- 1Department of Biological Sciences, Purdue University, West Lafayette, IN, United States
- 2Department of Life Sciences, Ben-Gurion University of the Negev, Beer-Sheva, Israel
- 3Department of Microbiology and Immunology, Bio21 Institute, The University of Melbourne, Melbourne, VIC, Australia
- 4Department of Ecology and Evolution, University of Chicago, Chicago, IL, United States
- 5Santa Fe Institute, Santa Fe, NM, United States
A corrigendum on
Frequency-dependent competition between strains imparts persistence to perturbations in a model of Plasmodium falciparum malaria transmission
by He, Q., Pilosof, S., Tiedje, K. E., Day, K. P., and Pascual, M. (2021). Front. Ecol. Evol. 9:633263. doi: 10.3389/fevo.2021.633263
In the original article, there was an error in the specific part of the code implementing migration between the global pool and the local population. This resulted in no migration after the initialization of the local population of parasites in the first 30 days. Thus, the simulations apply to a closed system and not to an open one with a regional intervention. The authors have corrected the description and discussion of the migration assumptions in the text (in sentences across four paragraphs), and in the diagrams of Figures 1A,B. The results apply to intervention in a closed population as now specified in the corrected text and Figure.
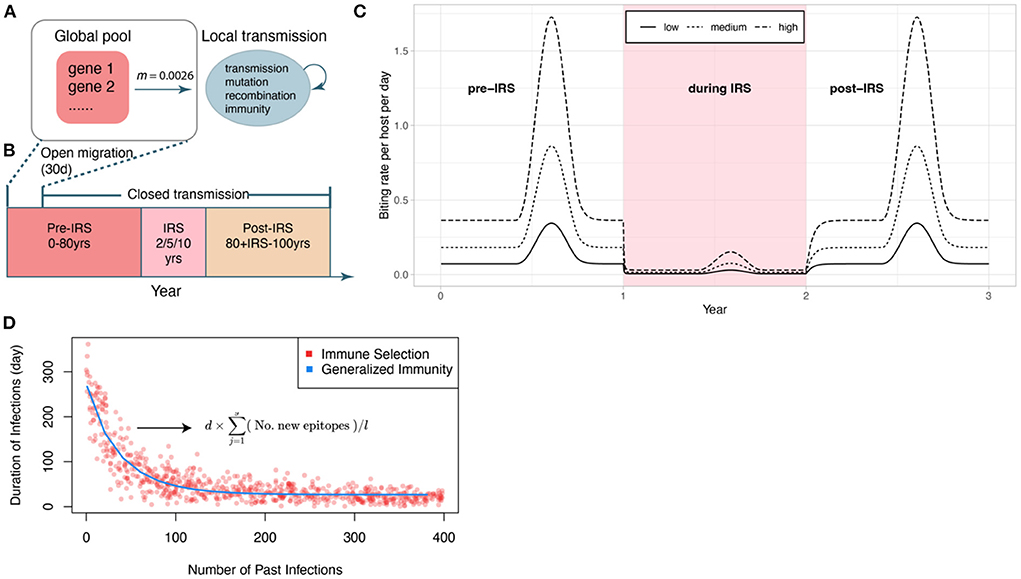
Figure 1. Schematic of the IRS experiment design. (A) Initial transmission events and migrant genomes in a local population (blue circle) are sourced from a global pool of genes (red square) (see Methods). The value of the migration rate is inferred from an empirical dataset for a high transmission region in Ghana (see Methods). Individual infections are tracked locally. We also track events of transmission, mutation of genes, recombination within and between genomes, and acquisition and loss of specific immune memory in hosts. (B) The period of open migration from the global pool lasts for 30 days to initialize the system. Each simulation follows three stages (after a burn-in period): a pre-IRS period during which the transmission in the local population reaches a stable state; an IRS period of 2, 5, or 10 years reducing transmission, and a post-IRS period when transmission rates go back to pre-IRS levels. (C) Three levels of transmission intensity (biting rates) are explored in the experiments (pre-/post-IRS, low: 44 bites per host per year; medium: 110 bites/h/y; high: 221 bites/h/y). (D) The two regimes (with and without NFDS) are ensured to be comparable by specifying the average duration of infection as a function of the number of previous infections in G with a curve fitted to the points generated under S. The expression for the duration of infection under S is given here, and its explanation can be found in the methods (2.2).
The corrected Figure 1 and legend appear below.
A correction has been made to Materials and Methods, “Agent-Based Model (ABM) and Model Setup,” paragraph 1. This paragraph previously stated:
“Malaria transmission and intervention are modeled using an agent-based, discrete-event, continuous-time stochastic system described in detail in He et al. (2018) and Pilosof et al. (2019). Here, we briefly describe the agent-based model (ABM), with an emphasis on the specific implementation of the regional intervention scheme that constitutes the press perturbation.”
The corrected paragraph appears below:
“Malaria transmission and intervention are modeled using an agent-based, discrete-event, continuous-time stochastic system described in detail in He et al. (2018) and Pilosof et al. (2019). Here, we briefly describe the agent-based model (ABM), with an emphasis on the specific implementation of intervention in a closed system that constitutes the press perturbation.”
A correction has been made to Materials and Methods, “Agent-Based Model (ABM) and Model Setup,” paragraph 4. The paragraph previously stated:
“The main modification to the model for this work is how the global pool interacts with local transmission. First, instead of remaining static, the global gene pool in this implementation updates its gene composition at the same mutation rate as that of the local population. Specifically, new genes are generated at a rate equal to the product of local parasite population size and the per-allele mutation rate. Once a new gene is generated, the old gene that it mutates from is removed from the global gene pool. Genomes migrate from the global pool to the local population (Figure 1A). The number of migrant genomes increases in wet seasons and decreases in dry seasons (see details in section Estimation of Migration Rate). Interventions are assumed to be applied at the regional level so that prevalence of the disease is the same across the region, including the local population (Figures 1B,C). Therefore, the proportion of infectious bites from migration is kept the same as that of local transmission.”
The corrected paragraph appears below:
“The main modification to the model for this work is how the local transmission is sourced from the global pool. After the initiation of local transmission, genomes continue to migrate from the global pool to the local population (Figure 1A) for the first 30 days (Figure 1B). Afterward, migration is discontinued, and local transmission fluctuates with the seasonality (Figure 1C).”
A correction has been made to Results, “Prevalence Recovers to a Lower Level Than Pre-intervention Under NFDS Because of Diversity Loss,” paragraph 1. The paragraph previously stated:
“We focus next on the recovery from intervention in terms of both antigenic diversity and prevalence. As expected, both antigenic diversity and prevalence decrease with intervention, and longer IRS leads to a higher reduction in antigenic diversity than the shorter 2-year IRS. Post-intervention, diversity settles on a new equilibrium, that is lower than the pre-intervention one (Figure 5). Although new genes should be strongly preferred under S (but not G), their generation from mutation, ectopic recombination, and immigration, is likely too slow to rebuild locally to pre-intervention levels given the parameters we use. Moreover, the intervention was implemented to also affect the var gene diversity of the regional pool, hindering regeneration of diversity via migration. We touch upon this point further in the Discussion.”
The corrected paragraph appears below:
“We focus next on the recovery from intervention in terms of both antigenic diversity and prevalence. As expected, both antigenic diversity and prevalence decrease with intervention, and longer IRS leads to a higher reduction in antigenic diversity than the shorter 2-year IRS. Post-intervention, diversity settles on a new equilibrium, that is lower than the pre-intervention one (Figure 5). Although new genes should be strongly preferred under S (but not G), their generation from mutation and ectopic recombination is likely to be too slow to rebuild diversity locally to pre-intervention levels given parameter values. We discuss the conditions of a closed system with no migrant genomes during intervention in the Discussion.”
A correction has been made to Discussion, paragraph 3. The paragraph previously stated:
“Although the strength of NFDS is quickly restored to maintain low similarity between strains, the var gene diversity in our simulations slowly rebounds but does not recover to pre-intervention levels. In part, this is the consequence of a regional intervention where the processes generating new genes are not fast enough to rebuild the original local and regional diversity. The time scale at which new genes can be generated and the selective advantage they represent will be critical parameters determining the speed of diversity recovery, but also whether it can practically rebuild to the original pre-intervention levels. This aspect of the system's recovery will be investigated in future work, in light of the recently introduced concept of a threshold for the accumulation of antigenic diversity, we named the innovation number Rdiv, whose critical value is one (He and Pascual, 2021). Here, we have considered parameter ranges representative of those in nature, although the values of ectopic recombination rates have been measured only in vitro (Claessens et al., 2014).”
The corrected paragraph appears below:
“Although the strength of NFDS is quickly restored to maintain low similarity between strains, the var gene diversity in our simulations slowly rebounds but does not recover to pre-intervention levels. In part, this is the consequence of a closed system where the processes generating new genes are not fast enough to rebuild the original local diversity. The time scale at which new genes can be generated and their selective advantage will be critical parameters determining the speed at which diversity recovers, and whether it can practically rebuild to the original pre-intervention levels. This aspect of the system's recovery will be investigated in future work by opening the local population to migration from the regional pool, and also in light of the recently introduced concept of a threshold for the accumulation of antigenic diversity, we named the innovation number Rdiv, whose critical value is one (He and Pascual, 2021). Here, we have considered parameter ranges representative of those in nature, although the values of ectopic recombination rates have been measured only in vitro (Claessens et al., 2014). We expect the results to hold in systems with weak immigration during and after intervention, where the intervention efforts have been applied regionally.”
A correction has been made to Discussion, paragraph 5. The paragraph preciously stated:
“Our model considers a local population embedded within a regional pool that provides a source of genetic variation. This approach is a first step toward developing a more comprehensive theory because in nature malaria is transmitted within and between local human populations, effectively creating a metapopulation of parasites. Therefore, processes that operate on metapopulations, such as dispersal and source-sink dynamics, may influence both the assembly of parasite populations and their stability in addition to local selection. For our purpose, this metapopulation context is particularly relevant in creating a vast pool of genetic variation as documented for endemic regions over larger spatial scales (Day et al., 2017; Tonkin-Hill, 2020) than those of local transmission, and longer temporal scales than those of the intervention we implement here. Addressing metapopulation dynamics explicitly for this highly diverse system would be however computationally extensive. Our approach relies on the initial compromise of a global pool typical of many assembly models in ecology.”
The corrected paragraph appears below:
“Our model considers a local population embedded within a regional pool that provides the source of initial genetic variation. This approach is a first step toward developing a more comprehensive theory because in nature malaria is transmitted within and between local human populations, effectively creating a metapopulation of parasites. Therefore, processes that operate on metapopulations, such as dispersal and source-sink dynamics, may influence both the assembly of parasite populations and their stability in addition to local selection. For our purpose, this metapopulation context is particularly relevant in creating a vast pool of genetic variation as documented for endemic regions over larger spatial scales (Day et al., 2017; Tonkin-Hill, 2020) than those of local transmission, and longer temporal scales than those of the intervention we implement here. Addressing metapopulation dynamics explicitly for this highly diverse system would be however computationally extensive. A potential compromise will be that of an open population with continuous migration from a global pool typical of many assembly models in ecology.”
The authors apologize for these errors and state that this does not change the scientific conclusions of the article. The original article has been updated.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Claessens, A., Hamilton, W. L., Kekre, M., Otto, T. D., Faizullabhoy, A., Rayner, J. C., et al. (2014). Generation of antigenic diversity in Plasmodium falciparum by structured rearrangement of var genes during mitosis. PLoS Genet. 10:e1004812. doi: 10.1371/journal.pgen.1004812
Day, K. P., Artzy-Randrup, Y., Tiedje, K. E., Rougeron, V., Chen, D. S., Rask, T. S., et al. (2017). Evidence of strain structure in Plasmodium falciparum var gene repertoires in children from Gabon, West Africa. Proc. Natl. Acad. Sci. U.S.A. 114, E4103–E4111. doi: 10.1073/pnas.1613018114
He, Q., and Pascual, M. (2021). An antigenic diversification threshold for falciparum malaria transmission at high endemicity. PLoS Comput. Biol. 17:e1008729. doi: 10.1371/journal.pcbi.1008729
He, Q., Pilosof, S., Tiedje, K. E., Ruybal-Pesántez, S., Artzy-Randrup, Y., Baskerville, E. B., et al. (2018). Networks of genetic similarity reveal non-neutral processes shape strain structure in Plasmodium falciparum. Nat. Commun. 9:1817. doi: 10.1038/s41467-018-04219-3
Pilosof, S., He, Q., Tiedje, K. E., Ruybal-Pesántez, S., Day, K. P., and Pascual, M. (2019). Competition for hosts modulates vast antigenic diversity to generate persistent strain structure in Plasmodium falciparum. PLoS Biol. 17:e3000336. doi: 10.1371/journal.pbio.3000336
Keywords: strain diversity, stabilizing competition, stochastic assembly, persistence, malaria and antigenic diversity, negative frequency-dependent selection, agent-based model, var genes
Citation: He Q, Pilosof S, Tiedje KE, Day KP and Pascual M (2022) Corrigendum: Frequency-dependent competition between strains imparts persistence to perturbations in a model of Plasmodium falciparum malaria transmission. Front. Ecol. Evol. 10:971161. doi: 10.3389/fevo.2022.971161
Received: 16 June 2022; Accepted: 20 July 2022;
Published: 04 August 2022.
Edited and reviewed by: Luís Borda-de-Água, Centro de Investigacao em Biodiversidade e Recursos Geneticos (CIBIO-InBIO), Portugal
Copyright © 2022 He, Pilosof, Tiedje, Day and Pascual. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mercedes Pascual, cGFzY3VhbG1tQHVjaGljYWdvLmVkdQ==
†These authors have contributed equally to this work
‡ORCID: Qixin He https://orcid.org/0000-0003-1696-8203
Shai Pilosof https://orcid.org/0000-0003-0430-5568
Kathryn E. Tiedje https://orcid.org/0000-0003-3305-0533
Karen P. Day https://orcid.org/0000-0002-6115-6135
Mercedes Pascual https://orcid.org/0000-0003-3575-7233
 Qixin He
Qixin He Shai Pilosof
Shai Pilosof Kathryn E. Tiedje
Kathryn E. Tiedje Karen P. Day
Karen P. Day Mercedes Pascual
Mercedes Pascual