- 1Department of Biological Sciences, Macquarie University, Sydney, NSW, Australia
- 2Centre for Superbug Solutions, Institute for Molecular Bioscience, The University of Queensland, St. Lucia, QLD, Australia
- 3Department of Physiology, Anatomy and Microbiology, La Trobe University, Melbourne, VIC, Australia
The global rise of antimicrobial resistance (AMR) phenotypes is an exemplar for rapid evolutionary response. Resistance arises as a consequence of humanity’s widespread and largely indiscriminate use of antimicrobial compounds. However, some features of this crisis remain perplexing. The remarkably widespread and rapid rise of diverse, novel and effective resistance phenotypes is in stark contrast to the apparent paucity of antimicrobial producers in the global microbiota. From the viewpoint of evolutionary theory, it should be possible to use selection coefficients to examine these phenomena. In this work we introduce an elaboration on the selection coefficient s termed selective efficiency, considering the genetic, metabolic, ecological and evolutionary impacts that accompany selective phenotypes. We then demonstrate the utility of the selective efficiency concept using AMR and antimicrobial production phenotypes as ‘worked examples’ of the concept. In accomplishing this objective, we also put forward cogent hypotheses to explain currently puzzling aspects of the AMR crisis. Finally, we extend the selective efficiency concept into a consideration of the ongoing management of the AMR crisis.
Introduction
The rise of antimicrobial resistance (AMR) is an acknowledged medical crisis for humanity (Michael et al., 2014; World Health Organization (WHO), 2014; Contained and Controlled, The UK’s 20-Year Vision for Antimicrobial Resistance, 2019). Increasing resistance to antimicrobials in general, and antibiotics in particular makes some diseases untreatable. The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) are of particular concern but a diverse range of other microorganisms such as M. tuberculosis and Candida auris have also acquired increasingly comprehensive suites of resistance determinants. The scope of this problem will continue to expand into the future.
A resistance phenotype can be inherited from progenitors, generated within a cell through mutation, or alternatively can be conferred by capture of genes from external sources in one of the various mechanisms of horizontal gene transfer (HGT). Many of the horizontally acquired genes that confer resistance are ancient (Recchia and Hall, 1997; Perron et al., 2015). These mobile resistance genes are part of the vast library of genes capable of conferring adaptive phenotypes, and form the floating genome shared amongst the microbiota (Shapiro, 2010; Aminov, 2011; Ghaly et al., 2019). These genes can move between individuals of the same species and between the cells of different species. The antimicrobial resistance genes acquired by human pathogens represent a tiny fraction of the floating genome (Shapiro, 2010; Aminov, 2011).
Resistance genes are ancient, and most likely arose in response to the production of equally ancient antimicrobials by other organisms (Aminov, 2009). In their native context, it is thought that antimicrobials were used briefly, and at low concentrations, possibly as a signaling mechanism to surrounding organisms (Rutherford and Bassler, 2012). The majority of the antimicrobials and antibiotics used by humans are ultimately derived from such natural products, with penicillin, vancomycin, streptomycin, tetracycline, chloroquine, and artemisinin being but a few examples. In contrast to the low concentrations and short-term diffusion of these compounds in their original context, the widespread, intense and persistent application of such materials has provoked an evolutionary response in the global proliferation and diversification of resistance phenotypes (Michael et al., 2014; Contained and Controlled, The UK’s 20-Year Vision for Antimicrobial Resistance, 2019).
These evolutionary responses are mirrored by the evolution of resistance to herbicides, insecticides, rodenticides, filaricides and other materials designed to kill or control other organisms. Clearly, while the antimicrobial resistance crisis is the particular focus of this work, in a wider sense, human attempts to control other organisms has sweeping and potentially long-lasting impacts on global ecology and on the passage of evolution. Understanding the dynamics of such evolutionary responses is key to managing the various crises in resistance.
The molecular mechanisms by which the AMR crisis has been generated are increasingly well understood. Novel phenotypes are generated through mutation and by acquisition of novel genes via the process of horizontal gene transfer. This dissemination within and between classes of microbe through the processes of both vertical and horizontal gene transfer continues to be an area of fruitful study (Gillings, 2014). These novel phenotypes then proliferate through the application of an applied selective pressure such as antimicrobials. However, the wider dynamics of the processes whereby AMR phenotypes both rapidly diversify and proliferate in microbial communities and across the globe so creating the current crisis, are less well understood. In particular, there are a number of outstanding questions that have led to the perspectives outlined in this paper. These are:
(i) Resistance genes, including recently arisen variants, appear to be present and persistent across the globe, rather than just being localized in areas where their cognate antimicrobial is used by humans.
(ii) In contrast, antimicrobial production phenotypes, at least those that produce antimicrobials that are lethal to one or more pathogens and yet do not unduly injure a human patient and so are considered to be therapeutically useful, appear to be rare (Silver, 2011; Ribeiro da Cunha et al., 2019). Indeed, the current therapeutic armamentarium is limited to approximately 12 main and some minor chemical skeletons derived from even fewer biological sources, despite decades of intense searching.
(iii) As the AMR crisis has progressed, the time taken for resistance phenotypes to appear in clinical settings is decreasing (Fairhurst and Dondorp, 2009; Das et al., 2018).
It is widely acknowledged that the AMR crisis is, at its heart, an evolutionary and ecological problem. Understanding and responding to this crisis will require an evolutionary and ecological perspective that has currently been lacking. Darwin’s apparently simple insight concerned the “survival of the fittest through the possession of advantageous heritable characters” and is central to modern evolutionary theory (Darwin, 1909). Natural selection posits that the possession of gene variants that generate phenotypes that confer a selective advantage in terms of improved reproduction. Over time, this results in those genes and the organisms that carry them becoming more common in populations. Exactly how successful such genes and their resulting phenotypes are in providing a selective advantage is measured by the Selection coefficient s. Typically, s is measured as the relative abundance of a phenotype bearing strain, as compared with a wild type strain over time (Thurman and Barrett, 2016). Such measurements have been taken of a wide range of organisms in many different environments over the years since the concept of s was initially put forward (Lande and Arnold, 1983). However, in the measurement of s, much variability has been seen (Dykhuizen, 2016). As the parameter, s has been primarily used to retrospectively assess differences between the “fitness” of various phenotypes in comparison to a “wildtype” reference in an experimental setting, it has limited ability to prospectively assess the ongoing ecological/evolutionary outcome of a particular phenotype in the environment. In order to extend the utility of this measurement we have arrived at an elaboration of the concept of the Selection coefficient. This elaboration, termed “Selective efficiency,” considers various factors contributing to or detracting from the “fitness” provided by a phenotype, and their changing interactions over time. Selective efficiency therefore provides an indicator, with which s might be determined prognostically.
The four additional parameters contributing to s, may be addressed separately, so allowing both the individual and overall examination of factors influencing fitness and so forming a cost-benefit comparison of the production of a phenotype. That is, we considered that the benefit to an organism from the presence of a selective phenotype in its increased survival and reproduction, is offset by the costs to the organism of providing that phenotype. These selective costs are subdivided into genetic, metabolic, ecological and evolutionary factors. Because both the advantages and costs of a phenotype may vary according to the context in which it acts, the calculation of selective efficiency is summed over a period of time. In approximating s, the summed selective efficiency of a strain demonstrating a particular phenotype is compared to the growth of a comparable wild type strain grown under equivalent conditions over the same number of generations to give the expanded estimate of s. In order to illustrate these concepts concisely Figure 1 shows the development of the selective efficiency concept and its factors and their relationship to the selection coefficients.
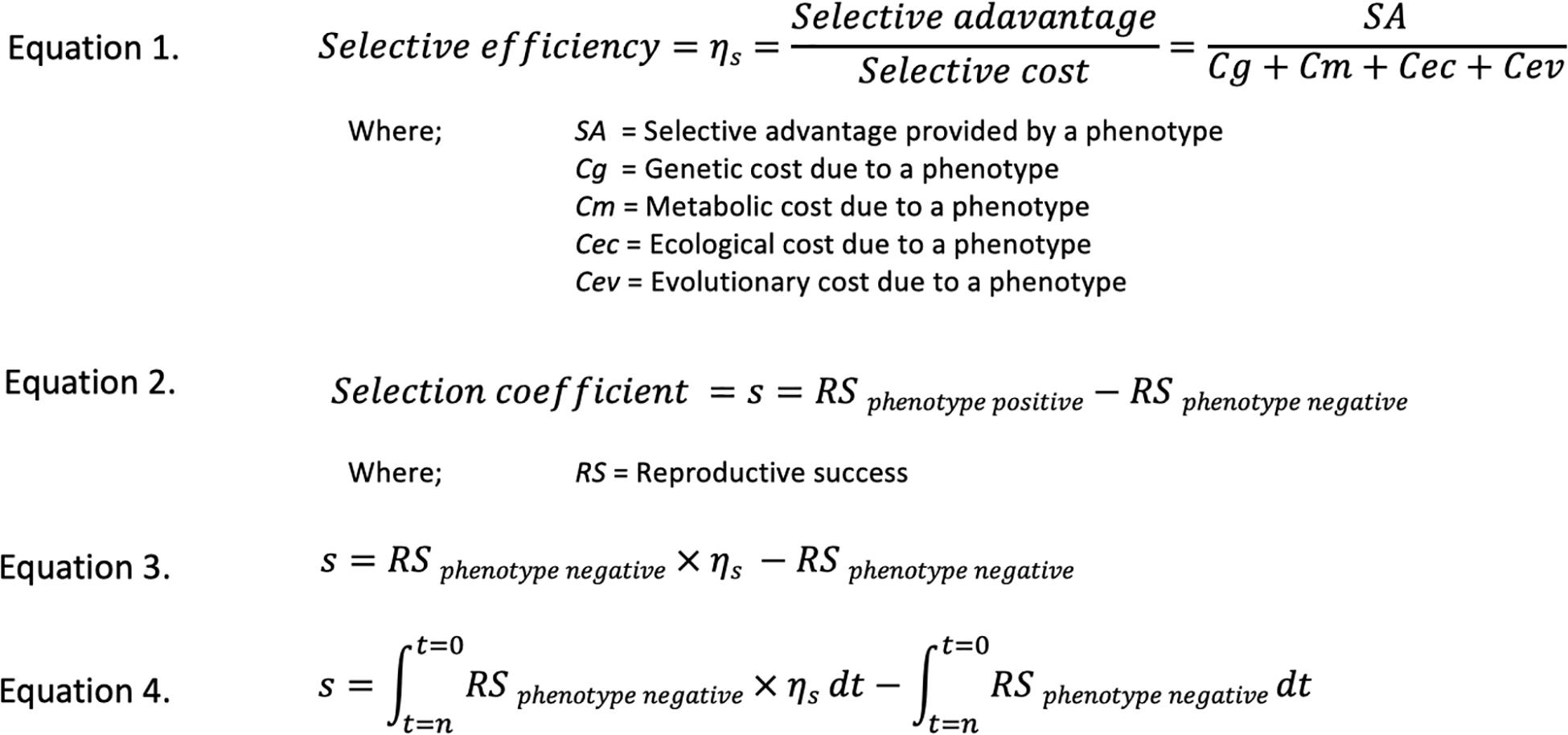
Figure 1. An explanation of the Selective efficiency concept and how it relates to the selection coefficient s using hypothetical equations. Selective efficiency provides a benefit/cost assessment of a selective phenotype and so expands the utility of the selective coefficient s as shown in these equations. Equation 1 shows the components of selective efficiency. Equation 2 shows the components of the selection coefficient s. Equation 3 shows the amalgamation of selective efficiency as represented by Equation 1, into Equation 2. Equation 4 shows the integration of Equation 3 with respect to time, from time 0 to time n, to account for possible time variant changes in selective efficiency. While not used in this work, it should be pointed out that the various factors involved in the selective efficiency concept may be determined experimentally. In essence, this may be accomplished in a microbial model, consisting of two mutants, one with the genes responsible for the phenotype in question removed and another with those genes still present but prevented from expression. These mutants are grown in parallel with both phenotype expressing and wild type organisms in the presence and absence of stressors to which the phenotype is relevant. The relevant values may then be determined by comparing the growth rates of various strains.
Using this concept as a metaphorical lens, we consider the considerable body of information published regarding both AMR phenotypes and Antimicrobial production both as phenotype and stressor imposed by humanity.
Antimicrobial Resistance
Antimicrobial resistance as a class of phenotype generally appears to provide an advantage while exacting a minimal toll upon the host organism and its environment. Perhaps the most elegant example is seen in Mycobacterium tuberculosis (Mtb) as well as in other similarly efficient micro-organisms (Szumowski et al., 2012; da Silva et al., 2016). Amongst other responses, these organisms up-regulate existing biochemical pathways (membrane efflux pumps) to export an antimicrobial agent that has entered the cell, so reducing or eliminating the threat (da Silva et al., 2016). Conferring up-regulation of existing membrane efflux pumps does not generally require the presence of additional genes in the host genome, but can be the result of small changes in the mechanisms controlling the expression of existing genes. Consequently, the genetic burden of such a phenotype is minimal. Similarly, the biochemical cost of this phenotype may be very low if the only cost is the additional energy required to increase the actuation of existing efflux pumps. Should the phenotype be provided through the provision of additional membrane efflux pumps in the cellular membrane, then an additional metabolic cost in the expression of the required genes will be incurred. Alternatively, should the phenotype be both conditional to the presence of an antimicrobial and reversible in its absence, then the metabolic costs of this form of antimicrobial resistance are negligible when the host is not experiencing an antimicrobial induced selective pressure.
Because the antimicrobial agent is merely exported from the cell and not necessarily changed by its passage, there is no net change on the surrounding environment due to the action of this phenotype. So, while the introduction of an antimicrobial may have large scale and lasting effects on the surrounding ecology, the response by Mtb imposes only a minimal burden on the cell and has no wider impact on other organisms. So, within the framework of selective efficiency, this is a very efficient phenotype. Because it provides significant advantage, at minimal “cost,” it is more likely to be maintained within host lineages for prolonged periods even in the absence of a stressor.
While the above example shows an extreme case of selective efficiency, other means of conferring AMR are similarly efficient. Many, such as the bla family of genes that code for variants of the beta lactamase enzyme and provide resistance to penicillin and other related antibiotics do so through the expression of just this one gene (Ur Rahman et al., 2018). While the burden of replicating and expressing this gene may be higher than that of an up-regulated efflux pump deleterious interference with other metabolic processes appears to be absent. Because beta lactamase is an enzymatic protein and so may not remain functional for extended periods of time and is also not exported, it does not materially alter the surrounding environment should it exit the cell. Hence, the presence of the bla gene does not have a direct effect upon the surrounding ecology beyond the preferential survival of organisms containing this gene in the presence of an antimicrobial stressor.
Many other types of antimicrobial resistance are also provided by single genes (Gillings, 2014). Interestingly, the small genetic constitution of such vectors of antimicrobial resistance contribute to their mobility both within and between organisms. Many AMR genes exist as mobile gene cassettes associated with HGT mechanisms such as integrons, plasmids and transposons and so may be readily mobilized within and between different types of microbe. However, some few examples of larger multi-gene operons also exist, such as vanA, the 7 gene structure providing one example of resistance to vancomycin (Ligozzi et al., 1998; Williams and Bardsley, 1999). The added necessity of maintaining not only the integrity of these multiple gene resistance operons, but also their spatial relationship upon a piece of horizontally transferred DNA may be one factor in the slower dissemination of this form of resistance. Additionally, the burden of expressing multiple additional enzymes and creating the finished altered peptidoglycan structure, may impose a higher fitness cost when expressed. However, conditional expression has been shown to reduce the fitness cost of vanB vancomycin resistance in enterococci (Foucault et al., 2010). Similarly, conditional expression of the vanA operon in VRE, may help to explain the currently increasing prevalence of VRE (Ramadhan and Hegedus, 2005).
In short, AMR phenotypes are typically selectively efficient. This efficiency may explain both the persistence, widespread dissemination and long-term maintenance of AMR phenotypes within the microbial world.
Additionally, it has been seen that AMR phenotypes are often provided by the various processes of HGT and many AMR phenotypes are often associated with class one integrons (Jackson et al., 2011). While integrons serve to mobilize selective gene cassettes they themselves are not intrinsically mobile. However, class one integrons have been noted as often being enclosed within mobilizing structures such as transposons (Tn21) which in turn may be carried upon mobile plasmids (Gillings, 2014). These nested mechanisms allow not only AMR genes carried as integron associated gene cassettes to be mobilized individually but also can transmit the mobilizing mechanisms carrying them as well. So, where a selective pressure acts to increase the abundance of AMR genes the same selective pressure will also increase the abundance of the associated mobilizing mechanisms within the community. This means, that with the resulting wider availability of HGT mechanisms within a microbial community the time taken to disseminate novel selective responses to new and existing antimicrobials will be reduced. This may well be a contributing factor in the observation that AMR seems to arise ever more quickly as both new and existing antimicrobials are introduced clinically.
Antimicrobial Production
Antimicrobials are molecules that adversely influence the survival of microbes. At one extreme, toxic substances such as hydrogen peroxide can cause extreme oxidative stress that can be lethal to virtually all exposed organisms and along with other antiseptics is a commonly used antimicrobial in clinical practice. Antibiotics are a more finely tuned group of antimicrobials that affect only specific groups of microbes and are generally not detrimental to humans although side effects such as nephrotoxicity can be dose-limiting. Most of the twelve major and various additional minor classes of antibiotic currently known originate in products produced in living organisms, presumably as a phenotype to gain selective advantage within their environment (Aminov, 2009). A good example of such an advantage is the production of penicillin by Penicillium spp. mold which is typically secreted from the cell and produces an “exclusion zone” around the organism. Observation of this competitor-free zone on agar media resulted in the discovery of penicillin by Fleming and arguably heralded the beginning of the antibiotic era in human medicine (Fleming, 1945).
However, in the microbial world, antibiotics such as these that are suitable for therapeutic use, exist at the lethal end of a spectrum of effect. This spectrum includes a variety of selective mechanisms whereby an organism can signal rather than lethally influence surrounding organisms and still ultimately derive an advantage. One school of thought suggests that the native function of native antimicrobials in general and antibiotics in particular is as an intercellular signaling mechanism akin to quorum sensing (Rutherford and Bassler, 2012). In this context, an antibiotic used in occasional non-lethal doses would selectively signal only susceptible organisms. This would limit the impact on the wider ecology of the use of an antibiotic while decreasing the cost to the producing cell. In this context, we now consider the selective efficiency of antibiotics considered to be useful therapeutically.
Genetic Costs
The majority of antibiotics currently used for therapeutic purposes are not protein molecules or protein derivatives. Consequently, they require a specific metabolic pathway for their production rather than a more direct protein production mechanism. Even the nearest protein analogs; the cyclic peptide derivatives daptomycin, polymyxins B and E, and vancomycin, are assembled on non-ribosomal gene clusters and require modified amino acid components (Williams and Bardsley, 1999). These additional pathways, whether they be derived from an existing mechanism or a completely novel addition to the genome, require the maintenance of additional genes and their appropriate expression mechanisms and so add a genetic load to the organism.
Additionally, the production of lethal materials within the cell presupposes the presence of either an intrinsic or acquired resistance to the produced antibiotic. This inherent resistance may involve yet further genes along with their attendant expression mechanisms. This multiplicity of genes required for a cell to survivably produce antibiotics and the genetic load it entails is in distinct contrast to the often elegantly simple single gene resistance mechanisms seen in the microbial world. It may also be conjectured given the ubiquitous nature of horizontal gene transfer, that the resistance mechanism inherent in the antibiotic producing cell, may itself form the basis for widespread resistance when spread by HGT to potential victims. If this were to be the case, then any long-term benefit to a strain provided by the intense production of antibiotics is inevitably limited by the eventual transmission of its own inherent immunity. This limitation is additional to the rise of other forms of resistance through mutation or pre-existing resistance genes in the surrounding ecology (Mazel, 2004).
Metabolic Costs
The production of antibiotics is a significant impost on the host cell. Because antibiotics of the type currently seen as therapeutically useful are typically non-protein, their production requires not only enzyme catalysis, but also additional raw materials and energy for their production. Since antibiotics are typically exported from the cell, the requirement for raw materials, their import, processing and subsequent export requires a significantly more cellular resources as compared to relatively passive resistance mechanisms. Additionally, the necessity of providing innate immunity may well add to the biochemical cost of antibiotic production. To minimize the selective pressure imposed by this larger use of resources, the production of an antibiotic that is lethal in very low concentrations and or else is produced only under specific circumstances may be favored. However, the production of such extravagantly lethal compounds may have a proportionately greater ecological effect when exported from the cell.
Ecological and Evolutionary Costs
The secretion of an antibiotic into the surrounding environment has a variety of ecological impacts. In the short term the presence of a lethal antibiotic kills or otherwise inhibits the growth of surrounding susceptible organisms. This reduces the number of organisms competing for available resources and also provides additional resource materials from killed cells, so benefiting the antibiotic producer as well as other fortuitously resistant organisms that may be present. But, in so doing, the application of an antibiotic results in an immediate increase in the proportion of organisms resistant to that antibiotic in the local environment, so producing a sudden shift in local community structure. These remaining organisms will be the original antibiotic producer and other organisms expressing innate or acquired resistance. This sudden shift in community structure may have ecological effects beyond an increase in local antibiotic resistance phenotypes. One possibility is that the marked reduction in community diversity may have wider impacts on local ecological resilience and hence the ability to recover from sudden perturbations caused by environmental challenges beyond the application of antibiotics (Peterson et al., 1998; Gunderson et al., 2012).
The impact of an antibiotic on the community in which it is present is also related to its toxicity. Secreted antibiotics create a concentration gradient centered on the producing organism as they diffuse into the environment around the producing cells. Close to the source, an antibiotic may be lethal to all except the producing organism. At increasing ranges and at consequently lower concentrations, the antibiotic may not be immediately lethal to other organisms. Such organisms, while stressed, may yet survive to pass on their ability to cope with the applied antibiotic. Because of the increased availability of resources closer to the source of the antibiotic as well as the reduction in the number and types of competing organisms present, selection for increasing resistance to the applied antibiotic will be exacerbated.
Where an antibiotic is more lethal at a given concentration or else is produced in higher concentrations, the number of organisms at the periphery of a survivable range is increased quadratically. The larger the number of organisms that can tolerate an applied antibiotic at any one time, the more opportunity there is for the random processes of mutation and or HGT to introduce or otherwise hone resistance phenotypes. Similarly, production of a “broad-spectrum” antibiotic that affects a larger proportion of the surrounding microbiota and will necessarily produce a faster evolutionary response from the surrounding ecology than a more specifically targeted antibiotic. Further, because of microbial movement in the environment, the longer an antibiotic is applied at a given concentration, the more likely it is for increasingly diverse populations to be exposed at survivable concentrations and hence develop increasingly efficient and diverse resistance phenotypes.
The focused selective pressure exerted by the application of antibiotics results in the evolution and dissemination of increasingly efficient resistance phenotypes in the local ecology. Further, it has been demonstrated that various mechanisms that promote selective responses such as the generalized SOS response and upregulation of various HGT processes are seen at antibiotic concentrations well below “minimum inhibitory concentration” (MIC) (Gullberg et al., 2011; Chow et al., 2015; Mira et al., 2015). Consequently, not only is it likely that the incidence of existing and novel phenotypes responding to the applied challenge will be increased, but it is also likely that other complementary mechanisms such as the ability to horizontally transfer genes will also be selected for. The increased availability of resistance phenotypes combined with the increased incidence of HGT capable phenotypes in the local community may result in a faster community response to further challenges with existing or novel antibiotics (Smalla et al., 2018).
In short, antibiotic production places a load on the producing cell. Additionally, the ecological and evolutionary consequences of antimicrobial production when used in lethal concentrations results in a selective advantage that is necessarily short lived for the clonal line producing it. Clearly the selective efficiency of such a phenotype, while beneficial in the short term, is ultimately very low. Low selective efficiency, particularly where a short-term gain is outweighed by long term consequences, is likely to impact the diversity and abundance of such phenotypes ultimately producing only a few different examples, even though they may be widely distributed in the global biota, such as the variations of streptomycin which appear to be virtually ubiquitous (Aminov, 2009).
The search for antibiotics deemed useful therapeutically, despite intense global effort has produced only a few distinct chemical backbones over the last 70 years. The exponentially decreasing number of novel therapeutic antibiotics is of great concern to the currently exponentially increasing human population. This apparent paucity of particularly lethal antibiotics may well reflect the selective inefficiency of such phenotypes as discussed above.
The current therapeutic antibiotic armamentarium is currently limited, despite our skills at modifying existing antibiotics to increase their lethality, durability and spectrum of targets. The search for new antibiotics thus far has been both geographically widespread and prolonged and so qualifies as a representative sampling of the global biota. Consideration of this historical data suggests that it is unlikely that multitudes of new classes of antibiotics with characteristics currently deemed therapeutically useful remain to be discovered in the global biota. Nevertheless, it should also be recognized that the search has not yet been exhaustive and that environments, niches and their contributing organisms remain to be examined. Indeed, that large fraction of the microbiota invisible to conventional culture-based techniques and only recently accessible through DNA based techniques, yet offers a significant field of exploration. So, it seems likely that novel natural antibiotics still remain to be discovered and the field of completely synthetic antimicrobial development remains available. However, given the rapid and concerted rise of resistance to existing antibiotics, measures to not only to develop but also preserve the effectiveness of novel antibiotics are of great importance.
Discussion
The elaboration on the selection coefficient provides a framework within which working hypotheses may be constructed. These hypotheses, as discussed in the foregoing section adequately explain some of the observed phenomena associated with the AMR crisis. While in no way being rigorous proofs, these hypotheses are sufficiently robust that their extension to wider aspects of the AMR crisis can be considered:
Antimicrobial Resistance Phenotypes Arise Quickly and Are Persistent
It is a small conceptual leap to suppose that selectively efficient phenotypes are comparatively slowly lost from the community metagenome in the absence of a stressor and proliferate and disseminate rapidly in its presence. Where the phenotype is provided by a single gene, is expressed conditionally, and does not have detrimental epistatic or pleiotropic effects in its expression, the cost to the organism for its carriage is minor. When such genes are mobile and so are dispersed amongst individuals in a microbial community, the cost of their maintenance in the wider metagenome against even a rare need is trivial. Antibiotic resistance in particular and antimicrobial resistance more widely appears to satisfy these criteria. Integron associated resistance gene cassettes are long established in the global microbiota as a source of diversity (Recchia and Hall, 1997; Perron et al., 2015). Such mobile genes further diversify and proliferate in the presence of a stressor (Gillings, 2014). Further, integron associated gene cassettes are both conditionally expressed and increasingly mobilized in the presence of a stressor (Michael and Labbate, 2010; Hocquet et al., 2012; Smalla et al., 2018). Consequently, both variant and completely novel resistance phenotypes have been observed to arise rapidly in response to the use of both derivatised and new antimicrobials (Guerin et al., 2009; Cambray et al., 2011). Indeed, the penicillin resistance conferring bla gene has currently diversified into a clade of over a hundred similar genes (Ur Rahman et al., 2018). Such diversification is a further indicator of not only the strength of the selective pressure applied, but also the efficiency with which such single gene responses can be generated.
The observed anthropic outcome of such selectively efficient characteristics has been the increasingly rapid origin and global dissemination of increasingly diverse antimicrobial resistance phenotypes. Given such a flexible and efficient system, it is likely that antimicrobial resistance is and will continue to be a permanent feature of health management (Michael et al., 2014). So, rather than seeking to eradicate resistance as a phenomenon, or perhaps more reasonably just trying to circumvent individual antimicrobial resistance phenotypes, the development of management strategies to both slow its rise and limit its incidence both locally and globally is additionally indicated (Michael et al., 2014). These measures may include but are not limited to global antimicrobial stewardship to utilize existing antimicrobials appropriately, monitoring of resistance phenotypes proactively and also the development and application of particular antimicrobials that may not provoke a rapid rise of resistance (Guerin et al., 2009; Baquero et al., 2011; Cambray et al., 2011; Michael et al., 2014).
Antimicrobials Suitable for Therapeutic Use Appear to Be Rare
The search for naturally occurring antimicrobials to be used therapeutically has been intensely pursued for nearly a centuary and has encompassed hundreds of thousands of samples taken from virtually every environmental niche on the planet (Ribeiro da Cunha et al., 2019). The search attempts to isolate compounds that will either inhibit or kill a broad range of pathogenic organisms, while administered at low concentrations, all while minimizing the detrimental “off-target” impact on the patient as much as possible (Ribeiro da Cunha et al., 2019). This search has uncovered few types of suitable antibiotic amongst the wider group of antimicrobials and even fewer specifically anti-filarial, anti-malarial, and anti-fungal compounds. The observation that only a few qualifying antimicrobials have been found despite an intensive search and that the same antimicrobials are repeatedly found suggests that the overall diversity of such comparatively lethal compounds is low (Silver, 2011; Cavera et al., 2015; Ribeiro da Cunha et al., 2019; Roope et al., 2019).
Further, antimicrobials for therapeutic use are often chemically modified during their development for therapeutic applications. This derivatization, quite apart from producing multiple patentable products from a single antimicrobial backbone, produces compounds that are better tolerated and more available to human patients, as well as compounds that are often more lethal to the pathogen in lower doses, broader spectrum in their effect and longer lasting in the environment. In contrast, the less than maximally lethal capability of native antimicrobials might be surprising if it were supposed that lethality was the aspect of these compounds being selected for. Given that such modifications are often chemically easy to produce, and hence that it is likely that they will also have been produced enzymatically in the course of evolution, it seems likely that the “native” antimicrobial represents a snapshot of the dynamic balance between short-term gain to the organism and long-term inefficiency to the clonal line. So, it may be assumed that such a dynamic balance will result in the evolution in the “natural environment” not only of such “moderated” antimicrobials, but also various alternative strategies by which individual antimicrobial producing organisms may survive in a competitive community. Such strategies may include the production of antimicrobials that are less lethal, are more specific to their target and which are used sporadically and in limited amounts. An examination of these and other strategies undoubtedly present in the global microbiota may yield alternate approaches to the current lethal broad-spectrum antimicrobials being developed for therapeutic use.
Potential Strategies
Antimicrobials in general and antibiotics in particular have had a profound positive effect upon human health and agriculture. Antibiotic therapy has alleviated or prevented infectious disease in many millions of human lives since its widespread introduction less than 80 years ago. As has been seen in the preceding discussion, unfortunately and perhaps inadvertently, the ways in which these marvelous materials have been used has catalyzed the evolutionary problem of AMR we now face. From the point of view of optimizing the clinical outcome to a patient suffering an infectious disease while limiting undesirable impacts, an ideal antibiotic therapy would target only those pathogenic organisms within a patient responsible for the disease being treated and only for the duration required to achieve the desired clinical result. No other organisms associated with the patient or the patient themselves would be adversely affected. Similarly, no organisms in the wider environment be they pathogenic or not, would be impacted by the application of the antibiotic. Unfortunately, for many reasons, such a finely focused treatment regimen is rarely, if ever achieved. The fact that “non-combatant” organisms both within the patient and in the wider environment are affected is the source of the AMR crisis as described in the body of this paper. Clearly, strategies that allow for the implementation of both existing and novel antibiotic use in a manner that is more “selectively efficient” and hence closer to the idealized therapy could be considered.
Strategies such as maintaining or improving the human immune system through vaccination and various other techniques such as the application of ecological control through phage therapy are useful. Nevertheless, there remains an imperative for the development of new antimicrobial therapies (Spellberg et al., 2004; Allen et al., 2014; Blaskovich et al., 2017; Baptista et al., 2018; Gupta et al., 2019; Roope et al., 2019). Clearly though, new antimicrobials or alternate therapies that display traits that lessen the wider impact of their use will be desirable. Such characteristics might include, greater specificity to particular pathogenic organisms (Doern, 2018; van Belkum et al., 2018). In addition, a shorter therapeutic half-life and the production of metabolites with little or no antimicrobial activity would be advantageous. It is notable that some currently available antimicrobials such as vancomycin with a typical metabolic half-life of 4–6 h, have the least diversity and a comparatively low incidence of resistance phenotypes despite their long-term usage. Similarly, fosfomycin is a decades-old antibiotic which has a short half-life (2.4 h) and is still used for urinary tract infections with renewed interest in additional applications (Ito et al., 2017). Finally, by limiting the duration of therapy to the minimum required to achieve a clinical result would also assist (Llewelyn et al., 2017). The requirement for the brief, targeted application of antimicrobial therapies obviously necessitates better and more frequently used diagnostic techniques for their implementation and would almost certainly have further consequences for their widespread application. But despite such procedural encumbrances compared to current practice, the long-term benefit is clear.
It should be noted that in the microbial world, strategies apart from widely lethal secreted antimicrobials that display some or all of the desired characteristics have been seen to be both effective and increasingly common. These include but are not limited to CRISPR, toxin-antitoxin systems and antimicrobial peptides (AMP’s) and many others. While most of these are not yet close to therapeutic use, some do show promise: AMP’s are both diverse and widely seen amongst macroscopic organisms and so, would appear to offer an efficient strategy in combatting microbial predation. These polypeptides often appear to function in a manner analogous to some antimicrobials in that similar host biochemical processes are targeted. With this apparent similarity, reasons why AMP’s are both diverse and relatively common while therapeutic antimicrobials apparently are not, are worth considering (Baquero et al., 2011; Cambray et al., 2011).
Antimicrobial peptides’s are relatively short protein sequences and so do not require a multiplicity of genes and may not require extensive post-translational processing in their production. So, both the genetic load as well as the metabolic cost of their production is minimized to the producing organism. Similarly, given that they are proteolytically susceptible, AMP’s are labile in the extracellular environment. This limits their field of action to either the intracellular milieu or else to the close proximity of the producing cell. Consequently, AMP’s cannot affect pathogenic organisms at great distance from the producing cell and so their ecological and evolutionary impact is comparatively limited, so also limiting the rise of compensating resistance. Where an AMP is deployed only within a multicellular organism, due to its rapid degradation its exposure to the wider microbial environment is also limited. This limited exposure and the selective efficiency it implies would be further enhanced by preventing all invading organisms from surviving encounters with an AMP so suggesting that AMP’s specifically and highly lethal to a pathogen, when combined with appropriate patient delivery and monitoring strategies, might be an attractive solution to the current dearth of novel antimicrobials. While the prospect of clinical AMP’s to address the AMR crisis is hopeful, significant challenges in developing AMP’s for therapeutic use have been encountered. These include but are not limited to developing AMP’s with a sufficiently broad therapeutic index and providing appropriate delivery systems that will protect a selectively efficient therapeutic AMP during storage and patient administration. Nevertheless, AMP’s as well as other selectively efficient microbial defense mechanisms indicate possible ways in which the human AMR crisis may yet be managed.
Conclusion
By separately considering the genetic, metabolic, ecological and evolutionary costs associated with a selective phenotype, a useful elaboration on the concept of the Selection coefficient may be arrived at. In the qualitative example considered here of the complex interplay of antimicrobial production and resistance phenotypes at the heart of the AMR crisis, useful hypotheses regarding the dynamics of the problem are considered. In addition, using this framework of both ecological and evolutionary considerations, potential strategies to mitigate and manage resistant microbial infections can be developed.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
CM conceived the work and wrote the manuscript. MG, MB, and AF contributed discussions, added significant concepts, and edited the work. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Allen, H. K., Trachsel, J., Looft, T., and Casey, T. A. (2014). Finding alternatives to antibiotics”. Ann. N. Y. Acad. Sci. 1323, 91–100. doi: 10.1111/nyas.12468
Aminov, R. I. (2009). The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 11, 2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158.
Baptista, P. V., McCusker, M. P., Carvalho, A., Ferreira, D. A., Mohan, N. M., Martins, M., et al. (2018). Nano-strategies to fight multidrug resistant bacteria“a battle of the titans”. Front. Microbiol. 9:1441. doi: 10.3389/fmicb.2018.01441
Baquero, F., Coque, T. M., and De La Cruz, F. (2011). Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob. Agents Chemother. 55, 3649–3660. doi: 10.1128/aac.00013-11
Blaskovich, M. A. T., Butler, M. S., and Cooper, M. A. (2017). Polishing the tarnished silver bullet: the quest for new antibiotics. Essays Biochem. 61, 103–114. doi: 10.1042/ebc20160077
Cambray, G., Sanchez-Alberola, N., Campoy, S., Guerin, É, Da Re, S., González-Zorn, B., et al. (2011). Prevalence of SOS mediated control of integron integrase expression as an adaptive trait of chromosomal and mobile integrons. Mobile DNA 2:6. doi: 10.1186/1759-8753-2-6
Cavera, V. L., Arthur, T. D., Kashtanov, D., and Chikindas, M. L. (2015). Bacteriocins and their position in the next wave of conventional antibiotics. Int. J. Antimicrob. Agents 46, 494–501. doi: 10.1016/j.ijantimicag.2015.07.011
Chow, L., Waldron, L., and Gillings, M. (2015). Potential impacts of aquatic pollutants: sub-clinical antibiotic concentrations induce genome changes and promote antibiotic resistance. Front. Microbiol. 6:803.
Contained and Controlled, The UK’s 20-Year Vision for Antimicrobial Resistance (2019). Available online at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/773065/uk-20-year-vision-for-antimicrobial-resistance.pdf
da Silva, P. E. A., Machado, D., Ramos, D., Couto, I., Von Groll, A., and Viveiros, M. (2016). “Efflux pumps in mycobacteria: antimicrobial resistance, physiological functions, and role in pathogenicity,” in Efflux-Mediated Antimicrobial Resistance in Bacteria, eds X. Z. Li, C. Elkins, and H. Zgurskaya (Cham), 527–559. doi: 10.1007/978-3-319-39658-3_21
Das, S., Saha, B., Hati, A. K., and Roy, S. (2018). Evidence of artemisinin-resistant Plasmodium falciparum malaria in Eastern India. New Engl. J. Med. 379, 1962–1964. doi: 10.1056/nejmc1713777
Doern, C. D. (2018). The slow march toward rapid phenotypic antimicrobial susceptibility testing: are we there yet? J. Clin. Microbiol. 56:e1999-17.
Dykhuizen, D. (2016). Thoughts toward a theory of natural selection: the importance of microbial experimental evolution. Cold Spring Harbor Perspect. Biol. 8:a018044. doi: 10.1101/cshperspect.a018044
Fairhurst, R. M., and Dondorp, A. M. (2009). Artemisinin resistance in Plasmodium falciparum malaria. New Engl. J. Med. 361, 455–467.
Fleming, A. (1945). Penicillin. Nobel Lecture. https://www.nobelprize.org/nobel_prizes/medicine/laureates/1945/fleming-lecture.pdf (accessed May 1, 2018).
Foucault, M. L., Depardieu, F., Courvalin, P., and Grillot-Courvalin, C. (2010). Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc. Natl. Acad. Sci. U.S.A. 107, 16964–16969. doi: 10.1073/pnas.1006855107
Ghaly, T. M., Jl Geoghegan, J., Alroy, M. R., and Gillings. (2019). High diversity and rapid spatial turnover of integron gene cassettes in soil. Environ. Microbiol. 21, 1567–1574. doi: 10.1111/1462-2920.14551
Gillings, M. R. (2014). Integrons: past, present, and future. Microbiol. Mol. Biol. Rev. 78, 257–277. doi: 10.1128/mmbr.00056-13
Guerin, É, Cambray, G., Sanchez-Alberola, N., Campoy, S., Erill, I., Da Re, S., et al. (2009). The SOS response controls integron recombination. Science 324, 1034–1034. doi: 10.1126/science.1172914
Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158. doi: 10.1371/journal.ppat.1002158
Gunderson, L. H., Allen, C. R., and Holling, C. S. (eds) (2012). Foundations of Ecological Resilience. Washington, D.C: Island Press.
Gupta, A., Mumtaz, S., Li, C. H., Hussain, I., and Rotello, V. M. (2019). Combatting antibiotic-resistant bacteria using nanomaterials. Chem. Soc. Rev. 48, 415–427. doi: 10.1039/c7cs00748e
Hocquet, D., Llanes, C., Thouverez, M., Kulasekara, H. D., Bertrand, X., Plésiat, P., et al. (2012). Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog. 8:e1002778. doi: 10.1371/journal.ppat.1002778
Ito, R., Mustapha, M. M., Tomich, A. D., Callaghan, J. D., McElheny, C. L., Mettus, R. T., et al. (2017). Widespread fosfomycin resistance in Gram-negative bacteria attributable to the chromosomal fosA gene. MBio 8, e749–e717.
Jackson, R. W., Vinatzer, B., Arnold, D. L., Dorus, S., and Murillo, J. (2011). The influence of the accessory genome on bacterial pathogen evolution. Mobile Genet. Elements 1, 55–65. doi: 10.4161/mge.1.1.16432
Lande, R., and Arnold, S. J. (1983). The measurement of selection on correlated characters. Evolution 37, 1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x
Ligozzi, M., Lo Cascio, G., and Fontana, R. (1998). vanA gene cluster in a vancomycin-resistant clinical isolate of Bacillus circulans. Antimicrob. Agents Chemother. 42, 2055–2059. doi: 10.1128/aac.42.8.2055
Llewelyn, M. J., Fitzpatrick, J. M., Darwin, E., SarahTonkin-Crine, Gorton, C., Paul, J., et al. (2017). The antibiotic course has had its day. Bmj 358:j3418. doi: 10.1136/bmj.j3418
Mazel, D. (2004). Integrons and the origin of antibiotic resistance gene cassettes. ASM News Am. Soc. Microbiol. 70, 520–525.
Michael, C. A., and Labbate, M. (2010). Gene cassette transcription in a large integron-associated array. BMC Genet. 11:82.
Michael, C. A., Dominey-Howes, D., and Labbate, M. (2014). The antimicrobial resistance crisis: causes, consequences, and management. Front. Public Health 2:145.
Mira, P. M., Meza, J. C., Nandipati, A., and Barlow, M. (2015). Adaptive landscapes of resistance genes change as antibiotic concentrations change. Mol. Biol. Evol. 32, 2707–2715. doi: 10.1093/molbev/msv146
Perron, G. G., Whyte, L., Turnbaugh, P. J., Goordial, J., Hanage, W. P., Dantas, G., et al. (2015). Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS One 10:e0069533. doi: 10.1371/journal.pone.0069533
Peterson, G., Allen, C. R., and Holling, C. S. (1998). Ecological resilience, biodiversity, and scale. Ecosystems 1, 6–18. doi: 10.1007/s100219900002
Ramadhan, A. A., and Hegedus, E. (2005). Survivability of vancomycin resistant enterococci and fitness cost of vancomycin resistance acquisition. J. Clin. Pathol. 58, 744–746. doi: 10.1136/jcp.2004.024091
Recchia, G. D., and Hall, R. M. (1997). Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 5, 389–394. doi: 10.1016/s0966-842x(97)01123-2
Ribeiro da Cunha, B., Fonseca, L. P., and Calado, C. R. C. (2019). Antibiotic discovery: where have we come from, where do we go? Antibiotics 8:45. doi: 10.3390/antibiotics8020045
Roope, L. S. J., Smith, R. D., Pouwels, K. B., Buchanan, J., Abel, L., Eibich, P., et al. (2019). The challenge of antimicrobial resistance: what economics can contribute. Science 364:eaau4679. doi: 10.1126/science.aau4679
Rutherford, S. T., and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427
Shapiro, J. A. (2010). Mobile DNA and evolution in the 21st century. Mob. DNA 1:4. doi: 10.1186/1759-8753-1-4
Smalla, K., Cook, K., Djordjevic, S. P., Klümper, U., and Gillings, M. (2018). Environmental dimensions of antibiotic resistance: assessment of basic science gaps. FEMS Microbiol. Ecol. 94:fiy195.
Spellberg, B., Powers, J. H., Brass, E. P., Miller, L. G., and Edwards, J. E. Jr. (2004). Trends in antimicrobial drug development: implications for the future. Clin. Infect. Dis. 38, 1279–1286. doi: 10.1086/420937
Szumowski, J. D., Adams, K. N., Edelstein, P. H., and Ramakrishnan, L. (2012). Antimicrobial efflux pumps and Mycobacterium tuberculosis drug tolerance: evolutionary considerations. Curr. Top Microbiol. Immunol. 374, 81–108. doi: 10.1007/82_2012_300
Thurman, T. J., and Barrett, R. D. H. (2016). The genetic consequences of selection in natural populations. Mol. Ecol. 25, 1429–1144.
Ur Rahman, S., Ali, T., Ali, I., Khan, N. A., Han, B., and Gao, J. (2018). The growing genetic and functional diversity of extended spectrum beta lactamases. BioMed Res. Int. 2018:9519718.
van Belkum, A., Bachmann, T. T., Lüdke, G., Lisby, J. G., Kahlmeter, G., Mohess, A., et al. (2018). Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 17, 51–62.
Williams, D. H., and Bardsley, B. (1999). The vancomycin group of antibiotics and the fight against resistant bacteria. Angewandte Chemie Int. Ed. 38, 1172–1193. doi: 10.1002/(sici)1521-3773(19990503)38:9<1172::aid-anie1172>3.0.co;2-c
World Health Organization (WHO) (2014). Antimicrobial Resistance: Global Report on Surveillance. Available online at: http://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf;jsessionid=16CE7CCA1D8B3D4E1EB4854BAE6C124B?sequence=1 (accessed February 5, 2014).
Keywords: evolutionary theory, antimicrobial, antibiotic, resistance, selective efficiency
Citation: Michael CA, Gillings MR, Blaskovich MAT and Franks AE (2021) The Antimicrobial Resistance Crisis: An Inadvertent, Unfortunate but Nevertheless Informative Experiment in Evolutionary Biology. Front. Ecol. Evol. 9:692674. doi: 10.3389/fevo.2021.692674
Received: 14 April 2021; Accepted: 01 June 2021;
Published: 01 July 2021.
Edited by:
Miguel Arenas, University of Vigo, SpainReviewed by:
Jan Zrimec, Chalmers University of Technology, SwedenSaúl Huitzil, National Autonomous University of Mexico, Mexico
Copyright © 2021 Michael, Gillings, Blaskovich and Franks. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carolyn A. Michael, Y2Fyb2x5bi5taWNoYWVsQG1xLmVkdS5hdQ==
†ORCID: Carolyn A. Michaell, orcid.org/0000-0002-3864-3380; Michael R. Gillings, orcid.org/0000-0002-4043-4351; Mark A. T. Blaskovich, orcid.org/0000-0001-9447-2292; Ashley E. Franks, orcid.org/0000-0003-1664-6060
 Carolyn A. Michael
Carolyn A. Michael Michael R. Gillings
Michael R. Gillings Mark A. T. Blaskovich
Mark A. T. Blaskovich Ashley E. Franks
Ashley E. Franks