Abstract
Mediterranean coastal lagoons are environmentally important ecosystems whose conservation has been challenged due to anthropogenic impacts that promoted the expansion of non-indigenous and, sometimes, invasive species. Therefore, it is crucial to inventory biodiversity in these areas for the development of strategies of conservation and management. Classical methods used for biodiversity surveys and detection of non-native species may be unsuccessful for the detection and identification of species in early development stages such as cryptic, microscopic, elusive, and new coming species at low population density. The development of metabarcoding techniques in the last decade offers new opportunities for reliable biodiversity surveillance and facilitates early detection of nuisance species. The objective of this study was to analyze the species occurring in the protected coastal lagoon Canet-Saint Nazaire using a simple sampling protocol based on water samples and environmental DNA (eDNA) metabarcoding with a single barcode (cytochrome c oxidase subunit I [COI] gene). Two invasive species (Polydora cornuta and Acartia tonsa), two polychaete bioindicators of pollution (Hediste diversicolor and Capitella capitata), and one alga that produces harmful algal blooms were detected from only 6 L of water, indicating environmental degradation in the lagoon despite its protected status. These results demonstrate the importance of COI as single barcode together with eDNA as an ecological early warning system and suggest the need for environmental restoration in this lagoon.
Introduction
Coastal lagoons occupy 13% of the coastal area worldwide (Barnes, 1980) and are among the marine habitats showing the highest biological productivity, by providing diverse habitat types for many species, nursery areas, and feeding grounds for marine and estuarine fishes (Pérez-Ruzafa et al., 2011). They are distinctive ecosystems because they are shallow coastal water bodies separated from the ocean by a barrier and connected intermittently to the sea (Kjerfve, 1994). They also support important fisheries and allow for intensive aquaculture exploitation (Cataudella et al., 2015). Despite their environmental and economic importance, and protected status, lagoons suffer from several threats derived from human activities such as the effects of climate change, pollution, eutrophication, and the introduction of non-indigenous species (NIS) (Reizopoulou et al., 1996; Chapman, 2012). Human activities, such as the increases in maritime traffic and the opening of the Suez Canal, have facilitated NIS introductions in the Mediterranean Sea (Katsanevakis et al., 2014). In coastal lagoons, NIS settlement is facilitated by their naturally stressful conditions, pollution, loss of native species, and intense shipping in the numerous nearby harbors (Ruiz et al., 2000; Crooks et al., 2011). NIS can become invasive and affect both native species and economic activities in the area (Galil, 2007).
Together with NIS, eutrophication plays an important role in the degradation of these estuarine systems. Inefficient wastewater management and intense agricultural activities in the catchment area of the lagoons can increase the concentration of nutrients (Carlier et al., 2008). The consequent eutrophic state facilitates the uncontrolled growth of organisms such as dinoflagellates, diatoms, and cyanobacteria that produce harmful algal blooms (HABs, also referred as red tides) (Anderson et al., 2002). The increasing algal bloom episodes is not only favored by eutrophication but also by climate change and the introduction of new strains by ballast water (Hallegraeff, 1993; Moore et al., 2008). HABs have several effects on the environment. The mere growth and following decay of organic material cause anoxic conditions, leading to the death of aquatic life. Some HABs also produce toxins that affect both marine life and human health even at low densities (Sellner et al., 2003). In humans, these toxins can cause different types of poisoning (diarrheal, neurotoxic, and paralytic). The most frequent are diarrheic shellfish poisoning (DSP), mainly due to toxic strains of Prorocentrum spp. and Dinophysis spp. (Yasumoto et al., 1980; Bravo et al., 2001), and paralytic shellfish poisoning (PSP) predominantly linked to toxic strains of Alexandrium spp. (Anderson et al., 2012).
The correct management and conservation of coastal areas requires efficient assessment of their biodiversity and detection and identification of species that may be of environmental concern, such as pollution indicators and NIS. For this purpose, species surveys are carried out periodically, generally consisting of manual sampling and visual identification of sampled specimens by taxonomical experts. Such a classical morphological analysis requires high sampling effort and might be inefficient in the detection of some species, for example those that are in early development stages, cryptic species, or microorganisms (Ficetola et al., 2008). For NIS, their early detection is crucial to prevent their establishment and dispersion because eradication or control are more efficient when the species are at low density soon after introduction (Gozlan et al., 2010). However, low population densities require greater survey efforts for detection, and many newly established populations may go unnoticed (Blanchet, 2012). Classical approaches for phytoplankton identification are often considered time consuming while requiring expertise in taxonomical identification. Identifying dinoflagellates represent a greater challenge due to high morphological similarities and a lack of unique characters between different species (Lin et al., 2009). DNA metabarcoding is a rapidly evolving method for assessing biodiversity that exceeds the limitations of traditional methods. It combines the use of environmental DNA (eDNA) and next-generation sequencing (NGS) allowing the detection of species from single cells in an environmental sample (e.g., soil or water) (Taberlet et al., 2012). Although eDNA metabarcoding was found to be less sensitive than a targeted monitoring approach for generating detailed specific distribution data (Furlan et al., 2015; Ardura et al., 2016; Bylemand et al., 2019), it has the ability to provide baseline information on biodiversity patterns (e.g., cryptic species and novel incursions of invasive species) (Blackman et al., 2017), capturing a great part of the aquatic community diversity (Zaiko et al., 2015). Borrell et al. (2017) have proposed the use of metabarcoding as an early alert method for the detection of invasive species in ports. They used two metabarcodes (cytochrome oxidase I and 18S rRNA genes) on 3 L of water from each sampling point. For the need of easy methods for alert of environmental disturbances, here, we will test a similar but simplified approach for the exploration of nuisance species, including invasive species and HABs.
We apply metabarcoding (eDNA) for the detection of aquatic nuisance organisms in the coastal lagoon Canet-Saint Nazaire, situated in the French Mediterranean coast. Previous studies developed in this lagoon using a rapid assessment of invertebrate species and barcoding (Ardura and Planes, 2017) suggested a high level of degradation and vulnerability of the lagoon. However, in that study, small species like HABs and organisms in early development stages could not be recognized. In the present investigation, eDNA was extracted from water samples from the canal that connects the lagoon to the open sea, and a region of the cytochrome c oxidase subunit I (COI) gene was used as a single metabarcode for species identification, since it has been used as a barcode of high-resolution power (Hebert et al., 2003a) and reference databases for aquatic organisms are more complete for this gene than other barcodes (Weigand et al., 2019). The principal aim of this study was focused on the detection of species that may reveal a risk for the conservation status and environmental health of the lagoon.
Materials and Methods
Study Area
Sampling was conducted in the canal that connects the Canet-Saint Nazaire Lagoon to the open sea (Figure 1). This lagoon is a special protection area (SPA) for birds within the Natura 2000 network, and it is one of the many coastal lagoons spanning the French Mediterranean coastline. It is a semiclosed system, with its principal water sources coming from two rivers and streams and, to a lesser extent, a canal connected to the sea. This, along with seasonal variations in temperature, drought periods, and sudden and intense rainfalls, causes important changes in its depth and salinity that varies from 13.2 to 35.6 during the year (Vouvé et al., 2014). Besides this, the lagoon suffers from intense pollution, eutrophication, and sediment filling due to human activities in its catchment area (agriculture, water treatment, and tourism) and scarce water exchange with the sea (Carlier et al., 2008; Souchu et al., 2010). Within 4 km of the lagoon, Canet port is located, which is a popular location for recreational boating with space for 977 vessels up to 24 m in length (Portbooker website, 2018). The presence of this harbor, along with the lagoon’s brackish water, makes Canet-Saint Nazaire extremely vulnerable to introduction of non-native species (Paavola et al., 2005; Ardura and Planes, 2017).
FIGURE 1

Lagoon of Canet-Saint Nazaire (below) and port of Canet (above) indicating the sampling site (Google Maps, 2018).
Sampling
Water samples of the lagoon were taken in the proximity of the canal (42°39′25.19″N 3° 1′39.30″E). Six liters of surface water was collected in three sterile bottles of 2 L (three replicates) on November–December 2016. All the water from each bottle was vacuum filtered through several filters of different material and pore size to collect all DNA from the sample without clogging the filters. Bottles are sterile, and cross-contamination is not expected because the sampling is done in unique point. First, the whole volume of each sample was filtered through 10-μm nylon filters; then, each sample was divided in four and filtered through 0.8-μm polyethersulfone (PES) filters. Finally, the whole volume of each sample was pooled together and filtered through 0.2-μm PES filters. In total, 18 filters (3 replicates × 6 filters each one) were obtained, which were thereafter preserved with 96% ethanol until eDNA extraction.
DNA and Bioinformatics Analysis
DNA was extracted from the filters using PowerWater® DNA Isolation Kit (MOBIO Laboratories, EE. UU.) following the manufacturer’s extraction protocol. In addition, ethanol was centrifuged, and the pellet, with precipitated DNA, was added in the extracting process. A negative control of pure water was added at this step to monitor contamination during the extraction process. In addition, bovine serum albumin (BSA) was added to the PCR reactions to increase PCR yields from low-purity templates and to avoid, as much as possible, the effect of inhibitors present in the water. Extracted DNA from all the filters was sent to Macrogen (Seoul, South Korea) were it was quantified and sequenced. The eDNA was quantified by a fluorescence-based method Victor 3 (Picogreen, Invitrogen). The modified universal COI primers mlCOIintF/jgHCO2198 (Leray et al., 2013) were used for PCR amplification of a fragment of ∼313 bp within the mitochondrial gene coding for the COI. The primers were modified to include Illumina sequencing adapters and sample-specific dual indices (i5 and i7) following the Illumina (2013) protocol in which the conditions for the amplicon PCR were changed for the ones described by Leray et al. (2013). The PCR reactions were undertaken by Macrogen, following the protocol Illumina (2013). After constructing the library, it was charged into the platform Illumina MiSeq v3 reagents that generates paired-end sequences (2 × 301). Adapters and indices were removed from the raw data along with reads <36 bp using Scythe and Buffalo software, respectively.
The trimmed data were received from Macrogen in Fastaq format and were further processed using the platform Qiime1 (Caporaso et al., 2010). All the sequence reads were assessed for quality by applying a Phred quality score threshold of 20 and were filtered by length (200 bp ≤ reads ≤400 bp) from the downstream analysis.
The paired-end reads from each sample were merged when they presented an overlapping region of at least 100 bp and <15% of differences within this region. This step is necessary to merge forward and reverse reads. Some errors often appear in the Illumina data as the merging is not always 100%, due to sequencing length and PCR errors. For this, one of the thresholds in the merging algorithm must be the percentage of coincidence within the merging region to avoid artifact sequences when merging them and to improve the following assignment against reference database.
For the taxonomic assignation, Basic Local Alignment Search Tool (BLAST) alignment was performed against NCBI database1 of COI sequences (obtained in 09/2017) filtered for environmental reads and using as threshold criteria: maximum E value = 1 × 10–50 and minimum percent identity = 97.0, which is enough in most of the cases for species identification from COI barcode (Hebert et al., 2003b). Some reads may have multiple BLAST hits at a 100% identity to different species, or the best BLAST hit have no species-level reference available. In those cases, the operational taxonomic unit (OTU) was assigned to a genus level.
OTU tables, a list of OTUs obtained for each sample and the number of sequences assigned to them, were constructed, clustering reads with a 100% identity between them and maintaining all assigned sequences including singletons to retain maximum sensitivity for species detection. The removal of singletons is usually employed to eliminate false positives as proposed by Scott et al. (2018), in the context of species survival, NIS early detection, or marine biosecurity surveillance; a false negative is most costly than a false positive (von Ammon et al., 2018). Sequences of organisms without relevance for the study (e.g., human, insects, terrestrial plants, etc.) were removed from the dataset (Figure 2). Generally, chimera formation is not extensive, although some chimeras can be found. In addition, one sequence per OTU was BLASTed manually to verify the reliability of the pipeline parameters and discard the presence of chimeras (Nilsson et al., 2012). The taxonomic information from the remaining OTUs was checked against the World Register of Marine Species2 and AlgaeBase3. The number of reads per OTU, as a proxy for species abundance, was used to generate OTU rarefaction curves using Vegan package in R software (Oksanen et al., 2013).
FIGURE 2
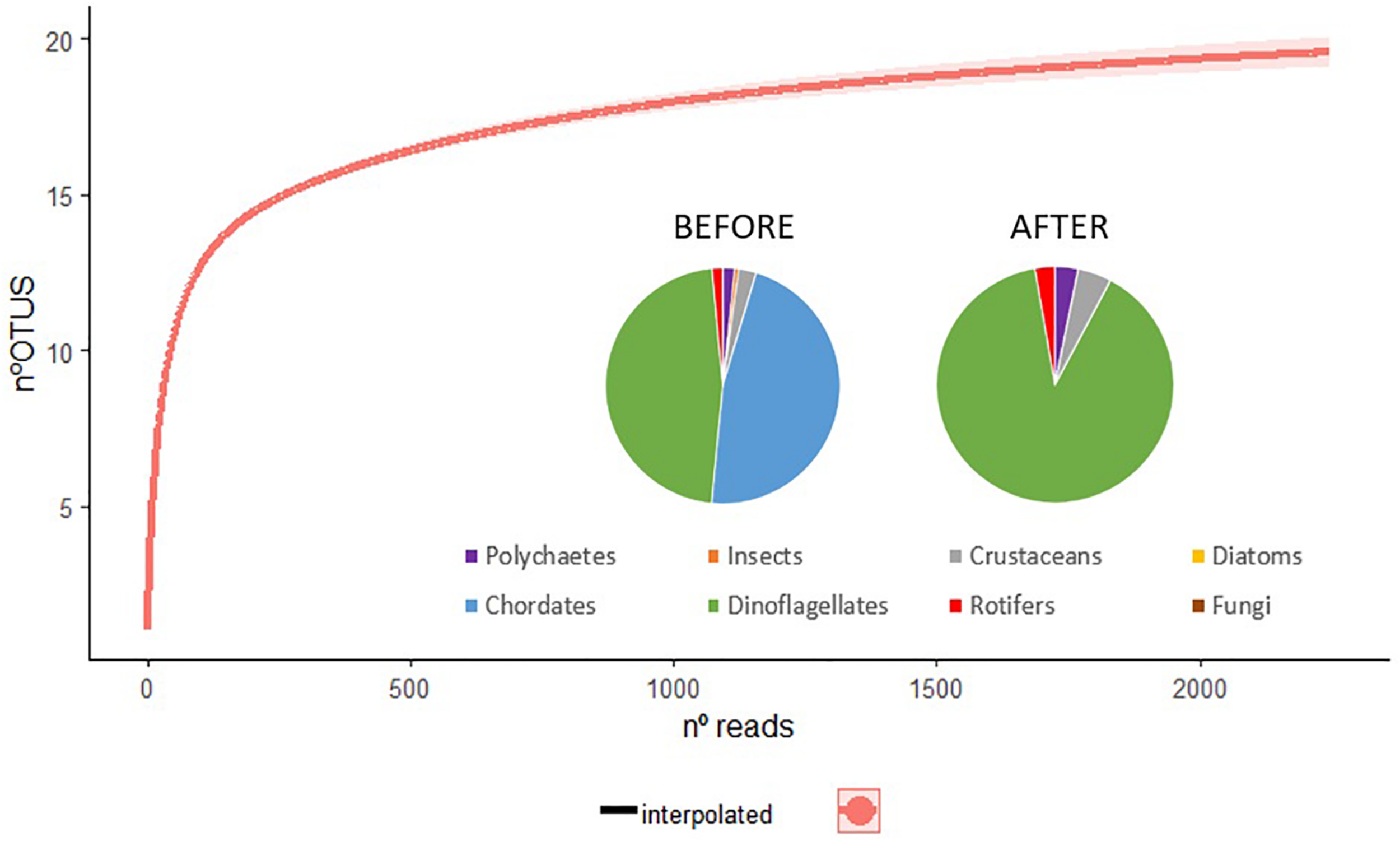
Rarefaction curve plotting the number of reads by the number of operational taxonomic units (OTUs) for the sample analyzed using high-throughput sequencing. Interpolated line in red. Graphics with the proportion of different taxonomical groups detected in the lagoon before and after expert check (eliminating the reads assigned to terrestrial and exclusive marine species).
Results
The minimum concentration of the extracted eDNA was 0.076 ng/μl. A total of 581,786 raw paired-end reads were sequenced from positive PCR amplicons (average read length, 301 bp). Following initial filtering, a total of 328,220 reads (56.4%) were retained for downstream analysis (average read length, 269 bp). After merging, and a second quality filtering, 239,842 sequences remained (41.2%), of which 98,282 matched a correct reference sequence from the database (16.9%; average read length, 359 bp) (Table 1).
TABLE 1
| GenBank ID | Phylum | Class | Species | 0.2 μ mesh size | 0.8 μ mesh size | 10 μ mesh size |
| KP254366.1 | Annelida | Polychaeta | Capitella capitata | 0 | 0 | 3 |
| KR916843.1 | Annelida | Polychaeta | Hediste diversicolor | 0 | 0 | 4 |
| AB636160.1 | Annelida | Polychaeta | Polydora cornuta | 0 | 0 | 1,619 |
| KM578774.1 | Arthropoda | Insecta | Attagenus gobicola | 0 | 0 | 2 |
| KJ962220.1 | Arthropoda | Insecta | Dermestes lardarius | 7 | 0 | 289 |
| HQ824544.1 | Arthropoda | Insecta | Dichotomius nisus | 1 | 0 | 1 |
| AF253029.1 | Arthropoda | Insecta | Ochlerotatus detritus | 0 | 0 | 125 |
| KM578800.1 | Arthropoda | Insecta | Trogoderma yunnaeunsis | 0 | 0 | 107 |
| KC287363.1 | Arthropoda | Maxillopoda | Acartia tonsa | 0 | 0 | 1 |
| KP136566.1 | Arthropoda | Maxillopoda | Cyclopoida environmental sample | 0 | 0 | 2 |
| KF297550.1 | Arthropoda | Maxillopoda | Perforatus perforatus | 0 | 2,324 | 0 |
| DQ059772.1 | Arthropoda | Maxillopoda | Sacculina carcini | 0 | 0 | 46 |
| FJ590523.1 | Ascomycota | Dothideomycetes | Cladosporium bruhnei | 28 | 0 | 0 |
| GQ844253.1 | Bacillariophyta | Bacillariophyceae | Cylindrotheca fusiformis | 0 | 0 | 1 |
| HF563534.1 | Bacillariophyta | Bacillariophyceae | Haslea crucigera | 0 | 0 | 9 |
| FJ519930.1 | Chordata | Chondrichthyes | Galeocerdo cuvier | 0 | 0 | 9 |
| AP008737.1 | Chordata | Mammalia | Homo sapiens | 20,333 | 0 | 25,756 |
| GQ501113.1 | Dinoflagellata | Dinophyceae | Akashiwo sanguinea | 0 | 0 | 670 |
| GQ501119.1 | Dinoflagellata | Dinophyceae | Alexandrium affine | 0 | 0 | 84 |
| GQ501128.1 | Dinoflagellata | Dinophyceae | Alexandrium catenella | 0 | 0 | 210 |
| GQ501141.1 | Dinoflagellata | Dinophyceae | Alexandrium lusitanicum | 0 | 0 | 1 |
| GQ501157.1 | Dinoflagellata | Dinophyceae | Alexandrium minutum | 0 | 0 | 667 |
| GQ501159.1 | Dinoflagellata | Dinophyceae | Alexandrium ostenfeldii | 0 | 0 | 64 |
| GQ501142.1 | Dinoflagellata | Dinophyceae | Alexandrium sp. RFS-2009a | 0 | 0 | 1 |
| GQ914937.1 | Dinoflagellata | Dinophyceae | Azadinium obesum | 0 | 0 | 1,104 |
| KJ503235.1 | Dinoflagellata | Dinophyceae | Dinophysis acuminata | 0 | 0 | 1,046 |
| GQ501853.1 | Dinoflagellata | Dinophyceae | Dinophysis sp. PL9-13 | 0 | 0 | 76 |
| GQ501849.1 | Dinoflagellata | Dinophyceae | Dinophysis sp. PL9-3 | 0 | 0 | 7 |
| GQ501848.1 | Dinoflagellata | Dinophyceae | Dinophysis sp. PL9-4 | 0 | 0 | 1 |
| GQ501217.1 | Dinoflagellata | Dinophyceae | Gonyaulax sp. AC551 | 0 | 0 | 3 |
| GQ501250.1 | Dinoflagellata | Dinophyceae | Karenia brevis | 0 | 0 | 9 |
| GQ501256.1 | Dinoflagellata | Dinophyceae | Karlodinium veneficum | 0 | 0 | 125 |
| GQ501233.1 | Dinoflagellata | Dinophyceae | Lepidodinium chlorophorum | 0 | 0 | 95 |
| GQ501243.1 | Dinoflagellata | Dinophyceae | Levanderina fissa | 0 | 0 | 4,751 |
| GQ501269.1 | Dinoflagellata | Dinophyceae | Peridinium inconspicuum | 0 | 0 | 51 |
| GQ502055.1 | Dinoflagellata | Dinophyceae | Peridinium sp. ES18-106 | 0 | 0 | 1 |
| GQ502043.1 | Dinoflagellata | Dinophyceae | Peridinium sp. ES18-111 | 0 | 0 | 5 |
| GQ502054.1 | Dinoflagellata | Dinophyceae | Peridinium sp. ES18-113 | 0 | 0 | 42 |
| GQ502044.1 | Dinoflagellata | Dinophyceae | Peridinium sp. ES18-128 | 0 | 0 | 1 |
| GQ502074.1 | Dinoflagellata | Dinophyceae | Prorocentrum micans | 0 | 0 | 1,137 |
| GQ502077.1 | Dinoflagellata | Dinophyceae | Prorocentrum sp. ES11-87 | 0 | 0 | 1 |
| GQ502071.1 | Dinoflagellata | Dinophyceae | Prorocentrum sp. ES18-118 | 0 | 0 | 6 |
| GQ502090.1 | Dinoflagellata | Dinophyceae | Prorocentrum sp. ES5-50 | 0 | 0 | 1 |
| GQ501301.1 | Dinoflagellata | Dinophyceae | Protoceratium reticulatum | 0 | 0 | 14 |
| GQ502107.1 | Dinoflagellata | Dinophyceae | Protoperidinium cf. depressum | 0 | 0 | 5,448 |
| GQ501312.1 | Dinoflagellata | Dinophyceae | Scrippsiella cf. precaria | 0 | 0 | 540 |
| GQ501317.1 | Dinoflagellata | Dinophyceae | Scrippsiella precaria | 0 | 0 | 2 |
| GQ501320.1 | Dinoflagellata | Dinophyceae | Scrippsiella sp. CS-297 | 0 | 0 | 4,620 |
| GQ501314.1 | Dinoflagellata | Dinophyceae | Scrippsiella sp. DINOB785-08 | 0 | 0 | 44 |
| GQ501322.1 | Dinoflagellata | Dinophyceae | Scrippsiella sweeneyae | 0 | 0 | 7 |
| GQ501325.1 | Dinoflagellata | Dinophyceae | Scrippsiella trochoidea | 0 | 0 | 144 |
| GQ501400.1 | Dinoflagellata | Dinophyceae | Thoracosphaera heimii | 0 | 0 | 1,215 |
| GQ501756.1 | Dinoflagellata | Dinophyceae | Uncultured dinoflagellate | 913 | 0 | 22,512 |
| GQ501539.1 | Dinoflagellata | Dinophyceae | Uncultured Peridinium | 0 | 0 | 168 |
| GQ501810.1 | Dinoflagellata | Dinophyceae | Uncultured Prorocentrum | 0 | 0 | 6 |
| GQ501436.1 | Dinoflagellata | Dinophyceae | Uncultured Thecadinium | 0 | 0 | 421 |
| JN809389.1 | Rotifera | Monogononta | Testudinella clypeata | 0 | 0 | 1,402 |
| Total per filter | 21,282 | 2,324 | 74,676 | |||
| Total | 98,282 |
Sequences matched a correct reference sequence from the GenBank database per filter, after merging and quality filtering.
The rarefaction curve of sequences indicated that the sample reached a plateau (Figure 2). This figure represents three 2-L superficial water samples, which were then grouped for the purpose of downstream analysis, hence the single plotted line. This indicates that, in terms of the number of sequences generated, our sequencing had enough depth to detect most of the possible species present within the sample.
After the application of expert check, eliminating the reads assigned to terrestrial and exclusive marine species, 51,624 sequences were left assigned to aquatic organisms at least at class level (Table 1). Of these reads, 10.5% were identified to a species level, while the majority was assigned to a genus or higher level. The presence of singletons may prevent reaching the asymptote in the curve of accumulation of species. In this case, the rare species (singletons and doublets) represent 11.1% (3 of the 27 OTU obtained), which did not impede to reach the asymptote, indicating that, in a number of sequences generated, the sample was adequately sized (Figure 2).
Regarding the biota profile captured from NGS after expert check, the assigned sequences were clustered in 27 different OTUs of which 17 belonged to the Class Dinophyceae (89.5% of the reads), while the rest was divided into four different classes by sequences number order: crustaceans, polychaetes, diatoms, and rotifers (Figure 2).
Considering only the sequences assigned at least at a genus level (28,029 in total), 25 taxa were identified, 10 of them at a species level. The genus with more reads was Protoperidinium (19.4%), followed by Scrippsiella (19.1%), and Levanderina (16.9%) (Table 1), pointing out at a clear dominance of Dinophyceae in the water samples analyzed not only in the number of taxa but also in abundance of reads. The following taxon in abundance of reads was the acorn barnacle Perforatus perforatus (8.3% of reads), then the whip mudworm Polydora cornuta (5.8%). Other two polychaetes, Capitella capitata and Hediste diversicolor, and another barnacle (Sacculina carcini) were represented by less reads: 3, 4, and 46, respectively. One rotifer accounting for 5% of the reads was identified: Testudinella clypeata. Diatoms were represented by only two species, Haslea crucigera and Cylindrotheca fusiformis, being together <0.04% of the reads. The copepod Acartia tonsa was also found at very low abundance in number of reads, which is only one read (Table 1).
One half of the species detected from metabarcoding in the analyzed lagoon can be considered nuisance species or indicators of bad environmental quality4 (Table 2). The three polychaetes are bioindicators of pollution. The polychaete P. cornuta and the copepod A. tonsa are considered invasive species in the Mediterranean Sea. Finally, the dinoflagellate Akashiwo sanguinea forms algal blooms (Table 2).
TABLE 2
| Species | Environmental threat | References |
| Polydora cornuta | Invasive species, pollution bioindicator | Dean, 2008; Zenetos et al., 2012 |
| Hediste diversicolor | Pollution bioindicator | Dean, 2008 |
| Capitella capitata | Pollution bioindicator | Dean, 2008 |
| Acartia tonsa | Invasive species | David et al., 2007 |
| Akashiwo sanguinea | Harmful algal blooms | Jones et al., 2017 |
Assessment of environmental threats from the species found in Canet-Saint Nazaire lagoon.
Moreover, seven of the dinoflagellate genera are known to contain numerous species that are considered HABs because they produce toxins and/or form algal blooms, being cataloged in the HAB list of Intergovernmental Oceanographic Commission (IOC)-Unesco (Moestrup et al., 2009). According to the Harmful Algal Events Database (HAEDAT)4, the genera Alexandrium (13 species) and Dynophysis (10 species) found in our study (Table 1) are responsible for most algal blooms in the area. The other five genera of dinoflagellates found in our study and listed as HABs were Karenia (10 species), Prorocentrum (12 species), Karlodinium (6 species), Protoceratium (1 species), and Azadinium (3 species) (Table 1).
Discussion
The present study, based on a single metabarcode and 6 L of surface water from only one sampling point from one lagoon of 600 ha, did reveal bioindicators of three different environmental or biosecurity threats in Canet-Saint Nazaire lagoon: pollution (three bioindicator species), biological invasions (two species), and harmful algal blooms (one species). Despite extreme methodological simplicity, the resource-efficient methodology employed in this study was capable of obtaining important environmental information. Although it is known that DNA remains in the environment for prolonged periods especially in cold conditions and that individuals may not be present at the time of sampling, these results confirm the utility of COI as an eDNA metabarcoding tool for early alert of biological risks as proposed by Borrell et al. (2017) for biological invasions in ports, expanding the applications to pollution assessments and HABs.
The results indicate a poor ecological status for the lagoon Canet-Saint Nazaire. The three polychaete species were found to have been previously used as pollution indicators due to their ability to survive in highly polluted waters, with wide ranges of temperature and salinity, hypoxia, and eutrophication states (Surugiu, 2005; Dean, 2008; Maranho et al., 2014). P. cornuta is also considered an invasive species in the Mediterranean due to its ability to change the composition and abundance of native species, replacing native species like C. capitata (Cinar et al., 2005; Çinar et al., 2012; Zenetos et al., 2012). It was first discovered in the Mediterranean from polluted sediments in Valencia’s harbor (Spain) (Tena et al., 1991); however, it now has a wider distribution. Its larval phase can survive transport in ship ballast water, and adults may form dense settlements through fouling of ship hulls facilitating its spread (Radashevsky and Selifonova, 2013). On the other hand, the invasive F. enigmaticus that was found in a previous survey in this lagoon (Ardura and Planes, 2017) could have facilitated the introduction and establishment of P. cornuta in the lagoon as reported in other locations (Read and Gordon, 1991; Heiman et al., 2008).
Acartia tonsa deserves additional attention, since it is a neritic copepod commonly found in the western Atlantic, the Pacific, and the Indian Ocean, especially in estuarine and coastal waters. It was first described in the Mediterranean in 1985 (Gaudy and Viñas, 1985) and has been previously reported in the Gulf of Lion in Berre Lagoon (Cervetto et al., 1999). It is considered an invasive species due to its ability to replace native species of copepods and its propensity to spread to new areas through ballast water (David et al., 2007).
On the other hand, despite having a single sampling point with only 6 L of water and forming only a limited description of the diversity of species in the whole ecosystem; the data set shows a great diversity of dinoflagellates in the sampling point analyzed. Most reads (89.5%) and OTUs (60.7%) detected were assigned to the class Dinophycea. This could be explained by the sampling protocol, since the samples were taken from the lagoon surface where a greater number of these organism are expected. In any case, this is a signal of the lagoon’s poor ecological state and high eutrophication and nitrogen and phosphorus levels that facilitate the proliferation of these organisms (Carlier et al., 2008; Heisler et al., 2008). Moreover, several dinoflagellates can produce resistance cysts, allowing them to survive during harsher periods in the lagoon (Sellner et al., 2003). However, the lack of variability in the COI region used for the species identification does not allow differences between species or strains (native or not) from the same genus to be resolved, making it impossible to detect species that can cause harm to the ecosystem. Several species of the genera detected (e.g., Alexandrium, Dinophysis, Karenia, or Prorocentrum) are known to cause HABs that could harm the lagoon ecosystem (Sellner et al., 2003). A. sanguinea (only species in the genus Akashiwo) blooms have been reported to produce powerful surfactant-like proteins that can coat bird plumage and collapse their feathers, causing loss of waterproofing and thermal insulation and even mass mortality (Jones et al., 2017). This could have serious effects in the lagoon because it is located along one of Europe’s principal migration routes, and since March 2006, it has been classified as a SPA (Ardura and Planes, 2017). Further analysis with more specific identification tools would be necessary to assess the risks to the lagoon, but the data obtained in this study should be taken as warning signal. In Thau Lagoon, 100 km away from Canet, a toxic strain from Alexandrium catenella originating in the West Pacific was probably introduced from ballast water released in Séte’s harbor that is connected to the lagoon (Lilly et al., 2002).
The current study has some limitations, likely because the sampling was limited to only one point and 6 L of water taken from the surface; thus, benthic organisms are unlikely detected. One limitation could be a relatively modest number of reads recovered (0.5 million reads). The presence of inhibitors due to the polluted state of the lagoon cannot be discarded as an additional explanation for this, since pollution can inhibit the PCR amplification of eDNA (Jane et al., 2015). The lack of detection of some invasive macroscopic species detected by Ardura and Planes (2017) in the lagoon (the bivalves Abra sp. and Cerastoderma glaucum and the tubeworm Ficopomatus enigmaticus) could be explained by the different sampling methods employed in the two studies. The species C. glaucum occurs in the bottom and is very scarce in the lagoon (in the previous survey, more sampling points were analyzed and only one individual for was obtained). Abra sp. and F. enigmaticus, which were abundant in the lagoon, were not seen where the water samples were taken in the present study; thus, it is possible that their DNA was simply absent from our water samples. Additional explanations could be very degraded DNA (Barnes et al., 2014) and/or lack of universality of the primers that may not anneal equally well in all taxa (Wilcox et al., 2013). In Ardura and Planes (2017), two different barcodes were amplified, e.g., COI (Geller et al., 2013) and 16S rRNA (Palumbi, 1996), but high-quality sequences for F. enigmaticus and Abra sp. were obtained only for the 16S rRNA marker, while in the present study, only COI primers were employed.
The number of detected OTUs in metabarcoding samples tends to be higher than in the visual and barcoding analysis, and effectively, the number of species detected here was much higher than in the previous work by Ardura and Planes (2017). False positives in metabarcoding due to incorrect assignations during the bioinformatic analysis (Ficetola et al., 2015) could be reasonably discarded since all the OTUs detected in this study were BLASTed manually to avoid incorrect assignations. On the other hand, it is possible to detect species that are not alive in the sampling area but whose DNA could come from the sea or from neighboring rivers (Pochon et al., 2017). If it was the case, they would still be around and be able to reach the lagoon eventually. In general, and despite possible errors and the obvious limitations of the simplified sampling scheme, the metabarcoding technique used in this study allowed the successful completion of a general assessment of the lagoon’s environmental state from a list of nuisance and indicator species. Despite this, the data should be considered qualitative rather than empirical. Although eDNA concentration and number of sequences yielded from NGS are positively correlated with biomass or population density, the accurate conversion of genome abundances to cell numbers and estimates of absolute abundance are still imprecise (Kelly et al., 2014; Bonk et al., 2018). A greater number of reads assigned to a specific OTU do not necessarily imply a greater abundance (Thomas et al., 2016).
This study provides new results from a new case study that support the applicability of the COI barcode as a metabarcoding tool using eDNA (e.g., Thomsen and Willerslev, 2015; Zaiko et al., 2015; Borrell et al., 2017; Pochon et al., 2017; Forster et al., 2019), showing in this particular case its usefulness to detect HAB-causing organisms. However, although it is useful to assign some organisms at species level, such as the genus Akashiwo, in most cases, this assignment is not possible because this small fragment, 300 bp, is not informative enough.
The simple sampling scheme in this study, with only one-point sample and three replicates, provides a proof of concept for the use of single samples as an approximation of ecological status. It is important to remark that the study trials a simplified sampling method to remark the value that small sample numbers can have on remote locations for future studies: even one sample with three replicates is informative. However, the limitations associated to collect small sample numbers should not be forgotten, bringing attention to considerations that must be had when analyzing NGS data collected from one sample. In these cases, the data must be analyzed as proof of concept. Further research with additional samples (at different depths and situational periods) would be necessary to build a more complete survey of the study area. Moreover, larger COI fragment or different molecular markers and the enlargement and improvement of barcode databases (Ardura, 2018) are also necessary and will provide a better representation of the ecosystem’s biodiversity.
On the other hand, new methodological tools are been developed to improve biological surveys, such as the use of environmental RNA (eRNA) (Pochon et al., 2017). Compared to eDNA, eRNA degrades more rapidly in the marine environment (typically hours to days, Thomsen et al., 2012; Sassoubre et al., 2016) and is therefore considered a better proxy for detecting living biota. However, susceptibility of RNA makes it difficult to work with. The collection of RNA samples requires dedicated sampling protocols, more careful preservation, and storage. There is also additional processing time and costs associated with isolation and reverse transcription of RNA (Laroche et al., 2016), making it more expensive and challenging and thus a less attractive molecule to work with (Zaiko et al., 2018).
As a final remark, the presence of invasive species, bioindicators of contamination, and organisms that can cause damage to the ecosystem in this single sampling point suggests a high level of degradation and vulnerability of the lagoon. This is a protected area under the Natura 2000 network, and the results may question the effectiveness of the current conservation programs, indicating that COI and metabarcoding can play an important role in monitoring the progress of conservation efforts.
Statements
Data availability statement
The Fastaq files are available in GenBank, reference (PRJNA549294).
Author contributions
AA: conception and design. SP: sampling. MS-M and AA: laboratory analysis. MS-M: NGS analysis. EG-V and AA: statistical analysis. AA and EG-V: writing the article. Finally, all authors revised the manuscript.
Funding
This study has been supported by the Spanish Ministry of Economy and Competitiveness Grant CGL2016-79209-R and the project FUO-201-17 supported by the Centre National de la Recherche Scientifique: “eDNA in Mediterranean lagoons.” This is a contribution from the Marine Observatory of Asturias (OMA), Asturias University Institute of Biotechnology (IUBA), and the Spanish Research Group of Excellence ARENA. AA holds a Juan de la Cierva-reincorporation Grant from the Spanish Ministry.
Acknowledgments
The authors thank Martin Desmalades for helping with the sampling tasks.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
1.^ https://www.ncbi.nlm.nih.gov/
2.^ http://www.marinespecies.org/
4.^ http://haedat.iode.org/ accessed on August 2019
References
1
Anderson D. M. Alpermann T. J. Cembella A. D. Collos Y. Masseret E. Montresor M. (2012). The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health.Harmful Algae1410–35. 10.1016/j.hal.2011.10.012
2
Anderson D. M. Glibert P. M. Burkholder J. M. (2002). Harmful algal blooms and eutrophication: nutrient sources, composition, and consequences.Estuaries25704–726. 10.1007/bf02804901
3
Ardura A. (2018). Species-specific markers for early detection of marine invertebrate invaders through eDNA methods: gaps and priorities in GenBank as database example.J. Nat. Conserv.4751–57. 10.1016/j.jnc.2018.11.005
4
Ardura A. Planes S. (2017). Rapid assessment of non-indigenous species in the era of the eDNA barcoding: a mediterranean case study.Estuar.Coast. Shelf Sci.18881–87. 10.1016/j.ecss.2017.02.004
5
Ardura A. Zaiko A. Borrell Y. Samuiloviene A. Garcia-Vazquez E. (2016). Novel tools for early detection of a global aquatic invasive, the zebra mussel Dreissena polymorpha.Aquat. Conserv.27165–176. 10.1002/aqc.2655
6
Barnes M. A. Turner C. R. Jerde C. L. Renshaw M. A. Chadderton W. L. Lodge D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems.Environ. Sci. Technol.481819–1827. 10.1021/es404734p
7
Barnes R. S. K. (1980). Coastal lagoons, Vol. 1. Cambridge: CUP Archive.
8
Blackman R. C. Constable D. Hahn C. Sheard A. Durkota J. (2017). Detection of a new non-native freshwater species by DNA metabarcoding of environmental samples – first record of Gammarus fossarum in the UK.Aquat. Invasion.12177–189. 10.3391/ai.2017.12.2.06
9
Blanchet S. (2012). The use of molecular tools in invasion biology: an emphasis on freshwater ecosystems.Fish. Manag Ecol.19120–132. 10.1111/j.1365-2400.2011.00832.x
10
Bonk F. Popp D. Harms H. Centler F. (2018). PCR-based quantification of taxa-specific abundances in microbial communities: quantifying and avoiding common pitfalls.J. Microbiol. Methods153139–147. 10.1016/j.mimet.2018.09.015
11
Borrell Y. J. Miralles L. Do Huu H. Mohammed-Geba K. Garcia-Vazquez E. (2017). DNA in a bottle—Rapid metabarcoding survey for early alerts of invasive species in ports.PLoS One12:e0183347. 10.1371/journal.pone.0183347
12
Bravo I. Fernández M. L. Ramilo I. Martınez A. (2001). Toxin composition of the toxic dinoflagellate Prorocentrum lima isolated from different locations along the Galician coast (NW Spain).Toxicon391537–1545. 10.1016/s0041-0101(01)00126-x
13
Bylemand J. Gleeson D. M. Duncan R. P. Hardy C. M. Furlan E. M. (2019). A performance evaluation of targeted eDNA and eDNA metabarcoding analyses for freshwater fishes.Environ. DNA1402–414. 10.1002/edn3.41
14
Caporaso J. G. Kuczynski J. Stombaugh J. Bittinger K. Bushman F. D. Costello E. K. et al (2010). QIIME allows analysis of high-throughput community sequencing data.Nat. Methods7335–336. 10.1038/nmeth.f.303
15
Carlier A. Riera P. Amouroux J. M. Bodiou J. Y. Desmalades M. Grémare A. (2008). Food web structure of two Mediterranean lagoons under varying degree of eutrophication.J. Sea Res.60264–275. 10.1016/j.seares.2008.10.006
16
Cataudella S. Crosetti D. Massa F. (2015). “Mediterranean coastal lagoons: sustainable management and interactions among aquaculture, capture fisheries and the environment,” in General Fisheries Commission for the Mediterranean.Rome: FAO.
17
Cervetto G. Gaudy R. Pagano M. (1999). Influence of salinity on the distribution of Acartia tonsa (Copepoda, Calanoida).J. Exp. Mar. Biol Ecol.23933–45. 10.1016/s0022-0981(99)00023-4
18
Chapman P. M. (2012). Management of coastal lagoons under climate change. Estuarine, Coastal and Shelf Science, 110, 32-35. g data.Nature Methods7:335.
19
Cinar M. E. Ergen Z. Dagli E. Petersen M. E. (2005). Alien species of spionid polychaetes (Streblospio gynobranchiata and Polydora cornuta) in Izmir Bay, eastern Mediterranean.J. Mar. Biol. Assoc. UK.85821–827. 10.1017/s0025315405011768
20
Çinar M. E. Katagan T. Öztürk B. Bakir K. Dagli E. (2012). Spatio-temporal distributions of zoobenthos in soft substratum of Izmir Bay (Aegean Sea, eastern Mediterranean), with special emphasis on alien species and ecological quality status.J. Mar. Biol. Assoc. UK921457–1477. 10.1017/s0025315412000264
21
Crooks J. A. Chang A. L. Ruiz G. M. (2011). Aquatic pollution increases the relative success of invasive species.Biol. Invasions13165–176. 10.1007/s10530-010-9799-3
22
David M. Gollasch S. Cabrini M. Perkoviè M. Bošnjak D. Virgilio D. (2007). Results from the first ballast water sampling study in the Mediterranean Sea–the Port of Koper study.Mar. Pollut. Bull.5453–65. 10.1016/j.marpolbul.2006.08.041
23
Dean H. K. (2008). The use of polychaetes (Annelida) as indicator species of marine pollution: a review.Rev. Biol. Trop.5611–38.
24
Ficetola G. F. Miaud C. Pompanon F. Taberlet P. (2008). Species detection using environmental DNA from water samples.Biol. Lett.4423–425. 10.1098/rsbl.2008.0118
25
Ficetola G. F. Pansu J. Bonin A. Coissac E. Giguet−Covex C. De Barba M. et al (2015). Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data.Mol. Ecol. Resour.15543–556. 10.1111/1755-0998.12338
26
Forster D. Filker S. Kochems R. Breiner H.-W. Cordier T. Pawlowski J. et al (2019). A comparison of different ciliate metabarcode genes as bioindicators for environmental impact assessments of salmon aquaculture.Eukaryot. Microbiol.66294–308. 10.1111/jeu.12670
27
Furlan E. M. Gleeson D. Hardy C. M. Duncan R. I. P. (2015). A framework for estimating the sensitivity of eDNA surveys.Mol. Ecol. Resour.16641–654. 10.1111/1755-0998.12483
28
Galil B. S. (2007). Loss or gain? Invasive aliens and biodiversity in the Mediterranean Sea.Mar. Pollut. Bull.55314–322. 10.1016/j.marpolbul.2006.11.008
29
Gaudy R. Viñas M. D. (1985). Première signalisation en Méditerranée du copépode pélagique Acartia tonsa.Rapp. Comm. Int. Mer Médit.29227–229.
30
Geller J. Meyer C. Parker M. Hawk H. (2013). Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all−taxa biotic surveys.Mol. Ecol. Resour.13851–861. 10.1111/1755-0998.12138
31
Gozlan R. E. Britton J. R. Cowx I. Copp G. H. (2010). Current knowledge on non−native freshwater fish introductions.J Fish Biol.76751–786. 10.1111/j.1095-8649.2010.02566.x
32
Hallegraeff G. M. (1993). A review of harmful algal blooms and their apparent global increase.Phycologia3279–99. 10.1186/1476-069X-7-S2-S4
33
Hebert P. D. Cywinska A. Ball S. L. (2003a). Biological identifications through DNA barcodes.Proc. R. Soc. Lon. B Biol. Sci.270313–321. 10.1098/rspb.2002.2218
34
Hebert P. D. Ratnasingham S. de Waard J. R. (2003b). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species.Proc. R. Soc. Lon. B Biol. Sci.270(Suppl. 1), S96–S99.
35
Heiman K. W. Vidargas N. Micheli F. (2008). Non-native habitat as home for non-native species: comparison of communities associated with invasive tubeworm and native oyster reefs.Aquati. Biol.247–56. 10.3354/ab00034
36
Heisler J. Glibert P. M. Burkholder J. M. Anderson D. M. Cochlan W. Dennison W. C. et al (2008). Eutrophication and harmful algal blooms: a scientific consensus.Harmful Algae83–13. 10.1016/j.hal.2008.08.006
37
Illumina (2013). “16S metagenomic sequencing library preparation,” in Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System, (Illumina, CA: Illumina), 1–28.
38
Jane S. F. Wilcox T. M. McKelvey K. S. Young M. K. Schwartz M. K. Lowe W. H. et al (2015). Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams.Mol. Ecol. Resour.15216–227. 10.1111/1755-0998.12285
39
Jones T. Parrish J. K. Punt A. E. Trainer V. L. Kudela R. Lang J. et al (2017). Mass mortality of marine birds in the Northeast Pacific caused by Akashiwo sanguinea.Mar. Ecol. Progr. Ser.579111–127. 10.3354/meps12253
40
Katsanevakis S. Coll M. Piroddi C. Steenbeek J. Ben Rais Lasram F. Zenetos A. et al (2014). Invading the mediterranean sea: biodiversity patterns shaped by human activities.Front. Mar. Sci.1:32. 10.3389/fmars.2014.00032
41
Kelly R. P. Port J. A. Yamahara K. M. Crowder L. B. (2014). Using environmental DNA to census marine fishes in a large mesocosm.PLoS One9:e86175. 10.1371/journal.pone.0086175.t001
42
Kjerfve B. (ed.) (1994). Coastal lagoon processes, Vol. 60. Berlin: Elsevier.
43
Laroche O. Wood S. A. Tremblay L. A. Ellis J. I. Lejzerowicz F. Pawlowski J. et al (2016). First evaluation of foraminiferal metabarcoding for monitoring environmental impact from an offshore oil drilling site.Mar. Environ. Res.120225–235. 10.1016/j.marenvres.2016.08.009
44
Leray M. Yang J. Y. Meyer C. P. Mills S. C. Agudelo N. Ranwez V. et al (2013). A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents.Front. Zool.10:34. 10.1186/1742-9994-10-34
45
Lilly E. L. Kulis D. M. Gentien P. Anderson D. M. (2002). Paralytic shellfish poisoning toxins in France linked to a human-introduced strain of Alexandrium catenella from the western Pacific: evidence from DNA and toxin analysis.J. Plankton Res.24443–452. 10.1093/plankt/24.5.443
46
Lin S. Zhang H. Hou Y. Zhuang Y. Miranda L. (2009). High-level diversity of dinoflagellates in the natural environment, revealed by assessment of mitochondrial cox1 and cob genes for dinoflagellate DNA barcoding.Appl. Environ. Microbiol.751279–1290. 10.1128/AEM.01578-08
47
Maranho L. A. Baena-Nogueras R. M. Lara-Martín P. A. DelValls T. A. Martín-Díaz M. L. (2014). Bioavailability, oxidative stress, neurotoxicity and genotoxicity of pharmaceuticals bound to marine sediments. The use of the polychaete Hediste diversicolor as bioindicator species.Environ. Res.134353–365. 10.1016/j.envres.2014.08.014
48
Moestrup Ø Akselmann R. Fraga S. Hoppenrath M. Iwataki M. Komárek J. et al (eds) (2009). IOC-UNESCO Taxonomic Reference List of Harmful Micro Algae. Avaliable at: http://www.marinespecies.org/hab(accessed December 16, 2018).
49
Moore S. K. Trainer V. L. Mantua N. J. Parker M. S. Laws E. A. Backer L. C. et al (2008). Impacts of climate variability and future climate change on harmful algal blooms and human health.Environ. Health7(Suppl. 2):S4.
50
Nilsson R. H. Tedersoo L. Abarenkov K. Ryberg M. Kristiansson E. Hartmann M. et al (2012). Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences.MycoKeys437–63. 10.3897/mycokeys.4.3606
51
Oksanen J. Blanchet F. Kindt R. Legendre P. Minchin R. O’Hara R. et al (2013). Vegan: Community Ecology Package. R Package Version. 2.0–10.
52
Paavola M. Olenin S. Leppäkoski E. (2005). Are invasive species most successful in habitats of low native species richness across European brackish water seas?Estuar.Coast. Shelf Sci.64738–750. 10.1016/j.ecss.2005.03.021
53
Palumbi S. R. (1996). “Nucleic acids II: the polymerase chain reaction,” in Molecular Systematics, edsHillisD. M.MoritzC.MableB. K. (Sunderland: Sinauer Associates, Inc.), 205–247.
54
Pérez-Ruzafa A. Marcos C. Pérez-Ruzafa I. M. (2011). Mediterranean coastal lagoons in an ecosystem and aquatic resources management context.Phys,. Chem. Earth Parts A/B/C36160–166. 10.1016/j.pce.2010.04.013
55
Pochon X. Zaiko A. Fletcher L. M. Laroche O. Wood S. A. (2017). Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications.PLoS One12:e0187636. 10.1371/journal.pone.0187636
56
Radashevsky V. I. Selifonova Z. P. (2013). Records of Polydora cornuta and Streblospio gynobranchiata (Annelida, Spionidae) from the Black Sea.Mediterr. Mar. Sci.14261–269.
57
Read G. B. Gordon D. P. (1991). Adventive occurrence of the fouling serpulid Ficopomatus enigmaticus (Polychaeta) in New Zealand.N. Z. J Mar. Freshwa. Res.25269–273. 10.1080/00288330.1991.9516478
58
Reizopoulou S. Thessalou-Legaki M. Nicolaidou A. (1996). Assessment of distubance in Mediterranean lagoons: an evaluation of methods.Mar. Biol.125189–197. 10.1007/bf00350773
59
Ruiz G. M. Fofonoff P. W. Carlton J. T. Wonham M. J. Hines A. H. (2000). Invasion of coastal marine communities in North America: apparent patterns, processes, and biases.Annu. Rev. Ecol. Syst.31481–531. 10.1146/annurev.ecolsys.31.1.481
60
Sassoubre L. M. Yamahara K. M. Gardner L. D. Block B. A. Boehm A. B. (2016). Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish.Environ. Sci. Technol.5010456–10464. 10.1021/acs.est.6b03114
61
Scott R. Zhan A. Brown E. A. Chain F. J. J. Cristescu M. E. Gras R. et al (2018). Optimization and performance testing of a sequence processing pipeline applied to detection of nonindigenous species.Evol. Appl.11891–905. 10.1111/eva.12604
62
Sellner K. G. Doucette G. J. Kirkpatrick G. J. (2003). Harmful algal blooms: causes, impacts and detection.J. Ind. Microbiol. Biotechnol.30383–406. 10.1007/s10295-003-0074-9
63
Souchu P. Bec B. Smith V. H. Laugier T. Fiandrino A. Benau L. et al (2010). Patterns in nutrient limitation and chlorophyll a along an anthropogenic eutrophication gradient in French Mediterranean coastal lagoons.Can. J. Fish Aquat. Sci.67743–753. 10.1139/f10-018
64
Surugiu V. (2005). The use of polychaetes as indicators of eutrophication and organic enrichment of coastal waters: a study case–Romanian Black Sea Coast. Analele Ştiinţifice ale Universităţii “Al.I. Cuza”. Iaşi, s.Biol. Anim.5155–62.
65
Taberlet P. Coissac E. Pompanon F. Brochmann C. Willerslev E. (2012). Towards next−generation biodiversity assessment using DNA metabarcoding.Mol. Ecol.212045–2050. 10.1111/j.1365-294X.2012.05470.x
66
Tena J. Capaccioni-Azzati R. Porras R. Torres-Gavilá F. J. (1991). Cuatro especies de poliquetos nuevas para las costas mediterráneas españolas en los sedimentos del antepuerto de Valencia.Miscel⋅ Zool.1529–41.
67
Thomas A. C. Deagle B. E. Eveson J. P. Harsch C. H. Trites A. W. (2016). Quantitative DNA metabarcoding: improved estimates of species proportional biomass using correction factors derived from control material.Mol. Ecol. Resour.16714–726. 10.1111/1755-0998.12490
68
Thomsen P. F. Willerslev E. (2015). Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity.Biol. Conserv.1834–18. 10.1016/j.biocon.2014.11.019
69
Thomsen P. T. Kielgast J. Iversen L. Møller P. R. Rasmussen M. Willerslev E. (2012). Detection of a diverse marine fish fauna using environmental DNA from seawater samples.PLoS One7:e41732. 10.1371/journal.pone.0041732
70
von Ammon U. Wood S. A. Larocge O. Zaiko A. Tait L. Lavery S. et al (2018). Combining morpho-taxonomy and metabarcoding enhances the detection of non-indigenous marine pests in biofouling communities.Sci. Rep.816290. 10.1038/s41598-018-34541-1
71
Vouvé F. Buscail R. Aubert D. Labadie P. Chevreuil M. Canal C. et al (2014). Bages-Sigean and Canet-St Nazaire lagoons (France): physico-chemical characteristics and contaminant concentrations (Cu, Cd, PCBs and PBDEs) as environmental quality of water and sediment.Environ. Sci. Pollut. Res.213005–3020. 10.1007/s11356-013-2229-1
72
Weigand A. Beermann A. J. Èiampor F. Costa F. O. Csabai Z. Duarte S. et al (2019). DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work.Sci. Total Environ.678499–524. 10.1016/j.scitotenv.2019.04.247
73
Wilcox T. M. McKelvey K. S. Young M. K. Jane S. F. Lowe W. H. Whiteley A. R. et al (2013). Robust detection of rare species using environmental DNA: the importance of primer specificity.PLoS One8:e59520. 10.1371/journal.pone.0059520
74
Yasumoto T. Oshima Y. Fukuyo Y. Oguri H. Igarashi T. Fujita N. (1980). Identification of Dinophysis fortii as the causative organism of diarrhetic shellfish poisoning.Bull. Japan. Soc. Sci. Fish.461405–1411. 10.2331/suisan.46.1405
75
Zaiko A. Pochon X. Garcia-Vazquez E. Olenin S. Wood S. A. (2018). Advantages and limitations of environmental DNA/RNA tools for marine biosecurity: management and surveillance of non-indigenous species.Front. Mar. Sci.5:322. 10.3389/fmars.2018.00322
76
Zaiko A. Samuiloviene A. Ardura A. Garcia-Vazquez E. (2015). Metabarcoding approach for nonindigenous species surveillance in marine coastal waters.Mar. Pollut. Bull.10053–59. 10.1016/j.marpolbul.2015.09.030
77
Zenetos A. Gofas S. Morri C. Rosso A. Violanti D. García Raso J. E. et al (2012). Alien species in the mediterranean Sea by 2012,” in a contribution to the application of european union’s marine strategy framework directive (MSFD), Part2. introduction trends and pathways.Mediterr. Mar. Sci.13328–352. 10.12681/mms.327
Summary
Keywords
COI “barcode, ” next generation sequencing, Canet lagoon, Nature 2000 areas, conservation
Citation
Suarez-Menendez M, Planes S, Garcia-Vazquez E and Ardura A (2020) Early Alert of Biological Risk in a Coastal Lagoon Through eDNA Metabarcoding. Front. Ecol. Evol. 8:9. doi: 10.3389/fevo.2020.00009
Received
03 October 2019
Accepted
15 January 2020
Published
28 February 2020
Volume
8 - 2020
Edited by
David S. Thaler, Universität Basel, Switzerland
Reviewed by
Alejandro Trujillo Gonzalez, University of Canberra, Australia; Rebecca Barr Simmons, University of North Dakota, United States
Updates
Copyright
© 2020 Suarez-Menendez, Planes, Garcia-Vazquez and Ardura.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alba Ardura, arduraalba@uniovi.es
This article was submitted to Phylogenetics, Phylogenomics, and Systematics, a section of the journal Frontiers in Ecology and Evolution
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.