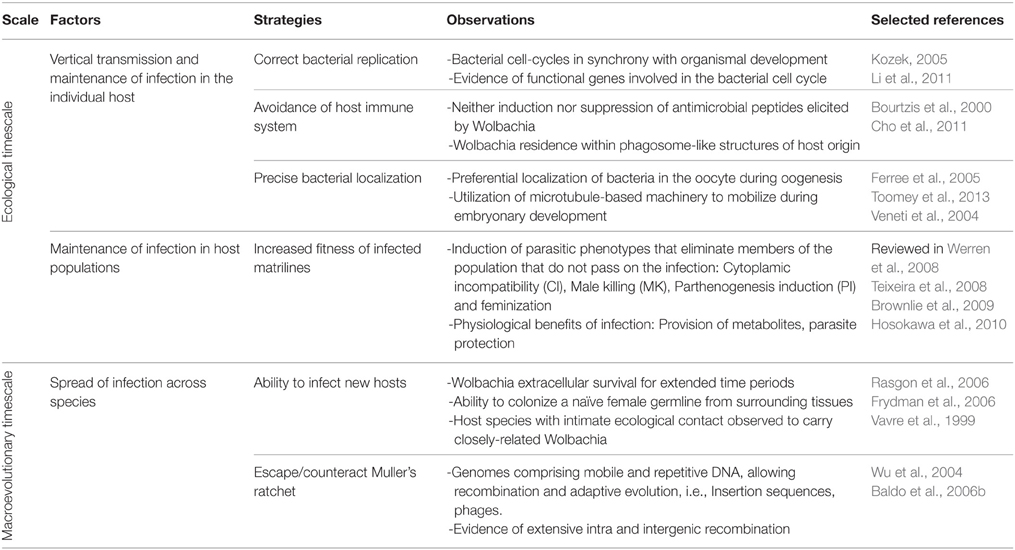
- The Ballard Lab, School of Biotechnology and Biomolecular Sciences, The University of New South Wales, Sydney, NSW, Australia
Wolbachia are intracellular, maternally inherited bacteria with an impressive history of adaptation to intracellular lifestyles. Instead of adapting to a single host lineage, Wolbachia evolved ways to jump across host species and establish relatively stable associations maintained through vertical transmission. Wolbachia are capable of manipulating the reproduction of infected hosts in a remarkable way. Traditionally, such reproductive manipulations have been regarded as the general mechanism by which Wolbachia spread through host populations. Recent evidence suggests that Wolbachia-host interactions are more complex than previously thought and may be driven by the onset and resolution of conflicts of interest. Here, we discuss how reproductive manipulation phenotypes may be transient. As the host adapts to infection, manipulation phenotypes attenuate and the continuity of the symbioses may rely on the physiological advantages Wolbachia may confer to their host. For facultative symbionts, such benefits are likely to be dependent on the environment. Here, we also review evidence that supports the view of environment-dependent facultative mutualism as a stable evolutionary outcome of Wolbachia infections beside extinction and obligate symbioses. Finally, our current understanding of the biology of mitochondria and Wolbachia unravels remarkable parallels in the way they interact with the nuclear genome. Great insights into both the Wolbachia and mitochondrial research fields can be revealed if such fields are considered to be overlapping, rather than independent from each other.
Introduction
With the publication of On the origins of mitosing cells in 1967, Lynn (Margulus) Sagan revived the long-standing but unpopular idea that essential components of eukaryotic cells such as mitochondria and plastids are derived from bacteria that some ancestral cell had engulfed (Sagan, 1967). Besides providing a robust explanation of how modern eukaryotic cells came to be, Margulis's idea showed that evolutionary change may occur with the acquisition of genomes through endosymbiosis; the cohabitation of nonrelated partners where one of them lives in the body of the other. The discovery of organelle DNA as a carrier of essential information for correct organelle function not only proved Margulis's predictions correct, but also showed that the genetic information of modern eukaryotic cells is compartmentalized into cytoplasmic and nuclear partitions (Mounolou and Lacroute, 2005). This implies that the highly coordinated work of cytoplasmic and nuclear genomes required for the assembly of the major energy-producing molecular machineries of eukaryotes is the product of two billion years of cyto-nuclear coevolution (Rand et al., 2004).
Mitochondria are not the only examples of genomes of disparate origin that coevolve with the host genome inside the cell. Intracellular endosymbioses have occurred repeatedly in nature and are found in a large variety of animals, plants, and other life forms. These organisms are known to play essential roles in aspects of their host ecology such as nutrition, reproduction, and pathogen resistance. It was once assumed that, because of the symbiont reliance on host reproduction for transmission, associations with endosymbiotic bacteria evolved toward beneficial symbioses. Although there are many mutualistic heritable symbionts, there are also those that appear to behave selfishly, favoring their transmission at the cost of the host nuclear genomes. Paramount among these so-called “reproductive parasites” is Wolbachia pipientis (hereafter Wolbachia), a heterogeneous group of intracellular bacteria of insect and other invertebrates.
Wolbachia are maternally inherited endosymbionts that have intrigued biologists since their discovery in 1924 (Hertig and Wolbach, 1924), and are among the most common life forms on earth (see Table 1). Being mostly non-essential from the host perspective, Wolbachia rely on their capacity to alter the host phenotype in order to spread (Moran et al., 2008). Such phenotypic alterations generally result in the reduction of reproductive chances of non-infective individuals, giving a relative advantage to individuals that pass the symbiont to their offspring. Maternal transmission implies that such manipulations, although beneficial for the infected matrilines, are harmful for the paternal lineages, clearly exemplifying the conflicts of interest that arise between cytoplasmic and nuclear genomes (Hurst, 1992; Rand et al., 2004). Such sexual asymmetry in favor of females is similar in principle to that between mitochondrial and nuclear genomes, where mitochondrial DNA (mtDNA) mutations can be maintained when favorable in females, even if they are harmful in males (Gemmell et al., 2004). Wolbachia-host associations therefore provide an opportunity to explore the coevolutionary processes that concur with intracellular lifestyles (Sicard et al., 2014) with new knowledge of Wolbachia biology coming from bacterial genome analysis (e.g., Wu et al., 2004; Ellegaard et al., 2013; Nikoh et al., 2014; Ramírez-Puebla et al., 2015).
The aim of this review is to discuss the expanding body of evidence that highlight Wolbachia as a powerful source of evolutionary innovation for many invertebrates (Duron and Hurst, 2013), with special attention to the roles Wolbachia play beyond reproductive manipulations. The first section summarizes the aspects intrinsic to Wolbachia biology that contribute to their evolutionary success across and within host species. The second section explores how the adaptation of the host to Wolbachia infection influences the evolutionary trajectories of the symbiosis. The final section of this review explores interesting parallels in the evolution of Wolbachia and the most successful animal endosymbiont on earth, the mitochondria.
Wolbachia: The Biology of a Master Manipulator
Wolbachia bacteria are a clear example of evolutionary success (Merçot and Poinsot, 2009). Estimates suggest that they infect more than 65% of all insect species (Hilgenboecker et al., 2008), but they are also widespread and common in other invertebrates such as arachnids, crustaceans, and nematodes (Werren et al., 2008). It is perplexing, however, why some other invertebrates such as mollusc's do not appear to be infected. The often discordant phylogenies between Wolbachia and their hosts (O'Neill et al., 1992; Heath et al., 1999; Vavre et al., 1999; Werren and Windsor, 2000) reveal extensive Wolbachia horizontal transmission over evolutionary time, which offers an explanation for the wide range of Wolbachia infections found among terrestrial invertebrates (Werren and Windsor, 2000). Nonetheless, lateral Wolbachia transmissions appear to be rare at ecological timescales. Therefore, the ample distribution and maintenance of Wolbachia infections in insects is likely to be a product of evolutionary processes that occur along very different timescales: in the longer term, Wolbachia retain the capacity to establish new infections through lateral transmission, while in the shorter term they maximize the reproductive fitness of infective matrilines. This section summarizes the aspects of the biology of Wolbachia that may contribute to their success in spreading both horizontally and vertically.
Wolbachia Across the Species Barrier: Horizontal Transmission
Wolbachia evolved from an ancient clade of alphaproteobacteria that comprises other obligatory intracellular organisms, such as Rickettsia, Ehrlichia, Anaplasma, Midichloria (Weinert et al., 2009) and the extinct ancestor of the modern mitochondria (Thrash et al., 2011). Wolbachia are a monophyletic group composed of at least eight different supergroups (A–H), where C and D are exclusively nematode symbionts, and supergroups A and B comprise the majority of arthropod infections (Glowska et al., 2015). Although the lack of suitable outgroups has prevented a satisfactory resolution of their phylogeny (Lo et al., 2007), estimates employing the base substitution rates of the ftzZ and 16S rRNA genes of super groups A, B, C and D suggest that their separation may have occurred approximately 100 million years ago (Werren et al., 1995). Considering that the split between nematodes and arthropods is approximately five times longer, lateral transmission between host phyla or independent acquisition of infection from a third party are a plausible origins for Wolbachia-invertebrate symbioses (Bandi et al., 1998). Furthermore, recent evidence suggests major nemathode-arthropode switches have occurred more than once (Glowska et al., 2015).
From an evolutionary perspective, lateral transmission of Wolbachia is a frequent and ongoing process (Duron and Hurst, 2013). The key pieces of evidence in this regard are: lack of congruence between host and symbiont phylogenies (O'Neill et al., 1992; Stouthamer et al., 1999), closely related Wolbachia present in taxonomically unrelated hosts (Baldo et al., 2006b; Raychoudhury et al., 2009) and disparate Wolbachia types present in the same host (Vavre et al., 1999). The astonishingly widespread distribution of Wolbachia across insects and other invertebrates highlights the importance of determining how Wolbachia crosses the species barrier.
Wolbachia: From One Species to Another
Although the processes involved in the establishment of new infections through lateral transmission are not thoroughly characterized, there is compelling evidence that suggests such events are frequent. First, Wolbachia are able to survive in extracellular environments for extended time periods. Rasgon et al. (2006) observed that, after a week of maintenance on a cell-free media, Wolbachia from Aedes albopictus lived and retained the capacity to invade cells and establish stable infections. Such ability would enhance the chance of the bacteria to colonize cells from an extracellular environment. Second, stable infections are routinely established in the laboratory through microinjections between different host taxa (Braig et al., 1994), which requires Wolbachia to successfully colonize a new female's germline from surrounding somatic tissues. In fact, Wolbachia's intrinsic ability to target and colonize stem cell niches in the ovary has been well demonstrated in Drosophila (Frydman et al., 2006; see Section The Egg as the Route to the Next Generation). Third, closely related Wolbachia infecting unrelated host species with intimate ecological contact have been observed, which suggests ecological routes for the symbiont to be laterally transmitted. Parasitoid wasps, whose larval development occurs within the body of other insects, have been observed to be especially susceptible to Wolbachia infection. European parasitoid wasps display double, and even triple, Wolbachia infections of bacterial types that closely resemble those of the insects they parasitize (Vavre et al., 1999). Another route of Wolbachia lateral transfer has been described in species of terrestrial isopods, where haemolymph contact between infected and non-infected individuals is sufficient to transfer the bacteria between species (Rigaud and Juchault, 1995).
Wolbachia Genomics and the Ability to Retain an Infectious Capacity
Intracellular lifestyles and maternal inheritance mean that bacterial endosymbionts are subject to environmental and population dynamics that drastically differ from those of a free-living organism. First, there is a reduction of the effective population size, owing to the constraints of living within host cells and tissues. Second, strong bottlenecks may occur during the passage of the symbiont from mother to offspring. Third, there is a much-reduced opportunity of horizontal gene transmission due to low intracellular bacterial diversity. Finally, there are relaxed constraints on gene function due to the metabolite-rich environment of the cytoplasm (Reviewed in Toft and Andersson, 2010). These conditions not only reduce genetic variability, but also lower the efficiency of purifying selection to eliminate slightly deleterious mutations. Increased chances of fixation of such mutations (Muller's ratchet) may ultimately lead to loss of gene function and genomic size reduction (Moran, 1996; Dale and Moran, 2006). Indeed, ancient obligate symbioses almost invariably show massive genome reduction, often retaining between 10 and 20% of the genes of their free-living counterparts (Moran, 2003; Dale and Moran, 2006). Their genomes also appear to be purged of pseudogenes, phage sequences and other mobile genetic elements, which led to extraordinary genetic stability (Tamas et al., 2002). Evidence suggests that in these ancient genomic associations, the symbiont has surrendered control of their genetic functions to the host, even in aspects related to DNA replication and gene expression (Moran et al., 2008).
Despite a history of adaptation to an intracellular lifestyle of tens of millions of years, Wolbachia seem to maintain autonomy in processes that involve host immune evasion and establishment of stable de novo infections (Table 1). This reflects an important conundrum in the evolutionary history of these organisms: how can Wolbachia be “generalists in host use” (Baldo et al., 2006b) despite the erosive genomic processes that come with host restriction? The analysis of Wolbachia genomes has provided important clues in this regard. Although their small genome (1–1.7 Mbp) agrees with a reductive trend (Werren et al., 2008), Wolbachia genomes comprise large segments of mobile and repetitive DNA, not a common trait among ancient, vertically-transmitted endosymbionts. The acquisition and preservation of these repetitive and mobile elements is hypothesized to play crucial roles in the evolution of Wolbachia (Wu et al., 2004). The following is a brief outline of how these genomic traits may contribute to Wolbachia adaptation.
Insertion sequences are exceedingly abundant in Wolbachia but almost completely absent from other intracellular obligate symbionts. Moran and Plague (2004) propose that during the early stages of adaptation to an intracellular lifestyle, reduced purifying selection fails to purge slightly deleterious insertions of mobile elements in the genome, which increases the chances of inactivation of functional genes and contributes to the processes of genome erosion. Although in most cases insertion events are predicted to be deleterious, they may induce adaptive evolutionary change by inactivating metabolically redundant genes. This creates regions of homology through the bacterial chromosomes for intra-chromosomal recombination, changing patterns of gene expression by carrying promoters to other regions in the chromosome.
Other important mobile elements found in the genome of Wolbachia are bacteriophage sequences. Although bacteriophages are a significant force of genomic evolution in free-living bacteria, they are notably absent in the genomes of ancient obligate endosymbionts (Bordenstein and Reznikoff, 2005). In contrast, complete and truncated bacteriophage sequences have been described in many Wolbachia types (WO phages) and in some instances, functional phages have been observed to undergo lytic cycle (Masui et al., 2001). The base composition of WO phage DNA is similar to that of the Wolbachia chromosome, which suggests that such particles have been associated with Wolbachia for a long time (Masui et al., 2000). Furthermore, evidence suggests that there has been exchange of viral DNA between distant Wolbachia lineages, and that some phage regions undergo fast rates of recombination. Bordenstein and Wernegreen (2004) propose that the phage-mediated exchange of DNA between bacterial strains within the same intracellular environment constitutes an important source of genomic instability and could drive significant evolutionary change in Wolbachia genomes.
All the necessary machinery for homologous recombination is present in the genome of wMel, the Wolbachia strain of Drosophila melanogaster (Wu et al., 2004). Either the result of functional recombination machinery or a consequence of the multiple mobile elements in the genome, wMel has been shown to display extensive intra and intergenic recombination (Baldo et al., 2006a). This capacity to recombine likely constitutes a key mechanism for adaptation within an arthropod host, as advantageous alleles that arise through recombination could be rapidly fixed and spread through horizontal transmission. Moreover, horizontal gene transfer between distinct Wolbachia types also appears to be frequent (Baldo et al., 2005, 2006a; Duplouy et al., 2013). Such process may give rise to advantageous genetic variants and reduce the accumulation of mildly deleterious mutations due to Muller's ratchet (Raychoudhury et al., 2009).
The maintenance of the global Wolbachia pandemic depends on the rates of acquisition and loss of infection within species, relative to the horizontal transmission (Werren et al., 2008). Plausibly, those Wolbachia lineages that readily establish new infections and recombine with other types have a greater chance to counteract the erosive effects of intracellular lifestyles than those confined to a single host population for extended periods of time. The peculiar host-generalist lifestyle of Wolbachia may result from a balance between vertical transmission, host switching and recombination.
Wolbachia Adaptations to Vertical Transmission
From the host perspective, endosymbionts can be categorized as obligate (primary), which are necessary for host survival and reproduction, or facultative (secondary), which are non-essential. As opposed to primary symbionts, facultative endosymbionts such as most Wolbachia in insects are capable of engineering their own mechanisms of transmission to the next generation of hosts. Because they are generally not fixed in host populations, facultative symbionts also need to confer a reproductive advantage to the transmitting hosts in order to spread (Moran et al., 2008). Perhaps the most fascinating aspect of the biology of Wolbachia is the variety of phenotypes they are capable of exerting upon their host. Wolbachia have been found to be anything from essential endosymbionts to parasites that effectively kill non-infective individuals. This versatility undoubtedly reflects a long evolutionary history of adaptation to intracellular lifestyle. Although many aspects of the biology of Wolbachia related to vertical transmission remain elusive, recent cytological, genetic, and genomic studies have provided insight into the mechanisms that allow Wolbachia to be maintained in host populations as stable infections.
The Egg as the Route to the Next Generation
The passage of symbionts through the egg cytoplasm is the primary mode of Wolbachia transmission. Accurate transmission involves correct symbiont replication, avoidance of host defenses and precise distribution of bacteria in the germline of the host cells (Moran et al., 2008). Some of what is known about the mechanisms of Wolbachia cell division comes from the study of filarial nematodes, as bacterial replication offers potential targets for pharmacologic treatment against filariasis (Taylor et al., 2005). Ultrastuructural analyses of the worm Dirofilaria immitis suggest two mechanisms of Wolbachia cell division: binary fission (common to bacillary forms) and a more complex Chlamydia-like cycle, during which Wolbachia is pleomorphic (Kozek, 2005). This cycle is thought to increase the survival potential of the microorganism by producing more progeny than binary fission and operating in synchrony with the worm development (Kozek, 2005). The analysis of Wolbachia genomes has also offered clues to the molecular mechanisms of cell division. Exploration of the genome of Wolbachia in Brugia malayi (wBm) revealed the presence of core fts genes, which are essential for cytokinesis. These included the highly conserved ftsZ, whose translated product possesses the key residues and secondary structure required for correct enzymatic function (Li et al., 2011). The rapidly increasing Wolbachia genomes available from multiple host species will further contribute to the understanding of Wolbachia cell division and propagation.
The maintenance of adequate microbe titres within the intracellular environment also requires Wolbachia to effectively escape the immune system of the host. This ability is not negligible as insects are capable of mounting sophisticated humoral and cellular immune responses against intruders (Siozios et al., 2008). Experiments on Drosophila simulans showed that Wolbachia do not activate the production of gram-negative specific antimicrobial peptides, but when infected animals were challenged with Escherichia coli, another gram-negative bacterium, they displayed a normal ability to mount an immune response. This indicates that Wolbachia in flies neither induce nor suppress humoral immune responses (Bourtzis et al., 2000). In addition, electron microscopy studies have revealed that Wolbachia reside within vacuoles of host origin in the cytoplasm (Cho et al., 2011). These vacuoles are likely phagosome-like structures that, under normal circumstances, are involved in the processes of degradation of intracellular particles through lysosome fusion and phagolysosome formation. The extent at which Wolbachia interact with and modifies this vacuole to avoid degradation is unknown, but it is plausible that they secrete effector molecules that manipulate the host intracellular environment.
Maternal transmission means that Wolbachia not only have to survive and multiply within the host, but also needs to effectively infect the female germline. Wolbachia are known to preferentially localize within specific regions of the oocyte during oogenesis (Ferree et al., 2005; Frydman et al., 2006). Recent studies show that Wolbachia access the cytoplasm of the forming egg by two main routes. The first route is the direct passage of symbionts from the progenitor germline stem cells, which in some cases become infected during the embryonary development of the gonads. When infected germline stem cells divide, the differentiating daughter cells that ultimately form developed oocytes contain Wolbachia (Serbus et al., 2008). The second route is through stem-cell tissue tropism, where Wolbachia targets and colonizes two groups of stem cell niches in the ovary, the germline stem cell niche at the anterior end of the ovariole and the somatic stem cell niche at the germarium. Evidence suggests that transmission of Wolbachia from stem cell niches into the developing oocyte is conserved in Drosophila and may even be the most prevalent mechanism of egg colonization by Wolbachia (Frydman et al., 2006; Toomey et al., 2013).
It has been observed that the distribution of Wolbachia from early embryogenesis and until late gastrulation is established during late oogenesis. In D. simulans, factors intrinsic to different Wolbachia strains determine their embryonic localization, which can be posterior, anterior, or cortical. Posterior and anterior localizations resemble those of the axis-specification mRNAs oskar and bicoid, and as such, bacteria may rely on microtubule-based machinery for their mobilization through the embryo (Veneti et al., 2004). Intriguingly, only the posterior localization pattern directly targets Wolbachia to the site of germ cell formation (germ plasm), which indicates that strains with a different localization pattern must rely on other mechanisms to reach embryonic germ cells, such as the previously described stem cell niche tropism. This has important evolutionary implications as such mechanisms would allow both the passage of bacteria from mother to offspring and the establishment of stable infection of symbionts transmitted horizontally (Toomey et al., 2013).
Reproductive Manipulations and Other Phenotypic Effects of Wolbachia
Despite Wolbachia's adaptations to reach the egg, symbiont transmission fidelity is often imperfect, especially in natural populations (Turelli et al., 1992; Turelli and Hoffmann, 1995). Unless there are mechanisms in which Wolbachia can increase the fitness of the matriline they infect, the persistence of imperfectly transmitted infection is difficult to explain (Hoffmann et al., 1998).
One strategy would be to establish a mutualistic association with the host, as increase in host fitness would aid the symbiont spread. Although mutualistic Wolbachia are not rare, many Wolbachia utilize another strategy: to induce parasitic phenotypes that selectively eliminate members of their host population that do not transmit the infection. These phenotypes, generally regarded as reproductive manipulations, have their basis in Wolbachia maternal transmission (Table 1). Four reproductive manipulation phenotypes are described as caused by Wolbachia: cytoplasmic incompatibility (CI), male killing (MK), parthenogenesis induction (PI), and feminization.
CI arrests the embryonic development of crosses between infected males and females that do not harbor the same Wolbachia type. Evidence suggests that Wolbachia modify the sperm of infected males and subsequently rescue the modification when present in the egg. This favors infected females but reduces the fitness of infected males. MK-inducing Wolbachia kills the sons of infected females, giving a competitive advantage to the female larvae over their brothers. PI turns haploid males into diploid females in haplo-diploid species. Finally, feminization turns genetic males into females by Wolbachia-related hormonal abnormalities.
This variety of reproductive phenotypes is atypical compared to other microbial reproductive manipulators (Werren et al., 2008). Nevertheless, different manipulations often share important similarities at the cytological level. Specifically, CI, MK, and PI appear to alter cell division during embryonic development. CI-induced embryonic mortality is due to the delayed entry of the male pronucleous into the first embryonic mitotic division, which results in paternal delayed nuclear envelope breakdown, semi-condensed chromatin in metaphase, and chromosome bridges during telophase (Tram and Sullivan, 2002). These anomalies are plausibly the consequence of delayed activity of the key cell cycle kinase Cdk 1, brought about by the modifications inflicted by Wolbachia on the sperm DNA during spermatogenesis (Tram and Sullivan, 2002).
Despite considerable work, very little is known about the processes involved in sperm modification or their subsequent rescue in the egg. Analyses of Wolbachia genomes, however, have rendered some important clues on the matter. A genomic trait of Wolbachia that has received considerable attention is their unusually high number of regions coding for ankyrin (ANK) domains (Iturbe-Ormaetxe et al., 2005). This is relevant as proteins with ANK domains often participate in cell signaling, regulation of gene expression and cytoskeleton integrity in eukaryotic cells, leading to the hypothesis that ANK sequences may be involved in Wolbachia-host interactions (Wu et al., 2004; Iturbe-Ormaetxe et al., 2005). Of particular interest is the fact that in Drosophila, closely related Wolbachia strains that are incompatible to each other differ in the structure and expression of their ANK proteins (Iturbe-Ormaetxe et al., 2005). Moreover, in the mosquito Culex pipiens the only Wolbachia gene identified so far that displays sex specific expression corresponds to an ANK protein that curiously co-expresses with a WO phage gene (Duron et al., 2007). These observations not only reveal the strong link between ANK repeats and the mechanisms of symbiont-host interactions, but also suggest that their variation and evolution may be influenced by rearrangements mediated by mobile elements. Another piece of evidence on CI mechanisms comes from the mosquito C. pipiens, where Wolbachia seem to be implicated on the regulation of host cell cycle genes. Specifically, the Drosophila homolog grauzone in C. pipiens, a regulator of female meiosis, appear to be over-expressed in CI-Wolbachia infected animals, and the levels of up-regulation differed among incompatible lines (Pinto et al., 2013). The increasing availability of annotated Wolbachia genomic sequences and the ability to test the effect of Wolbachia genes by transfection into their host nuclear DNA will prove useful in the discovery of Wolbachia genes involved in reproductive manipulation genotypes.
The mechanisms by which Wolbachia induce sex distortion are yet to be understood. Similarly to CI-Wolbachia, PI-Wolbachia also induce cell cycle abnormalities. This reproductive phenotype has only been observed in animals with haplo-diploid sex determination, specifically, those in which unfertilized (haploid) eggs produce males and diploid (fertilized) eggs produce females (arrhenotoky). In the parasitoid wasp Leptopilina clavipes, infected wasps have normal meiosis but diploidy is restored by an unresolved anaphase in the first mitotic division, which turns would-be males into parthenogenic females (Pannebakker et al., 2004). Indeed, except for CI that may be considered a secondary effect of the infection, it has been argued that all other phenotypic effects caused by Wolbachia might be interconnected, and possibly all correlated to feminization (Reviewed in Negri, 2012).
Feminization induced by Wolbachia has been demonstrated in lepidopteran and hemipteran insects and spiders (Negri, 2012; Curry et al., 2015). In the butterfly Eurema hecabe, it has been demonstrated that the Wolbachia feminizing effect acts continuously throughout the larval development for the maintenance of the female phenotype (Narita et al., 2007). This suggests that the bacterium acts on insect sex differentiation rather than sex determination, and it has been proposed that ecdysteroids are the best candidate for such an interaction (Negri, 2012). Support for this hypothesis is provided by the observation of incomplete Wolbachia suppression by antibiotic treatments during lepidopteran larval stages (Narita et al., 2007). A role for the ecdysteroid titer in regulating sexual dimorphism, including sex specific wing development, has been proved in Lepidoptera (Lobbia et al., 2003) strengthening the hypothesis of a link between Wolbachia and ecdysteroid signaling.
Finally, MK-Wolbachia in Drosophila bifasciata induces abnormalities at different times during early development of the male embryo. In these flies, defective re-modeling, and segregation of chromatin and chromosome bridging results in male embryos dying before eclosion. Although not thoroughly elucidated, these phenotypes are speculated to occur in relation to the X chromosome, due to its involvement in sex determination (Riparbelli et al., 2012).
Although less studied, beneficial Wolbachia are not negligible. For example, Wolbachia symbioses are essential for disease-causing filarial nematodes such as Onchocerca volvulus and B. malayi. Beneficial effects of endosymbiont bacteria are not surprising, as they possess many metabolic and biosynthetic capabilities that animals lack (Moran et al., 2008). For example, recent evidence suggests different Wolbachia types are involved in the provisioning of purines, pyrimidines (Brownlie et al., 2007), vitamin B (Hosokawa et al., 2010), and heme groups (Brownlie et al., 2009) to the host, as well as their involvement in iron metabolism and protection against natural enemies (Hedges et al., 2008; Brownlie and Johnson, 2009) (Table 1). These Wolbachia-host interactions are the subject of a later section in this review.
Wolbachia-Host Coevolution
It was once believed that parasitic and mutualist associations evolved in opposite ways. Classic theories stated that, while host-parasite adaptation typically followed evolutionary arms races, mutualist associations evolved in ways that enhanced the fitness of both partners (Reviewed in Sachs et al., 2011). Other models, however, suggest that mutualism can be more accurately understood as reciprocal exploitations of partners that nonetheless result in net benefits to each partner (Herre et al., 1999). This opens the possibility that the evolutionary mechanisms underlying parasitism and mutualist were more similar than previously thought.
Ewald (1987) proposed that endosymbionts evolve along a parasitism-mutualism continuum depending on their fitness gains, and emphasized that the mode of symbiont transmission was a key determinant of parasitic or mutualistic evolutionary trajectories. Ewald predicted that the more intimately associated host and symbionts are, the stronger their tendency to evolve toward beneficial symbiosis and mutualism. Further development of Ewald's concepts led to a more elaborate conflict-of-interest perspective, which states that the stability of symbiotic associations depends on how well the reproductive interests of host and symbiont converge. Vertical transmission and lack of free-living stages of endosymbionts are recognized as powerful factors aligning the partners' needs and promoting long-term stability (Herre et al., 1999). This has led to the longstanding prediction that vertically transmitted endosymbionts evolve to become benign and establish stable mutualistic associations with their host (Weeks et al., 2007). This is not only logically appealing but has received important support from theoretical, ecological and molecular evidence. However, it does not account for the fact that Wolbachia infections are often lost from insect hosts before co-speciation can occur (Kremer et al., 2009). Therefore, a key question relating to Wolbachia evolution is: what are the evolutionary trajectories of Wolbachia symbiosis within a single host species? This section is dedicated to exploring this question.
Conflicts of Interests between Intimate Associated Partners
Parasitic Wolbachia have been described as selfish genetic elements: heritable units that spread despite the adverse effects they may cause on other genes of the organism they inhabit (Hurst and Werren, 2001). As any other cytoplasmic gene, Wolbachia are in conflict with the host nuclear genome over control of sex ratio. This is the case as sex biases in favor of females would increase the propagation of the cytoplasmic element in spite of nuclear autosomal genes (Hurst, 1992). Indeed, Wolbachia and other selfish entities can, and do, spread by altering the host sex ratio (Charlat et al., 2003). Feminization, MK and PI are common sex ratio distorting phenotypes that effectively increase the frequency of infection in host populations (Hurst, 1992). Although not sex distorters per se, CI-Wolbachia are also in conflict with the nuclear genes, as infected males have reduced chances to produce viable offspring while infection in females protects their eggs from CI-induced mortality (Rand et al., 2004).
Any organismal fitness reduction caused by the disproportionate spread of selfish genetic elements has negative effects on unlinked genomic regions. Selection on those genomic regions would therefore favor the spread of suppressors or modifiers capable of counteracting the effect of the selfish elements (Werren, 2011). For Wolbachia, this would mean that host adaptation to infection would tend to resist or counteract the effects of reproductive modifications (Figure 1). Following, we present some examples of the dynamics of reproductive manipulations and how host adaptation may counteract their parasitic effects.
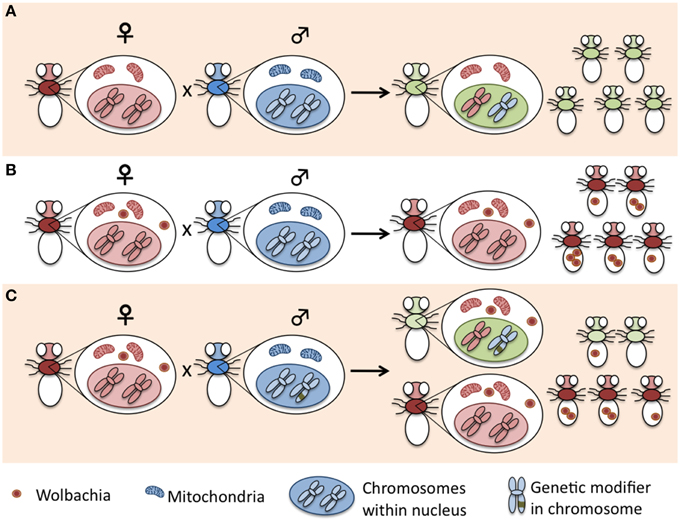
Figure 1. Reproductive-manipulator Wolbachia are selfish genetic elements in conflict with the host nuclear genome, as the disproportionate spread of Wolbachia adversely affects the inheritance of not-linked host genes. Selection on those genomic regions would therefore favor genetic modifiers that counteract the effects of the cytoplasmic selfish element. The figure is a cartoon representation of a sex-distorter Wolbachia in a generic insect population. (A) In a non-infected population, males and females pass on their nuclear genetic material. (B) Wolbachia disproportionately affect the spread of infected female's genes, rendering males less likely to pass their genetic material to the next generation. (C) As males become rare, those with genotypes that lower or suppress the sex-distortion phenotypes would have an increased reproductive success. Consequently, genes that supress or modify the sex-distortion would become common in the population.
Host Adaptation to Sex Distorter Wolbachia
MK, feminization, and PI result in an increase in the number of females in a population relative to males. The spread of these sex-distorting Wolbachia means that male individuals become increasingly rare, raising their reproductive success (Charlat et al., 2003). Therefore, unlinked nuclear genes that eliminate the infection or repress its parasitic effects would increase in frequency to restore the sex ratio to unity (Hurst, 1992). Of the three sex-distorting phenotypes, MK is perhaps the most harmful to host populations. Not only do host populations bear the costs of both mortality and failure to produce males, but the fitness compensation for female progeny is also low (Engelstädter and Hurst, 2009). Therefore, selection on MK suppressors would be predicted to be strong. The study by Hornett et al. (2009) on the butterfly Hypolimnas bolina clearly exemplify these dynamics in nature. Indo-Pacific populations of H. bolina are infected with the Wolbachia wBol1 strain. In Polynesian populations, this Wolbachia induces MK, but the same strain in Southeast Asian H. bolina is present in both sexes and infected females produce a 1:1 sex ratio. Crosses between the two populations demonstrated the presence of a dominant suppressor of the MK phenotype in insects from Thailand and Philippines that was absent from the populations of Polynesia. The authors estimated a very rapid spread of the suppressor gene from past populations, and suggested that MK will disappear from the Wolbachia-H. bolina association in the near future.
When the prevalence of Wolbachia in a host population is high, selection may favor the evolution of new sex determination systems to counteract the deleterious effects of sex-distorting Wolbachia. A notorious example of such adaptive change is presented by the woodlouse Armadillium vulgare (O'Neill et al., 1997). Normally, sex determination in these crustaceans is dictated by female heterogametic chromosomes (ZZ males and ZW females). In some populations, feminizing Wolbachia induced ZZ individuals to develop into females, leading to infection spread and female sex bias. Host genotypes that prevent Wolbachia transmission or suppress feminization, thus producing males, would have a high reproductive success and would tend to restore sex ratio balance. Fascinatingly, this seems to have resulted in the switch from the W chromosome to Wolbachia as sex determining factor in A. vulgare. Evidence suggests that sex determining Wolbachia may occur in other species besides A. vulgare (Charlat et al., 2003).
The genomic conflicts between PI-Wolbachia and the host genome, as well as their resolution, have particularly remarkable evolutionary consequences for species with haplo-diploid sex determination. As mentioned in Section Reproductive Manipulations and Other Phenotypic Effects of Wolbachia, sex determination in these organisms depends on the ploidy of the embryo; fertilized, diploid eggs hatch into diploid females while unfertilized eggs hatch into haploid males. Wolbachia infection turns unfertilized eggs into diploids, which would then hatch into females. This means that infected females produce daughters from both fertilization and parthenogenesis. Stouthamer et al. (2010) proposed that Wolbachia sex bias creates a selective advantage on females with “functional virginity alleles,” that is, alleles that decrease the rates of fertilization in order to favor male offspring. As these alleles spread and become fixed in the population, the capacity for sexual reproduction of the species is lost, ultimately rendering the host dependent on Wolbachia for reproduction. This model of evolution agrees with the observation that many wasp species that have fixed Wolbachia infections are completely parthenogenic, and that antibiotically cured females are no longer capable of sexual reproduction (Russell and Stouthamer, 2010).
Host Adaptation to Cytoplasmic Incompatibility Inducing-Wolbachia
In its simplest form, CI-Wolbachia induce embryonic mortality in crosses between infected males and uninfected females. In other words, CI-Wolbachia “utilize males to make uninfected females unviable” (Charlat et al., 2003). CI-Wolbachia pose opposite effects on each sex; infected females rescue their eggs from CI-inducing mortality, while infected males produce modified sperm that limit their successful matings with uninfected females. This also implies that costs and benefits are influenced by infection prevalence: at low prevalence, infection is a greater cost for males than it is a benefit to females; at high prevalence, infection is greatly advantageous for females and represents little cost to males (Turelli, 1994). Unlike sex-distorting Wolbachia, high prevalence of CI-Wolbachia produces little conflict with the host genome, as both females and males can readily transmit their genes to the next generation. Moreover, as resistance to infection is expected to be selected against, females are said to become addicted to CI-Wolbachia (Koehncke et al., 2009).
These models, although well supported by empirical data, cannot account for the recurrent losses of Wolbachia that are inferred from the incongruence of symbiont-insect phylogenies. It has been proposed the processes that result in Wolbachia extinction are likely related to the decline in CI penetrance (Engelstädter and Hurst, 2009), that is, when the number of eggs that hatch from incompatible crosses increase. A popular hypothesis suggests that it is the bacterium that loses the capacity to induce CI (Werren, 1997). This hypothesis assumes that the abilities of Wolbachia to induce CI (mod) and to rescue eggs from embryonic mortality (resc) are independent. In populations with high infection prevalence, the ability to modify sperm would be no longer necessary. As a consequence, mod− variants may arise and spread, as they would be fully compatible but do not invest in costly mod+ phenotypes. As the mod- variants spread, the ability to induce CI may be lost from the population and with time the endosymbiont may go extinct (Hurst and Mcvean, 1996).
It has also been proposed that attenuation of CI phenotypes is mediated by the spread of male-specific CI modifiers (Turelli, 1994; Koehncke et al., 2009). Using population genetics modeling, Koehncke et al. (2009) concluded that male-specific CI modifiers spread from fixed infections, even when the modifier bears a fitness cost to males. They also showed that the presence of an initial weak male-specific CI modifier eases the spread of subsequent modifiers, which may lead to the gradual elimination of CI. This theoretical finding has important implications for the evolutionary fate of Wolbachia; if symbioses break down after CI attenuation, the frequency of CI-suppressors would decline, once again making populations susceptible to CI-Wolbachia. Therefore, rather than stable, CI-Wolbachia symbioses, could be better understood as cycles of infection spread and loss.
Conflict Aftermath: Cooperation, Addiction, and Extinction
As long as there is conflict between Wolbachia and their host, evolution of resistance mechanisms is expected (Charlat et al., 2003). If adaptation of the host to Wolbachia infection leads to elimination of harmful manipulation phenotypes, does that mean that Wolbachia are inexorably destined to reach extinction? Are there any other factors besides reproductive manipulations that result in favorable selection of infected individuals? There is growing evidence indicating that Wolbachia may play important roles in host physiology, suggesting ecologically contingent benefits of infection (Engelstädter and Hurst, 2009; Duron and Hurst, 2013). Therefore, the importance of Wolbachia on the ecology and evolution of their hosts may go far beyond their ability to induce reproductive manipulations. This section is dedicated to summarizing these recent findings, and considers their implications in the evolutionary trajectories of the symbioses.
Wolbachia as Obligate Mutualists
Beneficial effects of Wolbachia symbiosis are particularly noticeable in organisms that depend on the symbiont for survival and/or reproduction. While Wolbachia are facultative in the vast majority of arthropod hosts, in filarial nematodes they are essential. Antibiotic elimination of Wolbachia results in infertility, inhibition of embryogenesis, arrested adult growth, and death of the worm (Taylor et al., 2005). Nematode Wolbachia display characteristics typical of other ancient primary endosymbionts: the distribution of Wolbachia in the body of the worms is highly specific, the host and symbiont phylogenies are congruent and genome reduction and elimination of repetitive regions relative to facultative Wolbachia. Analysis of the genome of Wolbachia wBm of B. malayi revealed the retention of metabolic pathways for the synthesis of riboflavin, flavin adenine dinucleotide, heme, and nucleotides, which are predicted to be the main metabolic contribution to the worm (Foster et al., 2005).
Obligate Wolbachia symbioses in insects are less common. Two examples that have caught the attention of evolutionary biologists involve the bedbug Cimex lectularius (Hosokawa et al., 2010; Nikoh et al., 2014) and the parasitic wasp Asobara tabida (Dedeine et al., 2001). In the bedbug C. lectularius, Wolbachia fullfils a role of dietary provisioner of vitamin B (Hosokawa et al., 2010) and biotin (Nikoh et al., 2014). Hosokawa and colleagues found that reduction or elimination of Wolbachia through antibiotic treatment rendered animals incapable of producing normally developing eggs, which could be restored by dietary supplementation of vitamin B. In Nikoh et al. (2014), authors sequenced the Wolbachia genome present in the bedbug wCle and found a full biotin-encoding operon capable of synthesizing the compound. Again, Wolbachia depletion produced unfit animals that would return to health after inclusion of the vitamin in their diet (Nikoh et al., 2014). The bedbug-Wolbachia association has characteristics of both primary and secondary endosymbioses, which suggest it is recent relative to other primary symbioses. Like other primary symbionts, Wolbachia in C. lectularius preferentially localizes in bacteriomes and female gonads. The genomic size of this Wolbachia strain, however, resembles that of facultative Wolbachia except in that it contains the biotin operon, which is thought to have been acquired by lateral transmission from an unrelated bacterium (Nikoh et al., 2014). It is speculated that the acquisition of Wolbachia may have influenced the evolution of the feeding habits of these animals (Hosokawa et al., 2010). Indeed, obligate endosymbionts seem to be very common among animals that feed on diets that are poor in specific essential nutrients, such as sap (Baumann and Moran, 1997).
As opposed to coadaptive processes (gradual, beneficial change symbiont, and host) shaping the evolution of Wolbachia-nematode and Wolbachia-bedbug associations, the obligate association with A. tabida is speculated to be the result of Wolbachia “hacking” into a very fundamental aspect of the wasp biology during a coevolutionary arms race (Aanen and Hoekstra, 2007). In insects, apoptosis is essential during the process of egg maturation, as it removes depleted nurse cells after they have transferred their cytoplasmic content to the oocyte (Cavaliere et al., 1998). In A. tabida, Wolbachia is necessary for normal progression of oogenesis, as aposymbiotic females show disproportionate apoptosis of nurse cells earlier than required, causing the abortion of egg development (Pannebakker et al., 2007). It has been hypothesized that during the early stages of the symbiosis, parasitic Wolbachia caused some degree of apoptotic inhibition that was harmful for gametogenesis. As a mechanism to recover ovarian functionality, the host may have responded by up-regulating apoptosis to a new level of functionality that compensated for the inhibitory effects of the symbiont. The spread and fixation of such a compensatory mechanism would have rendered the wasps incapable of producing their own eggs without Wolbachia. The evolutionary implications of these findings are crucial, as they show that obligate symbiosis may quickly evolve from parasitic associations through the spread of compensatory mutations, and that the gradual beneficial adaptation of coevolving partners is not strictly necessary for symbioses to become obligate.
Environment-Dependent Beneficial Effects of Wolbachia
Because of their drastic effects on the host biology, reproductive manipulations of facultative Wolbachia have been a major research focus. This, to some extent, has diverted attention from other phenotypic effects that may also be of great importance on the evolution of these symbioses (Iturbe-Ormaetxe and O'Neill, 2007). The last two decades of Wolbachia research have led to the discovery of multiple symbioses with largely or completely suppressed reproductive modifications that nonetheless appear at equilibrium (Iturbe-Ormaetxe and O'Neill, 2007). Models predict that, in order to persist, these associations require infected females to have a fecundity advantage that is independent from infection prevalence. In other words, these Wolbachia must confer fitness benefits to their host (O'Neill et al., 1997).
Any fitness benefit conferred by facultative symbionts is likely highly dependent on the environment and variable among host genotypes. If symbionts were uniformly beneficial, they would be expected to become fixated in the population (Moran et al., 2008). The symbiosis between D. melanogaster and wMel has been the subject of extensive analysis and has provided unparalleled insight into the phenotypic effects of infection other than reproductive modifications. The wMel-D. melanogaster symbiosis is geographically widespread and its incompatibility phenotypes are weak or absent. Hoffmann et al. (1998) investigated the dynamics of wMel in the field and found that the levels of CI were even weaker than in the laboratory, and maternal inheritance was imperfect. The observed stability of infection frequency in some populations led the authors to conclude that wMel likely provided fitness benefits to the flies, but such effects were not yet apparent. Another study by Fry and Rand (2002) evaluated the effects of wMel infection in laboratory maintained D. melanogaster, and found that fly lifespan was positively affected by infection, such effect being highly dependent on the nuclear background of the flies.
Sequencing of the wMel genome, the first available for Wolbachia, provided substantial indirect evidence for the plausible metabolic roles of wMel in flies. The presence of riboflavin and heme synthesis pathways (Wu et al., 2004), as well as evidence of positive selection in some of these genes (Brownlie et al., 2007), suggest that Wolbachia may provision the fly with these cofactors or some of their intermediates. Wolbachia may also represent an additional source of nucleotides, which could be beneficial in processes where high DNA replication is involved (Brownlie et al., 2007). Following, up on their genomic analyses of wMel, Brownlie et al. (2009) also found direct evidence on the beneficial role of Wolbachia in the iron metabolism of D. melanogaster. The authors investigated the effects of infection on flies exposed to different levels of dietary iron (Brownlie et al., 2009). Although Wolbachia appeared to have no effect on flies fed standard laboratory diets, flies raised on diets with a deficit or excess of iron greatly benefitted from the infection. Based on the seemingly low iron content of flies in nature, the authors concluded that Wolbachia contribution to the host iron metabolism is very likely an ecologically relevant trait.
Another fascinating effect of wMel is its capacity to confer protection against RNA viruses (Hedges et al., 2008; Teixeira et al., 2008). The presence of Wolbachia in fly tissues has been linked to a reduction in Drosophila C virus titres of up to 10,000-fold relative to uninfected flies, and correlates with extended lifespan. Moreover, wMel-related resistance is also effective against other RNA viruses and over different host genetic backgrounds (Teixeira et al., 2008). Importantly, new theoretical approaches suggest that protection against pathogens constitutes an important force driving the spread of facultative heritable symbionts (Reviewed in Haine, 2008). Models predict that wherever a virulent, horizontally transmitted pathogen infects a host population, a protective vertically transmitted endosymbiont would spread and greatly reduce the frequency of parasitised hosts at equilibrium. These three-way interactions would lead to the persistence of both pathogen and endosymbiont in the host population, but not to the fixation of the endosymbiont (Lively et al., 2005; Brownlie and Johnson, 2009). Empirical evidence on these symbiont-mediated protection dynamics is expanding. For example, Jaenike et al. (2010) documented a rapid spread of the vertically-transmitted Spiroplasma bacterium in American populations of Drosophila neotestacea and how such spread could be linked to their protective role against sterilizing parasitic nematodes. Therefore, Wolbachia-mediated protection against pathogens constitutes a very plausible mechanism of invasion of host populations besides or in addition to reproductive modifications (Fenton et al., 2011).
The expanding evidence on environment-dependent Wolbachia benefits allows for speculation of other evolutionary trajectories of Wolbachia beside fixation and extinction. Depending on the extent of host dependence on the microbe, Wolbachia could also stably persist as facultative symbionts that fullfil important but sporadic host needs. A study by Mondo et al. (2012) showed that the facultative symbioses between arbustal mycorrhizal fungi (Glomeromycota) and the bacterial Candidatus Glomeribacter gigasporarum have been evolutionary stable for 400 million years, almost twice the estimated time of the ancient obligate Buchnera-aphid association. This study suggests that such stability is due to a balance between vertical transmission, recombination and host switching. Sporadic horizontal transmission and recombination may counteract the erosive effects of genetic drift observed in non-recombining genomes (see Section Wolbachia Genomics and the Ability to Retain an Infectious Capacity), allowing organisms to maintain their infective capacity. Symbionts whose beneficial roles are restricted to particular environments would have an advantage if they retain the ability colonize new hosts. The authors of this study speculate that high environmental variability influences the symbiosis to be “locked” in a stable facultative state.
The many similarities between the Glomeribacter-Glomeromycota model and Wolbachia (sporadic horizontal transmission, recombination, environment dependent advantages) give some support to the hypothesis that Wolbachia could be stably maintained in host populations as facultative symbionts. The commonly observed lack of symbiont-host co-speciation, however, argues against Wolbachia symbioses being maintained for extended periods of evolutionary time. Plausibly, the immense environmental variability to which Wolbachia-arthropod associations are exposed means that there would be many situations where symbionts would not be necessary or would simply be too costly to maintain, therefore driving local symbiont extinctions. Indeed, population genomic analyses of Wolbachia in D. melanogaster indicate that this infection was globally established once some 8000 years ago and has been lost from multiple populations worldwide (Richardson et al., 2012).
Cardinium, is another symbiont causing similar reproductive alterations as Wolbachia (Giorgini et al., 2009; Ma et al., 2015). In the haplodiploid wasp Encarsia it has been shown that the Cardinium infection doesn't induce, as expected, thelytokous parthenogenesis, but feminization. In fact antibiotic treatment results in uninfected diploid male offspring, thus demonstrating that diploidy restoration is a necessary condition, but not sufficient, to elicit female development. In combination this result suggests that Cardinium is responsible for the feminization of the hymenopteran genetic males.
If Wolbachia play important ecological roles in their hosts, it is plausible that the local extinction of a given variant occurs through the replacement with new Wolbachia strains. An excellent example of this is presented by Kriesner et al. (2013) on the Australian coastal populations of D. simulans. Historically, Australian D. simulans harbored the non-CI inducing Wolbachia strain wAu, at frequencies lower than 0.3. At some point after 1994, the strong CI-inducer wRi was introduced to the continent. Between 2004 and 2012, wRi dramatically spread across the east coast completely replacing wAu and establishing itself at frequencies of over 0.9. A very important remark of this study was their suggestion that historical and recent invasions of wAu and wRi, respectively, cannot be fully explained by CI dynamics and must have been driven by fitness benefits conferred by Wolbachia, implying that Wolbachia may fill some ecological role in these populations. Interestingly, both wAu and wRi display antiviral protection phenotypes similar to those of wMel in D. melanogaster (Osborne et al., 2009). Whether this is the particular beneficial trait that allowed the spread of Wolbachia in Australian population of D. simulans remains unknown.
Our current understanding of Wolbachia no longer fits a model where parasitic and mutualistic associations are clear-cut (Zug and Hammerstein, 2015). We now know that there are Wolbachia-related phenotypes that are apparent under specific cellular and environmental circumstances and depend on the host genotypes (Correa and Ballard, 2014; Sicard et al., 2014). The study of Wolbachia infections in multiple environments and on carefully controlled host genetic backgrounds will continue to prove useful to our understanding of Wolbachia-host interactions.
Wolbachia, Mitochondria, and their Interactions with the Host and the Environment
As described in the previous section, Wolbachia induced phenotypes are often the result of complex interactions between the genotypes of both partners and the environment (Mouton et al., 2007). Interestingly, the current understanding of non-neutral mitochondrial genetic variations offers a very similar scenario, in which mtDNA mutants may have unequal fitness depending on the nuclear genetic background of the organism and the environment (Ballard and Pichaud, 2014). This section explores some of the evolutionary and ecological parallels between Wolbachia and mtDNA variants, and their implications in host adaptive processes.
Mitochondria and Wolbachia Origins
Although the idea of a prokaryotic origin of mitochondria can be traced to 1890 (Kutschera and Niklas, 2005), it was not until the discovery and sequencing of mtDNA that their bacterial ancestry became obvious. The retention of ribosomal RNA coding sequences, a universal trait of modern mitochondrial genomes (Gray, 2012), allowed mtDNA lineages to be traced to a single origin from an ancestral alphaproteobacterium of the order Rickettsiales (Ferla et al., 2013). Thus, mitochondria and Wolbachia not only share their lifestyles and mode of propagation but also their ancestry.
The adaptive processes that resulted in obligate intracellular lifestyles of mitochondria and Wolbachia, as well as their impact on host evolution, are arguably very different. A recent and relatively well-supported hypothesis on the origin of mitochondria states that their alphaproteobacterium ancestor (protomitochondrion) fused to or was engulfed by a highly complex archaeobacterium (instead of a basal amitochondrial eukaryote as classically believed). Under this “symbiogenic” hypothesis, the evolutionary novelty of eukaryotic cells emerged after or as a consequence of the establishment of this symbiosis (Koonin, 2010; Gray, 2012). Interestingly, such a scenario provides a plausible selective factor for the evolution of eukaryotic cellular compartmentalization. Hypothetically, the exposure of the archaeal genome to the DNA and translation products of the protomitochondrion may have negatively impacted the host gene expression, for which the separation of transcription and translation might have been adaptive (Koonin, 2010). Once in a compartmentalized cell, it has been argued that the conflicts of interest between cytoplasmic and nuclear genomes facilitated the evolution of other characteristic traits of eukaryotic cells such as sex, anisogamy and uniparental inheritance of cytoplasmic genes (Hurst, 1992; Law and Hutson, 1992). Although arguments against this symbiogenic hypothesis are yet to be addressed, the concept of the origin of modern eukaryotes being triggered by processes of conflict resolution between interacting genomes is intriguing.
The origin of Wolbachia is less clear. Their position within the Rickettsiales makes it reasonable to assume they originated from an ancestor with an intracellular lifestyle (Comandatore et al., 2013), more than100 million years ago. At present, it has not been possible to elucidate what was the first invertebrate lineage Wolbachia associated with, nor the nature of such association (mutualistic or parasitic). Considering their exclusive distribution within terrestrial invertebrates, it is plausible that Wolbachia emerged as symbionts of modern invertebrate forms. By the time the first Wolbachia associations appeared, invertebrate hosts had probably taken up most of the essential metabolic capacities from the protomitochondrion, which may have constrained the evolutionary novelty that the more derived Wolbachia alphaproteobacteria could have brought upon their hosts. Nonetheless, Wolbachia do confer novel physiologic abilities upon their hosts, enabling them to explore otherwise inadequate environments (Wernegreen, 2004; Duron and Hurst, 2013).
Genomic Reduction and Lateral Gene Transfer
Mitochondrial genomes are under the same population constrains of obligate endosymbionts (Burger and Lang, 2003; see Section Environment-Dependent Beneficial Effects of Wolbachia); consequently, they share many genomic traits such as extreme size reduction, genomic stability, and lack of repeat sequences. As a facultative symbiont, Wolbachia is said to possess a genome “in transition” (Moran et al., 2008). Knowledge on the current processes of genomic evolution in organisms like Wolbachia may contribute to the understanding of the early phases of mitochondrial evolution.
Genomic size reduction is perhaps the most consistent genomic consequence of endosymbiosis (Burger and Lang, 2003), which in mitochondria occurred through gene loss and transference to the nucleus (Rand et al., 2004). During eukaryotic evolution, such lateral gene transfer (LGT) was substantial, which partially accounts for the fact that nuclear genes now encode the vast majority of the mitochondrial proteome. In fact, the role of LGT goes beyond organelle function, and genes that formerly belonged to the protomitochondrion are now involved in other cellular processes (Gray, 2012).
Genomic reduction has occurred in Wolbachia, though to a lesser extent than in other ancient intracellular endosymbionts (Toft and Andersson, 2010). Interestingly, recent studies have shown that LGT also occurs from Wolbachia to host chromosomes. Wolbachia DNA fragments have been found in the genomes of various insect and nematode species. Hotopp et al. (2007) found traces of Wolbachia DNA fragments in six of 21 published genomes of various invertebrate taxa. Another study found that some transferred genes from wBm into the B. malayi genome seem to be transcribed in a way that is stage-specific during the development of the worm (Ioannidis et al., 2013). These results suggest that Wolbachia may indeed represent an important source of genomic innovation for their hosts. Considering the vast abundance of past and present Wolbachia associations, the evolutionary consequences of this phenomenon can be considerable.
Cyto-Nuclear Interactions and Compensatory Mutations
How similar are the processes of host adaptation to Wolbachia infection and mitochondrial genetic variants? It was once thought that genetic variation of mitochondrial genes was selectively neutral (Ballard and Kreitman, 1995). Such an assumption, however, has been weakened by increasing evidence on the differential fitness of mtDNA variants in host populations (Ballard and Whitlock, 2004; Ballard and Rand, 2005; Dowling et al., 2008).
It is now apparent that the coordinate assembly of mitochondrial and nuclear encoded proteins is necessary for the correct function the electron transport system. This intricate molecular machinery produces up to 90% of cellular energy through oxidative phosphorylation. Therefore, mutations in the mtDNA that result in protein conformational changes would be predicted to impact oxidative phosphorylation function, with consequences for cell energy homeostasis and overall organismal fitness (Ballard and Melvin, 2010; Horan et al., 2013). Because mtDNA is subjected to the effects of Muller's ratchet (Lynch and Blanchard, 1998; Rand et al., 2004), it has been suggested that selection favors responses in the nuclear genome that compensate for mildly deleterious mtDNA mutations to restore metabolic function (Dowling et al., 2008). The benefits of such compensatory mechanisms, limited to individuals with the mitochondrial mutation, are equivalent in principle to those that arise in response to negative effects of Wolbachia infection.
A simple prediction from this “compensatory mutation hypothesis” is that experimentally changing the native nuclear background of the cytoplasmic genetic elements (either Wolbachia or mitochondria) would result in the disruption of the compensatory phenotypes. For mtDNA, the disruption of mito-nuclear coadapted complexes would result in decreased host fitness. For CI-Wolbachia, it may result in increased CI levels of naive vs. adapted hosts. Experimental evidence supports these predictions. Laboratory crosses of the intertidal copepod Tigriopus californicus showed that third generation interpopulation hybrids displayed reduced fitness, subsequently restored by maternal (but not paternal) backcrosses. Moreover, lowered levels of ATP synthesis in the hybrid animals were restored by the re-introgression of the maternal nuclear background in these animals. These results clearly indicate the involvement of mito-nuclear interactions in ATP production and organismal fitness (Ellison and Burton, 2008). For Wolbachia, Poinsot et al. (1998) showed that the wMel induced weak CI in their native host D. melanogaster, but displayed very high levels of embryonic mortality upon transference to a naïve D. simulans background through microinjection.
Compensatory mutations are predicted to have important macroevolutionary consequences for their host. The T. californicus case exemplifies how mtDNA divergence between populations can drive differentiation in associated parts of the nuclear genome, which could lead to hybrid breakdown (Ellison and Burton, 2008). A study shows that Wolbachia variants may also drive reproductive isolation. Sympatric semispecies of the Neotropical Drosophila paulistorum require infection of semispecies-specific Wolbachia for viability. These obligate-mutualist Wolbachia are present at very low densities in host reproductive tissues. In semispecific hybrids, however, Wolbachia over-replication seems to cause pathogenic phenotypes such as embryonic inviability and male sterility, indicating semispecies-specific compensatory mechanisms against Wolbachia pathogenicity (Miller et al., 2010).
Sexual Antagonism and Mother's Curse
Similarly to Wolbachia, the maternal inheritance of mtDNA may represent an important asymmetry between the sexes, which may lead to significant fitness differences between males and females that harbor the same mtDNA type. The mother's curse hypothesis suggests that natural selection would fail to purge cytoplasmic genetic variants that are deleterious for males, provided they do not negatively affect female fitness (Gemmell et al., 2004). The deleterious effects of such mutants would be confined to male specific traits, and therefore very likely expressed as some degree of male infertility. A recent study examined the transcriptome of isogenic D. melanogaster flies with variable mtDNA and standardized nuclear background. The results of this study indicated striking effects of mtDNA mutations on the expression of male-specific genes (those related to testis and accessory gland function). Female-specific genes and those with identical function for both sexes were, however, only moderately affected by the mutation load. Compensatory effects that encompass widespread control of the nuclear transcriptome likely cause the discrepancy observed between the sexes (Innocenti et al., 2011). A follow-up study on these and other similarly constructed fly lines tested the effect of mtDNA mutations on sperm competitiveness, and found that male fertility indeed varies in vivo across mtDNA variants (Yee et al., 2013).
The asymmetric fitness effects produced by reproductive manipulations are discussed in Sections Reproductive Manipulations and Other Phenotypic Effects of Wolbachia and Conflicts of Interests Between Intimate Associated Partners. Aside from reproductive manipulations, Wolbachia has also been linked with reduction of sperm production (Snook et al., 2000) and sperm competitive ability (de Crespigny and Wedell, 2006) in D. simulans males. In CI-inducing Wolbachia, lower sperm competitiveness may undermine the effects of the reproductive manipulations and alter Wolbachia spread dynamics.
Cross-Talk, Trade-Offs, and Adaptation to the Local Environment
Life history traits are often negatively associated with each other (Zera and Harshman, 2001). When the fitness cost of a beneficial trait results in a detrimental change in another trait, it is said that a trade-off exists (Stearns, 1989). Understanding trade-offs is essential for the study of evolutionary processes, as they provide an explanation for the common occurrence of variability in life history traits (Zera and Harshman, 2001). Mutations that alter mito-nuclear molecular interactions may result in physiological trade-offs (Ballard and Pichaud, 2014). This is because the essential process of mitochondrial ATP synthesis produces by-products (in the form of reactive oxygen species) that lead to cellular oxidative damage, whose accumulated damaging effects are hypothesized to be causative of aging. Using D. simulans as a model organism, Ballard and Melvin (2011) found that reduced activity of the cytochrome oxidase complex due to a mutated nuclear encoded subunit resulted in flies having significantly higher reproduction at early life but reduced lifespan. The study showed that mutant flies up-regulated mitochondrial function as a whole in an attempt to compensate for suboptimal cytochrome oxidase complex activity, which resulted in efficient ATP synthesis but higher production of reactive oxygen species. The authors hypothesize that a retrograde response from a mildly dysfunctional electron transport system may lie at the heart of such a compensatory mechanism.
It has been argued that mutations that affect mitochondrial function could be positively selected if they provide benefits in specific environments. Pichaud et al. (2012) studied the mitochondrial performance of D. simulans with siII and siIII mitochondrial types and found that siII-harboring flies had a higher catalytic capacity, which may provide advantages in terms of intensity of aerobic activity, endurance, or both. Another study on similar fly cohorts showed that siII flies recovered faster from cold coma, while siIII were better at withstanding starvation, which may indicate that the improved catalytic capacity of siII flies comes at a cost (Ballard et al., 2007). Flies harboring siIII mitochondria are commonly found in sympatry with those harboring siII, with no indication of restricted gene flow between them (Ballard et al., 2002). This may be an indication that mtDNA genetic variation at the population level can occur if there are differential selective advantages for each mitochondrial type.
For Wolbachia-host associations, perhaps the most notorious trade-off is that between transmission fidelity and pathogenicity of infection, mediated by bacterial titres (McGraw et al., 2002). High Wolbachia titres may guarantee the transmission of microbes through the egg, but may result in pathogenicity for the host. Low symbiont density, on the other hand, may reduce the fitness costs of infection, albeit at a reduction in the number of infected host individuals in the next generation. If there is a fitness advantage for infected females, as in the case of those harboring CI-Wolbachia, host, and symbiont may act on the regulation of microbe titres to optimize the trade-off between the two parameters (Mouton et al., 2007). An exciting observation suggests a molecular mechanism by which Drosophila may modulate Wolbachia titres. Serbus et al. (2008) showed that the mRNP complex (mRNA and protein) formed by the transcript of the grk gene and its protein-binding partners has a cumulative, dose-sensitive impact on Wolbachia titre during oogenesis. The authors propose a model of interaction in which grk mRNP increases Wolbachia titres, which leads to a feed-back response that disrupts the function of the grk mRNP components, completing the regulatory loop. The involvement of cross-talk mechanisms in the regulation of symbiont/organelle function in response to environmental situations could represent an important hallmark in the long-term maintenance of stable cyto-nuclear associations. How widespread this mechanism is across Wolbachia-host symbioses is yet to be examined.
Wolbachia titres vary with environmental and physiological factors such as larval crowding (Wiwatanaratanabutr and Kittayapong, 2009), developmental and adult temperature (Mouton et al., 2006, 2007; Bordenstein and Bordenstein, 2011), age (Unckless et al., 2009; Tortosa et al., 2010), and diet (Ponton et al., 2015; Serbus et al., 2015). In some scenarios, such over-replication could be interpreted as the destabilization of the symbiosis where Wolbachia takes advantage of the host (McGraw et al., 2002; Rio et al., 2006). For example, the pathogenic wMelPop strain of Wolbachia is known to over-replicate in somatic tissues of D. melanogaster, producing abnormal fly phenotypes and shortening lifespan, which appear to be exacerbated at high temperatures (Min and Benzer, 1997; McGraw et al., 2002). In most cases, however, it is difficult to determine if Wolbachia titre is actively modulated in situations where higher bacterial densities are beneficial for the symbiosis. Osborne et al. (2012), for example, found important correlations between Wolbachia titres and antiviral protection for some naturally occurring Wolbachia variants in D. simulans. Wolbachia variants known to occur at low densities failed to confer antiviral protection.
Concluding Remarks
Wolbachia are a group of intracellular, maternally inherited bacteria with an impressive history of adaptation to intracellular lifestyles. Unlike other ancient obligate symbionts, Wolbachia are “generalists in host use.” This means that instead of adapting to a single host lineage, Wolbachia evolved ways to jump across host species and establish relatively stable associations in which they are maintained through vertical transmission. With every new established association, Wolbachia are in conflict with nuclear genes, often conferring a disproportionate advantage to infected females to allow infection to spread. The reproductive manipulations induced by Wolbachia, perhaps the clearest representations of conflicts between cytoplasmic and nuclear genomes, are thought to be the general mechanism by which Wolbachia spread through host populations.
Recent evidence, however, challenges the view of Wolbachia as a ruthless manipulator of the host reproductive biology and suggests an evolutionary scenario where Wolbachia-host interactions are driven by the onset and resolution of conflicts of interest. Here, we have highlighted a view in which reproductive manipulations are only transient phenotypes, attenuated as the host adapts to infection. After attenuation of reproductive phenotypes, the stability of the symbioses would rely on the physiological advantages Wolbachia may confer upon their host. The availability of multiple sequenced Wolbachia genomes has been an important step forward in the understanding of Wolbachia-induced phenotypes, as they reveal a whole biochemical and genetic repertoire from which the host can metabolically benefit from the infection. The recent discovery of various Wolbachia-induced beneficial phenotypes supports this line of thought.
Compensatory mutations (or genetic modifiers) are a hallmark of the adaptation of the host genome to Wolbachia and other cytoplasmic genetic elements in conflict, mitochondria among them. The evolutionary trajectories of Wolbachia symbioses after attenuation of reproductive manipulations would depend on the physiological benefits of infection, which for facultative symbionts are likely to be dependent on the environment. Here, we have also presented evidence that supports the view of environment-dependent facultative mutualism as a stable evolutionary outcome of Wolbachia infections beside extinction and obligate symbioses. Finally, our current understanding of the biology of mitochondria and Wolbachia unravels remarkable parallels in the way they interact with the nuclear genome. For example, we highlighted how Wolbachia and mtDNA polymorphisms may be maintained at equilibrium in host populations when they confer environment-specific fitness advantages. Great insights into both the Wolbachia and mitochondrial research fields can be revealed if these fields are considered to be overlapping, rather than independent from each other.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank John Janieke and Jeremy Brownlie for critical revision of this text. Members of the Ballard lab are also thanked for useful feedback on the manuscript draft.
References
Aanen, D. K., and Hoekstra, R. F. (2007). The evolution of obligate mutualism: if you can't beat'em, join'em. Trends Ecol. Evol. 22, 506–509. doi: 10.1016/j.tree.2007.08.007
Baldo, L., Bordenstein, S. R., Wernegreen, J. J., and Werren, J. H. (2006a). Widespread recombination throughout Wolbachia genomes. Mol. Biol. Evol. 23, 437–449. doi: 10.1093/molbev/msj049
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., Choudhury, R. R., et al. (2006b). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110. doi: 10.1128/AEM.00731-06
Baldo, L., Lo, N., and Werren, J. H. (2005). Mosaic nature of the Wolbachia surface protein. J. Bacteriol. 187, 5406–5418. doi: 10.1128/JB.187.15.5406-5418.2005
Ballard, J. W. O., Chernoff, B., and James, A. C. (2002). Divergence of mitochondrial DNA is not corroborated by nuclear DNA, morphology, or behavior in Drosophila simulans. Evolution 56, 527–545. doi: 10.1111/j.0014-3820.2002.tb01364.x
Ballard, J. W. O., and Kreitman, M. (1995). Is mitochondrial DNA a strictly neutral marker? Trends Ecol. Evol. 10, 485–488. doi: 10.1016/S0169-5347(00)89195-8
Ballard, J. W. O., and Melvin, R. G. (2010). Linking the mitochondrial genotype to the organismal phenotype. Mol. Ecol. 19, 1523–1539. doi: 10.1111/j.1365-294X.2010.04594.x
Ballard, J. W. O., and Melvin, R. G. (2011). Early life benefits and later life costs of a two amino acid deletion in Drosophila simulans. Evolution 65, 1400–1412. doi: 10.1111/j.1558-5646.2010.01209.x
Ballard, J. W. O., Melvin, R. G., Katewa, S. D., and Maas, K. (2007). Mitochondrial DNA variation associated with measurable differences in life-history traits and mitochondrial metabolism in Drosophila simulans. Evolution 61, 1735–1747. doi: 10.1111/j.1558-5646.2007.00133.x
Ballard, J. W. O., and Pichaud, N. (2014). Mitochondrial DNA: more than an evolutionary bystander. Funct. Ecol. 28, 218–231. doi: 10.1111/1365-2435.12177
Ballard, J. W. O., and Rand, D. M. (2005). The population biology of mitochondrial DNA and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36, 621–642. doi: 10.1146/annurev.ecolsys.36.091704.175513
Ballard, J. W. O., and Whitlock, M. C. (2004). The incomplete natural history of mitochondria. Mol. Ecol. 13, 729–744. doi: 10.1046/j.1365-294X.2003.02063.x
Bandi, C., Anderson, T. J., Genchi, C., and Blaxter, M. L. (1998). Phylogeny of Wolbachia in filarial nematodes. Proc. R. Soc. Lond. B Biol. Sci. 265, 2407–2413.
Baumann, P., and Moran, N. A. (1997). Non-cultivable microorganisms from symbiotic associations of insects and other hosts. Antonie Van Leeuwenhoek 72, 39–48.
Bordenstein, S. R., and Bordenstein, S. R. (2011). Temperature affects the tripartite interactions between bacteriophage WO, Wolbachia, and cytoplasmic incompatibility. PLoS ONE 6:e29106. doi: 10.1371/journal.pone.0029106
Bordenstein, S. R., and Reznikoff, W. S. (2005). Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 3, 688–699. doi: 10.1038/nrmicro1233
Bordenstein, S. R., and Wernegreen, J. J. (2004). Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21, 1981–1991. doi: 10.1093/molbev/msh211
Bourtzis, K., Pettigrew, M. M., and O'Neill, S. L. (2000). Wolbachia neither induces nor suppresses transcripts encoding antimicrobial peptides. Insect Mol. Biol. 9, 635–639. doi: 10.1046/j.1365-2583.2000.00224.x
Braig, H. R., Guzman, H., Tesh, R. B., and O'Neill, S. L. (1994). Replacement of the natural Wolbachia symbiont of Drosophila simulans with a mosquito counterpart. Nature 367, 453–455.
Brownlie, J. C., Adamski, M., Slatko, B., and McGraw, E. A. (2007). Diversifying selection and host adaptation in two endosymbiont genomes. BMC Evol. Biol. 7:68. doi: 10.1186/1471-2148-7-68
Brownlie, J. C., Cass, B. N., Riegler, M., Witsenburg, J. J., Iturbe-Ormaetxe, I., McGraw, E. A., et al. (2009). Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 5:e1000368. doi: 10.1371/journal.ppat.1000368
Brownlie, J. C., and Johnson, K. N. (2009). Symbiont-mediated protection in insect hosts. Trends Microbiol. 17, 348–354. doi: 10.1016/j.tim.2009.05.005
Burger, G., and Lang, B. F. (2003). Parallels in genome evolution in mitochondria and bacterial symbionts. IUBMB Life 55, 205–212. doi: 10.1080/1521654031000137380
Cavaliere, V., Taddei, C., and Gargiulo, G. (1998). Apoptosis of nurse cells at the late stages of oogenesis of Drosophila melanogaster. Dev. Genes Evol. 208, 106–112.
Charlat, S., Hurst, G. D. D., and Merçot, H. (2003). Evolutionary consequences of Wolbachia infections. Trends Genet. 19, 217–223. doi: 10.1016/S0168-9525(03)00024-6
Cho, K. O., Kim, G.-W., and Lee, O. K. (2011). Wolbachia bacteria reside in host Golgi-related vesicles whose position is regulated by polarity proteins. PLoS ONE 6:e22703. doi: 10.1371/journal.pone.0022703
Comandatore, F., Sassera, D., Montagna, M., Kumar, S., Koutsovoulos, G., Thomas, G., et al. (2013). Phylogenomics and analysis of shared genes suggest a single transition to mutualism in Wolbachia of nematodes. Genome Biol. 5, 1668–1674. doi: 10.1093/gbe/evt125
Correa, C. C., and Ballard, J. W. O. (2014). What can symbiont titres tell us about co-evolution of Wolbachia and their host? J. Invertebr. Pathol. 118, 20–27. doi: 10.1016/j.jip.2014.02.009
Curry, M. M., Paliulis, L. V., Welch, K. D., Harwood, J. D., and White, J. A. (2015). Multiple endosymbiont infections and reproductive manipulations in a linyphiid spider population. Heredity 115, 146–152. doi: 10.1038/hdy.2015.2
Dale, C., and Moran, N. A. (2006). Molecular interactions between bacterial symbionts and their hosts. Cell 126, 453–465. doi: 10.1016/j.cell.2006.07.014
de Crespigny, F. E. C., and Wedell, N. (2006). Wolbachia infection reduces sperm competitive ability in an insect. Proc. R. Soc. Lond. B Biol. Sci. 273, 1455–1458. doi: 10.1098/rspb.2006.3478
Dedeine, F., Vavre, F., Fleury, F., Loppin, B., Hochberg, M. E., and Boulétreau, M. (2001). Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc. Natl. Acad. Sci. U.S.A. 98, 6247–6252. doi: 10.1073/pnas.101304298
Dowling, D. K., Friberg, U., and Lindell, J. (2008). Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol. Evol. 23, 546–554. doi: 10.1016/j.tree.2008.05.011
Duplouy, A., Iturbe-Ormaetxe, I., Beatson, S. A., Szubert, J. M., Brownlie, J. C., McMeniman, C. J., et al. (2013). Draft genome sequence of the male-killing Wolbachia strain wBol1 reveals recent horizontal gene transfers from diverse sources. BMC Genomics 14:20. doi: 10.1186/1471-2164-14-20
Duron, O., Boureux, A., Echaubard, P., Berthomieu, A., Berticat, C., Fort, P., et al. (2007). Variability and expression of ankyrin domain genes in Wolbachia variants infecting the mosquito Culex pipiens. J. Bacteriol. 189, 4442–4448. doi: 10.1128/JB.00142-07
Duron, O., and Hurst, G. D. D. (2013). Arthropods and inherited bacteria: from counting the symbionts to understanding how symbionts count. BMC Biol. 11, 45–48. doi: 10.1186/1741-7007-11-45
Ellegaard, K. M., Klasson, L., Näslund, K., Bourtzis, K., and Andersson, S. G. E. (2013). Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet 9:e1003381. doi: 10.1371/journal.pgen.1003381
Ellison, C. K., and Burton, R. S. (2008). Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution 62, 631–638. doi: 10.1111/j.1558-5646.2007.00305.x
Engelstädter, J., and Hurst, G. D. D. (2009). The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 40, 127–149. doi: 10.1146/annurev.ecolsys.110308.120206
Ewald, P. W. (1987). Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N.Y. Acad. Sci. 503, 295–306.
Fenton, A., Johnson, K. N., Brownlie, J. C., and Hurst, G. D. D. (2011). Solving the Wolbachia paradox: Modelling the tripartite interaction between host, Wolbachia, and a natural enemy. Am. Nat. 178, 333–342. doi: 10.1086/661247
Ferla, M. P., Thrash, J. C., Giovannoni, S. J., and Patrick, W. M. (2013). New rRNA gene-based phylogenies of the alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PLoS ONE 8:e83383. doi: 10.1371/journal.pone.0083383
Ferree, P. M., Frydman, H. M., Li, J. M., Cao, J., Wieschaus, E., and Sullivan, W. (2005). Wolbachia utilizes host microtubules and dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 1:e14. doi: 10.1371/journal.ppat.0010014
Foster, J., Ganatra, M., Kamal, I., Ware, J., Makarova, K., Ivanova, N., et al. (2005). The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3:e121. doi: 10.1371/journal.pbio.0030121
Fry, A. J., and Rand, D. M. (2002). Wolbachia interactions that determine Drosophila melanogaster survival. Evolution 56, 1976–1981. doi: 10.1111/j.0014-3820.2002.tb00123.x
Frydman, H. M., Li, J. M., Robson, D. N., and Wieschaus, E. (2006). Somatic stem cell niche tropism in Wolbachia. Nature 441, 509–512. doi: 10.1038/nature04756
Gemmell, N. J., Metcalf, V. J., and Allendorf, F. W. (2004). Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol. Evol. 19, 238–244. doi: 10.1016/j.tree.2004.02.002
Giorgini, M., Monti, M. M., Caprio, E., Stouthamer, R., and Hunter, M. S. (2009). Feminization and the collapse of haplodiploidy in an asexual parasitoid wasp harboring the bacterial symbiont Cardinium. Heredity 102, 365–371. doi: 10.1038/hdy.2008.135
Glowska, E., Dragun-Damian, A., Dabert, M., and Gerth, M. (2015). New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect. Genet. Evol. 30, 140–146. doi: 10.1016/j.meegid.2014.12.019
Gray, M. W. (2012). Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 4:a011403. doi: 10.1101/cshperspect.a011403
Haine, E. R. (2008). Symbiont-mediated protection. Proc. R. Soc. Lond. B Biol. Sci. 275, 353–361. doi: 10.1098/rspb.2007.1211
Heath, B. D., Butcher, R. D., Whitfield, W. D., and Hubbard, S. F. (1999). Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr. Biol. 9, 313–316. doi: 10.1016/S0960-9822(99)80139-0
Hedges, L. M., Brownlie, J. C., O'Neill, S. L., and Johnson, K. N. (2008). Wolbachia and virus protection in insects. Science 322, 702–702. doi: 10.1126/science.1162418
Herre, E. A., Knowlton, N., Mueller, U. G., and Rehner, S. A. (1999). The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol. Evol. 14, 49–53. doi: 10.1016/S0169-5347(98)01529-8
Hertig, M., and Wolbach, S. B. (1924). Studies on rickettsia-like micro-organisms in insects. J. Med. Res. 44, 329–374.7.
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia? – A statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Hoffmann, A. A., Hercus, M., and Dagher, H. (1998). Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics 148, 221–231.
Horan, M. P., Rumbley, J. N., Melvin, R. G., Le Couteur, D. G., and Ballard, J. W. O. (2013). Quaternary protein modeling to predict the function of DNA variation found in human mitochondrial cytochrome c oxidase. J. Hum. Genet. 58, 127–134. doi: 10.1038/jhg.2012.144
Hornett, E. A., Charlat, S., Wedell, N., Jiggins, C. D., and Hurst, G. D. D. (2009). Rapidly shifting sex ratio across a species range. Curr. Biol. 19, 1628–1631. doi: 10.1016/j.cub.2009.07.071
Hosokawa, T., Koga, R., Kikuchi, Y., Meng, X. Y., and Fukatsu, T. (2010). Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. U.S.A. 107, 769–774. doi: 10.1073/pnas.0911476107
Hotopp, J. C. D., Clark, M. E., Oliveira, D. C. S. G., Foster, J. M., Fischer, P., Muñoz Torres, M. C., et al. (2007). Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756. doi: 10.1126/science.1142490
Hurst, G. D. D., and Werren, J. H. (2001). The role of selfish genetic elements in eukaryotic evolution. Nat. Rev. Genet. 2, 597–606. doi: 10.1038/35084545
Hurst, L. D. (1992). Intragenomic conflict as an evolutionary force. Proc. R. Soc. Lond. B Biol. Sci. 248, 135–140. doi: 10.1098/rspb.1992.0053
Hurst, L. D., and Mcvean, G. T. (1996). Clade selection, reversible evolution and the persistence of selfish elements: the evolutionary dynamics of cytoplasmic incompatibility. Proc. R. Soc. Lond. B Biol. Sci. 263, 97–104. doi: 10.1098/rspb.1996.0016
Innocenti, P., Morrow, E. H., and Dowling, D. K. (2011). Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science 332, 845–848. doi: 10.1126/science.1201157
Ioannidis, P., Johnston, K. L., Riley, D. R., Kumar, N., White, J. R., Olarte, K. T., et al. (2013). Extensively duplicated and transcriptionally active recent lateral gene transfer from a bacterial Wolbachia endosymbiont to its host filarial nematode Brugia malayi. BMC Genomics 14:639. doi: 10.1186/1471-2164-14-639
Iturbe-Ormaetxe, I., Burke, G. R., Riegler, M., and O'Neill, S. L. (2005). Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J. Bacteriol. 187, 5136–5145. doi: 10.1128/JB.187.15.5136-5145.2005
Iturbe-Ormaetxe, I., and O'Neill, S. L. (2007). Wolbachia-host interactions: connecting phenotype to genotype. Curr. Opin. Microbiol. 10, 221–224. doi: 10.1016/j.mib.2007.05.002
Jaenike, J., Unckless, R., Cockburn, S. N., Boelio, L. M., and Perlman, S. J. (2010). Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215. doi: 10.1126/science.1188235
Koehncke, A., Telschow, A., Werren, J. H., and Hammerstein, P. (2009). Life and death of an influential passenger: Wolbachia and the evolution of CI-modifiers by their hosts. PLoS ONE 4:e4425. doi: 10.1371/journal.pone.0004425
Koonin, E. V. (2010). The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11:209. doi: 10.1186/gb-2010-11-5-209
Kozek, W. J. (2005). What is new in the Wolbachia-Dirofilaria interaction? Vet. Parasitol. 133, 127–132. doi: 10.1016/j.vetpar.2005.02.005
Kremer, N., Charif, D., Henri, H., Bataille, M., Prévost, G., Kraaijeveld, K., et al. (2009). A new case of Wolbachia dependence in the genus Asobara: evidence for parthenogenesis induction in Asobara japonica. Heredity 103, 248–256. doi: 10.1038/hdy.2009.63
Kriesner, P., Hoffmann, A. A., Lee, S. F., Turelli, M., and Weeks, A. R. (2013). Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 9:e1003607. doi: 10.1371/journal.ppat.1003607
Kutschera, U., and Niklas, K. J. (2005). Endosymbiosis, cell evolution, and speciation. Theory Biosci. 124, 1–24. doi: 10.1016/j.thbio.2005.04.001
Law, R., and Hutson, V. (1992). Intracellular symbionts and the evolution of uniparental cytoplasmic inheritance. Proc. R. Soc. Lond. B Biol. Sci. 248, 69–77. doi: 10.1098/rspb.1992.0044
Li, Z., Garner, A. L., Gloeckner, C., Janda, K. D., and Carlow, C. K. (2011). Targeting the Wolbachia cell division protein FtsZ as a new approach for antifilarial therapy. PLoS Negl. Trop. Dis. 5:e1411. doi: 10.1371/journal.pntd.0001411
Lively, C. M., Clay, K., Wade, M. J., and Fuqua, C. (2005). Competitive co-existence of vertically and horizontally transmitted parasites. Evol. Ecol. Res. 7, 1183–1190.
Lo, N., Paraskevopoulos, C., Bourtzis, K., O'Neill, S. L., Werren, J. H., Bordenstein, S. R., et al. (2007). Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int. J. Syst. Evol. Microbiol. 57, 654–657. doi: 10.1099/ijs.0.64515-0
Lobbia, S., Niitsu, S., and Fujiwara, H. (2003). Female-specific wing degeneration caused by ecdysteroid in the tussock moth, Orgyia recens: hormonal and developmental regulation of sexual dimorphism. J. Insect Sci. 3, 1–7. doi: 10.1673/031.003.1101
Lynch, M., and Blanchard, J. L. (1998). Deleterious mutation accumulation in organelle genomes. Genetica 102, 29–39. doi: 10.1023/A:1017022522486
Ma, W.-J., Pannebakker, B. A., van de Zande, L., Schwander, T., Wertheim, B., and Beukeboom, L. W. (2015). Diploid males support a two-step mechanism of endosymbiont-induced thelytoky in a parasitoid wasp. BMC Evol. Biol. 15:84. doi: 10.1186/s12862-015-0370-9
Masui, S., Kamoda, S., Sasaki, T., and Ishikawa, H. (2000). Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 51, 491–497. doi: 10.1007/s002390010112
Masui, S., Kuroiwa, H., Sasaki, T., Inui, M., Kuroiwa, T., and Ishikawa, H. (2001). Bacteriophage, W. O., and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem. Biophys. Res. Commun. 283, 1099–1104. doi: 10.1006/bbrc.2001.4906
McGraw, E. A., Merritt, D. J., Droller, J. N., and O'Neill, S. L. (2002). Wolbachia density and virulence attenuation after transfer into a novel host. Proc. Natl. Acad. Sci. U.S.A. 99, 2918–2923. doi: 10.1073/pnas.052466499
Merçot, H., and Poinsot, D. (2009). Infection by Wolbachia: from passengers to residents. C. R. Biol. 332, 284–297. doi: 10.1016/j.crvi.2008.09.010
Miller, W. J., Ehrman, L., and Schneider, D. (2010). Infectious speciation revisited: impact of symbiont-depletion on female fitness and mating behavior of Drosophila paulistorum. PLoS Pathog. 6:e1001214. doi: 10.1371/journal.ppat.1001214
Min, K. T., and Benzer, S. (1997). Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. U.S.A. 94, 10792–10796. doi: 10.1073/pnas.94.20.10792
Mondo, S. J., Toomer, K. H., Morton, J. B., Lekberg, Y., and Pawlowska, P. E. (2012). Evolutionary stability in a 400-million-year-old heritable facultative mutualist. Evolution 66, 2564–2576. doi: 10.1111/j.1558-5646.2012.01611.x
Moran, N. A. (1996). Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U.S.A. 93, 2873–2878. doi: 10.1073/pnas.93.7.2873
Moran, N. A. (2003). Tracing the evolution of gene loss in obligate bacterial symbionts. Curr. Opin. Microbiol. 6, 512–518. doi: 10.1016/j.mib.2003.08.001
Moran, N. A., McCutcheon, J. P., and Nakabachi, A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. doi: 10.1146/annurev.genet.41.110306.130119
Moran, N. A., and Plague, G. R. (2004). Genomic changes following host restriction in bacteria. Curr. Opin. Genet. Dev. 14, 627–633. doi: 10.1016/j.gde.2004.09.003
Mounolou, J. C., and Lacroute, F. (2005). Mitochondrial DNA: an advance in eukaryotic cell biology in the 1960s. Biol. Cell 97, 743–748. doi: 10.1042/BC20040128
Mouton, L., Henri, H., Bouletreau, M., and Vavre, F. (2006). Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology 132, 49–56. doi: 10.1017/S0031182005008723
Mouton, L., Henri, H., Charif, D., Boulétreau, M., and Vavre, F. (2007). Interaction between host genotype and environmental conditions affects bacterial density in Wolbachia symbiosis. Biol. Lett. 3, 210–213. doi: 10.1098/rsbl.2006.0590
Narita, S., Kageyama, D., Nomura, M., and Fukatsu, T. (2007). Unexpected mechanism of symbiont-induced reversal of insect sex: feminizing Wolbachia continuously acts on the butterfly Eurema hecabe during larval development. Appl. Environ. Microbiol. 73, 4332–4341. doi: 10.1128/AEM.00145-07
Negri, I. (2012). Wolbachia as an “infectious” extrinsic factor manipulating host signaling pathways. Front. Endocrinol. 2:115. doi: 10.3389/fendo.2011.00115
Nikoh, N., Hosokawa, T., Moriyama, M., Oshima, K., Hattori, M., and Fakatsu, T. (2014). Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. U.S.A. 111, 10257–10262. doi: 10.1073/pnas.1409284111
O'Neill, S. L., Giordano, R., Colbert, A. M. E., Karr, T. L., and Robertson, H. M. (1992). 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. U.S.A. 89, 2699–2702.
O'Neill, S. L., Hoffmann, A. A., and Werren, J. H. (1997). Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. Oxford, U.K: Oxford University Press.
Osborne, S. E., Iturbe-Ormaetxe, I., Brownlie, J. C., O'Neill, S. L., and Johnson, K. N. (2012). Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans. Appl. Environ. Microbiol. 78, 6922–6929. doi: 10.1128/AEM.01727-12
Osborne, S. E., Leong, Y. S., O'Neill, S. L., and Johnson, K. N. (2009). Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 5:e1000656. doi: 10.1371/journal.ppat.1000656
Pannebakker, B. A., Loppin, B., Elemans, C. P. H., Humblot, L., and Vavre, F. (2007). Parasitic inhibition of cell death facilitates symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104, 213–215. doi: 10.1073/pnas.0607845104
Pannebakker, B. A., Pijnacker, L. P., Zwaan, B. J., and Beukeboom, L. W. (2004). Cytology of Wolbachia-induced parthenogenesis in Leptopilina clavipes (Hymenoptera: Figitidae). Genome 47, 299–303. doi: 10.1139/g03-137
Pichaud, N., Ballard, J. W. O., Tanguay, R. M., and Blier, P. U. (2012). Naturally occurring mitochondrial DNA haplotypes exhibit metabolic differences: insight into functional properties of mitochondria. Evolution 66, 3189–3197. doi: 10.1111/j.1558-5646.2012.01683.x
Pinto, S. B., Stainton, K., Harris, S., Kambris, Z., Sutton, E. R., Bonsall, M. B., et al. (2013). Transcriptional regulation of Culex pipiens mosquitoes by Wolbachia influences cytoplasmic incompatibility. PLoS Pathog. 9:e1003647. doi: 10.1371/journal.ppat.1003647
Poinsot, D., Bourtzis, K., Markakis, G., Savakis, C., and Merçot, H. (1998). Wolbachia transfer from Drosophila melanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics 150, 227–237.
Ponton, F., Wilson, K., Holmes, A., Raubenheimer, D., Robinson, K. L., and Simpson, S. J. (2015). Macronutrients mediate the functional relationship between Drosophila and Wolbachia. Proc. Biol. Sci. 282:20142029. doi: 10.1098/rspb.2014.2029
Ramírez-Puebla, S. T., Servín-Garcidueñas, L. E., Ormeño-Orrillo, E., Vera-Ponce de León, A., Rosenblueth, M., Delaye, L., et al. (2015). Species in Wolbachia? Proposal for the designation of ‘Candidatus Wolbachia bourtzisii’, ‘Candidatus Wolbachia onchocercicola', ‘Candidatus Wolbachia blaxteri’, ‘Candidatus Wolbachia brugii’, ‘Candidatus Wolbachia taylori’, ‘Candidatus Wolbachia collembolicola’ and ‘Candidatus Wolbachia multihospitum’ for the different species within Wolbachia supergroups. Syst. Appl. Microbiol. 38, 390–399. doi: 10.1016/j.syapm.2015.05.005
Rand, D. M., Haney, R. A., and Fry, A. J. (2004). Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19, 645–653. doi: 10.1016/j.tree.2004.10.003
Rasgon, J. L., Gamston, C. E., and Ren, X. (2006). Survival of Wolbachia pipientis in cell-free medium. Appl. Environ. Microbiol. 72, 6934–6937. doi: 10.1128/AEM.01673-06
Raychoudhury, R., Baldo, L., Oliveira, D. C., and Werren, J. H. (2009). Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 63, 165–183. doi: 10.1111/j.1558-5646.2008.00533.x
Richardson, M. F., Weinert, L. A., Welch, J. J., Linheiro, R. S., Magwire, M. M., and Jiggins, F. M. (2012). Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet. 8:e1003129. doi: 10.1371/journal.pgen.1003129
Rigaud, T., and Juchault, P. (1995). Success and failure of horizontal transfers of feminizing Wolbachia endosymbionts in woodlice. J. Evol. Biol. 8, 249–255. doi: 10.1046/j.1420-9101.1995.8020249.x
Rio, R. V. M., Wu, Y. N., Filardo, G., and Aksov, S. (2006). Dynamics of multiple symbiont density regulation during host development: Tsetse fly and its microbial flora. Proc. R. Soc. Lond. B Biol. Sci. 273, 805–814. doi: 10.1098/rspb.2005.3399
Riparbelli, M. G., Girodano, R., Ueyama, M., and Callaini, G. (2012). Wolbachia-mediated male killing is associated with defective chromatin remodeling. PLoS ONE 7:e30045. doi: 10.1371/journal.pone.0030045
Russell, J. E., and Stouthamer, R. (2010). The genetics and evolution of obligate reproductive parasitism in Trichogramma pretiosum infected with parthenogenesis-inducing Wolbachia. Heredity 106, 58–67. doi: 10.1038/hdy.2010.48
Sachs, J. L., Essenberg, C. J., and Turcotte, M. M. (2011). New paradigms for the evolution of beneficial infections. Trends Ecol. Evol. 26, 202–209. doi: 10.1016/j.tree.2011.01.010
Sagan, L. (1967). On the origin of mitosing cells. J. Theor. Biol. 14, 255–274. doi: 10.1016/0022-5193(67)90079-3
Serbus, L. R., Casper-Lindley, C., Landmann, F., and Sullivan, W. (2008). The genetics and cell biology of Wolbachia-host interactions. Annu. Rev. Genet. 42, 683–707. doi: 10.1146/annurev.genet.41.110306.130354
Serbus, L. R., White, P. M., Silva, J. P., Rabe, A., Teixeira, L., Albertson, R., et al. (2015). The impact of host diet on Wolbachia titer in Drosophila. PLoS Pathog. 11:e1004777. doi: 10.1371/journal.ppat.1004777
Sicard, M., Dittmer, J., Grève, P., Bouchon, D., and Braquart−Varnier, C. (2014). A host as an ecosystem: Wolbachia coping with environmental constraints. Environ. Microbiol. 16, 3583–3607. doi: 10.1111/1462-2920.12573
Siozios, S., Sapountzis, P., Ioannidis, P., and Bourtzis, K. (2008). Wolbachia symbiosis and insect immune response. Insect Sci. 15, 89–100. doi: 10.1111/j.1744-7917.2008.00189.x
Snook, R. R., Cleland, S. Y., Wolfner, M. F., and Karr, T. L. (2000). Offsetting effects of Wolbachia infection and heat shock on sperm production in Drosophila simulans: analyses of fecundity, fertility and accessory gland proteins. Genetics 155, 167–178.
Stearns, S. C. (1989). Trade-offs in life-history evolution. Funct. Ecol. 3, 259–268. doi: 10.2307/2389364
Stouthamer, R., Breeuwer, J. A. J., and Hurst, G. D. D. (1999). Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102. doi: 10.1146/annurev.micro.53.1.71
Stouthamer, R., Russell, J. E., Vavre, F., and Nunney, L. (2010). Intragenomic conflict in populations infected by parthenogenesis inducing Wolbachia ends with irreversible loss of sexual reproduction. BMC Evol. Biol. 10:229. doi: 10.1186/1471-2148-10-229
Tamas, I., Klasson, L., Canbäck, B., Näslund, A. K., Eriksson, A.-S., Wernegreen, J. J., et al. (2002). 50 million years of genomic stasis in endosymbiotic bacteria. Science 296, 2376–2379. doi: 10.1126/science.1071278
Taylor, M. J., Bandi, C., and Hoerauf, A. (2005). Wolbachia bacterial endosymbionts of filarial nematodes. Adv. Parasitol. 60, 245–284. doi: 10.1016/S0065-308X(05)60004-8
Teixeira, L., Ferreira, Á., and Ashburner, M. (2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6:e1000002. doi: 10.1371/journal.pbio.1000002
Thrash, J. C., Boyd, A., Huggett, M. J., Grote, J., Carini, P., Yoder, R. J., et al. (2011). Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 1:13. doi: 10.1038/srep00013
Toft, C., and Andersson, S. G. E. (2010). Evolutionary microbial genomics: insights into bacterial host adaptation. Nat. Rev. Genet. 11, 465–475. doi: 10.1038/nrg2798
Toomey, M. E., Panaram, K., Fast, E. M., Beatty, C., and Frydman, H. M. (2013). Evolutionarily conserved Wolbachia-encoded factors control pattern of stem-cell niche tropism in Drosophila ovaries and favor infection. Proc. Natl. Acad. Sci. U.S.A. 110, 10788–10793. doi: 10.1073/pnas.1301524110
Tortosa, P., Charlat, S., Labbé, P., Dehecq, J.-S., Barré, H., and Weill, M. (2010). Wolbachia age-sex-specific density in Aedes albopictus: a host evolutionary response to cytoplasmic incompatibility? PLoS ONE 5:e9700. doi: 10.1371/journal.pone.0009700
Tram, U., and Sullivan, W. (2002). Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 296, 1124–1126. doi: 10.1126/science.1070536
Turelli, M. (1994). Evolution of incompatibility-inducing microbes and their hosts. Evolution 48, 1500–1513. doi: 10.2307/2410244
Turelli, M., and Hoffmann, A. A. (1995). Cytoplasmic incompatibility in Drosophila simulans: Dynamics and parameter estimates from natural populations. Genetics 140, 1319–1338.
Turelli, M., Hoffmann, A. A., and McKechnie, S. W. (1992). Dynamics of cytoplasmic incompatibility and mtDNA variation in natural Drosophila simulans populations. Genetics 132, 713–723.
Unckless, R. L., Boelio, L. M., Herren, J. K., and Jaenike, J. (2009). Wolbachia as populations within individual insects: causes and consequences of density variation in natural populations. Proc. R. Soc. Lond. B Biol. Sci. 276, 2805–2811. doi: 10.1098/rspb.2009.0287
Vavre, F., Fleury, F., Lepetit, D., Fouillet, P., and Boulétreau, M. (1999). Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 16, 1711–1723. doi: 10.1093/oxfordjournals.molbev.a026084
Veneti, Z., Clark, M. E., Karr, T. L., Savakis, C., and Bourtzis, K. (2004). Heads or tails: host-parasite interactions in the Drosophila-Wolbachia system. Appl. Environ. Microbiol. 70, 5366–5372. doi: 10.1128/AEM.70.9.5366-5372.2004
Weeks, A. R., Turelli, M., Harcombe, W. R., Reynolds, K. T., and Hoffmann, A. A. (2007). From parasite to mutualist: Rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 5:e114. doi: 10.1371/journal.pbio.0050114
Weinert, L. A., Werren, J. H., Aebi, A., Stone, G. N., and Jiggins, F. M. (2009). Evolution and diversity of Rickettsia bacteria. BMC Biol. 7, 6–19. doi: 10.1186/1741-7007-7-6
Wernegreen, J. J. (2004). Endosymbiosis: lessons in conflict resolution. PLoS Biol. 2:e68. doi: 10.1371/journal.pbio.0020068
Werren, J. H. (1997). Biology of Wolbachia. Annu. Rev. Entomol. 42, 587–609. doi: 10.1146/annurev.ento.42.1.587
Werren, J. H. (2011). Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. U.S.A. 108, 10863–10870. doi: 10.1073/pnas.1102343108
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Werren, J. H., and Windsor, D. M. (2000). Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. Lond. B Biol. Sci. 267, 1277–1285. doi: 10.1098/rspb.2000.1139
Werren, J. H., Zhang, W., and Guo, L. R. (1995). Evolution and phylogeny of Wolbachia: Reproductive parasites of arthropods. Proc. R. Soc. Lond. B Biol. Sci. 261, 55–63. doi: 10.1098/rspb.1995.0117
Wiwatanaratanabutr, I., and Kittayapong, P. (2009). Effects of crowding and temperature on Wolbachia infection density among life cycle stages of Aedes albopictus. J. Invertebr. Pathol. 102, 220–224. doi: 10.1016/j.jip.2009.08.009
Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., et al. (2004). Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:e69. doi: 10.1371/journal.pbio.0020069
Yee, W. K. W., Sutton, K. L., and Dowling, D. K. (2013). In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr. Biol. 23, R55–R56. doi: 10.1016/j.cub.2012.12.002
Zera, A. J., and Harshman, L. G. (2001). The physiology of life history trade-offs in animals. Annu. Rev. Ecol. Syst. 32, 95–126. doi: 10.1146/annurev.ecolsys.32.081501.114006
Keywords: Wolbachia, conflict of interest, cytoplasmic genomes, mutualism, parasitism
Citation: Correa CC and Ballard JWO (2016) Wolbachia Associations with Insects: Winning or Losing Against a Master Manipulator. Front. Ecol. Evol. 3:153. doi: 10.3389/fevo.2015.00153
Received: 29 June 2015; Accepted: 18 December 2015;
Published: 19 January 2016.
Edited by:
Sampath S. Kumar, Bangalore University, IndiaReviewed by:
Kirk Matthew Schnorr, Novozymes, DenmarkViktoria Shcherbakova, Institute of Biochemistry and Physiology of Microorganisms Russian Academy of Sciences, Russia
Ilaria Negri, Koiné - Consulenze Ambientali S.n.c., Italy
Copyright © 2016 Correa and Ballard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. W. O. Ballard, dy5iYWxsYXJkQHVuc3cuZWR1LmF1
 Claudia C. Correa
Claudia C. Correa J. W. O. Ballard
J. W. O. Ballard