- 1Institute of Natural and Mathematical Sciences, Massey University, Auckland, New Zealand
- 2Institute for Applied Ecology, University of Canberra, Canberra, ACT, Australia
Despite decades of research, empirical support for the “compatible genes” and “good genes” hypotheses as explanations for adaptive female extra-pair mating remains discordant. One largely un-tested theoretical prediction that could explain equivocal findings is that mating for compatible genes benefits should reduce selection for good genes. However, this prediction does not consider demographic parameters, such as social structuring, that can indirectly influence extra-pair paternity (EPP) outcomes. Drawing on evidence from a previous study, we re-evaluate this hypothesis whilst considering social structuring in a population of tui, Prosthemadera novaeseelandiae, a socially monogamous passerine. Previous research has found possible evidence for mating for good genes because male ornament size correlates with EPP success in this population. Here, we test whether non-random inbreeding of social-pairs indirectly provides the opportunity to gain compatibility benefits from EPP, and thus whether selection for compatible genes and good genes can operate simultaneously. We found that (1) social mates were more closely related than expected through random mating, (2) extra-pair males (EPMs) were less closely related to females than were the females' within-pair mates, and (3) genetically dissimilar males sired offspring with faster growth rates. These results demonstrate that females gain compatible genes benefits from EPP. However, contrary to the compatible genes hypothesis, females in highly-related pairs did not engage more frequently in extra-pair mating. Together with previous research investigating female mate choice in this population, our findings suggest that social pairings between relatives provide a pathway for females to gain compatible genes benefits whilst engaging in extra-pair mating for good genes. This study provides evidence that some fitness benefits from EPP arise automatically through non-random social mating rather than through direct selection on extra-pair mate choice. We argue that when variance in compatibility benefits is an outcome of population structuring, compatible genes benefits need not be gained at the expense of good genes benefits.
Introduction
The adaptive function of female multiple mating in socially monogamous species has been variously debated over the past few decades (Westneat et al., 1990; Birkhead and Møller, 1992; Westneat and Stewart, 2003), however so far no obvious conclusion has been reached (Wan et al., 2013). Proposed hypotheses include non-adaptive mechanisms, for example pleiotropic gene effects such as the outcome of selection on males (Forstmeier et al., 2011), or convenience polyandry where females submit to extra-pair copulations to avoid harassment (Forstmeier et al., 2014). Of the adaptive hypotheses proposed to explain female extra-pair mating behavior, the “good genes” and “compatible genes” hypotheses in which females gain indirect fitness benefits have been the most extensively examined (reviewed in Griffith et al., 2002; Neff and Pitcher, 2005; Akçay and Roughgarden, 2007; Griffith and Immler, 2009; Prokop et al., 2012).
At the genic level, the effects of good genes and compatible genes differ in their functional mechanism. Under selection for good genes, a male's genetic quality is additive and conveyed through genetic covariance with phenotypic indicator traits (Zahavi, 1975; Hamilton and Zuk, 1982; Kirkpatrick and Ryan, 1991; Andersson, 1994; Kirkpatrick, 1996; Kokko et al., 2002, 2003). Therefore, a trait indicating good genes benefits can be used as proxy for genetic quality, and hence for the heritable fitness benefits (in the form of either increased viability or attractiveness) that are conferred to offspring by the mate choice decisions of all individuals regardless of their own genotype. Under the good genes hypothesis, females are therefore predicted to allocate paternity toward males with extreme forms of the trait. In contrast, under the compatible genes hypothesis, not all females stand to derive the same benefits from mating with a particular male (Neff and Pitcher, 2005; Puurtinen et al., 2009). Instead, it is the compatibility of male and female genotypes that act, through either heterozygote overdominance or avoidance of the deleterious effects of inbreeding, to produce genetically superior offspring (Zeh and Zeh, 1996, 1997; Jennions, 1997; Tregenza and Wedell, 2000). Inbreeding depression is a consequence of intragenomic conflict caused by increased shared genetic similarity by common descent and lowered functional genome-wide heterozygosity (Coulson et al., 1998). These genetic consequences have the potential to influence fitness through their detrimental effects on offspring birth weights, growth, and survival (Keller and Waller, 2002). Thus, the compatible genes hypothesis predicts that paternity should be biased toward genetically dissimilar males.
Theory predicts that good genes and compatible genes benefits are not mutually exclusive and therefore that females should be selected to exploit both types of benefits in mate choice (Colegrave et al., 2002; Neff and Pitcher, 2005; Puurtinen et al., 2005, 2009). Yet few empirical studies have demonstrated simultaneous optimization of both types of benefit (but see Foerster et al., 2003). As a possible explanation, Colegrave et al. (2002) hypothesized that when selection for both good genes and compatible genes operates in a population, unless there are large numbers of males of equally high additive genetic quality available to all females, selection for compatibility will weaken directional sexual selection for good genes. It has therefore been argued that females cannot simultaneously select the most ornamented and most genetically compatible male, and that female optimization of both good genes and compatible genes benefits can only operate via plasticity in mate choice in relation to the relative fitness advantages of each strategy (e.g., Roberts and Gosling, 2003; Oh and Badyaev, 2006; Mays et al., 2008). This antagonistic mechanism of selection could explain the so far equivocal support for the good genes and compatible genes hypotheses (reviewed in Griffith et al., 2002; Mays and Hill, 2004; Akçay and Roughgarden, 2007; Mays et al., 2008; Forstmeier et al., 2014).
Because extra-pair paternity (EPP) is purported to be a mechanism for socially monogamous females to circumvent the negative impacts of social inbreeding (Birkhead and Møller, 1998), compatible genes theory predicts that females should allocate paternity to genetically dissimilar extra-pair males (EPMs; Pusey and Wolf, 1996; Keller and Waller, 2002; Wheelwright et al., 2006; Kempenaers, 2007). Although the compatible genes hypothesis is emerging as an important component of EPP through its influence on offspring viability (Blomqvist et al., 2002; Masters et al., 2003; Richardson et al., 2005; Dowling and Mulder, 2006; Freeman-Gallant et al., 2006; Cohas et al., 2007; Rubenstein, 2007; Brouwer et al., 2011; Arct et al., 2015) the exact selective mechanism through which it acts remains unclear (reviewed in Griffith and Immler, 2009). Potential mechanisms can arise through pre- or post-copulatory (i.e., cryptic female choice; Eberhard, 1991; Colegrave et al., 2002; Birkhead and Pizzari, 2002) mate choice, or through sperm selection in which the sperm of more compatible males are more likely to fertilize an egg (Wilson et al., 1997; Zeh and Zeh, 1997; Birkhead and Møller, 1998; Tregenza and Wedell, 2000). These processes may operate disparately in different animal species. For example, while sophisticated olfactory mechanisms present in mammals, as well as visual and acoustic learning in birds with cooperative breeding systems, commonly facilitate the kin recognition mechanism necessary for female choice based on genetic compatibility (e.g., Pusey and Wolf, 1996; Mateo, 2003; Boulet et al., 2009; Leclaire et al., 2013), the role of pre- or post-copulatory female choice for genetic compatibility in socially monogamous birds remains unconvincing because there is scarce empirical support for the recognition of unfamiliar kin mechanism required for such precise copulatory control (Jamieson et al., 2009; but see Arct et al., 2010).
However, patterns in EPP can also be indirectly affected by demographic parameters such as population social structure; although to date this has been rarely considered in the EPP literature (Reid et al., 2015). Arguably the most common example of population structuring occurs when individuals mate non-randomly with respect to relatedness. In several socially monogamous species, studies have shown that individuals pair with relatives more frequently than expected through random mating (e.g., Foerster et al., 2006; Szulkin et al., 2009; Brekke et al., 2012). If related social-pairs then engage in extra-pair mating, compatible genes benefits could be gained automatically without specific mate choice for more genetically compatible partners, simply because of the high likelihood that the EPM would be less-related to the female than the within-pair male (WPM).
Recently, Reid et al. (2015) demonstrated that female song sparrows, Melospiza melodia, paired to related social males can lower offspring coefficients of inbreeding simply by engaging in random extra-pair mating. This finding suggests that social structuring by itself has the potential to influence the fitness outcomes of EPP without the need for pre- or post-copulatory kin recognition mechanisms. However, Reid et al. (2015) did not include any sexually selected traits in their analyses. This information is important to consider because social structuring could also have potential consequences for the predicted opposing interaction of selection for good genes and compatible genes. For example, in a non-randomly inbreeding population, females engaging in EPP for good genes benefits also stand to gain compatibility benefits as an automatic outcome of extra-pair mating. Under these circumstances, antagonistic selection between good genes and compatible genes effects would be minimized. Therefore, social structuring could provide a pathway through which the benefits of compatible genes and good genes could operate simultaneously. We test this hypothesis by investigating population social structure and its effect on the fitness outcomes of EPP in a naturally occurring population of the New Zealand tui, Prosthemadera novaeseelandiae.
Tui are socially monogamous but have a very high rate of EPP (57% of all offspring in 72% of all broods), and therefore the potential exists for females to gain significant indirect benefits from extra-pair mating (Wells et al., 2015). A previous study on a population of this honeyeater (Meliphagidae), revealed that both within-pair and EPP success are positively related to the size of a male's white throat feather ornament, suggesting mating preference for good genes (Wells et al., 2015). However, mate choice for genetic compatibility could also be an important adaptation to avoid inbreeding in this species because natal philopatry is common in both sexes and is exacerbated by competition for breeding territories near nectar resources (Stewart, 1980; Bergquist, 1985; Stewart and Craig, 1985).
In this study, we use offspring growth rates to quantify potential fitness benefits for genetic compatibility gained from extra-pair mating in this same population of tui, and examine how social structure can influence these potential benefits by increasing the occurrence of non-random social mating. To investigate population social structure, we first determine whether social-pairs are more genetically related than expected through random mating. We then test whether within-pair and extra-pair genetic relatedness influences male paternity success in addition to male ornamentation. If EPP functions as an inbreeding avoidance mechanism, females paired to related social partners are predicted to engage in extra-pair copulations and EPP should be biased toward genetically dissimilar males. Finally we test for increased offspring performance as a predicted fitness outcome of mating with genetically dissimilar EPMs (Zeh and Zeh, 1996; Puurtinen et al., 2009). Because growth rates are an important component of offspring performance (Lindstrom, 1999) that are associated with non-additive genetic quality (Keller and Waller, 2002; Kempenaers, 2007; Pitcher and Neff, 2007; Dziminski et al., 2008; Rosivall et al., 2009), we specifically examine the effect of genetic relatedness of biological parents on offspring growth rates, and compare within-pair and extra-pair maternal half-siblings to determine whether genetically dissimilar males provide compatibility benefits to females.
Methods
Field Methods
This study was conducted on a naturally breeding population of tui at Tawharanui Regional Park (36°22′S, 174°50′E), on the North Island of New Zealand. The large and open nature of this population (~1,000 pairs; Wells, unpublished data) means that we were unable to sample all individuals in the population, and thus poses a potential issue with regards to missing data on pairwise relatedness and identification of potential extra-pair sires. To mitigate this problem, we focussed our sampling effort on two main areas of the park (Ecology Bush and Jones Bay) separated by 2.5 km. We captured 390 adult tui using mist-nets between the years 2009 and 2012. Birds were color-banded for individual identification and blood samples were taken for DNA analysis. During the breeding seasons (Oct-Feb) of 2009–2012 we searched for nests belonging to banded pairs. Social pair-bonds were assessed by observation of territorial defense around a nest, mate guarding, song matching between pairs, and nestling provisioning. During this period we also caught as many un-banded neighboring males as possible to enable identification of potential EPMs. Blood samples were collected from 152 nestlings from 53 broods; 11 broods in 2009, 23 in 2010, 15 in 2011, and four in 2012.
In 2010 and 2011 twenty broods (N = 56 nestlings) with known hatching dates were followed through to fledging in order to track nestling growth. Nestlings were weighed at three stages of the nestling phase; day 0–1, 4–6, and 9–11 (Figure 1). For eight nestlings from three nests, measurements from days 15 to 16 were also available and were included in the analysis. Sampling was conducted at the same time of day to reduce diurnal variation in nestling mass. Nestlings were ringed with elastic cotton bands on the first measurement to allow future identification. On the second measurement they were provisionally banded with a single plastic color band until they could be permanently color-banded on the third measurement. Blood samples for paternity analysis were taken during the second measurement. This is study was approved and carried out in accordance with the recommendations of the Auckland Council, the New Zealand Department of Conservation (Research permit #37088_FAU) and the Massey University Animal Ethics Committee. The protocol was approved by the Massey University Animal Ethics Committee (permit #MUAEC09-39).
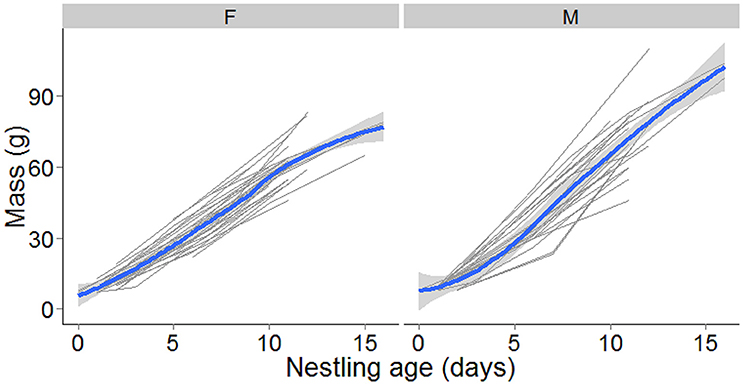
Figure 1. Raw data showing individual fledged nestlings mass in grams with nestling age for female (left plot, N = 26) and male (right plot, N = 22) nestlings. A locally weighted regression (lowess) line (in blue) with 95 % confidence interval shading has been added to show the overall trend with nestling age.
Paternity Analysis and Genetic Relatedness
Fourteen microsatellites were developed for the tui (Wells et al., 2013) and twelve of these were used to investigate paternity using the program CERVUS v.3.0.3 (Marshall et al., 1998; see Wells et al., 2015 for complete details of paternity analyses). Within-pair males (WPMs) were excluded as being the genetic father if they (a) mismatched at one or more loci within the mother-offspring dyad that could not be attributed to the presence of a low frequency null allele or (b) mismatched at three or more loci where null alleles could be present, but were unlikely due to their low frequencies, and if also (c) the WPM did not receive a positive LOD score (multilocus log-likelihood ratio score). Extra-pair males (EPMs) were assigned to a nestling only if they mismatched at zero loci within the mother-offspring dyad.
In order to calculate pairwise genetic relatedness estimates, the most appropriate estimator for the microsatellite dataset was chosen using COANCESTRY v.1.0.1.1 (Wang, 2011). This is because different marker-based relatedness estimators may perform better on different sets of loci due to variation between population composition and allele frequencies (van de Casteele et al., 2001). COANCESTRY estimates the performance of seven different relatedness estimators conditional on the degree of correlation of each estimator to simulated relatedness values based on allelic information from markers from the real dataset. TrioML, a maximum likelihood estimator (Wang, 2007) displayed the highest correlation to the true values (r = 0.96). Pairwise TrioML values for the Tawharanui population were subsequently calculated in COANCESTRY and used as an estimate of genetic relatedness in all further analyses. TrioML uses the genotype of a third individual as a reference in estimating the pairwise genetic relatedness between the two focal individuals, reducing the chance that genes identical in state are mistaken for genes that are identical by descent (IBD). This estimator calculates the probabilities of the IBD coefficients between two individuals X and Y which are then used to estimate relatedness according to the equation rXY = 2θXY where θXY is the coancestry coefficient defined by the IBD coefficients probabilities (Wang, 2007).
Pairwise TrioML values displayed a non-normal distribution with an overrepresentation of zero values (Figure S1). We therefore used a randomization procedure to investigate whether social pairings occur at random with respect to genetic relatedness. Two-tailed significance was approximated using 10,000 Monte Carlo resamplings. For each permutation the mean expected genetic relatedness was calculated by randomly selecting one male per female. This was repeated 10,000 times to give an approximated null distribution of relatedness values. The observed mean relatedness of social pairs was compared with this distribution and the null hypothesis of random mating was rejected if the observed mean relatedness was below or above the 2.5th and 97.5th percentiles of the null distribution respectively. Because the two areas of the park do not differ in their mean pairwise relatedness of either social pairs (randomization test: p = 0.66) or random male-female pairs within an area (randomization test: p = 0.15), the randomization procedure was performed for both areas together. However, only pairwise genetic relatedness values between females and their potential mates (i.e., males from the same area) were included in the analysis. We conducted this analysis for all years combined because (1) there is a high level of site philopatry across years in this population meaning that the pool of available males was temporally stable, (2) there is no significant difference in relatedness of social pairs between years (randomization test: p = 0.75, Figure S2), and (3) we had limited data on social pairings in some years. Because randomization tests assume independence between observations, if we had data on more than one pairing of a female (N = 21), we included data from only the first social pairing of that female in the analysis. To test the effect of eliminating these nests, and also to test whether a potential deviation from random mating occurs in only some of the observed years, we verified our results with all pairings using mixed-effects model analysis which can account for the non-independence of observations (Pinheiro and Bates, 2000). We examined the probability of male-female pairs from the same area being a social pair using a generalized linear mixed-effects model (GLMM) with a logit link function and binomial error distribution. We input as the binary response variable whether a pair was a social pair or not. Year and the pairwise genetic relatedness of the pair were included as fixed effects, and female identity as a random effect to control for the non-independence of observations. There is likely to have been some minor changes in individual composition of the population between years due to the migration of a small proportion of the breeding population. However, because tui exhibit significant natal and breeding site philopatry (Stewart, 1980; Bergquist, 1985) this was expected to affect only a small proportion of the sampled subpopulations. Therefore, for our analysis we considered that all males from the same area were available for each year.
Effect of Genetic Relatedness on Paternity Success
A previous analysis of this population found that paternity success is correlated to male ornamentation and body size (Wells et al., 2015). Here, we examine whether paternity success is also related to the genetic relatedness of the parents. GLMMs with a binomial error distribution and logit link function were used to test two measures of within-pair paternity success; the probability (as a binary response) of EPP occurring in a nest, and the proportion of extra-pair young (EPY) in a nest with brood size as the binomial denominator. To evaluate if WPMs that are closely related to the female are more likely to be cuckolded we included a predictor specifying the TrioML genetic relatedness value of the social pair. A random intercept for female identity was included to account for the non-independence of multiple broods from the same female (Pinheiro and Bates, 2000). Year was also included as a random effect to control for any annual variation in EPP (Figure S3).
To investigate whether EPP is biased toward EPMs that are genetically dissimilar to the female, we compared the genetic relatedness of females to their corresponding WPMs and EPMs in broods where both partners could be identified (N = 28). To account for some males being EPMs for more than one female and some broods having more than one EPM, a GLMM with a binomial error distribution and logit link function was used (Stiratelli et al., 1984; Anderson and Aitkin, 1985). The probability of a male being an EPM was modeled as the binary response variable (WPM = 0, EPM = 1), and the TrioML genetic relatedness value of the pair was included as a predictor. Female identity and year were included as random effects.
Effect of Parental Genetic Relatedness on Offspring Growth
To test the hypothesis that compatible genes benefits are gained from mate choice, we investigated offspring performance as a function of parental genetic compatibility. Specifically, we examined whether parental genetic relatedness predicted the pre-fledging offspring growth rates in mass. Nestling mass and tarsus length (i.e., skeletal size) are highly correlated in this population (Pearson's r = 0.98). Therefore, we only provide tests of offspring growth rates in mass. We chose mass rather than skeletal size as a measure of offspring growth because skeletal size is often heritable in birds (e.g., Alatalo and Lundberg, 1986), and therefore we posited that offspring skeletal size might simply be a function of parental size, rather than a genetic compatibility fitness proxy due to the relatedness of the parents. Only offspring for which we knew the identity of the true genetic father were included in the analysis. We also a priori only included offspring which survived to fledging (N = 38). Only fledged offspring were used because offspring that died before fledging includes nestlings with severely reduced growth rates (see Figure S4) that would have died due to causes such as differential nestling provisioning rates by parents when resources are short (i.e., brood reduction; Gebhardt-Henrich and Richner, 1998; e.g., Lack, 1947; Moreno-Rueda et al., 2007; Shizuka and Lyon, 2013), or differential male mortality in sexually size dimorphic species (e.g., Røskaft and Slagsvold, 1985). These factors are expected to have much more severe effects on growth rates (and hence on survival) than the subtle effects of parental relatedness. Thus, by including only fledglings in the analysis we expected that we would be more likely to detect any potential effect of relatedness on performance, free from additional confounding effects on survival. However, to verify the robustness of our results, we also performed all analyses investigating offspring growth on the full dataset of all offspring (N = 46), but also included a factor variable specifying whether or not an offspring survived to fledging (see Supplementary Material). This model specifically tests the effect of relatedness on all offspring whilst accounting for the reduced growth rates of non-fledged offspring.
To obtain a mean growth rate for each offspring we first used a linear mixed-effects model (LMM) to model offspring mass as a function of age (Table S1). To account for significant differences in mass due to sex we fitted sex-specific growth rates by including fixed effects for offspring age, offspring sex, and their interaction. We suppressed a random intercept for each nestling due to the low variation in hatching size, and over-parameterization identified by convergence errors when a random intercept was specified. We included random slopes for female identity, and for each nestling identity nested within female identity. These account for both multiple unbalanced repeated measures from each nestling, as well as non-independence of nestlings from the same female (Pinheiro and Bates, 2000). From this model, we extracted an individual nestling's residual mean growth rate from the model's random slope coefficients. These values were then used as the response variable in the LMM investigating the effect of parental relatedness on offspring growth rates. We included the genetic parents' relatedness values as a predictor and specified random intercepts for female identity and year.
Comparison of Within-Pair and Extra-Pair Young
If females gain genetic benefits from extra-pair mating, EPY should be fitter than within-pair young (WPY) in mixed paternity broods. To test this, we examined whether EPY possessed faster mass growth rates than their WPY maternal half-siblings. We fitted a LMM with offspring residual mean mass growth rates as the response variable and specified a factor variable indicating the nestling's extra-pair status (0 = WPY, 1 = EPY). We included a random intercept for female identity to compare maternal half-siblings (Stiratelli et al., 1984; Anderson and Aitkin, 1985). However, half (7 of 13) of the broods with EPY contained 100% EPP, leaving few mixed paternity broods to test this hypothesis. Therefore, we also conducted this analysis using data from all broods containing EPY (including nests with 100% EPP) and compared the results. Again, we also re-ran these models including all offspring and specifying a factor variable for whether an offspring fledged or not.
Due to our small sample sizes for some tests, we used the R package simr to conduct post-hoc power analyses for significant predictors to assess the power of our models to be able to detect the effect sizes that we observed. Tests were run for 1000 simulations. All analyses were conducted in R v.2.15.1 (R Development Core Team, 2011), using the lme4 package (Bates et al., 2015) for all mixed-model analyses. Significance for mixed-models was assessed using likelihood ratio tests (LRT) between nested models.
Results
Paternity Analysis
EPP was present in 72% (38 of 53) of all the broods and EPY accounted for 57% (87 of 152) of all offspring studied. Five out of the 28 broods in which the EPMs could be identified contained offspring from two EPMs. For the subset of nests that we had data on growth rates (N = 20), EPP occurred in 65% of all nests, and EPY accounted for 71% of all offspring.
Genetic Relatedness of Social Pairs
Pairwise relatedness between social pairs was often high, with 36% of all pairings being between close kin (equivalent to mating between first cousins or closer, r ≥ 0.125). Eight of the 13 pairs for which we had information on their pairings from multiple years maintained pair-bonds for at least 2 years. All four of the highly-related pairs for which we had information on their pairings from multiple years maintained pair-bonds for at least 2 years, and one of these was the only pair known to maintain a pair-bond for all 4 years of this study. Results from the mixed-model analysis indicate that social pairs are more closely related than expected by random mating (β = 4.517 ± 0.839, χ2 = 22.335, p < 0.001). The randomization test result demonstrated a similar outcome, however this analysis was not quite statistically significant (mean genetic relatedness of social pairs = 0.09 standard deviation (SD) 0.14, mean genetic relatedness of random pairs = 0.05 SD 0.09, p = 0.061). There was no significant difference in the mean genetic relatedness of pairs between years (p = 0.94).
Effect of Genetic Relatedness on Paternity Success
The genetic similarity of the social-pair did not significantly predict either the probability of a male being cuckolded (β = −0.447 ± 3.337, N = 35, χ2 = 0.017, p = 0.90), or the proportion of EPY in his brood (β = −2.000 ± 2.184, N = 35, χ2 = 0.748, p = 0.39). A comparison of WPMs and EPMs showed that EPMs were significantly less-related to the female than the WPMs they cuckolded (β = −3.051 ± 1.482, N = 28, χ2 = 4.567, p = 0.033, power = 52.5%). However, this relationship appears to be driven by the social pairings of close kin (Figure 2).
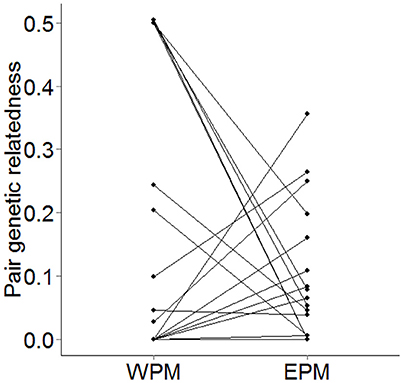
Figure 2. Paired scatterplot showing the pairwise genetic relatedness of WPMs and their corresponding EPMs to the female from each brood (N = 28). Where two EPMs within a brood occurred, the mean pairwise genetic relatedness of the two EPMs was plotted.
Effect of Parental Genetic Relatedness on Offspring Growth
When offspring mean mass growth rates were examined, less-related genetic parents produced offspring that exhibited significantly faster growth rates than more highly-related parents (β = −0.476 ± 0.234, N = 38, χ2 = 4.035, p = 0.044; Figure 3). The power for this effect size was 42.1%. When offspring that did not fledge, and a fixed effect taking into account the fact that non-fledged offspring have reduced growth rates, are included in the model, the effect of parental relatedness was highly significant (Table S2). As predicted, non-fledged offspring had significantly lower growth rates than fledged offspring (Table S2). We also tested the effect of relatedness whilst accounting for whether an offspring was a WPY or EPY by including an additional categorical variable specifying the nestling's extra-pair status (Table S3). Although we were limited by sample size for this test, both the relatedness predictor and the EP status predictor remained almost significant in the expected direction, and this test showed that while accounting for genetic relatedness of the parents, EPY tended to have faster growth rates than WPY (Table S3).
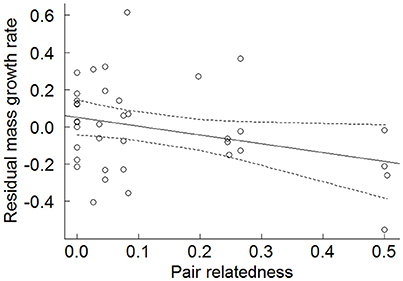
Figure 3. Effect of genetic relatedness of the biological parents on an offspring's residual mass growth rate (N = 38). Residual growth rates were extracted from the random slopes of a LMM of nestling mass on nestling age and represent the degree to which a certain nestling's growth rate is above or below the mean growth rate for that sex. Points are raw data and solid line is the fitted values output from a LMM with 95% confidence intervals (dashed lines).
Comparison of Within-Pair and Extra-Pair Young
EPY possessed significantly faster growth rates in mass (β = 0.173 ± 0.081, N = 13 nests, χ2 = 4.527, p = 0.033, power = 55.1%) than WPY. However, because 7 out of the 13 nests contained 100% EPP we consequently analyzed only mixed-paternity nests. Although this test lacked power (46.8%), EPY still tended to have faster growth rates than WPY (Figure 4, β = 0.106 ± 0.058, N = 6 nests, χ2 = 3.343, p = 0.067). Including all offspring in these models, with a variable specifying whether or not an offspring survived to fledging, did not change these results (Tables S4, S5).
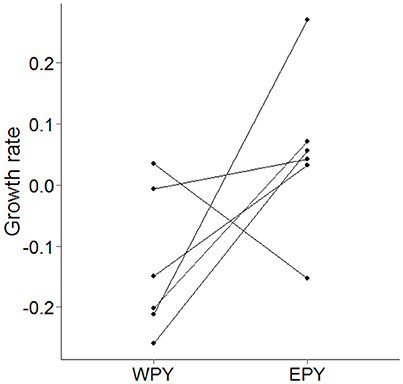
Figure 4. Difference in residual growth rates in mass between WPY and EPY maternal half-siblings in mixed paternity broods (N = 19 nestlings in 6 brood comparisons). Where multiple comparisons of half-siblings within broods existed, growth rates within each class have been averaged.
Discussion
Theory predicts that compatible genes selection should be greatest in populations where the cost of incompatibility is high (Pusey and Wolf, 1996; Colegrave et al., 2002; Michalczyk et al., 2011). This would occur, for example, in inbred populations where inbreeding depression can have detrimental effects on offspring performance (Keller and Waller, 2002). In our study, this prediction is borne out by several lines of evidence. First, social pairings between kin in tui are relatively common and occur more frequently than expected by chance, increasing the risk of inbreeding depression. Second, the offspring of genetically dissimilar biological parents had faster growth rates than the offspring of highly-related pairs, suggesting that inbreeding can reduce fitness under some circumstances. Third, EPMs were significantly less-related to the female than were the WPMs they cuckolded, EPY generally possessed faster growth rates than WPY, and EPY tended to have faster growth rates than their WPY half-siblings, which suggest that extra-pair mating helps females to circumvent the costs of social inbreeding. Together these findings provide empirical evidence that extra-pair mating in tui can provide compatible genes benefits.
The demonstrated adverse effect of inbreeding on offspring fitness in this study suggests that the high rate of EPP in tui could, at least in part, be an inbreeding avoidance mechanism that has evolved in response to significant natal philopatry (Birkhead and Møller, 1998; Foerster et al., 2006). Yet, we found no evidence that social inbreeding was compensated for by a facultative increase in frequency of EPP. Our findings are thus congruent with studies in other passerines in which females mated with genetically dissimilar EPMs and yet the genetic similarity of the social-pair did not predict the occurrence of EPP (Foerster et al., 2006; Suter et al., 2007; Brekke et al., 2012). Comparably, two of these studies also found some support for non-random social inbreeding (Foerster et al., 2006; Brekke et al., 2012). In our study, this was primarily driven by the social pairings of some highly-related individuals. We therefore argue that although female tui gain compatible genes benefits from EPP, this mating pattern is not a consequence of direct selection on pre- or post-copulatory female extra-pair mate choice (Fossøy et al., 2008) but is rather the automatic consequence of within-pair mating decisions. In support of this hypothesis, if highly-related pairs are removed from our analysis, extra-pair mating appears to be random with respect to genetic relatedness. Nevertheless, this mating pattern does not preclude that EPP in tui evolved as an inbreeding avoidance mechanism, because the social structuring of pairs with high genetic relatedness negates the requirement for female extra-pair mate choice based on genetic compatibility in order for EPP to be adaptive.
Indeed, mate choice based on relatedness would require the existence of sophisticated recognition of unfamiliar kin mechanisms. Although kin recognition by phenotype matching is common among mammals with refined olfactory mechanisms (e.g., Pusey and Wolf, 1996; Mateo, 2003; Boulet et al., 2009; Leclaire et al., 2013) and in cooperative breeders where visual or acoustic cues may be important in kin selection (Komdeur and Hatchwell, 1999; Hatchwell et al., 2001; Cornwallis et al., 2009; McDonald and Wright, 2011), few studies have definitively identified the methods through which socially monogamous birds can assess relatedness (Jamieson et al., 2009). Consequently, significant doubt still surrounds the ability of females to select mates based on genetic compatibility through either pre- or post-copulatory mechanisms (see Griffith, 2007 for a review). Instead, the EPP success of genetically dissimilar tui males likely derives from their superior post-copulatory fertilization success (Tregenza and Wedell, 2000; Griffith and Immler, 2009). Critically, this system does not require an active female preference or facultative manipulation of paternity. For example, under the “genetically loaded raffle” model (Ball and Parker, 2003) females may mate with many different males but only sperm from the most compatible males achieve fertilization (Wilson et al., 1997; Birkhead, 1998; Birkhead and Pizzari, 2002; Bernasconi et al., 2004; Griffith and Immler, 2009; Pryke et al., 2010). Foerster et al. (2006) similarly argued that sperm selection was responsible for the EPP success of genetically dissimilar blue tit, Cyanistes caeruleus, males. The tui system could be considered a specific form of the genetically loaded raffle driven by population social structure; if females are paired to highly-related social males, any chosen EPM should achieve significant paternity success because of his high likelihood of having more compatible sperm.
Previous studies have shown that population social structuring can influence rates of EPP in some species (e.g., Cohas and Allainé, 2009). However, the extent to which social structure influences the fitness outcomes of extra-pair mating has been little researched. To our knowledge, our study is the first to demonstrate that females increase offspring fitness by gaining compatible genes benefits from EPP simply as a result of population social structuring. Our findings are similar to those of a recent study (Reid et al., 2015) which found that female song sparrows, M. melodia, alter offspring inbreeding coefficients through random extra-pair mating due to their propensity to pair with related social partners more frequently than expected by random mating alone. These two studies demonstrate that population social structure can influence the fitness consequences of extra-pair mating without the overt need for an evolutionary mechanism based on kin recognition. However, Reid et al. (2015) did not include any aspect of sexually selected traits in their study, therefore it was unclear how selection for good genes might be affected. Although we do not include a measure of good genes selection here, this was the focus of another study on this tui population during the same time period (Wells et al., 2015). In contrast to the findings of Reid et al. (2015), the study found that extra-pair mating in this population of tui is not random: EPMs are more ornamented than their WPM counterparts, and males with smaller throat ornaments are more likely to be cuckolded (Wells et al., 2015). Theory predicts that females cannot gain good genes and compatible genes benefits from the same male (Mays et al., 2008), and that when these two mating strategies coexist in a population, selection for one benefit will weaken selection for the other (Colegrave et al., 2002). However, this may not be the case when compatibility benefits are the outcome of social structure, as we argue here. Most tui broods containing EPP are sired by only one EPM (Wells et al., 2015). Therefore, it is unlikely that female tui select different EPMs for good genes and compatible genes benefits, as has been demonstrated in blue tits (Foerster et al., 2003). Instead, any females paired to relatives that seek extra-pair copulations with more ornamented EPMs also stand to gain compatible genes benefits because of the higher likelihood that the chosen EPM will be more genetically dissimilar to her than the WPM. However, we stress that our finding that EPY have faster growth rates than WPY does not alone distinguish whether this is due to good genes or compatible genes benefits, or indeed whether it is a result of both processes, and examination of the fitness outcomes of good genes matings is needed to confirm this. Nevertheless, our findings highlight a potential pathway through which EPP in tui can provide the opportunity to simultaneously gain good genes and compatible genes benefits. Thus, population social structuring can facilitate a means by which selection for good genes and compatible genes can operate through EPP with little opposing selection.
The relatively stable pair-bonds and non-random social mating with respect to relatedness in tui then raises the question exactly what benefits, if any, being paired to a relative might provide (reviewed in Kokko and Ots, 2006)? One possibility is that inclusive fitness benefits gained from inbreeding and/or the costs of avoiding inbreeding may outweigh any cost of inbreeding depression (Lehmann and Perrin, 2003; Parker, 2006; Szulkin et al., 2013; Lehtonen and Kokko, 2015). Alternatively, individuals may not actively choose related social partners, yet inbreeding may provide females with some correlated benefit. For example, females may pair with relatives if local males are more likely than immigrants to procure high quality territories. This hypothesis is particularly relevant to tui, in which natal philopatry (Stewart, 1980; Bergquist, 1985; Stewart and Craig, 1985) exacerbated by social aggregations and competition for territories occurring at nectar resources at the beginning of the breeding season (Stewart, 1980), could render such a strategy adaptive.
Like most studies on a large, open population, our study has some limitations. First, our sample sizes, and hence the power of our tests, in this study are small, and should be replicated with a larger dataset. Second, due to the correlative nature of this study, we are unable to differentiate between the effects of relatedness itself on offspring fitness and between potential confounding factors such as age or condition. For example, individuals of low genetic quality or condition may experience limited breeding opportunities and may therefore be more likely to mate with relatives. Third, we cannot rule out the possibility that the greater fitness observed in the young of less related parents is the result of maternal effects. For example, females may preferentially invest more in extra-pair offspring that are deemed to be of greater reproductive value (Magrath et al., 2009; Tschirren et al., 2012). This is a possibility in our study because females choose EPMs based on phenotypic quality in this population. Finally, in including only fledged offspring in our tests we may have inadvertently created a bias, or reduced the power of our tests by reducing our sample size. However, the outcome of this was deemed to be preferable to potentially reporting results based on offspring that had extremely low growth rates that were unrelated to parental relatedness.
In conclusion, our study provides valuable insight into the effect that population social structure can have on the fitness outcomes of extra-pair mating in a large and naturally occurring population. Whether cryptic female choice or sperm selection is responsible for the demonstrated EPP success of genetically dissimilar males is outside the scope of the present study. Irrespective of the exact selective mechanism, our findings demonstrate that females gain compatible genes benefits from extra-pair mating, but that social inbreeding rather than active extra-pair mate choice is responsible. Moreover, in our study, females from highly-related pairs engaging in extra-pair mating for good genes benefits gained genetic compatibility benefits as an additional aside. Thus contrary to theoretical predictions (Colegrave et al., 2002; Mays et al., 2008) we argue that selection for compatible genes need not always weaken selection for good genes when genetic compatibility is an outcome of population social structuring. Such multiple genetic benefits could compensate for any costs of female extra-pair behavior and extra-pair mating could therefore be adaptive for females under such circumstances.
Author Contributions
SW and WJ conceived of and designed the study. SW and WJ collected field data. DG provided funding for molecular work. SW analyzed the data with input from DG and BJ. SW, JD, and WJ interpreted the data. SW wrote the manuscript with contributions from JD and WJ.
Funding
This study was funded by Landcare Research, New Zealand, and Auckland Council, New Zealand. SW was supported by a Commonwealth Doctoral Scholarship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ana Ramón-Laca, Diana Prada, and Robyn Howitt for advice on molecular work; many volunteers for assistance in the field; and Tim Lovegrove, Matt Maitland, and Morris Puckett at Auckland Council for logistical support in the field.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fevo.2017.00018/full#supplementary-material
References
Akçay, E., and Roughgarden, J. (2007). Extra-pair paternity in birds: review of the genetic benefits. Evol. Ecol. Res. 9, 855–868.
Alatalo, R. V., and Lundberg, A. (1986). Heritability and selection on tarsus length in the pied flycatcher (Ficedula hypoleuca). Evolution 40, 574–583. doi: 10.2307/2408578
Anderson, D. A., and Aitkin, M. (1985). Variance component models with binary response: interviewer variability. J. R. Stat. Soc. Ser. B 47, 203–210.
Arct, A., Drobniak, S. M., and Cichoń, M. (2015). Genetic similarity between mates predicts extrapair paternity—a meta-analysis of bird studies. Behav. Ecol. 26, 959–968. doi: 10.1093/beheco/arv004
Arct, A., Rutkowska, J., Martyka, R., Drobniak, S. M., and Cichoń, M. (2010). Kin recognition and adjustment of reproductive effort in zebra finches. Biol. Lett. 6, 762–764. doi: 10.1098/rsbl.2010.0417
Ball, M., and Parker, G. (2003). Sperm competition games: sperm selection by females. J. Theor. Biol. 224, 27–42. doi: 10.1016/S0022-5193(03)00118-8
Bates, D., Mächler, M., Bolker, B. M., and Walker, S. C. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01
Bergquist, C. A. (1985). Movements of groups of tui (Prosthemadera novaeseelandiae) in winter and settlement of juvenile tui in summer. N. Z. J. Zool. 12, 569–571. doi: 10.1080/03014223.1985.10428305
Bernasconi, G., Ashman, T.-L., Birkhead, T., Bishop, J., Grossniklaus, U., Kubli, E., et al. (2004). Evolutionary ecology of the prezygotic stage. Science 303, 971–975. doi: 10.1126/science.1092180
Birkhead, T. R. (1998). Cryptic female choice: criteria for establishing female sperm choice. Evolution 52, 1212–1218. doi: 10.2307/2411251
Birkhead, T. R., and Møller, A. P. (1992). Sperm Competition in Birds: Evolutionary Causes and Consequences. London: Academic Press.
Birkhead, T. R., and Møller, A. P. (1998). Sperm Competition and Sexual Selection. London: Academic Press.
Birkhead, T. R., and Pizzari, T. (2002). Postcopulatory sexual selection. Nat. Rev. Genet. 3, 262–273. doi: 10.1038/nrg774
Blomqvist, D., Andersson, M. B., Küpper, C., Cuthill, I. C., Kis, J., Lanctot, R. B., et al. (2002). Genetic similarity between mates and extra-pair parentage in three species of shorebirds. Nature 419, 613–615. doi: 10.1038/nature01104
Boulet, M., Charpentier, M. J., and Drea, C. M. (2009). Decoding an olfactory mechanism of kin recognition and inbreeding avoidance in a primate. BMC Evol. Biol. 9:281. doi: 10.1186/1471-2148-9-281
Brekke, P., Wang, J., Bennett, P. M., Cassey, P., Dawson, D. A., Horsburgh, G. J., et al. (2012). Postcopulatory mechanisms of inbreeding avoidance in the island endemic hihi (Notiomystis cincta). Behav. Ecol. 23, 278–284. doi: 10.1093/beheco/arr183
Brouwer, L., van de Pol, M., Atema, E., and Cockburn, A. (2011). Strategic promiscuity helps avoid inbreeding at multiple levels in a cooperative breeder where both sexes are philopatric. Mol. Ecol. 20, 4796–4807. doi: 10.1111/j.1365-294X.2011.05325.x
Cohas, A., and Allainé, D. (2009). Social structure influences extra-pair paternity in socially monogamous mammals. Biol. Lett. 5, 313–316. doi: 10.1098/rsbl.2008.0760
Cohas, A., Yoccoz, N. G., and Allaine, D. (2007). Extra-pair paternity in alpine marmots, Marmota marmota: genetic quality and genetic diversity effects. Behav. Ecol. Sociobiol. 61, 1081–1092. doi: 10.1007/s00265-006-0341-7
Colegrave, N., Kotiaho, J. S., and Tomkins, J. L. (2002). Mate choice or polyandry: reconciling genetic compatibility and good genes sexual selection. Evol. Ecol. Res. 4, 911–917.
Cornwallis, C. K., West, S. A., and Griffin, A. S. (2009). Routes to indirect fitness in cooperatively breeding vertebrates: kin discrimination and limited dispersal. J. Evol. Biol. 22, 2445–2457. doi: 10.1111/j.1420-9101.2009.01853.x
Coulson, T. N., Pemberton, J. M., Albon, S. D., Beaumont, M., Marshall, T. C., Slate, J., et al. (1998). Microsatellites reveal heterosis in red deer. Proc. R. Soc. B Biol. Sci. 265, 489–495. doi: 10.1098/rspb.1998.0321
Dowling, D. K., and Mulder, R. A. (2006). Combined influence of maternal and paternal quality on sex allocation in red-capped robins. J. Evol. Biol. 19, 440–449. doi: 10.1111/j.1420-9101.2005.01017.x
Dziminski, M. A., Roberts, J. D., and Simmons, L. W. (2008). Fitness consequences of parental compatibility in the frog Crinia georgiana. Evolution 62, 879–886. doi: 10.1111/j.1558-5646.2008.00328.x
Eberhard, W. G. (1991). Copulatory courtship and cryptic female choice in insects. Biol. Rev. 66, 1–31. doi: 10.1111/j.1469-185X.1991.tb01133.x
Foerster, K., Delhey, K., Johnsen, A., Lifjeld, J. T., and Kempenaers, B. (2003). Females increase offspring heterozygosity and fitness through extra-pair matings. Nature 425, 714–717. doi: 10.1038/nature01969
Foerster, K., Valcu, M., Johnsen, A., and Kempenaers, B. (2006). A spatial genetic structure and effects of relatedness on mate choice in a wild bird population. Mol. Ecol. 15, 4555–4567. doi: 10.1111/j.1365-294X.2006.03091.x
Forstmeier, W., Martin, K., Bolund, E., Schielzeth, H., and Kempenaers, B. (2011). Female extrapair mating behavior can evolve via indirect selection on males. Proc. Natl. Acad. Sci. U.S.A. 108, 10608–10613. doi: 10.1073/pnas.1103195108
Forstmeier, W., Nakagawa, S., Griffith, S. C., and Kempenaers, B. (2014). Female extra-pair mating: adaptation or genetic constraint. Trends Ecol. Evol. 29, 456–464. doi: 10.1016/j.tree.2014.05.005
Fossøy, F., Johnsen, A., and Lifjeld, J. T. (2008). Multiple genetic benefits of female promiscuity in a socially monogamous passerine. Evolution 62, 145–156. doi: 10.1111/j.1558-5646.2007.00284.x
Freeman-Gallant, C. R., Wheelwright, N. T., Meiklejohn, K. E., and Sollecito, S. V. (2006). Genetic similarity, extrapair paternity, and offspring quality in Savannah sparrows (Passerculus sandwichensis). Behav. Ecol. 17, 952–958. doi: 10.1093/beheco/arl031
Gebhardt-Henrich, S., and Richner, H. (1998). “Causes of growth variation and its consequences for fitness,” in Avian Growth and Development: Evolution within the Altricial-Precocial Spectrum, eds J. M. Starck and R. E. Ricklefs (Oxford: Oxford University Press), 324–339.
Griffith, S. C. (2007). The evolution of infidelity in socially monogamous passerines: neglected components of direct and indirect selection. Am. Nat. 169, 274–281. doi: 10.1086/510601
Griffith, S. C., and Immler, S. (2009). Female infidelity and genetic compatibility in birds: the role of the genetically loaded raffle in understanding the function of extrapair paternity. J. Avian Biol. 40, 97–101. doi: 10.1111/j.1600-048X.2009.04562.x
Griffith, S. C., Owens, I. P. F., and Thuman, K. A. (2002). Extra pair paternity in birds: a review of interspecific variation and adaptive function. Mol. Ecol. 11, 2195–2212. doi: 10.1046/j.1365-294X.2002.01613.x
Hamilton, W. D., and Zuk, M. (1982). Heritable true fitness and bright birds - a role for parasites. Science 218, 384–387. doi: 10.1126/science.7123238
Hatchwell, B. J., Ross, D. J., Fowlie, M. K., and McGowan, A. (2001). Kin discrimination in cooperatively breeding long-tailed tits. Proc. R. Soc. Lond. B Biol. Sci. 268, 885–890. doi: 10.1098/rspb.2001.1598
Jamieson, I. G., Taylor, S. S., Tracy, L. N., Kokko, H., and Armstrong, D. P. (2009). Why some species of birds do not avoid inbreeding: insights from New Zealand robins and saddlebacks. Behav. Ecol. 20, 575–584. doi: 10.1093/beheco/arp034
Jennions, M. D. (1997). Female promiscuity and genetic incompatibility. Trends Ecol. Evol. 12, 251–253. doi: 10.1016/S0169-5347(97)01128-2
Keller, L. F., and Waller, D. M. (2002). Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241. doi: 10.1016/S0169-5347(02)02489-8
Kempenaers, B. (2007). Mate choice and genetic quality: a review of the heterozygosity theory. Adv. Study Behav. 37, 189–278. doi: 10.1016/S0065-3454(07)37005-8
Kirkpatrick, M. (1996). Good genes and direct selection in evolution of mating preferences. Evolution 50, 2125–2140. doi: 10.2307/2410684
Kirkpatrick, M., and Ryan, M. J. (1991). The evolution of mating preferences and the paradox of the lek. Nature 350, 33–38. doi: 10.1038/350033a0
Kokko, H., and Ots, I. (2006). When not to avoid inbreeding. Evolution 60, 467–475. doi: 10.1111/j.0014-3820.2006.tb01128.x
Kokko, H., Brooks, R., Jennions, M. D., and Morley, J. (2003). The evolution of mate choice and mating biases. Proc. R. Soc. B Biol. Sci. 270, 653–664. doi: 10.1098/rspb.2002.2235
Kokko, H., Brooks, R., McNamara, J. M., and Houston, A. I. (2002). The sexual selection continuum. Proc. R. Soc. B Biol. Sci. 269, 1331–1340. doi: 10.1098/rspb.2002.2020
Komdeur, J., and Hatchwell, B. J. (1999). Kin recognition: function and mechanism in avian societies. Trends Ecol. Evol. 14, 237–241. doi: 10.1016/S0169-5347(98)01573-0
Lack, D. (1947). The significance of clutch-size. IBIS 89, 302–352. doi: 10.1111/j.1474-919X.1947.tb04155.x
Leclaire, S., Nielsen, J. F., Thavarajah, N. K., Manser, M., and Clutton-Brock, T. H. (2013). Odour-based kin discrimination in the cooperatively breeding meerkat. Biol. Lett. 9:20121054. doi: 10.1098/rsbl.2012.1054
Lehmann, L., and Perrin, N. (2003). Inbreeding avoidance through kin recognition: choosy females boost male dispersal. Am. Nat. 162, 638–652. doi: 10.1086/378823
Lehtonen, J., and Kokko, H. (2015). Why inclusive fitness can make it adaptive to produce less fit extra-pair offspring. Proc. R. Soc. Lond. B Biol. Sci. 282:20142716. doi: 10.1098/rspb.2014.2716
Lindstrom, J. (1999). Early development and fitness in birds and mammals. Trends Ecol. Evol. 14, 343–348. doi: 10.1016/S0169-5347(99)01639-0
Magrath, M. J. L., Vedder, O., van der Velde, M., and Komdeur, J. (2009). Maternal effects contribute to the superior performance of extra-pair offspring. Curr. Biol. 19, 792–797. doi: 10.1016/j.cub.2009.03.068
Marshall, T. C., Slate, J., Kruuk, L. E. B., and Pemberton, J. M. (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 7, 639–655. doi: 10.1046/j.1365-294x.1998.00374.x
Masters, B. S., Hicks, B. G., Johnson, L. S., and Erb, L. A. (2003). Genotype and extra-pair paternity in the house wren: a rare-male effect? Proc. R. Soc. B Biol. Sci. 270, 1393–1397. doi: 10.1098/rspb.2003.2380
Mateo, J. M. (2003). Kin recognition in ground squirrels and other rodents. J. Mammal. 84, 1163–1181. doi: 10.1644/BLe-011
Mays, H. L., Albrecht, T., Liu, M., and Hill, G. E. (2008). Female choice for genetic complementarity in birds: a review. Genetica 134, 147–158. doi: 10.1007/s10709-007-9219-5
Mays, H. L., and Hill, G. E. (2004). Choosing mates: good genes versus genes that are a good fit. Trends Ecol. Evol. 19, 554–559. doi: 10.1016/j.tree.2004.07.018
McDonald, P. G., and Wright, J. (2011). Bell miner provisioning calls are more similar among relatives and are used by helpers at the nest to bias their effort towards kin. Proc. R. Soc. Lond. B Biol. Sci. 278, 3403–3411. doi: 10.1098/rspb.2011.0307
Michalczyk, Ł., Millard, A. L., Martin, O. Y., Lumley, A. J., Emerson, B. C., Chapman, T., et al. (2011). Inbreeding promotes female promiscuity. Science 333, 1739–1742. doi: 10.1126/science.1207314
Moreno-Rueda, G., Soler, M., Soler, J. J., Martínez, J. G., and Pérez-Contreras, T. (2007). Rules of food allocation between nestlings of the black-billed magpie Pica pica, a species showing brood reduction. Ardeola 54, 15–25.
Neff, B. D., and Pitcher, T. E. (2005). Genetic quality and sexual selection: an integrated framework for good genes and compatible genes. Mol. Ecol. 14, 19–38. doi: 10.1111/j.1365-294X.2004.02395.x
Oh, K. P., and Badyaev, A. V. (2006). Adaptive genetic complementarity in mate choice coexists with selection for elaborate sexual traits. Proc. R. Soc. B Biol. Sci. 273, 1913–1919. doi: 10.1098/rspb.2006.3528
Parker, G. (2006). Sexual conflict over mating and fertilization: an overview. Philos. Trans. R. Soc. B Biol. Sci. 361, 235–259. doi: 10.1098/rstb.2005.1785
Pinheiro, J., and Bates, D. M. (2000). Mixed-Effects Models in S and S-PLUS. New York, NY: Springer.
Pitcher, T. E., and Neff, B. D. (2007). Genetic quality and offspring performance in Chinook salmon: implications for supportive breeding. Conserv. Genet. 8, 607–616. doi: 10.1007/s10592-006-9204-z
Prokop, Z. M., Michalczyk, Ł., Drobniak, S. M., Herdegen, M., and Radwan, J. (2012). Meta-analysis suggests choosy females get sexy sons more than “good genes.” Evolution 66, 2665–2673. doi: 10.1111/j.1558-5646.2012.01654.x
Pryke, S. R., Rollins, L. A., and Griffith, S. C. (2010). Females use multiple mating and genetically loaded sperm competition to target compatible genes. Science 329, 964–967. doi: 10.1126/science.1192407
Pusey, A., and Wolf, M. (1996). Inbreeding avoidance in animals. Trends Ecol. Evol. 11, 201–206. doi: 10.1016/0169-5347(96)10028-8
Puurtinen, M., Ketola, T., and Kotiaho, J. S. (2005). Genetic compatibility and sexual selection. Trends Ecol. Evol. 20, 157–158. doi: 10.1016/j.tree.2005.02.005
Puurtinen, M., Ketola, T., and Kotiaho, J. S. (2009). The good-genes and compatible-genes benefits of mate choice. Am. Nat. 174, 741–752. doi: 10.1086/606024
R Development Core Team (2011). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reid, J. M., Duthie, A. B., Wolak, M. E., and Arcese, P. (2015). Demographic mechanisms of inbreeding adjustment through extra-pair reproduction. J. Anim. Ecol. 84, 1029–1040. doi: 10.1111/1365-2656.12340
Richardson, D. S., Komdeur, J., Burke, T., and von Schantz, T. (2005). MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc. R. Soc. B Biol. Sci. 272, 759–767. doi: 10.1098/rspb.2004.3028
Roberts, S. C., and Gosling, L. M. (2003). Genetic similarity and quality interact in mate choice decisions by female mice. Nat. Genet. 35, 103–106. doi: 10.1038/ng1231
Rosivall, B., Szöllösi, E., Hasselquist, D., and Török, J. (2009). Effects of extrapair paternity and sex on nestling growth and condition in the collared flycatcher, Ficedula albicollis. Anim. Behav. 77, 611–617. doi: 10.1016/j.anbehav.2008.11.009
Røskaft, E., and Slagsvold, T. (1985). Differential mortality of male and female offspring in experimentally manipulated broods of the rook. J. Anim. Ecol. 54, 261–266. doi: 10.2307/4636
Rubenstein, D. R. (2007). Female extrapair mate choice in a cooperative breeder: trading sex for help and increasing offspring heterozygosity. Proc. R. Soc. B Biol. Sci. 274, 1895–1903. doi: 10.1098/rspb.2007.0424
Shizuka, D., and Lyon, B. E. (2013). Family dynamics through time: brood reduction followed by parental compensation with aggression and favouritism. Ecol. Lett. 16, 315–322. doi: 10.1111/ele.12040
Stewart, A. M. (1980). The Social Organisation and Foraging Ecology of the Tui, Prosthemadera Novaeseelandiae. Ph.D. Doctoral, University of Auckland.
Stewart, A. M., and Craig, J. L. (1985). Movements, status, access to nectar, and spatial-organization of the tui. N. Z. J. Zool. 12, 649–656. doi: 10.1080/03014223.1985.10428315
Stiratelli, R., Laird, N., and Ware, J. H. (1984). Random-effects models for serial observations with binary response. Biometrics 40, 961–971. doi: 10.2307/2531147
Suter, S. M., Keiser, M., Feignoux, R., and Meyer, D. R. (2007). Reed bunting females increase fitness through extra-pair mating with genetically dissimilar males. Proc. R. Soc. B Biol. Sci. 274, 2865–2871. doi: 10.1098/rspb.2007.0799
Szulkin, M., Stopher, K. V., Pemberton, J. M., and Reid, J. M. (2013). Inbreeding avoidance, tolerance, or preference in animals? Trends Ecol. Evol. 28, 205–211. doi: 10.1016/j.tree.2012.10.016
Szulkin, M., Zelazowski, P., Nicholson, G., and Sheldon, B. C. (2009). Inbreeding avoidance under different null models of random mating in the great tit. J. Anim. Ecol. 78, 778–788. doi: 10.1111/j.1365-2656.2009.01544.x
Tregenza, T., and Wedell, N. (2000). Genetic compatibility, mate choice and patterns of parentage: invited review. Mol. Ecol. 9, 1013–1027. doi: 10.1046/j.1365-294x.2000.00964.x
Tschirren, B., Postma, E., Rutstein, A. N., and Griffith, S. C. (2012). When mothers make sons sexy: maternal effects contribute to the increased sexual attractiveness of extra-pair offspring. Proc. R. Soc. B Biol. Sci. 279, 1233–1240. doi: 10.1098/rspb.2011.1543
van de Casteele, T., Galbusera, P., and Matthysen, E. (2001). A comparison of microsatellite-based pairwise relatedness estimators. Mol. Ecol. 10, 1539–1549. doi: 10.1046/j.1365-294X.2001.01288.x
Wan, D., Chang, P., and Yin, J. (2013). Causes of extra-pair paternity and its inter-specific variation in socially monogamous birds. Acta Ecol. Sin. 33, 158–166. doi: 10.1016/j.chnaes.2013.03.006
Wang, J. (2007). Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet. Res. 89, 135–153. doi: 10.1017/S0016672307008798
Wang, J. L. (2011). COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 11, 141–145. doi: 10.1111/j.1755-0998.2010.02885.x
Wells, S. J., Ji, W., Baillie, S. M., and Gleeson, D. (2013). Characterisation and cross-amplification of fourteen microsatellite loci for the endemic New Zealand tui (Meliphagidae), Prosthemadera novaeseelandiae. Conserv. Genet. Resour. 5, 113–116. doi: 10.1007/s12686-012-9745-z
Wells, S. J., Ji, W., Dale, J., Jones, B., and Gleeson, D. (2015). Male size predicts extrapair paternity in a socially monogamous bird with extreme sexual size dimorphism. Behav. Ecol. 26, 200–206. doi: 10.1093/beheco/aru173
Westneat, D. F., and Stewart, I. R. K. (2003). Extra-pair paternity in birds: causes, correlates, and conflict. Annu. Rev. Ecol. Evol. Syst. 34, 365–396. doi: 10.1146/annurev.ecolsys.34.011802.132439
Westneat, D. F., Sherman, P. W., and Morton, M. L. (1990). “The ecology and evolution of extra-pair copulations in birds,” in Current Ornithology, ed D. M. Power (New York, NY: Plenum Press), 331–369.
Wheelwright, N. T., Freeman-Gallant, C. R., and Mauck, R. A. (2006). Asymmetrical incest avoidance in the choice of social and genetic mates. Anim. Behav. 71, 631–639. doi: 10.1016/j.anbehav.2005.06.012
Wilson, N., Tubman, S. C., Eady, P. E., and Robertson, G. W. (1997). Female genotype affects male success in sperm competition. Proc. R. Soc. Lond. B Biol. Sci. 264, 1491–1495. doi: 10.1098/rspb.1997.0206
Zahavi, A. (1975). Mate selection—A selection for a handicap. J. Theor. Biol. 53, 205–214. doi: 10.1016/0022-5193(75)90111-3
Zeh, J. A., and Zeh, D. W. (1996). The evolution of polyandry I: intragenomic conflict and genetic incompatibility. Proc. R. Soc. B Biol. Sci. 263, 1711–1717. doi: 10.1098/rspb.1996.0250
Keywords: social structure, mating systems, extra-pair paternity, compatible genes, good genes, inbreeding
Citation: Wells SJ, Ji W, Gleeson D, Jones B and Dale J (2017) Population Social Structure Facilitates Indirect Fitness Benefits from Extra-Pair Mating. Front. Ecol. Evol. 5:18. doi: 10.3389/fevo.2017.00018
Received: 10 November 2016; Accepted: 13 March 2017;
Published: 28 March 2017.
Edited by:
Peter Schausberger, University of Vienna, AustriaReviewed by:
Keith Tarvin, Oberlin College, USARyan Germain, University of Aberdeen, UK
Aneta Izabela Arct, Jagiellonian University, Poland
Copyright © 2017 Wells, Ji, Gleeson, Jones and Dale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah J. Wells, cy5qLndlbGxzQG1hc3NleS5hYy5ueg==
 Sarah J. Wells
Sarah J. Wells Weihong Ji
Weihong Ji Dianne Gleeson2
Dianne Gleeson2 James Dale
James Dale