- Department of Environmental and Public Health Sciences, College of Medicine, University of Cincinnati, Cincinnati, OH, United States
Mucosal-associated invariant T (MAIT) cells are protective against tuberculous and non-tuberculous mycobacterial infections with poorly understood mechanisms. Despite an innate-like nature, MAIT cell responses remain heterogeneous in bacterial infections. To comprehensively characterize MAIT activation programs responding to different bacteria, we stimulated MAIT cells with E. coli to compare with Bacillus Calmette-Guérin (BCG), which remains the only licensed vaccine and a feasible tool for investigating anti-mycobacterial immunity in humans. Upon sequencing mRNA from the activated and inactivated CD8+ MAIT cells, results demonstrated the altered MAIT cell gene profiles by each bacterium with upregulated expression of activation markers, transcription factors, cytokines, and cytolytic mediators crucial in anti-mycobacterial responses. Compared with E. coli, BCG altered more MAIT cell genes to enhance cell survival and cytolysis. Flow cytometry analyses similarly displayed a more upregulated protein expression of B-cell lymphoma 2 and T-box transcription factor Eomesodermin in BCG compared to E.coli stimulations. Thus, the transcriptomic program and protein expression of MAIT cells together displayed enhanced pro-survival and cytotoxic programs in response to BCG stimulation, supporting BCG induces cell-mediated effector responses of MAIT cells to fight mycobacterial infections.
Introduction
An effective T cell response leads to a low lifetime risk of developing active tuberculosis and a successful vaccine mostly aims to boost protective T cell responses (Cooper, 2009). Conventional T cells are essential for maintaining a low bacterial load, as shown using antibody depletion or adoptive transfer of CD4+ or CD8+ T cells in mice (Flynn and Chan, 2001; Behar, 2013), increased risk of active tuberculosis with reduced CD4+ T cell frequency in HIV co-infection of humans (Cooper, 2009; Prezzemolo et al., 2014), and loss of protection against tuberculosis in primates with CD8+ T cell depletion (Chen et al., 2009). However, anti-mycobacterial roles of CD8+ T cells in humans remain poorly characterized, and weak peptide-specific CD8+ T cell responses have been shown in multiple human vaccine trials (Tameris et al., 2013). Recent findings demonstrated that a large portion of mycobacterial-reactive CD8+ T cells in humans are not conventional T cells but mucosal-associated invariant T (MAIT) cells (Gold et al., 2010; Le Bourhis et al., 2010). Unlike conventional T cells that are activated by the peptide antigens presented by major histocompatibility complex (MHC) or human leukocyte antigens (HLA), MAIT cells are activated by non-peptidic small metabolite antigens presented by MHC class I-related protein 1 (MR1) (Huang et al., 2005; Hansen et al., 2007; Huang et al., 2008; Huang et al., 2009; Gold et al., 2010; Le Bourhis et al., 2010; Chua et al., 2011; Young et al., 2013; Huang, 2016; Huang and Moody, 2016; Sharma et al., 2020). Different from the highly variable MHC or HLA proteins for the presentation of eminently divergent peptide antigens, MR1 is monomorphic in humans for the presentation of conserved metabolite antigens such as bacterial riboflavin metabolites, defining the innate-like nature of MAIT cell responses in a donor-unrestricted manner. Stimulation with mycobacterial-infected antigen-presenting cells can strongly enhance the expression of proinflammatory cytokines, surface activation markers, and cytolytic molecules of MAIT cells (Gold et al., 2010; Le Bourhis et al., 2010; Sharma et al., 2020) with poorly known mechanisms, to further bridge the innate and late onset of adaptive immune responses (Meierovics et al., 2013; Huang, 2016; Ioannidis et al., 2020).
MAIT cells are protective against multiple non-tuberculous mycobacterial and M. tuberculosis infections (Le Bourhis et al., 2010; Rossjohn et al., 2015; Huang, 2016). Specifically, MAIT cell overexpression in mice inhibits the growth of non-tuberculous M. abscessus (Le Bourhis et al., 2010) and M. bovis (Chua et al., 2012; Sakala et al., 2015), and partially suppresses M. tuberculosis infections (Sakai et al., 2021). In contrast, MR1-knockout mice display a higher load of M. abscessus (Le Bourhis et al., 2010), M. bovis (Chua et al., 2012; Sakala et al., 2015), and M. tuberculosis in vivo (Sakai et al., 2021). These recent findings highlighted MAIT cells as promising targets to induce immune protection against non-tuberculous and tuberculous mycobacterial infections. In the meantime, the M. bovis strain Bacillus Calmette-Guerin (BCG) activates MAIT cells rapidly (Chua et al., 2012; Sharma et al., 2020) and is the licensed vaccine against tuberculosis (Raviglione et al., 1995; Oettinger et al., 1999; Li J. et al., 2021). Although BCG vaccination of newborns or toddlers protects children or young adults from pulmonary tuberculosis, its efficacy in higher ages is usually compromised by various factors, including environmental mycobacteria infection (McShane et al., 2012; Kumar, 2021) or an extended period after vaccination (Michelsen et al., 2014; Katelaris et al., 2020). Conventional T cell responses have been a focus for interpreting anti-mycobacterial immunity. BCG vaccination induces antigen-specific CD4+ T cell responses critical for regulating cellular and humoral immunity (McShane et al., 2012; Soares et al., 2013). However, CD8+ T cell responses to BCG vaccination remain poorly characterized, and peptide-specific CD8+ T cells appear at a much smaller scale than CD4+ T cells (Murray et al., 2006). This deficit can be likely explained by a high percentage of mycobacterial-specific CD8+ T cell clones that have been recently characterized as mucosal-associated invariant T (MAIT) cells (Gold et al., 2010; Le Bourhis et al., 2010), which feature innate-like activation kinetics different from conventional CD8+ T cells.
It remains unclear how MAIT cells are stimulated by mycobacteria and develop unique transcriptomic programs to elicit anti-mycobacterial immunity. As known, bacterial activation of MAIT cells is mainly mediated by MR1 presentation of bacterial metabolites (Le Bourhis et al., 2010; Huang, 2016). BCG, E. coli (Le Bourhis et al., 2010; Sakala et al., 2015; Sharma et al., 2020), or Salmonella Typhimurium (Chen et al., 2017) are expected to provide riboflavin precursor (Kjer-Nielsen et al., 2012; Corbett et al., 2014; Harriff et al., 2018) or other metabolites to be presented by MR1 for MAIT cell activation. Further, MAIT cell activation is depleted by the anti-MR1 antibody blockade of the interaction between the bacterial antigen-loaded MR1 protein and T cell receptor (TCR) (Sharma et al., 2020). The human myelogenous cell line K562 with a defective expression of human leukocyte antigens (HLA) (Lozzio and Lozzio, 1975; Andersson et al., 1979; Roder et al., 1979; Koeffler and Golde, 1980; Li et al., 2019) has been widely used in various studies for testing antigen-specific T cell activation (Escobar et al., 2008; de Jong et al., 2010; de Jong et al., 2014; Sharma et al., 2020; Goodman et al., 2022) and as an ideal antigen-presenting cell for MAIT cell activation upon the overexpression of MR1 protein (Sharma et al., 2020). Similarly, MR1-dependent antigen presentation has been further demonstrated by an impaired anti-bacterial MAIT cell response in MR1 knockout mice (Le Bourhis et al., 2010; Sakala et al., 2015; Chen et al., 2017). Therefore, the co-culture of hMR1-expressing cells with MAIT cells provides a feasible model to investigate human MAIT cell transcriptomes upon the stimulation of MR1-mediated presentation of different bacterial antigens. The comparative MAIT cell transcriptomes stimulated by mycobacteria versus extracellular bacteria are expected to provide MAIT cell activation features and pathways potentially crucial for fighting mycobacterial infections. Recent MAIT cell transcriptomes, including single-cell transcriptomes, have differentiated various MAIT cell subsets in mice (Chandra et al., 2021; Tao et al., 2021) and humans (Vorkas et al., 2022). Moreover, human MAIT cell stimulation through signaling, cytokines, anti-CD3/CD28, or bacterial infections display transcriptomes associated with tissue repair (Hinks et al., 2019; Leng et al., 2019; Chandra et al., 2021; Tao et al., 2021; Vorkas et al., 2022), polyfunctional effector functions (Koay et al., 2019; Lamichhane et al., 2019; Salou et al., 2019; Lee et al., 2020), and innate-like activation programs (Sharma et al., 2020). However, MAIT cells stimulated by MR1 antigen presentation with different bacterial infections or stimulations display heterogeneous responses that have been considered as pathogen selectivity, labeled with diverse sequences of TCRβ chain and an invariant α chain, and attributed to potentially different antigens from various bacteria (Reantragoon et al., 2013; Gold et al., 2014; Jiang et al., 2014; Lepore et al., 2014; Sakala et al., 2015; Meermeier et al., 2016). Differential human MAIT cell response stimulated by BCG vs. E. coli is a representative example (Jiang et al., 2014) to further understand the mechanisms contributing to MAIT cell responses to different bacteria. It remains unknown whether mycobacteria stimulate MAIT cell transcriptomes and pathways that are associated with anti-mycobacterial immunity in comparison to extracellular bacteria E. coli. Therefore, we profiled the E. coli-stimulated MAIT cell transcriptomes to compare with the previously obtained BCG-stimulated MAIT transcriptomes (Sharma et al., 2020). Results demonstrated an enhanced program of pro-survival and cytolytic MAIT cell responses, particularly in BCG stimulation, supporting cell-mediated responses to mycobacterial infection.
Materials and methods
MAIT cells were activated by bacterial-incubated antigen-presenting cells and sorted for transcriptomic analyses with a summary of analyses provided below and the stepwise procedures detailed in the Supplementary Materials. We used human HLA-defective myelogenous leukemia cell line K562 (K562.hMR1) as antigen-presenting cells for bacterial infection and MAIT cell stimulation. Bacterial infection used Listeria monocytogenes (L. monocytogenes), Escherichia coli (E. coli), Mycobacterium bovis (M. bovis), and avirulent Mycobacterium tuberculosis (M. tuberculosis). Bacterial-incubated K562.hMR1 cells were co-cultured with anti-Vα7.2-enriched primary human MAIT cells from the blood samples of de-identified healthy donors following the Institutional Review Board (IRB)-approved protocol. The activated MAIT cells were gated on Vα7.2+CD161+CD4-CD8+ cells to sort CD69+CD26++ activated and CD69+/-CD26+/- inactivated CD8+ MAIT cells for transcriptomic profiling using an Illumina sequencing platform and flow cytometry analyses as we reported (Sharma et al., 2020). In this study, we profiled MAIT cell transcriptomes upon incubating with MAIT-stimulatory E. coli and non-stimulatory Listeria (accession # pending). The E. coli-activated MAIT cell transcriptomes were analyzed in comparison with previously obtained raw RANseq data from BCG-activated MAIT cells (accession # GSE124381), considering the baseline activities of inactivated CD69+/-CD26+/- CD8+ MAIT cells in the identical and inter-bacterial incubation (Sharma et al., 2020). The obtained transcriptomic data were analyzed using edgeR program for differentially expressed genes (DEG) with calculated p-values, Toppcluster program for gene clustering with Bonferroni correction of p-values, Cytoscape for pathway demonstration, and Gene Set Enrichment Analysis (GSEA) program with enrichment scores and nominal p-values for expression profile comparison. Statistical analyses of flow cytometry results used a pairwise t-test for the directional difference between E. coli and BCG stimulations. Detailed materials and methods are described in the Supplementary Materials.
Results
Differentially expressed genes (DEG) of MAIT cells upon BCG and E. coli stimulations
As various pathogens provide different immune stimuli (Kauffman et al., 2018; Sakai et al., 2021), the determination of differential MAIT transcriptomes stimulated by BCG versus E. coli facilitates characterizing anti-mycobacterial immunity of MAIT cells. We have shown that BCG and E. coli activated MAIT cells from human blood (Sharma et al., 2020), providing an in vitro MAIT cell activation model for intracellular vs. extracellular bacterial stimulation. In this model, human myelogenous leukemia cell line K562 with hMR1 overexpression (K562.hMR1) was incubated with BCG or E. coli overnight, washed, and co-cultured with MAIT cells, which were pre-enriched from the peripheral blood of healthy donors using magnetic bead-conjugated anti-Vα7.2 antibody. Upon overnight stimulation with bacterial-incubated K562.hMR1, CD8+ MAIT cells as the major peripheral MAIT cell population in humans were gated on Vα7.2+CD161+CD4-CD8+ (Figure S1A) to sort the activated (CD69+CD26++) and inactivated (CD69+/-CD26+/-) MAIT cell subsets using flow cytometry (Figures 1A, S1B) (Gold et al., 2010; Sharma et al., 2020) for total RNA extraction and mRNA profiling. Identification and separation of activated MAIT cells are based on the expression of CD69 and CD26 upregulated upon the incubation of stimulatory bacteria, but not non-stimulatory Listeria or bacterial-free condition (Sharma et al., 2020) (Figure S1B). It has been noted that E. coli and BCG activate MAIT cells through MR1 presentation of bacterial metabolite antigens to interact with MAIT cell TCR (Le Bourhis et al., 2010; Sakala et al., 2015; Chen et al., 2017; Sharma et al., 2020). Recent studies have also identified riboflavin precursor metabolites from E. coli, Salmonella, and Mycobacterium smegmatis for MAIT cell activation (Kjer-Nielsen et al., 2012; Corbett et al., 2014; Harriff et al., 2018), but not from non-stimulatory bacteria Listeria because of a defective expression of riboflavin synthesis enzymes in Listeria (Le Bourhis et al., 2010; Kjer-Nielsen et al., 2012; Gutierrez-Preciado et al., 2015). Further, the upregulation of CD69+CD26++ on activated MAIT cells upon bacterial stimulation can be largely blocked by anti-MR1 antibody, supporting the dependence of MR1-mediated bacterial antigen presentation for MAIT cell activation (Sharma et al., 2020). Therefore, the validated combinatory marker CD69+CD26++ was used to label the activated primary human MAIT cells stimulated by MR1-dependent presentation of bacterial antigens (Sharma et al., 2020). Upon RNAseq profiling, gene identities were assigned by aligning the detected sequences with the human genome in GenBank database. We used a robust algorithm of edgeR package in R for multi-factorial comparisons (McCarthy et al., 2012) to generate differentially expressed genes (DEGs) from inactivated MAIT cell subsets and from activated vs. inactivated MAIT cell subsets.
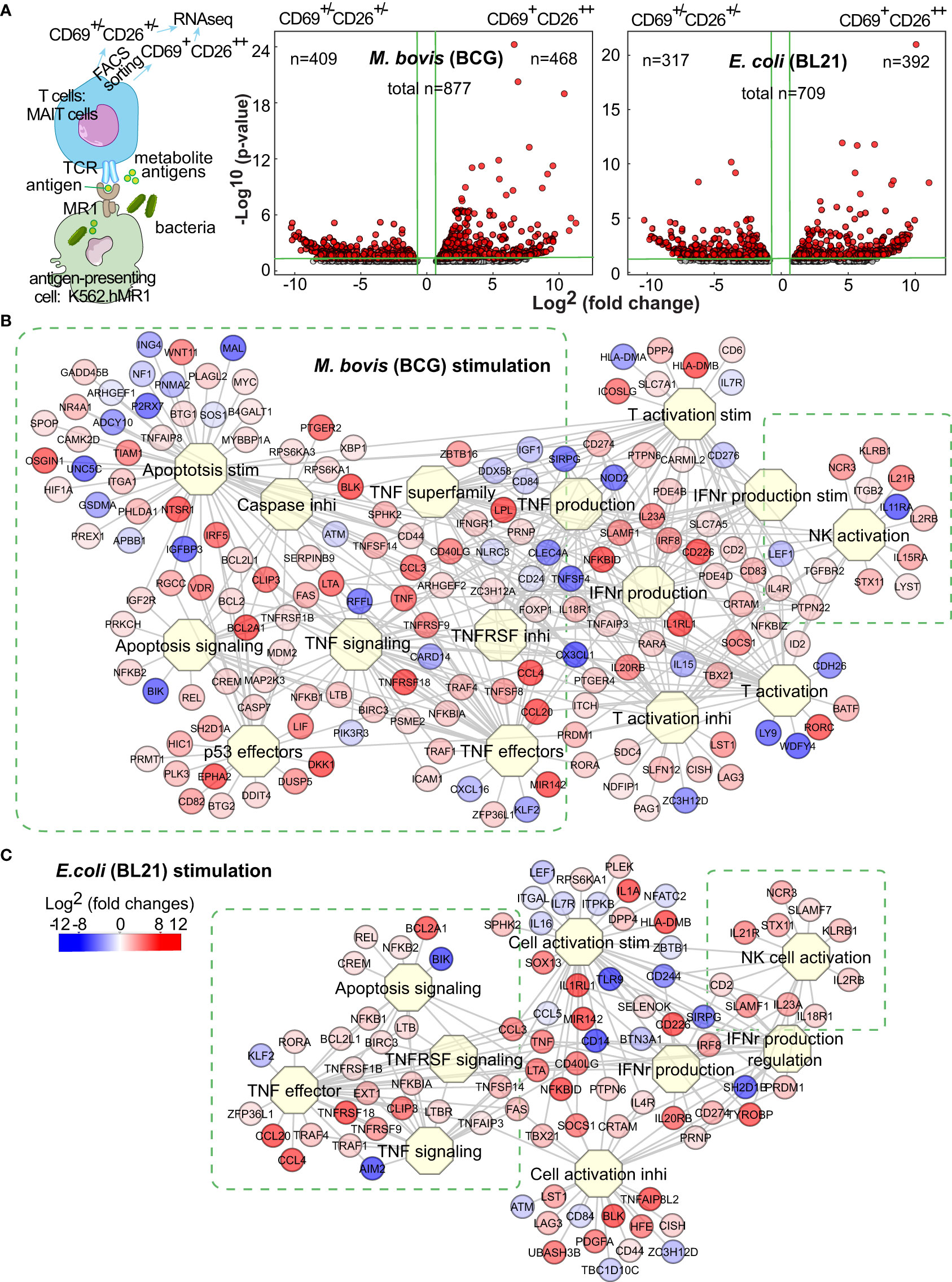
Figure 1 BCG and E.coli stimulate different MAIT cell transcriptomes. Anti-Vα7.2-enriched MAIT cells from blood of healthy donors were co-cultured overnight with bacterial-infected K562.hMR1 cells together with anti-CD28 antibody, then gated on Vα7.2+CD161+CD4-CD8+ cells, and further sorted into activated versus inactivated MAIT cells labeled with CD69+CD26++ and CD69+/-CD26+/-, respectively (A, Figure S1). The sorted activated vs. inactivated MAIT cells were used for RNA purification and next-generation sequencing. Based on the sequencing results, differentially expressed genes (DEGs) were identified using the edgeR package in R program and shown in Volcano plots as red dots. On the plots, vertical green lines label two-fold changes in intensity counts, and horizontal green lines label p-values of 0.05. For activated MAIT cells between BCG and E. coli stimulations, the horizontal green line label a p-value of 0.1 (A). DEGs were used to search biological processes and pathways through the Toppcluster program. Results show multiple functional clusters associated with cell proliferation, apoptosis, cytokine regulation, and cell activation upon BCG and E. coli stimulations (B, C).
To define a baseline comparator for activated MAIT cells, we generated DEGs for the inactivated MAIT cells with a CD69+/-CD26+/- phenotype upon the incubation of MAIT-stimulatory BCG or E. coli vs. the non-stimulatory Listeria (Figure S1C). Both volcano plots suggested the heterogeneity of baseline reactivity with a large number of DEGs between Listeria and BCG or E. coli incubations (Figure S1C) and the potential impact of MR1- or TCR-independent factors from different bacteria, such as pathogen-associated molecular patterns (PAMP). These bacterial factors independent of antigen presentation likely contributed to high numbers of DEGs with inter-bacterial comparison for activated vs. inactivated MAIT cells (Figure S1D). Further examining DEGs in inactivated MAIT cells (Figure S1C), we found many genes for regulating downstream reactivities, including effector molecules, cytokine receptors, signaling molecules, and exhaustion markers, remained unaltered or minimally altered, although some genes but not proteins for encoding surface markers (e.g., CD69, CD161) were higher in the inactivated MAIT cells from Listeria vs. BCG incubation. This heterogeneity of baseline inactivated MAIT cells from Listeria incubation is likely due to a broader CD26 expression similar to that in bacterial-free condition (Figure S1B), and likely comparable with a portion of pre-activated MAIT cells potentially induced by commensal bacteria in uninfected mice or healthy humans, unlike naïve conventional T cells (Kawachi et al., 2006; Gold et al., 2010; Le Bourhis et al., 2010). Moreover, the upregulated genes from the inactivated MAIT cells upon Listeria vs. BCG incubation were analyzed using Enrichr epigenetics enrichment tools (https://maayanlab.cloud/Enrichr/) (Kuleshov et al., 2016) and enriched in the top hit gene sets from thymus tissues or T cells with DNA methylation or histone modification, supporting potential epigenetic regulation in Listeria incubation. Thus, inactivated MAIT cells with different bacterial incubation remain heterogeneous. In contrast, MAIT cell subsets from an identically treated condition yielded more homogeneous inactivated MAIT cells to allow accurate analysis and association of altered genes with MR1-dependent MAIT cell activation (Figure 1A), because these activated vs. inactivated MAIT subsets responded to the same bacterium and stimulation, underwent identical incubation, staining, and FACS sorting process, and minimized the technical variation. Thus, we focused on the comparative transcriptomes of activated vs. inactivated MAIT cells from the identically treated samples to defined DEGs with a four-fold difference of intensity counts (Figure 1A). Several hundred upregulated genes from activated MAIT cells upon BCG and E. coli stimulations were shown with volcano plots (Figure 1A), in comparison with the inactivated MAIT cells identically stimulated and processed.
Different gene clusters of MAIT cells upon BCG and E. coli stimulations
To determine the gene clusters differentially stimulated by BCG vs. E. coli, we used Toppcluster (https://toppcluster.cchmc.org/) (Eisen et al., 1998; Chen et al., 2007) to search DEGs associated with various biological processes and pathways. Although multiple pathways in cytokine production and cell activation are similar (Figure S1E), gene clusters involving cell survival, death, and cytolysis differ between BCG and E. coli stimulations (Figure S1E, Figures 1B,C). Overall, BCG stimulation altered more gene clusters in regulating apoptosis and cell life in comparison to E. coli stimulation (Figure S1E, Figures 1B,C), such as tumor necrosis factor α (TNFα) stimulation and signaling, caspase (CASP)-induced apoptosis, and anti-proliferative effects of p53 (Figure 1B). These gene clusters include the intrinsic anti-apoptotic genes represented by upregulated B-cell lymphoma 2 (BCL2) and the counteracting molecules such as downregulated Bcl-2 interacting killer gene (BIK) but upregulated BLK (Hegde et al., 1998). E. coli altered less number of genes in the clusters regarding cell proliferation or apoptosis (Figure 1C). As MAIT cells are cytotoxic T cells, both bacteria upregulated the expression of GNLY (granulysin) and PRF1 (perforin) (Leeansyah et al., 2015) to induce the cytolytic effect of bacterial-infected cells (Figures 1B,C). Activated MAIT and natural killer NK cells also co-expressed multiple genes, including KLRB1 (CD161, Killer cell lectin-like receptor subfamily B, member 1) (Lanier et al., 1994) as an MAIT cell marker (Ussher et al., 2014), NCR3 (NKp30) a natural cytotoxicity triggering receptor to interact with CD3ξ (CD247) for NK cell differentiation (Siewiera et al., 2015), SLAMF1 (Signaling lymphocytic activation molecule 1, CD150) as an activation marker (Sharma et al., 2015), ID2 (DNA-binding protein inhibitor ID-2) a transcriptional regulatory protein constitutively expressed in NK cells (Li ZY. et al., 2021), together with genes LYST (lysosomal trafficking regulator) and STX11 (Syntaxin-11) regulating endocytic functions (Valdez et al., 1999; Gil-Krzewska et al., 2016). These data support the overall enhanced cell survival and cytotoxicity of MAIT cells upon BCG and E. coli stimulations, respectively. It was reasonable to expect that a direct comparison of the activated MAIT cells between BCG and E. coli stimulation generated less degree of difference in gene intensity counts and less altered gene clusters associated with cell reactivity (Figure S1F), although thresholds were set more aggressively to potentially visualize this critical difference. However, specific pathway analyses are needed to further understand gene interaction for regulating MAIT cell survival, death, and effector response.
Enriched pro-survival pathways of MAIT cells upon BCG and E. coli stimulations
We used the Cytoscape program to search the network that comprehensively depicts various recently reported pathway mechanisms (labeled by Roman numerals) for regulating cell survival and death (Figures 2A; S2). The intrinsic pathways include (i) anti-apoptotic BCL2 family gene members (Czabotar et al., 2014), (ii) pro-apoptotic BCL2 family gene members (Marsden and Strasser, 2003), and other intrinsic factors acting through BCL2 family genes for cell growth regulation, such as (iii) MDM2 (mouse double minute 2)-p53 (tumor suppressor gene) and pro-oncogene MYC-p53 counteraction (Wu and Prives, 2018) and (iv) insulin-like growth factor 1 (IGF1) signaling. The extrinsic pathways include TNF signals to mediate (v) proliferative nuclear factor-kappa B (NF-kB), (vi) proliferative mitogen-activated protein kinase (MAPK) pathways (Webster and Vucic, 2020), and (vii) necrotic receptor-interacting protein kinase (RIPK) signaling (Petersen et al., 2015), (viii) FAS-mediated apoptosis (Yamada et al., 2017), (ix) caspase-mediated apoptosis and pyroptosis (Wallach et al., 2014), and (x) interferon-regulatory factor 5 (IRF5)-promoted apoptosis associated with type I interferons (Hu and Barnes, 2009).
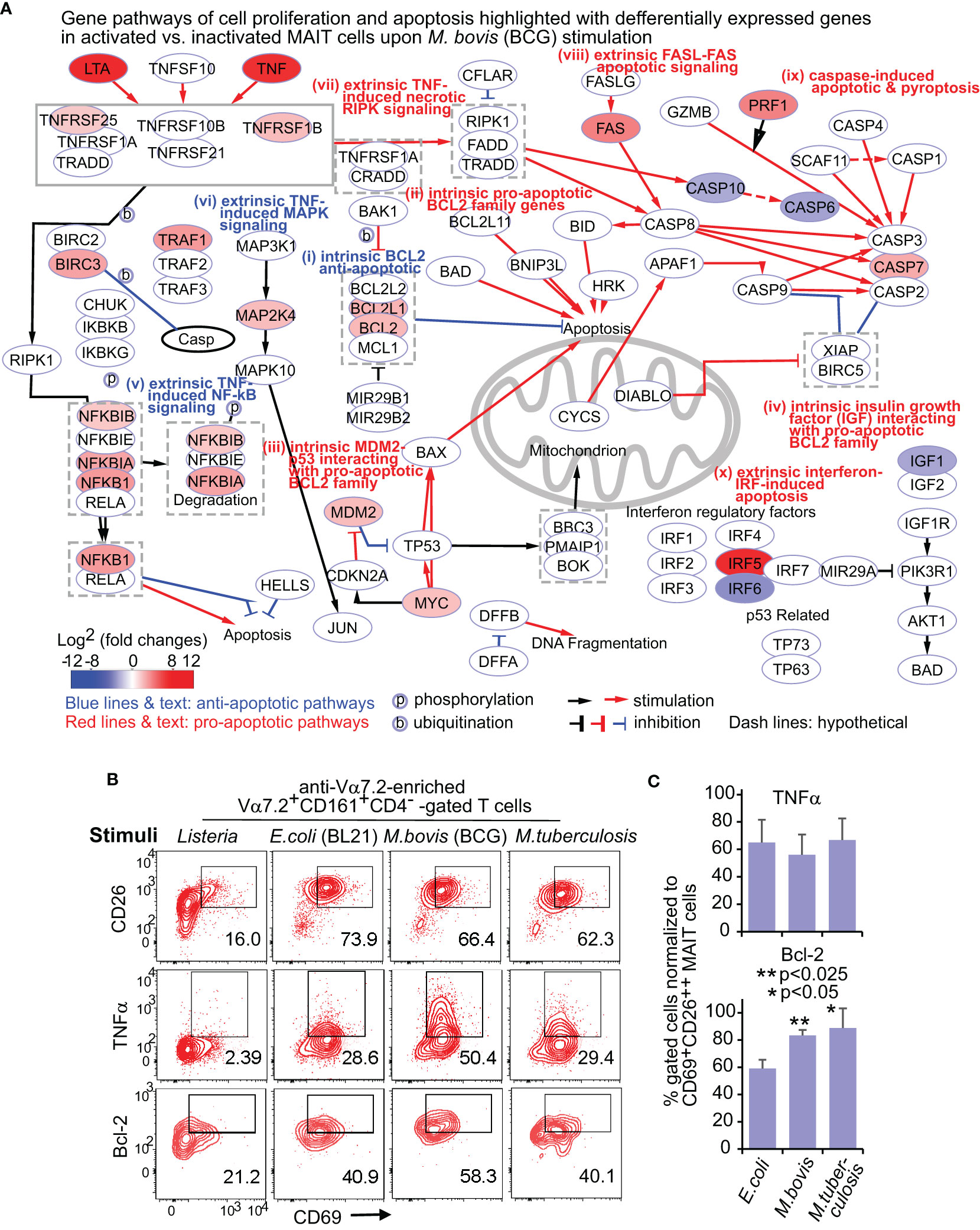
Figure 2 BCG stimulate differential gene and protein expression of MAIT cells in proliferation and apoptosis pathways. DEGs between activated (CD69+CD26++) subsets versus inactivated (CD69+/-CD26+/-) subsets of CD8+ MAIT cells upon BCG stimulation were annotated in pathways of cell proliferation and apoptosis (A). Protein expression of key cytokine TNFα and key regulatory protein Bcl-2 were confirmed with the percentage of CD69+TNFα+, and CD69+Bcl-2+ subsets in comparison to the CD69+CD26++ subset of CD8+ MAIT cells detected by flow cytometry (B). The gating strategy is shown in Figure S1. The percentage of CD69+TNFα+ or CD69+Bcl-2+ subsets of CD8+ MAIT cells was normalized with the percentage of activated CD69+CD26++ subsets of CD8+ MAIT cells. Plots show the means of these normalized percentages and standard errors from multiple donors. A pairwise t-test was used to determine the significance of differences between BCG and E. coli stimulations (C).
DEGs between activated versus inactivated CD8+ MAIT cells in BCG stimulation are expected to regulate cell activation, growth or death, cytokines, and cytotoxicity (Figure 2A). In the anti-apoptotic BCL2 gene family (i), the expression of BCL2, BCL2L1 (BCL-XL), BCL2A1 (BFL1), and BLK was enhanced. Favoring cell survival, the expression of pro-apoptotic BCL2 gene family (ii) was unaltered, including BID, BIM (BCL2L11), BAD, and pro-apoptotic “effectors” BAX and BAK1 (Marsden and Strasser, 2003). In MDM2-p53 and MYC-p53 counteractions (iii), enhanced MDM2 and MYC expression could inhibit the pro-apoptotic effect of p53 and stimulate cell growth (Eischen et al., 1999). Further, the reduced expression of insulin-like growth factor 1 (IGF1) gene (iv) was associated with a lower activity of the pro-apoptotic protein BAD, further supporting an anti-apoptotic effect of BCG stimulation. Together, enhanced expression of anti-apoptotic genes and unaltered or reduced expression of pro-apoptotic genes in various BCL2 gene family members and interacting pathways supported a pro-survival effect of MAIT cells in BCG stimulation (Figure 2A).
In extrinsic pathways altered by BCG stimulation (Figure 2A), the enhanced TNF (TNFα) and lymphotoxin (LTA) cytokines function as both effector molecules in anti-bacterial responses and self-feedback stimuli for regulating cell growth and death. TNF-induced proliferative nuclear factor κB (NF-κB) (v) and mitogen-activated protein kinase (MAPK) (vi) pathways supported a pro-survival effect. BCG induced a lower expression of caspase gene expression (CASP10 and CASP6) following TNF and necrotic RIPK1 signaling (vii), also supporting a pro-survival effect. However, a higher expression of the FAS gene (viii) involved in the self-recognition of FAS ligand could lead to FAS-mediated apoptosis (Yamada et al., 2017). Further, enhanced caspase 7 (CASP7) and upstream perforin 1 (PRF1) expression involved activation-mediated apoptosis and pyroptosis (Wallach et al., 2014). Enhanced expression of IRF5 (x) could promote the apoptosis associated with type I interferons (Hu and Barnes, 2009). In addition, the expression of TNF receptor 2 (TNFR2) also named TNF receptor superfamily 1B (TNFRSF1B), TNF receptor-associated factor 1 (TRAF1), and baculoviral IAP repeat containing 3 (BIRC3 or cIAP2) were enhanced to play an anti-apoptotic role (Silke and Brink, 2010). Overall, enhanced gene expression was associated with the extrinsic pathways promoting cell survival, with a balance of pro-apoptotic signals for self-activated control of cell growth and death in BCG stimulation (Figure 2A).
Although E. coli changed the expression of many above genes also altered in BCG stimulation, the expression of anti-apoptotic genes BCL2L1 (BCL-XL), TNFRSF25 (APO3 or DR3), MAP2K4, MDM2, IRF5, IRF6, IGF1, and the pro-apoptotic gene CASP7 were unaltered, while the MDM2 suppressor (CDKN2A) and FAS ligand (FASLG) were upregulated (Figure S2A). This difference suggested a more apoptotic effect with E. coli than BCG stimulation. We further directly compared the gene expression of the activated CD8+ MAIT cell subset upon BCG vs. E. coli stimulation. The higher expression of intrinsic BIM (BCL2L11) and CASP4 genes in BCG stimulation compared with E. coli stimulation likely suggest a higher baseline gene expression in BCG stimulation for regulating cell survival and apoptosis (Figure S2B). Results also showed a higher expression of BCL2 and BIRC3 genes, but a lower expression of CDKN2A (cyclin-dependent kinase inhibitor 2A) and IRF6 genes in BCG stimulation, supporting an overall pro-survival transcriptomic program.
Flow cytometry detected a more enhanced Bcl-2 expression upon BCG vs. E. coli stimulation
To confirm the protein expression of key upregulated genes for regulating cell growth or death, we applied flow cytometry to test the expression of TNFα and Bcl-2 proteins in E. coli, BCG, and M. tuberculosis stimulations. We similarly gated on the CD8+ MAIT cells (Vα7.2+CD161+CD4-CD8+ cells) as in Figure S1 and used the negative control L. monocytogenes (Figure 2B), which is absent of active riboflavin B metabolic pathway for providing an MAIT cell antigen. Because CD69+CD26++ was used as a combinatory marker to label the total activated MAIT cells (Sharma et al., 2020), we determined % CD69+TNFα+ or CD69+Bcl-2+ cells over CD69+CD26++ CD8+ MAIT cells (Figure 2C). This similar % of CD69+CD26++ CD8+ MAIT cells supported highly comparable conditions between BCG vs. E. coli stimulation, allowing further examining relative differences of various gene and protein expression. Results showed a higher percentage of Bcl-2-expressing CD8+ MAIT cells in mycobacterial than E. coli stimulations, supporting the enhanced Bcl-2 protein expression. Listeria incubation did not enhance CD69+CD26++ CD8+ MAIT cells and was not included in statistical analyses. TNF protein expression was more variable among donors and not statistically significant for BCG vs. E. coli comparison.
Enriched cytolytic pathways of MAIT cells upon BCG and E. coli stimulation
Upon bacterial stimulations, multiple genes involved in the cytolysis of MAIT cells were generally upregulated in BCG stimulation (Figure 3A). Beyond the upregulated genes for T cell activation, such as CD8A (CD8α), CD247 (CD3ξ), and CD69, cytokine receptor expression was also enhanced, including IL2RG (common γ chain), IL2RB (interleukin 2 receptor β subunit), and IL15RA (interleukin 15 receptor α subunit). Regarding cytotoxicity, multiple receptors, costimulatory molecules, signaling molecules, and transcription factors were upregulated in bacterial stimulations. CD161, a C-type lectin-like receptor encoded by the upregulated KLRB1, labels the maturation and cytotoxicity of NK cells (Lanier et al., 1994; Konjevic et al., 2009; Kurioka et al., 2018). The upregulated KLRG1 gene encodes a co-inhibitory receptor predominantly on late-differentiated effector and memory CD8+ T and NK cells (Nakamura et al., 2009). The enhanced NKG7 gene encodes natural killer cell granule protein 7, which is a regulator of lymphocyte granule exocytosis and inflammation, such as CD107a (Lamp1 or lysosomal-associated membrane protein-1) translocation to the cell surface for target cell killing (Ng et al., 2020). Multiple genes encoding effector molecules, such as FAS, perforin (PRF1), and granulysin (GNLY), were enhanced differentially in BCG stimulations (Figure 3A) for the cytolysis of the infected targeted cells (Lettau and Janssen, 2021). E. coli stimulation similarly upregulated cytotoxic genes (Figure S3A), with alteration of multiple genes at a lower degree compared with BCG stimulation (Figure S3B). Eomesodermin (EOMES) was interestingly more upregulated in activated MAIT cells in BCG than E. coli stimulation, indicating a higher basal expression of EOMES gene in BCG stimulation to promote cytolytic responses.
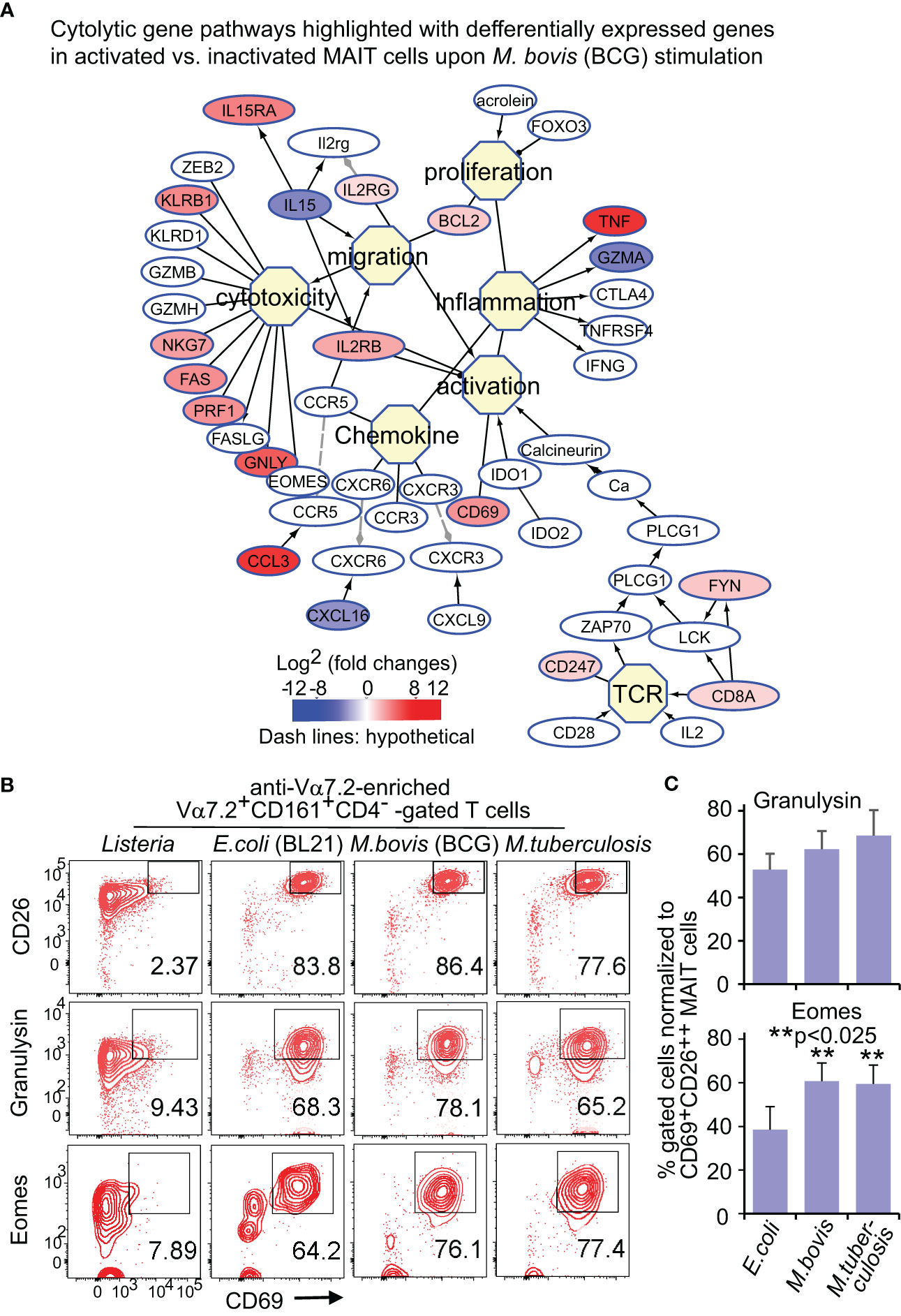
Figure 3 BCG and E.coli stimulate differential gene and protein expression of MAIT cells in cytotoxic pathways. Similar to Figure 2, DEGs between activated (CD69+CD26++) subsets versus inactivated (CD69+/-CD26+/-) subsets of CD8+ MAIT cells upon BCG stimulation were annotated in pathways of cytotoxic T cell responses (A). Protein expression of key effect molecule granulysin and key transcription factor Eomes were confirmed with the percentage of CD69+granulysin+ or CD69+Eomes+ subsets in comparison to the CD69+CD26++ subset of CD8+ MAIT cells detected by flow cytometry (B). The gating strategy is shown in Figure S1 as well. The percentage of CD69+granulysin+ or CD69+Eomes+ subsets of CD8+ MAIT cells were normalized with the percentage of activated CD69+CD26++ subsets of CD8+ MAIT cells. Plots show the means of these normalized percentages and standard error from multiple donors. A pairwise t-test was used to determine the significance of differences between BCG and E. coli stimulations (C).
Flow cytometry detected a more enhanced Eomes expression upon BCG than E. coli stimulation
For protein expression, flow cytometry was used to detect granulysin and Eomes as described in Figure 2. Data were similarly analyzed by normalizing % CD69+Eomes+ CD8+ MAIT cells with CD69+CD26++ CD8+ MAIT and showed more enhanced % CD69+Eomes+ CD8+ MAIT cells in BCG stimulation compared with E. coli, from one donor (Figure 3B) and multiple donors (Figure 3C). Together, both gene and protein expression demonstrated upregulated cytolytic molecules to fight intracellular mycobacterial infections.
MAIT transcriptomes stimulated by BCG are more comparable with conventional CD8+ T cells in intracellular microbial infections
To comprehensively compare BCG-stimulated MAIT transcriptomes with other cellular transcriptomes under bacterial or other stimulatory conditions, we performed gene set enrichment analyses (GSEA) for MAIT cell transcriptomes against MSigDB gene expression databases (http://software.broadinstitute.org/gsea/msigdb). The enriched gene sets were cut off with a significant nominal p-value (<0.05) and ranked by the normalized enrichment scores. GSEA analyses generally resulted in around two thousands of such reported gene sets, including various gene sets from conventional CD8+ T cells, CD4+ T cells, NKT, NK cells, macrophages, dendritic cells, and many other cell types under different stimulation conditions from different biological hosts. Enrichment plots displayed three and one representative gene sets that were selected out of the top twenty ranked gene sets enriched with activated or inactivated MAIT phenotypes, respectively (Figure 4). Heatmaps displayed the representative MAIT cell genes from both activated and inactivated MAIT subsets and from the Rank-Ordered List based on the top running enrichment scores (ES) of the tested genes in the targeted gene sets (Figure 4). As a result, gene enrichment of MAIT cell transcriptomes in BCG stimulation demonstrated a similar upregulation in activated conventional memory CD8+ T cells with intracellular bacterial infection and in PBMCs from infants at ten weeks after BCG vaccination at birth, or a similar downregulation in PBMCs of patients with sepsis, but reversely altered in bystander activated CD4+ T cells independent of antigen-presentation (Figure 4A). In contrast, MAIT cell genes in E. coli stimulation showed reversely altered in viral peptide-activated CD8+ T cells, bystander activated CD4+ T cells independent of antigens, lipopolysaccharide-stimulated dendritic cells, and IL15-stimulated NK cells (Figure 4B). Overall, MAIT cell gene profiles in BCG stimulation were comparable with memory CD8+ T cells or PBMCs responding to intracellular bacterial infections. In contrast, MAIT cell gene profiles in E.coli stimulation showed a different or reversed association. Results supported similar gene expression of MAIT and other CD8+ T cells in response to intracellular pathogens.

Figure 4 Gene set enrichment analyses (GSEA) support that BCG-stimulated MAIT cell transcriptomes are comparable with CD8+ T cells in intracellular microbial infections. BCG-stimulated DEG genes of MAIT cells were analyzed using the GSEA program to identify comparable gene sets with similarly enriched genes from different cell types under various stimulation conditions. The representative gene sets with T cells or other immune cells were selected from the top 20 out of over 4000 enriched gene sets. The representative core-enriched MAIT cell genes shown in heatmaps were selected based on the top running enrichment scores (ES) of the enriched genes (A). E. coli-stimulated DEG genes of MAIT cells were similarly analyzed using the GSEA program and shown with representative enrichment plots and enriched genes (B).
Discussion
Extracellular growing E. coli can secrete intermediate metabolites ribityllumazine and ribityluracil from riboflavin biosynthetic pathways to the culture supernatant (Kjer-Nielsen et al., 2012; Corbett et al., 2014; Harriff et al., 2018). These metabolites function as agonist antigens to be loaded to MR1 protein and presented on antigen-presenting cells for MAIT cell activation (McWilliam et al., 2020). Intracellular bacteria such as BCG and M. tuberculosis provide antigens for MAIT cell activation, likely involving endocytic compartments for antigen loading and presentation (Huang et al., 2008; Harriff et al., 2016; Huber et al., 2020; Sharma et al., 2020). Our MAIT gene profiles in BCG stimulation showed similar alterations to memory CD8+ T cells or PBMCs with intracellular bacterial infections, whereas MAIT cell gene profiles in E. coli stimulation showed reversal association with CD8+ T cells in intracellular bacterial infections. Transcriptomic analyses on intracellular mycobacterial infections have been mostly focused on macrophages (Nalpas et al., 2015) or monocytes (Kong et al., 2021) for understanding host-pathogen interaction and blood cells from tuberculosis patients. It remains poorly understood how T cell transcriptomes are altered in BCG vaccination or stimulation in a manner dependent on bacterial antigen presentation. As MAIT cells are protective against mycobacterial infections, including M. tuberculosis (Sakai et al., 2021), M. abscessus (Le Bourhis et al., 2010), and M. bovis (Chua et al., 2012; Sakala et al., 2015), this protection likely attributes to MAIT cell responses against intracellular bacterial growth (Le Bourhis et al., 2010; Chua et al., 2012; Sakala et al., 2015; Sakai et al., 2021). Indeed, MAIT cell transcriptomes stimulated by BCG in this study demonstrated pro-survival and cytolytic programs that crucially contribute to the immunity against mycobacterial infections. In the meantime, results discriminate differential MAIT cell transcriptomes responding to intracellular M. bovis BCG strain versus extracellular bacteria E. coli, suggesting genetic pathways regulating T cell immunity to fight intracellular bacterial infections.
Activated MAIT cells are expected to display pathogen selectivity and ligand discrimination, which have been recently characterized in MAIT cell responses to different bacteria (Reantragoon et al., 2013; Gold et al., 2014; Lepore et al., 2014; Meermeier et al., 2016). MAIT cells express an invariant T cell receptor α chain (TCRα) to recognize conserved antigens in contrast to conventional T cells (Nikolich-Zugich et al., 2004; Logunova et al., 2020). However, the β chain (TCRβ) expresses variable sequences in responses to different bacteria, such as E. coli and Salmonella Typhimurium that produce typical MAIT cell agonist antigens ribityllumazine and ribityluracil metabolites (Kjer-Nielsen et al., 2012; Corbett et al., 2014; Harriff et al., 2018), versus Streptococcus pyogenes that likely generate other unknown MAIT cell antigens (Meermeier et al., 2016). Our chemical purification of bacterial metabolites from BCG also supported the presence of alternative agonists different from the agonists derived from E. coli (Corbett et al., 2014). We adapted the HLA-defective myelogenous cell line K562 (Lozzio and Lozzio, 1975; Andersson et al., 1979; Roder et al., 1979; Koeffler and Golde, 1980; Li et al., 2019), widely used as antigen-presentation cells for various T cell activation (Escobar et al., 2008; de Jong et al., 2010; de Jong et al., 2014; Sharma et al., 2020; Goodman et al., 2022), with human MR1 overexpression to present bacterial metabolite antigens for primary MAIT cell activation. Downstream effects of differential pathogen stimulation and variable TCRβ chains (Reantragoon et al., 2013; Gold et al., 2014; Lepore et al., 2014; Meermeier et al., 2016) are expected to induce different MAIT cell transcriptomes that serve as predictors to link stimuli with effector responses. Comparative MAIT cell transcriptomes in this study depicted multifaceted programs regarding MAIT cell activation, survival, apoptosis, and cytolysis, in addition to various cytokine production in BCG vs. E.coli stimulation. More specifically, BCG stimulated a higher MAIT cell expression level of EOMES and BCL2 genes and proteins to mediate cytotoxic, cytokine production, and pro-survival programs (Figures 1–3), which are essential in fighting intracellular mycobacterial infections (Kim et al., 2015; Sharma et al., 2020).
Various DEGs of MAIT cells involved in cell survival and death pathways are astonishing. Although our clustering analyses suggested a broad alteration of these genes, pathway analyses comprehensively displayed altered gene expression in over ten intrinsic and extrinsic gene pathways for regulating cell survival, apoptosis, necrosis, and pyroptosis. Results demonstrated an anti-apoptotic gene expression of MAIT cells in four intrinsic pathways and a balanced survival vs. apoptotic gene expression in extrinsic pathways upon BCG stimulation. In various intrinsic pathways, anti-apoptotic BCL2 gene family members were enhanced, and other intrinsic factors interacting with BCL2 gene family members also showed an expression pattern favoring cell survival, such as upregulated MDM2 and MYC expression for a counteraction of p53 protein in BCG stimulation to facilitate MAIT cell proliferation instead of apoptosis or senescence (Wu and Prives, 2018). Multiple extrinsic pathways appear to balance the pro-apoptotic and anti-apoptotic pathways to play crucial roles in MAIT cell homeostasis. The extrinsic signals, including TNFα, LTα, and FAS ligand gene expression enhanced in BCG or E. coli stimulation, are common cytokines mediating anti-bacterial effector responses by interacting with corresponding receptors on other effector cells, such as mycobacterial-infected macrophages. If a bacterial infection is under control, these cytokines likely turn back and display a self-feedback control by interacting with their receptors on MAIT cells, leading to homeostatic control of MAIT cell growth by apoptosis or cell death. As both producers and targets of TNF cytokines, T cells can induce the positive feedback of proliferative responses and negative feedback of T cell apoptosis or regulatory T cell differentiation (Locksley et al., 2001; Mehta et al., 2018). TNF production upon BCG stimulation enhanced MAIT cell proliferative pathways mediated by NF-kB and MAPK signaling, but inhibited the necrotic signaling mediated by RIPK1 (receptor-interacting protein or RIP family of serine/threonine protein kinase 1) with reduced downstream caspase expression. TNF effect could be more complex by stimulating a signaling complex of TNF receptor-associated factors (TRAF) and TNFR2 (encoded by TNFRSF1B) proteins. For example, BCG enhanced TRAF1 and cellular inhibitor of apoptosis 2 (cIAP2 or BIRC3) to activate NF-κB signaling and prevent TNF-induced apoptosis (Silke and Brink, 2010). However, self-feedback regulation mediated by FAS in BCG stimulation could induce an apoptotic effect. Cell death signals initiated by an upstream cytotoxic molecule FAS mediate the clearance of bacterial-infected cells such as macrophages through MHC class I-restriction by conventional T cells (Kagi et al., 1994) and MR1 restriction by MAIT cells (Boulouis et al., 2020). FAS protein signals, together with the antigen-stimulated TCR signaling, will induce caspase 8-mediated apoptosis and provide feedback control (Bouillet and O’Reilly, 2009; Yamada et al., 2017). In parallel, perforin upregulation occurs with high expression of Tbet in conventional CD8+ T cells as reported (Makedonas et al., 2010) and in MAIT cells as we shown (Figure 1C). Cytokines IFN stimulate IRF family members together to fight microbial infections, but high IRF5 expression mediating TRAIL (TNF-related apoptosis-inducing ligand) receptors- or death receptor-induced apoptosis functions as negative feedback control (Hu and Barnes, 2009; Fabie et al., 2018). Moreover, lower IGF1 gene expression in the IGF (insulin-like growth factor) signaling pathway is contrasting with higher IRF5 expression, leading to less apoptotic effect in BCG stimulation (Figure 2A). Therefore, extrinsic pathways to maintain homeostasis often cross-regulate anti-microbial responses of MAIT cells in fighting infections.
Cytotoxicity-regulatory genes, including natural killer cell group 7 (NKG7), GNLY, EOMES, TNF, and FAS, were more enhanced in BCG stimulation than E. coli. Granulysin, perforin, and TNF function as effector molecules to mediate the cytolytic process of targeted cells. NKG7 interestingly optimizes the exocytosis of lytic granules for the perforin-dependent but not Fas ligand-mediated cytolytic pathway (Morikawa et al., 2021). Eomes is an important transcription factor critical for the formation of effector and memory CD8+ T cells (Kaech and Cui, 2012), by mediating the expression of many essential effector molecules, such as perforin, granzymes, and IFNγ (Intlekofer et al., 2005; Banerjee et al., 2010; Pipkin et al., 2010), promoting memory T cell differentiation by inhibiting apoptosis (Banerjee et al., 2010; Kavazovic et al., 2020), and regulating the exhaustion of highly activated CD8+ T cells (Li et al., 2018). Enhanced expression of TBX21, EOMES, and IL2RB (CD122) in MAIT cells in this study reflects the similarity of MAIT cells to memory CD8+ T cells or NK cells regarding cytolytic effector responses (Olson et al., 2013). Moreover, cytolytic responses are controlled by multiple different transcription factors, including ID2, Tbet, and Eomes, such as in BCG stimulation, leading to the differentiation of memory, cytolytic, even exhaustion phenotypes.
In this study, we specifically focused on comparing the activated MAIT cells labeled by high CD69 and CD26 expression (CD69+CD26++) with inactivated MAIT cells in BCG and E. coli stimulations to demonstrate MAIT cell transcriptomes dependent on MR1-mediated antigen presentation. Beyond dissecting the transcriptomes of MAIT cell subsets activated by different bacteria, future studies can include bacterial-free controls to assess the global effect of bacterial stimulation on the overall transcriptome. Technically, transcriptomic pathway analyses of T cell responses serve as a tool to integrate stimuli from different sources and facilitate understanding genetic pathways for developing protective anti-bacterial immunity. Consequently, the comparative transcriptomes of MAIT cells in BCG stimulation provided genetic pathways for regulating pro-survival, memory, cytolysis, and exhaustion to elicit anti-mycobacterial MAIT cell immune responses.
Data availability statement
The data presented in the study are deposited in the GEO repository of the NIH NCBI website (https://www.ncbi.nlm.nih.gov/geo/). Accession GSE228089 is for MAIT cell transcriptomes at E. coli and Listeria incubation conditions. Accession GSE124381 is for MAIT cell transcriptomes at BCG and anti-CD3 stimulation conditions.
Author contributions
All authors reviewed the manuscript. MS: perform assays; LN: initial transcriptomic data analyses; XZ: RNA sample preparation and sequencing; SH: study design, data analyses, and manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from National Institute of Allergy and Infectious Diseases (AI115358), American Lung Association (IA-629987), and National Institute of Environmental Health Sciences (ES006096).
Acknowledgments
The authors thank Branch Moody for mycobacterial BCG and H37Ra strains, Robert Giulitto for coordinating human blood samples, human blood donors in the Hoxworth Blood Center. The cell graphic elements in Figure 1A were licensed from Adobe Stock.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1134119/full#supplementary-material
Supplementary Figure 1 | Gating of activated versus inactivated CD8+ MAIT cells for RNA-seq analyses. CD8+ MAIT cells were gated on Vα7.2+CD161+CD4-CD8+ as Vα7.2+CD161+ gating has been used in multiple studies to detect MAIT cells (13, 29-32), especially the bacterial-activated MAIT cells (13, 33, 34) (A). % CD69+CD26++ CD8+ MAIT cells and CD69+/-CD26+/- cells are annotated, showing the cell populations sorted for RNA-sequencing and the strategy for gating activated MAIT cells (B). To determine whether the CD69+/-CD26+/- MAIT subsets at different bacterial incubation conditions show similar background response, we determined DEGs of CD69+/-CD26+/- inactivated MAIT cells between Listeria and BCG, or between Listeria and E. coli, suggesting high heterogeneity (C). High numbers of DEGs also occur with the activated MAIT subset (CD69+CD26++) upon BCG or E. coli incubations and inactivated MAIT subset (CD69+/-CD26+/-) with Listeria incubation (D). DEGs from the activated vs. inactivated MAIT cells responding to identical bacterial stimulations, the BCG and E. coli stimulation, were more clustered, respectively, to show genes associated with MAIT cell activation and survival (E). DEGs of activated MAIT cells from the direct comparison upon BCG vs. E. coli stimulation are expected to have a fewer number of genes and show more narrow clusters associated with MAIT cell reactivities (F).
Supplementary Figure 2 | BCG and E.coli stimulate differential gene expression of MAIT cells in cell proliferation and apoptosis pathways. Similar to , DEGs between activated (CD69+CD26++) subsets versus inactivated (CD69+/-CD26+/-) subsets of CD8+ MAIT cells upon E. coli stimulation were annotated in pathways of cell proliferation and apoptosis (A). DEGs of activated MAIT cells between BCG and E.coli stimulation are also annotated in pathways of cell proliferation and apoptosis (B).
Supplementary Figure 3 | BCG and E.coli stimulate differential gene expression of MAIT cells in cytotoxic pathways. Similar to , DEGs between activated (CD69+CD26++) subsets versus inactivated (CD69+/-CD26+/-) subsets of CD8+ MAIT cells upon E. coli stimulation were annotated in pathways of cell activation and cytotoxicity (A). DEGs of activated MAIT cells between BCG and E.coli stimulation were annotated in pathways of cytotoxic T cell responses (B).
Abbreviations
MAIT, mucosal-associated invariant T; MHC, major histocompatibility complex; MR1, MHC-related protein 1; CD, cluster of differentiation; NK, natural killer; TNFα, tumor necrosis factor α; IFNγ, interferon γ; TCR, T cell receptor; HLA, human leukocyte antigens; PBMCs, peripheral blood mononuclear cells; BCG, Bacillus Calmette-Guerin; M. bovis, Mycobacterium bovis; M. tuberculosis, Mycobacterium tuberculosis; E. coli, Escherichia coli; L. monocytogenes, Listeria monocytogenes; Vα7.2, variable segment 7.2 of the T cell receptor alpha chain; APC/Cy7, Allophycocyanin/Cyanine7; PE, phycoerythrin; DEGs, differentially expressed genes; Eomes, Eomesodermin; Bcl-2, B-cell lymphoma 2.
References
Andersson, L. C., Nilsson, K., Gahmberg, C. G. (1979). K562–a human erythroleukemic cell line. Int. J. Cancer 23 (2), 143–147. doi: 10.1002/ijc.2910230202
Banerjee, A., Gordon, S. M., Intlekofer, A. M., Paley, M. A., Mooney, E. C., Lindsten, T., et al. (2010). Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185 (9), 4988–4992. doi: 10.4049/jimmunol.1002042
Behar, S. M. (2013). Antigen-specific CD8(+) T cells and protective immunity to tuberculosis. Adv. Exp. Med. Biol. 783, 141–163. doi: 10.1007/978-1-4614-6111-1_8
Bouillet, P., O’Reilly, L. A. (2009). CD95, BIM and T cell homeostasis. Nat. Rev. Immunol. 9 (7), 514–519. doi: 10.1038/nri2570
Boulouis, C., Sia, W. R., Gulam, M. Y., Teo, J. Q. M., Png, Y. T., Phan, T. K., et al. (2020). Human MAIT cell cytolytic effector proteins synergize to overcome carbapenem resistance in escherichia coli. PloS Biol. 18 (6), e3000644. doi: 10.1371/journal.pbio.3000644
Chandra, S., Ascui, G., Riffelmacher, T., Chawla, A., Ramirez-Suastegui, C., Castelan, V. C., et al. (2021). Transcriptomes and metabolism define mouse and human MAIT cell heterogeneity. bioRxiv. doi: 10.1101/2021.12.20.473182
Chen, C. Y., Huang, D., Wang, R. C., Shen, L., Zeng, G., Yao, S., et al. (2009). A critical role for CD8 T cells in a nonhuman primate model of tuberculosis. PloS Pathog. 5 (4), e1000392. doi: 10.1371/journal.ppat.1000392
Chen, Z., Wang, H., D’Souza, C., Sun, S., Kostenko, L., Eckle, S. B., et al. (2017). Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. 10 (1), 58–68. doi: 10.1038/mi.2016.39
Chen, J., Xu, H., Aronow, B. J., Jegga, A. G. (2007). Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinf. 8, 392. doi: 10.1186/1471-2105-8-392
Chua, W. J., Kim, S., Myers, N., Huang, S., Yu, L., Fremont, D. H., et al. (2011). Endogenous MHC-related protein 1 is transiently expressed on the plasma membrane in a conformation that activates mucosal-associated invariant T cells. J. Immunol. 186 (8), 4744–4750. doi: 10.4049/jimmunol.1003254
Chua, W. J., Truscott, S. M., Eickhoff, C. S., Blazevic, A., Hoft, D. F., Hansen, T. H. (2012). Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect. Immun. 80 (9), 3256–3267. doi: 10.1128/IAI.00279-12
Cooper, A. M. (2009). Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 27, 393–422. doi: 10.1146/annurev.immunol.021908.132703
Corbett, A. J., Eckle, S. B., Birkinshaw, R. W., Liu, L., Patel, O., Mahony, J., et al. (2014). T-Cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509 (7500), 361–365. doi: 10.1038/nature13160
Czabotar, P. E., Lessene, G., Strasser, A., Adams, J. M. (2014). Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 15 (1), 49–63. doi: 10.1038/nrm3722
de Jong, A., Cheng, T. Y., Huang, S., Gras, S., Birkinshaw, R. W., Kasmar, A. G., et al. (2014). CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat. Immunol. 15 (2), 177–185. doi: 10.1038/ni.2790
de Jong, A., Pena-Cruz, V., Cheng, T. Y., Clark, R. A., Van Rhijn, I., Moody, D. B. (2010). CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nat. Immunol. 11 (12), 1102–1109. doi: 10.1038/ni.1956
Eischen, C. M., Weber, J. D., Roussel, M. F., Sherr, C. J., Cleveland, J. L. (1999). Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in myc-induced lymphomagenesis. Genes Dev. 13 (20), 2658–2669. doi: 10.1101/gad.13.20.2658
Eisen, M. B., Spellman, P. T., Brown, P. O., Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95 (25), 14863–14868. doi: 10.1073/pnas.95.25.14863
Escobar, H., Crockett, D. K., Reyes-Vargas, E., Baena, A., Rockwood, A. L., Jensen, P. E., et al. (2008). Large Scale mass spectrometric profiling of peptides eluted from HLA molecules reveals n-terminal-extended peptide motifs. J. Immunol. 181 (7), 4874–4882. doi: 10.4049/jimmunol.181.7.4874
Fabie, A., Mai, L. T., Dagenais-Lussier, X., Hammami, A., van Grevenynghe, J., Stager, S. (2018). IRF-5 promotes cell death in CD4 T cells during chronic infection. Cell Rep. 24 (5), 1163–1175. doi: 10.1016/j.celrep.2018.06.107
Flynn, J. L., Chan, J. (2001). Immunology of tuberculosis. Annu. Rev. Immunol. 19, 93–129. doi: 10.1146/annurev.immunol.19.1.93
Gil-Krzewska, A., Wood, S. M., Murakami, Y., Nguyen, V., Chiang, S. C. C., Cullinane, A. R., et al. (2016). Chediak-higashi syndrome: Lysosomal trafficking regulator domains regulate exocytosis of lytic granules but not cytokine secretion by natural killer cells. J. Allergy Clin. Immunol. 137 (4), 1165–1177. doi: 10.1016/j.jaci.2015.08.039
Gold, M. C., Cerri, S., Smyk-Pearson, S., Cansler, M. E., Vogt, T. M., Delepine, J., et al. (2010). Human mucosal associated invariant T cells detect bacterially infected cells. PloS Biol. 8 (6), e1000407. doi: 10.1371/journal.pbio.1000407
Gold, M. C., McLaren, J. E., Reistetter, J. A., Smyk-Pearson, S., Ladell, K., Swarbrick, G. M., et al. (2014). MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J. Exp. Med. 211 (8), 1601–1610. doi: 10.1084/jem.20140507
Goodman, D. B., Azimi, C. S., Kearns, K., Talbot, A., Garakani, K., Garcia, J., et al. (2022). Pooled screening of CAR T cells identifies diverse immune signaling domains for next-generation immunotherapies. Sci. Transl. Med. 14 (670), eabm1463. doi: 10.1126/scitranslmed.abm1463
Gutierrez-Preciado, A., Torres, A. G., Merino, E., Bonomi, H. R., Goldbaum, F. A., Garcia-Angulo, V. A. (2015). Extensive identification of bacterial riboflavin transporters and their distribution across bacterial species. PloS One 10 (5), e0126124. doi: 10.1371/journal.pone.0126124
Hansen, T. H., Huang, S., Arnold, P. L., Fremont, D. H. (2007). Patterns of nonclassical MHC antigen presentation. Nat. Immunol. 8 (6), 563–568. doi: 10.1038/ni1475
Harriff, M. J., Karamooz, E., Burr, A., Grant, W. F., Canfield, E. T., Sorensen, M. L., et al. (2016). Endosomal MR1 trafficking plays a key role in presentation of mycobacterium tuberculosis ligands to MAIT cells. PloS Pathog. 12 (3), e1005524. doi: 10.1371/journal.ppat.1005524
Harriff, M. J., McMurtrey, C., Froyd, C. A., Jin, H., Cansler, M., Null, M., et al. (2018). MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci. Immunol. 3 (25). doi: 10.1126/sciimmunol.aao2556
Hegde, R., Srinivasula, S. M., Ahmad, M., Fernandes-Alnemri, T., Alnemri, E. S. (1998). Blk, a BH3-containing mouse protein that interacts with bcl-2 and bcl-xL, is a potent death agonist. J. Biol. Chem. 273 (14), 7783–7786. doi: 10.1074/jbc.273.14.7783
Hinks, T. S. C., Marchi, E., Jabeen, M., Olshansky, M., Kurioka, A., Pediongco, T. J., et al. (2019). Activation and In vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 28 (12), 3249–3262 e3245. doi: 10.1016/j.celrep.2019.07.039
Hu, G., Barnes, B. J. (2009). IRF-5 is a mediator of the death receptor-induced apoptotic signaling pathway. J. Biol. Chem. 284 (5), 2767–2777. doi: 10.1074/jbc.M804744200
Huang, S. (2016). Targeting innate-like T cells in tuberculosis. Front. Immunol. 7. doi: 10.3389/fimmu.2016.00594
Huang, S., Gilfillan, S., Cella, M., Miley, M. J., Lantz, O., Lybarger, L., et al. (2005). Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J. Biol. Chem. 280 (22), 21183–21193. doi: 10.1074/jbc.M501087200
Huang, S., Gilfillan, S., Kim, S., Thompson, B., Wang, X., Sant, A. J., et al. (2008). MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J. Exp. Med. 205 (5), 1201–1211. doi: 10.1084/jem.20072579
Huang, S., Martin, E., Kim, S., Yu, L., Soudais, C., Fremont, D. H., et al. (2009). MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc. Natl. Acad. Sci. U.S.A. 106 (20), 8290–8295. doi: 10.1073/pnas.0903196106
Huang, S., Moody, D. B. (2016). Donor-unrestricted T cells in the human CD1 system. Immunogenetics 68 (8), 577–596. doi: 10.1007/s00251-016-0942-x
Huber, M. E., Kurapova, R., Heisler, C. M., Karamooz, E., Tafesse, F. G., Harriff, M. J. (2020). Rab6 regulates recycling and retrograde trafficking of MR1 molecules. Sci. Rep. 10 (1), 20778. doi: 10.1038/s41598-020-77563-4
Intlekofer, A. M., Takemoto, N., Wherry, E. J., Longworth, S. A., Northrup, J. T., Palanivel, V. R., et al. (2005). Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6 (12), 1236–1244. doi: 10.1038/ni1268
Ioannidis, M., Cerundolo, V., Salio, M. (2020). The immune modulating properties of mucosal-associated invariant T cells. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01556
Jiang, J., Wang, X., An, H., Yang, B., Cao, Z., Liu, Y., et al. (2014). MAIT cell function is modulated by PD-1 signaling in patients with active tuberculosis. Am. J. Respir. Crit. Care Med. 190 (3), 329–339. doi: 10.1164/rccm.201401-0106OC
Kaech, S. M., Cui, W. (2012). Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12 (11), 749–761. doi: 10.1038/nri3307
Kagi, D., Vignaux, F., Ledermann, B., Burki, K., Depraetere, V., Nagata, S., et al. (1994). Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science 265 (5171), 528–530. doi: 10.1126/science.7518614
Katelaris, A. L., Jackson, C., Southern, J., Gupta, R. K., Drobniewski, F., Lalvani, A., et al. (2020). Effectiveness of BCG vaccination against mycobacterium tuberculosis infection in adults: A cross-sectional analysis of a UK-based cohort. J. Infect. Dis. 221 (1), 146–155. doi: 10.1093/infdis/jiz430
Kauffman, K. D., Sallin, M. A., Hoft, S. G., Sakai, S., Moore, R., Wilder-Kofie, T., et al. (2018). Limited pulmonary mucosal-associated invariant T cell accumulation and activation during mycobacterium tuberculosis infection in rhesus macaques. Infect. Immun. 18 (6), e3000644. doi: 10.1128/IAI.00431-18
Kavazovic, I., Han, H., Balzaretti, G., Slinger, E., Lemmermann, N. A. W., Ten Brinke, A., et al. (2020). Eomes broadens the scope of CD8 T-cell memory by inhibiting apoptosis in cells of low affinity. PloS Biol. 18 (3), e3000648. doi: 10.1371/journal.pbio.3000648
Kawachi, I., Maldonado, J., Strader, C., Gilfillan, S. (2006). MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J. Immunol. 176 (3), 1618–1627. doi: 10.4049/jimmunol.176.3.1618
Kim, J. S., Kang, M. J., Kim, W. S., Han, S. J., Kim, H. M., Kim, H. W., et al. (2015). Essential engagement of toll-like receptor 2 in initiation of early protective Th1 response against rough variants of mycobacterium abscessus. Infect. Immun. 83 (4), 1556–1567. doi: 10.1128/IAI.02853-14
Kjer-Nielsen, L., Patel, O., Corbett, A. J., Le Nours, J., Meehan, B., Liu, L., et al. (2012). MR1 presents microbial vitamin b metabolites to MAIT cells. Nature 491, 717-723. doi: 10.1038/nature11605
Koay, H. F., Su, S., Amann-Zalcenstein, D., Daley, S. R., Comerford, I., Miosge, L., et al. (2019). A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci. Immunol. 4 (41), eaay6039. doi: 10.1126/sciimmunol.aay6039
Koeffler, H. P., Golde, D. W. (1980). Human myeloid leukemia cell lines: a review. Blood 56 (3), 344–350. doi: 10.1182/blood.V56.3.344.344
Kong, L., Moorlag, S., Lefkovith, A., Li, B., Matzaraki, V., van Emst, L., et al. (2021). Single-cell transcriptomic profiles reveal changes associated with BCG-induced trained immunity and protective effects in circulating monocytes. Cell Rep. 37 (7), 110028. doi: 10.1016/j.celrep.2021.110028
Konjevic, G., Martinovic, K. M., Vuletic, A., Jurisic, V., Spuzic, I. (2009). Distribution of several activating and inhibitory receptors on CD3(-)CD16(+) NK cells and their correlation with NK cell function in healthy individuals. J. Membrane Biol. 230 (3), 113–123. doi: 10.1007/s00232-009-9191-3
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97. doi: 10.1093/nar/gkw377
Kumar, P. (2021). A perspective on the success and failure of BCG. Front. Immunol. 12. doi: 10.3389/fimmu.2021.778028
Kurioka, A., Cosgrove, C., Simoni, Y., van Wilgenburg, B., Geremia, A., Bjorkander, S., et al. (2018). CD161 defines a functionally distinct subset of pro-inflammatory natural killer cells. Front. Immunol. 9. doi: 10.3389/fimmu.2018.00486
Lamichhane, R., Schneider, M., de la Harpe, S. M., Harrop, T. W. R., Hannaway, R. F., Dearden, P. K., et al. (2019). TCR- or cytokine-activated CD8(+) mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. 28 (12), 3061–306+. doi: 10.1016/j.celrep.2019.08.054
Lanier, L. L., Chang, C., Phillips, J. H. (1994). Human NKR-P1A. a disulfide-linked homodimer of the c-type lectin superfamily expressed by a subset of NK and T lymphocytes. J. Immunol. 153 (6), 2417–2428.
Le Bourhis, L., Martin, E., Peguillet, I., Guihot, A., Froux, N., Core, M., et al. (2010). Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11 (8), 701–708. doi: 10.1038/ni.1890
Lee, M., Lee, E., Han, S. K., Choi, Y. H., Kwon, D. I., Choi, H., et al. (2020). Single-cell RNA sequencing identifies shared differentiation paths of mouse thymic innate T cells. Nat. Commun. 11 (1), 4367. doi: 10.1038/s41467-020-18155-8
Leeansyah, E., Svard, J., Dias, J., Buggert, M., Nystrom, J., Quigley, M. F., et al. (2015). Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PloS Pathog. 11 (8), e1005072. doi: 10.1371/journal.ppat.1005072
Leng, T., Akther, H. D., Hackstein, C. P., Powell, K., King, T., Friedrich, M., et al. (2019). TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. 28 (12), 3077–3091 e3075. doi: 10.1016/j.celrep.2019.08.050
Lepore, M., Kalinicenko, A., Colone, A., Paleja, B., Singhal, A., Tschumi, A., et al. (2014). Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nat. Commun. 5, 3866. doi: 10.1038/ncomms4866
Lettau, M., Janssen, O. (2021). Intra- and extracellular effector vesicles from human T and NK cells: Same-same, but different? Front. Immunol. 12. doi: 10.3389/fimmu.2021.804895
Li, G., Bethune, M. T., Wong, S., Joglekar, A. V., Leonard, M. T., Wang, J. K., et al. (2019). T Cell antigen discovery via trogocytosis. Nat. Methods 16 (2), 183–190. doi: 10.1038/s41592-018-0305-7
Li, J., He, Y., Hao, J., Ni, L., Dong, C. (2018). High levels of eomes promote exhaustion of anti-tumor CD8(+) T cells. Front. Immunol. 9, 2981. doi: 10.3389/fimmu.2018.02981
Li, Z. Y., Morman, R. E., Hegermiller, E., Sun, M., Bartom, E. T., Maienschein-Cline, M., et al. (2021). The transcriptional repressor ID2 supports natural killer cell maturation by controlling TCF1 amplitude. J. Exp. Med. 218 (6), e20202032. doi: 10.1084/jem.20202032
Li, J., Zhan, L., Qin, C. (2021). The double-sided effects of mycobacterium bovis bacillus calmette-guerin vaccine. NPJ Vaccines 6 (1)14. doi: 10.1038/s41541-020-00278-0
Locksley, R. M., Killeen, N., Lenardo, M. J. (2001). The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104 (4), 487–501. doi: 10.1016/s0092-8674(01)00237-9
Logunova, N. N., Kriukova, V. V., Shelyakin, P. V., Egorov, E. S., Pereverzeva, A., Bozhanova, N. G., et al. (2020). MHC-II alleles shape the CDR3 repertoires of conventional and regulatory naive CD4(+) T cells. Proc. Natl. Acad. Sci. U.S.A. 117 (24), 13659–13669. doi: 10.1073/pnas.2003170117
Lozzio, C. B., Lozzio, B. B. (1975). Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 45 (3), 321–334. doi: 10.1182/blood.V45.3.321.321
Makedonas, G., Hutnick, N., Haney, D., Amick, A. C., Gardner, J., Cosma, G., et al. (2010). Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8 T cells. PloS Pathog. 6 (3), e1000798. doi: 10.1371/journal.ppat.1000798
Marsden, V. S., Strasser, A. (2003). Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annu. Rev. Immunol. 21, 71–105. doi: 10.1146/annurev.immunol.21.120601.141029
McCarthy, D. J., Chen, Y., Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40 (10), 4288–4297. doi: 10.1093/nar/gks042
McShane, H., Jacobs, W. R., Fine, P. E., Reed, S. G., McMurray, D. N., Behr, M., et al. (2012). BCG: myths, realities, and the need for alternative vaccine strategies. Tuberculosis (Edinb) 92 (3), 283–288. doi: 10.1016/j.tube.2011.12.003
McWilliam, H. E. G., Mak, J. Y. W., Awad, W., Zorkau, M., Cruz-Gomez, S., Lim, H. J., et al. (2020). Endoplasmic reticulum chaperones stabilize ligand-receptive MR1 molecules for efficient presentation of metabolite antigens. Proc. Natl. Acad. Sci. U.S.A. 117 (40), 24974–24985. doi: 10.1073/pnas.2011260117
Meermeier, E. W., Laugel, B. F., Sewell, A. K., Corbett, A. J., Rossjohn, J., McCluskey, J., et al. (2016). Human TRAV1-2-negative MR1-restricted T cells detect s. pyogenes and alternatives to MAIT riboflavin-based antigens. Nat. Commun. 7, 12506. doi: 10.1038/ncomms12506
Mehta, A. K., Gracias, D. T., Croft, M. (2018). TNF activity and T cells. Cytokine 101, 14–18. doi: 10.1016/j.cyto.2016.08.003
Meierovics, A., Yankelevich, W. J., Cowley, S. C. (2013). MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 110 (33), E3119–E3128. doi: 10.1073/pnas.1302799110
Michelsen, S. W., Soborg, B., Koch, A., Carstensen, L., Hoff, S. T., Agger, E. M., et al. (2014). The effectiveness of BCG vaccination in preventing mycobacterium tuberculosis infection and disease in Greenland. Thorax 69 (9), 851–856. doi: 10.1136/thoraxjnl-2014-205688
Morikawa, Y., Murakami, M., Kondo, H., Nemoto, N., Iwabuchi, K., Eshima, K. (2021). Natural killer cell group 7 sequence in cytotoxic cells optimizes exocytosis of lytic granules essential for the perforin-dependent, but not fas ligand-dependent, cytolytic pathway. Immunohorizons 5 (4), 234–245. doi: 10.4049/immunohorizons.2100029
Murray, R. A., Mansoor, N., Harbacheuski, R., Soler, J., Davids, V., Soares, A., et al. (2006). Bacillus calmette guerin vaccination of human newborns induces a specific, functional CD8+ T cell response. J. Immunol. 177 (8), 5647–5651. doi: 10.4049/jimmunol.177.8.5647
Nakamura, S., Kuroki, K., Ohki, I., Sasaki, K., Kajikawa, M., Maruyama, T., et al. (2009). Molecular basis for e-cadherin recognition by killer cell lectin-like receptor G1 (KLRG1). J. Biol. Chem. 284 (40), 27327–27335. doi: 10.1074/jbc.M109.038802
Nalpas, N. C., Magee, D. A., Conlon, K. M., Browne, J. A., Healy, C., McLoughlin, K. E., et al. (2015). RNA Sequencing provides exquisite insight into the manipulation of the alveolar macrophage by tubercle bacilli. Sci. Rep. 5, 13629. doi: 10.1038/srep13629
Ng, S. S., De Labastida Rivera, F., Yan, J., Corvino, D., Das, I., Zhang, P., et al. (2020). The NK cell granule protein NKG7 regulates cytotoxic granule exocytosis and inflammation. Nat. Immunol. 21 (10), 1205–1218. doi: 10.1038/s41590-020-0758-6
Nikolich-Zugich, J., Slifka, M. K., Messaoudi, I. (2004). The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 4 (2), 123–132. doi: 10.1038/nri1292
Oettinger, T., Jorgensen, M., Ladefoged, A., Haslov, K., Andersen, P. (1999). Development of the mycobacterium bovis BCG vaccine: review of the historical and biochemical evidence for a genealogical tree. Tuber Lung Dis. 79 (4), 243–250. doi: 10.1054/tuld.1999.0206
Olson, J. A., McDonald-Hyman, C., Jameson, S. C., Hamilton, S. E. (2013). Effector-like CD8(+) T cells in the memory population mediate potent protective immunity. Immunity 38 (6), 1250–1260. doi: 10.1016/j.immuni.2013.05.009
Petersen, S. L., Chen, T. T., Lawrence, D. A., Marsters, S. A., Gonzalvez, F., Ashkenazi, A. (2015). TRAF2 is a biologically important necroptosis suppressor. Cell Death Differ 22 (11), 1846–1857. doi: 10.1038/cdd.2015.35
Pipkin, M. E., Sacks, J. A., Cruz-Guilloty, F., Lichtenheld, M. G., Bevan, M. J., Rao, A. (2010). Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32 (1), 79–90. doi: 10.1016/j.immuni.2009.11.012
Prezzemolo, T., Guggino, G., La Manna, M. P., Di Liberto, D., Dieli, F., Caccamo, N. (2014). Functional signatures of human CD4 and CD8 T cell responses to mycobacterium tuberculosis. Front. Immunol. 5. doi: 10.3389/fimmu.2014.00180
Raviglione, M. C., Snider, D. E., Jr., Kochi, A. (1995). Global epidemiology of tuberculosis. morbidity and mortality of a worldwide epidemic. Jama 273 (3), 220–226. doi: 10.1001/jama.1995.03520270054031
Reantragoon, R., Corbett, A. J., Sakala, I. G., Gherardin, N. A., Furness, J. B., Chen, Z., et al. (2013). Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210 (11), 2305–2320. doi: 10.1084/jem.20130958
Roder, J. C., Ahrlund-Richter, L., Jondal, M. (1979). Target-effector interaction in the human and murine natural killer system: specificity and xenogeneic reactivity of the solubilized natural killer-target structure complex and its loss in a somatic cell hybrid. J. Exp. Med. 150 (3), 471–481. doi: 10.1084/jem.150.3.471
Rossjohn, J., Gras, S., Miles, J. J., Turner, S. J., Godfrey, D. I., McCluskey, J. (2015). T Cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200. doi: 10.1146/annurev-immunol-032414-112334
Sakai, S., Kauffman, K. D., Oh, S., Nelson, C. E., Barry, C. E., 3rd, Barber, D. L. (2021). MAIT cell-directed therapy of mycobacterium tuberculosis infection. Mucosal Immunol. 14 (1), 199–208. doi: 10.1038/s41385-020-0332-4
Sakala, I. G., Kjer-Nielsen, L., Eickhoff, C. S., Wang, X., Blazevic, A., Liu, L., et al. (2015). Functional heterogeneity and antimycobacterial effects of mouse mucosal-associated invariant T cells specific for riboflavin metabolites. J. Immunol. 195 (2), 587–601. doi: 10.4049/jimmunol.1402545
Salou, M., Legoux, F., Gilet, J., Darbois, A., du Halgouet, A., Alonso, R., et al. (2019). A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J. Exp. Med. 216 (1), 133–151. doi: 10.1084/jem.20181483
Sharma, P. K., Wong, E. B., Napier, R. J., Bishai, W. R., Ndung’u, T., Kasprowicz, V. O., et al. (2015). High expression of CD26 accurately identifies human bacteria-reactive MR1-restricted MAIT cells. Immunology 145 (3), 443–453. doi: 10.1111/imm.12461
Sharma, M., Zhang, S., Niu, L., Lewinsohn, D. M., Zhang, X., Huang, S. (2020). Mucosal-associated invariant T cells develop an innate-like transcriptomic program in anti-mycobacterial responses. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01136
Siewiera, J., Gouilly, J., Hocine, H. R., Cartron, G., Levy, C., Al-Daccak, R., et al. (2015). Natural cytotoxicity receptor splice variants orchestrate the distinct functions of human natural killer cell subtypes. Nat. Commun. 6, 10183. doi: 10.1038/ncomms10183
Silke, J., Brink, R. (2010). Regulation of TNFRSF and innate immune signalling complexes by TRAFs and cIAPs. Cell Death Differ 17 (1), 35–45. doi: 10.1038/cdd.2009.114
Soares, A. P., Kwong Chung, C. K., Choice, T., Hughes, E. J., Jacobs, G., van Rensburg, E. J., et al. (2013). Longitudinal changes in CD4(+) T-cell memory responses induced by BCG vaccination of newborns. J. Infect. Dis. 207 (7), 1084–1094. doi: 10.1093/infdis/jis941
Tameris, M. D., Hatherill, M., Landry, B. S., Scriba, T. J., Snowden, M. A., Lockhart, S., et al. (2013). Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 381 (9871), 1021–1028. doi: 10.1016/S0140-6736(13)60177-4
Tao, H., Pan, Y., Chu, S., Li, L., Xie, J., Wang, P., et al. (2021). Differential controls of MAIT cell effector polarization by mTORC1/mTORC2 via integrating cytokine and costimulatory signals. Nat. Commun. 12 (1), 2029. doi: 10.1038/s41467-021-22162-8
Ussher, J. E., Bilton, M., Attwod, E., Shadwell, J., Richardson, R., de Lara, C., et al. (2014). CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur. J. Immunol. 44 (1), 195–203. doi: 10.1002/eji.201343509
Valdez, A. C., Cabaniols, J. P., Brown, M. J., Roche, P. A. (1999). Syntaxin 11 is associated with SNAP-23 on late endosomes and the trans-golgi network. J. Cell Sci. 112 (Pt 6), 845–854. doi: 10.1242/jcs.112.6.845
Vorkas, C. K., Krishna, C., Li, K., Aube, J., Fitzgerald, D. W., Mazutis, L., et al. (2022). Single-cell transcriptional profiling reveals signatures of helper, effector, and regulatory MAIT cells during homeostasis and activation. J. Immunol. 208 (5), 1042–1056. doi: 10.4049/jimmunol.2100522
Wallach, D., Kang, T. B., Kovalenko, A. (2014). Concepts of tissue injury and cell death in inflammation: a historical perspective. Nat. Rev. Immunol. 14 (1), 51–59. doi: 10.1038/nri3561
Webster, J. D., Vucic, D. (2020). The balance of TNF mediated pathways regulates inflammatory cell death signaling in healthy and diseased tissues. Front. Cell Dev. Biol. 8. doi: 10.3389/fcell.2020.00365
Wu, D., Prives, C. (2018). Relevance of the p53-MDM2 axis to aging. Cell Death Differ 25 (1), 169–179. doi: 10.1038/cdd.2017.187
Yamada, A., Arakaki, R., Saito, M., Kudo, Y., Ishimaru, N. (2017). Dual role of Fas/FasL-mediated signal in peripheral immune tolerance. Front. Immunol. 8. doi: 10.3389/fimmu.2017.00403
Keywords: transcriptome, MHC-related protein 1 (MR1), Bacillus Calmette-Guérin (BCG), mucosal-associated invariant T (MAIT) cells, Mycobacterium tuberculosis (M. tuberculosis)
Citation: Sharma M, Niu L, Zhang X and Huang S (2023) Comparative transcriptomes reveal pro-survival and cytotoxic programs of mucosal-associated invariant T cells upon Bacillus Calmette–Guérin stimulation. Front. Cell. Infect. Microbiol. 13:1134119. doi: 10.3389/fcimb.2023.1134119
Received: 29 December 2022; Accepted: 15 March 2023;
Published: 06 April 2023.
Edited by:
Alexander S. Apt, Central Tuberculosis Research Institute (RAMS), RussiaReviewed by:
Marianna Orlova, McGill University Health Center, CanadaMatt Johansen, University of Technology Sydney, Australia
Copyright © 2023 Sharma, Niu, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouxiong Huang, U2hvdXhpb25nLmh1YW5nQHVjLmVkdQ==
 Manju Sharma
Manju Sharma Liang Niu
Liang Niu Xiang Zhang
Xiang Zhang Shouxiong Huang
Shouxiong Huang