- 1Chongqing Key Laboratory of Vector Insects, College of Life Sciences, Chongqing Normal University, Chongqing, China
- 2Key Laboratory of Zoological Systematics and Evolution, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 3International College, University of Chinese Academy of Sciences, Beijing, China
- 4College of Biological Sciences, University of Chinese Academy of Sciences, Beijing, China
- 5State Key Laboratory of Integrated Pest Management, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 6College of Life Sciences, Chongqing Normal University, Chongqing, China
To meet the pollination need of economic crops, Osmia excavata has been successfully used to improve the pollination efficiency of Rosaceae and Brassicaceae plants. As a widely used pollinator of economic crops, a systematic study of flower-visiting species and diversities of O. excavata stocked in China was not found. To investigate the foraging pollen species and diversities of O. excavata, beebread from 20 experimental plots in China was collected by the trap-nesting method and analyzed by DNA metabarcoding technology. A total of 26 pollen plants in 14 genera and nine families were identified. A further analysis showed that the richness and abundance of the wild flowering plants in orchards and farmlands were lower than those in the nearby semi-natural habitats. The favorite pollen comes from economic crops apple and rape and wild flowering plants Juncus interior, Rosa gymnocarpa, and Rosa laevigata. Through a diversity index analysis, it was found that the Anhui region has the highest pollen plant diversity, while the Liaoning region has the lowest. Our results can provide a basis for flower-visiting species and diversities of O. excavata.
Introduction
Pollinators are of great importance in ecosystems and contribute about $136 billion annually to agricultural systems in China (Ouyang et al., 2019). Due to the intensification and expansion of agriculture, the diversity of pollinators has been declining rapidly for decades, which has led to the loss, fragmentation, and degradation of pollinator habitats and a severe decline in global pollination (Foley et al., 2011; Féon et al., 2013; Belsky and Joshi, 2019). In the past 50 years, the dependence of global agriculture on pollinators has increased by more than 300%, while the abundance of pollinators worldwide has continued to decline (Potts et al., 2016). Previous studies have shown that planting of pollen plants in pollinator habitats can significantly increase the number and species of pollinators (Jönsson et al., 2015; M’Gonigle et al., 2016; Warzecha et al., 2018; Williams and Lonsdorf, 2018; Nichols et al., 2019). Therefore, it is necessary to clarify how to choose the best plants for different pollinators.
The internal transcribed spacer II (ITS2) barcode based on metabarcoding method can identify pollen plant species from mixed pollen and shows great potential in the analysis of beebread ingredients (Pornon et al., 2016; De-Vere et al., 2017; Lucas et al., 2018). Compared with traditional palynology, the molecular method not only has a higher sensitivity for the identification of mixed pollen but also effectively reduces time and cost (Lang et al., 2018; Tremblay et al., 2019). The ITS2 region has proved to be a core barcode for pollen identification (Richardson et al., 2015; Gous et al., 2019; Tremblay et al., 2019). The barcode combination of rbcL, trnH-psbA, and ITS2 successfully identified the main pollen plants, Vitex negundo and Vitex quinata, in the beebread of Megachile strupigera by He et al. (2020). Therefore, DNA metabarcode not only helps to identify species and determine the relative abundance of pollen plants in beebread but is also used to construct pollination networks, monitor plant pathogens, assess species diversity, and predict biodiversity patterns (Huang et al., 2020).
To meet the pollination demand of economic crops, pollinators with commercial potential have been found and developed (Ryder et al., 2020). Bumblebees have been successfully used as pollinators for eggplant, tomato, peach, and pear (Wu et al., 2006). Megachile rotundata was used for the pollination of alfalfa (Xu et al., 2009). In orchards, the bee of genus Osmia was considered as efficient pollinators of some Rosaceae plants, such as apricot, plum, cherry, peach, pear, and apple (Parker, 1981; Torchio et al., 1987; Wei et al., 1991; Xu et al., 1994). Importantly, Osmia bee can be artificially stocked due to its advantages such as easy management, low cost, and high pollination efficiency (Wei and Zhao, 1995; Wei et al., 2000b; Dai, 2004). In north and northwest of China, there are mainly five species of Osmia (O. excavata, O. cornifrons, O. taurus, O. jacoti, and O. pedicornis) that play important roles in enhancing pollination, increasing fruit diameter and the number of seeds per fruit, and decreasing the percentage of asymmetrical fruit (Wei et al., 2002). For O. excavata, it has many advantages, such as low-temperature tolerance, long daily activity time, high flower-visiting efficiency, centralized pollination range, and easy management (Wei and Yuan, 1997). Therefore, it was considered as an excellent and large-scale stocking pollinator. The pollination of O. excavata can create more than $27.8, $9.4, and $3.0 billion for cherries, apples, and rapes planting, respectively, each year (Liu et al., 2018, 2019a,b). Pollination can significantly improve the fruit set rate and fruit quality, and it has broad application prospects in the future of fruit planting industry (He and Zhou, 2009).
However, the short flowering period of fruit trees limits the nutritional requirements of O. excavata during the foraging period (Yan et al., 2018). Because of this limitation, survival and large-scale stocking of O. excavata face great challenges. A previous study showed that the diversity of pollen plants is the main factor affecting the diversity of wild bee species (Kovács-Hostyánszki et al., 2017). Therefore, the effective solution is to grow other pollen plants in the orchard. This will not only improve the biodiversity of plantations but also provide adequate food sources for bees (Boyle et al., 2020). So, it is necessary to understand the foraging preferences of bees. In this study, beebread of O. excavata was collected by trap-nesting from 20 experimental plots in China. The species and abundance of plants were determined by ITS2 barcode based on metabarcoding method, and the dominant plants that meet the nutrition needs of bees were further analyzed.
Materials and Methods
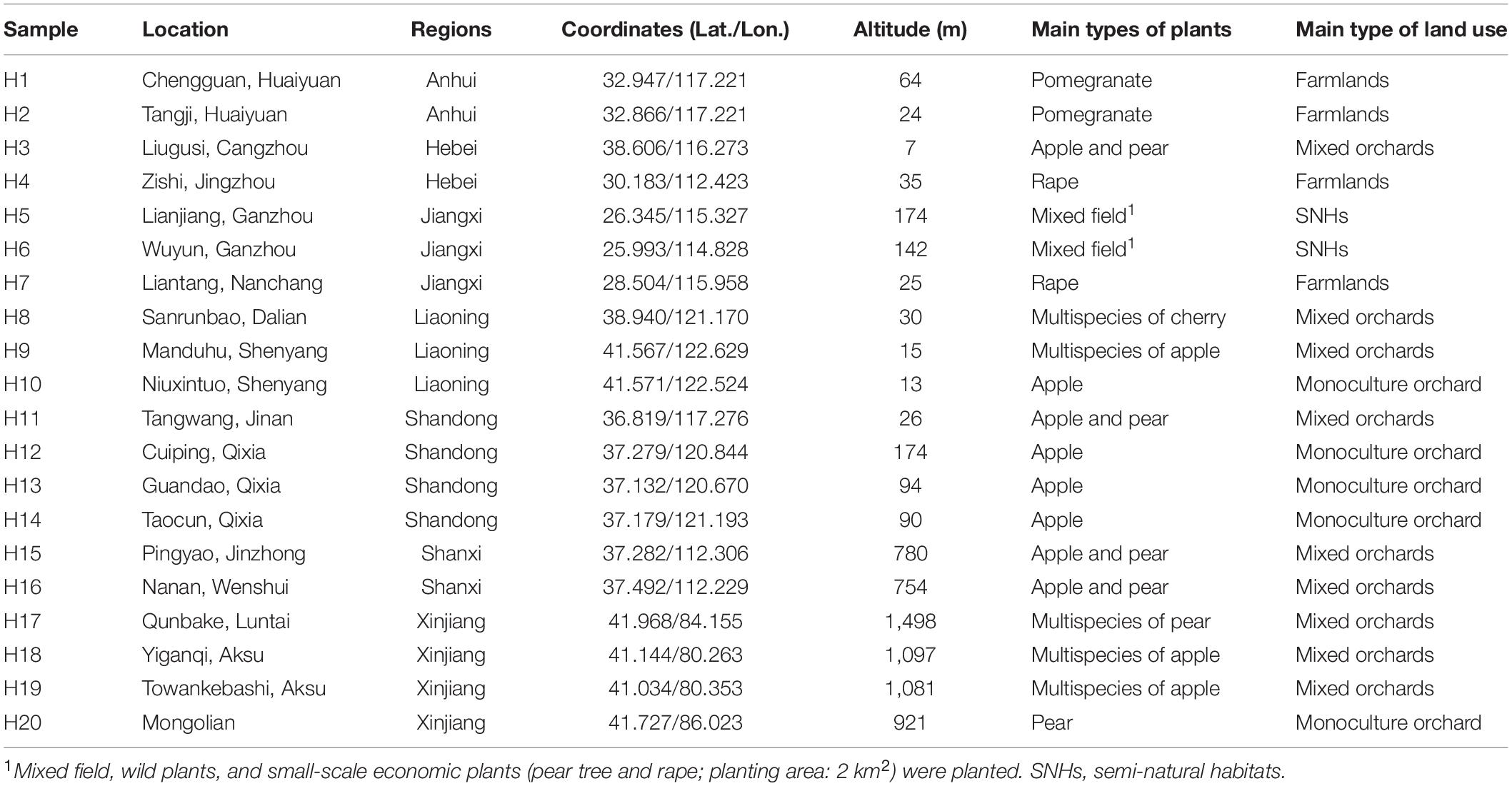
Sample Collection and DNA Extraction
Osmia excavata can build a nest in trap nests made by reed tubes and make beebread to breed its offspring (Wang et al., 2001; Huang et al., 2013). According to the main activity period (March–April) of O. excavata, trap nests were placed in similar habitats (e.g., flower composition, competitors, and predators, etc.) in each experimental plot. Trap nests of O. excavata were collected from 20 regions of China (March–May 2020), covering three types of land: orchards (monoculture or mixed-species plantation), farmlands, and semi-natural habitats (SNHs; agroforestry ecotone). Detailed geographic information of each sampling plot and surrounding main plant types are listed in Table 1. In the laboratory, mixed pollens within five groups of beebread from the trap nests of each site were stored at –80°C for later use. Mixed pollens were extracted using the E.Z.N.A.® Soil DNA Kit according to the manufacturer’s protocol (Omega Bio-Tek, Norcross, GA, United States). The concentration and purity of purified DNA were determined by a NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc., United States).
PCR Amplification and Sequencing
The ITS2 region was amplified using the forward primer ITSS2F: 5′ATGCGATACTTGGTGTGAAT3′ and the reverse primer ITSS4R: 5′TCCTCCGCTTATTGATATGC 3′. PCR reactions were performed in a 20-μl system, containing 4 μl FastPfu Buffer (5×), 2 μl dNTPs (2.5 mM/each), 0.8 μl each primer (5 μM), 0.4 μl FastPfu Polymerase (5 U/μl), 2 μl template DNA (5 ng/μl), and 10 μl ddH2O. All reactions were performed in an ABI GeneAmp® 9700 (Applied Biosystems, Carlsbad, CA, United States) with the following parameters: an initial denaturation at 95°C for 5 min, followed by 29 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and a final extension at 72°C for 10 min. PCR products were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States). The DNA concentrations were measured with a Qubit® 3.0 fluorometer (Life Invitrogen, Waltham, MA, United States) and then paired-end sequenced (2 × 250 bp) on an Illumina MiSeq platform (Illumina, San Diego, CA, United States) at the Biozeron Biological Technology Co. Ltd. (Shanghai, China).
Data Analysis
Raw sequencing data were filtered to delete the tail-base of reads (Q < 20) using Trimmomatic v0.38 (Bolger et al., 2014). The chimeric sequences were removed and effective sequences clustered into operational taxonomic units (OTUs; ≥97% similarity) by Usearch v10 (Edgar, 2013). Based on the Unite database v8.2,1 the OTU representative sequences were analyzed taxonomically to obtain the community composition of each sample using the RDP classifier Bayes algorithm with default parameters (Wang et al., 2007).
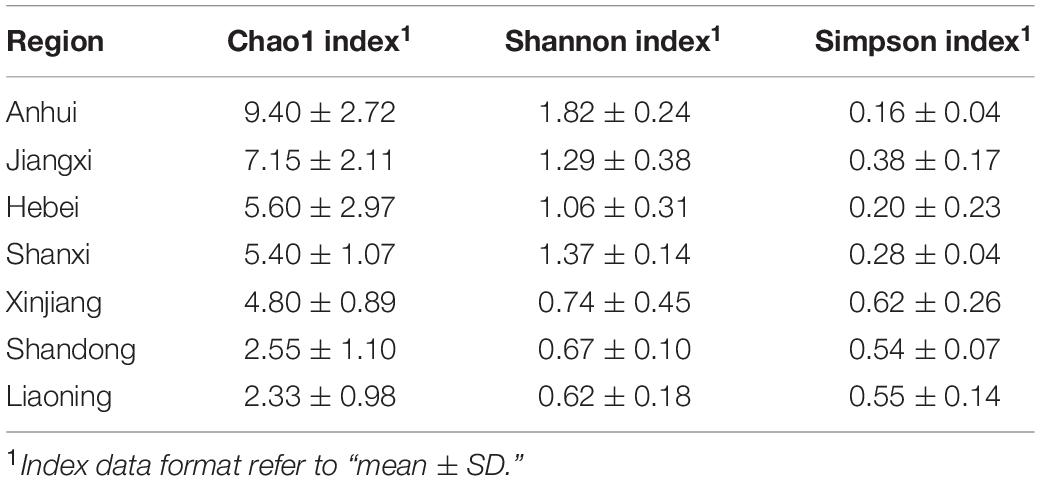
Alpha diversity indexes (Shannon, Simpson, and Chao1) were calculated using Mothur v1.30 (Schloss et al., 2009). The Fisher’s least significant difference (LSD) in SPSS v25.0 (SPSS, Inc., Chicago, IL, United States) was used to analyze the differences of diversities of beebread between different land types and experimental plots. Species accumulation curves (SACs), the abundance, and composition of beebread were prepared and visualized by the “ggplot2” package (Hadley, 2016) and the “pheatmap” package (Raivo, 2019).
Results
Sequencing and Assembly
A total of 4,563,930 clean ITS2 sequences from all experimental plots were obtained with an average length of 335 ± 9 bp and clustered into 64 OTUs (Supplementary Table 1). A statistical analysis showed that the SACs tend to be flat, indicating that the sample sizes reach the conditions that species are fully discovered. The species richness in the environment should not increase significantly with the increase of sample sizes (Supplementary Figure 1).
Species and Abundance Analysis of Pollen Plants
Based on OTU representative sequences, a total of nine families (14 genera and 26 species) were identified from all samples combined (Supplementary Table 1). The clustering results of plant species showed that the rape and apple pollens were rich in four samples (H1, H2, H4, and H7) and 11 samples (H3, H9–H16, H18, and H19), respectively. Wild plant pollens were dominated in samples H2 (Juncus interior and Papaver rhoeas), H5 (J. interior, Rosa gymnocarpa, Rosa laevigata, and Rosa primula), H6 (R. gymnocarpa, R. laevigata, and R. primula), H7 (J. interior), and H16 (Prinsepia sinensis; Figure 1 and Supplementary Table 2). A further analysis revealed that plant species in beebread was dominated by economic crops, including apple (Malus domestica, Malus pumila, and Malus sp.), pear (Pyrus betulifolia, Pyrus communis, Pyrus cordata, and Pyrus spinosa), plum (Prunus brachypoda), cherry (Cerasus avium), and rape (Brassica juncea and Brassica sp.). In addition, nine types of wild plants were detected in beebread. Among them, the abundance of J. interior, R. gymnocarpa, and R. laevigata was higher than others (Figure 1 and Supplementary Table 2).
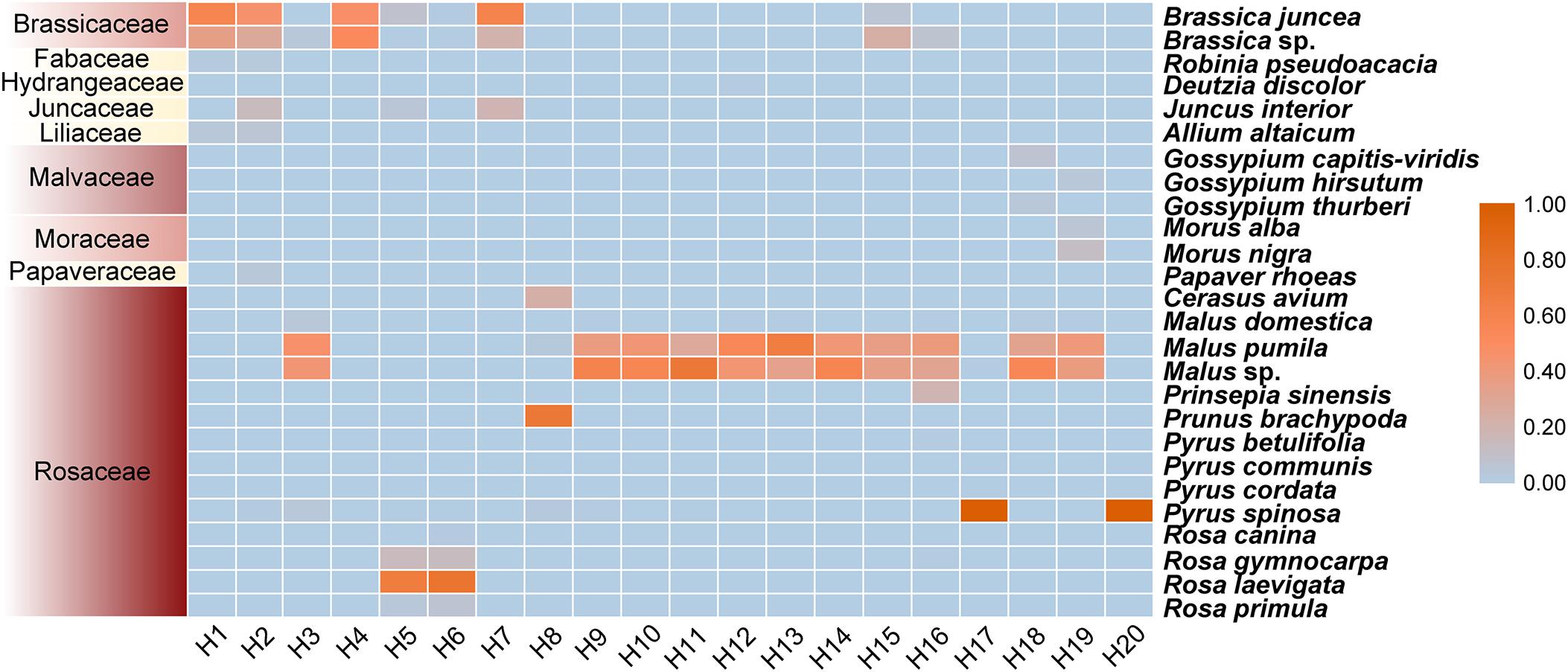
Figure 1. Composition and relative abundance of pollen plants. The left and right of the Y-axis and the X-axis represent the family and species of pollen plants and sampling plot, respectively. The legend in the upper right represents the proportional abundance of pollen plants.
Impacts of Different Types of Land on the Composition of Beebread
The composition and abundance of pollen in beebread from different types of land use were compared (Table 1). The results showed that economic crop pollens mainly came from the single- or mixed-fruit plantations and intensive farmland, and wild plant pollens mainly came from the agroforestry ecotone (Figure 2A). Pollen plants in beebread from semi-natural habitat (SNH; agroforestry ecotone) samples have the highest species abundance (4.7 ± 0.95, n = 10), then followed by that from mixed plantations (3.33 ± 0.76, n = 45), farmlands (3.05 ± 0.94, n = 20), and single-fruit plantations (2.08 ± 0.56, n = 25; Figure 2B). A further analysis found that the species abundance of pollen plants in beebread from farmlands and mixed-fruit plantations did not differ significantly (p = 0.36), but there were significant differences between other experimental plots (p < 0.05; Supplementary Table 3).
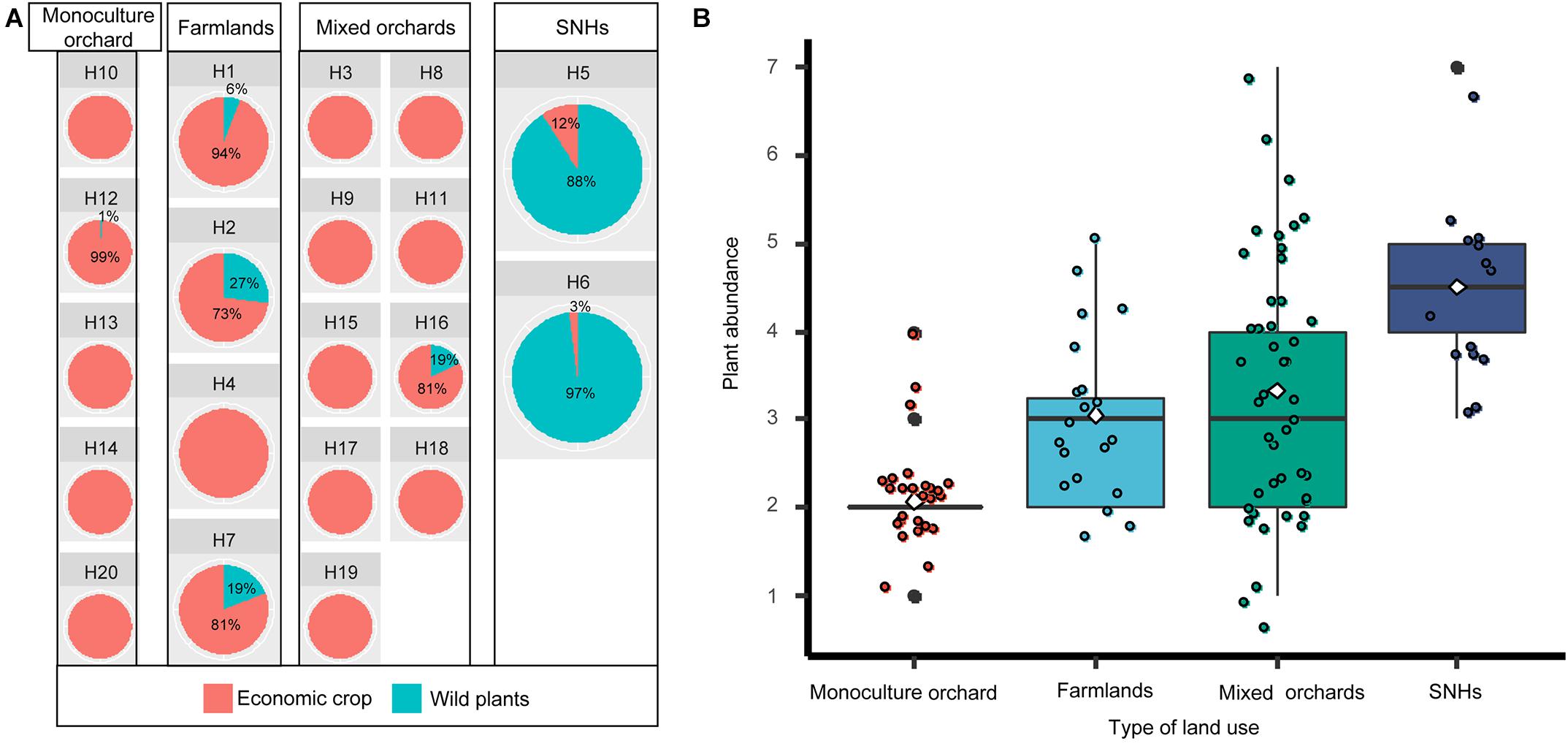
Figure 2. The composition (A) and abundance (B) of beebread in different types of land use. (A) The proportion of economic crops and wild plants in each experimental plot. (B) Plant abundance under different land-use types. The X-axis and Y-axis represent land-use types and plant abundance, respectively.
Diversity of Pollen Plants in Beebread
To better understand the richness and diversity of pollen plants in beebread, 20 experimental plots were divided into seven regions according to the province where O. excavata is attracted (Table 1). The results showed that the Chao1 index of Anhui (9.40 ± 2.72, n = 10) is the highest, then followed by that of Jiangxi (7.15 ± 2.11, n = 20), Hebei (5.60 ± 2.97, n = 5), Shanxi (5.40 ± 1.07, n = 10), Xinjiang (4.80 ± 0.89, n = 20), Shandong (2.55 ± 1.10, n = 20), and Liaoning (2.33 ± 0.98, n = 15; Table 2 and Supplementary Table 4). The Shannon index of Anhui (1.82 ± 0.24, n = 10), Shanxi (1.37 ± 0.14, n = 10), Jiangxi (1.29 ± 0.38, n = 20), and Hebei (1.06 ± 0.31, n = 5) is significantly higher than that of Xinjiang (0.74 ± 0.45, n = 20), Shandong (0.67 ± 0.10, n = 20), and Liaoning (0.62 ± 0.18, n = 15; all p < 0.05), which indicated that the diversity of pollen plants in Anhui, Shanxi, Jiangxi, and Hebei is high (Table 2 and Supplementary Table 4). A large Chao1 and Shannon indexes or a small Simpson index can explain the high species richness and diversity in beebread. Therefore, it can be concluded that the abundance of pollen plants in the beebread from Anhui was significantly higher than that in other regions. In general, the richness and diversity of pollen plants in different regions can be divided into three levels from high to low: Anhui → Hebei, Shanxi, and Jiangxi → Xinjiang, Shandong, and Liaoning (Table 2).
Discussion
Wild bees are important pollinators for economic crops and wild plants. They rely on different landscapes to obtain flowering resources and nesting habitats (Kratschmer et al., 2019). In this study, 20 experimental plots in China were set up to collect the beebread of O. excavata. A total of 26 pollen plant species were identified, belonging to 14 genera of nine families and including 17 economic crops and nine wild plant species (Robinia pseudoacacia, Deutzia discolor, J. interior, P. rhoeas, P. sinensis, Rosa canina, R. gymnocarpa, R. laevigata, and R. primula). Based on the composition and relative abundance of pollens (Figures 1, 2), O. excavata is an effective pollinator for economic crops (B. juncea, Brassica sp., M. pumila, Malus sp., P. brachypoda, and P. spinosa).
Previous studies indicated that the larvae of O. excavata released at the early flowering stages of economic crops will grow into effective pollinators, especially Rosaceae and Brassicaceae plants (Yang et al., 1997; Wei et al., 2000a; Hawkins et al., 2015; Gresty et al., 2018). However, the flowering period (7–15 days; Supplementary Table 5) (Lech et al., 2008; Liu et al., 2008, 2010; Wang et al., 2012; Lu et al., 2013; Lin et al., 2019; Zhang et al., 2019, 2020; Li et al., 2020) of most economic crops is short and cannot cover the entire adult stage of O. excavata (40–55 days; Wei et al., 2002; Men et al., 2018). Through the observation of professional beekeepers and our field work on the stocking of O. excavata, when O. excavata had no diverse food sources except economic crops, this gap seriously affects the bees to reserve beebread for their offspring, which causes a low recovery rate after artificial stocking. The number of reduced populations needs to be supplemented every year, excluding the situation of unnatural death, human activities, and population migration.
Although when the beebread from H5/H6 experimental plots were collected, the economic crops (pear and rape trees) in the plantation were in the late flowering stage, which led to the lack of pollen resources, O. excavata could still obtain pollens from wild plants (J. interior, R. gymnocarpa, R. laevigata, and R. primula; Figure 1). This means that in order to reduce the impact of the lack of pollen resources of economic crops, O. excavata can use the pollen of wild plants as their main source of nutrition for meeting survival needs. Plantations (orchards and farmlands) were dominated by a single type of economic crops and lack of flowering resources of wild plants (Figure 2A). A further analysis revealed that more wild plants in beebread were from SHNs (Figure 2A). Papanikolaou et al. (2017) found that increasing the richness of plants in the habitat would significantly affect the abundance and diversity of bees. When the pollen resources of economic crops cannot meet the needs of the entire adult stage of O. excavata, wild flowering plants can be planted in plantations or introduced into SNHs. The transplanting of wild flowering plants into the plantation helps to attract O. excavata and increase food sources. Ideal type of wild flowering plants can be selected from nine wild plants identified in beebread, especially J. interior, R. laevigata, and R. gymnocarpa.
Piko et al. (2021) concluded that the mixed flower field model can effectively provide attractive food resources, thereby greatly increasing the abundance and diversity of bumblebee. Bihaly et al. (2020) also emphasized that planting a variety of flowering herbaceous plants on a small scale could significantly increase the number of bee nests in the orchard. In response to concerns about the food source of the O. excavata, combing the previous results, a conjecture is proposed: a mixed flower field model suitable for O. excavata (MFF-OE model). We can plant a variety of favorite wild flowering plants around the plantation to meet the needs of pollen resources during the entire adult stage of O. excavata. However, the model needs to be further explored in terms of variety allocation, optimum planting area selection, and planting scale, etc. The comprehensive analysis of Chao1 and Shannon indexes reflected lower species richness and diversity in Liaoning. The Liaoning region can be used as a pilot for the exploration and implementation of the MFF-OE model. Taken together, we look forward that the implementation of the MFF-OE model can effectively solve the shortage of pollen resources of economic crops.
Conclusion
Osmia excavata has been successfully used for pollination in commercial orchards because it can significantly improve fruit setting rate and fruit quality. In this study, beebread of O. excavata was collected from various habitats by trap-nesting and analyzed by DNA metabarcoding technology. A total 26 pollen plants in 14 genera and nine families were identified, including 17 economic crops and nine wild plant species. The abundances of wild flowering plants in the plantation were lower than those in SNHs. According to the composition and abundance of pollen plants, O. excavata is regarded as an effective pollinator for economic crops in Rosaceae and Brassicaceae from the molecular level.
Data Availability Statement
The sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/. The associated BioProject, SRA, and BioSample numbers are PRJNA742556, SRR15011441, and SAMN19967841, respectively.
Ethics Statement
All experiments procedures for this study complied with the current animal ethics guidelines and did not involve any protected animals.
Author Contributions
AL and DH were responsible for the conceptualization and supervision of the study. HL, FD, and YH were responsible for the methodology and writing – original draft preparation. HL, FD, YL, KZ, and HZ were responsible for the software. ZZ, CZ, AL, and DH performed the validation. HL and FD conducted the formal analysis and visualization. FD, YL, KZ, HZ, and DH conducted the investigation. FD and DH were responsible for the resources. HL, FD, YL, KZ, HZ, and DH were responsible for data curation. YH, ZZ, CZ, AL, and DH were responsible for writing – review and editing. DH was responsible for project administration. HL, ZZ, AL, and DH were responsible for funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Program of Ministry of Science and Technology of China (2018FY100405); the Research and Innovation Projects of Graduate Students of Chongqing Normal University (YKC20037); the National Natural Science Foundation of China (31970484 and 32070465); and the Key Laboratory of Animal Evolution and Systematics, Chinese Academy of Sciences (E052G21305).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to the editor and reviewers for helpful comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.730549/full#supplementary-material
Footnotes
References
Belsky, J., and Joshi, N. K. (2019). Impact of biotic and abiotic stressors on managed and feral bees. Insects 10, 1–42. doi: 10.3390/insects10080233
Bihaly, ÁD., Kovács−Hostyánszki, A., Szalai, M., and Sárospataki, M. (2020). Nesting activity of cavity−nesting bees and wasps is lower in small−scale apple orchards compared to nearby semi−natural habitats. Agr. Forest Entomol. 23, 49–58. doi: 10.1111/afe.12403
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyle, N. K., Artz, D. R., Lundin, O., Ward, K., Picklum, D., Wardell, G. I., et al. (2020). Wildflower plantings promote blue orchard bee, Osmia lignaria (Hymenoptera: megachilidae), reproduction in California almond orchards. Ecol. Evol. 10, 3189–3199. doi: 10.1002/ece3.5952
Dai, Y. G. (2004). The advantage of Osmia pollination for fruit trees of Rosaceae. Apicul. Sci. Technol. 2:48.
De-Vere, N., Jones, L. E., Gilmore, T., Moscrop, J., Lowe, A., Smith, D., et al. (2017). Using DNA metabarcoding to investigate honey bee foraging reveals limited flower use despite high floral availability. Sci. Rep. 7:42838. doi: 10.1038/srep42838
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Féon, V. L., Burel, F., Chifflet, R., Henry, M., Ricroch, A., Vaissière, B. E., et al. (2013). Solitary bee abundance and species richness in dynamic agricultural landscapes. Agr. Ecosyst. Environ. 166, 94–101. doi: 10.1016/j.agee.2011.06.020
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478, 337–342. doi: 10.1038/nature10452
Gous, A., Swanevelder, D. Z. H., Eardley, C. D., and Willows-Munro, S. (2019). Plant-pollinator interactions over time: pollen metabarcoding from bees in a historic collection. Evol. Appl. 12, 187–197. doi: 10.1111/eva.12707
Gresty, C. E. A., Clare, E., Devey, D. S., Cowan, R. S., Csiba, L., Malakasi, P., et al. (2018). Flower preferences and pollen transport networks for cavity−nesting solitary bees: implications for the design of agri-environment schemes. Ecol. Evol. 8, 7574–7587. doi: 10.1002/ece3.4234
Hawkins, J., de Vere, N., Griffith, A., Ford, C. R., Allainguillaume, J., Hegarty, M. J., et al. (2015). Using DNA metabarcoding to identify the floral composition of honey: a new tool for investigating honey bee foraging preferences. PLoS One 10:e0134735. doi: 10.1371/journal.pone.0134735
He, B., Gu, Z. Y., Li, H. Y., and Huang, D. Y. (2020). Analysis of species and diversities in the pollen plants of Megachile strupigera (Hymenoptera: megachilidae) by DNA barcoding. Acta Ecol. Sin. 40, 2122–2129. doi: 10.5846/stxb201809021867
He, W. Z., and Zhou, W. R. (2009). Study on the pollination effect of Osmia jacoti, Apis mellifera and artificial pollination on apple. Apicul. China 60, 9–11.
Huang, D. Y., He, B., Yu, J. F., Gu, P., Peng, F., Huang, G. N., et al. (2013). Nesting biology of Euodynerus nipanicus Schulthess (Hymenoptera: vespidae). J. Environ. Entomol. 35, 783–791.
Huang, D. Y., Huang, S. C., Li, H. Y., Dou, F. Y., Kou, R. M., Li, Y., et al. (2020). Identification methods of nectar or pollen plants for pollinators. J. Chongqing Normal U. (Nat. Sci.) 37, 1–8.
Jönsson, A. M., Ekroos, J., Dänhardt, J., Andersson, G. K., Olsson, O., and Smith, H. G. (2015). Sown flower strips in southern Sweden increase abundances of wild bees and hoverflies in the wider landscape. Biol. Conserv. 184, 51–58. doi: 10.1016/j.biocon.2014.12.027
Kovács-Hostyánszki, A., Espíndola, A., Vanbergen, A. J., Settele, J., Kremen, C., and Dicks, L. V. (2017). Ecological intensification to mitigate impacts of conventional intensive land use on pollinators and pollination. Ecol. Lett. 20, 673–689. doi: 10.1111/ele.12762
Kratschmer, S., Petroviæ, B., Curto, M., Meimberg, H., and Pachinger, B. (2019). Pollen availability for the Horned mason bee (Osmia cornuta) in regions of different land use and landscape structures. Ecol. Entomol. 45, 525–537. doi: 10.1111/een.12823
Lang, D., Tang, M., and Zhou, X. (2018). Qualitative and quantitative molecular construction of plant-pollinator network: application and prospective. Biodivers. Sci. 26, 445–456. doi: 10.17520/biods.2018058
Lech, W., Malodobry, M., Dziedzic, E., Bieniasz, M., and Doniec, S. (2008). Biology of sweet cherry flowering. J. Fruit Ornam. Plant Res. 16, 189–199.
Li, W. J., Huang, W. W., Li, Q., Sun, Q. Q., and Luo, Y. (2020). Research on the forecast method of rape florescence in the Yangtze River basin. Chin. J. Agr. Res. Reg. Planning 41, 101–108.
Lin, X., Hou, K. Q., Chen, L. A., Wei, Z. P., and Fang, Q. Y. (2019). Introduction performance of 8 citrus cultivars in eastern fujian province. Southeast Horticul. 7, 15–18.
Liu, H. T., Wang, J. S., Liu, W. J., Li, C. Y., and Guo, G. Z. (2008). Study on the characteristics of apricot florescence. Chin. Agr. Sci. Bull. 24, 171–175.
Liu, L., Li, L. L., Ouyang, F., Li, C., Yu, Y., Qu, C. H., et al. (2019a). Fruit-setting and yield increase for apple pollination by Osmia excavata Alfken and evaluation of economic value in Shandong province. Apicul. China 70, 65–68.
Liu, L., Li, L. L., Ouyang, F., Li, C., Yu, Y., Qu, C. H., et al. (2019b). Fruit-setting, yield increase and economic value evaluation for cherry pollination by Osmia excavata Alfken in Shandong province. Shandong Agr. Sci. 51, 125–128.
Liu, L., Li, L. L., Ouyang, F., Li, L. L., Qu, C. H., Li, C., et al. (2018). Economic value of the pollination services provided by Osmia excavata Alfken to rape seed crops. Chin. J. Appl. Entomol. 55, 1016–1022.
Liu, Y. N., He, W. L., Li, Y. L., Bo, Q. F., Liang, Y., and Zhang, T. (2010). A study on the risk index design of agricultural insurance on apple florescence freezing injury in Shaanxi fruit zone. Chin. J. Agrometeorol. 31, 125–129.
Lu, X. H., Yang, Y. M., Zhu, Z. Y., Wu, Q. S., Liu, S. H., and Sun, G. M. (2013). Study on florescence characteristics of pineapple. South China Fruits 42, 66–68.
Lucas, A., Bodger, O., Brosi, B. J., Ford, C. R., Forman, D. W., Greig, C., et al. (2018). Generalisation and specialisation in hoverfly (Syrphidae) grassland pollen transport networks revealed by DNA metabarcoding. J. Animal Ecol. 87, 1008–1021. doi: 10.1111/1365-2656.12828
Men, X. Y., Li, L. L., Lu, Z. B., Ouyang, F., Liu, L., Xu, H., et al. (2018). Biological characteristics and pollination service of Mason bee. Chin. J. Appl. Entomol. 55, 973–983.
M’Gonigle, L. K., Ponisio, L. C., Cutler, K., and Kremen, C. (2016). Habitat restoration promotes pollinator persistence and colonization in intensively managed agriculture. Ecol. Appl. 25, 1557–1565. doi: 10.1890/14-1863.1
Nichols, R. N., Goulson, D., and Holland, J. M. (2019). The best wildflowers for wild bees. J. Insect Conserv. 23, 819–830. doi: 10.1007/s10841-019-00180-8
Ouyang, F., Wang, L., Yan, Z., Men, X. Y., and Ge, F. (2019). Evaluation of insect pollination and service value in China’s agricultural ecosystems. Acta Ecol. Sin. 39, 131–145. doi: 10.5846/stxb201809172030
Papanikolaou, A. D., Ingolf, K., Frenzel, M., Kuhlmann, M., Poschlod, P., Potts, S. G., et al. (2017). Wild bee and floral diversity co-vary in response to the direct and indirect impacts of land use. Ecosphere 8:e02008. doi: 10.1002/ecs2.2008
Parker, F. D. (1981). A candidate red clover pollinator Osmia coerulescens (L.). J. Apicult. Res. 20, 62–65. doi: 10.1080/00218839.1981.11100474
Piko, J., Keller, A., Geppert, C., Batáry, P., and Hass, A. L. (2021). Effects of three flower field types on bumblebees and their pollen diets. Basic Appl. Ecol. 52, 95–108. doi: 10.1016/j.baae.2021.02.005
Pornon, A., Escaravage, N., Burrus, M., Holota, H., Khimoun, A., Mariette, J., et al. (2016). Using metabarcoding to reveal and quantify plant-pollinator interactions. Sci. Rep. 6:27282. doi: 10.1038/srep27282
Potts, S. G., Imperatriz-Fonseca, V., Ngo, H. T., Báldi, A., Bartuska, A., Baste, I. A., et al. (2016). The Assessment Report on Pollinators, Pollination and Food Production: Summary for Policymakers. Bonn: IPBES.
Richardson, R. T., Lin, C. H., Sponsler, D. B., Quijia, J. O., Goodell, K., and Johnson, R. M. (2015). Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Appl. Plant Sci. 3:1400066. doi: 10.3732/apps.1400066
Ryder, J. T., Cherrill, A., Prew, R., Shaw, J., Thorbek, P., and Walters, K. F. A. (2020). Impact of enhanced Osmia bicornis (Hymenoptera: megachilidae) populations on pollination and fruit quality in commercial sweet cherry (Prunus avium L.) orchards. J. Apicul. Res. 59, 77–87.
Schloss, P., Westcott, S., Ryabin, T., Hall, J., Hartmann, M., Hollister, E., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Torchio, P. F., Asensio, E., and Thorp, R. W. (1987). Introduction of the European bee, Osmia cornuta, into California almond orchards (Hymenoptera: megachilidae). Environ. Entomol. 58, 42–52. doi: 10.1093/ee/16.3.664
Tremblay, ÉD., Duceppe, M. O., Thurston, G. B., Gagnon, M. C., Côté, M. J., and Bilodeau, G. J. (2019). High−resolution biomonitoring of plant pathogens and plant species using metabarcoding of pollen pellet contents collected from a honey bee hive. Environ. DNA 1, 155–175. doi: 10.1002/edn3.17
Wang, F. H., Guo, Z. H., Xu, X. L., and Chen, C. G. (2001). The main influence factors about retrieving of osmia and its improvement mechanism. Beijing Forest Sci. 4, 29–30. doi: 10.1201/9780367801618-7
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267.
Wang, X., Wang, J. Z., Zhao, B. Y., Cao, D., and Zhang, S. L. (2012). Research on the changes of temperature and humidity and phenophase of pear between plastic tunnel and open field. Chin. Agr. Sci. Bull. 28, 201–206.
Warzecha, D., Diekötter, T., Wolters, V., Jauker, F., Didham, R., and Batáry, P. (2018). Attractiveness of wildflower mixtures for wild bees and hoverflies depends on some key plant species. Insect Conserv. Diver. 11, 32–41.
Wei, S. G., Wang, R., Smirle, M. J., and Xu, H. L. (2002). Release of Osmia excavata and Osmia jacoti (Hymenoptera: megachilidae) for apple pollination. Can. Entomol. 134, 369–380. doi: 10.4039/Ent134369-3
Wei, S. G., Wei, S. L., Wang, R., Zhou, W. R., and Xu, Z. G. (1991). Morphological and biological study of fruit pollinator Osmia cornifrons. Kun chong Zhi shi 28, 106–108.
Wei, S. G., and Zhao, L. Y. (1995). Attention should be paid to the use of Osmia to pollinate fruit trees. Northern Fruits 2, 15–16.
Wei, Y. P., and Yuan, F. (1997). Applying trap-nesting to collection Osmia jacoti for fruit pollinating. Shaanxi J. Agr. Sci. 5, 46–47.
Wei, Y. P., Yuan, F., and Zhang, Y. L. (2000a). Flower visiting habits and the essential number of Osmia excavata Alfken for economic apple production. Acta U. Agric. Boreali Occidentalis 28, 76–79.
Wei, Y. P., Yuan, F., Zhang, Y. L., and Wang, Y. H. (2000b). The reproductive characteristics of Osmia excavata Alfken. Acta Agric. Boreali Occidentalis Sin. 3, 35–38.
Williams, N. M., and Lonsdorf, E. V. (2018). Selecting cost-effective plant mixes to support pollinators. Biol. Conserv. 217, 195–202.
Wu, J., An, J. D., Li, J. L., Huang, J. X., Guo, Z. B., and Tong, Y. M. (2006). Research and application of bumblebee in China. Proc. 7th Symposium Chin. Beekeeping Soc. Pollination Committee 11, 124–136.
Xu, H. L., Wu, Y. R., Zhou, W. R., Wei, S. G., and Wang, T. (1994). Biological Study on Pollinators of fruit trees——Osima jacoti, Osima excavate. J. Fruit Sci. 3, 153–156.
Xu, H. L., Yang, J. W., and Sun, J. R. (2009). Current status on the study of wild bee-pollinators and conservation strategies in China. Acta Phytophyl. Sin. 36, 371–376.
Yan, Z., Wang, L. N., Men, X. Y., Xiao, Y. L., Feng, G. E., and Ouyang, F. (2018). Effect of maintaining surrounding habitat near apple orchards on Osmia excavata (Hymenoptera: Megachilidae). Chinese J. Appl. Entomol. 55, 1007–1015.
Yang, L. L., Xu, H. L., and Wu, Y. R. (1997). Comparative studies of nesting and foraging behavior and pollination ecology of Osmia excavata Alfken and O. jacoti Cockerel in apple orchards (Hymenoptera: megachilidae). Acta Ecol. Sin. 17, 1–6.
Zhang, L., Yang, M. F., Liu, Y., and Ji, C. R. (2019). Effects of spring temperature change on flowering period of flat peach in shihezi. Heilongjiang Agric. Sci. 12, 42–45.
Keywords: Osmia excavata, metabarcoding method, pollen plants, diversity, species conservation
Citation: Lu H, Dou F, Hao Y, Li Y, Zhang K, Zhang H, Zhou Z, Zhu C, Huang D and Luo A (2021) Metabarcoding Analysis of Pollen Species Foraged by Osmia excavata Alfken (Hymenoptera: Megachilidae) in China. Front. Ecol. Evol. 9:730549. doi: 10.3389/fevo.2021.730549
Received: 25 June 2021; Accepted: 30 July 2021;
Published: 01 September 2021.
Edited by:
Shu-Jun Wei, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Xiangqun Yuan, Northwest A&F University, ChinaHuanli Xu, China Agricultural University, China
Xingyuan Men, Institute of Plant Protection, Shandong Academy of Agricultural Sciences, China
Copyright © 2021 Lu, Dou, Hao, Li, Zhang, Zhang, Zhou, Zhu, Huang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dunyuan Huang, MjAxNzAwNTRAY3FudS5lZHUuY24=; Arong Luo, bHVvYXJAaW96LmFjLmNu
†These authors have contributed equally to this work and share first authorship
 Huanhuan Lu
Huanhuan Lu Feiyue Dou1†
Feiyue Dou1† Chaodong Zhu
Chaodong Zhu