- 1Molecular Phytobacteriology Laboratory, Infectious Disease Research Center, KRIBB, Daejeon, South Korea
- 2University of Science and Technology, Daejeon, South Korea
- 3Department of Plant Physiology, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
Infestation of plants with the phloem-feeding whitefly Bemisia tabaci modulates root microbiota and both local and systemic immunity against microbial pathogens. Specifically, aboveground whitefly infestation suppresses pathogen propagation and symptom development caused by the soil-borne pathogens Agrobacterium tumefaciens and Ralstonia solanacearum in the root system through systemic signal transduction. Therefore, we hypothesized that secreted protein(s)/non-protein factors from whitefly saliva (referred to as candidate effectors) might function as insect determinants that activate systemic acquired resistance (SAR) in the host plant. Here, we intensively screened a cDNA library constructed from mRNA from whitefly feeding on Nicotiana benthamiana leaves and selected three candidate effectors 2G4, 2G5, and 6A10, that appear to reduce disease development caused by the aboveground pathogen Pseudomonas syringae pv. tabaci and the soil-borne pathogen R. solanacearum. Transient expression of the three candidate effector cDNAs in leaves primed the expression of SAR marker genes NbPR1a and NbPR2 in local and systemic leaves against P. syringae pv. tabaci, while leaf infiltration with 2G4 or 6A10 cDNA elicited strong defense priming of SAR markers following drench application of R. solanacearum on plant roots. In silico and qRT-PCR analyses revealed the presence of 2G5 and 6A10 transcripts in insect salivary glands. This is the first report of whitefly effectors that prime SAR against aboveground and belowground bacterial pathogens.
Introduction
Plants are constantly exposed to diverse insect pests and microbial pathogens (Agrios, 2005). To protect from these enemies, immune responses, including chemical and physical defense mechanisms, are activated in local and systemic plant tissues (Koornneef and Pieterse, 2008; Dangl et al., 2013). Plants have developed a sophisticated immune system against insect herbivory. Compared with our understanding of plant defensive responses against chewing insects, little is known about plant responses to phloem-feeding insects in the order Hemiptera (Van Oosten et al., 2008; Walling, 2008; Rodriguez-Medina et al., 2011; van Dam and Heil, 2011; Louis and Shah, 2013; Pitino and Hogenhout, 2013; Rao et al., 2013; VanDoorn et al., 2015). The detailed mechanisms of plant responses to this group of insects, such as aphids and whitefly (Bemisia tabaci Genn.), have only recently begun to be uncovered due to the small size and limited genetic and physiological information about these insects (Louis and Shah, 2013; Pitino and Hogenhout, 2013; VanDoorn et al., 2015).
For instance, aboveground (leaf) whitefly infestation increases plant immunity against soil-borne plant pathogens, indicating that systemic plant signaling is activated and translocated from leaf to root. Infestation of pepper leaves by whitefly increases systemic resistance against the soil-borne pathogen Ralstonia solanacearum (Yang et al., 2011; Lee et al., 2012). Further investigation revealed that infestation with this insect leads to the recruitment of beneficial rhizosphere bacterial species, which act as a biological trigger to elicit plant systemic defense responses against subsequent whitefly attack (Murphy et al., 2003). More recently, whitefly infestation was found to reduce Agrobacterium tumefaciens mediated crown gall formation on stems and roots (Song et al., 2015). Transcriptome and virus-induced gene silencing analyses demonstrated that whitefly-induced salicylic acid (SA) signaling attenuates Agrobacterium T-DNA transformation and gall formation. Root exudates that were collected from tobacco contained approximately 2.5-fold higher SA levels when whiteflies had infested leaf tissues (aboveground) compared to the uninfested control. Intriguingly, whitefly-elicited plant immunity in pepper activates both SA- and jasmonic acid (JA)-related gene expression in aboveground and belowground tissue, indicating that SA- and JA-dependent pathways are activated from leaf to root in response to whitefly feeding on leaves (Park and Ryu, 2014). Further investigation involving the fine-tuning of these signaling pathways following whitefly infestation using virus-induced gene knockdown of SA- and JA-responsive and biosynthesis genes revealed that SA is a major player in whitefly feeding-dependent signaling (Lazebnik et al., 2014; Song et al., 2015).
The whitefly determinant that confers resistance against soil borne pathogens in systemic plant tissues is still unknown. Our understanding of insect-mediated changes in the activation of plant immune responses is limited due to the lack of information on whitefly determinants that suppress or induce plant immune responses. To fill this knowledge gap, most studies on insect factors that modulate plant immunity have focused on the suppression of insect resistance rather than the induction of plant resistance responses such as systemic acquired resistance (SAR) (Kempema et al., 2007; Cooper et al., 2010; Su et al., 2012, 2015). The effector proteins from pathogenic bacteria and fungi induce and suppress plant immunity via a well-known process described by the zigzag theory (Jones and Dangl, 2006). Effectors are a group of proteins that translocate from microbes such as bacteria, fungi, and nematodes to host plants and animals (Elzinga and Jander, 2013). The major function of effectors is to modulate host immune responses though interactions with their counterpart proteins in the host plant. The outcomes of these interactions include effector-triggered susceptibility (ETS) and effector-triggered immunity (ETI) or immune reactions in the target plant, which occur in a protein-dependent manner.
Unlike microbial effectors, insect effectors have not been intensively studied. Recent studies explored effector proteins, primarily from sucking insects, and their role in plant immunity (Elzinga and Jander, 2013). Hemipteran and dipteran insect species, including phloem-feeding aphids and whiteflies, secrete certain proteins and translocate them into the cytosol of the host cell through their stylets (Kaloshian and Walling, 2015). These effectors play important roles in suppressing plant defense responses and helping the insect overcome plant immunity. The aphid effectors Coo2 and Armet, which were identified through transcriptome analysis of aphid glands, increase insect survival and host colonization (Mutti et al., 2006; Wang et al., 2015a). Another aphid effector, SHP (structure sheath protein), is primarily expressed in saliva and functions as a virulence factor. Interestingly SHP does not share any sequence homology with proteins from other insects, suggesting that it would be a good target for RNA interference-mediated insect control in SHP dsRNA-overexpressing transgenic plants (Abdellatef et al., 2015; Will and Vilcinskas, 2015). ACE2 (angiotensin-converting enzyme 2), SSGPs (secreted salivary gland proteins), and Mp10, Mp55, Me10, and Me23 are also candidates for this technique (Elzinga and Jander, 2013; Wang et al., 2015b; Zhao et al., 2015). However, the functions and molecular roles of effectors from whitefly have only recently been explored. Whitefly saliva is thought to contain proteins that modulate plant defense responses and facilitate feeding. Secreted whitefly laccase 1 (LAC1) and small RNAs have been identified and are thought to help the insect overcome plant immunity responses (Yang et al., 2017; van Kleeff et al., 2016).
In the current study, to extend our understanding of plant-microbe-insect tritrophic interactions, we focused on the following: (1) establishing a high-throughput screening system to screen whitefly effectors that elicit plant immune responses against aboveground virulent pathogens, (2) evaluating aboveground effector-mediated plant SAR against soil-borne pathogens, and (3) characterizing the identified effectors and confirming expression in the salivary gland. We used whitefly as a model insect and two microbial pathogens as model pathogens, including Pseudomonas syringae on local and systemic leaves (aboveground) and the soil-borne pathogen Ralstonia solanacearum on the plant root system (belowground).
The objective of the current study was to identify candidate whitefly effectors that activate plant immunity, as revealed by the suppression of symptom development caused by virulent Pseudomonas syringae pv. tabaci or attenuation of the hypersensitive response (HR, a plant programmed cell death response) caused by avirulent Pseudomonas syringae pv. syringae. We hypothesized that pre-infiltration of Nicotiana benthamiana leaves with candidate whitefly effectors would delay or totally suppress lesion formation caused by the two P. syringae pathovars in the overlapping regions of leaves after cross-infiltration. Finally, we validated three putative effectors identified from high-throughput screening of an Agrobacterium tumefaciens-mediated transient expression system as candidate effectors that elicit plant systemic immunity (SAR) against a soil-borne pathogen Ralstonia solanacearum, prime plant SAR marker gene expression on root and confirmed their localization in silico. This study represents the first demonstration of whitefly effectors that trigger SAR against aboveground and belowground bacterial pathogens.
Materials and Methods
Insect Culture and Plant Growth Conditions
Whiteflies (Bemisia tabaci) were grown and maintained in the KRIBB Greenhouse Facility in Daejeon, South Korea, as described previously (Yang et al., 2011; Park and Ryu, 2014). The whitefly was identified as Q biotype (data now shown). N. benthamiana was used as the model system, as described previously (Song et al., 2016). N. benthamiana seeds were surface-sterilized with 6% sodium hypochlorite, washed four times with sterile distilled water, and incubated in a growth chamber at 25 ± 2°C under fluorescent lights (light: dark 12: 12 h; c. 7000 lux light intensity). Seedlings were individually grown in plastic pots 9 cm in diameter at 25 ± 2°C under fluorescent lights in a controlled environment growth room (12 h/12 h day/night cycle, 7000 lux light intensity). Three-week-old N. benthamiana plants were used in the experiments. The experiments were repeated three times with 10 replications (one plant per replication and three leaves per plant).
Whitefly cDNA Library Construction
Whiteflies were collected from N. benthamiana leaves at mid-day and quickly ground in liquid nitrogen for RNA isolation using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and RNase-free DNase I (Promega). Purified total RNA was reverse-transcribed to cDNA using a Cloneminer II cDNA Library Construction Kit (Invitrogen) following the manufacturer's instructions. The cDNA was inserted into the pDONR222 Gateway vector. The initial titer of the library was determined through colony counts via plating on LB plates containing kanamycin (50 μg/mL). The average insert size was analyzed by restriction enzyme digestion with BsrG1 (New England Biolabs). The initial cDNA fragments were transferred to the pK7WG2 vector using LR recombinase (Invitrogen), and transformed into A. tumefaciens LBA4404 cells by electroporation and used for transgene expression in N. benthamiana.
5′ Rapid Amplification of cDNA Ends (RACE)
Sense cDNA was synthesized via 5′ rapid amplification of cDNA ends (RACE) adapter (5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA-3′) ligation with a First choice RLM-RACE Kit (Ambion). Antisense cDNA was synthesized by PCR using the 5′ RACE adaptor outer primer following the manufacturer's instructions. The amplified double-stranded cDNA was cloned into the pGEM-T Easy vector (Promega).
Intensive Whitefly Effector Screening via a Cross-Infiltration Assay
Agrobacterium-mediated gene transfer was conducted as described elsewhere with minor modifications (Win et al., 2011). The A. tumefaciens GV2260 culture was pelleted by centrifugation for 5 min at 4,000 rpm at room temperature (RT), and the cell pellet was washed three times with distilled water. The cells were re-suspended in Agro-induction medium (10 mM MgCl2, 150 μM acetosyringone, pH 5.6). The concentration of the suspension was adjusted to OD600 = 0.1 prior to infiltration. To identify effectors from a total cDNA library prepared from whitefly during plant infestation, we reasoned that Agrobacterium tumefaciens-mediated transient transformation should delay or totally suppress lesion formation caused by Pseudomonas syringae pv. tabaci (Pta) and P. syringae pv. syringae (Psy) in the overlapping regions of leaves after cross-infiltration. Agro-infiltration assays were performed on the middle leaves of 3-week-old N. benthamiana plants. The two P. syringae pathovars (Pta and Psy) were selected on solid King's B medium containing 100 μg/mL rifampicin at 30°C for 2 days, scraped off the plates, and re-suspended in 10 mM MgCl2 (King and Zeevaart, 1974; Song et al., 2015). The negative control was empty vector (pK7WG2). For Pta symptom evaluation, leaves were infiltrated with Pta (OD600 = 0.01) 3 days after agro-infiltration. To visualize HR symptoms, leaves were infiltrated with Psy (OD600 = 0.1) 3 days after agro-infiltration. The HR is normally apparent 24 h after infiltration.
P. syringae pv. tabaci Pathogenesis Assay
To investigate the impact of the effectors in detail, symptom development and bacterial numbers in local and systemic leaves at day were measured on days 0, 3, and 5 after pathogen challenge (Figure 2A). The foliar parts of 3-week-old N. benthamiana seedlings were infiltrated with 2 mL of a 106–107 cfu/mL suspension of A. tumefaciens. The positive control was 0.5 mM BTH (Syngenta, Durham, NC, USA), which elicits SAR to bacterial pathogens. The negative control was empty vector. Pta was selected on solid King's B medium containing 100 μg/mL rifampicin at 30°C for 2 days, scraped off the plates, and re-suspended in 10 mM MgCl2 (King and Zeevaart, 1974; Song et al., 2015). For Pta symptom evaluation, leaves were infiltrated with Pta (OD600 = 0.01) at 3 days after agro-infiltration. To investigate the population size of Pta in leaves, Pta cells were counted at 0, 3, and 5 days after pathogen inoculation. Leaf discs (1 cm diameter) were ground in 10 mM MgCl2, and serial dilutions of bacterial solution were spread onto selection medium (King's B agar medium containing 100 μg/mL rifampicin) and incubated for 2 days in a 30°C growth chamber.
R. solanacearum Pathogenesis Assay
Ralstonia solanacearum was grown on Casamino acid-Peptone-Glucose (CPG) at 30°C for 1 day. The R. solanacearum culture was pelleted at RT for 5 min at 4,000 rpm and re-suspended in 10 mM MgCl2 (Song et al., 2015). A freshly prepared 50 mL aliquot of R. solanacearum suspension at OD600 = 1 was used to drench the roots of N. benthamiana seedlings at 3 days after leaf infiltration with whitefly cDNA clones 2G4, 2G5, and 6A10 and empty vector (EV) (Chandrasekaran et al., 2016). The severity of R. solanacearum symptoms was scored on a scale of 0–10 as follows: 0, no leaves wilted; 1, 1–5% of leaves wilted; 2, 6–20% of leaves wilted; 3, 21–35% of leaves wilted; 4, 36–50% of leaves wilted; 5, 51–65% of leaves wilted; 6, 66–80% of leaves wilted; 7, 81–95% of leaves wilted; 8, 96–100% of leaves wilted but stems intact; 9, 96–100% of leaves wilted and stems broken; and 10, 100% of leaves wilted and stems broken (Song et al., 2016; Figure 4A). The total number of R. solanacearum cells in the rhizosphere was counted at 10 days after drench application. Whole roots were collected from each plant without soil particles, placed in a flask containing 200 mL of sterilized distilled water, and incubated with shaking for 30 min at 30°C. The liquid from the flask was serially diluted and spread onto CPG agar plates. To test the effect of the candidate whitefly effectors on plant growth rates, root weights were measured 10 days after pathogen challenge as described previously (Yang et al., 2009). The experiment was repeated three times with 10 replications. Preparation of graphs were performed using the R studio (R-Studio, Boston, MA, USA).
GUS Staining
Histochemical GUS staining was performed as described previously (Brown et al., 2003). Three plants treated with candidate effectors were collected for GUS staining on days 0, 3, 5, and 7. The positive control was 0.5 mM BTH (Syngentay Crop Protection Inc., Greensboro, NC. USA), and the negative control was empty vector. Leaves were immersed in staining solution (2 mM X-Gluc in N,N-dimethyl formamide, 100 mM NaH2PO4, 10 mM Na2EDTA, 0.5 mM ferrocyanide, and 0.1% Triton X-100, pH 7.0) and incubated at 37°C overnight in the dark. The leaves were cleared of chlorophyll by treating them with 70% ethanol after staining at RT for 24 h. Stained samples were observed and photographed with a digital camera (Sony, Park Ridge, NJ, USA).
Extraction of Plant RNA, cDNA Synthesis, and Quantitative RT-PCR
For the leaf pathogen (P. syringae) experiment, following agro-infiltration, leaf tissue was collected 0, 12, and 24 h after agro-infiltration and used for total RNA isolation. Following inoculation with Pta, the leaf tissue was harvested at 0, 24, and 48 h after inoculation and used for total RNA isolation. Following agro-infiltration, root tissue was collected 0, 1, and 2 days after agro-infiltration and used for total RNA isolation. For the soil-borne pathogen, following inoculation with R. solanacearum, root tissue was harvested at 0, 1, and 2 days after inoculation and used for total RNA isolation. To validate candidate effector production in whitefly, whitefly adults were collected from N. benthamiana leaves at mid-day and used for total RNA isolation. Total RNA was isolated from tobacco leaf tissues using Trizol (Invitrogen) according to the manufacturer's instructions and as described in our previous study (Lee et al., 2012). First-strand cDNA synthesis was performed with 1 μg of DNase-treated total RNA, oligo-dT primers, and Moloney murine leukemia virus reverse transcriptase (Enzynomics, Daejeon, Korea). The qRT-PCR reaction mixtures consisted of cDNA, iQ™ SYBR® Green Supermix (Bio-Rad Inc., Hercules, CA, USA), and 10 pM each primer. The cycling parameters were as follows: initial polymerase activation for 3 min at 95°C, followed by 45 cycles of 30 s at 95°C, 60 s at 60°C, and 30 s at 72°C. Relative transcript levels were calculated using the 2-ΔΔ CT method. The reference genes were NbACT mRNA (GenBank accession no. U60489) in tobacco plants, and BtACT mRNA (GenBank accession no. AF071908) in whitefly. For convenient comparisons, the expression levels were presented as fold change relative to those of the control (where empty vector or Lac1 is equal to 1). Gene specific primer sequences are listed in Table S1.
RNA Isolation, De Novo Assembly and Relative Expression Levels
Whiteflies (B. tabaci MEAM1_UvA) were reared on cucumber plants (Cumumis sativus, Ventura, RijkZwaan, the Netherlands) in a climatized chamber (Snijders, Tilburg, the Netherlands; 28°C, 16H light 150 μE m−2s−1, RH75%) as previously described (Bleeker et al., 2011).
The samples for RNA sequencing were obtained as follows: whitefly eggs and nymphs (1st, 2nd, and 3rd instar) were removed, between 10 a.m. and 6 p.m., from the cucumber leaf using an insect pin and transferred directly into 100% acetone for storage at RT. For whole body (n = 1), thorax (n = 2), and abdomen (n = 1) samples the whiteflies were collected at 9 AM by aspiration and transferred to 100% acetone at RT. The whiteflies were halved with a surgical knife while submerged in 100% acetone to obtain the thorax and abdomen samples. Salivary glands (n = 1) and midguts (n = 1) were dissected from adult whiteflies as described previously (Kliot et al., 2014).
RNA isolation was performed using the RNeasy mini kit from Qiagen (www.qiagen.com) according to manufacturer's protocol. RNA integrity was examined using the 2200 TapeStation System with Agilent RNA ScreenTapes (Agilent). RNA with RIN values greater than 7.4 were used for Illumina RNA sequencing (www.illumina.com, HiSeq 2000) except for salivary glands (RIN 5.4) and midgut (RIN 5.8). The Illumina reads were cleaned from adapter and ambiguous sequences by Trimmomatic 0.32 software (usadellab.org) (Bolger et al., 2014). The clean reads of thorax, abdomen, eggs and whole body (±20 million per sample) were used for de novo assembly using Trinity software (r20140313) [https://github.com/trinityrnaseq/trinityrnaseq/wiki; (Grabherr et al., 2011; Haas et al., 2013)] using the default settings. Trinity (r20140313) was used to realign reads to the de novo transcriptome contigs and to calculate the fragments per kilobase transcript length per million fragments mapped (FPKM) using RSEM (http://deweylab.github.io/RSEM; Li and Dewey, 2011), after which an normalization (Trimmed Mean of M) was performed across all whitefly samples using the abundance_estimates_to_matrix.pl script (Li and Dewey, 2011; Haas et al., 2013). The RNA-seq data are deposited at the European Nucleotide Archive (https://www.ebi.ac.uk/ena) which is a mirror site of NCBI (the project number: PRJEB26594).
Statistical Analysis
Data were subjected to analysis of variance using JMP software ver. 4.0 (SAS Institute Inc., Cary, NC, USA; www.sas.com). The significance of biological or chemical treatment effects was determined by the magnitude of the F-value at P = 0·05. When a significant F-value was obtained for treatments, separation of means was accomplished using Fisher's protected least significant difference (LSD) test at P < 0.05. The results of repeated trials of each experiment outlined above were similar. Hence, one representative trial of each experiment is reported.
Results
High-Throughput System Design and Identification of Potential Effectors
We developed a new screening method to assess the attenuation or suppression of the HR or symptom development caused two P. syringae pathovars (Pta and Psy) in the overlapping regions of N. benthamiana leaves after cross-infiltration with candidate whitefly effectors. In the first screening with usingthe two P. syringae pathovars in the overlapping regions of leaves after cross-infiltration, we selected 24 and 9 clones after Pta and Psy infiltration, respectively (Figures 1A,B). Of the 893 clones in the cDNA library, we ultimately selected three cDNA clones, 2G4, 2G5, and 6A10, due to their clear suppressive effects on lesion formation (Table S2).
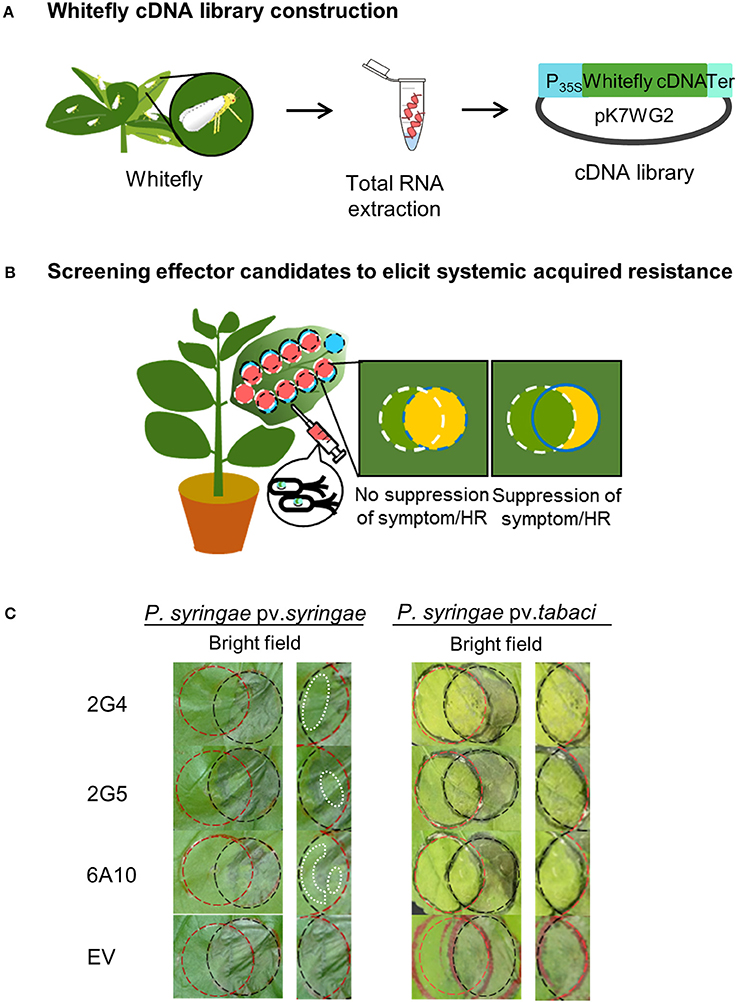
Figure 1. Screening system to identify candidate whitefly effectors that induce plant immunity. (A) Whitefly cDNA library construction. Extraction of total RNA from whitefly during infestation of Nicotiana benthamiana leaves. The cDNA library from whitefly mRNA was constructed in Agrobacterium tumefaciens using a binary vector system. (B) Screening candidate effectors that elicit systemic acquired resistance against virulent Pseudomonas syringae pv. tabaci and avirulent (a non-host pathogen) Pseudomonas syringae pv. syringae. To identify whitefly effectors that activate plant pathogen immunity, cross-circle infiltration was performed with cDNA clones and pathogens, and the suppression of disease symptoms caused by Pta and the hypersensitive response (HR) caused by Psy at 1–5 days after pathogen infiltration were evaluated. (C) Suppression of the HR and symptoms by whitefly cDNA clones 2G4, 2G5, and 6A10. The white dotted lines in the left panel indicate the suppressed HR. The empty vector (EV) treatment showed no inhibition of the HR and symptom development in the intersecting area. The experiment was repeated three times with similar results.
Effector-Mediated Plant Immunity Against the Aboveground Pathogen P. syringae pv. tabaci
We evaluated whether the three candidate effectors would elicit plant immunity in local or systemic tissues of N. benthamiana. We infiltrated each whitefly cDNA clone into one half of a leaf and the vector control into the other half. At 3 days after infiltration, we challenged the plants with Pta and measured bacterial numbers on days 0, 3, and 5. First, we confirmed the inhibition of symptom development by Pta using the overlay method after infiltration of the candidate effectors into whole leaves (Figures 2B,C). Plants pretreated with the three candidate whitefly effectors, 2G4, 2G5, and 6A10, showed significantly (P < 0.05) fewer (10-fold) bacteria number on days 3 and 5 than the empty vector control in local leaves (Figure 2F). On day 5 after pathogen challenge in leaves infiltrated with the three candidate effectors, the number of bacteria was not statistically different among systemic leaves while they differed compared to empty vector control on day 3 (Figure 2G), and the number of bacteria was similar to that of the BTH-pretreated positive control in local leaves (Figures 2F,G). The symptom in local and systemic leaves at day 5 after pathogen challenge are presented (Figures 2D,E).
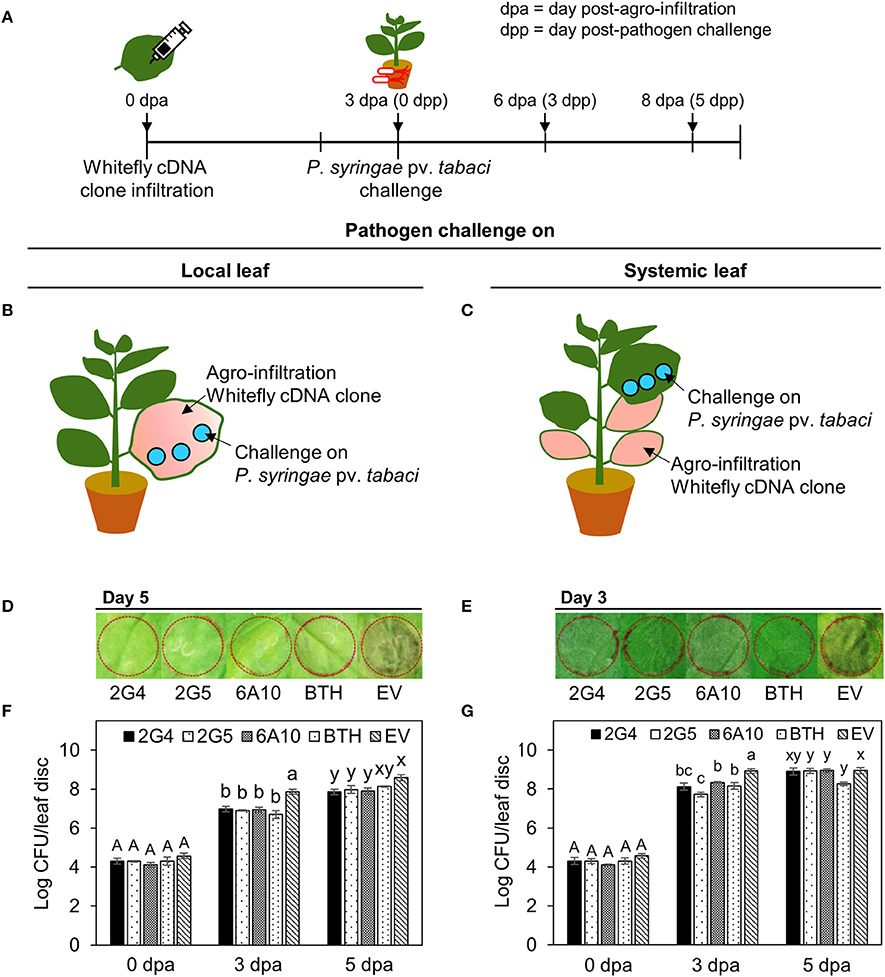
Figure 2. Systemic acquired resistance against P. syringae pv. tabaci elicited by candidate whitefly effectors on local and systemic tobacco leaves. (A) Schematic diagram of the experimental design for investigating SAR against Pta after leaf infiltration with whitefly cDNA clones 2G4, 2G5, and 6A10. (B) SAR against Pta in a local tobacco leaf. A suspension of Pta at OD600 = 0.001 was infiltrated into a tobacco leaf that had been pre-infiltrated with whitefly cDNA clones 2G4, 2G5, and 6A10 throughout the leaf at 3 days before pathogen challenge. (C) SAR against Pta on a systemic tobacco leaf. A suspension of Pta at OD600 = 0.001 was infiltrated into a whole tobacco leaf that had been pre-infiltrated with whitefly cDNA clones 2G4, 2G5, and 6A10 at 3 days before pathogen challenge using a needleless syringe. (D) Disease symptom development at 5 days after pathogen challenge on local leaf. (E) Disease symptom development at 3 days after pathogen challenge on systemic leaf. (F) Bacterial population size of local leaf were measured at 0, 3, and 5 days after Pta infiltration with a needleless syringe. (G) Bacterial population size of systemic leaf were measured at 0, 3, and 5 days after Pta infiltration. Bars represent the mean value ± SEM (N = 10). Infiltration with 1 mM BTH and Agrobacterium empty vector (EV) suspension was used as a positive and negative control, respectively. Different letters (a, b, and c; x and y) within day indicate statistically significant differences (P < 0.05).
Candidate Whitefly Effector-Elicited SAR Marker Gene Expression
We evaluated the short- and long-term elicitation of SAR by quantifying the expression of SAR marker genes NbPR1a and NbPR2 at 0, 12, and 24 h and 0, 3, 5, and 7 days after infiltration with the candidate effector cDNAs (Figures 3A,B). To quantify the early expression of SAR marker genes, we evaluated the effects of pretreatment with the three candidate effectors, 2G4, 2G5, and 6A10, which induced the early expression of marker genes both locally and systemically (Figure 3B). Of the three clones, 2G4 had the strongest effects at 24 h to a level similar with that of positive control BTH treatment in local leaves (Figure 3B, left panel), while all three clones increased NbPR2 expression, and clones 2G4 and 6A10 more strongly increased NbPR1a expression in systemic leaves compared with the control (Figure 3B, right panel). At the same time, the transcriptional level of NbPR1a and NbPR2 on the local leaf infiltrated with clone 2G5 was similar with 6A10 but higher than empty vector control (Figure 3B, left panel).
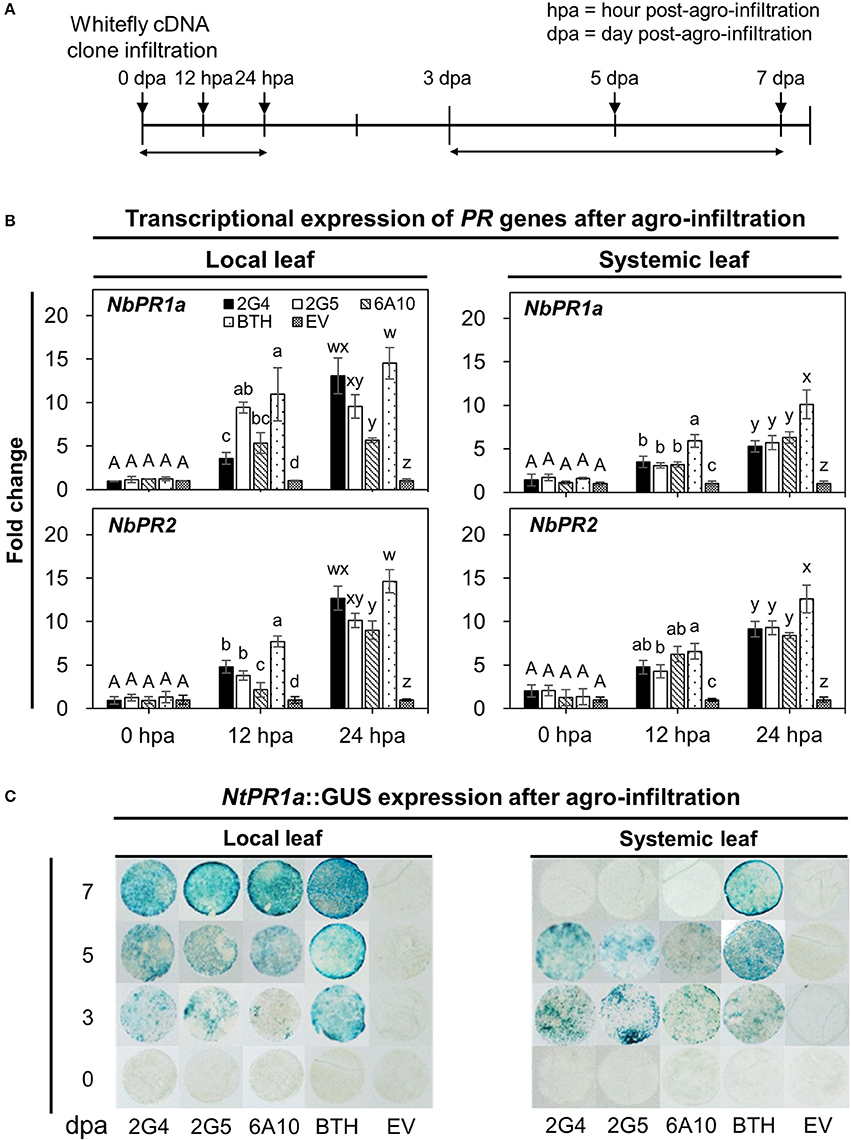
Figure 3. Whitefly effector-mediated early and late expression of SAR marker genes in N. benthamiana. (A) Schematic diagram of the experimental design for investigating the expression of marker genes (NbPR1a, NbPR2) and the staining of leaf disc from NtPR1a::GUS transgenic tobacco after leaf infiltration with whitefly cDNA clones 2G4, 2G5, and 6A10 and empty vector (EV). The samples used for the early gene expression were local and systemic leaves after whitefly cDNA clone leaf-infiltration at 0, 12, and 24 h post-Agrobacterium infiltration (hpa). The samples for GUS staining to late expression of a SAR marker gene were leaf disc collected at 3, 5, and 7 days post-Agrobacterium infiltration (dpa). (B) Quantification of defense priming of SAR marker genes NbPR1a and NbPR2 in plants at 0, 12, and 24 h after inoculation with whitefly cDNA clones 2G4, 2G5, and 6A10 on local (left panel) and systemic (right panel) leaves by qRT-PCR. Transcription is shown relative to empty vector (expression level = 1) with the NbActin gene as an internal reference. Bars represent the mean value ± SEM (N = 10). Infiltration with a suspension of 1 mM BTH and Agrobacterium with empty vector (EV) was used as a positive and negative control, respectively. Different letters (a, b, c, and d; w, x, y, and z) within hpa indicate statistically significant differences (P < 0.05). (C) NtPR1a gene expression pattern following infiltration with candidate effectors using NtPR1a::GUS transgenic Nicotiana tabacum plants. GUS staining was conducted by incubating leaf tissues (N = 30) in X-gluc solution at 0, 3, 5, and 7 days post-Agrobacterium infiltration with whitefly cDNA clones 2G4, 2G5, and 6A10 and empty vector. Infiltration with 1 mM BTH suspension was used as a positive control.
Next, to quantify the long-term expression of SAR marker genes, we evaluated their expression at 0, 3, 5, and 7 days after agro-infiltration. The activation of local and systemic plant immune responses was confirmed by examining the expression of NtPR1a::GUS, a representative SAR biomarker gene for plant immunity in tobacco (Figure 3C). GUS expression in tobacco leaves infiltrated with the three candidate effectors was first detected on day 3 and reached a maximized level on day 7 in local leaves, whereas the expression of this gene changed little in leaves pretreated with 1 mM BTH on days 3 to 7 (Figure 3C, left panel). The systemic expression of NtPR1a induced by the three whitefly effectors was detected only on day 3 and 5 but not on day 7 (Figure 3C, right panel). Maximum expression of NtPR1a in the positive BTH-treated control was detected on day 7 in local leaves and day 5 in systemic leaves (Figure 3C).
Whitefly Effector Expression in Aboveground Plant Tissues Activates Immunity Against the Soil-Borne Pathogen R. solanacearum
One important characteristic of plant immunity is “defense priming” (Song et al., 2015). Strong defense priming is generally detected at an early time point after pathogen challenge. To evaluate candidate effector-mediated SAR and defense priming against the soil-borne pathogen R. solanacearum, we measured bacterial wilt symptoms at 10 days after drench application of a 108 cfu/mL R. solanacearum suspension at 3 days after leaf infiltration with the three candidate effector cDNAs. Disease severity and the pathogen population were significantly reduced by 2G4 and 6A10 cDNA treatment (Figures 4B,C). We detected a 24, 12, and 27% reduction in symptom development in tobacco plants treated by leaf infiltration with 2G4, 2G5, and 6A10 cDNA, respectively (Figure 4B). The root fresh weight was 20% higher in plants pre-infiltrated with 6A10 than in the control (Figure 4D). By contrast, the bacterial number and root fresh weight in plants treated with 2G5 cDNA did not differ from those in the control (Figures 4C,D). However, the bacterial number in root system of plants treated with 2G4 cDNA was statistically lower than control treatment (Figure 4C).
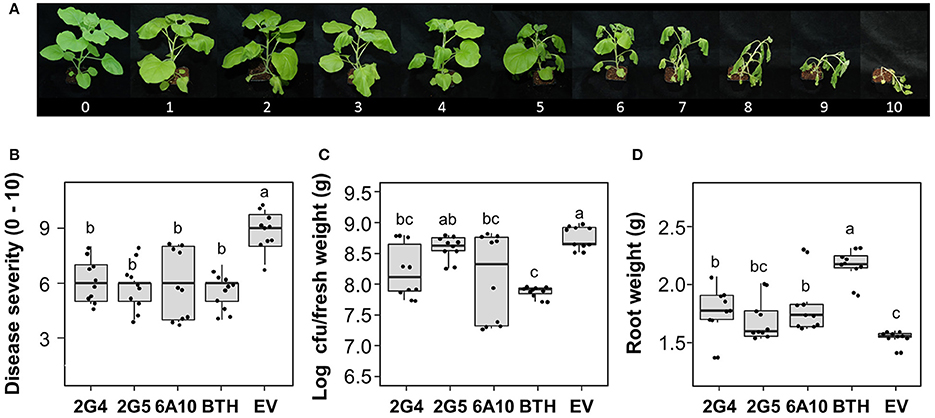
Figure 4. Whitefly effector-mediated SAR against the soil-borne pathogen Ralstonia solanacearum. A freshly prepared 50 mL aliquot of R. solanacearum suspension at OD600 = 1 was used to drench the roots of N. benthamiana seedlings at 3 days after leaf infiltration with whitefly cDNA clones 2G4, 2G5, and 6A10 and empty vector (EV). The disease severity (0–100) was measured at 10 days after pathogen challenge. (A) Disease scale (0–10). The disease severity of bacterial wilt caused by R. solanacearum was scored from 0 to 10 as follows: 0, no leaves wilted; 1, 1–5% of leaves wilted; 2, 6–20% of leaves wilted; 3, 21–35% of leaves wilted; 4, 36–50% of leaves wilted; 5, 51–65% of leaves wilted; 6, 66–80% of leaves wilted; 7, 81–95% of leaves wilted; 8, 96–100% of leaves wilted but stems intact; 9, 96–100% of leaves wilted and stems broken; and 10, 100% of leaves wilted and stems broken. (B) Quantification of disease severity, (C) pathogen population size, and (D) root fresh weight at 10 days after drench application of an R. solanacearum suspension at OD600 = 0.01 on 3 days after leaf infiltration with whitefly cDNA clones 2G4, 2G5, and 6A10 and empty vector. Infiltration with 1 mM BTH suspension in tobacco leaves was used as a positive control. Different letters indicate statistically significant differences (P < 0.05). Error bars represent mean ± maximum and minimum values (N = 10).
To obtain further confirmation of candidate effector-mediated SAR against R. solanacearum, we performed two qRT-PCR experiments to evaluate the transcriptional expression of SAR marker genes NbPR1a and NbPR2 in roots on days 0, 1, and 2 after cDNA infiltration and pathogen challenge (Figures 5A,B). First, we evaluated transcript levels of the two SAR marker genes after cDNA infiltration without pathogen challenge (Figure 5B, left panel). Compared with the control, the expression of NbPR1a was significantly different under all treatments on day 1. No significant difference was detected on day 2 across 2G4 and 6A10 treatments (Figure 5B, left and above panel). The maximum expression of NbPR2 was detected in 2G4-, 2G5-, and 6A10-treated plants at 2 days (2 dpa) after leaf infiltration than in the control (Figure 5B, left panel). The expression of these genes was not different compared to control at the pathogen inoculation time point (3 dpa and 0 dpp) when R. solanacearum drench-applied 3 d after clone infiltration on the leaves (0 d for pathogen challenge in the roots) (Figure 5B, right panel).
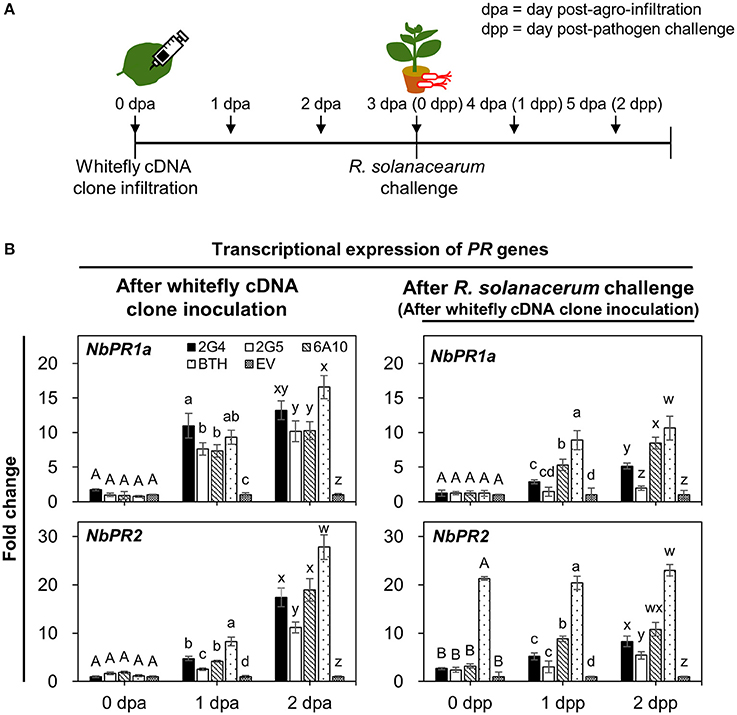
Figure 5. Whitefly effector candidate-mediated defense priming of SAR marker genes in roots after challenge with Ralstonia solanacearum. (A) Schematic diagram of the experimental design for investigating the expression of SAR marker genes NbPR1a and NbPR2 after leaf infiltration with whitefly cDNA clones 2G4, 2G5, and 6A10 into three whole leaves and on days 0, 1, and 2 after R. solanacearum challenge. The samples used to investigate whitefly effector candidate-mediated direct systemic activation of SAR marker genes were root tissues collected after leaf infiltration with whitefly cDNA at 0, 1, and 2 days post-Agrobacterium infiltration (dpa). The samples for the defense priming experiment were root tissues collected after pathogen challenge at 0, 1, and 2 day post-pathogen challenge (dpp). (B) Quantification of the defense priming of SAR marker genes NbPR1a and NbPR2 in roots at 0, 1, and 2 days after inoculation with whitefly cDNA clones 2G4, 2G5, and 6A10 (left panel) and after challenge with R. solanacearum (right panel). Transcription is shown relative to empty vector (expression level = 1) with the NbActin gene as an internal reference. Bars represent the mean value ± SEM (N = 10). Infiltration with 1 mM BTH and Agrobacterium empty vector (EV) suspension was used as a positive and negative control, respectively. Different letters (A, B, a, b, c, and d; w, x, y, and z) within dpa and dpp indicate statistically significant differences (P < 0.05).
Second, to evaluate the defense priming of SAR marker genes NbPR1a and NbPR2, we measured their expression at 0, 1, and 2 days after drench application of R. solanacearum on roots (Figure 5B, right panel). In plants subjected to leaf infiltration with 2G4 and 6A10 cDNA, NbPR1a and NbPR2 were upregulated compared with the control at 2 days after pathogen challenge (Figure 5B, right panel). By contrast, pre-infiltration with 2G5 cDNA did not prime the expression of the two marker genes. The positive control treatment, 1 mM BTH, significantly increased NbPR1a and NbPR2 expression after both direct infiltration and pathogen drench treatment (Figure 5B, right panel).
In Silico Analysis of Effector Expression in Whitefly
When whiteflies feed on phloem, they first produce saliva (Jiang et al., 1999; Jiang and Walker, 2003). This saliva, like the saliva of other herbivores, is thought to contain effector molecules produced by the salivary glands (Bos et al., 2010; Villarroel et al., 2016). From the de novo assembled RNA-seq data, the contig N50 number, median contig length, average contig length, and total assembled bases were 2445, 448, 1089.85, and 94604597 respectively. To validate candidate effector production in whitefly, we first performed qRT-PCR analysis to measure the expression levels of the three candidate genes from whitefly fed on tobacco (Figure 6A). All three candidates were expressed in whole whitefly adults (Figure 6A). Second, to confirm expression of the whitefly effectors in salivary glands, we generated RNA sequencing (RNA seq) libraries from whitefly salivary glands, thorax (salivary gland enriched), midgut, abdomen (midgut enriched), nymphs, and eggs from whiteflies collected from cucumber (Figure 6B).
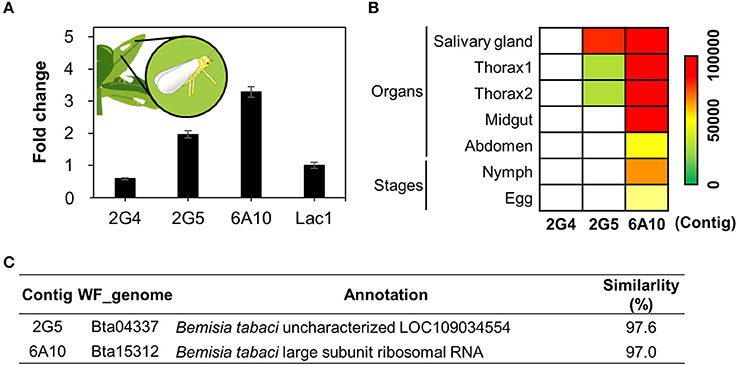
Figure 6. Validation of candidate whitefly effectors via in silico and qRT-PCR analysis. (A) Transcript levels of candidate effector 2G4, 2G5, and 6A10 in whitefly feeding on tobacco as determined by qRT-PCR. The whitefly effector Lac1 was used as a positive control. The expression levels were presented as relative values compared to Lac1(expression level = 1). (B) Relative expression levels of the candidate effectors 2G4, 2G5, and 6A10 in different organs during whitefly feeding on cucumber. Colors indicate normalized expression levels within the whitefly RNAseq samples of salivary gland, thorax (salivary gland enriched), midgut, abdomen, 1st−3rd instar (nymphs) and eggs. (C) Annotation of the three candidate effector 2G4, 2G5, and 6A10. The detailed methodology is described in the Materials and Methods.
Transcripts of 6A10 and 2G5 were detected in both whitelfy salivary gland and thorax tissue, indicating that they might be transferred in to plant tissue (Figure 6B). The 2G5 effector shows high expression in salivary glands and thorax compared to midgut, abdomen, nymph and egg. However, 2G4 expression is overall low (Figures 6A,B). These data indicate that 6A10 and 2G5 are indeed expressed in salivary glands, pointing to the possibility that they are transferred into plant tissue (Figure 6B). However, we did not detect any mRNA of 2G4 in the whitefly organs nor nymph/egg (Figure 6B). The candidate effector cDNAs encode proteins annotated as follows: 2G5, an unknown protein, and 6A10, large subunit ribosomal RNA (Figure 6A). The expression levels of these genes did not significantly differ from that of the positive control, Lac1 (Figure 6A), encoding a recently identified whitefly effector (Yang et al., 2017). Collectively, of the three clones, only clones 2G5 and 6A10 represent solid candidate whitefly effectors.
Discussion
Our study demonstrates that transient expression of putative whitefly effector cDNA induces plant systemic resistance against the soil-borne pathogen R. solanacearum, as well as the air-borne (aboveground) pathogen syringae pv. tabaci (Pta), in N. benthamiana. These findings add to our previous finding that whitefly infestation elicits plant immunity in the root system through signal transduction from aboveground to belowground plant parts (Yang et al., 2011). Crosstalk between hormone signaling pathways is often detected in plants infested by chewing and phloem-sucking insects. In contrast to JA/ET-dependent signaling elicited by chewing insects, the infestation of Arabidopsis with sucking insects such as whitefly increases the expression of marker genes for the SA-response pathway (Park and Ryu, 2014).
In this study, we designed a new high-throughput screening protocol for isolating putative whitefly effectors that are translocated to the plant and activate systemic plant immunity. We performed Agrobacterium-mediated transient transformation of N. benthamiana leaf tissues with cDNA prepared from total RNA extracted from whitefly during infestation at mid-day (Figure 1A). Cross-inoculation with whitefly cDNA and virulent/avirulent pathogens allowed us to detect the induction of plant resistance responses (Figures 1B,C). The three selected candidate effectors suppressed avirulent pathogen-mediated HR responses and virulent pathogen-mediated symptom development. Indeed, we previously demonstrated bacterial effector-induced suppression of both the HR and pathogen-mediated symptom development, and transcriptome analysis revealed that whitefly infestation induced the expression of a large portion of a set of genes for plant immunity in leaves and roots (Park and Ryu, 2014). However, the identity of the determinants from whitefly that elicit SA signaling from leaf to root was previously unclear.
Our current results describing SA-responsive gene expression induced by candidate effectors in leaves are in agreement with previous investigations of SA marker genes in pepper and tobacco (Yang et al., 2011; Song et al., 2015). The induction of SA-responsive genes detected in the current study corresponded to the induction of plant immunity against Pta (Figure 2). More interestingly, the strength of whitefly effector-mediated SAR was more obvious in local leaves (transiently expressing elicitor) than in systemic (distal) leaves. The infiltration of all three candidate effectors was sufficient to attenuate pathogen growth in the intracellular spaces of local but not systemic leaves on day 5, but bacterial numbers were reduced on day 3 in both local and systemic leaves (Figure 2). The results of defense-related gene expression analysis support the differential induction of SAR in local vs. systemic leaves (Figure 3). The defense priming of SA-responsive biomarker genes NbPR1a and NbPR2 was weaker in systemic vs. local leaves (Figure 3A). More importantly, NbPR1a expression was not detected systemically at 7 days after cDNA infiltration in the face of pathogen challenge (Figure 3B).
As expected, systemic plant immunity was induced by transient expression of the candidate whitefly effectors. Of the three candidates, 2G5 and 6A10 cDNA suppressed bacterial wilt symptom caused by R. solanacearum (Figure 4B). This decrease in 2G5 treatment severity could not be explained by defense priming of SA-responsive genes NbPR1a and NbPR2 (Figure 5B). Pre-infiltration with 2G5 cDNA did not prime the response of the two SAR marker genes to R. solanacearum challenge at 1 dpp in roots (Figure 5B). These results might be due to the weak activation of defense priming by clone 2G5 (Figure 5B). The induction of SAR by clone 2G5 cannot be dependent of SA signaling but can be dependent some other defense signaling such as jasmonic acid or ethylene signaling (Hase et al., 2008; Baichoo and Jaufeerally-Fakim, 2017; Liu et al., 2017). In contrast, the pre-infiltration with 6A10 cDNA elicited defense priming in similar level of BTH treatment used as a positive control (Figure 5B). A previous study also showed strong defense priming when plants activated the SAR response (Song et al., 2015). Overall, these results represent the first demonstration that a single whitefly effector elicits SAR against aboveground and belowground microbial pathogens through systemic signal transduction.
The candidate effector cDNAs are annotated as encoding large subunit ribosomal RNA (6A10) and an unknown protein (2G5). Bioinformatics analysis revealed the presence of 2G5 and 6A10 in whitefly salivary glands. The identification of 6A10 as a large subunit ribosomal RNA of B. tabaci deserves further study. There are many examples of the secretion of effector proteins that modulate plant immunity in saliva from hemipterans including aphids and whitefly (Atkins et al., 2011; Will et al., 2013; Sharma et al., 2014; Su et al., 2015; Peng et al., 2016; Villarroel et al., 2016). For instance, the effector proteins C002, Mp1, and Mp2 from aphid promote fecundity, whereas Mp10 and Mp42 decrease fecundity (Bos et al., 2010; Pitino and Hogenhout, 2013).
We do not fully understand how whitefly rRNAs such as 6A10 are translocated to plant cells to elicit SAR. Besides protein effectors, non-protein salivary factors can also act as effectors (Su et al., 2015). Bacterial rRNAs and plant DNA were recently shown to elicit SAR (Bhat and Ryu, 2016; Lee et al., 2016). Destroying the structure of bacterial rRNA via sonication and RNase treatment greatly reduced its effect on inducing SAR, indicating that certain (structural or sequence) signatures of rRNA are required for full SAR elicitation. While this signature has not been identified, bacterial rRNA was successfully detected in plant cells. It appears that the plant recognizes whitefly rRNA as a non-self-molecular pattern. Also, three small RNAs from whitefly were detected in tomato leaf tissue through sequencing tomato phloem RNA from whitefly-infested plants and the nymphs themselves, and the translocation of these RNAs was confirmed using stem-loop qRT-PCR (van Kleeff et al., 2016). The translocation of sRNA has also been observed during plant-fungus interactions. The gray mold fungal pathogen Botrytis cinerea delivers its sRNA and suppresses host defense responses through silencing host mRNAs related to defense signaling (Weiberg et al., 2013). Like non-coding sRNAs that function as effectors from whitefly, the non-coding rRNA identified in the current study appears to function as a trigger of SAR, a notion that is currently under investigation.
In conclusion, we revealed a new function for whitefly effectors, i.e., eliciting systemic immunity from aboveground to belowground plant parts. Adding to our previous discovery of whitefly-mediated SAR against aboveground and soil-borne plant pathogens, in the current study, we demonstrated that treatment with candidate whitefly effectors alone was sufficient to elicit plant immunity against microbial pathogens in local and systemic tissues (Figures 7A,B). In silico and qRT-PCR analyses confirmed that the candidate effectors were expressed. Both 2G5 and 6A10 were expressed in salivary glands and could be translocated into the host plant, resulting in defense priming of SAR-related marker genes, even in distal tissues. To our knowledge, this is the first report of whitefly effector-mediated induction of plant systemic immunity.
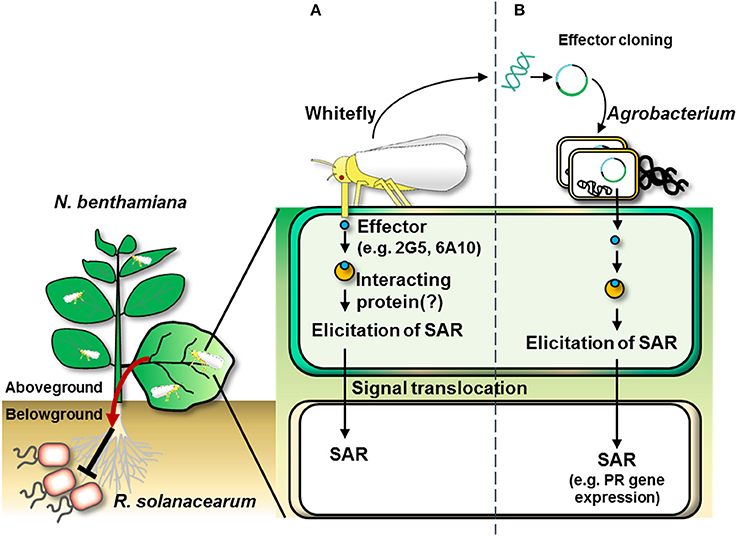
Figure 7. Overall scheme of whitefly effector-induced plant SAR against aboveground and belowground pathogens. (A) The infestation of whitefly on the leaf delivers effectors into the plant. The whitefly effectors might interact with plant partner proteins to induce plant systemic immunity (referred to as “systemic acquired resistance”) as indicated by the transcriptional activation of pathogenesis-related genes (e.g., NbPR1a and NbPR2). (B) Whitefly effector cloning. Agrobacterium-mediated transient expression of cDNAs of candidate effectors derived from total RNA from whitefly elicits SAR in N. benthamiana.
Author Contributions
C-MR designed the study. H-RL, SL, and SP performed experiments. PvK performed the RNA-seq sampling and bioinformatic analysis. RC supervised PvK. C-MR contributed to scientific discussions that guided the project's direction. C-MR and H-RL wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ji Hyun Lee to provide the disease scale of bacterial wilt. This research was supported by the grants from the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Initiative Program, the Agenda Project (Agenda Project No. PJ012814) of the Rural Development Administration, and by the Advanced Biomass R&D Center (ABC) of the Global Frontier Project funded by the Ministry of Science and ICT (ABC-2015M3A6A2065697), South Korea for C-MR. PvK was supported by the Netherlands Organization for Scientific Research (NWO grant 848.13.001).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00090/full#supplementary-material
References
Abdellatef, E., Will, T., Koch, A., Imani, J., Vilcinskas, A., and Kogel, K. H. (2015). Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol. J. 13, 849–857. doi: 10.1111/pbi.12322
Agrios, G. N. (2005). Plant Pathology, 5th Edn. San Diego, CA: Academic Press; Department of Plant Pathology, University of Florida.
Atkins, C. A., Smith, P. M., and Rodriguez-Medina, C. (2011). Macromolecules in phloem exudates—a review. Protoplasma 248, 165–172. doi: 10.1007/s00709-010-0236-3
Baichoo, Z., and Jaufeerally-Fakim, Y. (2017). Ralstonia solanacearum upregulates marker genes of the salicylic acid and ethylene signaling pathways but not those of the jasmonic acid pathway in leaflets of Solanum lines during early stage of infection. Eur. J. Plant Pathol. 147, 615–625. doi: 10.1007/s10658-016-1030-7
Bhat, A., and Ryu, C. M. (2016). Plant perceptions of extracellular DNA and RNA. Mol. Plant. 9, 956–958. doi: 10.1016/j.molp.2016.05.014
Bleeker, P. M., Diergaarde, P. J., Ament, K., Schütz, S., Johne, B., Dijkink, J., et al. (2011). Tomato-produced 7-epizingiberene and R-curcumene act as repellents to whiteflies. Phytochemistry 72, 68–73. doi: 10.1016/j.phytochem.2010.10.014
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bos, J. I., Prince, D., Pitino, M., Maffei, M. E., Win, J., and Hogenhout, S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6:e1001216. doi: 10.1371/journal.pgen.1001216
Brown, R. L., Kazan, K., McGrath, K. C., Maclean, D. J., and Manners, J. M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1. 2 gene of Arabidopsis. Plant Physiol. 132, 1020–1032. doi: 10.1104/pp.102.017814
Chandrasekaran, M., Subramanian, D., Yoon, E., Kwon, T., and Chun, S. C. (2016). Meta-analysis reveals that the genus Pseudomonas can be a better choice of biological control agent against bacterial wilt disease caused by Ralstonia solanacearum. Plant Pathol. J. 32, 216. doi: 10.5423/PPJ.OA.11.2015.0235
Cooper, W. R., Dillwith, J. W., and Puterka, G. J. (2010). Salivary proteins of russian wheat aphid (Hemiptera: Aphididae). Environ. Entomol. 39, 223–231. doi: 10.1603/EN09079
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Elzinga, D. A., and Jander, G. (2013). The role of protein effectors in plant–aphid interactions. Curr. Opin. Plant Biol. 16, 451–456. doi: 10.1016/j.pbi.2013.06.018
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hase, S., Takahashi, S., Takenaka, S., Nakaho, K., Arie, T., Seo, S., et al. (2008). Involvement of jasmonic acid signalling in bacterial wilt disease resistance induced by biocontrol agent Pythium oligandrum in tomato. Plant Pathol. 57, 870–876. doi: 10.1111/j.1365-3059.2008.01858.x
Jiang, Y. X., Lei, H., Collar, J. L., Martin, B., Muñiz, M., and Fereres, A. (1999). Probing and feeding behavior of two distinct biotypes of Bemisia tabaci (Homoptera: Aleyrodidae) on tomato plants. J. Econ. Entomol. 92, 357–366. doi: 10.1093/jee/92.2.357
Jiang, Y. X., and Walker, G. P. (2003). Electrical penetration graphs of the nymphal stage of Bemisia argentifolii. Entomol. Exp. Appl. 109, 101–111. doi: 10.1046/j.1570-7458.2003.00093.x
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323. doi: 10.1038/nature05286
Kaloshian, I., and Walling, L. L. (2015). “Plant immunity: connecting the dots between microbial and hemipteran immune responses,” in Management of Insect Pests to Agriculture; Lessons Learned From Deciphering Their Genome, Transcriptome and Proteome, eds H. Czosnek and M. Ghanim (Cham: Springer), 217–243. doi: 10.1007/978-3-319-24049-7_9
Kempema, L. A., Cui, X., Holzer, F. M., and Walling, L. L. (2007). Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 143, 849–865. doi: 10.1104/pp.106.090662
King, R. W., and Zeevaart, J. (1974). Enhancement of phloem exudation from cut petioles by chelating agents. Plant Physiol. 53, 96–103. doi: 10.1104/pp.53.1.96
Kliot, A., Kontsedalov, S., Lebedev, G., Brumin, M., Cathrin, P. B., Marubayashi, J. M., et al. (2014). Fluorescence in situ hybridizations (FISH) for the localization of viruses and endosymbiotic bacteria in plant and insect tissues. J. Vis. Exp. e51030. doi: 10.3791/51030
Koornneef, A., and Pieterse, C. M. (2008). Cross talk in defense signaling. Plant Physiol. 146, 839–844. doi: 10.1104/pp.107.112029
Lazebnik, J., Frago, E., Dicke, M., and van Loon, J. J. (2014). Phytohormone mediation of interactions between herbivores and plant pathogens. J. Chem. Ecol. 40, 730–741. doi: 10.1007/s10886-014-0480-7
Lee, B., Lee, S., and Ryu, C. M. (2012). Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Ann. Bot. 110, 281–290. doi: 10.1093/aob/mcs055
Lee, B., Park, Y. S., Lee, S., Song, G. C., and Ryu, C. M. (2016). Bacterial RNAs activate innate immunity in Arabidopsis. New Phytol. 209, 785–797. doi: 10.1111/nph.13717
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Liu, Q., Liu, Y., Tang, Y., Chen, J., and Ding, W. (2017). Overexpression of NtWRKY50 increases resistance to Ralstonia solanacearum and alters salicylic acid and jasmonic acid production in Tobacco. Front. Plant Sci. 8:1710. doi: 10.3389/fpls.2017.01710
Louis, J., and Shah, J. (2013). Arabidopsis thaliana—Myzus persicae interaction: shaping the understanding of plant defense against phloem-feeding aphids. Front. Plant Sci. 4:213. doi: 10.3389/fpls.2013.00213
Murphy, J. F., Reddy, M. S., Ryu, C. M., Kloepper, J. W., and Li, R. (2003). Rhizobacteria-mediated growth promotion of tomato leads to protection against Cucumber mosaic virus. Phytopathology 93, 1301–1307. doi: 10.1094/PHYTO.2003.93.10.1301
Mutti, N. S., Park, Y., Reese, J. C., and Reeck, G. R. (2006). RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J. Insect Sci. 6, 1–7. doi: 10.1673/031.006.3801
Park, Y. S., and Ryu, C. M. (2014). Understanding cross-communication between aboveground and belowground tissues via transcriptome analysis of a sucking insect whitefly-infested pepper plants. Biochem. Biophys. Res. Commun. 443, 272–277. doi: 10.1016/j.bbrc.2013.11.105
Peng, H. C., Mantelin, S., Hicks, G. R., Takken, F. L., and Kaloshian, I. (2016). The conformation of the plasma membrane-localized SlSERK1-Mi-1.2 complex is altered by a potato aphid derived effector. Plant Physiol. 171, 2211–2222. doi: 10.1104/pp.16.00295
Pitino, M., and Hogenhout, S. A. (2013). Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant Microbe. Interact. 26, 130–139. doi: 10.1094/MPMI-07-12-0172-FI
Rao, S. A., Carolan, J. C., and Wilkinson, T. L. (2013). Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PLoS ONE 8:e57413. doi: 10.1371/journal.pone.0057413
Rodriguez-Medina, C., Atkins, C. A., Mann, A. J., Jordan, M. E., and Smith, P. M. (2011). Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.). BMC Plant Biol. 11:36. doi: 10.1186/1471-2229-11-36
Sharma, A., Khan, A. N., Subrahmanyam, S., Raman, A., Taylor, G. S., and Fletcher, M. J. (2014). Salivary proteins of plant-feeding hemipteroids–implication in phytophagy. Bull. Entomol. Res. 104, 117–136. doi: 10.1017/S0007485313000618
Song, G. C., Lee, S., Hong, J., Choi, H. K., Hong, G. H., Bae, D. W., et al. (2015). Aboveground insect infestation attenuates belowground Agrobacterium-mediated genetic transformation. New Phytol. 207, 148–158. doi: 10.1111/nph.13324
Song, G. C., Sim, H.-J., Kim, S.-G., and Ryu, C.-M. (2016). Root-mediated signal transmission of systemic acquired resistance against above-ground and below-ground pathogens. Ann. Bot. 118, 821–831. doi: 10.1093/aob/mcw152
Su, Q., Oliver, K. M., Xie, W., Wu, Q., Wang, S., and Zhang, Y. (2015). The whitefly-associated facultative symbiont Hamiltonella defensa suppresses induced plant defences in tomato. Funct. Ecol. 29, 1007–1018. doi: 10.1111/1365-2435.12405
Su, Y. L., Li, J. M., Li, M., Luan, J. B., Ye, X. D., Wang, X. W., et al. (2012). Transcriptomic analysis of the salivary glands of an invasive whitefly. PLoS ONE 7:e39303. doi: 10.1371/journal.pone.0039303
van Dam, N. M., and Heil, M. (2011). Multitrophic interactions below and above ground: en route to the next level. J. Ecol. 99, 77–88. doi: 10.1111/j.1365-2745.2010.01761.x
VanDoorn, A., de Vries, M., Kant, M. R., and Schuurink, R. C. (2015). Whiteflies glycosylate salicylic acid and secrete the conjugate via their honeydew. J. Chem. Ecol. 41, 52–58. doi: 10.1007/s10886-014-0543-9
van Kleeff, P. J., Galland, M., Schuurink, R. C., and Bleeker, P. M. (2016). Small RNAs from Bemisia tabaci are transferred to Solanum lycopersicum phloem during feeding. Front. Plant Sci. 7:1759. doi: 10.3389/fpls.2016.01759
Van Oosten, V. R., Bodenhausen, N., Reymond, P., Van Pelt, J. A., Van Loon, L. C., Dicke, M., et al. (2008). Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis. Mol. Plant Microbe. Interact. 21, 919–930. doi: 10.1094/MPMI-21-7-0919
Villarroel, C. A., Jonckheere, W., Alba, J. M., Glas, J. J., Dermauw, W., Haring, M. A., et al. (2016). Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 86, 119–131. doi: 10.1111/tpj.13152
Walling, L. L. (2008). Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 146, 859–866. doi: 10.1104/pp.107.113142
Wang, W., Dai, H., Zhang, Y., Chandrasekar, R., Luo, L., Hiromasa, Y., et al. (2015a). Armet is an effector protein mediating aphid–plant interactions. FASEB J. 29, 2032–2045. doi: 10.1096/fj.14-266023
Wang, W., Luo, L., Lu, H., Chen, S., Kang, L., and Cui, F. (2015b). Angiotensin-converting enzymes modulate aphid–plant interactions. Sci. Rep. 5:8885. doi: 10.1038/srep08885
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Will, T., Furch, A. C., and Zimmermann, M. R. (2013). How phloem-feeding insects face the challenge of phloem-located defenses. Front. Plant Sci. 4:336. doi: 10.3389/fpls.2013.00336
Will, T., and Vilcinskas, A. (2015). The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Biol. 57, 34–40. doi: 10.1016/j.ibmb.2014.12.005
Win, J., Kamoun, S., and Jones, A. M. (2011). Purification of effector–target protein complexes via transient expression in Nicotiana benthamiana. Methods Mol. Biol. 712, 181–194. doi: 10.1007/978-1-61737-998-7_15
Yang, C. H., Guo, J. Y., Chu, D., Ding, T. B., Wei, K. K., Cheng, D. F., et al. (2017). Secretory laccase 1 in Bemisia tabaci MED is involved in whitefly-plant interaction. Sci. Rep. 7, 3623. doi: 10.1038/s41598-017-03765-y
Yang, J. W., Yi, H. S., Kim, H., Lee, B., Lee, S., Ghim, S. Y., et al. (2011). Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below-ground microflora. J. Ecol. 99, 46–56. doi: 10.1111/j.1365-2745.2010.01756.x
Yang, J.-W., Yu, S.-H., and Ryu, C.-M. (2009). Priming of defense-related genes confers root-colonizing bacilli-elicited induced systemic resistance in pepper. Plant Pathology J. 25, 389–399. doi: 10.5423/PPJ.2009.25.4.389
Keywords: whitefly, effector, Nicotiana benthamiana, Ralstonia solanacearum, transient expression
Citation: Lee H-R, Lee S, Park S, van Kleeff PJM, Schuurink RC and Ryu C-M (2018) Transient Expression of Whitefly Effectors in Nicotiana benthamiana Leaves Activates Systemic Immunity Against the Leaf Pathogen Pseudomonas syringae and Soil-Borne Pathogen Ralstonia solanacearum. Front. Ecol. Evol. 6:90. doi: 10.3389/fevo.2018.00090
Received: 13 March 2018; Accepted: 06 June 2018;
Published: 06 July 2018.
Edited by:
Ana Pineda, Netherlands Institute of Ecology (NIOO-KNAW), NetherlandsReviewed by:
Ioannis Stringlis, Utrecht University, NetherlandsFabio Cortesi, The University of Queensland, Australia
Copyright © 2018 Lee, Lee, Park, van Kleeff, Schuurink and Ryu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Choong-Min Ryu, Y21yeXVAa3JpYmIucmUua3I=
 Hae-Ran Lee1
Hae-Ran Lee1 Seyeon Park
Seyeon Park Paula J. M. van Kleeff
Paula J. M. van Kleeff Robert C. Schuurink
Robert C. Schuurink Choong-Min Ryu
Choong-Min Ryu