- 1Biodiversity, Evolution and Adaptation, Natural Environment Research Council, British Antarctic Survey, Cambridge, UK
- 2Museum Victoria, Melbourne, VIC, Australia
- 3Área de Zoología, Facultad de Ciencias, Universidad de Extremadura, Badajoz, Spain
The Southern Ocean is anomalously rich in benthos. This biodiversity is native, mostly endemic and perceived to be uniquely threatened from climate- and anthropogenically- mediated invasions. Major international scientific effort throughout the last decade has revealed more connectivity than expected between fauna north and south of the worlds strongest marine barrier—the Polar Front (the strongest jet of the Antarctic Circumpolar Current). To date though, no research has demonstrated any radiations of marine taxa out from the Southern Ocean, except at abyssal depths (where conditions differ much less). Our phylogeographic investigation including mitochondrial sequences of 135 specimens of one of the most ubiquitous and abundant clades at high southern latitudes, the ophiuroids (brittlestars), shows that one of them, Ophiura lymani, has gone against the flow. Remarkably our phylogenetic and coalescent analyses suggest that O. lymani has successfully invaded the South American shelf from Antarctica at least three times, in recent (Pleistocene) radiation. Many previous studies have demonstrated links within clades across the Polar Front this is the first in which northwards directional movement of a shelf-restricted species is the only convincing explanation. Rapid, recent, regional warming is likely to facilitate multiple range shift invasions into the Southern Ocean, whereas movement of cold adapted fauna (considered highly stenothermal) out of the Antarctic to warmer shelves has, until now, seemed highly unlikely.
Introduction
Marine animals inhabiting continental shelves can disperse widely over ecological time scales due to extensive shelf connectivity, current regimes, and reasonably consistent ocean chemistry (Paulay and Meyer, 2006). Nevertheless, some local endemism occurs in most continental shelf regions (Sanford and Kelly, 2011). Temperature is a strong predictor of biogeographic regions (Belanger et al., 2012) and studies demonstrate that given a warming climate many marine shelf species track pole-wards and into deeper, cooler habitats (Norberg et al., 2012; Pinsky et al., 2013; Hiddink et al., 2015). Shelf areas within the Southern Ocean are unusually isolated compared with those north of the Southern Ocean. They are surrounded by deep seas (>2000 m depth), the strongest oceanic current on the planet, the Antarctic Circumpolar Current (ACC) (Whitworth, 1980; Orsi et al., 1995), and encircled by the Polar Front, a narrow jet within the ACC, across which there are steep gradients in temperature and salinity. These isolating conditions, established in the Oligocene (Barker and Thomas, 2004), have left a signature of high endemicity of the Southern Ocean shelf fauna (Dell, 1972; Clarke and Crame, 1989; Griffiths et al., 2009).
Over the ~24 million years since the opening of the Drake Passage, the Miocene Antarctic benthos has had to respond to the cooling conditions by either adapting in situ or range shifts into appropriate environmental conditions. This for instance could have resulted in species ranges being restricted to the northern continental shelves of South Georgia and South America, while Antarctic shelf populations went extinct (Clarke and Crame, 1989). The onset of glaciation caused extinction in most, but not all, of the terrestrial flora and fauna (Convey et al., 2008), but in the more constant marine environment, much of the fauna has adapted successfully and has become comparably rich (Clarke and Johnston, 2003). There has long been debate regarding the origins of the Antarctic fauna and how much movement there has been of fauna into and out of the Antarctic realm over time since its isolation (Hedgpeth, 1969; Dell, 1972; Clarke and Crame, 1989; Clarke et al., 2005; Barnes et al., 2006; Griffiths et al., 2009). Although there is clearly biogeographic evidence of a South American influence in the Southern Ocean fauna (Griffiths et al., 2009), phylogenetic studies suggest that octopus (Strugnell et al., 2008), and pleurobranch gastropods (Göbbeler and Klussmann-Kolb, 2010) have radiated out of the Antarctic, possibly assisted by the initiation of the formation of Antarctic bottom water, around the time of the Oligocene cooling. There is also molecular evidence suggesting that some Antarctic species and their northern lineages diverged much more recently in the Pliocene or Pliestocene (Page and Linse, 2002; Hunter and Halanych, 2008; González-Wevar et al., 2010; Krabbe et al., 2010; Poulin et al., 2014), while several species are currently thought to be distributed on either side of the Polar Front indicating recurrent gene flow (see Sands et al., 2013 Supplementary Material for ophiuroid examples).
The recently observed increase in Southern Ocean surface temperatures (Meredith and King, 2005; Whitehouse et al., 2008; Turner et al., 2014) is suggested to have serious implications for Southern Ocean biota. The fauna is widely considered to be cold adapted with no option of migration except to the deep sea (Peck, 2005), which in itself requires specific adaptations for survival (Brown and Thatje, 2015). Warming of the Southern Ocean is also likely to facilitate invasion of species from more northern regions. This is most likely to be observed first on the more northern shelf regions of the Southern Ocean, especially the large micro-continental shelf regions of South Georgia and Shag Rocks (Barnes et al., 2009; Hogg et al., 2011; Morley et al., 2014) and the even larger but less studied waters of the Kerguelen Plateau. Despite being located south of the Polar Front, South Georgia is eastwards of the tip of South America and directly affected by the ACC flowing across from the South American Shelf. The benthic fauna of the South Georgia shelf has been described as “Antarctic” (Hogg et al., 2011), although biogeographic work indicates that there is substantial South American influence (Griffiths et al., 2009). There are several shelf-restricted brittle star (Ophiuroidea) species that are located on both the Patagonian and South Georgian shelves (Sands et al., 2013). Given the strong ACC with the potential to promote gene flow we expect a pattern consistent with the South American influence hypothesis that predicts (1) decreasing genetic diversity from the Patagonian Shelf to Antarctic Shelf; (2) older lineages i.e., more basal clades in a phylogeny, will consist of Patagonian Shelf specimens and (3) tests for gene flow reveal asymmetrical migration in the direction of the ACC from the Patagonian shelf population to South Georgia. Using a fragment of the mitochondrial cytochrome c oxidase subunit 1 gene (CO1) amplified from collections of the brittle star Ophiura lymani and its sister species Ophiura carinifera, acquired throughout their ranges around the Southern Ocean, we tested these expectations under the hypothesis of South American origin against the alternative hypothesis of Antarctic origin.
Materials and Methods
Our approach was to first test for genetic structure within our samples using phylogenetic methods. We then conducted tests using coalescent techniques to best identify the processes that underlying the phylogenetic patterns identified.
Specimens of O. lymani and O. carinifera were collected on several international expeditions (JR 144, JR179, JR230, PS77, ANDEEP, ICEFISH, CEAMARC, and MacRidge 2) using Agassiz Trawls. Sampling locations are illustrated in Figure 1. Samples were sorted immediately on collection and stored in 96% ethanol. Individuals were identified morphologically to species level.
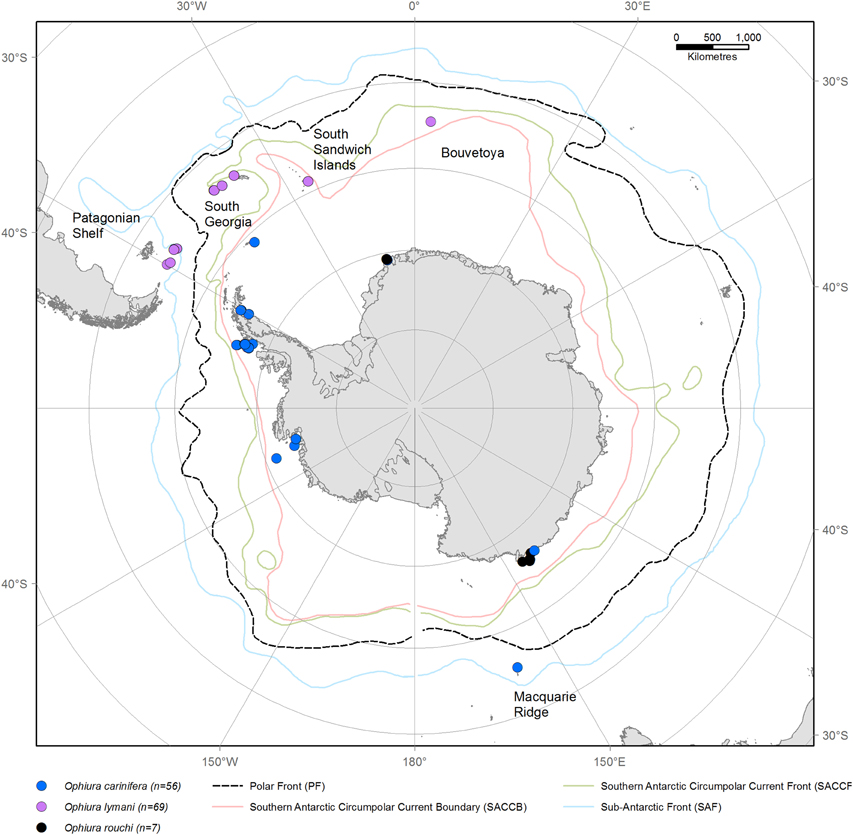
Figure 1. Map of Antarctica and the Southern Ocean showing sampling locations of Ophiura lymani (purple) and Ophiura carinifera (blue) and Ophiura rouchi (green). The approximate positions of the fronts defining the ACC are indicated with the Polar Front indicated in a black dashed line.
Tissue samples taken from arm tips were used for DNA extraction. DNA extraction and sequencing of partial cytochrome c oxidase subunit 1 (CO1) was carried out by the Canadian Center for DNA Barcoding using the primers LCOech1aF1 and HCO2198 (Folmer et al., 1994), with sequences and metadata stored and made publically available on the Barcode of Life Data Systems (Ratnasingham and Hebert, 2007). Forward and reverse trace files were downloaded from the database and assembled and edited in CodonCode Aligner V5.0.1. Sequences were translated to ensure an open reading frame, and aligned using the MUSCLE (Edgar, 2004) alignment process. We included sequences from Ophiura rouchi as the most reasonable outgroup and to stabilize more basal nodes (Felsenstein, 1981) we included two further outgroups, Ophiocten dubium and Ophioperla koehleri.
Descriptive statistics produced using DnaSP v5.10.01 (Librado and Rozas, 2009) and Arlequin v3.5 (Excoffier et al., 2005). Departures from drift mutation equilibrium were tested by comparing estimates of nucleotide diversity with segregating sites—Tajima's D (Tajima, 1989)—and by assessing observed vs. expected haplotype diversity—Fu's Fs (Fu, 1996). Both these statistics are affected by population size changes as well as selection. Significance was assessed from 1000 coalescent simulations in DnaSP. We conducted mismatch analysis (Rogers and Harpending, 1992) to test whether the frequency distribution of pairwise differences conformed with the random (ragged) distribution expected under equilibrium or whether the distribution appeared non-random as expected under past population change. P-values were generated by conducting 1000 bootstrap pseudoreplicates in Arlequin. Within and among clade distances were calculated in MEGA v5.2.2 (Tamura et al., 2011). Base compositional heterogeneity was examined using a match paired test in SeqVis v1.5 (Ho et al., 2006).
To visualize the change of effective size through time in each population, Bayesian Skyline (Drummond et al., 2005) and Bayesian Skyride plots (Minin et al., 2008) were produced in BEAST v1.8.1(Drummond and Rambaut, 2007) using coalescent Skyline and Gaussian Markov random field Bayesian Sky Ride tree priors respectively, and a mutation rate of 2.48 × 10 −8 (see below). Ten million generations of MCMC produced 10,000 trees. TRACER v1.6 (Rambaut et al., 2014) was used to check diagnostics of convergence and effective sample sizes of parameters, as well as to analyse the trees and produce the plots.
To determine the relationships among O. lymani populations and the closely related High Antarctic species O. carinifera, we produced a Maximum Likelihood estimate of phylogeny using RAxML v8.0.26 (Stamatakis, 2014) and a Bayesian estimate using the program MrBayes v3.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). In order to account for violations of assumptions of stationarity and heterogeneity we used JModeltest v2 (Darriba et al., 2012) and used AIC to determine that GTR+I+G was the most suitable substitution model. As proportion of invariant sites (I) is closely correlated with the gamma distribution (+G) we used the GTR+G option in RAxML, with 10 replicate runs on both a codon partitioned dataset and a non-partitioned dataset. We ran 1000 “thorough” (as opposed to the “rapid” option) bootstrap pseudoreplicates to estimate node support. A codon partitioned dataset was used in our Bayesian analysis to better account for variation in substitution rates across sites and lineages. Each partition was set to estimate all parameters (nst = 6) effectively allowing a GTR model with a gamma distribution. Phylogenies were estimated using four replicates of four heated chains over 108 generations with a 25% burnin that ensured trees were sampled only once split frequencies fell below 0.01. Convergence and effective sample size estimates of parameters were checked using TRACER v1.6. An alternative visualization of the relationships among haplotypes of O. lymani sampled from the Patagonian shelf and the South Georgia/Shag Rocks shelf areas was generated in the form of a statistical parsimony network (Templeton et al., 1992) using the package TCS v1.21 (Clement et al., 2000). From this reduced dataset we constructed a Bayesian phylogeny as described above to be used as the gene tree estimate in coalescent simulations.
Species delimitation was explored using popular analytic techniques. Automatic barcode gap discovery (Puillandre et al., 2012) is a distance-based technique that explores a predicted “gap” in the frequencies of pairwise distances indicating the discrete change in intra- and interspecific evolutionary processes. Parameters Pmin (lowest expected distance to include a barcode gap), Pmax (maximum expected distance to include a barcode gap), X (relative width of the barcode gap), and distance correction (JC69, K2P, SimpleP) were adjusted to best encompass the range of variation in the data (Pmin = 0.001, Pmax, X = 0.1, K2P distance). The resulting number of species groups was estimated after the stabilization of recursive partitioning of the data. The general mixed Yule coalescent (GMYC) method is a phylogenetic technique (Pons et al., 2006) that was implemented in the R package SPLITS (Fujisawa and Barraclough, 2013) using an ultrametric tree produced in the software BEAST v1.8.1 (Drummond and Rambaut, 2007) as described below.
Where results of non-monophyly between regions were observed we used a test of the cost of deep coalescence (cDC) events required to explain the data compared with that of distribution of coalescent trees simulated from the original gene tree. As the coalescent process is stochastic, any pattern inferred by phylogeny may be chance—rather than process—driven. In the software package Mesquite v3.01 (Maddison and Maddison, 2014) we constructed a species tree specifying a topology representing two populations: Patagonian shelf and South Georgian shelf. The Bayesian gene tree estimated from the reduced dataset was forced into the species tree and the cDC recorded. The same gene tree was used as a starting point to simulate 10,000 coalescent trees, each one forced to fit the species tree and the number of deep coalescent events noted. The cDC from the gene tree was compared to a distribution of the cDC from the simulations.
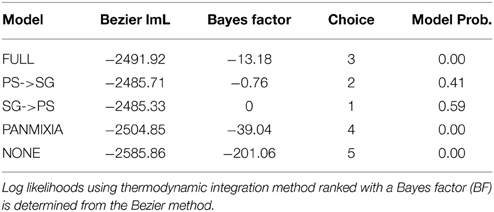
The likelihood of the exchange of migrants between the Patagonian shelf and South Georgia shelf was tested in Migrate v3.6.4 (Beerli, 2009) by examining the posterior distribution of migration estimates from an unrestricted model allowing full symmetrical migration, and then by ranking competing migration models using thermodynamic integration with a Bayes factor test (Beerli and Palczewski, 2010). The models examined were symmetrical migration (PS<−>SG), asymmetrical migration (PS->SG), (PS<-SG), panmixia (PS = SG) and no migration (PS-x-SG). Prior to running these models, Migrate-n settings were optimized using a full model (symmetrical migration) to ensure priors were appropriate and runs would converge. Final run conditions were 10 replicate long chains each with four heated chains with temperatures 1, 1.5, 3, 106. Each chain was run for 106 generations, sampling every 100 generations.
The timing of divergence events was estimated using a strict molecular clock in BEAST v1.8.1 (Drummond and Rambaut, 2007). We allowed a GTR + G model on partitioned codon sites and a Yule process speciation prior. A lognormal prior was used for “clock rate” such that the median reflected the mutation rate per lineage per year (2.48 × 10 −8) as determined by Naughton et al. (2014) and 95% distribution between 1.2 × 10 −8 and 8.7 × 10 −8 thus covering a broad range of uncertainty. Sixteen test runs of 108 generations were used to test and adjust priors and their effects on convergence, mixing and parameter estimates.
All data and meta-data used in this study has been made publically available on BOLD Systems (www.boldsystems.org) under the project code DS-MS001 (DOI: dx.doi.org/10.5883/DS-MS001) and on GenBank with accession numbers KR861569-KR86164.
Results
The Samples
A total of 69 O. lymani specimens were collected; 33 from the Patagonian shelf, 31 from the South Georgia/Shag Rocks shelf, three from South Sandwich Islands and two close to the remote Bouvetoya. Morphological examination of all specimens supported a single morpho-species.
O. carinifera, the sister taxon to O. lymani, is clearly identifiable by strongly elevated and granulated disc plates and radial shields (flattened without granules in O. lymani), and narrow dorsal arm plates with a serrated keel (wide without serrated rest in O. lymani). We included 56 specimens sampled from around the Antarctic continental shelf—from the Amundsen Sea, Bellingshausen Sea, Larsen shelf, South Orkney shelf and the south-east region of theWeddell Sea, east Antarctica near the Mertz Glacier and a specimen from the Macquarie Ridge. The Macquarie Ridge specimen (OPHAN310) was morphologically distinct from all others having a more flattened disc, similar to O. lymani and genetically divergent from all other O. carinifera, appearing to be the sole representative of a sister clade (Figure 2).
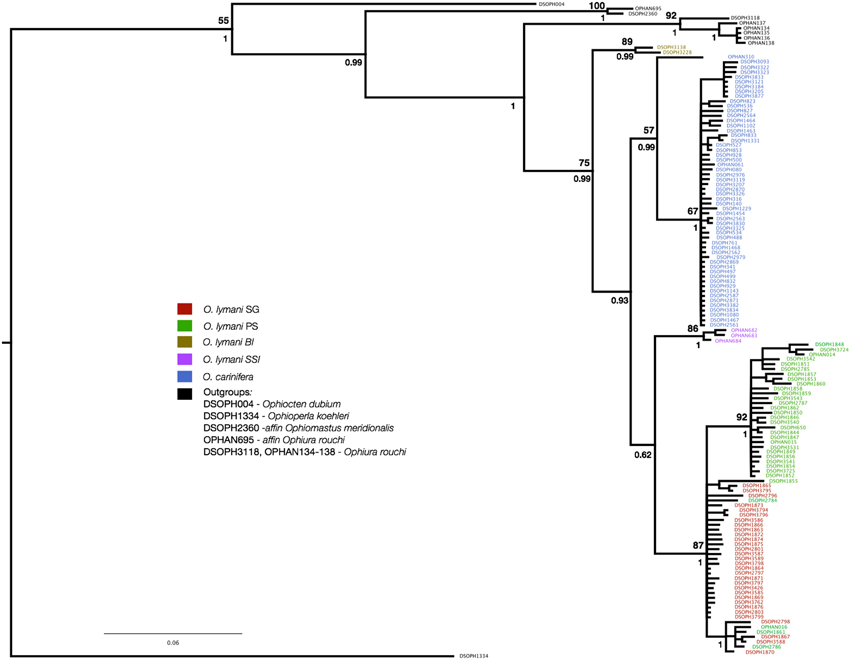
Figure 2. Bayesian consensus phylogeny of partitioned CO1 sequences derived from Ophiura lymani from South Georgia shelf (red) Patagonian shelf (green), South Sandwich Islands (purple) and Bouvetoya (mustard). Ophiura carinifera samples are blue, Ophiura rouchi samples are black. Posterior probabilities are given under the line by the associated adjacent node and bootstrap values are given above the line. No value was given if bootstrap was less than 50%. The identifiers on the tips are BOLD system accession numbers for the individual sequences.
Molecular Diversity
Our alignment of 132 sequences consisted of 658 bases, of which 138 were variable and 97 were parsimony informative. As expected with mitochondrial sequence there was a bias toward Adenosine and Thymine residues (GC = 39.2%).
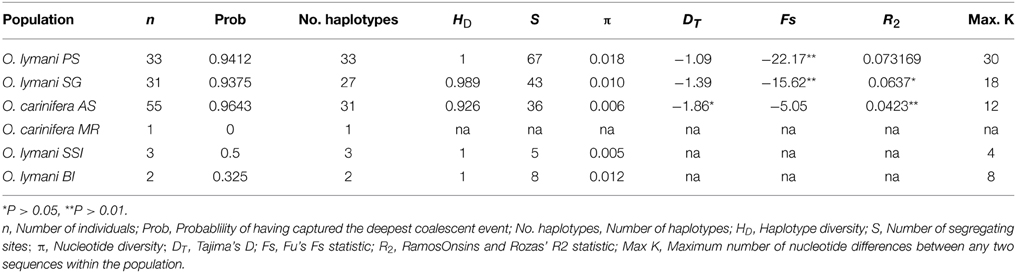
Haplotype diversity (HD) was exceptionally high in all populations of O. lymani and O. carinifera (Table 1). Tajima's D was negative, although not significantly different from zero in O. lymani. Significant negative values as seen in O. carinifera may indicate either purifying selection or population size change. Fu's Fs was also negative as to be expected given the high haplotype diversity. Fu's Fs was highly significant in the O. lymani populations from the Patagonian shelf and South Georgian shelf, both of which have very high haplotype diversity. Significant R2-values also suggest population expansion, but only in O. carinifera and the South Georgia population of O. lymani. The frequency distribution of pairwise differences under a constant population is expected to be largely random, whereas the expectation under population growth is for a smooth curve. Although non-significant, the two O. lymani populations, particularly the South Georgia population, appeared to more closely fit the expectations of population expansion (Figure 3). Coalescent-based demographic reconstruction using Bayesian Skyline and Skyride plots indicated a population increase over the past few glacial cycles in all three populations (Figure 4). The Skyline plots show a sharp increase in population size starting 100+ ka, whereas smoothing algorithms in the Skyride plots indicate a steady increase over time.
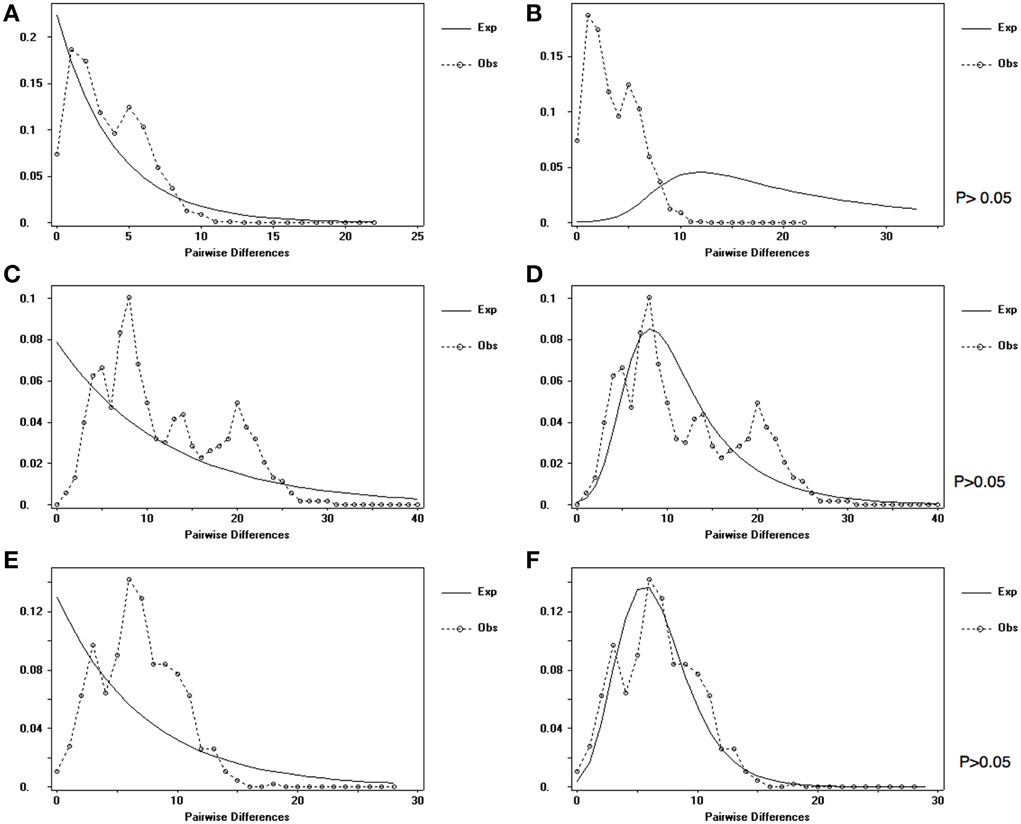
Figure 3. The expected (dotted line) and observed (solid line) distribution of pairwise frequencies between haplotypes under the expectation of constant size (A,C,E) and exponential growth (B,D,F) for O. carinifera (A,B), O. lymani from the Patagonian Shelf (C,D) and O. lymani from the South Georgia shelf (E,F).
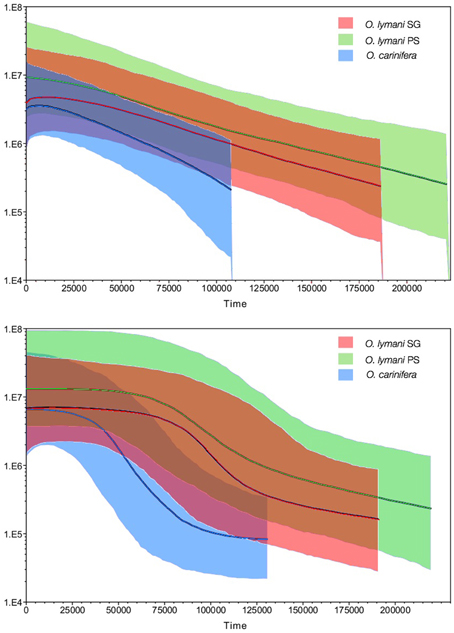
Figure 4. Bayesian SkyRide (top) and SkyLine (bottom) plots indicating change in population size over time in O. carinifera (blue), O. lymani from the Patagonian Shelf (green) and the South Georgia shelf (red).
Phylogenetic Reconstruction
Phylogenetic reconstruction identified strongly supported clades of geographically defined populations of O. lymani, with the exception of five individuals sampled from the Patagonian shelf that grouped with the South Georgian shelf individuals, and a single Antarctic-wide clade including two lineages of O. carinifera (Figure 2). The low support of the node containing the South Sandwich Islands, South Georgia and Patagonian Shelf clades (pp. 0.62) persisted despite trialing various plausible evolutionary models and partitioning schemes. Low support for nodes should not always be interpreted as phylogenetic failure, but may rather reflect a genuine (hard) polytomy due to a radiation or series of speciation events between which there was insufficient time to accumulate mutational variation to permit accurate reconstruction (Hoelzer and Meinick, 1994).
The statistical parsimony network highlighted five individuals sampled from the Patagonian shelf that either originated from the South Georgia clade, or are retained ancient lineages indicative of incomplete lineage sorting (Figure 5). In order to determine the likelihood of these lineages being retained, we determined the cost of deep coalescence (cDC) required to explain the The distribution of cDC of the simulated trees had a mean of 48 (41–55, 95% CI). The cDC estimated from the Bayesian phylogeny was 4, well outside of this distribution, indicating that chance alone is very unlikely to explain the low number of deep coalescences required to produce this species tree from the gene tree.
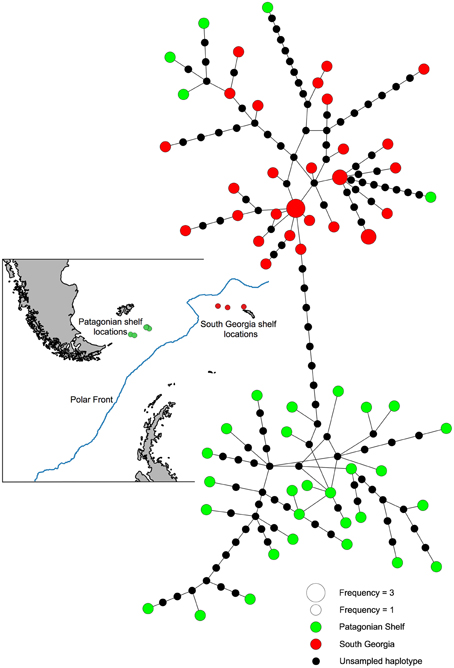
Figure 5. Statistical Parsimony network indicating sampling locations and genetic relationships among Patagonian (green) and South Georgian (red) Ophiura lymani haplotypes.
Species Delimitation
Genetic distances between populations were considerably greater than within-population distances, and generally greater than are expected for conspecific populations (Table 2). Visual inspection suggests that clades representing O. lymani from Bouvet, the South Sandwich Islands and the Patagonian shelf/South Georgia are likely to be three distinct species, considering that the clearly morphologically distinct O. carinifera is grouped within the pan O. lymani clade. Depending on the relative gap width setting (X), ABGD identified these groupings, as well as OPHAN310 (O. carinifera from Macquarie Ridge) and DSOPH1855 (O. lymani from Patagonian shelf grouped into the South Georgia clade), implied to be two distinct species when X is set to 1. For larger values of X, ABGD failed to stabilize.
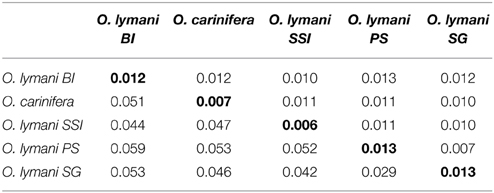
Table 2. Average pairwise distances between clades (below diagonal), associated standard error estimates (above diagonal) and within clades distances (along the diagonal).
The phylogenetic method was less conservative: with the likelihood test finding the GMYC model a significant improvement over the null model (HGMYC 505.08 v H0 492.12, P < 0.0001). The test identified 14 entities (CI 9–24) and 11 clusters (CI 6–16).
Gene Flow
Visual inspection of the phylogenies suggested distinct geographically distinct clades with the exception of five individuals from the Patagonian shelf that are more closely related to South Georgia haplotypes than other Patagonian haplotypes. As coalescent simulations suggest this pattern is non-random, we investigated migration between these regions.
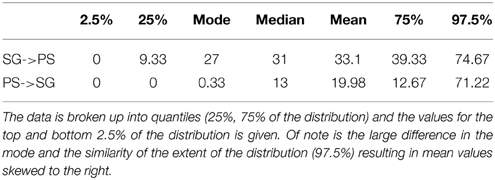
The full model, allowing co-estimation of population size variation and free migration, indicates that migration is asymmetrical from South Georgia to the Patagonian shelf (Table 3). The posterior distribution of migration rates has clear modes: at 0.3 for migration from Patagonia to South Georgia, and 27 for South Georgia to Patagonia (Figure 6). A comparative exploration of migration scenarios strongly supports asymmetrical migration scenarios over symmetrical migration, free migration (panmixia) or no migration (Table 4). The scenario of migration from South Georgia to the Patagonian shelf was favored (probability of 0.59) and, although support for this model was only marginal compared with migration from the Patagonian shelf to South Georgia (probability of 0.41), it is hard to reconcile the pattern visualized in Figure 5 with migration from the Patagonian shelf as no “Patagonian shelf”-like haplotypes were found around South Georgia. This seemingly marginal result lies at least partly with the incompatibility of the distributions being compared (Figure 6). Given that the posterior distribution must be positive, and the large amount of variation expected in a single marker analysis, even a “true” value of 0 will have a wide distribution and positive mean. In this case the distribution of 97% of the data was similar for both scenarios, producing a high mean (19.98) for the PS->SG migration estimate (Table 3) even though the mode (highest posterior estimate) was near 0, compared with SG->PS where the mode (27) was much closer to the mean (33.1). Low confidence due to wide distribution of the posterior is largely down to the use of a single marker.
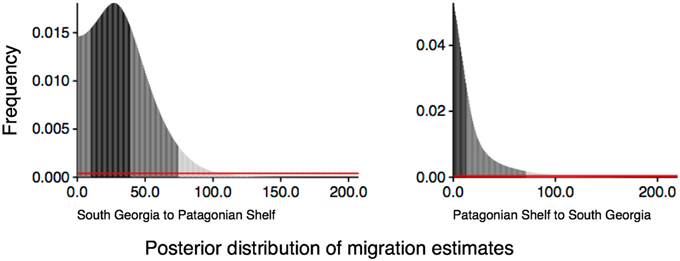
Figure 6. The distribution of the posterior migration estimates from the analysis of the full migration model allowing migration between South Georgia to Patagonian shelf and Patagonian shelf to South Georgia.
Timing of Divergences
The root height of the tree was estimated as 8.3 million years ago (ma) (5.77, 11.07 95% HPD). The main radiation of the O. lymani/O. carinifera clades (nodes B–D, Figure 7) was inferred to be around 2.5 ma. The most recent common ancestor of the Patagonian shelf and South Georgia shelf populations (node G) was estimated at around 1.5 ma. A similar time frame was estimated for the common ancestor of the Antarctic O. carinifera and its Macquarie Ridge relative (node E). The two groups of Patagonian shelf O. lymani that cluster within the South Georgia clade indicate that there were at least two separate migration events, one around 0.75 ma (node J) and the second around 0.22 ma (node L) although the time dependency of the molecular clock would affect these date estimates strongly and they should be considered to be more recent.
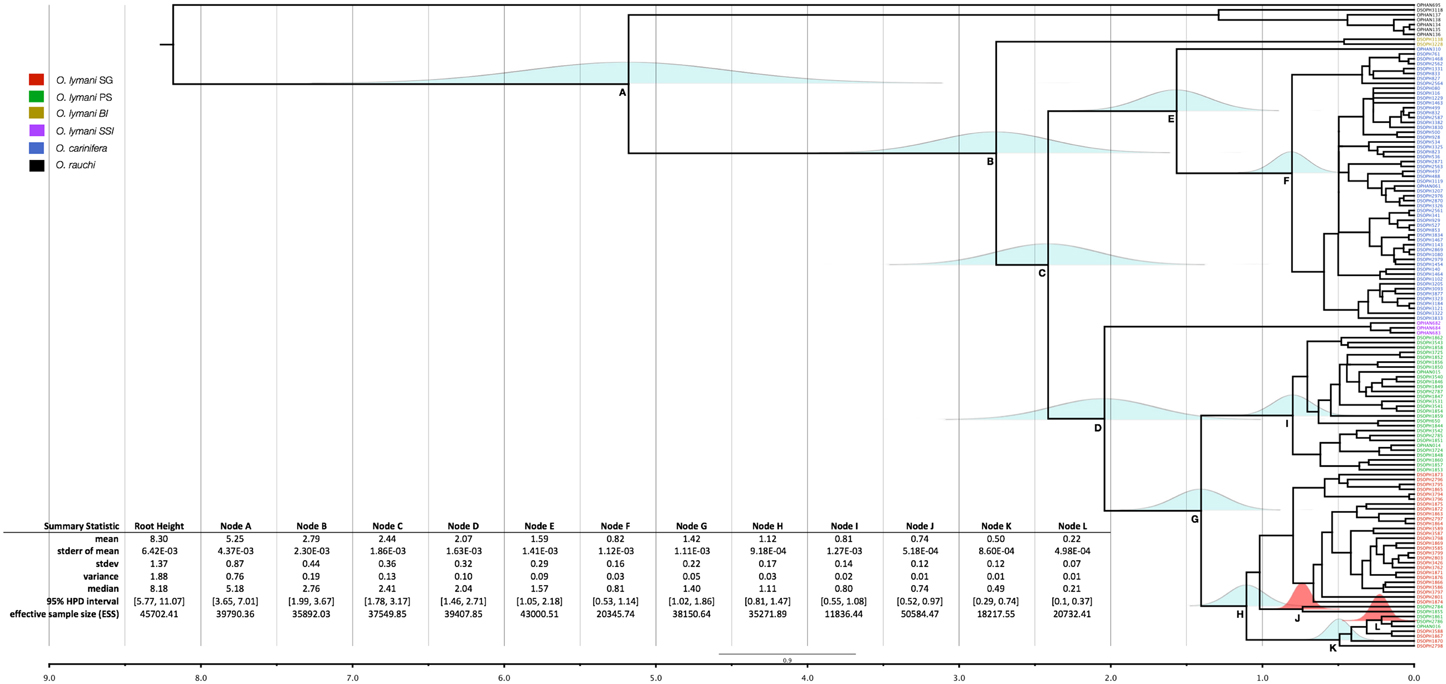
Figure 7. Bayesian chronogram of partitioned CO1 sequences derived from Ophiura lymani from South Georgia shelf (red) Patagonian shelf (green), South Sandwich Islands (purple) and Bouvetoya (mustard). Ophiura carinifera samples are blue, Ophiura rouchi samples are black. The distribution of uncertainty of node placement is indicated around each of the main nodes in light blue. The distribution of uncertainty around the nodes indicating migration events from South Georgia to Patagonian shelf are indicated in red. The scale bar is in millions of years.
Discussion
The works of Hedgpeth (1969) and Griffiths et al. (2009) provide a biogeographic hypothesis in which the ACC strongly affect the distribution of Southern Ocean benthos by facilitating gene flow over vast geographic distances, and yet inhibiting gene flow particularly in and out of Antarctica. These processes have resulted in many species with circumpolar distributions, and assemblages that indicate a South American component, particularly at South Georgia. Our expectations under the South American input hypothesis were that there would be (1) appreciably more genetic diversity and (2) deeper coalescent times in the Patagonian Shelf populations, and (3) indication of asymmetrical movement along the direction of the ACC. However, our phylogeographic examination of O. lymani and O. carinifera demonstrated (1) very high diversity in all populations, (2) deeper coalescence times for the South Georgia population than any other, and (3) a mitochondrial signal of asymmetrical migration that appears to go against the ACC from South Georgia to the Patagonian shelf. Importantly, it appears that there are several independently evolving lineages of O. lymani, with the diverging Patagonian shelf lineage breaking out of the Antarctic realm and thereby rejecting the South American input hypothesis in this case.
Crypsis
The populations of Bouvetoya and South Sandwich Islands are as genetically distinct from each other, and the Patagonian shelf and South Georgia populations, as each is also from O. carinifera, a clearly different morpho-species. Cryptic or unrecognized species (Janosik and Halanych, 2010) are commonly identified by mitochondrial DNA studies of benthic invertebrates (For Antarctic examples see Linse et al., 2007; Leese and Held, 2008; Krabbe et al., 2010; Arango et al., 2011; Havermans et al., 2011; Hemery et al., 2012). There is considerable debate as to whether or not biological species should be delimited based solely on mitochondrial DNA sequence data (DeSalle et al., 2005; Taylor and Harris, 2012; Zhang et al., 2013). In this case, based on clear genetic clades that correspond to discrete geographic areas, distinct morphology, and prior evidence that CO1 reliably distinguishes ophiuroid species (Ward et al., 2008), we can assume that O. lymani could be conservatively divided into a Bouvetoya species, a South Sandwich Island species and a diverging Patagonian shelf/South Georgia species.
Against the Flow
It appears counter intuitive that the most closely related populations are separated by what should be an intense barrier to gene flow. Similarly, despite currents that could facilitate movement from Patagonia to South Georgia, or South Georgia to the South Sandwich Islands or Bouvetoya, there is no evidence of movement in this direction or that currents themselves are responsible for population structuring. The only evidence of migration is from South Georgia to the Patagonian shelf, against the ACC and across a gradient between two water masses that strongly differ in temperature and salinity (Orsi et al., 1995).
There are three possible mechanisms for dispersal: larval transport, active adult dispersal or passive adult dispersal. There is no published literature regarding the reproductive biology of O. lymani or O. carinifera. However, lack of obvious genetic structure of O. carinifera haplotypes around Antarctic continental shelves and O. lymani haplotypes between Shag Rocks and South Georgia shelf regions indicate moderate dispersal is possible. The narrow but deep gap between South Georgia and Shag Rock shelf fragments is known to inhibit gene flow in more mobile species (Allcock et al., 1997; Strugnell et al., 2012), and yet is observed in O. lymani, suggesting that O. lymani may have a more mobile life stage. Transport of larvae in ships ballast water is a possible mechanism, although the genetic distances of the haplotypes suggest the timing of the movement substantially predates shipping in the area.
Dispersal could occur by moving down slope and along abyssal sea floor. Such a deep sea link has been used to explain the close genetic connectivity between sub-Antarctic and Antarctic morphotypes of the echinoid Sterechinus (Díaz et al., 2011). The limits of adult O. lymani/carinifera dispersal are unknown. They are moderately large (20+ mm disc size) and muscular, and certainly able to move across the sea floor (Sands, personal observation). Their depth range is unknown, largely due to the rarity of deep sampling events in the region. In the collections available to this study, O. lymani and O. carinifera were only recovered in the shallower trawls (194–723 and 153–556 m depth, respectively), and absent from deeper trawls (20 trawls between 1000 and 3213 m) indicating that they prefer shelf and upper slope depths. Therefore, while dispersal of adults remains possible, it appears unlikely.
Rafting, where individuals are attached to or entangled in kelp, pumice or other buoyant substrata and drift from one area to another, is a third possible mechanism. Drifting kelp is common in the Southern Ocean (Fraser et al., 2009) and is a known dispersal mechanism for a variety of benthos (Barnes et al., 2006; Leese et al., 2010). However, rafting from South Georgia to Patagonian shelf would require either a reversal of the ACC or an almost complete circumpolar journey that, if sufficiently frequent to result in detectable gene flow, would be considerably higher in other closer shelf areas more convincingly in the path of the ACC and associated eddies, such as South Sandwich Islands and Bouvetoya.
Two other scenarios may explain the apparent migration pattern between South Georgia and Patagonia but without the need to invoke migration against the flow of the ACC. One is vicariance as a result of continental drift with retained ancestral lineages. Our results suggest that the most recent common ancestor of the clade (Node B, 2.8 ma) considerably post-dates the estimated opening of the North Scotia Ridge sufficient to allow the northward flow of the ACC around 17 Ma (Eagles, 2010), and coalescent analyses indicate that retained ancestral lineages are an unlikely explanation. A second scenario is that both populations are supplied by an un-sampled source as in the case of the limpet Nacella magellanica, where samples from the Atlantic South American coast and Falkland Islands show two highly divergent lineages, but also more closely related lineages and one shared haplotype. Sampling the Pacific Patagonian shelf revealed that the shared haplotype between the Atlantic population and the Falkland Islands population was the most frequent haplotype from the Pacific region (González-Wevar et al., 2012). We hope that further sampling opportunities will open up to test this possibility.
Out of Antarctica
A clear pattern in our data is the Antarctic or sub-Antarctic origin of the O. lymani / O. carinifera clade. The change in morphology from O. lymani type to O. carinifera type occurred between 2.5 and 1.6 ma, and was maintained on the Antarctic continental shelf from around 0.8 ma. Movement out of the sub Antarctic occurred between 1.4 and 0.8 ma, with two subsequent migration events 0.74 and 0.22 ma. Poulin et al. (2014) summarized the divergences of several taxa across the Drake Passage and found that divergences generally occurred around 4–5 ma, although the brittle star Astrotoma agassizi and the sea star Odontaster validus had more recent divergences of 1.5–1 ma using a 3.5% mutation rate (Hunter and Halanych, 2008; Janosik et al., 2011) which, if standardized to the mutation rate used here, would be in line with our results. Although there is some evidence of benthic groups radiating out of the Antarctic during the Tertiary (Strugnell et al., 2008; Göbbeler and Klussmann-Kolb, 2010; González-Wevar et al., 2010), and there are populations of some benthic species either side of the Polar Front (see Supplementary Tables in Sands et al., 2013), to our knowledge this is the first evidence strongly suggesting Quaternary asymmetrical movement out of the Antarctic and into warmer waters. At a time when regional warming is causing a detectable shift in range distributions toward the poles (Thomas, 2010), and there is an expectation that indigenous Antarctic cold-adapted species may be threatened by the warming marine environment (Peck, 2005), the example of O. lymani shows that at least one species has had the ability to migrate and adapt to warmer temperature regimes over time.
Caveats and Interpretation Limits
Although evidence for crypsis, asymmetrical migration and radiation out of Antarctica appears compelling, there are caveats that apply to the current results and interpretations. The DNA sequence marker cytochrome c oxidase subunit 1 is widely used and heavily relied on in species identification and biodiversity studies. However, we emphasize that it is a single line of evidence and despite our best attempts at controlling for stochastic or confounding processes, without other gene sequences we cannot be overly confident that our gene trees reflect the species tree. Similarly, our mutation rates are best estimates, and the process they are derived from (the closing of the Isthmus of Panama) is in itself a best estimate, so the dates and error distributions provided here should be treated with great caution. The analyses here assume mutation drift equilibrium and selective neutrality, neither of which are safe assumptions in real populations. Finally, the sampling, although the best available, is still limited due to the enormous time and expense of carrying out surveys in the Southern Ocean and remote southern areas of other oceans. Perhaps the most significant sample related limitation for this study is the geopolitical boundaries on the South American continent, which have prevented wide sampling of the Patagonian shelf. However, with these caveats in mind, we can still interpret clear patterns of diversification within the Southern Ocean and what appears to be a recent movement from of lineages from South Georgia to the Patagonian Shelf.
Conclusions
We have found evidence that the ophiuroid O. lymani has radiated in the Southern Ocean, with deep sea between isolated lineages likely to be the mechanism maintaining distinct populations or cryptic species. Our results indicate occasional migration over evolutionary time against the prevailing ACC and across the perceived barrier of the Polar Front. We provide evidence that O. lymani is a species complex of which O. carinifera is an element, with the South Georgia population highly divergent from other geographic populations in the Southern Ocean, and the likely source of an invasion into the cool temperate seas of South America. These results and hypotheses may cause a rethink in our general view of Antarctic biogeography, but will require further testing including better South American sampling, replication of markers, and extension to other taxa.
Author Contributions
CS conceived the study, collected the majority of the specimens, acquired funding for the molecular sequencing, conducted the analyses and wrote the manuscript. TO provided additional specimens and sequences, helped shape the manuscript. DB assisted in the collections both financially and practically, contributed to the text and provided intellectual input throughout the project. RL identified the majority of the specimens, provided unique and relevant observations and contributed to the production of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Captains and crews of the RRS James Clarke Ross (JR144) and RV Polarstern (PS77) for their commitment to excellent science discovery, and the science leaders of the cruises that contributed to the collection of this material. Thanks to Elena Zaikina for assisting RL, Rachel Downey, Camille Moreau and Huw Griffiths for assistance in sorting material at BAS. Thanks also to Olivia Martin-Sanchez who assisted with the GIS data. Jennifer Jackson, Elise Biersma, Peter Convey and Will Goodall-Copestake provided useful discussion throughout the analysis and writing process. Funding for the sequencing service was supported by Census of Antarctic Marine Life. CS was supported by an Antarctic Science Bursary, a SCAR MarBIN grant and a SynTax award scheme supported by BBSRC and NERC.
References
Allcock, A. L., Brierley, A. S., Thorpe, J. P., and Rodhouse, P. G. (1997). Restricted gene flow and evolutionary divergence between geographically separated populations of the Antarctic octopus. Pareledone turqueti. Mar. Biol. 129, 97–102. doi: 10.1007/s002270050150
Arango, C. P., Soler-Membrives, A., and Miller, K. J. (2011). Genetic differentiation in the circum—Antarctic sea spider Nymphon australe (Pycnogonida; Nymphonidae). Deep Sea Res. Part II Top. Stud. Oceanogr. 58, 212–219. doi: 10.1016/j.dsr2.2010.05.019
Barker, P. F., and Thomas, E. (2004). Origin, signature and palaeoclimatic influence of the Antarctic Circumpolar Current. Earth Sci. Rev. 66, 143–162. doi: 10.1016/j.earscirev.2003.10.003
Barnes, D. K. A., Griffiths, H. J., and Kaiser, S. (2009). Geographic range shift responses to climate change by Antarctic benthos: where we should look. Mar. Ecol. Prog. Ser. 393, 13–26. doi: 10.3354/meps08246
Barnes, D. K. A., Hodgson, D. A., Convey, P., Allen, C. S., and Clarke, A. (2006). Incursion and excursion of Antarctic biota: past, present and future. Glob. Ecol. Biogeogr. 15, 121–142. doi: 10.1111/j.1466-822X.2006.00216.x
Beerli, P. (2009). How to use MIGRATE or why are Markov chain Monte Carlo programs difficult to use. Popul. Genet. Anim. Conserv. 17, 42–79.
Beerli, P., and Palczewski, M. (2010). Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 185, 313–326. doi: 10.1534/genetics.109.112532
Belanger, C. L., Jablonski, D., Roy, K., Berke, S. K., Krug, A. Z., and Valentine, J. W. (2012). Global environmental predictors of benthic marine biogeographic structure. Proc. Natl. Acad. Sci. U.S.A. 109, 14046–14051. doi: 10.1073/pnas.1212381109
Brown, A., and Thatje, S. (2015). The effects of changing climate on faunal depth distributions determine winners and losers. Glob. Change Biol. 21, 173–180. doi: 10.1111/gcb.12680
Clarke, A., Barnes, D. K. A., and Hodgson, D. A. (2005). How isolated is Antarctica? Trends Ecol. Evol. 20, 1–3. doi: 10.1016/j.tree.2004.10.004
Clarke, A., and Crame, J. A. (1989). The origin of the Southern Ocean marine fauna. Geol. Soc. Lond. Spec. Publ. 47, 253–268. doi: 10.1144/GSL.SP.1989.047.01.19
Clarke, A., and Johnston, N. M. (2003). Antarctic marine benthic diversity. Oceanogr. Mar. Biol. Annu. Rev. 41, 47–114.
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Convey, P., Gibson, J. A. E., Hillenbrand, C.-D., Hodgson, D. A., Pugh, P. J. A., Smellie, J. L., et al. (2008). Antarctic terrestrial life - challenging the history of the frozen continent? Biol. Rev. 83, 103–117. doi: 10.1111/j.1469-185X.2008.00034.x
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Dell, R. K. (1972). Antarctic benthos. Adv. Mar. Biol. 10, 1–216. doi: 10.1016/s0065-2881(08)60416-2
DeSalle, R., Egan, M. G., and Siddall, M. (2005). The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philos. Trans. R. Soc. B Biol. Sci. 360, 1905–1916. doi: 10.1098/rstb.2005.1722
Díaz, A., Féral, J.-P., David, B., Saucède, T., and Poulin, E. (2011). Evolutionary pathways among shallow and deep-sea echinoids of the genus Sterechinus in the Southern Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 58, 205–211. doi: 10.1016/j.dsr2.2010.10.012
Drummond, A. J., Rambaut, A., Shapiro, B., and Pybus, O. G. (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192. doi: 10.1093/molbev/msi103
Drummond, A., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Eagles, G. (2010). South Georgia and Gondwana's Pacific margin: lost in translation? J. South Am. Earth Sci. 30, 65–70. doi: 10.1016/j.jsames.2010.04.004
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Excoffier, L., Laval, L. G., and Schneider, S. (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinformatics Online 1, 47–50.
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Fraser, C. I., Hay, C. H., Spencer, H. G., and Waters, J. M. (2009). Genetic and morphological analyses of the southern bull kelp Durvillaea antarctica (Phaeophyceae: Durvillaeales) in New Zealand reveal cryptic species. J. Phycol. 45, 436–443. doi: 10.1111/j.1529-8817.2009.00658.x
Fu, Y. X. (1996). New statistical tests of neutrality for DNA samples from a population. Genetics 143, 557–570.
Fujisawa, T., and Barraclough, T. G. (2013). Delimiting species using single-locus data and the generalized mixed Yule Coalescent Approach: a revised method and evaluation on simulated data sets. Syst. Biol. 62, 707–724. doi: 10.1093/sysbio/syt033
Göbbeler, K., and Klussmann-Kolb, A. (2010). Out of Antarctica? – New insights into the phylogeny and biogeography of the Pleurobranchomorpha (Mollusca, Gastropoda). Mol. Phylogenet. Evol. 55, 996–1007. doi: 10.1016/j.ympev.2009.11.027
González-Wevar, C. A., Hüne, M., Cañete, J. I., Mansilla, A., Nakano, T., and Poulin, E. (2012). Towards a model of postglacial biogeography in shallow marine species along the Patagonian Province: lessons from the limpet Nacella magellanica (Gmelin, 1791). BMC Evol. Biol. 12:139. doi: 10.1186/1471-2148-12-139
González-Wevar, C. A., Nakano, T., Cañete, J. I., and Poulin, E. (2010). Molecular phylogeny and historical biogeography of Nacella (Patellogastropoda: Nacellidae) in the Southern Ocean. Mol. Phylogenet. Evol. 56, 115–124. doi: 10.1016/j.ympev.2010.02.001
Griffiths, H. J., Barnes, D. K. A., and Linse, K. (2009). Towards a generalized biogeography of the Southern Ocean benthos. J. Biogeogr. 36, 162–177. doi: 10.1111/j.1365-2699.2008.01979.x
Havermans, C., Nagy, Z. T., Sonet, G., De Broyer, C., and Martin, P. (2011). DNA barcoding reveals new insights into the diversity of Antarctic species of Orchomene sensu lato (Crustacea: Amphipoda: Lysianassoidea). Deep Sea Res. Part II Top. Stud. Oceanogr. 58, 230–241. doi: 10.1016/j.dsr2.2010.09.028
Hedgpeth, J. W. (1969). “Introduction to Antarctic zoogeography,” in Distribution of Selected Groups of Marine Invertebrates in Waters South of 35° S. Antarctic Map Folio Series, eds V. C. Bushnell and J. W. Hedgpeth (New York, NY: American Geographical Society), 1–29.
Hemery, L. G., Eléaume, M., Roussel, V., Améziane, N., Gallut, C., Steinke, D., et al. (2012). Comprehensive sampling reveals circumpolarity and sympatry in seven mitochondrial lineages of the Southern Ocean crinoid species Promachocrinus kerguelensis (Echinodermata). Mol. Ecol. 21, 2502–2518. doi: 10.1111/j.1365-294X.2012.05512.x
Hiddink, J. G., Burrows, M. T., and García Molinos, J. (2015). Temperature tracking by North Sea benthic invertebrates in response to climate change. Glob. Change Biol. 21, 117–129. doi: 10.1111/gcb.12726
Ho, J. W. K., Adams, C. E., Lew, J. B., Matthews, T. J., Ng, C. C., Shahabi-Sirjani, A., et al. (2006). SeqVis: visualization of compositional heterogeneity in large alignments of nucleotides. Bioinformatics 22, 2162–2163. doi: 10.1093/bioinformatics/btl283
Hoelzer, G. A., and Meinick, D. J. (1994). Patterns of speciation and limits to phylogenetic resolution. Trends Ecol. Evol. 9, 104–107. doi: 10.1016/0169-5347(94)90207-0
Hogg, O. T., Barnes, D. K. A., and Griffiths, H. J. (2011). Highly diverse, poorly studied and uniquely threatened by climate change: an assessment of marine biodiversity on South Georgia's continental shelf. PLoS ONE 6:e19795. doi: 10.1371/journal.pone.0019795
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hunter, R. L., and Halanych, K. M. (2008). Evaluating connectivity in the brooding brittle star Astrotoma agassizii across the Drake Passage in the Southern Ocean. J. Hered. 99, 137–148. doi: 10.1093/jhered/esm119
Janosik, A. M., and Halanych, K. M. (2010). Unrecognized Antarctic biodiversity: a case study of the genus Odontaster (Odontasteridae; Asteroidea). Integr. Comp. Biol. 50, 981–992. doi: 10.1093/icb/icq119
Janosik, A. M., Mahon, A. R., and Halanych, K. M. (2011). Evolutionary history of Southern Ocean Odontaster sea star species (Odontasteridae; Asteroidea). Polar Biol. 34, 575–586. doi: 10.1007/s00300-010-0916-7
Krabbe, K., Leese, F., Mayer, C., Tollrian, R., and Held, C. (2010). Cryptic mitochondrial lineages in the widespread pycnogonid Colossendeis megalonyx Hoek, 1881 from Antarctic and Subantarctic waters. Polar Biol. 33, 281–292. doi: 10.1007/s00300-009-0703-5
Leese, F., Agrawal, S., and Held, C. (2010). Long-distance island hopping without dispersal stages: transportation across major zoogeographic barriers in a Southern Ocean isopod. Naturwissenschaften 97, 583–594. doi: 10.1007/s00114-010-0674-y
Leese, F., and Held, C. (2008). Identification and characterization of microsatellites from the Antarctic isopod Ceratoserolis trilobitoides: nuclear evidence for cryptic species. Conserv. Genet. 9, 1369–1372. doi: 10.1007/s10592-007-9491-z
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Linse, K., Cope, T., Lörz, A.-N., and Sands, C. (2007). Is the Scotia Sea a centre of Antarctic marine diversification? Some evidence of cryptic speciation in the circum-Antarctic bivalve Lissarca notorcadensis (Arcoidea: Philobryidae). Polar Biol. 30, 1059–1068. doi: 10.1007/s00300-007-0265-3
Maddison, W. P., and Maddison, D. R. (2014). Mesquite: A Modular System for Evolutionary Analysis. Version 3.01. Available online at: mesquiteproject.org
Meredith, M. P., and King, J. C. (2005). Rapid climate change in the ocean west of the Antarctic Peninsula during the second half of the 20th century. Geophys. Res. Lett. 32, L19604. doi: 10.1029/2005GL024042
Minin, V. N., Bloomquist, E. W., and Suchard, M. A. (2008). Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25, 1459–1471. doi: 10.1093/molbev/msn090
Morley, S. A., Belchier, M., Sands, C., Barnes, D. K. A., and Peck, L. S. (2014). Geographic isolation and physiological mechanisms underpinning species distributions at the range limit hotspot of South Georgia. Rev. Fish Biol. Fish. 24, 485–492. doi: 10.1007/s11160-013-9308-8
Naughton, K. M., O'Hara, T. D., Appleton, B., and Cisternas, P. A. (2014). Antitropical distributions and species delimitation in a group of ophiocomid brittle stars (Echinodermata: Ophiuroidea: Ophiocomidae). Mol. Phylogenet. Evol. 78, 232–244. doi: 10.1016/j.ympev.2014.05.020
Norberg, J., Urban, M. C., Vellend, M., Klausmeier, C. A., and Loeuille, N. (2012). Eco-evolutionary responses of biodiversity to climate change. Nat. Clim. Change 2, 747–751. doi: 10.1038/nclimate1588
Orsi, A. H., Whitworth, T. III, and Nowlin, W. D. Jr. (1995). On the meridional extent and fronts of the Antarctic Circumpolar Current. Deep Sea Res. Part Oceanogr. Res. Pap. 42, 641–673. doi: 10.1016/0967-0637(95)00021-W
Page, T., and Linse, K. (2002). More evidence of speciation and dispersal across the Antarctic Polar Front through molecular systematics of Southern Ocean Limatula (Bivalvia: Limidae). Polar Biol. 25, 818–826. doi: 10.1007/s00300-002-0414-7
Paulay, G., and Meyer, C. (2006). Dispersal and divergence across the greatest ocean region: do larvae matter? Integr. Comp. Biol. 46, 269–281. doi: 10.1093/icb/icj027
Peck, L. (2005). Prospects for surviving climate change in Antarctic aquatic species. Front. Zool. 2:9. doi: 10.1186/1742-9994-2-9
Pinsky, M. L., Worm, B., Fogarty, M. J., Sarmiento, J. L., and Levin, S. A. (2013). Marine taxa track local climate velocities. Science 341, 1239–1242. doi: 10.1126/science.1239352
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., Hazell, S., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609. doi: 10.1080/10635150600852011
Poulin, E., González-Wevar, C., Díaz, A., Gérard, K., and Hüne, M. (2014). Divergence between Antarctic and South American marine invertebrates: what molecular biology tells us about Scotia Arc geodynamics and the intensification of the Antarctic Circumpolar Current. Glob. Planet. Change 123, 392–399. doi: 10.1016/j.gloplacha.2014.07.017
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
Rambaut, A., Suchard, M. A., Xie, D., and Drummond, A. (2014). Tracer v1.6. Available online at: http://beast.bio.ed.ac.uk/Tracer
Ratnasingham, S., and Hebert, P. D. N. (2007). Bold: the barcode of life data system (http://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Rogers, A. R., and Harpending, H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569.
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sands, C. J., Griffiths, H. J., Downey, R. V., Barnes, D. K. A., Linse, K., and Martín-Ledo, R. (2013). Observations of the ophiuroids from the West Antarctic sector of the Southern Ocean. Antarct. Sci. 25, 3–10. doi: 10.1017/S0954102012000612
Sanford, E., and Kelly, M. W. (2011). Local adaptation in marine invertebrates. Annu. Rev. Mar. Sci. 3, 509–535. doi: 10.1146/annurev-marine-120709-142756
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Strugnell, J. M., Rogers, A. D., Prodohl, P. A., Collins, M. A., and Allcock, A. L. (2008). The thermohaline expressway: the Southern Ocean as a centre of origin for deep-sea octopuses. Cladistics 24, 853–860. doi: 10.1111/j.1096-0031.2008.00234.x
Strugnell, J. M., Watts, P. C., Smith, P. J., and Allcock, A. L. (2012). Persistent genetic signatures of historic climatic events in an Antarctic octopus. Mol. Ecol. 21, 2775–2787. doi: 10.1111/j.1365-294X.2012.05572.x
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Taylor, H. R., and Harris, W. E. (2012). An emergent science on the brink of irrelevance: a review of the past 8 years of DNA barcoding. Mol. Ecol. Resour. 12, 377–388. doi: 10.1111/j.1755-0998.2012.03119.x
Templeton, A. R., Crandall, K. A., and Sing, C. F. (1992). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonucleaase mapping and DNA sequence data. III. Cladogram estimation. Genetics 132, 619–633.
Thomas, C. D. (2010). Climate, climate change and range boundaries. Divers. Distrib. 16, 488–495. doi: 10.1111/j.1472-4642.2010.00642.x
Turner, J., Barrand, N. E., Bracegirdle, T. J., Convey, P., Hodgson, D. A., Jarvis, M., et al. (2014). Antarctic climate change and the environment: an update. Polar Rec. 50, 237–259. doi: 10.1017/S0032247413000296
Ward, R. D., Holms, B. H., and O'Hara, T. D. (2008). DNA barcoding discriminates echinoderm species. Mol. Ecol. Resour. 8, 1202–1211. doi: 10.1111/j.1755-0998.2008.02332.x
Whitehouse, M. J., Meredith, M. P., Rothery, P., Atkinson, A., Ward, P., and Korb, R. E. (2008). Rapid warming of the ocean around South Georgia, Southern Ocean, during the 20th century: forcings, characteristics and implications for lower trophic levels. Deep Sea Res. Part Oceanogr. Res. Pap. 55, 1218–1228. doi: 10.1016/j.dsr.2008.06.002
Whitworth, T. III. (1980). Zonation and geostrophic flow of the Antarctic circumpolar current at Drake Passage. Deep Sea Res. Part Oceanogr. Res. Pap. 27, 497–507. doi: 10.1016/0198-0149(80)90036-9
Keywords: adaptation, climate change, cryptic species, Ophiuroidea, range expansion, radiation, phylogeography, Pleistocene
Citation: Sands CJ, O'Hara TD, Barnes DKA and Martín-Ledo R (2015) Against the flow: evidence of multiple recent invasions of warmer continental shelf waters by a Southern Ocean brittle star. Front. Ecol. Evol. 3:63. doi: 10.3389/fevo.2015.00063
Received: 01 April 2015; Accepted: 03 June 2015;
Published: 22 June 2015.
Edited by:
Christian C. Figueroa, Universidad de Talca, ChileReviewed by:
Florian Leese, Ruhr University Bochum, GermanyPilar Haye Molina, Universidad Católica del Norte, Chile
Copyright © 2015 Sands, O'Hara, Barnes and Martín-Ledo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chester J. Sands, British Antarctic Survey, High Cross, Madingley Road, Cambridge, Cambridgeshire, CB3 0ET, UK,Y2pzYW5AYmFzLmFjLnVr
 Chester J. Sands
Chester J. Sands Timothy D. O'Hara2
Timothy D. O'Hara2 Rafael Martín-Ledo
Rafael Martín-Ledo