- 1Department of Veterinary Medicine, College of Veterinary Medicine, National Pingtung University of Science and Technology, Pingtung, Taiwan
- 2Southern Taiwan Fish Disease Centre, College of Veterinary Medicine, National Pingtung University of Science and Technology, Pingtung, Taiwan
- 3International Degree Program of Ornamental Fish Technology and Aquatic Animal Health, International College, National Pingtung University of Science and Technology, Pingtung, Taiwan
- 4Research Center for Animal Biologics, National Pingtung University of Science and Technology, Pingtung, Taiwan
Transcriptome analysis is a powerful tool that enables a deep understanding of complicated physiological pathways, including immune responses. RNA sequencing (RNA-Seq)-based transcriptome analysis and various bioinformatics tools have also been used to study non-model animals, including aquaculture species for which reference genomes are not available. Rapid developments in these techniques have not only accelerated investigations into the process of pathogenic infection and defense strategies in fish, but also used to identify immunity-related genes in fish. These findings will contribute to fish immunotherapy for the prevention and treatment of bacterial infections through the design of more specific and effective immune stimulants, adjuvants, and vaccines. Until now, there has been little information regarding the universality and diversity of immune reactions against pathogenic infection in fish. Therefore, one of the aims of this paper is to introduce the RNA-Seq technique for examination of immune responses in pathogen-infected fish. This review also aims to highlight comparative studies of immune responses against bacteria, based on our previous findings in largemouth bass (Micropterus salmoides) against Nocardia seriolae, gray mullet (Mugil cephalus) against Lactococcus garvieae, orange-spotted grouper (Epinephelus coioides) against Vibrio harveyi, and koi carp (Cyprinus carpio) against Aeromonas sobria, using RNA-seq techniques. We demonstrated that only 39 differentially expressed genes (DEGs) were present in all species. However, the number of specific DEGs in each species was relatively higher than that of common DEGs; 493 DEGs in largemouth bass against N. seriolae, 819 DEGs in mullets against L. garvieae, 909 in groupers against V. harveyi, and 1471 in carps against A. sobria. The DEGs in different fish species were also representative of specific immune-related pathways. The results of this study will enhance our understanding of the immune responses of fish, and will aid in the development of effective vaccines, therapies, and disease-resistant strains.
Introduction
Transcriptome analysis is used to study principal pathways of development, cellular fate, physiology, activity, and disease progression. RNA sequencing (RNA-Seq) is a modern technology for transcriptome profiling that uses next-generation sequencing (NGS). Advancements in bioinformatics has significantly supported RNA-Seq technology to accelerate the knowledge on transcriptomes (1).
In the aquaculture field, there is wide utility for RNA-Seq in various applications, such as understanding the development of embryo and larvae, toxicology, environmental stress, effect of dietary conditions, and discovery of novel transcripts (2–4). In addition, RNA-Seq has been used in many studies of fish immunology (5, 6). Pathogenic infection is a major concern for maintaining economic sustainability in natural and farmed fish; it results in high mortality and economical loss in aquaculture. An innate immunity is a front line of host defense, producing effectors that directly to the pathogen and attack it. An adaptive immune system is also present in teleost, including humoral and cellular mechanisms. To reduce disease outbreaks, it is essential to understand the immune mechanisms in fish during pathogenic infections. This knowledge will support the development of effective vaccines and adjuvants against pathogens. However, it has not been reported that the universality and diversity of immune reactions against pathogenic infection in fish.
In this paper, we firstly introduce the RNA-Seq technique and current knowledge for investigations of immune responses in pathogen-infected fish. This review also aims to highlight comparative studies of fish immune responses against bacteria based on our previous studies that we demonstrated the transcriptome of bacteria infected fish, largemouth bass (Micropterus salmoides) against Nocardia seriolae, gray mullet (Mugil cephalus) against Lactococcus garvieae, orange-spotted grouper (Epinephelus coioides) against Vibrio harveyi, and koi carp (Cyprinus carpio) against Aeromonas sobria.
Advantage of RNA-seq Analysis in Fish Aquaculture
Since the 2000s, hybridization-based microarray has been used to examine fish immunology in aquacultures; some early examples included Japanese flounder (Paralichthys olivaceus) (7), rainbow trout (Oncorhynchus mykiss) (8), and Atlantic salmon (Salmo salar) (9). Although species-specific probes should be designed, microarray technology could provide us with high throughput gene expression data. For transcriptome analysis, series analysis of gene expression (SAGE) and cap analysis gene expression (CAGE) have also been utilized. SAGE and CAGE, which are tag-based technology, are more precise; however, the number of genes that can be analyzed at one time is lower as compared with that of microarrays. Recently, the number of reports that uses RNA-Seq in aquaculture studies has rapidly increased. For the complete detail of RNA-Seq methodology, please refer the nicely reviews focused on aquaculture field (3, 6). The advantage of RNA-Seq is that it could determine expression levels of low-level transcripts as well as each splice variant isoforms. Current focus in aquaculture fish research is to examine organisms that do not process reference gene sequences; unigenes are obtained via de novo assembly using Trinity or similar programs without requiring reference gene sequences (10). RNA-Seq could provide novel transcript sequences, thereby expanding our current list of known transcripts in fish.
Annotation, Enrichment Analysis, and Pathway Analysis Using de novo Assembly Data
Although RNA-Seq technology could be applied to non-model animals, there are problems associated with the functional annotation and enrichment analysis of transcripts data. Transcripts sequences following assembly are usually searched via several databases, such as NCBI nucleotide sequences (NT), NCBI non-redundant protein (NR), Clusters of Orthologous Groups (COGs) (11), Kyoto Encyclopedia of Genes and Genomes (KEGG) (12, 13), gene ontology (GO) (14), and InterPro annotation (15). In our previous study on orange-spotted grouper (Epinephelus coioides), a total of 79,128 unigenes were identified and aligned with each database; 58,926 (74.47%) in NT, 43,576 (55.07%) in NR, 14,750 (18.64%) in COG, 34711 (43.87%) in KEGG, and 4232 (5.35%) in GO (16). The number of genes aligned with existing genes in the database was the highest in NT, while gene alignment was the lowest in GO. Differences in the number of aligned genes between the databases were similar to those found in other aquaculture studies (17–19). Although the GO database could provide enrichment analysis and pathway analysis, due to low gene alignment, results may be limited to generalized conclusions. There are programs that can convert gene IDs from one database to those of another database. For example, DAVID and ID Converter Systems are able to change gene IDs from NT to that of GO. However, these systems are not very useful in genes of aquaculture species. Currently, the KEGG database shows a relatively high number of aligned genes, which allows for enrichment analysis of aquaculture species. To obtain generalized conclusions using transcriptome data, it is essential that systems are developed to describe non-model organisms. There are databases now under construction that contain transcriptome information of aquaculture species. In the European common carp (Cyprinus carpio), a wide range of data on tissue-specific gene expression and translation (20) has been presented. These datasets will allow us to investigate immune responses in aquaculture species via transcriptome analysis.
Immune Responses Against Pathogens in Teleost Using RNA-seq
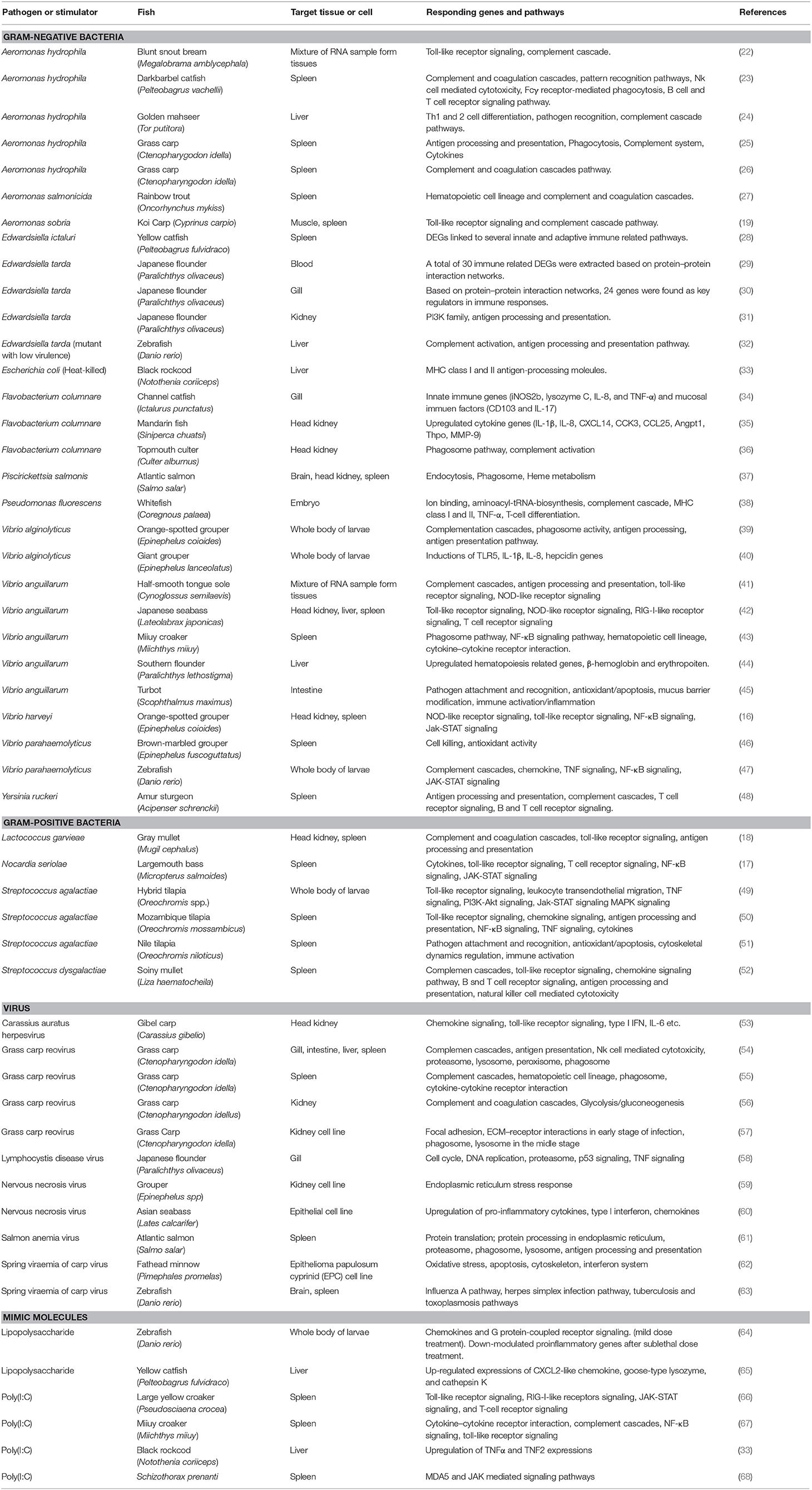
Although fish are constantly exposed to risk of microorganism pathogens, fish could keep in shape to act immune mechanisms against pathogens. In a first line of immune response, fish are protected by non-specific humoral factors including growth inhibiting substances (transferrin and antiproteases), lysins (lysozyme, C-reactive protein, and bactericidal peptides), and making a link with non-specific phagocyte responses. Second, fish produce antibody constitutes for a specific humoral defense inhibiting bacterial adherence and invasion of non-phagocytic host cells and counteracting toxins from bacterial (21). After developing the technology of molecular biology, the immune-related gene functions and responses against pathogens have been one of the major topic in fish immunology field. To investigate the expression pattern of immune-associated genes, real-time PCR is usually performed. However, this method is expensive and not recommended for a genome-wide survey of gene expression. As RNA-Seq could provide us with quantitative data on transcript expression levels, this technique has been commonly used to identify genes that respond to pathogenic conditions during exogenous challenge. Table 1 lists transcriptome analysis studies that examined immune regulations in teleost. In this review, we will also focus on studies that investigated immune responses to pathogen or their mimic molecules, using RNA-Seq analysis.
Aeromonus hydrophila is a Gram-negative bacterium, and causes a wide spectrum of diseases in vertebrates (69, 70). It is a major pathogen in aquaculture farms, and leads to high mortalities and economic losses worldwide (71, 72). In blunt snout bream (Megalobrama amblycephala), RNA-Seq analysis was conducted with RNA from several tissues, and 238 differentially expressed unigenes were identified in infected fish (22). In grass carp (Ctenopharygodon idella), 2121 DEGs were identified in spleens of A. hydrophila (6 hpi)-infected fish, some of which were involved in phagocytosis, the complement system, and cytokine production (25). Using transcriptome analysis, another study showed that A. hydrophila infected grass carp exhibited 2992 DEGs in the spleen, which were associated with the complement and coagulation cascades (26). In golden mahseer (Tor putitora), DEGs in A. hydrophila-infected livers were mainly associated with Th1/2 cell differentiation pathways, as well as in pathogen recognition and complement system (24).
Flavobacterium columnare is a Gram-negative bacterium, and causes columnaris in freshwater fish (73). This disease induces pathological changes, and damages epidermal tissues, gills, and the skin (74). In channel catfish (Ictalurus punctatus), the transcript profile of F. columnare-infected gills was examined using RNA-Seq to investigate differences in susceptibility to F. columnare (34). In resistant fish, the expression level of innate immune-associated genes (iNOS2b, lysozyme C, IL-8, and TNFα) was found to be elevated. In susceptible fish, the expression of secreted mucin forms, mucosal immune factors (CD103 and IL-17a), and rhamnose-binding lectin (34) was upregulated. The transcriptomic profiles of F. columnare-infected and non-infected mandarin fish (Siniperca chuatsi) have been reported using the head kidney F. columnare-infected and non-infected group (35). The results indicated that 1019 genes were differentially expressed between the two groups, of which 27 were immune-related (35). A similar study using the head kidney F. columnare-infected topmouth culter (Culter alburnus) (36) was also conducted. A total of 4037 DEGs (1217 upregulated and 2820 downregulated genes) were identified, and were found to be involved in phagosome formation, carbohydrate metabolism, amino acid metabolism, and lipid metabolism (36).
Streptococcus agalactiae, a Gram-positive round bacterium, is a harmful aquaculture pathogen that leads to enormous economic losses in various teleost (75–78). Transcriptome analysis of hybrid tilapia (Oreochromis spp.) after S. agalactiae infection was conducted, and results indicated that DEGs are mainly involved in immune-related pathways, especially Toll-like receptor signaling and leukocyte transendothelial migration (49). Moreover, time-course expression profile of genes suggested that induction of the NADPH oxidase complex and piscidin is mediated by Toll-like receptor pathways (49). Another research group conducted RNA-Seq analysis in tilapia (Oreochromis niloticus) spleens following S. agalactiae infections (51). A total of 2822 DEGs were detected, many of which were involved in pathogen attachment and recognition, antioxidant/apoptosis, cytoskeletal rearrangement, and immune activation (51). Wang et al. (50) focused on the relation between temperature and bacterial infection. They showed that temperature influences mRNA profiles of the spleen in tilapia during S. agalactiae infections. In addition, it was suggested that DEGs are involved in immune responses and oxygen related metabolisms (50).
Vibrio alginolyticus is a halophilic Gram-negative bacterium that causes septicemias, ulcers, exophthalmia, and corneal opaqueness in marine fish worldwide (79, 80). Transcriptome analysis in larvae of orange-spotted grouper (Epinephelus coioides) revealed that the expression of genes involved in the complement pathway and antimicrobial peptides is enhanced upon V. alginolyticus infection (39). In addition, transcriptome profiles of giant grouper (Epinephelus lanceolatus) larvae infected with Vibrio alginolyticus suggested that TLR5 signaling induces secretion of several cytokines (IL-1β and IL-8) (40).
Diversity of Immune Responses Among Species and Pathogens
In the previous section, we introduced various RNA-seq analyses conducted in fish with bacterial infections. We have also previously published four research papers that conducted transcriptome analysis on infected fish, namely largemouth bass (Micropterus salmoides) against Nocardia seriolae (17), gray mullet (Mugil cephalus) against Lactococcus garvieae (18), orange-spotted grouper (Epinephelus coioides) against Vibrio harveyi (16), and koi carp (Cyprinus carpio) against Aeromonas sobria (19). Based on the transcriptome data from these reports, we gained a deeper understanding of immune responses to bacterial infections. However, there is little information regarding the universality and diversity of immune reactions of fish against pathogenic infections. Here, we investigated specific genes and pathways that are involved in each bacterial infection in various fish species. In this study, we used DEGs (transcripts from spleen at 1 dpi with log2 > 1 or < −1 between infected and control group) with KEGG-annotations. We first identified overlapping and specific genes that were up- or down- regulated in each species. Venn diagrams (Figure 1) showed that only 39 DEGs (25 up-regulated and 14 down regulated) were involved in all species. The number of specific DEGs in each species was relatively higher than that of common DEGs; 493 DEGs (167 up-regulated and 326 down regulated) were found in largemouth bass against N. seriolae, 819 DEGs (291 up-regulated and 528 down regulated) were found in mullets against L. garvieae, 909 DEGs (601 up-regulated and 308 down regulated) were found in groupers (Epinephelus coioides) against V. harveyi, and 1471 DEGs (1,001 up-regulated and 470 down regulated) were found in carps against A. sobria (Figure 1).
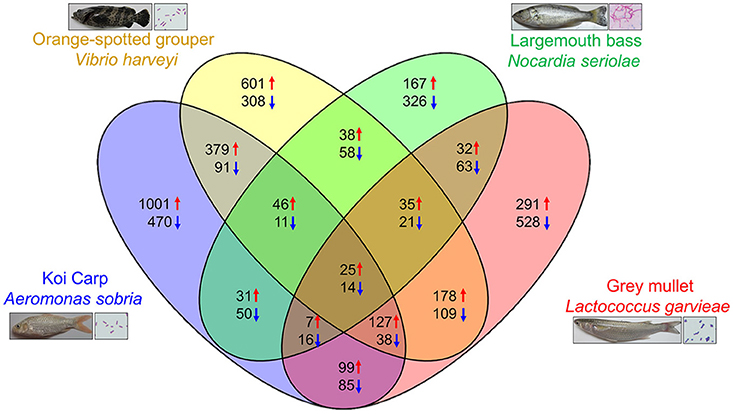
Figure 1. Venn diagrams showing overlaps of up and down regulated genes among each fish with bacterial challenge. The numbers indicate up (red arrow) and down (blue arrow) regulated genes in each categories.
Of the common DEGs, we found several immune-related genes that were upregulated, including C4 (complement component 4), CCL19 (C-C motif chemokine 19), and SOCS1 (suppressor of cytokine signaling 1) (Table S1). The complement system is an important innate immune system that functions to detect pathogenic infections in both vertebrates and invertebrates. C4 is an important part of the classical and lectin pathways, which form enzymes C3 and C5 convertases (81, 82). CCL19, a CC chemokine that is expressed in lymphoid organs, manages the migration of antigen presenting cells and lymphocytes (83). In teleost, there are also various reports that investigated potential chemokine genes and their chemotactic activity (84–86). SOCS1 is a regulator of JAK/STAT signaling, and is induced by type I interferon (IFN) and IFN-γ via binding and blocking of JAK2 activation (87). It has been reported that SOCS1 acts as an inhibitor of IFN-mediated signaling in Atlantic salmon (Salmo salar) (88). From other reports of the transcriptome analysis (Table 1), complement system, JAK/STAT signaling and chemokine systems are also commonly appeared in responding pathways to bacterial infections. Therefore, it is suggested that these genes contribute to early immune responses following bacterial infections (within 24 h).
Nocardia seriolae is a filamentous Gram-positive bacterium that causes nocardiosis with high mortality in many fish species in Japan, Taiwan and Japan. The infected fish showed a lethal granulomatous disease of the skin, muscle, spleen, kidney, and liver tissues (89). Unlike other bacterial species from our previous studies, N. seriolae is an intercellular bacteria. To determine specific DEGs elicited by N. seriolae infections, we performed functional enrichment of the KEGG pathway for specific up-regulated genes in largemouth bass. As shown in Table S2, specific upregulated genes were assigned to 11 KEGG pathways; based on the enrichment analysis. From the enrichment analysis, Notch signaling pathway was focused and illustrated using expression levels of RNA-seq data from all four fish species (Figure 2A). Results indicated that Notch1 and HES1 (hairy and enhancer of split 1) were specifically upregulated in largemouth bass against N. seriolae. The Notch and HES1 axis present in hematopoietic cells and stroma of the thymus plays an important role in T cell development (90, 91). Additionally, in the “cytokine-cytokine receptor interaction” pathway, IL12RB1 (interleukin 12 receptor β-1) and IL12RB2 (interleukin 12 receptor β2) in largemouth bass against N. seriolae were upregulated. IL12B is a ligand of IL12RBs, and is highly, but not specifically expressed, in largemouth bass (Figure 2B). IL-12, a heterodimetric cytokine consisting of p35 and p40 subunits, is a key regulator of T helper 1 development (Th1), which promotes cellular immunity against intracellular pathogens. In Amberjack (Seriola dumerili), administrated recombinant IL-12 and formalin-killed N. seriolae showed the higher survival rates after challenged with N. seriolae, compared to vehicle and FKC only groups (92). These pathways promote immune reactions against N. seriolae during early stages of the infection, and are candidates for infection prevention and adjuvants in fish.
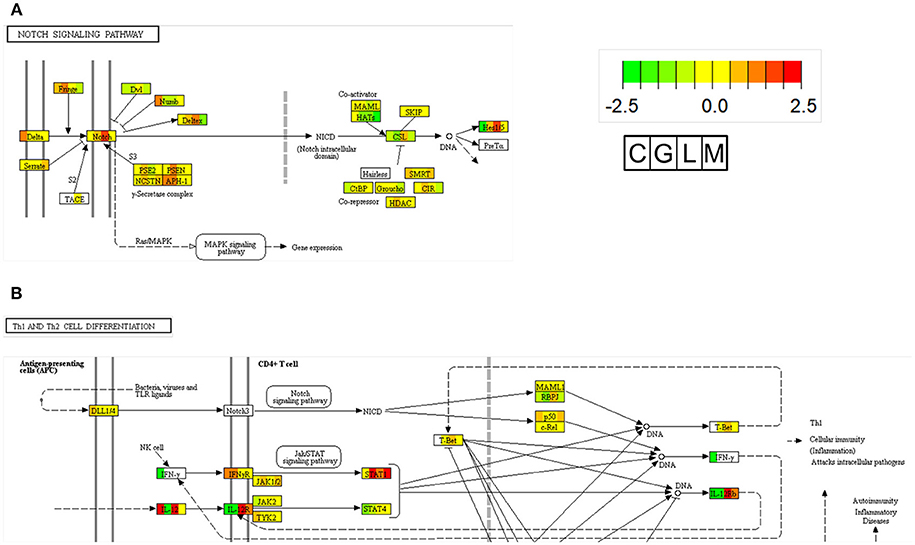
Figure 2. Pathway map of Notch signaling (A) and Th1 differentiation (B) in KEGG. In each gene boxes, the gene expression levels are shown in 4 fish (C, Carp; G, Grouper; L, Largemouth bass; M, Mullet) spleen 1 day after infection with A. sobria, V. harveyi, N. seriolae, and L. garvieae, respectively, when compared to the control group. The lower expression levels of genes are shown in green, and the higher expression levels of genes are shown in red. Undetected genes are shown by white coloring (see color legend in figure).
Aeromonas sobria is a Gram-negative, motile, rod-shaped bacterium that has been isolated from many diseased fish (93–95). In the spleen and head kidney of disease fish, necrotized spleen cells and hemorrhagic pulps were observed (93). From the extracted data of specific up-regulated genes (1001 genes) in koi carp against A sobria (Figure 1), we performed functional enrichment of the KEGG pathway for specific up-regulated genes. As shown in Table S3, specific upregulated genes of koi carp against A. sobria are associated with 45 KEGG pathways. As shown in Figure S1, regulation of the actin cytoskeleton was activated during A. sobria infection. In addition, CXCL12 (C-X-C motif chemokine 12) and CXCR4 (C-X-C motif chemokine receptor 4) in were also up-regulated in koi carps during A sobria infections (Figure S1). The CXCL12-CXCR4 axis modulates various immune functions, such as induction of hematopoiesis and accumulation of immune cells in inflamed tissues (96). Therefore, the CXCL12-CXCR4 axis may function in reorganization of hematopoiesis in injured tissues during A. sobria infections.
Vibrio harveyi is one of the major photogenes of a luminescent Gram-negative bacterium, which impacts to wide range of aquaculture species (97–99). The 601 specific upregulated genes in orange-spotted grouper against Vibrio harveyi (Figure 1) were assigned to 8 KEGG pathways (Table S4). We focused on the ErbB signaling pathway, and found that expression of TGFα (transforming growth factor α) and its receptor, ERBB1 (epidermal growth factor receptor), were upregulated (Figure S2). Previous studies have shown that TGFα promotes the expression and activity of TLR5 and TLR9 in skin keratinocytes (100). In our previous study, expressions of TRL5 and its downstream genes in the spleen were found to be enhanced 2 days following V. harveyi infections (16). While the immunological function of TGFα in the spleen of fish is unclear, we hypothesize that TGFα is a key regulator for prevention of V. harveyi infection in fish.
Lactococcus garvieae is a Gram-positive, facultative anaerobic, non-motile bacterium, and affects freshwater and marine cultured fish species worldwide (101, 102). Functional enrichment analysis of the KEGG pathway was performed to determine specific upregulated genes (291 genes) in gray mullets against L. garvieae (Figure 1). Specific upregulated genes were mapped to 10 KEGG pathways during L. garvieae infection in gray mullets (Table S5). Results indicated that the IL-17 signaling pathway is clearly enhanced during the infection, as illustrated by Figure S3. IL-17 is composed of six ligands (IL-17A to F), and plays critical roles in inflammatory responses and host defenses during invasion by extracellular pathogens. Binding of IL-17s to its five perspective receptors (IL-17Rs; IL-17RA to E), induces inflammatory and immune responses (103). In vitro study, exogenous IL-17A induced bacterial clearance in F. tularensis LVS- live vaccine strain infected cells (104). Additionally, in mice model, it has been reported that in vivo administration of IL-17A moderately delays time of death from lethal infection of Francisella tularensis live vaccine strain (105). While we did not detect expression of IL-17 ligands in this study, we found that expressions of IL-17RB, IL17RC, and IL-17RE were up-regulated in gray mullets infected with L. garvieae. There are studies that aimed to identify and characterize IL-17 and IL17Rs in fish (106, 107). However, functional differentiations of teleost IL-17s and these receptors remain elusive. Our findings on the expression pattern of IL17Rs will provide useful models that can be used to investigate immune functions of IL17s in teleost.
A. sobria and V. harveyi are classified to Gram-negative bacteria. Therefore, we approached to find the immune-related genes and pathways using commonly DEG (379 upregulated genes and 91 downregulated genes) of koi carp against A. sobria and orange-spotted grouper against V. harveyi (Figure 1). However, any immune-related pathways were not assigned by KEGG enrichments analysis. TLR4, which is the pathogen recognized receptor for the Gram-negative bacteria specific lipopolysaccharide, is not highly expression in the spleen of koi carp against A. sobria and orange-spotted grouper against V. harveyi. While, we could find the up-regulated immune-related gene of these two species, such as pattern recognition receptors (TLR6 and TLR5), cytokines and chemokines (CSF3 and CCL21), lysosome related genes (LYPLA3 and SLC11A1) and caspase recruitment domain-containing protein (Card) 9 (Table S6). N. seriolae and L. garvieae are classified to Gram-positive bacteria. Therefore, we investigated to commonly immune-related genes and pathways in Gram-positive bacteria using commonly DEGs (32 upregulated genes and 63 downregulated genes) of largemouth bass against N. seriolae and gray mullet against L. garvieae (Figure 1). Up-regulated immune-related genes of these two species were identified, such as IL-6, TNF Receptor Superfamily Member 11b (TNFRSF11B), interferon regulatory factors (IRF4 and IRF8) and CD83 (Table S7). Although it is unclear the pathways to induce these up-regulated genes, these immune related genes may become the marker and immune factors in Gram-negative or positive bacterial infection.
Conclusion and Future Perspectives
In this review, we first introduced applications of the RNA-Seq technology in aquaculture studies. The RNA-Seq technology has allowed us to identify many novel genes, and to investigate the expression patterns at various conditions in non-model teleost. Therefore, findings based on this technology have accelerated research in the aquaculture field. Additionally, high throughput quantification by RNA-seq could be used to identify pathogen, and to evaluate the efficacy of vaccines and adjuvants against pathogen in vivo. We also summarized current knowledge on immune responses to pathogenic challenges via RNA-Seq in teleost. In this study, we could identify the specific pathway in each fish against bacteria species, Noth1 signaling and IL-12 signaling pathway in largemouth bass against N. seriolae, CXCL12 and CXCR4 signaling pathway in koi carp against A. sobria, TGFα signaling pathway in orange-spotted grouper against V. harveyi, and IL-17 signaling pathway in gray mullet against L. garvieae. These types of studies are increasing, and have enormously aided in our understanding of pathogenic strategies and immune defense systems in aquaculture fish. However, there remains certain limitations of RNA-Seq analysis in aquaculture species. Additionally, the RNA-seq technology could be used to expand existing datasets on splicing variants in mRNA and SNP. Currently, differences in immune response against different pathogens are not well-described. In this study, we attempted to investigate both species-specific and common immune related genes that are up-regulated during bacterial infections based on our previous RNA-seq data. Secondary use of RNA-seq datasets may be essential for preparation of future RNA-seq studies in aquaculture species, which can further deepen our understanding of specific immune functions against pathogens. In aquaculture field, these deep and particular understanding of immune response against each pathogens will provide us to more accurate diagnosis of disease and develop a more effective vaccine and adjuvant of each pathogens.
Author Contributions
SM analyzed transcriptome data and wrote the paper. P-CW and S-CC reviewed the paper.
Funding
This research was funded by National Science Council, Taiwan, grant number NSC 104-2622-B-020-002-CC1 and MOST 107-2313-B-020−012 -MY3.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Genomics Bioscience Technology Co. Ltd. (Taipei, Taiwan) for assistance with transcriptome analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00153/full#supplementary-material
Figure S1. Pathway map of Regulation of actin cytoskeleton in KEGG. In each gene boxes, the gene expression levels are shown in 4 fish (C, Carp; G, Grouper; L, Largemouth bass; M, Mullet) spleen 1 day after infection with A. sobria, V. harveyi, N. seriolae, and L. garvieae, respectively, when compared to the control group. The lower expression levels of genes are shown in green, and the higher expression levels of genes are shown in red. Undetected genes are shown by white coloring (see color legend in figure).
Figure S2. Pathway map of ErbB signaling pathway in KEGG. In each gene boxes, the gene expression levels are shown in 4 fish (C, Carp; G, Grouper; L, Largemouth bass; M, Mullet) spleen 1 day after infection with A. sobria, V. harveyi, N. seriolae, and L. garvieae, respectively, when compared to the control group. The lower expression levels of genes are shown in green, and the higher expression levels of genes are shown in red. Undetected genes are shown by white coloring (see color legend in figure).
Figure S3. Pathway map of Il-17 signaling pathway in KEGG. In each gene boxes, the gene expression levels are shown in 4 fish (C, Carp; G, Grouper; L, Largemouth bass; M, Mullet) spleen 1 day after infection with A. sobria, V. harveyi, N. seriolae, and L. garvieae, respectively, when compared to the control group. The lower expression levels of genes are shown in green, and the higher expression levels of genes are shown in red. Undetected genes are shown by white coloring (see color legend in figure).
Table S1. Expression levels of commonly up or down regulated genes among 4 fish infected with bacteria. Each expression data indicated fold-change (log2) of bacterial infection/control groups.
Table S2. Functional enrichment KEGG pathway for specific up-regulated genes of largemouth bass against N. seriolae.
Table S3. Functional enrichment KEGG pathway for specific up-regulated genes of koi carp against A. sobria.
Table S4. Functional enrichment KEGG pathway for specific up-regulated genes of orange-spotted grouper against V. harveyi.
Table S5. Functional enrichment KEGG pathway for specific up-regulated genes of gray mullet against Lactococcus garvieae.
Table S6. Expression levels of commonly up regulated immune-related genes among koi carp against A. sobria and orange-spotted grouper against V. harveyi. Each expression data indicated fold-change (log2) of bacterial infection/control groups.
Table S7. Expression levels of commonly up regulated immune-related genes among largemouth bass against N. seriolae and gray mullet against L. garvieae. Each expression data indicated fold-change (log2) of bacterial infection/control groups.
References
1. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. (2009) 10:57–63. doi: 10.1038/nrg2484
2. Qian X, Ba Y, Zhuang Q, Zhong G. RNA-Seq technology and its application in fish transcriptomics. OMICS (2014) 18:98–110. doi: 10.1089/omi.2013.0110
3. Martin SA, Dehler CE, Krol E. Transcriptomic responses in the fish intestine. Dev Comp Immunol. (2016) 64:103–17. doi: 10.1016/j.dci.2016.03.014
4. Jacobson G, Muncaster S, Mensink K, Forlenza M, Elliot N, Broomfield G, et al. Omics and cytokine discovery in fish: presenting the Yellowtail kingfish (Seriola lalandi) as a case study. Dev Comp Immunol. (2017) 75:63–76. doi: 10.1016/j.dci.2017.04.001
5. Petit J, David L, Dirks R, Wiegertjes GF. Genomic and transcriptomic approaches to study immunology in cyprinids: what is next? Dev Comp Immunol. (2017) 75:48–62. doi: 10.1016/j.dci.2017.02.022
6. Sudhagar A, Kumar G, El-Matbouli M. Transcriptome analysis based on RNA-Seq in understanding pathogenic mechanisms of diseases and the immune system of fish: a comprehensive review. Int J Mol Sci. (2018) 19:245. doi: 10.3390/ijms19010245
7. Byon JY, Ohira T, Hirono I, Aoki T. Use of a cDNA microarray to study immunity against viral hemorrhagic septicemia (VHS) in Japanese flounder (Paralichthys olivaceus) following DNA vaccination. Fish Shellfish Immunol. (2005) 18:135–47. doi: 10.1016/j.fsi.2004.06.008
8. MacKenzie S, Iliev D, Liarte C, Koskinen H, Planas JV, Goetz FW, et al. Transcriptional analysis of LPS-stimulated activation of trout (Oncorhynchus mykiss) monocyte/macrophage cells in primary culture treated with cortisol. Mol Immunol. (2006) 43:1340–8. doi: 10.1016/j.molimm.2005.09.005
9. Martin SA, Blaney SC, Houlihan DF, Secombes CJ. Transcriptome response following administration of a live bacterial vaccine in Atlantic salmon (Salmo salar). Mol Immunol. (2006) 43:1900–11. doi: 10.1016/j.molimm.2005.10.007
10. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. (2011) 29:644–52. doi: 10.1038/nbt.1883
11. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
12. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. (1999) 27:29–34. doi: 10.1093/nar/27.1.29
13. Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. (2016) 44:D457–62. doi: 10.1093/nar/gkv1070
14. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics (2005) 21:3674–6. doi: 10.1093/bioinformatics/bti610
15. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, et al. InterProScan: protein domains identifier. Nucleic Acids Res. (2005) 33:W116–20. doi: 10.1093/nar/gki442
16. Maekawa S, Byadgi O, Chen YC, Aoki T, Takeyama H, Yoshida T, et al. Transcriptome analysis of immune response against Vibrio harveyi infection in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. (2017) 70:628–37. doi: 10.1016/j.fsi.2017.09.052
17. Byadgi O, Chen CW, Wang PC, Tsai MA, Chen SC. De novo transcriptome analysis of differential functional gene expression in largemouth bass (Micropterus salmoides) after challenge with Nocardia seriolae. Int J Mol Sci. (2016) 17:1315. doi: 10.3390/ijms17081315
18. Byadgi O, Chen YC, Barnes AC, Tsai MA, Wang PC, Chen SC. Transcriptome analysis of grey mullet (Mugil cephalus) after challenge with Lactococcus garvieae. Fish Shellfish Immunol. (2016) 58:593–603. doi: 10.1016/j.fsi.2016.10.006
19. Byadgi O, Chen YC, Maekawa S, Wang PC, Chen SC. Immune-Related functional differential gene expression in Koi carp (Cyprinus carpio) after Challenge with Aeromonas sobria. Int J Mol Sci. (2018) 19:2107. doi: 10.3390/ijms19072107
20. Kolder IC, van der Plas-Duivesteijn SJ, Tan G, Wiegertjes GF, Forlenza M, Guler AT, et al. A full-body transcriptome and proteome resource for the European common carp. BMC Genomics (2016) 17:701. doi: 10.1186/s12864-016-3038-y
21. Ellis AE. Immunity to bacteria in fish. Fish Shellfish Immunol. (1999) 9:291–308. doi: 10.1006/fsim.1998.0192
22. Tran NT, Gao ZX, Zhao HH, Yi SK, Chen BX, Zhao YH, et al. Transcriptome analysis and microsatellite discovery in the blunt snout bream (Megalobrama amblycephala) after challenge with Aeromonas hydrophila. Fish Shellfish Immunol. (2015) 45:72–82. doi: 10.1016/j.fsi.2015.01.034
23. Qin C, Gong Q, Wen Z, Yuan D, Shao T, Wang J, et al. Transcriptome analysis of the spleen of the darkbarbel catfish Pelteobagrus vachellii in response to Aeromonas hydrophila infection. Fish Shellfish Immunol. (2017) 70:498–506. doi: 10.1016/j.fsi.2017.09.042
24. Kumar R, Sahoo PK, Barat A. Transcriptome profiling and expression analysis of immune responsive genes in the liver of Golden mahseer (Tor putitora) challenged with Aeromonas hydrophila. Fish Shellfish Immunol. (2017) 67:655–66. doi: 10.1016/j.fsi.2017.06.053
25. Yang Y, Yu H, Li H, Wang A. Transcriptome profiling of grass carp (Ctenopharyngodon idellus) infected with Aeromonas hydrophila. Fish Shellfish Immunol. (2016) 51:329–36. doi: 10.1016/j.fsi.2016.02.035
26. Dang Y, Xu X, Shen Y, Hu M, Zhang M, Li L, et al. Transcriptome analysis of the innate immunity-related complement system in spleen tissue of Ctenopharyngodon idella Infected with Aeromonas hydrophila. PLoS ONE (2016) 11:e0157413. doi: 10.1371/journal.pone.0157413
27. Long M, Zhao J, Li T, Tafalla C, Zhang Q, Wang X, et al. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J Proteomics (2015) 122:41–54. doi: 10.1016/j.jprot.2015.03.031
28. Zhu R, Liu XX, Lv X, Li SY, Li YD, Yu XJ, et al. Deciphering transcriptome profile of the yellow catfish (Pelteobagrus fulvidraco) in response to Edwardsiella ictaluri. Fish Shellfish Immunol. (2017) 70:593–608. doi: 10.1016/j.fsi.2017.08.040
29. Li Z, Liu X, Liu J, Zhang K, Yu H, He Y, et al. Transcriptome profiling based on protein-protein interaction networks provides a core set of genes for understanding blood immune response mechanisms against Edwardsiella tarda infection in Japanese flounder (Paralichthys olivaceus). Dev Comp Immunol. (2018) 78:100–13. doi: 10.1016/j.dci.2017.09.013
30. Li Z, Liu X, Cheng J, He Y, Wang X, Wang Z, et al. Transcriptome profiling provides gene resources for understanding gill immune responses in Japanese flounder (Paralichthys olivaceus) challenged with Edwardsiella tarda. Fish Shellfish Immunol. (2018) 72:593–603. doi: 10.1016/j.fsi.2017.11.041
31. Liu X, Li Z, Wu W, Liu Y, Liu J, He Y, et al. Sequencing-based network analysis provides a core set of gene resource for understanding kidney immune response against Edwardsiella tarda infection in Japanese flounder. Fish Shellfish Immunol. (2017) 67:643–54. doi: 10.1016/j.fsi.2017.06.051
32. Yang D, Liu Q, Yang M, Wu H, Wang Q, Xiao J, et al. RNA-seq liver transcriptome analysis reveals an activated MHC-I pathway and an inhibited MHC-II pathway at the early stage of vaccine immunization in zebrafish. BMC Genomics (2012) 13:319. doi: 10.1186/1471-2164-13-319
33. Ahn DH, Kang S, Park H. Transcriptome analysis of immune response genes induced by pathogen agonists in the Antarctic bullhead notothen Notothenia coriiceps. Fish Shellfish Immunol. (2016) 55:315–22. doi: 10.1016/j.fsi.2016.06.004
34. Peatman E, Li C, Peterson BC, Straus DL, Farmer BD, Beck BH. Basal polarization of the mucosal compartment in Flavobacterium columnare susceptible and resistant channel catfish (Ictalurus punctatus). Mol Immunol. (2013) 56:317–27. doi: 10.1016/j.molimm.2013.04.014
35. Zhou W, Zhang Y, Wen Y, Ji W, Zhou Y, Ji Y, et al. Analysis of the transcriptomic profilings of Mandarin fish (Siniperca chuatsi) infected with Flavobacterium columnare with an emphasis on immune responses. Fish Shellfish Immunol. (2015) 43:111–9. doi: 10.1016/j.fsi.2014.12.006
36. Zhao L, Tu J, Zhang Y, Wang J, Yang L, Wang W, et al. Transcriptomic analysis of the head kidney of Topmouth culter (Culter alburnus) infected with Flavobacterium columnare with an emphasis on phagosome pathway. Fish Shellfish Immunol. (2016) 57:413–8. doi: 10.1016/j.fsi.2016.09.001
37. Valenzuela-Miranda D, Gallardo-Escarate C. Novel insights into the response of Atlantic salmon (Salmo salar) to Piscirickettsia salmonis: interplay of coding genes and lncRNAs during bacterial infection. Fish Shellfish Immunol. (2016) 59:427–38. doi: 10.1016/j.fsi.2016.11.001
38. Wilkins LG, Clark ES, Farinelli L, Wedekind C, Fumagalli L. Embryonic gene expression of Coregonus palaea (whitefish) under pathogen stress as analyzed by high-throughput RNA-sequencing. Fish Shellfish Immunol. (2015) 47:130–40. doi: 10.1016/j.fsi.2015.08.035
39. Wang YD, Huang SJ, Chou HN, Liao WL, Gong HY, Chen JY. Transcriptome analysis of the effect of Vibrio alginolyticus infection on the innate immunity-related complement pathway in Epinephelus coioides. BMC Genomics (2014) 15:1102. doi: 10.1186/1471-2164-15-1102
40. Wang YD, Wang YH, Hui CF, Chen JY. Transcriptome analysis of the effect of Vibrio alginolyticus infection on the innate immunity-related TLR5-mediated induction of cytokines in Epinephelus lanceolatus. Fish Shellfish Immunol. (2016) 52:31–43. doi: 10.1016/j.fsi.2016.03.013
41. Zhang X, Wang S, Chen S, Chen Y, Liu Y, Shao C, et al. Transcriptome analysis revealed changes of multiple genes involved in immunity in Cynoglossus semilaevis during Vibrio anguillarum infection. Fish Shellfish Immunol. (2015) 43:209–18. doi: 10.1016/j.fsi.2014.11.018
42. Zhao C, Fu M, Wang C, Jiao Z, Qiu L. RNA-Seq analysis of immune-relevant genes in Lateolabrax japonicus during Vibrio anguillarum infection. Fish Shellfish Immunol. (2016) 52:57–64. doi: 10.1016/j.fsi.2016.02.032
43. Chu Q, Song W, Cui J, Xu T. Genome-guided transcriptome analysis of miiuy croaker provides insights into pattern recognition receptors and cytokines in response to Vibrio anguillarum. Dev Comp Immunol. (2017) 73:72–8. doi: 10.1016/j.dci.2017.03.009
44. Bayha KM, Ortell N, Ryan CN, Griffitt KJ, Krasnec M, Sena J, et al. Crude oil impairs immune function and increases susceptibility to pathogenic bacteria in southern flounder. PLoS ONE (2017) 12:e0176559. doi: 10.1371/journal.pone.0176559
45. Gao C, Fu Q, Su B, Zhou S, Liu F, Song L, et al. Transcriptomic profiling revealed the signatures of intestinal barrier alteration and pathogen entry in turbot (Scophthalmus maximus) following Vibrio anguillarum challenge. Dev Comp Immunol. (2016) 65:159–68. doi: 10.1016/j.dci.2016.07.007
46. Low CF, Mariana NS, Maha A, Chee HY, Fatimah MY. Identification of immune response-related genes and signalling pathways in spleen of Vibrio parahaemolyticus-infected Epinephelus fuscoguttatus (Forskal) by next-generation sequencing. J Fish Dis. (2016) 39:389–94. doi: 10.1111/jfd.12359
47. Zhang Q, Ji C, Ren J, Dong X, Zu Y, Jia L, et al. Differential transcriptome analysis of zebrafish (Danio rerio) larvae challenged by Vibrio parahaemolyticus. J Fish Dis. (2018) 41:1049–62. doi: 10.1111/jfd.12796
48. Li S, Zhang Y, Cao Y, Wang D, Liu H, Lu T. Trancriptome profiles of Amur sturgeon spleen in response to Yersinia ruckeri infection. Fish Shellfish Immunol. (2017) 70:451–60. doi: 10.1016/j.fsi.2017.09.033
49. Ken CF, Chen CN, Ting CH, Pan CY, Chen JY. Transcriptome analysis of hybrid tilapia (Oreochromis spp.) with Streptococcus agalactiae infection identifies Toll-like receptor pathway-mediated induction of NADPH oxidase complex and piscidins as primary immune-related responses. Fish Shellfish Immunol. (2017) 70:106–20. doi: 10.1016/j.fsi.2017.08.041
50. Wang L, Liu P, Wan ZY, Huang SQ, Wen YF, Lin G, et al. RNA-Seq revealed the impairment of immune defence of tilapia against the infection of Streptococcus agalactiae with simulated climate warming. Fish Shellfish Immunol. (2016) 55:679–89. doi: 10.1016/j.fsi.2016.06.058
51. Zhu J, Fu Q, Ao Q, Tan Y, Luo Y, Jiang H, et al. Transcriptomic profiling analysis of tilapia (Oreochromis niloticus) following Streptococcus agalactiae challenge. Fish Shellfish Immunol. (2017) 62:202–12. doi: 10.1016/j.fsi.2017.01.023
52. Qi Z, Wu P, Zhang Q, Wei Y, Wang Z, Qiu M, et al. Transcriptome analysis of soiny mullet (Liza haematocheila) spleen in response to Streptococcus dysgalactiae. Fish Shellfish Immunol. (2016) 49:194–204. doi: 10.1016/j.fsi.2015.12.029
53. Gao FX, Wang Y, Zhang QY, Mou CY, Li Z, Deng YS, et al. Distinct herpesvirus resistances and immune responses of three gynogenetic clones of gibel carp revealed by comprehensive transcriptomes. BMC Genomics (2017) 18:561. doi: 10.1186/s12864-017-3945-6
54. Shi M, Huang R, Du F, Pei Y, Liao L, Zhu Z, et al. RNA-seq profiles from grass carp tissues after reovirus (GCRV) infection based on singular and modular enrichment analyses. Mol Immunol. (2014) 61:44–53. doi: 10.1016/j.molimm.2014.05.004
55. Xu BH, Zhong L, Liu QL, Xiao TY, Su JM, Chen KJ, et al. Characterization of grass carp spleen transcriptome during GCRV infection. Genet Mol Res. (2016) 15. doi: 10.4238/gmr.15026650
56. He L, Zhang A, Pei Y, Chu P, Li Y, Huang R, et al. Differences in responses of grass carp to different types of grass carp reovirus (GCRV) and the mechanism of hemorrhage revealed by transcriptome sequencing. BMC Genomics (2017) 18:452. doi: 10.1186/s12864-017-3824-1
57. Chen G, He L, Luo L, Huang R, Liao L, Li Y, et al. Transcriptomics sequencing provides insights into understanding the mechanism of grass carp reovirus infection. Int J Mol Sci. (2018) 19:488. doi: 10.3390/ijms19020488
58. Wu R, Sheng X, Tang X, Xing J, Zhan W. Transcriptome analysis of flounder (Paralichthys olivaceus) gill in response to Lymphocystis Disease Virus (LCDV) infection: novel insights into fish defense mechanisms. Int J Mol Sci. (2018) 19:160. doi: 10.3390/ijms19010160
59. Lu MW, Ngou FH, Chao YM, Lai YS, Chen NY, Lee FY, et al. Transcriptome characterization and gene expression of Epinephelus spp in endoplasmic reticulum stress-related pathway during betanodavirus infection in vitro. BMC Genomics (2012) 13:651. doi: 10.1186/1471-2164-13-651
60. Liu P, Wang L, Kwang J, Yue GH, Wong SM. Transcriptome analysis of genes responding to NNV infection in Asian seabass epithelial cells. Fish Shellfish Immunol. (2016) 54:342–52. doi: 10.1016/j.fsi.2016.04.029
61. Dettleff P, Moen T, Santi N, Martinez V. Transcriptomic analysis of spleen infected with infectious salmon anemia virus reveals distinct pattern of viral replication on resistant and susceptible Atlantic salmon (Salmo salar). Fish Shellfish Immunol. (2017) 61:187–93. doi: 10.1016/j.fsi.2017.01.005
62. Yuan J, Yang Y, Nie H, Li L, Gu W, Lin L, et al. Transcriptome analysis of epithelioma papulosum cyprini cells after SVCV infection. BMC Genomics (2014) 15:935. doi: 10.1186/1471-2164-15-935
63. Wang Y, Zhang H, Lu Y, Wang F, Liu L, Liu J, et al. Comparative transcriptome analysis of zebrafish (Danio rerio) brain and spleen infected with spring viremia of carp virus (SVCV). Fish Shellfish Immunol. (2017) 69:35–45. doi: 10.1016/j.fsi.2017.07.055
64. Dios S, Balseiro P, Costa MM, Romero A, Boltana S, Roher N, et al. The involvement of cholesterol in sepsis and tolerance to lipopolysaccharide highlighted by the transcriptome analysis of zebrafish (Danio rerio). Zebrafish (2014) 11:421–33. doi: 10.1089/zeb.2014.0995
65. Liu QN, Xin ZZ, Liu Y, Zhang DZ, Jiang SH, Chai XY, et al. De novo transcriptome assembly and analysis of differential gene expression following lipopolysaccharide challenge in Pelteobagrus fulvidraco. Fish Shellfish Immunol (2018) 73:84–91. doi: 10.1016/j.fsi.2017.11.045
66. Mu Y, Li M, Ding F, Ding Y, Ao J, Hu S, et al. De novo characterization of the spleen transcriptome of the large yellow croaker (Pseudosciaena crocea) and analysis of the immune relevant genes and pathways involved in the antiviral response. PLoS ONE (2014) 9:e97471. doi: 10.1371/journal.pone.0097471
67. Chu Q, Gao Y, Xu G, Wu C, Xu T. Transcriptome comparative analysis revealed poly(I:C) activated RIG-I/MDA5-mediated signaling pathway in miiuy croaker. Fish Shellfish Immunol. (2015) 47:168–74. doi: 10.1016/j.fsi.2015.08.032
68. Du X, Li Y, Li D, Lian F, Yang S, Wu J, et al. Transcriptome profiling of spleen provides insights into the antiviral mechanism in Schizothorax prenanti after poly (I: C) challenge. Fish Shellfish Immunol. (2017) 62:13–23. doi: 10.1016/j.fsi.2017.01.004
69. Cascon A, Yugueros J, Temprano A, Sanchez M, Hernanz C, Luengo JM, et al. A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect Immun. (2000) 68:3233–41. doi: 10.1128/IAI.68.6.3233-3241.2000
70. Gong YX, Zhu B, Liu GL, Liu L, Ling F, Wang GX, et al. Single-walled carbon nanotubes as delivery vehicles enhance the immunoprotective effects of a recombinant vaccine against Aeromonas hydrophila. Fish Shellfish Immunol. (2015) 42:213–20. doi: 10.1016/j.fsi.2014.11.004
71. Kozinska A. Dominant pathogenic species of mesophilic aeromonads isolated from diseased and healthy fish cultured in Poland. J Fish Dis. (2007) 30:293–301. doi: 10.1111/j.1365-2761.2007.00813.x
72. Hossain MJ, Sun D, McGarey DJ, Wrenn S, Alexander LM, Martino ME, et al. An Asian origin of virulent Aeromonas hydrophila responsible for disease epidemics in United States-farmed catfish. MBio (2014) 5:e00848–00814. doi: 10.1128/mBio.00848-14
73. Decostere A. Flavobacterium columnare infections in fish: the agent and its adhesion to the gill tissue. Verh K Acad Geneeskd Belg. (2002) 64:421–30.
74. Wakabayashi H. Effect of environmental conditions on the infectivity of Flexibacter columnaris to fish. J Fish Dis. (1991) 14:279–90. doi: 10.1111/j.1365-2761.1991.tb00825.x
75. Duremdez R, Al-Marzouk A, Qasem JA, Al-Harbi A, Gharabally H. Isolation of Streptococcus agalactiae from cultured silver pomfret, Pampus argenteus (Euphrasen), in Kuwait. J Fish Dis. (2004) 27:307–10. doi: 10.1111/j.1365-2761.2004.00538.x
76. Jafar QA, Sameer AZ, Salwa AM, Samee AA, Ahmed AM, Al-Sharifi F. Molecular investigation of Streptococcus agalactiae isolates from environmental samples and fish specimens during a massive fish kill in Kuwait Bay. Pak J Biol Sci. (2008) 11:2500–4. doi: 10.3923/pjbs.2008.2500.2504
77. Amal MN, Zamri-Saad M, Iftikhar AR, Siti-Zahrah A, Aziel S, Fahmi S. An outbreak of Streptococcus agalactiae infection in cage-cultured golden pompano, Trachinotus blochii (Lacepede), in Malaysia. J Fish Dis. (2012) 35:849–52. doi: 10.1111/j.1365-2761.2012.01443.x
78. Bowater RO, Forbes-Faulkner J, Anderson IG, Condon K, Robinson B, Kong F, et al. Natural outbreak of Streptococcus agalactiae (GBS) infection in wild giant Queensland grouper, Epinephelus lanceolatus (Bloch), and other wild fish in northern Queensland, Australia. J Fish Dis. (2012) 35:173–86. doi: 10.1111/j.1365-2761.2011.01332.x
79. Cai SH, Wu ZH, Jian JC, Lu YS. Cloning and expression of the gene encoding an extracellular alkaline serine protease from Vibrio alginolyticus strain HY9901, the causative agent of vibriosis in Lutjanus erythopterus (Bloch). J Fish Dis. (2007) 30:493–500. doi: 10.1111/j.1365-2761.2007.00835.x
80. Liang HY, Wu ZH, Jian JC, Huang YC. Protection of red snapper (Lutjanus sanguineus) against Vibrio alginolyticus with a DNA vaccine containing flagellin flaA gene. Lett Appl Microbiol. (2011) 52:156–61. doi: 10.1111/j.1472-765X.2010.02981.x
81. Muller-Eberhard HJ. Molecular organization and function of the complement system. Annu Rev Biochem. (1988) 57:321–47. doi: 10.1146/annurev.bi.57.070188.001541
82. Boshra H, Gelman AE, Sunyer JO. Structural and functional characterization of complement C4 and C1s-like molecules in teleost fish: insights into the evolution of classical and alternative pathways. J Immunol. (2004) 173:349–59. doi: 10.4049/jimmunol.173.1.349
83. Takamura K, Fukuyama S, Nagatake T, Kim DY, Kawamura A, Kawauchi H, et al. Regulatory role of lymphoid chemokine CCL19 and CCL21 in the control of allergic rhinitis. J Immunol. (2007) 179:5897–906. doi: 10.4049/jimmunol.179.9.5897
84. Khattiya R, Ohira T, Hirono I, Aoki T. Identification of a novel Japanese flounder (Paralichthys olivaceus) CC chemokine gene and an analysis of its function. Immunogenetics (2004) 55:763–9. doi: 10.1007/s00251-003-0638-x
85. Li YX, Sun JS, Sun L. An inflammatory CC chemokine of Cynoglossus semilaevis is involved in immune defense against bacterial infection. Fish Shellfish Immunol. (2011) 31:446–52. doi: 10.1016/j.fsi.2011.06.017
86. Chen C, Hu YH, Xiao ZZ, Sun L. SmCCL19, a CC chemokine of turbot Scophthalmus maximus, induces leukocyte trafficking and promotes anti-viral and anti-bacterial defense. Fish Shellfish Immunol. (2013) 35:1677–82. doi: 10.1016/j.fsi.2013.08.020
87. Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, et al. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. (1999) 18:1309–20. doi: 10.1093/emboj/18.5.1309
88. Sobhkhez M, Joensen LL, Tollersrud LG, Strandskog G, Thim HL, Jorgensen JB. A conserved inhibitory role of suppressor of cytokine signaling 1 (SOCS1) in salmon antiviral immunity. Dev Comp Immunol. (2017) 67:66–76. doi: 10.1016/j.dci.2016.11.001
89. Maekawa S, Yoshida T, Wang PC, Chen SC. Current knowledge of nocardiosis in teleost fish. J Fish Dis. (2018) 41:413–9. doi: 10.1111/jfd.12782
90. Wong GW, Knowles GC, Mak TW, Ferrando AA, Zuniga-Pflucker JC. HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRbeta-selected mouse thymocytes. Blood (2012) 120:1439–48. doi: 10.1182/blood-2011-12-395319
91. Rani A, Greenlaw R, Smith RA, Galustian C. HES1 in immunity and cancer. Cytokine Growth Factor Rev. (2016) 30:113–7. doi: 10.1016/j.cytogfr.2016.03.010
92. Matsumoto M, Araki K, Hayashi K, Takeuchi Y, Shiozaki K, Suetake H, et al. Adjuvant effect of recombinant interleukin-12 in the Nocardiosis formalin-killed vaccine of the amberjack Seriola dumerili. Fish Shellfish Immunol. (2017) 67:263–9. doi: 10.1016/j.fsi.2017.06.025
93. Yu J, Koo BH, Kim DH, Kim DW, Park SW. Aeromonas sobria infection in farmed mud loach (Misgurnus mizolepis) in Korea, a bacteriological survey. Iran J Vet Res. (2015) 16:194–201.
94. Dar GH, Dar SA, Kamili AN, Chishti MZ, Ahmad F. Detection and characterization of potentially pathogenic Aeromonas sobria isolated from fish Hypophthalmichthys molitrix (Cypriniformes: Cyprinidae). Microb Pathog. (2016) 91:136–40. doi: 10.1016/j.micpath.2015.10.017
95. Zhu M, Wang XR, Li J, Li GY, Liu ZP, Mo ZL. Identification and virulence properties of Aeromonas veronii bv. sobria isolates causing an ulcerative syndrome of loach Misgurnus anguillicaudatus. J Fish Dis (2016) 39:777–81. doi: 10.1111/jfd.12413
96. Kucia M, Jankowski K, Reca R, Wysoczynski M, Bandura L, Allendorf DJ, et al. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol Histol. (2004) 35:233–45. doi: 10.1023/B:HIJO.0000032355.66152.b8
97. Zhang X, Austin B. Pathogenicity of Vibrio harveyi to salmonids. J Fish Dis. (2000) 23:93–102. doi: 10.1046/j.1365-2761.2000.00214.x
98. Lee K, Liu P, Chuang W. Pathogenesis of gastroenteritis caused by Vibrio carchariae in cultured marine fish. Marine Biotechnology (2002) 4:267–77. doi: 10.1007/s10126-002-0018-9
99. Pujalte M, Sitjà-Bobadilla A, Macián M, Belloch C, Alvarez-Pellitero P, Pérez-Sánchez J, et al. Virulence and molecular typing of Vibrio harveyi strains isolated from cultured dentex, gilthead sea bream and European sea bass. Syst Appl Microbiol. (2003) 26:284–92. doi: 10.1078/072320203322346146
100. Miller LS, Sorensen OE, Liu PT, Jalian HR, Eshtiaghpour D, Behmanesh BE, et al. TGF-alpha regulates TLR expression and function on epidermal keratinocytes. J Immunol. (2005) 174:6137–43. doi: 10.4049/jimmunol.174.10.6137
101. Vendrell D, Balcazar JL, Ruiz-Zarzuela I, de Blas I, Girones O, Muzquiz JL. Lactococcus garvieae in fish: a review. Comp Immunol Microbiol Infect Dis. (2006) 29:177–98. doi: 10.1016/j.cimid.2006.06.003
102. Meyburgh CM, Bragg RR, Boucher CE. Lactococcus garvieae: an emerging bacterial pathogen of fish. Dis Aquat Org. (2017) 123:67–79. doi: 10.3354/dao03083
103. Moseley TA, Haudenschild DR, Rose L, Reddi AH. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. (2003) 14:155–74. doi: 10.1016/S1359-6101(03)00002-9
104. Lin Y, Ritchea S, Logar A, Slight S, Messmer M, Rangel-Moreno J, et al. Interleukin-17 is required for T helper 1 cell immunity and host resistance to the intracellular pathogen Francisella tularensis. Immunity (2009) 31:799–810. doi: 10.1016/j.immuni.2009.08.025
105. Markel G, Bar-Haim E, Zahavy E, Cohen H, Cohen O, Shafferman A, et al. The involvement of IL-17A in the murine response to sub-lethal inhalational infection with Francisella tularensis. PLoS ONE (2010) 5:e11176. doi: 10.1371/journal.pone.0011176
106. Kono T, Korenaga H, Sakai M. Genomics of fish IL-17 ligand and receptors: a review. Fish Shellfish Immunol. (2011) 31:635–43. doi: 10.1016/j.fsi.2010.11.028
Keywords: transcriptome, RNA-Seq, immune response, fish disease, bacteria
Citation: Maekawa S, Wang P-C and Chen S-C (2019) Comparative Study of Immune Reaction Against Bacterial Infection From Transcriptome Analysis. Front. Immunol. 10:153. doi: 10.3389/fimmu.2019.00153
Received: 29 October 2018; Accepted: 17 January 2019;
Published: 05 February 2019.
Edited by:
Hetron Mweemba Munang'andu, Norwegian University of Life Sciences, NorwayReviewed by:
Ming-Wei Lu, National Taiwan Ocean University, TaiwanSyarul Nataqain Baharum, National University of Malaysia, Malaysia
Copyright © 2019 Maekawa, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Chi Wang, cGM5MjEwMDNAZ21haWwuY29t
Shih-Chu Chen, c2NjaGVuQG1haWwubnB1c3QuZWR1LnR3
 Shun Maekawa
Shun Maekawa Pei-Chi Wang
Pei-Chi Wang Shih-Chu Chen
Shih-Chu Chen