Van Endert P. Amigorena S. (2003). ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature425 (6956), 397–402. doi: 10.1038/nature01911
64
Hakansson A. P. Orihuela C. J. Bogaert D. (2018). Bacterial-host interactions: Physiology and pathophysiology of respiratory infection. Physiol. Rev.98 (2), 781–811. doi: 10.1152/physrev.00040.2016
65
Harriff M. J. Burgdorf S. Kurts C. Wiertz E. J.H.J. Lewinsohn D. A. Lewinsohn D. M. . (2013). TAP mediates import of mycobacterium tuberculosis-derived peptides into phagosomes and facilitates loading onto HLA-I. PloS One8 (11), e79571. doi: 10.1371/journal.pone.0079571
66
Harriff M. J. Cansler M. E. Toren K. G. Canfield E. T. Kwak S. Gold M. C. et al . (2014). Human lung epithelial cells contain mycobacterium tuberculosis in a late endosomal vacuole and are efficiently recognized by CD8+ T cells. PloS One9 (5), e97515. doi: 10.1371/journal.pone.0097515
67
Harriff M. J. Karamooz E. Burr A. Grant W. F. Canfield E. T. Sorensen M. L. et al . (2016). Endosomal MR1 trafficking plays a key role in presentation of mycobacterium tuberculosis ligands to MAIT cells. PloS Pathog.12 (3), e1005524. doi: 10.1371/journal.ppat.1005524
68
Harriff M. J. McMurtrey C. Froyd C. A. Jin H. Cansler M. Null M. et al . (2018). MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci. Immunol.3 (25), 24 doi: 10.1126/sciimmunol.aao2556
69
Hoft D. F. Blazevic A. Selimovic A. Turan A. Tennant J. Abate G. et al . (2016). Safety and immunogenicity of the recombinant BCG vaccine AERAS-422 in healthy BCG-naive adults: A randomized, active-controlled, first-in-human phase 1 trial. EBioMedicine7, 278–286. doi: 10.1016/j.ebiom.2016.04.010
70
Hoft D. F. Brown R. M. Roodman S. T. (1998). Bacille calmette-guérin vaccination enhances human gamma delta T cell responsiveness to mycobacteria suggestive of a memory-like phenotype. J. Immunol. (Baltimore Md. : 1950)161 (2), 1045–1054. doi: 10.4049/jimmunol.161.2.1045
71
Hossain M. M. Norazmi M. N. (2013). Pattern recognition receptors and cytokines in mycobacterium tuberculosis infection–the double-edged sword? BioMed. Res. Int.2013, 18. doi: 10.1155/2013/179174
72
Howcroft T. K. Raval A. Weissman J. D. Gegonne A. Singer D. S. . (2003). Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression. Mol. Cell. Biol.23 (10), 3377–3391. doi: 10.1128/mcb.23.10.3377-3391.2003
73
Hu D. Wu J. Wang W. Mu M. Zhao R. Xu X. et al . (2015). Autophagy regulation revealed by SapM-induced block of autophagosome-lysosome fusion via binding RAB7. Biochem. Biophys. Res. Commun.461 (2), 401–407. doi: 10.1016/j.bbrc.2015.04.051
74
Huang D. D. Bao L. (2016). Mycobacterium tuberculosis EspB protein suppresses interferon-γ-induced autophagy in murine macrophages. J. Microbiology Immunol. Infection49 (6), 859–865. doi: 10.1016/j.jmii.2014.11.008
75
Humphreys I. R. Sebastian S. (2018). Novel viral vectors in infectious diseases. Immunology153 (1), 1–9. doi: 10.1111/imm.12829
76
Huppa J. B. Axmann M. Mortelmaier M. A. Lillemeier B. F. Newell E. W. Brameshuber M. et al . (2010). TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature463 (7283), 963–967. doi: 10.1038/nature08746
77
James C. A. Yu K. K.Q. Mayer-Blackwell K. Fiore-Gartland A. Smith M. T. Layton E. D. et al et al . (2022). Durable expansion of TCR-δ meta-clonotypes after BCG revaccination in humans. Front. Immunol.13. doi: 10.3389/fimmu.2022.834757
78
Jamwal S. Midha M. K. Verma H. N. Basu A. Rao K. V.S. Manivel V. et al . (2013). Characterizing virulence-specific perturbations in the mitochondrial function of macrophages infected with mycobacterium tuberculosis. Sci. Rep.3, 1328. doi: 10.1038/srep01328
79
Jamwal S. V. Mehrotra P. Singh A. Siddiqui Z. Basu A. Rao K. V.S. et al . (2016). Mycobacterial escape from macrophage phagosomes to the cytoplasm represents an alternate adaptation mechanism. Sci. Rep.6, 9. doi: 10.1038/srep23089
80
Jayawardena-Wolf J. Benlagha K. Chiu Y. H. Mehr R. Bendelac A. . (2001). CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity15 (6), 897–908. doi: 10.1016/S1074-7613(01)00240-0
81
Jiang J. Natarajan K. Margulies D. H. (2019). MHC molecules, T cell receptors, natural killer cell receptors, and viral immunoevasins-key elements of adaptive and innate immunity. Adv. Exp. Med. Biol.1172, 21–62. doi: 10.1007/978-981-13-9367-9_2
82
Joly E. Mucke L. Oldstone M. B. (1991). Viral persistence in neurons explained by lack of major histocompatibility class I expression. Sci. (New York N.Y.)253 (5025), 1283–1285. doi: 10.1126/science.1891717
83
Joosten S. A. Van Meijgaarden K. E. Van Weeren P. C. Kazi F. Geluk A. Savage N. D. L. et al . (2010). Mycobacterium tuberculosis peptides presented by HLA-e molecules are targets for human CD8+ T-cells with cytotoxic as well as regulatory activity. PloS Pathog.6 (2), e1000782. doi: 10.1371/journal.ppat.1000782
84
Kawka M. Brzostek A. Dzitko K. Kryczka J. Bednarek R. Płocińska R. et al . (2021). Mycobacterium tuberculosis binds human serum amyloid a, and the interaction modulates the colonization of human macrophages and the transcriptional response of the pathogen. Cells10 (5), 26. doi: 10.3390/cells10051264
85
Kim K. H. An D. R. Song J. Yoon J. Y. Kim H. S. Yoon H. J. et al . (2012). Mycobacterium tuberculosis eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc. Natl. Acad. Sci. United States America109 (20), 7729–7734. doi: 10.1073/pnas.1120251109
86
Kim W. S. Kim J. S. Cha S. Bin Kim H. Kwon K. W. Kim S. J. et al . (2016). Mycobacterium tuberculosis Rv3628 drives th1-type T cell immunity via TLR2-mediated activation of dendritic cells and displays vaccine potential against the hyper-virulent beijing K strain. Oncotarget7 (18), 24962–24982. doi: 10.18632/oncotarget.8771
87
Kimmey J. M. Stallings C. L. (2016). Bacterial pathogens versus autophagy: Implications for therapeutic interventions. Trends Mol. Med.22 (12), 1060–1076. doi: 10.1016/j.molmed.2016.10.008
88
Köster S. Upadhyay S. Chandra P. Papavinasasundaram K. Yang G. Hassan A. et al . (2017). Mycobacterium tuberculosis is protected from NADPH oxidase and LC3-associated phagocytosis by the LCP protein CpsA. Proc. Natl. Acad. Sci. United States America114 (41), E8711–E8720. doi: 10.1073/pnas.1707792114
89
Koh K. W. Lehming N. Seah G. T. (2009). Degradation-resistant protein domains limit host cell processing and immune detection of mycobacteria. Mol. Immunol.46 (7), 1312–1318. doi: 10.1016/j.molimm.2008.11.008
90
Kwon K. W. Lee A. Larsen S. E. Baldwin S. L. Coler R. N. Reed S. G. et al . (2019). Long-term protective efficacy with a BCG-prime ID93/GLA-SE boost regimen against the hyper-virulent mycobacterium tuberculosis strain K in a mouse model. Sci. Rep.9 (1), 15560. doi: 10.1038/s41598-019-52146-0
91
Layre E. (2020). Trafficking of mycobacterium tuberculosis envelope components and release within extracellular vesicles: Host-pathogen interactions beyond the wall. Front. Immunol.11. doi: 10.3389/fimmu.2020.01230
92
Leddon S. A. Sant A. J. (2010). Generation of MHC class II-peptide ligands for CD4 T-cell allorecognition of MHC class II molecules. Curr. Opin. Organ Transplant.15 (4), 505–511. doi: 10.1097/MOT.0b013e32833bfc5c
93
Lella R. K. Sharma C. (2007). Eis (enhanced intracellular survival) protein of mycobacterium tuberculosis disturbs the cross regulation of T-cells. J. Biol. Chem.282 (26), 18671–18675. doi: 10.1074/jbc.C600280200
94
Lerner T. R. Carvalho-Wodarz C. D.S. Repnik U. Russell M. R.G. Borel S. DIedrich C. R. et al et al . (2016). Lymphatic endothelial cells are a replicative niche for mycobacterium tuberculosis. J. Clin. Invest.126 (3), 1093–1108. doi: 10.1172/JCI83379
95
Lewinsohn D. A. Heinzel A. S. Gardner J. M. Zhu L. Alderson M. R. Lewinsohn D. M. et al . (2003). Mycobacterium tuberculosis-specific CD8+ T cells preferentially recognize heavily infected cells. Am. J. Respir. Crit. Care Med.168 (11), 1346–1352. doi: 10.1164/rccm.200306-837OC
96
Lewinsohn D. A. Swarbrick G. M. Park B. Cansler M. E. Null M. D. Toren K. G. et al . (2017). Comprehensive definition of human immunodominant CD8 antigens in tuberculosis. NPJ Vaccines2, 11. doi: 10.1038/s41541-017-0008-6
97
Lewinsohn D. M. Swarbrick G. M. Cansler M. E. Null M. D. Rajaraman V. Frieder M. M. et al . (2013). Human mycobacterium tuberculosis CD8 T cell Antigens/Epitopes identified by a proteomic peptide library. PloS One8 (6), e67016. doi: 10.1371/journal.pone.0067016
98
Lipworth S. Hammond R. J.H. Baron V. O. Hu Y. Coates A. Gillespie S. H. . (2016). Defining dormancy in mycobacterial disease. Tuberculosis (Edinburgh Scotland)99, 131–142. doi: 10.1016/j.tube.2016.05.006
99
Liu Q. Via L. E. Luo T. Liang L. Liu X. Wu S. et al . (2015). Within patient microevolution of mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci. Rep.5, 17507. doi: 10.1038/srep17507
100
Long Q. Ji L. Wang H. Xie J. . (2010). Riboflavin biosynthetic and regulatory factors as potential novel anti-infective drug targets: Perspective. Chem. Biol. Drug Design75 (4), 339–347. doi: 10.1111/j.1747-0285.2010.00946.x
101
Lopez K. Iwany S. K. Suliman S. Reijneveld J. F. Ocampo T. A. Jimenez J. et al . (2020). CD1b tetramers broadly detect T cells that correlate with mycobacterial exposure but not tuberculosis disease state. Front. Immunol.11. doi: 10.3389/fimmu.2020.00199
102
Lopes R. L. Borges T. J. Zanin R. F. Bonorino C. . (2016). IL-10 is required for polarization of macrophages to M2-like phenotype by mycobacterial DnaK (heat shock protein 70). Cytokine85, 123–129. doi: 10.1016/j.cyto.2016.06.018
103
López-Jiménez A. T. Cardenal-Muñoz E. Leuba F. Gerstenmaier L. Barisch C. Hagedorn M. et al . (2018). The ESCRT and autophagy machineries cooperate to repair ESX-1-dependent damage at the mycobacterium-containing vacuole but have opposite impact on containing the infection. PloS Pathog.14 (12), 29. doi: 10.1371/JOURNAL.PPAT.1007501
104
Loxton A. G. Knaul J. K. Grode L. Gutschmidt A. Meller C. Eisele B. et al . (2017). Safety and immunogenicity of the recombinant mycobacterium bovis BCG vaccine VPM1002 in HIV-unexposed newborn infants in south Africa. Clin. Vaccine Immunol.24 (2), e00439-16. doi: 10.1128/CVI.00439-16
105
Lundegaard C. Lund O. Buus S. Nielsen M. (2010). Major histocompatibility complex class I binding predictions as a tool in epitope discovery. Immunology130 (3), 309–318. doi: 10.1111/J.1365-2567.2010.03300.X
106
Ly D. Moody D. B. (2014). The CD1 size problem: lipid antigens, ligands, and scaffolds. Cell. Mol. Life Sci.71 (16), 3069–3079. doi: 10.1007/S00018-014-1603-6
107
Mahmood A. Srivastava S. Tripathi S. Ansari M. A. Owais M. Arora A. et al . (2011). Molecular characterization of secretory proteins Rv3619c and Rv3620c from mycobacterium tuberculosis H37Rv. FEBS J.278 (2), 341–353. doi: 10.1111/j.1742-4658.2010.07958.x
108
McShane H. Pathan A. A. Sander C. R. Goonetilleke N. P. Fletcher H. A. Hill A. V. S. (2005). Boosting BCG with MVA85A: the first candidate subunit vaccine for tuberculosis in clinical trials. Tuberculosis (Edinburgh Scotland)85 (1–2), 47–52. doi: 10.1016/j.tube.2004.09.015
109
Mehaffy C. Belisle J. T. Dobos K. M. (2019). Mycobacteria and their sweet proteins: An overview of protein glycosylation and lipoglycosylation in m. tuberculosis. Tuberculosis115, 1–13. doi: 10.1016/j.tube.2019.01.001
110
Millington K. A. Fortune S. M. Low J. Garces A. Hingley-Wilson S. M. Wickremasinghe M. et al . (2011). Rv3615c is a highly immunodominant RD1 (Region of difference 1)-dependent secreted antigen specific for mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. United States America108 (14), 5730–5735. doi: 10.1073/pnas.1015153108
111
Montealegre S. Van Endert P. M. (2019). Endocytic recycling of MHC class I molecules in non-professional antigen presenting and dendritic cells. Front. Immunol.9 (JAN). doi: 10.3389/FIMMU.2018.03098
112
Moodley A. Fatoba A. Okpeku M. Emmanuel Chiliza T. Blessing Cedric Simelane M. Pooe O. J. . (2022). Reverse vaccinology approach to design a multi-epitope vaccine construct based on the mycobacterium tuberculosis biomarker PE_PGRS17. Immunologic Res.70 (4), 501–517. doi: 10.1007/s12026-022-09284-x
113
Moody D. B. Besra G. S. Wilson I. A. Porcelli S. A. . (1999). The molecular basis of CD1-mediated presentation of lipid antigens. Immunol. Rev.172, 285–296. doi: 10.1111/J.1600-065X.1999.TB01373.X
114
Moody D. B. Reinhold B. B. Guy M. R. Beckman E. M. Frederique D. E. Furlong S. T. et al . (1997). Structural requirements for glycolipid antigen recognition by CD1b- restricted T cells. Science278 (5336), 283–286. doi: 10.1126/science.278.5336.283
115
Moody D. B. Young D. C. Cheng T. Y. Rosat J. P. Roura-Mir C. O’Connor P. B. et al . (2004). T Cell activation by lipopeptide antigens. Sci. (New York N.Y.)303 (5657), 527–531. doi: 10.1126/SCIENCE.1089353
116
Moody D. B. Suliman S. (2017). CD1: From molecules to diseases. F1000Research6, 10. doi: 10.12688/F1000RESEARCH.12178.1
117
Moscoso J. Serrano-Vela J. I. Pacheco R. Arnaiz-Villena A. . (2006). HLA-G, -e and -f: Allelism, function and evolution. Transplant. Immunol.17 (1), 61–64. doi: 10.1016/j.trim.2006.09.010
118
Moss C. X. Villadangos J. A. Watts C. (2005). Destructive potential of the aspartyl protease cathepsin d in MHC class II-restricted antigen processing. Eur. J. Immunol.35 (12), 3442–3451. doi: 10.1002/eji.200535320
119
Mukamolova G. V. Turapov O. A. Young D. I. Kaprelyants A. S. Kell D. B. Young et al . (2002). A family of autocrine growth factors in mycobacterium tuberculosis. Mol. Microbiol.46 (3), 623–635. doi: 10.1046/j.1365-2958.2002.03184.x
120
Murima P. Zimmermann M. Chopra T. Pojer F. Fonti G. Dal Peraro M. et al . (2016). A rheostat mechanism governs the bifurcation of carbon flux in mycobacteria. Nat. Commun.7, 12527. doi: 10.1038/ncomms12527
121
Nair S. Ramaswamy P. A. Ghosh S. Joshi D. C. Pathak N. Siddiqui I. et al . (2009). The PPE18 of mycobacterium tuberculosis interacts with TLR2 and activates IL-10 induction in macrophage. J. Immunol.183 (10), 6269–6281. doi: 10.4049/jimmunol.0901367
122
Ndiaye B. P. Thienemann F. Ota M. Landry B. S. Camara M. Dieye S. et al . (2015). Safety, immunogenicity, and efficacy of the candidate tuberculosis vaccine MVA85A in healthy adults infected with HIV-1: a randomised, placebo-controlled, phase 2 trial. Lancet Respir. Med.3 (3), 190–200. doi: 10.1016/S2213-2600(15)00037-5
123
Neumann H. Schmidt H. Cavalié A. Jenne D. Wekerle H. . (1997). Major histocompatibility complex (MHC) class I gene expression in single neurons of the central nervous system: differential regulation by interferon (IFN)-gamma and tumor necrosis factor (TNF)-alpha. J. Exp. Med.185 (2), 305–316. doi: 10.1084/JEM.185.2.305
124
Nguyen B. N. Peterson B. N. Portnoy D. A. (2019). Listeriolysin O: A phagosome-specific cytolysin revisited. Cell. Microbiol.21 (3), e12988. doi: 10.1111/cmi.12988
125
Nieuwenhuizen N. E. Kulkarni P. S. Shaligram U. Cotton M. F. Rentsch C. A. Eisele B. et al . (2017). The recombinant bacille calmette-guerin vaccine VPM1002: Ready for clinical efficacy testing. Front. Immunol.8. doi: 10.3389/fimmu.2017.01147
126
Nieweg O. E. Tanis P. J. Kroon B. B. R. (2001). The definition of a sentinel node. Ann. Surg. Oncol.8 (6), 538–541. doi: 10.1007/S10434-001-0538-Y
127
Noss E. H. Pai R. K. Sellati T. J. Radolf J. D. Belisle J. Golenbock D. T. et al . (2001). Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of mycobacterium tuberculosis. J. Immunol.167 (2), 910–918. doi: 10.4049/jimmunol.167.2.910
128
O’Callaghan C. A. Bell J. I. (1998). Structure and function of the human MHC class ib molecules HLA-e, HLA-f and HLA-G. Immunol. Rev.163, 129–138. doi: 10.1111/J.1600-065X.1998.TB01192.X
129
O’Callaghan C. A. Tormo J. Willcox B. E. Braud V. M. Jakobsen B. K. Stuart D. I. et al . (1998). Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-e. Mol. Cell1 (4), 531–541. doi: 10.1016/s1097-2765(00)80053-2
130
Orr M. T. Ireton G. C. Beebe E. A. Huang P.-W. D. Reese V. A. Argilla D. et al . (2014). Immune subdominant antigens as vaccine candidates against mycobacterium tuberculosis. J. Immunol. (Baltimore Md. : 1950)193 (6), 2911–2918. doi: 10.4049/jimmunol.1401103
131
Pathni A. Özçelikkale A. Rey-Suarez I. Li L. Davis S. Rogers N. et al . (2022). Cytotoxic T lymphocyte activation signals modulate cytoskeletal dynamics and mechanical force generation. Front. Immunol.13. doi: 10.3389/fimmu.2022.779888
132
Pattanaik K. P. Ganguli G. Naik S. K. Sonawane A. . (2021). Mycobacterium tuberculosis EsxL induces TNF-α secretion through activation of TLR2 dependent MAPK and NF-κB pathways. Mol. Immunol.130, 133–141. doi: 10.1016/j.molimm.2020.11.020
133
Paulson T. (2013). A mortal foe. Nature502 (7470). doi: 10.1038/502S2a
134
Penn B. H. . (2018). An mtb-human protein-protein interaction map identifies a switch between host antiviral and antibacterial responses. Mol. Cell71 (4), 637–648.e5. doi: 10.1016/j.molcel.2018.07.010
135
Penn-Nicholson A. Tameris M. Smit E. Day T. A. Musvosvi M. Jayashankar L. et al . (2018). Safety and immunogenicity of the novel tuberculosis vaccine ID93 + GLA-SE in BCG-vaccinated healthy adults in south Africa: a randomised, double- blind, placebo-controlled phase 1 trial. Lancet Respir. Med.6 (4), 287–298. doi: 10.1016/S2213-2600(18)30077-8
136
Perreault C. (2010). The origin and role of MHC class I-associated self-peptides. Prog. Mol. Biol. Trans. Sci.92 (C), 41–60. doi: 10.1016/S1877-1173(10)92003-6
137
Pires D. Marques J. Pombo J. P. Carmo N. Bettencourt P. Neyrolles O. et al . (2016). Role of cathepsins in mycobacterium tuberculosis survival in human macrophages. Sci. Rep.6, 32247. doi: 10.1038/srep32247
138
Pomaznoy M. Kuan R. Lindvall M. Burel J. G. Seumois G. Vijayanand P. et al . (2020). Quantitative and qualitative perturbations of CD8(+) MAITs in healthy mycobacterium tuberculosis-infected individuals. ImmunoHorizons4 (6), 292–307. doi: 10.4049/immunohorizons.2000031
139
Portal-Celhay C. Tufariello J. M. Srivastava S. Zahra A. Klevorn T. Grace P. S. et al . (2016). Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4(+) T-cell activation. Nat. Microbiol.2, 16232. doi: 10.1038/nmicrobiol.2016.232
140
Prados-Rosales R. Baena A. Martinez L. R. Luque-Garcia J. Kalscheuer R. Veeraraghavan U. et al . (2011). Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Invest.121 (4), 1471–1483. doi: 10.1172/JCI44261
141
Provine N. M. Klenerman P. (2020). MAIT cells in health and disease. Annu. Rev. Immunol.38, 203–228. doi: 10.1146/annurev-immunol-080719-015428
142
Purcell A. W. Ramarathinam S. H. Ternette N. (2019). Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc.14 (6), 1687–1707. doi: 10.1038/s41596-019-0133-y
143
Rahman M. A. Sobia P. Dwivedi V. P. Bhawsar A. Singh D. K. Sharma P. et al . (2015). Mycobacterium tuberculosis TlyA protein negatively regulates T helper (Th) 1 and Th17 differentiation and promotes tuberculosis pathogenesis. J. Biol. Chem.290 (23), 14407–14417. doi: 10.1074/jbc.M115.653600
144
Rastogi S. Briken V. (2022). Interaction of mycobacteria with host cell inflammasomes. Front. Immunol.13. doi: 10.3389/FIMMU.2022.791136
145
Riaño F. Arroyo L. París S. Rojas M. Friggen A. H. Van Meijgaarden K. E. et al . (2012). T Cell responses to DosR and rpf proteins in actively and latently infected individuals from Colombia. Tuberculosis92 (2), 148–159. doi: 10.1016/j.tube.2011.12.005
146
Riegert P. Wanner V. Bahram S. (1998). Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J. Immunol. (Baltimore Md. : 1950)161 (8), 4066–4077. doi: 10.4049/jimmunol.161.8.4066
147
Roberts E. A. Chua J. Kyei G. B. Deretic V. . (2006). Higher order rab programming in phagolysosome biogenesis. J. Cell Biol.174 (7), 923–929. doi: 10.1083/jcb.200603026
148
Robinson J. Guethlein L. A. Cereb N. Yang S. Y. Norman P. J. Marsh S. G.E. et al . (2017). Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-a, -b and -c alleles. PloS Genet.13 (6), e1006862. doi: 10.1371/journal.pgen.1006862
149
Rodo M. J. Rozot V. Nemes E. Dintwe O. Hatherill M. Little F. et al . (2019). A comparison of antigen-specific T cell responses induced by six novel tuberculosis vaccine candidates. PloS Pathog.15 (3), e1007643. doi: 10.1371/journal.ppat.1007643
150
Römisch K. Roberts E.A. Chua J. (2017). A case for Sec61 channel involvement in ERAD. Trends Biochem. Sci.42 (3), 171–179. doi: 10.1016/j.tibs.2016.10.005
151
Rothchild A. C. Jayaraman P. Nunes-Alves C. Behar S. M. . (2014). iNKT cell production of GM-CSF controls mycobacterium tuberculosis. PloS Pathog.10 (1), e1003805. doi: 10.1371/JOURNAL.PPAT.1003805
152
Rozot V. Nemes E. Geldenhuys H. Musvosvi M. Toefy A. Rantangee F. et al . (2020). Multidimensional analyses reveal modulation of adaptive and innate immune subsets by tuberculosis vaccines. Commun. Biol.3 (1), 563. doi: 10.1038/s42003-020-01288-3
153
Rozot V. Vigano S. Mazza-Stalder J. Idrizi E. Day C. L. Perreau M. et al . (2013). Mycobacterium tuberculosis-specific CD8+ T cells are functionally and phenotypically different between latent infection and active disease. Eur. J. Immunol.43 (6), 1568–1577. doi: 10.1002/eji.201243262
154
Sánchez A. Espinosa P. García T. Mancilla R. . (2012). The 19 kDa mycobacterium tuberculosis lipoprotein (lpqh) induces macrophage apoptosis through extrinsic and intrinsic pathways: A role for the mitochondrial apoptosis-inducing factor. Clin. Dev. Immunol.2012, 950503. doi: 10.1155/2012/950503
155
Saiga H. Nieuwenhuizen N. Gengenbacher M. Koehler A.-B. Schuerer S. Moura- Alves P. et al . (2015). The recombinant BCG ΔureC::hly vaccine targets the AIM2 inflammasome to induce autophagy and inflammation. J. Infect. Dis.211 (11), 1831–1841. doi: 10.1093/infdis/jiu675
156
Saini N. K. Baena A. Ng T. W. Venkataswamy M. M. Kennedy S. C. Kunnath- Velayudhan S. et al . (2016). Suppression of autophagy and antigen presentation by mycobacterium tuberculosis PE_PGRS47. Nat. Microbiol.1 (9), 16133. doi: 10.1038/nmicrobiol.2016.133
157
Sakai S. Kauffman K. D. Oh S. Nelson C. E. Barry C. E. 3rd Barber D. L. et al . (2021). MAIT cell-directed therapy of mycobacterium tuberculosis infection. Mucosal Immunol.14 (1), 199–208. doi: 10.1038/s41385-020-0332-4
158
Samuel L. P. Song C. H. Wei J. Roberts E. A. Dahl J. L. Barry C. E. et al . (2007). Expression, production and release of the eis protein by mycobacterium tuberculosis during infection of macrophages and its effect on cytokine secretion. Microbiology153 (2), 529–540. doi: 10.1099/mic.0.2006/002642-0
159
Schrambach S. Ardizzone M. Leymarie V. Sibilia J. Bahram S. . (2007). In vivo expression pattern of MICA and MICB and its relevance to auto-immunity and cancer. PloS One2 (6), e518. doi: 10.1371/journal.pone.0000518
160
Segura J. M. Guillaume P. Mark S. Dojcinovic D. Johannsen A. Bosshard G. et al . (2008). Increased mobility of major histocompatibility complex i-peptide complexes decreases the sensitivity of antigen recognition. J. Biol. Chem.283 (35), 24254–24263. doi: 10.1074/jbc.M803549200
161
Sette A. Sidney J. (1999). Nine major HLA class I supertypes account for the vast preponderance of HLA-a and -b polymorphism. Immunogenetics50 (3–4), 201–212. doi: 10.1007/S002510050594
162
Sharma T. Alam A. Ehtram A. Rani A. Grover S. Ehtesham N. Z. et al . (2022). The mycobacterium tuberculosis PE_PGRS protein family acts as an immunological decoy to subvert host immune response. Int. J. Mol. Sci.23 (1), 17. doi: 10.3390/IJMS23010525
163
Shi J. Zhang H. Fang L. Xi Y. Zhou Y. Luo R. et al . (2014). A novel firefly luciferase biosensor enhances the detection of apoptosis induced by ESAT-6 family proteins of mycobacterium tuberculosis. Biochem. Biophys. Res. Commun.452 (4), 1046–1053. doi: 10.1016/j.bbrc.2014.09.047
164
Shi S. Ehrt S. (2006). Dihydrolipoamide acyltransferase is critical for mycobacterium tuberculosis pathogenesis. Infection Immun.74 (1), 56–63. doi: 10.1128/IAI.74.1.56-63.2006
165
Sillé F. C. M. Martin C. Jayaraman P. Rothchild A. Fortune S. Besra G. S. et al . (2011). Requirement for invariant chain in macrophages for mycobacterium tuberculosis replication and CD1d antigen presentation. Infection Immun.79 (8), 3053–3063. doi: 10.1128/IAI.01108-10
166
Singh P. Katoch V. M. Mohanty K. K. Chauhan D. S. . (2016). Analysis of expression profile of mce operon genes (mce1, mce2, mce3 operon) in different mycobacterium tuberculosis isolates at different growth phases. Indian J. Med. Res.143 (April), 487–494. doi: 10.4103/0971-5916.184305
167
Smaill F. Jeyanathan M. Smieja M. Medina M. F. Thanthrige-Don N. Zganiacz A. et al . (2013). A human type 5 adenovirus-based tuberculosis vaccine induces robust T cell responses in humans despite preexisting anti-adenovirus immunity. Sci. Trans. Med.5 (205), 205ra134. doi: 10.1126/scitranslmed.3006843
168
Sperisen P. Schmid C. D. Bucher P. Zilian O. . (2005). Stealth proteins: In silico identification of a novel protein family rendering bacterial pathogens invisible to host immune defense. PloS Comput. Biol.1 (6), 492–499. doi: 10.1371/journal.pcbi.0010063
169
Stephenson L. Byard R. W. (2020). An atlas overview of characteristic features of tuberculosis that may be encountered at autopsy. Forensic science medicine Pathol.16 (1), 143–151. doi: 10.1007/S12024-019-00161-Y
170
Stewart G. R. Wilkinson K. A. Newton S. M. Sullivan S. M. Neyrolles O. Wain J. R. et al . (2005). Effect of deletion or overexpression of the 19-kilodalton lipoprotein Rv3763 on the innate response to mycobacterium tuberculosis. Infection Immun.73 (10), 6831–6837. doi: 10.1128/IAI.73.10.6831-6837.2005
171
Stylianou E. Griffiths K. L. Poyntz H. C. Harrington-Kandt R. Dicks M. D. Stockdale L. et al . (2015). Improvement of BCG protective efficacy with a novel chimpanzee adenovirus and a modified vaccinia Ankara virus both expressing Ag85A. Vaccine33 (48), 6800–6808. doi: 10.1016/j.vaccine.2015.10.017
172
Stylianou E. Harrington-Kandt R. Beglov J. Bull N. Pinpathomrat N. Swarbrick G. M. et al . (2018). Identification and evaluation of novel protective antigens for the development of a candidate tuberculosis subunit vaccine. Infection Immun.86 (7), e00014-18. doi: 10.1128/IAI.00014-18
173
Su H. Peng B. Zhang Z. Liu Z. Zhang Z. . (2019). The mycobacterium tuberculosis glycoprotein Rv1016c protein inhibits dendritic cell maturation, and impairs Th1 /Th17 responses during mycobacteria infection. Mol. Immunol.109, 58–70. doi: 10.1016/j.molimm.2019.02.021
174
Suliman S. Gela A. Mendelsohn S. C. Iwany S. K. Tamara K. L. Mabwe S. et al . (2020). Peripheral blood mucosal-associated invariant T cells in tuberculosis patients and healthy mycobacterium tuberculosis-exposed controls. J. Infect. Dis.222 (6), 995–1007. doi: 10.1093/infdis/jiaa173
175
Sunita Singhvi N. Singh Y. Shukla P. . (2020). Computational approaches in epitope design using DNA binding proteins as vaccine candidate in mycobacterium tuberculosis. Infection Genet. Evol.83, 7. doi: 10.1016/j.meegid.2020.104357
176
Tang S. T. van Meijgaarden K. E. Caccamo N. Guggino G. Klein M. R. van Weeren P. et al . (2011). Genome-based in silico identification of new mycobacterium tuberculosis antigens activating polyfunctional CD8+ T cells in human tuberculosis. J. Immunol. (Baltimore Md. : 1950)186 (2), 1068–1080. doi: 10.4049/jimmunol.1002212
177
Tantivitayakul P. Juthayothin T. Ruangchai W. Smittipat N. Disratthakit A. Mahasirimongkol S. et al . (2020). Identification and in silico functional prediction of lineage-specific SNPs distributed in DosR-related proteins and resuscitation-promoting factor proteins of mycobacterium tuberculosis. Heliyon6 (12), e05744–e05744. doi: 10.1016/j.heliyon.2020.e05744
178
Teitelbaum R. Cammer M. Maitland M. L. Freitag N. E. Condeelis J. Bloom B. R. . (1999). Mycobacterial infection of macrophages results in membrane-permeable phagosomes. Proc. Natl. Acad. Sci. United States America96 (26), 15190. doi: 10.1073/PNAS.96.26.15190
179
ten Broeke T. Wubbolts R. Stoorvogel W. (2013). MHC class II antigen presentation by dendritic cells regulated through endosomal sorting. Cold Spring Harbor Perspect. Biol.5 (12), a016873. doi: 10.1101/cshperspect.a016873
180
Tiwari B. Ramakrishnan U. M. Raghunand T. R. (2015). The mycobacterium tuberculosis protein pair PE9 (Rv1088)-PE10 (Rv1089) forms heterodimers and induces macrophage apoptosis through toll-like receptor 4. Cell. Microbiol.17 (11), 1653–1669. doi: 10.1111/cmi.12462
181
Tiwari B. Soory A. Raghunand T. R. (2014). An immunomodulatory role for the mycobacterium tuberculosis region of difference 1 locus proteins PE35 (Rv3872) and PPE68 (Rv3873). FEBS J.281 (6), 1556–1570. doi: 10.1111/febs.12723
182
Tobian A. A. R. Potter N. S. Ramachandra L. Pai R. K. Convery M. Boom W. H. et al (2003). Alternate class I MHC antigen processing is inhibited by toll-like receptor signaling pathogen-associated molecular patterns: Mycobacterium tuberculosis 19-kDa Lipoprotein, CpG DNA, and Lipopolysaccharide and lipopolysaccharide. J. Immunol.171 (3), 1413–1422. doi: 10.4049/jimmunol.171.3.1413
183
Torrelles J. B. Schlesinger L. S. (2017). Integrating lung physiology, immunology, and tuberculosis. Trends Microbiol.25 (8), 688–697. doi: 10.1016/j.tim.2017.03.007
184
Trost B. Lucchese G. Stufano A. Bickis M. Kusalik A. Kanduc D. et al . (2010). No human protein is exempt from bacterial motifs, not even one. Self/nonself1 (4), 328–334. doi: 10.4161/self.1.4.13315
185
Tundup S. Mohareer K. Hasnain S. E. (2014). Mycobacterium tuberculosis PE25/PPE41 protein complex induces necrosis in macrophages: Role in virulence and disease reactivation? FEBS Open Bio4, 822–828. doi: 10.1016/j.fob.2014.09.001
186
Turing A. M. (1990). The chemical basis of morphogenesis. Bull. Math. Biol.52 (1–2), 153–197. doi: 10.1007/BF02459572
187
Udgata A. Qureshi R. Mukhopadhyay S. (2016). Transduction of functionally contrasting signals by two mycobacterial PPE proteins downstream of TLR2 receptors. J. Immunol.197 (5), 1776–1787. doi: 10.4049/jimmunol.1501816
188
Underhill D. M. Ozinsky A. Smith K. D. Aderem A. . (1999). Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. United States America96 (25), 14459–14463. doi: 10.1073/pnas.96.25.14459
189
Uzhachenko R. V. Shanker A. (2019). CD8(+) T lymphocyte and NK cell network: Circuitry in the cytotoxic domain of immunity. Front. Immunol.10. doi: 10.3389/fimmu.2019.01906
190
van Crevel R. Ottenhoff T. H. M. van der Meer J. W. M. (2003). Innate immunity to mycobacterium tuberculosis. Adv. Exp. Med. Biol.531, 241–247. doi: 10.1007/978-1-4615-0059-9_20
191
van Meijgaarden K. E. Haks M. C. Caccamo N. Dieli F. Ottenhoff T. H.M. Joosten S. A. . (2015). Human CD8+ T-cells recognizing peptides from mycobacterium tuberculosis (Mtb) presented by HLA-e have an unorthodox Th2-like, multifunctional, mtb inhibitory phenotype and represent a novel human T-cell subset. PloS Pathog.11 (3), e1004671. doi: 10.1371/journal.ppat.1004671
192
Van Rhijn I. Young D. C. De Jong A. Vazquez J. Cheng T.-Y. Talekar R. et al . (2009). CD1c bypasses lysosomes to present a lipopeptide antigen with 12 amino acids. J. Exp. Med.206 (6), 1409–1422. doi: 10.1084/jem.20082480
193
Vázquez C. L. Lerner T. R. Kasmapour B. Pei G. Gronow A. Bianco M. V. et al . (2014). Experimental selection of long-term intracellular mycobacteria. Cell. Microbiol.16 (9), 1425–1440. doi: 10.1111/cmi.12303
194
Vergne I. Chua J. Lee H. H. Lucas M. Belisle J. Deretic V. et al . (2005). Mechanism of phagolysosome biogenesis block by viable mycobacterium tuberculosis. Proc. Natl. Acad. Sci. United States America102 (11), 4033–4038. doi: 10.1073/pnas.0409716102
195
Vico T. Youssif C. Zare F. Comalada M. Sebastian C. Lloberas J. et al . (2022). GM-CSF protects macrophages from DNA damage by inducing differentiation. Cells11 (6), 20. doi: 10.3390/cells11060935
196
Vordermeier H. M. Hewinson R. G. Wilkinson R. J. Wilkinson K. A. Gideon H. P. Young D.B. et al . (2012). Conserved immune recognition hierarchy of mycobacterial PE/PPE proteins during infection in natural hosts. PloS One7 (8), e40890. doi: 10.1371/JOURNAL.PONE.0040890
197
Walker K. B. Guo M. Guo Y. Poecheim J. Velmurugan K. Schrager L. K. et al . (2016). “Novel approaches to preclinical research and TB vaccine development,” in Tuberculosis (Edinburgh, Scotland) (Scotland), ppS12–ppS15. doi: 10.1016/j.tube.2016.05.012
198
Wang J. Teng J. L. L. Zhao D. Ge P. Li B. Woo P. C.Y. et al . (2016). The ubiquitin ligase TRIM27 functions as a host restriction factor antagonized by mycobacterium tuberculosis PtpA during mycobacterial infection. Sci. Rep.6, 13. doi: 10.1038/srep34827
199
WHO (2017) 2017, GLobal tuberculosis report. Available at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports.
200
WHO (2020) Global tuberculosis report 2020. Available at: https://www.who.int/publications/i/item/9789240013131.
201
WHO Key Facts (2022). Available at: https://www.who.int/news-room/fact-sheets/detail/tuberculosis.
202
Wilkie M. Satti I. Minhinnick A. Harris S. Riste M. Ramon R. L. et al . (2020). A phase I trial evaluating the safety and immunogenicity of a candidate tuberculosis vaccination regimen, ChAdOx1 85A prime - MVA85A boost in healthy UK adults. Vaccine38 (4), 779–789. doi: 10.1016/j.vaccine.2019.10.102
203
Willcox C. R. Pitard V. Netzer S. Couzi L. Salim M. Silberzahn T. et al . (2012). Cytomegalovirus and tumor stress surveillance by binding of a human γδ T cell antigen receptor to endothelial protein c receptor. Nat. Immunol.13 (9), 872–879. doi: 10.1038/ni.2394
204
Witherden D. A. Havran W. L. (2012). “EPCR: a stress trigger for γδ T cells,” in Nature immunology (United States), pp 812–pp 814. doi: 10.1038/ni.2398
205
Wong D. Bach H. Sun J. Hmama Z. Av-Gay Y. . (2011). Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc. Natl. Acad. Sci. United States America108 (48), 19371–19376. doi: 10.1073/pnas.1109201108
206
Wong E. B. Gold M. C. Meermeier E. W. Xulu B. Z. Khuzwayo S. Sullivan Z. A. et al . (2019). TRAV1-2+ CD8+ T-cells including oligoconal expansions of MAIT cells are enriched in the airways in human tuberculosis. Commun. Biol.2 (1), 13. doi: 10.1038/s42003-019-0442-2
207
Wu C. Li Z. Fu X. Yu S. Lao S. Yang B. et al . (2015). Antigen-specific human NKT cells from tuberculosis patients produce IL-21 to help b cells for the production of immunoglobulins. Oncotarget6 (30), 28633–28645. doi: 10.18632/oncotarget.5764
208
Xu Y. Yang E. Huang Q. Ni W. Kong C. Liu G. et al . (2015). PPE57 induces activation of macrophages and drives Th1-type immune responses through TLR2. J. Mol. Med.93 (6), 645–662. doi: 10.1007/s00109-014-1243-1
209
Yang J. D. Mott D. Sutiwisesak R. Lu Y.-J. Raso F. Stowell B. et al . (2018). Mycobacterium tuberculosis-specific CD4+ and CD8+ T cells differ in their capacity to recognize infected macrophages. PloS Pathog.14 (5), e1007060. doi: 10.1371/journal.ppat.1007060
210
Yaseen I. Kaur P. Nandicoori V. K. Khosla S. . (2015). Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3. Nat. Commun.6, 13. doi: 10.1038/ncomms9922
211
Zhen Y. Radulovic M. Vietri M. Stenmark H. . (2021). Sealing holes in cellular membranes. EMBO J.40 (7), e106922. doi: 10.15252/EMBJ.2020106922
Summary
Keywords
Mycobacterium tuberculosis , antigen processing, MHC-I, vaccine, host-pathogen interactions, cytotoxic T cells
Citation
Witt KD (2023) Role of MHC class I pathways in Mycobacterium tuberculosis antigen presentation. Front. Cell. Infect. Microbiol. 13:1107884. doi: 10.3389/fcimb.2023.1107884
Received
25 November 2022
Accepted
23 February 2023
Published
15 March 2023
Volume
13 - 2023
Edited by
Edwin Leeansyah, Tsinghua University, China
Reviewed by
Fei Han, Tsinghua University, China; Shouxiong Huang, University of Cincinnati, United States
Updates
Copyright
© 2023 Witt.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karolina D. Witt, karolina.witt@ndm.ox.ac.uk
This article was submitted to Molecular Bacterial Pathogenesis, a section of the journal Frontiers in Cellular and Infection Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.