- 1Key Laboratory of Jilin Province for Zoonosis Prevention and Control, Changchun Veterinary Research Institute, Chinese Academy of Agriculture Sciences, Changchun, Jilin, China
- 2China-Japan Union Hospital, Jilin University, Changchun, Jilin, China
Backgroud: The emergence of carbapenem-resistant Pseudomonas aeruginosa (CRPA) co-producing KPC-2 and VIM-2 has increased the healthcare threats.
Results: In this study, a CRPA strain 18102011, was isolated from the bile of a burn patient in ICU of China. Its whole genome was sequenced via the PacBio platform. The molecular characteristics of the genome were analyzed to assess the genetic environment of the carbapenemase genes blaKPC-2 and blaVIM-2. Antimicrobial susceptibility, plasmid stability, bacterial growth curves, and plasmid conjugation were measured. Strain 18102011 exhibited a resistant pattern to all 23 antibiotics tested, which could be defined as a pan-drug resistant P. aeruginosa strain. Two plasmids were identified in this strain: the IncpRBL16 mega-plasmid pP2011–1 carrying blaVIM-2 and the IncP6 plasmid pP2011–2 carrying blaKPC-2. blaVIM-2 was located in the region of In2057 (a novel class 1 integron) that was inserted into pP2011-1, and the expression of the blaVIM-2 gene was increased by the PcWTGN-10 promoter located at the 5’-CS. For the blaKPC-2 gene, the core module Tn3-ISKpn27-blaKPC-ΔISKpn6 served as the blaKPC-2 platform in pP2011-2, and the expression of the blaKPC-2 gene was achieved via the P1 promoter located downstream of ISKpn27. This expression pattern resulted in MICs of 4,096 μg/mL of imipenem for both strain 18102011 and its transconjugant D2011. Both plasmids were stable in strain 18102011 and could be co-transferred to other strains.
Conclusion: This study raised concerns regarding the high stability and non-inferior fitness of blaKPC-2-blaVIM-2-CRPA, shed light on its genomic characteristics, and underscored the importance of continued surveillance of CRPA.
Introduction
Pseudomonas aeruginosa accounts for 10-15% of nosocomial infections worldwide (Blanc et al., 1998). It can attach to the surfaces of medical instruments through biofilm formation, facilitating its spread within hospitals, particularly in the ICU (Cornaglia et al., 2000; Li et al., 2022). Most ICU patients use broad-spectrum antimicrobials to treat bacterial infections, and the prolonged use of antibiotics to eradicate bacterial infections is commonly practiced (Pang et al., 2019). However, the acquisition of antibiotic resistance genes by mobile genetic elements (e.g., plasmids, transposons, integrative elements, and conjugative elements), combined with their transfer among bacterial strains, leads to the development of multidrug-resistant P. aeruginosa strains in patients (De Vos et al., 1997; Partridge et al., 2018; Botelho et al., 2019; Allameh et al., 2020; Mohammadnejad et al., 2023). Such infections are associated with high mortality rates, ranging from 18 to 61% (Kang et al., 2003). Although carbapenems are the most important antibiotics for treating their P. aeruginosa infections, the bacteria can be resistant to them due in part to the acquisition of carbapenemase genes, complicating treatment (Partridge et al., 2018; Botelho et al., 2019). Some strains even exhibit pan-drug resistance, rendering subsequent treatments with drugs such as colistin and amikacin ineffective, which creates a therapeutic dilemma. Consequently, antimicrobial resistance has become a global health challenge that threatens many medical achievements of the last century and causing serious harm to health system outcomes.
In this study, P. aeruginosa strain 18102011 was isolated from the bile of a burn patient in a hospital ICU in 2018 (Changchun, China). The strain was genetically investigated to evaluate the genetic mechanism of its drug resistance. The whole-genome sequence of the strain was generated, and its molecular characteristics were subsequently investigated. The strain belonged to multi-locus sequence typing (ST2374) and serotype O4. Furthermore, the strain co-harbored the blaKPC-2 and blaVIM-2 genes, enabling it to resist carbapenems. The MICs of imipenem and meropenem (carbapenem) were both >256 μg/mL. Additionally, it was a pan-drug resistant P. aeruginosa that was resistant to 23 antibiotics, including amikacin, colistin, and fosfomycin. Further genetic analyses were applied to the plasmids pP2011–1 carrying blaVIM-2, and pP2011–2 carrying blaKPC-2 to characterize their genetic environments. The transcriptional expression of resistance genes blaKPC-2 and blaVIM-2 changed after imipenem exposure. Moreover, the fitness cost, loss of resistance genes under serially passage, and horizontal transmission ability of the blaKPC-2 and blaVIM-2 genes in bacteria were analyzed. These results provide a deeper understanding of the acquisition of drug resistance genes in P. aeruginosa.
Materials and methods
Identification of blaKPC-2-blaVIM-2-CRPA
A CRPA obtained from bile collected from a patient in the intensive care unit (ICU) of a public Chinese hospital in 2018, exhibited resistance to both imipenem and meropenem. This isolate was forwarded to our laboratory for species identification and antimicrobial susceptibility testing via the BD Phoenix-100 system, using Escherichia coli ATCC25922 as the quality control for susceptibility testing (The antibiotics tested were shown in Supplementary Table S1). Additionally, the isolate was stored at -80°C for potential future use. The experimental protocols were approved by the Ethics Committee of the Jilin University (JDKQ202316EC).
To ensure the accuracy of our findings, the MICs of imipenem and meropenem were determined by E-test. Furthermore, the MICs for other antibiotics that have not been tested in the BD Phoenix-100 system, such as colistin, ceftazidime-avibactam, and fosfomycin, which were commonly used to treat carbapenem-resistant bacteria, were also ascertained using E-test (The antibiotics tested were shown in Supplementary Table S2). And the quality control strain was E. coli ATCC25922. Drug-resistance levels, including resistance, intermediary, and sensitivity thresholds, were assessed by the guidelines established by the Clinical and Laboratory Standards Institute.
It was subject to polymerase chain reaction (PCR) and Sanger sequencing and targets the oprL gene (which was specific to P. aeruginosa), which encoded a peptidoglycan-associated lipoprotein (De Vos et al., 1997). Additionally, PCR assays were performed to detect the presence of carbapenemase genes, including blaIMP, blaSPM, blaAIM, blaVIM, blaGIM, blaBIC, blaSIM, blaNDM, blaDIM, blaKPC and blaOXA-48 (Poirel et al., 2011) (The primer sequences and PCR conditions were listed in Supplementary Table S3).
Sequencing and genome sequence assembly
Bacterial genomic DNA was isolated using the UltraClean microbial DNA extraction kit (Qiagen, Germany) and sequenced via a PacBio RS II sequencer (Pacific Biosciences, USA). The sequence reads were de novo assembled using the SMARTdenovo assembler (http://github.com/ruanjue/smartdenovo). The assembly results were corrected based on sequencing data through three rounds of error correction by using the Racon software (version 1.4.13). Subsequently, three rounds of error correction were performed using the Pilon software (version 1.22) with second-generation reads, yielding the final assembly results.
Genome annotation and comparison
Precise bacterial species identification was evaluated using pair-wise ANI (http://www.ezbiocloud.net/tools/ani) analysis between the genomes generated in this study and the P. aeruginosa reference genome sequence PAO1 (GenBank ID: NC_002516.2). A ≥95% ANI cut-off was used to define bacterial species (Yoon et al., 2017). The PAst (https://cge.food.dtu.dk/services/PAst/) server was used to perform O-antigen classification. In addition, RAST 2.0 (Brettin et al., 2015) and BLASTP/BLASTN (Boratyn et al., 2013) searches were used to predict open reading frames (ORFs), whereas comparisons with the ResFinder 4.0 (Bortolaia et al., 2020) (https://cge.cbs.dtu.dk/services/ResFinder/), CARD (https://card.mcmaster.ca) (Alcock et al., 2023) and VFDB (Liu et al., 2022) (http://www.mgc.ac.cn/VFs/) databases were used to identify acquired resistance genes and virulence genes. In addition, the ISfinder (Varani et al., 2011) (https://www-is.biotoul.fr/; Lastest Database Update 2021-9-21), TnCentral (https://tncentral.ncc.unesp.br), INTEGRAL (Moura et al., 2009) (http://integrall.bio.ua.pt/), and ICEberg 2.0 (Liu et al., 2019) (http://db-mml.sjtu.edu.cn/ICEberg/) platforms were used to identify mobile elements. The online database BPROM (Cassiano and Silva-Rocha, 2020) was used for promoter prediction. Pairwise sequence comparisons were conducted using BLASTN searches. Functional analysis of proteins in families and domain prediction was conducted using the InterPro (https://www.ebi.ac.uk/interpro) database. Gene organization diagrams were drawn in Inkscape 1.0 (http://inkscape.org/en/).
Multilocus sequence typing (ST) was conducted by evaluating gene sequence data (including seven conserved housekeeping genes: acsA, aroE, gtaA, mutL, nuoD, ppsA, and trpE) with the pubMLST platform (https://pubmlst.org/).
Conjugation experiments
Plasmid transferability was tested using E. coli EC600 (rifampicin resistant) and strain 18102011 as the recipient strain and donor strain, respectively. Both the donor and recipient strains were cultured separately overnight at 37°C. Adjust the bacterial concentrations of both recipient strain EC600 and the donor strain 18102011 to 109 CFU/mL. The donor and recipient strains were then subsequently mixed at a 1:1 ratio. The mixture was then spotted onto a 1cm2 hydrophilic nylon membrane filter (Millipore; 0.45 µm pore size), which was placed on an LB agar plate and incubated at 37°C for 6 h to initiate mating. After incubation, cells were recovered from the filter and serially diluted from 10–1 to 10-9, with three parallel replicates per dilution. The dilution was spotted onto LB agar (containing 80 µg/mL rifampicin and 4 µg/mL imipenem) and LB agar (containing 80 µg/mL rifampicin) to select the carbapenem-resistant E. coli transconjugants and recipient strain E. coli EC600, respectively. The entire experimental procedure was repeated three times. The conjugation transfer efficiency between strain 18102011 and EC600 was calculated using the Equation (1.1). Antimicrobial susceptibility of transconjugants was determined using the BD Phoenix-100 system, while the MIC of imipenem for both strain 18102011 and its transconjugant D2011 was determined by the broth dilution method.
Bacterial growth curve assay
In studies assessing plasmid fitness burden, it’s standard and scientifically appropriate to compare an isogenic pair. To specifically determine the fitness cost associated with the presence of plasmids harboring blaKPC-2 and blaVIM-2 genes in strain 18102011, bacterial growth curves were constructed for both transconjugant D2011 (carrying blaKPC-2 and blaVIM-2) of strain 18102011 and recipient strain E. coli EC600. The overnight culture of strain D2011 and EC600 were diluted to 0.5 McFarland standard and subsequently diluted 1:100 in antibiotic-free LB broth. Over the next 12 hours, the optical density (OD600) of each culture was monitored at 2-hour intervals via a NanoPhotometer N60 (Implen, Germany). The data represent the mean ± SEM of two independent experiments. Statistical differences were determined by two-tailed t-test (*p< 0.05).
Plasmid stability testing
Strain 18102011 was grown at 37°C in a shaking incubator set to 200 rpm and serially passaged for 10 days, with each passage diluted 1:500 in antibiotic-free LB broth. After 5 and 10 days, the cultures were serially diluted and plated on antibiotic-free LB agar. Fifty single colonies were randomly selected for detection of the blaKPC-2 and blaVIM-2 genes by PCR on the 5th and 10th day, respectively, and were used as markers to reflect the loss of their plasmid. The data represent the mean ± SEM of two independent experiments. Statistical differences were determined by two-tailed t-test (*p< 0.05).
RNA preparation and transcriptome sequencing
Bacterial genomic RNA (before and after the addition of imipenem) was isolated using the RNAprep Pure Cell/Bacteria Kit RNAprep Pure (TianGen, China). Starting with total RNA, the mRNA was purified by rRNA depletion. And cDNA libraries were constructed using mRNA. Transcriptome sequencing was performed on triplicate samples, which was carried out by Illumina NovaSeq. Using HTSeq v0.6.1, the number of reads mapped to each gene was counted. FPKM values for each gene were then calculated based on gene length and the corresponding read counts. FPKM, which stands for Fragments Per Kilobase of transcript sequence per Million base pairs sequenced, accounts for both sequencing depth and gene length, making it a widely used method for estimating gene expression levels. The DESeq R package (1.18.0) was employed to determine differential expression of strain 18102011 carrying plasmids before and after the addition of imipenem, using a model based on the negative binomial distribution. Using the Equstion (1.2), we calculate the fold change in expression levels of the resistance genes (blaKPC-2 and blaVIM-2), conjugative transfer genes, and replicon genes, before and after antibiotic treatment. The resulting p-values were adjusted using the Benjamini-Hochberg method to control the false discovery rate. Genes with an adjusted p-value<0.05, identified by DESeq, were classified as differentially expressed.
Quantitative real-time polymerase chain reaction
All primers and probes for blaKPC-2 gene, blaVIM-2 gene, and repA genes (IncpRBL16 and IncP6) were designed using Primer 5.0 software, based on the sequences from strain 18102011 carrying plasmid 1 (CP116229) and plasmid 2 (CP116230) (The primer sequences are listed in Supplementary Table S4). DNA contamination was removed from RNA samples using the 4x gDNA wiper rMix (reaction conditions: 42°C for 2 min). cDNA was then synthesized using the used 5x HiScripII qRT SuperMIx II. The reverse transcription program included incubation at 50 °C for 15 min, followed by incubation at 85 °C for 5 s. Quantitative real-time polymerase (qPCR) analysis specifically targeted four key genetic elements. These included the blaKPC-2 gene and blaVIM-2 gene responsible for encoding KPC-2 and VIM-2 carbapenemases respectively. The analysis also targeted the IncpRBL16 repA and IncP6 repA genes, which encode essential replication initiation proteins for plasmid 1 and plasmid 2 (The qPCR conditions were listed in Supplementary Table S5). The experiment was conducted with three technical replicates using three-fold serial dilutions of cDNA. The entire experimental procedure was repeated three times.
Nucleotide sequence accession numbers
The complete sequences of strain 18102011, plasmid pP2011-1, and plasmid pP2011–2 have been submitted to GenBank under the accession numbers CP116228, CP116229, and CP116230, respectively. The transcriptome data of strain 18102011 and its plasmids before and after the addition of imipenem have been uploaded to the China National Center for Bioinformatics (CNCB) (http://ngdc.cncb.ac.cn/gsub/) under accession number CRA023298.
Results and discussion
Identification and antimicrobial resistance profile of strain 18102011
After strain 18102011 was cultured overnight at 37°C on Brain Heart Infusion (BHI) agar containing an imipenem concentration of 4 μg/mL, distinct colonies with smooth and regular edges, measuring approximately 0.5 mm in diameter, were observed. These colonies exhibited non-fusion growth, and pyocyanin production, and were devoid of metallic sheens. Subsequently, strain 18102011 was identified as P. aeruginosa using the BD Phoenix-100 automated identification system and sequencing of the oprL gene, a marker specific to this species. Following this identification, a comprehensive analysis of its drug resistance spectrum was conducted, revealing resistance to all evaluated antibiotics (Supplementary Tables S1, S2).
Genomic characterization of the P. aeruginosa strain 18102011
To characterize the genome of the bacterium thoroughly, we employed Single Molecule Real-Time (SMRT) sequencing. The genome sequencing data revealed a 6.6 Mb chromosome in strain 18102011 (GenBank ID: CP116228), exhibiting a GC content of 66.2%. Additionally, the presence of two plasmids was detected in this bacterium. This strain’s genome revealed an ANI value of ≥95% with the reference strain P. aeruginosa PAO1 (GenBank ID: NC_002516.2), verifying the species information for this bacterium again.
The molecular genetic characteristics of the bacterium were analyzed to determine whether it was a member of a potential epidemic clone group. The isolate belonged to the multi-locus sequence type ST2374 and serotype O4 based on MLST and PAst identification, respectively. As of March 2024, the pbMLST database contained two total strains of ST2374 P. aeruginosa, including strain SX69 from a sputum origin in China and strain 127Gr isolated from soft tissue in Belarus in 2016. More than twenty serotypes of P. aeruginosa were known, of which serotype O4 was not known as a multidrug-resistant serotype (Nasrin et al., 2022). However, serotype switching from O4 to the multidrug-resistant serotype O12 had been observed under specific conditions (Thrane et al., 2015).
Three virulence genes were detected on the chromosome of strain 18102011: the phospholipase C (PLC) gene (plcH) and exotoxin genes (exoS and exoT). PLC was a thermolabile hemolysin that degrades phospholipid surfactants and reduces surface tension, thus preventing alveoli from collapsing completely when air leaves them during breathing (Khan et al., 2021; Wang et al., 2021). The ExoS and ExoT proteins could be secreted via the type III secretion system (T3SS) and could disrupt the cytoskeleton, induce host cell rounding, disrupt intercellular tight junctions, prevent wound healing, and inhibit bacterial internalization into epithelial cells and macrophages (Rao et al., 2021; Jouault et al., 2022). Consistent with previous reports, the isolate harboring exoS-like sequences did not contain exoU-like sequences (Feltman et al., 2001). In this study, in addition to the exoU gene encoding ExoU (a T3SS effector), the exoA gene encoding Exotoxin A (ExoA), which was also a strong pathogenic virulence factor such as ExoU, was not identified (Jørgensen et al., 2005; Foulkes et al., 2019; Morgan et al., 2021). Additionally, the exoY gene encoding ExoY (another T3SS effector) was also not detected in this study. It might exhibit greater cytotoxicity to epithelial cells than strains secreting active ExoY, as ExoY had been found to possibly play a protective role at certain stages of bacterial infection, either to facilitate host colonization or to establish and/or maintain chronic infection in the host (Silistre et al., 2021). Thus, this strain might cause cellular damage in immunocompromised patients, but it didn’t cause acute toxicity.
According to the Resfinder database, CARD database, and PCR results, the bacteria carried the acquired genes blaVIM-2 and blaKPC-2 simultaneously. Furthermore, through the Resfinder platform, other acquired resistance genes were identified in strain 18102011, including aminoglycoside resistance genes (aph(3’)-IIb, aac(6’)-Ib-cr, ant(2’’)-Ia), β-lactam resistance genes (blaOXA-396, blaPAO, blaPER-1), a sulfonamide resistance gene (sul1), a chloramphenicol resistance gene (catB7), a quinolone resistance gene (qnrVC6), and a fosfomycin resistance gene (fosA). These resistance genes were inserted into chromosomes and plasmids by mobile elements. Plasmid-mediated drug resistance genes could lead to their rapid spread among bacteria. Pan-resistance P. aeruginosa co-carrying blaVIM-2 and blaKPC-2 genes was first reported in Colombia and had subsequently spread in this country, harboring many resistance genes (Correa et al., 2015; Rada et al., 2021), which was not common in China.
Overview of plasmid pP2011–1 carrying blaVIM-2
The bacteria carried two plasmids, which were named pP2011–1 and pP2011-2, in this study. The mega-plasmid pP2011-1 (GenBank ID: CP116229) carrying blaVIM-2 was 474 kb and exhibited a GC content of 56.9%. The plasmid harbored a repA (replication initiation) gene sharing ≥96% nucleotide identity to repAIncpRBL16. The IncpRBL16 plasmid was first reported in the mega-plasmid p12969-DIM (GenBank ID: KU130294) of strain P. aeruginosa 12969 (Sun et al., 2016). As of March 2024, there were many genomes containing repAIncpRBL16-like sequences in the GenBank database, all of which belonged to Pseudomonas spp. However, the pP2011–1 plasmid had a low genetic identified with the previously reported IncpRBL16 family plasmid (Jiang et al., 2020; Dong et al., 2022), and there were large structural variations. The IncpRBL16 family plasmids carried by bacteria may have co-evolved with their chromosomes to maintain their presence in the host bacteria.
As shown in Figure 1, the backbone of plasmid pP2011–1 contained genes for partitioning (parB2-parAB), conjugal transfer (cpl and tivF), chemotaxis (che), pilus assembly (pil), and tellurium resistance (ter), in addition to repAIncpRBL16 (Figure 1). The plasmid also contained 3 novel recombination sites in addition to the common recombination site 2 for IncpRBL16 family plasmids (Jiang et al., 2020). ISPa75 (IS66 family) and fosA formed the accessory module 1 region that was inserted between two hypothetical proteins of the tellurium resistance region. In addition, dnaK and yaeT, along with several phage integrase genes and stress protein genes, form the accessory module 2 region that was inserted into the region of the catalytic subunit of Pol V (UmuC), which was a major recombination site of IncpRBL16 family plasmids. During conjugation, plasmids were transferred as single-stranded DNA, which in turn activates the bacterial SOS stress response. The SOS response coordinated the expression of dozens of bacterial genes involved in DNA repair and cell cycle control, and it was known to fuel bacterial evolvability through an increase in recombination and mutagenesis (Rodríguez-Beltrán et al., 2021). The accessory module 3 region comprised heavy-metal efflux protein genes (merE, merD, merA, merP, merT, and merR), Tn6001 (Tn3 family), two copies of ISCR1 (IS91 family), blaPER-1, and qnrVC6, and a novel class 1 integron In2057 carrying ant(2’’)-IIa, blaVIM-2 and aac(6’)-Ib-cr, which was inserted between the hypothetical protein genes that were 708 bp and 1,257 bp in length. Finally, the accessory module 4 region included ISPa141 (IS30 family), ISPa61 (ISL3 family), ISPst3 (IS21 family), ISPa60 (ISAs1 family), Tn4662a (Tn3 family), and Tn5046.1 (Tn3 family), which were inserted between the hypothetical protein and phage protein genes.
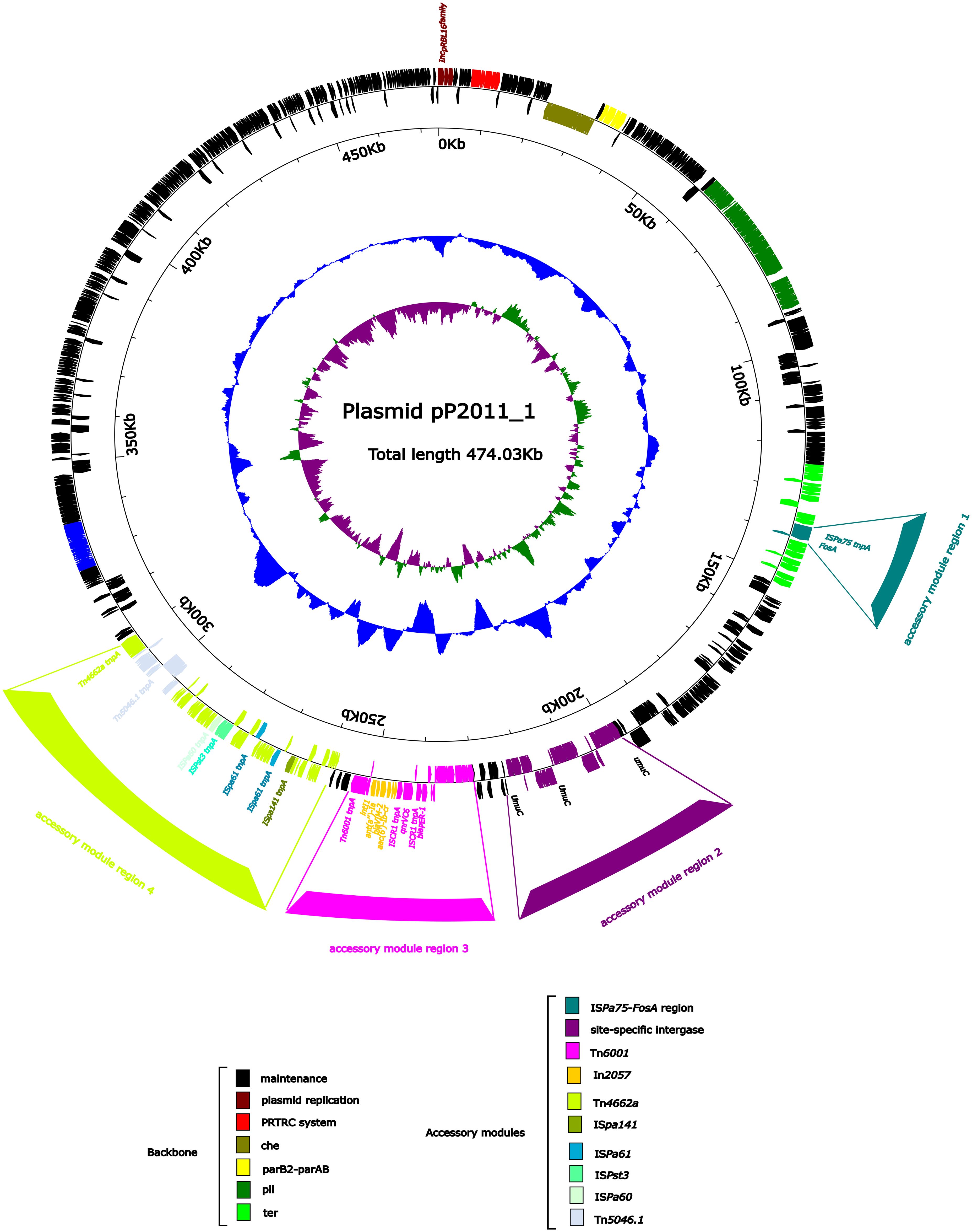
Figure 1. Annotation of the plasmid pP2011-1. The circles represented (from outside to inside) the following: (1) Genome functional annotation. The backbone region was black, the plasmid replicon was brown, the PRTRC system was red, the che region was dark green, the parB2-parAB region was yellow, the pil region was lighter green, and the ter region was bright green. The ISPa75 and fosA genes form the accessory module 1 region (dark cyan), whereas the dnaK and yaeT genes, along with several phage integrase genes and stress protein genes, form the accessory module 2 region (purple), which was inserted into the UmuC region (purple). The accessory module 3 region (pink) comprises Tn6011 (pink) and In2057 (orange). The accessory module 4 region (green-yellow) comprises ISpa141 (olive), ISpa61 (dark blue), ISpst3 (turquoise), ISPa60 (honeydew), Tn4662a (green-yellow) and Tn5046.1 (alice blue); (2) GC skew calculated as [(G-C)/(G+C)]; and (3) GC content.
Overview of plasmid pP2011–2 carrying blaKPC-2
As shown in Figure 2, the pP2011–2 plasmid (GenBank ID: CP116230) carrying blaKPC-2 was 40 kb in length and exhibited a GC content of 58.1%. The plasmid harbored a repA gene sharing ≥96% nucleotide identity to repAIncp6. The IncP6 plasmid was first identified in pRms149 (GenBank ID: GCA_019400855.1) of the P. aeruginosa JX05 strain in 2005 (Haines et al., 2005). According to the GenBank database, this replicon sequence could be found in Enterobacteriaceae, P. aeruginosa, and Aeromonas strains. The backbone of the repAIncp6 family plasmid included repAIncp6, genes for partitioning (parABC), and a mobilization region (mobABCDE), in addition to two accessory modules in this study. The accessory module 1 region comprised Tn5563a (Tn3 family) and ISPa19, whereas accessory module 2 includes ISEc33-Tn3-ISApu2-ISApu2-orf7-ISKpn27-blaKPC-2-ΔISKpn6-korC-orf6-klcA-ΔrepB. Typical transposons Tn4401 and Tn1722, both members of Tn3 family, have been demonstrated to mobilize the blaKPC-2 at high transposition frequencies (Song et al., 2024). KPC-1/KPC-2 was first identified in Klebsiella pneumoniae in 2001 (Yigit et al., 2001). While KPC carried by Enterobacteriaceae, it has also been detected in non-Enterobacteriaceae due to horizontal gene transfer, such as P. aeruginosa. Tn4401 is a removable element that commonly carries the blaKPC-2 gene in Europe, Brazil, and the United States. In Asia, blaKPC-2 is mainly located on different variants of Tn1722-like transposons (Song et al., 2024). Zhang DF et al. found that there were three divergent forms of blaKPC-2 transposon unit in K. pneumoniae, including Tn1721-blaKPC-2 transposon, IS26-Tn1721-blaKPC-2 transposon and IS26-blaKPC-2 transposon (Zhang et al., 2022). The IncP6 plasmid (p10265-KPC) was first characterized in 2016, different from the Tn1722/Tn1721-like unit transposons (pKP048 from K. pneumoniae), the ISApu1-orf7-ISApu2 structure truncates the Tn3 transposase and inserts a truncated blaTEM-1 gene downstream of ISKpn27 (Dai et al., 2016). In this study, the distinction lines in the fact that p10265-KPC has a truncated blaTEM-1 gene inserted between ISKpn27 and the blaKPC-2 gene (Dai et al., 2016).
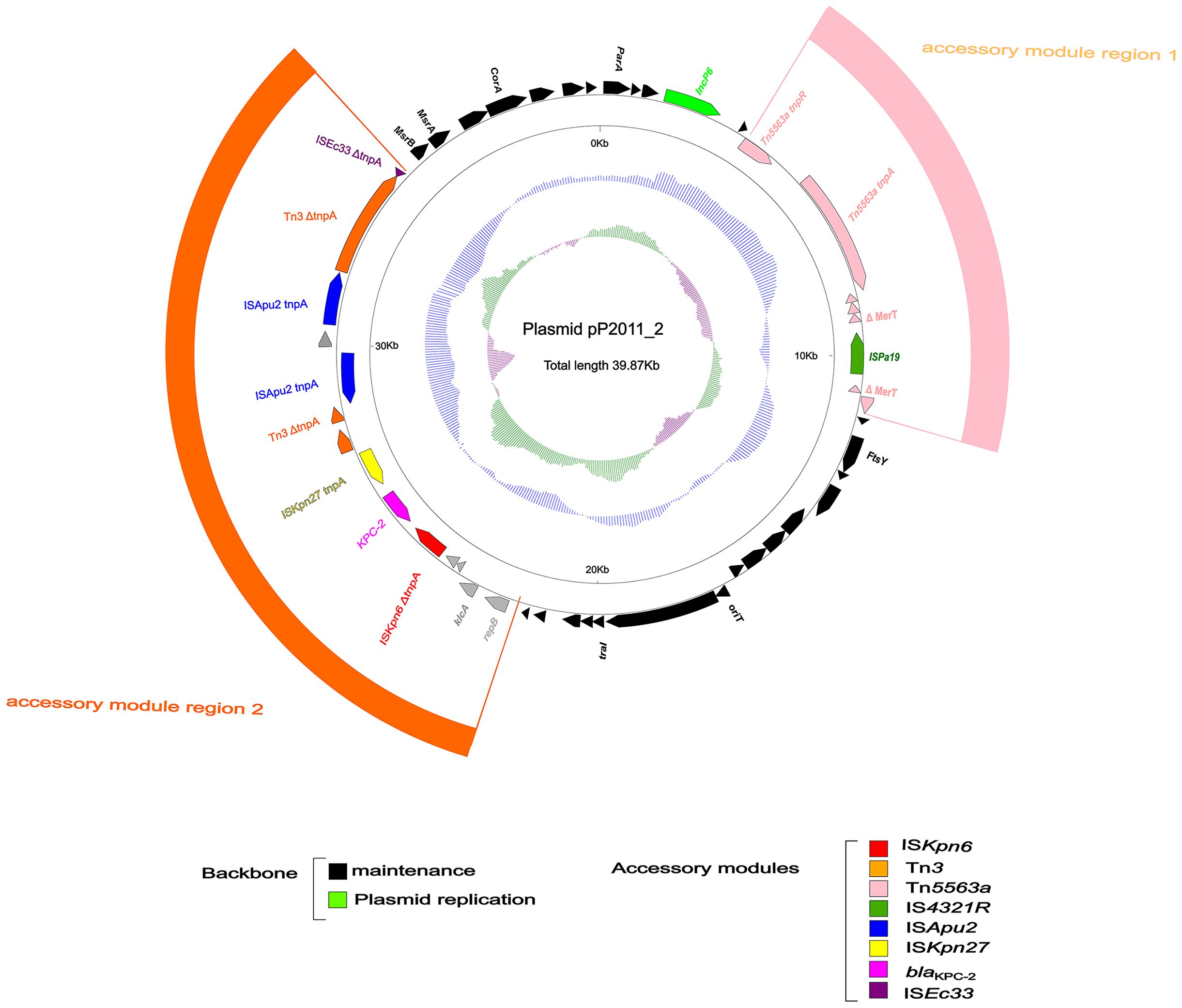
Figure 2. Annotation of plasmid pP2011-2. The circles represented (from outside to inside) the following: (1) Genome functional annotation. The backbone region was black, in which the plasmid replicon was green. Tn5563a (pink) and ISPa19 (sea-green) comprise the accessory module 1 region (pink), whereas the module 2 region (orange) comprises ISKpn6 (red), blaKPC-2 (magenta), ISKpn27 (yellow), Tn3 (orange), ISApu2 (blue), and ISEc33 (purple); (2) the GC skew was calculated as [(G-C)/(G+C)]; and (3) the GC content.
Conjugative transfer and resistance phenotype dissemination of blaVIM-2 and blaKPC-2-encoding plasmids
To evaluate the transferability of plasmids harboring blaKPC-2 and blaVIM-2, conjugation assays were performed by co-culturing the strain 18102011 with E. coli EC600. Conjugants grown on dilution gradients of 10-1, 10-2, and 10-3. And recipient bacteria can grow on all dilution gradients from 10–1 to 10–9 on a medium containing only rifampicin. The conjugation transfer efficiency was calculated to be 10–6 using the Equation (1.1). Because the IncP6 plasmid lacked a conjugation-transfer region and only had a mobile region, the plasmid could only carry out horizontal transfer under the synergistic action of the conjugation plasmid IncpRBL16 (Coluzzi et al., 2022). In this study, all suspected transconjugants were picked from the culture medium at 103 dilution gradients, with number of 19, 15, and 21 respectively (corresponding to three parallel replicates). A single carbapenem-resistant gene, namely blaKPC-2 or blaVIM-2, was not found by conjugation assays. Although the MICs of transconjugant D2011 for many antibiotics tested decreased significantly, indicating susceptibility, it was still resistant to imipenem and meropenem (Supplementary Tables S1, S2). The MICs of strain 18102011 and its transconjugant D2011 against imipenem were determined by broth method, both of which were 4,096 μg/mL. Therefore, the main resistance mechanism of strain 18102011 to carbapenem antibiotics was plasmid-mediated blaVIM-2 and blaKPC-2 genes, resulting in a stable carbapenem-resistant transmission phenotype.
Plasmid stability and fitness cost analysis of blaKPC-2 and blaVIM-2-co-harboring strain 18102022
The impact of the acquisition of both plasmids carrying blaKPC-2 and blaVIM-2 on the biological fitness cost was evaluated. Notably, the OD600 of recipient strain E. coli EC600 was significantly lower than that of the transconjugant D2011 (carrying blaKPC-2 and blaVIM-2) in 12h. Taken together, these microbiological characteristics indicated that strain 18102011 could carry both the blaKPC-2 and blaVIM-2 genes without compromising their fitness and maintaining them stably over time (Figure 3a). Furthermore, during serial passage in the laboratory for 10 days to evaluate the stability of plasmids carrying blaKPC-2 and blaVIM-2 from strain 1810211. Interestingly, both the blaKPC-2 and blaVIM-2 plasmids were present in 50 colonies (100%, 50/50) on the 5th and 10th day, respectively, indicating high stability (Figure 3b, PCR identification of blaKPC-2 and blaVIM-2 genes of the strain D2011 on 5th and 10th day were shown in Supplementary Figure S1). This increased our concern about blaKPC-2-blaVIM-2-CRPA, as it posed not only significant challenges in treatment but also a remarkable ability to stably maintain these plasmids without apparent fitness costs, making it difficult to retrogress.
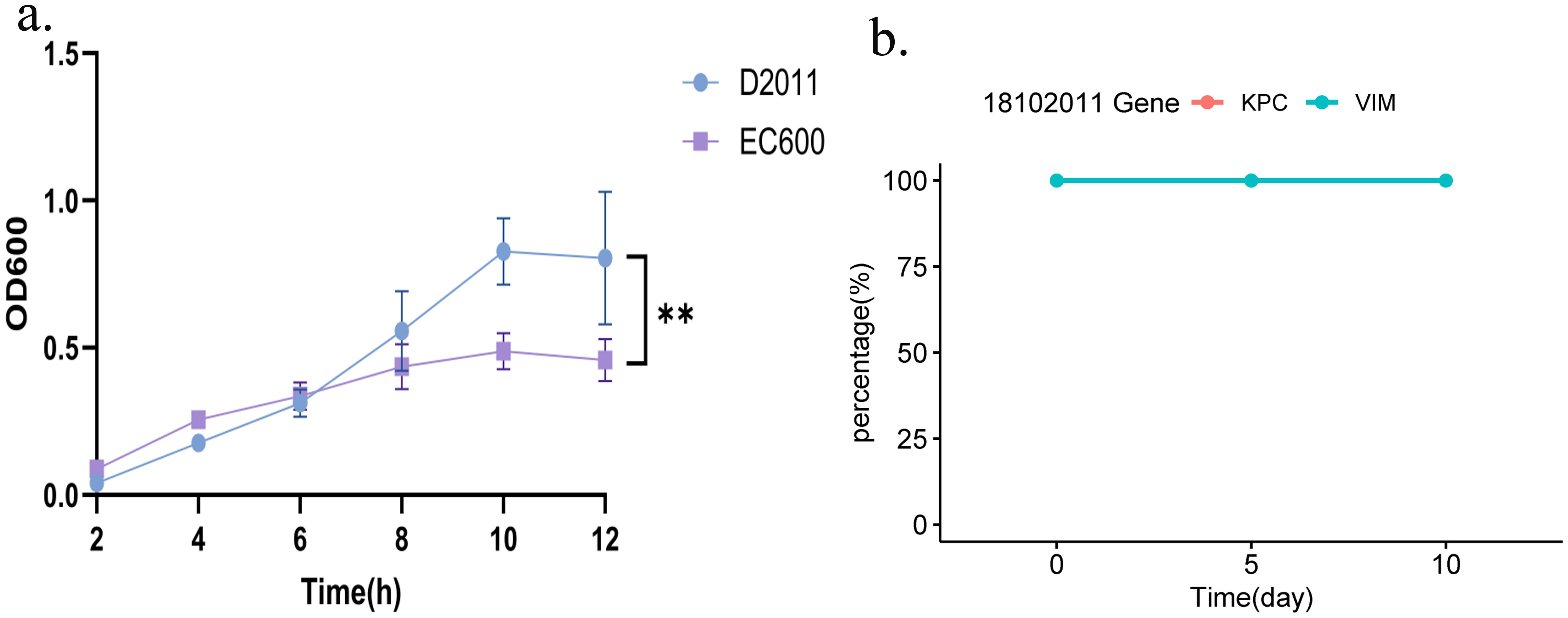
Figure 3. Growth curves of strain D2011 and stability of the blaKPC-2 or blaVIM-2 gene. (a) Growth curve of strain D2011 carrying the blaKPC-2 and blaVIM-2 genes and E. coli EC600. (b) Stability of the blaKPC-2 and blaVIM-2 genes during the 10-day serial passage. p< 0.05 *(significant), p< 0.01 ** (highly significant).
Transcriptional regulation of blaVIM-2 and blaKPC-2 under carbapenem pressure
The integron In2057-blaVIM-2 sequence was uploaded into the BPROM database, revealing only one promoter with the -10/-35 region (AGCTTACCA/TGTCCA) located within In2057’s integrase. This promoter sequence matched the PcWTGN-10 promoter sequence identified in the research by Nesvera J et al., driving the expression of the downstream gene cassette (Nesvera et al., 1998). It had been reported that the TGN-10 motif increased the PcW promoter strength efficiency 15-fold (Jové et al., 2010). Therefore, this promoter might have increased the expression of a series of downstream gene cassettes in this study, including blaVIM-2. This might be one of the reasons for the high resistance of bacteria to carbapenem.
The ISKpn27-blaKPC-2 sequence was uploaded into the BPROM database. Only one promoter with the -10/-35 region (ATGTAA/GGATTA) was identified between the ISKpn27 and blaKPC-2 genes. This promoter sequence was consistent with the P1 promoter sequence commonly found in ISKpn7-blaKPC-2, which drove the expression of the downstream blaKPC-2 gene (Huang et al., 2019; Huang et al., 2020).
Through transcriptome analysis of the expression of the blaVIM-2 gene carried by plasmid pP2011–1 and the blaKPC-2 gene carried by plasmid pP2011-2, it was found that both genes were up-regulated when imipenem was added to the culture medium (Figure 4, the data for Figure 4 can be found in Supplementary Table S6). However, after the addition of imipenem, the expression level of blaVIM-2 significantly increased, which might be related to the regulatory effect of its upstream strong promoter (Figures 4, 5, the data for Figure 4 can be found in Supplementary Table S7). Additionally, under the influence of imipenem, the expression levels of some other genes on the plasmid also changed. For example, the expression levels of plasmid conjugative transfer gene (tra), the mobilization genes (mob, excluding mobA), and the repA gene on the IncpRBL16 plasmid all showed increase, although this increase was not significant (Figures 4, 5). The low levels of read counts (FPKM) for the replicon genes repA (located on the IncpRBL16 plasmids) and the conjugative transfer gene (traD) indicated their low expression levels (Supplementary Table S6). Since the repA gene is a key factor for initiating plasmid replication, its low expression leads to a reduction in plasmid copy number. This could be a strategy employed by the IncpRBL16 plasmid to decrease its fitness cost to the host bacterium by maintaining a lower copy number (Figure 4).
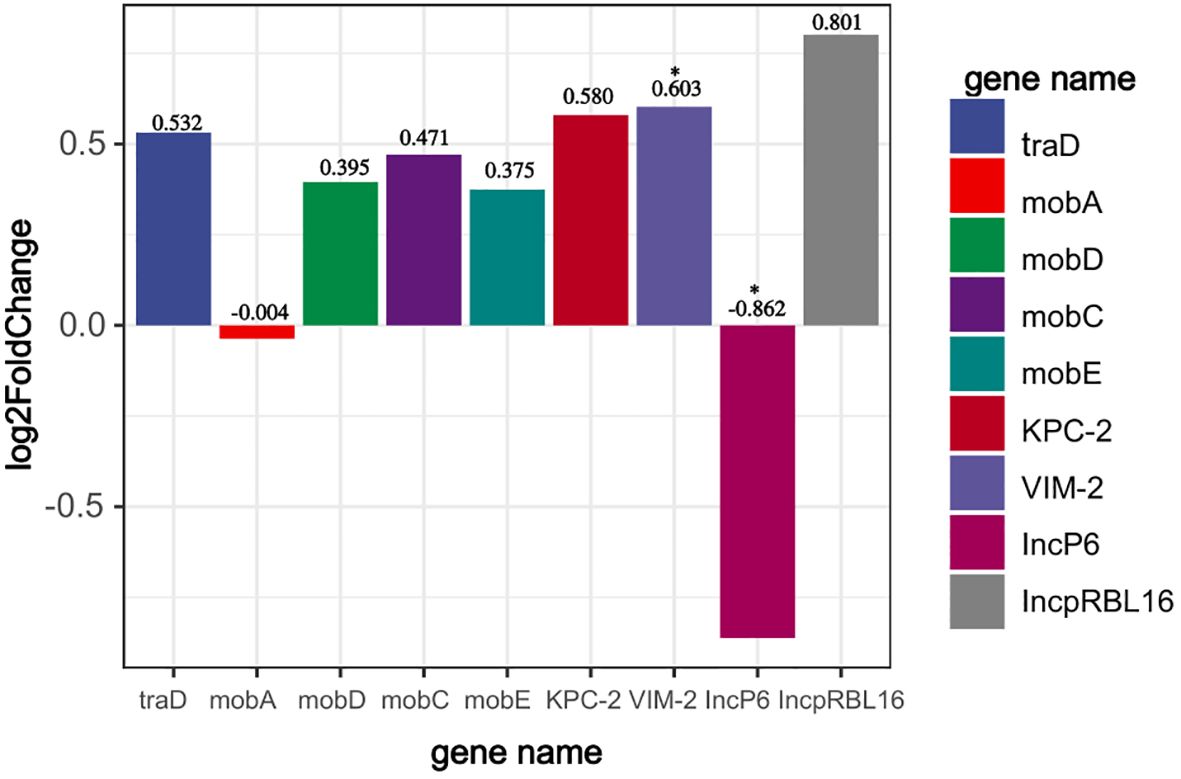
Figure 4. Read counts of the IncpRBL16 (repA), IncP6 (repA), blaVIM-2, blaKPC-2, conjugative transfer genes (tra), and mobile genetic genes (mob) in strain 18102011. The horizontal axis represents gene name, whereas the vertical axis represents change in read counts (FPKM) of genes after imipenem addition (expressed as log2FoldChange). p<0.05 *(significant), p<0.01 **(highly significant).

Figure 5. Results of qPCR identification of the IncpRBL16 (repA), IncP6 (repA), blaVIM-2, and blaKPC-2 in strain 18102011. The horizontal axis represents group classifications (IncP6; IncpRBL16; blaKPC-2; blaVIM-2), whereas the vertical axis represents amplification fold. p<0.05 *(significant), p<0.01 **(highly significant), p>0.05 ns (not significant).
Conclusion
Currently, there were relatively few reports on pan-resistant P. aeruginosa co-carrying blaVIM-2 and blaKPC-2 genes in China, which were mostly concentrated in Columbia. The limited existing reports also lacked an analysis of the fitness costs associated with the plasmids carried by these bacteria. In this study, the presence of blaKPC-2 and blaVIM-2 plasmids didn’t cause fitness costs to the growth of the host bacteria. Since the IncP6 plasmid lacked a conjugation transfer region, its horizontal transfer required the assistance of IncpRBL16 conjugation plasmids for co-transfer into E. coli EC600. Through horizontal plasmid transfer, high-level resistance to imipenem was stably maintained within the bacterial population. However, there were differences in the composition of the cell membranes between P. aeruginosa and E. coli, causing the pan-drug resistant phenotype to not be shared between the donor and recipient bacteria through plasmid conjugation transfer. As the treatment of pan-drug resistant bacteria was extremely challenging, their epidemiological and molecular genetics across the world should be closely monitored.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: The complete sequences of strain 18102011, plasmid pP2011-1, and plasmid pP2011-2 have been submitted to GenBank under the accession numbers CP116228, CP116229, and CP116230, respectively. The transcriptome data of strain 18102011 and its plasmids before and after the addition of imipenem have been uploaded to the China National Center for Bioinformatics (CNCB) (http://ngdc.cncb.ac.cn/gsub/) under accession number CRA023298.
Author contributions
LZ: Conceptualization, Investigation, Writing – original draft, Writing – review & editing, Data curation, Validation, Visualization, Methodology. ZW: Writing – original draft. GL: Writing – original draft, Supervision. XZ: Supervision, Validation, Writing – original draft. JJ: Writing – original draft, Supervision, Validation. SS: Project administration, Writing – original draft. YS: Writing – original draft, Resources, Project administration, Funding acquisition. XJ: Writing – original draft, Project administration. BJ: Project administration, Writing – original draft. LWZ: Conceptualization, Resources, Funding acquisition, Methodology, Supervision, Writing – review & editing. XG: Methodology, Investigation, Data curation, Funding acquisition, Resources, Project administration, Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Funding for the study design, data collection, data generation, and publication costs was provided by Jilin Provincial Department of Finance (jcsz2023481-1) and the National Science and Natural Science Foundation of China (Grant agreement 31872486).
Acknowledgments
We are grateful to the members of the China-Japan Union Hospital, Jilin University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1617614/full#supplementary-material
Supplementary Figure 1 | PCR identification of blaKPC-2 and blaVIM-2 genes of strain 18102011 on the 5th and 10th day. (a) PCR identification of blaKPC-2 and blaVIM-2 genes of strain 18102011 on the 5th day; (b) PCR identification of blaKPC-2 and blaVIM-2 genes of strain 18102011 on the 10th day; (c) PCR identification of blaKPC-2 and blaVIM-2 genes of strain 18102011 on the 5th day (repeat); (d) PCR identification of blaKPC-2 and blaVIM-2 genes of strain 18102011 on the 10th day (repeat).
References
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–d699. doi: 10.1093/nar/gkac920
Allameh, S. F., Nemati, S., Ghalehtaki, R., Mohammadnejad, E., Aghili, S. M., Khajavirad, N., et al. (2020). Clinical characteristics and outcomes of 905 COVID-19 patients admitted to imam khomeini hospital complex in the capital city of tehran, Iran. Arch. Iran Med. 23, 766–775. doi: 10.34172/aim.2020.102
Blanc, D. S., Petignat, C., Janin, B., Bille, J., and Francioli, P. (1998). Frequency and molecular diversity of Pseudomonas aeruginosa upon admission and during hospitalization: a prospective epidemiologic study. Clin. Microbiol. Infect. 4, 242–247. doi: 10.1111/j.1469-0691.1998.tb00051.x
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Botelho, J., Grosso, F., and Peixe, L. (2019). Antibiotic resistance in Pseudomonas aeruginosa - Mechanisms, epidemiology and evolution. Drug Resist. Updat 44, 100640. doi: 10.1016/j.drup.2019.07.002
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/srep08365
Cassiano, M. H. A. and Silva-Rocha, R. (2020). Benchmarking bacterial promoter prediction tools: potentialities and limitations. mSystems 5. doi: 10.1128/mSystems.00439-20
Coluzzi, C., Garcillán-Barcia, M. P., de la Cruz, F., and Rocha, E. P. C. (2022). Evolution of plasmid mobility: origin and fate of conjugative and nonconjugative plasmids. Mol. Biol. Evol. 39. doi: 10.1093/molbev/msac115
Cornaglia, G., Mazzariol, A., Lauretti, L., Rossolini, G. M., and Fontana, R. (2000). Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-beta-lactamase. Clin. Infect. Dis. 31, 1119–1125. doi: 10.1086/317448
Correa, A., Del Campo, R., Perenguez, M., Blanco, V. M., Rodríguez-Baños, M., Perez, F., et al. (2015). Dissemination of high-risk clones of extensively drug-resistant Pseudomonas aeruginosa in Colombia. Antimicrob. Agents Chemother. 59, 2421–2425. doi: 10.1128/AAC.03926-14
Dai, X., Zhou, D., Xiong, W., Feng, J., Luo, W., Luo, G., et al. (2016). The IncP-6 Plasmid p10265-KPC from Pseudomonas aeruginosa Carries a Novel ΔISEc33-Associated blaKPC-2 Gene Cluster. Front. Microbiol. 7, 310. doi: 10.3389/fmicb.2016.00310
De Vos, D., Lim, A., Jr., Pirnay, J. P., Struelens, M., Vandenvelde, C., Duinslaeger, L., et al. (1997). Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J. Clin. Microbiol. 35, 1295–1299. doi: 10.1128/jcm.35.6.1295-1299.1997
Dong, N., Liu, C., Hu, Y., Lu, J., Zeng, Y., Chen, G., et al. (2022). Emergence of an Extensive Drug Resistant Pseudomonas aeruginosa Strain of Chicken Origin Carrying bla(IMP-45), tet(X6), and tmexCD3-toprJ3 on an Inc(pRBL16) Plasmid. Microbiol. Spectr 10, e0228322. doi: 10.1128/spectrum.02283-22
Feltman, H., Schulert, G., Khan, S., Jain, M., Peterson, L., and Hauser, A. R. (2001). Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiol. (Reading) 147, 2659–2669. doi: 10.1099/00221287-147-10-2659
Foulkes, D. M., McLean, K., Haneef, A. S., Fernig, D. G., Winstanley, C., Berry, N., et al. (2019). Pseudomonas aeruginosa toxin exoU as a therapeutic target in the treatment of bacterial infections. Microorganisms 7. doi: 10.3390/microorganisms7120707
Haines, A. S., Jones, K., Cheung, M., and Thomas, C. M. (2005). The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187, 4728–4738. doi: 10.1128/JB.187.14.4728-4738.2005
Huang, J., Hu, X., Zhao, Y., Shi, Y., Ding, H., Wu, R., et al. (2019). Comparative analysis of bla(KPC) expression in tn4401 transposons and the tn3-tn4401 chimera. Antimicrob. Agents Chemother. 63:e02434-18. doi: 10.1128/AAC.02434-18
Huang, J., Hu, X., Zhao, Y., Shi, Y., Ding, H., Xv, J., et al. (2020). Genetic factors associated with enhanced bla(KPC) expression in tn3/tn4401 chimeras. Antimicrob. Agents Chemother. 64. :e01836-19. doi: 10.1128/AAC.01836-19
Jiang, X., Yin, Z., Yuan, M., Cheng, Q., Hu, L., Xu, Y., et al. (2020). Plasmids of novel incompatibility group IncpRBL16 from Pseudomonas species. J. Antimicrob. Chemother. 75, 2093–2100. doi: 10.1093/jac/dkaa143
Jørgensen, R., Merrill, A. R., Yates, S. P., Marquez, V. E., Schwan, A. L., Boesen, T., et al. (2005). Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature 436, 979–984. doi: 10.1038/nature03871
Jouault, A., Saliba, A. M., and Touqui, L. (2022). Modulation of the immune response by the Pseudomonas aeruginosa type-III secretion system. Front. Cell Infect. Microbiol. 12, 1064010. doi: 10.3389/fcimb.2022.1064010
Jové, T., Da Re, S., Denis, F., Mazel, D., and Ploy, M. C. (2010). Inverse correlation between promoter strength and excision activity in class 1 integrons. PloS Genet. 6, e1000793. doi: 10.1371/journal.pgen.1000793
Kang, C. I., Kim, S. H., Kim, H. B., Park, S. W., Choe, Y. J., Oh, M. D., et al. (2003). Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin. Infect. Dis. 37, 745–751. doi: 10.1086/377200
Khan, M., Willcox, M. D. P., Rice, S. A., Sharma, S., and Stapleton, F. (2021). Development of antibiotic resistance in the ocular Pseudomonas aeruginosa clone ST308 over twenty years. Exp. Eye Res. 205, 108504. doi: 10.1016/j.exer.2021.108504
Li, X., Gu, N., Huang, T. Y., Zhong, F., and Peng, G. (2022). Pseudomonas aeruginosa: A typical biofilm forming pathogen and an emerging but underestimated pathogen in food processing. Front. Microbiol. 13, 1114199. doi: 10.3389/fmicb.2022.1114199
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–d665. doi: 10.1093/nar/gky1123
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–d917. doi: 10.1093/nar/gkab1107
Mohammadnejad, E., Seifi, A., Ghanei Gheshlagh, R., Aliramezani, A., Fattah Ghazi, S., Salehi, M., et al. (2023). Determine phenotypical patterns of resistance to antibiotics in COVID-19 patients with associated bacterial infection: largest medical center in Iran. Iran J. Microbiol. 15, 336–342. doi: 10.18502/ijm.v15i3.12893
Morgan, R. N., Saleh, S. E., Aboshanab, K. M., and Farrag, H. A. (2021). ADP-ribosyl transferase activity and gamma radiation cytotoxicity of Pseudomonas aeruginosa exotoxin A. AMB Express 11, 173. doi: 10.1186/s13568-021-01332-3
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nasrin, S., Hegerle, N., Sen, S., Nkeze, J., Sen, S., Permala-Booth, J., et al. (2022). Distribution of serotypes and antibiotic resistance of invasive Pseudomonas aeruginosa in a multi-country collection. BMC Microbiol. 22, 13. doi: 10.1186/s12866-021-02427-4
Nesvera, J., Hochmannová, J., and Pátek, M. (1998). An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol. Lett. 169, 391–395. doi: 10.1111/j.1574-6968.1998.tb13345.x
Pang, Z., Raudonis, R., Glick, B. R., Lin, T. J., and Cheng, Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31. doi: 10.1128/CMR.00088-17
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Rada, A. M., de la Cadena, E., Agudelo, C. A., Pallares, C., Restrepo, E., Correa, A., et al. (2021). Genetic Diversity of Multidrug-Resistant Pseudomonas aeruginosa Isolates Carrying bla (VIM-2) and bla (KPC-2) Genes That Spread on Different Genetic Environment in Colombia. Front. Microbiol. 12, 663020. doi: 10.3389/fmicb.2021.663020
Rao, L., de la Rosa, I., Xu, Y., Sha, Y., Bhattacharya, A., Holtzman, M. J., et al. (2021). Pseudomonas aeruginosa survives in epithelia by ExoS-mediated inhibition of autophagy and mTOR. EMBO Rep. 22, e50613. doi: 10.15252/embr.202050613
Rodríguez-Beltrán, J., DelaFuente, J., León-Sampedro, R., MacLean, R. C., and San Millán, Á. (2021). Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 19, 347–359. doi: 10.1038/s41579-020-00497-1
Silistre, H., Raoux-Barbot, D., Mancinelli, F., Sangouard, F., Dupin, A., Belyy, A., et al. (2021). Prevalence of exoY activity in pseudomonas aeruginosa reference panel strains and impact on cytotoxicity in epithelial cells. Front. Microbiol. 12, 666097. doi: 10.3389/fmicb.2021.666097
Song, J. M., Long, H. B., Ye, M., Yang, B. R., Wu, G. J., He, H. C., et al. (2024). Genomic characterization of a bla (KPC-2)-producing IncM2 plasmid harboring transposon ΔTn6296 in Klebsiella michiganensis. Front. Cell Infect. Microbiol. 14, 1492700. doi: 10.3389/fcimb.2024.1492700
Sun, F., Zhou, D., Wang, Q., Feng, J., Feng, W., Luo, W., et al. (2016). Genetic characterization of a novel blaDIM-2-carrying megaplasmid p12969-DIM from clinical Pseudomonas putida. J. Antimicrob. Chemother. 71, 909–912. doi: 10.1093/jac/dkv426
Thrane, S. W., Taylor, V. L., Freschi, L., Kukavica-Ibrulj, I., Boyle, B., Laroche, J., et al. (2015). The Widespread Multidrug-Resistant Serotype O12 Pseudomonas aeruginosa Clone Emerged through Concomitant Horizontal Transfer of Serotype Antigen and Antibiotic Resistance Gene Clusters. mBio 6, e01396–e01315. doi: 10.1128/mBio.01396-15
Varani, A. M., Siguier, P., Gourbeyre, E., Charneau, V., and Chandler, M. (2011). ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12, R30. doi: 10.1186/gb-2011-12-3-r30
Wang, J., Chen, Y. L., Li, Y. K., Chen, D. K., He, J. F., and Yao, N. (2021). Functions of sphingolipids in pathogenesis during host-pathogen interactions. Front. Microbiol. 12, 701041. doi: 10.3389/fmicb.2021.701041
Yigit, H., Queenan, A. M., Anderson, G. J., Domenech-Sanchez, A., Biddle, J. W., Steward, C. D., et al. (2001). Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Keywords: carbapenem-resistant Pseudomonas aeruginosa, blaKPC-2, blaVIM-2, plasmid, fitness cost
Citation: Zheng L, Wang Z, Zhang X, Lu G, Jing J, Sun S, Sun Y, Ji X, Jiang B, Zhu L and Guo X (2025) Genomic features and fitness cost of co-existence of blaKPC-2 and blaVIM-2 plasmids in ICU-derived pan-drug resistant Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 15:1617614. doi: 10.3389/fcimb.2025.1617614
Received: 24 April 2025; Accepted: 14 July 2025;
Published: 27 August 2025.
Edited by:
Valerio Baldelli, University of Milan, ItalyReviewed by:
Dao-Feng Zhang, Hohai University, ChinaRhishita Chourashi, University of Maryland, United States
Shijun Sun, First Affiliated Hospital of Zhengzhou University, China
Copyright © 2025 Zheng, Wang, Zhang, Lu, Jing, Sun, Sun, Ji, Jiang, Zhu and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Zheng, emwwNTA1MTRAMTYzLmNvbQ==; Lingwei Zhu, bGluZ3dlaXpAMTI2LmNvbQ==; Xuejun Guo, eHVlanVuZzIwMjFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Lin Zheng
Lin Zheng Zixian Wang1†
Zixian Wang1† Xuejun Guo
Xuejun Guo