- 1Department of Biology, University of Pennsylvania, Philadelphia, PA, USA
- 2Department of Biology, Indiana University, Bloomington, IN, USA
Kin selection theory has always been explicitly genetic and has long been invoked to explain the evolution of the sterile worker caste in the social insects. However, sociogenomic studies of the evolution and genomic basis of social insect caste have been largely disconnected from kin selection theory and other related genetic theories of social evolution. Two previous population genetic models make testable predictions for patterns of sequence diversity for genes shaped indirectly by kin selection compared to genes shaped directly by natural selection, but there is some confusion in the literature regarding the predicted effects of kin selection and sex-limited expression on molecular evolution. We review the previous models and then use a simple parental effect model to clarify that the two factors, kin selection and sex-limited expression, are distinct and each has a separate effect on the expected patterns of molecular evolution. We further build on the previous models to show how categories of genes in social insect genomes with diverse combinations of fitness effects (direct, parental, sib, and offspring) and patterns of sex-limitation are predicted to evolve. We discuss how caste- and sex-specific transcriptomic profiling, coupled with population genomic data, can be used to identify different functional categories of genes and subsequently test whether observed patterns of molecular evolution fit theoretical predictions.
Introduction
Kin selection theory has always had an explicitly genetic focus. In first developing kin selection theory, Hamilton (1964) was motivated to explain how natural selection could favor an allele associated with altruism, which by definition entails a direct fitness cost to altruists expressing the allele as well as an indirect fitness benefit to social partners. Hamilton (1964) demonstrated that such an allele can spread when the beneficiaries of altruism also tend to bear the altruism allele, such as when altruists and beneficiaries are genetically related (see also Williams and Williams, 1957). Hamilton's Rule describes how the allele associated with altruism can experience a net positive fitness effect, and thus be adaptive, only when the indirect fitness benefit weighted by relatedness outweighs the direct fitness cost (Hamilton, 1964).
Kin selection theory has also had a special focus on explaining the evolution of eusociality, i.e., the evolution of sterile castes (Hamilton, 1964, 1972). Indeed, the evolution of a sterile worker caste provides the textbook example of the evolution of reproductive altruism by kin selection. Furthermore, the development and application of kin selection theory to social insect traits (e.g., caste, sex ratio) has been touted as providing some of the best testable predictions and quantitative support for these predictions in all of evolutionary biology (Queller and Strassmann, 1998; Abbot et al., 2011).
It would seem natural then for kin selection and related social evolution theory to be used to motivate and provide testable predictions for the explosion of recent transcriptomic and comparative genomic social insect studies of the evolution of caste and eusociality. Surprisingly, with relatively few exceptions (Hall and Goodisman, 2012; Thompson et al., 2013; Vojvodic et al., 2015; Ronai et al., 2016; Sobotka et al., 2016), this is not the case. The majority of sociogenomics research from the past decade has been divorced from and not motivated by kin selection theory or other related forms of established social evolution theory (e.g., indirect genetic effects theory, Moore et al., 1997; Wolf et al., 1998; Bijma et al., 2007; Bijma and Wade, 2008; Linksvayer and Wade, 2009; Mcglothlin et al., 2010). Instead, concepts originating in evolutionary developmental biology have motivated most sociogenomic studies (Toth and Robinson, 2007). For example, sociogenomic studies have often sought to test the hypothesis that highly conserved “gene toolkits” underlie social novelty in diverse lineages (e.g., Toth et al., 2010; Woodard et al., 2011; Berens et al., 2015; Morandin et al., 2016), or alternatively, that the evolution of social innovations involves rapidly evolving “novel genes” (Johnson and Tsutsui, 2011; Feldmeyer et al., 2014; Sumner, 2014; Jasper et al., 2015).
How can kin selection theory be touted as providing some of the best testable predictions of all evolutionary biology on the one hand, and yet, on the other hand, not be used by sociogenomic researchers studying the very genetic phenomena that initially spurred Hamilton (1964) to develop kin selection theory? This trend suggests to us that, either established theory is perceived to be inappropriate or inadequate for generating and testing predictions with sociogenomic datasets, or that its theoretical predictions are not yet testable with available datasets.
Here, we start by reviewing two previous population genetic models that build on kin selection theory/indirect genetic effects theory and make testable predictions for sociogenomic datasets, in particular for social insects (Linksvayer and Wade, 2009; Hall and Goodisman, 2012). While these studies have been described as providing conflicting results (Hall and Goodisman, 2012), we show that they are actually complementary. We use a simple parental effects, population genetic model to clarify that indirect fitness effects (i.e., kin selection) and sex-limited gene expression are distinct causal factors that influence expected patterns of molecular evolution. We extend the previous models of Linksvayer and Wade (2009) and Hall and Goodisman (2012) to show more generally how genes in the genomes of social insects (and more broadly, other social species) are expected on theoretical grounds to fit into several categories. The categories are the result of different patterns of gene expression and combinations of direct and indirect fitness effects, which determine the expected rates of molecular evolution for each category. Finally, we describe how empiricists can use RNA sequencing studies to categorize genes based on their expression patterns and inferred fitness effects, in order to test whether observed patterns of molecular evolution fit theoretical predictions. While we hope to also motivate further theory (and we briefly discuss promising routes), our target audience is mainly empiricists, because, as described above, we believe there is very strong potential for a tighter linkage between social evolution theory and sociogenomic data. The recent interface of population genetic models and genomic data for topics related to the evolution of sex-limited expression and sexual dimorphism provide a nice example of the promise of such a link between theory and genomic data (e.g., Connallon and Clark, 2010; Meisel et al., 2012; Griffin et al., 2013; Kirkpatrick and Guerrero, 2014; Wilkinson et al., 2015; Dapper and Wade, 2016).
Although, our results are general and applicable to loci with diverse fitness effects in diverse social organisms, we focus on the eusocial insects, since sociogenomic research in social insects continues to be a very rapidly growing field (e.g., Ferreira et al., 2013; Simola et al., 2013; Berens et al., 2015; Harpur et al., 2014; Roux et al., 2014; Jasper et al., 2015; Kapheim et al., 2015; Mikheyev and Linksvayer, 2015; Patalano et al., 2015; Smith et al., 2015; Vojvodic et al., 2015; Morandin et al., 2016).
Comparison of Two Previous Population Genetic Models of Molecular Evolution in Social Insect Genomes
Linksvayer and Wade (2009) sought to make sociogenomic predictions broadly applicable to species with diverse social effects, but focused their discussion on the social insects. Specifically, they extended previous maternal effects theory (Wade, 1998) to study how indirect fitness effects, resulting from genes expressed in relatives, are expected to affect rates of molecular evolution and the resulting patterns of sequence variation within and between populations. They modeled how the fitness of new reproductives (i.e., new queens and males) depends directly on genes expressed in these individuals, as well as indirectly on genes expressed in the mother (i.e., the queen) and in their siblings (i.e., workers).
Their main conclusion was that “all else being equal, selection acts less effectively on loci with indirect social effects on fitness than on loci with direct effects on fitness” (Linksvayer and Wade, 2009). This conclusion can be understood as a straightforward restatement of Hamilton's (1964) basic insight that indirect fitness effects must be weighted by relatedness, i.e., a factor between 0 and 1, when compared with direct fitness effects (note that the effect of relatedness on rates of evolution has also long been recognized in maternal effect quantitative genetic models; e.g., Dickerson, 1947; Willham, 1963; Cheverud, 1984; Lynch, 1987). Linksvayer and Wade (2009) mentioned in passing that some genes with direct fitness effects are likely only expressed in one sex, e.g., queen-expressed genes influencing egg-laying rate.
Hall and Goodisman (2012) sought to build on Linksvayer and Wade (2009) by focusing on the effects of female-limited expression, because reproductive and sterile females (i.e., queens and workers) are the main actors in hymenopteran insect societies. Specifically, they modeled how the queen's own genotype (“queen effects”) and the genotype of her offspring workers (“worker effects”) affect the colony-level output of new queens and males that are offspring of the queen and her mate(s) (following Linksvayer and Wade, 2009; Hall and Goodisman, 2012 considered colonies with a single queen and one to multiple mates). They ascribed this fitness to the queen and her mate(s).
Despite the seeming close overlap of the two models and their results, Hall and Goodisman (2012) make a point of emphasizing that their results are “in sharp contrast” (p. 2081) to the results of Linksvayer and Wade (2009), explaining further: “This result is in contrast to the primary conclusion of Linksvayer and Wade (2009), who noted that selection is less efficient for loci with indirect social effects regardless of queen mate number. The difference between our results and theirs is caused by our assumption of sex-limited selection” (Hall and Goodisman, 2012, p. 2086). This statement begs the question: do the results of Hall and Goodisman (2012) refute the main conclusion of Linksvayer and Wade (2009) that genes shaped by indirect selection experience relaxed adaptive molecular evolution compared to genes shaped by direct selection? Or, is it correct to state that all-else-equal, indirect selection is always expected to be effectively weaker than direct selection (consistent with previous authors, including Hamilton, 1964), but, in some cases, the effects of sex-limited gene expression can combine with the effects of indirect selection, so that some categories of genes experiencing kin selection can show similar expected rates of molecular evolution as some categories of genes experiencing direct, albeit sex-limited, selection?
In the next section, we use a simple parental effects model to demonstrate how sex-limited expression and indirect fitness effects (i.e., kin selection) act separately as distinct causal factors to shape patterns of molecular evolution. This model and the subsequent models illustrate that the original main conclusion of Linksvayer and Wade (2009) is general, but indeed, other causal factors such as sex-limitation or other forms of conditional expression are also expected to shape patterns of molecular evolution (Whitlock and Wade, 1995; Van Dyken and Wade, 2010). But first, we explain further how the models of Linksvayer and Wade (2009) and Hall and Goodisman (2012) can be seen to overlap and to be complementary to each other. After presenting the parental effects model, we develop a more general model that includes and goes beyond all of the effects considered by the two previous models.
The two models apply to two different but overlapping forms of selection, both likely to be important for social insects (Figure 1). Linksvayer and Wade (2009) modeled how the fitness of new queens and males depends on direct, maternal, and sib effects. This situation can be considered as a form of viability selection on the traits of new queens and males. For example, genes expressed in developing larvae, the mother queen, and sibling workers may affect caste fate or the viability, and subsequent fecundity of new queens and males. In contrast, Hall and Goodisman (2012) modeled how the fitness of a mature queen and her mate(s) depend on queen- and worker-expressed genes. Here, “queen effects” are direct effects of genes expressed in the queen's genome on queen fecundity, and “worker effects” are most clearly understood as indirect effects of genes expressed in the workers' genomes on their parents' fecundity. These genes have offspring effects (as opposed to sib, parental, or direct effects). This situation of direct and offspring effects can be considered as a form of fecundity selection on traits related to the fecundity of mature queens. For example, genes expressed in a mature queen and her offspring workers that influence queen egg-laying rate naturally fit these categories of direct and offspring fitness effects, respectively. As illustrated in Figure 1, the traits and fitnesses of all individuals in insect societies are likely influenced by all of these types of effects. Indeed, more generally, genes expressed in all species, in particular those living in family-based groups, can have diverse fitness effects, including direct, parental, sibling, and offspring effects (Lynch, 1987; Linksvayer and Wade, 2005; Linksvayer, 2006; Drown and Wade, 2014).
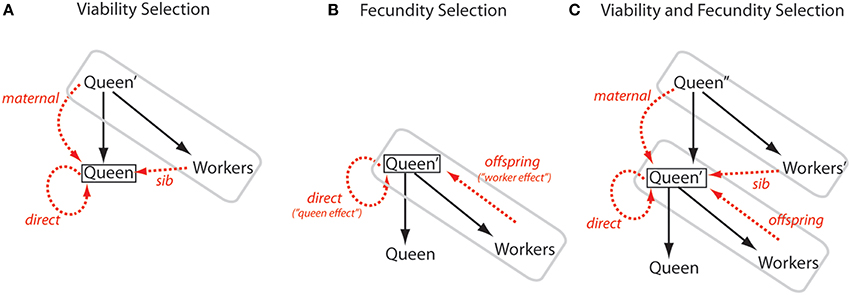
Figure 1. Diagram of alternate ways to model the direct and indirect fitness effects in social insect colonies. The queen and her offspring workers that make up a colony unit are enclosed by a gray box; solid black arrows represent direct reproduction of a queen to produce new queens or new workers; a black box encloses the focal queen whose fitness is being considered; dashed red arrows represent direct and indirect effects on the focal queen's fitness; and individuals from the previous one or two generations are marked with a single or double quotation marks, respectively. Each model considers the fitness of either a reproductively active queen with an established colony (i.e., within a gray box), or the fitness of a new queen. Males are excluded for clarity. (A) Viability Selection. Linksvayer and Wade (2009) modeled how a queen's fitness can depend directly on its own genes (direct arrow) and indirectly on its mother's genes (maternal arrow) and its sib's genes (sib arrow). Such selection could act on the traits of virgin or mated queens that affect their survival, or could act on traits that later affect queen fecundity. (B) Fecundity Selection. The fitness of established queens may primarily depend on colony-level production of new queens and males. This output may depend mainly on the queen's own genes (direct arrow) and on her worker offspring's genes (offspring arrow). Hall and Goodisman's (2012) model can be understood to be such a case, where the direct effect is called a “queen effect” and the indirect offspring effect is called a “worker effect.” This pattern of selection can shape queen traits related to fecundity but could also affect traits relating to the survival of established queens. (C) Viability and Fecundity Selection. Realistically, both viability and fecundity selection shape queen traits, and all types of direct and indirect effects can shape the traits of queens and other members of insect societies. In all cases, the change in allele frequency at a locus with a single type of fitness effect will depend on the relatedness between the individual expressing the locus and the individual that expresses its fitness effects (here the focal queen).
As a side note to readers attempting to compare the details of the two models: Without careful inspection, it may be unclear how the results of Hall and Goodisman (2012) (e.g., their Table 2) are related to the results of Linksvayer and Wade (2009), their later steps, and our models below. Both models start with tables describing the link between genotype and fitness (e.g., their Table 1; note the use of q vs. p to describe the frequency of the alternate allele), but Hall and Goodisman (2012) take some extra steps before calculating the expected per generation change in the frequency of a new allele with a fitness effect relative to the wildtype. As they explain, they present the leading eigenvalue, λ, of the stability matrix for the introduction of the new allele, and its approximation, instead of directly presenting the Δp equations (see their Table 2), where Δp ~ (λ−1)p. Subsequently, like Linksvayer and Wade (2009), they calculate fixation probabilities of each category of genes in their model. Hall and Goodisman (2012) then go one further step to calculate the adaptive substitution rate, which is Δp, weighted in proportion to the number of new adaptive alleles that arise per generation, 2 and 3/2 Nμ, for diploids and haplodiploids, respectively. In our models below, we use the traditional approach of directly calculating Δp equations based on the relationship between genotype and fitness, and then assume weak selection (Charlesworth and Charlesworth, 2010, p. 90–91), because such calculations are more transparent (see Supplementary Material) and also produce the same result.
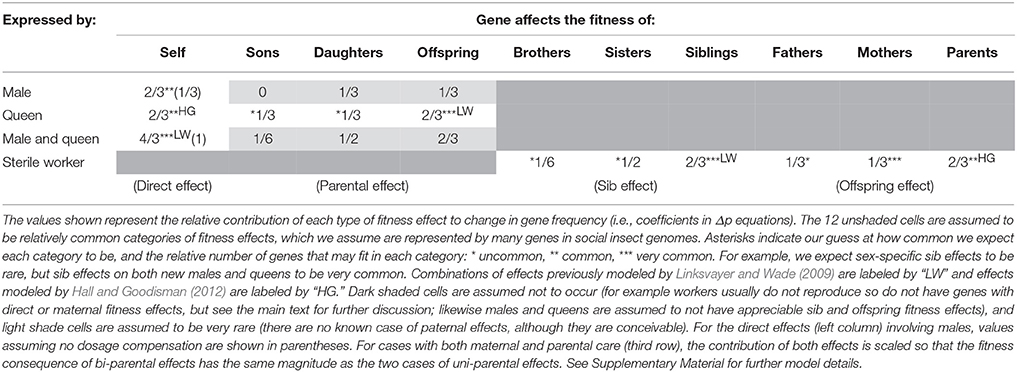
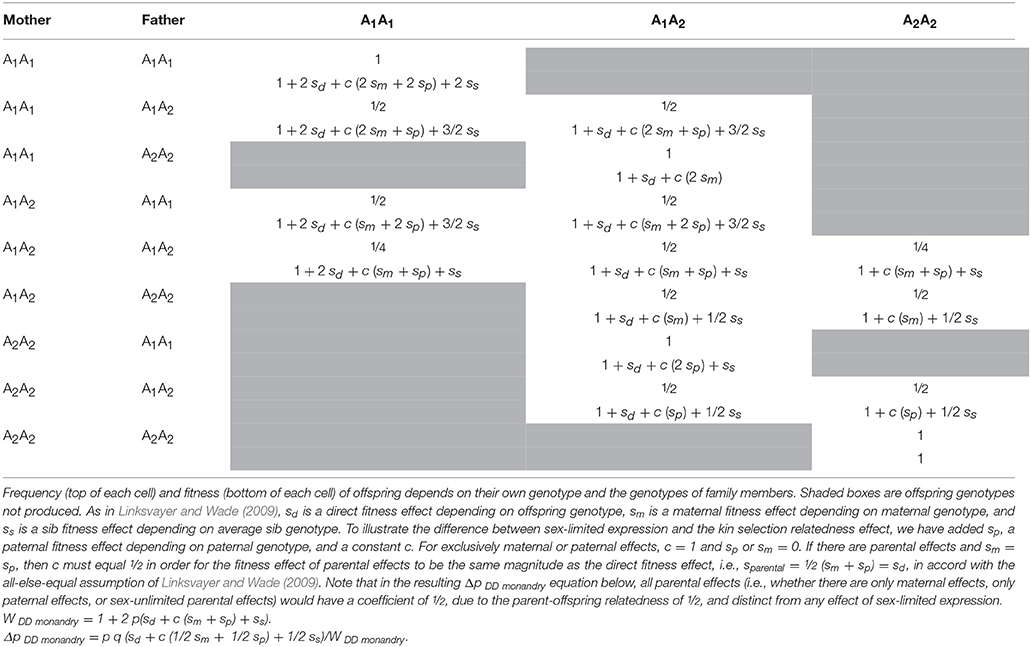
Table 2. In a haplodiploid colony with a single, singly-mated queen, the relative effective strength of selection experienced by genes with each combination of fitness effect and sex-limitation.
Parental Effect Model Clarifies the Distinct Effects of Kin Selection and Sex-Limitation
First consider a single diploid locus with a sex-limited direct effect, sd♀, on female fitness (note, here we assume diploidy for simplicity, but for the rest of the paper we model haplodiploid loci). This gene is expected to experience half the effective strength of selection as another direct effect locus with a similar magnitude of fitness effect that is expressed by both sexes (Whitlock and Wade, 1995). Half of the gene copies, those in males, are not expressed and therefore have no fitness effects for natural selection to act upon. Mean fitness, W, under this model is the average of the mean fitnesses across the two sexes, 1 in males and 1 + 2sd♀p in females, for an average of 1 + sd♀p. The gene frequency change, Δp, with this model equals (sdpq/2W), where sd♀is the direct fitness effect on females depending on their genotype, and p and q are the frequencies of the A1 and wild type alleles, respectively.
Now imagine a maternal effect locus that is expressed by mothers and indirectly influences offspring fitness regardless of offspring genotype by an amount sm for each A1 allele carried by the mother (Wade, 1998). This too can be considered to be a gene whose expression is limited to females (i.e., moms), but its evolution is different. Mean fitness, W, under this model is 1 + 2smpA1. Note that here there are twice as many deaths for the same gene frequency as there are in the female-limited direct fitness case, but here the deaths are less effective at changing gene frequency, because the individuals dying are not dying because of the expression of their own genes but rather owing to the effects of expression of their mother's genes. The gene frequency change, ΔpA1, with this model equals (smpq/2W) (Wade, 1998). Although both models have the selection coefficient discounted by the factor of ½, the reason for the discounting is different for each model. In the female-limited direct fitness case, females die because of their genotype, while males of the same genotype enjoy normal fitness. The factor of ½ appears because of averaging across the two sexes. In the maternal fitness effect case, the factor of ½ appears because offspring die as a result of mother's genotype and the regression from offspring death to maternal genotype is ½. That is, the maternal effect locus indirectly affects offspring fitness and the parent-offspring relatedness coefficient is ½. More generally, whenever a locus expressed in one individual affects its relative's fitness (e.g., a sib effect locus), the effective strength of selection experienced by the locus depends on the relatedness between social partners (Linksvayer and Wade, 2009).
To further illustrate that the effects of sex-limited expression and kin selection are distinct, consider the case of a parental effect locus that is expressed in both parents and indirectly influences offspring fitness. As shown in Table 1, the parental effect locus will still experience half the effective strength of selection compared to a direct effect locus that is expressed in both sexes and has the same magnitude of fitness effect. More precisely, in practice, the effect of one or both parents' expressing the parental effect on offspring fitness will depend on specific life history assumptions in the model (Table 1). If the magnitude of the fitness effect on offspring is not reduced when the parental effect locus is expressed by only one sex, for example if single parents can provide as much care as both parents together, then the expected effective strength of selection operating on the maternal effect locus will still be half that of the direct effect locus. If, on the other hand, sex-limited expression at the parental effect locus means that the magnitude of the indirect effect on offspring fitness is half as strong as when the parental effect locus is expressed by both parents, the effective strength of selection experienced by the parental effect locus will be one fourth that of the direct effect locus (i.e., ½ * ½). The first halving is caused by kin selection and the second by sex-limited expression.
Note that certain factors differentially affect sex-limited direct effect loci and kin selection loci with indirect fitness effects, further illustrating that kin selection and sex limitation are distinct. For example, sex ratio affects expected allele frequency change for sex-limited direct effect loci but not for maternal effect loci. In contrast, inbreeding has a larger effect on allele frequency change for parental effect loci (inbreeding acts to increase parent-offspring relatedness) compared to direct effect loci (Wade, 2000; Wolf and Wade, 2016).
These simple cases illustrate that the effects of sex-limited expression and indirect fitness effects are distinct and can be additive, depending on the underlying biology. These models also illustrate that it is possible to formulate models where selection experienced by direct effect loci and indirect loci are or appear equivalent because of additional differences in sex-limited expression (e.g., when the direct effect locus is sex-limited and the indirect effect locus is expressed in both sexes) (Hall and Goodisman, 2012). As we discuss below, such formulation is likely relevant to specific loci and biological scenarios, however ultimately it can obscure the fact that the causal factors underlying selection and gene frequency change are distinct.
We note that, in addition to sex-limited expression described in detail above, genes with indirect fitness effects may also have sex-limited fitness effects. For example, regardless whether a parentally-expressed locus has sex-limited expression (e.g., only maternal or both maternal and paternal), its fitness effects may be sex-limited (e.g., only affecting the fitness of daughters/sons, rather than all offspring regardless of sex). Thus, genes with indirect effects can have sex-limited expression as well as sex-limited fitness effects. In contrast, the effects of sex-limited expression and fitness effects are exactly overlapping for direct effect genes because the same individual expresses the gene as well as its fitness effects.
A More Complete Model of Molecular Evolution for Genes with Diverse Fitness Effects and Patterns of Sex-Limitation
The model of Linksvayer and Wade (2009) explicitly considers three distinct types of genes with distinct fitness effects (direct, maternal, and sib effects) on the fitness of new males and new queens, under conditions of monandry and polyandry. The model of Hall and Goodisman (2012) similarly explicitly considers two different types of genes with distinct fitness effects: queen-specific direct fitness effects, and offspring effects on the fitness of queens and her mate(s). Altogether, these combine to five distinct categories of genes. There are many more conceivable types of genes with distinct patterns of fitness effects and sex-limitation, for example, genes with male-specific direct effects, queen-specific offspring effects, sex-specific maternal effects, or sex-specific sib effects.
Following the approach of Linksvayer and Wade (2009), we modeled all possible combinations of direct, parental, sib, and offspring fitness effects with all types of sex-limitation. We calculated the per generation change in allele frequency (Δp) associated with each fitness effect (see Supplementary Material). Tables 2, 3 shows the resulting coefficients for each fitness effect in the Δp equations associated with each combination of fitness effect and sex limitation, for monandrous and polyandrous colonies, respectively. These coefficients reflect the effective strength of selection, and relative rates of molecular evolution, experienced by genes with these particular effects. We show 12 different categories of effects (and corresponding categories of genes), marked with at least one asterisk, likely to play roles in the biology of social insects. In particular, we have highlighted seven categories, marked with two or three asterisks, that we think are likely to be especially common, and together likely represent the majority of genes in social insect genomes.
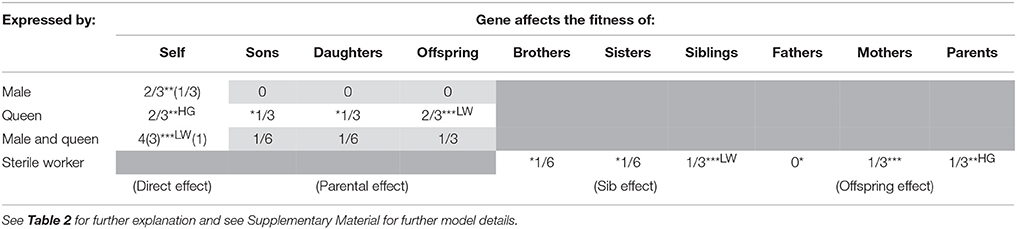
Table 3. In a haplodiploid colony with a single, multiply-mated queen, the relative effective strength of selection experienced by genes with each combination of fitness effect and sex-limitation.
Social insect adult workers are expected to simultaneously affect the fitness of their mother, through offspring effects (Hall and Goodisman, 2012), and the fitness of their younger reproductive siblings (new males and queens), through sib effects (Linksvayer and Wade, 2009). For example, by collecting food and defending the colony, workers increase both the survival and fecundity of their mother as well as the likelihood of survival and future fecundity of their younger siblings. However, inspection of Tables 2, 3 reveals that allele frequency change due to sib effects and offspring effects are different, unless considering average effects across both sexes (this may be why Hall and Goodisman, 2012 stated in passing that they could have equivalently modeled their offspring effects as sib effects). This makes sense, because for haplodiploids, offspring-parent relatedness is not always the same as relatedness between sibs. Consider two specific cases likely to be very common: worker genes that affect the fitness of new queens (a sib effect) and worker genes that affect the fitness of the mother queen (an offspring effect). The Δp coefficients for these fitness effects in monandrous colonies are 1/2 and 1/3, respectively (Table 2). The relative fitness effect of worker genes on their mother queen vs. sibs will determine which coefficient more strongly shapes rates of molecular evolution for these genes. For example with monandry, compared to worker genes with both sib and offspring effects, female-limited direct effect genes (coefficient 2/3), are expected to experience selection that is 1.3–2 times as effective, leading to stronger purifying selection and higher rates of adaptive divergence for these “queen genes” compared to “worker genes.” For a detailed discussion of the subtle issue of when fitness should be ascribed to parents vs. offspring, see Wolf and Wade (2001).
Besides providing predictions for worker-associated vs. queen-associated genes, how can the theoretical predictions presented in Tables 2, 3 be useful to empiricists? One way is by examining the relative ranking of gene categories to identify which types of genes, all-else-equal, are predicted to experience the effectively strongest to most relaxed selection: Genes with direct effects on both males and queens are expected to experience the strongest effective selection (Tables 2, 3). With complete dosage compensation, genes with male-specific and queen-specific direct effects are expected to experience similar strengths of effective selection (Whitlock and Wade, 1995), although with no dosage compensation (as modeled by Linksvayer and Wade, 2009), genes with male-specific direct effects will experience relaxed selection relative to genes with queen-specific direct effects (Tables 2, 3). Next in rank are several categories: genes with maternal effects on both sons and daughters, genes with sib effects on both brothers and sisters, and genes with offspring effects on both mothers and fathers. Genes expected to experience the most relaxed selection are those with sex-limited indirect fitness effects, in particular worker-expressed genes that only affect the fitness of brothers are expected to experience the lowest effective selection for monandrous colonies (Table 2). Only male-expressed genes with son-specific fitness effects are predicted to experience more relaxed selection (i.e., a 0 coefficient for this fitness effect, indicating that these genes would not experience selection). This coefficient is 0 because haplodiploid fathers and sons are unrelated (haploidploid males do not have fathers), so that male-specific paternal effects cannot be shaped by kin selection. However, it is difficult to imagine a paternal effect gene with male-specific effects in the first place. For polyandrous species, worker-expressed genes that only affect their father's fitness also would not experience kin selection (0 coefficient; Table 3), because with a large number of mates, the average worker in the colony is unrelated to any particular male parent.
Note that we did not model worker-expressed genes with direct fitness effects, because workers are usually assumed to be functionally sterile (Tables 2, 3). But in fact, workers in the large majority of haplodiploid social insect species can lay unfertilized, male-destined eggs in some situations (e.g., after the death of their mother queen). (Note that for species with workers that lay haploid eggs, the coefficients for direct effects and maternal effects on sons are the same as for queens). Genes exclusively expressed by workers in these species may then have direct fitness effects, and may be more strongly shaped by direct selection than kin selection. There are only a handful of social insect lineages (i.e., ant genera such as Solenopsis, Monomorium, Wasmannia) where workers are obligately sterile (Hölldobler and Wilson, 1990). These ant genera present particularly strong systems to study patterns of molecular evolution for genes expected to be shaped by direct versus kin selection, because genes exclusively expressed by obligately sterile workers can only be shaped by kin selection.
Inspection of Tables 2, 3 can also help explain why Hall and Goodisman (2012) rejected Linksvayer and Wade's (2009) general conclusion that genes with indirect fitness effects are expected to experience relaxed selection compared to genes with direct fitness effects. Hall and Goodisman (2012) modeled classes of direct and indirect effect genes that happen to have equivalent expected rates of molecular evolution (worker genes with offspring effects on both their parents, and queen genes with direct fitness effects). Inspection of Tables 2, 3 illustrates that more generally, genes with indirect fitness effects do experience relaxed selection compared to genes with direct fitness effects. When they do not, it is simply because the degree of relaxation of selection due to relatedness is the same as the degree of relaxation due to sex-limitation.
Promising Empirical Approaches to Test Predicted Patterns of Molecular Evolution in Social Insect Genomes
While it may be tempting to treat the two previous models (and also the more general model detailed above) as alternative hypotheses with distinct predictions, we do not recommend this approach because the two models are relevant to different categories of genes in social insect genomes. As illustrated in Tables 2, 3, there are a variety of types of combinations of fitness effects and patterns of sex limitation, putatively representing categories of genes that are likely to exist in social insect genomes. We recommend that empiricists seek to identify genes with distinct patterns of sex- and caste-specific expression that correspond to the categories in Tables 2, 3. Declining costs of RNA sequencing mean that extensive information about the context of expression across life stages, sexes, and castes should help to categorize genes within a genome as being associated with sex-limited, stage-limited, caste-limited, etc. expression. For example, genes that are significantly upregulated in worker tissues, queen tissues, or male tissues, are likely to have mainly sib/offspring, direct/maternal, and direct fitness effects, respectively. Determining whether, for example queen-associated genes mainly have direct fitness effects or maternal fitness effects will require further study of time- and tissue-specific gene expression patterns. For example, it is possible to identify sets of worker-associated genes that are functionally linked to sib care behaviors and have indirect effects on larval development (Linksvayer, 2015; Vojvodic et al., 2015). After identifying categories of genes with different combinations of sex-limitation and direct and indirect fitness effects, the models described above can be used as predictions for relative rates of molecular evolution.
We note that all of the models described above assume that the magnitude of selection associated with each type of fitness effect is the same (i.e., the all-else-equal assumption described in detail by Linksvayer and Wade, 2009). That is, for example, the strength of selection on worker traits and queen traits is assumed to be the same. The models describe how the effective strength of selection, and corresponding rate of molecular evolution, experienced by genes in each category vary. This variation in effective strength of selection experienced by genes results from differences in relatedness among social partners that modulate the effective strength of kin selection, and patterns of sex-limitation that modulate the proportion of individuals (and genomes) in the population that experience selection. When this all-else-equal assumption does not hold, all bets are off. For example, if, as a result of the natural history of a particular ant species, worker-associated traits have a much stronger effect on colony productivity than queen-associated traits, the underlying worker-associated genes may experience elevated rates of molecular evolution relative to the queen-associated genes, despite the fact that worker genes experience kin selection and queen genes experience direct selection. That is, the effective weakening of selection experienced by loci with indirect fitness effects can be offset (or further exacerbated) by the different magnitudes of selection. Thus, the patterns shown in Tables 2, 3 can be understood as null expectations in the absence of other forces. As the categories of identified genes become smaller, mean rates of molecular evolution are more likely to be influenced by unique characteristics of individual genes, and not fit the all-else equal assumption of the models. Of course, in contrast, when categories become too large, they will just represent the average genome attributes.
All of the models described above assume that genes more-or-less have a single type of fitness consequence. As described in Linksvayer and Wade (2009), genes are more realistically likely to be pleiotropic, and this may be especially true in social insect genomes (Linksvayer, 2015; Mullen and Thompson, 2015). For example, while studies looking for differentially-expressed genes between queens and workers at a single life history stage (e.g., adults) often find hundreds or even thousands of genes, studies that consider multiple life history stages find fewer (Morandin et al., 2015; Smith et al., 2015). Moreover, very few genes seem to be exclusively expressed in only queens or workers (Smith et al., 2015). Future empirical studies will have to clarify exactly what level of differential expression is functionally important. Given these caveats, it is perhaps surprising that empirical studies consistently find evidence for different rates of molecular evolution for genes that are significantly upregulated in for example workers, vs. queens.
Most past studies have relied on comparing relative rates of divergence between species at non-synonymous and synonymous sites (dN/dS). As discussed in detail by Helanterä and Uller (2014), such divergence can reflect a variety of processes, including relaxed selective constraint (i.e., relaxed purifying selection) or elevated positive selection. Differentiating between these alternatives is difficult (Helanterä and Uller, 2014). Furthermore, in some cases, relaxed selection within populations could lead to increased segregating variation, which could later lead to elevated rates of divergence among populations associated with local adaptation (Linksvayer and Wade, 2009). Population genomic datasets should have more potential to partition adaptive molecular evolution (i.e., positive selection) from signatures of relaxed selective constraint (i.e., purifying selection). For example, using McDonald-Kreitman type tests (Mcdonald and Kreitman, 1991), it is possible to differentiate between genes experiencing positive versus negative selection, and it is possible to estimate the proportion of genes experiencing adaptive molecular divergence (Smith and Eyre-Walker, 2002; Stoletzki and Eyre-Walker, 2011).
Interestingly, some recent studies indicate that worker-biased genes may experience elevated rates of molecular evolution relative to queen-biased genes (e.g., Feldmeyer et al., 2014; Harpur et al., 2014; Vojvodic et al., 2015). This could be due to relaxed purifying selection (which fits the predictions described above) or elevated positive selection (which does not fit the predictions described above). The Harpur et al. (2014) honey bee study uses a population genomic dataset with McDonald-Kreitman type tests to suggest higher rates of adaptive divergence (i.e., elevated positive selection) for worker-associated genes (see also Vojvodic et al., 2015). In some species, worker traits may simply be experiencing much stronger selection than queen traits, so that even though worker traits evolve predominantly through indirect selection, worker-associated genes may experience similar or even higher rates of adaptive molecular evolution. For species such as honey bees, it may often be the case that worker traits experience much stronger selection than queen traits. Honey bee queens are always accompanied by a large number of workers (i.e., dependent colony foundation; Winston, 1987), so that selection on many queen traits (except traits such as developmental rate) may be relaxed relative to queen traits in species with independent colony foundation.
In addition to comparing rates of molecular evolution between different categories of genes within a genome, it is also possible to try to study how rates of molecular evolution differ for lineages with different mating systems or social structures that could affect relatedness patterns and hence the relative strength of direct vs. kin selection. For example, Hall and Goodisman (2012) compared available estimates of molecular evolution between genes upregulated in adult queens versus adult workers for the honey bee Apis mellifera, which has multiple mating, and the fire ant Solenopsis invicta, which has single mating, under the hypothesis that the relative strength of kin selection is reduced with multiple mating. However, tremendous life history differences between these lineages, including dependent versus independent colony foundation, likely strongly differentially shapes patterns of selection for queen and worker traits in these lineages. These life history differences may swamp out any effects of direct versus indirect selection on observed patterns of molecular evolution. Future comparative studies should sample more extensively and use imethods accounting for phylogeny to try to control for the effects of variation in life history on patterns of molecular evolution (e.g., Morandin et al., 2016).
Conclusions and Future Directions
The general lack of interface between established social evolution theory and recent sociogenomic empirical studies is somewhat surprising given the degree to which kin selection theory in particular has been embraced by social insect biologists in the past. This is likely in part due to a lack of detailed information available about the functional roles that particular genes play. As described above, emerging RNA sequencing data sets can be used to identify genes with putative direct and indirect fitness effects, for example first by identifying genes associated with queen vs. worker development and function. A perceived lack of simple testable predictions may well also be a problem. To help clear up this issue, we have further developed our previous model to provide testable predictions for relative rates of molecular evolution for genes with different patterns of direct and indirect fitness effects and sex-limitation. We also reiterate our main previous prediction (Linksvayer and Wade, 2009): all-else-equal, genes with indirect vs. direct selection should experience relaxed adaptive molecular evolution. This can be understood as a null expectation for genomic patterns of molecular evolution in species with social fitness effects. For example, when sets of genes upregulated in obligately sterile workers have higher rates of adaptive molecular evolution compared to sets of genes upregulated in queens, something else must be going on. In this case, the all-else-equal assumption is likely incorrect, and selection on worker traits may be stronger, overcoming the dampening relatedness effect. Overall, we encourage researchers to use emerging sociogenomic datasets to interface with longstanding social evolution models that explicitly model and consider the special genetic and evolutionary features of social traits.
Promising future directions for further developing theory include incorporating life history details, together with direct and indirect selection to models of trait evolution and resulting molecular evolution. Additionally, recent studies indicate that genes associated with social traits may exist in distinct gene regulatory contexts (Molodtsova et al., 2014; Jasper et al., 2015; Mikheyev and Linksvayer, 2015; Morandin et al., 2016) and models explicitly considering the evolution of the components of gene regulatory networks involved in social interactions can lead to further testable predictions for genes associated with social traits relative to the rest of the genome (Akcay et al., 2015).
Author Contributions
TL and MW developed the model and wrote the paper.
Funding
TL was funded by the NSF award IOS-1452520.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Alexander Mikheyev for providing comments on an earlier version of the manuscript, and to Luke Holman and the two reviewers for comments that helped improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2016.00065
References
Abbot, P., Abe, J., Alcock, J., Alizon, S., Alpedrinha, J. A. C., Andersson, M., et al. (2011). Inclusive fitness theory and eusociality. Nature 471, E1–E4. doi: 10.1038/nature09831
Akcay, E., Linksvayer, T. A., and Van Cleve, J. (2015). Bridging social evolution theory and emerging empirical approaches to social behavior. Curr. Opin. Behav. Sci. 6, 59–64. doi: 10.1016/j.cobeha.2015.09.002
Berens, A. J., Hunt, J. H., and Toth, A. L. (2015). Comparative transcriptomics of convergent evolution: different genes but conserved pathways underlie caste phenotypes across lineagse of eusocial insects. Mol. Biol. Evol. 32, 690–703. doi: 10.1093/molbev/msu330
Bijma, P., Muir, W. A., and Van Arendonk, J. A. M. (2007). Multilevel selection 1: quantitative genetics of inheritance and response to selection. Genetics 175, 277–288. doi: 10.1534/genetics.106.062711
Bijma, P., and Wade, M. J. (2008). The joint effects of kin, multilevel selection and indirect genetic effects on response to genetic selection. J. Evol. Biol. 21, 1175–1188. doi: 10.1111/j.1420-9101.2008.01550.x
Charlesworth, B., and Charlesworth, D. (2010). Elements of Evolutionary Genetics. Greenwood Village, CO: Roberts and Company Publishers.
Cheverud, J. M. (1984). Evolution by kin selection: a quantitative genetic model illustrated by maternal performance in mice. Evolution 38, 766–777. doi: 10.2307/2408388
Connallon, T., and Clark, A. G. (2010). Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution 64, 3417–3442. doi: 10.1111/j.1558-5646.2010.01136.x
Dapper, A. L., and Wade, M. J. (2016). The evolution of sperm competition genes: the effect of mating system on levels of genetic variation within and between species. Evolution 70, 502–511. doi: 10.1111/evo.12848
Dickerson, G. E. (1947). Composition of hog carcasses as influenced by heritable differences in rate and economy of gain. Iowa Agric. Exp. Station Res. Bull. 354, 489–524.
Drown, D. M., and Wade, M. J. (2014). Runaway coevolution: adaptation to heritable and nonheritable environments. Evolution 68, 3039–3046. doi: 10.1111/evo.12470
Feldmeyer, B., Elsner, D., and Foitzik, S. (2014). Gene expression patterns associated with caste and reproductive status in ants: worker-specific genes are more derived than queen-specific ones. Mol. Ecol. 23, 151–161. doi: 10.1111/mec.12490
Ferreira, P. G., Patalano, S., Chauhan, R., Ffrench-Constant, R., Gabaldon, T., Guigo, R., et al. (2013). Transcriptome analyses of primitively eusocial wasps reveal novel insights into the evolution of sociality and the origin of alternative phenotypes. Genome Biol. 14:R20. doi: 10.1186/gb-2013-14-2-r20
Griffin, R. M., Dean, R., Grace, J. L., Rydén, P., and Friberg, U. (2013). The shared genome is a pervasive constraint on the evolution of sex-biased gene expression. Mol. Biol. Evol. 30, 2168–2176. doi: 10.1093/molbev/mst121
Hall, D. W., and Goodisman, M. A. (2012). The effects of kin selection on rates of molecular evolution in social insects. Evolution 66, 2080–2093. doi: 10.1111/j.1558-5646.2012.01602.x
Hamilton, W. D. (1964). The genetical evolution of social behaviour, I. J. Theor. Biol. 7, 1–16. doi: 10.1016/0022-5193(64)90038-4
Hamilton, W. D. (1972). Altruism and related phenomena, mainly in social insects. Annu. Rev. Ecol. Syst. 3, 193–232. doi: 10.1146/annurev.es.03.110172.001205
Harpur, B. A., Kent, C. F., Molodtsova, D., Lebon, J. M. D., Alqarni, A. S., Owayss, A. A., et al. (2014). Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc. Natl. Acad. Sci. U.S.A. 111, 2614–2619. doi: 10.1073/pnas.1315506111
Helanterä, H., and Uller, T. (2014). Neutral and adaptive explanations for an association between caste-biased gene expression and rate of sequence evolution. Front. Genet. 5:297. doi: 10.3389/fgene.2014.00297
Jasper, W. C., Linksvayer, T. A., Atallah, J., Friedman, D., Chiu, J. C., and Johnson, B. R. (2015). Large scale coding sequence change underlies the evolution of post-developmental novelty in honey bees. Mol. Biol. Evol. 32, 334–346. doi: 10.1093/molbev/msu292
Johnson, B. R., and Tsutsui, N. D. (2011). Taxonomically restricted genes are associated with the evolution of sociality in the honey bee. BMC Genomics 12:164. doi: 10.1186/1471-2164-12-164
Kapheim, K. M., Pan, H., Li, C., Salzberg, S. L., Puiu, D., Magoc, T., et al. (2015). Genomic signatures of evolutionary transitions from solitary to group living. Science 348, 1139–1143. doi: 10.1126/science.aaa4788
Kirkpatrick, M., and Guerrero, R. F. (2014). Signatures of sex-antagonistic selection on recombining sex chromosomes. Genetics 197, 531–541. doi: 10.1534/genetics.113.156026
Linksvayer, T. A., and Wade, M. J. (2005). The evolutionary origin and elaboration of sociality in the aculeate Hymenoptera: maternal effects, sib-social effects, and heterochrony. Q. Rev. Biol. 80, 317–336. doi: 10.1086/432266
Linksvayer, T. A., and Wade, M. J. (2009). Genes with social effects are expected to harbor more sequence variation within and between species. Evolution 63, 1685–1696. doi: 10.1111/j.1558-5646.2009.00670.x
Linksvayer, T. A. (2006). Direct, maternal, and sibsocial genetic effects on individual and colony traits in an ant. Evolution 60, 2552–2561. doi: 10.1554/06-011.1
Linksvayer, T. A. (2015). The molecular and evolutionary genetic implications of being truly social for the social insects. Adv. Insect Phys. 48, 271–292. doi: 10.1016/bs.aiip.2014.12.003
Lynch, M. (1987). Evolution of intrafamilial interactions. Proc. Natl. Acad. Sci. U.S.A. 84, 8507–8511. doi: 10.1073/pnas.84.23.8507
Mcdonald, J. H., and Kreitman, M. (1991). Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654. doi: 10.1038/351652a0
Mcglothlin, J. W., Moore, A. J., Wolf, J. B., and Brodie, E. D. (2010). Interacting phenotypes and the evolutionary process. III. Social evolution. Evolution 64, 2558–2574. doi: 10.1111/j.1558-5646.2010.01012.x
Meisel, R. P., Malone, J. H., and Clark, A. G. (2012). Faster-X evolution of gene expression in Drosophila. PLoS Genet. 8:e1003013. doi: 10.1371/journal.pgen.1003013
Mikheyev, A. S., and Linksvayer, T. A. (2015). Genes associated with ant social behavior show distinct transcriptional and evolutionary patterns. Elife 4:e04775. doi: 10.7554/eLife.04775
Molodtsova, D., Harpur, B. A., Kent, C. F., Seevananthan, K., and Zayed, A. (2014). Pleiotropy constrains the evolution of protein but not regulatory sequences in a transcription regulatory network influencing complex social behaviors. Front Genet. 5:431. doi: 10.3389/fgene.2014.00431
Moore, A. J., Brodie, E. D. III, and Wolf, J. B. (1997). Interacting phenotypes and the evolutionary process.1. Direct and indirect genetic effects of social interactions. Evolution 51, 1352–1362. doi: 10.2307/2411187
Morandin, C., Dhaygude, K., Paviala, J., Trontti, K., Wheat, C., and Helanterä, H. (2015). Caste-biases in gene expression are specific to developmental stage in the ant Formica exsecta. J. Evol. Biol. 28, 1705–1718. doi: 10.1111/jeb.12691
Morandin, C., Tin, M. M. Y., Abril, S., Gómez, C., Pontieri, L., Schiøtt, M., et al. (2016). Comparative transcriptomics reveals the conserved building blocks involved in parallel evolution of diverse phenotypic traits in ants. Genome Biol. 17, 1–19. doi: 10.1186/s13059-016-0902-7
Mullen, E. K., and Thompson, G. J. (2015). Chapter ten-understanding honey bee worker self-sacrifice: a conceptual–empirical Framework. Adv. Insect Phys. 48, 325–354. doi: 10.1016/bs.aiip.2014.12.002
Patalano, S., Vlasova, A., Wyatt, C., Ewels, P., Camara, F., Ferreira, P. G., et al. (2015). Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl. Acad. Sci. U.S.A. 112, 13970–13975. doi: 10.1073/pnas.1515937112
Queller, D. C., and Strassmann, J. E. (1998). Kin selection and social insects. Bioscience 48, 165–175. doi: 10.2307/1313262
Ronai, I., Oldroyd, B. P., Barton, D. A., Cabanes, G., Lim, J., and Vergoz, V. (2016). Anarchy is a molecular signature of worker sterility in the honey bee. Mol. Biol. Evol. 33, 134–142. doi: 10.1093/molbev/msv202
Roux, J., Privman, E., Moretti, S., Daub, J. T., Robinson-Rechavi, M., and Keller, L. (2014). Patterns of positive selection in seven ant genomes. Mol. Biol. Evol. 31, 1661–1685. doi: 10.1093/molbev/msu141
Simola, D. F., Wissler, L., Donahue, G., Waterhouse, R. M., Helmkampf, M., and Roux, J. (2013). Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 23, 1235–1247. doi: 10.1101/gr.155408.113
Smith, C. R., Helms Cahan, S., Kemena, C., Brady, S. G., Yang, W., Bornberg-Bauer, E., et al. (2015). How do genomes create novel phenotypes? Insights from the loss of the worker caste in ant social parasites. Mol. Biol. Evol. 32, 2919–2931. doi: 10.1093/molbev/msv165
Smith, N. G., and Eyre-Walker, A. (2002). Adaptive protein evolution in Drosophila. Nature 415, 1022–1024. doi: 10.1038/4151022a
Sobotka, J. A., Daley, M., Chandrasekaran, S., Rubin, B. D., and Thompson, G. J. (2016). Structure and function of gene regulatory networks associated with worker sterility in honeybees. Ecol. Evol. 6, 1692–1701. doi: 10.1002/ece3.1997
Stoletzki, N., and Eyre-Walker, A. (2011). Estimation of the Neutrality Index. Mol. Biol. Evol. 28, 63–70. doi: 10.1093/molbev/msq249
Sumner, S. (2014). The importance of genomic novelty in social evolution. Mol. Ecol. 23, 26–28. doi: 10.1111/mec.12580
Thompson, G. J., Hurd, P. L., and Crespi, B. J. (2013). Genes underlying altruism. Biol. Lett. 9:20130395. doi: 10.1098/rsbl.2013.0395
Toth, A. L., and Robinson, G. E. (2007). Evo-devo and the evolution of social behavior. Trends in Genetics 23, 334–341. doi: 10.1016/j.tig.2007.05.001
Toth, A. L., Varala, K., Henshaw, M. T., Rodriguez-Zas, S. L., Hudson, M. E., and Robinson, G. E. (2010). Brain transcriptomic analysis in paper wasps identifies genes associated with behaviour across social insect lineages. Proc. R. Soci. B Biol. Sci. 277, 2139–2148. doi: 10.1098/rspb.2010.0090
Van Dyken, J. D., and Wade, M. J. (2010). The genetic signature of conditional expression. Genetics 184, 557–570. doi: 10.1534/genetics.109.110163
Vojvodic, S., Johnson, B. R., Harpur, B., Kent, C., Zayed, A., Anderson, K. E., et al. (2015). The transcriptomic and evolutionary signature of social interactions regulating honey bee caste development. Ecol. Evol. 5, 4759–4807. doi: 10.1002/ece3.1720
Wade, M. J. (1998). “The evolutionary genetics of maternal effects,” in Maternal Effects As Adaptations, eds T. A. Mousseau and C. W. Fox (New York, NY: Oxford University Press), 5–21.
Wade, M. J. (2000). Opposing levels of selection can cause neutrality: mating patterns and maternal-fetal interactions. Evolution 54, 290–292. doi: 10.1111/j.0014-3820.2000.tb00029.x
Whitlock, M. C., and Wade, M. J. (1995). Speciation: founder events and their effects on X-linked and autosomal genes. Am. Nat. 145, 676–685. doi: 10.1086/285762
Wilkinson, G. S., Breden, F., Mank, J. E., Ritchie, M. G., Higginson, A. D., Radwan, J., et al. (2015). The locus of sexual selection: moving sexual selection studies into the post-genomics era. J. Evol. Biol. 28, 739–755. doi: 10.1111/jeb.12621
Willham, R. L. (1963). The covariance between relatives for characters composed of components contributed by related individuals. Biometrics 19, 18–27. doi: 10.2307/2527570
Williams, G. C., and Williams, D. C. (1957). Natural selection of individually harmful social adaptations among sibs with special reference to social insects. Evolution, 11, 32–39. doi: 10.2307/2405809
Wolf, J. B., Brodie, E. D. III, Cheverud, J. M., Moore, A. J., and Wade, M. J. (1998). Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13, 64–69. doi: 10.1016/S0169-5347(97)01233-0
Wolf, J. B., and Wade, M. J. (2001). On the assignment of fitness to parents and offspring: whose fitness is it and when does it matter? J. Evol. Biol. 14, 347–356. doi: 10.1046/j.1420-9101.2001.00277.x
Wolf, J. B., and Wade, M. J. (2016). Evolutionary genetics of maternal effects. Evolution. 70, 827–839. doi: 10.1111/evo.12905
Keywords: maternal effects, eusociality, social evolution, indirect genetic effects, kin selection, sociogenomics
Citation: Linksvayer TA and Wade MJ (2016) Theoretical Predictions for Sociogenomic Data: The Effects of Kin Selection and Sex-Limited Expression on the Evolution of Social Insect Genomes. Front. Ecol. Evol. 4:65. doi: 10.3389/fevo.2016.00065
Received: 31 March 2016; Accepted: 27 May 2016;
Published: 10 June 2016.
Edited by:
Miriam H. Richards, Brock University, CanadaReviewed by:
Luke Holman, University of Melbourne, AustraliaDavid Queller, Washington University in St. Louis, USA
Copyright © 2016 Linksvayer and Wade. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Timothy A. Linksvayer, dGxpbmtzQHNhcy51cGVubi5lZHU=
 Timothy A. Linksvayer
Timothy A. Linksvayer Michael J. Wade
Michael J. Wade