- 1Division of Comparative Medicine, Massachusetts Institute of Technology, Cambridge, MA, United States
- 2Island Veterinary Services, George Town, Cayman Islands
- 3Department of the Environment, George Town, Cayman Islands
- 4Wildlife Conservation Society, Zoological Health Program, Bronx, NY, United States
Blue iguanas (Cyclura lewisi) are endangered reptiles found only on Grand Cayman. Previously, DNA for a novel Helicobacter species GCBI1 was detected in sick and dead iguanas. In the current study, fecal and cloacal swab samples were obtained from 25 iguanas. Through molecular and microbiological techniques, a novel Helicobacter species was cultured from feces and characterized, for whom we propose the name Helicobacter cyclurae. This novel helicobacter had a prevalence of 56% by PCR and 20% by culture in samples analyzed. The type strain MIT 16-1353 was catalase, oxidase, and gamma-glutamyl transpeptidase positive. By electron microscopy, H. cyclurae has a curved rod morphology and a single sheathed polar flagellum. Phylogenetic analysis using 16S rRNA, gyrB, and hsp60 indicated that these strains were most closely related to Helicobacter sp. 12502256-12 previously isolated from lizards. H. cyclurae has a 1.91-Mb genome with a GC content of 33.37%. There were 1,969 genes with four notable virulence genes: high temperature requirement-A protein-secreted serine protease, gamma-glutamyl transpeptidase, fibronectin/fibrinogen binding protein, and neutrophil-activating protein. Whole-genome phylogeny, average nucleotide identity, and digital DNA–DNA hybridization analysis confirmed that H. cyclurae is a novel species, and the first helicobacter cultured and characterized from blue iguanas.
Introduction
The Cayman Islands are a group of Caribbean islands composed of the Grand Cayman, Little Cayman, and Cayman Brac. These islands are inhabited with diverse fauna and flora including numerous orchids and animals such as the Cayman Brac Parrot and dwarf boa. The Grand Cayman iguana, Cyclura lewisi, a blue colored reptile is indigenous only to Grand Cayman. It is currently listed as Endangered under the US Endangered Species Act. They have been threatened to the point of extinction when in the past their numbers dwindled to fewer than 20 animals in the wild due to predation from feral species, vehicular accidents, and habitat loss (Goodman and Burton, 2005). Due to the collective efforts of the Wildlife Conservation Society and local partners in Grand Cayman, the number of this species in the wild has drastically improved.
Helicobacter is a genus of bacteria that are known to cause diseases in humans and a variety of animal species. Specifically, enterohepatic Helicobacter species (EHS) colonize the gastrointestinal tract and are associated with gastroenteritis and hepatitis in humans and animals. Importantly, several of these Helicobacter species cause bacteremia and systemic diseases (Fox, 2002; Stacy and Wellehan, 2010; Shen et al., 2017). Reptiles are also colonized by Helicobacter spp., with one report finding a prevalence of 4.8 and 39.1% by culture and PCR, respectively, and more specifically 7.4 and 30.7% for animals of the suborder Lacertilia (Gilbert et al., 2014). In a recent study, two free-ranging blue iguanas located at the Grand Cayman Queen Elizabeth II Botanic Park (QEIIBP) in 2015 developed hind limb paralysis and septicemia with spiral-shaped bacteria noted on blood smears (Conley et al., 2021). PCR results, using specific Helicobacter species primers, from the blood and tissue of these animals were positive for a novel Helicobacter species, provisionally designated Helicobacter sp. GCBI1. The novel helicobacter, by 16S rRNA analysis, is 95.1% similar to a helicobacter identified in a Greek tortoise, and phylogenetic analysis of Helicobacter sp. GCBI1 suggested that it is an EHS as it grouped with other EHS in the chelonian clade, Helicobacter spp. taxon 2 (Gilbert et al., 2014). Examination and clinical testing of affected animals along with responsive treatment for helicobacter infection was suggestive that Helicobacter sp. GCBI1 was the causative agent of disease in Grand Cayman blue iguanas. Retrospective analysis from a mortality event of green iguanas a year prior and screening of healthy free-ranging green iguanas from QEIIBP also found this helicobacter by qPCR (Conley et al., 2021). In this paper, we describe, by utilizing molecular and microbiological methodology, the isolation of a novel Helicobacter species, H. cyclurae, that is distinct from previously described Helicobacter sp. GCBI1, from feces and cloacal swabs in Grand Cayman blue iguanas.
Materials and Methods
Animals
A total of 25 blue iguanas were screened for helicobacter. For 17 animals, fecal samples were collected; for eight animals, feces were not available, so eight cloacal swab samples were collected instead. Collected samples were placed in freeze media (20% glycerol in Brucella broth) and kept at −80°C prior to processing.
Bacterial Isolation and Characterization
Feces were homogenized in freeze media; the freeze media containing fecal material and cloacal swab suspensions were directly placed on CVA (cefoperazone, vancomycin, and amphotericin) plates or filtered through 0.65-μm filters onto tryptic soy agar plates with 5% sheep blood agar plates (Remel Laboratories, Lenexus, KS) and incubated under microaerobic conditions at 37°C with a gas mixture of N2, CO2, and H2 (80:10:10). Detailed biochemical characterization analysis was performed on five individual isolates using a RapIDTM NH System (Remel Laboratories, Lenexus, KS). Biochemical characterization of urease, catalase, and oxidase production; sensitivity to nalidixic acid and cephalothin; and growth in the presence of 1% glycine were analyzed as previously described by our laboratory (Shen et al., 2005, 2017) and following the guideline of minimal standards for describing new species belonging to the families Campylobacteraceae and Helicobacteraceae (On et al., 2017; Shen et al., 2020).
DNA Extraction, PCR, and Sequence Analysis for 16S rRNA, gyrB, and hsp60 Genes
A High Pure PCR Template Preparation Kit (Roche Molecular Biochemicals, Indianapolis, IN) was used for bacterial DNA extraction, and the QIAamp DNA Stool Mini Kit (Qiagen, Germantown, MD) was used for fecal DNA extraction. The nearly full 16S rRNA sequence of five strains was amplified with primer 9F (5′-GAG TTT GAT YCT GGC TCA G-3′) and 1541R (5′-AAG GAGGTG WTC CAR CC-3′). Helicobacter genus-specific primers which amplify a 1.2-kb product from 16S rRNA gene were used for PCR for fecal and cloacal swab samples (Fox et al., 1998). Primers HSP60AF (5′-GCT AAT CCT ATT GAA GTG AAA AGA GGN ATG GAY AA-3′) and HSP60DR (5′-CAC TAA GGT AGT TAA AGC TTC CCC TTC DAT RTC YT-3′) were used to amplify the hsp60 gene (Inglis et al., 2006); primers specific for Helicobacter species iguana isolates gyrB-igu-F (5′-GAT ACT TAT AAA GTT TCC GG-3′) and gyrB-igu-R (5′-CAA ATT CCT TAT CAA TTC CGC A-3′) were used to amplify the gryB gene. PCR amplifications were performed using the Expand High Fidelity PCR System (Roche Molecular Biochemicals, Indianapolis, IN). The following conditions were used for amplification: 35 cycles of denaturation at 94°C for 1 min, annealing at 55–58°C for 1 min, and elongation at 72°C for 1.5 min, followed by an elongation step of 7 min at 72°C. The PCR products from bacterial isolates and fecal samples were directly sequenced, using a commercial sequencing facility. Sequences were compared directly with the NCBI GenBank nucleotide database by BLAST search. Phylogenetic trees were constructed by the neighbor-joining method with the Lasergene software package (DNASTAR Madison WI) based on the comparison of genes from other Helicobacter species. Bootstrap values (>75%) were based on 1,000 replications.
Draft Genome Sequencing and Comparative Analysis
Genomic DNA from the type strain MIT 16-1353 isolated from the feces of a blue iguana was extracted using the MasterPure Complete DNA and RNA Purification Kit (Epicenter) following the manufacturer’s protocol for bacterial cell samples. DNA libraries were prepared using NextraXT for sequencing of 2 × 150 paired-end reads by Illumina MiSeq. Raw sequenced reads were decontaminated of adapter sequences and quality trimmed to a Phred quality score (Q) ≥ 10 using BBDuk from the BBMap package version 37.17 (BBMap, SourceForge, Fairfax, VA). Decontaminated reads were then assembled into contigs with SPAdes followed by genome annotation with RAST, with both services hosted by PATRIC (Wattam et al., 2017). Through OrthoFinder (Emms and Kelly, 2019), concatenated core gene sequences were determined followed by MAFFT for multi-sequence alignment and FastTree to infer approximately maximum-likelihood phylogenetic trees. Orthogroups, determined using OrthoFinder, were further analyzed to identify shared and unique gene between isolate MIT 16-1353 from the blue iguana and the genomes of other helicobacters isolated from reptile hosts. Average nucleotide identities (ANI) were calculated with pyani (Pritchard et al., 2016). Genomes with an ANI >95% were considered the same species. Digital DNA–DNA hybridization (dDDH) percentages were calculated using the Genome-to-Genome Distance Calculator (GGDC) (Meier-Kolthoff et al., 2013). Genomes with a dDDH >70% were considered the same species. Virulence factor gene homologs were identified by BLASTP analysis of genome annotation against the virulence factor database (VFDB) (Liu et al., 2019). Homolog genes were considered to have sequence identity ≥50% and sequence coverage ≥ 95%.
Electron Microscopy
Isolate MIT 16-1353 was examined by electron microscopy. Cells grown on blood agar plate for 48 h were gently suspended in PBS at a concentration of about 108 cells/ml. The sample was negatively stained with 1% (w/v) phosphotungstic acid (pH 6.5) for 20–30 s. Specimens were examined with a JEOL model 2100F transmission electron microscope.
Results
Helicobacter Isolation and Characterization
Helicobacter-like organisms were isolated from 4 of the 17 fecal samples and one of the eight cloacal swab samples. These helicobacter isolates were compared with other Helicobacter species to determine differences in biochemical characteristics (Table 1). They were gram negative and grew at 28°C and 37°C but not at 42°C or with 1% glycine. The bacterium appeared on 5% sheep blood agar plates as a single colony, 2–3 mm in diameter, mucoid, and clear or grayish in color. These isolates of the Helicobacter species had strong catalase, oxidase, and gamma-glutamyl transpeptidase activity and displayed resistance to nalidixic acid and cephalothin. There was no nitrate reduction or urease activity nor hydrolyzing activity of indoxyl acetate in these isolates; variation on alkaline phosphatase was noted among isolates.
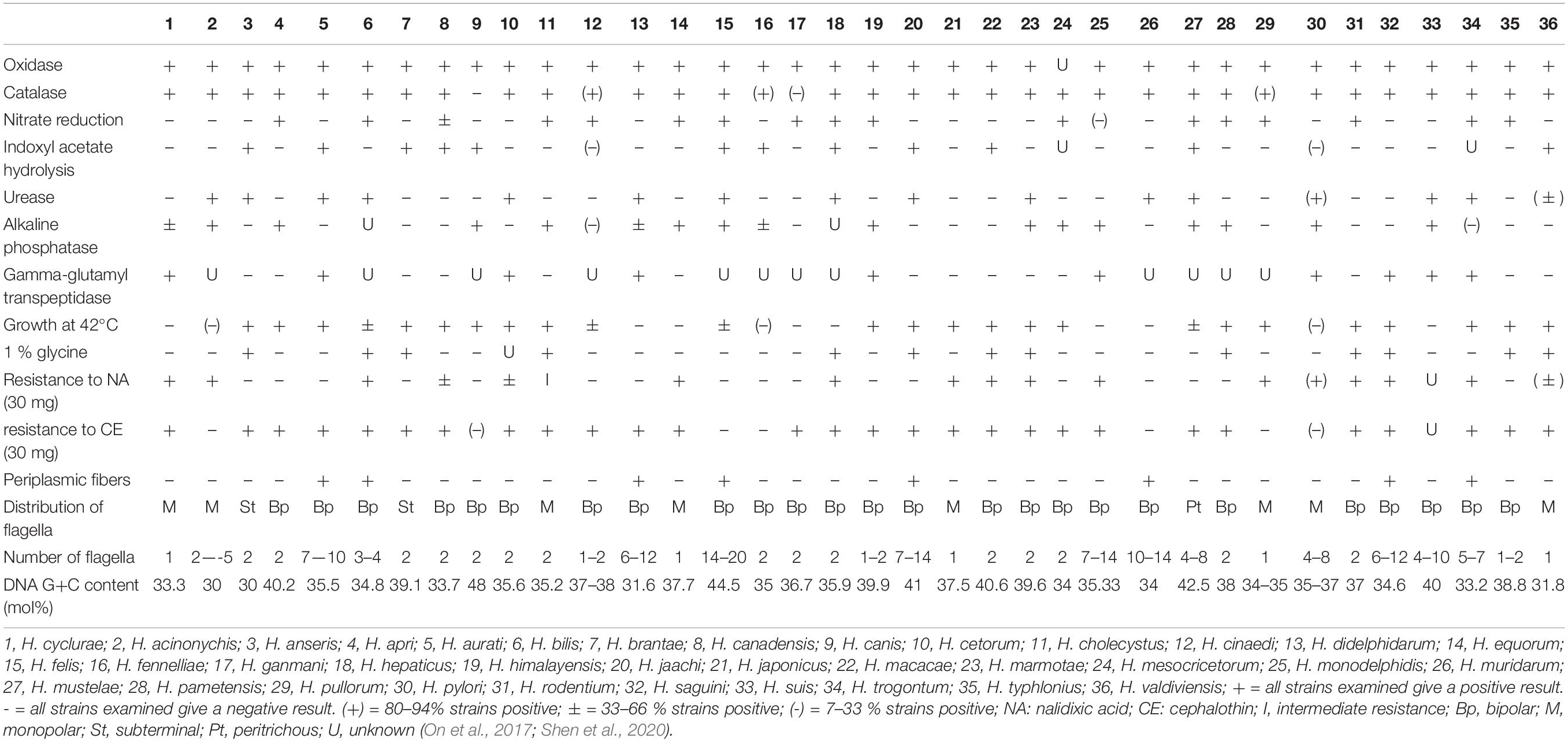
Table 1. Phenotypic characteristics that differentiate H. cyclurae from other EHS and selected gastric Helicobacter species.
PCR and Phylogenetic Analysis
Ten fecal samples and four cloacal swab samples from 25 animals were helicobacter PCR positive using genus 16S rRNA-specific primers. Helicobacter species isolated from four fecal samples and one cloacal swab sample (see above) had over 99% 16S rRNA sequences similarity with each other. Its 16S rRNA sequence clustered with other Helicobacter species that have been isolated from lizards. The most closely related 16S rRNA sequence was Helicobacter sp. 12S02256-12 (KJ081208) with 98% similarity (Figure 1A; Gilbert et al., 2014). Sequences from PCR products of nine fecal or cloacal swab samples which were culture-negative had similar sequences with the novel cultured Helicobacter sp. The 16S rRNA sequences of this novel Helicobacter species were different from the Helicobacter sp. GCBI1, which was identified by PCR in the blood and tissues of blue iguanas; these two Helicobacters only shared 96% sequence identity (Figure 1A; Conley et al., 2021).
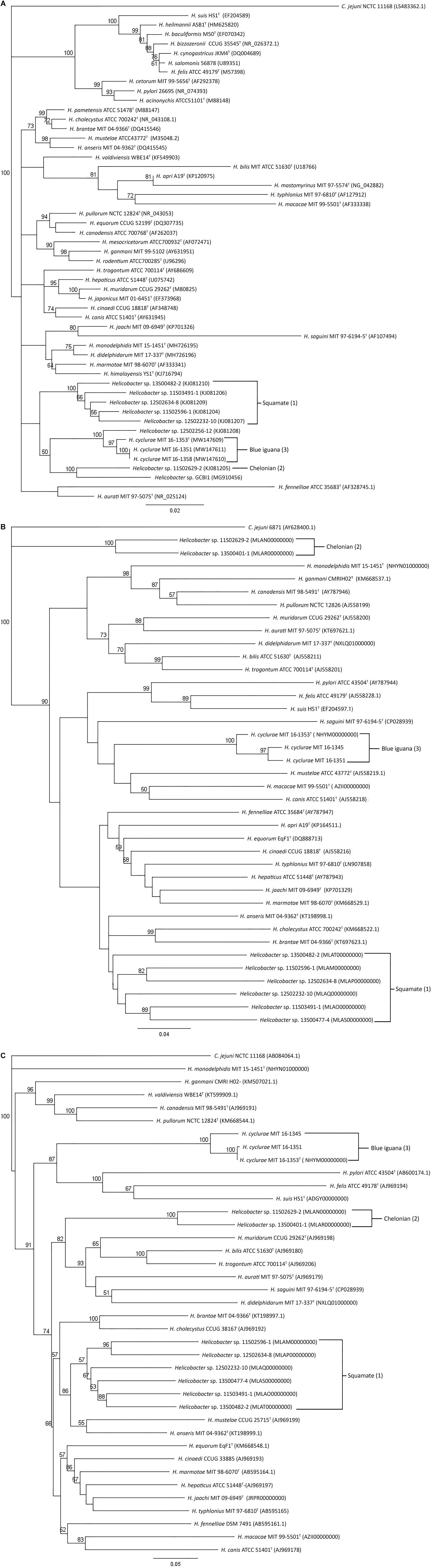
Figure 1. Phylogenetic analysis of 16S rRNA, (A) hsp60, (B) and gyrB (C) gene sequences. Neighbor-joining trees were based on the comparison of genes from different Helicobacter species. C. jejuni is used as an outgroup. Bootstrap values (>50%) based on 1,000 replications are indicated. GenBank accession numbers (in parentheses) are provided for each strain. The scale bar indicates branch length as number of substitutions per nucleotide. (1) Reptiles in the order of Chelonian: turtles and tortoises. (2) Reptiles in the order of Squamata: snakes and lizards. (3) Blue iguana: belongs to the order of Squamata.
The housekeeping gene sequences for heat shock protein 60 (hsp60) and DNA gyrase subunit B (gyrB) have been used for classification of phylogenetic relationships among Helicobacter species (Mikkonen et al., 2004; Hannula and Hanninen, 2007). Phylogenetic trees based on the partial gyrB and hsp60 gene sequences of the novel Helicobacter species are presented in Figures 1B,C. The three H. cyclurae isolates shared 94–99% sequence identity within the species in their gyrB but only had 72% sequence identity with the gyrB gene sequence of closely related species, Helicobacter anseris (Fox et al., 2006). For hsp60 gene analysis, the three H. cyclurae isolates shared 96–97% sequence identity and had 78% sequence identity with the hsp60 gene of the most closely related species, Helicobacter brantae (Fox et al., 2006).
Whole-Genome Analysis
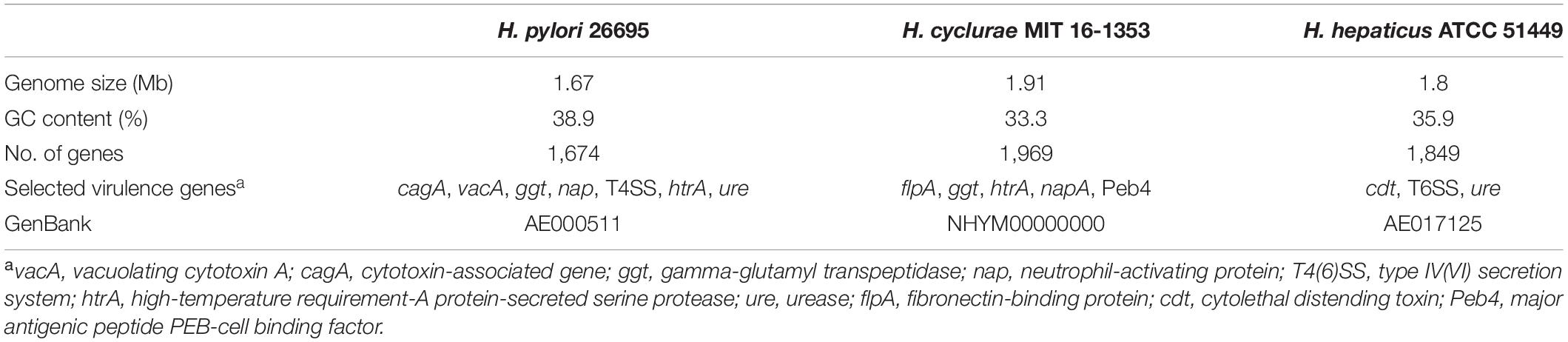
Whole-genome sequencing of the type strain, MIT 16-1353, indicated that H. cyclurae has a genome size of 1.91 Mb with a GC content of 33.3% (Figure 2) and contains 1,969 genes (Table 2). A whole-genome phylogenetic tree constructed from the multi-sequence alignment of concatenated core genes confirms the iguana helicobacter is distinct from other known Helicobacter species, including the species isolated from reptiles described by Gilbert et al. (2017). Instead, H. cyclurae was most closely placed with Helicobacter monodelphidis MIT 15–1451 isolated from captive opossums with cloacal prolapse (Shen et al., 2020). ANI and dDDH comparisons between the iguana isolate genome and other Helicobacter species were below 95 and 70%, respectively, supporting the finding that the iguana isolate is a novel Helicobacter species (Table 3). The type strain, MIT 16-1353, has been submitted to GenBank under accession number NHYM00000000.
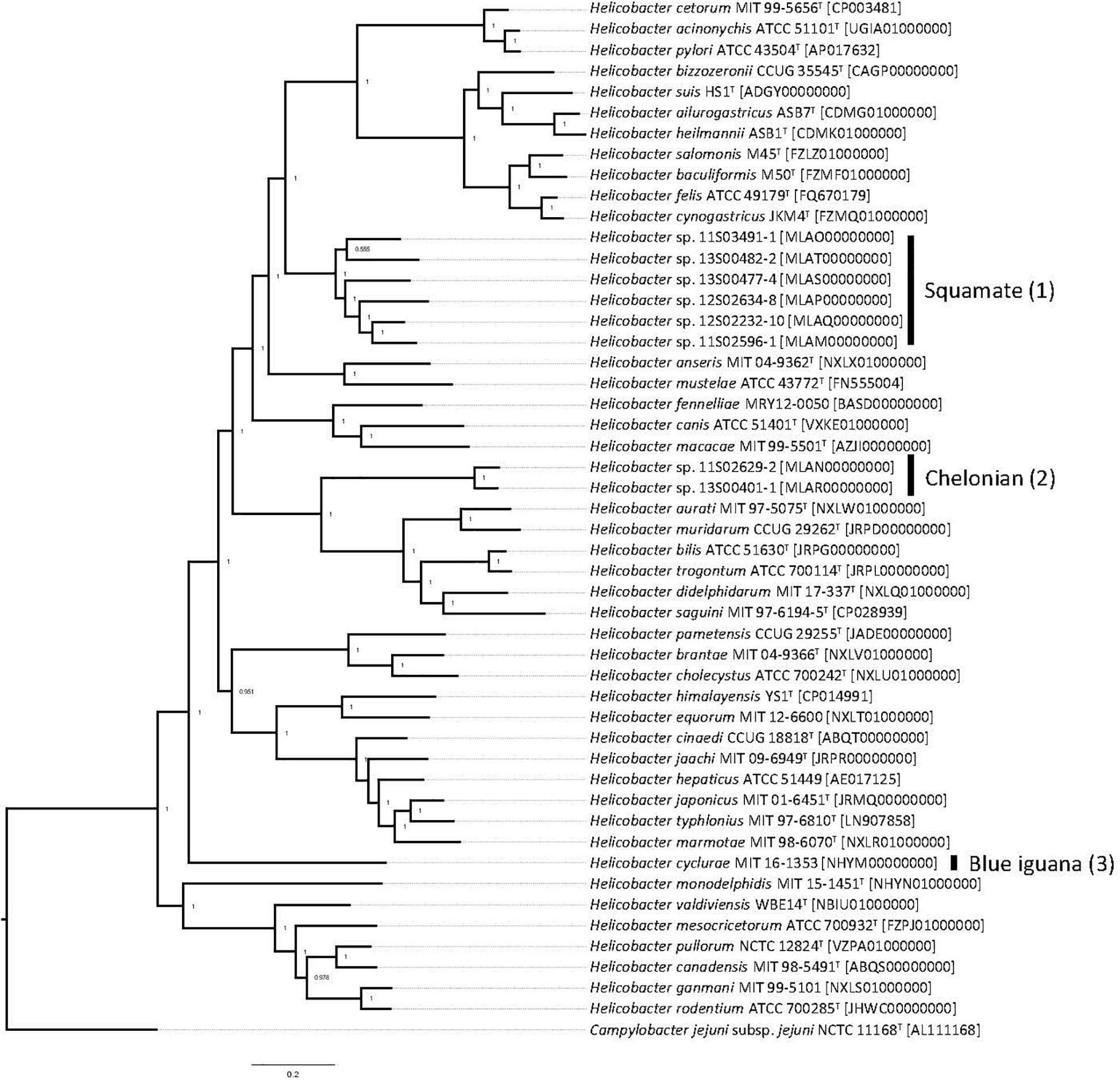
Figure 2. Pan-genomic phylogenetic tree. Genomes for Helicobacter species were accessed from public databases. Type strains were used when available, and GenBank accession numbers for each genome are shown in brackets. The pan-genomic phylogenetic tree was created from the multi-sequence alignment of concatenated core gene sequences. Local support values, which indicate the reliability of each branch in the tree (0–1, lowest to highest support), were calculated by FastTree by resampling the site likelihoods 1,000 times using the Shimodaira–Hasegawa test. The scale bar indicates branch length as number of substitutions per amino acid. (1) Reptiles in the order of Chelonian: turtles and tortoises. (2) Reptiles in the order of Squamata: snakes and lizards. (3) Blue iguana: belongs to the order of Squamata.

Table 3. ANI and dDDH values for Helicobacter cyclurae MIT 16-1353 [NHYM00000000] vs. other Helicobacter species genomes.
In agreement with the whole-genome phylogenetic relationship, H. cyclurae shared more orthogroups (i.e., homologous genes) with the Helicobacter species isolated from chelonians (Helicobacter spp. 11S02629-2 and 13S00401-1) compared to squamates (Helicobacter spp. 13S00482-2, 11S03491-1, 13S00477-4, 12S02634-8, 12S02232-10, and 11S02596-1) (Figure 3). More than 25% of orthogroups for H. cyclurae were not found in these other reptile-associated helicobacter genomes. The unique genes harbored by H. cyclurae included metabolic and functional pathways for amino acid synthesis, transporters, and oxidative phosphorylation (Figure 4). However, the H. cyclurae genome was deficient for several genes related to lipid and lipopolysaccharide biosynthesis as well as other metabolic and functional pathways compared to the unique orthogroups belonging to the chelonian and squamate helicobacters (Figure 4). These included glutamyl-tRNA amidotransferase subunit A, putative methyltransferase YcgJ, and the putative tricarballylate catabolism locus tcuRABC, which are present in the other reptile genomes described by Gilbert et al. (2017).
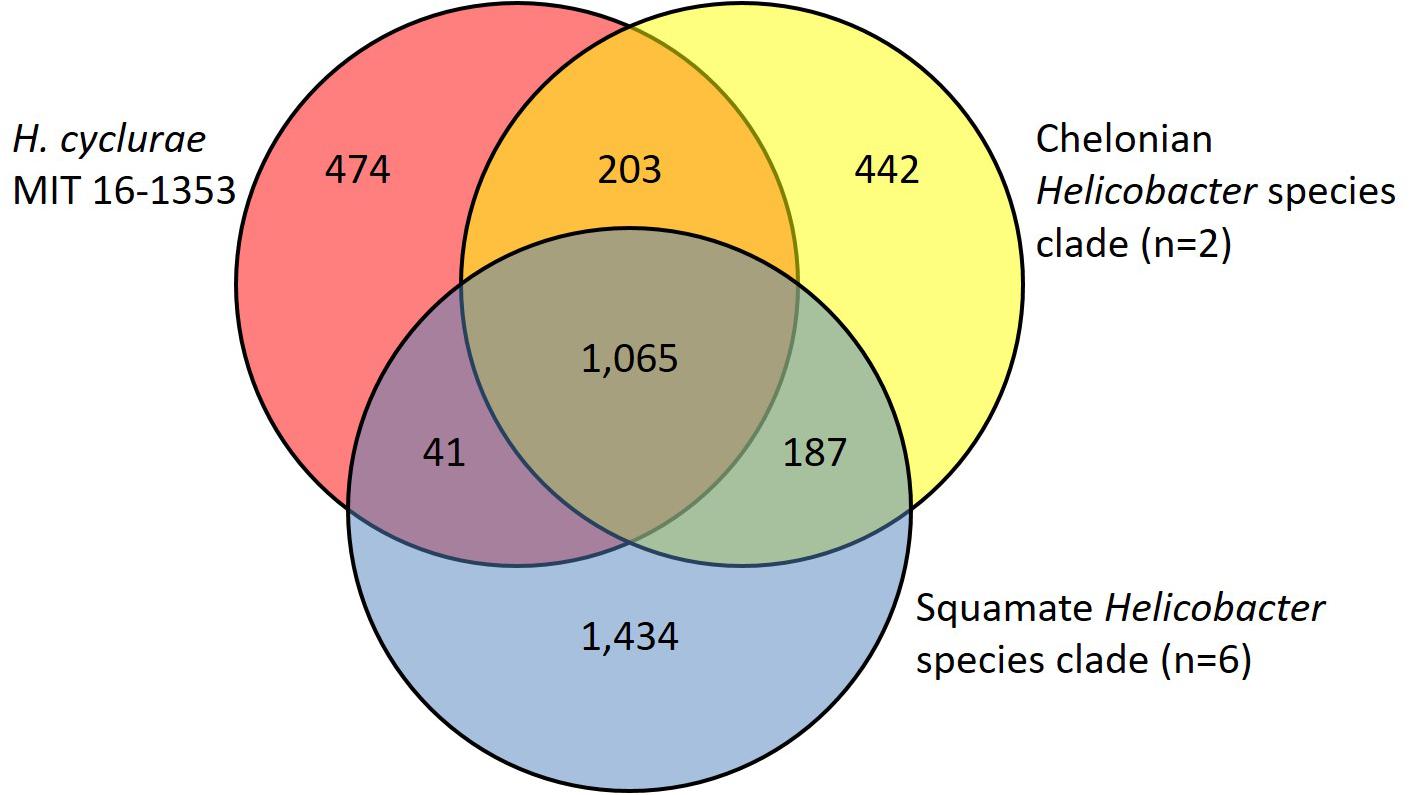
Figure 3. Venn diagram showing shared and unique orthogroups among genomes from Helicobacter species isolated from reptilian hosts.
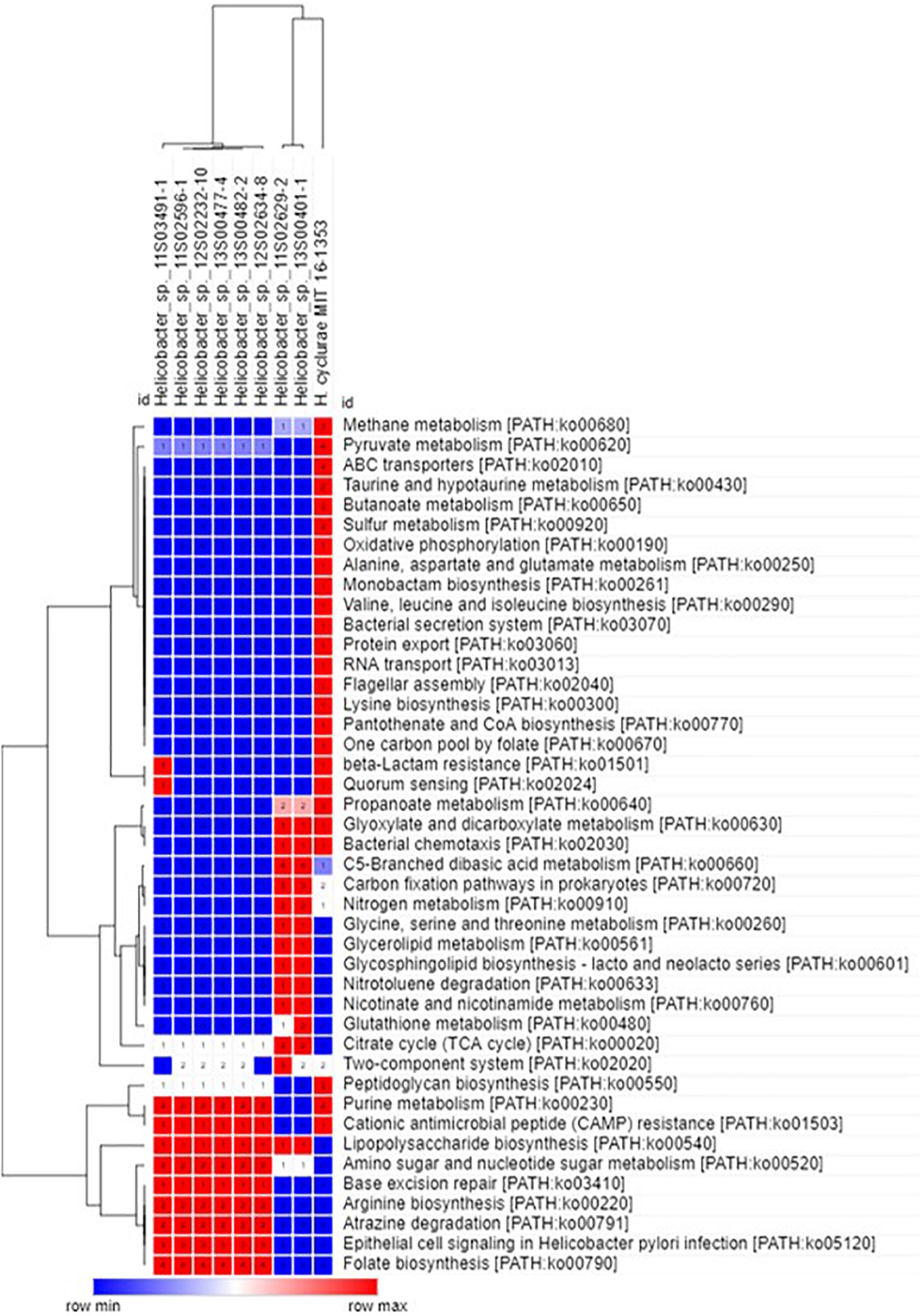
Figure 4. Hierarchal clustering heatmap of unique orthogroups and associated KEGG metabolic/functional pathways from Helicobacter species isolated from reptilian hosts.
Virulence factor profiles were distinct between H. cyclurae and other reptile-associated helicobacters. All helicobacter genomes, including H. cyclurae, encoded high-temperature requirement-A protein-secreted serine protease (htrA), gamma-glutamyl transpeptidase (ggt), and neutrophil activating protein (napA). Homologous sequences to the major antigenic peptide PEB-cell binding factor (Peb4) from Campylobacter jejuni ssp. jejuni NCTC 11168 were present in all reptile-associated helicobacters except strain 13S00401-1 isolated from a chelonian host. Unlike most other reptile-associated helicobacter genomes, H. cyclurae did not harbor a homologous sequence to dupA, hopZ, or campylobacter invasion antigen B (ciaB) gene. Similar to Helicobacter sp. 11S02629-2 (chelonian), H. cyclurae has a homologous sequence to the C. jejuni virulence factor fibronectin-binding protein gene (flpA). Interestingly, both chelonian-associated helicobacters contain the cdtABC operon for cytolethal distending toxin, while H. cyclurae or the helicobacters from squamate hosts did not. H. cyclurae and both chelonian isolates did not have genes for the alpha and beta urease subunits, which were present in the squamates-associated helicobacter genomes. Together, these results support that H. cyclurae has unique genetic, metabolic, and virulence profiles compared to helicobacter isolated from other reptile hosts.
Electron Microscopy
In order to visualize and describe the morphology of H. cyclurae, electron microscopy was performed. H. cyclurae MIT 16-1353 is 2–3 μm in length and 0.3–0.5 μm in width and are curved with a smooth surface and a single sheathed polar flagellum (Figure 5).
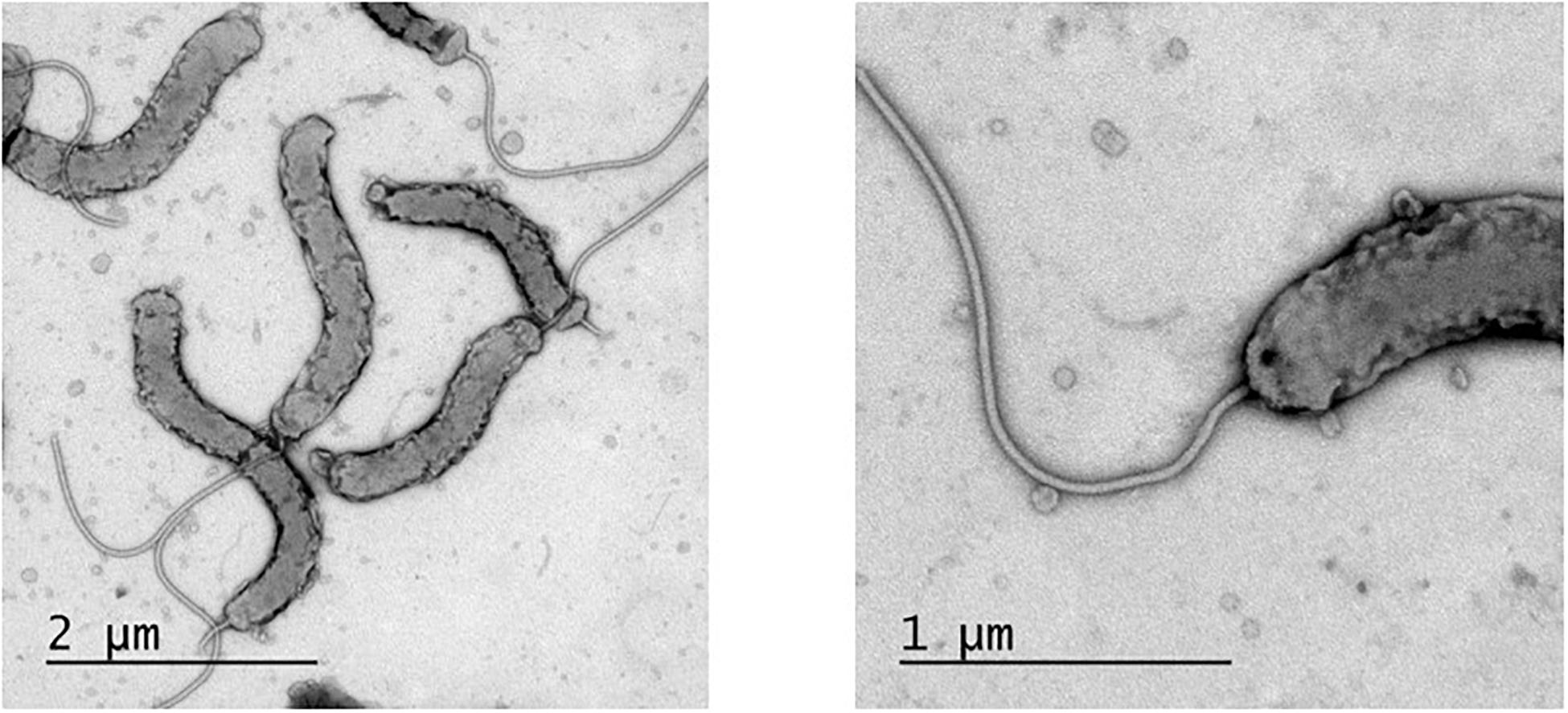
Figure 5. Transmission electron micrograph of Helicobacter cyclurae MIT 16-1353. Measured 2–3 μm in length and 0.3–0.5 μm in width and with a single polar sheathed flagellum.
Discussion
In this study, we isolated the first novel Helicobacter species, H. cyclurae, from fecal samples and cloacal swabs of blue iguanas inhabiting Grand Cayman. The novel helicobacter was characterized by biochemical and whole-genome sequencing; 16S rRNA, gyrB, and hsp60 gene sequencing; and ultrastructural characterization, following the guidelines described by On et al. (2017) pertaining to the minimal standards for describing new Helicobacter spp. (Shen et al., 2020). According to these guidelines, these phenotypic and genotypic characterizations are necessary to effectively and unambiguously distinguish a new taxon from extant species and subspecies and determine phylogeny in the genus. This was particularly important for supporting that H. cyclurae is a novel species, considering helicobacters have been previously isolated from reptilian hosts (Gilbert et al., 2014). A single phenotypic and/or genotypic method is insufficient to make accurate taxonomic determinations, and as demonstrated in our recent publication describing two novel Helicobacter species isolated from opossums, the aggregate of phenotypic and genotypic data is appropriate instead (Shen et al., 2020).
Biochemically and phenotypically, H. cyclurae was compared to other Helicobacter species listed in Table 1. All of the isolates were oxidase and catalase positive, had gamma-glutamyl transpeptidase activity, did not have urease activity nor hydrolyze indoxyl acetate, did not reduce nitrate to nitrite, and did not grow in 1% glycine or at 42°C. Phylogenetic analysis of the 16S RNA gene indicated this novel helicobacter is most closely related to a helicobacter sequenced from a Lacertilia, Helicobacter sp. 12S02256-12 (Gilbert et al., 2014). H. cyclurae is different from Helicobacter sp. GCBI1 identified by PCR in a study of septic blue iguanas, which was most closely related to Helicobacter sp. 11S02629-2 isolated from tortoises (Gilbert et al., 2014).
Interestingly, we observed that the Helicobacter species from chelonians, squamates, and blue iguanas each formed distinct clusters in our phylogenetic analyses; however, the phylogenetic topology differed depending on whether 16S rRNA, hsp60, and gyrB genes were used. This is an example of how a single gene is insufficient to ensure accurate species classification and phylogenetic placement, which has been previously appreciated for causing discordant phylogenies for Helicobacter species (Dewhirst et al., 2005). Therefore, to complement our single-gene phylogenetic analyses, we also performed whole-genome sequence analysis, which is the most robust determination of phylogenetic and taxonomic classification for prokaryotes. Whole-genome phylogenetic analysis of core genes indicated that H. cyclurae was most similar to H. monodelphidis MIT 15-1451 isolated from captive opossums with cloacal prolapse (Shen et al., 2020). Together, these analyses show how the phylogenetic placement of Helicobacter species does not appear to be dependent on the host and that H. cyclurae is phylogenetically divergent from other helicobacters isolated from reptiles.
Conventional DNA–DNA hybridization (DDH) has been used to differentiate known vs. novel bacterial species, including Helicobacter species, based on a threshold of 70% similarity (i.e., species that are ≥ 70% similar are the same species). ANI and dDDH are in silico nucleotide-level similarity indexes analogous to conventional DDH that use whole-genome sequence data instead. ANI and dDDH similarity of ∼95 and ∼70%, respectively, are the equivalent thresholds to conventional DDH. Therefore, ANI and dDDH analyses of H. cyclurae against Helicobacter species, including those from reptiles, were below these ∼95 and ∼70% similarity thresholds, supporting our other phenotypic and genotypic characterizations that H. cyclurae is a novel species.
Similar to H. pylori, this novel helicobacter does have the virulence factors high-temperature requirement-A protein-secreted serine protease (htrA) and gamma-glutamyl transpeptidase (ggt). Additionally, it has a virulence factor fibronectin/fibrinogen binding protein (fbps) that is not present in H. pylori but identified in Helicobacter bilis; this virulence property is capable of providing invasive properties in host cells. Unlike Helicobacter hepaticus, the prototypical EHS H. cyclurae lacks urease and cytolethal distending toxin genes. While morphology alone cannot be used to differentiate helicobacters, electron microscopy allows description of the cell morphology and size, the number and arrangement of flagella, and the presence or absence of flagellar sheaths and periplasmic fibers, all of which can vary substantially between different Helicobacter species. Transmission electron microscopy illustrates that H. cyclurae has a single polar sheathed flagellum similar to Helicobacter cholecystus (Owen, 1998; Whary and Fox, 2004).
The 56% prevalence of H. cyclurae in the blue iguanas surveyed in our study is higher than what has been previously reported in lizards (Gilbert et al., 2014). Also, our culture and PCR results indicated that H. cyclurae was more commonly identified in the feces and cloacal samples from clinical normal animals in our study compared with Helicobacter sp. GCBI1 identified in clinically ill iguanas in blood and tissue samples (Conley et al., 2021). In our previous study, clinically ill blue iguanas who developed lethargy, weakness, inappetence, and septicemia had spiral-shaped bacteria identified on peripheral blood smears (Conley et al., 2021). Through 16S rRNA gene-specific primers, Helicobacter spp. were detected in 11/19 blood or tissue samples of iguanas tested. Whereas PCR utilizing Helicobacter sp. GCBI1-specific primers identified Helicobacter sp. GCBI1 in 7/19 of these animals, no helicobacters were cultured from the blood and tissue samples of these animals (Conley et al., 2021). One of the animals which had Helicobacter sp. GCBI1 identified in its blood also had H. cyclurae isolated from its feces, indicating that more than one Helicobacter species may be present in the same iguana. It is hypothesized that Helicobacter GCBI1 might have originated from an invasive species, the green iguana (Iguana iguana), that is competing for habitat and resources with blue iguanas (Conley et al., 2021). Ongoing screening of green iguanas for these two novel Helicobacter species will be important in elucidating Helicobacter species colonization dynamics in iguanas inhabiting Grand Cayman (Popescu, 2018).
Conclusion
In conclusion, using the criteria for characterizing Helicobacter species (On et al., 2017; Shen et al., 2020), we have described the isolation of a novel helicobacter, H. cyclurae, from blue iguanas, an endangered species from Grand Cayman. To support the recovery effort of the blue iguana population, it is important to identify and understand the mechanisms whereby newly recognized microorganisms contribute to diseases. Further studies are needed to identify if H. cyclurae can be pathogenic under certain circumstances in blue iguanas.
Description of Helicobacter cyclurae sp. Nov.
H. cyclurae sp. nov. (cy.clu’rae. N.L. gen. n. cyclurae of the blue iguana Cyclura). The organism is motile; cells are slightly curved, 2–3 μm long and 0.3–0.5 μm wide, with a monopolar sheathed flagellum. The bacteria are gram negative and non-sporulating. The organism under microaerobic conditions grows slowly at 28 and 37°C, but not at 42°C. It appears on solid agar as single colonies. The bacterium is oxidase, catalase, and gamma-glutamyl transpeptidase positive and has variable alkaline phosphatase activity, but urease, indoxyl acetate hydrolysis, and nitrate reduction are negative. It did not grow on 1% glycine and is resistant to nalidixic acid and cephalothin. The type strain MIT 16-1353T isolated from the feces of a blue iguana in Grand Cayman has been deposited in the Belgian Coordinated Collections of Microorganisms (BCCM) as LMG 31270 and in The National Collection of Type Cultures as NCTC 14190. It has a DNA G+C content of 33.27 mol%, and its genome size is ∼1.91 Mb. The 16S rRNA and the whole-genome sequence of the type strain have been deposited in GenBank under accession numbers MW147609 and NHYM00000000.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, NHYM00000000; https://www.ncbi.nlm.nih.gov/genbank/, MW147609.
Ethics Statement
Ethical review and approval was not required for the animal study because IACUC approval for this project was not required as WCS institutional requirements for IACUC review do not include field projects that take place outside of our facilities. However, sampling, capture, and release of wild reptiles, collecting dead animals found in the environment, or sampling after euthanasia are all standard techniques and procedures for clinical and pathology examinations or investigations. The welfare of animals included in this study was considered throughout their care, with analgesics used as deemed necessary. Animals that survived infection were released back into the environment where they were found, either in the captive setting or free-ranging within the QEIIBP. No animals were housed for continued research purposes. Within the Cayman Islands regulatory framework, the project operates under the terms of a protected species permit issued to the BIRP by the National Conservation Council. Blood, tissue, and fecal samples were collected by or under the direction of licensed veterinarians. Cayman Islands CITES export permits 2014/KY/000674, 2014/KY/000687, 2014/KY/000686, 2014/KY/000690, 2014/KY/000689, 2015/KY/000777, 2015/KY/000808, 2015/KY/000809, 2016/KY/000817, 2017/KY/000874, and 2017/KY/000923; and United States CITES import permits 15US033594/9, 16US0333594/9, and 17US033594/9 were used for sample export and import.
Author Contributions
JF and PC designed and supervised the study. NC, ZS, and SK processed samples for helicobacter isolation and characterization. AM performed and analyzed the whole-genome sequence. IP and FB collected and organized the iguana samples. NC, ZS, AM, and JF analyzed and interpreted the data, and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the NIH grants T32-OD010978, P30-ES002109, and R01-OD011141 (to JF), the Wildlife Conservation Society’s Zoological Health Program, the Cayman Islands Government, the National Trust for the Cayman Islands, and the Derald H Ruttenberg Memorial Fund (to PC). The funders played no role in the study or in the preparation of article or decision to publish.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Aharon Oren of The Hebrew University of Jerusalem for providing nomenclature expertise in naming of these novel Helicobacter species, and we thank Parisa Zarringhalam for assistance with manuscript preparation. We are grateful for the assistance of Veterinary Technicians from the Wildlife Conservation Society and the staff and volunteers of the Blue Iguana Recovery Program and Cayman Islands Department of the Environment and the St. Matthew’s University School of Veterinary Medicine for laboratory support.
References
Conley, K. J., Seimon, T. A., Popescu, I. S., Wellehan, J. F. X. Jr., Fox, J. G., Shen, Z., et al. (2021). Systemic Helicobacter infection and associated mortalities in endangered Grand Cayman blue iguanas (Cyclura lewisi) and introduced green Iguanas (Iguana iguana). PLoS One 16:e0247010. doi: 10.1371/journal.pone.0247010
Dewhirst, F. E., Shen, Z., Scimeca, M. S., Stokes, L. N., Boumenna, T., Chen, T., et al. (2005). Discordant 16S and 23S rRNA gene phylogenies for the genus Helicobacter: implications for phylogenetic inference and systematics. J. Bacteriol. 187, 6106–6118. doi: 10.1128/JB.187.17.6106-6118.2005
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. doi: 10.1186/s13059-019-1832-y
Fox, J. G. (2002). The non-H pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 50, 273–283. doi: 10.1136/gut.50.2.273
Fox, J. G., Dewhirst, F. E., Shen, Z., Feng, Y., Taylor, N. S., Paster, B. J., et al. (1998). Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114, 755–763. doi: 10.1016/s0016-5085(98)70589-x
Fox, J. G., Taylor, N. S., Howe, S., Tidd, M., Xu, S., Paster, B. J., et al. (2006). Helicobacter anseris sp. nov. and Helicobacter brantae sp. nov., isolated from feces of resident Canada geese in the greater Boston area. Appl. Environ. Microbiol. 72, 4633–4637. doi: 10.1128/AEM.02876-05
Gilbert, M. J., Duim, B., Timmerman, A. J., Zomer, A. L., and Wagenaar, J. A. (2017). Whole genome-based phylogeny of reptile-associated Helicobacter indicates independent niche adaptation followed by diversification in a poikilothermic host. Sci. Rep. 7:8387. doi: 10.1038/s41598-017-09091-7
Gilbert, M. J., Kik, M., Timmerman, A. J., Severs, T. T., Kusters, J. G., Duim, B., et al. (2014). Occurrence, diversity, and host association of intestinal Campylobacter, Arcobacter, and Helicobacter in reptiles. PLoS One 9:e101599. doi: 10.1371/journal.pone.0101599
Goodman, R., and Burton, F. (2005). Natural history notes:Cyclura lewisi (Grand Cayman Blue Iguana): hatchlings. Herpetol. Rev. 36:176.
Hannula, M., and Hanninen, M. L. (2007). Phylogenetic analysis of Helicobacter species based on partial gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 57(Pt 3), 444–449. doi: 10.1099/ijs.0.64462-0
Inglis, G. D., McConville, M., and de Jong, A. (2006). Atypical Helicobacter canadensis strains associated with swine. Appl. Environ. Microbiol. 72, 4464–4471. doi: 10.1128/AEM.02843-05
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Goker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Mikkonen, T. P., Karenlampi, R. I., and Hanninen, M. L. (2004). Phylogenetic analysis of gastric and enterohepatic Helicobacter species based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 54(Pt 3), 753–758. doi: 10.1099/ijs.0.02839-0
On, S. L. W., Miller, W. G., Houf, K., Fox, J. G., and Vandamme, P. (2017). Minimal standards for describing new species belonging to the families Campylobacteraceae and Helicobacteraceae: Campylobacter, Arcobacter, Helicobacter and Wolinella spp. Int. J. Syst. Evol. Microbiol. 67, 5296–5311. doi: 10.1099/ijsem.0.002255
Owen, R. J. (1998). Helicobacter–species classification and identification. Br. Med. Bull. 54, 17–30. doi: 10.1093/oxfordjournals.bmb.a011667
Popescu, I. (2018). Investigations into Green Iguana (Iguana iguana) as a Potential Carrier for a Novel Strain of Helicobacter spp, Pathogenic for the Endangered Blue Iguana (Cyclura lewisi). Edinburgh: The University of Edinburgh.
Pritchard, L., Glover, R. H., Humphris, S., Elphinstoneb, J. G., and Toth, I. K. (2016). Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8, 12–24. doi: 10.1039/c5ay02550h
Shen, Z., Batac, F., Mannion, A., Miller, M. A., Bakthavatchalu, V., Ho, C., et al. (2017). Novel urease-negative Helicobacter sp. ‘H. enhydrae sp. nov.’ isolated from inflamed gastric tissue of southern sea otters. Dis. Aquat. Organ. 123, 1–11. doi: 10.3354/dao03082
Shen, Z., Mannion, A., Lin, M., Esmail, M., Bakthavatchalu, V., Yang, S., et al. (2020). Helicobacter monodelphidis sp. nov. and Helicobacter didelphidarum sp. nov., isolated from a colony of gray short-tailed opossums (Monodelphis domestica) with endemic cloacal prolapses. Int. J. Syst. Evol. Microbiol. 70, 6032–6043. doi: 10.1099/ijsem.0.004424
Shen, Z., Xu, S., Dewhirst, F. E., Paster, B. J., Pena, J. A., Modlin, I. M., et al. (2005). A novel enterohepatic Helicobacter species ‘Helicobacter mastomyrinus’ isolated from the liver and intestine of rodents. Helicobacter 10, 59–70. doi: 10.1111/j.1523-5378.2005.00292.x
Stacy, B. A., and Wellehan, J. F. Jr. (2010). Fatal septicemia caused by Helicobacter infection in a pancake tortoise (Malacochersus tornieri). J. Vet. Diagn. Invest. 22, 660–662. doi: 10.1177/104063871002200431
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017). Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 45, D535–D542. doi: 10.1093/nar/gkw1017
Keywords: Helicobacter, Cyclura lewisi, blue iguana, novel species, Grand Cayman, endangered, reptiles
Citation: Chan N, Shen Z, Mannion A, Kurnick S, Popescu IS, Burton FJ, Calle PP and Fox JG (2021) Helicobacter cyclurae sp. Nov., Isolated From Endangered Blue Iguanas (Cyclura lewisi). Front. Ecol. Evol. 9:676682. doi: 10.3389/fevo.2021.676682
Received: 08 March 2021; Accepted: 05 August 2021;
Published: 14 September 2021.
Edited by:
Enrique P. Lessa, Universidad de la República, UruguayReviewed by:
Gregorio Iraola, Institut Pasteur de Montevideo, UruguayRodney L. Honeycutt, Pepperdine University, United States
Copyright © 2021 Chan, Shen, Mannion, Kurnick, Popescu, Burton, Calle and Fox. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James G. Fox, amdmb3hAbWl0LmVkdQ==
 Nathan Chan
Nathan Chan Zeli Shen1
Zeli Shen1 Anthony Mannion
Anthony Mannion Ioana S. Popescu
Ioana S. Popescu Paul P. Calle
Paul P. Calle James G. Fox
James G. Fox