- 1Nordcee, Department of Biology, University of Southern Denmark, Odense, Denmark
- 2Department of Engineering, University of Aarhus, Aarhus, Denmark
Direct electron uptake by prokaryotes is a recently described mechanism with a potential application for energy and CO2 storage into value added chemicals. Members of Methanosarcinales, an environmentally and biotechnologically relevant group of methanogens, were previously shown to retrieve electrons from an extracellular electrogenic partner performing Direct Interspecies Electron Transfer (DIET) and were therefore proposed to be electroactive. However, their intrinsic electroactivity has never been examined. In this study, we tested two methanogens belonging to the genus Methanosarcina, M. barkeri, and M. horonobensis, regarding their ability to accept electrons directly from insoluble electron donors like other cells, conductive particles and electrodes. Both methanogens were able to retrieve electrons from Geobacter metallireducens via DIET. Furthermore, DIET was also stimulated upon addition of electrically conductive granular activated carbon (GAC) when each was co-cultured with G. metallireducens. However, when provided with a cathode poised at −400 mV (vs. SHE), only M. barkeri could perform electromethanogenesis. In contrast, the strict hydrogenotrophic methanogen, Methanobacterium formicicum, did not produce methane regardless of the type of insoluble electron donor provided (Geobacter cells, GAC or electrodes). A comparison of functional gene categories between the two Methanosarcina showed differences regarding energy metabolism, which could explain dissimilarities concerning electromethanogenesis at fixed potentials. We suggest that these dissimilarities are minimized in the presence of an electrogenic DIET partner (e.g., Geobacter), which can modulate its surface redox potentials by adjusting the expression of electroactive surface proteins.
Introduction
Extracellular electron uptake by methanogens may impact carbon turnover in electron-acceptor limited environments (Morris et al., 2013). In these environments, thermodynamically challenging processes become possible due to syntrophic interactions between bacteria and archaea. A syntrophic interaction requires a bacterium, which oxidizes organics to interspecies-transferrable molecules. Moreover, syntrophy requires a partner methanogen to scavenge the transferable molecules. For decades, we have assumed that interspecies-transferrable molecules were either H2 or formate (Stams and Plugge, 2009). We now know that some species can also transfer electrons directly (Lovley, 2017). During direct interspecies electron transfer (DIET), species like Geobacter oxidize ethanol according to Reaction (1), only in the presence of methanogens like Methanosarcina, which scavenge reducing equivalents (H+ and e−) and acetate (Rotaru et al., 2014a,b) (Figure 1).
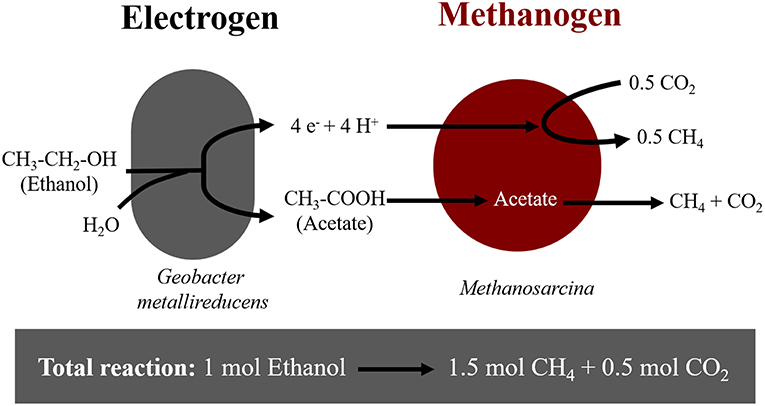
Figure 1. Diagram of electron flow during DIET interactions between Geobacter metallireducens and Methanosarcina-species provided with ethanol as sole electron donor.
In DIET co-cultures, only those Geobacter species producing high current densities, met the energetic needs of their DIET partners (Rotaru et al., 2015). For this purpose, Geobacter up-regulates the expression of redox active and conductive proteins (outer membrane c-type cytochromes and pili) (Shrestha et al., 2013; Holmes et al., 2018b). Geobacter's requirement for outer-surface proteins during DIET was confirmed earlier with gene-deletion studies (Rotaru et al., 2014a,b). Thus, if Geobacter lacked the ability to produce e.g., pili it was incapable of DIET.
Although we understand how Geobacter releases electrons outside their cells during DIET, the way Methanosarcinales retrieve DIET-electrons is poorly understood. A glimpse at this mechanism was provided in a recent comparative transcriptomic study (Holmes et al., 2018b). In this study, the transcriptomes of DIET co-cultures (G. metallireducens—Methanosarcina barkeri) were compared to those of co-cultures performing interspecies H2-transfer (Pelobacter carbinolicus—M. barkeri). During DIET, M. barkeri had higher expression of membrane-bound redox-active proteins like cupredoxins, thioredoxins, pyrroloquinoline, and quinone-, cytochrome-, or Fe-S containing proteins (Holmes et al., 2018b). Still, the exact mechanism of electron uptake by Methanosarcina has not been validated and warrants further investigation.
Moreover, Methanosarcina can also retrieve electrons from electrically conductive particles charged by Geobacter oxidizing organics (Shrestha and Rotaru, 2014). In effect, DIET is accelerated by electrically conductive particles/minerals perhaps because they replace conductive and redox active surface proteins diminishing cellular energy expenditure required to overexpress such surface constituents (Liu et al., 2012). For instance, co-cultures of G. metallireducens and M. barkeri were stimulated at the addition of conductive particles, such as GAC (Liu et al., 2012), carbon cloth (Chen et al., 2014a), biochar (Chen et al., 2014b), or magnetite (Wang et al., 2018). On the other hand, the addition of non-conductive materials did not stimulate DIET (Chen et al., 2014a; Rotaru et al., 2018a). In addition, conductive particles appear to play a significant role in interspecies interactions from natural and artificial ecosystems such as sediments, soils, rice paddies or anaerobic digesters (Holmes et al., 2017a; Rotaru et al., 2018b; Ye et al., 2018; Zhang L. et al., 2018a). In these cases, the addition of conductive particles enriched for DIET-related Methanosarcinales (Zheng et al., 2015; Dang et al., 2016; Holmes et al., 2018b; Rotaru et al., 2018a). However, exceptions were observed since occasionally conductive particles enriched for H2-utilizing methanogens of the genus Methanospirillum (Lee et al., 2016) or Methanobacterium (Zhuang et al., 2015; Lin et al., 2017).
Since methanogens retrieve extracellular electrons from cells or conductive particles to reduce CO2 to methane, it was expected that they could also retrieve electrons from a poised electrode via electromethanogenesis. Nevertheless, electromethanogenesis was only verified with H2-utilizing methanogens like Methanobacterium palustre (Cheng et al., 2009). Yet, H2-utilizers were incapable of DIET (Rotaru et al., 2014b). Conversely, it is unknown if Methanosarcinales, which are capable of DIET, are also capable of electron uptake from a cathode. Our objective was to investigate the ability to carry electromethanogenesis and DIET in two Methanosarcina species. We have shown that both Methanosarcina species grew by DIET, however only M. barkeri performed methanogenesis on the cathode at −400 mV (vs. SHE). This indicates that extracellular electron-uptake routes from cathodes or other cells might differ between Methanosarcina species.
Materials and Methods
Microorganism Strains and Cultivation Conditions
Methanosarcina barkeri MS (DSM 800) and Methanosarcina horonobensis HB-1 (DSM 21571) were purchased from the German culture collection DSMZ while Methanobacterium formicicum (NBRC 100475) was from the Japanese culture collection NBRC. Geobacter metallireducens GS-15 was sent to us by Dr. Sabrina Beckmann from the University of New South Wales, Australia.
Routine cultivation was performed under strictly anaerobic conditions in serum bottles sealed with butyl rubber stoppers and incubated statically at 37°C. All the microorganisms had been adapted to grow in DSMZ medium 120c with the following modifications: 1 g/L NaCl, 0.5 g/L yeast, and no tryptone (Rotaru et al., 2014a). During incubations in co-cultures or for electrochemical experiments, sulfide, and yeast extract was omitted. When grown in pure cultures, Methanosarcina species were provided with 30 mM acetate and 20 mM methanol as methanogenic substrates, while M. formicicum was provided with 150 kPa of H2: CO2 (80:20) in the headspace. G. metallireducens was routinely grown with 20 mM ethanol and 56 mM ferric citrate. All media and cultures were prepared and kept under an N2: CO2 (80:20) atmosphere.
The co-cultures of Geobacter and methanogens were initiated with 0.5 mL of G. metallireducens and 1 mL of acetate-grown Methanosarcina-species or H2-grown M. formicicum inoculated into 8.5 ml of the media prepared as above. The starting cell numbers for the methanogens in co-cultures were approximately 2.6 × 106 cells/mL, 8.2 × 106 cells/mL and 6.7 × 105 cells/mL for M. barkeri, M. formicicum, and M. horonobensis, respectively. The starting cell numbers for G. metallireducens in co-cultures were circa 1.5 × 105. Incubations were carried out in 20 ml pressure vials. For the co-cultures ethanol (10 mM) was added as an electron donor and CO2 was the sole electron acceptor. When noted, sterile granular activated carbon (GAC) was added at a concentration of 25 g/L and prepared as described before (Rotaru et al., 2018a).
Electrochemical Setup and Measurements
All bioelectrochemical incubations were carried with a modified DSMZ 120c medium (see above) in the absence of sulfide and yeast extract. The pH of this medium in the bioelectrochemical reactors was set to 6.5. We used bioelectrochemical reactors with a standard dual chamber configuration as shown in Figure S1. Two-chamber glass bottles were purchased from Adams & Chittenden Scientific Glass (USA) with side ports fitted with butyl septa to allow for medium transfer, sampling, and the introduction of a reference electrode. Each chamber of the reactors had a total volume of 650 ml with a flange diameter of 40 mm and the chambers were separated by a Nafion™ N117 proton exchange membrane (Ion Power) held by an O-ring seal with a knuckle clamp.
Both the working and counter electrodes were made of graphite (Mersen MI Corp., Greenville USA) with dimensions of 2.5 × 7.5 × 1.2 cm thus a total projected surface area of 61.5 cm2. The working and counter electrodes were coupled to titanium wires, which pierced through rubber stoppers fitted into the main opening of each chamber. A 2 cm deep and 2 mm wide hole was drilled on the short side of the electrode and a 12.5 cm long; 2 mm wide titanium rod (Alfa-Aesar, DE) was inserted and sealed from the outside with biocompatible non-conductive epoxy. Electrodes with a resistance of <10 Ω were used to ensure proper electrical connections. The assembled electrodes were introduced into the chamber via the main opening and 2 mm-wide holes were drilled in the black rubber stopper to allow access of the titanium rod. After autoclaving the reactors, sterile medium was transferred into the reactors anaerobically and aseptically. Sterile (bleach and ethanol series) reference electrodes were lodged through a side port in the working electrode chamber at a distance of about 1 cm from the surface of the working electrode. After lodging the electrodes, degassing with N2: CO2 (80:20) for circa 30 min in each reactor chamber ensured anaerobic conditions. When the pre-cultures were in mid-exponential phase, they were inoculated (20%) into fresh medium in the cathodic chamber following sterile anoxic techniques to a final volume of 550 ml leaving a headspace of approximately 100 mL in each chamber. The approximate cell numbers at the time of inoculation into the electrochemical reactors for M. barkeri, M. formicicum, and M. horonobensis were 2.6 × 107 cells/mL, 8.2 × 107 cells/mL, and 6.7 × 106 cells/mL respectively. Cell counts were done with microscopic examination using DAPI (1 μg/mL) stained cells.
The reference electrodes used were leak-free Ag/AgCl reference electrodes (3.4M KCl) (CMA Microdialysis, Sweden), which are 242 mV above the standard hydrogen electrode (SHE) according to the manufacturer and our own measurements against a Hydroflex® reference electrode used as NHE (normal hydrogen electrode). The difference between NHE and SHE is experimentally negligible (Smith and Stevenson, 2007). All potentials in this paper from here onwards are reported vs. SHE by adjusting accordingly from the Ag/AgCl reference electrodes values. Cathode poising and electrochemical measurements were carried with a multichannel potentiostat (MultiEmstat, Palmsens, NL) operated by the Multitrace software (Palmsens, NL).
Analytical Measurements and Calculations
Headspace samples for CH4 and H2 analysis were taken with hypodermic needles and kept in airtight exetainers until measurement. Methane (CH4) and hydrogen gas (H2) were measured on a Trace 1300 gas chromatograph (Thermo-Scientific) with a TracePLOT™ TG-BOND Msieve 5A column and a thermal conductivity detector (TCD). The carrier gas was argon at a flow rate of 25 mL/min. The injector, oven and detector temperatures were 150, 70, and 200°C respectively. The detection limit for CH4 and H2 was ca. 5 μM for both. The concentration unit was converted to molarity by using the ideal gas law (p × V = n × R × T) under standard conditions, where p = 1 atm, V is the volume of the gaseous phase (L), n is amount of gas (mol), R is the gas constant (0.08205 atm × L/ mol × K) and T = 298.15 K. For ethanol detection, 0.5 mL samples were filtered (0.2 μm pore size) into appropriate sampling vials and were heated for 5 min. at 60°C. The headspace gas was then passed through the Trace 1300 gas chromatograph (Thermo-Scientific) with a TRACE™ TR-Wax column and detected by a flame ionization detector (FID). Nitrogen gas at a flow of 1 mL/min was used as the carrier and the injector, oven, and detectors were kept at 220, 40, and 230°C respectively. Short-chained volatile fatty acids (VFA) were analyzed with a Dionex™ ICS-1500 Ion Chromatography system, using a Dionex™ IonPac™ AS22 IC Column and a mixture of 1.4 mM NaHCO3 and 4.5 mM Na2CO3 as the eluent fitted with an electron capture detector (ECD) at 30 mA.
Genome Comparison
Genomes for all tested microorganisms were available at the JGI integrated microbial genomes and microbiomes. Functional category comparisons and pairwise average nucleotide identity (ANI) were determined using the IMG/M- “Compare Genomes” tools. The IMG genome IDs of the studied M. barkeri, M. horonobensis, and M. formicicum used were 2630968729, 2627854269 and 2645727909, respectively. The gene functions were analyzed from the annotated names of all the protein-coding genes retrieved from the National Center for Biotechnology Information (NCBI) database. The accession numbers used were NZ_CP009528, NZ_CP009516, and NZ_LN515531 for M. barkeri, M. horonobensis, and M. formicicum, respectively. To scan for the cytochrome motif (CxxCH) through all the genomes, we used a pattern-matching Web-application (Seiler et al., 2006) in addition to manual search.
Results and Discussion
It was previously shown that two Methanosarcinales-methanogens, Methanosarcina barkeri and Methanothrix harundinacea grew via DIET whereas strict H2-utilizing methanogens did not (Rotaru et al., 2014a,b). Here we show that M. barkeri could retrieve electrons not only from an exoelectrogen but also from an electrode poised at −400 mV (non-H2 generating conditions) to carry electromethanogenesis. As expected, the H2-utilizing methanogen M. formicicum did not carry electromethanogenesis under this condition. We tested an environmentally relevant Methanosarcina, M. horonobensis for extracellular electron uptake from cells and electrodes, and we observed that it could only retrieve electrons from exoelectrogenic Geobacter and from granular activated carbon but not from electrodes.
Methanosarcina barkeri
M. barkeri grows in co-culture with G. metallireducens via DIET, and the interaction could be accelerated by electrically conductive particles (Rotaru et al., 2014a, Figure S2). This was anticipated because G. metallireducens, a respiratory organism, is incapable of substrate fermentation and consequent H2 production according to previous physiological tests (Cord-Ruwisch et al., 1998) and genetic investigations (Aklujkar et al., 2009). Since H2 could not be generated by G. metallireducens, a strict H2-utilizer like M. formicicum was rendered incapable of an interspecies association based on H2-transfer with this bacterium (Rotaru et al., 2014b). However, more recent studies indicated that conductive carbon nanotubes stimulated methanogenesis by M. formicicum (Salvador et al., 2017). This implied that M. formicicum might be encouraged by the presence of conductive particle to interact syntrophically with Geobacter. Therefore, we tested if conductive GAC aids M. formicicum to establish a syntrophic association with G. metallireducens. This was not the case since co-cultures of M. formicicum and G. metallireducens did not generate methane regardless of the presence or absence of conductive particles, over the course of 120 days (Figure 2).
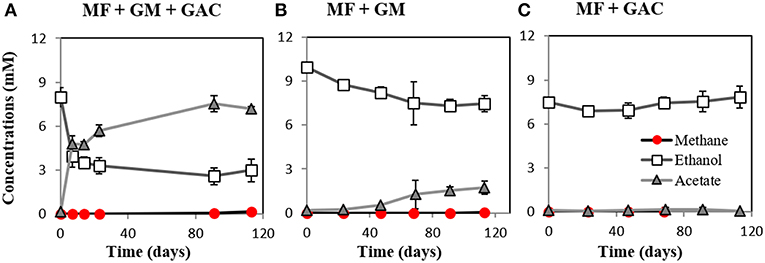
Figure 2. Co-cultures experiments with M. formicicum and G. metallireducens (n = 3). M. formicicum did not produce methane when incubated with G. metallireducens (A) in the presence of or (B) in the absence of GAC. (C) Alone, M. formicicum could not utilize ethanol or produce methane in the presence of GAC. MF, Methanobacterium formicicum; GM, Geobacter metallireducens; GAC, granular activated carbon.
During DIET, the extracellular electron transfer machinery of G. metallireducens plays a crucial role in Geobacter-Methanosarcina interactions, indicating that Geobacter releases extracellular electrons for the methanogen to use. Therefore, we suspected that Methanosarcina might also be able to directly retrieve extracellular electrons from electrodes to do electromethanogenesis.
In this study, we tested for the first time if M. barkeri could retrieve electrons directly from an electrode poised at −400 mV. Indeed, M. barkeri produced significantly more methane (4.4 ± 0.33 mM; p < 0.001) (Figure 3) when provided with an applied potential at the cathode, in contrast to open circuit controls without an applied potential (1.3 ± 0.33 mM) (Figure S3). The background methane in control reactors resulted from carry-over substrates, once this was subtracted, the additional methane produced by M. barkeri in poised reactors (3.1 ± 0.34 mM) could be solely credited to electricity. Moreover, the highest rate of methane production was observed when current density profiles indicated the highest current draw by M. barkeri (Figure 3).
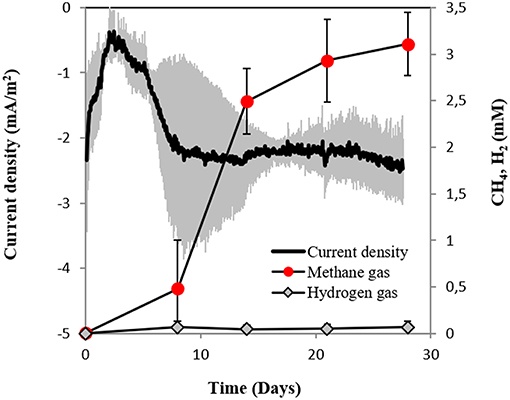
Figure 3. Current consumption and gas production in triplicate M. barkeri cultures provided with a poised cathode at −400 mV (vs. SHE) as sole electron donor.
There are two possible scenarios for M. barkeri growing successfully at a cathode poised at −400 mV:
1. It may use low concentrations of H2 generated electrochemically at the cathode, or
2. It retrieves electrons directly via an unknown mechanism
To determine abiotic electrochemical H2 evolution we (i) verified for H2 accumulation over a month of incubation and (ii) verified the threshold for H2-evolution by linear sweep voltammetry at the beginning and the end of the incubation. H2 did not accumulate over a month of incubation in six independent abiotic controls (Figure 4A). Linear sweep voltammetry profiles indicated that in our media the threshold for H2-evolution was below −700 mV (Figure 4B). This was in agreement with previous studies determining electrochemical H2-evolution under physiological conditions on a graphite electrode, which was below −400 mV due to high overpotentials (Cheng et al., 2009; Mitov et al., 2012; Batlle-Vilanova et al., 2014; Beese-Vasbender et al., 2015).
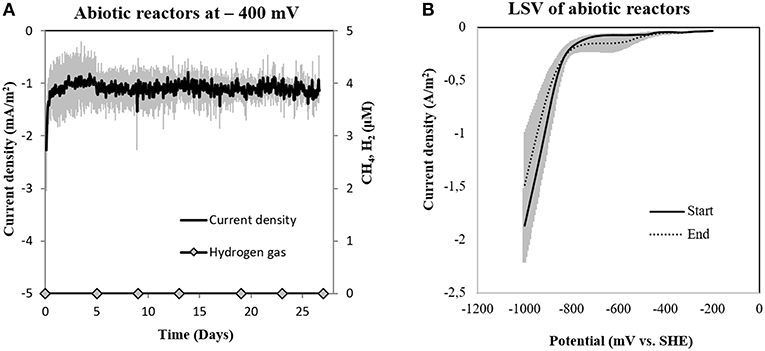
Figure 4. (A) Current consumption and gas production in four abiotic reactors at −400 mV (vs. SHE) and (B) Linear Sweep Voltammetry (LSV) of abiotic reactors at the start and end of the experiment (n = 3).
On the other hand, in reactors inoculated with M. barkeri, the detected H2 stabilized at 0.065 ± 0.02 mM, similar to concentrations observed for co-cultures of M. barkeri with (0.077 ± 0.03 mM) or without conductive particles (0.076 ± 0.06 mM) and in pure culture (0.068 mM). This is supported by previous research, which demonstrated H2-cycling (H2-production and H2-uptake) in M. barkeri (Kulkarni et al., 2009, 2018; Mand et al., 2018). The cellular-evolved H2 is well above the H2-uptake threshold for M. barkeri (296–376 nM) (Lovley, 1985; Kral et al., 1998) possibly because in these cultures there is an alternative, competitive electron donor.
Secondly, if H2 evolved electrochemically to concentrations under the detection limit (which was not the case, see above), we anticipated that a sensitive hydrogenotrophic methanogen could effectively reclaim low concentrations of electrochemical H2, draw current and produce methane. To test this hypothesis we used a highly effective H2-utilizing methanogen—M. formicicum, which has a low H2 uptake threshold of approximately 6 nM (Lovley, 1985). However, when M. formicicum was incubated in electrochemical reactors, neither H2, methane nor current draw was observed at −400 mV (Figure 5) indicating that methanogenesis from H2 could not occur at this potential. In addition, to ensure that the growth of M. formicicum was unrestrained by the poised electrode, we carried control incubations at −400 mV with extrinsic H2 as substrate. M. formicicum was unaffected by a poised electrode since it produced methane from the extrinsic H2 in an electrochemical setup (Figure S4).
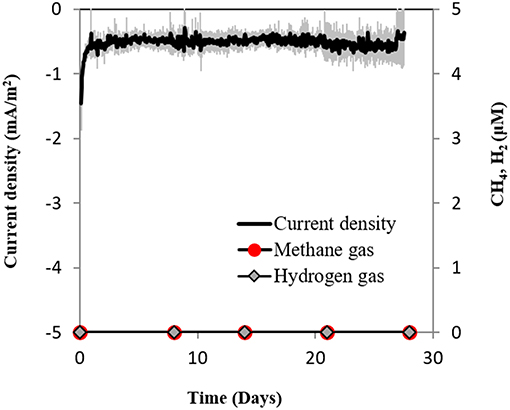
Figure 5. Current consumption and gas production in triplicate M. formicicum cultures provided with a cathode poised at −400 mV (vs. SHE) as sole electron donor.
As electrochemical H2 was unlikely in our electrochemical setup, according to cumulative gas-detection analyses, electrochemical tests, and tests with a highly effective H2-utilizer, we confer that M. barkeri is likely to retrieve electrons directly from the electrode.
Methanosarcina horonobensis
The distribution of extracellular electron uptake to other species of the order Methanosarcinales has not been explored, with the exception of M. barkeri and M. harudinacea. M. barkeri and M. harundinacea species have been isolated from and associated with anaerobic wastewater treatment (Bryant and Boone, 1987; Ma et al., 2006; De Vrieze et al., 2012). We were interested to see if other environmentally relevant Methanosarcina species had similar electron-uptake properties. We focused on Methanosarcina horonobensis because of its provenience and consistent association with deep aquifers (Shimizu et al., 2010; Holmes et al., 2018a).
Here we showed that M. horonobensis did establish successful syntrophic associations with G. metallireducens with or without conductive particles as an electrical conduit (Figure 6). Theoretically, G. metallireducens oxidizes ethanol to acetate only if they could use the methanogen as an electron acceptor (Reaction 1). The acetate is then further disproportionated by the acetoclastic methanogen to produce methane and CO2 (Reactions 2, 3). During DIET we expect the conversion of 1 mol ethanol to 1.5 mol methane according to Reactions 1–3 (Figure 1). As predicted, in the G. metallireducens—M. horonobensis co-cultures, the syntroph oxidized 8.8 ± 0.4 mM ethanol providing the reducing equivalents (directly and via acetate) to generate 13.1 ± 0.8 mM CH4 by the methanogen. These co-cultures achieved stoichiometric recoveries of 98.5 ± 3.3 %. Similar recoveries (109 ± 18.5 %) were also observed at the addition of conductive particles. Single species controls with GAC showed that ethanol could not be converted to methane by the methanogen or the syntroph alone (Figure 6C and Figure S5). However, similar to previous reports (Zhang P. et al., 2018b), Geobacter could partially convert ethanol to acetate using GAC as insoluble electron acceptor (Figure S5; Van Der Zee et al., 2003; Zhang P. et al., 2018b), likely until it reaches its maximum capacitance of 40 F/g (Zhang et al., 2009). Co-cultures of G. metallireducens and M. horonobensis could not carry interspecies H2 transfer because G. metallireducens is a strict respiratory microorganism which cannot ferment ethanol to generate H2 (Shrestha et al., 2013) and because M. horonobensis is unable to use H2 as electron donor for their metabolism (Shimizu et al., 2010).
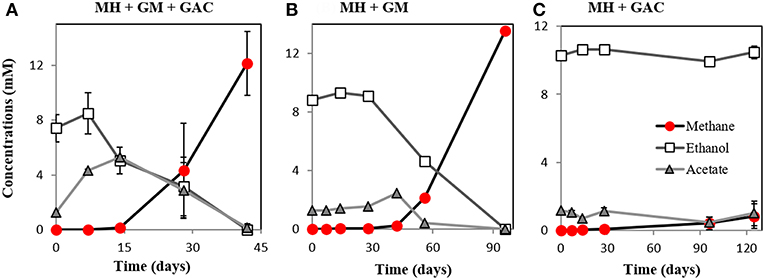
Figure 6. Co-culture experiments with M. horonobensis and G. metallireducens. M. horonobensis established successful co-cultures with G. metallireducens as apparent from ethanol utilization and methane production in the presence (A, n = 3) or absence of GAC (B, n = 1; see replication in Figure S6). Alone, M. horonobensis could not utilize ethanol or produce methane in the presence of GAC (C, n = 3). MH, Methanosarcina horonobensis; GM, Geobacter metallireducens; GAC, granular activated carbon.
Surprisingly, M. horonobensis, which could grow by DIET, was incapable of electromethanogenesis (Figure 7). Thus, we compared the genomes of the two Methanosarcina, M. horonobensis, and M. barkeri to further explain why they were both capable of DIET, but showed dissimilar activities on cathodes at −400 mV.
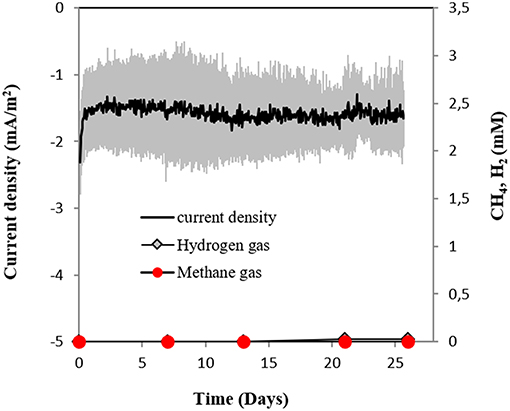
Figure 7. Current consumption and gas production in cultures of M. horonobensis (n = 3) provided with a cathode poised at −400 mV (vs. SHE) as sole electron donor.
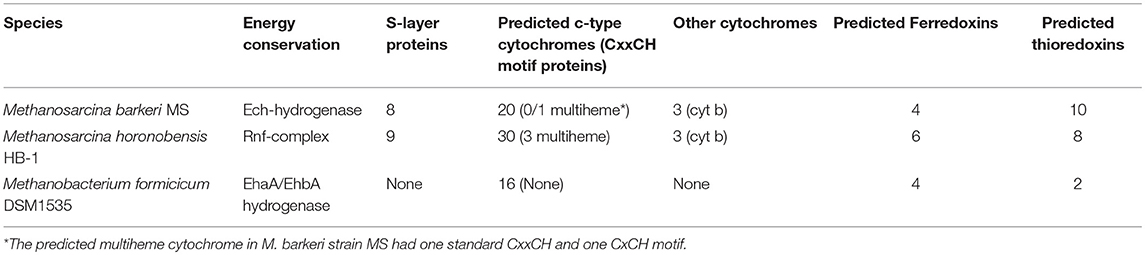
The main difference between the genomes of M. barkeri and M. horonobensis, was related to their energy metabolism (Table 1). M. barkeri utilizes an energy-converting hydrogenase (Ech) (Kulkarni et al., 2018), which couples the reduction of protons with ferredoxin (Fdx−) to the production of a proton motive force according to the reaction: Fdx− (red) + 2H+ → Fdx (ox) + H2 + ΔμH+/ ΔμNa+ (Thauer et al., 2008). M. horonobensis does not have the Ech (Table 1). An alternative to Ech is the Na+-pumping Rnf complex described biochemically in M. acetivorans (Schlegel et al., 2012; Suharti et al., 2014), and predicted via genome mining in M. thermophila (Wang et al., 2011) and ANME-2 archaea (Wang et al., 2014). Since we could not find any Ech in the genome of M. horonobensis, we screened for the genes encoding an Rnf-complex. In M. horonobensis, we found all eight representative Rnf-genes (including the cytochrome subunit and Rnf A to G; MSHOH_3554 to 3561), which showed 65–91% protein identity to their M. acetivorans counterparts (MA_0658 to 0665).
Both Ech and Rnf contain Fe-S centers (Welte and Deppenmeier, 2014), however, the Rnf complex has an accompanying c-type cytochrome (Suharti et al., 2014) possibly influencing the overall redox-chemistry on the cell surface. We presume that differences in surface redox chemistry will impact how different Methanosarcina interact with extracellular electron donors. Thus, electromethanogenesis at a set potential of −400 mV is unlikely to match the redox requirements of each type of Methanosarcina. On the other hand, in co-cultures, Geobacter may coordinate its cytochrome expression to match the redox potential of the partner methanogen, who plays the role of a terminal electron acceptor. This is supported by previous studies showing Geobacter modulates their cell-surface proteins to match the electron acceptor provided (Ishii et al., 2018; Otero et al., 2018).
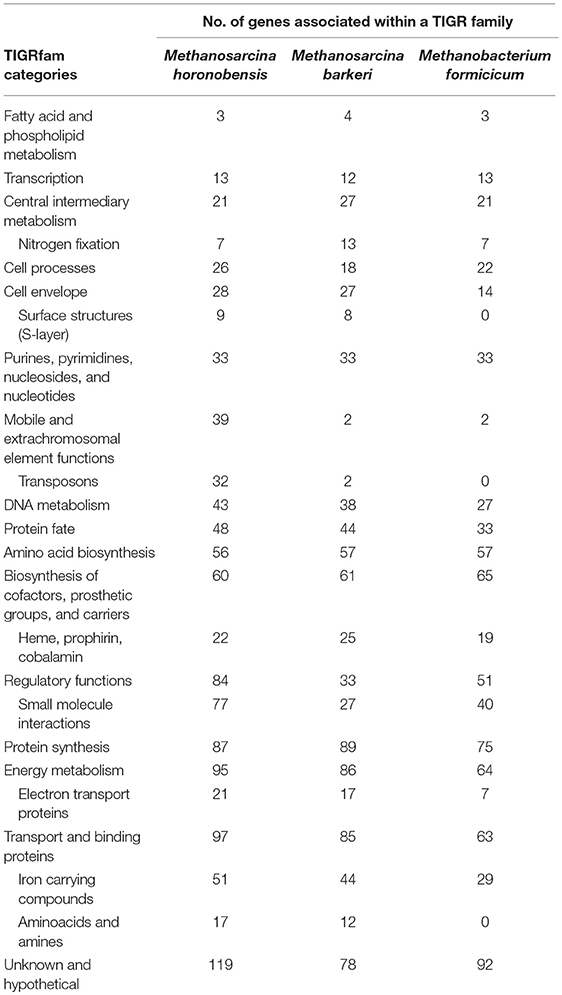
When contrasting the genomes of the two Methanosarcina species we also observed significant differences regarding nitrogen fixation, mobile elements, and sensing/chemotaxis proteins (Table 2). As such, compared to M. horonobensis, M. barkeri encodes for more N2-fixation proteins (86%). Compared to M. barkeri, M. horonobensis encodes for more small-molecule-interaction proteins such as redox-sensing and chemotaxis proteins (185%) and mobile elements than M. barkeri (16-fold increase) (Table 2). The exact role of these proteins in extracellular electron uptake by these Methanosarcinas is unknown and warrants further investigation.
Furthermore, to determine why Methanosarcina could do DIET, but not Methanobacterium, we compared the genomes of the two Methanosarcina species with that of M. formicicum (Table 2). In contrast to the Methanobacterium, both Methanosarcina species encode in their genomes three times the amount of genes for electron transport proteins and circa 50% more genes for cell surface and transport proteins (Table 2). Especially, outer surface S-layer proteins were only present in the two Methanosarcina (Table 2). S-layer proteins were previously suggested to play a role in extracellular electron transfer in Methanosarcina related ANME-2, which carry anaerobic methane oxidation syntrophically (McGlynn et al., 2015; McGlynn, 2017; Timmers et al., 2017). Future gene-expression and deletion studies could shed light on the possible role of S-layer proteins in DIET-interactions.
Conclusion
Three methanogens were investigated for their ability to do extracellular electron uptake from (1) a cathode at −400 mV, (2) directly from an electrogenic-DIET partner, or (3) from a DIET-partner, but mediated by conductive particles. Only M. barkeri was able to carry out all three forms of extracellular electron uptake, making this the first observation of a Methanosarcina in pure culture performing electromethanogenesis. The conditions in our abiotic electrochemical controls did not lead to H2- evolution at −400 mV, according to electrochemical and analytical tests. Therefore, under these conditions, it was impossible to sustain a methanogen with high H2-affinity, like M. formicicum. Besides M. formicicum was incapable to retrieve electrons directly from the electrode or from a DIET partner (direct or via conductive particles). In this study, we also demonstrated that another Methanosarcina, M. hornobensis performed DIET with Geobacter (direct or via conductive particles). However, surprisingly, M. horonobensis was incapable of electromethanogenesis. We screened the genomes of the two Methanosarcina and identified differences (e.g., energy metabolism), which could lead to phenotypic variability and thus contrasting electromethanogenesis-ability. Compared to M. barkeri, M. horonobensis is a better candidate for understanding electron uptake from a DIET syntrophic partner. This is because unlike M. barkeri, M. horonobensis does not utilize H2, and it grows as single cells on freshwater media, which is ideal for genetic studies.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
MY and A-ER conceived the study with support from BT and LO. MY performed all experiments with support from OS-W. MY analyzed the data with support from A-ER. MY wrote the manuscript with help from A-ER. All authors contributed to drafting and editing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Innovationsfond grant number 4106-00017 funded this work. We would like to thank Lasse Ørum Smidt and Heidi Grøn Jensen for lab assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2019.00029/full#supplementary-material
References
Aklujkar, M., Krushkal, J., Dibartolo, G., Lapidus, A., Land, M. L., and Lovley, D. R. (2009). The genome sequence of Geobacter metallireducens: features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 9, 1–22. doi: 10.1186/1471-2180-9-109
Batlle-Vilanova, P., Puig, S., Gonzalez-Olmos, R., Vilajeliu-Pons, A., Bañeras, L., Balaguer, M. D., et al. (2014). Assessment of biotic and abiotic graphite cathodes for hydrogen production in microbial electrolysis cells. Int. J. Hydrogen Energy 39, 1297–1305. doi: 10.1016/j.ijhydene.2013.11.017
Beese-Vasbender, P. F., Grote, J.-P., Garrelfs, J., Stratmann, M., and Mayrhofer, K. J. J. (2015). Selective microbial electrosynthesis of methane by a pure culture of a marine lithoautotrophic archaeon. Bioelectrochemistry 102, 50–55. doi: 10.1016/j.bioelechem.2014.11.004
Bryant, M. P., and Boone, D. R. (1987). Emended Description of Strain MST(DSM 800T), the Type Strain of Methanosarcina barkeri. Int. J. Syst. Bacteriol. 37, 169–170. doi: 10.1099/00207713-37-2-169
Chen, S., Rotaru, A.-E., Liu, F., Philips, J., Woodard, T. L., Nevin, K. P., et al. (2014a). Carbon cloth stimulates direct interspecies electron transfer in syntrophic co-cultures. Bioresour. Technol. 173, 82–86. doi: 10.1016/j.biortech.2014.09.009
Chen, S., Rotaru, A.-E., Shrestha, P. M., Malvankar, N. S., Liu, F., Fan, W., et al. (2014b). Promoting interspecies electron transfer with biochar. Sci. Rep. 4, 1–7. doi: 10.1038/srep05019
Cheng, S., Xing, D., Call, D. F., and Logan, B. E. (2009). Direct biological conversion of electrical current into methane by electromethanogenesis. Environ. Sci. Technol. 43, 3953–3958. doi: 10.1021/es803531g
Cord-Ruwisch, R., Lovley, D. R., and Schink, B. (1998). Growth of Geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl. Environ. Microbiol. 64, 2232–2236.
Dang, Y., Holmes, D. E., Zhao, Z., Woodard, T. L., Zhang, Y., Sun, D., et al. (2016). Enhancing anaerobic digestion of complex organic waste with carbon-based conductive materials. Bioresour. Technol. 220, 516–522. doi: 10.1016/j.biortech.2016.08.114
De Vrieze, J., Hennebel, T., Boon, N., and Verstraete, W. (2012). Methanosarcina: the rediscovered methanogen for heavy duty biomethanation. Bioresour. Technol. 112, 1–9. doi: 10.1016/j.biortech.2012.02.079
Holmes, D. E., Orelana, R., Giloteaux, L., Wang, L.-Y., Shrestha, P. M., Williams, K., et al. (2018a). Potential for methanosarcina to contribute to uranium reduction during acetate-promoted groundwater bioremediation. Microb. Ecol. 76, 660–7. doi: 10.1007/s00248-018-1165-5
Holmes, D. E., Rotaru, A.-E., Ueki, T., Shrestha, P. M., Ferry, J. G., and Lovley, D. R. (2018b). Electron and proton flux for carbon dioxide reduction in methanosarcina barkeri during direct interspecies electron transfer. Front. Microbiol. 9:3109. doi: 10.3389/fmicb.2018.03109
Holmes, D. E., Shrestha, P. M., Walker, D. J. F., Dang, Y., Nevin, K. P., Woodard, T. L., et al. (2017a). Metatranscriptomic evidence for direct interspecies electron transfer between geobacter and methanothrix species in methanogenic rice paddy soils. Appl. Environ. Microbiol. 83:AEM.00223-17. doi: 10.1128/AEM.00223-17
Ishii, S., Suzuki, S., Tenney, A., Nealson, K. H., and Bretschger, O. (2018). Comparative metatranscriptomics reveals extracellular electron transfer pathways conferring microbial adaptivity to surface redox potential changes. ISME J. 12, 2844–2863. doi: 10.1038/s41396-018-0238-2
Kral, T. A., Brink, K. M., Miller, S. L., and McKay, C. P. (1998). Hydrogen consumptions by methanogens on the early earth. Orig. Life Evol. Biosph. 28, 311–319. doi: 10.1023/A:1006552412928
Kulkarni, G., Kridelbaugh, D. M., Guss, A. M., and Metcalf, W. W. (2009). Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc. Natl. Acad. Sci. U.S.A. 106, 15915–15920. doi: 10.1073/pnas.0905914106
Kulkarni, G., Mand, T. D., and Metcalf, W. W. (2018). Energy conservation via hydrogen cycling in the methanogenic archaeon Methanosarcina barkeri. MBio 9, 1–10. doi: 10.1128/mBio.01256-18
Lee, J. Y., Lee, S. H., and Park, H. D. (2016). Enrichment of specific electro-active microorganisms and enhancement of methane production by adding granular activated carbon in anaerobic reactors. Bioresour. Technol. 205, 205–212. doi: 10.1016/j.biortech.2016.01.054
Lin, Q., Fang, X., Ho, A., Li, J., Yan, X., Tu, B., et al. (2017). Different substrate regimes determine transcriptional profiles and gene co-expression in Methanosarcina barkeri (DSM 800). Appl. Microbiol. Biotechnol. 101(19):7303–16. doi: 10.1007/s00253-017-8457-4
Liu, F., Rotaru, A.-E., Shrestha, P. M., Malvankar, N. S., Nevin, K. P., and Lovley, D. R. (2012). Promoting direct interspecies electron transfer with activated carbon. Energy Environ. Sci. 5:8982. doi: 10.1039/c2ee22459c
Lovley, D. R. (1985). Minimum threshold for hydrogen metabolism in methanogenic bacteria. Appl. Environ. Microbiol. 49, 1530–1531.
Lovley, D. R. (2017). Syntrophy goes electric: direct interspecies electron transfer. Annu. Rev. Microbiol. 71, annurev-micro-030117-020420. doi: 10.1146/annurev-micro-030117-020420
Ma, K., Liu, X., and Dong, X. (2006). Methanosaeta harundinacea sp. nov., a novel acetate-scavenging methanogen isolated from a UASB reactor. Int. J. Syst. Evol. Microbiol. 56, 127–131. doi: 10.1099/ijs.0.63887-0
Mand, T. D., Kulkarni, G., and Metcalf, W. W. (2018). Genetic, biochemical, and molecular characterization of methanosarcina barkeri mutants lacking three distinct classes of hydrogenase. J. Bacteriol. 200, e00342–e00318. doi: 10.1128/JB.00342-18
McGlynn, S. E. (2017). Energy metabolism during anaerobic methane oxidation in ANME archaea. Microbes Environ. 32, 5–13. doi: 10.1264/jsme2.ME16166
McGlynn, S. E., Chadwick, G. L., Kempes, C. P., and Orphan, V. J. (2015). Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535. doi: 10.1038/nature15512
Mitov, M., Chorbadzhiyska, E., Rashkov, R., and Hubenova, Y. (2012). Novel nanostructured electrocatalysts for hydrogen evolution reaction in neutral and weak acidic solutions. Int. J. Hydrogen Energy 37, 16522–16526. doi: 10.1016/J.IJHYDENE.2012.02.102
Morris, B. E. L., Henneberger, R., Huber, H., and Moissl-Eichinger, C. (2013). Microbial syntrophy: interaction for the common good. FEMS Microbiol. Rev. 37, 384–406. doi: 10.1111/1574-6976.12019
Otero, F. J., Chan, C. H., and Bond, D. R. (2018). Identification of different putative outer membrane electron conduits necessary for Fe(III) Citrate, Fe(III) Oxide, Mn(IV) Oxide, or electrode reduction by geobacter sulfurreducens downloaded from. J. Bacteriol. 200, 347–365. doi: 10.1128/JB.00347-18
Rotaru, A.-E., Calabrese, F., Stryhanyuk, H., Musat, F., Shrestha, P. M., Weber, H. S., et al. (2018a). Conductive particles enable syntrophic acetate oxidation between geobacter and methanosarcina from coastal sediments. MBio 9, 1–14. doi: 10.1128/mBio.00226-18
Rotaru, A.-E., Posth, N. R., Miracle, M. R., Vicente, E., Cox, R. P., Thompson, J., et al. (2018b). Interspecies interactions mediated by conductive minerals in the sediments of the ferruginous Lake La Cruz, Spain. Limnetica 38, 1–30. doi: 10.1101/366542
Rotaru, A.-E., Shrestha, P. M., Liu, F., Markovaite, B., Chen, S., Nevin, K. P., et al. (2014a). Direct interspecies electron transfer between geobacter metallireducens and Methanosarcina barkeri. Appl. Environ. Microbiol. 80, 4599–4605. doi: 10.1128/AEM.00895-14
Rotaru, A.-E., Shrestha, P. M., Liu, F., Shrestha, M., Shrestha, D., Embree, M., et al. (2014b). A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ. Sci. 7, 408–415. doi: 10.1039/C3EE42189A
Rotaru, A.-E., Woodard, T. L., Nevin, K. P., and Lovley, D. R. (2015). Link between capacity for current production and syntrophic growth in Geobacter species. Front. Microbiol. 6:744. doi: 10.3389/fmicb.2015.00744
Salvador, A. F., Martins, G., Melle-Franco, M., Serpa, R., Stams, A. J. M., Cavaleiro, A. J., et al. (2017). Carbon nanotubes accelerate methane production in pure cultures of methanogens and in a syntrophic coculture. Environ. Microbiol. 19, 2727–2739. doi: 10.1111/1462-2920.13774
Schlegel, K., Welte, C., Deppenmeier, U., and Müller, V. (2012). Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 279, 4444–4452. doi: 10.1111/febs.12031
Seiler, M., Mehrle, A., Poustka, A., and Wiemann, S. (2006). The 3of5 web application for complex and comprehensive pattern matching in protein sequences. BMC Bioinform. 7:144. doi: 10.1186/1471-2105-7-144
Shimizu, S., Upadhye, R., Ishijima, Y., and Naganuma, T. (2010). Methanosarcina horonobensis sp. nov., a methanogenic archaeon isolated from a deep subsurface Miocene formation. Int. J. Syst. Evol. Microbiol. 61, 2503–2507. doi: 10.1099/ijs.0.028548-0
Shrestha, P. M., and Rotaru, A.-E. (2014). Plugging in or going wireless: strategies for interspecies electron transfer. Front. Microbiol. 5:237. doi: 10.3389/fmicb.2014.00237
Shrestha, P. M., Rotaru, A.-E., Aklujkar, M., Liu, F., Shrestha, M., Summers, Z. M., et al. (2013). Syntrophic growth with direct interspecies electron transfer as the primary mechanism for energy exchange. Environ. Microbiol. Rep. 5, 904–910. doi: 10.1111/1758-2229.12093
Smith, T. J., and Stevenson, K. J. (2007). “Reference electrodes,” in Handbook of Electrochemistry ed C. G. Zoski (Las Cruces: Elsevier B.V.), p. 73–110.
Stams, A. J. M., and Plugge, C. M. (2009). Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7, 568–577. doi: 10.1038/nrmicro2166
Suharti, S., Wang, M., De Vries, S., and Ferry, J. G. (2014). Characterization of the RnfB and RnfG subunits of the Rnf complex from the archaeon Methanosarcina acetivorans. PLoS ONE 9:97966. doi: 10.1371/journal.pone.0097966
Thauer, R. K., Kaster, A.-K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. doi: 10.1038/nrmicro1931
Timmers, P. H. A., Welte, C. U., Koehorst, J. J., Plugge, C. M., Jetten, M. S. M., and Stams, A. J. M. (2017). Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237
Van Der Zee, F. P., Bisschops, I. A. E., Lettinga, G., and Field, J. A. (2003). Activated carbon as an electron acceptor and redox mediator during the anaerobic biotransformation of azo dyes. Environ. Sci. Technol. 37, 402–408. doi: 10.1021/es025885o
Wang, F. P., Zhang, Y., Chen, Y., He, Y., Qi, J., Hinrichs, K.-U., et al. (2014). Methanotrophic archaea possessing diverging methane-oxidizing and electron-Transporting pathways. ISME J. 8, 1069–1078. doi: 10.1038/ismej.2013.212
Wang, M., Tomb, J.-F., and Ferry, J. G. (2011). Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol. 11:165. doi: 10.1186/1471-2180-11-165
Wang, O., Zheng, S., Wang, B., Wang, W., and Liu, F. (2018). Necessity of electrically conductive pili for methanogenesis with magnetite stimulation. PeerJ. 6:e4541. doi: 10.7717/peerj.4541
Welte, C. U., and Deppenmeier, U. (2014). Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta Bioenerg. 1837, 1130–1147. doi: 10.1016/j.bbabio.2013.12.002
Ye, Q., Zhang, Z., Huang, Y., Fang, T., Cui, Q., He, C., et al. (2018). Enhancing electron transfer by magnetite during phenanthrene anaerobic methanogenic degradation. Int. Biodeterior. Biodegrad. 129, 109–116. doi: 10.1016/j.ibiod.2018.01.012
Zhang, L., Zhang, J., and Loh, K. C. (2018a). Activated carbon enhanced anaerobic digestion of food waste–Laboratory-scale and Pilot-scale operation. Waste Manag. 75, 270–279. doi: 10.1016/j.wasman.2018.02.020
Zhang, P., Zheng, S., Liu, J., Wang, B., Liu, F., and Feng, Y. (2018b). Surface properties of activated sludge-derived biochar determine the facilitating effects on Geobacter co-cultures. Water Res. 142, 441–451. doi: 10.1016/j.watres.2018.05.058
Zhang, Y., Feng, H., Wu, X., Wang, L., Zhang, A., Xia, T., Zhang, L., et al. (2009). Progress of electrochemical capacitor electrode materials: a review. Int. J. Hydrogen Energy 34, 4889–4899. doi: 10.1016/j.ijhydene.2009.04.005
Zheng, S., Zhang, H., Li, Y., Zhang, H., Wang, O., Zhang, J., et al. (2015). Co-occurrence of Methanosarcina mazei and Geobacteraceae in an iron (III)-reducing enrichment culture. Front. Microbiol. 6:941. doi: 10.3389/fmicb.2015.00941
Keywords: extracellular electron uptake, methanogen, Methanosarcina, direct interspecies electron transfer (DIET), electromethanogenesis, GAC (Granular Activated Carbon), Geobacter
Citation: Yee MO, Snoeyenbos-West OL, Thamdrup B, Ottosen LDM and Rotaru A-E (2019) Extracellular Electron Uptake by Two Methanosarcina Species. Front. Energy Res. 7:29. doi: 10.3389/fenrg.2019.00029
Received: 31 October 2018; Accepted: 05 March 2019;
Published: 02 April 2019.
Edited by:
S. Venkata Mohan, Indian Institute of Chemical Technology (CSIR), IndiaReviewed by:
Sunil A. Patil, Indian Institute of Science Education and Research Mohali, IndiaSarah Glaven, United States Naval Research Laboratory, United States
Copyright © 2019 Yee, Snoeyenbos-West, Thamdrup, Ottosen and Rotaru. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amelia-Elena Rotaru, YXJvdGFydUBiaW9sb2d5LnNkdS5kaw==
†Present Address: Oona L. Snoeyenbos-West, Department of Microbiology and Molecular Genetics, Michigan State University, East Lansing, MI, United States
 Mon Oo Yee
Mon Oo Yee Oona L. Snoeyenbos-West
Oona L. Snoeyenbos-West Bo Thamdrup
Bo Thamdrup Lars D. M. Ottosen
Lars D. M. Ottosen Amelia-Elena Rotaru
Amelia-Elena Rotaru