- 1Key Laboratory of Animal Disease and Human Health of Sichuan Province, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2The Humanities Laboratory for Ecological Civilization and Environmental Governance of Zhejiang Province, Institute of Virology, Wenzhou University, Wenzhou, China
- 3Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China
- 4Wenzhou Municipal Center for Disease Control and Prevention, Wenzhou Municipal Institute of Health Supervision, Wenzhou, China
- 5Wenzhou Polytechnic, Wenzhou, China
Chikungunya virus (CHIKV) infection is often linked to acute interstitial nephritis (AIN) in fatal cases. Given the global spread of CHIKV and the lack of targeted antiviral treatments, there is an urgent need for effective therapeutic strategies against CHIKV-induced AIN. This study explored the therapeutic potential of salidroside (Sal) using an integrative approach involving network pharmacology, molecular docking and in vitro validation. Network pharmacology analysis identified 18 overlapping targets between Sal and AIN, including TNF, IL6 and AKT1. Molecular docking revealed strong binding affinities between Sal and key pathway proteins (Vina scores < –6), notably TNF, IL6 and BCL2. In vitro assays using CHIKV-infected 293T cells demonstrated that Sal (7.8125–2000 μM) enhanced cell viability by 8.9–25.9%, with the greatest effect observed at 1000 μM, without significantly altering viral replication. Mechanism analysis using the KEGG and FerrDB databases implicated apoptosis and ferroptosis in CHIKV-induced AIN pathogenesis. RT-qPCR analysis confirmed that Sal significantly downregulated ferroptosis-related genes (IL-6, IL-1β, SIRT1, PARP1, HMOX1) and apoptosis-associated markers (Bax, TNF-α, PARP1) in infected cells. Consistent with these findings, molecular docking demonstrated that Sal binds strongly to the ferroptosis-related protein GPX4 (Vina score: –6.3) and the apoptosis regulator NFKB1 (Vina score: –6.0). These results suggest that Sal is a promising therapeutic candidate for the treatment of CHIKV-induced AIN.
1 Introduction
Chikungunya virus (CHIKV) is a mosquito-borne arbovirus belonging to the genus Alphavirus (family Togaviridae) (Soumahoro et al., 2009). It is primarily transmitted to humans via infected Aedes aegypti and Aedes albopictus mosquitoes (Rougeron et al., 2015). The viral genome is a ~12 kb positive-sense single-stranded RNA that encodes four non-structural proteins (nsP1–nsP4) involved in replication, and three structural proteins (C, E1 and E2) essential for viral entry and assembly (Silva and Dermody, 2017).
CHIKV is endemic in more than half of the world’s regions, with over four million infections reported annually (Staples et al., 2009; Rougeron et al., 2015). Clinically, infection manifests as an acute febrile illness characterised by high fever, severe polyarthralgia, myalgia, headache and rash (Vairo et al., 2019). Although most patients recover within one to two weeks, approximately 30–40% develop chronic arthralgia that can persist for months or even years, contributing to significant morbidity and economic burden in endemic areas (Vu and Jungkind, 2017; Martins et al., 2020). The pathogenesis of CHIKV involves complex virus-host interactions. The virus infects dendritic cells, macrophages, fibroblasts and endothelial cells, eliciting a pronounced inflammatory response characterised by elevated levels of TNF-α, IL-6 and IL-1β (Mathew et al., 2017). Recent studies suggest that CHIKV can cause systemic complications, including renal dysfunction. In fatal cases, acute interstitial nephritis (AIN) is frequently observed and is believed to result from direct viral infection of renal cells, cytokine storm-induced injury and oxidative stress-mediated apoptosis (Joubert et al., 2012; Mercado et al., 2018; Aurore et al., 2021; Morel et al., 2024). However, no targeted therapies are currently available for treating AIN in CHIKV-infected individuals.
Salidroside (Sal), a phenylethanoid glycoside derived from Rhodiola rosea, possesses anti-inflammatory, antioxidant and immunomodulatory properties. It reduces oxidative stress by activating the Nrf2/HO-1 pathway and suppresses inflammation via inhibition of NF-κB and MAPK signalling pathways (Wang et al., 2019; Zhang et al., 2021). Moreover, Sal has demonstrated renoprotective effects in various disease models (Liu et al., 2024).
This study aims to evaluate the therapeutic potential of Sal against CHIKV-induced AIN using an integrative strategy. First, network pharmacology will be employed to identify key molecular targets and pathways implicated in CHIKV-associated AIN. Next, molecular docking will assess the binding interactions between Sal and critical host proteins. Finally, in vitro experiments using CHIKV-infected 293T cells will be conducted to investigate the cytoprotective effects of Sal and elucidate its underlying mechanisms.
2 Materials and methods
2.1 Cell culture, viral strain and compounds
293T and BHK-21 cell lines (China Center for Type Culture Collection, CCTCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/mL penicillin and 100 μg/mL streptomycin (Thermo Fisher Scientific). Cells were maintained at 37 °C in a humidified incubator with 5% CO2.
The CHIKV strain (GenBank: MT933041.1; provided by the Changchun Veterinary Research Institute) was stored at −80°C. Sal (MedChemExpress; CAS No. 10338-51-9; purity ≥99.9%) was dissolved in sterile distilled H2O to prepare a 100 mM stock solution.
2.2 Network pharmacology-based analysis
2.2.1 Targets acquisition
A total of 706 CHIKV-related targets were obtained from our previous study (Xin et al., 2025). Targets associated with acute interstitial nephritis (AIN) were retrieved from DisGeNET (https://disgenet.com), GeneCards (https://www.genecards.org) and OMIM (https://www.omim.org).
Sal-associated targets were identified by querying ‘Salidroside’ and ‘10338-51-9’ in TCMSPS (https://old.tcmsp-e.com/tcmsp.php), CTD (https://ctdbase.org), Swiss Target Prediction (http://www.swisstargetprediction.ch/), Herb (https://herb.ac.cn), with results filtered for Homo sapiens. All identified targets were consolidated and duplicate entries were removed.
2.2.2 Disease and disease-compound intersection target analysis
Overlapping targets between CHIKV-induced AIN and Sal were identified. Protein–protein interaction (PPI) networks were constructed using the STRING database (Version 12.0; confidence score ≥0.9; species: Homo sapiens) and visualised in Cytoscape (Version 3.9.1). Core targets were ranked using four topological algorithms implemented via the CytoNCA plugin in Cytoscape. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted on the intersection targets.
2.2.3 Molecular docking validation
The three-dimensional structures of target proteins were retrieved exclusively from the PDB (https://www.rcsb.org), focusing on Homo sapiens with (< 3 A) X-ray crystallography structures.
The two-dimensional structure of Sal was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Structure-based blind docking was performed using CB-DOCK2 (http://cadd.labshare.cn/cb-dock2/php/index.php). Docking results were evaluated based on binding affinity, with particular emphasis on complexes exhibiting the lowest Vina scores.
2.3 Experimental validation
2.3.1 Cytotoxicity assay
The 50% cytotoxic concentration (CC50) of Sal was determined in 293T cells seeded in 96-well plates. Cells were treated with varying Sal concentrations (15.625 to 4000 μM) in DMEM supplemented with 2% FBS for 48 h. Cell viability was assessed using the CCK-8 assay (Vazyme, A311-01, China).
2.3.2 Antiviral activity
293T cells were infected with CHIKV (MOI 0.01) in the presence of different Sal concentrations. After 24 h of incubation, cell viability was measured using the CCK-8 assay. The percentage of viability was calculated using the formula: [(As-Ab)/(Ac-Ab)] × 100, where As, Ab and Ac represent the sample, blank and control absorbances, respectively.
2.3.3 Viral replication
CHIKV-infected 293T cells (MOI 0.01) were treated with Sal (250–1000 μM) or left untreated as controls. After 24 h, viral RNA levels in the culture supernatants were quantified by RT-qPCR. Viral infectivity was further assessed by plaque assay on BHK-21 cells. Primer sequences are listed in Supplementary Table 1.
2.3.4 Proliferation assay
293T cells at 30% confluence were treated with Sal at concentrations ranging from 7.8125 to 2000 μM, or left untreated as controls and incubated for 48 h. Cell viability was subsequently assessed using the CCK-8 assay.
2.4 Analysis of ferroptosis and apoptosis
To investigate the involvement of apoptosis in CHIKV-induced AIN, apoptosis-related genes enriched in the KEGG pathway were randomly selected and analysed using RT-qPCR across treatment groups.
Ferroptosis targets from FerrDB (http://www.zhounan.org/ferrdb/current/) and compared with CHIKV-AIN and Sal-associated targets using Venn analysis. Core ferroptosis-related targets were identified using Cytoscape (v3.9.1). Molecular docking and RT-qPCR were employed to validate the predicted targets. Primer sequences are provided in Supplementary Table 1.
2.5 Statistical analysis
Statistical analyses were conducted using GraphPad Prism (v9.5). Unpaired t-tests were used for comparisons between two groups, and one-way ANOVA was applied for multiple group comparisons. A p-value < 0.05 was considered statistically significant.
3 Results
3.1 CHIKV-induced AIN involves multiple pathogenic pathways
The workflow for identifying targets associated with CHIKV-induced acute interstitial nephritis (AIN) is shown in Figure 1A. Venn analysis revealed 250 overlapping targets between 719 CHIKV-related and 1,435 AIN-related targets (Figure 1B). Core targets were ranked using four topological algorithms (Betweenness, Closeness, Degree and Eigenvector) within Cytoscape 3.9.1 and the CytoNCA plugin, identifying TNF, IL6 and IL1B as the top three nodes (Figure 1C). GO enrichment analysis indicated that these targets are primarily involved in inflammatory and immune responses, as well as the positive regulation of cell proliferation (Figure 1D). KEGG pathway analysis identified ferroptosis, the PI3K-Akt signalling pathway and apoptosis as key pathways implicated in CHIKV-induced AIN (Figure 1E).
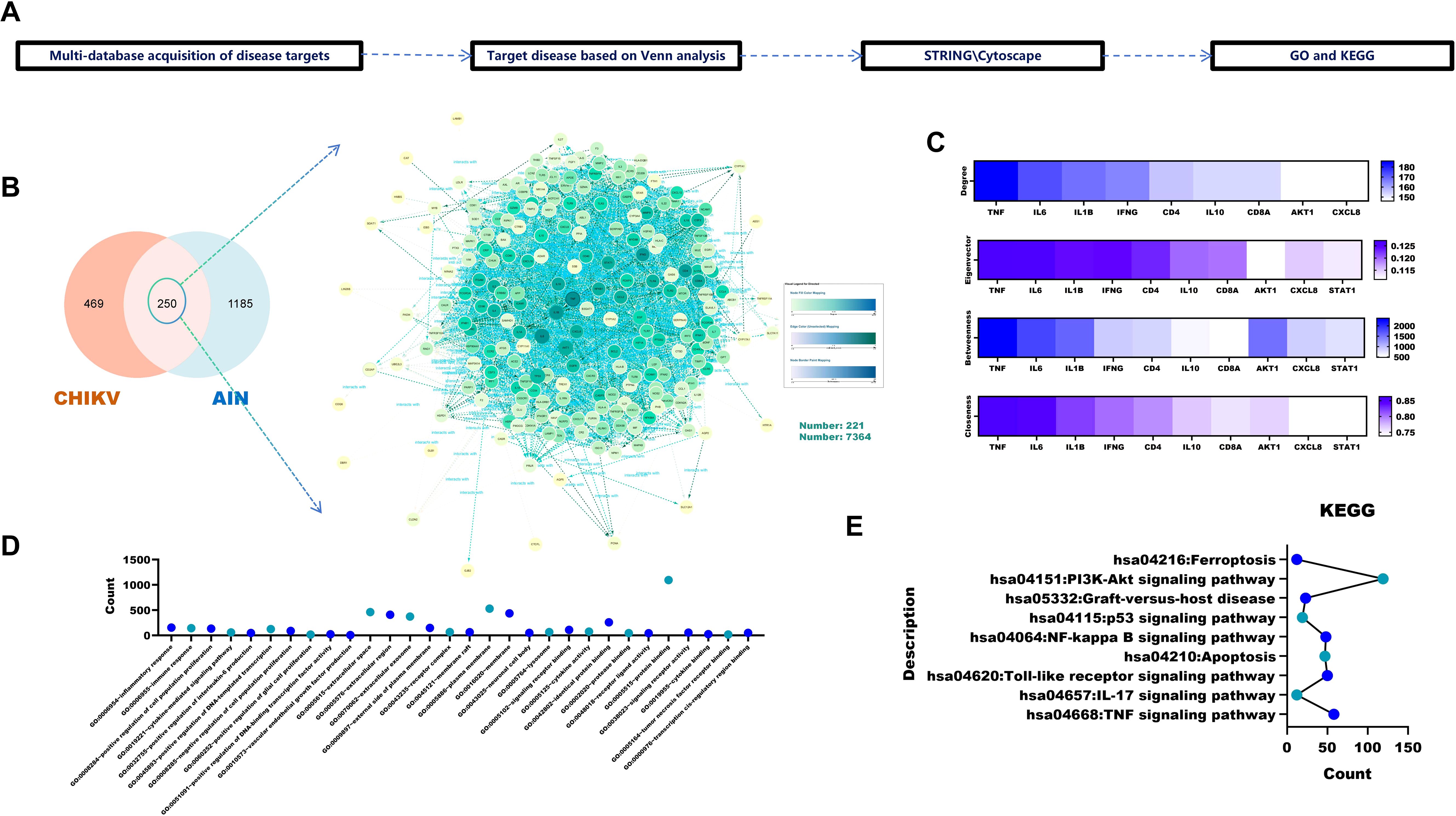
Figure 1. Pathogenic pathways and targets of CHIKV-induced AIN. (A) Acquisition and analysis of CHIKV-induced AIN targets. (B) Identification of CHIKV-induced targets via Venn analysis. (C) Analysis of core disease targets using four algorithms in Cytoscape 3.9.1 (D) GO analysis of core targets. (E) KEGG analysis of the core targets.
3.2 Sal has potential anti-AIN effects against CHIKV
To evaluate the potential role of Sal in treating CHIKV-induced AIN, network pharmacology analysis was conducted (Figure 2A). Venn analysis of 124 Sal-associated targets and 250 disease targets identified 18 overlapping targets, including TNF, IL6 and AKT1 (Figure 2B).
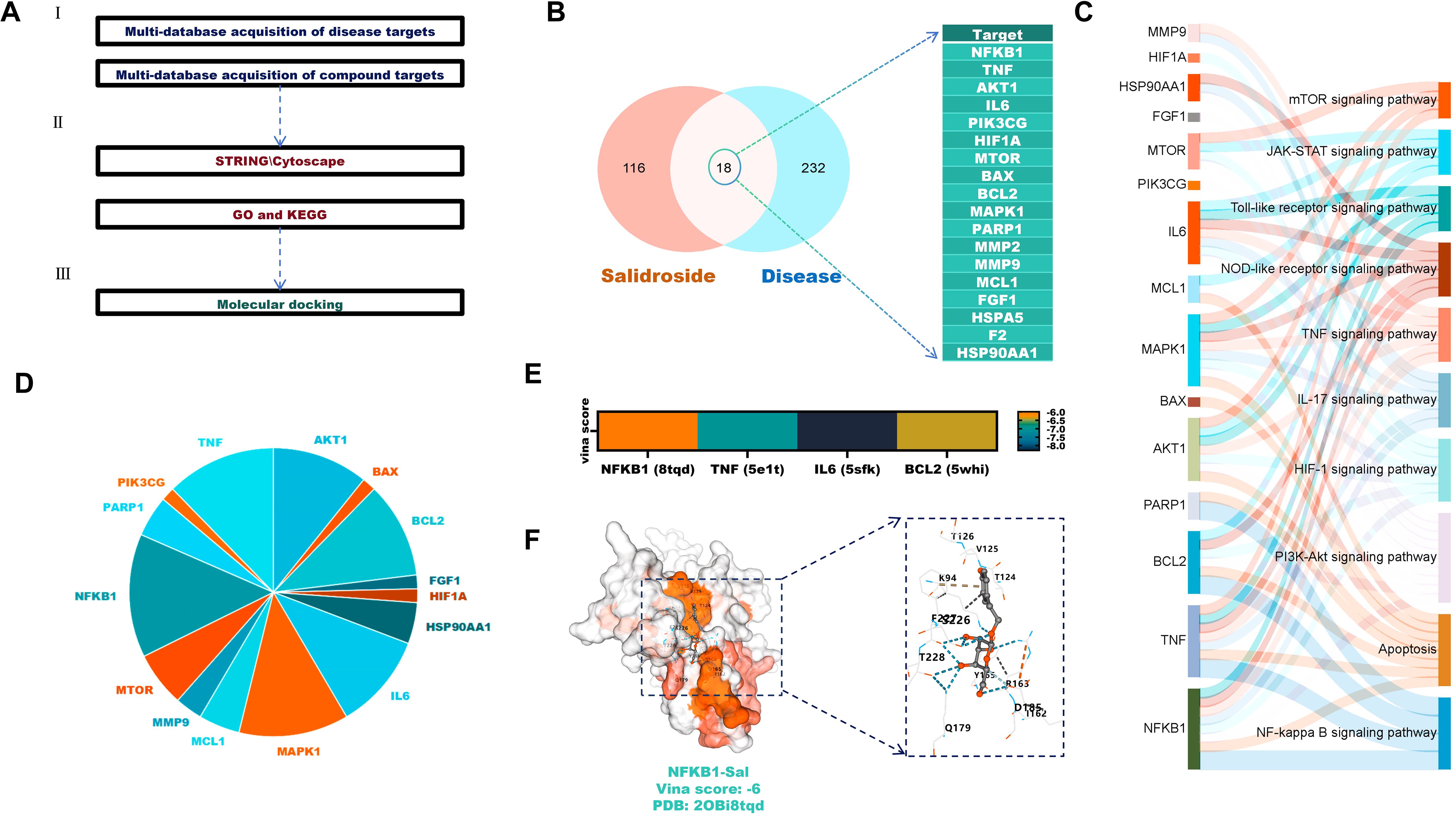
Figure 2. Anti-AIN potential of Sal against CHIKV. (A) Analysis of Sal’s disease-targeting effects (B) Venn analysis identifies overlapping targets between Sal and the disease. (C) KEGG analysis of overlapping targets. (D) Frequency analysis of the overlapping targets in KEGG. (E) Molecular docking verification. (F) Visualisation of the molecular docking between Sal and NFKB1 proteins.
KEGG enrichment analysis showed that these targets are primarily involved in the PI3K-Akt, apoptosis and NF-κB signalling pathways (Figure 2C). Frequency analysis highlighted NFKB1, TNF, IL6 and BCL2 as the most frequently involved targets (Figure 2D). Molecular docking confirmed strong binding affinities between Sal and these proteins, with Vina scores below -6 (Figures 2E, F).
3.3 Sal enhances the viability of CHIKV-infected 293T cells
The chemical structure of Sal is presented in Figure 3A. Cytotoxicity assays indicated that Sal was non-toxic to 293T cells at concentrations up to 2,000 μM (Figure 3B). Moreover, Sal (7.8125 μM–2,000 μM) promoted cell proliferation in a dose-dependent manner (Figure 3C).
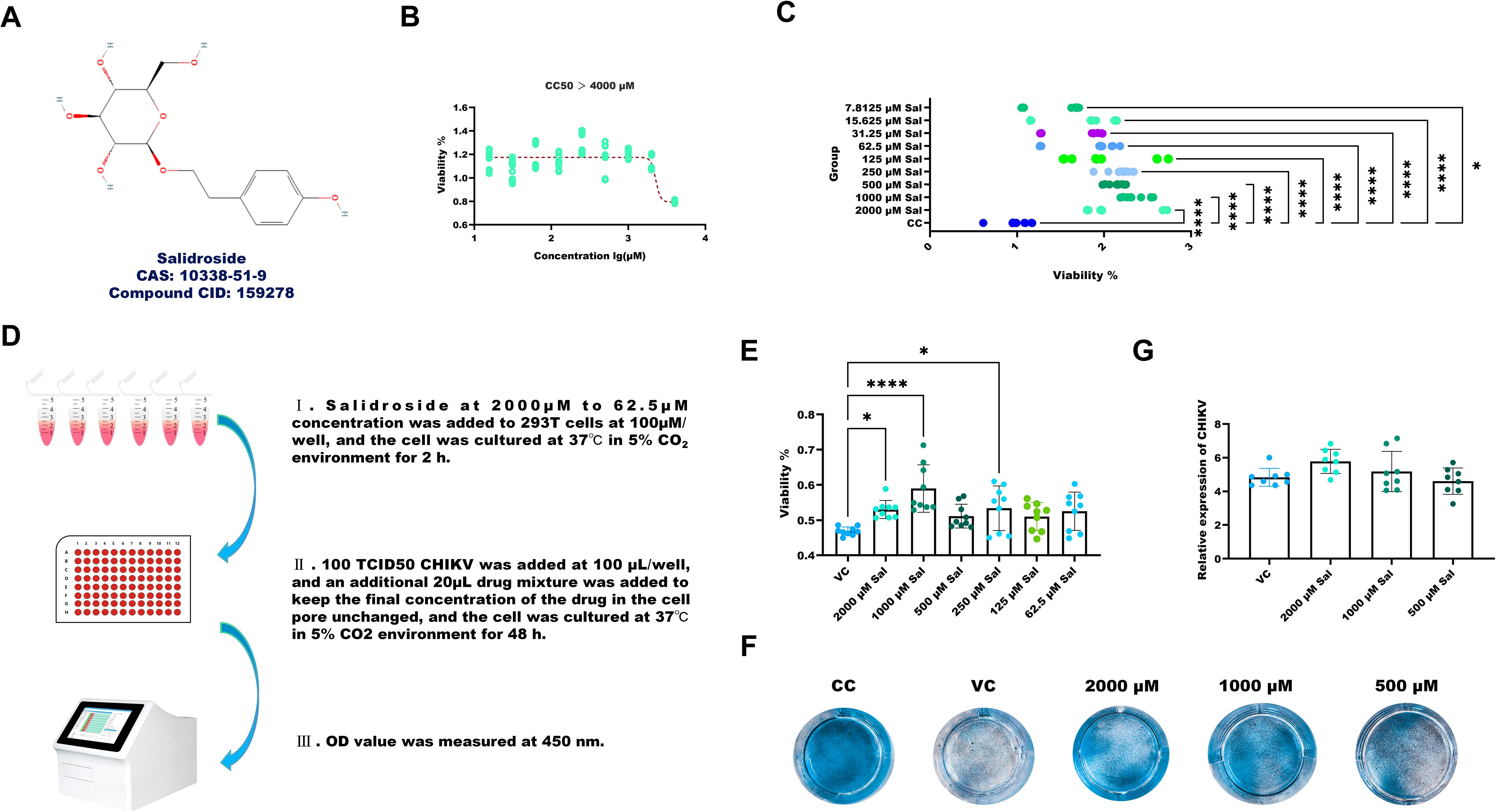
Figure 3. Sal enhances the viability of CHIKV-infected 293T cells. (A) Chemical structure of Sal. (B) Determination of CC50. (C) The results of Sal promoting the viability of 293T cells. (D) Experimental design for assessing the protective effects of Sal in CHIKV-infected 293T cells. (E) The results of Sal inhibiting CHIKV-induced cytotoxicity in 293T cells. (F) Crystal violet staining results. (G) Determination of CHIKV copy number in the supernatant of CHIKV-infected 293T cells treated with 1000 to 250 μM Sal. VC, Virus infection control; CC, Cell control. All values represent the mean ± SD. Compared with VC or CC, ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
The experimental setup for assessing the protective effects of Sal in CHIKV-infected 293T cells is illustrated in Figure 3D. Treatment with Sal (250–2,000 μM) significantly improved the viability of infected cells, as measured by CCK-8 assay (Figure 3E) and further supported by crystal violet staining (Figure 3F). However, RT-qPCR analysis showed that Sal did not reduce CHIKV RNA levels, indicating that its protective effects are not due to inhibition of viral replication (Figure 3G).
3.4 The anti-CHIKV-induced AIN effect of Sal is associated with ferroptosis and apoptosis
Venn analysis identified 109 overlapping targets between CHIKV-induced AIN and ferroptosis-related genes (Figure 4A). Core target analysis using Cytoscape 3.9.1 and the CytoNCA plugin identified TP53, IL6 and STAT3 as the top three key regulators (Figures 4B, C). Notably, 12 out of 18 (66.7%) predicted Sal targets against CHIKV-induced AIN overlapped with ferroptosis-associated genes (Figure 4D).
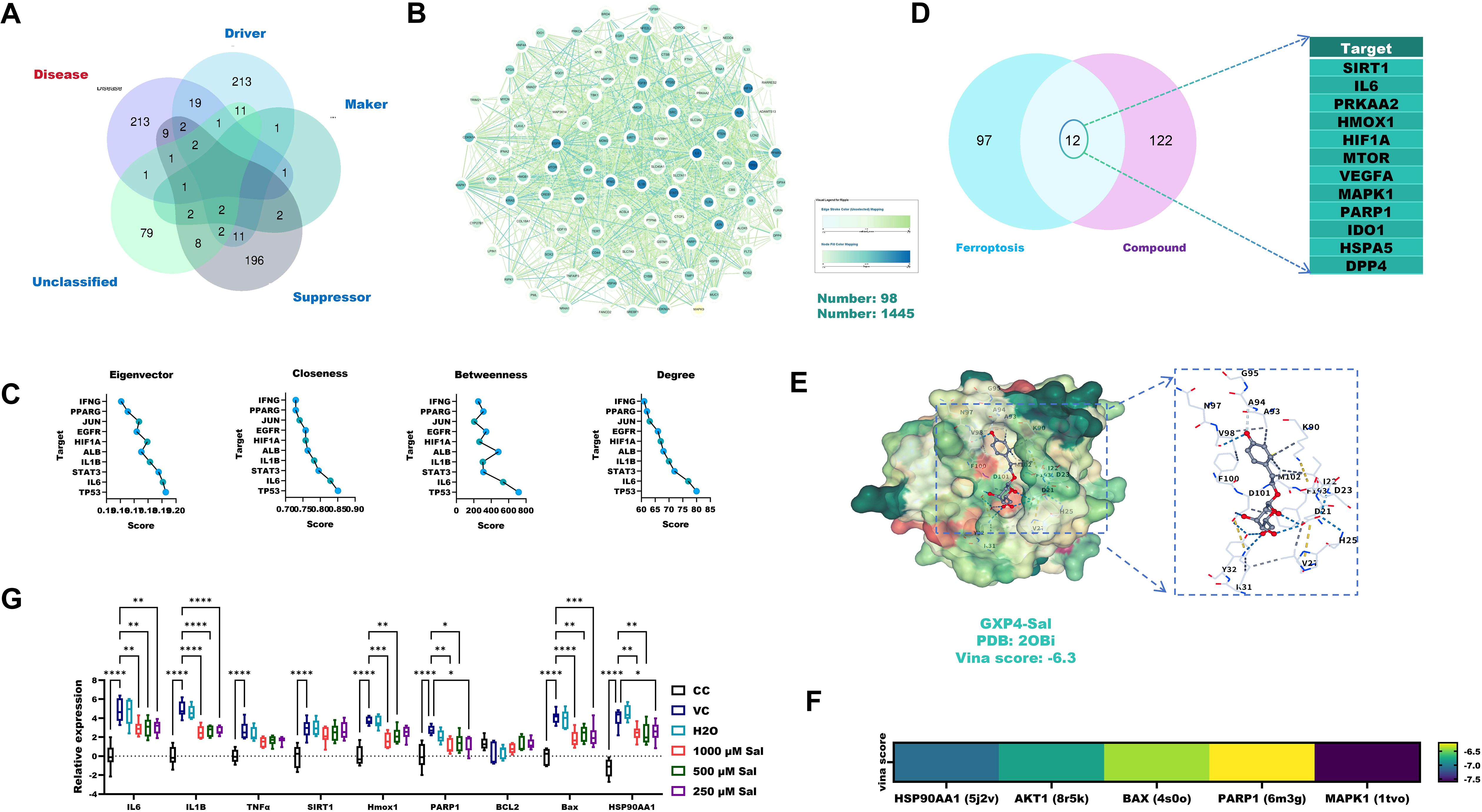
Figure 4. The anti-CHIKV-induced AIN effect of Sal is associated with ferroptosis and apoptosis. (A) Venn analysis identified overlapping targets between CHIKV-induced AIN and ferroptosis-related targets. (B) PPI analysis of overlapping targets. (C) Core target analysis of overlapping targets using Cytoscape 3.9.1 and CytoNCA. (D) Predicted targets of Sal against ferroptosis-related targets of CHIKV-induced AIN. (E) Molecular docking details between Sal and GPX4 proteins. (F) Molecular docking verification. (G) RT-qPCR verification. VC, Virus infection control; CC, Cell control. All values represent the mean ± SD. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
Molecular docking analysis revealed a strong binding affinity between Sal and GPX4 (Vina score: –6.3) (Figure 4E). Sal also exhibited high binding affinities (Vina scores < –6.5) with additional key targets, including HSP90AA1, AKT1, BAX, PARP1 and MAPK1 (Figure 4F). RT-qPCR analysis confirmed that Sal (250–1000 μM) significantly downregulated the expression of ferroptosis-related genes (IL-6, IL-1β, SIRT1, PARP1, HMOX1), apoptosis-associated markers (BAX, TNF-α, PARP1) and HSP90AA1 in CHIKV-infected 293T cells (Figure 4G).
4 Discussion
Renal impairment is a common and clinically significant complication in severe cases of Chikungunya virus (CHIKV) infection, with acute interstitial nephritis (AIN) identified as the predominant pathological manifestation (Mercado et al., 2018). Although the precise mechanisms underlying CHIKV-induced AIN remain incompletely understood, existing evidence implicates an excessive inflammatory response, particularly the upregulation of pro-inflammatory cytokines such as IL-1β, IL-6 and TNF-α during the acute phase of infection (Ng et al., 2009). Our findings support this hypothesis. Through network topological analysis, IL1B, IL6 and TNF emerged as central molecular targets associated with CHIKV-induced AIN. This prediction was validated in our in vitro model, where CHIKV-infected 293T cells exhibited significant transcriptional upregulation of IL-1β, IL-6 and TNF-α. Bioinformatic analysis further revealed that these cytokines are functionally interconnected with key signalling pathways, including NF-κB, PI3K-Akt and ferroptosis, suggesting a synergistic role in promoting renal injury.
Given the well-documented anti-inflammatory and renoprotective properties of Sal (Pu et al., 2020), we hypothesised that it might exert therapeutic effects against CHIKV-induced AIN. This was evaluated using our established 293T CHIKV infection model. Sal treatment (7.8–2000 μM) significantly improved cell viability in infected cells, with the most pronounced protective effect observed at 1,000 μM. Integrating network pharmacology and experimental data, we identified HSP90AA1 as a key target of Sal. RT-qPCR analysis confirmed that Sal (1000 μM) significantly downregulated HSP90AA1 expression in CHIKV-infected cells. As HSP90AA1 is known to stabilise the CHIKV non-structural protein NSP2 and facilitate viral replication (Nam et al., 2021), its inhibition was expected to reduce viral load. However, contrary to this expectation, quantitative RT-qPCR showed no significant difference in viral RNA levels between Sal-treated and untreated cells. This apparent paradox suggests that while Sal may disrupt HSP90AA1-mediated support of viral replication, compensatory mechanisms might maintain viral propagation. These could involve alternative host pathways that offset the antiviral impact of HSP90AA1 inhibition.
Several mechanisms may explain this unexpected finding. First, our network pharmacology analysis indicated that Sal is involved in the PI3K/Akt signalling pathway, which has been shown to facilitate CHIKV replication (Thaa et al., 2015). This is particularly noteworthy, as previous studies have demonstrated Sal’s ability to activate PI3K/Akt signalling to promote cell survival and tissue repair in various pathological contexts (Hou et al., 2023). By activating this pathway, Sal may inadvertently create a cellular environment conducive to CHIKV replication. However, the effect of Sal on the PI3K/Akt pathway appears to be context-dependent. For instance, in CoCl2-induced HT22 cells, Sal enhances mitochondrial function via activation of the PI3K-Akt-MAPK pathway (Hou et al., 2023). Conversely, in human gastric cancer AGS cells, Sal promotes apoptosis and protective autophagy through the PI3K/Akt/mTOR axis (Rong et al., 2020). Therefore, its precise impact on PI3K/Akt signalling in CHIKV-infected 293T cells requires further experimental validation. Second, our study confirms Sal’s anti-apoptotic effects. RT-qPCR analysis demonstrated that Sal significantly downregulated the expression of key apoptotic markers, including TNF-α, PARP1 and BAX. Molecular docking provided structural support for this activity, revealing a strong binding affinity between Sal and NFKB1 (Vina score: -6.0 kcal/mol). In addition, although CHIKV infection has not been widely associated with ferroptosis, our integrated analysis identified 109 CHIKV-AIN-related targets linked to ferroptosis pathways. Experimental validation showed that Sal downregulated several ferroptosis-associated genes in CHIKV-infected 293T cells, including IL-6, HMOX1 and SIRT1. Docking analysis also confirmed Sal’s interaction with the ferroptosis regulator GPX4 (Vina score: -6.3 kcal/mol). By simultaneously inhibiting apoptosis and ferroptosis, Sal may inadvertently promote the survival of infected cells, potentially allowing CHIKV more time to replicate. This interpretation aligns with previous reports suggesting that pharmacological inhibition of apoptosis can enhance CHIKV production by prolonging host cell viability (Nghia et al., 2020). In this context, the extended survival of infected cells could compensate for the reduced replication efficiency resulting from HSP90AA1 downregulation, thereby sustaining overall viral output.
In conclusion, this study suggests that Sal holds promise as a potential therapeutic agent for CHIKV-induced AIN. However, as these findings are based solely on in vitro experiments, further in vivo investigations are necessary to validate its efficacy and elucidate its mechanisms of action in a physiological context.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
SC: Writing – review & editing, Methodology, Software, Writing – original draft. JX: Software, Methodology, Writing – original draft, Writing – review & editing. TZ: Methodology, Writing – review & editing, Software, Writing – original draft. YZ: Writing – review & editing, Investigation, Writing – original draft. CJ: Writing – original draft, Investigation, Writing – review & editing. LK: Writing – original draft, Investigation, Writing – review & editing. XZ: Investigation, Writing – original draft, Writing – review & editing. HZ: Resources, Writing – review & editing, Writing – original draft, Validation. WW: Writing – original draft, Resources, Writing – review & editing, Supervision. XL: Writing – review & editing, Supervision, Writing – original draft, Resources.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Wenzhou Scientific Research Project (S20240001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1623860/full#supplementary-material
Supplementary Table 1 | Primer pairs used for target validation.
References
Aurore, A. C., Couderc, T., Dueymes, J. M., Deligny, C., Lecuit, M., Molinié, V., et al. (2021). The clinicopathological spectrum of kidney lesions in chikungunya fever: A report of 5 cases with kidney biopsy. Am. J. Kidney Dis. 78, 902–906. doi: 10.1053/j.ajkd.2021.04.012
Hou, Y., Zhang, Y., Jiang, S., Xie, N., Zhang, Y., Meng, X., et al. (2023). Salidroside intensifies mitochondrial function of CoCl2-damaged HT22 cells by stimulating PI3K-AKT-MAPK signaling pathway. Phytomedicine. 109, 154568. doi: 10.1016/j.phymed.2022.154568
Joubert, P. E., Werneke, S., de la Calle, C., Guivel-Benhassine, F., Giodini, A., Peduto, L., et al. (2012). Chikungunya-induced cell death is limited by ER and oxidative stress-induced autophagy. Autophagy. 8, 1261–1263. doi: 10.4161/auto.20751
Liu, Q., Chen, J., Zeng, A., and Song, L. (2024). Pharmacological functions of salidroside in renal diseases: facts and perspectives. Front. Pharmacol. 14. doi: 10.3389/fphar.2023.1309598
Martins, D. O. S., Santos, I. A., de Oliveira, D. M., Grosche, V. R., and Jardim, A. C. G. (2020). Antivirals against chikungunya virus: is the solution in nature? Viruses 12, 272. doi: 10.3390/v12030272
Mathew, A. J., Ganapati, A., Kabeerdoss, J., Nair, A., Gupta, N., Chebbi, P., et al. (2017). Chikungunya infection: a global public health menace. Curr. Allergy Asthma Rep. 17, 13. doi: 10.1007/s11882-017-0680-7
Mercado, M., Acosta-Reyes, J., Parra, E., Guzmán, L., Beltrán, M., Gasque, P., et al. (2018). Renal involvement in fatal cases of chikungunya virus infection. J. Clin. Virol. 103, 16–18. doi: 10.1016/j.jcv.2018.03.009
Morel, Z., Martínez, T., Galeano, F., Coronel, J., Quintero, L., Jimenez, R., et al. (2024). Cytokine storm in Chikungunya: Can we call it multisystem inflammatory syndrome associated with Chikungunya? Reumatol Clin. (Engl Ed) 20, 223–225. doi: 10.1016/j.reumae.2024.04.005
Nam, S., Ga, Y. J., Lee, J. Y., Hwang, W. Y., Jung, E., Shin, J. S., et al. (2021). Radicicol inhibits chikungunya virus replication by targeting nonstructural protein 2. Antimicrob. Agents Chemother. 65, e0013521. doi: 10.1128/AAC.00135-21
Ng, L. F. P., Chow, A., Sun, Y.-J., Kwek, D. J. C., Lim, P.-L., Dimatatac, F., et al. (2009). IL-1beta, IL-6, and RANTES as biomarkers of chikungunya severity. PloS One 4, e4261. doi: 10.1371/journal.pone.0004261
Nghia, V. X., Giang, N. V., Canh, N. X., Ha, N. H., Duong, N. T., Hoang, N. H., et al. (2020). Stimulation of dendritic cell functional maturation by capsid protein from chikungunya virus. Iran J. Basic Med. Sci. 23, 1268–1274. doi: 10.22038/IJBMS.2020.40386.9558
Pu, W. L., Zhang, M. Y., Bai, R. Y., Sun, L. K., Li, W. H., Yu, Y. L., et al. (2020). Anti-inflammatory effects of Rhodiola rosea L.: A review. BioMed. Pharmacother. 121, 109552. doi: 10.1016/j.biopha.2019.109552
Rong, L., Li, Z., Leng, X., Li, H., Ma, Y., Chen, Y., et al. (2020). Salidroside induces apoptosis and protective autophagy in human gastric cancer AGS cells through the PI3K/Akt/mTOR pathway. BioMed. Pharmacother. 122, 109726. doi: 10.1016/j.biopha.2019.109726
Rougeron, V., Sam, I. C., Caron, M., Nkoghe, D., Leroy, E., and Roques, P. (2015). Chikungunya, a paradigm of neglected tropical disease that emerged to be a new health global risk. J. Clin. Virol. 64, 144–152. doi: 10.1016/j.jcv.2014.08.032
Silva, L. A. and Dermody, T. S. (2017). Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Invest. 127, 737–749. doi: 10.1172/JCI84417
Soumahoro, M. K., Gérardin, P., Boëlle, P. Y., Perrau, J., Fianu, A., Pouchot, J., et al. (2009). Impact of Chikungunya virus infection on health status and quality of life: a retrospective cohort study. PloS One 4, e7800. doi: 10.1371/journal.pone.0007800
Staples, J. E., Breiman, R. F., and Powers, A. M. (2009). Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin. Infect. Dis. 49, 942–948. doi: 10.1086/605496
Thaa, B., Biasiotto, R., Eng, K., Neuvonen, M., Götte, B., Rheinemann, L., et al. (2015). Differential phosphatidylinositol-3-kinase-akt-mTOR activation by semliki forest and chikungunya viruses is dependent on nsP3 and connected to replication complex internalization. J. Virol. 89, 11420–11437. doi: 10.1128/JVI.01579-15
Vairo, F., Haider, N., Kock, R., Ntoumi, F., Ippolito, G., and Zumla, A. (2019). Chikungunya: epidemiology, pathogenesis, clinical features, management, and prevention. Infect. Dis. Clin. North Am. 33, 1003–1025. doi: 10.1016/j.idc.2019.08.006
Vu, D. M. and Jungkind, D. (2017). Angelle desiree laBeaud. Chikungunya virus. Clin. Lab. Med. 37, 371–382. doi: 10.1016/j.cll.2017.01.008
Wang, J., Pan, Y., Cao, Y., Zhou, W., and Lu, J. (2019). Salidroside regulates the expressions of IL-6 and defensins in LPS-activated intestinal epithelial cells through NF-κB/MAPK and STAT3 pathways. Iran J. Basic Med. Sci. 22, 31–37. doi: 10.22038/ijbms.2018.26994.6602
Xin, J., Song, X., Zheng, H., Li, W., Qin, Y., Wang, W., et al. (2025). Exploring the antiviral potential of shikimic acid against Chikungunya virus through network pharmacology, molecular docking, and In vitro experiments. Front. Vet. Sci. 12. doi: 10.3389/fvets.2025.1524812
Zhang, Q., Liu, J., Duan, H., Li, R., Peng, W., and Wu, C. (2021). Activation of Nrf2/HO-1 signaling: An important molecular mechanism of herbal medicine in the treatment of atherosclerosis via the protection of vascular endothelial cells from oxidative stress. J. Adv. Res. 34, 43–63. doi: 10.1016/j.jare.2021.06.023
Keywords: CHIKV, salidroside, network pharmacology, molecular docking, ferroptosis, apoptosis
Citation: Cheng S, Xin J, Zhang T, Zhang Y, Ji C, Kang L, Zhu X, Zhang H, Wang W and Liao X (2025) Assessing the potential impact of salidroside on Chikungunya virus-induced acute interstitial nephritis via network pharmacology, molecular docking and in vitro experiments. Front. Cell. Infect. Microbiol. 15:1623860. doi: 10.3389/fcimb.2025.1623860
Received: 06 May 2025; Accepted: 30 June 2025;
Published: 22 July 2025.
Edited by:
He Zhang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Changqing Yu, Yibin Vocational and Technical College, ChinaYuancheng Zhou, Sichuan Animal Science Academy, China
Mixia Cao, Anhui Science and Technology University, China
Copyright © 2025 Cheng, Xin, Zhang, Zhang, Ji, Kang, Zhu, Zhang, Wang and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinfei Liao, eGluZmVpX2xpYW9AMTI2LmNvbQ==; Wei Wang, d3dreTExMDFAMTI2LmNvbQ==; He Zhang, aGV6aGFuZ3ZzQDEyNi5jb20=; Xiangyu Zhu, emh1eGlhbmd5dTAwQDEyNi5jb20=
†These authors share first authorship
 Sheng Cheng
Sheng Cheng Jialiang Xin
Jialiang Xin Tianran Zhang
Tianran Zhang Yulin Zhang2,3†
Yulin Zhang2,3† Xiangyu Zhu
Xiangyu Zhu He Zhang
He Zhang Wei Wang
Wei Wang Xinfei Liao
Xinfei Liao