- 1Shanghai Key Laboratory of Forensic Medicine, Key Lab of Forensic Science, Ministry of Justice, Academy of Forensic Sciences, Shanghai, China
- 2Department of Forensic Medicine, Guizhou Medical University, Guiyang, China
- 3Department of Anthropology and Ethnology, Institute of Anthropology, School of Sociology and Anthropology, Xiamen University, Xiamen, China
- 4State Key Laboratory of Cellular Stress Biology, School of Life Sciences, Xiamen University, Xiamen, China
- 5State Key Laboratory of Marine Environmental Science, Xiamen University, Xiamen, China
- 6Institute of Artificial Intelligence, Xiamen University, Xiamen, China
Insertion-deletion (Indel) serves as one of the important markers in forensic personal identification and parentage testing, especially for cases with degraded samples. However, the genetic diversity and forensic features in ethnolinguistically diverse southwestern Chinese populations remain to be explored. Sui, one Tai-Kadai-speaking population residing in Guizhou, has a complex genetic history based on linguistic, historic, and anthropological evidence. In this study, we genotyped 30 Indels from 511 Guizhou Sui individuals and obtained approximately 700,000 genome-wide single-nucleotide polymorphisms (SNPs) in 15 representative Sui individuals to comprehensively characterize the genetic diversity, forensic characteristics, and genomic landscape of Guizhou Sui people. The estimated forensic statistically allele frequency spectrum and parameters demonstrated that this Indels panel was polymorphic and informative in Tai-Kadai populations in southern China. Results of principal component analysis (PCA), STRUCTURE, and phylogenetic trees showed that Guizhou Sui had a close genetic relationship with geographically close Tai-Kadai and Hmong-Mien people. Furthermore, genomic analysis based on the Fst and f4-statistics further suggested the genetic affinity within southern Chinese Tai-Kadai-speaking populations and a close relationship with geographically adjoining Guizhou populations. Admixture models based on the ADMIXTURE, f4, three-way qpAdm, and ALDER results demonstrated the interaction between the common ancestor for Tai-Kadai/Austronesian, Hmong-Mien, and Austroasiatic speaking populations played a significant role in the formation of modern Tai-Kadai people. We observed a sex-biased influence in Sui people by finding that the dominant Y chromosomal type was a Hmong-Mien specific lineage O2a2a1a2a1a2-N5 but the mtDNA lineages were commonly found in Tai-Kadai populations. The additional southward expansion of millet farmers in the Yellow River Basin has impacted the gene pool of southern populations including Tai-Kadai. The whole-genome sequencing in the future will shed more light on the finer genetic profile of Guizhou populations.
Introduction
Southwest China has always been regarded as a multi-ethnolinguistic region, comprising diverse language families, namely, Sino-Tibetan, Tai-Kadai, Hmong-Mien, and Austroasiatic. There was evidence that southwest China was characterized by significantly different genetic lineages from southern and northern East Asia (represented by Qihe and Bianbian, respectively) (Wang T. et al., 2021). The population continuity from earlier Longlin and bidirectional gene flow between southwest and southeast China played a formative role in the demographic history of this region. Furthermore, southwest China was a vital channel of population migration between Southeast Asia and southern China since the Neolithic Age. The ancestry related to Hoabinhian hunter-gatherers extending from Southeast Asia into southern China was detected in Neolithic Baojianshan samples, while Mainland Southeast Asians from Neolithic Age and Bronze Age showed a genetic affinity with modern southern Chinese Tai-Kadai and Hmong-Mien people (Lipson et al., 2018; Mccoll et al., 2018; Wang T. et al., 2021). Besides, the genetic makeup of ancient and modern southern Chinese was influenced by the southward migration of northern millet farming populations throughout history (Wen et al., 2004; Wang et al., 2014, 2022; Wang C. C. et al., 2021; Wang T. et al., 2021; Yao et al., 2017; Guo et al., 2019b; 2022; Xu et al., 2019; Liu Y. et al., 2020; Ning et al., 2020; Yang et al., 2020; He et al., 2021).
The Sui people in southwest China belonging to the Tai-Kadai language family has a unique and ancient culture–“Sui Shu”. However, the origin of the Sui people is relatively controversial currently, and there are several hypotheses as follows: (1) Sui is an ancient indigenous group in southern China for 6,000 years; (2) Sui is a branch of Baiyue people who had inhabited southern China before the Han dynasty; (3) Sui originated from the Sui River Basin, gradually migrated to southern China, and integrated into the Baiyue tribes. In summary, the origin of the Sui population is still in dispute and needs to be further investigated.
The resolution of previous studies focused on the Tai-Kadai populations including Sui people was quite limited due to the low density of genetic markers used: the study analyzed 15 autosomal STRs of 2,022 unrelated individuals from five ethnic groups including Sui, Han, Gelao, Jing, and Zhuang from Guangxi and Guangdong and found that Zhuang and Gelao were genetically closer with Han when comparing with Sui and Jing people (Yang et al., 2013). A study using 23 Y-STR loci of 200 unrelated Sui individuals from Guizhou showed that Sui people were genetically distantly related to northern Chinese populations such as northern Han, Manchu, Hui, Mongolian, Japanese, and Koreans (Ji et al., 2017). The analysis by 19 X-STRs of 400 unrelated individuals from Guizhou Sui suggested that the Sui had greater genetic similarity with the Tai-Kadai-speaking populations and southern Han Chinese (Guo et al., 2019a). From the analyses of uniparental lineages, Sui possessed high frequencies of southern Chinese specific paternal Y-chromosome lineage O1a-M119 and also maternal mitochondrial haplogroups, such as B, M7, F, and R (Li et al., 2007, 2008). Bin et al. (2021) reported genome-wide SNPs of 68 Sui individuals from seven indigenous populations in Guizhou Province and Guangxi in Southwest China and found that Sui people showed a strong genetic affinity with populations from southern China and Southeast Asia, especially Tai-Kadai and Hmong-Mien-speaking populations, which supported the hypothesis that Sui people came from southern China originally.
Although several reports explored the genetic relationship of Guizhou Sui by autosomal and X chromosomal STRs, STR may not be the best marker for population genetic structure, origin, and immigration analysis due to the high mutation rate of 10–3 to 10–2 (Slooten and Ricciardi, 2013). Insertion and deletions (Indels) are abundant in the human genome with a low mutation rate and are beginning to be used in forensic, anthropological, and population genetic studies (Mills et al., 2006). The Investigator® DIPplex kit with 30 InDels is a sophisticated assay that is used to investigate the allele frequency distribution among the worldwide ethnically and linguistically diverse populations in a large number of genetic studies (Turrina et al., 2011). However, only a few studies reported these data from Chinese ethnic groups.
In summary, the Guizhou Sui population is an ancient ethnic group with a unique culture. However, the available genetic information on Sui people is largely limited to only autosomal and X chromosomal STRs (Ji et al., 2017; Guo et al., 2019a). Therefore, we genotyped 30 InDels from 511 Guizhou Sui unrelated individuals and 700,000 genome-wide SNPs from 15 Sui individuals to thoroughly explore the genetic origin, admixture history, and forensic characteristics of the Guizhou Sui population.
Materials and Methods
Sample Collection, DNA Extraction, and Quantification
A total of 511 blood samples were collected using the FTA card, and 15 saliva samples were collected using the DNA Genotek Kit (WeGene, Shenzhen, China) from unrelated individuals of the Guizhou Sui population with written informed consent, and the Geographical distribution of Guizhou Sui was shown in Supplementary Figure 1A. Our study and sample collection were reviewed and approved by the Medical Ethics Committee of Guizhou Medical University and followed the recommendations provided by the revised Helsinki Declaration of 2000. The participants included are all with official Sui recognition and non-consanguineous marriages over three generations. Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany), and bloodstain samples were extracted using the Chelex-100 (Walsh et al., 1991), quantified using the Nanodrop-2000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States).
PCR Amplification, Capillary Electrophoresis, and Array Genotyping
A total of 30 InDel markers were amplified on the GeneAmp PCR System 9700 Thermal Cycler (Thermo Fisher Scientific, Wilmington, MA, United States) using the Investigator DIPplex Commercial Kit (Qiagen, Hilden, Germany) and 10 μl reaction volume, which included 0.24 μl multi Taq2 DNA polymerase, 1.0 μl template DNA, 2 μl primer mix, and 2 μl reaction mix, appropriated ddH2O. PCR condition was followed by 4 min at 94 °C, 30 cycles of 94 °C for the 30 s, 61 °C for 120 s, and 72 °C for 75 s, and a final extension at 68 °C for 60 min. The PCR product was separated on Applied Biosystems 3500 Genetic Analyzer (Thermo Fisher Scientific, Wilmington, MA, United States) with HiDi Formamide (Thermo Fisher Scientific, Wilmington, MA, United States) and DNA size standard 550 (BTO) (Qiagen, Hilden, Germany) and genotyped according to GeneMapper ID-X version 1.3 software (Thermo Fisher Scientific, Wilmington, MA, United States). The ddH2O and 9948 (Promega, Madison, WI, United States) were used as negative control and positive control, respectively. A total of 690,193 SNPs were genotyped on the Illumina WeGene Arrays at the WeGene genotyping center, Shenzhen.
Reference Datasets and Data Merging
All the allelic frequency data of 30 InDel markers from 100 populations, including African, (Northern and Southern) American, European, and Asian, were collected as the dataset f. Since the genotype data could not be found in some populations in their reports, we sorted out the genotype data from 52 populations as the dataset g, including an African, 3 European, 7 American, 41 Asian populations, and Guizhou Sui (Fondevila et al., 2012; Friis et al., 2012; Kis et al., 2012; Larue et al., 2012; Akhteruzzaman et al., 2013; Carvalho and Pinheiro, 2013; da silva et al., 2013; Martin et al., 2013; Kim et al., 2014; Ferreira Palha et al., 2015; Hefke et al., 2015; Meng et al., 2015; Guo et al., 2016, 2018; Martinez-Cortes et al., 2016; Mei et al., 2016; Du et al., 2017, 2019; Kong et al., 2017; Li et al., 2018, 2019; Ma et al., 2018; Chen et al., 2019; He et al., 2019a,c; Jian et al., 2019; Liu Y. et al., 2020). The genome-wide SNP data of the Sui population were merged with the reference dataset including 1240 k, Human Origin, and so on (Patterson et al., 2012; Lipson et al., 2018; Mccoll et al., 2018; Liu D. et al., 2020; Ning et al., 2020; Yang et al., 2020; Wang C. C. et al., 2021) to generate two combined datasets covering 189,177 and 71,989 SNPs for subsequent analyses. The 1240 k harbored more SNPs than the Human Origin dataset; nevertheless, the Human Origin dataset included more reference peoples than 1240 k, such as Maonan, Tibetan, Dong_Guizhou, Han, and others from China.
Statistical Analysis
Allelic frequency distribution and corresponding forensic statistical parameters of 30 Indels including the power of discrimination (PD), probability of exclusion (PE), probability of match (PM), and the polymorphism information content (PIC) were calculated using the online tool of STRAF (Gouy and Zieger, 2017) (STR analysis for forensics). Linkage disequilibrium (LD), Hardy–Weinberg equilibrium (HWE), observed heterozygosity (Ho), and expected heterozygosity (He) were analyzed using Arlequin software version 3.5 (Excoffier and Lischer, 2010). Nei’s and DA genetic distance between Guizhou Sui and other 99 reference populations were calculated according to PHYLIP software version 3.52 (Shimada and Nishida, 2017) and the DISPAN program based on the dataset f. The principal component analyses (PCAs) were performed using MVSP software version 3.22 (Kovach, 2007) among the 100 populations, and the results were shown by the heat map and scatter diagram according to R Statistical software version 3.0.2. The phylogenetic tree was conducted based on DA genetic distance using a neighbor-joining algorithm implemented in Mega 7.0 (Kumar et al., 2016). The analysis of ancestral components for individuals in each population was performed using STRUCTURE version 2.3.4.21 (Evanno et al., 2005) based on the dataset g, with the parameters of running 15 replicates from K = 2 to K = 8 with 10,000 burn-ins and 10,000 MCMC.
Genomic-Based Statistical Analysis
We performed the population genetic analysis including PCA (Patterson et al., 2006) and ADMIXTURE after filtering strong LD by Plink with parameters “–indep-200 25 0.4” (Alexander et al., 2009; Chang et al., 2015), f statistics (Patterson et al., 2012), qpWave, and qpAdm (Patterson et al., 2012; Haak et al., 2015). We calculated outgroup-f3(Sui_Sandu, Populations; Mbuti) to measure the shared genetic drifts between the Sui population and reference groups and computed admixture-f3-statistics in the form of f3(Source1, Source2; Sui_Sandu populations) using the qp3pop program with default parameters in ADMIXTOOLS to explore the potential existing admixture signals. We further estimated the f4-statistics values in the forms of f4(Mbuti, X; Sui_Sandu, Tai-Kadai/Guizhou) and f4(ancient1, ancient2; Sui_Sandu, Mbuti) using the qpDstat program to study genetic affinity, continuity, and admixture using the default block jackknife. We estimated that pairwise genetic distance by Fst using the smartpca program of EIGENSOFT with fstonly was YES and inbreed was YES parameter. Then, we also inferred a rooted maximum likelihood tree using TreeMix software (Pickrell and Pritchard, 2012). We used qpWave as implemented in ADMIXTOOLS to explore the minimum number of potential ancestral sources of the Sui population and used qpAdm to quantify the proportions of ancestral populations. We finally used ALDER (Loh et al., 2013) to estimate the time of admixtures with 28 years as one generation length.
Results
Genetic Diversity, Allele Frequencies, and Forensic Efficiency Parameters Based on the 30 InDels
The genotypes of 30 InDels are all listed in Supplementary Table 1. The results of HWE analysis showed that after Bonferroni correction (p < 0.05/30), 30 InDels were all in HWE. The deeper the red color in LD analysis represents the stronger the linkage degree (Supplementary Figure 1B). No sites were found with LD when considering r2 > 0.8 as the threshold. When using the p-value to measure the LD, the result showed no LD between every two loci after Bonferroni correction (p < 0.05/30) (Supplementary Table 2).
The insertion allelic frequencies ranged from 0.0939 (HLD39) to 0.9207 (HLD118) (Supplementary Table 3). We found that the values of forensic parameters HO, PD, PE, PIC, and TPI of loci HLD118, HLD64, HLD39, HLD99, HLD111, HLD84, HLD122, HLD114, HLD81, and HLD67 were lower than that of other InDel sites, indicating a low level of genetic diversity in these loci in Guizhou Sui population, which is similar to other Asian ethnic groups reported by previous studies (Guo et al., 2018; He et al., 2019c; Zhang et al., 2019). The combined power of discrimination (CPD) of 30 InDels was 0.999999999983, suggesting this assay could satisfy the requirement of individual identification for the Guizhou Sui population, while the cumulative probability of exclusion (CPE) was only 0.9986, indicating that those 30 InDels could only be used as a supplement system for parentage testing.
Genetic Structure and Genetic Affinity Explorations Among 99 Worldwide Populations via the DIPplex System
We collected the frequency data of the 30 InDels of the Investigator® DIPplex system from 99 published global reference populations (Supplementary Figure 2). These populations were mainly divided into four branches, namely, the African populations, the East Asian populations, the American populations, and the European populations, demonstrating that these InDels showed excellent population differentiation power among different intercontinental populations. The 30 InDels were roughly clustered into three branches: cluster A contained six loci HLD118, HLD67, HLD84, HLD99, HLD64, and HLD81, while cluster B included five loci HLD114, HLD128, HLD111, HLD39, and HLD122, and the remaining loci were clustered into a large branch C. The frequencies of the five loci included in branch B showed the most significant differences among these 100 populations, which should be more advantageous to be used as ancestral information markers for inferring ethnic groups. Guizhou Sui clustered with the Tai-Kadai and Hmong-Mien speaking populations in southern China, and its allelic frequencies showed no significant difference from other East Asian ethnic groups.
To investigate the genetic relationships between Guizhou Sui and other reference populations, we performed a genetic analysis based on 30 InDel markers. The PCA analysis based on the frequency data of 30 InDels of 100 populations in Dataset f showed that populations from each continent clustered together and Sui clustered with East Asian populations (Figure 1A). When removing European, African, American, and South Asian populations, three genetic clusters of East Asian populations were observed as follows: high-altitude Tibetans, low-altitude northern populations, and southern populations. Sui samples clustered with the Tai-Kadai populations and were most closely to Gelao_Guangxi, Zhuang_Guangxi, and Vietnamese belonging to Austroasiatic people (Figure 1B). We then calculated the DA and Nei’s genetic distance (Supplementary Tables 4, 5 and Supplementary Figures 3, 4) and constructed a phylogenetic tree (Figure 2). We found that population clustering was consistent with geographic and linguistic similarity, and Sui had the closest genetic relationship with other Tai-Kadai-speaking groups and geographical neighbors.
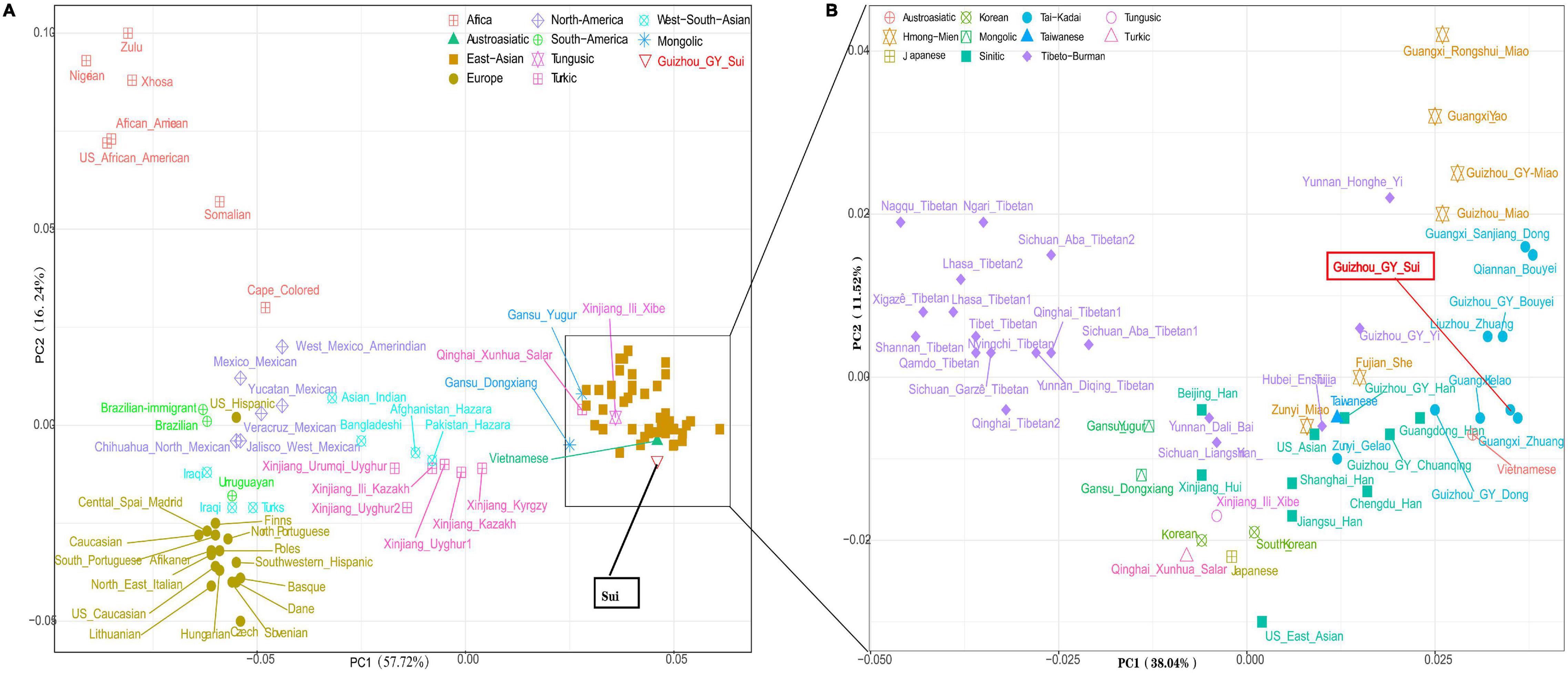
Figure 1. The patterns of genetic relationship between Guizhou Sui and global reference populations based on the InDel low-density dataset. (A) The first two components of principal component analysis (PCA) results between the Guizhou Sui and 99 worldwide reference populations. (B) PCA-based East Asian populations showed the closer genetic Sui and other Tai-Kadai.
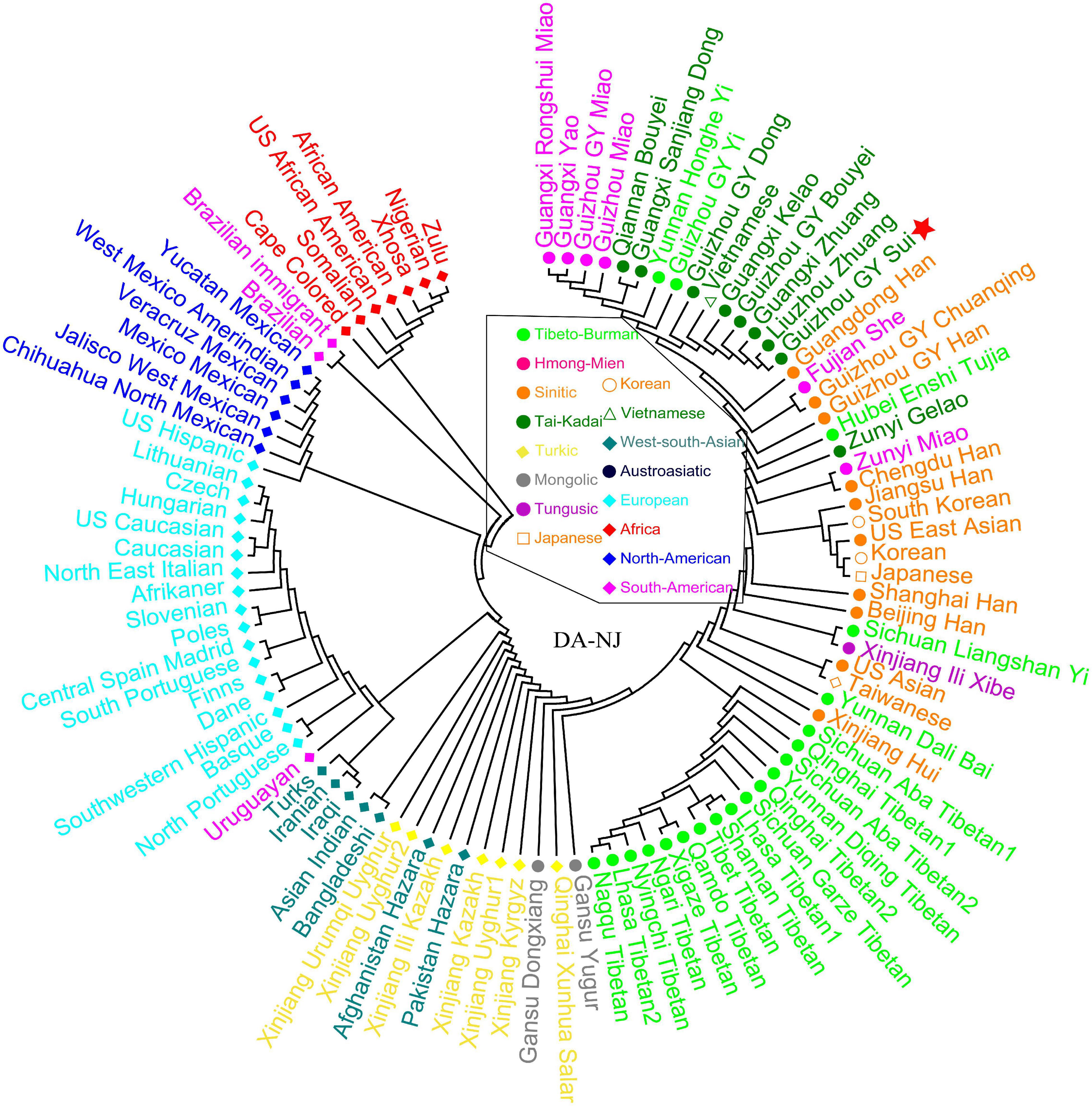
Figure 2. Neighbor-joining polygenetic tree constructed on the DA genetic distance among Guizhou Sui and 99 worldwide reference populations.
We further performed ancestral component analysis using STRUCTURE (Supplementary Figure 5) according to the model-based algorithm and hypothetic ancestry source populations from 2 to 8 using a dataset with 30 InDel genotype data of Guizhou Sui and 51 populations from different continents. We observed the lowest CV errors at K = 4 with four ancestral components in East Asian populations. Most East Asians possessed two ancestral components except for Hui, Kazakh, Dongxiang, and Yugur due to West Eurasian admixture (Wang C. C. et al., 2021). There was a subtle difference in ancestral component composition between Tai-Kadai and Hmong-Mien speaking populations. The ancestral component composition of the Guizhou Sui was similar to Hmong-Mien speakers such as Miao, Yao, and She and some Tai-Kadai-speaking groups such as Gelao_Guangxi and Zhuang.
Population Genomic Analyses Revealed the Genetic Affinity of Sui Based on Genome-Wide Data
We generated genome-wide data from 15 Sui individuals, and first conducted PCA based on genome-wide data to explore the population genetic structure across East and Southeast Asia. We projected published ancient individuals onto PC plots (Figure 3A). Tai-Kadai-speaking populations clustered at the southern end of the south-north cline and overlapped with some Austronesian individuals. Ancient populations of southern China, such as Late Neolithic Xitoucun samples from Fujian and Iron Age Hanben samples from Taiwan, partly overlapped with the modern Tai-Kadai populations, documenting a long-term genetic continuity. In addition, mainland Southeast Asians from Bronze Age ancient populations to modern Vietnamese such as Muong/Kinh also had a closer genetic relationship with Tai-Kadai speakers. The 15 Sui individuals clustered tightly with other Tai-Kadai-speaking populations.
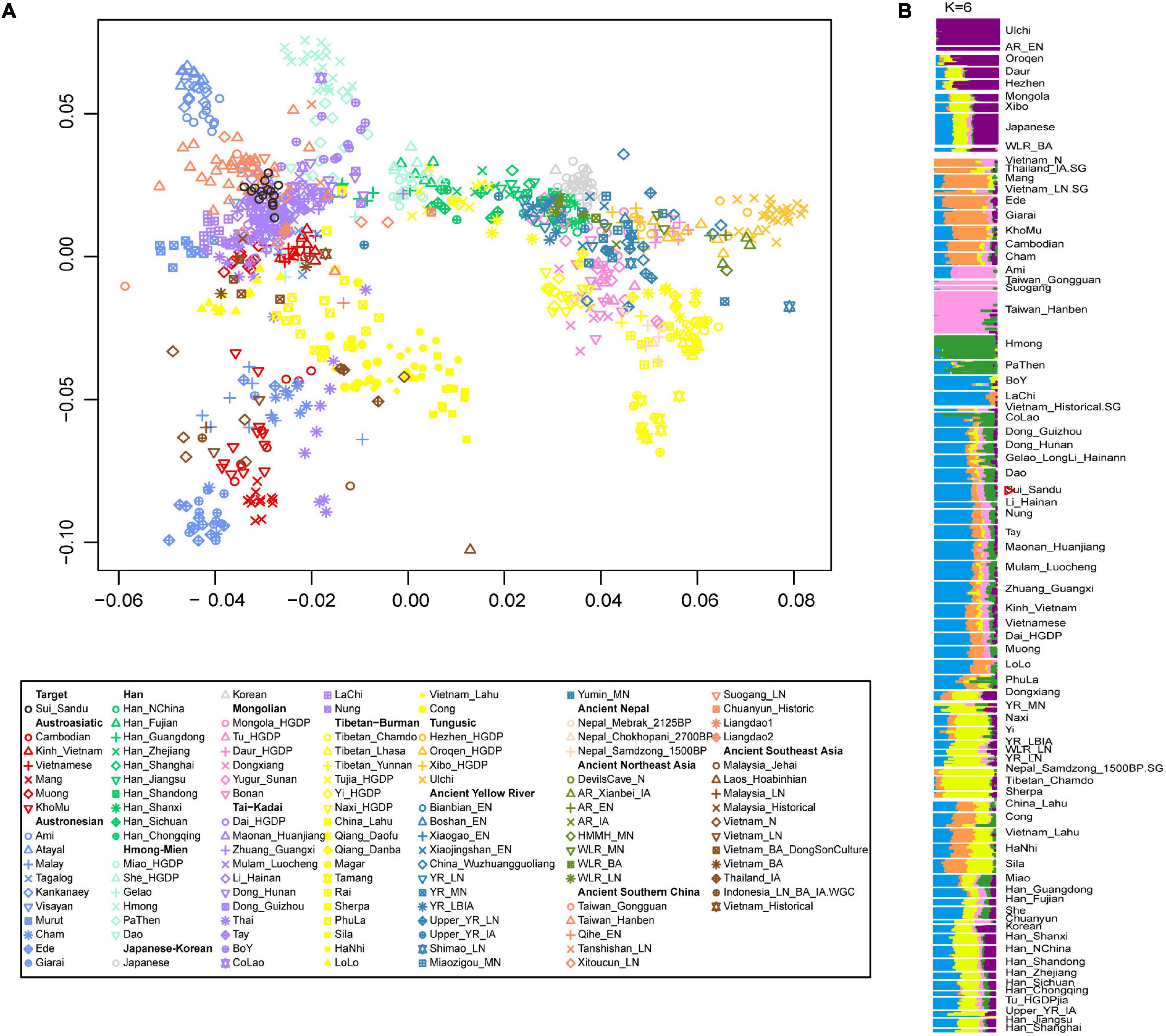
Figure 3. The population structure of modern and ancient populations in Asia based on genome-wide data. (A) PCA results showed an overview population relationship between modern populations and ancient populations. (B) ADMIXTURE results (the lowest CV errors K = 6): ancestral components among Guizhou Sui and modern and ancient populations in Asia.
The outgroup f3 statistics (Figure 4) showed that the Sui population had the closest genetic affinity with Tai-Kadai-speaking groups and also shared more genetic drift with Hmong-Mien groups in Vietnam and with some Austronesian groups such as Ami and Kankanaey. When compared with ancient populations, Sui shared most alleles with ancient Taiwanese and Southeast Asians such as Malaysia_Historical, Vietnam_BA_NuiNap, and Neolithic Vanuatu (Lipson et al., 2020), followed by Yellow River farming populations from the Late Neolithic to Iron Age. The phylogenetic relationships between the studied Sui population and modern East and Southeast Asian populations were further confirmed by a Treemix-based phylogenetic tree (Supplementary Figure 6). We observed southern populations including Austronesian, Austroasiatic, Hmong-Mien, and Tai-Kadai speaking groups clustered together as a southern branch. Sui first clustered with Maonan of Guangxi in China and then with Nung of Vietnam.
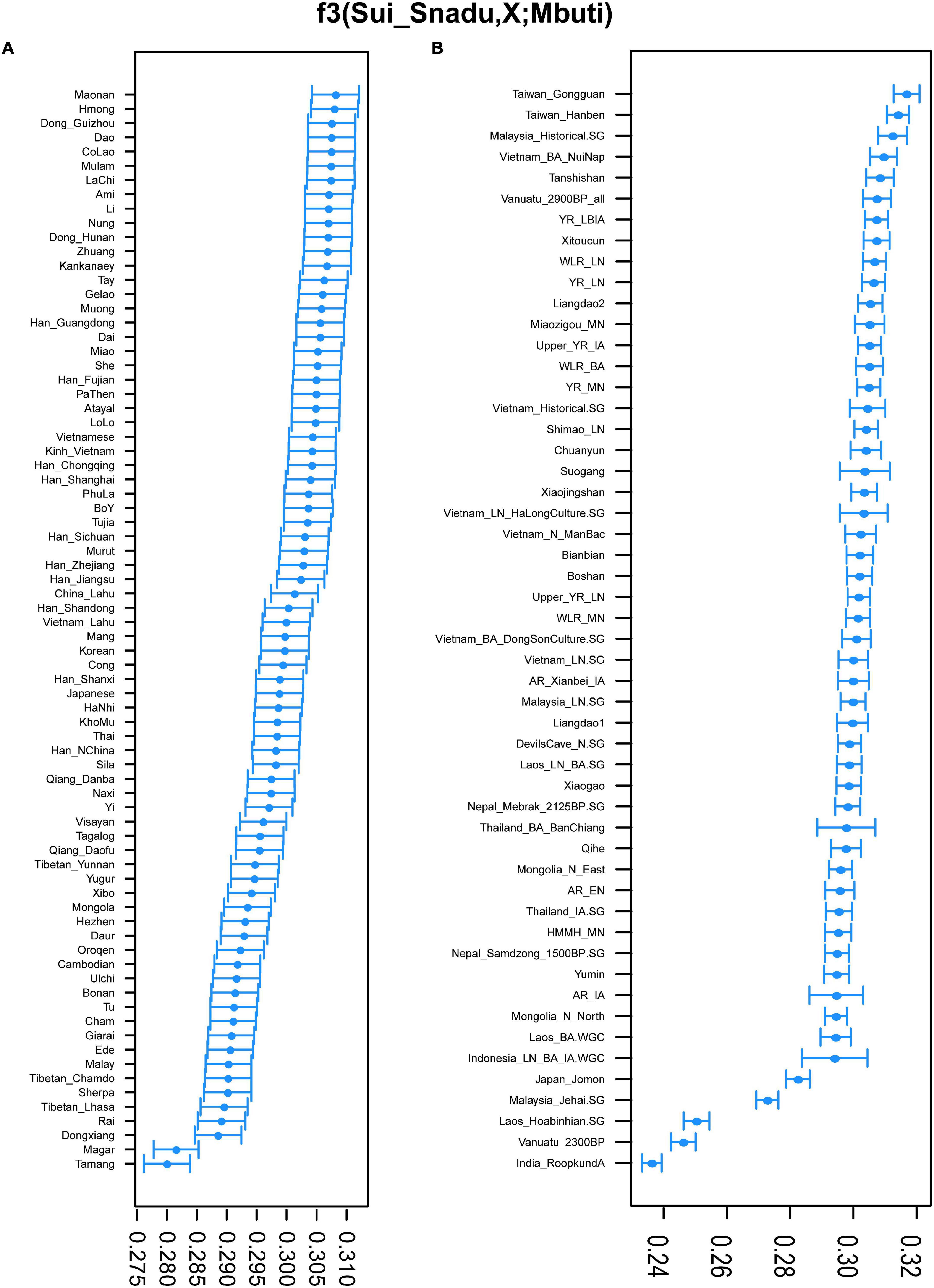
Figure 4. The shared genetic drift between Sui and modern/ancient populations in Asia showed a closer affinity of Sui and Tai-Kadai/Hmong-Mien populations.
We also conducted Wright’s fixation index Fst among 87 modern and ancient populations (Supplementary Table 7), and Sui possessed the smallest genetic distance with Zhuang and Dong_Guizhou (Fst = 0.003), followed by Dong_Hunan and Maonan (Fst = 0.004). Overall, Sui showed a close genetic relationship first with Tai-Kadai-speaking groups and then with Austroasiatic-speaking groups such as Kinh, Muong, Vietnamese, and southern Han. The closer genetic affinity of Kinh/Vietnamese/southern Han with Tai-Kadai groups was also shown in other Chinese Tai-Kadai speakers, indicating genetic influence between Tai-Kadai groups and Vietnamese Austroasiatic groups and southern Han. We also found a smaller genetic distance between Sui and ancient millet farming populations in the Yellow River basin from Late Neolithic Age onward.
We further performed four population statistics to study the genetic relationship of Sui_Sandu with other Tai-Kadai-speaking groups in the form of f4(Mbuti, X; Sui_Sandu, CoLao/Dai/Dong_Guizhou/LaChi/Li/Maonan/Mulam/Zhuang) (Supplementary Table 7). Sui showed relatively genetic homogeneity with other Tai-Kadai populations as reflected in non-significant f statistics (|Z| < 2), except for Dai who was genetically influenced by Austroasiatic speakers. Notably, we observed significant allele sharing with Hmong in Sui_Sandu as reflected in significantly negative statistics in the form of f4(Mbuti, Hmong; Sui_Sandu, LaChi/Li/Maonan/Mulam/Zhuang). When the Z-value was less than negative 3 in the form f4(Hmong-Mien/Austronesians/Austroasiatic, Hmong-Mien/Austronesians/Austroasiatic; Sui_Sandu, Mbuti) (Figure 5), the levels of allele sharing with Sui were roughly similar in the Hmong-Mien population, Austronesians in and adjacent to Taiwan including Ami, Atayal, and Kankanaey, and Austroasiatic speaking including Kinh, Vietnamese, and Muong. In ancient populations from the Neolithic to Iron Age, we found that later populations related to Austronesians in Taiwan shared more alleles with Sui than the earlier populations in southern China, and we observed the same pattern in using ancient Southeast Asians and northern populations in the form f4(Taiwan_Hanben/Taiwan_Gongguan, ancients; Sui_Sandu, Mbuti) with the Z-value was more than 3 (Supplementary Figure 6). Millet farmers in the Yellow River Basin since Late Neolithic also showed a closer genetic affinity with Sui in the form f4(YR_LN/YR_LBIA, ancients; Sui_Sandu, Mbuti) with the Z-value was more than 3. Late Neolithic southern Chinese populations and Southeast Asian populations since the Neolithic Age shared roughly equal levels of genetic affinity with Sui in the formf4 (Xitoucun/Tanshishan, Neolithic/Bronze Age/Historical Southeast Asian population; Sui_Sandu, Mbuti) with the Z-value was less than 2 (Supplementary Figure 7). When the Z-value was less than negative 2 in the form (f4(Laos_Hoabinhian, Vietnam_N_ManBac/Vietnam_LN/Vietnam_BA_NuiNap/Vietna m_BA_DongSonCulture/Vietnam_LN_HaLong Culture; Sui_ Sandu, Mbuti), and f4(Vietnam_N_ManBac, Vietnam_LN; Sui_Sandu, Mbuti) (Supplementary Figure 6), these results suggesting the genetic influx from Sui flowed into Southeast Asia could be detected in Neolithic Vietnamese. Furthermore, we explored the genetic relationship between the Sui and other populations in Guizhou by f4 statistics in form of f4(Mbuti, X; Sui_Sandu, Manchu/Mongolian/Dong) (Supplementary Table 8). The result suggested that Sui showed a similar genetic affinity pattern with Dong as mentioned above, and Sui had a significantly different affinity pattern with Tungusic and Mongolic speaking groups in Guizhou since Sui shared more alleles with southern populations compared with Manchu and Mongolian.
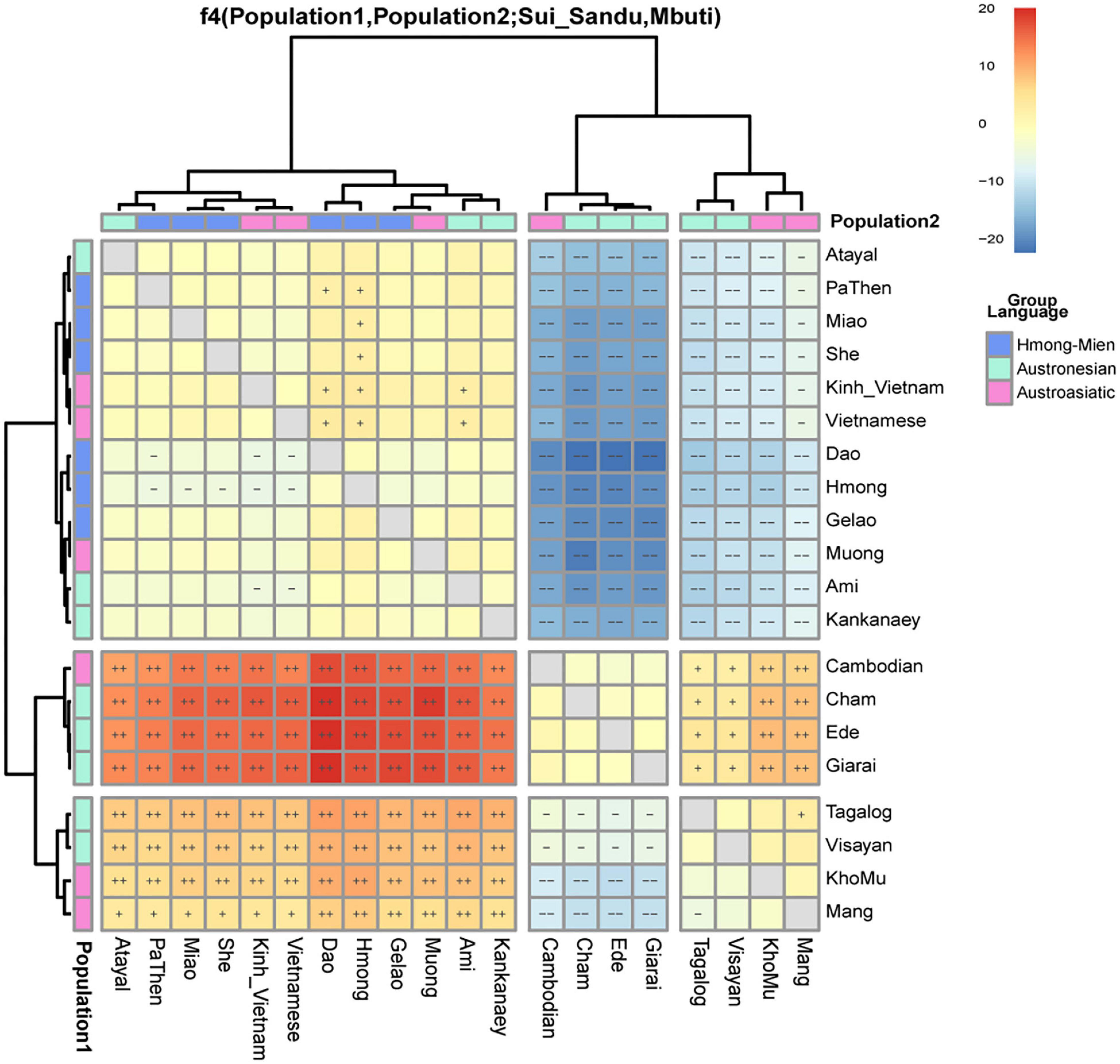
Figure 5. Genetic affinity between Guizhou Sui and southern populations shown in f4(population1, population2; Sui_Sandu, Mbuti). Significant Z scores were labeled by “++” “– –” (Z > 6, Z < –6, respectively), and Z-values with 3 < Z < 6, –6 < Z < –3 were labeled by “+,” “–,” respectively.
Ancestral Origins and Genetic History Reconstruction of Sui From Modern/Ancient DNA Perspectives
We co-analyzed Sui samples with modern and ancient East and Southeast Asians using ADMIXTURE to infer the population clustering patterns (Figure 3B). We observed that the Sui population harbored four main ancestral components. The first dominant ancestral component was maximized in Lachi/Boy which were the Sino-Tibetan group, the second ancestral component was enriched in Hmong-Mien speaking groups represented by Hmong, the third component was related to Austroasiatic-speaking populations represented by Malaysia_LN, and the fourth component was related to Austronesian-speaking populations represented by Taiwan_Hanben. We also detected a small proportion of northern ancestral components represented by ancient samples from Nepal and Yellow River Basin, present-day Ulchi from northeast Asia. Interestingly, the genetic composition of some Hmong-Mien groups was similar to that of Tai-Kadai populations, such as Dao, which was consistent with previous studies that showed there have been extensive interactions between the Hmong-Mien and Tai-Kadai speaking groups (Liu Y. et al., 2020; Macholdt et al., 2020).
To find out the potential genetic sources of Sui_Sandu, we then performed f3 statistics in the form of f3 (source1, source2; Sui_Sandu) using 147 populations from Asia including 91 modern populations and 46 ancient populations (Supplementary Table 9). After excluding ancient source pairs with overlapping SNPs of less than 1,000 (a total of 139 samples), we just found 179 source pairs performing admixture signals when the f3 value was negative (−2.2 < Z < 0). But the numbers of overlapping SNPs in generating those negative results were small, implying that there might be no genetic admixture in the Sui population in recent times due to the geographic isolation, or it is a false-negative signal caused by the strong genetic drift of Sui.
The Z-value was less than negative 2 in the form f4(Mbuti, YR_LN; Sui_Sandu, Southern ancient Chinese/Southeast Asian), and the Z-value was less than negative 3 in the form f4(Mbuti, Taiwan_Hanben; Sui_Sandu, YR/Northeast Asian/Southeast Asian/Nepal)) and f4(Mbuti, ancient Southeast Asian; Sui_Sandu, YR/Northeast Asian), indicating that the allele frequencies of Sui people were in-between those of northern and southern Chinese populations (Supplementary Table 10). To quantitatively explore the plausible admixture models of Sui and other Tai-Kadai groups and estimate the corresponding ancestral proportion, we used qpWave/qpAdm statistics to infer the minimum number of ancestry and proportion estimation (Supplementary Table 11). The qpWave results showed that the best two-way or three-way admixture models could be used to explain the genetic formation of the Sui people (χ2 p > 0.05). The Tai-Kadai populations could be modeled as the admixture of southern East Asian and northern East Asian populations. We used qpAdm to model the proportions of potential ancestral sources. We found that Tai-Kadai populations descended from Atayal-related ancestry spanning from ∼61.4 to ∼85% and Late Neolithic millet farmers from Yellow River belonging to Longshan Culture-related ancestry spanning ∼38.6–∼15% (0.045 < χ2 p < 0.19), which was in accordance with the historical record of the southward expansion of agriculturists from the Yellow River Basin. Sui_Sandu was suggested to harbor ∼75.2% Atayal-related ancestry and ∼34.8% YN_LN-related ancestry (χ2 p = 0.0498197467). A more complex three-way model of YR_LN + Atayal + Vietnam_LN fitted all populations (χ2 p > 0.05) but with a large standard error for Sui_Sandu, Maonan, and Dong_Guizhou. When we replaced ancient Southeast Asians with modern Vietnamese Austroasiatic speakers by Mang, Tai-Kadai populations could be modeled as 9.4–34.5% YR_LN + 56.7–74.5% Atayal + 8.8–19.5% Mang, suggesting that Southeast Asian-related ancestry contributed to the gene pool of Tai-Kadai populations. Therefore, we inferred that the interaction of rice farmers in the Yangtze River and earlier Austroasiatic populations and millet farmers in the Yellow River Basin influenced the formation of Tai-Kadai.
We further used the ALDER method based on weighted LD statistics to estimate the date when northern, southern Chinese, or Southeast Asian-related ancestry was introduced into the gene pool of the Sui population. We explored a total of 1,539 pairwise two-reference groups and found that two pairs give a reasonable admixture date of Guizhou Sui (Supplementary Table 12). Our results showed the time for Southeast Asian admixture in Sui spanning from 49.65 ± 23.78 generations ago corresponding to 1,390 ± 666 BP in the Hmong-Ede model to 42.59 ± 12.46 generations ago corresponding to 1,193 ± 349 BP in the Hmong-Cambodian model. We also used one-reference mode and detected more recent admixture events in Sui including the northern admixture at approximately ∼31.7 ± 17.0 generations using Dongxiang as a single reference and the admixture with other Tai-Kadai speakers at approximately 79.4 ± 20.3 generations ago using Li as a single reference and 87.6 ± 11.6 generations ago using Dai as a single reference.
The Uniparental Genetic Profile of Guizhou Sui
We obtained uniparental Y chromosomal and mtDNA haplotypes for 15 individuals, of whom 7 were men (Supplementary Table 13). From the maternal perspective, Sui individuals had 10 mitochondrial haplotypes with frequencies ranging from 0.067 in F3a1 to 0.2 in M7b1a1a3, in which M7b was the dominant lineage. The maternal profile was similar to that of the Tai-Kadai populations in southern China, but we also detected a northwestern lineage D4 in Sui samples (Li et al., 2007).
We found 3 different paternal Y chromosomal lineages with frequencies ranging from 0.143 of O1a1a1b2-Z23404 and C2c-F9851 to 0.714 of O2a2a1a2a1a2-N5 in Sui samples. The haplogroup O2a2a1a2a1a2-N5 is a lineage prevalently found in Hmong-Mien-speaking populations (Xia et al., 2019), documenting the paternal genetic affinity of Sui and Hmong-Mien people.
Discussion
Guizhou is not only a multiethnic province located in the hinterland of southwest China but also a multi-language region comprising Hmong-Mien, Tai-Kadai, and Sinitic-Tibetan languages. The region was regarded as an indispensable resource for exploring genetic diversity, origin, migration, and mixing of various ethnic groups in China. However, the genetic origin, admixture, and forensic characteristics of Tai-Kadai are still unclear and need to be comprehensively explored. Therefore, we provided 30 InDels from 511 unrelated individuals of Sui in Guizhou and also generated genome-wide SNP data and merged with all publicly available modern and ancient genomes to reconstruct the Sui population history of Guizhou in southwest China and shed light on the formation of the people.
First, we observed that the majority of 30 InDel loci were highly polymorphic in the Guizhou Sui population and can be used for individual identification in Chinese populations, but we also noted that the following 11 low polymorphic loci: HLD118, HLD64, HLD39, HLD99, HLD111, HLD84, HLD122, HLD114, HLD81, and HLD6710, were not recommended to be included in the InDel system for forensic use in Chinese populations. In addition, the InDel system can be widely used as a powerful tool for individual identification based on CPD and CPE, but could only be used as a supplement for paternity testing due to the low CPE.
The origin of Sui has been relatively controversial since the description of historical documents was indistinct. What is more, the genetic relationship and admixture history of the Sui population are not well studied due to the sparse sampling. In this study, we first used forensic high polymorphic and informative InDel markers to explore the genetic relationship between Sui and East Asian populations. We found that the allelic frequencies of 30 InDels showed significant differences among intercontinental populations and the genetic markers’ frequency characteristics in Sui people were similar to other Chinese groups. Sui had a close genetic relationship with the Tai-Kadai-speaking groups in the surrounding area. Second, through the comprehensive genome-wide SNP analysis, our findings clearly showed that the genetic formation of Sui people has been influenced by surrounding populations, including Tibetan-Burman, Hmong-Mien, Austronesian, and Austroasiatic speaking groups (He et al., 2020). We observed that the Tai-Kadai populations have the closest genetic affinity with Hmong-Mien people. The extensive interaction between Tai-Kadai and Hmong-Mien has also been shown in the analyses of uniparental and InDel markers and also in linguistics (Chen et al., 2007, 2018; Blench, 2017; He et al., 2019b; Huang et al., 2020; Macholdt et al., 2020). According to linguistic information and historic documents, Hmong-Mien and Tai-Kadai language families both originated in southern China and then expanded southward into mainland Southeast Asia approximately two millenniums ago (Edmondson and Gregerson, 2007; Bellwood and Ness, 2015). We also detected the gene flow from Tai-Kadai-related populations into Southeast Asians since the Neolithic Age in the negative f4(Laos_Hoabinhian, Neolithic to present-day Southeast Asians; Sui_Sandu, Mbuti), which is consistent with previous genomic studies (Lipson et al., 2018; Huang et al., 2020; Liu D. et al., 2020). We found that the Tai-Kadai people had a high proportion of Austroasiatic-related ancestry at a proportion of 8.8–19.5% using Mang as the genetic source, indicating the prosperous communications between ancient Tai-Kadai and Austroasiatic populations. The admixture between Tai-Kadai, Hmong-Mien, and Austronesian/Austroasiatic was consistent with the bidirectional interactions between populations in southwest China and Southeast Asia. The admixture times among ancient Tai-Kadai, Austronesian, and Austroasiatic speakers were estimated at approximately 2,600–1,400 years ago. For populations in Guizhou, the studied Sui was genetically similar to the previously studied Tai-Kadai populations in Guizhou, such as Sui, Bouyei, and Dong. These populations could be modeled as the admixtures of Neolithic Yellow River farmers (15–40%) and ancient coastal southern East Asians (45–85%) (Bin et al., 2021; Wang C. C. et al., 2021). Interestingly, we found that the dominant paternal Y chromosomal lineage of our studied Sui samples derived most likely from Hmong-Mien people, but the mtDNA profile was similar to that of the Tai-Kadai populations in southern China, documenting a possible sex-biased admixture in the formation of the Guizhou Sui people.
Our findings also showed a phylogenetic relationship between present-day Tai-Kadai and Austronesian speaking populations, which corroborated previous genetic study about the affinity between Tai-Kadai and Austronesian (Wang C. C. et al., 2021) and was also compatible with the hypothesis of “the common origin of Austronesian-Tai-Kadai” in linguistics (Sagart, 2004). Meanwhile, the genomic analysis of ancient populations in Fujian Province and Taiwan supported that the origin of the Austronesian language family can trace back to southeastern China (Yang et al., 2020; Wang C. C. et al., 2021). We observed a close connection between ancient populations in southeastern China and Tai-Kadai populations, which indicated that the common origin place of Austronesian-Tai-Kadai-speaking groups was probably southern China (Huang et al., 2020). According to historic documents, southern China was used to be a region where the ancient Baiyue tribes inhabited, and the Baiyue tribes gradually developed into Austronesian and Tai-Kadai groups (Lee, 2011). The Yangtze River rice farmers were hypothesized to contribute to the gene pool of Austronesian, Tai-Kadai, and Austroasiatic speakers (Lipson et al., 2018; Mccoll et al., 2018). The formation of Tai-Kadai speakers has also been influenced by the southward expansion of northern populations, such as the millet farmers of Neolithic Yangshao Culture and Longshan Culture in the Yellow River Basin (Yang et al., 2020).
Conclusion
We used both low-density forensic InDels and high-density genome-wide SNPs in this study to infer the genetic characteristics of Guizhou Tai-Kadai-speaking Sui people. The 30 InDel loci were found forensically informative in Guizhou Sui and can be used for individual identification. Furthermore, population genetic landscape and demographic history reconstruction based on genome-wide SNP data showed that Sui people were genetically similar to other Tai-Kadai groups. We found that Tai-Kadai populations had a genetic affinity with Hmong-Mien, Austroasiatic Kinh, Muang, and modern and ancient Austronesian-related populations in southern China, suggesting that the interactions among Tai-Kadai/Austronesian, Hmong-Mien, and Austroasiatic speaking populations in southern China during the past 2,000 years played a crucial part in the formation of modern Tai-Kadai. We found a possible sex-biased admixture in the formation of the Guizhou Sui people with the dominant paternal Y chromosomal lineage deriving most likely from Hmong-Mien-speaking people. The additional southward expansion of millet farmers from the Yellow River Basin has impacted the gene pool of southern populations including Tai-Kadai-speaking people.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical Committee on Clinical Trials of Drugs of the Affiliated Hospital of Guizhou Medical University (2020 Ethics Approval No. 85). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
C-CW and JH designed this study. MY, ZR, HaZ, QW, YL, HoZ, JJ, and JC collected the samples and conducted the experiment. XY wrote the manuscript. XY, GH, and JG analyzed the results. C-CW modified the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by the Guizhou Scientific Support Project, Qian Science Support (2021) General 448; Guizhou Province Education Department, Characteristic Region Project, Qian Education KY No. (2021)065; Guizhou “Hundred” High-Level Innovative Talent Project, Qian Science Platform Talents (2020)6012; Guizhou Scientific Support Project, Qian Science Support (2020) 4Y057; Guizhou Science Project, Qian Science Foundation (2020) 1Y353; Guizhou Scientific Support Project, Qian Science Support (2019) 2825; Guizhou Scientific Cultivation Project, Qian Science Platform Talent (2018) 5779-X; and Guizhou Engineering Technology Research Center Project, Qian High-Tech of Development and Reform Commission No. (2016) 1345, the Major Project of National Social Science Foundation of China granted to C-CW (21&ZD285) and (20&ZD248), the National Natural Science Foundation of China (31801040), the “Double First Class University Plan” key construction project of Xiamen University (the origin and evolution of East Asian populations and the spread of Chinese civilization, 0310/X2106027), Major Project of Marxist Theoretical Research and Construction Project (2021MZD014), European Research Council (ERC) grant (ERC-2019-ADG-883700-TRAM), and Nanqiang Outstanding Young Talents Program of Xiamen University (X2123302).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
S. Fang and Z. Xu from the Information and Network Center of Xiamen University are acknowledged for their help with high-performance computing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.844761/full#supplementary-material
Supplementary Figure 1 | (A) Geographical distribution of Guizhou Sui in this study. (B) The linkage disequilibrium patterns among the 30 InDel loci of the Guizhou Sui population. (C,D) Allele frequencies and corresponding forensic parameters of the 30 Indels in the Guizhou Sui population.
Supplementary Figure 2 | The heat map on the basis of the deletion allelic frequency distributions for Guizhou Sui and 99 worldwide reference populations.
Supplementary Figure 3 | The heat map of pairwise Nei’s genetic distances among Guizhou Sui and 99 worldwide reference populations.
Supplementary Figure 4 | The heat map of pairwise DA genetic distances among Guizhou Sui and 99 worldwide reference populations.
Supplementary Figure 5 | The results of ancestry component analysis on the 30 InDels with k ranging from 2 to 8 of the Guizhou Sui and 51 worldwide reference populations by STRUCTURE.
Supplementary Figure 6 | The phylogenetic relationships between the Guizhou Sui population and modern Asian populations based on Treemix.
Supplementary Figure 7 | The genetic affinity between Guizhou Sui and ancient populations in Asia. Significant Z scores were labeled by “++” “– –” (Z > 6, Z < −6, respectively), and Z-values with 3 < Z < 6, −6 < Z < −3 were labeled by “+,” “–,” respectively.
References
Akhteruzzaman, S., Das, S. A., Hosen, I., and Ferdous, A. (2013). Genetic polymorphism of 30 InDel markers for forensic use in Bangladeshi population. Forensic Sci. Int. Genet. Suppl. Ser. 4, e348–e349.
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Blench, R. M. (2017). “Origins of Ethnolinguistic Identity in Southeast Asia,” in Handbook of East and Southeast Asian Archaeology, eds J. Habu, P. V. Lape, and J. W. Olsen (New York, NY: Springer), 733–753. doi: 10.1007/978-1-4939-6521-2_42
Bin, X., Wang, R., Huang, Y., Wei, R., Zhu, K., Yang, X., et al. (2021). Genomic insight into the population structure and admixture history of tai-kadai-speaking sui people in Southwest China. Front. Genet. 12:735084. doi: 10.3389/fgene.2021.735084
Carvalho, A., and Pinheiro, M. F. (2013). Population data of 30 insertion/delection polymorphisms from a sample taken in the North of Portugal. Int. J. Legal Med. 127, 65–67. doi: 10.1007/s00414-012-0693-7
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/s13742-015-0047-8
Chen, P., He, G., Zou, X., Zhang, X., Li, J., Wang, Z., et al. (2018). Genetic diversities and phylogenetic analyses of three Chinese main ethnic groups in southwest China: a Y-Chromosomal STR study. Sci. Rep. 8:15339. doi: 10.1038/s41598-018-33751-x
Chen, P., Luo, L., Gao, H., Wu, J., Wang, Y., He, G., et al. (2019). Forensic performance of 30 InDels included in the Investigator DIPplex system in Miao population and comprehensive genetic relationship in China. Int. J. Legal Med. 133, 1389–1392. doi: 10.1007/s00414-019-02057-6
Chen, S., Ren, X., Liu, Y., Hu, Q., Hong, W., and Xu, A. (2007). Human leukocyte antigen class I polymorphism in Miao. Bouyei, and Shui ethnic minorities of Guizhou, China. Hum. Immunol. 68, 928–933. doi: 10.1016/j.humimm.2007.09.006
da silva, C. V., Matos, S., Costa, H. A., Morais, P., Dos Santos, R. M., Espinheira, R., et al. (2013). Genetic portrait of south Portugal population with InDel markers. Forensic Sci. Int. Genet. 7, e101–e103. doi: 10.1016/j.fsigen.2013.03.009
Du, W., Feng, C., Yao, T., Xiao, C., Huang, H., Wu, W., et al. (2019). Genetic variation and forensic efficiency of 30 indels for three ethnic groups in Guangxi: relationships with other populations. PeerJ 7:e6861. doi: 10.7717/peerj.6861
Du, W., Peng, Z., Feng, C., Zhu, B., Wang, B., Yue, W., et al. (2017). Forensic efficiency and genetic variation of 30 InDels in Vietnamese and Nigerian populations. Oncotarget 8, 88934–88940. doi: 10.18632/oncotarget.21494
Edmondson, J. A., and Gregerson, K. J. (2007). The languages of Vietnam: mosaics and expansions. Lang. Linguist. Compass 1, 727–749. doi: 10.1111/j.1749-818x.2007.00033.x
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Ferreira Palha, T. J. B., Ribeiro Rodrigues, E. M., Cavalcante, G. C., Marrero, A., De Souza, I. R., Seki Uehara, C. J., et al. (2015). Population genetic analysis of insertion-deletion polymorphisms in a Brazilian population using the Investigator DIPplex kit. Forensic Sci. Int. Genet. 19, 10–14. doi: 10.1016/j.fsigen.2015.03.015
Fondevila, M., Phillips, C., Santos, C., Pereira, R., Gusmao, L., Carracedo, A., et al. (2012). Forensic performance of two insertion-deletion marker assays. Int. J. Legal Med. 126, 725–737. doi: 10.1007/s00414-012-0721-7
Friis, S. L., Borsting, C., Rockenbauer, E., Poulsen, L., Fredslund, S. F., Tomas, C., et al. (2012). Typing of 30 insertion/deletions in Danes using the first commercial indel kit–Mentype(R) DIPplex. Forensic Sci. Int. Genet. 6, e72–e74. doi: 10.1016/j.fsigen.2011.08.002
Gouy, A., and Zieger, M. (2017). STRAF-A convenient online tool for STR data evaluation in forensic genetics. Forensic Sci. Int. Genet. 30, 148–151. doi: 10.1016/j.fsigen.2017.07.007
Guo, J., Xu, B., Li, L., He, G., Zhang, H., Cheng, H. Z., et al. (2019b). Paternal Y chromosomal genotyping reveals multiple large-scale admixtures in the formation of Lolo-Burmese-speaking populations in southwest China. Ann. Hum. Biol. 46, 581–588. doi: 10.1080/03014460.2019.1698655
Guo, J., Ji, J., He, G., Ren, Z., Zhang, H., Wang, Q., et al. (2019a). Genetic structure and forensic characterisation of 19 X-chromosomal STR loci in Guizhou Sui population. Ann. Hum. Biol. 46, 246–253. doi: 10.1080/03014460.2019.1623911
Guo, J., Wang, W., Zhao, K., Li, G., He, G., Zhao, J., et al. (2022). Genomic insights into Neolithic farming-related migrations in the junction of east and southeast Asia. Am. J. Biol. Anthropol. 177, 328–342. doi: 10.1002/ajpa.24434
Guo, Y., Chen, C., Jin, X., Cui, W., Wei, Y., Wang, H., et al. (2018). Autosomal DIPs for population genetic structure and differentiation analyses of Chinese Xinjiang Kyrgyz ethnic group. Sci. Rep. 8:11054. doi: 10.1038/s41598-018-29010-8
Guo, Y., Shen, C., Meng, H., Dong, Q., Kong, T., Yang, C., et al. (2016). Population Differentiations and Phylogenetic Analysis of Tibet and Qinghai Tibetan Groups Based on 30 InDel Loci. DNA Cell Biol. 35, 787–794. doi: 10.1089/dna.2016.3395
Haak, W., Lazaridis, I., Patterson, N., Rohland, N., Mallick, S., Llamas, B., et al. (2015). Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211. doi: 10.1038/nature14317
He, G., Ren, Z., Guo, J., Zhang, F., Zou, X., Zhang, H., et al. (2019a). Population genetics, diversity and forensic characteristics of Tai-Kadai-speaking Bouyei revealed by insertion/deletions markers. Mol. Genet. Genomics 294, 1343–1357 doi: 10.1007/s00438-019-01584-6
He, G., Wang, M., Zou, X., Chen, P., Wang, Z., Liu, Y., et al. (2021). Peopling history of the tibetan plateau and multiple waves of admixture of tibetans inferred from both ancient and modern genome-wide data. Front. Genet. 12:725243. doi: 10.3389/fgene.2021.725243
He, G., Wang, Z., Guo, J., Wang, M., Zou, X., Tang, R., et al. (2020). Inferring the population history of Tai-Kadai-speaking people and southernmost Han Chinese on Hainan Island by genome-wide array genotyping. Eur. J. Hum. Genet. 28, 1111–1123. doi: 10.1038/s41431-020-0599-7
He, G., Wang, Z., Zou, X., Wang, M., Liu, J., Wang, S., et al. (2019b). Tai-Kadai-speaking Gelao population: Forensic features, genetic diversity and population structure. Forensic Sci. Int. Genet. 40, e231–e239. doi: 10.1016/j.fsigen.2019.03.013
He, G., Ren, Z., Guo, J., Zhang, F., Zou, X., Zhang, H., Wang, Q., Ji, J., Yang, M., Zhang, Z., Zhang, J., Nabijiang, Y., Huang, J., and Wang, C. C. (2019c). “Population genetics, diversity and forensic characteristics of Tai-Kadai-speaking Bouyei revealed by insertion/deletions markers.” Mol. Genet Genomics 294(5): 1343–1357. doi: 10.1007/s00438-019-01584-6
Hefke, G., Davison, S., and D’amato, M. E. (2015). Forensic performance of Investigator DIPplex indels genotyping kit in native, immigrant, and admixed populations in South Africa. Electrophoresis 36, 3018–3025. doi: 10.1002/elps.201500243
Huang, X., Xia, Z.-Y., Bin, X., He, G., Guo, J., Lin, C., et al. (2020). Genomic insights into the demographic history of Southern Chinese. bioRxiv [Preprint]. doi: 10.1101/2020.11.08.373225
Ji, J., Ren, Z., Zhang, H., Wang, Q., Wang, J., Kong, Z., et al. (2017). Genetic profile of 23 Y chromosomal STR loci in Guizhou Shui population, southwest China. Forensic Sci. Int. Genet. 28, e16–e17. doi: 10.1016/j.fsigen.2017.01.010
Jian, H., Wang, L., Wang, H., Bai, X., Lv, M., and Liang, W. (2019). Population genetic analysis of 30 insertion-deletion (INDEL) loci in a Qinghai Tibetan group using the Investigator DIPplex Kit. Int. J. Legal Med. 133, 1039–1041. doi: 10.1007/s00414-018-1954-x
Kim, E. H., Lee, H. Y., Yang, I. S., Yang, W. I., and Shin, K. J. (2014). Population data for 30 insertion-deletion markers in a Korean population. Int. J. Legal Med. 128, 51–52. doi: 10.1007/s00414-013-0851-6
Kis, Z., Zalan, A., Volgyi, A., Kozma, Z., Domjan, L., and Pamjav, H. (2012). Genome deletion and insertion polymorphisms (DIPs) in the Hungarian population. Forensic Sci. Int. Genet. 6, e125–e126. doi: 10.1016/j.fsigen.2011.09.004
Kong, T., Chen, Y., Guo, Y., Wei, Y., Jin, X., Xie, T., et al. (2017). Autosomal InDel polymorphisms for population genetic structure and differentiation analysis of Chinese Kazak ethnic group. Oncotarget 8, 56651–56658. doi: 10.18632/oncotarget.17838
Kovach, W. L. (2007). MVSP-A MultiVariate Statistical Package for Windows, ver. 3.1. Pentraeth: Kovach Computing Services.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larue, B. L., Ge, J., King, J. L., and Budowle, B. (2012). A validation study of the Qiagen Investigator DIPplex(R) kit; an INDEL-based assay for human identification. Int. J. Legal Med. 126, 533–540. doi: 10.1007/s00414-012-0667-9
Lee, H. (2011). Some remarks on reconstructing the prehistoric linguistic relationships of the tai-kadai language family and its putative linguistic affiliations with other language families. Int. Rev. Soc. Sci. Hum. 2, 163–175.
Li, H., Cai, X., Winograd-Cort, E. R., Wen, B., Cheng, X., Qin, Z., et al. (2007). Mitochondrial DNA diversity and population differentiation in southern East Asia. Am. J. Phys. Anthropol. 134, 481–488. doi: 10.1002/ajpa.20690
Li, H., Wen, B., Chen, S. J., Su, B., Pramoonjago, P., Liu, Y., et al. (2008). Paternal genetic affinity between Western Austronesians and Daic populations. BMC Evol. Biol. 8:146. doi: 10.1186/1471-2148-8-146
Li, L., Ye, Y., Song, F., Wang, Z., and Hou, Y. (2019). Genetic structure and forensic parameters of 30 InDels for human identification purposes in 10 Tibetan populations of China. Forensic Sci. Int. Genet. 40, e219–e227. doi: 10.1016/j.fsigen.2019.02.002
Li, Z., Xu, J., Chen, P., Yin, C., Hu, L., Huang, H., et al. (2018). Forensic efficiency and genetic divergence of 30 autosomal InDels in Chinese Han population from Jiangsu province. Forensic Sci. Int. Genet. 37, e17–e19. doi: 10.1016/j.fsigen.2018.08.006
Lipson, M., Cheronet, O., Mallick, S., Rohland, N., Oxenham, M., Pietrusewsky, M., et al. (2018). Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science 361, 92–95. doi: 10.1126/science.aat3188
Lipson, M., Spriggs, M., Valentin, F., Bedford, S., Shing, R., Zinger, W., et al. (2020). Three phases of ancient migration shaped the ancestry of human populations in Vanuatu. Curr. Biol. 30, 4846–4856.e6. doi: 10.1016/j.cub.2020.09.035
Liu, D., Duong, N. T., Ton, N. D., Van Phong, N., Pakendorf, B., Van Hai, N., et al. (2020). Extensive ethnolinguistic diversity in vietnam reflects multiple sources of genetic diversity. Mol. Biol. Evol. 37, 2503–2519. doi: 10.1093/molbev/msaa099
Liu, Y., Zhang, H., He, G., Ren, Z., Zhang, H., Wang, Q., et al. (2020). Forensic features and population genetic structure of dong, yi, han, and chuanqing human populations in Southwest China inferred from insertion/deletion markers. Front. Genet. 11:360. doi: 10.3389/fgene.2020.00360
Loh, P. R., Lipson, M., Patterson, N., Moorjani, P., Pickrell, J. K., Reich, D., et al. (2013). Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193, 1233–1254. doi: 10.1534/genetics.112.147330
Ma, R., Shen, C., Wei, Y., Jin, X., Guo, Y., Mu, Y., et al. (2018). Genetic differentiation and forensic efficiency evaluation for Chinese Salar ethnic minority based on a 5-dye multiplex insertion and deletion panel. Gene 660, 41–50. doi: 10.1016/j.gene.2018.03.058
Macholdt, E., Arias, L., Duong, N. T., Ton, N. D., Van Phong, N., Schroder, R., et al. (2020). The paternal and maternal genetic history of Vietnamese populations. Eur. J. Hum. Genet. 28, 636–645. doi: 10.1038/s41431-019-0557-4
Martin, P., Garcia, O., Heinrichs, B., Yurrebaso, I., Aguirre, A., and Alonso, A. (2013). Population genetic data of 30 autosomal indels in Central Spain and the Basque Country populations. Forensic Sci. Int. Genet. 7, e27–e30. doi: 10.1016/j.fsigen.2012.10.003
Martinez-Cortes, G., Garcia-Aceves, M., Favela-Mendoza, A. F., Munoz-Valle, J. F., Velarde-Felix, J. S., and Rangel-Villalobos, H. (2016). Forensic parameters of the Investigator DIPplex kit (Qiagen) in six Mexican populations. Int. J. Legal Med. 130, 683–685. doi: 10.1007/s00414-015-1242-y
Mccoll, H., Racimo, F., Vinner, L., Demeter, F., Gakuhari, T., Moreno-Mayar, J. V., et al. (2018). The prehistoric peopling of Southeast Asia. Science 361, 88–92. doi: 10.1126/science.aat3628
Mei, T., Shen, C. M., Liu, Y. S., Meng, H. T., Zhang, Y. D., Guo, Y. X., et al. (2016). Population genetic structure analysis and forensic evaluation of Xinjiang Uigur ethnic group on genomic deletion and insertion polymorphisms. SpringerPlus 5:1087. doi: 10.1186/s40064-016-2730-3
Meng, H. T., Zhang, Y. D., Shen, C. M., Yuan, G. L., Yang, C. H., Jin, R., et al. (2015). Genetic polymorphism analyses of 30 InDels in Chinese Xibe ethnic group and its population genetic differentiations with other groups. Sci. Rep. 5:8260. doi: 10.1038/srep08260
Mills, R. E., Luttig, C. T., Larkins, C. E., Beauchamp, A., Tsui, C., Pittard, W. S., et al. (2006). An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 16, 1182–1190. doi: 10.1101/gr.4565806
Ning, C., Li, T., Wang, K., Zhang, F., Li, T., Wu, X., et al. (2020). Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun. 11:2700. doi: 10.1038/s41467-020-16557-2
Patterson, N., Moorjani, P., Luo, Y., Mallick, S., Rohland, N., Zhan, Y., et al. (2012). Ancient admixture in human history. Genetics 192, 1065–1093. doi: 10.1534/genetics.112.145037
Patterson, N., Price, A. L., and Reich, D. (2006). Population structure and eigenanalysis. PLoS Genet. 2:e190. doi: 10.1371/journal.pgen.0020190
Pickrell, J. K., and Pritchard, J. K. (2012). Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. doi: 10.1371/journal.pgen.1002967
Sagart, L. (2004). The higher phylogeny of Austronesian and the position of Tai-Kadai. Ocean. Linguist. 43, 411–444. doi: 10.1353/ol.2005.0012
Shimada, M. K., and Nishida, T. (2017). A modification of the PHYLIP program: a solution for the redundant cluster problem, and an implementation of an automatic bootstrapping on trees inferred from original data. Mol. Phylogenet. Evol. 109, 409–414. doi: 10.1016/j.ympev.2017.02.012
Slooten, K., and Ricciardi, F. (2013). Estimation of mutation probabilities for autosomal STR markers. Forensic Sci. Int. Genet. 7, 337–344. doi: 10.1016/j.fsigen.2013.01.006
Turrina, S., Filippini, G., and De Leo, D. (2011). Forensic evaluation of the Investigator DIPplex typing system. Forensic Sci. Int. Genet. Suppl. Ser. 3, e331–e332.
Walsh, P., Metzger, D., and Higuchi, R. (1991). Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10, 506–513.
Wang, C. C., Wang, L. X., Shrestha, R., Zhang, M., Huang, X. Y., Hu, K., et al. (2014). Genetic structure of Qiangic populations residing in the western Sichuan corridor. PLoS One 9:e103772. doi: 10.1371/journal.pone.0103772
Wang, T., Wang, W., Xie, G., Li, Z., Fan, X., Yang, Q., et al. (2021). Human population history at the crossroads of East and Southeast Asia since 11,000 years ago. Cell 184, 3829–3841.e21. doi: 10.1016/j.cell.2021.05.018
Wang, C. C., Yeh, H. Y., Popov, A. N., Zhang, H. Q., Matsumura, H., Sirak, K., et al. (2021). Genomic insights into the formation of human populations in East Asia. Nature 591, 413–419. doi: 10.1038/s41586-021-03336-2
Wang, M., Du, W., Tang, R., Liu, Y., Zou, X., Yuan, D., et al. (2022). Genomic history and forensic characteristics of Sherpa highlanders on the Tibetan Plateau inferred from high-resolution InDel panel and genome-wide SNPs. Forensic Sci. Int. Genet. 56:102633. doi: 10.1016/j.fsigen.2021.102633
Wen, B., Li, H., Lu, D., Song, X., Zhang, F., He, Y., et al. (2004). Genetic evidence supports demic diffusion of Han culture. Nature 431, 302–305. doi: 10.1038/nature02878
Xia, Z. Y., Yan, S., Wang, C. C., Zheng, H. X., Zhang, F., Liu, Y. C., et al. (2019). Inland-coastal bifurcation of Southern East Asians Revealed by Hmong-mien genomic history. bioRxiv [Preprint]. doi: 10.1101/730903
Xu, B., Guo, J., Huang, Y., Chen, X., Deng, X., and Wang, C. C. (2019). The paternal genetic structure of Jingpo and Dai in southwest China. Ann. Hum. Biol. 46, 279–283. doi: 10.1080/03014460.2019.1624821
Yang, L., Zhao, Y., Liu, C., Chan, D. W., Chan, M., and He, M. (2013). Allele frequencies of 15 STRs in five ethnic groups (Han, Gelao, Jing, Shui and Zhuang) in South China. Forensic Sci. Int. Genet. 7, e9–e14. doi: 10.1016/j.fsigen.2012.10.009
Yang, M. A., Fan, X., Sun, B., Chen, C., Lang, J., Ko, Y. C., et al. (2020). Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 369, 282–288. doi: 10.1126/science.aba0909
Yao, H. B., Tang, S., Yao, X., Yeh, H. Y., Zhang, W., Xie, Z., et al. (2017). The genetic admixture in Tibetan-Yi Corridor. Am. J. Phys. Anthropol. 164, 522–532. doi: 10.1002/ajpa.23291
Keywords: Sui, InDel, genome-wide data, genetic profile, demographic history
Citation: Yang M, Yang X, Ren Z, He G, Zhang H, Wang Q, Liu Y, Zhang H, Ji J, Chen J, Guo J, Huang J and Wang C-C (2022) Genetic Admixture History and Forensic Characteristics of Guizhou Sui People Inferred From Autosomal Insertion/Deletion and Genome-Wide Single-Nucleotide Polymorphisms. Front. Ecol. Evol. 10:844761. doi: 10.3389/fevo.2022.844761
Received: 28 December 2021; Accepted: 04 April 2022;
Published: 16 May 2022.
Edited by:
Lian Deng, Fudan University, ChinaReviewed by:
Yungang He, Fudan University, ChinaHaoliang Fan, Southern Medical University, China
Weian Du, Third Affiliated Hospital of Sun Yat-sen University, China
Catarina Xavier, Medical University of Innsbruck, Austria
Copyright © 2022 Yang, Yang, Ren, He, Zhang, Wang, Liu, Zhang, Ji, Chen, Guo, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Huang, bW1tX2hqQDEyNi5jb20=; Chuan-Chao Wang, d2FuZ0B4bXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Meiqing Yang
Meiqing Yang Xiaomin Yang
Xiaomin Yang Zheng Ren1†
Zheng Ren1† Guanglin He
Guanglin He Jing Chen
Jing Chen Jiang Huang
Jiang Huang Chuan-Chao Wang
Chuan-Chao Wang