- Institute of Loess Plateau, Shanxi University, Taiyuan, China
Ecological restoration of mining areas has mainly focused on the succession dynamics of vegetation and the fate of microbial communities remains poorly understood. We examined changes in soil characteristics and plant and microbial communities with increasing reclamation period in an open coal mine. Bacterial, archaeal and fungal communities were assessed by tag-encoded 454 pyrosequencing. At the phylum level, Proteobacteria, Crenarchaeota, and Ascomycota had the highest detected relative abundance within bacteria, archaea, and fungi, respectively. Partial regressions and canonical correspondence analysis demonstrated that vegetation played a major role in bacterial and archaeal diversity and assemblies, and soil characteristics, especially nitrogen, were important for fungal diversity and assemblies. Spearman rank correlation indicated that bacterial and archaeal communities showed synergistic succession with plants; whereas, fungal communities showed no such pattern. Overall, our data suggest that there are different drivers of bacterial, archaeal and fungal succession during secondary succession in a reclaimed open mine.
Introduction
Microorganisms are key drivers of global biogeochemical cycles and are critical for the development, abundance, and diversity of aboveground plant communities (Falkowski et al., 2008; Herzberger et al., 2015). Studies of microbial succession are increasingly important, and have been explored in a variety of environments and over different timescales (Williams et al., 2013; Dini-Andreote et al., 2014; Zhao et al., 2014). Previous studies have shown that soil microbial community succession is a long and non-monotonic process (López-Lozano et al., 2013; Mastrogianni et al., 2014). Lozano et al. (2014) showed that microbial community succession is closely correlated with multiple ecosystem functions. However, the links between below-ground and above-ground succession processes are poorly understood.
Soil factors and vegetation are important determinants of microbial community composition and could determine the trajectory of ecosystem development (Yarwood et al., 2015). Dimitriu et al. (2010) showed that reclamation-mediated effects on microbial properties are mainly attributable to changes in abiotic properties such as soil pH; however, the composition of soil bacterial and fungal communities in forest soil is largely determined by dominant trees (Urbanová et al., 2015). Both parent material (sandstone and siltstone) and nutrient addition (including nitrogen and phosphorus fertilization) affected the rate of succession because of the differences in resource availability (Knelman et al., 2014; Yarwood et al., 2015). In severely degraded soil, microbial community structure and generation were mediated by soil aggregation via the protection afforded by soil organic carbon (Zhang et al., 2010). Microbial succession in the volcanic deter of Mount Fuji was initially strongly affected by the pioneer herbaceous plants and subsequent invasion of shrubs (Yoshitake et al., 2013).
Bacteria and fungi demonstrate different succession patterns (Schmidt et al., 2014). Banning et al. (2011) showed that the bacterial community successional trend became more similar to those of the surrounding non-mined forest with increased rehabilitation age, but fungi did not show a similar successional trajectory. Yannarell et al. (2014) found that both bacterial and fungal communities demonstrated significant variation along transects spanning the prairie–shrub–forest continuum, but their predominant patterns were different. Cutler et al. (2014) showed that the composition of plant communities was significant for fungal communities, but less relevant for bacterial communities during primary succession on an 850-year chronosequence of lava flows. In retreating glaciers, the presence of plants was important in bacterial successional dynamics, but played a minor role in those of fungi (Brown and Jumpponen, 2014). Soil substrates were the major drivers of archaeal community succession across a receding glacier foreland (Nicol et al., 2005; Zumsteg et al., 2012). In addition, the major soil factors that were significantly correlated with bacterial, archaeal, and fungal communities were different (Zumsteg et al., 2012; Li et al., 2013a).
High-throughput sequencing technologies have expanded our understanding of microbial community assembly across environmental gradients and during succession, and ecological hypotheses have been tested and developed using microbes as indicators (Nemergut et al., 2013). Dini-Andreote et al. (2015) found that soil bacterial community composition was initially subject to stochastic effects, but there was a progressive increase in deterministic selection as succession proceeded in a salt marsh chronosequence succession. Schmidt et al. (2014) found a strong deterministic community assembly pattern for bacteria but not for fungi during primary succession in the foreland of a receding glacier. Temporal niche partitioning was the dominant mechanism of bacterial community assembly in the initial succession (Dini-Andreote et al., 2014).
The Antaibao mining area, with a documented management history of over 30 years, is located in the east of the Loess Plateau, China. The existence of gradients in vegetation succession patterns and soil chemical characteristics has been clearly shown in these rehabilitation sites (Zhao et al., 2012, 2013). Our objective in the current study was to examine how soil microbial communities develop and assemble with reclamation processes in an open coal mine. A pyrosequencing-based approach was used to determine bacterial, archaeal, and fungal phylogenetic composition in reclaimed soils following reclamation of a period between 2 and 30 years.
Materials and Methods
Study Site Description
Our research was conducted at the Antaibao opencast coal mining area in Plateau Loess (39°23′−39°23′ N; 112°11′−112°30′ E), China. The climate is terrestrial temperate, and the area experiences monsoons. The altitude is 1300–1400 m. Annual average precipitation is about 450 mm, and annual average air temperature is about 6.2°C. The frost-free season ranges in length from 115 to 130 days.
Study Plots Survey and Soil Sampling
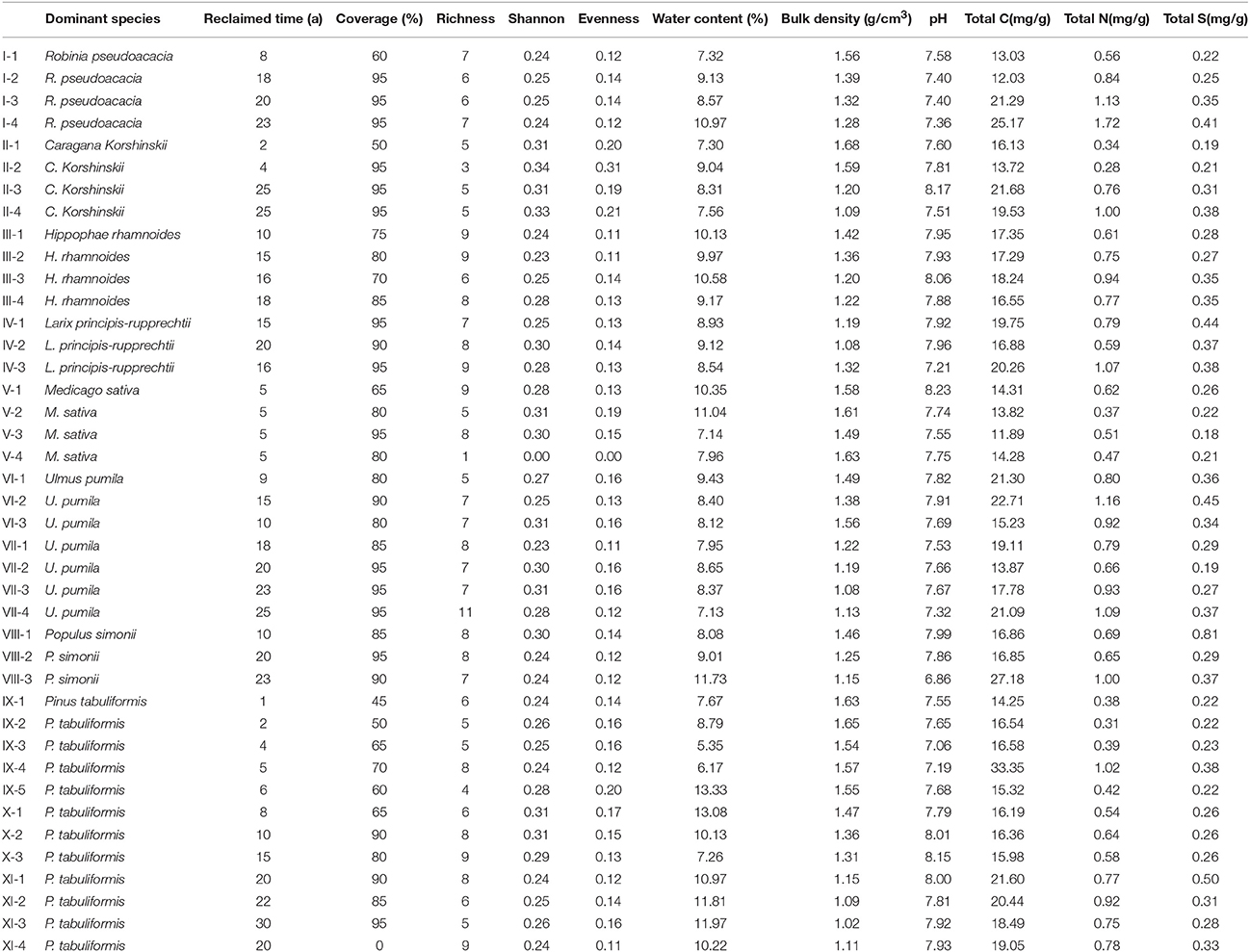
To study the characteristics of the various plant communities, quadrats of dimensions 20 × 20 m, 4 × 4 m, and 1 × 1 m were established in forest, scrubland, and grassland communities. We recorded the cover, height, diameter at breast height, and the individual number for each tree species and the cover and height for shrubs and herbs at 41 rehabilitation sites (Figure S1). The sites were classified into 11 groups (Groups I-XI) using TWINSPAN analysis (Zhang, 2005).
The upper 10 cm of soil was collected from six random locations at each site and mixed into a single bulk sample in July, 2013. Soil samples were sieved to 2 mm and homogenized. Subsamples for microbial analysis were stored at −80°C, and the remainder was air dried for chemical analyses.
Soil Chemical Analysis
Soil bulk density (BD) was obtained using the gravimetric method. Soil pH in distilled water was measured using a glass combination electrode at a soil: solution ratio of 1:5 after 1 h. Soil total carbon (C), nitrogen (N) and sulfur (S) were analyzed using a Vario MACRO cube (Elemental Analyzer, Germany). Soil and plant characteristics are given in Table 1.
RNA Genetic Pyrosequencing
DNA was extracted from 0.5 g soil using the Ultra-clean TM soil DNA Isolation Kits (MoBio Laboratory, USA) according the manufacturer's protocol. Soil microbial rRNA gene were amplified using primer sets containing the Roche 454 pyrosequencing adaptors (underlined), which followed bacterial primer set (5′-CGTATCGCCTCCCTCGCGCCATCAG-barcode-AGA GTTTGATCMTGGCTCAG-3′ and 5′-CTATGCGCCTTGCCAGCCCGCTCAG-GTATTACCGCGGCTGCTGGC AC-3′) (Justé et al., 2008), archaeal set (5′-TTTTCTATGCGCCTTGCCAGCCCGCT-CAGCAGCMGCCGCGGTA A-3′ and 5′-CGTATCGCCTCCCTCGCGCCATCAG-barcode-GGCCATGCACCWCCT CTC-3′) (Kolganova et al., 2002), and fungal set (5′-CGTATCGCCTCCCTCGCGCCATCAG-barcode-CA GTAGTCATATGCTTGTCTC-3′ and 5′-CTATGCGCCTTGCCAGCCCGCTCAG-GCTGCTGGCACCAGACTT GC-3′) (Costa et al., 2006). There were 7-, 10-, and 10-base barcodes for primers of bacteria, archaea, and fungi, respectively. To amplify the rRNA genes, 50 μl of PCR reactions were carried out according to the following reaction mixture: 5 μl 10 × PCR buffer (MgCl2, 2 mM), 0.5 μl dNTPs (10 mM), 0.5 μl Plantium Taq (5 U/μl), 1 μl BSA (1 μl/μg), 10 ng genomic DNA, and 1 μl of each primer (50 mM).
Bacterial 16S rRNA gene PCR reactions were performed in the following program: 3 min at 94°C; 30 cycles of 45 s at 94°C, 45 s at 54°C and 60 s at 72°C, followed by a final extension period of 5 min at 72°C. Archaea followed the program: 94°C: 3 min; 5 cycles of 20 s at 94°C, 20 s at 45°C, 60 s at 72°C; 25 cycles of 45 s at 94°C, 45 s at 58°C, 60 s at 72°C; 5 min at 72°C. The 18S rRNA gene PCR program for fungi was: 94°C: 5 min; 30 cycles of 30 s at 94°C, 30 s at 56°C, 60 s at 72°C; 5 min at 72°C. The PCR products were separated by 1% agarose gel electrophoresis and purified with Wizard® SV Gel and PCR Clean-Up System (Promega, USA). High-throughput sequencing was determined using a Roche GS-FLX 454 pyrosequencer (Roche, Basel, Switzerland) at Sangon Biotech (Shanghai) Co., Ltd.
All raw sequence processing was conducted using QIIME pipeline (Caporaso et al., 2010). Sequences were trimmed if they were shorter than 300 bp and quality scores lower than 25. Sequences were clustered into operational taxonomic units (OTUs) using UCLUST, with a 97% identity threshold (Edgar, 2010). The sequences comprising each OTU were aligned using the Ribosomal Database Project (RDP, http://rdp.cme.msu.edu).
Data Analysis
Plant diversity indices for each quadrat were calculated following Li et al. (2013a). Microbial diversity including Shannon, ACE and Chao1 indices were determined using the Mothur software (Schloss et al., 2009) (http://www.mothur.org). A one-way analysis of variance and Duncan test were used to examine the significant effects on microbial diversity indices (Benes and Carpenter, 2015). Spearman rank correlation was used to examine the association between plant species pairs and microbial genus pairs (Li et al., 2013a). Pearson linear correlations were calculated to determine whether there were significant correlations between the environmental factors and microbial properties.
We conducted partial regressions to correlate the vegetative effects (including reclamation period and plant coverage, richness, and evenness) with soil characteristics. We established a pure model only containing plant or soil variables, and a full model containing both plant and soil variables. The total variation of microbial diversity was partitioned into independent, co-varying, and unexplained components (Wang et al., 2011).
A matrix of important values for plants and matrices of genus ratios for bacteria, archaea, fungi and total microbe were used as the basis for community analysis. The total microbial matrix was established using the following formula: Matrix (total microbe) = (4 × ratios(Bacteria-genus) + 3 × ratios(Archaea-genus) + 3 × ratios(Fungi-genus))/10 (Li et al., 2013a). Environmental and biotic matrixes were subjected to canonical correspondence analysis (CCA) to examine the significant environmental factors on biotic compositions. We used the Monte Carlo permutation to test the significant level between species and environmental data in CANOCO software (version 4.5) (Ter Braak and Smilauer, 2002).
Results
Microbial Diversity
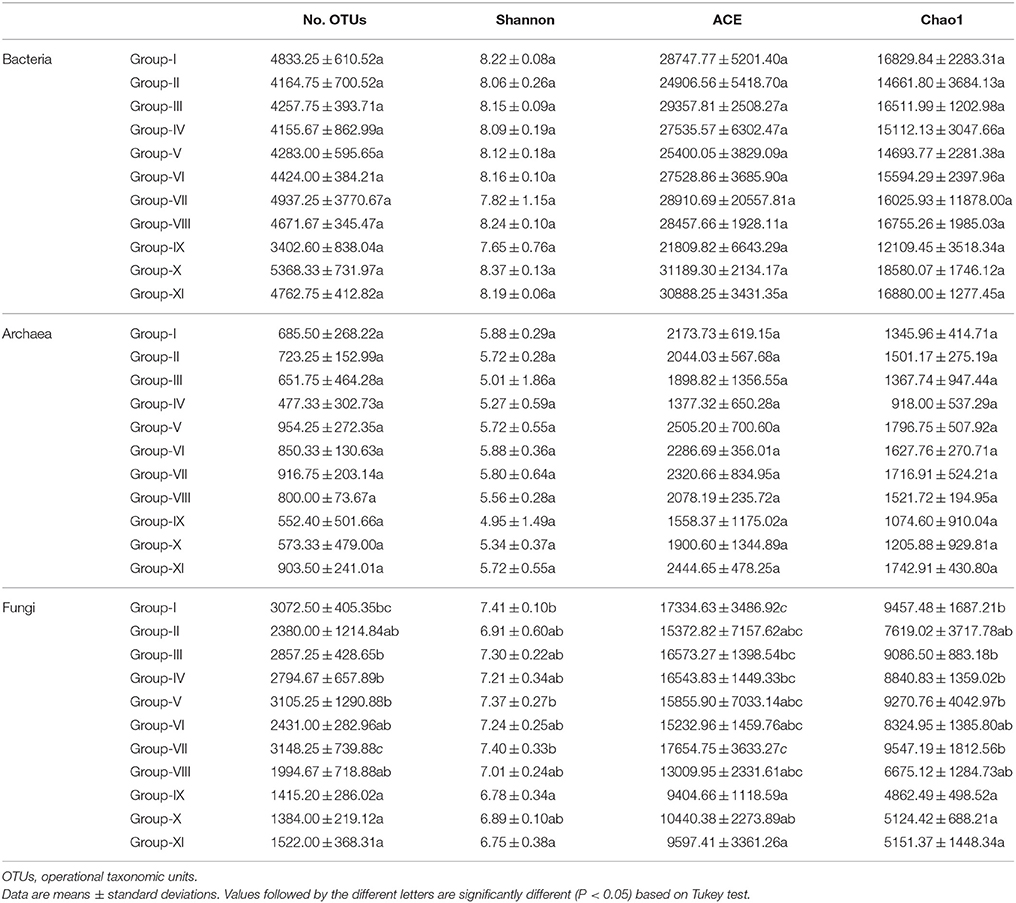
In the 41 study sites, there were 181,827, 30,205, and 97,230 high-quality sequences for bacteria, archaea and fungi, respectively (Figure S2). There were significant differences between the 11 groups in fugal diversity indices, but not in bacterial and archaeal indices (Table 2). Significantly low fungal diversity indices (P < 0.05) were found in Pinus tabuliformis plantations (Groups IX, X, and XI), and the significantly high fungal diversity indices (P < 0.05) were found in Robinia pseudoacacia (Group I) and Ulmus pumila (Group-VII) with the longer reclamation period.
Microbial Taxonomic Distribution and Affiliation
Bacterial rarefied sequences were affiliated to 33 phyla, and 12 phyla presenting sequences abundances were above 1% (Figure 1A). The highest relative abundant sequences were Proteobacteria at the study sites, except for group VIII, in which Acidobacteria was the highest. Other bacterial phyla with average abundance < 0.1% were not universal for each plot (not listed). At class level, the highest relative abundances were affiliated to Actinobacteria in most study sites. Alphaproteobacteria dominated among Proteobacteria sequences.
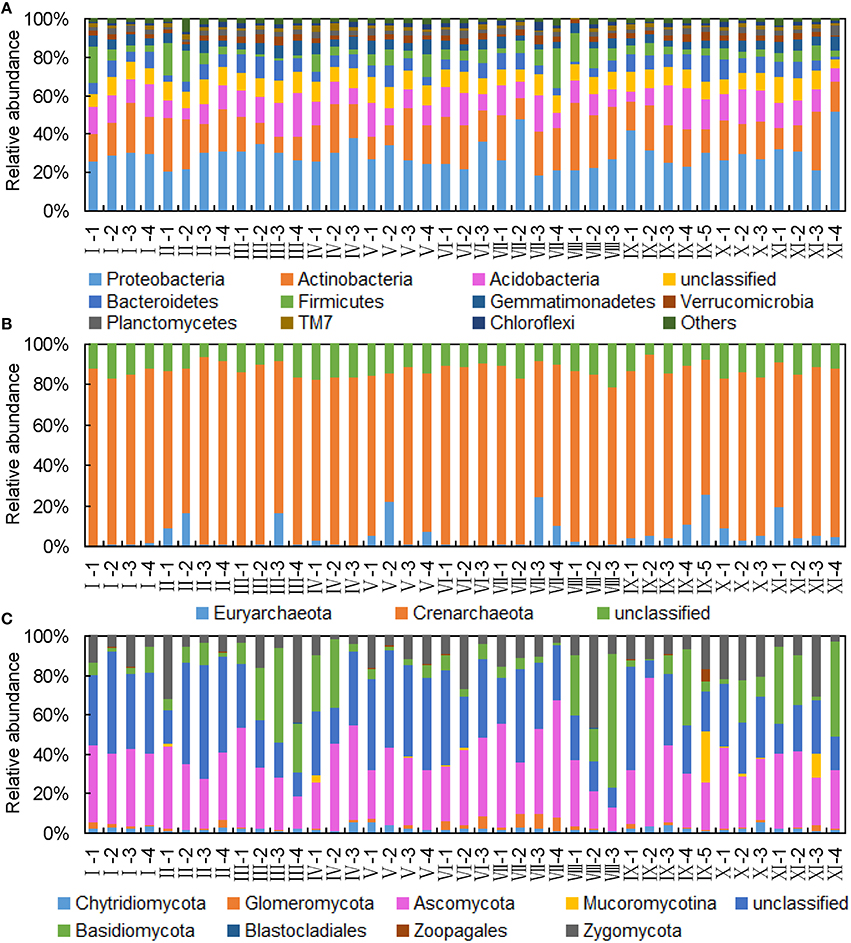
Figure 1. Relative abundance of different microbial phyla in the reclaimed mining area. (A) Bacterial phyla accounting for >1%. (B) Archaeal phyla. (C) Fungal phyla.
Classified archaeal sequences were Crenarchaeota and Euryarchaeota, and Crenarchaeota accounted for the majority (Figure 1B). Euryarchaeota sequences were classified as Thermoplasmata, Methanobacteria, Methanomicrobia, Archaeoglobi, and Methanopyri, and only Thermoplasmata occurred across all samples. At genus level, sequences showed that Fervidicoccus, with the highest relative abundance, was common to every sample.
The relative sequences of unclassified fungal sequences varied among study sites (8.85–57.55%) (Figure 1C). The dominant fungal phyla were Ascomycota, Basidiomycota, and Zygomycota. At class level, Agaricomycetes had the highest ratios of the classified sequences. Fungal classified sequences were affiliated to 28 classes, 88 orders, 177 families, and 463 genera.
Spearman Rank Correlation Test
The results of the Spearman rank correlation test of plants and soil microbes are given in Table 3. The lowest and highest plant species-pair ratios of positive to negative association were observed in the shortest and longest reclaimed-time P. tabuliformis plantations (i.e., Group IX and XI), respectively. Both plant and bacteria ratios were significantly correlated with reclaimed time (P < 0.05), and the Pearson correlation coefficients were 0.700 and 0.630, respectively. There was a positive relationship between plant and bacteria ratios (r = 0.641, P < 0.05). The genus-pair ratios of positive and negative association for archaea were higher than those for bacteria and fungi, except in Groups II and III. Fungal genus-pair ratios retained a relatively low level in most samples.
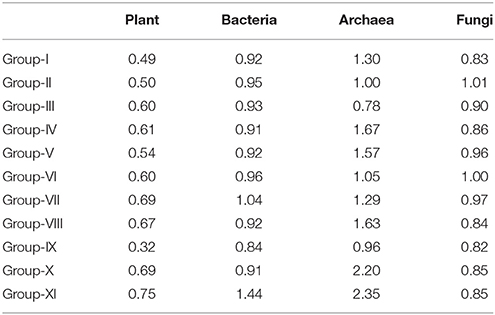
Table 3. The species/genus-pair ratios of positive to negative association for plant and microbe in the reclaimed mining area.
Relationship between Microbial Community and Environmental Factors
Partial regressions indicated that vegetation and soil independently accounted for 12.3–28.0% and 2.7–5.2% of variation in bacterial diversity indices, respectively, and their joint effects accounted for 7.4–41.2%. The independent effects of vegetation and soil on archaeal diversity variation increased; however, their joint effects decreased. For fungal diversity indices, soil independently explained more (17.7–26.2%) than vegetation (6.3–7.9%). The highest residual variations were associated with archaeal diversity indices, and the lowest were associated with fungal indices (Figure 2).
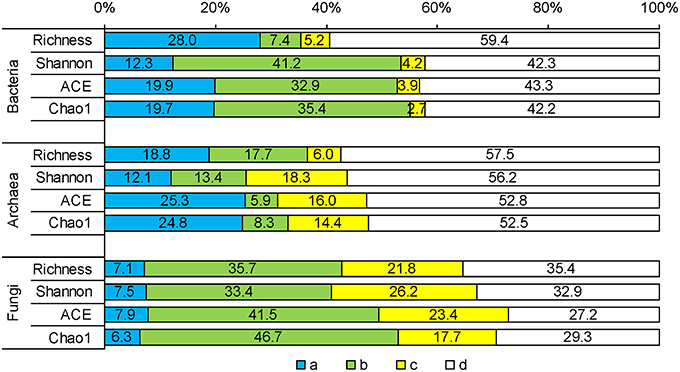
Figure 2. Comparisons between effects of vegetation and the effects of soil properties on microbial diversity indices by partial regressions. The variation of microbial diversity indices is partitioned into (a) the independent components of plant and (c) soil properties, (b) the covarying component and (d) residual variation.
The first two axes accounted for 54.9, 63.3, 42.6, and 45.2% of correlations between environmental variables and bacterial, archaeal, fungal and total microbial community compositions, respectively (Figure 3). The year after reclamation significantly influenced bacterial and archaeal compositions (Figures 3A,B). Fungal compositions were significantly affected by soil N (P = 0.002), soil C (P = 0.024), and water content (P = 0.014) (Figure 3C). Bulk density was strongly related to total microbial compositions (Figure 3D).
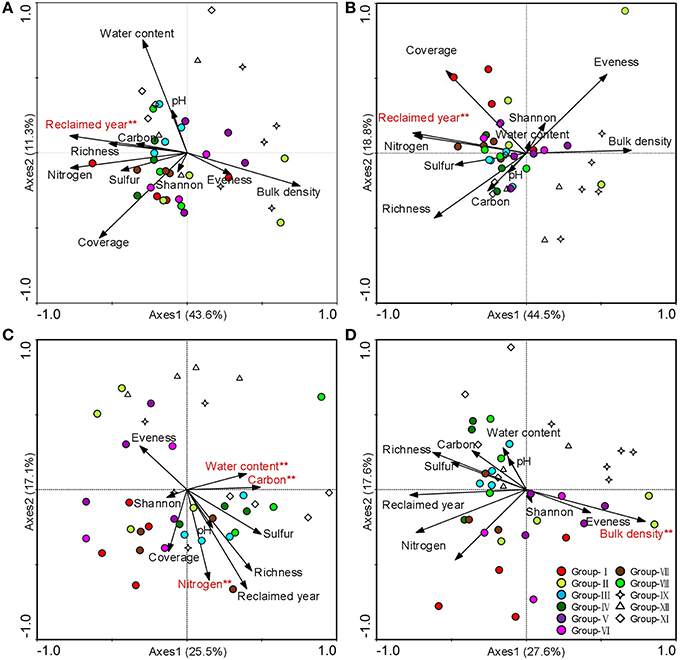
Figure 3. CCA ordination biplot of 41 quadrats and environmental factors for microbial communities in the reclaimed mining area. (A) Bacteria, (B) Archaea, (C) Fungi, and (D) Total microbe. ** Significant environmental factor at the 0.01 level (Monte Carlo permutation test).
At the bacterial phylum level, the relative abundances of Proteobacteria, Acidobacteria, and Planctomycetes positively correlated with reclamation period, water content, and total N, and negatively correlated with water content. However, the opposite correlations were demonstrated between the relative abundance of Firmicutes and these environmental factors (Table 4). Crenarchaeota positively correlated with total N. For fungal phyla, the significant correlations were largely associated with soil characteristics, except for Blastocladiales, which was negatively correlated with plant richness.
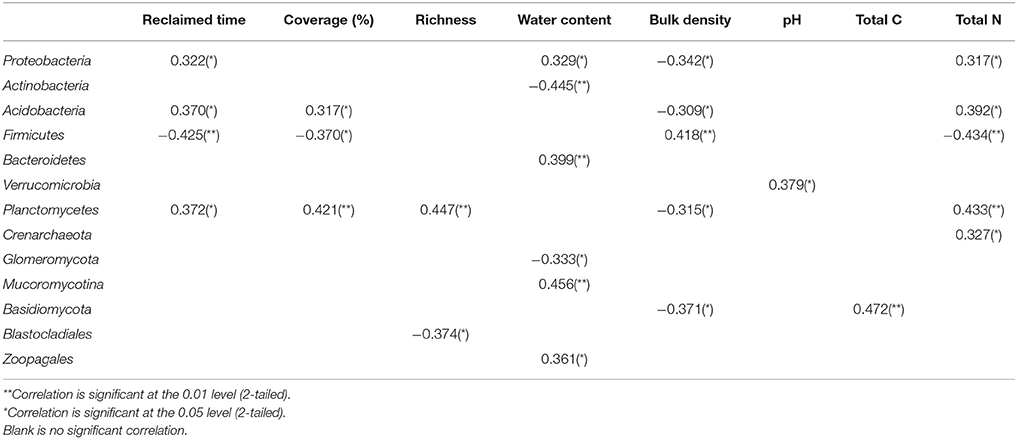
Table 4. The Pearson correlation coefficients for the linear regressions between environmental factors and microbial phylum-level relative abundance.
Discussion
Soil C (r = 0.375, P < 0.05) and N (r = 0.652, P < 0.01) were significantly correlated with reclamation period, which supports previous studies on an increase in organic matter along the successional stage (Zhao et al., 2013; Lozano et al., 2014). pH gradually approximated to neutral with soil carbon improvement, and was negatively correlated with soil C (r = −0.328, P < 0.05). Litter inputs, and root and microbial exudates reduced pH, and the optimal pH was beneficial for aboveground and belowground biomass accumulations (Putten et al., 2013; Lozano et al., 2014). This indicates that plant and soil mutually promote the rehabilitation process.
We assessed the succession of soil microbial communities after mining reclamation through 454- pyrosequencing. Fungal diversity indices significantly changed in response to reclamation vegetation and time, but bacterial and archaeal indices were only slightly influenced by reclamation characteristics (Table 2). A potential explanation for this pattern is that bacteria and archaea are less likely to be dispersal limited than fungi (Wilkinson et al., 2012; Brown and Jumpponen, 2014). Another possible reason for the three domains of microbes demonstrating different responses is that bacteria and archaea have a broader range of physiologies than fungi (Nemergut et al., 2013). The strong correlations between soil N content with fungal ACE (r = 0.480, P < 0.01) and Chao1 (r = 0.425, P < 0.01) indices support the suggestion that fungi are more dependent than bacteria or archaea on C and N sources prior to any significant organic matter build-up as a result of succession (Schmidt et al., 2014). Li et al. (2013b) found that the key competition for nutrients between vegetation and microbes took place during the initial rehabilitation period, which supports our suggestion that soil nutrients represent a limitation for microbial communities in reclamation succession.
Spearman rank pairwise correlations (Table 3), and partial regressions (Figure 2) and CCA analysis (Figure 3) demonstrated that reclamation vegetation played major roles in bacterial and archaeal diversity, compositions and succession, and soil properties produced strong effects on those of fungi. In retreating glacier soils, the presence of plants was important for structuring bacterial communities and played a minor role in fungal communities (Brown and Jumpponen, 2014). In forest ecosystems, the effect of trees on both bacterial and fungal communities was stronger than that of soil properties and explained a large proportion of the variation in community composition (Urbanová et al., 2015). Furthermore, a strong influence of wood type was also found on fungal community and composition during decay in a forest soil (Prewitt et al., 2014). The differences between studies suggest that the roles of soil properties on fungal communities might largely depend on the soil nutrient level. Soil nutrients would be limiting factors for fungal communities in poor reclaimed and glacier soil; whereas, changes to soil factors would produce little effect on fungi in fertile forest soils.
Archaea are known to be able to live under extreme conditions; therefore, it is easily understood that archaeal diversity changed little with reclamation period (Table 2). Li et al. (2013a) also found that in the Loess Plateau of China, archaeal abundance and diversity did not change with restoration of abandoned land. However, the decline or increase in diversity took place along a successional gradient in a receding glacier foreland (Nicol et al., 2005; Zumsteg et al., 2012). In the current study, archaea at high taxonomic ranks, such as the phylum or class level, displayed ecological coherence (Figure 1B), but archaea compositions at family or genus level were visibly different. According to archaeal genus composition, we found that archaeal compositions significantly evolved with reclamation period (Figure 3B), and the genus level pairwise association of archaea was positively correlated with plant species pairwise association (Table 3). Crenarchaeota are often found in plant rhizospheres (Timonen and Bomberg, 2009), so Crenarchaeota-dominated archaeal communities were more susceptible to reclamation of vegetation than soil properties (Figures 1B, 2). At the landscape scale, the variation in archaea was also more strongly related to the changes in vegetation type, rather than soil properties (Nielsen et al., 2010). Crenarchaeota are influenced by soil organic matter (Zinger et al., 2011), and Crenarchaeota increased with the enhancement of soil total N (Table 4). This may partly originate from the shift from a Euryarchaeota-dominated to a Crenarchaeota-dominated archaeal community with chronosequence succession in a receding glacier (Zumsteg et al., 2012).
Firmicutes, including many endospore formers including Bacillus and Clostrium, are advantageous traits under poor nutrient and dry conditions. Therefore, it would be better to understand that Frimicutes decreased with increased reclamation period and enhancement of soil N (Table 4). Shrestha et al. (2007) reported that Betaproteobacteria and Gammaproteobacteria were typical at the early stage, while Alphaproteobacteria and Actinobacteria prevailed in late succession. We also found that Proteobacteria was significantly correlated with reclamation period (Table 4); however, the classes of Proteobacteria displayed no distinct succession pattern. Negative and positive correlations between soil content and relative abundance of Actinobacteria and Bacteroidetes were found in this study, respectively, which supports results from the Ödenwinkelkees glacier foreland across the chronosequence (Philippot et al., 2011). This phenomenon might be because Actinobacteria and Bacteroidetes prefer arid and wet environments, respectively. Kuramae et al. (2011) found that under low nutrient conditions, the combination of soil available phosphorus and ammonia nitrogen produced a positive effect on a number of OTUs of Planctomycetes. In the current study, we found that the relative abundance of Planctomycetes was positively correlated with reclamation period, plant cover and richness, and soil N.
Plant species pairwise ratios significantly correlated with bacterial genus pairwise ratios, which suggested that plant and bacterial communities displayed a similar pattern along the reclaimed succession; however, fungal succession showed a dissimilar pattern. In a dry environment, microbial succession may lag behind plant succession, but plant and microbes demonstrate a similar succession pattern (Lozano et al., 2014). Previous studies reported that bacteria and fungi evolved along different trajectories under various conditions, such as secondary succession in a restored ecosystem (Banning et al., 2011; Li et al., 2013a), shrub encroachment (Yannarell et al., 2014), and the primary succession of lava flows (Cutler et al., 2014) and glacier soils (Brown and Jumpponen, 2014). The different succession patterns were determined by the range of microbial physiologies, dispersal ability, and assembly pattern (Schmidt et al., 2014).
Conclusions
Our study demonstrates that bacterial, archaeal and fungal communities are dynamic along a secondary succession in a reclaimed mining area. Soil C and N significantly improved with reclamation period. Reclamation scenarios and times produced more significant effects on fungal diversity indices than on bacterial and archaeal indices. Vegetation had stronger effects on the variations in bacterial and archaeal diversity, and a lower effect on the variations in fungal diversity than soil characteristics. Reclamation period was significant for bacterial and archaeal assemblies, and soil N, C and water content significantly influenced fungal assembly. Bacterial and archaeal succession followed plant succession, but fungi did not. Taken together, our data highlight that fungal successional dynamics distinctly differ from bacterial and archaeal successional dynamics.
Author Contributions
JL and JC took part in vegetation survey and soil sampling. FL contributed to measuring soil microbial diversity. JL analyzed data and wrote paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the Natural Science Foundation of China (41271530 and 41201374).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2016.00032
Figure S1. The location of Study sites.
Figure S2. Soil microbial richness rarefaction curves in the reclaimed mining area. (A) Bacteria. (B) Archaea. (C) Fungi.
References
Banning, N. C., Gleeson, D. B., Grigg, A. H., Grant, C. D., Andersen, G. L., Brodie, E. L., et al. (2011). Soil microbial community successional patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 77, 6158–6164. doi: 10.1128/AEM.00764-11
Benes, K. M., and Carpenter, R. C. (2015). Kelp canopy facilitates understory algal assemblage via competitive release during early stages of secondary succession. Ecology 96, 241–251. doi: 10.1890/14-0076.1
Brown, S. P., and Jumpponen, A. (2014). Contrasting primary successional trajectories of fungi and bacteria in retreating glacier soils. Mol. Ecol. 23, 481–497. doi: 10.1111/mec.12487
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Costa, R., Götz, M., Mrotzek, N., Lottmann, J., Berg, G., and Smalla, K. (2006). Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56, 236–249. doi: 10.1111/j.1574-6941.2005.00026.x
Cutler, N. A., Chaput, D. L., and van der Gast, C. J. (2014). Long-term changes in soil microbial communities during primary succession. Soil Biol. Biochem. 69, 359–370. doi: 10.1016/j.soilbio.2013.11.022
Dimitriu, P. A., Prescott, C. E., Quideau, S. A., and Grayston, S. J. (2010). Impact of reclamation of surface-mined boreal forest soils on microbial community composition and function. Soil Biol. Biochem. 42, 2289–2297. doi: 10.1016/j.soilbio.2010.09.001
Dini-Andreote, F., de Cássia Pereira e Silva, M., Triadó-Margarit, X., Casamayor, E. O., van Elsas, J. D., and Salles, J. F. (2014). Dynamics of bacterial community succession in a salt marsh chronosequence: evidences for temporal niche partitioning. ISME J. 8, 1989–2001. doi: 10.1038/ismej.2014.54
Dini-Andreote, F., Stegen, J. C., van Elsas, J. D., and Salles, J. F. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. U.S.A. 112, E1326–E1332. doi: 10.1073/pnas.1414261112
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Herzberger, A. J., Meiners, S. J., Towey, J. B., Butts, P. A., and Armstrong, D. L. (2015). Plant–microbe interactions change along a tallgrass prairie restoration chronosequence. Restor. Ecol. 23, 220–227. doi: 10.1111/rec.12165
Justé, A., Thomma, B., and Lievens, B. (2008). Recent advances in molecular techniques to study microbial communities in food-associated matrices and processes. Food Microbiol. 25, 745–761. doi: 10.1016/j.fm.2008.04.009
Knelman, J. E., Schmidt, S. K., Lynch, R. C., Darcy, J. L., Castle, S. C., Cleveland, C. C., et al. (2014). Nutrient addition dramatically accelerates microbial community succession. PLoS ONE 9:e102609. doi: 10.1371/journal.pone.0102609
Kolganova, T., Kuznetsov, B. B., and Tourova, T. (2002). Designing and testing oligonucleotide primers for amplification and sequencing of archaeal 16S rRNA genes. Microbiology 71, 243–246. doi: 10.1023/A:1015122926687
Kuramae, E., Gamper, H., van Veen, J., and Kowalchuk, G. (2011). Soil and plant factors driving the community of soil-borne microorganisms across chronosequences of secondary succession of chalk grasslands with a neutral pH. FEMS Microbiol. Ecol. 77, 285–294. doi: 10.1111/j.1574-6941.2011.01110.x
Li, J.-J., Zheng, Y.-M., Yan, J.-X., Li, H.-J., and He, J.-Z. (2013a). Succession of plant and soil microbial communities with restoration of abandoned land in the Loess Plateau, China. J. Soils Sediments 13, 760–769. doi: 10.1007/s11368-013-0652-z
Li, J.-J., Zheng, Y., Yan, J., Li, H., Wang, X., He, J., et al. (2013b). Effects of different regeneration scenarios and fertilizer treatments on soil microbial ecology in reclaimed opencast mining areas on the loess plateau, China. PLoS ONE 8:e63275. doi: 10.1371/journal.pone.0063275
López-Lozano, N. E., Heidelberg, K. B., Nelson, W. C., García-Oliva, F., Eguiarte, L. E., and Souza, V. (2013). Microbial secondary succession in soil microcosms of a desert oasis in the Cuatro Cienegas Basin, Mexico. Peer J. 1, e47. doi: 10.7717/peerj.47
Lozano, Y. M., Hortal, S., Armas, C., and Pugnaire, F. I. (2014). Interactions among soil, plants, and microorganisms drive secondary succession in a dry environment. Soil Biol. Biochem. 78, 298–306. doi: 10.1016/j.soilbio.2014.08.007
Mastrogianni, A., Papatheodorou, E. M., Monokrousos, N., Menkissoglu-Spiroudi, U., and Stamou, G. P. (2014). Reclamation of lignite mine areas with Triticum aestivum: the dynamics of soil functions and microbial communities. Appl. Soil Ecol. 80, 51–59. doi: 10.1016/j.apsoil.2014.03.009
Nemergut, D. R., Schmidt, S. K., Fukami, T., O'Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/MMBR.00051-12
Nicol, G. W., Tscherko, D., Embley, T. M., and Prosser, J. I. (2005). Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ. Microbiol. 7, 337–347. doi: 10.1111/j.1462-2920.2005.00698.x
Nielsen, U. N., Osler, G. H., Campbell, C. D., Burslem, D. F., and van der Wal, R. (2010). The influence of vegetation type, soil properties and precipitation on the composition of soil mite and microbial communities at the landscape scale. J. Biogeogr. 37, 1317–1328. doi: 10.1111/j.1365-2699.2010.02281.x
Philippot, L., Tscherko, D., Bru, D., and Kandeler, E. (2011). Distribution of high bacterial taxa across the chronosequence of two alpine glacier forelands. Microb. Ecol. 61, 303–312. doi: 10.1007/s00248-010-9754-y
Prewitt, L., Kang, Y., Kakumanu, M. L., and Williams, M. (2014). Fungal and bacterial community succession differs for three wood types during decay in a forest soil. Microb. Ecol. 68, 212–221. doi: 10.1007/s00248-014-0396-3
Putten, W. H., Bardgett, R. D., Bever, J. D., Bezemer, T. M., Casper, B. B., Fukami, T., et al. (2013). Plant–soil feedbacks: the past, the present and future challenges. J. Ecol. 101, 265–276. doi: 10.1111/1365-2745.12054
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schmidt, S. K., Nemergut, D. R., Darcy, J. L., and Lynch, R. (2014). Do bacterial and fungal communities assemble differently during primary succession? Mol. Ecol. 23, 254–258. doi: 10.1111/mec.12589
Shrestha, P. M., Noll, M., and Liesack, W. (2007). Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ. Microbiol. 9, 2464–2474. doi: 10.1111/j.1462-2920.2007.01364.x
Ter Braak, C. J., and Smilauer, P. (2002). “CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (version 4.5)”. Available online at: www.canoco.com
Timonen, S., and Bomberg, M. (2009). Archaea in dry soil environments. Phytochemistry Reviews 8, 505–518. doi: 10.1007/s11101-009-9137-5
Urbanová, M., Šnajdr, J., and Baldrian, P. (2015). Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 84, 53–64. doi: 10.1016/j.soilbio.2015.02.011
Wang, Z., Fang, J., Tang, Z., and Lin, X. (2011). Patterns, determinants and models of woody plant diversity in China. Proc. R. Soc. Lond. B 278, 2122–2132. doi: 10.1098/rspb.2010.1897
Wilkinson, D. M., Koumoutsaris, S., Mitchell, E. A., and Bey, I. (2012). Modelling the effect of size on the aerial dispersal of microorganisms. J. Biogeogr. 39, 89–97. doi: 10.1111/j.1365-2699.2011.02569.x
Williams, M. A., Jangid, K., Shanmugam, S. G., and Whitman, W. B. (2013). Bacterial communities in soil mimic patterns of vegetative succession and ecosystem climax but are resilient to change between seasons. Soil Biol. Biochem. 57, 749–757. doi: 10.1016/j.soilbio.2012.08.023
Yannarell, A. C., Menning, S. E., and Beck, A. M. (2014). Influence of shrub encroachment on the soil microbial community composition of remnant hill prairies. Microb. Ecol. 67, 897–906. doi: 10.1007/s00248-014-0369-6
Yarwood, S., Wick, A., Williams, M., and Daniels, W. L. (2015). Parent material and vegetation influence soil microbial community structure following 30-years of rock weathering and pedogenesis. Microb. Ecol. 69, 383–394. doi: 10.1007/s00248-014-0523-1
Yoshitake, S., Fujiyoshi, M., Watanabe, K., Masuzawa, T., Nakatsubo, T., and Koizumi, H. (2013). Successional changes in the soil microbial community along a vegetation development sequence in a subalpine volcanic desert on Mount Fuji, Japan. Plant Soil 364, 261–272. doi: 10.1007/s11104-012-1348-7
Zhang, B., Deng, H., Wang, H.-L., Yin, R., Hallett, P. D., Griffiths, B. S., et al. (2010). Does microbial habitat or community structure drive the functional stability of microbes to stresses following re-vegetation of a severely degraded soil? Soil Biol. Biochem. 42, 850–859. doi: 10.1016/j.soilbio.2010.02.004
Zhang, J.-T. (2005). Succession analysis of plant communities in abandoned croplands in the eastern Loess Plateau of China. J. Arid Environ. 63, 458–474. doi: 10.1016/j.jaridenv.2005.03.027
Zhao, J., Li, S., He, X., Liu, L., and Wang, K. (2014). The Soil Biota composition along a progressive succession of secondary vegetation in a karst area. PLoS ONE 9:e112436. doi: 10.1371/journal.pone.0112436
Zhao, Z., Bai, Z., Zhang, Z., Guo, D., Li, J., Xu, Z., et al. (2012). Population structure and spatial distributions patterns of 17 years old plantation in a reclaimed spoil of Pingshuo opencast mine, China. Ecol. Eng. 44, 147–151. doi: 10.1016/j.ecoleng.2012.03.018
Zhao, Z., Shahrour, I., Bai, Z., Fan, W., Feng, L., and Li, H. (2013). Soils development in opencast coal mine spoils reclaimed for 1–13 years in the West-Northern Loess Plateau of China. Eur. J. Soil Biol. 55, 40–46. doi: 10.1016/j.ejsobi.2012.08.006
Zinger, L., Lejon, D. P., Baptist, F., Bouasria, A., Aubert, S., Geremia, R. A., et al. (2011). Contrasting diversity patterns of crenarchaeal, bacterial and fungal soil communities in an alpine landscape. PLoS ONE 6:e19950. doi: 10.1371/journal.pone.0019950
Keywords: bacteria, archaea, fungi, 454 pyrosequencing, microbial succession, reclaimed mine
Citation: Li J, Liu F and Chen J (2016) The Effects of Various Land Reclamation Scenarios on the Succession of Soil Bacteria, Archaea, and Fungi Over the Short and Long Term. Front. Ecol. Evol. 4:32. doi: 10.3389/fevo.2016.00032
Received: 29 December 2015; Accepted: 17 March 2016;
Published: 31 March 2016.
Edited by:
Eric Altermann, AgResearch Ltd, New ZealandReviewed by:
Steven Singer, Lawrence Berkeley National Laboratory, USAAmy Pruden, Virginia Tech, USA
Copyright © 2016 Li, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junjian Li, bGlqdW5qaWFuQHN4dS5lZHUuY24=
 Junjian Li
Junjian Li Feng Liu
Feng Liu