Abstract
The detection of female-released species-specific sex pheromones in moths is mediated by the pheromone receptors that are expressed in the sensory neurons in the olfactory sensilla of conspecific male antennae. Since the pioneering studies on the tobacco budworm Heliothis virescens and the silkworm Bombyx mori a decade ago, genes encoding pheromone receptors have been identified from a number of moth species. Pheromone receptor genes constitute a specialized olfactory receptor subfamily that shares sequence homology. In most cases the pheromone receptor genes are more abundantly expressed in male antennae, and the expression is confined to the neurons in the long sensilla trichodea, which are responsible for pheromone sensing. Both highly specific and more broadly tuned pheromone receptors have been described in various moth species. We review the advances in moth pheromone receptor studies over the past decade, including the methods used in receptor gene isolation and functional characterization, the different ligand profiles of the identified receptors, and the evolution of this multigene family.
Introduction
Mate-finding behavior, mediated by species-specific sex pheromones, is important in mate recognition in moths. Moth sex pheromones are normally released by adult females during “calling” behavior and tracked by the conspecific males over a long distance. Based on their chemical properties, moth sex pheromones are classified into two major types, Type I sex pheromones comprising C10-C18 straight chain fatty alcohols and corresponding acetates and aldehydes, and Type II sex pheromones including long-chain polyunsaturated hydrocarbons and the corresponding epoxides (Millar, 2000; Ando et al., 2004).
The reception of these chemical signals is conducted by specialized pheromone receptors (PRs) expressed in specific olfactory sensory neurons (OSNs) in antennal sensilla. As members of the insect olfactory receptor (OR) family, PRs possess a seven-transmembrane structure and form heteromeric ligand-gated non-selective ion channels with the olfactory co-receptor Orco (Sato et al., 2008). The pheromones are solubilized and transported by pheromone binding proteins (PBPs) through the lymph around the dendrite of the OSNs, and activate the PR/Orco complex (Vogt, 2005). In Drosophila, the presence of sensory neuron membrane proteins (SNMPs) is required for proper pheromone-evoked response (Benton et al., 2007). Recent studies indicated that in moth pheromone detection system, SNMPs might contribute to the sensitivity (Pregitzer et al., 2014), or rapid activation and termination of pheromone-induced activity (Li et al., 2014).
Following the pioneering studies on odorant receptors in the vinegar fly, Drosophila melanogaster (Clyne et al., 1999; Vosshall et al., 1999), moth PR genes were initially discovered from two intensively studied species, the tobacco budworm Heliothis virescens and the silkworm Bombyx mori (Krieger et al., 2004, 2005; Sakurai et al., 2004; Nakagawa et al., 2005; Große-Wilde et al., 2007). Since then, a number of PR genes have been identified from various moth species. In this review, we summarize the progress to date in the isolation and functional characterization of moth PRs, to enable a discussion on the evolution of PR function.
Moth PR gene sequences and expression pattern
In H. virescens, the genomic database was BLAST analyzed with candidate chemosensory receptor genes from D. melanogaster and the malaria mosquito Anopheles gambiae, combined with screening of antennal cDNA libraries with specific probes (Krieger et al., 2004). In B. mori, different cloning strategies were used in two independent studies. BmorOR1 was identified by differential screening of a male antennal cDNA library (Sakurai et al., 2004), whereas more candidate PR genes were identified by the method used in H. virescens (Krieger et al., 2005). The sequence homology found in PRs from these two species made it possible to explore new PR genes using degenerate PCR. This approach turned out to be an efficient strategy in various moth species, including the diamondback moth Plutella xylostella, the armyworm moth Mythimna separata, and the cucumber moth Diaphania indica (Mitsuno et al., 2008), the cotton bollworm Helicoverpa armigera and the tobacco budworm Helicoverpa assulta (Zhang, 2010; Zhang et al., 2010), the European corn borer Ostrinia nubilalis and related Ostrinia species (Miura et al., 2010; Wanner et al., 2010), the navel orangeworm Amyelois transitella (Xu et al., 2012), the beet armyworm Spodoptera exigua (Liu et al., 2013a), and the turnip moth Agrotis segetum (Zhang and Löfstedt, 2013). The identified PRs cluster in a single lineage, forming a specialized subfamily of olfactory receptors.
The alignment of hitherto known moth PR sequences shows a relatively conserved C-terminal region that contains three highly conserved motifs (Figure 1A). Motif 1 has a signature sequence L-(L/M)-(L/V)-(E/Q)-C-(S/T/A); motif 2 contains the signature sequences (Q/G/T)-(Q/E/L)-L-(I/V)-(Q/L/E) and P-W-(E/Q/D); and motif 3 contains the signature sequence (I/V)-(L/I)-(K/R)-(T/A)-(S/T). These motifs provide useful sites for designing degenerate primers to isolate new PR genes. From the functional perspective, the significance of these motifs has not been fully investigated. Previous studies on BmorOR1 in silkworm showed that site-directed mutagenesis of the residue E in the signature sequence L-(L/M)-(L/V)-(E/Q)-C-(S/T/A) or P-W-(E/Q/D) caused functional alterations in the odor-evoked cation channel activity, indicating an essential role of the residues in keeping the PR/Orco complex channel activity (Nakagawa et al., 2012). Further mutagenesis studies will help to define the roles of the other residues in these motifs.
Figure 1
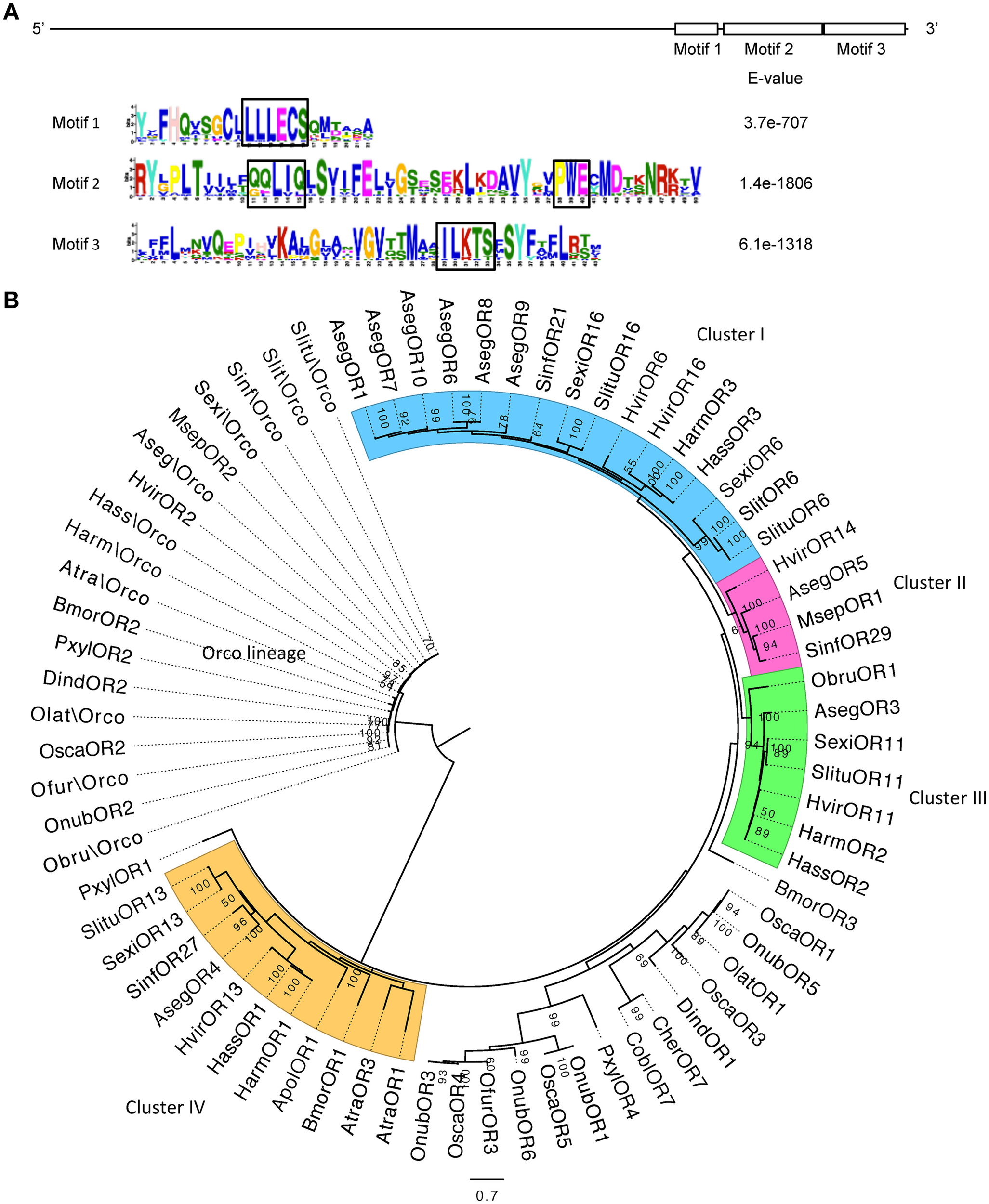
The motif sequences and phylogeny of moth pheromone receptor genes. (A) The upper bar indicates the location of the three motifs on the PR sequences. The lower shows the sequences of the three motifs and respective E-values. The signature sequences in the motifs are boxed in black. (B) The evolutionary history was inferred with MEGA6 by using the Maximum Likelihood method based on the LG model (Le and Gascuel, 2008; Tamura et al., 2013). The tree with the highest log likelihood (−22599.7) is shown. Support values above 50% are labeled next to the branches, which were derived from 100 bootstrap replicates. Initial tree for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 2.6982)]. The tree was rooted with the Orco lineage. Color coding indicates the four different orthologous clusters.
In recent years, RNA sequencing of moth antennal transcriptomes has become a powerful alternative to degenerate PCR when exploring the repertoire of genes coding for olfactory receptors (Montagné et al., 2015). For example, 2 out of 47 ORs of the tobacco hornworm Manduca sexta, 5 out of 43 ORs in the codling moth Cydia pomonella, and 4 out of 47 ORs in the cotton leafworm Spodopetra littoralis were found belonging to the PR subfamily based on the respective transcriptome data (Große-Wilde et al., 2011; Bengtsson et al., 2012; Poivet et al., 2013).
In addition, the expression levels of PR genes may provide clues to receptor function, which can be assessed by in situ hybridization and quantitative PCR (Krieger et al., 2005; Wanner et al., 2010; Zhang et al., 2010), or directly from the RNA-seq data. The latter makes it more convenient to compare the expression levels of many target genes in different tissues. In general, the expression level of PR genes is higher in male antennae than in female antennae, and the expression is confined to neurons located in the long sensilla trichodea (Krieger et al., 2005), which are known to be responsive to moth sex pheromones (Schneider, 1974).
Functional assays of moth PRs
Different heterologous expression systems have been used to characterize moth PR gene function during the past decade (Table 1). The first moth PR, BmorOR1 was deorphanized from B. mori using the Xenopus oocyte expression system (Sakurai et al., 2004; Nakagawa et al., 2005), which, since then has been most commonly used in moth PR studies (Table 1 and references therein). In short, the complementary RNAs (cRNAs) of a candidate PR gene and Orco gene are co-injected into the oocytes of the African clawed frog, Xenopus laevis, where the target receptors are efficiently and faithfully translated, assembled and inserted into the plasma membrane. The oocytes are subsequently incubated and perfused with respective pheromone compounds diluted in buffers. During the perfusion the stimulated inward currents conferred by the PR/Orco heteromeric complex are recorded under the two-electrode voltage clamp (TEVC) at a certain holding potential. The PR ligand profiles obtained from this system agree well with the properties of the olfactory neurons identified by in vivo electrophysiological studies, which makes it possible to hypothetically assign the PR genes to corresponding neurons in the sensilla (Miura et al., 2010; Wang et al., 2011; Zhang and Löfstedt, 2013).
Table 1
| Species | Family | Heterologous expressiona | Genesb | Ligands | References |
|---|---|---|---|---|---|
| Bombyx mori | Bombycidae | Oocytes with G protein | BmorOR1 | E10,Z12-16:OH** | Sakurai et al., 2004 |
| Oocytes | BmorOR1 | E10,Z12-16:OH** | Nakagawa et al., 2005 | ||
| BmorOR3 | E10,Z12-16:Ald | ||||
| HEK293 with G protein | BmorOR1 | E10,Z12-16:OH** (in the presence of PBP) | Große-Wilde et al., 2006 | ||
| BmorOR3 | E10,Z12-16:Ald | ||||
| DmelOr22a empty neuron | BmorOR1 | E10,Z12-16:OH** | Syed et al., 2006 | ||
| DmelOr67d empty neuron | BmorOR1 | E10,Z12-16:OH** | Syed et al., 2010 | ||
| Cell-free (giant vesicles) | BmorOR1 | E10,Z12-16:OH** | Hamada et al., 2014 | ||
| Heliothis virescens | Noctuidae | HEK293 with G protein | HvirOR13 | Z11-16:Ald** (in the presence of PBP2) | Große-Wilde et al., 2007 |
| HvirOR14 | Z11-16:OAc, Z11-16:Ald, Z9-16:Ald, Z9-14:Ald* | ||||
| HvirOR16 | Z9-14:Ald, Z11-16:OH | ||||
| Oocytes | HvirOR6 | Z9-14:Ald* | Wang et al., 2011 | ||
| HvirOR11 | – | ||||
| HvirOR13 | Z11-16:Ald** | ||||
| HvirOR14 | Z11-16:OAcc | ||||
| HvirOR15 | – | ||||
| HvirOR16 | Z11-16:OHc | ||||
| Plutella xylostella | Plutellidae | Oocytes | PxylOR1 | Z11-16:Ald** | Mitsuno et al., 2008 |
| Oocytes | PxylOR4 | Z9,E12-14:OAc, Z9-14:OAc | Sun et al., 2013 | ||
| Mythimna separata | Noctuidae | Oocytes | MsepOR1 | Z11-16:OAc** | Mitsuno et al., 2008 |
| Diaphania indica | Crambidae | Oocytes | DindOR1 | E11-16:Ald** | Mitsuno et al., 2008 |
| Antheraea polyphemus | Saturniidae | HEK293 with G protein | ApolOR1 | E6,Z11-16:Ald* (only specific at low concentration, in the presence of PBP2) | Forstner et al., 2009 |
| Ostrinia latipennis | Crambidae | Oocytes | OlatOR1 | E11-14:OH** | Miura et al., 2009 |
| Ostrinia scapulalis (E type) | Crambidae | Oocytes | OscaOR1 | E11-14:OH | Miura et al., 2009 |
| Oocytes | OscaOR3 | Broadly tuned to Z11-14:OAc*, E11-14:OAc**, Z12-14:OAc, E12-!4:OAc, and Z9-14:OAcc | Miura et al., 2010 | ||
| OscaOR4 | E11-14:OAc | ||||
| OscaOR5 | Marginal responses to a few pheromone components | ||||
| OscaOR6 | – | ||||
| OscaOR7 | – | ||||
| OscaOR8 | – | ||||
| Ostrinia nubilalis (Z races) | Crambidae | Oocytes | OnubOR1 | Broadly tuned to Z11-14:OAc**, E11-14:OAc*, Z12-14:OAc, E12-14:OAc and Z9-14:OAcc, more sensitive to E12-!4:OAc | Wanner et al., 2010 |
| OnubOR3 | Broadly tuned to Z11-14:OAc, E11-14:OAc, Z12-14:OAc, E12-!4:OAc and Z9-14:OAc | ||||
| OnubOR4 | – | ||||
| OnubOR5 | Broadly tuned to Z11-14:OAc, E11-14:OAc, Z12-14:OAc, E12-!4:OAc and Z9-14:OAc | ||||
| Ostrinia furnacalis | Crambidae | Oocytes | OfurOR3 | Preferentially responsive to E12-14:OAc** and Z12-14:OAc** | Leary et al., 2012 |
| Helicoverpa armigera | Noctuidae | Oocytes | HarmOR1 | Z11-16:Ald** | Zhang, 2010 |
| HarmOR2 | – | ||||
| HarmOR3 | Z11-16:OHc, Z9-14:Ald | ||||
| Oocytes | HarmOR13 | Z11-16:Ald** | Liu et al., 2013b | ||
| HarmOR6 | Z9-14:Ald, Z9-16:Ald* | ||||
| HarmOR16 | Z11-16:OH, Z9-14:Ald | ||||
| HarmOR11 | – | ||||
| HarmOR14 | – | ||||
| HarmOR15 | – | ||||
| Helicoverpa assulta | Noctuidae | Oocytes | HassOR1 | Z11-16:Ald* | Zhang, 2010 |
| HassOR2 | – | ||||
| HassOR3 | Z9-14:Ald, Z11-16:OH | ||||
| Sf9 | HassOR13 | Z11-16:Ald* | Xu et al., 2015 | ||
| Amyelois transitella | Pyralidae | Oocytes | AtraOR1 | Z11,Z13-16:Ald** | Xu et al., 2012 |
| AtraOR3 | Z11-16:Ald, Z9,Z11-14OFor | ||||
| Spodoptera littoralis | Noctuidae | DmelOr67d empty neuron | SlitOR6 | Z9,E12-14:OAc* | Montagné et al., 2012 |
| Agrotis segetum | Noctuidae | Oocytes | AsegOR1 | Z5-10:OAc, Z7-12:OAc, Z8-12:OAcc | Zhang and Löfstedt, 2013 |
| AsegOR3 | Low sensitivity to all the tested compounds | ||||
| AsegOR4 | Z7-12:OAc** | ||||
| AsegOR5 | Z9-14:OAc* | ||||
| AsegOR6 | Z5-10:OHc, Z5-10:OAc | ||||
| AsegOR7 | Similar to AsegOR1, lower sensitivity | ||||
| AsegOR8 | Similar to AsegOR6, lower sensitivity | ||||
| AsegOR9 | Z5-10:OAc* | ||||
| AsegOR10 | Minor responses to Z9-14:OAc, Z5-10:OAc | ||||
| Spodoptera exigua | Noctuidae | Oocytes | SexiOR6 | – | Liu et al., 2013a |
| SexiOR11 | – | ||||
| SexiOR13 | Z9,E12-14:OAc**, Z9-14:OAc* | ||||
| SexiOR16 | Z9-14:OH* | ||||
| Sesamia inferens | Noctuidae | Oocytes | SinfOR21 | Z11-16:OH* | Zhang et al., 2014 |
| SinfOR29 | Z11-16:OAc** | ||||
| SinfOR27 | Z9,E12-14:OAc | ||||
| Ctenopseustis obliquana | Tortricidae | HEK293 | CoblOR7 | Z8-14:OAc** | Steinwender et al., 2015 |
| Ctenopseustis herana | Tortricidae | HEK293 | CherOR7 | Z8-14:OAc*, Z7-14:OAc** | Steinwender et al., 2015 |
| Spodoptera litura | Noctuidae | Oocytes | SlituOR6 | Z9,E12-14:OAc* | Zhang et al., 2015b |
| SlituOR11 | – | ||||
| SlituOR13 | Z9-14:OAc*, Z9,E12-14:OAc | ||||
| SlituOR16 | Broadly tuned with the largest response to Z9-14:OHc | ||||
| Operophtera brumata | Geometridae | Oocytes | ObruOR1 | 1,Z3,Z6,Z9-19:H | Zhang et al., 2015a |
Functionally identified PR genes in lepidopteran species.
Except for when specified differently in this column, the PRs were co-expressed with the respective Orco.
HarmOR13, HarmOR11, HarmOR16 in Liu et al. (2013b) are equivalent genes to HarmOR1, HarmOR2 and HarmOR3 in Zhang et al. (2010), respectively; HassOR13 in Xu et al. (2015) is equivalent to HassOR1 in Zhang et al. (2010).
Behavioral antagonist to corresponding species.
Minor pheromone components of corresponding species.
Major pheromone component of corresponding species.
– No response to the tested compounds was observed.
Pheromone compounds are abbreviated in a standard way including (in order) geometry of the double bond, position of unsaturation, chain length followed by a colon and functionality. For example, E10,Z12-16:OH, (10E,12Z)-hexadecadienol; E10,Z12-16:Ald, (10E,12Z)-hexadecadienal; Z11-16:OAc, (11Z)-hexadecenyl acetate; and 1,Z3,Z6,Z9-19:H, (1,3Z,6Z,9Z)-nonadecatetraene.
Another in vitro gene expression system using human embryonic kidney 293 (HEK293) cells was also applied in moth PR functional assays, in which the PRs and Gα proteins are co-expressed in the cells (Große-Wilde et al., 2006, 2007; Forstner et al., 2009), because PRs were previously assumed to be canonical G protein-coupled receptors (GPCR). The coupling of these exogenous proteins elicits an increase in the level of intracellular Ca2+ upon pheromone stimulation, which can be monitored by calcium imaging. To improve the response specificity of the transfected HEK293 cells, the matching PBPs were required in above studies. Recently, a functional assay using modified HEK293 cell lines co-expressing PRs with Orco instead of Gα proteins, but in the absence of PBPs was reported (Steinwender et al., 2015), following a previously described protocol for OR study (Corcoran et al., 2014).
The Drosophila..empty neuron.. has been employed as an in vivo heterologous expression system in moth PR functional assays. Firstly, the flies are genetically modified by replacing an endogenous OR gene with a candidate moth PR gene in corresponding Drosophila OR-expressing neurons. The antennae of the flies are then stimulated by moth pheromone compounds and the evoked neuronal responses are recorded by single-sensillum recording. The ab3A neurons that host the endogenous Drosophila DmelOr22a gene were initially used to express B. mori PRs (Syed et al., 2006). However, the T1 neurons that host the DmelOr67d gene and respond to the Drosophila pheromone cis-vaccenyl acetate (Ha and Smith, 2006) were later found to functionally express moth PR genes more efficiently (Kurtovic et al., 2007; Syed et al., 2010; Montagné et al., 2012). A likely explanation is that the T1 neurons are equipped with necessary components such as SNMP1, which is required for the sensing of sex pheromones in Drosophila (Benton et al., 2007).
More recently, a cell-free expression system involving in situ protein synthesis has been reported (Hamada et al., 2014). In this study BmorOR1 was co-expressed with Bmor\Orco in giant vesicles and excited in the presence of the ligand bombykol (10E,12Z)-hexadecadienol, as shown by patch-clamp recording.
To what extent the different assays give similar results is currently not known, when it comes to specificity and sensitivity, but the bulk of available data (Table 1) have been collected using the Xenopus oocyte expression system as mentioned above.
Ligand profiles of moth PRs
The specific PRs
A number of PRs are specifically responsive to a single pheromone compound, which in most cases is the major pheromone component for the species in question. The specificity of these PRs confer on them the ability to distinguish compounds sharing very similar chemical structures, including: (1) analogs with different fatty chain lengths, e.g., AsegOR9, AsegOR4, and AsegOR5 in A. segetum, which are specifically tuned to the pheromone components (5Z)-decenyl, (7Z)-dodecenyl, and (9Z)-tetradecenyl acetates, respectively (Zhang and Löfstedt, 2013); (2) compounds with the same molecular skeletons but different oxygen-containing functional groups, e.g., BmorOR1 and BmorOR3 in B. mori specifically tuned to the sex pheromone components bombykol and its oxidized form bombykal (10E, 12Z)-hexadecadienal, respectively (Nakagawa et al., 2005); (3) stereoisomeric pheromone compounds with different geometry and/or position of the double bond(s), e.g., OnubOR6 in the European corn borer O. nubilalis Z strain tuned to (11Z)-tetradecenyl acetate, but not to (11E)-tetradecenyl acetate (Wanner et al., 2010), OscarOR4 in O. scapulalis tuned to (11E)-tetradecenyl acetate rather than (12E)-tetradecenyl acetate (Miura et al., 2010), and SlitOR6 in S. littoralis tuned to (9Z,12E)-tetradecadienyl acetate, but not to (9Z,11E)-tetradecadienyl acetate (Montagné et al., 2012).
The broadly tuned PRs
In addition to the above-mentioned specific receptors that are tuned to the major pheromone components in respective species, some PRs have broader response spectra. For example, OscaOR3 from O. scapulalis responds not only to the conspecific pheromone components (11E)- and (11Z)-tetradecenyl acetates, but also to those from closely related species, such as (9Z)-, (12E)-, and (12Z)-tetradecenyl acetates (Miura et al., 2010); OnubOR1, OnubOR3, and OnubOR5 from O. nubilalis also respond to all the five tetradecenyl acetate isomers mentioned above (Wanner et al., 2010); and similarly, SexiOR16 from S. exigua shows broad activity to multiple sex pheromone components (Liu et al., 2013a).
PR responses to behavioral antagonists
Behavioral antagonism mediated by pheromone-like compounds may provide a mechanism for pheromone specificity and prevent cross-attraction between sympatric species and hence reproductive isolation. These compounds can be used as pheromone components in one species, but have antagonistic effects in sibling species (Linn and Roelofs, 1995; Cardé and Haynes, 2004; Linn et al., 2007). The receptors for the behavioral antagonists are also found in the PR subfamily. In H. virescens, HvirOR16 and HvirOR14 are specifically responsive to the behavioral antagonists, (11Z)-hexadecenol and (11Z)-hexadecenyl acetate, respectively (Wang et al., 2011). In some other species, however, the broadly tuned receptors may respond to both their own pheromone compounds and the interspecific behavioral antagonists. For example, the above mentioned OscaOR3 in O. scapulalis responds not only to the conspecific pheromone components (11E)- and (11Z)-tetradecenyl acetates, but also to (9Z)-tetradecenyl acetate, a behavioral antagonist in O. scapulalis but pheromone component in the closely related species O. zaguliaevi and O. zealis (Miura et al., 2010). In S. litura, in addition to the modest responses to three conspecific sex pheromone components and an analog, SlituOR16 showed the strongest response to (9Z)-tetradecenol, a behavioral antagonist in S. litura, but a sex pheromone component in S. exigua (Zhang et al., 2015b). In A. segetum, AsegOR1 responds to both the behavioral antagonist (8Z)-dodecenyl acetate and the sex pheromone components (5Z)-decenyl and (7Z)-dodecenyl acetates; similarly, AsegOR6 responds to both (5Z)-decenol, another behavioral antagonist, and the pheromone compound (5Z)-decenyl acetate (Zhang and Löfstedt, 2013). The fact that a receptor can respond to both a behavioral agonist and an antagonist might simply because these compounds share similar chemical structures. However, when both agonists and antagonists are perceived, the behavioral outcome might be an olfactory antagonistic balance (Baker, 2008) that depends on the glomerular projection of OSNs and the integration of the information from different receptors in the central nervous system (CNS).
The “ligand unknown” receptors
Among all the moth PRs investigated to date, there is a cluster of orthologous PRs, for which the ligands remain unknown (see Cluster III in Figure 1B). The ratio of nonsynonymous to synonymous substitutions (dN/dS value) in this cluster is considerably lower than the other clades, indicating strong purifying selection on the whole cluster, and possibly a conserved function for these receptors (Zhang et al., 2015a). Previous hypotheses of the function of these receptors focused on structurally related pheromone compounds, behavioral antagonists or the degradation products of the major sex pheromone component (Baker, 2009; Krieger et al., 2009). However, these assumptions have not yet received any support from functional analyses. Most recently, our study on pheromone reception in the winter moth, Operophtera brumata (Geometridae) has shown that the receptor ObruOR1 in this ligand-unknown cluster is specifically tuned to a tetraene (1,3Z,6Z,9Z)-nonadecatetraene, the single component sex pheromone of this species (Roelofs et al., 1982). Similarly, our subsequent functional characterization of another member of Cluster III, AsegOR3 from the noctuid moth A. segetum showed the strongest response to a triene, in this case (3Z,6Z,9Z)-tricosatriene (Zhang et al., 2015a). These results suggest that members in this cluster may all respond to Type II polyene pheromones.
The evolution of moth PRs
As mentioned above, the co-existence of specific and more broadly tuned PRs in moths might be a common phenomenon. The highly specific PRs play essential roles in the accurate perception of conspecific pheromones in the presence of structurally similar compounds in the surroundings, ensuring effective mate recognition. On the other hand, following the asymmetric tracking hypothesis, males (the signal receivers) are under stronger selective pressures than females, and a subset of receptors with a broader response spectrum may serve as a preadaptation to be able to track variation in female-released pheromone signals (Phelan, 1997; Heckel, 2010; Wanner et al., 2010; Zhang and Löfstedt, 2013).
Some broadly tuned PRs are responsive to the behavioral antagonists. In this case a nonspecific neuron tuned to several antagonists might be sufficient to abort the flight toward the source (Takanashi et al., 2006), and the corresponding receptors may maintain a broad tuning profile instead of evolving specificity for a specific antagonist. Alternatively, as was recently found in O. nubilalis, a single OSN that respond to different behavioral antagonists may co-express multiple receptors. This might be another strategy for the moths to broaden the antagonism and increase the specificity of pheromone detection (Koutroumpa et al., 2014).
The phylogeny of the identified moth PRs reveals several apparent orthologous clusters (Cluster I–IV in Figure 1B) mainly expanded in the noctuids but also contain several genes from Bombycidae, Saturniidae, Geometridae, and Pyralidae. There are also some less defined clades expanded in the crambids, which contain PRs from Plutellidae and Tortricidae as well. Identification of PR genes from more Crambidae species may contribute to the recognition of orthologous clusters in these clades. PRs within the same orthologous cluster may respond to the same ligand, e.g., the HvOR13, HarmOR1, HassOR1, and AtraOR3 in Cluster IV are all specifically tuned to (11Z)-hexadecenal. However, the ligand profile of a candidate PR cannot be predicted simply by its orthology with known receptors. In clusters that have strong selective pressure indicated by a low dN/dS value, the PRs' ligand profiles tend to be conserved, whereas clusters with a high dN/dS value are relaxed from evolutionary constraint, thus have more divergent ligand profiles. In some species, paralogous PRs and their ligand profiles are more divergent compared to orthologous PRs (Zhang and Löfstedt, 2013). Because of the limited data of functionally characterized PRs, these patterns are put forward as hypotheses to be tested rather than conclusions.
In general, moth PR genes are under strong selective pressure to ensure the species-specific communication. It remains a conundrum how the moth PR functional diversity evolves under stabilizing selection. Gene duplication, which was suggested as an important mechanism for the diversification of olfactory receptors (Nei et al., 2008; Sánchez-Gracia et al., 2009), might also apply in PRs. Some closely related PR genes form a tightly linked cluster of duplicated genes as indicated by genetic mapping (Gould et al., 2010), and the PR paralogs arisen in the duplication events are under relaxed constraint, allowing the differentiation of their ligand preference (Zhang and Löfstedt, 2013). Another possible mechanism might be that the common ancestor of current orthologous PRs was broadly tuned, and later selected to respond specifically to certain pheromone compounds in different species.
Future research on moth PRs
With the facility of transcriptome sequencing, it is now straightforward to obtain the sequences of candidate PRs. Since most of the PRs identified to date are from noctuid species that normally use fatty acyl alcohol, aldehyde and acetate pheromone compounds, it would be interesting to broaden the search to explore the PRs tuned to other type of pheromones, such as the Type II long chain polyenes and epoxides, or the short chain ketones and secondary alcohols that are used as pheromones in more basal lepidopteran families (Löfstedt and Kozlov, 1997).
The mechanisms underlying ligand selectivity within a receptor still remain largely unclear. Determination of the key amino acids in the ligand-binding region may help to clarify what determines specificity. Comparison of orthologous PRs with different pheromone specificities, or with the same ligand specificity in evolutionary distant species, as well as mutagenesis of the sites under positive selection (Leary et al., 2012) will help to identify the amino acids of importance to the receptor-ligand interaction. Solution of the crystal structures of pheromone receptors, a major challenge due to the technical difficulties of working with membrane proteins, may ultimately provide the information necessary to test hypotheses concerning the relationship between receptor sequence and specificity, as well as the interaction between PR and the co-receptor.
The transduction of sex pheromone signals has been intensively investigated since the early days of pheromone research and remains a hotspot of current research effort on PRs. Research has focused on the formation of the heteromeric ligand-gated non-selective ion channels through the combination of Orco and PRs (e.g., Nakagawa et al., 2005; Wicher et al., 2008), the binding and transport of the target sex pheromone components to the OSN's dendrites (Vogt, 2005; Sato et al., 2008), as well as the close association of PRs and SNMPs (Benton et al., 2007; Jin et al., 2008; Li et al., 2014; Pregitzer et al., 2014). Progress on these fundamental questions will greatly enrich our understanding of the working mechanism of moth PRs.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Statements
Acknowledgments
The authors thank the two reviewers for their constructive comments, Jürgen Krieger from Martin Luther University of Halle-Wittenberg, Richard Newcomb in The New Zealand Institute for Plant & Food Research, and Hong-Lei Wang from Lund University for their comments on this manuscript. This work was supported by the Swedish Research Councils, the Carl Trygger Foundation, and the Birgit and Sven Håkan Ohlsson foundation (to CL), as well as the Royal Physiographic Society in Lund (to DDZ).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AndoT.InomateS. I.YamamotoM. (2004). Lepidopteran sex pheromones in The Chemistry of Pheromones and Other Semiochemicals: Vol. 239, Topics in Current Chemistry, ed SchulzS. (New York, NY: Springer), 51–96. 10.1007/b95449
2
BakerT. C. (2008). Balanced olfactory antagonism as a concept for understanding evolutionary shifts in moth sex pheromone blends. J. Chem. Ecol.34, 971–981. 10.1007/s10886-008-9468-5
3
BakerT. C. (2009). Nearest neural neighbors: moth sex pheromone receptors HR11 and HR13. Chem. Senses34, 465–468. 10.1093/chemse/bjp025
4
BengtssonJ. M.TronaF.MontagnéN.AnforaG.IgnellR.WitzgallP.et al. (2012). Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE7:e31620. 10.1371/journal.pone.0031620
5
BentonR.VanniceK. S.VosshallL. B. (2007). An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature450, 289–293. 10.1038/nature06328
6
CardéR. T.HaynesK. F. (2004). Structure of the pheromone communication channel in moths, in Advances in Insect Chemical Ecology, ed CardéR. T.MillarJ. G. (Cambridge: Cambridge University Press), 283–332. 10.1017/cbo9780511542664.009
7
ClyneP. J.WarrC. G.FreemanM. R.LessingD.KimJ.CarlsonJ. R. (1999). A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron22, 327–338. 10.1016/S0896-6273(00)81093-4
8
CorcoranJ. A.JordanM. D.CarraherC.NewcombR. D. (2014). A novel method to study insect olfactory receptor function using HEK293 cells. Insect Biochem. Mol. Biol.54, 22–32. 10.1016/j.ibmb.2014.08.005
9
ForstnerM.BreerH.KriegerJ. (2009). A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int. J. Biol. Sci.5, 745–757. 10.7150/ijbs.5.745
10
GouldF.EstockM.HillierN. K.PowellB.GrootA. T.WardC. M.et al. (2010). Sexual isolation of male moths explained by a single pheromone response QTL containing four receptor genes. Proc. Natl. Acad. Sci. U.S.A.107, 8660–8665. 10.1073/pnas.0910945107
11
Große-WildeE.GohlT.BouchéE.BreerH.KriegerJ. (2007). Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci.25, 2364–2373. 10.1111/j.1460-9568.2007.05512.x
12
Große-WildeE.KueblerL. S.BucksS.VogelH.WicherD.HanssonB. S. (2011). Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. U.S.A. 108, 7449–7454. 10.1073/pnas.1017963108
13
Große-WildeE.SvatošA.KriegerJ. (2006). A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses31, 547–555. 10.1093/chemse/bjj059
14
HaT. S.SmithD. P. (2006). A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci.26, 8727–8733. 10.1523/JNEUROSCI.0876-06.2006
15
HamadaS.TabuchiM.ToyotaT.SakuraiT.HosoiT.NomotoT.et al. (2014). Giant vesicles functionally expressing membrane receptors for an insect pheromone. Chem. Commun. 50, 2958–2961. 10.1039/c3cc48216b
16
HeckelD. G. (2010). Smells like a new species: gene duplication at the periphery. Proc. Natl. Acad. Sci. U.S.A.107, 9481–9482. 10.1073/pnas.1004511107
17
JinX.HaT. S.SmithD. P. (2008). SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. U.S.A.105, 10996–11001. 10.1073/pnas.0803309105
18
KoutroumpaF. A.KárpátiZ.MonsempesC.HillS. R.HanssonB. S.Jacquin-JolyE.et al. (2014). Shifts in sensory neuron identity parallel differences in pheromone preference in the European corn borer. Front. Ecol. Evol.2:65. 10.3389/fevo.2014.00065
19
KriegerJ.GondesenI.ForstnerM.GohlT.DewerY.BreerH. (2009). HR11 and HR13 receptor-expressing neurons are housed together in pheromone-responsive sensilla trichodea of male Heliothis virescens. Chem. Senses34, 469–477. 10.1093/chemse/bjp012
20
KriegerJ.Große-WildeE.GohlT.BreerH. (2005). Candidate pheromone receptors of the silkmoth Bombyx mori. Eur. J. Neurosci. 21, 2167–2176. 10.1111/j.1460-9568.2005.04058.x
21
KriegerJ.Große-WildeE.GohlT.DewerY. M.RamingK.BreerH. (2004). Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Natl. Acad. Sci. U.S.A.101, 11845–11850. 10.1073/pnas.0403052101
22
KurtovicA.WidmerA.DicksonB. J. (2007). A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature446, 542–546. 10.1038/nature05672
23
LeS. Q.GascuelO. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol.25, 1307–1320. 10.1093/molbev/msn067
24
LearyG. P.AllenJ. E.BungerP. L.LuginbillJ. B.LinnC. E.Jr.MacallisterI. E.et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A.109, 14081–14086. 10.1073/pnas.1204661109
25
LiZ.NiJ. D.HuangJ.MontellC. (2014). Requirement for Drosophila SNMP1 for rapid activation and termination of pheromone-induced activity. PLoS Genet.10:e1004600. 10.1371/journal.pgen.1004600
26
LinnC. E.Jr.DomingueM. J.MustoC. J.BakerT. C.RoelofsW. L. (2007). Support for (Z)-11-hexadecanal as a pheromone antagonist in Ostrinia nubilalis: flight tunnel and single sensillum studies with a New York population. J. Chem. Ecol.33, 909–921. 10.1007/s10886-007-9268-3
27
LinnC. E.RoelofsW. L. (1995). Pheromone communication in the moths and its role in the speciation process in Speciation and the Recognition Concept: Theory and Application, ed LambertD.SpencerH. (Baltimore, MD: Johns Hopkins University Press), 263–300.
28
LiuC.LiuY.WalkerW. B.DongS.WangG. (2013a). Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hübner). Insect Biochem. Mol. Biol.43, 747–754. 10.1016/j.ibmb.2013.05.009
29
LiuY.LiuC.LinK.WangG. (2013b). Functional specificity of sex pheromone receptors in the cotton bollworm Helicoverpa armigera. PLoS ONE8:e62094. 10.1371/journal.pone.0062094
30
LöfstedtC.KozlovM. (1997). A phylogenetic analysis of pheromone communication in primitive moths, in Insect Pheromone Research: New Directions, eds CardéR. T.MinkA. K. (New York, NY: Chapman & Hall), 473–489. 10.1007/978-1-4615-6371-6_41
31
MillarJ. G. (2000). Polyene hydrocarbons and epoxides: a second major class of lepidopteran sex attractant pheromones. Annu. Rev. Entomol. 45, 575–604. 10.1146/annurev.ento.45.1.575
32
MitsunoH.SakuraiT.MuraiM.YasudaT.KugimiyaS.OzawaR.et al. (2008). Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci.28, 893–902. 10.1111/j.1460-9568.2008.06429.x
33
MiuraN.NakagawaT.TatsukiS.TouharaK.IshikawaY. (2009). A male-specific odorant receptor conserved through the evolution of sex pheromones in Ostrinia moth species. Int. J. Biol. Sci.5, 319–330. 10.7150/ijbs.5.319
34
MiuraN.NakagawaT.TouharaK.IshikawaY. (2010). Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. Insect Biochem. Mol. Biol.40, 64–73. 10.1016/j.ibmb.2009.12.011
35
MontagnéN.ChertempsT.BrigaudI.FrançoisA.FrançoisM. C.de FouchierA.et al. (2012). Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci.36, 2588–2596. 10.1111/j.1460-9568.2012.08183.x
36
MontagnéN.de FouchierA.NewcombR. D.Jacquin-JolyE. (2015). Advances in the identification and characterization of olfactory receptors in insects. Prog. Mol. Biol. Transl. Sci. 130, 55–80. 10.1016/bs.pmbts.2014.11.003
37
NakagawaT.PellegrinoM.SatoK.VosshallL. B.TouharaK. (2012). Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. PLoS ONE7:e32372. 10.1371/journal.pone.0032372
38
NakagawaT.SakuraiT.NishiokaT.TouharaK. (2005). Insect sex pheromone signals mediated by specific combinations of olfactory receptors. Science307, 1638–1642. 10.1126/science.1106267
39
NeiM.NiimuraY.NozawaM. (2008). The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet.9, 951–963. 10.1038/nrg2480
40
PhelanP. L. (1997). Genetics and phylogenetics in the evolution of sex pheromones, in Insect Pheromone Research: New Directions, eds CardéR. T.MinksA. K. (New York, NY: Chapman & Hall), 563–579.
41
PoivetE.GallotA.MontagneN.GlaserN.LegeaiF.Jacquin-JolyE. (2013). A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PLoS ONE8:e60263. 10.1371/journal.pone.0060263
42
PregitzerP.GreschistaM.BreerH.KriegerJ. (2014). The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol. Biol.23, 733–742. 10.1111/imb.12119
43
RoelofsW. L.HillA. S.LinnC. E.MeinwaldJ.JainS. C.HerbertH. J.et al. (1982). Sex pheromone of the winter moth, a geometrid with unusually low temperature precopulatory responses. Science217, 657–659. 10.1126/science.217.4560.657
44
SakuraiT.NakagawaT.MitsunoH.MoriH.EndoY.TanoueS.et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A.101, 16653–16658. 10.1073/pnas.0407596101
45
Sánchez-GraciaA.VieiraF. G.RozasJ. (2009). Molecular evolution of the major chemosensory gene families in insects. Heredity103, 208–216. 10.1038/hdy.2009.55
46
SatoK.PellegrinoM.NakagawaT.VosshallL. B.TouharaK. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature452, 1002–1006. 10.1038/nature06850
47
SchneiderD. (1974). The sex-attractant receptor of moths. Sci. Am.231, 28–35. 10.1038/scientificamerican0774-28
48
SteinwenderB.ThrimawithanaA. H.CrowhurstR. N.NewcombR. D. (2015). Pheromone receptor evolution in the cryptic leafroller species, Ctenopseustis obliquana and C. herana. J. Mol. Evol.80, 42–56. 10.1007/s00239-014-9650-z
49
SunM.LiuY.WalkerW. B.LiuC.LinK.GuS.et al. (2013). Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth, Plutella xyllostella. PLoS ONE8:e62098. 10.1371/journal.pone.0062098
50
SyedZ.IshidaY.TaylorK.KimbrellD. A.LealW. S. (2006). Pheromone reception in fruit flies expressing a moth's odorant receptor. Proc. Natl. Acad. Sci. U.S.A.103, 16538–16543. 10.1073/pnas.0607874103
51
SyedZ.KoppA.KimbrellD. A.LealW. S. (2010). Bombykol receptors in the silkworm moth and the fruit fly. Proc. Natl. Acad. Sci. U.S.A.107, 9436–9439. 10.1073/pnas.1003881107
52
TakanashiT.IshikawaY.AndersonP.HuangY.LöfstedtC.TatsukiS.et al. (2006). Unusual response characteristics of pheromone-specific olfactory receptor neurons in the Asian corn borer moth, Ostrinia furnacalis. J. Exp. Biol.209, 4946–4956. 10.1242/jeb.02587
53
TamuraK.StecherG.PetersonD.FilipskiA.KumarS. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol.30, 2725–2729. 10.1093/molbev/mst197
54
VogtR. G. (2005). Molecular basis of pheromone detection in insects, in Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology, Vol. 3, eds GilbertL. I.IatrouK.GillS. S. (Oxford: Elsevier Pergamon), 753–803.
55
VosshallL. B.AmreinH.MorozovP. S.RzhetskyA.AxelR. (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell96, 725–736. 10.1016/S0092-8674(00)80582-6
56
WangG.VásquezG. M.SchalC.ZwiebelL. J.GouldF. (2011). Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol. Biol.20, 125–133. 10.1111/j.1365-2583.2010.01045.x
57
WannerK. W.NicholsA. S.AllenJ. E.BungerP. L.GarczynskiS. F.LinnC. E.et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE5:e8685. 10.1371/journal.pone.0008685
58
WicherD.SchäferR.BauernfeindR.StensmyrM. C.HellerR.HeinemannS. H.et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature452, 1007–1011. 10.1038/nature06861
59
XuP.GarczynskiS. F.AtunguluE.SyedZ.ChooY. M.VidalD. M.et al. (2012). Moth sex pheromone receptors and deceitful parapheromones. PLoS ONE7:e41653. 10.1371/journal.pone.0041653
60
XuW.PapanicolaouA.LiuN. Y.DongS. L.AndersonA. (2015). Chemosensory receptor genes in the oriental tobacco budworm Helicoverpa assulta. Insect Mol. Biol.24, 253–263. 10.1111/imb.12153
61
ZhangD. D. (2010). Cloning And Functional Identification Of Pheromone Receptor Genes in Two Sibling Moth Species, Helicoverpa Armigera And Helicoverpa Assulta. Ph. D. thesis, Institute of Zoology, Chinese Academy of Sciences, Beijing.
62
ZhangD. D.LöfstedtC. (2013). Functional evolution of a multigene family: orthologous and paralogous pheromone receptor genes in the turnip moth, Agrotis segetum. PLoS ONE8:e77345. 10.1371/journal.pone.0077345
63
ZhangD. D.WangH. L.SchultzeA.FroßH.FranckeW.KriegerJ.et al. (2015a). Functional characterization of a receptor for a Type II sex pheromone in the winter moth, Operophtera brumata (Lepidoptera: Geometridae), The 31st Annual Meeting of the International Society of Chemical Ecology (Stockholm), 81.
64
ZhangD. D.ZhuK. Y.WangC. Z. (2010). Sequencing and characterization of six cDNAs putatively encoding three pairs of pheromone receptors in two sibling species, Helicoverpa armigera and Helicoverpa assulta. J. Insect. Physiol.56, 586–593. 10.1016/j.jinsphys.2009.12.002
65
ZhangJ.YanS.LiuY.Jacquin-JolyE.DongS.WangG. (2015b). Identification and functional characterization of sex pheromone receptors in the common cutworm (Spodoptera litura). Chem. Senses40, 7–16. 10.1093/chemse/bju052
66
ZhangY. N.ZhangJ.YanS. W.ChangH. T.LiuY.WangG. R.et al. (2014). Functional characterization of sex pheromone receptors in the purple stem borer, Sesamia inferens (Walker). Insect Mol. Biol.23, 611–620. 10.1111/imb.12109
Summary
Keywords
pheromone receptor, lepidoptera, cloning, functional characterization, evolution
Citation
Zhang D-D and Löfstedt C (2015) Moth pheromone receptors: gene sequences, function, and evolution. Front. Ecol. Evol. 3:105. doi: 10.3389/fevo.2015.00105
Received
20 May 2015
Accepted
24 August 2015
Published
15 September 2015
Volume
3 - 2015
Edited by
Sharon Rose Hill, Swedish University of Agricultural Sciences, Sweden
Reviewed by
Nicolas Montagné, Université Pierre et Marie Curie, France; Patricia Nagnan-Le Meillour, Institut National de la Recherche Agronomique, France
Updates
Copyright
© 2015 Zhang and Löfstedt.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan-Dan Zhang, Department of Biology, Lund University, Sölvegatan 37, SE-223 62 Lund, Sweden dan-dan.zhang@biol.lu.se
This article was submitted to Chemical Ecology, a section of the journal Frontiers in Ecology and Evolution
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.