- 1Jilin Collaborative Innovation Center for Antibody Engineering, Jilin Medical University, Jilin, China
- 2Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Changchun, China
- 3Institute of Virology, Wenzhou University, Wenzhou, China
Introduction: The newly emerged Omicron subvariants demonstrate resistance to current therapeutic antibodies and an enhanced ability to evade the vaccine-induced immune responses. Among them, JN.1 sublineages are considered highly immune-evasive, underscoring the urgent need for broadly protective vaccines. Ferritin nanoparticles, with their unique hollow nanocage structure, provide an efficient antigen-display platform for next-generation vaccine development.
Methods: Based on the previously constructed Delta-6P-S recombinant protein vaccine with broad-spectrum protective effects, this study optimized the S protein structure displayed on the surface of ferritin nanoparticles by comparing the immune responses induced in C57BL/6J mice.
Results: Delta-4S1158 nanoparticles, containing a truncated S-6P structure with four additional mutation sites, elicited robust S-specific immunoglobulin G (IgG), potent neutralizing antibodies, and a Th2-biased T-cell response in C57BL/6J mice, demonstrating favorable immunogenicity and safety. The JN.1-4S1158 nanoparticles, based on this structural design, induced a strong cross-neutralizing antibody response in C57BL/6J mice and conferred effective protection against Omicron BA.5, XBB, and JN.1 variants. Vaccinated mice exhibited significantly reduced viral genomic loads in trachea and lung tissues compared to controls, with no infectious virus detected. Lung tissue pathology was minimal in vaccinated mice.
Conclusion: The JN.1-4S1158 nanoparticle vaccine demonstrates broad-spectrum protective effects against Omicron subvariants and shows potential for further development. It also provides a basis for the development of a universal SARS-CoV-2 vaccine.
1 Introduction
Although the emergency phase of the COVID-19 pandemic has been declared over by the World Health Organization (Wang et al., 2024), SARS-CoV-2 continues to evolve and spread (Wang and Guo, 2023). Since the emergence of the Omicron variant, it has spread rapidly worldwide, giving rise to numerous subvariants that have caused widespread infection (Fahrbach et al., 2025; Wasim et al., 2025). Vaccination has long been an effective strategy to combat infectious disease outbreaks (Mambelli and de Araujo, 2025; Moore et al., 2025). Although various vaccine platforms have achieved different degrees of immune protection, they also exhibit inherent limitations (Pischel et al., 2024; Widyaningsih and Hakim, 2024).
Advances in SARS-CoV-2 structural biology and gene delivery technologies offer new opportunities to develop vaccines capable of conferring broader protection against diverse variants (Chen and Farzan, 2025; Holmes, 2024). Recent studies increasingly demonstrate that nanoparticle-mediated antigen delivery systems can significantly enhance immune responses compared with monomeric antigens (Chen et al., 2025; Sun et al., 2025; Wu et al., 2025). Additionally, nanoparticle vaccines have a favorable safety profile and can stimulate robust humoral and cellular immune responses, particularly strong Th2-biased immunity (Belghith et al., 2025; Chiba et al., 2024; Lin et al., 2025). Among these platforms, ferritin-based nanoparticles show particular promise as vaccine carriers (Cao et al., 2024; Chen et al., 2024). Ferritin is a naturally occurring spherical nanocage composed of 24 symmetrically arranged subunits with a conserved structure across species (Ahmadivand and Fux, 2024; Lucignano and Ferraro, 2024). Studies have shown that ferritin nanoparticle vaccines induce T follicular helper cell responses that are three to four times higher than those of traditional subunit vaccines, while maintaining an excellent safety profile (Wang et al., 2023; Widge and Hofstetter, 2023). Therefore, ferritin nanoparticle vaccines represent a promising design strategy with substantial research potential (Yang et al., 2024).
We previously reported a candidate vaccine, Delta-6P-S, with broad-spectrum protective effects (Feng et al., 2023). This immunogen incorporates a T4 fibrin trimerization tag and six proline substitutions, stabilizing the spike protein in its pre-fusion conformation. It exhibited excellent immunogenicity and cross-neutralization activity against wild-type, Beta, Delta, and Omicron variants in C57BL/6J mice and golden hamsters. It protected the hamsters against Delta and Omicron challenges. In the present study, we designed structural variants based on this immunogen, displayed them on the surface of ferritin nanoparticles, and evaluated the induced immune response in C57BL/6J mice. After identifying the spike protein structure most suitable for ferritin display, we developed a nanoparticle vaccine, “JN.1-4S1158”, based on the Omicron JN.1 spike sequence. This vaccine demonstrated strong immunogenicity and cross-neutralization activity against Omicron BA.5, XBB, and JN.1 variants in C57BL/6J mice and conferred protection against challenge with these variants.
2 Materials and methods
2.1 Cell culture
CHO-S cells were purchased from Thermo Fisher Scientific and cultured in ExpiCHO Expression Medium (Thermo Fisher Scientific, USA). Vero-E6 cells were cryopreserved in our laboratory and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific, USA) and 1% penicillin–streptomycin. The cells were maintained in a 37°C incubator.
2.2 Animals and viruses
Specific pathogen-free (SPF) 6-week-old female C57BL/6J mice (18–20 g) were obtained from Beijing Charles River Laboratory Animal Technology Co., Ltd. The authentic Delta (CSTR.16698.06.NPRC6.CCPM-B-V-049-2105-6), BA.5 (SARS-CoV-2 strain Omicron CoV/human/CHN_CVRI-12/2022), XBB (SARS-CoV-2 strain Omicron CoV/human/CHN_CVRI-10/2023), and JN.1 (SARS-CoV-2 strain Omicron CoV/human/CHN_CVRI-05/2024) strains were isolated from patients with COVID-19. All virus experiments were conducted in a Biosafety Level 3 laboratory with standard operating procedures.
2.3 Design and preparation of ferritin nanoparticles
The Helicobacter pylori ferritin sequence was obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). The spike protein sequences of Delta and Omicron JN.1 variants were designed according to mutation sites reported by the World Health Organization (https://www.who.int/). Six proline substitutions were introduced at residues 817, 892, 899, 942, 986, and 987. Four additional mutations (P1143S, F1148I, Y1155I, and F1156H) were also introduced. The furin cleavage site (682-RRAR-686) was replaced with 682-GSAS-686. The construct also contained an N-terminal Kozak sequence and signal peptide, and a C-terminal T4 fibritin trimerization tag fused to H. pylori ferritin. Ferritin nanoparticles were expressed in CHO-S cells cultured in ExpiCHO Expression Medium at 37°C with agitation at 125 rpm in a humidified atmosphere containing 8% CO2. Nanoparticles were purified by affinity chromatography using Strep-Tactin®XT (IBA Life Sciences, Germany) with a gravity-flow column. The target proteins were eluted with Strep-Tactin®XT Elution Buffer (IBA Life Sciences, Germany) and resuspended in phosphate-buffered saline (PBS) for storage at −80°C.
2.4 Western blot
Ferritin nanoparticle samples were mixed with an appropriate amount of 5× loading buffer, heated at 100°C for 10min, and separated on a 7.5% polyacrylamide Tris-glycine gel for 2h at 80V. Then, the immunoblotting was transferred to the NC membrane for 90min at 100V. The membrane was sealed overnight at 4°C in TBS and 0.1% Tween 20 (TBST) with 5% bovine serum albumin (BSA) and then incubated at room temperature with a 1:3,000 dilution of the RBD pAb and strep-tag mAb (Thermo Fisher Scientific, USA) for 2.5h. After four washes with TBST, the membranes were incubated for 1h at room temperature with the horseradish peroxidase (HRP)-labeled goat anti-mouse immunoglobulin G (IgG) (Beyotime, China, 1:5,000 dilution). The membranes were developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, USA), and images were acquired with Amersham Imager 600 (General Electric Company, USA).
2.5 Immunization of C57BL/6J mice
To compare immune responses induced by different nanoparticle designs, 25 female C57BL/6J mice were divided into five groups (n = 5). Mice were immunized with three doses of Delta-6P-S, Delta-S1208, Delta-S1158, or Delta-4S1158 nanoparticle vaccines at weeks 3, 6, and 9. Control groups received alum alone. Tail-vein blood was collected every 3 weeks, and spleens were harvested 3 weeks after the final immunization.
To evaluate the immunogenicity of JN.1-4S1158, 30 female C57BL/6J mice were divided into two groups (n = 15). Mice in the immunized group received three doses of JN.1-4S1158 nanoparticles at weeks 3, 6, and 9, while controls received alum. Tail-vein blood was collected every 3 weeks.
2.6 Omicron variant challenge in C57BL/6J mice
Three groups of JN.1-4S1158-immunized and three groups of alum-treated C57BL/6J mice (n=5 per group) were anesthetized with 3% isoflurane inhalation and inoculated intranasally with 100 μL of 104.75 TCID50/mL Omicron BA.5, XBB, and JN.1 variants. Mice were euthanized by cervical dislocation 4 days post-challenge (dpi). Lungs and trachea were harvested for measurement of viral titers and gene copies at 4 dpi. Lung pathology was compared between vaccinated and control groups.
2.7 ELISA
The S-specific IgG antibody ELISA (enzyme-linked immunosorbent assay) was performed as described earlier. Plates coated with spike proteins were used (Sangon Biotech, China). Serial dilutions of serum were added for incubation for 50min at 37°C. After washing, HRP‐conjugated goat anti-mouse IgG antibodies (ZSGB-BIO, China) diluted 1:10,000 with PBS were added and incubated for 30min at 37°C. TMB Single-Component Substrate solution was used for color development. The reaction was terminated using 2% H2SO4, and the optical density (OD) was measured at 450 nm.
2.8 Neutralization of Omicron live virus and determination of TCID50
Neutralization assays were performed with Omicron BA.5, XBB, and JN.1 variants. Diluted sera were incubated with 100 TCID50 of live Omicron virus at 37°C for 1h, then added to Vero-E6 cells (5 × 103/well). Cultures were incubated for 4 days, and cytopathic effects (CPEs) were assessed microscopically. Neutralization titers were reported as the highest serum dilution achieving 50% neutralization.
For TCID50 determination, Vero-E6 cells (5 × 10³/well) were seeded in 96-well plates overnight. Serially diluted tissue homogenates (100 µL/well) were added and incubated for 4 days. CPE was recorded using a microscope.
2.9 Quantitative real-time PCR
The HiScript II U+ One Step qRT-PCR (quantitative real-time polymerase chain reaction) Probe Kit (Vazyme, China) was used for qPCR of viral RNA detection. Primers and probes targeting the N gene and sgE gene of SARS-CoV-2 were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Viral gene loads were quantified by TaqMan qRT-PCR as previously described (Li et al., 2022).
2.10 Flow cytometry
To analyze T-cell response (CD3+CD4+ and CD3+CD8+) and cytokine production, 2 × 106 splenic lymphocytes from each mouse were stimulated in 24-well plates with 10 μg/well SARS-CoV-2 spike peptide pools (Sino Biological, China) for 12h. Cells were stained on ice with surface antibodies, including PE/Cyanine7 anti-mouse CD3ϵ Antibody (BioLegend, USA), FITC anti-mouse CD4 Antibody (BioLegend, USA), and APC anti-mouse CD8a Antibody (BioLegend, USA). After washing, cells were fixed and permeabilized (BD, USA) for 30min, washed in 1×Perm/Wash™ buffer, and stained intracellularly with PE anti-mouse IFN-γ Antibody (BioLegend, USA) and PE anti-mouse IL-4 Antibody (BioLegend, USA). Data were acquired on a Beckman flow cytometer.
2.11 Statistical analysis
GraphPad Prism v.8.0.2 was used for all statistical analyses, and intergroup differences were compared by multiple t-tests. Antibody titer data were log-transformed before analysis. Correlations were assessed by Spearman rank-correlation tests.
3 Results
3.1 Design, expression, and purification of ferritin nanoparticles
Based on the structure of the previously developed “Delta-6P-S” candidate vaccine (stabilized in the pre-fusion conformation), we designed four immunogens for evaluation: (1) “Delta-6P-S” (reference candidate); (2) Delta-S1208: the original design linked to H. pylori ferritin; (3) Delta-S1158: derived from Delta-S1208 by truncating the S protein, removing the helix region, while retaining the T4 fibritin trimer tag and six proline mutations; and (4) Delta-4S1158: derived from Delta-S1158 with four additional amino acid substitutions (P1143S, F1148I, Y1155I, and F1156H), as described by Joyce et al. (2021) (Figure 1A). The sequences were codon-optimized, cloned into the pSN vector, transfected into CHO-S, and purified by Strep-Tactin®XT affinity chromatography. Western blot analysis with RBD-pAb and Strep-tag mAb confirmed protein expression. The purified ferritin nanoparticles migrated at 180 kDa (Figures 1B–D). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) showed a single band (Figures 1E–G). Transmission electron microscopy revealed nanoparticles with uniform size and morphology (Figures 2A–C).
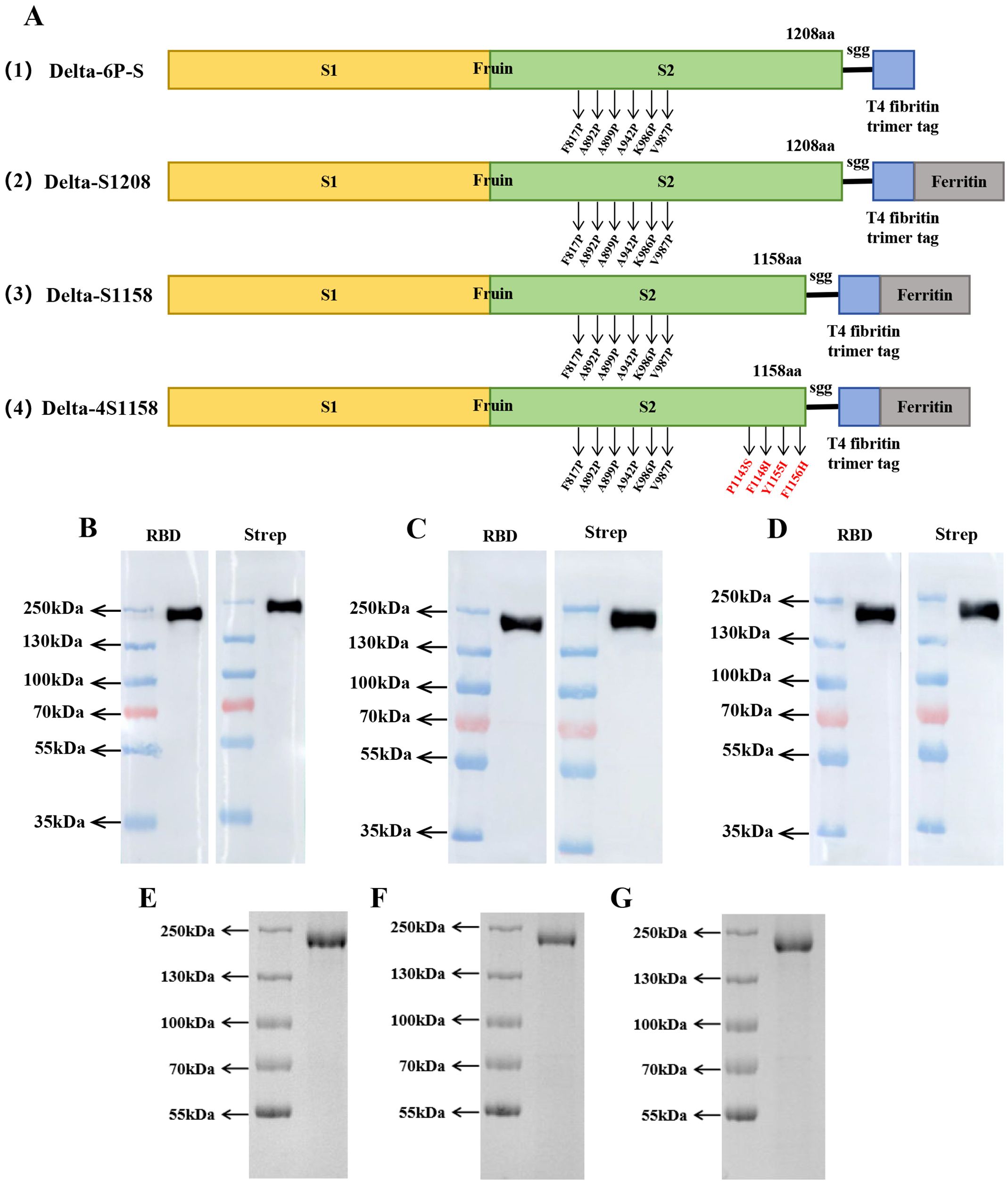
Figure 1. Immunogenic design and purification of ferritin nanoparticles. (A) Delta-6P-S recombinant protein, Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticle design and schematic diagram. (B–D) Characterization of Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticles by Western blot. (E–G) SDS-PAGE gel showing Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticle bands.
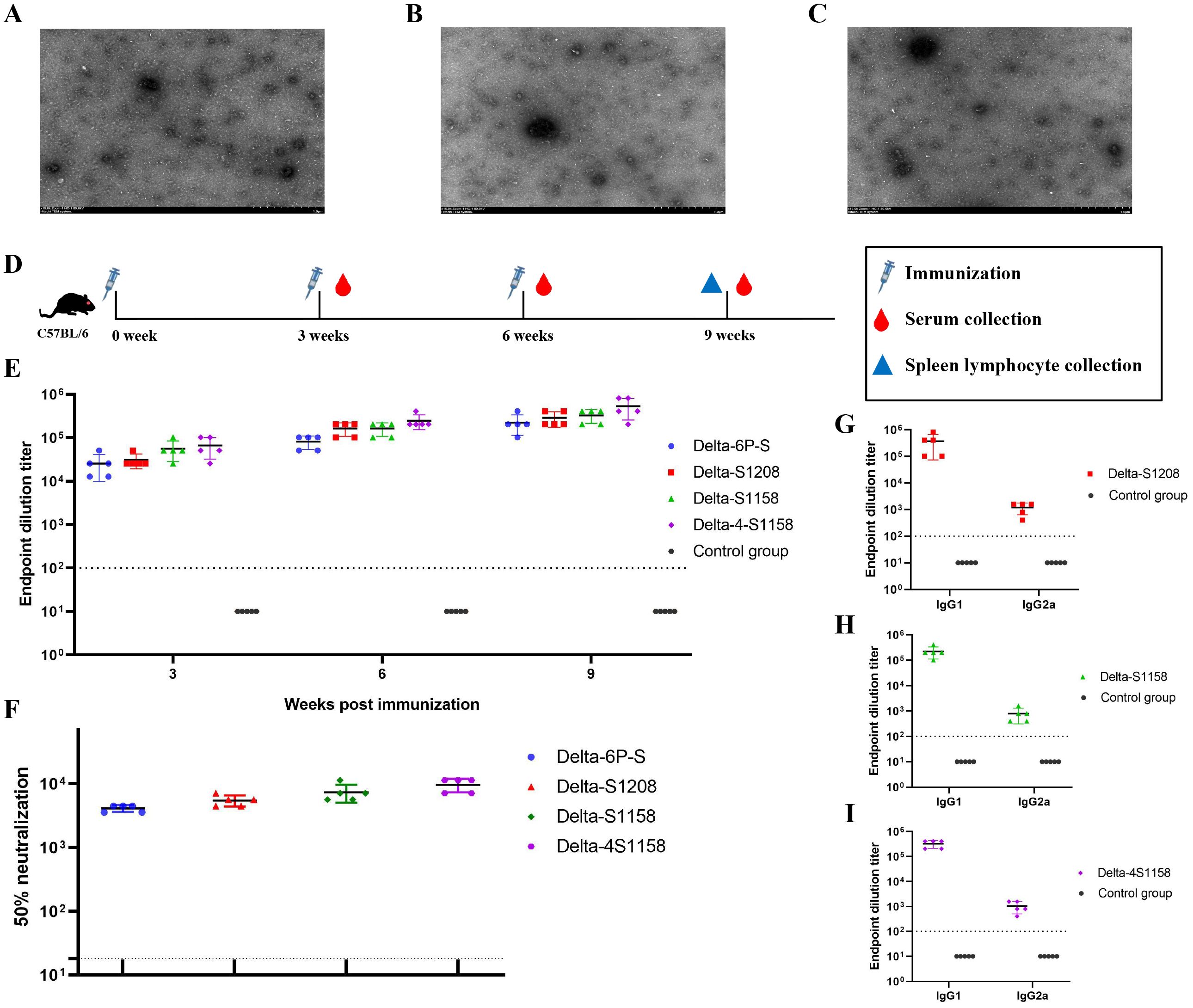
Figure 2. Electron microscopy observation of Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticles and induction of immune response in C57BL/6J mice. (A–C) Negative staining electron microscopy images of Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticles. Scale bar = 1 μm. (D) Immunization and evaluation procedures of Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticles in C57BL/6J mice. (E) The S-specific IgG antibodies of Delta-6P-S recombinant protein, Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticles in serum at 3, 6, and 9 weeks post-immunization (n = 5). (F) The 50% neutralization titers of Delta-6P-S recombinant protein, Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticle groups against the Delta variant (n=5). (G–I) Antibody subclass detection of Delta-S1208, Delta-S1158, and Delta-4S1158 nanoparticle groups. Individual animal values are indicated by colored symbols. Source data are provided as a Source Data file, and the data are presented as mean values ± SEM. The horizontal dashed line indicates the lower limit of detection (LLOD).
3.2 Immune responses induced by differently designed ferritin nanoparticles in C57BL/6J mice
To compare the immunogenicity of structurally distinct nanoparticles, C57BL/6J mice (5 groups, n=5) were immunized at 3-week intervals. Serum S-specific IgG titers were measured (Figure 2D). High antibody levels were detected after the first dose, with geometric mean titers (GMTs) ranging from 1:25,600 to 1:66,560. After the third booster, GMTs were 1:225,280 (Delta-6P-S), 1:286,720 (Delta-S1208), 1:327,680 (Delta-S1158), and 1:532,480 (Delta-4S1158) (Figure 2E). The control group exhibited background levels of the signal (Figure 2E). Neutralizing antibody titers against the Delta variant followed a similar trend. The Delta-4S1158 group reached an NT50 of 1:9,757, which was 2.34-fold higher than Delta-6P-S, 1.76-fold higher than Delta-S1208, and 1.31-fold higher than Delta-S1158 (Figure 2F). Antibody subclasses were predominantly IgG1, consistent with a Th2-biased response (Figures 2G–I). Thus, immunization with Delta-4S1158 nanoparticles induced a higher level of immune response in C57BL/6J mice. The spleen lymphocytes were detected by flow cytometry. Flow cytometry showed significantly increased frequencies of CD3+CD4+ and CD3+CD8+ T cells compared with controls (p<0.05; Figures 3B, C). Intracellular cytokine staining revealed higher proportions of CD3+CD4+IFN-γ+ and CD3+CD4+IL-4+ T cells in immunized mice (p<0.05), indicating enhanced Th2-type responses (Figures 3E, F, H, I). A preliminary in vivo safety assessment showed no apparent adverse effects in immunized mice (Supplementary Figure S1).
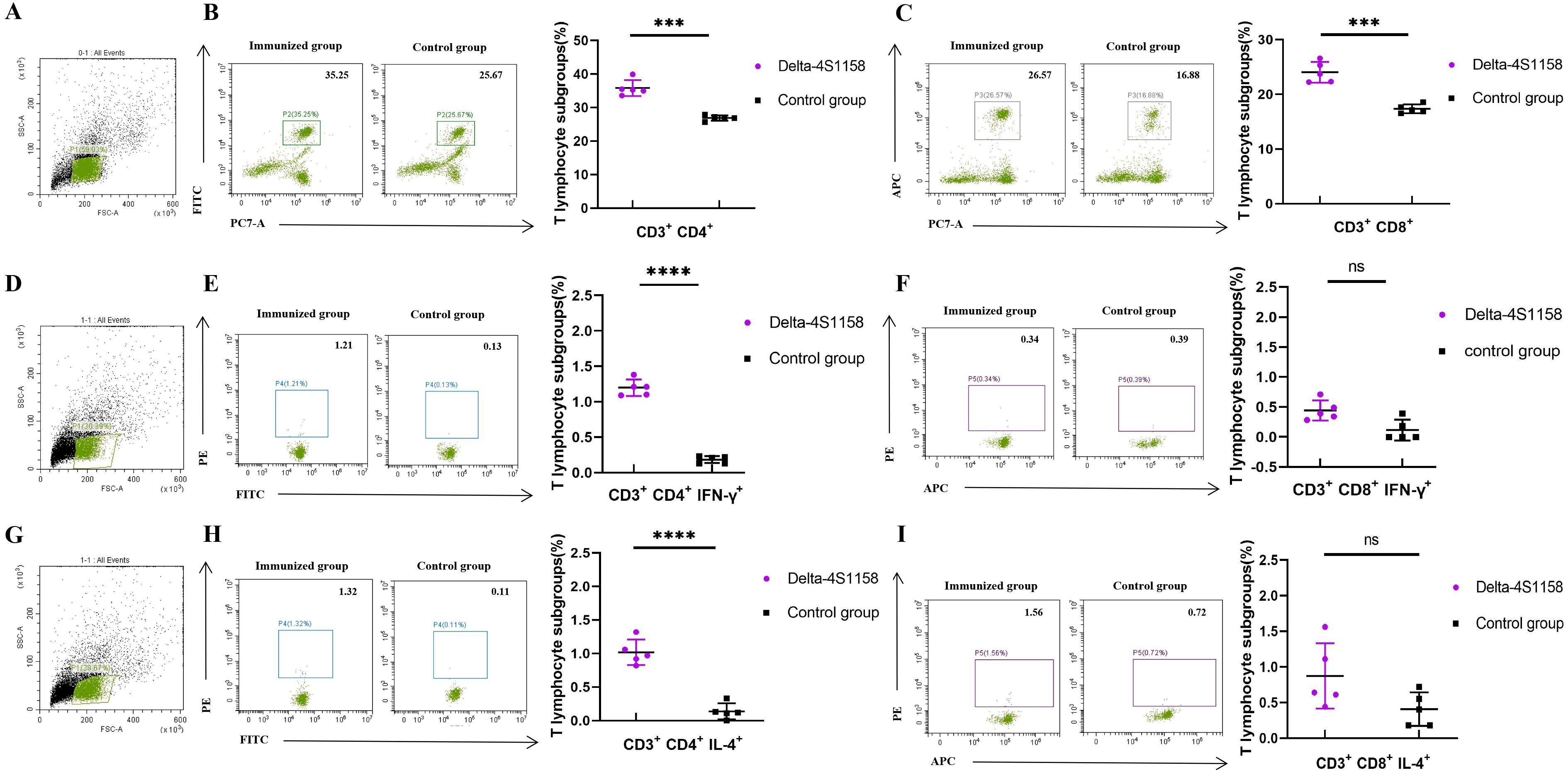
Figure 3. Flow cytometry detection of mouse spleen lymphocytes in the Delta-4S1158 nanoparticle group. Representative plots from each group are shown. (A, D, G) Gating strategy of flow cytometry. (B, C) Delta-4S1158 nanoparticles induced T lymphocyte differentiation in C57BL/6J mice. (E, F, H, I) Delta-4S1158 nanoparticles induced intracellular cytokines in T lymphocytes of C57BL/6J mice. Individual animal values are indicated by colored symbols, and the data are presented as mean values ± SEM.
3.3 Design, expression, purification, and immune response induced in C57BL/6J mice by JN.1-4S1158
To address emerging Omicron variants, we developed JN.1-4S1158, based on the Delta-4S1158 design but incorporating the Omicron JN.1 S protein sequence with six proline substitutions and the four additional stabilizing mutations (P1143S, F1148I, Y1155I, and F1156H) (Figure 4A). Expression in CHO-S cells and purification were confirmed by Western blot, which showed a band at ~180 kDa (Figure 4B). SDS-PAGE revealed a single band (Figure 4C), and electron microscopy confirmed nanoparticle formation (Figure 4D). Mice (2 groups, n=15) were immunized on a prime-boost regimen. On week 3, the GMT of S-specific IgG was 1:63,146, rising to 1:477,867 by week 9 (Figure 4F). The control group showed background levels. Neutralization assays demonstrated cross-protection: NT50s were 1:4,811 (BA.5), 1:5,493 (XBB), and 1:8,449 (JN.1) (Figure 4G).
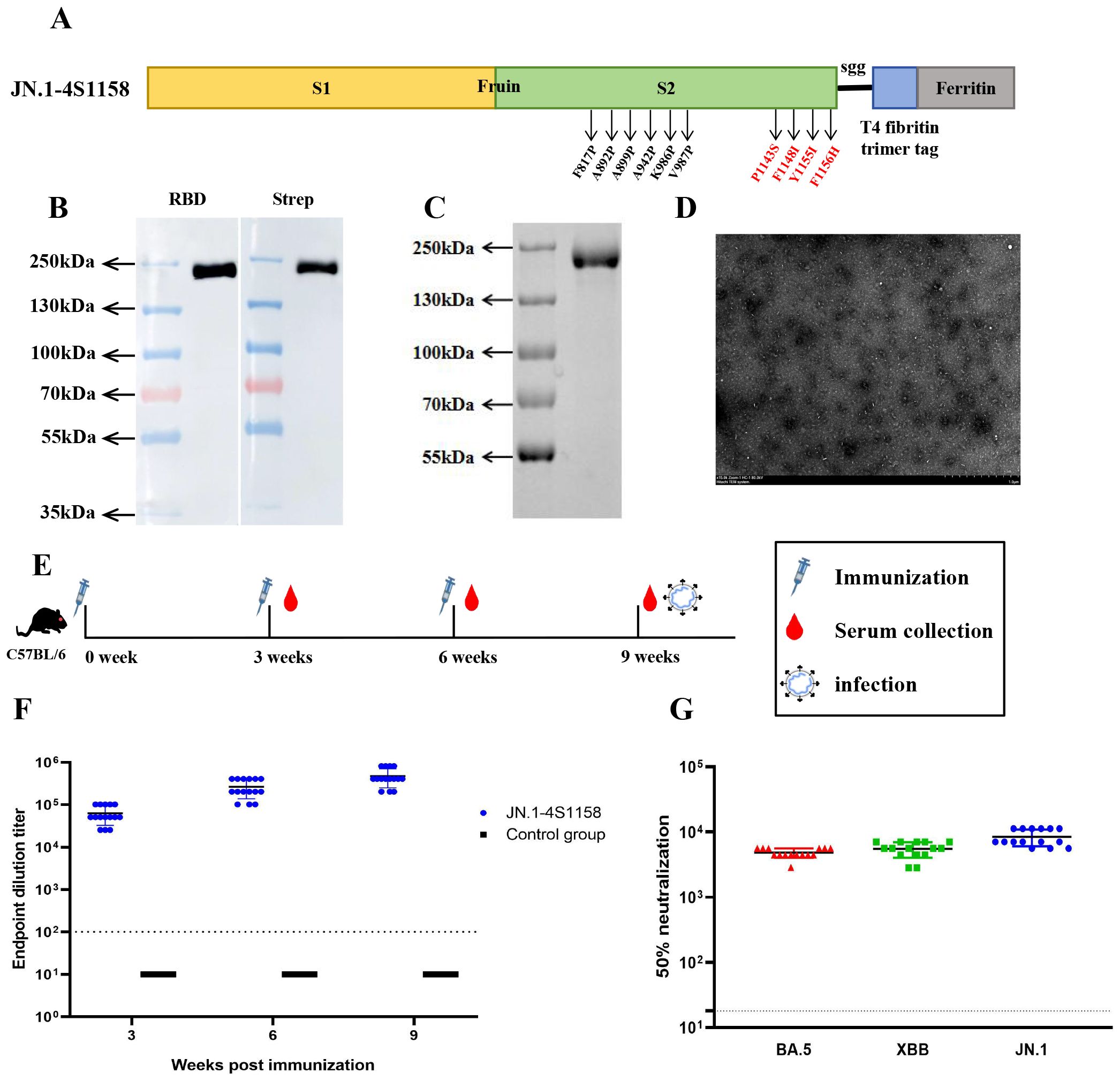
Figure 4. Immunogen design and purification of JN.1-4S1158 nanoparticles and induced immune response in C57BL/6J mice. (A) JN.1-4S1158 nanoparticle design and schematic diagram: six proline mutations and four additional mutation sites. (B) Characterization of JN.1-4S1158 nanoparticles by Western blot. (C) SDS-PAGE gel showing the JN.1-4S1158 nanoparticle band. (D) Negative staining electron microscopy images of JN.1-4S1158 nanoparticles. Scale bar = 1 μm. (E) Immunization and evaluation procedures of JN.1-4S1158 nanoparticles in C57BL/6J mice. (F) The S-specific IgG antibodies of JN.1-4S1158 nanoparticles in serum at 3, 6, and 9 weeks post-immunization (n = 15). (G) The 50% neutralization titers of JN.1-4S1158 nanoparticles against different Omicron variants (n = 15). Individual animal values are indicated by colored symbols, and the data are presented as mean values ± SEM. Source data are provided as a Source Data file. The horizontal dashed line indicates the lower limit of detection (LLOD).
3.4 JN.1-4S1158 protection against Omicron variants in C57BL/6J mice
To evaluate protection, immunized and control mice (n = 5 per group) were challenged intranasally with Omicron BA.5, XBB, or JN.1 (100 μL, 104.75 TCID50/mL). In JN.1-challenged mice, lung viral RNA levels were 106.22 copies/g compared with 1010.82 in controls (p<0.05; Figures 5K, L). Tracheal viral loads were 107.71 vs. 109.74 copies/g, respectively (p<0.05; Figures 5I, J). Infectious virus was undetectable in the lungs and trachea of immunized mice but present at high levels in controls (Figures 6C, F). Similar protection was observed against BA.5 and XBB (Figures 5A–H, 6A–D). Histopathological analysis revealed that control mice exhibited hemorrhage, inflammatory infiltration, and alveolar wall thickening, whereas vaccinated mice showed minimal to no lung pathology (Figure 7). In summary, JN.1-4S1158 nanoparticles induced robust antibody and T-cell responses and protected against multiple Omicron variants in C577BL/6J mice.
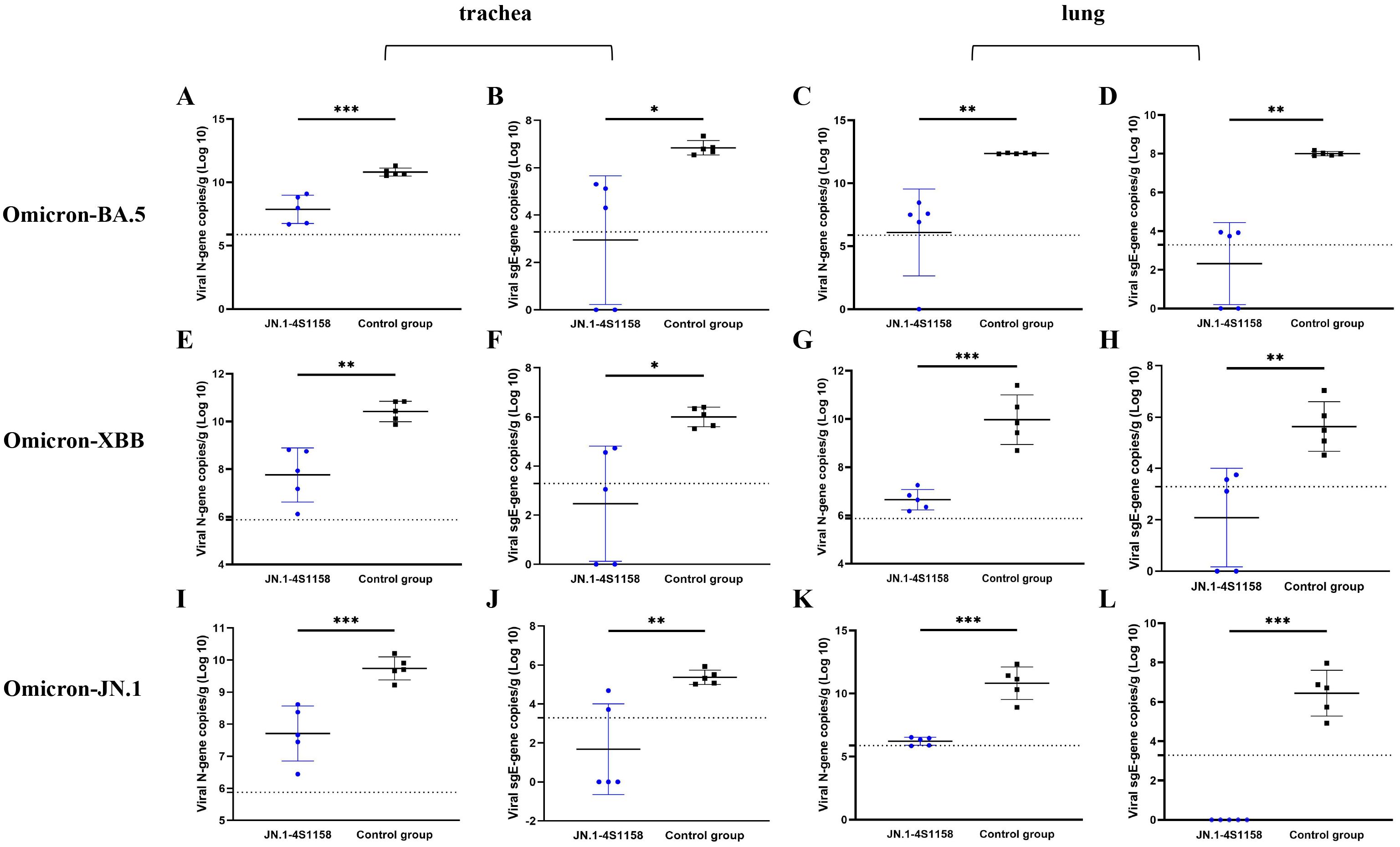
Figure 5. Viral N gene and sgE gene copies of trachea and lungs in C57BL/6J mice at 4 dpi. (A, C, E, G, I, K) Comparison of viral gene copy measurements of qPCR targeting viral N gene from tracheas (A, E, I) and lungs (C, G, K) between the immunized groups and the control groups after challenge with Omicron BA.5, XBB, and JN.1 variants at 4 dpi. (B, D, F, H, J, L) Comparison of viral gene copy measurements of qPCR targeting viral N gene from tracheas (B, F, J) and lungs (D, H, L) between the immunized groups and the control groups after challenge with Omicron BA.5, XBB, and JN.1 variants at 4 dpi. Data of the JN.1-4S1158 nanoparticle groups are shown in blue. Data of the control groups are shown in black. Individual animal values are indicated by colored symbols. Comparisons were performed by multiple t-test; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. Source data are provided as a Source Data file. The horizontal dashed line indicates the lower limit of detection (LLOD).
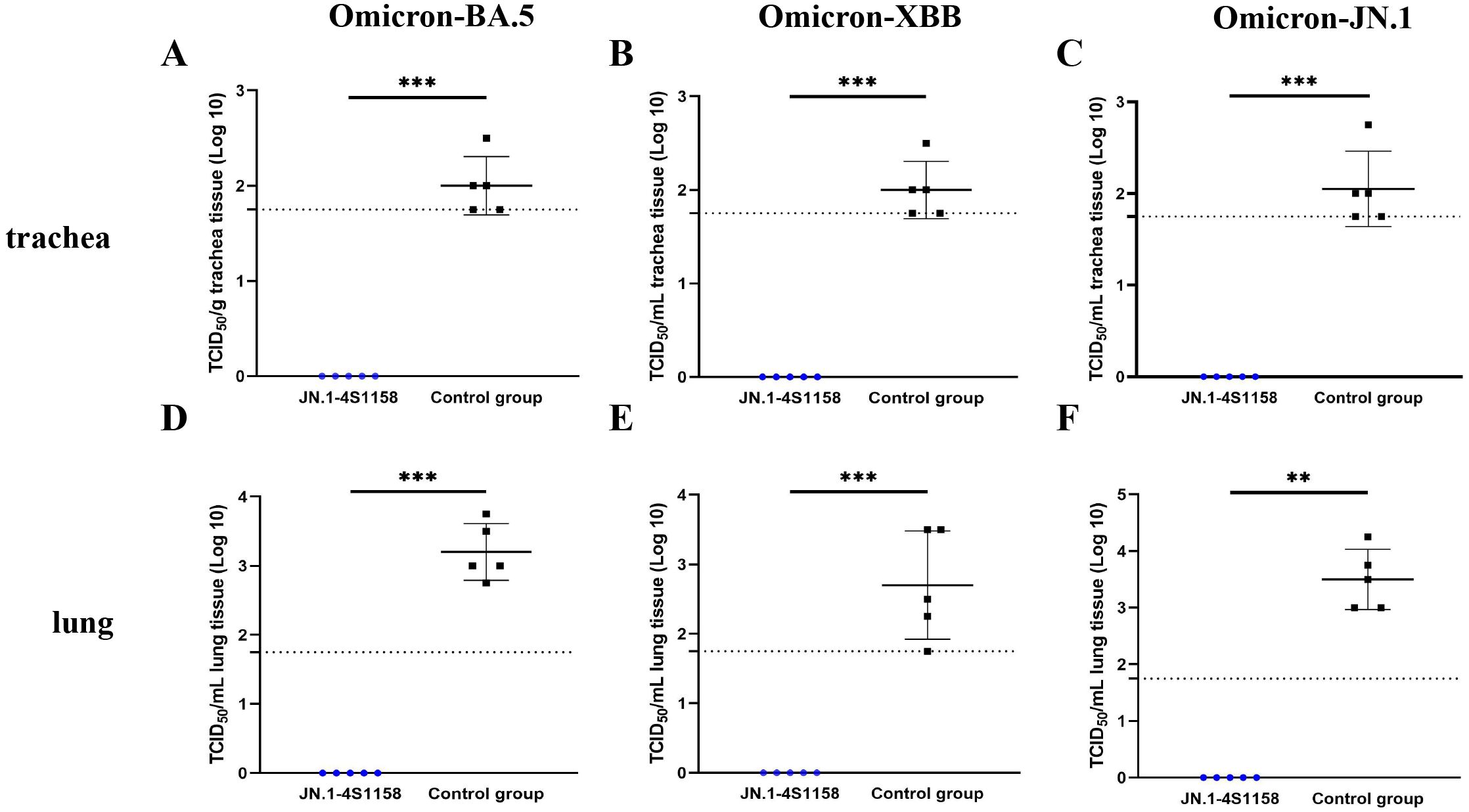
Figure 6. Protection of JN.1-4S1158 nanoparticles against Omicron variant infection in C57BL/6J mice. (A–F) Viral load in trachea (A–C) and lung (D–F) homogenates at 4 days post-Omicron BA.5, XBB, and JN.1 variant challenge. Bars represent the mean viral load (n = 5). Data of JN.1-4S1158 nanoparticle groups are presented in blue. Data from the control groups are presented in black. Individual animal values are indicated by colored symbols. Comparisons were performed by multiple t-test; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. Source data are provided as a Source Data file. The horizontal dashed line indicates the lower limit of detection (LLOD).
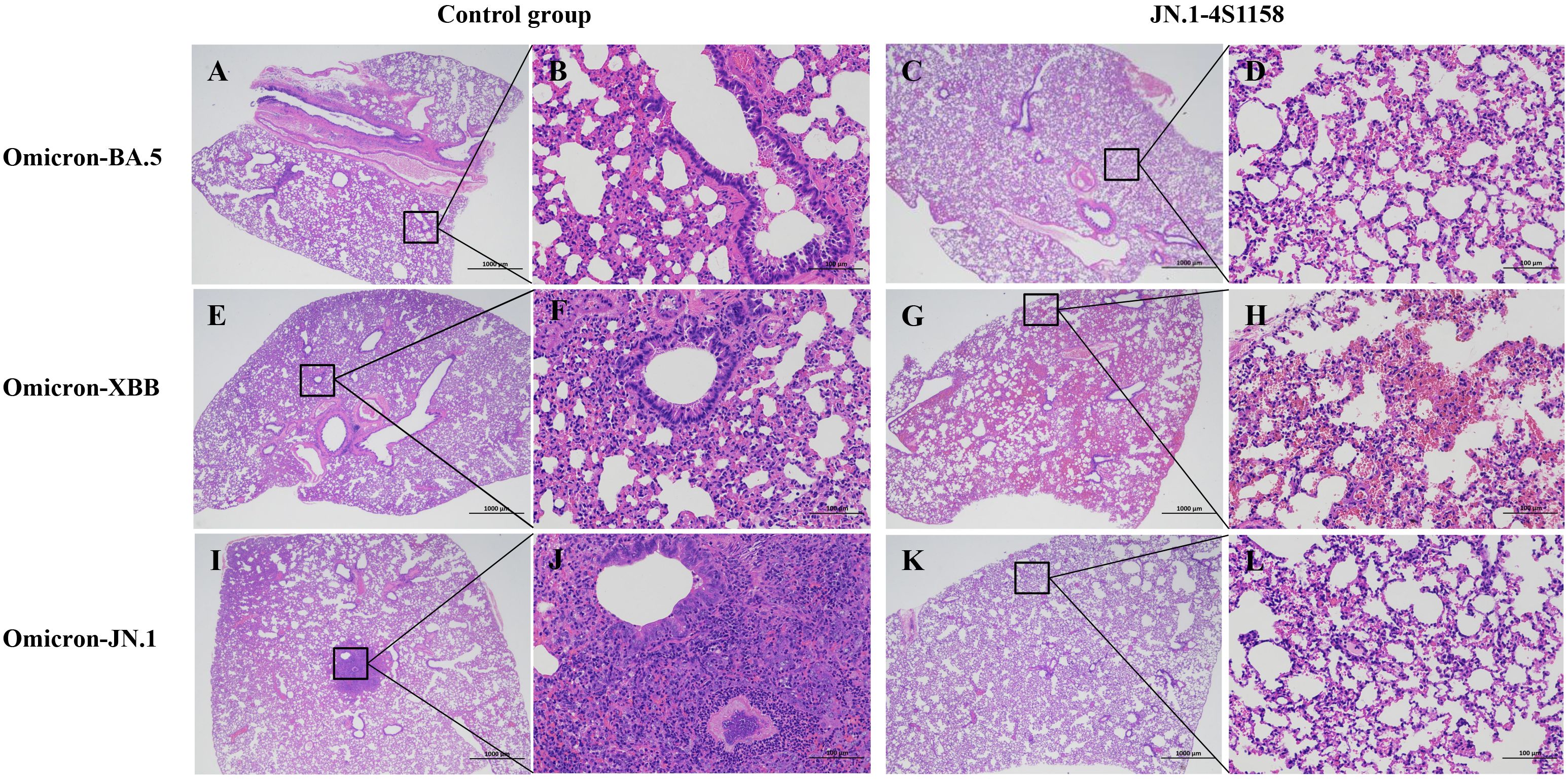
Figure 7. Pathological observation of lungs in C57BL/6J mice challenged with Omicron variants. Representative plots from each group are shown. (A, E, I) Lungs of C57BL/6J mice from control groups at 4 dpi. (A) Moderate alveolar wall thickening and inflammatory cell infiltration; scale bar = 1,000 μm. (E) Moderate alveolar wall thickening and inflammatory cell infiltration with bleeding; scale bar = 1,000 μm. (I) Moderate alveolar wall thickening and inflammatory cell infiltration with bleeding; scale bar = 1,000 μm. (B, F, J) Scale bar = 100 μm. (C, G, K) Lungs of C57BL/6J mice from JN.1-4S1158 nanoparticle groups at 4 dpi. (C) Mild alveolar wall thickening; scale bar = 1,000 μm. (G) Mild inflammatory cell infiltration with bleeding; scale bar = 1,000 μm. (K) Mild inflammatory cell infiltration; scale bar = 1,000 μm (D, H, J).
4 Discussion
The continued evolution of SARS-CoV-2 presents a significant challenge to global public health (Li et al., 2025). Large numbers of people remain infected with emerging Omicron variants (Giombini et al., 2025; Naveed Siddiqui et al., 2025). Currently, several vaccines have been developed for SARS-CoV-2, which can induce an excellent protective immune response; however, this protective effect is reduced against new variants (Saha and Ghosh Roy, 2025). Ferritin is an attractive vaccine platform because it is safe, stable, and can display antigens on its surface without disrupting the self-assembly of the nanoparticles. This versatility makes ferritin nanoparticles promising candidates for vaccine development and antigen delivery (Yang et al., 2024). While previous studies have demonstrated the strong immunogenicity of ferritin nanoparticle vaccines, relatively few have evaluated their efficacy against newer Omicron variants (Joyce and King, 2022; Wuertz and Barkei, 2021). In previous studies, we confirmed that the six-proline stabilized S-6P construct provided stronger protection than S-2P in mice and golden hamsters against SARS-CoV-2 infection (Feng et al., 2023). Other studies have further demonstrated that truncation of S-2P combined with four amino acid substitutions (P1143S, F1148I, Y1155I, and F1156H) enhances the immune response (Joyce et al., 2021). Building on this knowledge, we compared the immunogenicity of ferritin nanoparticles presenting S proteins with different structural designs in C57BL/6J mice and identified the optimal configuration. This led to the development of the “JN.1-4S1158” nanoparticle vaccine, which elicited strong immune responses and provided effective protection against Omicron BA.5, XBB, and JN.1 variants. No infectious virus was detected in the lungs or trachea of vaccinated animals, and levels of both the N and sgE genes were near the limit of detection, in contrast to the high viral burdens observed in control mice.
Since the receptor binding domain (RBD) of wild-type SARS-CoV-2 does not interact with mouse ACE2, hACE2 transgenic mouse models have commonly been used to test vaccine efficacy (Park et al., 2024; Thiede and Dick, 2024). Mutations such as N501Y expanded the host range by improving binding to mouse ACE2 (Halfmann and Iida, 2022; Wei et al., 2021). In this study, we used C57BL/6J mice to assess immunogenicity and cross-protection. We found that S-specific IgG titers increased significantly following immunization, with nanoparticle formulations eliciting higher antibody levels than monomeric proteins. Neutralizing antibody titers followed the same trend, consistent with the principle that multivalent antigen display enhances immunogenicity in line with prior vaccine studies (Sankhala et al., 2024; Wu et al., 2024). Among the designs tested, Delta-4S1158 nanoparticles induced the strongest immune response, confirming that this truncated S-6P design with four stabilizing mutations is well-suited for ferritin display.
The emergence of the Omicron JN.1 variant has raised concern due to its pronounced immune evasion compared with earlier variants (Kaku et al., 2024; Li et al., 2024). Although there is no clinical evidence that JN.1 causes more severe disease than other variants, mutations such as L455S in the RBD may enhance infectivity and broaden tissue tropism (Chang et al., 2025; Tian et al., 2025; Yang et al., 2024). In our study, the JN.1-4S1158 vaccine elicited high levels of cross-neutralizing antibodies against Omicron BA.5, XBB, and JN.1 variants. While neutralization titers against BA.5 and XBB were lower than those against JN.1, they remained within the range considered protective, consistent with findings from other recent studies (Ao et al., 2025; Yang et al., 2025). Challenge experiments confirmed these results: vaccinated mice showed no detectable infectious virus in the lungs or trachea, in contrast to high viral titers in controls. The ability of JN.1-4S1158 to confer cross-protection suggests that it may be effective against multiple Omicron sublineages, although its activity against other emerging variants requires further study. Concerns about antibody-dependent enhancement (ADE) have been raised in the context of SARS-CoV and MERS-CoV vaccines, where enhanced respiratory disease was reported in animal models (Karthik et al., 2020; Li et al., 2021). However, in preclinical models and large clinical trials of SARS-CoV-2 vaccines, such effects have not been observed (Gao and Bao, 2020; Wang et al., 2020). In our study, preliminary safety assessments revealed no evidence of pathology or adverse effects in vaccinated mice, supporting the biosafety of the ferritin nanoparticle platform.
In summary, the JN.1-4S1158 ferritin nanoparticle vaccine demonstrated strong preclinical efficacy against Omicron BA.5, XBB, and JN.1 variants in mice. These findings highlight the potential of ferritin-based vaccines as adaptable platforms for combating emerging SARS-CoV-2 variants. Further studies, including evaluation in additional animal models and eventual clinical trials, are warranted to validate safety, durability, and breadth of protection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Ethics Committee of Jilin Medical University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SF: Writing – review & editing, Writing – original draft. XY: Writing – review & editing. HW: Writing – review & editing. ZL: Writing – review & editing. SL: Writing – review & editing. HW: Writing – review & editing. ZH: Writing – review & editing. WW: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the following grants: the National Key Research and Development Program of China (grant number 2021YFC2302502) and the Science and Technology Planning Project of Jilin Province of China (grant number 20220204030YY). The funders did not play any role in the design, conclusions, or interpretation of the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1676592/full#supplementary-material
References
Ahmadivand, S. and Fux, R. (2024). Ferritin vaccine platform for animal and zoonotic viruses. Vaccines (Basel) 12, 1112. doi: 10.3390/vaccines12101112
Ao, D., Peng, D., He, C., Ye, C., Hong, W., Huang, X., et al. (2025). A promising mRNA vaccine derived from the JN.1 spike protein confers protective immunity against multiple emerged Omicron variants. Mol. Biomed. 6, 13. doi: 10.1186/s43556-025-00258-7
Belghith, A. A., Cotter, C. A., Ignacio, M. A., Earl, P. L., Hills, R. A., and Howarth, M. R. (2025). Mpox multiprotein virus-like nanoparticle vaccine induces neutralizing and protective antibodies in mice and non-human primates. Nat. Commun. 16, 4726. doi: 10.1038/s41467-025-59826-8
Cao, S., Ma, D., and Ji, S. (2024). Self-assembled ferritin nanoparticles for delivery of antigens and development of vaccines: from structure and property to applications. Molecules 29, 4221. doi: 10.3390/molecules29174221
Chang, S., Shin, J., Park, S., Park, H., Kim, J. H., Kim, T. W., et al. (2025). Continuous tracking for effective tackling: Ad5/35 platform-based JN1 lineage vaccines development in response to evolving SARS-CoV-2 variants. J. Med. Virol. 97, e70206. doi: 10.1002/jmv.70206
Chen, B. and Farzan, M. (2025). SARS-CoV-2 spike protein: structure, viral entry and variants. Nat. Rev. Microbiol. 23, 455–468. doi: 10.1038/s41579-025-01185-8
Chen, T., Gao, Y., Chen, X., Dong, Y., Wang, S., Huang, Q., et al. (2025). Self-assembling nanoparticle vaccine elicits a robust protective immune response against avian influenza H5N6 virus in chickens. Int. J. Biol. Macromol 287, 138405. doi: 10.1016/j.ijbiomac.2024.138405
Chen, S., Zhang, X., Yao, Y., Wang, S., Li, K., Zhang, B., et al. (2024). Ferritin nanoparticle-based Nipah virus glycoprotein vaccines elicit potent protective immune responses in mice and hamsters. Virol. Sin. 39, 909–916. doi: 10.1016/j.virs.2024.09.005
Chiba, S., Maemura, T., Loeffler, K., Frey, S. J., Gu, C., and Biswas, A. (2024). Single immunization with an influenza hemagglutinin nanoparticle-based vaccine elicits durable protective immunity. Bioeng. Transl. Med. 9, e10689. doi: 10.1002/btm2.10689
Fahrbach, K., Cichewicz, A., Chu, H., Di Fusco, M., Burnett, H., Volkman, H. R., et al. (2025). Comparative effectiveness of Omicron XBB 1.5-adapted COVID-19 vaccines: a systematic literature review and network meta-analysis. Expert Rev. Vaccines 24, 416–432. doi: 10.1080/14760584.2025.2505754
Feng, S., Fan, Z., Zhou, K., Ma, S., Liang, M., Zhang, H., et al. (2023). Subunit vaccine raised against the SARS-CoV-2 spike of Delta and Omicron variants. J. Med. Virol. 95, e29160. doi: 10.1002/jmv.29160
Gao, Q. and Bao, L. (2020). Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369, 77–81. doi: 10.1126/science.abc1932
Giombini, E., Schiavoni, I., Ambrosio, L., Lo Presti, A., Di Martino, A., Fiore, S., et al. (2025). JN.1 variants circulating in Italy from October 2023 to April 2024: genetic diversity and immune recognition. BMC Infect. Dis. 25, 291. doi: 10.1186/s12879-025-10685-0
Halfmann, P. J. and Iida, S. (2022). SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603, 687–692. doi: 10.1038/s41586-022-04441-6
Holmes, E. C. (2024). The emergence and evolution of SARS-CoV-2. Annu. Rev. Virol. 11, 21–42. doi: 10.1146/annurev-virology-093022-013037
Joyce, M. G., Chen, W. H., Sankhala, R. S., Hajduczki, A., Thomas, P. V., Choe, M., et al. (2021). SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 37, 110143. doi: 10.1016/j.celrep.2021.110143
Joyce, M. G. and King, H. A. D. (2022). A SARS-CoV-2 ferritin nanoparticle vaccine elicits protective immune responses in nonhuman primates. Sci. Transl. Med. 14, eabi5735. doi: 10.1126/scitranslmed.abi5735
Kaku, Y., Okumura, K., Padilla-Blanco, M., Kosugi, Y., Uriu, K., Hinay, A. A., Jr., et al. (2024). Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect. Dis. 24, e82. doi: 10.1016/s1473-3099(23)00813-7
Karthik, K., Senthilkumar, T. M. A., Udhayavel, S., and Raj, G. D. (2020). Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum. Vaccin Immunother. 16, 3055–3060. doi: 10.1080/21645515.2020.1796425
Li, P., Faraone, J. N., Hsu, C. C., Chamblee, M., Liu, Y., Zheng, Y. M., et al. (2025). Neutralization and spike stability of JN.1-derived LB.1, KP.2.3, KP.3, and KP.3.1.1 subvariants. mBio 16, e0046425. doi: 10.1128/mbio.00464-25
Li, Z. X., Feng, S., Zhang, H., Zhuang, X. Y., Shang, C., Sun, S. Y., et al. (2022). Immunogenicity and protective efficacy of a DNA vaccine inducing optimal expression of the SARS-CoV-2 S gene in hACE2 mice. Arch. Virol. 167, 2519–2528. doi: 10.1007/s00705-022-05562-z
Li, D., Luan, N., Li, J., Zhao, H., Zhang, Y., Long, R., et al. (2021). Waning antibodies from inactivated SARS-CoV-2 vaccination offer protection against infection without antibody-enhanced immunopathology in rhesus macaque pneumonia models. Emerg. Microbes Infect. 10, 2194–2198. doi: 10.1080/22221751.2021.2002670
Li, L., Shi, K., Gu, Y., Xu, Z., Shu, C., Li, D., et al. (2024). Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants. Structure 32, 1055–1067.e1056. doi: 10.1016/j.str.2024.06.012
Lin, G., Tang, Y. L., Fu, Z., Chen, R., Liu, Y., Liu, Z., et al. (2025). Enhancing protective immunity against SARS-CoV-2 with a self-amplifying RNA lipid nanoparticle vaccine. J. Control Release 378, 250–265. doi: 10.1016/j.jconrel.2024.12.003
Lucignano, R. and Ferraro, G. (2024). Bioactive molecules delivery through ferritin nanoparticles: sum up of current loading methods. Molecules 29, 4045. doi: 10.3390/molecules29174045
Mambelli, F. and de Araujo, A. (2025). An update on anti-COVID-19 vaccines and the challenges to protect against new SARS-CoV-2 variants. Pathogens 14, 23. doi: 10.3390/pathogens14010023
Moore, M., Anderson, L., Schiffer, J. T., Matrajt, L., and Dimitrov, D. (2025). Durability of COVID-19 vaccine and infection induced immunity: A systematic review and meta-regression analysis. Vaccine 54, 126966. doi: 10.1016/j.vaccine.2025.126966
Naveed Siddiqui, A., Musharaf, I., and Gulumbe, B. H. (2025). The JN.1 variant of COVID-19: immune evasion, transmissibility, and implications for global health. Ther. Adv. Infect. Dis. 12, 20499361251314763. doi: 10.1177/20499361251314763
Park, U., Lee, J. H., Kim, U., and Jeon, K. (2024). A humanized ACE2 mouse model recapitulating age- and sex-dependent immunopathogenesis of COVID-19. J. Med. Virol. 96, e29915. doi: 10.1002/jmv.29915
Pischel, L., Martini, B. A., Yu, N., Cacesse, D., Tracy, M., Kharbanda, K., et al. (2024). Vaccine effectiveness of 3rd generation mpox vaccines against mpox and disease severity: A systematic review and meta-analysis. Vaccine 42, 126053. doi: 10.1016/j.vaccine.2024.06.021
Saha, A. and Ghosh Roy, S. (2025). Beyond the pandemic era: recent advances and efficacy of SARS-CoV-2 vaccines against emerging variants of concern. Vaccines (Basel) 13, 424. doi: 10.3390/vaccines13040424
Sankhala, R. S., Lal, K. G., and Jensen, J. L. (2024). Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques. Nat. Commun. 15, 200. doi: 10.1038/s41467-023-44265-0
Sun, Y., Gao, Y., Su, T., Zhang, L., Zhou, H., Zhang, J., et al. (2025). Nanoparticle vaccine triggers interferon-gamma production and confers protective immunity against porcine reproductive and respiratory syndrome virus. ACS Nano 19, 852–870. doi: 10.1021/acsnano.4c12212
Thiede, J. M. and Dick, J. K. (2024). Human ACE2 gene replacement mice support SARS-CoV-2 viral replication and nonlethal disease progression. ImmunoHorizons 8, 712–720. doi: 10.4049/immunohorizons.2400030
Tian, J., Shang, B., Zhang, J., and Guo, Y. (2025). T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots. Nat. Immunol. 26, 265–278. doi: 10.1038/s41590-024-02051-0
Wang, Q. and Guo, Y. (2023). Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature 624, 639–644. doi: 10.1038/s41586-023-06750-w
Wang, Q., Guo, Y., Bowen, A., Mellis, I. A., Valdez, R., Gherasim, C., et al. (2024). XBB.1.5 monovalent mRNA vaccine booster elicits robust neutralizing antibodies against XBB subvariants and JN.1. Cell Host Microbe 32, 315–321.e313. doi: 10.1016/j.chom.2024.01.014
Wang, C., Liu, Q., and Huang, X. (2023). Ferritin nanocages: a versatile platform for nanozyme design. J. Mater. Chem. B 11, 4153–4170. doi: 10.1039/d3tb00192j
Wang, H., Zhang, Y., Huang, B., Deng, W., Quan, Y., Wang, W., et al. (2020). Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell 182, 713–721.e719. doi: 10.1016/j.cell.2020.06.008
Wasim, R., Sumaiya, and Ahmad, A. (2025). Across-the-board review on Omicron SARS-CoV-2 variant. Inflammopharmacology 33, 1–10. doi: 10.1007/s10787-024-01627-4
Wei, C., Shan, K. J., Wang, W., Zhang, S., Huan, Q., and Qian, W. (2021). Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J. Genet. Genomics 48, 1111–1121. doi: 10.1016/j.jgg.2021.12.003
Widge, A. T. and Hofstetter, A. R. (2023). An influenza hemagglutinin stem nanoparticle vaccine induces cross-group 1 neutralizing antibodies in healthy adults. Sci. Transl. Med. 15, eade4790. doi: 10.1126/scitranslmed.ade4790
Widyaningsih, S. A. and Hakim, M. S. (2024). COVID-19 pandemic and its impact on vaccine hesitancy: A review. Oman Med. J. 39, e646. doi: 10.5001/omj.2024.103
Wu, X., Li, W., Rong, H., Pan, J., Zhang, X., Hu, Q., et al. (2024). A nanoparticle vaccine displaying conserved epitopes of the preexisting neutralizing antibody confers broad protection against SARS-CoV-2 variants. ACS Nano 18, 17749–17763. doi: 10.1021/acsnano.4c03075
Wu, H., Weng, R., Li, J., Huang, Z., Tie, X., Li, J., et al. (2025). Self-Assembling protein nanoparticle platform for multivalent antigen delivery in vaccine development. Int. J. Pharm. 676, 125597. doi: 10.1016/j.ijpharm.2025.125597
Wuertz, K. M. and Barkei, E. K. (2021). A SARS-CoV-2 spike ferritin nanoparticle vaccine protects hamsters against Alpha and Beta virus variant challenge. NPJ Vaccines 6, 129. doi: 10.1038/s41541-021-00392-7
Yang, H., Guo, H., Wang, A., Cao, L., Fan, Q., Jiang, J., et al. (2024). Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants. Nat. Commun. 15, 7715. doi: 10.1038/s41467-024-51973-8
Yang, J., He, X., and Shi, H. (2025). Recombinant XBB.1.5 boosters induce robust neutralization against KP.2- and KP.3-included JN.1 sublineages. Signal Transduct. Target. Ther. 10, 47. doi: 10.1038/s41392-025-02139-5
Keywords: immune-evasive, ferritin nanoparticles, immunogenicity, cross-neutralizing, Omicron
Citation: Feng S, Yu X-Y, Wang H-T, Li Z-X, Li S-Z, Wang H-Y, Ha Z and Wang W (2025) Ferritin nanoparticle vaccine displaying optimized spike protein confers broad protection against Omicron subvariants. Front. Cell. Infect. Microbiol. 15:1676592. doi: 10.3389/fcimb.2025.1676592
Received: 30 July 2025; Accepted: 17 October 2025;
Published: 04 November 2025.
Edited by:
He Zhang, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2025 Feng, Yu, Wang, Li, Li, Wang, Ha and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuo Ha, aGF6aHVvMTYzQDE2My5jb20=; Wei Wang, d3dreTExMDFAMTI2LmNvbQ==; Sheng Feng, ZmVuZ3NoZW5nMTk5NDA1MDRAMTYzLmNvbQ==
 Sheng Feng
Sheng Feng Xiao-Yang Yu
Xiao-Yang Yu Hai-Tong Wang1
Hai-Tong Wang1 Zhuo-Xin Li
Zhuo-Xin Li Hui-Yan Wang
Hui-Yan Wang Zhuo Ha
Zhuo Ha Wei Wang
Wei Wang