Abstract
Phenoloxidase (PO)–catalyzed melanization is a vital immune response in insects for defense against pathogen infection. This process is mediated by clip domain serine proteases and regulated by members of the serpin superfamily. We here revealed that the infection of Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) significantly inhibited the PO activity in Ostrinia furnacalis hemolymph and induced the expression of O. furnacalis serpin–4. Addition of recombinant serpin-4 protein to O. furnacalis hemolymph resulted in a great increase of AcMNPV copies. Serpin-4 significantly suppressed the PO activity and the amidase activity in cleaving colorimetric substrate IEARpNA (IEARase activity) of hemolymph. Further experiments indicated it formed covalent complexes with three serine proteases (SP1, SP13 and SP105) and prevented them from cleaving their cognate downstream proteases in vitro. Altogether, O. furnacalis melanization restricted AcMNPV replication and serpin-4 facilitated AcMNPV infection by inhibiting serine proteases, SP1, SP13, and SP105 which were all involved in the melanization response.
Introduction
Insects inevitably encounter various pathogens including viruses during their lifetime, but they still survive in a microbe–rich natural environment (1). This is mainly attributed to the powerful innate immune system in insects, especially in the case most insects lack a typical adaptive immune system (2, 3). Among insect innate immune responses, melanization is a prominent humoral reaction and leads to the synthesis of melanin (4). A number of studies have shown that melanization, combined with other immune mechanisms such as antimicrobial peptide (AMP) production, provides defense against bacteria (5), fungi (6, 7), and parasites (8). Some research revealed that melanization was also effective in resistance against viruses. For example, the melanized tracheal epidermis limited the spread of Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) to hemocytes and other tissues in resistant Helicoverpa zea larvae (9). The melanized plasma in Heliothis virescens accounted for the inactivation of H. zea single capsid nucleopolyhedrovirus (HzSNPV) in vitro (10). Hemolymph melanization in Lepidoptera was correlated with antiviral activity against Microplitis demolitor bracovirus (MdBV), Lymantria dispar MNPV and Helicoverpa armigera nucleopolyhedrovirus (HearNPV) (11, 12).
The melanization reaction is mediated by multiple activating and regulating factors. During insect melanization, a series of serine proteases (SPs) are sequentially activated upon infection and culminate in the activation of prophenoloxidase activating proteinase (PAP). Activated PAP further converts inactive prophenoloxidase (PPO) to phenoloxidase (PO) (13). The resulting active phenoloxidase catalyzes the oxidation of phenols to quinones which spontaneously polymerize to form melanin (14, 15). This process is strictly regulated by members of the serine protease inhibitor (serpin) superfamily through targeting at specific serine protease(s) (16). Serpins adopt a canonical fold of three β–sheets and up to nine α–helices with a reactive center loop (RCL) exposed at the surface (17, 18). When a serpin interacts with its target serine protease, it is cleaved at the scissile bond in the RCL by the target serine protease and subsequently forms a covalent complex with the target serine protease, which is therefore irreversibly inhibited (18, 19). Many serpins have been reported to regulate insect melanization, such as SRPN1 and SRPN2 in Aedes agypti (20), SRPN2 in Anopheles gambiae (21), serpin–5, –6, –15 and –32 in Bombyx mori (22–25), serpin–5 and serpin–9 in Helicoverpa armigera (26), serpin–1, serpin–3, serpin–4, serpin–5, serpin–6, serpin–7, serpin–9, serpin–12 and serpin–13 in Manduca sexta (27–30) and SPN40, SPN55 and SPN48 in Tenebrio molitor (31).
The inhibition of melanization by serpins has been reported to affect the antibacterial and antiparasitic responses (23, 32, 33). Recent studies revealed that serpins also participated in the antiviral processes in insects. For example, in H. armigera, suppression of melanization by serpin–5 and serpin–9 promoted the baculovirus infection (26). In B. mori, depletion of serpin-2 resulted in the decrease of the number of BmNPV genomic DNA copies (34). Expression of a viral serpin Hesp018 increased the virulence of baculovirus in infected Trichoplusia ni larvae, possibly due to its ability to inhibit the activity of host protease involved in PPO activation (35). Comparing with the understanding of the role of serpin in antibacterial response, knowledge about its function in insect antiviral reaction is relatively lacking.
The Asian corn borer, Ostrinia furnacalis (Guenée), is an important agricultural pest in many regions of Asia and causes great economic losses (36). The strategy suppressing O. furnacalis larvae by the natural enemy such as entomopathogenic virus or fungi has been proposed. Our previous work illuminated partly the molecular and biochemical mechanisms involved in interaction between O. furnacalis and entomopathogenic fungi Beauveria bassiana (37). Four serine proteases, SP1, SP7, SP13 and SP105 mediated the melanization in O. furnacalis upon the infection of B. bassiana (38–40). However, knowledge about the crosstalk between O. furnacalis and entomopathogenic virus is very incomplete. In this study, we investigated the relationship between AcMNPV infection and O. furnacalis melanization, and discovered that O. furnacalis serpin-4 facilitated AcMNPV infection by inhibiting its target serine proteases, SP1, SP13, and SP105 which were all involved in the melanization response.
Materials and Methods
Insect Rearing and AcMNPV Preparation
Asian corn borers, O. furnacalis (Guenée) larvae were reared on an artificial diet at 28°C under a relative humidity of 70–90% and a photoperiod of 16 h of light and 8 h dark. AcMNPV was purchased from Henan Jiyuan Baiyun Industry Co., Ltd. and dissolved in phosphate–buffered saline (PBS).
Examination of gDNA Copies of AcMNPV in Infected O. furnacalis Larvae
To explore the replication process of AcMNPV in its host O. furnacalis larvae, each fifth–instar day 0 larvae were injected with 1 μL of AcMNPV (2.5 × 104 polyhedral inclusion body (PIB)/μL) or PBS as a control (2 larvae/treatment). Each treatment was performed 3 times individually. After 1, 6, 12 and 18 h, the total genomic DNA (gDNA) was individually extracted with the Genomic DNA Extraction Kit Ver.5.0 (TaKaRa, Japan) following the manufacturer’s instructions. Specific primers (Table S1) were designed to amplify AcMNPV ODV–e56 which encoded an occlusion–derived virus–specific envelope protein (41). O. furnacalis rpL8 was used to normalize the expression of ODV–e56. qRT–PCR was performed on an Applied Biosystems 7500 Real Time System (Life Technologies™) using SuperReal PreMix Plus (SYBRGreen) (TIANGEN, Beijing, China) with gDNA as a template, according to the manufacturer’s instructions. The thermal cycling conditions for qRT–PCR were 95°C for 15 min followed by 40 cycles of 95°C for 10 s, 60°C for 30 s and 72°C for 32 s to generate a melting curve. Each qRT–PCR experiment was performed in 3 biological replicates. The relative viral gDNA expression was calculated using the 2–ΔΔCt method.
Analysis of PO Activity and Expression of Serpin in AcMNPV–Infected O. furncalis Larvae
To check whether the replication of AcMNPV in O. furnacalis was affected by the melanization response of O. furnacalis, we injected 1 μL of AcMNPV at different concentrations (2.5 × 103, 2.5 × 104 and 6 × 104 PIB/μL) into the hemocoel of O. furnacalis fifth instar day 0 larvae. Injection of 1 μL of sterile PBS was used as a control. At 1, 3, 6, 9, 12 and 18 h post infection (hpi), 1 µL of hemolymph was collected from individual larva and incubated with 9 µL of saline buffer (20 mM Tris, 150 mM NaCl, pH 8.0) at room temperature for 10 min. Then, PO activity of the reaction mixture was measured using dopamine as the substrate. One unit of PO activity was defined as the amount of enzyme producing an increase in absorbance (ΔA490) of 0.001 per min.
To check whether the replication of AcMNPV in O. furnacalis was related to the expression of serpins which are known inhibitors of insect melanization response, we injected 1 µL of AcMNPV suspension (2.5 × 104 PIB/μL, sterile PBS as a control) into fifth instar day 0 larvae. Three hours later, total RNA was isolated from the whole body of each larva (3 larvae/each group) with TRIzol reagent (TIANGEN, Beijing, China) following the manufacturer’s instructions. One microgram of total RNA from each larva was converted into first–strand cDNA using a FastQuant RT Kit (TIANGEN, Beijing, China) following the manufacturer’s protocol. The cDNA products were diluted 10–fold for use as templates in qRT-PCR. Specific primers were designed based on the cDNA sequences from assembled O. furnacalis transcriptome (37) and listed in Table S1. qRT–PCR was performed as described above.
Cloning and Expression Profile Analysis of O. furnacalis Serpin–4
Based on the data from “Material and methods 2.3”, O. furnacalis serpin-4 (37) was selected for further cloning and characterization. Specific primers (Table S1) were designed for the amplification of full–length cDNA encompassing the entire reading frame with cDNA from O. furnacalis larvae as a template. The products were cloned into the pMD19–T vector, and the nucleotide sequences were confirmed by DNA sequencing. To investigate the transcriptional changes of serpin–4 during the various developmental stages, total RNA was individually prepared from three different stages including egg, larva, and pupa. To determine the expression patterns of serpin–4 in different tissues, total RNA samples were isolated separately from the heads, guts, fat bodies, and hemocytes of day 1 fifth–instar larvae. The synthesis of first–strand cDNA and qRT–PCR analyses were performed as described above.
Production of Recombinant Serpin–4 and GFP Proteins and Preparation of Antiserum Against Serpin–4
To produce recombinant serpin–4 (rserpin-4), a cDNA fragment encoding mature serpin–4 was amplified by PCR using the specific primers listed in Table S1. The forward primer included an Nco I site, which provided the start codon, followed by one codon for a glycine residue and six codons for histidine residues. The reverse primer contained a stop codon and a Not I site. The PCR products were ligated into the pMD19–T vector and then digested with Nco I and Not I (TaKaRa, Japan). The digested product was subcloned into the same restriction sites of the expression vector pET28a (Novagen). After sequence confirmation, the resulting serpin–4/pET28a plasmids and gifted GFP/pET28a plasmids were used to transform E. coli BL21 (DE3) cells, respectively. For recombinant protein expression, a single clone was incubated at 37°C in LB medium containing 50 μg/mL kanamycin. When the OD600 of the culture reached 0.8, isopropyl β–D–thiogalactoside was added to a final concentration of 0.1 mM, and recombinant protein was expressed for 13 h at 22°C and 220 rpm. The bacteria were harvested by centrifugation at 3,000 × g for 30 min and resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl and 5 mM imidazole, pH 8.0). The bacteria cells were lysed by sonication, and a cleared clear lysate was obtained by centrifugation. The soluble rserpin–4 or rGFP in the supernatant was purified as described previously (29). Two milligram of the purified serpin–4 was used as an antigen to produce a rabbit polyclonal antiserum (Beijing CoWin Bioscience Co., Ltd). The remaining recombinant protein was stored at –80°C for further use.
The other recombinant proteins, including O. furnacalis PPO2, mutated proSP1 (proSP1Xa), wild type proSP13, mutated proSP13 (proSP13Xa), and mutated proSP105 (proSP105Xa) were successfully obtained in our previous work (38, 39, 42). In proSP1Xa and proSP13Xa, the cleavage activation site was changed to IEGR to permit its activation by bovine Factor Xa which was commercially available (43).
Effect of Serpin–4 on AcMNPV Replication and Hemolymph Melanization
To investigate the effect of serpin–4 on AcMNPV replication in O. furnacalis plasma (cell–free hemolymph), AcMNPV (1.25×104 PIB) was mixed with 5 μL of plasma from day 0 fifth instar larvae plus 5 μL of rSerpin–4 (1.2 μg/μL) or 5 μL of rGFP (1.2 μg/μL), or 5 μL of 20 mM phenylthiourea (PTU, specific inhibitor of PO), respectively. Phosphate buffer (50 mM sodium phosphate, pH 6.5) was supplied to adjust the final volume of the reaction mixtures to 110 μL. After incubation for 1, 3 and 6 h at room temperature, the total viral gDNA in each sample was extracted as described above. The numbers of viral DNA copies in each mixture were quantified with a standard curve that was generated from a series of diluted plasmids containing the fragment encoding AcMNPV ODV–e56 (Figure S1) (41).
To investigate the effect of serpin–4 on melanization, nickel–nitrilotriacetic acid (Ni–NTA) agarose beads (Qiagen, Hilden, Germany) were incubated with recombinant serpin–4 overnight at 4°C (recombinant GFP was used as control). The coated Ni–NTA beads were washed three times with PBS and resuspended in PBS to approximately 100 beads/μL. Then, one microliters of resuspended Ni–NTA beads was added to a 24-well plate containing 10 μL of fresh hemolymph and 450 μL of Sf9 medium. After incubation for 2 h at room temperature, the melanization of Ni-NTA beads were observed under an inverted fluorescence microscope.
Analysis of Inhibition of Serpin–4 on PO Activity and Amidase Activity in O. furnacalis Hemolymph
To measure the inhibitory potential of serpin–4 on PO activity of O. furnacalis hemolymph, hemolymph was collected into a 1.5 mL microcentrifuge tube from the cut abdominal prolegs of fifth instar day 1 O. furnacalis larvae. Aliquots (1 μL) of hemolymph was incubated for 10 min at room temperature with 9 μL of purified recombinant serpin–4 at varying concentrations. The residual PO activity in the mixtures was measured as described above. Additionally, the amidase activity of the mixtures was measured using acetyl–Ile–Glu–Ala–Arg–p–nitroanilide (IEARpNA) as colorimetric substrate. Changes in absorbance at 405 nm were monitored in a microplate reader (Bio-Tek Instrument, Inc.). One unit of amidase activity was defined as the amount of enzyme producing an increase in absorbance (ΔA405) of 0.001 per min (39).
Analysis of Inhibition of Serpin–4 on SP1, SP13 and SP105
To investigate whether the inhibition of PO activity by serpin-4 was due to it inhibiting SP1, SP13, and SP105 which were all involved in PPO activation (38, 39), we firstly checked whether serpin-4 could form an SDS-stable complex with one of these three serine proteases. Recombinant proSP1Xa, proSP13Xa or proSP105Xa (200 ng) were activated by Factor Xa as described previously (38, 39), and then mixed with purified recombinant serpin–4 at molar ratio of 1:1 or 1:10 (proSPs:rserpin–4). In control samples, proSPs or Factor Xa was omitted. After incubation at 37°C for 15 min, the reaction mixtures were treated with 5 × SDS sample buffer containing dithiothreitol (DTT) at 95°C for 5 min and subjected to 10% SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblot analysis with mouse anti–His (1:2,000) or rabbit anti–serpin–4 (1:2,000) as the primary antibodies. Antibody binding was visualized using alkaline phosphate–conjugated horse anti–mouse (1:3,000) or goat anti–rabbit (1:3,000) and 5–bromo–4–chloro–3–indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) staining buffer containing 165 mg/mL BCIP and 330 mg/mL NBT in 100 mM Tris (pH 9.5), 150 mM NaCl, and 5 mM MgCl2.
Furthermore, we analyzed the inhibitory potential of serpin-4 on the activities of SP1, SP13, and SP105 cleaving the respective substrate. Factor Xa-activated SP1, SP13 or SP105 (200 ng) was mixed with serpin–4 at a molar ratio of 1:1 or 1:10 (proSPs:serpin–4). After incubation at 37°C for 15 min, 200 ng of proSP13 (for SP1’s cleavage) or OfPPO2 (for SP13 and SP105) was added to the reaction mixtures and incubated at 37°C for another 15 min. The mixtures were separated with 10% or 8% SDS–PAGE and subjected to immunoblot analysis using antiserum against the mouse anti–His (1:2,000) or rabbit anti–PPO2 (1:2,000).
Results
AcMNPV Infection Suppressed PO Activity in O. furnacalis Hemolymph and Induced Serpin–4 Expression
As a first step to investigate the interaction between entomopathogenic virus AcMNPV and the host O. furnacalis, we infected O. furnacalis larvae with AcMNPV and measured the viral gDNA copies 1, 6, 12, and 18 hours post infection (hpi). As shown in Figure 1A, the gDNA amounts of AcMNPV remained unchanged within 12 hours after infection, but increased significantly at 18 hpi. Meanwhile, we checked PO activities of O. furnacalis hemolymph after AcMNPV infection. PO activity kept unchanged within 3 hours after infection, and began to decrease significantly at 6 hpi. Until 18 hpi, PO activity was still suppressed significantly (Figure 1B, S2A). It suggested that AcMNPV infection reduced PO activity in O. furnacalis hemolymph, and this suppression might facilitate the viral replication in the host.
Figure 1
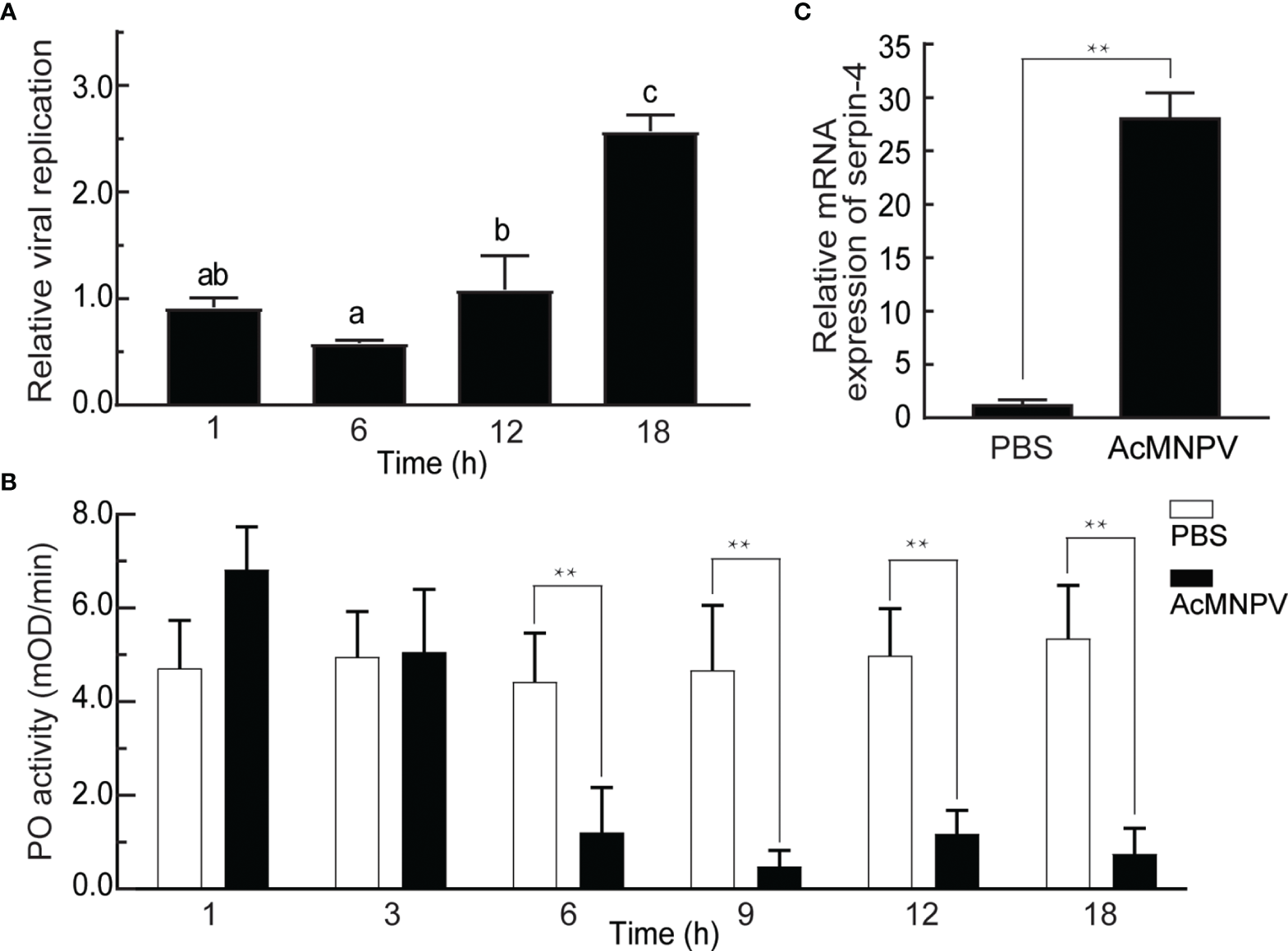
Relationship between AcMNPV infection and the innate immune response in O. furnacalis. (A) Analysis of viral replication after AcMNPV infection. Viral gDNA was extracted from larvae at 1/6/12/18 h after infection, and the relative viral amounts were determined by qRT–PCR. O. furnacalis ribosomal protein L8 (rpL8) was used as the internal control. The bars represented the mean ± S.D. (n = 3). Different marked letters indicated means that were significantly different (one–way ANOVA followed by Tukey’s multiple comparisons test, P < 0.05). (B) Analysis of PO activity after AcMNPV infection. Hemolymph (1 μL) collected from virus-infected or control larvae was incubated for 10 min at room temperature. PO activity was monitored using dopamine as substrate. The bars represented the mean ± S.D. (n = 3). Statistical significance was determined using Sidak’s multiple comparisons test (*P < 0.05, **P < 0.01). (C) Analysis of expression of serpin–4 after AcMNPV infection. Fifth–instar larvae were injected with AcMNPV. The transcript level of serpin–4 was assayed by qRT–PCR three hours later, and rpL8 was used as an internal standard. The bars represented the mean ± S.D. (n = 3). Asterisks indicated means that were significantly different (unpaired t test, two–tailed, **P < 0.01).
On the other hand, PPO activation was regulated by the serpin(s) in insects (44). We speculated that the decrease of PO activity upon AcMNPV infection was related to the serpin(s) in O. furnacalis, and then checked mRNA expression of several transcripts encoding potential serpins, including CL7904.Contig2 (serpin-3), CL9195.Contig5, and CL5354.Contig1 (serpin-6) (37). As shown in Figure 1C and S2B, the abundance of 3 transcripts all increased significantly in the larvae challenged with AcMNPV. The transcript level of CL9195.Contig5 had the largest increase, up to around 25 folds 3 h after AcMNPV infection. Therefore, we only focused on CL9195.Contig5 in the studies that followed. This transcript was named as serpin-4.
Molecular Cloning and Sequence Analysis of Serpin–4
The cDNA fragments encoding the entire coding region of serpin-4 were amplified by PCR using specific primers designed based on the cDNA sequence of CL9195.Contig5 from our previous O. furnacalis transciptome (37). The obtained cDNA sequence of serpin–4 (GenBankTM accession number ON323051) contained a 1,242–bp open reading frame. The conceptual protein deduced from nucleotide sequence consisted of 413 amino acid residues, including a predicted 18–residue secretion signal peptide. The calculated molecular weight and the theoretical isoelectric point of the mature protein was 44.7 kDa and 6.63, respectively (Figure S3). Phylogenetic analysis showed that O. furnacalis serpin–4 clustered together with B. mori serpin–4, M. sexta serpin–4, Operophtera brumata serpin–4 and Plutella xylostella serpin–4, with the bootstrap value of 100 (Figure S4). Among them, M. sexta serpin–4 has been verified experimentally to inhibit PPO activation by inhibiting at least 4 target serine proteases (27, 45). Therefore, we predicted that O. furnacalis serpin–4 could also function as an inhibitor of melanization response.
Expression Profiles of O. furnacalis Serpin–4
We analyzed the mRNA levels of O. furnacalis serpin–4 in various developmental stages or different tissues using qRT–PCR methods. As shown in Figure 2A, serpin–4 transcripts in eggs were significantly more than that in other developmental stages. The mRNA level of serpin–4 remained consistent in the third, fourth and fifth instar larvae, but was significantly higher than that in the first and second instar larvae and pupae. In different tissues, serpin–4 was expressed at significantly higher levels in fat bodies than in the head, gut and hemocytes (Figure 2B).
Figure 2
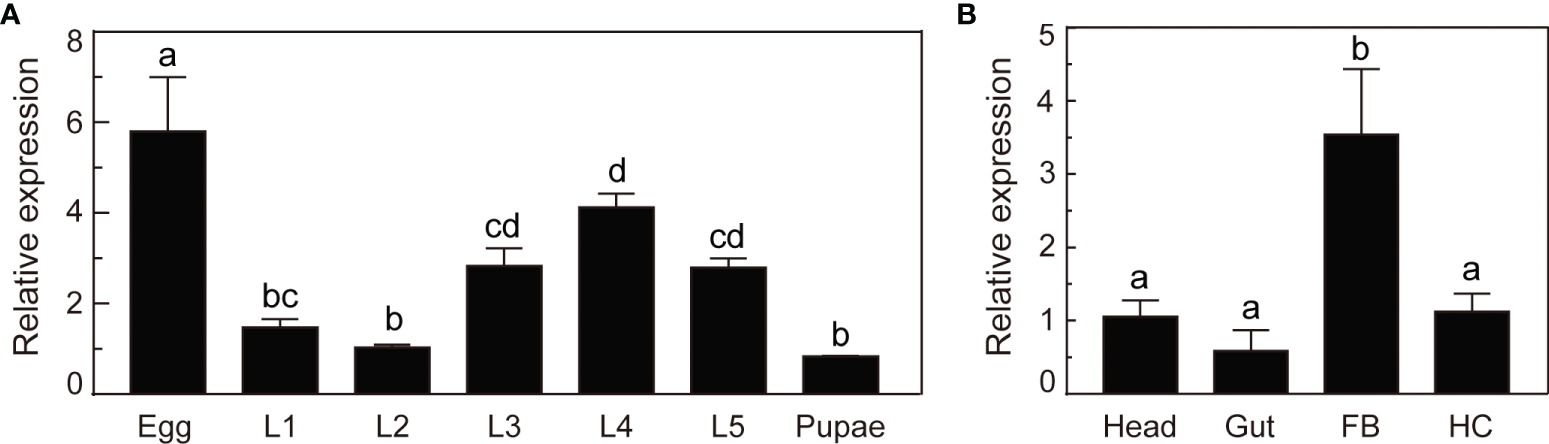
Expression profile analysis of O. furnacalis serpin–4. (A) qRT–PCR analysis of O. furnacalis serpin–4 transcripts at different stages of development. RNA was extracted from eggs, first–instar larvae (L1), second–instar larvae (L2), third–instar larvae (L3), fourth–instar larvae (L4), fifth–instar larvae (L5) and pupae. (B) qRT–PCR analysis of O. furnacalis serpin–4 transcripts in different tissues. RNA was extracted from the head, gut, fat body (FB) and hemocytes (HC). The bars represented the mean ± S.D. (n = 3). The rpL8 gene was used as an internal standard to indicate a consistent total mRNA amount. Bars labeled with different letters were significantly different (one–way ANOVA, followed by Tukey’s multiple comparisons test, P < 0.05).
Production and Purification of Recombinant Serpin–4 and GFP
In order to investigate the function of serpin–4, we produced the recombinant serpin–4 protein with an amino-terminal hexahistidine tag. SDS–PAGE analysis indicated that purified rSerpin–4 had an apparent molecular mass of 45 kDa, approximately consistent with that predicted based on its cDNA sequence (Figure 3A). Recombinant serpin–4 was clearly recognized by the antibodies against O. furnacalis serpin-4 and the commercial anti–His serum in immunoblotting analysis (Figure 3B). Additionally, we produced the recombinant GFP protein as a control. It had an apparent mass of approximately 34 kDa, and was recognized as a single band by anti–His serum (Figure 3).
Figure 3
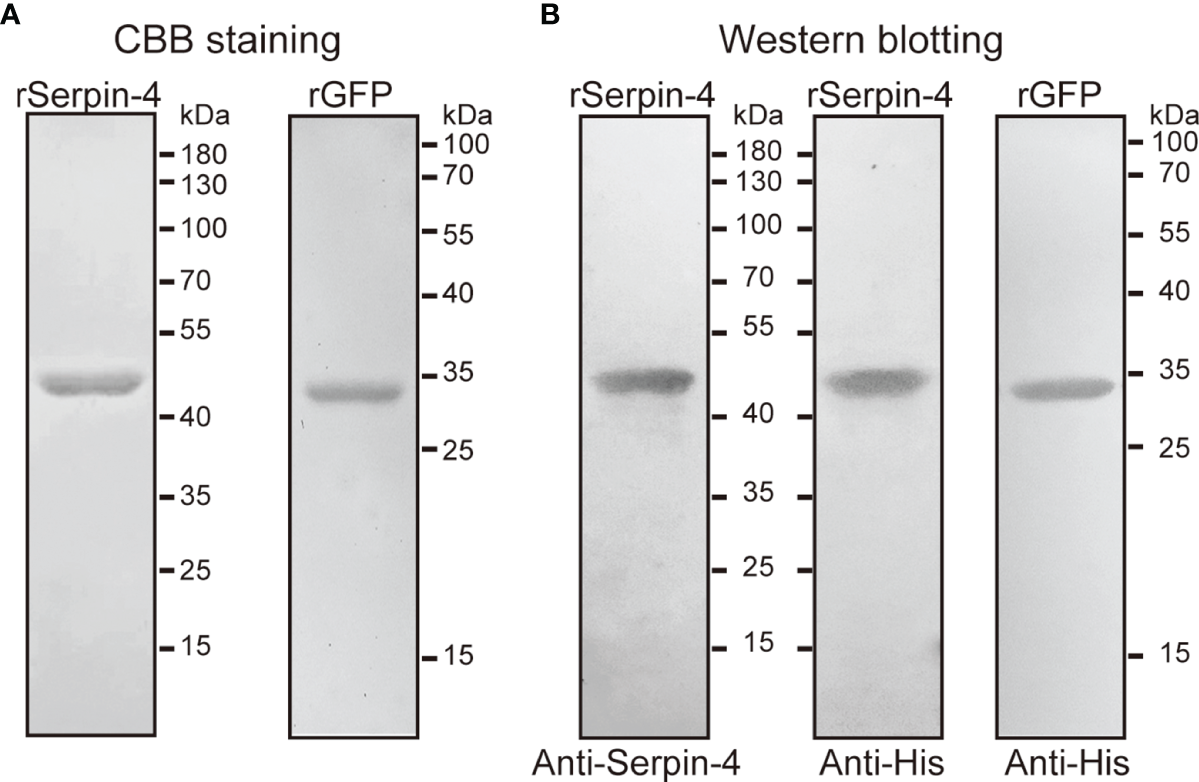
SDS–PAGE (A) and immunoblot analysis (B) of purified recombinant serpin–4 and GFP. The purified recombinant serpin–4 (250 ng) or recombinant GFP (250 ng) was treated with SDS sample buffer containing DTT, separated by 10% or 15% SDS–PAGE and subjected to Coomassie brilliant blue staining or immunoblotting with anti–His or anti–serpin–4 as primary antibodies.
Effects of Serpin–4 on AcMNPV Infection and O. furnacalis Melanization
To evaluate the effects of serpin-regulated hemolymph melanization on virus infection, we incubated AcMNPV with plasma only, or plasma plus rGFP, or plasma plus rSerpin–4, or plasma plus PTU (PTU blocks the melanization by specifically inhibiting PO) for 1, 3, and 6 h, and determined the viral gDNA copies. After 3 h incubation, the virus copies in the sample containing plasma only or plasma plus rGFP decreased to be nearly undetectable (Figure 4A). However, when PTU was incubated together with plasma and virus, the viral DNA copies was unchanged even after incubation for 6 h. Similar results were observed when virus was incubated with plasma together with the recombinant serpin-4 (Figure 4A). These results demonstrated that melanization could reduce the virus copies in vitro and serpin-4 worked like PTU to inhibit the melanization of hemolymph.
Figure 4
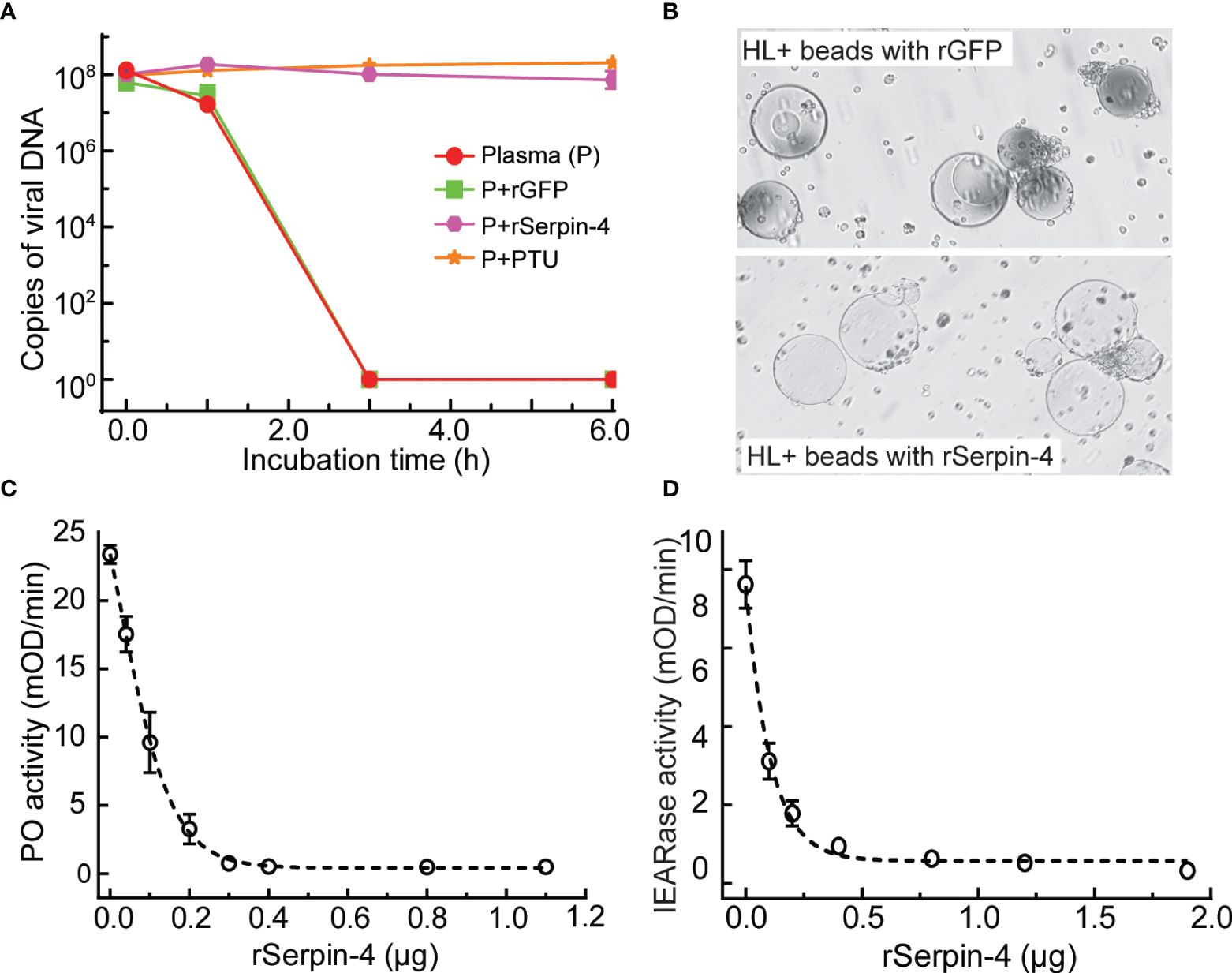
Inhibition analysis of melanization by O. furnacalis serpin–4. (A) Determination of gDNA copy numbers of AcMNPV in plasma incubated with inhibitors. AcMNPV was mixed with plasma, plasma plus PTU (P+PTU), plasma plus recombinant GFP (P+rGFP) or plasma plus recombinant serpin–4 (P+rSerpin–4). The mixtures were incubated at room temperature for 0, 1, 3, or 6 h, and the numbers of gDNA copies were determined by qRT–PCR. The data points represented the mean ± S.D. (n = 3). (B) Serpin–4 suppressed the melanization of Ni–NTA agarose beads. Ni–NTA agarose beads coated with recombinant GFP or serpin–4 were incubated with hemolymph from O. furnacalis larvae. The melanized beads were observed and photographed under a microscopy after 2 h of incubation. (C) Inhibition of PO activation by serpin–4. Hemolymph (1 μL) collected from fifth instar larvae was incubated for 10 min at room temperature with purified recombinant serpin–4 at different concentrations. PO activity was monitored using dopamine as substrate. The bars represented the mean ± S.D. (n = 3). (D) Inhibition of IEARase activation by serpin–4. Hemolymph (1 μL) collected from fifth-instar larvae was incubated with purified recombinant serpin–4 at different concentrations. The residual IEARase activity of hemolymph was plotted as the mean ± S.D. (n = 3).
We further performed in vitro experiments to test the potential of serpin-4 inhibiting melanization. We coated Ni-NTA agarose beads with recombinant serpin-4 or GFP (as a control), and then incubated with O. furnacalis hemolymph. After 2 h, GFP–coated beads turned black. However, the beads coated with rSerpin-4 had no change (Figure 4B).
Hemolymph melanization was accompanied by induced PO activity (20). Inhibition of melanization by serpin-4 inferred that it could suppress PO activity in the hemolymph. To test this hypothesis, we measured the PO activity after the incubation of O. furnacalis hemolymph with different amounts of recombinant serpin–4. Serpin–4 inhibited PPO activation in a concentration–dependent manner (Figure 4C). It blocked PPO activation by 50% at 10 μg/mL and 95% at 30 μg/mL. On the other hand, PPO activation was mediated by multiple serine proteases, some of which exhibit IEARase activity (cleaving after arginine residue in IEARpNA substrate). Thus, we examined the IEARase activity of hemolymph with the addition of different amounts of recombinant serpin–4. As the concentration of rSerpin-4 in the reaction mixtures increased, the IEARase activity gradually declined (Figure 4D). These results indicated that serpin–4 inhibited at least one serine protease in PPO activation cascade in O. furnacalis.
Formation of SDS–Stable Complexes Between rSerpin-4 and SP1, SP13 and SP105
In previous studies, we demonstrated that two serine proteases (SP13 and SP105) acted as prophenoloxidase-activating protease in PPO activation pathway in O. furnacalis, and proSP13 was cleaved and activated by another serine protease (SP1) (38, 39). To reveal which protease serpin–4 inhibited in blocking PPO activation, we firstly checked whether serpin–4 could form SDS–stable, high molecular weight complex with anyone of these three proteases because the formation of such a complex was a characteristic feature for serpin to inhibit its target protease (44).
The anti–His antiserum recognized purified proSP1Xa, proSP13Xa and proSP105Xa as approximately 43 kDa, 49 kDa and 50 kDa, respectively (Figure 5, circles in upper panels). After activation by Factor Xa, the bands representing the three zymogens disappeared (for proSP1Xa) or decreased in intensities (for proSP13Xa and proSP105Xa). Meanwhile, a new band with the apparent molecular weight of 34 kDa, 34 kDa and 35 kDa, appeared, which corresponded to the catalytic domain of proSP1Xa, proSP13Xa and proSP105Xa, respectively (Figure 5, asterisks in upper panels). When rSerpin–4 was mixed with Factor Xa alone or proSP1Xa/proSP13Xa/proSP105Xa zymogen, no change was observed. However, when rSerpin-4 was mixed with Factor Xa-activated SP1Xa, SP13Xa and SP105Xa, respectively, the band corresponding to the catalytic domain disappeared, and a new immunoreactive band at ~ 80 kDa (for SP1Xa) or ~90 kDa position (for SP13Xa and SP105Xa) was detected, which was the expected size of a serpin-4/SP complex (Figure 5, arrows in upper panel). This band with high molecular mass was more abundant when the molar ratio of serpin-4 to SPs increased from 1:1 to 10:1 (Figure 5, upper panel). Moreover, these complexes were also recognized by antibody against serpin–4 (Figure 5, arrows in lower panel). It indicated that serpin-4 could form a covalent complex with each of SP1, SP13, and SP105 in vitro.
Figure 5
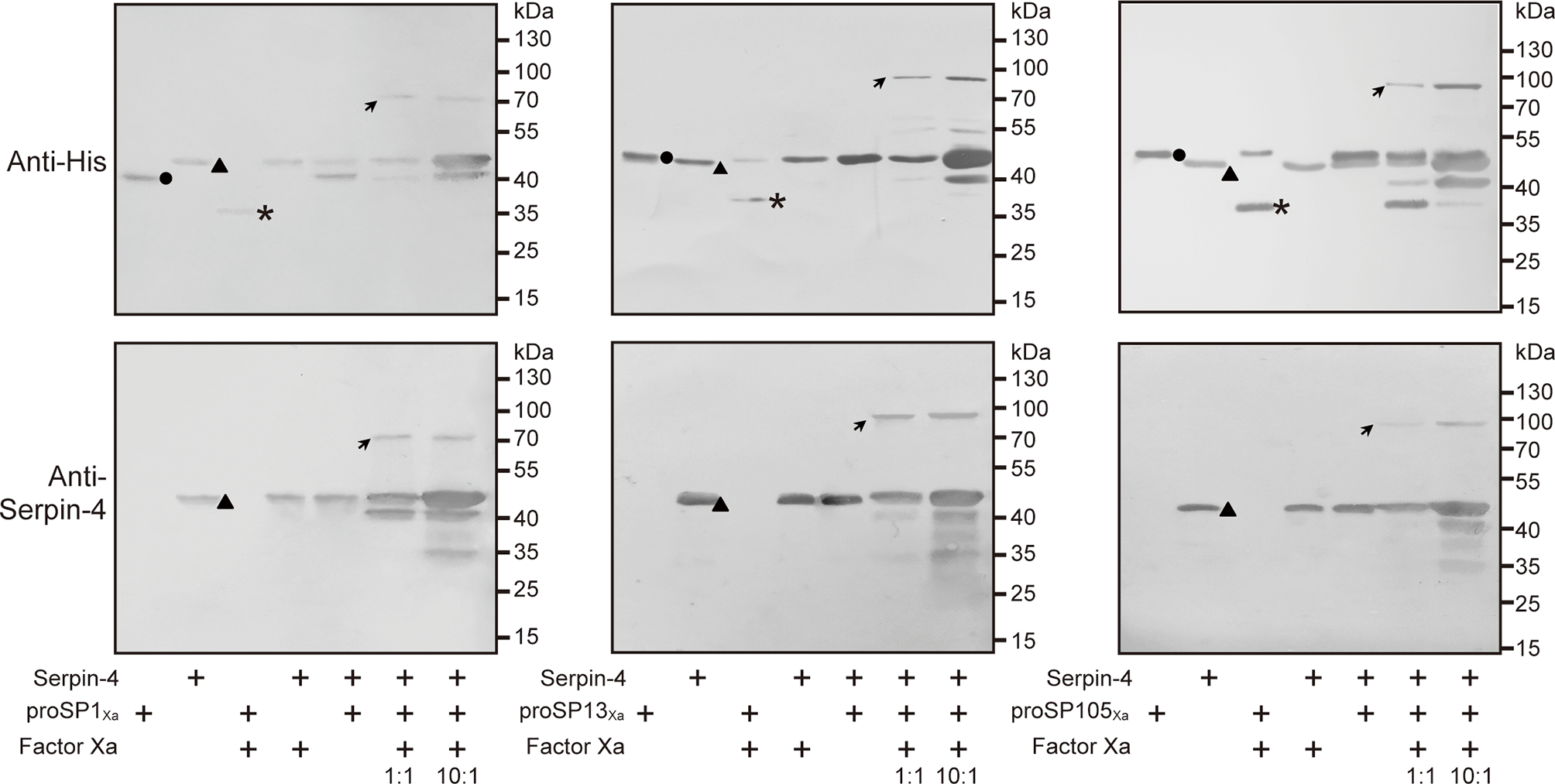
Detection of covalent complex formation between serpin-3 and three serine proteases. 200 ng of proSP1Xa, proSP13Xa or proSP105Xa was activated by Factor Xa, respectively, and incubated with purified serpin–4 at a molar ratio of 1:1 or 10:1 (serpin–4:SPXa) at 37°C for 15 min. The samples were subjected to 10% SDS–PAGE and immunoblot analysis using antiserum against His (upper panel) or O. furnacalis serpin–4 (lower panel) as primary antibodies. The sizes and positions of the molecular mass standards were indicated to the right of each blot. Circles, proSPXa; triangles, serpin–4; asterisks, catalytic domain of proSPXa; arrows, serpin–4/SP complex.
Serpin–4 Prevented SP1Xa, SP13Xa and SP105Xa From Cleaving Its Respective Downstream Substrate
Our previous work indicated that SP1, SP13, and SP105 could cleave O. furnacalis proSP13, PPO2, PPO2, respectively (38, 39). If SP1/SP13/SP105 could be inhibited by serpin-4, the cleavage of their respective substrate would be theoretically suppressed in the presence of serpin-4. To test this hypothesis, we incubated Factor Xa–activated SPs with their substrates (SP1Xa and proSP13; SP13Xa and PPO2; SP105Xa and PPO2) in the absence or presence of serpin–4. As shown in Figure 6A, when proSP13 was incubated with Factor Xa-activated SP1Xa, the ~49-kDa band corresponding to proSP13 zymogen (hollow square in Figure 6A) disappeared, and a ~34-kDa band corresponding to the cleaved catalytic domain of proSP13 showed up (solid square in Figure 6A). When Factor Xa-activated SP1Xa was pre-treated with serpin–4 before mixed with proSP13, proSP13 was clearly detected and the ~34-kDa band indicating the cleavage of proSP13 became faint. Pre-treatment of more serpin-4 resulted in stronger inhibition on the cleavage of proSP13 by SP1Xa (Figure 6A). Similarly, when Factor Xa-activated SP13Xa was pre-treated with serpin–4, the processing of PPO2 by SP13Xa was partly inhibited (Figure 6B and Figure S5A). Especially when Factor Xa-activated SP105Xa was pre-treated with serpin–4 at a molar ration of 1:10, all added PPO2 was recognized as ~80-kDa zymogen band and no cleaved PO2 was detected (Figure 6C and Figure S5B).
Figure 6
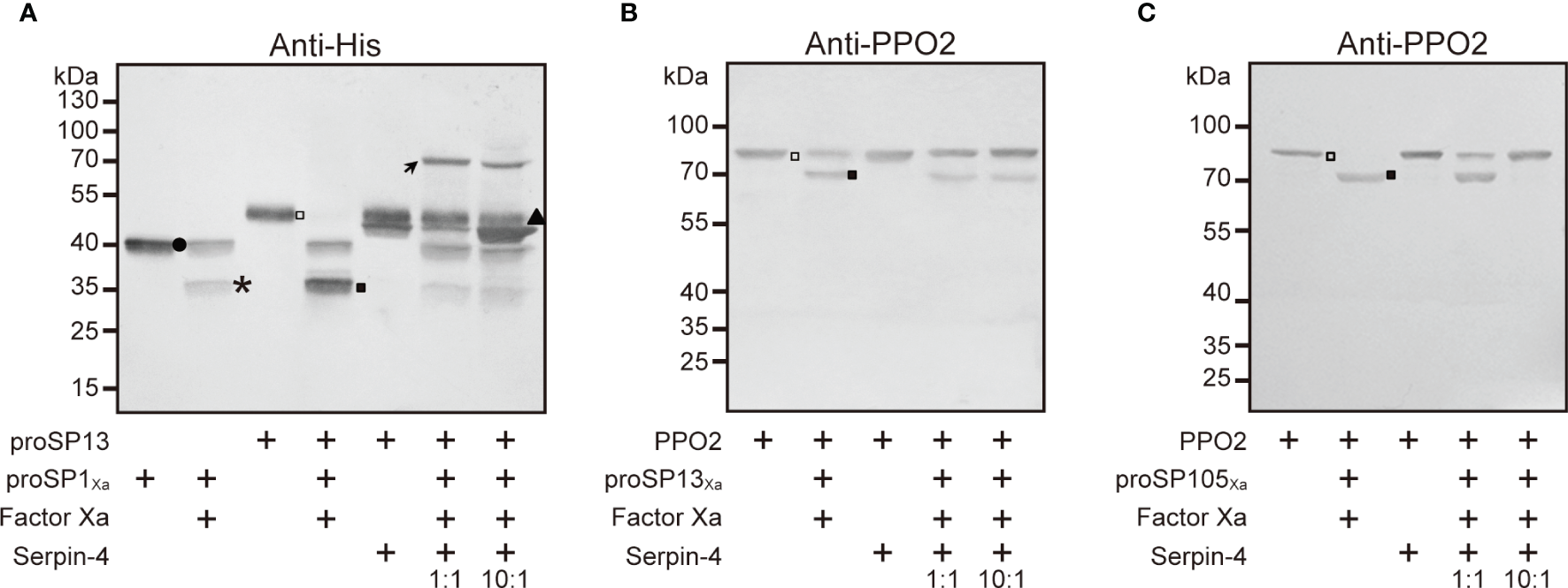
Serpin–4 prevented SP1 (A), SP13 (B) or SP105 (C) from cleaving its respective downstream protease. Factor Xa–activated SPXa (200 ng) was combined with a 1- or 10-fold molar excess of serpin–4 and then incubated with recombinant proSP13 (200 ng) or OfPPO2 (200 ng) at 37°C for 15 min. The mixtures were subjected to 10% or 7.5% SDS–PAGE and immunoblotting using antiserum against His or PPO2. Circle, proSP13Xa; asterisk, catalytic domain of proSP1Xa, proSP13Xa or proSP105Xa; triangle, serpin–4; hollow square, proSP13 or PPO2 zymogen; solid square, activated SP13 or PO2.
Discussion
The understanding of the immune interaction between entomopathogenic viruses and their insect hosts is incomplete. The insect, such as a serious pest O. furnacalis, employs its own immune response including melanization reaction to defend against the microbial infection. On the other hand, entomopathogenic virus, such as AcMNPV, suppresses O. furnacalis immunity and finally kills it. Comprehensive understanding of the biochemical mechanisms involved in the crosstalk between O. furnacalis and AcMNPV would improve the killing effects of AcMNPV, and further help to develop a new strategy on controlling O. furnacalis. Here, we investigated the interaction between AcMNPV infection and O. furnacalis melanization, and discovered that O. furnaclais serpin-4 facilitated AcMNPV infection by inhibiting the melanization. We further revealed serpin-4 performed the inhibitory function possibly by blocking its target proteases, SP1, SP13, and SP105.
Upon the viral infection, insects rely on several defenses including RNA interference (RNAi), Jak/STAT signaling pathway, apoptosis and autophagy to restrict viral replication and dissemination (46–48). Here we discovered that AcMNPV infection resulted in a significant decrease in PO activity of O. furnacalis hemolymph from 6 hpi to 18 hpi or possibly longer (Figure 1B). Meanwhile, viral copies decreased significantly when AcMNPV was incubated with O. furnacalis plasma only in vitro, but remained unchanged when incubated with plasma and PTU which inhibited the melanization of plasma (Figure 4A). It suggested hemolymph melanization in O. furnacalis was related to AcMNPV replication and infection. Similarly, upon AcMNPV infection, PO activity decreased and viral copies significantly increased in susceptible silkworm strains p50. Instead, PO activity increased and viral copies kept unchanged in resistant strain C108 (49). In Aedes albopictus - derived U4.4 cell, more cells were infected by Semliki Forest virus when PO activity of the conditioned medium was blocked (11, 50). 5, 6-dihydroxyindole (DHI), a reactive compound generated by PO, and its spontaneous oxidation products were active against viruses (51). It possibly explained why AcMNPV infection was associated with PO–catalyzed melanization in O. furnacalis.
On the other hand, PO–catalyzed melanization is mediated by a series of sequentially activated serine proteases and regulated by serpin superfamily (14). In this work, we identified a novel serpin transcript, O. furnacalis serpin-4, and illustrated that the melanization of the beads coated with recombinant serpin-4 were obviously restrained (Figure 4B). Recombinant serpin-4 protein inhibited PO activity and IEARase activity of O. furnacalis hemolymph in a concentration-dependent manner (Figures 4C, D). Furthermore, it was interesting that the number of AcMNPV copies significantly increased when the melanization of plasma was suppressed by serpin-4 (Figure 4A). It suggested AcMNPV replication was indeed associated with melanization, and as well inferred that O. furnacalis serpin-4 had the potential to inhibit the melanization. Similar results were found in other insects. Knockdown of serpin-5 or serpin-9 in H. armigera with RNAi significantly increased PO activity of hemolymph and dramatically reduced the number of HearNPV DNA copies (26). In B. mori, the copy numbers of viral genomic DNA also decreased in Bmserpin2-depleted hemolymph (34). Therefore, we concluded that the suppression of melanization caused by serpin responded to viral infection, and depletion of serpin might enhance the virulence of entomopathogenic virus. In our study, O. furnacalis serpin-3 and serpin-6 was also induced upon AcMNPV infection besides serpin-4 (Figure S2B). The function of serpin-6 was completely unknown so far. Serpin-3 has been clarified to regulate the melanization of O. furnacalis hemolymph (32). Future work would test whether serpin-3 could also facilitate virus infection by inhibiting melanization response. We further deciphered the mechanism of serpin-4 regulating O. furnacalis melanization reaction. Recombinant serpin–4 formed covalent complexes with three serine proteases (SP1, SP13 and SP105) which were all involved in melanization pathway (Figure 5) (38, 39). It was consistent with the characteristic feature of serpin in which it forms covalent complexes with its target protease(s) (44). Such serpin/protease regulatory unites were reported as serpin-12/HP14 (30), serpin-5/HP6 (28) and serpin-5/HP1 (27) in M. sexta, serpin-5/cSP4, serpin-9/cSP6, and serpin-9/cSP29 in H. armigera (26). Complex formation made serpin covalently linked to the target protease, which was therefore irreversibly inhibited (19). Here, we also observed serpin-4 strongly prevented SP1, SP13 and SP105 from cleaving their cognate downstream protease - proSP13, PPO2, and PPO2, respectively (Figure 6). Therefore, we speculated serpin-4 regulated the melanization of O. furnacalis hemolymph in this way, and further made for the virus infection. It is reasonable that insects are infected more easily by pathogens or viruses when its immune system is weakened by the negative inhibitors such as serpins. For example, expression of a serpin homologue (Hesp018) in AcMNPV increased the viral virulence and resulted in an increased mortality of infected Trichoplusia ni (35). In Galleria mellonella and Myzus persicae, serpin-expressing B. bassiana strain suppressed PO activation in host hemolymph and exhibited higher virulence (52).
For entomopathogenic microbes, they evolved various strategies to overcome host immunity, for example, inhibit the melanization reaction. Some produced antibiotics to suppress melanization. Entomopathogenic bacterium Photorhabdus luminescens released a small-molecule antibiotic to directly block PO activity (53). Some expressed its own viral protein to inhibit the host PAPs, for example, Egf1.0 and Egf1.5 produced in Microplitis demolitor bracovirus suppressed the processing of host PPO by PAP and SPH (54). Some expressed serpin-like protein to target at host serine protease, for example, parasitoid wasp Pteromalus puparum secreted serpin isoform PpS1V to inhibit host PPO activation by forming complexes with host PrHP8 and PrPAP1 (55). In our study, entomopathogenic virus AcMNPV employed host serpin-4 to suppress host melanization by inhibiting its potential target proteases SP1, SP13 and SP105. However, it is unknown how AcMNPN induced the expression of O. furnacalis serpin–4 and whether other serpin(s) also contributed to AcMNPV infection. More investigation is ongoing. The findings would provide a theoretical basis for better controlling agricultural pests with entomopathogenic virus.
Funding
This work was supported by National Key R&D Program of China (2019YFE0120400), grants 31672361 from the National Natural Science Foundation of China, and the Youth Program of Natural Science Foundation of Jiangsu Province (BK20190959).
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
CA and JJ contributed to the conception and design of the experiments. JJ, SZ, and DS performed the experiments. JJ and LW processed the data. JJ and CA wrote the article. All authors contributed to the article and approved the submitted version.
Acknowledgments
Asian corn borer (O. furnacalis (Guenée)) larvae were kindly provided by Kanglai He, Institute of Plant Protection, Chinese Academy of Agricultural Sciences. B. bassiana was gifted kindly by Weiguo Fang from Zhejiang University. The plasmid for the recombinant expression of GFP was provided by Zhichao Yan from Zhejiang University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.905357/full#supplementary-material
Supplementary Figure 1Standard curve for the quantification of viral copy numbers by qRT-PCR. The CT values were plotted as X-axis. The logarithm of viral copy numbers was indicated on Y-axis. The standard curve parameters calculation was shown above the curve.
Supplementary Figure 2Analysis of PO activity (A) and mRNA expression (B) after AcMNPV infection. (A) Hemolymph (1 μL) collected from O. furnacalis larvae infected by AcMNPV at different concentrations was incubated for 10 min at room temperature. PO activity was monitored using dopamine as a substrate. The bars represented mean ± S.D. (n = 3). Statistical significance was determined using Tukey’s multiple comparisons test (*P < 0.05, **P < 0.01). (B) Fifth–instar larvae were injected with AcMNPV. The transcript levels of O. furnacalis serpin–3, serpin–4 and serpin–6 were assayed by qRT–PCR. rpL8 was used as an internal standard to normalize the templates. The bars represented the mean ± S.D. (n = 3). Asterisks indicated means that were significantly different (unpaired t test, two–tailed, *P < 0.05, **P < 0.01).
Supplementary Figure 3Sequence analysis of serpin–4. The deduced amino acid sequence was shown below the nucleotide sequence of O. furnacalis serpin–4. The one–letter code for each amino acid was aligned with the second nucleotide of the corresponding codon. The stop codon was marked with an asterisk (*). The predicted secretion signal peptide was underlined and assigned negative numbers. Putative N–linked and O–linked glycosylation sites were heavily and lightly shaded, respectively. The potential RCL region was in the square box with the predicted P1 and P1’ residues in boldface. The predicted scissile peptide bond was indicated with “ǁ”.
Supplementary Figure 4Phylogenetic analysis of O. furnacalis serpin–4 and serpins from other insect species. The used amino acid sequences were from Ostrinia furnacalis (Of, red), Anopheles gambiae (Ag), Bombyx mori (Bm), Drosophila melanogaster (Dm), Manduca sexta (Ms), Operophtera brumata (Ob), Plutella xylostella (Px), Tenebrio molitor (Tm). GenBank accession numbers of this specific genes were given in parentheses. The clade that groups O. furnacalis serpin-4 with other serpin-4s was shaded in blue. The numbers at the nodes indicated the bootstrap values as percentages of 1000 repetitions.
Supplementary Figure 5Serpin–4 inhibited the cleavage of recombinant PPO2 by SP13 (A) and SP105 (B). Factor Xa–activated SP13Xa or SP105Xa (200 ng) was combined with a 1- or 10-fold molar excess of serpin–4 at 37°C for 15 min and then incubated with recombinant PPO2 (200 ng) at 37°C for another 15 min. The mixtures were subjected to 7.5% SDS–PAGE and immunoblotting using antiserum against His. Circle, proSP13Xa or proSP105Xa; asterisk, catalytic domain of proSP13Xa or proSP105Xa; triangle, serpin–4; hollow square, PPO2 zymogen; solid square, activated PO2.
Supplementary Table 1Oligonucleotides primers used in this study.
References
1
Shakeel M Xu X Mandal S Jin F . Role of Serine Protease Inhibitors in Insect-Host-Pathogen Interactions. Arch Insect Biochem (2019) 102(3):21556. doi: 10.1002/Arch.21556
2
Cherry S Silverman N . Host-Pathogen Interactions in Drosophila: New Tricks From an Old Friend. Nat Immunol (2006) 7(9):911–7. doi: 10.1038/Ni1388
3
Kingsolver MB Hardy RW . Making Connections in Insect Innate Immunity. Proc Natl Acad Sci U S A (2012) 109(46):18639–40. doi: 10.1073/pnas.1216736109
4
Sheehan G Garvey A Croke M Kavanagh K . Innate Humoral Immune Defences in Mammals and Insects: The Same, With Differences? Virulence (2018) 9(1):1625–39. doi: 10.1080/21505594.2018.1526531
5
Ferrandon D Imler J Hetru C Hoffmann JA . The Drosophila Systemic Immune Response: Sensing and Signalling During Bacterial and Fungal Infections. Nat Rev Immunol (2007) 7(11):862–74. doi: 10.1038/nri2194
6
Tawidian P Rhodes VL Michel K . Mosquito-Fungus Interactions and Antifungal Immunity. Insect Biochem Mol Biol (2019) 111:103182. doi: 10.1016/J.Ibmb.2019.103182
7
Zhang J Huang W Yuan C Lu Y Yang B Wang CY et al . Prophenoloxidase-Mediated Ex Vivo Immunity to Delay Fungal Infection After Insect Ecdysis. Front Immunol (2017) 8:1445. doi: 10.3389/fimmu.2017.01445
8
Severo MS Levashina EA . Mosquito Defenses Against Plasmodium Parasites. Curr Opin Insect Sci (2014) 3:30–6. doi: 10.1016/J.Cois.2014.07.007
9
Washburn JO Kirkpatrick BA Volkman LE . Insect Protection Against Viruses. Nature (1996) 383(6603):767. doi: 10.1038/383767a0
10
Shelby KS Popham HJ . Plasma Phenoloxidase of the Larval Tobacco Budworm, Heliothis Virescens, is Virucidal. J Insect Sci (2006) 6:1–12. doi: 10.1673/2006_06_13.1
11
Rodriguez-Andres J Rani S Varjak M Chase-Topping ME Beck MH Ferguson MC et al . Phenoloxidase Activity Acts as a Mosquito Innate Immune Response Against Infection With Semliki Forest Virus. PLoS Pathog (2012) 8(11):e1002977. doi: 10.1371/journal.ppat.1002977
12
Wang Q Yin M Yuan C Liu X Hu Z Zou Z et al . Identification of a Conserved Prophenoloxidase Activation Pathway in Cotton Bollworm Helicoverpa Armigera. Front Immunol (2020) 11:785. doi: 10.3389/fimmu.2020.00785
13
Veillard F Troxler L Reichhart JM . Drosophila Melanogaster Clip-Domain Serine Proteases: Structure, Function and Regulation. Biochimie (2016) 122:255–69. doi: 10.1016/J.Biochi.2015.10.007
14
Kan H Kim C Kwon H Park J Roh K Lee H et al . Molecular Control of Phenoloxidase-Induced Melanin Synthesis in an Insect. J Biol Chem (2008) 283(37):25316–23. doi: 10.1074/Jbc.M804364200
15
Sugumaran M Barek H . Critical Analysis of the Melanogenic Pathway in Insects and Higher Animals. Int J Mol Sci (2016) 17(10):1753. doi: 10.3390/Ijms17101753
16
Meekins DA Kanost MR Michel K . Serpins in Arthropod Biology. Semin Cell Dev Biol (2017) 62:105–19. doi: 10.1016/j.semcdb.2016.09.001
17
Silverman GA Bird PI Carrell RW Church FC Coughlin PB Gettins PG et al . The Serpins are an Expanding Superfamily of Structurally Similar But Functionally Diverse Proteins. Evolution, Mechanism of Inhibition, Novel Functions, and a Revised Nomenclature. J Biol Chem (2001) 276(36):33293–6. doi: 10.1074/Jbc.R100016200
18
Huntington JA . Serpin Structure, Function and Dysfunction. J Thromb Haemost (2011) 9(1):26–34. doi: 10.1111/J.1538-7836.2011.04360.X
19
Whisstock JC Bottomley SP . Molecular Gymnastics: Serpin Structure, Folding and Misfolding. Curr Opin Struc Biol (2006) 16(6):761–8. doi: 10.1016/j.sbi.2006.10.005
20
Zou Z Shin SW Alvarez KS Kokoza V Raikhel AS . Distinct Melanization Pathways in the Mosquito Aedes Aegypti. Immunity (2010) 32(1):41–53. doi: 10.1016/j.immuni.2009.11.011
21
Zhang X Li M El Moussawi L Saab S Zhang S Osta MA et al . CLIPB10 is a Terminal Protease in the Regulatory Network That Controls Melanization in the African Malaria Mosquito Anopheles Gambiae. Front Cell Infect Microbiol (2021) 10:585986. doi: 10.3389/fcimb.2020.585986
22
Liu D Wang L Yang L Qian C Wei G Dai L et al . Serpin-15 From Bombyx Mori Inhibits Prophenoloxidase Activation and Expression of Antimicrobial Peptides. Dev Comp Immunol (2015) 51(1):22–8. doi: 10.1016/J.Dci.2015.02.013
23
Li J Ma L Lin Z Zou Z Lu Z . Serpin-5 Regulates Prophenoloxidase Activation and Antimicrobial Peptide Pathways in the Silkworm, Bombyx Mori. Insect Biochem Mol Biol (2016) 73:27–37. doi: 10.1016/J.Ibmb.2016.04.003
24
Li B Yu H Ye C Ma Y Li X Fan T et al . Bombyx Mori Serpin6 Regulates Prophenoloxidase Activity and the Expression of Antimicrobial Proteins. Gene (2017) 610:64–70. doi: 10.1016/J.Gene.2017.02.011
25
Wang L Liu H Fu H Zhang L Guo P Xia Q et al . Silkworm Serpin32 Functions as a Negative-Regulator in Prophenoloxidase Activation. Dev Comp Immunol (2019) 91:123–31. doi: 10.1016/j.dci.2018.10.006
26
Yuan C Xing L Wang M Wang X Yin M Wang Q et al . Inhibition of Melanization by Serpin-5 and Serpin-9 Promotes Baculovirus Infection in Cotton Bollworm Helicoverpa Armigera. PLoS Pathog (2017) 13(9):e1006645. doi: 10.1371/Journal.Ppat.1006645
27
Tong Y Jiang H Kanost MR . Identification of Plasma Proteases Inhibited by Manduca Sexta Serpin-4 and Serpin-5 and Their Association With Components of the Prophenol Oxidase Activation Pathway. J Biol Chem (2005) 280(15):14932–42. doi: 10.1074/Jbc.M500532200
28
An C Kanost MR . Manduca Sexta Serpin-5 Regulates Prophenoloxidase Activation and the Toll Signaling Pathway by Inhibiting Hemolymph Proteinase Hp6. Insect Biochem Mol Biol (2010) 40(9):683–9. doi: 10.1016/J.Ibmb.2010.07.001
29
An C Ragan EJ Kanost MR . Serpin-1 Splicing Isoform J Inhibits the Prospätzle-Activating Proteinase HP8 to Regulate Expression of Antimicrobial Hemolymph Proteins in Manduca Sexta. Dev Comp Immunol (2011) 35:135–41. doi: 10.1016/J.Dci.2010.09.004
30
Wang Y Yang F Cao X Huang R Paskewitz S Hartson SD et al . Inhibition of Immune Pathway-Initiating Hemolymph Protease-14 by Manduca Sexta Serpin-12, a Conserved Mechanism for the Regulation of Melanization and Toll Activation in Insects. Insect Biochem Mol Biol (2020) 116:103261. doi: 10.1016/J.Ibmb.2019.103261
31
Jiang R Kim E Gong J Kwon H Kim C Ryu K et al . Three Pairs of Protease-Serpin Complexes Cooperatively Regulate the Insect Innate Immune Responses. J Biol Chem (2009) 284(51):35652–8. doi: 10.1074/jbc.M109.071001
32
Chu Y Zhou F Liu Y Hong F Wang G An C . Ostrinia Furnacalis Serpin-3 Regulates Melanization Cascade by Inhibiting a Prophenoloxidase-Activating Protease. Insect Biochem Mol Biol (2015) 61:53–61. doi: 10.1016/J.Ibmb.2015.03.007
33
Wang M Hu Z . Cross-Talking Between Baculoviruses and Host Insects Towards a Successful Infection. Philos Trans R Soc Lond B Biol Sci (2019) 374(1767):20180324. doi: 10.1098/Rstb.2018.0324
34
Toufeeq S Wang J Zhang SZ Li B Hu P Zhu LB et al . Bmserpin2 is Involved in BmNPV Infection by Suppressing Melanization in Bombyx Mori. Insects (2019) 10(11):399. doi: 10.3390/insects10110399
35
Ardisson-Araujo DMP Rohrmann GF Ribeiro BM Clem RJ . Functional Characterization of Hesp018, a Baculovirus-Encoded Serpin Gene. J Gen Virol (2015) 96(5):1150–60. doi: 10.1099/Vir.0.000041
36
Afidchao MM Musters CJ se Snoo GR . Asian Corn Borer (ACB) and Non-ACB Pests in GM Corn (Zea Mays L.) in the Philippines. Pest Manag Sci (2013) 69(7):792–801. doi: 10.1002/Ps.3471
37
Liu Y Shen D Zhou F Wang G An C . Identification of Immunity-Related Genes in Ostrinia Furnacalis Against Entomopathogenic Fungi by RNA-Seq Analysis. PLoS One (2014) 9(1):e86436. doi: 10.1371/Journal.Pone.0086436
38
Chu Y Liu Y Shen D Hong F Wang G An C . Serine Proteases SP1 and SP13 Mediate the Melanization Response of Asian Corn Borer, Ostrinia Furnacalis, Against Entomopathogenic Fungus Beauveria Bassiana. J Invertebr Pathol (2015) 128:64–72. doi: 10.1016/J.Jip.2015.02.010
39
Chu Y Hong F Liu Q An C . Serine Protease SP105 Activates Prophenoloxidase in Asian Corn Borer Melanization, and is Regulated by Serpin-3. Sci Rep (2017) 7(1):45256. doi: 10.1038/Srep45256
40
Zhang S Feng T Ji J Wang L An C . Serine Protease SP7 Cleaves Prophenoloxidase and is Regulated by Two Serpins in Ostrinia Furnacalis Melanization. Insect Biochem Mol Biol (2022) 141:103699. doi: 10.1016/j.ibmb.2021.103699
41
Li Z Blissard GW . Cellular VPS4 is Required for Efficient Entry and Egress of Budded Virions of Autographa Californica Multiple Nucleopolyhedrovirus. J Virol (2012) 86(1):459–72. doi: 10.1128/JVI.06049-11
42
Zhang S Hong F Song H Wang L Liu Q An C . Cloning, Expression, and Characterization of Prophenoloxidases From Asian Corn Borer, Ostrinia Furnacalis (Gunée). J Immunol Res (2016) 2016:1781803. doi: 10.1155/2016/1781803
43
An C Ishibashi J Ragan EJ Jiang H Kanost MR . Functions of Manduca Sexta Hemolymph Proteinases HP6 and HP8 in Two Innate Immune Pathways. J Biol Chem (2009) 284(29):19716–26. doi: 10.1074/jbc.M109.007112
44
Silverman GA Whisstock JC Bottomley SP Huntington JA Kaiserman D Luke CJ et al . Serpins Flex Their Muscle: I. Putting the Clamps on Proteolysis in Diverse Biological Systems. J Biol Chem (2010) 285(32):24299–305. doi: 10.1074/Jbc.R110.112771
45
Wang Y Yang F Cao X Zou Z Lu Z Kanost MR et al . Hemolymph Protease-5 Links the Melanization and Toll Immune Pathways in the Tobacco Hornworm, Manduca Sexta. Proc Natl Acad Sci USA (2020) 117(38):23581–7. doi: 10.1073/Pnas.2004761117
46
Jiang L Liu W Guo H Dang Y Cheng T Yang W et al . Distinct Functions of Bombyx Mori Peptidoglycan Recognition Protein 2 in Immune Responses to Bacteria and Viruses. Front Immunol (2019) 10:776. doi: 10.3389/fimmu.2019.00776
47
Mussabekova A Daeffler L Imler J . Innate and Intrinsic Antiviral Immunity in Drosophila. Cell Mol Life Sci (2017) 74(11):2039–54. doi: 10.1007/S00018-017-2453-9
48
Schuster S Miesen P van Rij RP . Antiviral RNAi in Insects and Mammals: Parallels and Differences. Viruses (2019) 11(5):448. doi: 10.3390/V11050448
49
Li T Wang X Qin S Sun X Wang S Li M . The Hemolymph Melanization Response is Related to Defence Against the AcMNPV Infection in Bombyx Mori. Arch Insect Biochem Physiol (2021) 108(1):e21764. doi: 10.1002/arch.21764
50
Ourth DD Renis HE . Antiviral Melanization Reaction of Heliothis Virescens Hemolymph Against DNA and RNA Viruses In Vitro. Comp Biochem Physiol B (1993) 105(3-4):719–23. doi: 10.1016/0305-0491(93)90111-h
51
Zhao P Lu Z Strand MR Jiang H . Antiviral, Anti-Parasitic, and Cytotoxic Effects of 5, 6-Dihydroxyindole (DHI), a Reactive Compound Generated by Phenoloxidase During Insect Immune Response. Insect Biochem Mol Biol (2011) 41(9):645–52. doi: 10.1016/J.Ibmb.2011.04.006
52
Yang L Keyhani NO Tang G Tian C Lu R Wang X et al . Expression of a Toll Signaling Regulator Serpin in a Mycoinsecticide for Increased Virulence. Appl Environ Microbiol (2014) 80(15):4531–9. doi: 10.1128/AEM.01197-14
53
Eleftherianos I Boundy S Joyce SA Aslam S Marshall JW Cox RJ et al . An Antibiotic Produced by an Insect-Pathogenic Bacterium Suppresses Host Defenses Through Phenoloxidase Inhibition. Proc Natl Acad Sci USA (2007) 104(7):2419–24. doi: 10.1073/pnas.0610525104
54
Lu Z Beck MH Strand MR . Egf1.5 is a Second Phenoloxidase Cascade Inhibitor Encoded by Microplitis Demolitor Bracovirus. Insect Biochem Mol Biol (2010) 40(7):497–505. doi: 10.1016/j.ibmb.2010.04.009
55
Yan Z Fang Q Liu Y Xiao S Yang L Wang F et al . A Venom Serpin Splicing Isoform of the Endoparasitoid Wasp Pteromalus Puparum Suppresses Host Prophenoloxidase Cascade by Forming Complexes With Host Hemolymph Proteinases. J Biol Chem (2017) 292(3):1038–51. doi: 10.1074/jbc.M116.739565
Summary
Keywords
serpin-4, baculovirus, melanization, inhibition, Ostrinia furnacalis
Citation
Ji J, Shen D, Zhang S, Wang L and An C (2022) Serpin–4 Facilitates Baculovirus Infection by Inhibiting Melanization in Asian Corn Borer, Ostrinia furnacalis (Guenée). Front. Immunol. 13:905357. doi: 10.3389/fimmu.2022.905357
Received
27 March 2022
Accepted
25 April 2022
Published
09 June 2022
Volume
13 - 2022
Edited by
Erjun Ling, Shanghai Institutes for Biological Sciences (CAS), China
Reviewed by
Kai Wu, Shangrao Normal University, China; Kang He, Zhejiang University, China
Updates
Copyright
© 2022 Ji, Shen, Zhang, Wang and An.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunju An, anchunju@cau.edu.cn
This article was submitted to Comparative Immunology, a section of the journal Frontiers in Immunology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.