- 1School of Biological Sciences, University of Auckland, Auckland, New Zealand
- 2Department of Anthropology, University of Auckland, Auckland, New Zealand
- 3Department of Statistics, University of Auckland, Auckland, New Zealand
Two species of invasive rats (Rattus norvegicus and R. rattus) arrived in New Zealand with Europeans in the mid to late eighteenth and nineteenth century respectively. They rapidly spread across the main islands of New Zealand and its offshore islands, displacing the historically introduced R. exulans. Today both species are widespread although the distribution of the sub-dominant R. norvegicus is patchy. Tissue samples were obtained from 425 R. rattus and 130 R. norvegicus across the New Zealand archipelago and neighboring islands. We sequenced a standard 545 base pair section of the mitochondrial D-loop in order to construct a modern phylogeography of the two species and to make inference on historical invasion pathways and spread across the country. We found limited diversity in R. norvegicus haplotypes, with two widespread haplotypes across New Zealand and its offshore islands most likely corresponding to two independent invasions, potentially with English and Chinese origins, respectively. In contrast we found widespread diversity in R. rattus haplotypes across New Zealand and its offshore islands, most likely corresponding to at least four independent invasions to the main North and South Islands, Great Barrier Island archipelago, and Stewart Island archipelago. The most common R. rattus haplogroup was found throughout New Zealand and many of its offshore islands, as well as neighboring islands in the Tasman Sea, and has been documented elsewhere across the Pacific, but with European origins. We also found both geographic partitioning and secondary invasions of haplotypes within the main North and South Island. In addition to distinct haplogroups differing by over three base pairs, which exhibit geographical partitioning suggestive of independent invasion events, for both species we also found instances of single base-pair differences within localities, elevating haplotype diversity. The geographical distribution of pelage color morphs also correlates with haplotype distribution, lending further support to the hypothesis and role of independent invasion events.
Introduction
Three species of rats have been introduced to the New Zealand archipelago: Rattus exulans (Pacific or Polynesian rat), R. norvegicus (Norway or brown rat) and R. rattus (ship or black rat). R. exulans was introduced by Polynesian settlers in the late thirteenth century (Wilmshurst et al., 2008), while R. norvegicus and R. rattus were introduced by European explorers and settlers in the mid to late eighteenth and nineteenth century respectively (Atkinson, 1973). Each species rapidly spread throughout the country, and in turn displaced those rat species which had arrived previous to it (Russell et al., 2014). As of the late twentieth century R. exulans was mostly restricted to remote parts of the main islands and a few offshore islands of New Zealand, R. norvegicus was distributed patchily across the main islands and some offshore islands of New Zealand, and R. rattus was distributed abundantly across the main islands and some offshore islands of New Zealand (Russell and Clout, 2004).
Since the middle of the twentieth century these rat species have been progressively eradicated from the uninhabited offshore islands of New Zealand (Clout and Russell, 2006; Russell and Broome, 2016), and since the start of the twenty-first century controlled widely across the main islands of New Zealand (Brown et al., 2015; Russell et al., 2015). The rat species of European origin (R. norvegicus and R. rattus) have well-documented dispersal capabilities over water and land (Russell et al., 2005; Abdelkrim et al., 2010), indeed R. norvegicus is particularly effective at dispersal by swimming and can cross water gaps of two kilometers, while R. rattus can cross gaps of hundreds of meters (Bassett et al., 2016). Population genetic studies have helped determine the putative origin of rats on islands (Robins et al., 2016), and whether rats discovered on islands following eradication are survivors or reinvaders (Russell et al., 2010). Such genetic studies have contributed to improved management of invasive rats across New Zealand.
The population genetics of house mice (Mus musculus) has been rigorously assessed across the New Zealand archipelago using a variety of genetic markers (Searle et al., 2009; King et al., 2016; Veale et al., 2018). The population genetics of R. exulans has also been assessed, as a proxy for Polynesian migration (Matisoo-Smith et al., 1998). However, to date, there has not been a comprehensive national survey of R. norvegicus and R. rattus genetic diversity across the entirety of the New Zealand archipelago. Regional studies of genetic diversity in New Zealand have been undertaken, particularly for island groups and adjacent coastlines (Russell et al., 2009, 2010; Abdelkrim et al., 2010; Fewster et al., 2011; Robins et al., 2016). Similar regional studies of R. norvegicus and R. rattus have also been undertaken internationally (Aplin et al., 2011; Song et al., 2014). Such molecular studies can have a variety of applications, including increasing knowledge of invasive rat taxonomy (Bastos et al., 2011; Conroy et al., 2013), evolutionary biology (Lack et al., 2012; Konečný et al., 2013,), ecology (Theuerkauf et al., 2015; Varudkar and Ramakrishnan, 2015), disease epidemiology (Tollenaere et al., 2012; Brouat et al., 2013; Richardson et al., 2017), management (Kaleme et al., 2011; Kajdacsi et al., 2013; Haniza et al., 2015) and historical biogeography (Tollenaere et al., 2010; Lack et al., 2013; López et al., 2013; Brouat et al., 2014; Colangelo et al., 2015; Berthier et al., 2016). In this work, the first comprehensive national survey of mitochondrial genetic diversity for R. norvegicus and R. rattus across the New Zealand archipelago and surrounding islands is undertaken.
Materials and Methods
Samples
Tissue samples were acquired from New Zealand and surrounding islands from 2001 to 2015 through research collections (i.e., regional studies), opportunistic collection (e.g., road kill), and requests to people or groups undertaking rat trapping (Supplementary Table 1). Tissue samples were provided as a by-product of pest control in accordance with the Conservation Act (1987) and thus were exempt from animal ethics committee approval under the New Zealand Animal Welfare Act (1999) 30B.1.b.iii, except where indicated in cited studies. Collections were made with a focus on representative geographic coverage of as many islands as possible on which either rat species was known to be or have been present, and for islands larger than 100,000 hectares representative coverage was sought across those islands (i.e., North, South, Stewart). As well as all outlying islands within the New Zealand Exclusive Economic Zone (Campbell, Chatham and Raoul Islands), samples were also obtained from Macquarie, Lord Howe and Norfolk Islands, and Port Jackson, Sydney (Australia). Tissue samples ranged in quality from freshly caught to degraded by some weeks, but previous work assured us that this did not affect the quality of DNA extraction for the level of molecular resolution we required. Tissue samples were geoindexed precisely with GPS or on large islands approximately to nearby landmarks (variation of a few kilometers). Rat species identification was not always known or accurate, but was invariably confirmed following molecular typing.
Sequencing
DNA was extracted using the DNeasy Tissue Kit (Qiagen), or the High Pure PCR Template Preparation Kit (Roche Diagnostics). The 585 bp amplicon of the D-Loop was amplified with the primers EGL4L and RJ3R (Robins et al., 2007). The reaction volume was 20 μL comprising: 10 mM Tris HCl pH 8.3; 50 mM KCl, 2.5 mM MgCl2, primers at 0.5 μM each, dNTPs at 0.15 mM each; 0.5 U of Taq polymerase, 1 μL of DNA template. The PCR (polymerase chain reaction) regime was an initial denaturation step of 94°C for 2 min; 35 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 1 min with a final extension step of 72°C for 5 min. PCR products were visualized and quantified, using a low mass ladder for comparison, on ethidium bromide stained 1% agarose gels. PCR products were purified with ExoSAP-IT (Affymetrix, Inc.). Sequencing was carried out at the Massey University Genome Service, Palmerston North, New Zealand using the BigDye Terminator version 3 sequencing kit, the GeneAmp PCR System 9700 and a capillary ABI3730 DNA analyser, all from Applied Biosystems.
Analysis
The raw sequences were trimmed, edited, aligned, and grouped into haplotypes using the software package SEQUENCHER (Gene Codes). The relationships among the haplotypes were estimated with a minimum spanning haplotype network (Bandelt et al., 1999) as implemented in PopART (http://popart.otago.ac.nz).
Results
A total of 23 unique haplotypes were found and lodged in GenBank (Table 1).
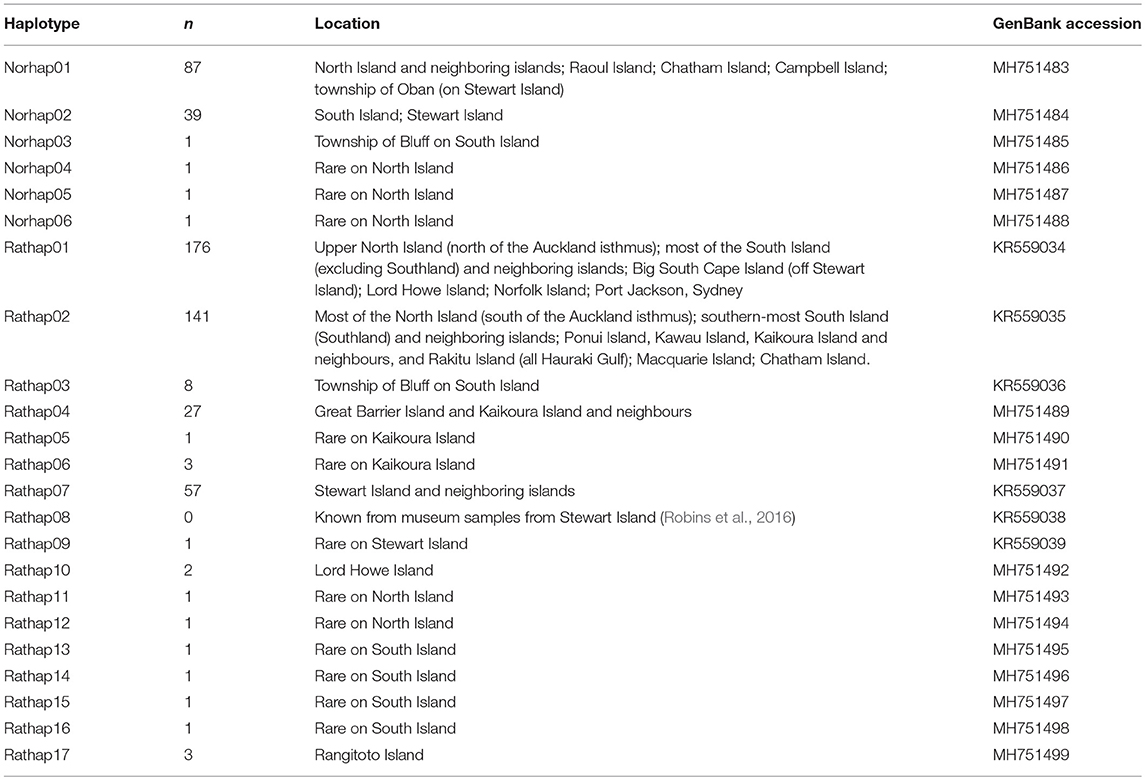
Table 1. The number of unique haplotypes across New Zealand and surrounding islands with GenBank accession numbers.
Rattus norvegicus
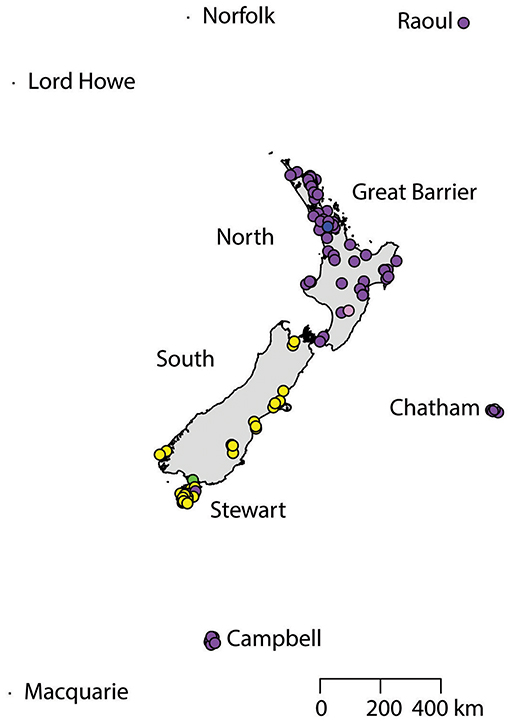
A total of 130 samples of R. norvegicus, from 24 islands, were included in our study. Notable absences included Kapiti and Mayor Islands (eradicated and no samples located). The aligned sequences, 545 nucleotides long, had differences in the base composition at 12 positions, and six haplotypes were found (Table 2). The relationships among the haplotypes are shown in Figure 1, and their distribution across New Zealand and surrounding islands in Figure 2.

Table 2. Differences in the base composition of R. norvegicus haplotypes from across the New Zealand archipelago.
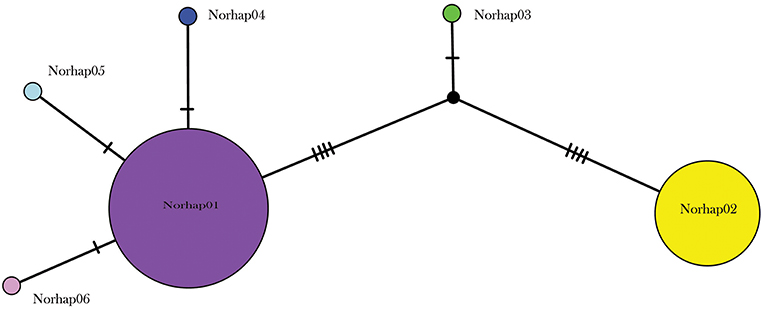
Figure 1. Median joining haplotype network for 130 R. norvegicus individuals from across the New Zealand archipelago.
Two geographically partitioned haplotypes were predominant. Norhap01 was found throughout the North Island and neighboring islands; Raoul Island; Chatham Island; and Campbell Island. The substantially different Norhap02 was found throughout the South Island and neighboring islands. Both Norhap01 and Norhap02 were found on Stewart Island, but Norhap01 was restricted to the port town of Oban. Forming a haplogroup with Norhap01, single instances of Norhap04, Norhap05 and Norhap06, singleton variations of Norhap01, were also found scattered throughout the North Island. The distinct haplotype Norhap03, intermediate between Norhap1 and Norhap02, was found in the port town of Bluff.
Rattus rattus
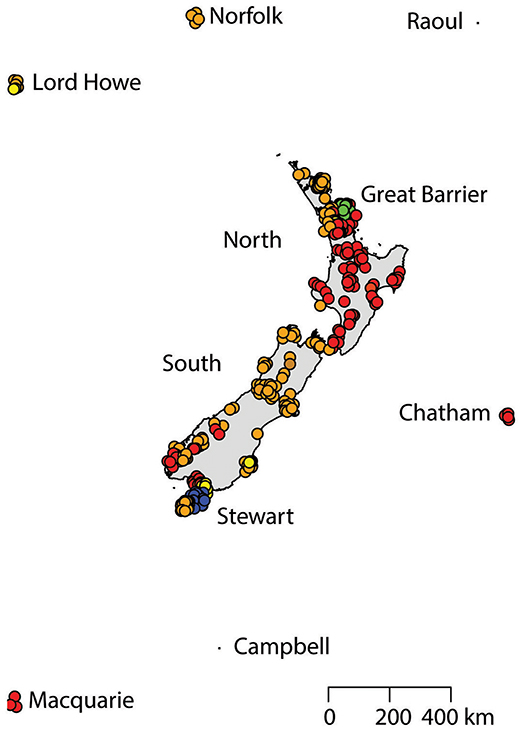
A total of 425 samples of R. rattus, from 31 islands, were included in our study. The only absence of note was Arapawa Island (extant but no samples located). The aligned sequences, 545 nucleotides long, had differences in the base composition at 17 positions, and 17 haplotypes were found (Table 3). The relationships among the haplotypes are shown in Figure 3 and their distribution across New Zealand and surrounding islands in Figures 4–6.
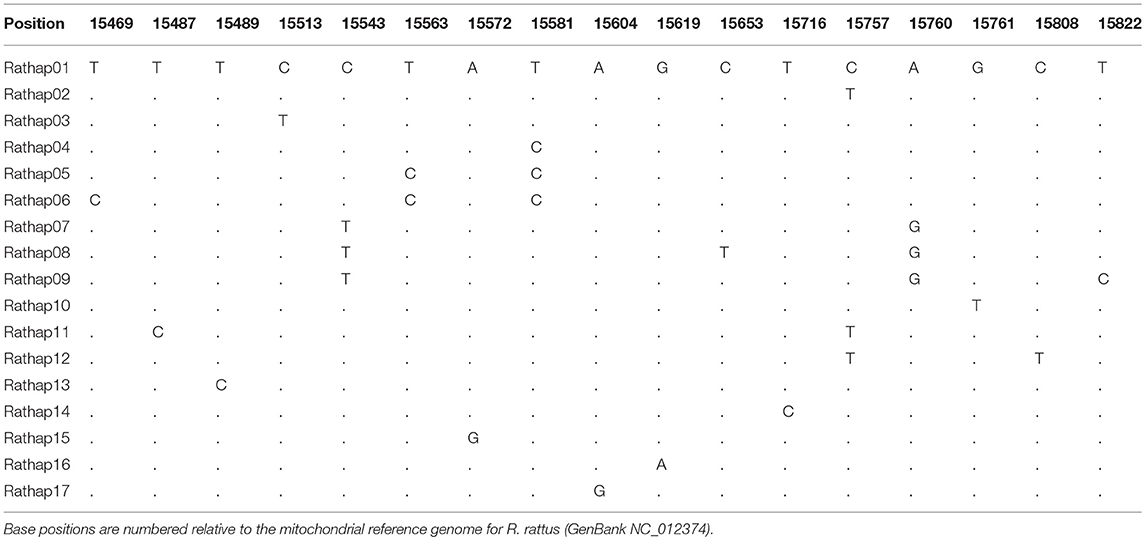
Table 3. Differences in the base composition of R. rattus haplotypes from across the New Zealand archipelago.
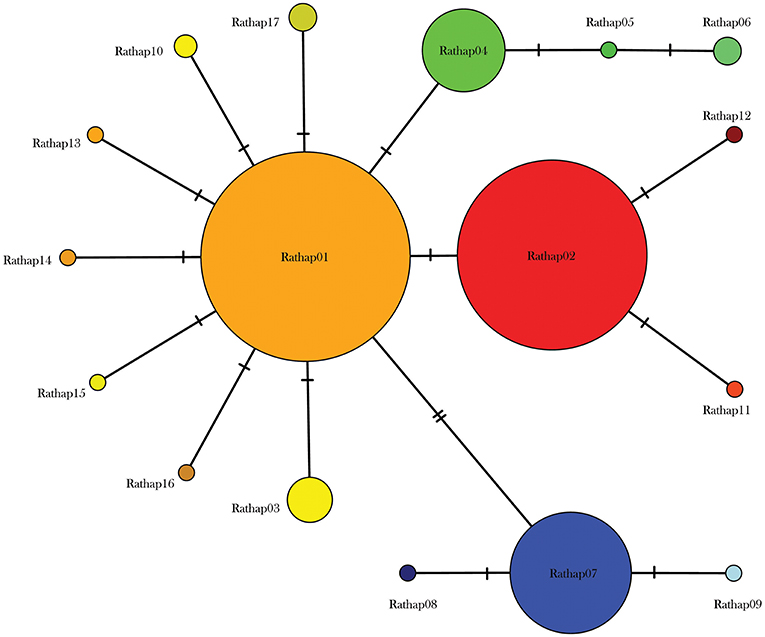
Figure 3. Median joining haplotype network for 425 R. rattus individuals from across the New Zealand archipelago.
Two geographically partitioned haplotypes were predominant. Rathap01 was found in the upper North Island (north of the Auckland isthmus); most of the South Island (excluding Southland) and neighboring islands; as well as Big South Cape Island (off Stewart Island); Lord Howe Island; Norfolk Island; and Port Jackson, Sydney. The closely related Rathap02 was found in most of the North Island (south of the Auckland isthmus); southern-most South Island (Southland) and neighboring islands; as well as Ponui Island, Kawau Island, Kaikoura Island and neighbours, and Rakitu Island (all Hauraki Gulf, Figure 5); Macquarie Island; and Chatham Island. Two other haplotypes were common. Rathap04 was found in Great Barrier Island and Kaikoura Island and neighbours, and Rathap07 was found in Stewart Island and neighboring islands (Figure 6).
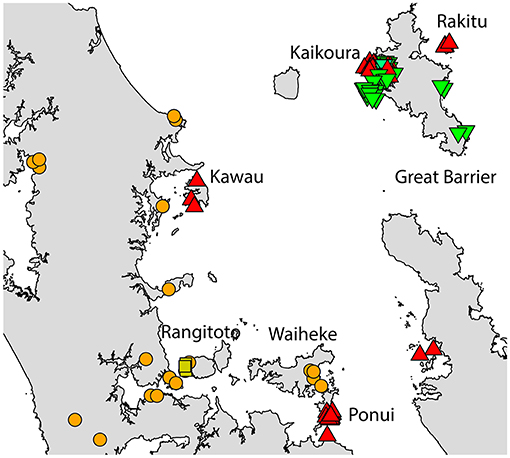
Figure 5. The distribution of R. rattus haplotypes across the Hauraki Gulf, Auckland.  = RatHap01,
= RatHap01,  = RatHap02,
= RatHap02,  = RatHap04, RatHap05 & RatHap06,
= RatHap04, RatHap05 & RatHap06,  = RatHap17.
= RatHap17.
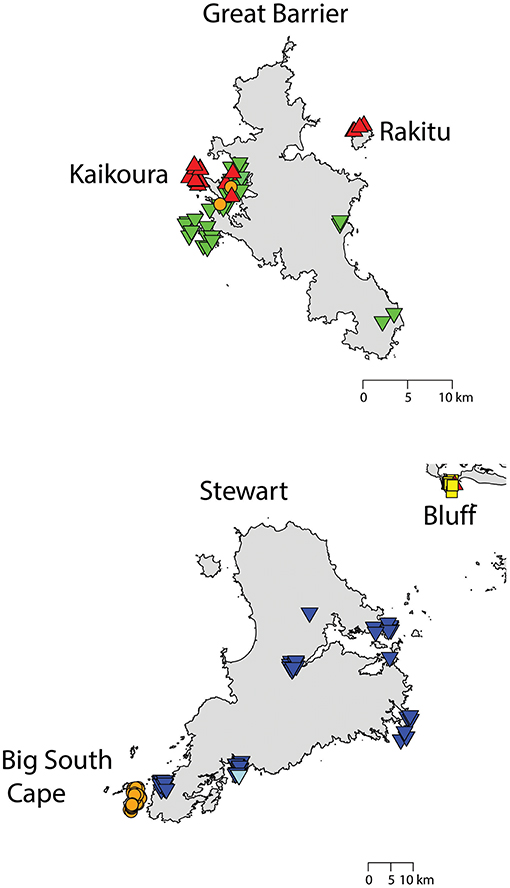
Figure 6. The distribution of R. rattus haplotypes on (top) Great Barrier Island and neighboring islands,  = RatHap01,
= RatHap01,  = RatHap02,
= RatHap02,  = RatHap04, RatHap05 & RatHap06; and (bottom) Stewart Island and neighboring islands,
= RatHap04, RatHap05 & RatHap06; and (bottom) Stewart Island and neighboring islands,  = RatHap01,
= RatHap01,  = RatHap02,
= RatHap02,  = RatHap03,
= RatHap03,  = RatHap07, RatHap08 & RatHap09.
= RatHap07, RatHap08 & RatHap09.
Forming a haplogroup with Rathap01, a few instances of Rathap17 and Rathap03, singleton variations of Rathap01, were found on Rangitoto Island and in the port town of Bluff. Single instances of Rathap13, Rathap14, Rathap15, and Rathap16, singleton variations of Rathap01, were also found scattered throughout the South Island. Rathap10, also a singleton variation of Rathap01, was found on Lord Howe Island. Forming a haplogroup with Rathap02, single instances of Rathap11 and Rathap12, singleton variations of Rathap02, were also found scattered throughout the North Island. Forming a haplogroup with Rathap04, a few instances of Rathap05 and Rathap06, singleton variations of Rathap04, were also found on Kaikoura Island. Forming a haplogroup with Rathap07, a single instance of Rathap09, a singleton variation of Rathap07, was also found on Stewart Island. We only had coverage of the area of the genome distinguishing RatHap09 from RatHap07 for 25% of samples from Stewart Island (the only location where either haplotype occurred). However, we had coverage of this area of the genome for 60% of samples in total, and did not find any further mutations. Nonetheless, RatHap09 might be under-represented in our results.
Discussion
The phylogeographies of both rat species introduced by Europeans exhibit marked geographic partitioning between distinct haplogroups, suggesting that more than one introduction of each species took place. Multiple independent introductions have also been inferred from mitochondrial DNA haplotypes of R. exulans and M. musculus in New Zealand (Matisoo-Smith et al., 1998; King et al., 2016).
From known historical records R. norvegicus was first observed in the North Island in 1772 and was widespread by the 1830s, while in the South Island the first observations were in the 1850s (Innes, 2005a), and was first observed in Stewart Island in the 1870s (Thomson, 1922). These records are in agreement with the two haplogroups identified, which are suggestive of at least two independent introductions. In keeping with the earlier North Island introduction date, single base-pair mutations from the dominant haplogroup were only detected in the North Island. Similar within-archipelago independent introductions of R. norvegicus have also been found in the Falklands Islands (Hingston et al., 2016). Large gaps in our coverage of the South Island for R. norvegicus correspond with the extent of alpine and beech forest distribution in New Zealand (Wardle, 1984), from where R. norvegicus is seemingly absent.
From known historical records R. rattus was only widespread in the North Island and Great Barrier Island after 1860, in the South Island after 1890, and was first observed on Stewart Island in 1911 (Atkinson, 1973). These records are in agreement with the four geographically partitioned haplogroups identified, which are suggestive of at least four independent introductions across the North Island, South Island, Great Barrier Island, and Stewart Island. In keeping with the similar introduction dates, single base-pair mutations from the dominant haplogroups were detected on all four islands. Together this evidence suggests invasion and spread were rapid across islands, and therefore a limited role for bridgehead effects from subsequent invasions (Bertelsmeier et al., 2018). Instead, incumbent advantage appears to have inhibited over-invasion by conspecifics (Waters et al., 2013).
For both species our findings most likely represent a minimum number of introductions, as additional introductions of an already-extant haplotype may also have taken place, perhaps explaining observations such as the dispersed geographic distributions of Rathap01 and Rathap02 across the North Island and South Island. Rare pre- or post-introduction mutations of single base pairs also cannot be unequivocally distinguished from additional introductions. Although there is good national coverage, sampling intensity was generally low within sites across the North Island and South Island. Thus, with rare haplotypes being detected, the results presented here should be interpreted cautiously (Muirhead et al., 2008).
Many of the haplotypes found in this study have been reported in other studies worldwide, but global representation is too patchy to support conclusive inference on historical invasion pathways to New Zealand. For R. norvegicus, Norhap01 was the most common haplotype found in modern samples from England (Haniza et al., 2015), while Norhap02 has only been reported in China (Liu et al., 2012). For R. rattus, Rathap01 has been found in modern relict island populations in England (Hingston et al., 2005) and across the Pacific Islands from New Guinea to French Polynesia (Robins et al., 2007). Closely related haplotypes to Rathap01 and Rathap02 have been found throughout the Mediterranean basin (Colangelo et al., 2015). Rathap04 has not been reported anywhere previously, while Rathap07 has been reported in New South Wales, Australia (Rowe et al., 2011). These links reaffirm the correspondence of R. rattus and R. norvegicus with European movements throughout the wider Pacific. The Asian link with Norhap02 might indicate a non-European origin for R. norvegicus in the South Island of New Zealand, as has also been inferred for introduced mice in the southern South Island (King et al., 2016).
Our study also detected evidence of ongoing contemporary movement of both rat species within New Zealand, particularly where we had comprehensive sampling on Great Barrier and Stewart Islands. A subset of data from this study has already been used to determine the putative distant origin of the R. rattus which invaded Big South Cape Island, off Stewart Island, in the 1960s (Robins et al., 2016). Stewart Island itself was probably colonized first by the South Island haplotype of R. norvegicus, and then more recently by the North Island haplotype restricted to the port town of Oban. Similarly, unique haplotypes were found for both R. norvegicus and R. rattus in the southern port town of Bluff, and had seemingly not spread far beyond this locality. Kaikoura Island and its neighboring islands (Nelson Island and Motuhaku Island), off the western coast of Great Barrier Island, had an unusual number of R. rattus haplotypes, with five haplotypes identified from 29 samples comprising a mix of the North Island and Great Barrier Island haplotypes, possibly indicative of recent long-distance transportation by boat. Invasive rats demonstrate a strong incumbent advantage both between and within species (Russell et al., 2014), so localized occurrence of a haplotype may indicate that this haplotype arrived subsequent to the initial invasion. This is especially suggestive for R. norvegicus, where the Bluff haplotype differs from all other sampled haplotypes by several base pairs, so is not a plausible in-situ mutation. Together, these results are suggestive that long-distance rat movements occurred at least well into the twentieth century.
The documented proportion of R. rattus pelage phenotypes in a region also appears to correspond to our haplotype distributions (Innes, 2005b). For example, higher proportions of the melanistic “rattus” (black) pelage occur in the South Island and upper North Island, corresponding with haplotype Rathap01, but are almost absent from Stewart Island, corresponding with Rathap07. The inheritance of pelage color is through known genetic markers unrelated to mtDNA haplotype (Kambe et al., 2011), but a geographical association between pelage color and haplogroup may arise due to the genetic makeup of different founding populations. Similarly, the rare occurrence in R. rattus of the Tyr25Phe mutation in VKorc1, which is associated with anti-coagulant resistance, also corresponds with the distribution of haplotype Rathap02 (Cowan et al., 2017).
Advances in molecular biology since this work was undertaken already allow higher resolution characterisation of individual genetic variability. Single nucleotide polymorphisms (SNPs) in particular facilitate deeper insight into the population genetics of invasive rats (e.g., Puckett et al., 2016). Results at a higher resolution from future genetic studies of invasive rats in New Zealand may help further distinguish independent introductions, and better facilitate characterisation of invasion spread across New Zealand. Such studies may be facilitated by the availability of high-throughput tools such as SNP chips developed for medical research on laboratory rats, once ascertainment bias has been accounted for. Ultimately, the availability of low-cost genomes for individual invasive rats will usher in the era of population genomics (Aitman et al., 2008).
Author Contributions
JCR and RF conceived the research. JCR and RF oversaw sample acquisition. JHR performed laboratory analyses. JHR and JCR undertook analysis. JCR wrote the manuscript with input from RF.
Funding
This work was funded by the 2012 Prime Minister's MacDiarmid Emerging Scientist Prize to JCR, and by the Royal Society of New Zealand Marsden Fund 03-UOA-117 to RF.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are exceptionally grateful to all people who contributed rat tissue samples for this project, in particular Phil Cowan, Grant Harper, Robyn Howitt, Kim King, and Steven Miller with assistance from Te Papa and Ecogene. Thanks also to the following people who kindly provided assistance collecting rat tissue samples: T. Agnew, S. Anderson, M. Aviss, M. Ayre, L. Baigent, P. Banks, D. Barnes, N. Barton, R. Blenk, M. Bowie, C. Brosnlian, P. Brown, T. Butcher, H. Chester, S. Collins, J. Cook, M. de Beurs, A. DeGraaf, J. Edgar, S. Foster, I. Gazzard, J. Gilbert, R. Goldring, N. Goldwater, R. Griffith, J. Guilbert, P. Hall, O. Hamilton, J. Hellyer, G. Heron, C. James, I. James, T. Kelly, E. King, M. King, S. Klaere, J. Lambie, J. Latham, W. Linklater, J. MacKay, J. Mather, E. Matisoo-Smith, P. McClelland, K. McMannas, S. McManus, P. McMurtrie, D. Merton, C. Mitchell, P. Mitchell, R. Molloy, H. Nathan, M. Ogle, R. Renwick, H. Ricardo, C. Roberts, S. Sawyer, L. Shapiro, R. Shaw, W. Shaw, G. Smith, K. Springer, I. Squires, N. Taylor, K. Taylor, R. Taylor, B. Thomas, P. Thomson, T. Thurley, A. Walker, R. Weaver, J. Welsh, R. Whittam, A. Williams, G. Wilson, M. Wylie, J. Zammit-Ross and any others we may have overlooked. Thanks to John Innes, Kim King and Rowley Taylor for discussions on the distribution and history of rats in New Zealand. This work was presented at the VIII Southern Connection Congress and 15th Rodens et Spatium: International Conference on Rodent Biology. Thanks to the editor and reviewers for feedback on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00048/full#supplementary-material
Supplementary Table 1. Samples including species, island, haplotype, and coordinates. *indicates museum specimen.
References
Abdelkrim, J., Byrom, A. E., and Gemmell, N. J. (2010). Fine-scale genetic structure of mainland invasive Rattus rattus populations: implications for restoration of forested conservation areas in New Zealand. Conserv. Genet. 11, 1953–1964. doi: 10.1007/s10592-010-0085-9
Aitman, T. J., Critser, J. K., Cuppen, E., Dominiczak, A., Fernandez-Suarez, X. M., Flint, J., et al. (2008). Progress and prospects in rat genetics: a community view. Nat. Genet. 40, 516–522. doi: 10.1038/ng.147
Aplin, K. P., Suzuki, H., Chinen, A. A., Chesser, R. T., Ten Have, J., Donnellan, S. C., et al. (2011). Multiple geographic origins of commensalism and complex dispersal history of black rats. PLoS ONE. 6:e26357. doi: 10.1371/journal.pone.0026357
Atkinson, I. A. E. (1973). Spread of the ship rat (Rattus r. rattus L.) in New Zealand. J. R. Soc. N. Z. 3, 457–472. doi: 10.1080/03036758.1973.10421869
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bassett, I. E., Cook, J., Buchanan, F., and Russell, J. C. (2016). Treasure islands: biosecurity in the Hauraki Gulf marine park. N. Z. J. Ecol. 40, 250–266. doi: 10.20417/nzjecol.40.28
Bastos, A. D., Nair, D., Taylor, P. J., Brettschneider, H., Kirsten, F., Mostert, E., et al. (2011). Genetic monitoring detects an overlooked cryptic species and reveals the diversity and distribution of three invasive Rattus congeners in South Africa. BMC. Genet. 12:26. doi: 10.1186/1471-2156-12-26
Bertelsmeier, C., Ollier, S., Liebhold, A. M., Brockerhoff, E. G., Ward, D., and Keller, L. (2018). Recurrent bridgehead effects accelerate global alien ant spread. Proc. Natl. Acad. Sci. U.S.A. 115, 5486–5491. doi: 10.1073/pnas.1801990115
Berthier, K., Garba, M., Leblois, R., Navascués, M., Tatard, C., Gauthier, P., et al. (2016). Black rat invasion of inland Sahel: insights from interviews and population genetics in south-western Niger. Biol. J. Linn. Soc. 119, 748–765. doi: 10.1111/bij.12836
Brouat, C., Rahelinirina, S., Loiseau, A., Rahalison, L., Rajerison, M., Laffly, D., et al. (2013). Plague circulation and population genetics of the reservoir Rattus rattus: the influence of topographic relief on the distribution of the disease within the Madagascan focus. PLoS Negl. Trop. Dis. 7:e2266. doi: 10.1371/journal.pntd.0002266
Brouat, C., Tollenaere, C., Estoup, A., Loiseau, A., Sommer, S., Soanandrasana, R., et al. (2014). Invasion genetics of a human commensal rodent: the black rat Rattus rattus in Madagascar. Mol. Ecol. 23, 4153–4167. doi: 10.1111/mec.12848
Brown, K., Elliott, G., Innes, J., and Kemp, J. (2015). Ship Rat, Stoat and Possum Control on Mainland New Zealand. An Overview of Techniques, Successes and Challenges. Wellington: Department of Conservation.
Clout, M. N., and Russell, J. C. (2006) “The eradication of mammals from New Zealand islands,” in Assessment Control of Biological Invasion Risks, eds F. Koike, M. N. Clout, M. Kawamichi, M. De Poorter, and K. Iwatsuki (Gland: IUCN; Kyoto: Shoukadoh Book Sellers), 127–141.
Colangelo, P., Abiadh, A., Aloise, G., Amori, G., Capizzi, D., Vasa, E., et al. (2015). Mitochondrial phylogeography of the black rat supports a single invasion of the western Mediterranean basin. Biol. Invasions 17, 1859–1868. doi: 10.1007/s10530-015-0842-2
Conroy, C. J., Rowe, K. C., Rowe, K. M., Kamath, P. L., Aplin, K. P., Hui, L., et al. (2013). Cryptic genetic diversity in Rattus of the San Francisco Bay region, California. Biol. Invasions 15, 741–758. doi: 10.1007/s10530-012-0323-9
Cowan, P. E., Gleeson, D. M., Howitt, R. L., Ramón-Laca, A., Esther, A., and Pelz, H. J. (2017). Vkorc1 sequencing suggests anticoagulant resistance in rats in New Zealand. Pest Manag. Sci. 73, 262–266. doi: 10.1002/ps.4304
Fewster, R. M., Miller, S. D., and Ritchie, J. (2011). “DNA profiling - a management tool for rat eradication,” in Island Invasives: Eradication and Management, eds C. R. Veitch, M. N. Clout, and D. R. Towns (Gland: IUCN), 430–435.
Haniza, M. Z., Adams, S., Jones, E. P., MacNicoll, A., Mallon, E. B., Smith, R. H., et al. (2015). Large-scale structure of brown rat (Rattus norvegicus) populations in England: effects on rodenticide resistance. PeerJ 3:e1458. doi: 10.7717/peerj.1458
Hingston, M., Goodman, S. M., Ganzhorn, J. U., and Sommer, S. (2005). Reconstruction of the colonization of southern Madagascar by introduced Rattus rattus. J. Biogeogr. 32, 1549–1559. doi: 10.1111/j.1365-2699.2005.01311.x
Hingston, M., Poncet, S., Passfield, K., Tabak, M. A., Gabriel, S. I., Piertney, S. B., et al. (2016). Phylogeography of Rattus norvegicus in the South Atlantic Ocean. Diversity 8:32. doi: 10.3390/d8040032
Innes, J. G. (2005a). “Norway rat,” in The Handbook of New Zealand Mammals. 2nd edition, ed C. M. King, (Melbourne, VIC: Oxford University Press), 174–187.
Innes, J. G. (2005b). “Ship rat,” in The Handbook of New Zealand Mammals. 2nd edition, ed C. M. King, (Melbourne, VIC: Oxford University Press), 187–203.
Kajdacsi, B., Costa, F., Hyseni, C., Porter, F., Brown, J., Rodrigues, G., et al. (2013). Urban population genetics of slum-dwelling rats (Rattus norvegicus) in Salvador, Brazil. Mol. Ecol. 22, 5056–5070. doi: 10.1111/mec.12455
Kaleme, P. K., Bates, J. M., Belesi, H. K., Bowie, R. C. K., Gambalemoke, M., Kerbis-Peterhans, J., et al. (2011). Origin and putative colonization routes for invasive rodent taxa in the Democratic Republic of Congo. Afr. Zool. 46, 133–145. doi: 10.1080/15627020.2011.11407486
Kambe, Y., Tanikawa, T., Matsumoto, Y., Tomozawa, M., Aplin, K. P., and Suzuki, H. (2011). Origin of agouti-melanistic polymorphism in wild black rats (Rattus rattus) inferred from Mc1r gene sequences. Zool. Sci. 28, 560–567. doi: 10.2108/zsj.28.560
King, C., Alexander, A., Chubb, T., Cursons, R., MacKay, J., McCormick, H., et al. (2016). What can the geographic distribution of mtDNA haplotypes tell us about the invasion of New Zealand by house mice Mus musculus? Biol. Invasions 18, 1551–1565. doi: 10.1007/s10530-016-1100-y
Konečný, A., Estoup, A., Duplantier, J. M., Bryja, J., Bâ, K., Galan, M., et al. (2013). Invasion genetics of the introduced black rat (Rattus rattus) in Senegal, West Africa. Mol. Ecol. 22, 286–300. doi: 10.1111/mec.12112
Lack, J. B., Greene, D. U., Conroy, C. J., Hamilton, M. J., Braun, J. K., Mares, M. A., et al. (2012). Invasion facilitates hybridization with introgression in the Rattus rattus species complex. Mol. Ecol. 21, 3545–3561. doi: 10.1111/j.1365-294X.2012.05620.x
Lack, J. B., Hamilton, M. J., Braun, J. K., Mares, M. A., and Van Den Bussche, R. A. (2013). Comparative phylogeography of invasive Rattus rattus and Rattus norvegicus in the US reveals distinct colonization histories and dispersal. Biol. Invasions 15, 1067–1087. doi: 10.1007/s10530-012-0351-5
Liu, J., Liu, D.-Y., Chen, W., Li, J.-L., Luo, F., Li, Q., et al. (2012). Genetic analysis of hantaviruses and their rodent hosts in central-south China. Virus Res. 163, 439–447. doi: 10.1016/j.virusres.2011.11.006
López, M., Foronda, P., Feliu, C., and Hernández, M. (2013). Genetic characterization of black rat (Rattus rattus) of the Canary Islands: origin and colonization. Biol. Invasions 15, 2367–2372. doi: 10.1007/s10530-013-0466-3
Matisoo-Smith, E., Roberts, R. M., Irwin, G. J., Allen, J. S., Penny, D., and Lambert, D. M. (1998). Patterns of prehistoric human mobility in polynesia indicated by mtDNA from the Pacific rat. Proc. Natl Acad. Sci. U.S.A. 95, 15145–15150. doi: 10.1073/pnas.95.25.15145
Muirhead, J. R., Gray, D. K., Kelly, D. W., Ellis, S. M., Heath, D. D., and MacIsaac, H. J. (2008). Identifying the source of species invasions: sampling intensity vs. genetic diversity. Mol. Ecol. 17, 1020–1035. doi: 10.1111/j.1365-294X.2008.03669.x
Puckett, E. E., Park, J., Combs, M., Blum, M. J., Bryant, J. E., Caccone, A., et al. (2016). Global population divergence and admixture of the brown rat (Rattus norvegicus). Proc. R. Soc. 283:20161762. doi: 10.1098/rspb.2016.1762
Richardson, J. L., Burak, M. K., Hernandez, C., Shirvell, J. M., Mariani, C., Carvalho-Pereira, T. S., et al. (2017). Using fine-scale spatial genetics of Norway rats to improve control efforts and reduce leptospirosis risk in urban slum environments. Evol. Appl. 10, 323–337. doi: 10.1111/eva.12449
Robins, J. H., Hingston, M., Matisoo-Smith, E., and Ross, H. A. (2007). Identifying Rattus species using mitochondrial DNA. Mol. Ecol. Resour. 7, 717–729. doi: 10.1111/j.1471-8286.2007.01752.x
Robins, J. H., Miller, S. D., Russell, J. C., Harper, G. A., and Fewster, R. M. (2016). Where did the rats of Big South Cape Island come from? N. Z. J. Ecol. 40, 229–234. doi: 10.20417/nzjecol.40.26
Rowe, K. C., Aplin, K. P., Baverstock, P. R., and Moritz, C. (2011). Recent and rapid speciation with limited morphological disparity in the genus Rattus. Syst. Biol. 60, 188–203. doi: 10.1093/sysbio/syq092
Russell, J. C., and Broome, K. G. (2016). Fifty years of rodent eradications in New Zealand: another decade of advances. N. Z. J. Ecol. 40, 197–204. doi: 10.20417/nzjecol.40.22
Russell, J. C., and Clout, M. N. (2004). Modelling the distribution and interaction of introduced rodents on New Zealand offshore islands. Global Ecol. Biogeograp. 13, 497–507. doi: 10.1111/j.1466-822X.2004.00124.x
Russell, J. C., Innes, J. G., Brown, P. H., and Byrom, A. E. (2015). Predator-free New Zealand: conservation country. BioScience 65, 520–525. doi: 10.1093/biosci/biv012
Russell, J. C., Mackay, J. W., and Abdelkrim, J. (2009). Insular pest control within a metapopulation context. Biol. Conserv. 142, 1404–1410. doi: 10.1016/j.biocon.2009.01.032
Russell, J. C., Miller, S. D., Harper, G. A., MacInnes, H. E., Wylie, M. J., and Fewster, R. M. (2010). Survivors or reinvaders? Using genetic assignment to identify invasive pests following eradication. Biol. Invasions 12, 1747–1757. doi: 10.1007/s10530-009-9586-1
Russell, J. C., Sataruddin, N. S., and Heard, A. D. (2014). Over-invasion by functionally equivalent invasive species. Ecology 95, 2268–2276. doi: 10.1890/13-1672.1
Russell, J. C., Towns, D. R., Anderson, S. H., and Clout, M. N. (2005). Intercepting the first rat ashore. Nature 437:1107. doi: 10.1038/4371107a
Searle, J. B., Jamieson, P. M., Gündüz, I., Stevens, M. I., Jones, E. P., Gemmill, C. E., et al. (2009). The diverse origins of New Zealand house mice. Proc. Biol. Sci. 276, 209–217. doi: 10.1098/rspb.2008.0959
Song, Y., Lan, Z., and Kohn, M. H. (2014). Mitochondrial DNA phylogeography of the Norway rat. PLoS ONE. 9:e88425. doi: 10.1371/journal.pone.0088425
Theuerkauf, J., Kuehn, R., Gula, R., Sztencel-Jabłonka, A., Jourdan, H., Taugamoa, A., et al. (2015). Invasion history affects genetic structure in island rat populations. J. Zool. 295, 197–205. doi: 10.1111/jzo.12206
Thomson, G. M. (1922). The Naturalization of Animals & Plants in New Zealand. London: Cambridge University Press.
Tollenaere, C., Brouat, C., Duplantier, J. M., Rahalison, L., Rahelinirina, S., Pascal, M., et al. (2010). Phylogeography of the introduced species Rattus rattus in the western Indian Ocean, with special emphasis on the colonization history of Madagascar. J. Biogeogr. 37, 398–410. doi: 10.1111/j.1365-2699.2009.02228.x
Tollenaere, C., Ivanova, S., Duplantier, J. M., Loiseau, A., Rahalison, L., Rahelinirina, S., et al. (2012). Contrasted patterns of selection on MHC-linked microsatellites in natural populations of the malagasy plague reservoir. PLoS ONE. 7:e32814. doi: 10.1371/journal.pone.0032814
Varudkar, A., and Ramakrishnan, U. (2015). Commensalism facilitates gene flow in mountains: a comparison between two Rattus species. Heredity 115, 253–261. doi: 10.1038/hdy.2015.34
Veale, A. J., Russell, J. C., and King, C. M. (2018). The genomic ancestry, landscape genetics and invasion history of introduced mice in New Zealand. R. Soc. Open Sci. 5:170879. doi: 10.1098/rsos.170879
Wardle, J. (1984). The New Zealand Beeches: Ecology, Utilisation and Management. Wellington: New Zealand Forest Service.
Waters, J. M., Fraser, C. I., and Hewitt, G. M. (2013). Founder takes all: density-dependent processes structure biodiversity. Trends Ecol. Evol. 28, 78–85. doi: 10.1016/j.tree.2012.08.024
Keywords: D-loop, genetics, island, mitochondrial DNA, rattus, rodent
Citation: Russell JC, Robins JH and Fewster RM (2019) Phylogeography of Invasive Rats in New Zealand. Front. Ecol. Evol. 7:48. doi: 10.3389/fevo.2019.00048
Received: 20 June 2018; Accepted: 07 February 2019;
Published: 08 March 2019.
Edited by:
Michael H. Parsons, Fordham University, United StatesReviewed by:
Jason Munshi-South, Fordham University, United StatesPeter Convey, British Antarctic Survey (BAS), United Kingdom
Copyright © 2019 Russell, Robins and Fewster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James C. Russell, ai5ydXNzZWxsQGF1Y2tsYW5kLmFjLm56
 James C. Russell
James C. Russell Judith H. Robins2
Judith H. Robins2 Rachel M. Fewster
Rachel M. Fewster