- 1Guangxi Key Laboratory for Forest Ecology and Conservation, College of Forestry, Guangxi University, Nanning, China
- 2Ministry of Education Key Laboratory for Biodiversity Science and Ecological Engineering, Institute of Biodiversity Science, Center of Evolutionary Biology, School of Life Sciences, Fudan University, Shanghai, China
- 3Key Laboratory of Environment Change and Resource Use, Beibu Gulf, Nanning Normal University, Nanning, China
Introduction: With 78 species, Sinocyclocheilus cavefish constitute the largest cavefish radiation in the world. They exhibit remarkable morphological diversity across three habitat types: surface (surface morphs, normal-eyed, variably colored), exclusively-cave-dwelling (stygobitic morphs, eyeless, depigmented), and intermediate between cave and surface (stygophilic morphs, micro-eyed, partially depigmented). Distinctive traits of Sinocyclocheilus include variations in eye and skin conditions associated with their habitat, despite the role of the skin in sensing environmental changes, its habitat correlates are less understood, compared to the well-studied eye conditions.
Methods: Here, we analyzed the correlation between Sinocyclocheilus skin morphology and its habitat, utilizing morphological and transcriptomics-based methods. We generated skin RNA-sequencing data for nine species and integrated those with existing data from five additional species. These 14 species represent the primary clades and major habitats of these cavefish.
Result: We identified 7,374 orthologous genes in the skin transcriptome data. Using a comparative transcriptomics approach, 1,348 differentially expressed genes (DEGs) were identified in the three morphotypes. GO and KEGG enrichment analyses suggested that these genes were mainly involved in energy metabolism, immunity and oxidative stress-related functions. Genes related to immune, apoptotic, and necrotic functions were identified through positive selection analysis of orthologous genes. The maximum likelihood phylogenetic tree, based on 1,369, single-copy orthologous genes of the species, was largely concordant with the currently established RAD-seq and mt-DNA based phylogenies. Species with higher cave dependence present lighter coloration, fewer dark blotches, and diminished scale morphology and coverage. We also found that differences in skin gene expression and positive selection effects may have contributed to the degradation of skin color and scales.
Discussion: Our study highlights the significance of habitat in shaping skin metabolism, pigmentation variation, and morphology while offering insights into the molecular mechanisms driving these habitat-specific adaptations in Sinocyclocheilus. These findings underscore the transcriptional variation in adapting to diverse environments and contribute to future studies on the evolution and ecology of cavefish.
1 Introduction
Cave-adapted organisms, particularly cavefish, make up a significant portion of vertebrates inhabiting caves. They present unique opportunities for evolutionary biologists to study the patterns of adaptation to new environments (Jeffery, 2001). Over 300 cavefish species have been discovered worldwide, evolving rapidly from their surface-dwelling ancestors, making them ideal for comparative analysis of adaptive processes (Borowsky, 2018; Fortune et al., 2020; Policarpo et al., 2021). The best known among these is the well-studied Astyanax mexicanus cavefish system, a single species that includes surface and cave-adapted populations representing two distinct morpho-habitat types (Gross, 2012; Loomis et al., 2019; Krishnan et al., 2020). Interestingly, phylogenetically distant groups display convergent stygomorphic traits, such as the loss of pigmentation, degeneration or loss of eyes, elongated appendages, lowered metabolism, and specialized sensory systems (Jeffery, 2019), providing opportunities for cave-adaptation related comparative studies (Protas and Jeffery, 2012; Stahl and Gross, 2017).
China’s Sinocyclocheilus cavefish, with 78 species, constitute the largest cavefish radiation in the world (Mao T. et al., 2022; Wen et al., 2022; Luo et al., 2023; Xu et al., 2023). Phylogenetic analyses based on mitochondrial and nuclear DNA have consistently resolved Sinocyclocheilus as a monophyletic genus, comprising of 4-6 main clades, that share a common normal-eyed surface dwelling ancestor (Zhao and Zhang, 2009; Jiang et al., 2019; Mao et al., 2021). Sinocyclocheilus species can be grouped into three categories based on habitat occupation: surface (living outside caves, SU), stygophilic (cave-associated habit, SP), and stygobitic (exclusively cave-dwelling habit, SB) species (Yang et al., 2016; Zhao et al., 2021; Zhou S. et al., 2022). The eye morphology and skin coloration of these fish strongly correlate with their habitats and hence, eye-condition can be used as a proxy to identify their habitat associations (Jiang et al., 2019; Mao et al., 2021; Zhao et al., 2021). SUs have normal eyes and yellow or charcoal gray coloration, while SBs lack eyes and have white-pink, translucent skin (Li et al., 2020; Mao et al., 2021). SPs usually have micro-eyes and variable coloration, although there are exceptions (Mao et al., 2021; Chen et al., 2022). There are several instances of independent evolution of SUs (in two clades) and SBs (Three Clades) within the diversification (Mao T. et al., 2022; Wen et al., 2022). So far, Sinocyclocheilus research has predominantly focused on eye regression, leaving the skin relatively understudied (Meng et al., 2013; Huang et al., 2019; Zhao et al., 2021).
Epidermal adaptations are essential for organisms conquering new environments (Jablonski and Chaplin, 2010; Ángeles Esteban, 2012). Sinocyclocheilus have repeatedly evolved both regressive and constructive skin traits as adaptation to cave environments. Regressive traits include: reduced pigmentation, reduction in scales and constructive features include increased fat accumulation and enhanced non-visual sensory abilities such as well-developed lateral lines and neuromast systems (Yoshizawa et al., 2014; Yang et al., 2016; Chen et al., 2022). Hence, the independent evolution of the morphotypes of Sinocyclocheilus provide an attractive system to investigate the molecular mechanisms of convergent adaptations of their skin at the molecular level.
Transcriptomics based tools provide an unprecedented opportunity to understand the gene expression profiles and patterns of genetic variation behind the independent evolution of these diverse cavefish morphotypes. At the broader scale, studies on Astyanax mexicanus cavefish and Sinocyclocheilus cavefish have shown how protein sequence alterations (transcriptome) play a role in eye morphology and color degeneration in cavefish (Hinaux et al., 2013; McGaugh et al., 2014; Torres-Paz et al., 2018; Huang et al., 2019; Li et al., 2020; Zhao et al., 2020). However, little is known about the mechanisms of scale degradation (Yang et al., 2016; Simon et al., 2017). The convergent evolution of skin coloration is largely driven by the similar sensory adaptations to similar light environments that evolved independently in each species (Moran et al., 2023). However, the relationship between scales and cave habitat is unclear (Zhao and Zhang, 2009). Therefore, it is also necessary to explore the morphological traits of the skin in an evolutionary comparative framework.
It is known that the habitat influences skin immunity, microbial composition, and host skin sensitivity (Scharsack et al., 2007; Peuß et al., 2020; Zhou S. et al., 2022). The maintenance of the health of an organism is a result of a dynamic interplay between the microbiota, host skin cells, and immune system, which work synergistically in a mutually beneficial manner (Austin, 2006; Belkaid and Hand, 2014; Ellison et al., 2021). In a study involving the three representative morpho-species, it has been shown that S. rhinoceros (SP) displayed the strongest innate immunity, which suggests this as a possible adaptation for greater habitat heterogeneity (Yang et al., 2016).
Caves present a challenging environment characterized by limited food resources and low diversity (Gibert and Deharveng, 2002; Lafferty, 2012). Under such conditions, skin functions may be significantly impacted, with effects observed in mucus composition and immune response. As a result, there may be an increased risk of harmful microbial infections and associated health-related issues (McCormick and Larson, 2008).
Multispecies transcriptomics offer new insights into the origins of adaptive phenotypes in cavefish (Stahl and Gross, 2017; Meng et al., 2018; Qi et al., 2018). It can also reveal plasticity or adaptive changes in habitats of related genes that may mediate energy metabolism (Riddle et al., 2018; Lam et al., 2022), immune regulation (Huang et al., 2016; Peuß et al., 2020), and oxidative stress (Krishnan et al., 2020). However, the broader extent of such responses in Sinocyclocheilus skin remains underexplored, especially regarding the diverse morphological variations and intriguing immune mechanisms observed in SPs.
To better understand the habitat correlations, morphotypes, and genetic basis of Sinocyclocheilus cavefish skin related adaptation, we conducted a comprehensive radiation-scale analysis. These incorporated representatives from the major clades and the three main habitat types. We envision that this approach will allow us to uncover subtle patterns pertaining to the complex interplay among gene regulation, physiological adaptations, and the unique constraints imposed by different habitats on the skin of these cavefish. Specifically, we focus on the following objectives: (1) investigating the shared and derived adaptive mechanisms of the skin of cavefish to diverse habitats; (2) examining the phylogenetic relationships among species based on transcriptomic analysis of their skin-related genes; and (3) elucidating the associations between variations in skin color, scale morphology, and their respective habitats, by integrating molecular mechanisms with observations of morphological traits of the skin.
2 Materials and methods
2.1 Sample collection
Animal care and experimental protocols were approved for this study by the Guangxi University Ethical Committee under the approval document (GXU-2021-126). To investigate the adaptive mechanism for cave-dwelling in the Sinocyclocheilus radiation, 9 species were collected from caves in Yunnan, Guizhou and Guangxi of China during 2019 – 2021 as a part of an ongoing phylogenomic study. Cavefish were transported alive to the laboratory in oxygenated plastic bags in a cooler box to both keep the temperatures low (18 – 20°C) and to provide darkness. We let them settle down prior to initiation of the experiments. Following morphological observations (mentioned below), they were anesthetized using 30 mg/L MS-222 (ethyl 3-aminobenzoate methanesulfonate; Sigma-Aldrich). The skin tissue was taken from the right side near the back of each individual in approximately 0.8 × 1.0 cm under sterile conditions. Three biological repeats were taken for each species. The tissue extraction was done in a DNA/RNA-free clean room. Biopsied tissues were placed in RNAlater and stored in an ultra-low temperature freezer at -83°C prior to further analysis.
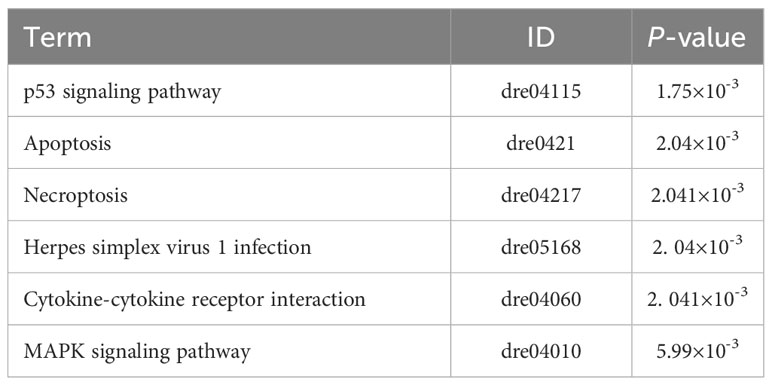
Given their rarity, sampling difficulty, and the need for a representation of the diversification, we enhanced taxon sampling. By adding skin transcriptomes for five additional Sinocyclocheilus species from previous studies and supplementing the morphological data with micrographs of Sinocyclocheilus qiubeiensis and Sinocyclocheilus tianlinensis (Yang et al., 2016; Li, 2019; Li et al. 2020). Overall, the dataset included species representing the four main clades in the context of Mao et al. (Mao et al., 2021), as well as the three main eye-types/habitats (normal-eyed/surface, SU; normal-eyed/stygophilic, SP; micro-eyed/stygophilic, SP; eyeless/stygobitic, SB), representative of the Sinocyclocheilus radiation (Figure 1; Supplementary Table S1). The eye-type in this study serves as a practical, easily observable feature that corresponds with the habitat occupation of these species.
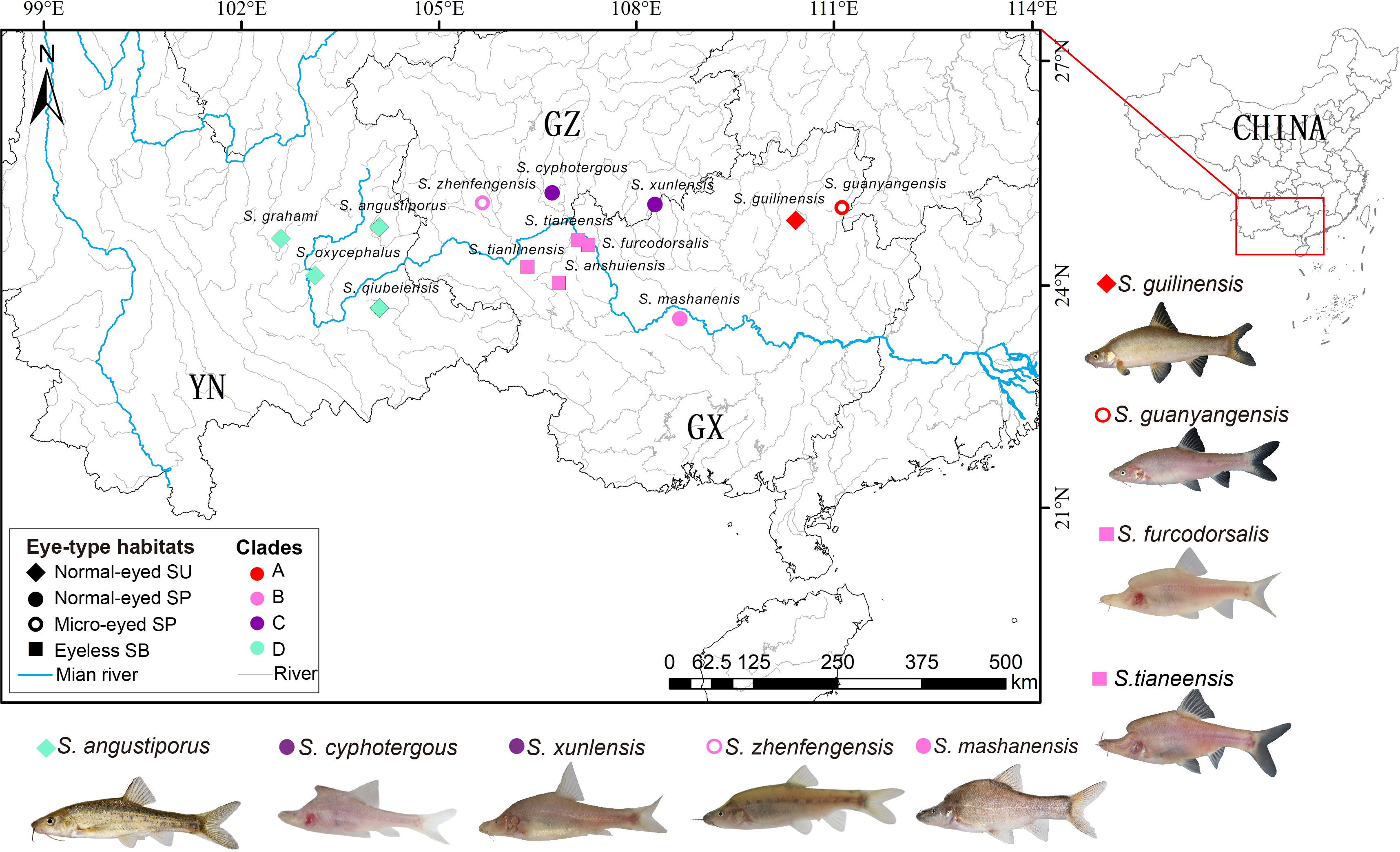
Figure 1 Geographic distribution of sampling sites, morphotypes and clade affiliations for the 14 species representative of the 4 major clades of the Sinocyclocheilus radiation. Symbols represent eye-types and habitats; color represents their clades.
2.2 RNA extraction and transcriptome sequencing
In order to generate transcription data for Sinocyclocheilus skin from the morphotypes, the total RNA was extracted from the 27 skin samples (9 species that we sampled) using Trizol methods; mRNA was captured from total RNA through Oligo (dT). To synthesize cDNA, fragmented mRNA was utilized as a template in the M-MuLV reverse transcriptase system. Random oligonucleotides acted as primers to initiate the synthesis of the first cDNA strand. RNaseH was then used to degrade the RNA strand, and dNTPs were added under the DNA polymerase I system to synthesize the second cDNA strand. The library was constructed using the NEBNext® Ultra™ RNA Library Prep Kit (Illumina, USA). The RNA quality and concentration were measured using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). The effective concentration of the library was again accurately quantified by qPCR to ensure the quality of the library. An Illumina NovaSeq 6000 was used to sequence the library, producing paired-end reads of 150 bp each. The library construction and sequencing were conducted at Beijing Novogene Bioinformatics Technology Co., Ltd.
2.3 Data preprocessing
In addition to our data, RNA-seq data for 5 species were obtained from two public databases. We downloaded transcriptome datasets for 3 species from the public DRYAD database: Sinocyclocheilus oxycephalus, Sinocyclocheilus tianlinensis, and Sinocyclocheilus qiubeiensis (https://datadryad.org/stash/share/t5cZIXoVUgyhpzEP6z-GN6xjc5EU3TvPPEwbdIo7siI), and transcriptome datasets for 2 species: Sinocyclocheilus grahami (NCBI: SRR2960332) and Sinocyclocheilus anshuiensis (NCBI: SRR2960751) from the NCBI database. The skin transcriptomes from the 9 species from our study and the 5 additional species were evaluated with FastQC v0.11.9 (Andrews, 2010). To ensure the accuracy and reliability of our data, we implemented a rigorous filtering process. This involved removing reads containing adapters, poly-N sequences, and those with low-quality scores. The resulting high-quality, clean reads formed the basis for all downstream analyses. The clean reads are deposited in the NCBI database and will be made public upon acceptance (NCBI SRA accession number: SUB13474193).
2.4 Transcriptome assembly, annotation and selection of orthogroups
We used Trinity v2.8.6 (Grabherr et al., 2011) to assemble clean reads for each species and extracted the longest transcripts (unigenes), to obtain single gene sequences. Subsequently, the CD-HIT v4.6.8 (Fu et al., 2012) was used with a 95% threshold to cluster sequences and eliminate redundancy in the final assembly. To predict the full open reading frames (ORFs) for each gene, we used TransDecoder (http://transdecoder.github.io/) default parameters. We then annotated the resulting protein sequences using BLASTP (Blast+ v2.6.0) with an E-value of 10-5 and using the UniProt database (v2022_05) (Mahram and Herbordt, 2015). We retained only UniProt germplasm from the top 10 hits for each species and selected the final UniProt ID based on the number of species that used it as the best match. Using OrthoFinder v1.1.2. (Emms and Kelly, 2015), we performed gene clustering while retaining only those containing at least one transcript per species. These filtering steps resulted in 7,374 annotated orthologous groups for downstream analysis of expression differences.
2.5 Construction of phylogenetic trees and screening of positive selection gene
For phylogenetic inference, we utilized single-copy gene families obtained from OrthoFinder, as previously described. We discarded gene families comprising sequences shorter than 200 amino acids. The retained amino acid sequence families were aligned with Muscle 5 (Edgar, 2022), with default parameters. To excise ambiguously aligned regions, we used Gblock v0.91b (Castresana, 2000). We conducted a maximum likelihood (ML) phylogenetic analysis on a refined set of 1,369 single-copy orthogroups using IQ-TREE 2. We selected the best-fit model, GTR+F+I+I+R3, which encompasses 72,820 distinct patterns, 77,156 parsimony-informative sites, 76,466 singleton sites, and 1,318,361 constant sites, to construct an unrooted phylogeny. To determine node support, we performed an ML bootstrap analysis using 1,000 pseudoreplicates over 102 iterations. (Minh et al., 2020). The resulting ML phylogenetic tree (best log likelihood score: -3,627,797.972) was visualized using Figtree (Rambaut, 2009). We explored two methods to root the tree. First, we used mid-point rooting, which places the root of the tree at the midpoint between the two most distant taxa, making the fewest assumptions about the evolutionary rates in different lineages. Second, we rooted the tree using a well-established early-merging Clade A species, S. guyangensis (Mao et al., 2021; Mao T. et al., 2022).
To understand the functional genes that may facilitate the adaptation of Sinocyclocheilus to its environment, orthologous of the skin of 14 species were tested for signals of positive selection. The dN, dS, and dN/dS values of the orthologous were calculated using the CodeML program of the PAML package (Yang, 2007). The CodeML parameter was set to “Runmode = 0, Model = 0”. Then, ω> 1 can be judged to have experienced positive selection pressure effects in this gene, and a total of 329 genes were screened. KEGG enrichment of PSGs was performed using KOBAS v2.0.12, and the PSGs in significantly enriched pathways were annotated based on the Evolutionary Genealogy of Genes: Nonsupervised Orthologous Groups (eggNog) database (Xie et al., 2011; Huerta-Cepas et al., 2016).
2.6 Differentially expressed gene analysis of orthogroups
To study how orthologous genes are expressed in three different habitats - SB, SP, and SU - we conducted a mapping analysis of high-quality reads of each species against its corresponding representative orthogroups obtained earlier; for this, we used Bowtie 2 v2.2.9 for direct homology filtering (default settings) (Langmead and Salzberg, 2012), and for gene expression levels analysis we used RSEM v1.2.26 (default setting) (Li and Dewey, 2011). To analyze the gene expression profiles of each species, we utilized the ggbiplot package to conduct a principal component analysis (PCA) following the method outlined by Yeung and Ruzzo (2001). The ggplot2 package was used to statistically generate stacked bar graphs with clustered trees reflecting the similarity between species and habitat groups, as well as information on the expression profiles of the orthologous genes for each species (Thiergart et al., 2020). We then identified differential expression in the orthogroups between species using edgeR and adjusted the resulting P-values through the application of the Benjamini and Hochberg method, which effectively estimates the false discovery rate (FDR). We considered differentially expressed genes to be those with absolute log2 fold change (|log2FC|) greater than 1 and FDR less than 0.05. These thresholds were chosen to determine statistical significance. Finally, GOseq and KOBAS v2.0.12 were used to perform GO enrichment analysis and KEGG enrichment analysis of differentially expressed orthogroups (Kanehisa et al., 2008; Young et al., 2010). P-values were obtained using a hypergeometric test and the significance term with an adjusted P-value threshold of 0.05. Using these methods, we identified significantly enriched biological pathways, including important biochemical metabolic and signal transduction pathways across the orthogroups.
2.7 Skin photography
Using a digital camera (Canon EOS 6D Mark II AF-A) set at a fixed distance of 0.3m from the tank and LUX = 45 for ambient light, we captured images with the following settings: Shutter speed: 1/250s; F/20; ISO 200. Next, the fish were returned to their aquarium system (pH: 7.0-8.0; temperature: 19 ± 1°C; dissolved oxygen: 8.5 mg/L) devoid of light to maintain their natural skin characteristics. Subsequently, we examined the pigmentation and scales in samples preserved in 70% ethanol using a Leica M165FC stereomicroscope.
2.8 Calculation of mean color, dark clustering and measurements of scales
To compare the body color differences of the Sinocyclocheilus species, we used the Color Summarizer v0.8 (http://mkweb.bcgsc.ca/colorsummarizer/analyze) to digitize the average color values of the skin. In order to ascertain the average red-green-blue (RGB) color component of the image, we employed a systematic approach. Firstly, a set of reference colors that had the least deviation from the image colors were identified, which allowed to discern variation in color with greater precision. Specifically, 5 reference colors denoted by “k” were selected. Subsequently, 5 representative color ratios from each sample taken. Finally, the average RGB color values were calculated from these ratios to obtain an overall RGB color component of the image. This procedure provided an accurate and reliable estimation of the RGB color values. This process was repeated three times separately for the three samples collected to obtain a representative color for each Sinocyclocheilus species. Our analysis focused on the 5 most representative color ratios in order to calculate the proportion of darker blotches. For instance, in a particular species, mean color values of RGB (250, 250, 250) were observed in the white areas while the darker black areas had color values closer to RGB (0, 0, 0). We postulated that a higher concentration of melanin deposition is associated with a decrease in RGB values towards RGB (0, 0, 0). The area and number of lateral line scales were counted for each sample using ImageJ (Abràmoff et al., 2004). The scale sizes were classified into three categories: no-scales, small-scales, and large-scales, and the degree of scale cover was classified as full-cover, no-cover, and partial-cover.
3 Results
3.1 Transcriptome sequencing data and identification of orthologous
To understand the molecular mechanisms underlying skin coloration in Sinocyclocheilus species, we generated high-quality transcriptome sequencing data from skin tissue samples of 9 distinct Sinocyclocheilus species (Supplementary Table S2). Following this, a de novo assembly was performed for sequences from the 14 species. Among them, S. oxycephalus (205,993) has the highest number of transcripts and S. grahami (80,118) has the lowest. The average transcript length post-assembly varied between 856 and 1,158 bps, while the N50 length ranged from 1,621 to 2,414 bps. The OrthoFinder identified a total of 384,146 orthologous genes in the transcripts. Overall, orthogroups accounted for 94.3% of the genes. To facilitate comparative analysis among the various species, we focused on the 7,374 orthologous genes present in all examined species. These orthologous gene families were clustered, and 1,369 single-copy orthologues genes were obtained after multiple sequence alignment and low quality pairwise pruning. These orthologous genes were subjected to quantitative comparative analysis.
3.2 Distinct habitats influence gene expression variation
A total of 1,348 differentially expressed genes (DEGs) were identified on the skins of Sinocyclocheilus species by conducting a pairwise comparison across the three habitats (Figure 2; Supplementary Table S3). When compared to SUs, SPs exhibited 200 up-regulated and 467 down-regulated genes; SBs displayed 292 up-regulated and 394 down-regulated genes. The majority of DEGs were up-regulated in SUs and the highest number of down-regulated DEGs was observed in SPs (Figure 2A). We also found more unique gene up-regulation in SP vs. SB, and that more of their shared genes were consistently down-regulated compared to SUs (SU vs. SP and SU vs. SB, Figure 2B). This suggested a significant impact of cave habitats on gene expression patterns.
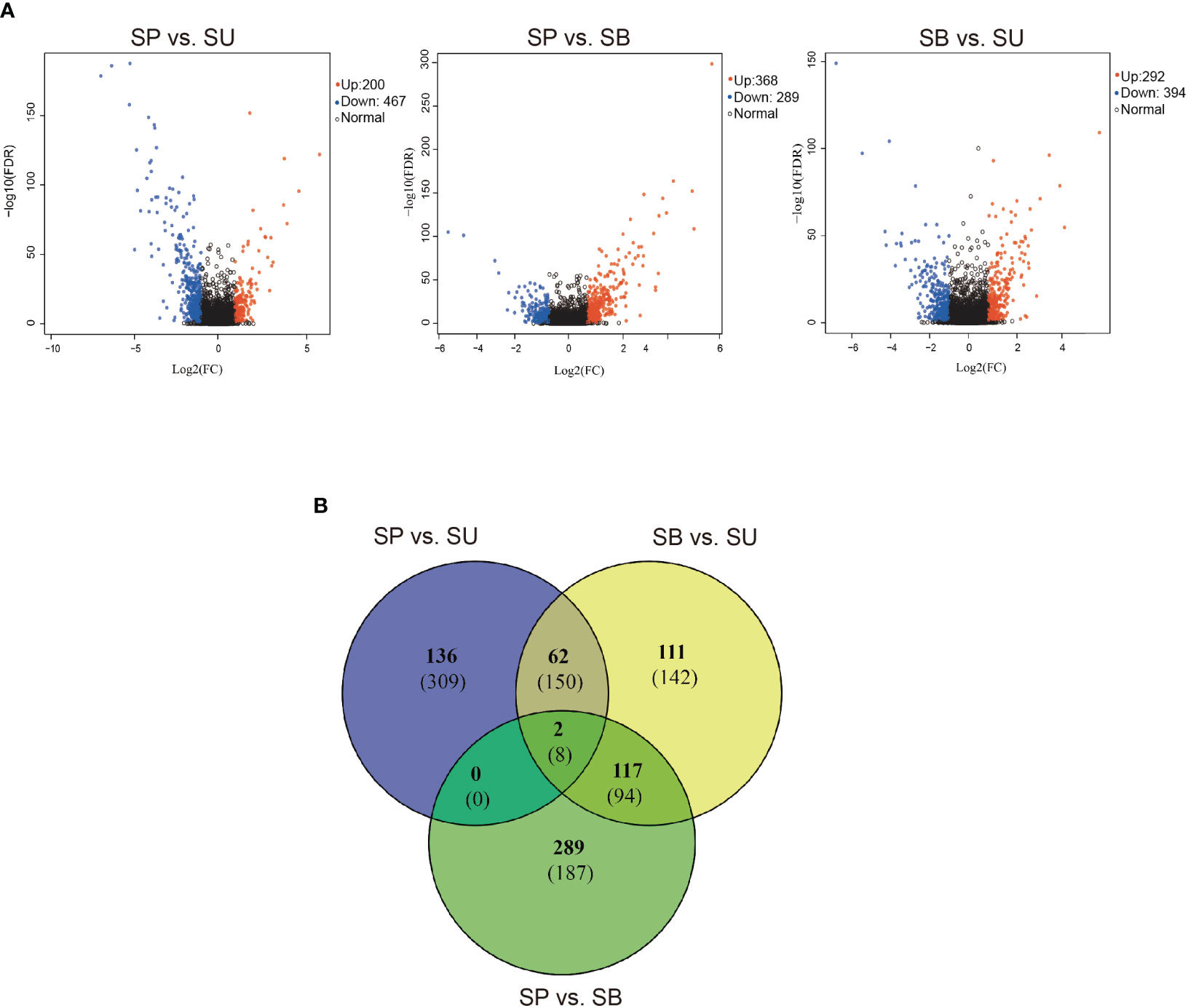
Figure 2 (A) Volcano plots of the distribution of DEGs between SU vs. SP, SP vs. SB and SU vs. SB. The x-axis shows log 2-fold change in gene expression. The y-axis shows -log 10 (P-value). The further away from 0 on the x-axis, the greater the change in expression, and the higher the y-axis, the greater the significance. Blue dots indicate up-regulation, red dots indicate down-regulation and black dots indicate no change in expression in the DEG. (B) Venn diagrams depict shared and unique variations in gene expression among the three main habitats (SU, SP and SB). The numbers in each section correspond to the number of DEGs from gene expression estimates. The number of up-regulated DEGs is listed at the top (in bold) and the number of down-regulated DEGs is listed at the bottom.
GO enrichment analysis of 1,348 DEGs in different eye-types/habitats identified 630, 565 and 799 significantly enriched GO terms in SU vs. SP, SP vs. SB and SU vs. SB, respectively (adjusted P<0.05) (Supplementary Table S4). These DEGs were involved in functional responses mainly related to stimulus responses, catalytic activity, multi-organism process, and immune system process (Figure 3A). We found that the co-enriched terms in these three groups were energy metabolism related (Supplementary Table S5). We also found two major categories of GO terms have been enriched in SU vs. cave dwellers (SP and SB). Stimulus response-related terms such as external biotic stimulus, bacterial, fungal, and viral reactions, and numerous immune-related terms. SU vs. SP was mainly related to cellular immunity, such as leukocyte mediated immunity (GO:0002444), regulation of macrophage derived foam cell differentiation (GO:0010743); while SU vs. SB increased the regulation of apoptosis, such as: positive regulation of MAPK cascade (GO:0043410), regulation of neuron apoptotic process (GO:0043523), regulation of epithelial cell apoptotic process (GO:1904035). Anyhow, this suggested that these observations may be due to the differences in the exogenous biological stimulation of surface water environment. Cave species seemed to have different immune strategies to these external stimuli.
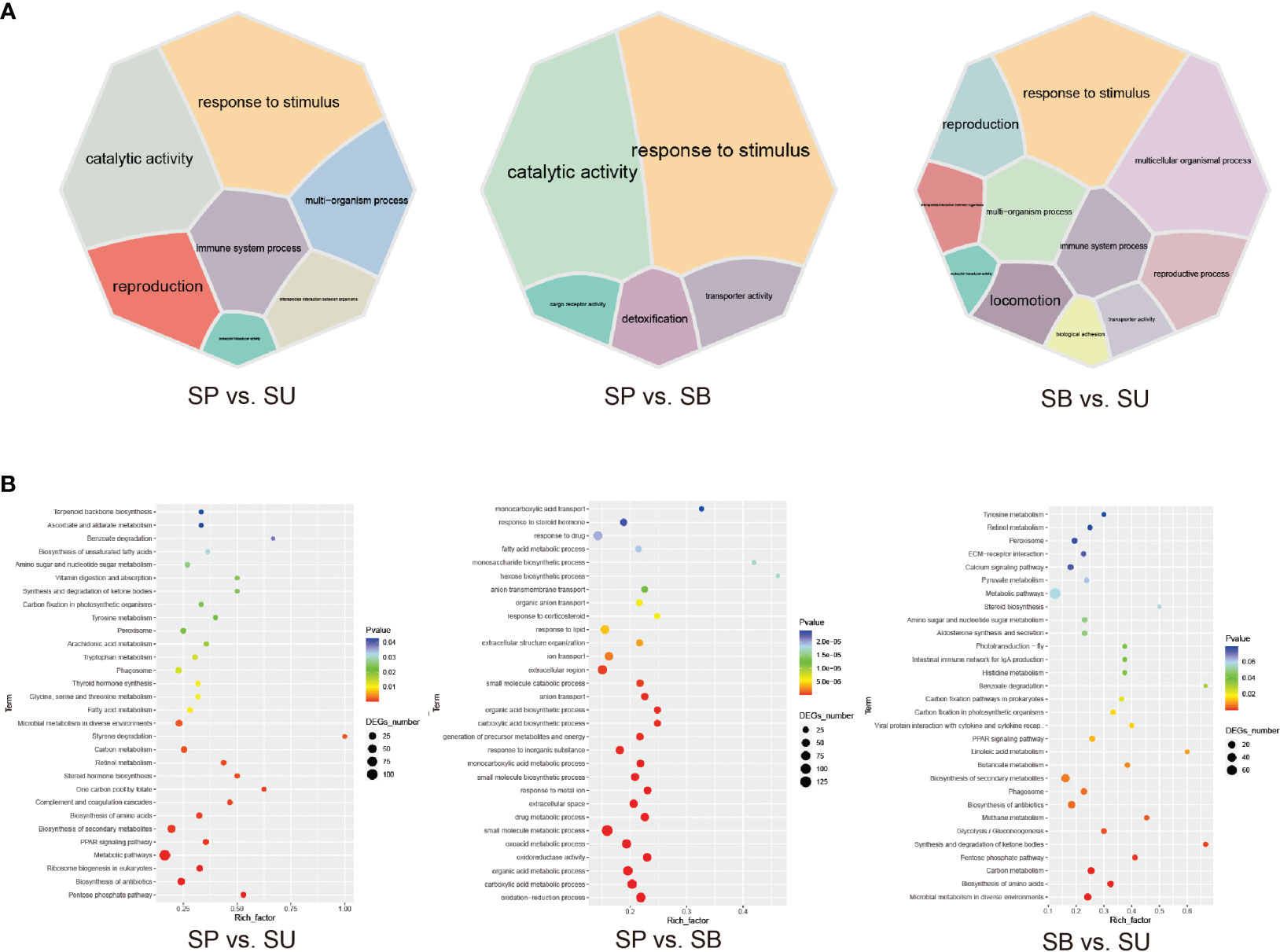
Figure 3 Enrichment maps for DEG among the three main habitats (SU, SP and SB). (A) Classification of GO terms significantly enriched in differentially expressed genes (DEGs). 7 GO categories in SU vs. SP; 5 GO categories in SP vs. SB, and 11 GO categories in SU vs. SB. Different GO categories are displayed in different colors, and the size of the module represents the number of DEGs associated with the corresponding functional item. (B) The top 30 KEGG pathways that are enriched in differentially expressed genes (DEGs), including SU vs. SP, SP vs. SB and SU vs. SB.
Genes associated with changes in oxygen levels showed differences in expression across different habitats, for example, response to hypoxia (GO:0001666), response to decreased oxygen levels (GO:0036293), cellular response to hypoxia (GO:0071456), etc. Interestingly, only in SPs and SBs can respiratory electron transport chain (GO:0022904), mitochondrial respirasome (GO:0005746), respiratory electron transport chain (GO:0022904), and mitochondrial respirasome (GO:0005746), respiratory chain complex (GO:0098803) and response to pH (GO:0009268) be identified (Supplementary Table S5). This may reflect the different cellular and mitochondrial respiration efficiency of cave dwellers.
The number of significantly enriched pathways in three groups was 30, 38, and 22, respectively, and the same pathways were mainly related to metabolisms, such as pentose phosphate pathway (ko00030), biosynthesis of amino acids (ko01130), Carbon metabolism (ko01200), PPAR signaling pathway (ko03320) and microbial metabolism in diverse environments (ko01120), etc. Secondly, immune response-related pathways were phagosome (ko04145) (adjusted P<0.05) (Figure 3B; Supplementary Table S6). This suggested that the strong influence of the habitat environment leads to differences in skin metabolism and skin microbial metabolism among different habitat populations. Compared to SPs, all DEGs (28) enriched in the PPAR signaling pathway and carbon metabolism pathway were up-regulated in SUs, followed by 16 DGEs in SBs. However, all DEGs (9) enriched oxidative phosphorylation (OXPHOS, ko00190) pathway and 5 DEGs enriched fatty acid degradation pathway (ko00071) were upregulated in SPs. This may indicate differences in the regulation of energy and mitochondrial metabolism between species in different habitats (Supplementary Table S7). These pathways in different eye-types/habitats showed that there were common molecular mechanisms to understand their habitual differences and evidence of genetic variation and transcriptional plasticity or adaptation in response to environmental change.
In particular, microbial immune-related pathways such as complement and coagulation cascades (ko04610), intestinal immune network for IgA production (ko04672), and viral protein interaction with cytokine and cytokine receptors (ko04061) found in SU vs. cave dwellers (SP and SB) (Supplementary Table S6). This may indicate differences in immune responses to microorganisms between SUs and cave dwellers. Moreover, hematopoietic cell lineage (ko04640) and ECM-receptor interaction (ko04512), and pathways associated with oxidative stress (OXPHOS, glutathione metabolism, cysteine and methionine metabolism, metabolism of xenobiotics by cytochrome P450, drug metabolism - cytochrome P450) exist only in SP vs. SB. This may indicate a widespread oxidative stress response of cave dwellers to stimuli of the cave environments.
We also found that phenylalanine, tyrosine and tryptophan biosynthesis pathway (ko00400), and especially the tyrosine metabolism pathway (ko00350) may be influenced by their habitats, and affected melanin differences (Supplementary Table S6).
3.3 Positive selection genes in the skin of 14 species
After analyzing the KEGG pathways of these 329 positive selection genes (PSGs), we found that genes under positive selection were most significantly enriched in six metabolic pathways (Table 1). They were associated with viral infection, signaling, and cell necrosis and apoptosis. A total of 18 PSGs were identified in these enriched pathways, among which the tumor protein p63 regulatory 1 (OG0018161) was under the strongest selection pressure (Supplementary Table S8). These genes may be involved in the adaptation process of Sinocyclocheilus to cave dwelling. We also found that Reticulocalbin 3, EF-hand calcium binding domain (OG0016528), S100 calcium binding protein U (OG0016522), and positive regulation of vitamin D (OG0016683) have all been implicated in calcium regulation.
3.4 Phylogeny based on orthologous genes
We filtered the clusters that comprised a single sequence from each of the 14 transcriptomes and retrieved 1,369 putative single-copy orthologous genes. We concatenated and aligned these genes into a supermatrix 1,471,983 informative sites for the 14 taxa. The two orthologous gene-based maximum likelihood trees, rooted using both the mid-point method and S. guanyangensis as the outgroup, showed two similar trees, each with five well-supported clades (Figure 4; Supplementary Figure S1). Three notable discrepancies with previous phylogenies were the positions of early diverging Sinocyclocheilus lineages: S. xunlensis, S. oxycephalus, and S. furcodorsalis. Our phylogenetic tree unambiguously showed that S. oxycephalus was a separate lineage, while S. xunlensis was recovered as the sister group of S. guilinensis; four species including S. furcodorsalis constituted a separate lineage.
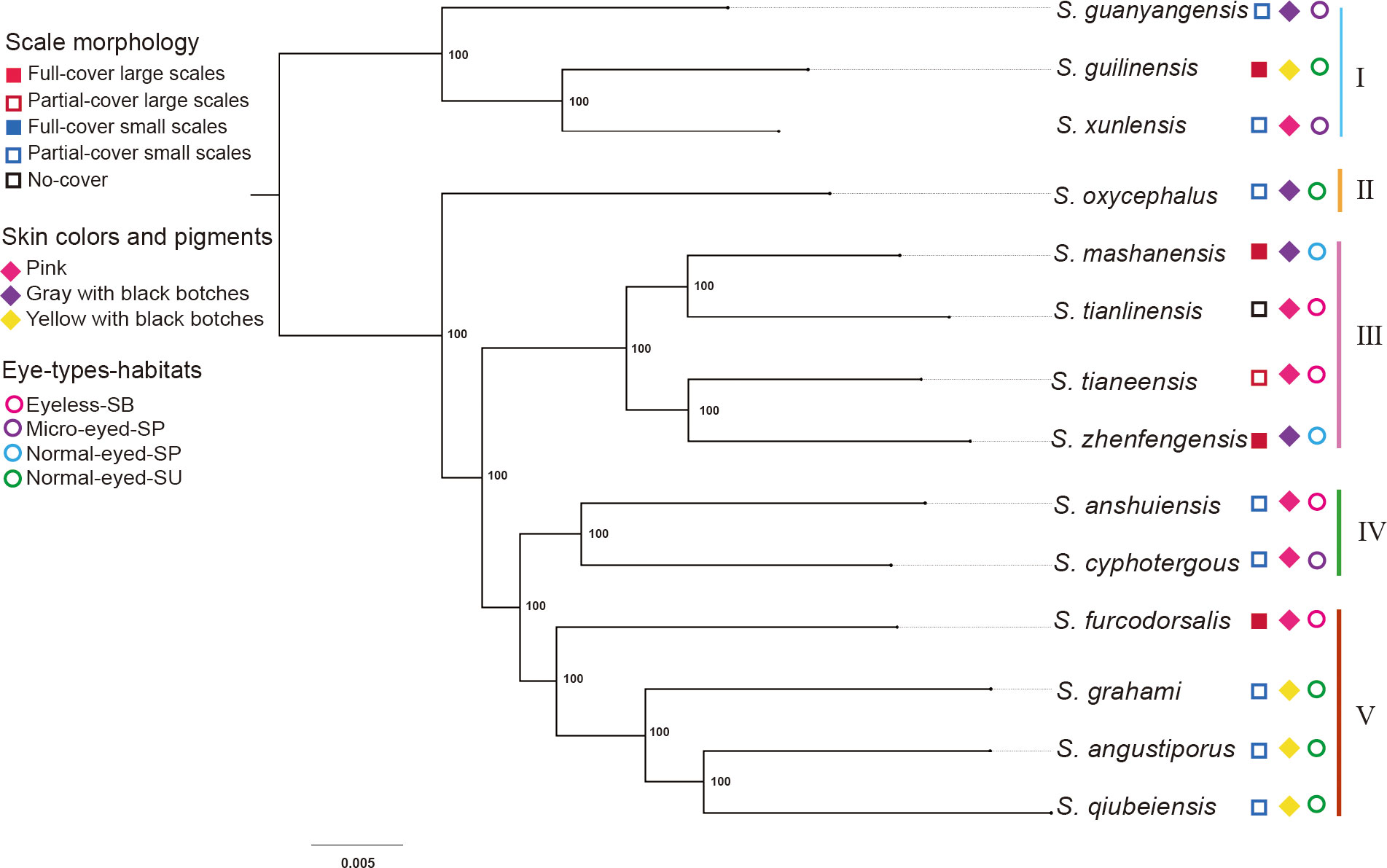
Figure 4 Phylogeny of 14 Sinocyclocheilus species with the main clades and Lineages designated by I-V. The maximum likelihood tree was derived from the series data of 1,369 single-copy orthologous genes; bootstrap support is displayed at nodes.
3.5 The skin color
The analysis of color variation in the 14 Sinocyclocheilus species highlighted a rich diversity in body coloration and patterns, with each species exhibiting unique colors, mainly in a combination of pinkish-white, gray, and yellow (Figure 5; Supplementary Figure S2).
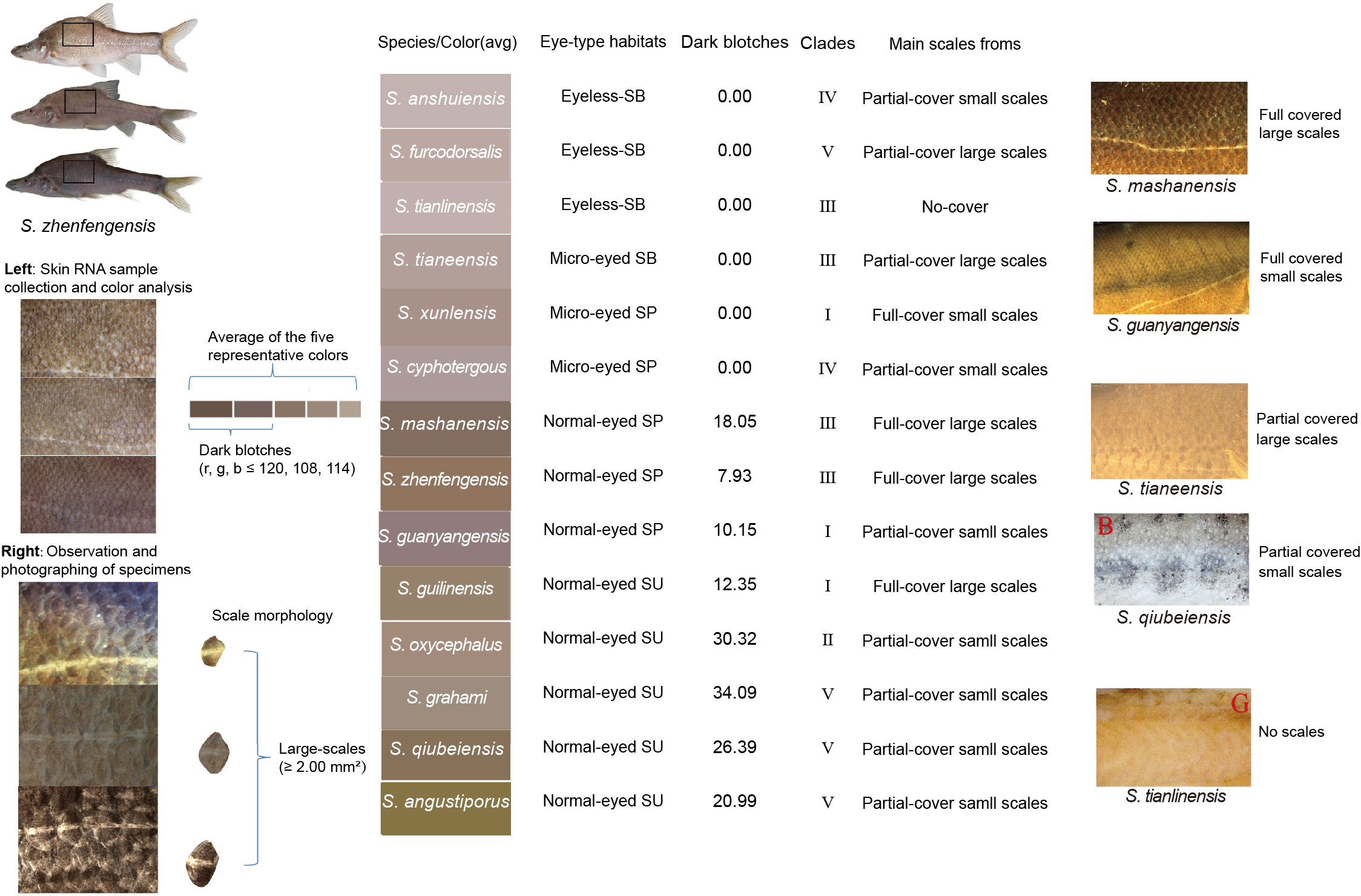
Figure 5 Color and scales characteristics of the 14 Sinocyclocheilus species. The micrographs of S. qiubeiensis and S. tianlinsis named "B" and "G", respectively, were obtained from a previous study (Li, 2019).
For every species, color attributes were assessed based on average skin color value and the proportion of dark blotches and distinct spots on the skin. Sinocyclocheilus tianlinensis had the highest skin RGB value (195, 181, 177), followed by S. anshuiensis (191, 179, 177) and S. furcodorsalis (188, 163, 155); S. grahami (138, 123, 111), S. oxycephalus (142, 116, 90), and S. qiubeiensis (142, 127, 104) displayed the lowest RGB values (Supplementary Table S9). The RGB values for black blotches in these species were under (120, 108, 114) with lower values signifying darker colors, such as dark blotches and spots (Figure 5; Supplementary Table S9).
Skin colors were associated with eye-types and habitats. Normal-eyed SUs had yellow or gray skin, while most SPs exhibited gray or pale gray tones. Unique spots were also indicative of eye-type and habitat groups; normal-eyed S. grahami (SU) had the highest proportion of black spots, succeeded by normal-eyed S. oxycephalus (SU) and normal-eyed S. qiubeiensis (SU). Eyeless SBs and micro-eyed S. xunlensis (SP), which displayed depigmented pink-white skin, had the lowest percentage of dark blotches (Figure 5; Supplementary Table S9). However, microscopic observations showed that SBs and S. xunlensis (SP) still had numerous tiny black blotches dispersed across their skin (Supplementary Figure S2); comparatively, eyeless SBs were pink with fewer pronounced dark blotches and distinct spots.
When examining color in the context of phylogeny, Clades I-V displayed various combinations of skin color and pigmentation relative to eye-types and habitats (Figure 5; Supplementary Figure S2). Notably, in Clade III and Clade IV, which contain the largest proportion of exclusive cave dwellers, SBs were pink with fewer dark blotches. Hence, the Sinocyclocheilus genus adapted to cave environments and evolved with convergent skin coloration in different evolutionary clades.
3.6 Scale characteristics
Microscopic examination of scale characteristics showed a marked reduction in scale size and coverage in 11 Sinocyclocheilus species except for three species (S. mashanensis: mean ± SD: 3.74 ± 1.36, S. zhenfengensis: mean ± SD: 2.93 ± 0.95, S. guilinensis: mean ± SD: 2.00 ±0.62), which exhibited complete coverage by large scales, and the mean number of their lateral line scales are respectively 49 ± 3, 43 ± 4 and 50±8. S. oxycephalus (mean ± SD: 0.17 ± 0.11), S. qiubeiensis (mean ± SD: 0.61 ± 0.28), and S. grahami (mean ± SD: 0.72 ± 0.40) had smaller scales, also with more lateral line scales (mean ± SD: 70 ± 3, 78 ± 3 and 72 ± 2), but most of the body scales were buried in the skin and disappeared (Figure 5; Supplementary Figure S2). We found that larger scales and fewer lateral line scales were mainly present in Clade III, IV while smaller scales and more lateral line scales species were mainly found in Clade V. The reduction of scale coverage can be found in all clades (I-V). This may indicate convergent evolution of scale reduction in Sinocyclocheilus.
Generally, SBs exhibited smaller and fewer scales than SPs. The scales of S. tianlinensis (SB) were absent altogether, and S. anshuiensis (SB) had only rare scales. These species in Clade III, Clade IV revealed a pattern of scales, characterized by a gradual reduction in scale size, decreased coverage and the number of lateral line scales, and eventual disappearance, which may be related to cave adaptation. Clade V species displayed variability in scale morphology and coverage, suggesting a chaotic pattern of scale evolution in Clade V unrelated to habitat. Here, the small scales were widely spaced (Figure 5).
3.7 Gene expression patterns and skin morphology
The PCA analysis and cluster analysis of the 14 Sinocyclocheilus species using expression data from 7,374 orthologous genes revealed that 13 species clustered together, separate from S. angustiporus (Figure 6A; Supplementary Table S10). In addition, micro-eyed SPs and eyeless SBs showed differences in gene expression compared with normal-eyed SUs (Figure 6B; Supplementary Table S10). This suggested that patterns of gene expression were similar between these species, but the effects of habitats can still be seen.
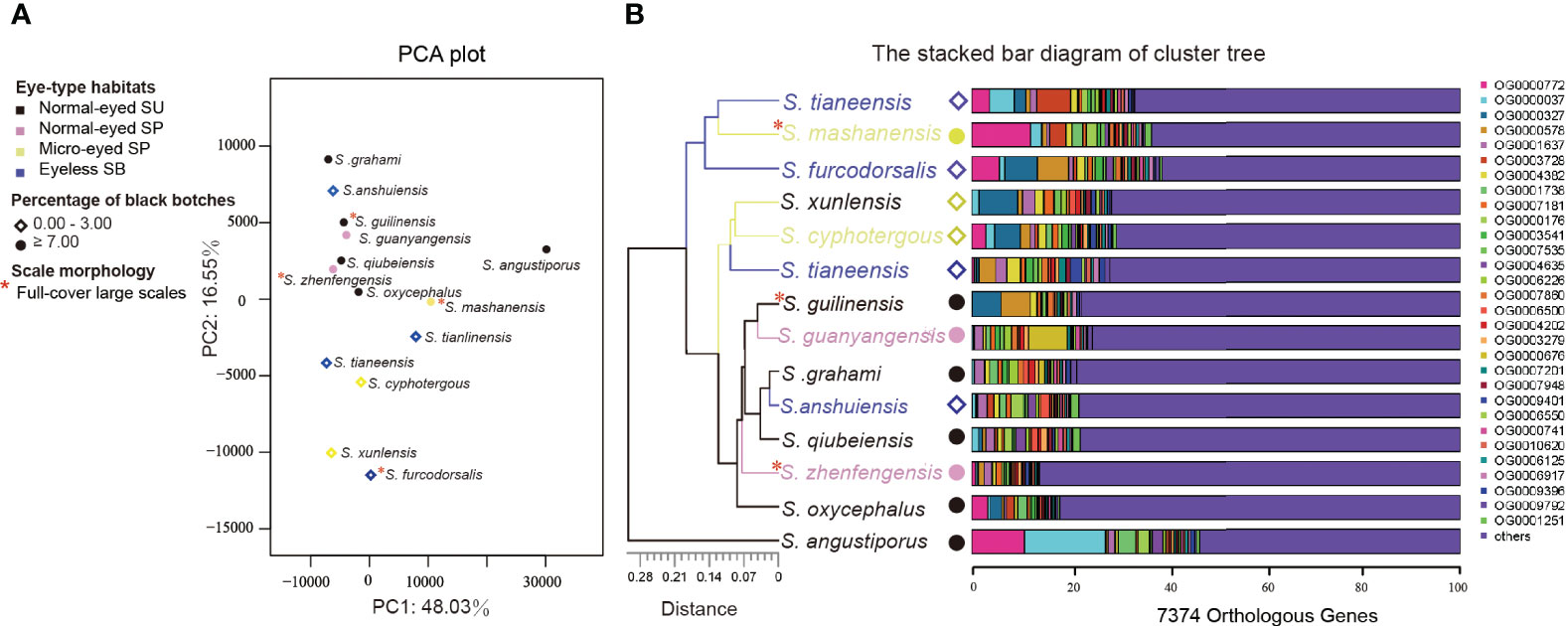
Figure 6 Gene expression clustering of the 14 Sinocyclocheilus species based on 7374 orthologous genes. (A) PCA plot showing the relationship between gene expression patterns and habitats/skin morphologies. Together, PC1 and PC2 explain 64.58% of the variability. (B) The stacked bar diagram of cluster tree showing the expression differences of the top 30 orthologous genes with expression changes in different species, and the relationship between these orthologous gene expression changes and habitats/skin morphology.
Interestingly, the second principal component (variance explained 16.55%) differentiated species with and without black blotches (Figure 6A). Moreover, S. tianeensis, S. cyphotergous and S. xunlensis with little black blotches were clustered in one branch, but expression patterns vary among species with full-cover large scales (Figure 6B). This suggested that changes in gene expression patterns may affect pigmentation similarly but not scales.
4 Discussion
Broadly, we investigated the skin-related morphology (color and scale characteristics), gene expression patterns, and functional enrichment of differentially expressed gene (DEGs) and positive selection genes (PSGs) among various morphotypes of Sinocyclocheilus species, representative of the phylogeny, living in three habitat types (surface, SU; stygophilic, SP; stygobitic, SB). Our results suggested that habitats may influence changes in color and scale characteristics, gene expression and their function, and hence a driver of skin evolution. Here we discuss our findings and their implications for understanding the broad scale patterns of adaptation of Sinocyclocheilus skin for cave-dwelling.
4.1 Possible adaptive mechanisms to different habitats
In our analysis of DEGs, we identified distinct patterns pertaining to metabolism, oxidative stress, and immune responses in various Sinocyclocheilus species. These patterns appear to be correlated with their specific habitats. Such results substantiate the presence of cave-environmental gradients within natural ecosystems. In line with our findings, several studies have pointed toward physiological and metabolic distinctions between surface dwelling and cave dwelling species (Yang et al., 2016; Stahl and Gross, 2017; Krishnan et al., 2020; Boggs and Gross, 2021; Medley et al., 2022).
These variances could be due to differences in habitat characteristics like light exposure, ambient oxygen levels, and nutrient resource availability. Furthermore, the effects of different experimental sampling methods on gene expression has also been shown (Tessler et al., 2020). The intricate interplay among these factors significantly intensifies the complexity of environmental gradients, making the prediction of gene expression responses more challenging (Passow et al., 2017).
While the analysis of coloration and large-scale gene expression patterns may provide some insights, they fall short of identifying the specific environmental factors driving convergent evolution in these habitats. A recent comprehensive genomic study encompassing 248 species of A. mexicanus from diverse habitats shed light on the underlying mechanisms for convergent evolution in cavefish (Moran et al., 2023). Likewise, changes in fish transcriptome levels have been linked to phenotypic responses to environmental extremes (Wang and Guo, 2019). However, solely relying on this approach provides insights that might be limited to the specific context of the study in question. A potentially more effective strategy would combine pinpointing and investigating differentially expressed genes (DEGs) with functional annotations based on a priori hypotheses. Such an integrated approach could provide deeper insights into the range of environmental stressors within a habitat and illuminate the evolutionary pathways leading to different phenotypes.
4.1.1 Metabolic differences among species across habitats
A large number of GO terms related to energy metabolism (e.g., lipid metabolism, fatty acid metabolism, carbohydrate metabolism, mitochondrial respiration, etc.) were enriched in the comparison of species from different habitats, which were further supported by common KEGG enrichment pathways, such as energy-related carbon metabolism, pentose phosphate pathway, PPAR signaling pathway, fatty acid metabolism, fatty acid degradation, and OXPHOS.
Most of these DEGs in energy metabolism-related pathways were up-regulated in SUs. This may indicate that SUs, that live in resource rich environments compared to cave dwellers, have a higher energy metabolic rates. In fact, the lowering of metabolism is a well-known feature of organisms living in resource poor cave-environments (Soares and Niemiller, 2020). For instance, Astyanax cave-morphs have a lower oxygen consumption and metabolic rate compared to their surface morphs (Moran et al., 2014; Boggs and Gross, 2021). Many of these genes were associated with glycolysis such as fructose-1, 6-diphosphatase (OG0001025), glyceraldehyde-3-phosphate dehydrogenase (OG0001738), 6-phosphogluconate dehydrogenase (OG0011314) and glucose-6-phosphate 1-dehydrogenase (OG0011750) (Okar and Lange, 1999; Randhawa et al., 2014). One of the reasons for this may be due to an abundance of UV light in surface habitats stimulating the skins of SUs leading to enhanced glycolysis (Randhawa et al., 2014).
The differences between cave dwellers (SP and SB) were mainly due to OXPHOS and fatty acid degradation related genes, where they were upregulated in SPs. In addition, under the condition of using fat as an energy source, the expression of the pyruvate dehydrogenase complex-related genes (OG0011863) was upregulated; this is known to contribute to the dynamic balance of glycolysis and tricarboxylic acid cycles (Gray et al., 2014; Pham et al., 2022). The enhanced mitochondrial activity in the skin of cave fish may reflect an increased allocation to detect the environment using non-visual sensory organs, such as lateral line organs, neuromasts and other detectors in the skin, which are enhanced in some cavefish, including in Sinocyclocheilus (Yoshizawa et al., 2010; Chen et al., 2022).
In contrast, SBs mainly have enhanced glycolytic processes, as well as enhanced expression of enzymes and proteins related to lipid synthesis and transport, such as fatty acid synthase (OG0001380), very long chain fatty acid elongation protein 6 (OG0010037), fatty acid-binding protein (OG0003744). We also found that troglomorphic traits, such as changes in lipid and energy metabolism, appeared to be linked to increased carbohydrate and fat synthesis processes in SPs and SBs, which promotes fat storage (Xiong, 2021; Lam et al., 2022). Overall, the reduced metabolism of nutrients and mitochondrial respiration in SBs may help Sinocyclocheilus lower their energy consumption, and increase energy storage, facilitating adaptation to a resource-depleted cave environment (Riddle et al., 2018).
4.1.2 Immune responses in species from different habitats
Fish skin is acutely sensitive to alterations in the aquatic environment, and it is noteworthy that DEGs identified within the skin are also responsive to environmental stimuli. Results of the enrichment analysis showed a wide variation in immune mechanisms but with general enrichment in biological processes pertinent to stimulus response, leukocyte proliferation, and apoptosis. Our results also agrees that fish skin has an immune function and highlight the critical role of macrophages during infection (Bangert et al., 2011). In addition to the significant co-enrichment of phagocytosis and inflammatory regulation pathways, we also found that species in different habitats had different immune responses, which may be related to the greater contribution of microbial stimulation and oxygen concentration. Enhanced cave microbial diversity and stimulation in surface water environments may have shaped stronger adaptive immunity in SUs. In contrast, nutrient limitation and reduced dissolved oxygen in cave water environments may contribute to increased susceptibility to pathogens and risk of inflammation to cave dwellers (SP and SB).
Compared with cave dwellers (SP, SB), more pro-inflammatory factors such as tumor necrosis factor, C-X-C chemokine, and complement factor related genes were up-regulated in SUs in immune-related pathways. Moreover, genes that were significantly upregulated in microbial immune-related pathways included: ATPase genes (V-type proton ATPase 116 kDa subunit a: OG0001307; V-type proton ATPase subunit B: OG0009257), which were involved in lysosomal function and autophagy flux. The expression of the vesicle-trafficking protein SEC22b-B (OG0002841) was also upregulated. These results may indicate an enhanced ability of macrophages to deliver in the skins of SUs, and, along with T cells, support the function of adaptive immunity. In addition, the vesicles were subsequently internalized by macrophages, playing a key role in inflammation resolution. Generally, the macrophages and adaptive immunity in the skins of SUs contributed to microbial resistance (Mohanty and Sahoo, 2010; Lü et al., 2012). The reason for this difference was that there were significant differences in microbial metabolism between different habitats in Sinocyclocheilus. we observed upregulation of inositol-3-phosphate synthase 1-A-like isoenzyme X2 (OG0003974) and UDP-N-acetylglucosamine pyrophosphorylase (UAP1, OG0008682), indicating increased eukaryotic growth and reproductive activity in skin microorganisms (Reynolds, 2009; Behr, 2011). The dynamics and intensity of viral replication may tend to weaken due to the lowering of temperature within the cave environment (Demory et al., 2017). In fact, it is generally known that caves are a biodiversity-depleted environment, including that of pathogens (Peuß et al., 2020). Thus, investment strategies for microbial immunity in cave-dwelling species may be lower compared to those of SUs.
There is a disparity between cave-dwelling species (SP, SB) with respect to the expression levels of genes involved in hypoxia (ECM-receptor interaction pathway and hematopoietic cell lines pathway) and inflammatory (complement and coagulation cascades pathway) related responses and pathways (Morikawa and Takubo, 2016; Chen et al., 2021). Inflammation plays an important role in the immune response, serving as a critical pathophysiological reaction of the organism to pathogenic invasion, tissue damage, and other stimuli. Hypoxia also emerges as a significant modulator of both inflammatory and immune responses (Zhao et al., 2016; Bhatti et al., 2017; Duan et al., 2022). In contrast to SPs, a large number of proinflammatory factors (such as complement factors, coagulation factors, and integrin) were up-regulated in SBs. This suggested that SBs had a stronger innate immune response to microbes. Interestingly, we found the least immune-associated GO between SBs and SPs, especially T-cellular immune-related. Thus, cavefish may reduce investment in immune cells, but enhance the sensitivity of the innate immune system as suggested by some previous studies as well (Mayer et al., 2016; Peuß et al., 2020). Hence, our findings suggest that SBs may employ adaptive strategies in cave environments that involve a reduction in T-cell mediated immune responses, but an enhancement in the regulation of innate immune defenses. Moreover, efficient resolution of inflammation and infection in the dermal tissues of SBs necessitates spontaneous apoptosis, induced by microenvironmental factors, to facilitate the clearance of abundant neutrophils at the site of infection.
4.1.3 Oxidative stress in species from different habitats
Though both inflammatory signaling and the mitochondrial electron transport chain (ETC) contribute to the generation of cellular reactive oxygen species (ROS), the mitochondria principally serve as the primary site of cellular ROS production. Additionally, they are the primary targets of a multitude of exogenous toxic effects stemming from environmental chemical agents and ROS themselves (Li et al., 2017). In our study, cave dwellers (SP, SB) were significantly enriched for most of the diverse antioxidant stress-related pathways. The lower dissolved oxygen concentrations and nutrients in the cave water environment, as demonstrated in previous studies, constitute a significant limiting factor (Riddle et al., 2018; Boggs and Gross, 2021). This could contribute to the oxidative stress observed in cave-dwelling species (SP, SB). Examination of genes in the anti-oxidative stress-related pathways of SUs and SBs revealed slight differences in expressions between them. Some of the minor differences were that SUs mainly upregulated genes in the peroxisome pathway, while SBs mainly enhanced genes in the P450 and glutathione metabolism pathway to enhance cellular antioxidant action and integrated detoxification. The increased activity of catalase in SUs contributes to mitochondrial respiration and enhances fatty acid oxidation and utilization (Yu et al., 2003). And cytochrome P450 may target SBs in their sensitivity to exogenous chemicals (including pesticides) in the cave environment, as well as the need for thermoregulation (Guengerich et al., 2016). In a previous study, cavefish showed increased levels of stress compared to surface fish, and the expression of genes involved in glutathione metabolism was also increased to prevent oxidative stress under prolonged fasting (Krishnan et al., 2020; Medley et al., 2022).
Interestingly, almost all oxidative stress-related genes in the peroxisome pathway and P450 pathway were down-regulated in SP skins, suggesting that they may have a stronger environmental oxidative stress resistance. It has been show that the down-regulation of stress-related genes in response to abiotic or biological changes in the environment is an indicator of stress-resistant populations and species (Bailey et al., 2017). Previous studies had shown the heterogeneity of SP environments may enhance their immune capacity (Yang et al., 2016). The extent to which environmental stressors influence appears to be lesser and is contingent upon the physiological strategies adopted by the species; furthermore, microevolutionary processes contribute to the augmentation of resistance of an organism (Sun et al., 2015).
SPs reduced tissue hypoxia by enhancing oxidative phosphorylation (OXPHOS), thereby enhancing mitochondrial respiration and ATP synthesis, thereby enhancing adaptation to hypoxia. Cytochrome c oxidase (COX) was a key enzyme in establishing a more efficient mitochondrial respiratory chain (MRC) to improve oxygen utilization under hypoxic conditions. This may help SPs maintain homeostasis of mitochondrial regulation of energy production, reactive oxygen species homeostasis, and cell death in the hypoxic microenvironment caused by skin immunization (Heather et al., 2012; Garvin et al., 2015). Our study demonstrated different hypoxic adaptation strategies in cave dwellers, implicating that alterations in mitochondrial respiration rate and enzymatic activity might play pivotal roles in the regulation and maintenance of redox homeostasis.
4.2 Transcriptional plasticity and convergent evolution
Our ML trees, based on 1,369 unique single-copy orthologous genes, showed some discordance when compared to recent mitochondrial gene (mt-DNA) and RAD-seq based studies (Zhao and Zhang, 2009; Mao et al., 2021; Mao T. et al., 2022; Wen et al., 2022; Xu et al., 2023). The robust node support in our phylogeny might be attributed to the abundance of genes under selection derived from transcriptome sequencing. However, this precision may have a downside: the unique genes associated with the transcriptome we used could potentially not be under neutral evolution, thereby causing the phylogeny of expressed genes important for the functions of the skin. Most notably, Clade I of our study contains species from three clades that employed mt-DNA/RAD-seq data. This was possibly due to shared skin traits. In addition, S. oxycephalus formed a distinct divergent lineage (II). The skin of these species had a distinctive gray color. And previous studies have shown that its skin is charcoal gray skin unlike other types (Li et al., 2020). Furthermore, we identified S. furcodorsalis, which was from clade B of previous studies placed in our Clade V, which contained predominantly species from clade D (which are mostly SUs) (Mao et al., 2021). This suggested that S. furcodorsalis may have a skin transcription profile similar to that of SUs. In fact, this species, despite being a cave-dweller, enigmatically had some of the most prominent scales. However, our phylogeny lacked high taxon sampling, which prevented us from making deeper inferences on transcriptional plasticity. However, this study highlights the significance and precision of transcriptomic data in understanding the evolutionary relationships among Sinocyclocheilus species.
It is known that the phenotypic adaptations of cave-dwelling in Sinocyclocheilus cavefish involve changes in eye types and color and pigmentation patterns (Meng et al., 2013; Li et al., 2020); our results from the context of skin morphology and gene expression confirm this. We found that eyeless SBs and some micro-eyed SPs without obvious black blotches showing similar expression patterns, even though they were in different clades (Figure 5). Existing literature has established that changes in skin color, driven primarily by adaptation to diverse habitats, serve as a major contributing factor to differential gene expression patterns (Hinaux et al., 2013; Stahl and Gross, 2015; Gross et al., 2016; Li et al., 2020). Skin color adaptations in fish are known to occur rapidly when they are introduced to novel environments (Leclercq et al., 2010; Nilsson Sköld et al., 2013).
The relationship between fish skin color and melanin content was examined, focusing on key genes involved in phenylalanine, tyrosine and tryptophan biosynthesis pathway, and especially the tyrosine metabolism (Figure 7). We found that the tyrosine metabolic pathway, influenced by habitat, played a role in determining melanosis in the skin of Sinocyclocheilus. Previous research has demonstrated that decreased phenylalanine-4-hydroxylase (PAH, OG: 0000390) activity can affect Try activity (Leandro et al., 2017), and inhibition of phenylalanine conversion to tyrosine will lead to reduced melanin production (Staudigl et al., 2011; Zhou et al., 2021). However, we found low expression of the gene encoding PAH in these species. This may be due to the lack of phenylalanine and protein catabolic inputs in the diet of cavefish (Borowsky, 2018). However, L-phenylalanine (L-Phe) is also an essential protein-producing amino acid, and it is important to avoid it being completely catabolic (Xiao et al., 2020). Therefore, in order to achieve the dual role of effective preservation and removal of excess L-Phe, this regulatory mechanism may be more favorable to the adaptation of their environment.
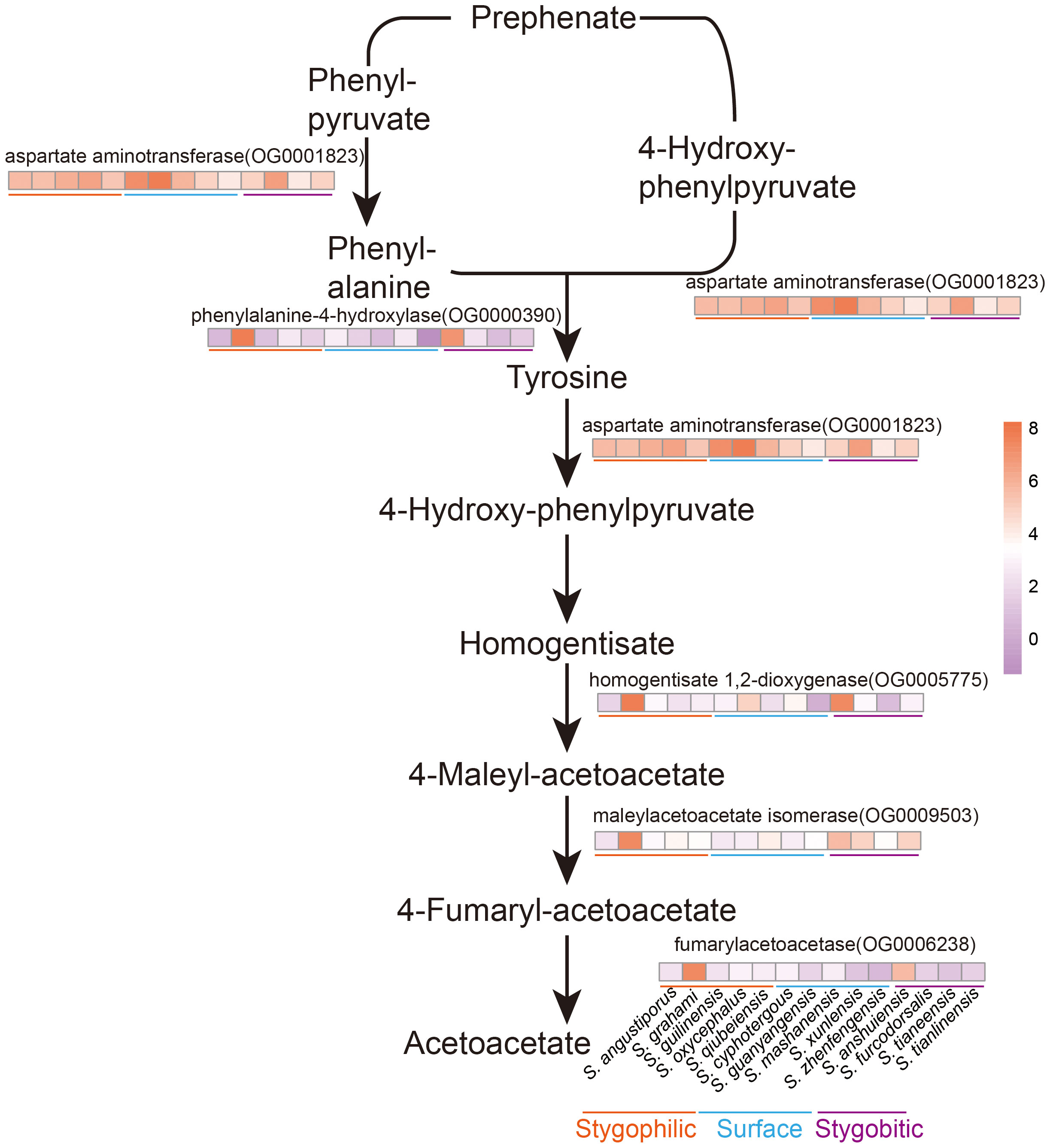
Figure 7 Flow of phenylalanine metabolism and tyrosine metabolism and heat map of related enzyme gene expression in 14 Sinocyclocheilus species. The darker the orange, higher the expression; the darker the purple, lower the expression.
Our analysis revealed that SBs had the fewest black blotches and the lowest expression of tyrosine metabolism-related enzymes: aspartate aminotransferase (AST, OG0001823), fumarylacetoacetase (FAH, OG0006238; Figures 5; 7). This indicated that phenylalanine and tyrosine metabolism was inhibited, and the final melanin synthesis was affected. Consistent with previous research, melanin synthesis is triggered by sunlight, which can offer benefits such as camouflage, protection from ultraviolet radiation, and a role in social signaling (Rzepka et al., 2016). Thus, whereas melanin synthesis is inhibited in SBs that inhabit lightless environments.
SPs such as S. guanyangensis and S. cyphotergous had elevated AST activity. The way these species produce melanin may be related to the accumulation of homogentisic acid (HGA). HGA oxidation leads to melanin-like pigmentation (Giustarini et al., 2012). Interestingly, cultivation of these species in our laboratory revealed that SPs seemed to exhibit an enhanced ability to adapt rapidly to fluctuating light conditions. This may enable them to adapt to the changing light environment between cave and semi-enclosed cave environments. Therefore, AST may have an important role in fish skin differentiation and variation through the melanogenesis pathway. The observed pigmentation differences among Sinocyclocheilus species may represent adaptations to their specific environments. Reduced pigmentation could be advantageous in the dark cave environment, while increased pigmentation may be more beneficial for survival in semi-enclosed and surface environments (Romero and Green, 2005; Soares and Niemiller, 2020).
Degeneration of scales was found in all clades in our study (Figure 4). It is not unexpected to find skin scale degradation prevalent in Sinocyclocheilus, as it is a trait seen in some other Carpiformes, unrelated to cave environments (Harris et al., 2008; Zhao and Zhang, 2009; Zhu et al., 2019). The scales of Sinocyclocheilus are known to be bony scales, and the posterior part of their scales consists of bone (Zhao and Zhang, 2009). We found that Reticulocalbin 3, EF-hand calcium binding domain (OG0016528), S100 calcium binding protein U (OG0016522), and Positive regulation of vitamin D receptor signaling pathway (OG0016683) were under positive selection, which may contribute to the calcium-bone homeostasis of fish skin (Schäfer and Heizmann, 1996; Bouillon and Suda, 2014; Girard et al., 2015). And PSGs are enriched in the MAPK signaling pathway, which is known to be associated with scale development, so the role of such genes involved in scale evolution is strong (Zhou Q. L. et al., 2022); there is also a strong association between scale regression and cave-dwelling. However, a deeper analysis is warranted to explore the connection between cave environments and scale features perse, i.e. a comparison between scaleless Sinocyclocheilus and scaleless other Carpiformes. Our research showcases the remarkable adaptability of cave-dwelling species to dynamic environments, offering insights into fish adaptation processes in response to anthropogenic alterations, such as habitat degradation and climate change. Sinocyclocheilus cavefish share few positive selections among their skin orthologous genes, and genes associated with the limited skin phenotype are more difficult to detect. The most likely plausible reason for this evolutionary phenomenon is the low rate of evolution of the cavefish genome and genome-wide relaxation of selection (Torres-Paz et al., 2018; Policarpo et al., 2020; Zhao et al., 2022). Our study demonstrates the remarkable adaptability of cave-dwelling species to dynamic environments and provides insights into the process of fish adaptation to anthropogenic changes such as habitat degradation and climate change.
PSGs in Sinocyclocheilus were enriched in herpes simplex virus 1 (HSV-1) infection, cytokine-cytokine receptor interactions, p53 signaling pathway, necrosis, and apoptosis related pathways. The current study has found that under prolonged environmental stress (e.g. hypoxia, prolonged hypothermia, starvation, etc.), facilitation and evolution of pathways such as p53 signaling pathway, MAPK signaling pathway, cytokine-cytokine receptor interaction, and apoptosis related pathways are prone to occur and echoes our study (Tong et al., 2021; Mao J. et al., 2022; Voskarides et al., 2022; Li et al., 2023). Our study highlighted that an important aspect of skin evolution is resistance to viral infection. These pathways promote coordination between cells and contribute to the clearance of pathogenic infections. Previous studies have shown that p53 plays a dual role in the replication of HSV-1 at different stages of infection (Sato and Tsurumi, 2013; Voskarides et al., 2022). Our study found that one of the p53 homologs: the Tumor protein p63 regulated 1 (OG0018161) was subject to the strongest positive selection (Aloni-Grinstein et al., 2018). Interestingly, it has been shown that HSV-1 manages to counteract this negative effect of p53 through the viral protein ICP22, which is able to bind p53 directly and eliminate its function (Sato and Tsurumi, 2013). Like many other viruses, herpesviruses their hosts co-evolved (Chawla et al., 2022). Fish skin is the first line of defense against viral and pathogenic microbial infections in the aquatic environment, and the adaptive evolution of immune and apoptosis-related genes may have facilitated the development of immune function in the skin of Sinocyclocheilus, enabling their ability to rapidly adapt to new environments and to rapidly occupy vacant ecological niches.
5 Conclusion
In conclusion, our study focused on investigating the genetic basis of environmental adaptation in Sinocyclocheilus cavefish, specifically exploring the influence of gene regulation on skin-related traits. Through a radiation-scale analysis involving representatives from major clades and different habitat types, we uncovered important findings regarding the adaptive mechanisms of these cave-dwelling fish.
We observed that different habitats exerted significant influences on color and scale characteristics, gene expression patterns, and functional enrichment of differentially expressed genes (DEGs) and positively selected genes (PSGs) in Sinocyclocheilus species. The functional enrichment analysis revealed distinct patterns in signaling mechanisms, oxidative stress, energy metabolism, and immune responses associated with different habitats.
Regarding metabolic differences among species across habitats, we found that species from surface habitats (SUs) exhibited higher energy metabolic rates, reflected in enriched energy metabolism pathways such as carbohydrate metabolism, amino acid metabolism and energy meta signaling mechanisms.
Conversely, stygophilic species (SPs) enhanced fat decomposition and mitochondrial respiration, while stygobitic species (SBs) showed enhanced glycolysis processes, lipid synthesis, and transport pathways, possibly as an adaptation to energy-poor cave environments.
The immune responses of species from different habitats also exhibited variations. SUs displayed the mechanism by which macrophages combine to adapt immune resistance to microbial infection, indicating a stronger response to microbial challenges. In contrast, cave dwellers showed differences in immune response patterns, with SBs exhibiting stronger innate immune responses to microbes. Additionally, oxidative stress-related pathways were enriched in cave dwellers, potentially as a response to the unique challenges of their low-oxygen cave environments.
The study of transcriptional plasticity and convergent evolution in Sinocyclocheilus cavefish revealed interesting findings. Skin color adaptations were associated with differential gene expression patterns, particularly in the tyrosine metabolism pathway, which influences melanin production. The presence or absence of scales in different species was also observed, with scale regression prevalent in cave-dwelling species. These observations highlight the significance of skin morphology and gene expression in understanding the evolutionary relationships among Sinocyclocheilus species.
Our research demonstrates the adaptability of cave-dwelling species to dynamic environments. The knowledge generated maybe useful also in predicting the processes of fish adaptation in response to habitat degradation and climate change. Even with the stabilization of key physiological functions and pathways observed in the skin transcriptomes across species from three different habitats and experiments, it is evident that the genetic mechanisms driving environmental adaptations—such as metabolic adjustments, immune responses, oxidative stress regulation, and certain phenotypic traits—shed light on the overarching patterns of adaptation in Sinocyclocheilus skin specific to cave-dwelling. Further investigations are warranted to deepen our knowledge of skin evolution and its adaptive significance in this unique group of cavefish.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal studies were approved by Guangxi University Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
XL: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. BC: Investigation, Methodology, Resources, Visualization, Writing – original draft, Writing – review & editing. TM: Formal Analysis, Investigation, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. YL: Investigation, Methodology, Resources, Validation, Writing – review & editing. JY: Investigation, Resources, Supervision, Validation, Writing – review & editing. MM: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by National Natural Science Foundation of China #32260333 and Guangxi University Startup funds to MM.
Acknowledgments
We thank the following individuals: Juntao Hu (Fudan University) and JY for suggestions to improve the paper; Shipeng Zhou and Dan Sun for assistance in the field.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author MM declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1264214/full#supplementary-material
References
Abràmoff M. D., Magalhães P. J., Ram S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11, 36–42. doi: 10.3233/ISU-1991-115-601
Aloni-Grinstein R., Charni-Natan M., Solomon H., Rotter V. (2018). p53 and the viral connection: back into the future (‡). Cancers (Basel) 10. doi: 10.3390/cancers10060178
Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Ángeles Esteban M. (2012). An overview of the immunological defenses in fish skin. Int. Scholarly Res. Notices 2012, 853470. doi: 10.5402/2012/853470
Austin B. (2006). The bacterial microflora of fish, revised. TheScientificWorldJOURNAL 6, 931–945. doi: 10.1100/tsw.2006.181
Bailey A., De Wit P., Thor P., Browman H. I., Bjelland R., Shema S., et al. (2017). Regulation of gene expression is associated with tolerance of the Arctic copepod Calanus glacialis to CO2-acidified sea water. Ecol. Evol. 7, 7145–7160. doi: 10.1002/ece3.3063
Bangert C., Brunner P. M., Stingl G. (2011). Immune functions of the skin. Clinics Dermatol. 29, 360–376. doi: 10.1016/j.clindermatol.2011.01.006
Behr J. B. (2011). Chitin synthase, a fungal glycosyltransferase that is a valuable antifungal target. Chimia 65, 49–49. doi: 10.2533/chimia.2011.49
Belkaid Y., Hand T. W. (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. doi: 10.1016/j.cell.2014.03.011
Bhatti J. S., Bhatti G. K., Reddy P. H. (2017). Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta (BBA)-Molecular Basis Dis. 1863, 1066–1077. doi: 10.1016/j.bbadis.2016.11.010
Boggs T., Gross J. (2021). Reduced oxygen as an environmental pressure in the evolution of the blind Mexican cavefish. Diversity 13, 26. doi: 10.3390/d13010026
Bouillon R., Suda T. (2014). Vitamin D: calcium and bone homeostasis during evolution. Bonekey Rep. 3, 480. doi: 10.1038/bonekey.2013.214
Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chawla K., Subramanian G., Rahman T., Fan S., Chakravarty S., Gujja S., et al. (2022). Autophagy in virus infection: A race between host immune response and viral antagonism. Immuno 2, 153–169. doi: 10.3390/immuno2010012
Chen B., Mao T. R., Liu Y. W., Dai W. A., Li X. L., Rajput A. P., et al. (2022). Sensory evolution in a cavefish radiation: patterns of neuromast distribution and associated behaviour in Sinocyclocheilus (Cypriniformes: Cyprinidae). Proc. R. Soc. B: Biol. Sci. 289, 20221641. doi: 10.1098/rspb.2022.1641
Chen G., Pang M., Yu X., Wang J., Tong J. (2021). Transcriptome sequencing provides insights into the mechanism of hypoxia adaption in bighead carp (Hypophthalmichthys nobilis). Comp. Biochem. Physiol. Part D: Genomics Proteomics 40, 100891. doi: 10.1016/j.cbd.2021.100891
Demory D., Arsenieff L., Simon N., Six C., Rigaut-Jalabert F., Marie D., et al. (2017). Temperature is a key factor in Micromonas–virus interactions. ISME J. 11, 601–612. doi: 10.1038/ismej.2016.160
Duan H., Jing L., Xiang J., Ju C., Wu Z., Liu J., et al. (2022). CD146 associates with Gp130 to control a macrophage pro-inflammatory program that regulates the metabolic response to obesity. Advanced Sci. 9, 2103719. doi: 10.1002/advs.202103719
Edgar R. C. (2022). Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 13, 6968. doi: 10.1038/s41467-022-34630-w
Ellison A. R., Wilcockson D., Cable J. (2021). Circadian dynamics of the teleost skin immune-microbiome interface. Microbiome 9, 1–18. doi: 10.1186/s40168-021-01160-4
Emms D. M., Kelly S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157. doi: 10.1186/s13059-015-0721-2
Fortune E. S., Andanar N., Madhav M., Jayakumar R. P., Cowan N. J., Bichuette M. E., et al. (2020). Spooky interaction at a distance in cave and surface dwelling electric fishes. Front. Integr. Neurosci. 14. doi: 10.3389/fnint.2020.561524
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Garvin M. R., Thorgaard G. H., Narum S. R. (2015). Differential Expression of Genes that Control Respiration Contribute to Thermal Adaptation in Redband Trout (Oncorhynchus mykiss gairdneri). Genome Biol. Evol. 7, 1404–1414. doi: 10.1093/gbe/evv078
Gibert J., Deharveng L. (2002). Subterranean Ecosystems: A Truncated Functional Biodiversity: This article emphasizes the truncated nature of subterranean biodiversity at both the bottom (no primary producers) and the top (very few strict predators) of food webs and discusses the implications of this truncation both from functional and evolutionary perspectives. BioScience 52, 473–481. doi: 10.1641/0006-3568(2002)052[0473:SEATFB]2.0.CO;2
Girard F., Venail J., Schwaller B., Celio M. R. (2015). The EF-hand Ca2+-binding protein super-family: A genome-wide analysis of gene expression patterns in the adult mouse brain. Neuroscience 294, 116–155. doi: 10.1016/j.neuroscience.2015.02.018
Giustarini D., Dalle-Donne I., Lorenzini S., Selvi E., Colombo G., Milzani A., et al. (2012). Protein thiolation index (PTI) as a biomarker of oxidative stress. Free Radical Biol. Med. 53, 907–915. doi: 10.1016/j.freeradbiomed.2012.06.022
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gray L. R., Tompkins S. C., Taylor E. B. (2014). Regulation of pyruvate metabolism and human disease. Cell Mol. Life Sci. 71, 2577–2604. doi: 10.1007/s00018-013-1539-2
Gross J. B. (2012). The complex origin of Astyanax cavefish. BMC evolutionary Biol. 12, 1–12. doi: 10.1186/1471-2148-12-105
Gross J. B., Powers A. K., Davis E. M., Kaplan S. A. (2016). A pleiotropic interaction between vision loss and hypermelanism in Astyanax mexicanus cave x surface hybrids. BMC Evol. Biol. 16, 145. doi: 10.1186/s12862-016-0716-y
Guengerich F. P., Waterman M. R., Egli M. (2016). Recent structural insights into cytochrome P450 function. Trends Pharmacol. Sci. 37, 625–640. doi: 10.1016/j.tips.2016.05.006
Harris M. P., Rohner N., Schwarz H., Perathoner S., Konstantinidis P., Nüsslein-Volhard C. (2008). Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PloS Genet. 4, e1000206. doi: 10.1371/journal.pgen.1000206
Heather L. C., Cole M. A., Tan J. J., Ambrose L. J. A., Pope S., Abd-Jamil A. H., et al. (2012). Metabolic adaptation to chronic hypoxia in cardiac mitochondria. Basic Res. Cardiol. 107, 268. doi: 10.1007/s00395-012-0268-2
Hinaux H., Poulain J., Da Silva C., Noirot C., Jeffery W. R., Casane D., et al. (2013). De novo sequencing of Astyanax mexicanus surface fish and Pachón cavefish transcriptomes reveals enrichment of mutations in cavefish putative eye genes. PloS One 8, e53553. doi: 10.1371/journal.pone.0053553
Huang Y., Chain F. J. J., Panchal M., Eizaguirre C., Kalbe M., Lenz T. L., et al. (2016). Transcriptome profiling of immune tissues reveals habitat-specific gene expression between lake and river sticklebacks. Wiley-Blackwell Online Open 25 (4), 943–958. doi: 10.1111/mec.13520
Huang Z., Titus T., Postlethwait J. H., Meng F. (2019). Eye Degeneration and Loss of otx5b Expression in the Cavefish Sinocyclocheilus tileihornes. J. Mol. Evol. 87, 199–208. doi: 10.1007/s00239-019-09901-8
Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Jablonski N. G., Chaplin G. (2010). Human skin pigmentation as an adaptation to UV radiation. Proc. Natl. Acad. Sci. 107, 8962–8968. doi: 10.1073/pnas.0914628107
Jeffery W. R. (2001). Cavefish as a model system in evolutionary developmental biology. Dev. Biol. 231, 1–12. doi: 10.1006/dbio.2000.0121
Jeffery W. R. (2019). “Chapter 12 - Astyanax mexicanus: A vertebrate model for evolution, adaptation, and development in caves,” in Encyclopedia of caves (Third edition). Eds. Culver D. C., White W. B., T. Pipan (London: Academic/Elsevier Press), 85–93. doi: 10.1016/B978-0-12-814124-3.00012-1
Jiang W. S., Li J., Lei X. Z., Wen Z. R., Han Y. Z., Yang J. X., et al. (2019). Sinocyclocheilus sanxiaensis, a new blind fish from the Three Gorges of Yangtze River provides insights into speciation of Chinese cavefish. Zoological Res. 40, 552. doi: 10.24272/j.issn.2095-8137.2019.065
Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Krishnan J., Persons J. L., Peuss R., Hassan H., Kenzior A., Xiong S., et al. (2020). Comparative transcriptome analysis of wild and lab populations of Astyanax mexicanus uncovers differential effects of environment and morphotype on gene expression. J. Exp. Zool B Mol. Dev. Evol. 334, 530–539. doi: 10.1002/jez.b.22933
Lam S. M., Li J., Sun H., Mao W., Lu Z., Zhao Q., et al. (2022). Quantitative lipidomics and spatial MS-Imaging uncovered neurological and systemic lipid metabolic pathways underlying troglomorphic adaptations in cave-dwelling fish. Mol. Biol. Evol. 39, msac050. doi: 10.1093/molbev/msac050
Lafferty K. (2012). Biodiversity loss decreases parasite diversity: theory and patterns. Philos. Trans. R Soc. Lond B Biol. 367, 2814–2827. doi: 10.1098/rstb.2012.0110
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leandro J., Stokka A. J., Teigen K., Andersen O. A., Flatmark T. (2017). Substituting Tyr138 in the active site loop of human phenylalanine hydroxylase affects catalysis and substrate activation. FEBS Open Bio 7, 1026–1036. doi: 10.1002/2211-5463.12243
Leclercq E., Taylor J. F., Migaud H. (2010). Morphological skin colour changes in teleosts. Fish Fisheries 11, 159–193. doi: 10.1111/j.1467-2979.2009.00346.x
Li C. Q., Chen H. Y., Zhao Y. C., Chen S. Y., Xiao H. (2020). Comparative transcriptomics reveals the molecular genetic basis of pigmentation loss in Sinocyclocheilus cavefishes. Ecol. Evol. 10, 14256–14271. doi: 10.1002/ece3.7024
Li C. Q. (2019). Exploring the genetic basis of differences in body color phenotypic traits in fishes of the genus Sinocyclocheilus based on transcriptome sequencing (in Chinese). (Yunnan: Yunnan University), 35–39.
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li X., Fang P., Yang W. Y., Chan K., Lavallee M., Xu K., et al. (2017). Mitochondrial ROS, uncoupled from ATP synthesis, determine endothelial activation for both physiological recruitment of patrolling cells and pathological recruitment of inflammatory cells. Can. J. Physiol. Pharmacol. 95, 247–252. doi: 10.1139/cjpp-2016-0515
Li X., Wang X., Yang C., Lin L., Yuan H., Lei F., et al. (2023). A de novo assembled genome of the Tibetan Partridge (Perdix hodgsoniae) and its high-altitude adaptation. Integr. Zoology 18, 225–236. doi: 10.1111/1749-4877.12673
Loomis C., Peuß R., Jaggard J. B., Wang Y., Mckinney S. A., Raftopoulos S. C., et al. (2019). An adult brain atlas reveals broad neuroanatomical changes in independently evolved populations of Mexican cavefish. Front. Neuroanat 13. doi: 10.3389/fnana.2019.00088
Lü A., Hu X., Xue J., Zhu J., Wang Y., Zhou G. (2012). Gene expression profiling in the skin of zebrafish infected with Citrobacter freundii. Fish Shellfish Immunol. 32, 273–283. doi: 10.1016/j.fsi.2011.11.016
Luo Q., Tang Q., Deng L., Duan Q., Zhang R. (2023). A new cavefish of Sinocyclocheilus (Teleostei: Cypriniformes: Cyprinidae) from the Nanpanjiang River in Guizhou, China. J. Fish Biol. 1–13. doi: 10.1111/jfb.15490
Mahram A., Herbordt M. C. (2015). NCBI BLASTP on high-performance reconfigurable computing systems. ACM Trans. Reconfigurable Technol. Syst. (TRETS) 7, 1–20. doi: 10.1145/2629691
Mao J., Huang X., Sun H., Jin X., Guan W., Xie J., et al. (2022). Transcriptome analysis provides insight into adaptive mechanisms of scallops under environmental stress. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.971796
Mao T. R., Liu Y. W., Meegaskumbura M., Yang J., Ellepola G., Senevirathne G., et al. (2021). Evolution in Sinocyclocheilus cavefish is marked by rate shifts, reversals and origin of novel traits. BMC Ecol. Evol. 21, 45. doi: 10.1186/s12862-021-01776-y
Mao T., Liu Y., Vasconcellos M. M., Pie M. R., Ellepola G., Fu C., et al. (2022). Evolving in the darkness: phylogenomics of Sinocyclocheilus cavefishes highlights recent diversification and cryptic diversity. Mol. Phylogenet. Evol. 168, 107400. doi: 10.1016/j.ympev.2022.107400
Mayer A., Mora T., Rivoire O., Walczak A. M. (2016). Diversity of immune strategies explained by adaptation to pathogen statistics. Proc. Natl. Acad. Sci. U.S.A. 113, 8630–8635. doi: 10.1073/pnas.1600663113
McCormick M. I., Larson J. K. (2008). Effect of hunger on the response to, and the production of, chemical alarm cues in a coral reef fish. Anim. Behav. 75, 1973–1980. doi: 10.1016/j.anbehav.2007.12.007
McGaugh S. E., Gross J. B., Aken B., Blin M., Borowsky R., Chalopin D., et al. (2014). The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 5, 5307. doi: 10.1038/ncomms6307
Medley J. K., Persons J., Biswas T., Olsen L., Peuß R., Krishnan J., et al. (2022). The metabolome of Mexican cavefish shows a convergent signature highlighting sugar, antioxidant, and Ageing-Related metabolites. Elife 11, e74539. doi: 10.7554/eLife.74539
Meng F., Braasch I., Phillips J. B., Lin X., Titus T., Zhang C., et al. (2013). Evolution of the eye transcriptome under constant darkness in Sinocyclocheilus cavefish. Mol. Biol. Evol. 30, 1527–1543. doi: 10.1093/molbev/mst079
Meng F. W., Zhao Y. H., Titus T., Zhang C. G., Postlethwait J. H. (2018). Brain of the blind: transcriptomics of the golden-line cavefish brain. Curr. Zoology 64, 765–773. doi: 10.1093/cz/zoy005
Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., Von Haeseler A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mohanty B. R., Sahoo P. K. (2010). Immune responses and expression profiles of some immune-related genes in Indian major carp, Labeo rohita to Edwardsiella tarda infection. Fish Shellfish Immunol. 28, 613–621. doi: 10.1016/j.fsi.2009.12.025
Moran R. L., Richards E. J., Ornelas-García C. P., Gross J. B., Donny A., Wiese J., et al. (2023). Selection-driven trait loss in independently evolved cavefish populations. Nat. Commun. 14, 2557. doi: 10.1038/s41467-023-37909-8
Moran D., Softley R., Warrant E. J. (2014). Eyeless Mexican cavefish save energy by eliminating the circadian rhythm in metabolism. PloS One 9, e107877. doi: 10.1371/journal.pone.0107877
Morikawa T., Takubo K. (2016). Hypoxia regulates the hematopoietic stem cell niche. Pflügers Archiv-European J. Physiol. 468, 13–22. doi: 10.1007/s00424-015-1743-z
Nilsson Sköld H., Aspengren S., Wallin M. (2013). Rapid color change in fish and amphibians–function, regulation, and emerging applications. Pigment Cell melanoma Res. 26, 29–38. doi: 10.1111/pcmr.12040
Okar D. A., Lange A. J. (1999). Fructose-2, 6-bisphosphate and control of carbohydrate metabolism in eukaryotes. Biofactors 10, 1–14. doi: 10.1002/biof.5520100101
Passow C. N., Brown A. P., Arias-Rodriguez L., Yee M. C., Sockell A., Schartl M., et al. (2017). Complexities of gene expression patterns in natural populations of an extremophile fish (Poecilia mexicana, Poeciliidae). Mol. Ecol. 26, 4211–4225. doi: 10.1111/mec.14198
Peuß R., Box A. C., Chen S., Wang Y., Tsuchiya D., Persons J. L., et al. (2020). Adaptation to low parasite abundance affects immune investment and immunopathological responses of cavefish. Nat. Ecol. Evol. 4, 1416–1430. doi: 10.1038/s41559-020-1234-2
Pham L., Komalavilas P., Eddie A. M., Thayer T. E., Greenwood D. L., Liu K. H., et al. (2022). Neutrophil trafficking to the site of infection requires Cpt1a-dependent fatty acid β-oxidation. Commun. Biol. 5, 1366. doi: 10.1038/s42003-022-04339-z
Policarpo M., Fumey J., Lafargeas P., Naquin D., Thermes C., Naville M., et al. (2021). Contrasting gene decay in subterranean vertebrates: insights from cavefishes and fossorial mammals. Mol. Biol. Evol. 38, 589–605. doi: 10.1093/molbev/msaa249
Policarpo M., Fumey J., Lafargeas P., Naquin D., Thermes C., Naville M., et al. (2020). Contrasting gene decay in subterranean vertebrates: insights from cavefishes and fossorial mammals. Mol. Biol. Evol. 38, 589–605. doi: 10.1093/molbev/msaa249
Protas M., Jeffery W. R. (2012). Evolution and development in cave animals: from fish to crustaceans. Wiley Interdiscip Rev. Dev. Biol. 1, 823–845. doi: 10.1002/wdev.61
Qi D., Chao Y., Wu R., Xia M., Chen Q., Zheng Z. (2018). Transcriptome analysis provides insights into the adaptive responses to hypoxia of a schizothoracine fish (Gymnocypris eckloni). Front. Physiol. 9. doi: 10.3389/fphys.2018.01326
Rambaut A. (2009). FigTree. Tree figure drawing tool. Available at: http://tree.bio.ed.ac.uk/software/figtree/.
Randhawa M., Sangar V., Tucker-Samaras S., Southall M. (2014). Metabolic signature of sun exposed skin suggests catabolic pathway overweighs anabolic pathway. PloS One 9, e90367. doi: 10.1371/journal.pone.0090367
Reynolds T. B. (2009). Strategies for acquiring the phospholipid metabolite inositol in pathogenic bacteria, fungi and protozoa: making it and taking it. Microbiology 155, 1386. doi: 10.1099/mic.0.025718-0
Riddle M. R., Aspiras A. C., Gaudenz K., Peuß R., Sung J. Y., Martineau B., et al. (2018). Insulin resistance in cavefish as an adaptation to a nutrient-limited environment. Nature 555, 647–651. doi: 10.1038/nature26136
Romero A., Green S. M. (2005). The end of regressive evolution: examining and interpreting the evidence from cave fishes. J. Fish Biol. 67, 3–32. doi: 10.1111/j.0022-1112.2005.00776.x
Rzepka Z., Buszman E., Beberok A., Wrześniok D. (2016). From tyrosine to melanin: Signaling pathways and factors regulating melanogenesis. Postepy Hig. Med. Dosw (Online) 70, 695–708. doi: 10.5604/17322693.1208033
Sato Y., Tsurumi T. (2013). Genome guardian p53 and viral infections. Rev. Med. Virol. 23, 213–220. doi: 10.1002/rmv.1738
Schäfer B. W., Heizmann C. W. (1996). The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem. Sci. 21, 134–140. doi: 10.1016/s0968-0004(96)80167-8
Scharsack J. P., Kalbe M., Harrod C., Rauch G. (2007). Habitat-specific adaptation of immune responses of stickleback (Gasterosteus aculeatus) lake and river ecotypes. Proc. R. Soc. B: Biol. Sci. 274, 1523–1532. doi: 10.1098/rspb.2007.0210
Simon V., Elleboode R., Mahé K., Legendre L., Ornelas-Garcia P., Espinasa L., et al. (2017). Comparing growth in surface and cave morphs of the species Astyanax mexicanus: insights from scales. EvoDevo 8, 1–13. doi: 10.1186/s13227-017-0086-6
Soares D., Niemiller M. L. (2020). Extreme adaptation in caves. Anatomical Rec. 303, 15–23. doi: 10.1002/ar.24044
Stahl B. A., Gross J. B. (2015). Alterations in Mc1r gene expression are associated with regressive pigmentation in. Astyanax cavefish. Dev. Genes Evol. 225, 367–375. doi: 10.1007/s00427-015-0517-0
Stahl B. A., Gross J. B. (2017). A comparative transcriptomic analysis of development in two Astyanax cavefish populations. J. Exp. Zoology Part B: Mol. Dev. Evol. 328, 515–532. doi: 10.1002/jez.b.22749
Staudigl M., Gersting S. W., Danecka M. K., Messing D. D., Woidy M., Pinkas D., et al. (2011). The interplay between genotype, metabolic state and cofactor treatment governs phenylalanine hydroxylase function and drug response. Hum. Mol. Genet. 20, 2628–2641. doi: 10.1093/hmg/ddr165
Sun Y., Liu W. Z., Liu T., Feng X., Yang N., Zhou H. F. (2015). Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Receptors Signal Transduction 35, 600–604. doi: 10.3109/10799893.2015.1030412
Tessler M., Brugler M. R., Burns J. A., Sinatra N. R., Vogt D. M., Varma A., et al. (2020). Ultra-gentle soft robotic fingers induce minimal transcriptomic response in a fragile marine animal. Curr. Biol. 30, R157–R158. doi: 10.1016/j.cub.2020.01.032
Thiergart T., Durán P., Ellis T., Vannier N., Garrido-Oter R., Kemen E., et al. (2020). Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nat. Ecol. Evol. 4, 122–131. doi: 10.1038/s41559-019-1063-3
Tong C., Li M., Tang Y., Zhao K. (2021). Genomic signature of shifts in selection and alkaline adaptation in highland fish. Genome Biol. Evol. 13. doi: 10.1093/gbe/evab086
Torres-Paz J., Hyacinthe C., Pierre C., Rétaux S. (2018). Towards an integrated approach to understand Mexican cavefish evolution. Biol. Lett. 14 (8), 20180101. doi: 10.1098/rsbl.2018.0101
Voskarides K., Koutsofti C., Pozova M. (2022). TP53 mutant versus wild-type zebrafish larvae under starvation stress: larvae can live up to 17 days post-fertilization without food. Zebrafish 19, 49–55. doi: 10.1089/zeb.2022.0003
Wang Y., Guo B. (2019). Adaption to extreme environments: a perspective from fish genomics. Rev. Fish Biol. Fisheries 29, 735–747. doi: 10.1007/s11160-019-09577-9
Wen H., Luo T., Wang Y., Wang S., Liu T., Xiao N., et al. (2022). Molecular phylogeny and historical biogeography of the cave fish genus Sinocyclocheilus (Cypriniformes: Cyprinidae) in Southwest China. Integr. Zoology 17, 311–325. doi: 10.1111/1749-4877.12624
Xiao W., Zou Z., Li D., Zhu J., Yue Y., Yang H. (2020). Effect of dietary phenylalanine level on growth performance, body composition, and biochemical parameters in plasma of juvenile hybrid tilapia, Oreochromis niloticus × Oreochromis aureus. J. World Aquaculture Soc. 51, 437–451. doi: 10.1111/jwas.12641
Xie C., Mao X. Z., Huang J. J., Ding Y., Wu J. M., Dong S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xiong S. (2021). How cavefish gain high body fat to adapt to food scarcity (United Kingdom: Open University).
Xu C., Luo T., Zhou J.-J., Wu L., Zhao X.-R., Yang H.-F., et al. (2023). Sinocyclocheilus longicornus (Cypriniformes, Cyprinidae), a new species of microphthalmic hypogean fish from Guizhou, Southwest China. ZooKeys 1141, 1–28. doi: 10.3897/zookeys.1141.91501
Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang J., Chen X., Bai J., Fang D., Qiu Y., Jiang W., et al. (2016). The Sinocyclocheilus cavefish genome provides insights into cave adaptation. BMC Biol. 14, 1–13. doi: 10.1186/s12915-015-0223-4
Yeung K. Y., Ruzzo W. L. (2001). Principal component analysis for clustering gene expression data. Bioinformatics 17, 763–774. doi: 10.1093/bioinformatics/17.9.763
Yoshizawa M., Goricki S., Soares D., Jeffery W. R. (2010). Evolution of a behavioral shift mediated by superficial neuromasts helps cavefish find food in darkness. Curr. Biol. 20, 1631–1636. doi: 10.1016/j.cub.2010.07.017
Yoshizawa M., Jeffery W. R., Van Netten S. M., Mchenry M. J. (2014). The sensitivity of lateral line receptors and their role in the behavior of Mexican blind cavefish (Astyanax mexicanus). J. Exp. Biol. 217, 886–895. doi: 10.1242/jeb.094599
Young M. D., Wakefield M. J., Smyth G. K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Yu S., Rao S., Reddy J. K. (2003). Peroxisome proliferator-activated receptors, fatty acid oxidation, steatohepatitis and hepatocarcinogenesis. Curr. Mol. Med. 3, 561–572. doi: 10.2174/1566524033479537
Zhao Y. C., Chen H. Y., Li C. Q., Chen S. Y., Xiao H. (2020). Comparative transcriptomics reveals the molecular genetic basis of cave adaptability in Sinocyclocheilus fish species. Front. Ecol. Evol. 8. doi: 10.3389/fevo.2020.589039
Zhao Y., Huang X., Ding T. W., Gong Z. (2016). Enhanced angiogenesis, hypoxia and neutrophil recruitment during Myc-induced liver tumorigenesis in zebrafish. Sci. Rep. 6, 31952. doi: 10.1038/srep31952
Zhao Y., Huang Z., Huang J., Zhang C., Meng F. (2021). Phylogenetic analysis and expression differences of eye-related genes in cavefish genus Sinocyclocheilus. Integr. Zool 16, 354–367. doi: 10.1111/1749-4877.12466
Zhao Q., Shao F., Li Y., Yi S. V., Peng Z. (2022). Novel genome sequence of Chinese cavefish (Triplophysa rosa) reveals pervasive relaxation of natural selection in cavefish genomes. Mol. Ecol. 31, 5831–5845. doi: 10.1111/mec.16700
Zhao Y. H., Zhang C. G. (2009). Endemic fishes of Sinocyclocheilus (Cypriniformes: Cyprinidae) in China−species diversity, cave adaptation, systematics and zoogeograph (Beijing: Science Press).
Zhou J., Luo J., Zeng Y., Xu L. (2021). Characterization of PAH gene mutations and analysis of genotype-phenotype correlation in patients with phenylalanine hydroxylase deficiency from Fujian province, Southeastern China. Mol. Biol. Rep. 49, 10409–10419. doi: 10.21203/rs.3.rs-1096859/v1
Zhou S., Rajput A. P., Mao T., Liu Y., Ellepola G., Herath J., et al (2022). Adapting to novel environments together: evolutionary and ecological correlates of the bacterial microbiome of the world’s largest cavefish diversification (Cyprinidae, Sinocyclocheilus). Front. Microbiol. 13. doi: 10.3389/fmicb.2022.823254
Zhou Q. L., Wang L. Y., Zhao X. L., Yang Y. S., Ma Q., Chen G. (2022). Effects of salinity acclimation on histological characteristics and miRNA expression profiles of scales in juvenile rainbow trout (Oncorhynchus mykiss). BMC Genomics 23, 300. doi: 10.1186/s12864-022-08531-7
Keywords: Sinocyclocheilus, radiation, skin adaptations, morphological diversity, habitat, comparative transcriptomics, pigmentation, phylogeny
Citation: Luo X, Chen B, Mao T, Liu Y, Yang J and Meegaskumbura M (2023) Skin transcriptomic correlates of cave-dwelling Sinocyclocheilus cavefish. Front. Ecol. Evol. 11:1264214. doi: 10.3389/fevo.2023.1264214
Received: 20 July 2023; Accepted: 04 October 2023;
Published: 25 October 2023.
Edited by:
Mark A. Elgar, The University of Melbourne, AustraliaReviewed by:
Tereza Manousaki, Hellenic Centre for Marine Research (HCMR), GreeceLiandong Yang, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Luo, Chen, Mao, Liu, Yang and Meegaskumbura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Madhava Meegaskumbura, bWFkaGF2YV9tQG1hYy5jb20=
 Xiayue Luo
Xiayue Luo Bing Chen
Bing Chen Tingru Mao
Tingru Mao Yewei Liu
Yewei Liu Jian Yang
Jian Yang Madhava Meegaskumbura
Madhava Meegaskumbura