- 1Department of Biology, Western University, London, ON, Canada
- 2Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland
The relatively simple communication, breeding, and egg-making systems that govern reproduction in female Drosophila retain homology to eusocial species in which these same systems are modified to the social condition. Despite having no parental care, division of labor, or subfertile caste, Drosophila may nonetheless offer a living test of certain sociobiological hypotheses framed around gene function. In this review, we make this case, and do so around the recent discovery that the non-social fly, Drosophila melanogaster, can respond to the ovary-suppressing queen pheromone of the honey bee Apis mellifera. Here, we first explain the sociobiological imperative to reconcile kin theory with molecular biology, and qualify a potential role for Drosophila. Then, we offer three applications for the fly-pheromone assay. First, the availability and accessibility of massive mutant libraries makes immediately feasible any number of open or targeted gene screens against the ovary-inhibiting response. The sheer tractability of Drosophila may therefore help to accelerate the search for genes in pheromone-responsive pathways that regulate female reproduction, including potentially any that are preserved with modification to regulate worker sterility in response to queen pheromones in eusocial taxa. Secondly, Drosophila's powerful Gal4/UAS expression system can complement the pheromone assay by driving target gene expression into living tissue, which could be well-applied to the functional testing of genes presumed to drive ovary activation or de-activation in the honey bee or other eusocial taxa. Finally, coupling Gal4 with UAS-RNAi lines can facilitate loss-of-function experiments against perception and response to the ovary inhibiting pheromone, and do so for large numbers of candidates in systematic fashion. Drosophila's utility as an adjunct to the field of insect sociobiology is not ideal, but retains surprising potential.
Introduction
Sociobiology is the evolutionary genetic study of social behavior (Crozier and Pamilo, 1996; Frank, 1998; Bourke, 2011). Its premise is simple: natural selection of gene variants can explain evolved aspects of social behavior. By adopting this gene's-eye-view of behavioral evolution we can begin to ask, how does a gene associated with a particular behavior change in frequency? For social phenotypes, whereby the trait is defined in terms of fitness consequences (West et al., 2007), this approach has proven useful—for example, by explaining how genes for altruism can evolve despite their cost to personal fitness.
Further, by incorporating the notion of environmental selection of genetic variants into its framework, sociobiology implicitly melds “nature” with “nurture” (Robinson, 2004; Crozier, 2008) and can advance our understanding of behavioral evolution on two fronts. First, it makes clear that differences in genotype (or environment) can explain individual differences in behavior, which opens up the prospect of finding the very genes (or environments) involved. Second, it questions the adaptive value of behavioral variants and their relative impact on fitness, which helps us to understand the ultimate function of behavior. The molecular “how” combined with the evolutionary “why” of sociobiology make this field as fascinating as it is contentious.
At a practical level, sociobiologists have been successful at adopting analytical tools developed within the social sciences—for example, longitudinal or cross-sectional studies, common garden or cross-fostering techniques, and population vs. individual level analyses of variance, etc.—are all commonly used in this field (Hughes, 1998). Moreover, sociobiologists have increasingly begun to deploy sophisticated molecular tools that are ultimately imported from the health sciences (Smith et al., 2008). These tools include genotype–phenotype association analyses, molecular screens, and statistical tests for determining the genetic effects on social traits. The field of sociobiology is therefore bourgeoning and has a history of assimilating seemingly disparate angles and ideas into its single unified conceptual framework that is Hamilton's (1964, 1972) inclusive fitness theory (Reeve, 2001; Lehmann and Keller, 2006; Abbot et al., 2011; Gardner et al., 2011; Bourke, 2014; Liao et al., 2015).
Inclusive fitness theory lends itself well to the evolutionary genetic study of behavior. For one, it is explicitly gene centric, which makes clear the primacy of gene-level thinking. This emphasis on the gene as the unit of selection has, in the past 50 years, re-shaped our understanding of how selection actually works (Dawkins, 1976; Herbers, 2013). Specifically, it generalizes the Darwinian understanding that selection tends to maximize direct fitness of individuals when, rather, it tends to optimize direct vs. indirect fitness trade-offs, the latter of which is measured through the focal individual's fitness effect on its non-descendent relatives. Darwin's original emphasis on direct fitness can therefore be regarded as a special case of Hamilton's more general theory of inclusive fitness (West and Gardner, 2013), which includes both direct and indirect fitness components.
When fitness is thus partitioned it is possible to clearly distinguish four different types of social behavior (West et al., 2007). For example, selfishness is recognized by a positive (direct) fitness effect to an individual actor with a correspondingly negative effect on one or more receivers who are, in effect, exploited. Cooperation, by contrast, is recognized from the positive fitness outcome for actor and receiver alike. Selfishness and cooperation are readily explained by positive fitness effects on the actor and do not invoke any indirect fitness effects. Altruism, however, is different. This type of social interaction is costly to the actor, and thus cannot evolve via direct benefit. Still, altruism has evolved and is most evident in the reproductively altruistic (even sterile) castes of the eusocial insects. Hamilton's insight has proven especially helpful to explain this type of altruism (Crozier and Crespi, 2000), which can evolve under conditions specified by Hamilton's Rule. The rule—so named by Charnov (1977)—is a heuristic statement that predicts altruism when the direct cost, c, of helping is small in relation to the direct fitness benefit, b, to the recipient of that help, provided the recipient is sufficiently likely, r, to carry copies of the causal gene. Or, rb > c.
Genes for Altruism
The independent evolution of eusociality featuring altruism across different spectra of life creates opportunities for comparative analyses (Crespi, 1996; Thorne and Traniello, 2003; Terrapon et al., 2014; Berens et al., 2015; Kapheim et al., 2015; Patalano et al., 2015; Rehan and Toth, 2015). On one hand, it is possible that each origin is a unique response to indirect selection on taxon-specific genes for socially altruistic phenotypes (or, more loosely, on taxon-specific “social genes;” Kapheim et al., 2015). If so, then we expect to see little homology between species for gene sets that regulate aspects of sociality. On the other hand, it is conceivable that the evolution and expression of altruistic work occasionally or regularly involves functionally related genes, or other genetic features like gene pathways, gene modules or gene networks (reviewed in Mikheyev and Linksvayer, 2015). To the extent that conserved genetic features are merely modified from selfish ancestries to now regulate complex social phenotypes, we can exploit comparative analyses to find them.
Finding these genes and their extended genotypes (modules, networks) may not be easy. As insightful as inclusive fitness theory is at highlighting the importance of gene-level thinking for understanding the evolution and expression of social traits, the identity or even qualities of the genes promoting social novelty, including altruism, remain mostly unknown (Thompson et al., 2013; Kapheim, 2016). We echo recent sentiment that there is every opportunity to couple gene-minded behavioral ecology and sociobiology with the practical tools of molecular biology (Robinson et al., 2005; Smith et al., 2008; Johnson and Linksvayer, 2010; Kapheim et al., 2015; Rehan and Toth, 2015). In principle, it should be possible to find real genes that specifically underlie reproductive altruism for a given taxon (Thompson et al., 2013; Akçay et al., 2015; Linksvayer, 2015). Why not? This fitness-defined trait has clearly evolved, and as such, must be underlain by gene variants that have been selected to regulate this behavior within social environments.
In this practical sense, genes for altruism are not just hypothetical variables in a conceptual formula—as in Hamilton's (1963) “g.” They are real molecular loci, even if they are pleiotropic, and otherwise known for relatively unspectacular metabolic, structural, nervous, developmental or regulatory functions (Bloch and Grozinger, 2011; Mullen and Thompson, 2015). In effect, a “gene for altruism,” or, a gene for any social trait, can be detected by the genotypic difference that explains the behavioral variant in a given environment—for example, a genotypic tendency for parental (selfish) vs. alloparental (altruistic) care at a mixed brood nest. For sociobiology this basic tenet of behavioral genetics should be readily applied through Hamilton's rule, which nicely phrases the conditional effect of genetic differences (r) within an environmental context (b, c). If so, it should be possible to find the genes that mediate the expression of reproductive altruism. Of course, the causal gene is not kin selected if its transmission is incidental, but rather is selected when the effect of altruism itself is to increase the direct fitness of related beneficiaries, over and above what their direct fitness would have otherwise been.
So where are these genes, and what qualities might they have? For one, we expect them to be differentially expressed between reproducing (selfish) and non-reproducing (altruistic) individuals. The honey bee (Apis mellifera) has proven to be a useful model in this regard. It is possible to experimentally manipulate the application of queen or brood pheromone, and with it the activation or de-activation of worker ovaries. From here, one can screen for genes that are functionally associated with the pheromone-induced onset of worker sterility and associated helping behavior that, together, represent the honey bee's instance of altruism. Early screens using microarrays uncovered suites of genes that correlated in their expression with worker ovary de-activation (reviewed by Mullen et al., 2014; Sobotka et al., 2016). The technology-driven generation of these candidates was an important step toward understanding the molecular basis of honey bee worker sterility (Lattorff and Moritz, 2013; Mullen and Thompson, 2015). But how do these genes respond to their environment and to each other to coordinate direct vs. indirect fitness trade-offs within individuals?
We expect genetic effects on social traits to be mediated through environmentally responsive networks (Schwander et al., 2010; Bendesky and Bargmann, 2011), and indeed, this idea is substantiated by empirical studies that situate socially-relevant gene sets within a transcriptional regulatory context (Grozinger and Robinson, 2007; Cardoen et al., 2011; Chandrasekaran et al., 2011; Molodtsova et al., 2014), including pheromone-responsive genes for honey bee worker sterility (Mullen et al., 2014; Sobotka et al., 2016). Multi-gene network studies are useful because they represent graphical molecular hypotheses about how sociality is coordinated at the molecular level (Kapheim, 2016). The validity of these and other molecular hypotheses for the coordination of worker sterility are, however, only beginning to be tested in vivo (Jarosch et al., 2011; Ronai et al., 2015), probably because it is not as easy to perturb known or presumed network nodes in honeybees or other eusocial taxa via strategic gene knock-down as it is for a fully tractable insect model like Drosophila melanogaster. Notwithstanding tremendous progress in the application of gene-transforming technologies to A. mellifera (Jarosch and Moritz, 2011; Scott et al., 2013; Schulte et al., 2014), we present a complementary approach that seeks to leverage more from Drosophila as a surrogate model in insect sociobiology, beyond the regular informatics comparisons to Drosophila gene databases.
Drosophila as a Non-social Model of Social Gene Discovery
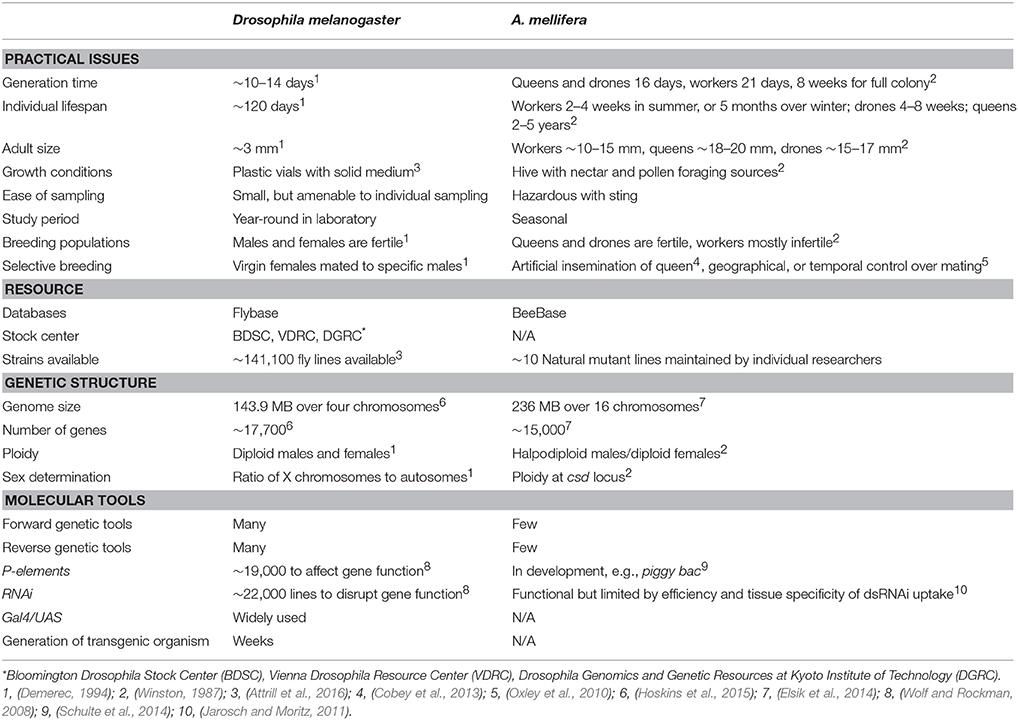
Genomic comparisons between species that differ in social structure help to highlight the genes involved in each social transition (Schwarz et al., 2007; Woodard et al., 2011; Kapheim et al., 2015; Rehan and Toth, 2015). But how far down the phylogenetic tree can you go? Despite ~350 million years of divergence between flies (Diptera) and the order containing bees (Hymenoptera), comparative studies using species of flies to model the genetics of complex behavior has proven useful and retains surprising potential (Sokolowski, 2001; Robinson and Ben-Shahar, 2002; Reaume and Sokolowski, 2011). For example, studies using fruit flies to elucidate the mechanism involved in honey bee worker sterility date back to the late 1960s; Sannasi (1969) found that adult female Drosophila reduced their ovary size in a manner comparable to queen-right worker bees when exposed to a component (9-ODA) of honeybee queen pheromone. So did the house fly Musca domestica (Nayer, 1963). Further, the same component reduces ovarian development in other insect species, including social ones like termites (Hrdý et al., 1960; Sannasi and George, 1972) and ants (Carlisle and Butler, 1956). It is clear that A. mellifera queen mandibular pheromone can potentially influence the reproductive phenotype of other insects, but the reason is unclear. We speculate that, despite its derived chemical composition (Oi et al., 2015), A. mellifera queen mandibular pheromone, like other Hymenopteran queen pheromones (Van Oystaeyen et al., 2014), likely evolved through modification of fertility signals already present in pre-social ancestors (Chapuisat, 2014; Oliveira et al., 2015; Peso et al., 2015) and thus may retain sufficient structural homology to extant pheromones to inadvertently stimulate the fertility pathways of these insects. Alas, despite lacking a social repertoire—no parental care, division of labor, or subfertile caste—Drosophila's conspicuous response to ovary-inhibiting queen bee pheromone, together with its base homology and genetic tractability (Table 1), open experimental possibilities to tease-out conserved gene functions in reproduction-related behaviors. This bee-to-fly approach will not help to directly uncover any role for genetic novelty in social evolution, which is clearly important (Johnson and Tsutsui, 2011), but will address its converse—the role for conserved mechanisms that are not unique to social taxa, just secondarily co-opted to perform within a social context. At any rate, Drosophila spp. do express mild social phenotypes related to feeding (Tinette et al., 2004) larval burrowing (Wu et al., 2003), mating (Villella and Hall, 2008), and nearest-neighbor spacing (Simon et al., 2012), among other traits that are potentially comparable to their highly social versions in eusocial taxa.
One reproduction-related trait from Drosophila that may prove relevant to comparative insect sociobiology is how females regulate their ovaries in response to environmental cues (Emerson et al., 2009). The ramping up and down of personal reproduction in females via ovary activation and de-activation is comparable to this same physiological process in worker honeybees, where workers de-activate their ovaries and adopt reproductively altruistic roles in response to queen signal (Winston, 1987). The potential for this comparison was realized in full by Camiletti et al. (2013) who built upon earlier findings (Nayer, 1963; Sannasi, 1969) to show that female Drosophila are responsive to a synthetic blend of ovary-inhibiting queen mandibular pheromone (QMP) components from A. mellifera. Virgin Oregon-R flies, among other Drosophila strains (Camiletti et al., 2014), treated with QMP develop smaller ovaries with fewer eggs than do un-treated controls, but otherwise have normal phenotypes. Remarkably, Drosophila females exposed to this Apis fertility signal respond in a manner that is comparable to the pheromone's normal effect on workers—that is, it renders them partially “sterile.” This unexpected finding opens new experimental possibilities whereby we can use QMP to manipulate ovary activation in Drosophila in a controlled and repeatable manner, and then use mutant lines, gene drivers and gene knock-downs, among other Drosophila resources, to experimentally infer the genes, pathways and neural circuits involved. To the extent that any of these genetic or genetically encoded factors are present and functional in Apis or other social insects, they become prime candidates for functional studies in the social taxa themselves.
Application of Drosophila Gene-finding Tools to Predict Apis Biology
The massive availability of Drosophila mutant lines via global stock centers offer an obvious entry-level test for probing the function of genes that have previously been implicated in Apis or other social insect reproduction. This classic “reverse genetic” approach has proven useful—for example, by testing how foraging (Camiletti et al., 2014) or Orco mutants respond to ovary-inhibiting pheromone in Drosophila. Early observations that Drosophila sitter (forS) and rover (forR) strains (Osborne, 1997) are behaviorally analogous, if not homologous, to nurse and forager worker castes (Ben-Shahar et al., 2002) led Camiletti et al. (2014) to test the sitter/rover response to QMP. Their prediction was similar to that of Ben-Shahar et al. (2002)—namely, that if nurse bees have higher reproductive potential than do forager bees (Fussnecker et al., 2011) then sitter flies should be more responsive to this ovary-inhibiting cue. Camiletti et al. (2014) showed that sitters are more responsive to QMP relative to rovers, and respond more-or-less as nurse bees normally would, with smaller ovaries that contain fewer eggs. This “social” response from flies to a bee pheromone underscores the notion that even non-social insects can express some responsiveness to social cues, and that this response varies with genotype. In general, the interspecific response is consistent with a conserved gene module affecting reproductive physiology. The differential response among fly strains in particular suggests that for is itself important for modulating the fly's pheromonal response.
Similarly, we predict that the fly's response to bee pheromone is likely mediated by olfaction, and if so it should be possible to use a pheromone assay on Drosophila mutant lines to test this prediction. The gene Orco encodes a major olfactory co-factor, and available homozygous loss-of-function mutants that are characterized by coding region deletions are effective at blocking a wide range of olfactory stimuli, to the point of rendering the Orco mutants essentially anosmic (Larsson et al., 2004; Steck et al., 2012). We predict therefore that the ovary-inhibiting response by wild-type Drosophila to Apis QMP will be muted in Orco mutant assays. If this prediction is up-held, any such lack-of-response would suggest that Orco—a 1-to-1 ortholog with the bee's AmOr2 (Robertson and Wanner, 2006)—is essential for the perception of QMP and its downstream effect on ovaries. This comparative test between Drosophila and Apis would provide functional evidence for a leading hypothesis in insect sociobiology—namely, for a deeply conserved reproductive regulatory system shared with modification between social and pre-social orders. Further, a functional demonstration from mutant screens that Orco is specifically required to perceive QMP would identify a clear ortholog (AmOr2) from the pheromone-responsive pathway that may regulate honeybee, or other social insect, worker sterility. But this, together with so many other similarly reasoned bee-fly hypotheses, remains to be tested.
Next, Drosophila's gene-driving Gal4/UAS system allows for targeted control of transgene expression in specific tissues, and even at specific developmental time points (Duffy, 2002; del Valle Rodríguez et al., 2012). Gal4 itself is a transcription factor that can be coupled with an endogenous promoter to direct transgene expression of UAS (Upstream Activating Sequence)-responder gene elements. The most widely used GAL4 drivers for tissues of reproductive interest include elav-Gal4 (for neural tissues; Yao and White, 1994), nos-Gal4 (for ovary tissues; Rørth, 1998), and FB-Gal4 (for fat body tissues; Grönke et al., 2003), among other driver lines, including those with ubiquitous expression (i.e., actin-Gal4; Wilder, 2000). When Gal4-individuals are mated with UAS-transgene construct partners—for example, elav-Gal4 × UAS-GFP—their offspring inherit both transgenes (elav-Gal4/UAS-GFP) and thus express the target gene, in this case a green fluorescent protein (GFP). The ability to keep the driver and responder components of this system separated in parental lines until combined in the F1 affords a level of control over gene expression that is not yet possible in any eusocial taxon. For example, it allows for the controlled expression of potentially toxic gene products under the regulation of UAS that will then only be expressed in progeny that also carry the Gal4 transgene, but remain unexpressed in stock populations. This may be of interest to bee researchers wishing to exploit the fly model to screen for toxicity of transgene products being vetted for control of pests and pathogens in an agricultural context (Malone and Pham-Delègue, 2001). It is also possible to use the fly Gal4/UAS system to induce gene expression at different developmental time points by, for example, using a heat shock-Gal4 driver (i.e., heat shock protein 70) to induce UAS-responder gene expression as individuals are heat shocked at, say, 37°C. One application here might be to shock-induce target gene expression at larval, pupal, or adult stages in the fly, and monitor the effects on reproductive or sexual behavior, ovary activation, or any other trait of interest, and do so as flies are exposed to QMP, royal jelly or other social cues. The fly-enabled targeted gene expression system, together with the newly reported bee-like effects from flies in response to social cues, could be adopted to fuel a series of creative experiments centered on gene function.
One sociobiological application is to drive genes into female flies that are predicted to induce social phenotypes in the honeybee or other social taxa, even if these genes are not normally present in the Drosophila genome. For example, Kamakura (2011) drove a gene encoding royalactin—a protein that induces the “royal” queen phenotype in developing A. mellifera larvae—into female D. melanogaster to monitor the gene's exogenous effect on growth and reproduction. Likewise, Ren and Hughes (2014) drove the gene encoding A. mellifera vitellogenin into Drosophila metabolic tissues to test its effect on lifespan and fecundity. Specifically they crossed a UAS-Apis Vg transgenic fly line with flies harboring the fat body S106-Gal4. These pioneering studies demonstrate the potential and feasibility of Drosophila for testing the crude function of genes related to social reproduction in a non-social insect model, with the understanding that the ultimate test remains forthcoming when similar technology eventually becomes available for the eusocial taxa themselves. Any results from the fly model must therefore be considered as provisional support for (or rejection of) the functional molecular hypothesis under consideration. Negative results in particular require careful interpretation of whether the model failed (was idiosyncratic) or the biological hypothesis is genuinely not supported.
Finally, the Gal4/UAS system can be coupled with RNAi to knock down “social” genes in Drosophila. This combination of targeted gene expression and gene silencing affords a higher measure of control over the timing and target of knock-down effects (Bakal, 2011). Depending on the availability of UAS-RNAi construct lines (Dietzl et al., 2007) one can drive the silencing of candidate genes or, systematically, whole gene families of functional groups to reveal which individual genes from a larger set are functionally required for the expression of a social or reproductive phenotype. For honey bee genes, the combined Gal4/UAS-RNAi canon could compliment cases where RNAi via injection, feeding or nanospray of dsRNA was inefficient or lacking in tissue specificity (Jarosch and Moritz, 2011; Li-Byarlay et al., 2013; Scott et al., 2013).
One application might be to use the Gal4/UAS-RNAi approach to uncover the subset of olfactory receptors (ORs) that specifically “tune” Orco to the perception of QMP. It should be possible to knock down individual ORs via Gal4-driven RNAi insertions that are readily available from the Vienna Drosophila RNAi Center (Dietzl et al., 2007). We envision that crossing UAS-RNAi males with virgin elav-Gal4; UAS-dcr2 females will generate F1s that express OR-specific knockdowns. From the recombinant offspring of each RNAi lines one could use the Camiletti et al. (2013, 2014) queen pheromone assay to systematically screen all of the ORs specific to Drosophila adults (~48; Laissue and Vosshall, 2008) to identify any receptors with a strong knock down effect—i.e., where knock-downs fail to show a worker-like phenotype upon exposure to QMP. Any candidates thus identified might therefore have some involvement in the fly's “normal” worker-like response to queen pheromone and, through any functional homology, some involvement in response to ovary inhibiting signal in the worker bee herself. This fly-to-bee comparison will be challenging owing to the evolutionary radiation of olfactory receptors in A. mellifera (Robertson and Wanner, 2006), but detailed genealogies of bee-fly olfactory receptor gene families (Zhou et al., 2012) provide a guide and will facilitate identification of the A. mellifera olfactory receptors that are most likely involved in the honey bee worker's altruistic response to queen pheromone. The validity of this Drosophila-Apis comparative approach will rest in part on future tests of this hypothesis.
What Next from the Drosophila Model?
Although, kin theory has long used a gene's-eye view to help model the evolution of eusocial breeding systems, molecular biology is only beginning to uncover the genes involved. Here, we highlight the role for one established genetic model to help accelerate our understanding of the genetic and molecular underpinnings of complex social behavior, including, potentially, the genes that underpin reproductive altruism in eusocial insects. We forecast several experiments in insect sociobiology using the queen pheromone assay described above and a Drosophila model. First, it should readily be possible to expand the comparative utility of the Camiletti et al. (2013) pheromone assay to test the response from male flies, and to test the response from other insect species that share a more recent ancestry with bees than does Drosophila. If female flies respond as (female) worker bees do, then do male flies respond as (male) drones do? Preliminary work in our lab at Western University (Canada) suggests that QMP does excite male Drosophila to orient toward and court conspecific females, much as it does to male Apis. What of other Drosophila species? If the response from Oregon-R and strains noted above is not strictly idiosyncratic then we predict other insects should respond to QMP. If so, then QMP could potentially be used to test the reproductive response in any number of taxa, including subsocial (Rehan et al., 2014) and primitively eusocial (Sadd et al., 2015) species that are emerging as alternate models from within the Hymenoptera. At present, we argue that Drosophila, despite its lack of eusocial phenotype, could play a useful role in social gene discovery—a major mandate of the field of social insect biology. There is obviously every reason to invest into non-Drosophilid models to advance insect sociobiology (Hasselmann et al., 2015; Wurm, 2015), but at the same time we can harness other tools that are currently available, where appropriate. We are also optimistic of on-going progress toward developing stable transgenic lines for social Hymenoptera through use of a germ-line targeted transposon called piggyBac (Schulte et al., 2014). The further development of this technique could eventually facilitate the implementation of tools like Gal4/UAS for direct use in Apis (Ben-Shahar, 2014) and other social taxa to reveal more of the genetic and molecular underside of sociobiology.
Author Contributions
AC and GT co-conceived the manuscript, drafted, and revised the manuscript together, and co-approved the final version for publication.
Funding
This work was funded by an Ontario Graduate Scholarship to AC and a Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery Grant to GT. We thank the Herbette Foundation (La Fondation Herbette) for facilitating GT's stay at the University of Lausanne.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
For help and discussion we thank all members of the Social Biology Group at Western University (Canada). We thank Luke Holman (University of Melbourne) and Jan Oettler (University Regensburg) for very helpful comments on an earlier draft of the manuscript.
References
Abbot, P., Abe, J., Alcock, J., Alizon, S., Alpedrinha, J. A., Andersson, M., et al. (2011). Inclusive fitness theory and eusociality. Nature 471, E1–E4. doi: 10.1038/nature09831
Akçay, E., Linksvayer, T. A., and van Cleve, J. (2015). Bridging social evolution theory and emerging empirical approaches to social behavior. Curr. Opin. Behav. Sci. 6, 59–64. doi: 10.1016/j.cobeha.2015.09.002
Attrill, H., Falls, K., Goodman, J. L., Millburn, G. H., Antonazzo, G., Rey, A. J., et al. (2016). FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 44, D786–D792. doi: 10.1093/nar/gkv1046
Bakal, C. (2011). Drosophila RNAi screening in a postgenomic world. Brief Funct. Genomics 10, 197–205. doi: 10.1093/bfgp/elr015
Bendesky, A., and Bargmann, C. I. (2011). Genetic contributions to behavioural diversity at the gene-environment interface. Nat. Rev. Genet. 12, 809–820. doi: 10.1038/nrg3065
Ben-Shahar, Y. (2014). A piggyBac route to transgenic honeybees. Proc. Natl. Acad. Sci. U.S.A. 111, 8708–8709. doi: 10.1073/pnas.1407876111
Ben-Shahar, Y., Robichon, A., Sokolowski, M. B., and Robinson, G. E. (2002). Influence of gene action across different time scales on behavior. Science 296, 741–744. doi: 10.1126/science.1069911
Berens, A. J., Hunt, J. H., and Toth, A. L. (2015). Comparative transcriptomics of convergent evolution: different genes but conserved pathways underlie caste phenotypes across lineages of eusocial insects. Mol. Biol. Evol. 32, 690–703. doi: 10.1093/molbev/msu330
Bloch, G., and Grozinger, C. M. (2011). Social molecular pathways and the evolution of bee societies. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 366, 2155–2170. doi: 10.1098/rstb.2010.0346
Bourke, A. F. G. (2014). Hamilton's rule and the causes of social evolution. Phil. Trans. R. Soc. B 369:20130362. doi: 10.1098/rstb.2013.0362
Camiletti, A. L., Awde, D. N., and Thompson, G. J. (2014). How flies respond to honey bee pheromone: the role of the foraging gene on reproductive response to queen mandibular pheromone. Naturwissenschaften 101, 25–31. doi: 10.1007/s00114-013-1125-3
Camiletti, A. L., Percival-Smith, A., and Thompson, G. J. (2013). Honey bee queen mandibular pheromone inhibits ovary development and fecundity in a fruit fly. Entomol. Exp. Appl. 147, 262–268. doi: 10.1111/eea.12071
Cardoen, D., Wenseleers, T., Ernst, U. R., Danneels, E. L., Laget, D., DE Graaf, D., et al. (2011). Genome-wide analysis of alternative reproductive phenotypes in honeybee workers. Mol. Ecol. 20, 4070–4084. doi: 10.1111/j.1365-294X.2011.05254.x
Carlisle, D., and Butler, C. (1956). The queen-substance of honeybees and the ovary inhibiting hormone of crustaceans. Nature 177, 276–277. doi: 10.1038/177276b0
Chandrasekaran, S., Ament, S. A., Eddy, J. A., Rodriguez-Zas, S. L., Schatz, B. R., Price, N. D., et al. (2011). Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc. Natl. Acad. Sci. U.S.A. 108, 18020–18025. doi: 10.1073/pnas.1114093108
Chapuisat, M. (2014). Smells like queen since the cretaceous. Science 343, 254–255. doi: 10.1126/science.1249285
Charnov, E. L. (1977). An elementary treatment of the genetical theory of kin selection. J. Theor. Biol. 66, 541–550. doi: 10.1016/0022-5193(77)90301-0
Cobey, S. W., Tarpy, D. R., and Woyke, J. (2013). Standard methods for instrumental insemination of Apis mellifera queens. J. Api. Res. 52, 18. doi: 10.3896/IBRA.1.52.4.09
Crespi, B. J. (1996). “Comparative analysis of the origins and losses of eusociality: causal mosaics and historical uniqueness,” in Phylogenies and the Comparative Method in Animal Behaviour, ed E. P. Martins (New York, NY: Oxford University Press), 253–287.
Crozier, R. H. (2008). Advanced eusociality, kin selection and male haploidy. Aust. J. Entomol. 47, 2–8. doi: 10.1111/j.1440-6055.2007.00621.x
Crozier, R. H., and Crespi, B. J. (2000). A life of insight - in memoriam William D. Hamilton (1936–2000). Insect Soc. 47, 297.
Crozier, R., and Pamilo, P. (1996). Evolution of Social Insect Colonies. Oxford: Oxford University Press.
del Valle Rodríguez, A., Didiano, D., and Desplan, C. (2012). Power tools for gene expression and clonal analysis in Drosophila. Nat. Methods 9, 47–55. doi: 10.1038/nmeth.1800
Demerec, M. (ed.). (1994). Biology of Drosophila. New York, NY: Cold Spring Harbor Laboratory Press.
Dietzl, G., Chen, D., Schnorrer, F., Su, K. C., Barinova, Y., Fellner, M., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. doi: 10.1038/nature05954
Duffy, J. B. (2002). GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34, 1–15. doi: 10.1002/gene.10150
Elsik, C. G., Worley, K. C., Bennett, A. K., Beye, M., Camara, F., Childers, C. P., et al. (2014). Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15:86. doi: 10.1186/1471-2164-15-86
Emerson, K. J., Uyemura, A. M., McDaniel, K. L., Schmidt, P. S., Bradshaw, W. E., and Holzapfel, C. M. (2009). Environmental control of ovarian dormancy in natural populations of Drosophila melanogaster. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 195, 825–829. doi: 10.1007/s00359-009-0460-5
Fussnecker, B. L., McKenzie, A. M., and Grozinger, C. M. (2011). cGMP modulates responses to queen mandibular pheromone in worker honey bees. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 197, 939–948. doi: 10.1007/s00359-011-0654-5
Gardner, A., West, S. A., and Wild, G. (2011). The genetical theory of kin selection. J. Evol. Biol. 24, 1020–1043. doi: 10.1111/j.1420-9101.2011.02236.x
Grönke, S., Beller, M., Fellert, S., Ramakrishnan, H., Jäckle, H., and Kühnlein, R. P. (2003). Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 13, 603–606. doi: 10.1016/S0960-9822(03)00175-1
Grozinger, C. M., and Robinson, G. E. (2007). Endocrine modulation of a pheromone-responsive gene in the honey bee brain. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 193, 461–470. doi: 10.1007/s00359-006-0202-x
Hamilton, W. D. (1963). The evolution of altruistic behavior. Am. Nat. 97, 354–356. doi: 10.1086/497114
Hamilton, W. D. (1964). The genetical evolution of social behaviour II. J Theor. Biol. 7, 17–52. doi: 10.1016/0022-5193(64)90039-6
Hamilton, W. D. (1972). Altruism and related phenomena, mainly in social insects. Annu. Rev. Ecol. Syst. 3, 193–232. doi: 10.1146/annurev.es.03.110172.001205
Hasselmann, M., Ferretti, L., and Zayed, A. (2015). Beyond fruit-flies: population genomic advances in non-Drosophila arthropods. Brief Funct. Genomics 14, 424–431. doi: 10.1093/bfgp/elv010
Herbers, J. M. (2013). 50 years on: the legacy of William Donald Hamilton. Biol. Lett. 9:20130792. doi: 10.1098/rsbl.2013.0792
Hoskins, R. A., Carlson, J. W., Wan, K. H., Park, S., Mendez, I., Galle, S. E., et al. (2015). The release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25, 445–458. doi: 10.1101/gr.185579.114
Hrdý, I., Novák, V. J. A., and Škrobal, D. (1960). “Influence of the queen inhibitory substance of honeybee on the development of supplementary reproductives in the termite Kalotermes flavicollis,” in The Ontogeny of Insects (Acta Symposii de Evolutione Insectorum, Prague, 1959), ed I. Hrdý (London: Academic Press), 172–174.
Hughes, C. (1998). Integrating molecular techniques with field methods in studies of social behavior: a revolution of results. Ecology 79, 383–399. doi: 10.1890/0012-9658(1998)079[0383:IMTWFM]2.0.CO;2
Jarosch, A., and Moritz, R. F. (2011). Systemic RNA-interference in the honeybee Apis mellifera: tissue dependent uptake of fluorescent siRNA after intra-abdominal application observed by laser-scanning microscopy. J. Insect Physiol. 57, 851–857. doi: 10.1016/j.jinsphys.2011.03.013
Jarosch, A., Stolle, E., Crewe, R. M., and Moritz, R. F. A. (2011). Alternative splicing of a single transcription factor drives selfish reproductive behavior in honeybee workers (Apis mellifera). Pro. Natl. Acad. Sci. U.S.A. 108, 15282–15287. doi: 10.1073/pnas.1109343108
Johnson, B. R., and Linksvayer, T. A. (2010). Deconstructing the superorganism: social physiology, groundplans, and sociogenomics. Q. Rev. Biol. 85, 57–79. doi: 10.1086/650290
Johnson, B. R., and Tsutsui, N. D. (2011). Taxonomically restricted genes are associated with the evolution of sociality in the honey bee. BMC Genomics 12:164. doi: 10.1186/1471-2164-12-164
Kamakura, M. (2011). Royalactin induces queen differentiation in honeybees. Nature 473, 478–483. doi: 10.1038/nature10093
Kapheim, K. M. (2016). Genomic sources of phenotypic novelty in the evolution of eusociality in insects. Curr. Opin. Insect Sci. 13, 24–32. doi: 10.1016/j.cois.2015.10.009
Kapheim, K. M., Pan, H., Li, C., Salzberg, S. L., Puiu, D., Magoc, T., et al. (2015). Genomic signatures of evolutionary transitions from solitary to group living. Science 348, 1139–1143. doi: 10.1126/science.aaa4788
Laissue, P. P., and Vosshall, L. B. (2008). The olfactory sensory map in Drosophila. Adv. Exp. Med. Biol. 628, 102–114. doi: 10.1007/978-0-387-78261-4_7
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Lattorff, H. M., and Moritz, R. F. (2013). Genetic underpinnings of division of labor in the honeybee (Apis mellifera). Trends Genet. 29, 641–648. doi: 10.1016/j.tig.2013.08.002
Lehmann, L., and Keller, L. (2006). The evolution of cooperation and altruism–a general framework and a classification of models. J. Evol. Biol. 19, 1365–1376. doi: 10.1111/j.1420-9101.2006.01119.x
Liao, X., Rong, S., and Queller, D. (2015). Relatedness, conflict, and the evolution of eusociality. PLoS Biol. 13:e1002098. doi: 10.1371/journal.pbio.1002098
Li-Byarlay, H., Li, Y., Stroud, H., Feng, S., Newman, T. C., Kaneda, M., et al. (2013). RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. U.S.A. 110, 12750–12755. doi: 10.1073/pnas.1310735110
Linksvayer, T. A. (2015). The molecular and evolutionary genetic implications of being truly social for the social insects. Adv. Insect Physiol. 48, 271–292. doi: 10.1016/bs.aiip.2014.12.003
Malone, L. A., and Pham-Delègue, M. H. (2001). Effects of transgene products on honey bees (Apis mellifera) and bumblebees (Bombus sp.). Apidologie 32, 287–304. doi: 10.1051/apido:2001130
Mikheyev, A. S., and Linksvayer, T. A. (2015). Genes associated with ant social behavior show distinct transcriptional and evolutionary patterns. eLife 4:e04775. doi: 10.7554/eLife.04775
Molodtsova, D., Harpur, B. A., Kent, C. F., Seevananthan, K., and Zayed, A. (2014). Pleiotropy constrains the evolution of protein but not regulatory sequences in a transcription regulatory network influencing complex social behaviors. Front. Genet. 5:431. doi: 10.3389/fgene.2014.00431
Mullen, E. K., Daley, M., Backx, A. G., and Thompson, G. J. (2014). Gene co-citation networks associated with worker sterility in honey bees. BMC Syst. Biol. 8:13. doi: 10.1186/1752-0509-8-38
Mullen, E. K., and Thompson, G. J. (2015). Understanding honey bee worker self-sacrifice: a conceptual–empirical framework. Adv. Insect Physiol. 48, 325–354. doi: 10.1016/bs.aiip.2014.12.002
Nayer, J. K. (1963). Effect of synthetic ‘queen substance’ (9-oxodec-trans-2-enoic acid) on ovary development of the house-fly, Musca domestica L. Nature 197, 923–924. doi: 10.1038/197923c0
Oi, C. A., van Zweden, J. S., Oliveira, R. C., Van Oystaeyen, A., Nascimento, F. S., and Wenseleers, T. (2015). The origin and evolution of social insect queen pheromones: novel hypotheses and outstanding problems. Bioessays 37, 808–821. doi: 10.1002/bies.201400180
Oliveira, R. C., Oi, C. A., do Nascimento, M. M. C., Vollet-Neto, A., Alves, D. A., Campos, M. C., et al. (2015). The origin and evolution of queen and fertility signals in corbiculate bees. BMC Evol. Biol. 15:254. doi: 10.1186/s12862-015-0509-8
Osborne, K. A. (1997). Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science 277, 834–836. doi: 10.1126/science.277.5327.834
Oxley, P. R., Hinhumpatch, P., Gloag, R., and Oldroyd, B. P. (2010). Genetic evaluation of a novel system for controlled mating of the honeybee, Apis mellifera. J. Hered. 101, 334–338. doi: 10.1093/jhered/esp112
Patalano, S., Vlasova, A., Wyatt, C., Ewels, P., Camara, F., Ferreira, P. G., et al. (2015). Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl. Acad. Sci. U.S.A. 112, 13970–13975. doi: 10.1073/pnas.1515937112
Peso, M., Elgar, M. A., and Barron, A. B. (2015). Pheromonal control: reconciling physiological mechanism with signalling theory. Biol. Rev. 90, 542–559. doi: 10.1111/brv.12123
Reaume, C. J., and Sokolowski, M. B. (2011). Conservation of gene function in behaviour. Philos. Trans. R Soc. Lond. B Biol. Sci. 366, 2100–2110. doi: 10.1098/rstb.2011.0028
Reeve, H. K. (2001). “In search of unified theories in sociobiology: help from social wasps,” in Model Systems in Behavioral Ecology: Integrating Conceptual, Theoretical and Empirical Approaches, ed L. A. Dugatkin (Princeton, NJ: Princeton University Press), 57–71.
Rehan, S. M., Berens, A. J., and Toth, A. L. (2014). At the brink of eusociality: transcriptomic correlates of worker behaviour in a small carpenter bee. BMC Evol. Biol. 14:260. doi: 10.1186/s12862-014-0260-6
Rehan, S. M., and Toth, A. L. (2015). Climbing the social ladder: the molecular evolution of sociality. Trends Ecol. Evol. 30, 426–433. doi: 10.1016/j.tree.2015.05.004
Ren, Y., and Hughes, K. A. (2014). Vitellogenin family gene expression does not increase Drosophila lifespan or fecundity. F1000Res. 3, 125. doi: 10.12688/f1000research.3975.1
Robertson, H., and Wanner, K. (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16, 1395–1403. doi: 10.1101/gr.5057506
Robinson, G. E. (2004). Beyond nature and nurture. Science 304, 397–399. doi: 10.1126/science.1095766
Robinson, G. E., and Ben-Shahar, Y. (2002). Social behavior and comparative genomics: new genes or new gene regulation? Genes Brain Behav. 1, 197–203. doi: 10.1034/j.1601-183X.2002.10401.x
Robinson, G. E., Grozinger, C. M., and Whitfield, C. W. (2005). Sociogenomics: social life in molecular terms. Nat. Rev. Genet. 6, 257–270. doi: 10.1038/nrg1575
Ronai, I., Oldroyd, B. P., Barton, D. A., Cabanes, G., Lim, J., and Vergoz, V. (2015). Anarchy is a molecular signature of worker sterility in the honey bee. Mol. Biol. Evol. 33, 134–142. doi: 10.1093/molbev/msv202
Rørth, P. (1998). Gal4 in the Drosophila female germline. Mech. Dev. 78, 113–118. doi: 10.1016/S0925-4773(98)00157-9
Sadd, B. M., Barribeau, S. M., Bloch, G., de Graaf, D. C., Dearden, P., Elsik, C. G., et al. (2015). The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16, 76. doi: 10.1186/s13059-015-0623-3
Sannasi, A. (1969). Inhibition of ovary development of the fruit-fly, Drosophila melanogaster by synthetic “queen substance.” Life Sci. 8, 785–789. doi: 10.1016/0024-3205(69)90016-2
Sannasi, A., and George, C. J. (1972). Termite queen substance: 9-oxodec-trans-2-enoic acid. Nature 237, 457–457. doi: 10.1038/237457a0
Schulte, C., Theilenberg, E., Müller-Borg, M., Gempe, T., and Beye, M. (2014). Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). Proc. Natl. Acad. Sci. U.S.A. 111, 9003–9008. doi: 10.1073/pnas.1402341111
Schwander, T., Lo, N., Beekman, M., Oldroyd, B. P., and Keller, L. (2010). Nature versus nurture in social insect caste differentiation. Trends Ecol. Evol. 25, 275–282. doi: 10.1016/j.tree.2009.12.001
Schwarz, M. P., Richards, M. H., and Danforth, B. N. (2007). Changing paradigms in insect social evolution: insights from halictine and allodapine bees. Annu. Rev. Entomol. 52, 127–150. doi: 10.1146/annurev.ento.51.110104.150950
Scott, J. G., Michel, K., Bartholomay, L. C., Siegfried, B. D., Hunter, W. B., Smagghe, G., et al. (2013). Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221. doi: 10.1016/j.jinsphys.2013.08.014
Simon, A. F., Chou, M. T., Salazar, E. D., Nicholson, T., Saini, N., Metchev, S., et al. (2012). A simple assay to study social behavior in Drosophila: measurement of social space within a group. Genes Brain Behav. 11, 243–252. doi: 10.1111/j.1601-183X.2011.00740.x
Smith, C. R., Toth, A. L., Suarez, A. V., and Robinson, G. E. (2008). Genetic and genomic analyses of the division of labour in insect societies. Nat. Rev. Genet. 9, 735–748. doi: 10.1038/nrg2429
Sobotka, J. A., Daley, M., Chandrasekaran, S., Rubin, B. D., and Thompson, G. J. (2016). Structure and function of gene regulatory networks associated with worker sterility in honeybees. Ecol. Evol. 6, 1692–1701. doi: 10.1002/ece3.1997
Sokolowski, M. B. (2001). Drosophila: genetics meets behaviour. Nat. Rev. Genet. 2, 879–890. doi: 10.1038/35098592
Steck, K., Veit, D., Grandy, R., Badia, S. B., Mathews, Z., Verschure, P., et al. (2012). A high-throughput behavioral paradigm for Drosophila olfaction - the flywalk. Sci. Rep. 2:361. doi: 10.1038/srep00361
Terrapon, N., Li, C., Robertson, H. M., Ji, L., Meng, X., Booth, W., et al. (2014). Molecular traces of alternative social organization in a termite genome. Nat. Commun. 5, 3636. doi: 10.1038/ncomms4636
Thompson, G. J., Hurd, P. L., and Crespi, B. J. (2013). Genes underlying altruism. Biol. Lett. 9:20130395. doi: 10.1098/rsbl.2013.0395
Thorne, B. L., and Traniello, J. F. (2003). Comparative social biology of basal taxa of ants and termites. Annu. Rev. Entomol. 48, 283–306. doi: 10.1146/annurev.ento.48.091801.112611
Tinette, S., Zhang, L., and Robichon, A. (2004). Cooperation between Drosophila flies in searching behavior. Genes Brain Behav. 3, 39–50. doi: 10.1046/j.1601-183x.2003.0046.x
Van Oystaeyen, A., Oliveira, R. C., Holman, L., van Zweden, J. S., Romero, C., Oi, C. A., et al. (2014). Conserved class of queen pheromones stops social insect workers from reproducing. Science 343, 287–290. doi: 10.1126/science.1244899
Villella, A., and Hall, J. C. (2008). Neurogenetics of courtship and mating in Drosophila. Adv. Genet. 62, 67–184. doi: 10.1016/S0065-2660(08)00603-2
West, S. A., and Gardner, A. (2013). Adaptation and inclusive fitness. Curr. Biol. 23, R577–R584. doi: 10.1016/j.cub.2013.05.031
West, S. A., Griffin, A. S., and Gardner, A. (2007). Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. J. Evol. Biol. 20, 415–432. doi: 10.1111/j.1420-9101.2006.01258.x
Wilder, E. L. (2000). “Ectopic expression in Drosophila,” in Developmental Biology Protocols, eds R. Tuan and C. Lo (Totowa, NJ: Humana Press), 9–14.
Wolf, M. J., and Rockman, H. A. (2008). Drosophila melanogaster as a model system for the genetics of postnatal cardiac function. Drug Discov. Today Dis. Models 5, 117–123. doi: 10.1016/j.ddmod.2009.02.002
Woodard, S. H., Fischman, B. J., Venkat, A., Hudson, M. E., Varala, K., Cameron, S. A., et al. (2011). Genes involved in convergent evolution of eusociality in bees. Proc. Natl. Acad. Sci. U.S.A. 108, 7472–7477. doi: 10.1073/pnas.1103457108
Wu, Q., Wen, T., Lee, G., Park, J. H., Cai, H. N., and Shen, P. (2003). Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 39, 147–161. doi: 10.1016/S0896-6273(03)00396-9
Wurm, Y. (2015). Arthropod genomics beyond fruit flies: bridging the gap between proximate and ultimate causation. Brief Funct. Genomics 14, 381–383. doi: 10.1093/bfgp/elv034
Yao, K. M., and White, K. (1994). Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. J. Neurochem. 63, 41–51. doi: 10.1046/j.1471-4159.1994.63010041.x
Keywords: insect sociobiology, functional genetics, Apis mellifera, mutations, GAL4-UAS
Citation: Camiletti AL and Thompson GJ (2016) Drosophila As a Genetically Tractable Model for Social Insect Behavior. Front. Ecol. Evol. 4:40. doi: 10.3389/fevo.2016.00040
Received: 01 January 2016; Accepted: 04 April 2016;
Published: 19 April 2016.
Edited by:
Heikki Helanterä, University of Helsinki, FinlandReviewed by:
Jan Oettler, University Regensburg, GermanyLuke Holman, University of Melbourne, Australia
Copyright © 2016 Camiletti and Thompson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alison L. Camiletti, YWNhbWlsZXRAdXdvLmNh
 Alison L. Camiletti
Alison L. Camiletti Graham J. Thompson
Graham J. Thompson