- 1Department of Microbiology, Anatomy, Physiology and Pharmacology, School of Agriculture, Biomedicine and Environment, La Trobe University, Melbourne, VIC, Australia
- 2Research Centre for Extracellular Vesicles, School of Agriculture, Biomedicine and Environment, La Trobe University, Melbourne, VIC, Australia
- 3ACTV Research Group, Centre for Oral Health Research, Royal Dental Hospital, Melbourne Dental School, The University of Melbourne, Melbourne, VIC, Australia
- 4Department of Biochemistry and Chemistry, School of Agriculture, Biomedicine and Environment, La Trobe University, Melbourne, VIC, Australia
- 5Institute for Health and Sport, Victoria University, Melbourne, VIC, Australia
The release of bacterial membrane vesicles (BMVs) has become recognized as a key mechanism used by both pathogenic and commensal bacteria to activate innate immune responses in the host and mediate immunity. Outer membrane vesicles (OMVs) produced by Gram-negative bacteria can harbor various immunogenic cargo that includes proteins, nucleic acids and peptidoglycan, and the composition of OMVs strongly influences their ability to activate host innate immune receptors. Although various Gram-negative pathogens can produce OMVs that are enriched in immunogenic cargo compared to their parent bacteria, the ability of OMVs produced by commensal organisms to be enriched with immunostimulatory contents is only recently becoming known. In this study, we investigated the cargo associated with OMVs produced by the intestinal commensal Bacteroides fragilis and determined their ability to activate host innate immune receptors. Analysis of B. fragilis OMVs revealed that they packaged various biological cargo including proteins, DNA, RNA, lipopolysaccharides (LPS) and peptidoglycan, and that this cargo could be enriched in OMVs compared to their parent bacteria. We visualized the entry of B. fragilis OMVs into intestinal epithelial cells, in addition to the ability of B. fragilis OMVs to transport bacterial RNA and peptidoglycan cargo into Caco-2 epithelial cells. Using HEK-Blue reporter cell lines, we identified that B. fragilis OMVs could activate host Toll-like receptors (TLR)-2, TLR4, TLR7 and nucleotide-binding oligomerization domain-containing protein 1 (NOD1), whereas B. fragilis bacteria could only induce the activation of TLR2. Overall, our data demonstrates that B. fragilis OMVs activate a broader range of host innate immune receptors compared to their parent bacteria due to their enrichment of biological cargo and their ability to transport this cargo directly into host epithelial cells. These findings indicate that the secretion of OMVs by B. fragilis may facilitate immune crosstalk with host epithelial cells at the gastrointestinal surface and suggests that OMVs produced by commensal bacteria may preferentially activate host innate immune receptors at the mucosal gastrointestinal tract.
Introduction
Bacterial membrane vesicles (BMVs) are nanoparticles released by both pathogenic and non-pathogenic bacteria as part of their normal growth. BMVs are referred to as outer membrane vesicles (OMVs) or membrane vesicles (MVs), if produced by Gram-negative or Gram-positive bacteria, respectively. BMVs contain a range of biological and immunogenic cargo originating from their parent bacteria which includes proteins (1), DNA (2), RNA (3), lipids (4) and peptidoglycan (5, 6), and also lipopolysaccharides (LPS) if produced by Gram-negative bacteria (7). In addition to containing a range of biological cargo, pathogen-derived BMVs can also harbor virulence effectors and immunostimulatory molecules derived from their parent bacteria, enabling them to enhance pathogenesis in the host (8). Due to the diverse range of biological cargo associated with BMVs, they can activate a wide range of host pattern recognition receptors (PRRs) which include Toll-like receptors (TLRs) at the host cell surface, or they can enter host cells and deliver their cargo to intracellular TLRs or nucleotide-binding oligomerisation domain-containing protein (NOD) receptors to mediate inflammation in the host (8). More recently, it has been demonstrated that BMVs produced by pathogenic bacteria may also be enriched in specific cargo compared to their parent bacteria which can include toxins (9), proteins (7), LPS (7), peptidoglycan (6), lipids (4), DNA (10), and RNA (11), and that the differential enrichment of cargo into pathogen-derived BMVs can enhance their immunostimulatory or immunomodulatory functions (12). Therefore, pathogen-derived BMVs are immunogenic, and can be enriched in cargo that facilitates pathogenesis independently of their parent bacteria.
In addition to pathogen-derived BMVs that can elicit immune responses in the host (8), commensal bacteria and their secreted BMVs can also be immunostimulatory or immunomodulatory in the host (13). Recently, the gut microbiota has emerged as a key player in regulating host immune responses, and one mechanism by which they do this is via the secretion of immunomodulatory BMVs (13). A large body of evidence now demonstrates that microbiota-derived BMVs contain diverse bacterial cargo including proteins (14–19), RNA (18, 20) and peptidoglycan (21), and that they can deliver their cargo to host cells to activate PRRs and drive immune responses (22–28). One important member of the gut microbiota is Bacteroides fragilis, which constitutes 1-2% of the normal intestinal microflora (29), and has the ability to modulate host immunity by mediating IL-10 production as a result of detection of its polysaccharide A (PSA) capsule by TLR2 (30–32). It also was identified that B. fragilis OMVs contain PSA and can modulate host immunity, as a result of TLR2 activation and the secretion of IL-10, to ultimately confer protection against colitis in murine models of disease (22, 25). B. fragilis OMVs can also contain proteins and lipid A (15), however, it is unclear whether other immunostimulatory bacterial components such as nucleic acids or peptidoglycan are also present in B. fragilis OMVs, and their ability to activate innate immune receptors remains unknown.
In this study, we examined the immunogenic cargo associated with B. fragilis OMVs and investigated their ability to enter host epithelial cells and activate PRRs. We showed that B. fragilis OMVs contain proteins, DNA, RNA, LPS and peptidoglycan. Additionally, we identified that B. fragilis OMVs could enter host intestinal epithelial cells and transport their peptidoglycan and RNA cargo intracellularly, rendering this cargo accessible to intracellular PRRs. Moreover, due to the cargo they packaged and their ability to enter host cells, we identified that B. fragilis OMVs were able to activate cell-surface receptors TLR2 and TLR4, as well as intracellular PRRs TLR7 and NOD1, in a dose-dependent manner. In comparison, B. fragilis bacteria were only able to activate TLR2 and did not activate any other PRRs examined. Collectively, our data demonstrates that B. fragilis OMVs are laden with potentially immunogenic cargo that enables them to activate a broader range of PRRs compared to their parent bacteria. These findings highlight the importance of OMV secretion by the commensal B. fragilis in maintaining intercellular communication at the mucosal epithelial cell surface.
Materials and methods
Bacterial culturing conditions
Bacteroides fragilis strain NCTC 9343 was cultured as previously described (33). Briefly, B. fragilis was cultured using Horse Blood Agar medium consisting of Blood Agar Base No. 2 (Oxoid, USA) supplemented with 8% (v/v) horse blood (Australian Ethical Biologicals, Australia), or using Brain Heart Infusion (BHI) broth (BD Biosciences, USA) supplemented with 5 μg/ml Hemin (Sigma-Aldrich, USA) with shaking at 120 rpm. Cultures were grown at 37°C in anaerobic conditions using an AnaeroGen 2.5L sachet (Oxoid, USA) and an AnaeroJar 2.5L anaerobic jar (Oxoid, USA).
Isolation of B. fragilis OMVs
B. fragilis OMVs were isolated using our established methods of OMV isolation (5, 6, 12, 34–36). Briefly, BHI broth was inoculated using an overnight B. fragilis culture at a starting optical density (O.D. 600 nm) of 0.05 and grown at 37°C with shaking for 16 h to stationary phase of growth (O.D. 600 nm of approximately 1.8-2.0) using anaerobic conditions. Bacteria were pelleted by centrifugation at 3, 800 × g for 1 h at 4°C, and the supernatant was subsequently filtered using a 0.22μm polyethersulfone (PES) filter (Nalgene, USA) to remove any remaining bacteria. OMVs contained within bacterial free supernatants were concentrated by tangential flow filtration using a VivaFlow 200 PES crossflow cassette with a 10 kDa molecular weight cut-off filter (Sartorius, Australia), and then pelleted by ultracentrifugation at 100, 000 × g for 2 h at 4°C using a P28S rotor in a CP100NX ultracentrifuge (Hitachi, Japan). The resulting OMV pellets were resuspended in Dulbecco’s phosphate-buffered saline (DPBS; Gibco, USA) and stored at -80°C until further purified.
Purification of B. fragilis OMVs
B. fragilis OMVs were purified by OptiPrep (60% iodixanol(v/v); Sigma-Aldrich, USA) density gradient ultracentrifugation as previously described (6, 12, 35, 36). In brief, OMV samples were adjusted to 45% (v/v) OptiPrep in 2ml DPBS and were then overlaid with a discontinuous OptiPrep gradient containing 2ml each of 40%, 35%, 30%, 25% and 20% OptiPrep (v/v) in DPBS. The OptiPrep gradient was subjected to ultracentrifugation at 100, 000 × g for 16 h at 4°C. Twelve fractions (1 ml each) were collected, each fraction was washed with 10 volumes of DPBS by ultracentrifugation at 100, 000 × g for 2 h at 4°C, and then resuspended in DPBS. Fractions 3 to 9 containing purified OMVs were pooled and washed using ultracentrifugation at 100,000 × g for 2 h at 4°C and the purity of OMV preparations was confirmed using Transmission electron microscopy (TEM). Purified OMVs were stored at -80°C until required.
Nanoparticle tracking analysis (NTA)
Quantification of purified OMVs was performed using ZetaView™ Nanoparticle Tracking Analysis (NTA; Particle Metrix, Germany) as previously described (6). Briefly, OMVs were diluted in DPBS to a concentration of 50 - 200 particles per field of view. NTA measurements of OMV samples were performed using a 488 nm 40 mW laser and CMOS camera by observing 11 cell positions at 25°C with 60 frames captured per position. Analysis was then performed using ZetaView software version 8.05.14 SP7 (minimum brightness: 30, maximum brightness: 255, minimum area: 5, maximum area: 1000, minimum trace length: 15). The average of three biological replicates was calculated and plotted as particle size versus particles per ml using GraphPad Prism v9.3.1.
Transmission electron microscopy (TEM)
TEM sample preparation was performed as previously described (5, 35). Briefly, OMVs were coated onto carbon‐coated 400 mesh copper grids (ProSciTech, Australia) for 10 min, fixed in 1% (w/v) glutaraldehyde (Sigma-Aldrich, USA) and negatively-stained with 2% (w/v) uranyl-acetate (ProSciTech, Australia). OMV samples were then coated with 2% (w/v) methyl-cellulose (Sigma-Aldrich, USA) in 0.4% (w/v) uranyl acetate. Samples were air dried and viewed using a JEM-2100 transmission electron microscope (JEOL, Japan) operated at 200 kV using a Valeta 4 MP CCD camera (Emsis, Germany).
Quantification of the cargo associated with B. fragilis OMVs and B. fragilis bacteria
The protein cargo associated with B. fragilis OMVs was quantified using Qubit protein assay (Invitrogen, USA) using a Qubit 3.0 Fluorometer, according to the manufacturer’s instructions.
OMV-associated DNA was quantified using Qubit broad-range DNA assay. Briefly, OMVs were incubated with 4U Turbo DNase (Invitrogen, USA) at 37°C for 1 h to degrade extra-vesicular DNA, according to the manufacturer’s rigorous DNA degradation protocol. Alternatively, OMVs were incubated at 37°C for 1 h with DPBS as a control. To confirm DNase activity, B. fragilis genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, USA) and incubated with DNase as a control. DNA associated with OMVs and controls was quantified using a Qubit 3.0 Fluorometer, according to the manufacturer’s instructions.
OMV associated RNA was quantified using the Qubit high-sensitivity assay. Briefly, OMVs were incubated with 10 pg/μl RNase A (Invitrogen, USA) at 37°C for 1 h as previously described (37), or OMVs were incubated with DPBS as a control. To confirm the efficiency of RNase, B. fragilis RNA was extracted using the Isolate II RNA Mini Kit (Bioline, UK) and incubated with RNase as a control. RNA in samples was quantified using a Qubit 3.0 Fluorometer, according to the manufacturer’s instructions.
OMV-associated peptidoglycan was quantified as described previously (5, 6, 38). Briefly, OMV samples and L-18 muramyldipeptide (MDP) standards (Invivogen, USA) were adjusted to a volume of 0.5 ml in 1M NaOH and incubated at 38°C for 30 min. Samples were then incubated with 0.5 ml of 0.5M H2SO4 and 5 ml concentrated H2SO4 at 95°C for 5 min. Samples were cooled immediately under running water, followed by the addition of 50 μl CuSO4 (4% w/v) and 100 μl of 1.5% (w/v) 4-phenylphenol (dissolved in 96% (v/v) ethanol) and incubated at 30°C for 30 min. Absorbance was measured at 560 nm using a spectrophotometer and the amount of peptidoglycan associated with B. fragilis OMVs was determined using the MDP standard curve.
LPS associated with OMVs was quantified using the Pierce Chromogenic Endotoxin Quant kit, according to the manufacturer’s instructions (Thermo Scientific, USA). Briefly, OMV samples and LPS standards were adjusted to a volume of 50 μl and incubated with 50 μl limulus amoebocyte lysate for 9 min. Chromogenic substrate solution was added and samples were incubated for 6 min at 37°C, then the reaction was stopped by the addition of 25% (v/v) acetic acid. Absorbance was measured at 405 nm using a CLARIOstar plate reader (BMG Labtech, Germany) and the amount of LPS associated with 108 OMVs was quantified using the standard curve (0.1-1.0 EU/ml), according to the manufacturer’s instructions. Each assay was performed in technical triplicate.
Detection of protein, LPS and peptidoglycan cargo associated with B. fragilis OMVs and B. fragilis bacteria
To detect proteins associated with either B. fragilis OMVs or B. fragilis bacteria, samples were boiled at 95°C for 5 min in 1x NuPAGE LDS sample buffer (Invitrogen, USA) and 1x NuPAGE Reducing Agent (Invitrogen, USA). Samples were normalized by an equivalent amount of protein and were separated by SDS-PAGE as previously described (6). Proteins associated with B. fragilis OMVs and B. fragilis bacteria were detected by staining SDS-PAGE gels using Sypro Ruby (Invitrogen, USA), according to the manufacturer’s instructions, and visualized at 560 nm using a ChemiDoc image system (Bio-Rad Laboratories, USA).
Peptidoglycan associated with B. fragilis OMVs and their parent bacteria was detected by Western immunoblot, as described previously (6). In brief, 10 μg of B. fragilis OMVs and B. fragilis bacterial samples were separated by SDS-PAGE, transferred to a 0.2 μm polyvinylidene difluoride membrane and then blocked using 5% (w/v) bovine serum albumin (BSA; Sigma-Aldrich, USA) in Tris-buffered saline containing 0.05% (v/v) Tween (TBS-T). The membrane was then incubated with an anti-peptidoglycan mouse monoclonal antibody (Bio-Rad Laboratories, USA; clone number 3F6B3, 1:1,000 dilution), washed and then incubated with goat anti-mouse IgG HRP antibody (Invitrogen, USA, 1:5,000 dilution). The membranes were then washed, developed using Clarity Western ECL Substrate (Bio-Rad Laboratories, USA) and imaged using a GE Amersham imager 600 (GE Life Sciences, UK).
To detect LPS associated with B. fragilis OMVs and their parent bacteria, samples (10 μg protein) were first incubated with proteinase K (10 μM; Invitrogen, USA), or DPBS as a control, for 90 min at 37°C. Samples were then resuspended in 1x NuPAGE LDS sample buffer and 1x NuPAGE Reducing Agent and separated by SDS-PAGE. Next, to visualize LPS, SDS-PAGE gels were stained using ProQ Emerald 300 LPS stain kit (Invitrogen, USA), according to the manufacturer’s instructions. Briefly, samples were fixed [50% methanol (v/v), 5% acetic acid (v/v)] for 90 min, oxidized using periodic acid containing 3% acetic acid (v/v) for 30 min, and stained with ProQ Emerald stain for 2 h. SDS-PAGE gels were then washed using 3% acetic acid (v/v), and LPS was visualized at 300 nm using a ChemiDoc image system (Bio-Rad Laboratories). To determine the amount of protein associated with these samples, SDS-PAGE gels were then counterstained using Sypro Ruby protein stain and visualized at 560 nm.
Cell culture and stimulations
Human intestinal epithelial cells (Caco-2) were routinely cultured as previously described (39). In brief, Caco-2 cells were cultured in high-glucose Dulbecco’s modified eagle medium (DMEM; Gibco, USA) supplemented with 10% (v/v) fetal calf serum (FCS; Gibco, USA), 1% (v/v) L-glutamine (Gibco, USA), 1% (v/v) penicillin-streptomycin (Gibco, USA), 1% non-essential amino acids (Gibco, USA) and 25mM HEPES (Gibco, USA). HEK-Blue null cells and HEK-Blue hTLR2, hTLR4, hTLR7, hTLR8, hTLR9, hNOD1 and hNOD2 cells (Invivogen, USA) were maintained in DMEM supplemented with 10% (v/v) FCS, 1% (v/v) L-glutamine, 1% (v/v) penicillin-streptomycin and selective antibiotics required for each individual cell line as described previously (6, 40). All cell lines were cultured at 37°C with 5% CO2.
To perform HEK-Blue assays, HEK-Blue cells were seeded in 96-well plates (Greiner, Germany) at a density of 1 × 105 cells per well in 200 μl culture media and cultured to approximately 80-90% confluence. B. fragilis bacteria were cultured for 16 h and washed with PBS, then added to HEK-Blue cells at an increasing multiplicity of infection (MOI) for 18 h. Alternatively, B. fragilis bacteria were heat-killed at 95°C for 45 min as previously described (41), before their addition to HEK-Blue cells at an increasing MOI for 18 h. Viable counts were performed on live and heat-killed B. fragilis bacteria by enumerating serial dilutions spread on horse blood agar and cultured overnight at 37°C using anaerobic conditions. Additionally, HEK-Blue cells were either stimulated with an increasing MOI of purified B. fragilis OMVs for 18 h, or not-stimulated as negative controls. Positive controls for each cell line included: 50 ng/ml Pam3CSK4 (Pam3CysSerLys4) for TLR2 cells (Invivogen, USA), 6.25 ng/ml LPS for TLR4 cells (Invivogen, USA), 1 pg/ml R848 (resiquimod) for TLR7 and TLR8 cells (Invivogen, USA), 5 nM CpG ODN for TLR9 cells (Invivogen, USA), 100ng/ml TriDap for NOD1 cells (Invivogen, USA) and 0.001 pg/ml L18‐MDP for NOD2 cells (Invivogen, USA). After 24 h, 20 μl of cell culture supernatant was transferred to a fresh 96-well plate and incubated with 180 μl of QUANTI-Blue solution (Invivogen, USA) at 37°C. SEAP activity was measured at 625 nm using a CLARIOstar plate reader (BMG Labtech, Germany).
MTT cell viability assay
The viability of HEK-Blue cells following 18 h stimulation with either live or heat-killed bacteria, or not-stimulated as controls, was determined using the MTT Cell Proliferation Kit (Abcam, UK), according to the manufacturer’s protocol. Briefly, HEK-Blue cell lines (Null, hTLR2, hTLR4, hTLR7 and hNOD1) were seeded at 1 × 105 cells per well in 96-well plates and stimulated with either live or heat-killed B. fragilis bacteria at an MOI of 1,000 for 18 h. Culture media was then replaced with DMEM containing 100 μg/ml gentamicin for 2 h. The media was then replaced with 50 μl MTT reagent in 50 μl DMEM for 3 h followed by adding 150 μl MTT solvent for 15 minutes with shaking. Absorbance was measured at 590 nm using a CLARIOstar plate reader (BMG Labtech, Germany).
Fluorescent labelling of OMVs and OMV-associated cargo
B. fragilis OMVs were labelled using Vybrant DiI (10 μM; Invitrogen, USA) as described previously (6, 10, 42–44). Briefly, OMVs were adjusted to 1.2 × 1012 OMVs per ml in 100 μl of DPBS and stained with DiI for 30 min at 37°C with gentle agitation. The RNA content of OMVs was labelled by incubating with Syto RNASelect (1μM; Invitrogen, USA) for 60 min with gentle agitation, as previously described (43–45). The peptidoglycan content of B. fragilis OMVs was labelled using BODIPY-FL vancomycin (4 ng/ml; Invitrogen, USA) and non-labelled vancomycin (4 ng/ml; Sigma-Aldrich, USA) for 20 min, as previously described (42). An equivalent amount of each fluorescent stain in DPBS (in the absence of OMVs) was used as a negative control. Excess DiI, Syto RNASelect or BODIPY-FL vancomycin dye were removed by washing OMVs and controls four times with 4 ml DPBS using a 10 kDa centrifugal filtration unit (Merck Millipore, Germany).
Examining OMV entry into host cells by confocal microscopy
To visualise OMV entry into host cells, Caco-2 cells were seeded on 18mm round coverslips (Marienfeld, Germany) in 12-well plates at a density of 3 × 105 cells per well in 1ml of media for 24 h. Caco-2 cells were stimulated with either DiI, BODIPY-FL or Syto RNASelect-labelled B. fragilis OMVs for 4 h at an MOI of 4 × 105 OMVs per cell, or each respective stain in DPBS as a control. Following incubation, cells were washed three times with DPBS, and extracellular fluorescence was quenched with 0.025% (v/v) Trypan blue as previously described (34). Cells were fixed using 4% paraformaldehyde (Sigma-Aldrich, USA) and blocked using 1% BSA (w/v) in DPBS. Cell nuclei and cellular actin were stained with 4’,6‐diamidino‐2‐phenylindole dilactate (DAPI; Merck, Germany) and Alexa Fluor 680 phalloidin (Invitrogen, USA), respectively. Samples were then mounted using VectaShield mounting medium (Vector Laboratories, USA) and imaged using a Zeiss 780 PicoQuant confocal microscope (Zeiss, Germany) using a 63x/1.4NA oil objective at 1024 × 1024 × 32 bit per channel. Image analysis was performed using Imaris x64 v9.5.0 (Bitplane, Switzerland). Three biological replicates of Caco-2 cells stimulated with OMVs, labelled with each individual stain, were examined. Three fields of view were imaged for each treatment containing a minimum of 10 cells per field of view.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software v9.3.1. Qubit quantification experiments were analysed using an unpaired t-test. HEK-Blue experiments were analysed using an unpaired t-test or one-way ANOVA with Dunnett’s multiple comparisons test, as indicated. Differences were considered statistically significant when *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Results
B. fragilis OMVs contain DNA, RNA, protein, LPS, and peptidoglycan
B. fragilis OMVs contain a range of cargo including polysaccharides (22), proteins (15, 16, 46) and LPS (15). However, it remains unclear if B. fragilis OMVs also contain other biological cargo including peptidoglycan and nucleic acids. B. fragilis OMVs were isolated and purified to examine their biological cargo composition, as well as their size and morphology. Examination of purified B. fragilis OMVs using transmission electron microscopy (TEM) and Nanoparticle Tracking Analysis (NTA) revealed that they were heterogeneous in size, with the predominant population of OMVs being approximately 135 nm in diameter (Figures 1A, B). Next, we quantified the amount of protein, DNA, RNA, peptidoglycan and LPS cargo associated with B. fragilis OMVs (Figures 1C–G). Having previously shown that there is variability in the quantification of OMV-associated cargo based on the type of protein assay used and the method used to normalise BMV number (35), we quantified the biological cargo associated with OMVs and represented the quantity of cargo per 109 OMVs (Figures 1C–G). Quantification of the cargo associated with 109 B. fragilis OMVs revealed that they contained protein, DNA and RNA (Figures 1C–E). Furthermore, degradation of extra-vesicular DNA using DNase, or RNA using RNase, revealed that the majority of DNA and RNA was protected from degradation and was therefore predominantly located within OMVs (Figures 1D, E). We also identified that B. fragilis OMVs contained peptidoglycan, with approximately 600 ng of peptidoglycan associated with 109 OMVs (Figure 1F), in addition to LPS with approximately 38 endotoxin units (EU) of LPS per 109 OMVs (Figure 1G). Collectively, these findings demonstrate that B. fragilis OMVs contain a diverse range of biological cargo including protein, DNA, RNA, peptidoglycan and LPS.
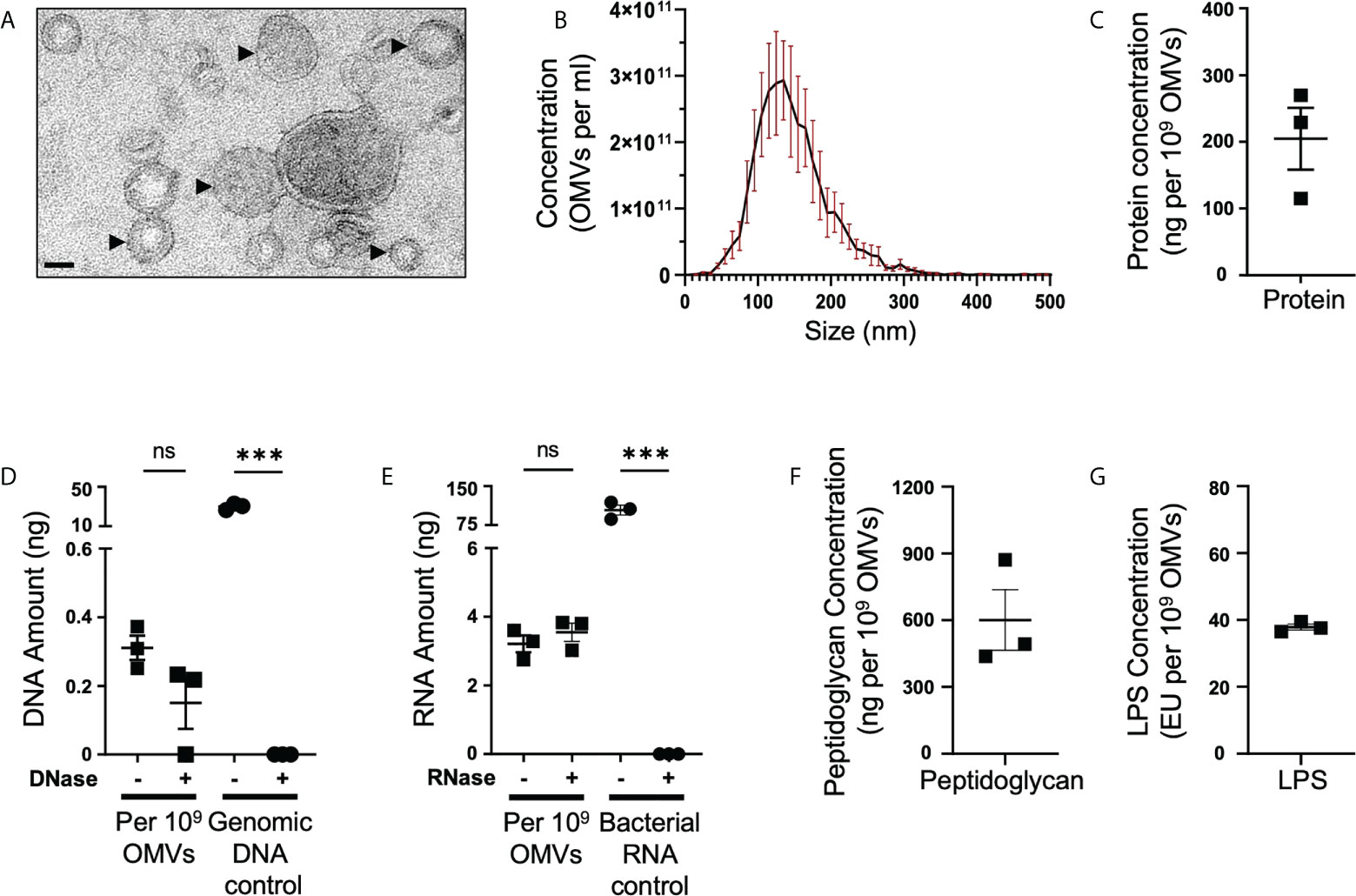
Figure 1 B fragilis OMVs are heterogenous in size and morphology, and harbour protein, DNA, RNA, peptidoglycan and LPS cargo. (A) Purified B. fragilis OMVs were visualized using transmission electron microscopy (TEM). OMVs are indicated by black arrows. Images are representative of three biological replicates (Scale bar = 100 nm). (B) The size distribution of B. fragilis OMVs was determined using ZetaView Nanoparticle Tracking Analysis. Data shows the mean (black line) ± SEM (red error bars) of three biological replicates. (C) Quantification of OMV-associated protein, per 109 B. fragilis OMVs, using Qubit fluorometric analysis. Data shows the mean ± SEM of three biological replicates. (D) Quantification of DNA associated with DNase-treated (+) or non-treated (-) B. fragilis OMVs using Qubit fluorometric analysis. B. fragilis genomic DNA (Genomic DNA control), either treated with DNase (+) or non-treated (-), was used as a control for DNase activity. Shown is the mean ± SEM of three biological replicates. ns, not significant, ***p < 0.001 (unpaired t-test). (E) Quantification of RNA associated with RNase-treated (+) or non-treated (-) B. fragilis OMVs using Qubit fluorometric analysis. B. fragilis bacterial RNA (Bacterial RNA control), either treated with RNase (+) or not-treated (-), was used as a control for RNase activity. Shown is the mean ± SEM of three biological replicates. ns, not significant, ***p < 0.001 (unpaired t-test). (F) Quantification of the peptidoglycan cargo associated with 109 B. fragilis OMVs. Shown is the mean ± SEM of three biological replicates. (G) LPS associated with 109 B. fragilis OMVs was quantified using the Pierce™ Chromogenic Endotoxin Quant kit. Data represents the mean ± SEM of three biological replicates.
B. fragilis OMVs have an altered cargo composition compared to their parent bacteria
We have previously demonstrated that BMVs produced by Gram-negative and Gram-positive bacteria contain cargo that differs from their parent bacterium, suggesting that bacteria can preferentially package or enrich biological cargo in their BMVs (6, 12). Furthermore, we showed that enrichment of biological cargo within BMVs can have a profound effect on their subsequent biological functions, when compared to their parent bacteria and to BMVs produced during different stages of bacterial growth, indicating that selective cargo packaging into BMVs regulates their functions (12). As B. fragilis OMVs contain protein, DNA, RNA, LPS and peptidoglycan, we next investigated if B. fragilis OMVs were enriched in biological cargo compared to their parent bacteria. Examination of the overall protein profiles of B. fragilis OMVs by SDS-PAGE revealed that a range of predominant protein bands, including those at approximately 125 kDa, 75 kDa, 30 kDa and 17 kDa, were enriched in B. fragilis OMVs compared to their parent bacteria (Figure 2A, arrows). Similarly, some protein bands present in B. fragilis bacteria, such as bands of approximately 60 and 45 kDa, were not equally prominent in OMVs (Figure 2A, stars), suggesting that there is selective cargo packaging of protein into B. fragilis OMVs.
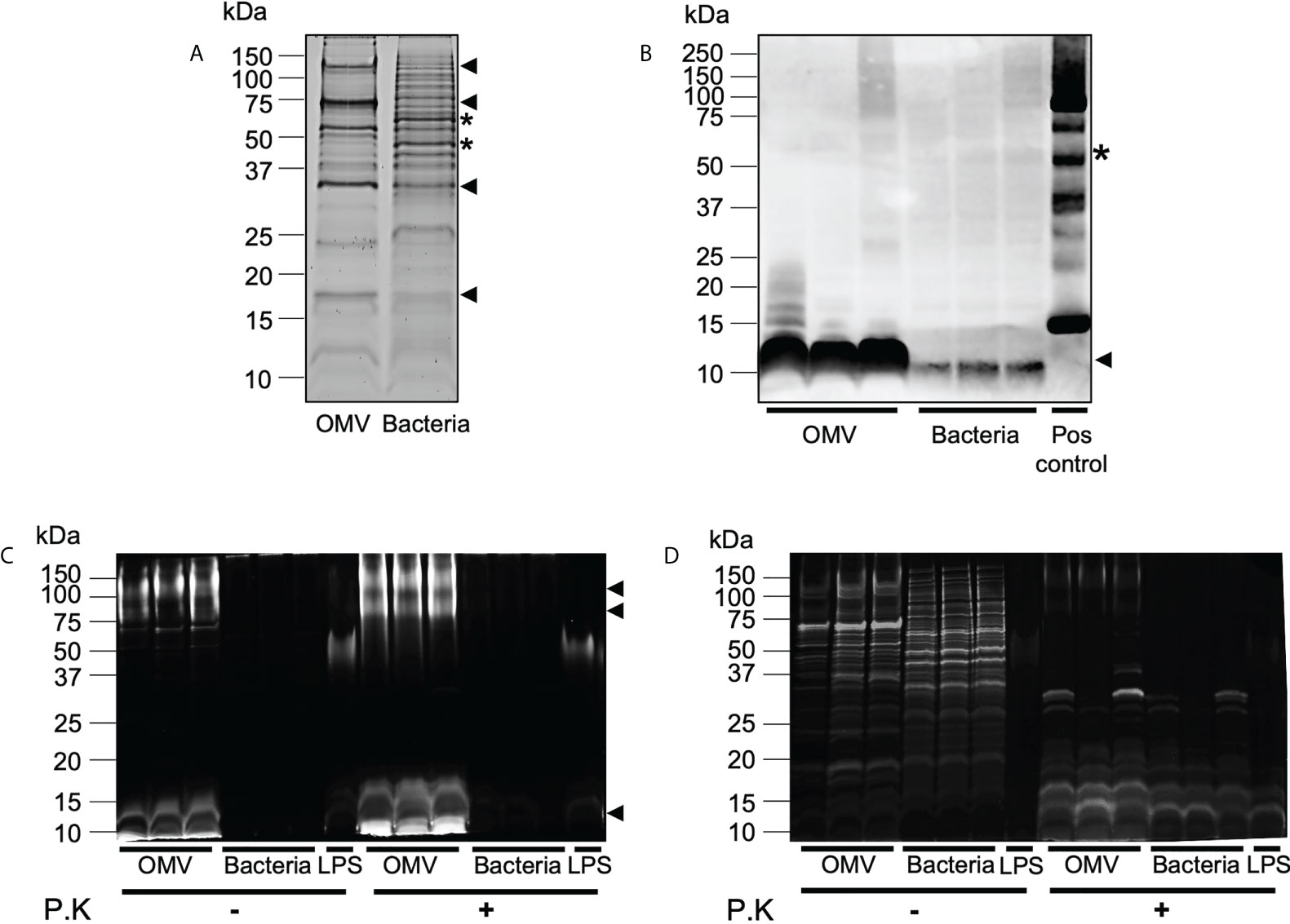
Figure 2 B. fragilis OMVs are enriched in protein, peptidoglycan and LPS cargo compared to their parent bacteria. (A) B. fragilis OMVs (10 μg) and B. fragilis bacteria (10 μg) were separated using SDS-PAGE and their protein cargo was stained using Sypro Ruby. Data is representative of three biological replicates. (B) The presence of peptidoglycan cargo associated with B. fragilis OMVs (10 μg) and B. fragilis bacteria (10 μg) was detected using Western immunoblot using an anti-peptidoglycan antibody. Peptidoglycan was used as a positive control (Pos control). Data shows three biological samples of B. fragilis OMVs and B. fragilis bacteria and is representative of three experiments. (C) B. fragilis OMVs and B. fragilis bacteria were treated with Proteinase K (P.K) (+) or not treated as controls (-), and their LPS cargo was detected by staining with ProQ Emerald 300 LPS stain. (D) ProQ Emerald-stained SDS-PAGE gels were counterstained with the protein-specific stain Sypro Ruby. Data in panel C and D represents three independent biological samples of B. fragilis OMVs and B. fragilis bacteria and is representative of three independent experiments. In all panels (A–D), black arrows represent cargo that is enriched in OMVs compared to bacteria, stars represent cargo that is less abundant in OMVs compared to bacteria.
We next detected the presence of peptidoglycan within OMVs and bacterial samples by Western immunoblot. Examination of an equivalent amount of B. fragilis OMVs and bacteria, normalized by protein amount, revealed that OMVs were enriched in peptidoglycan, evidenced by a prominent band of approximately 10 kDa, compared to their parent bacteria (Figure 2B). These differences in peptidoglycan profiles suggest differences in peptidoglycan packaging into OMVs compared to their parent bacteria, similar to what we have previously observed in MVs produced by the Gram-positive pathogen S. aureus (6).
Although it was previously reported that there were no structural differences between the lipid A LPS moiety of B. fragilis and their OMVs (15), the relative abundance of LPS cargo in B. fragilis OMVs compared to their parent bacteria remains unknown. To address this, we examined the LPS profiles of B. fragilis OMVs compared to their parent bacteria using SDS-PAGE, loaded with equivalent protein amounts, and visualized LPS using the ProQ Emerald stain (Figure 2C). LPS moieties of approximately 100 kDa, 75 kDa and 10 kDa were prominent in B. fragilis OMVs, however these moieties were less evident in B. fragilis bacterial samples, suggesting that LPS was enriched in OMVs compared to their parent bacteria (Figure 2C). The higher molecular weight staining (approximately 75 to 100 kDa) in the B. fragilis OMV samples suggests the presence of LPS. In contrast, the low molecular weight smear (10 kDa) may represent lipooligosaccharide (LOS) or lipid A components (Figure 2C), as described previously (40). Additionally, both high and low molecular weight smears were resistant to proteinase K degradation (Figure 2C) and were not visualized by Sypro Ruby staining (Figure 2D), validating that these bands were representative of LPS. Taken together, these data demonstrate that B. fragilis OMVs harbor a range of biological cargo that includes proteins, peptidoglycan, and LPS and that there are differences in the preferential packaging of these immunostimulatory cargo into OMVs compared to their parent bacteria, which may ultimately alter their ability to activate host PRRs.
B. fragilis OMVs activate TLR2 and TLR4 responses compared to their parent bacteria that can only activate TLR2
Having revealed that B. fragilis OMVs contain various biological cargo with distinct differences in their cargo composition compared to their parent bacteria (Figures 2A–D), we sought to determine if there was a difference in the ability of B. fragilis OMVs and their parent bacteria to activate host PRRs. To address this, HEK-Blue reporter cells that express either human TLR2 or TLR4, or control HEK-Blue null cells, were stimulated with an increasing dose of either B. fragilis OMVs, live B. fragilis bacteria, or heat killed B. fragilis bacteria as a control. First, we identified that B. fragilis OMVs as well as live or heat-killed bacteria were unable to induce the activation of HEK-Blue null cells (Figures 3A–D; Supplementary Figure 1A). However, both B. fragilis OMVs and their parent bacteria activated TLR2 in a dose-dependent manner (Figures 3A, B), consistent with previous reports (22, 32). OMV-mediated TLR2 activation occurred at an MOI as low as approximately 80 OMVs per cell (p < 0.01), suggesting that OMVs can readily activate TLR2 (Figure 3A). Next, we investigated whether B. fragilis and their OMVs could activate TLR4, and observed dose-dependent activation of TLR4 by B. fragilis OMVs, which occurred at MOI as low as approximately 625 OMVs per cell (p < 0.01) (Figure 3C). However, we did not observe any TLR4 activation in response to stimulation with live B. fragilis bacteria at any of the concentrations examined, suggesting that there is a difference in the TLR4 immunostimulatory abilities of B. fragilis OMVs compared to their parent bacteria (Figure 3D). Similarly, heat-killed B. fragilis bacteria also activated TLR2 but not TLR4-expressing HEK-Blue cells (Supplementary Figures 1B, C), identifying that the inability of B. fragilis to activate TLR4 was independent of bacterial viability or potential cytotoxic effects mediated by live bacteria on host cells. We also confirmed the viability of HEK-Blue null cells and TLR2 and TLR4 expressing HEK-Blue cells following stimulation with either live or heat killed B. fragilis bacteria at maximal MOI (Supplementary Figures 1D–F), validating that the lack of TLR4 activation in response to stimulation with live B. fragilis bacteria was not due to a decrease in HEK-Blue cell viability. Overall, these data demonstrate that B. fragilis OMVs activate both TLR2 and TLR4, whereas live and heat killed B. fragilis bacteria can only activate TLR2, suggesting that the immunogenic cargo of B. fragilis OMVs may enhance their ability to activate TLR4 compared to their parent bacteria.
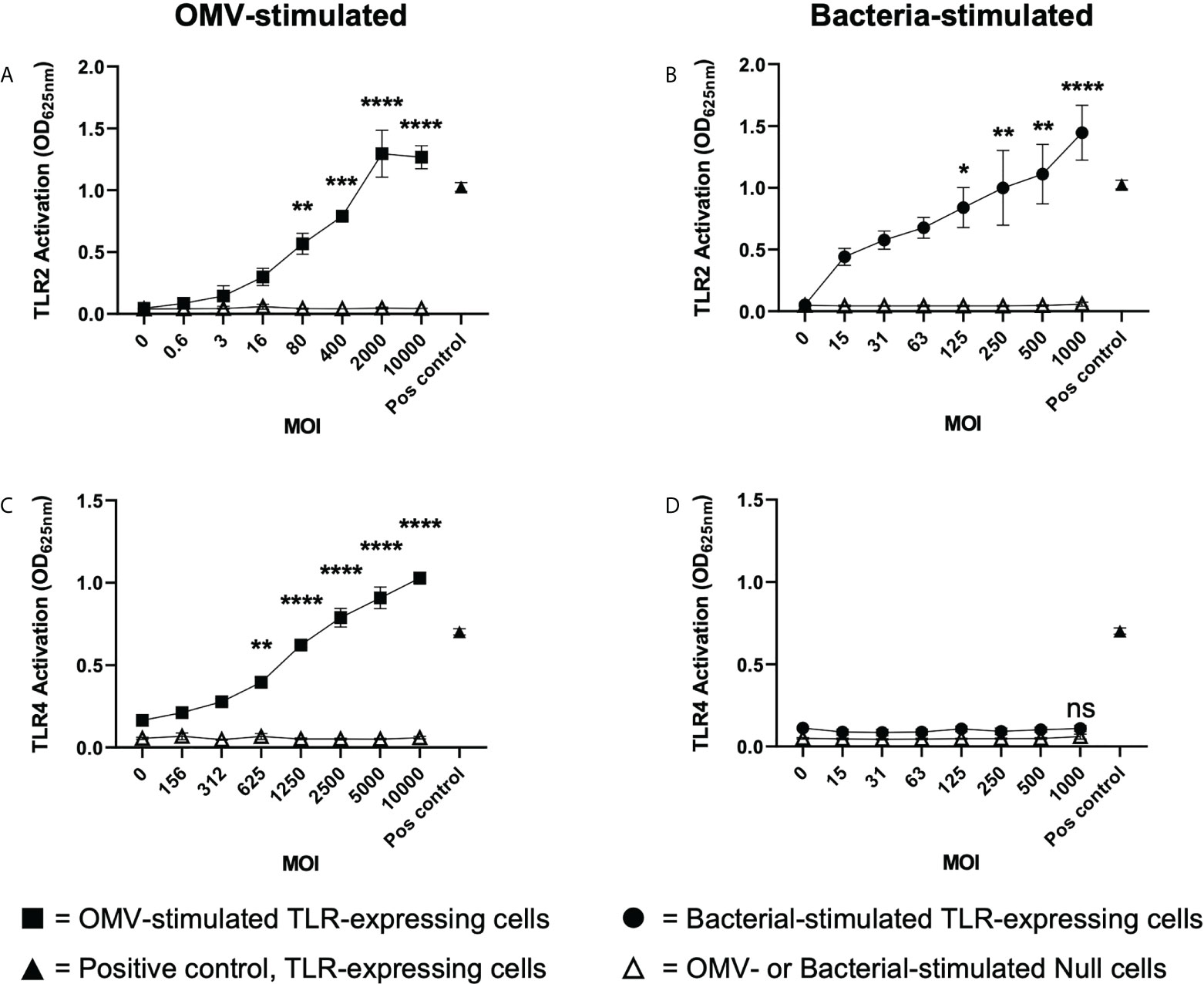
Figure 3 B. fragilis OMVs activate TLR2 and TLR4 compared to their parent bacteria which only activate TLR2. (A, B) TLR2 and (C, D) TLR4 expressing HEK-Blue cells were stimulated with either B. fragilis OMVs (A, C, squares) or B fragilis bacteria (B, D, circles) at an increasing multiplicity of infection (MOI) for 18 hours. In all panels, open triangles indicate stimulation of the HEK-Blue null cell line with either OMVs (A, C) or bacteria (B, D) in each assay, filled triangles indicate positive controls for each respective cell line. Data represents mean ± SEM of three biological replicates. ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (One-way ANOVA with Dunnett’s multiple comparisons test, compared to non-stimulated controls).
B. fragilis OMVs can enter host intestinal epithelial cells and deliver peptidoglycan and RNA intracellularly
We next investigated the ability of B. fragilis OMVs to enter and deliver their immunogenic cargo, including peptidoglycan and RNA, to host epithelial cells. First, we confirmed the ability of B. fragilis OMVs to enter human intestinal epithelial cells (Caco-2). To do this, DiI-labelled B. fragilis OMVs were incubated with Caco-2 cells for 4 hours, and the ability of OMVs to enter epithelial cells was determined by confocal microscopy (Figure 4A). Examination revealed that B. fragilis OMVs were capable of entering Caco-2 epithelial cells, and therefore able to deliver their immunogenic cargo intracellularly to host cells (Figure 4A).
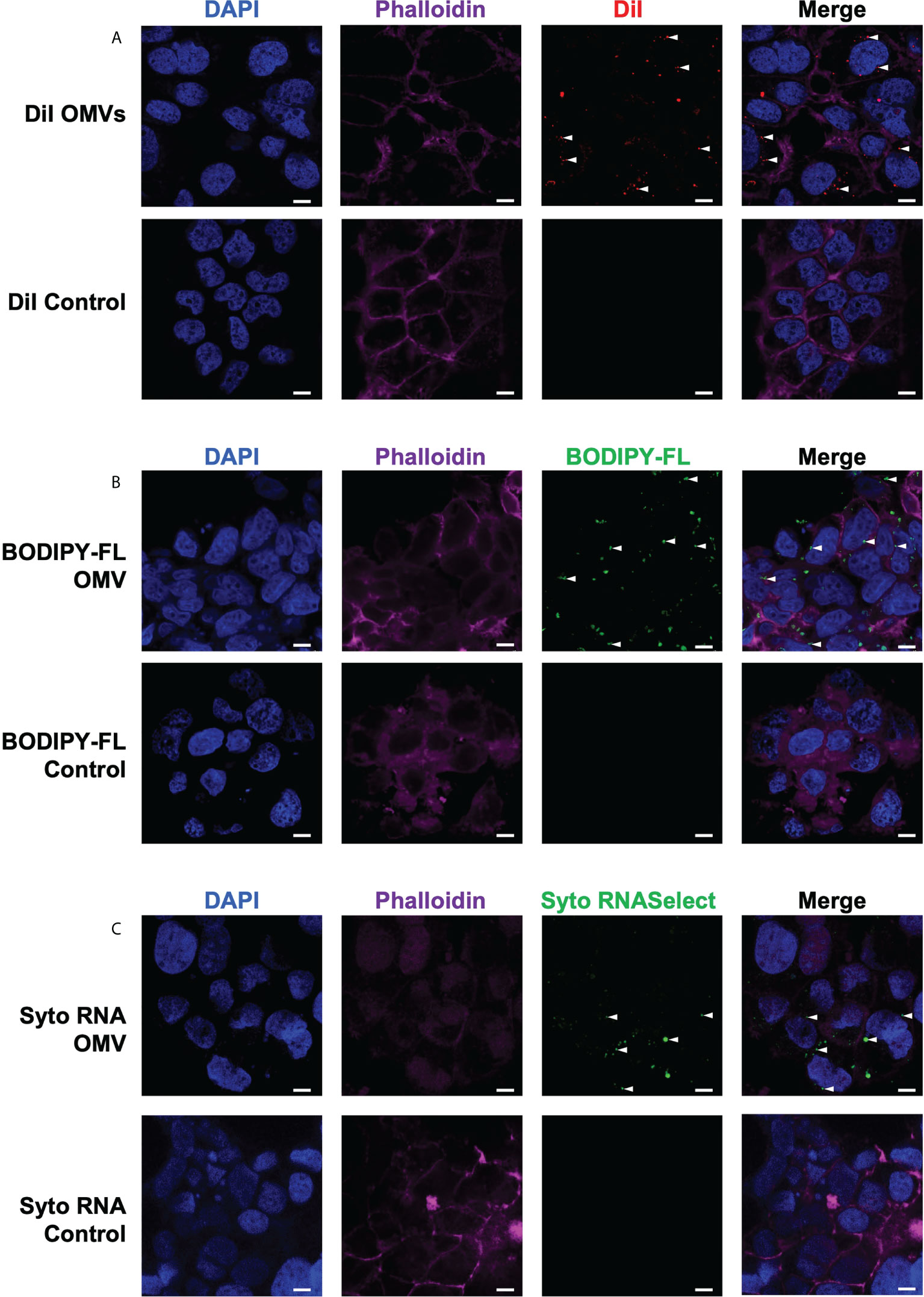
Figure 4 B fragilis OMVs enter host intestinal epithelial cells and transport their peptidoglycan and RNA cargo intracellularly. The (A) lipid (DiI; red), (B) peptidoglycan (BODIPY-FL vancomycin; green) and (C) RNA (Syto RNASelect; green) cargo associated with B. fragilis OMVs was fluorescently labelled. Fluorescently labelled B. fragilis OMVs were then incubated with Caco-2 cells for 4 hours, and OMV entry was visualized by confocal microscopy. Cell nuclei and cellular actin were visualized by staining with DAPI (blue) or Phalloidin (magenta). Intracellular B. fragilis OMVs are indicated by white arrows. DPBS containing each respective stain (Control) were incubated with Caco-2 cells to control for the non-specific uptake of each fluorescent stain by host cells. Images are representative of three biological replicates of Caco-2 cells stimulated with each type of fluorescently-labelled B. fragilis OMVs. Scale bar = 10 μm.
We have previously shown that Helicobacter pylori OMVs contain peptidoglycan, and that they can deliver their peptidoglycan cargo into host epithelial cells, resulting in the activation of the cytoplasmic host innate immune receptor NOD1 and the induction of an innate immune response (5, 42). As B. fragilis OMVs also contain peptidoglycan (Figures 1, 2), we next investigated the ability of B. fragilis OMVs to deliver their peptidoglycan cargo intracellularly to host epithelial cells. To do this, peptidoglycan associated with B. fragilis OMVs was fluorescently labelled using BODIPY-FL vancomycin, and the ability of fluorescently-labelled B. fragilis OMVs to enter Caco-2 cells was determined using confocal microscopy (Figure 4B). We found that BODIPY-FL-labelled B. fragilis OMVs entered Caco-2 epithelial cells and were therefore capable of delivering their fluorescently-labelled peptidoglycan intracellularly (Figure 4B).
Pathogen-derived BMVs can also mediate the intracellular delivery of bacterial RNA to host cells, resulting in the induction of innate immunity (6, 37, 44, 47, 48). However, the ability of commensal-derived BMVs to deliver RNA cargo into host cells remains to be elucidated. Therefore, we next examined the ability of B. fragilis OMVs to deliver their RNA cargo intracellularly to intestinal epithelial cells. To do this, RNA associated with B. fragilis OMVs was labelled using Syto RNASelect. B. fragilis OMVs containing fluorescently-labelled RNA were incubated with Caco-2 cells and examined by confocal microscopy, revealing that B. fragilis OMVs could deliver RNA intracellularly to Caco-2 cells (Figure 4C). As a control for the non-specific uptake of DiI, BODIPY-FL vancomycin or Syto RNASelect, Caco-2 cells were incubated with each respective stain resuspended in DPBS in the absence of OMVs and examined by confocal microscopy, revealing a lack of each fluorescent stain intracellularly (Figure 4, controls). These findings reveal that B. fragilis OMVs can enter host epithelial cells to deliver their cargo, that includes peptidoglycan and RNA.
B. fragilis OMVs activate NOD1 and TLR7 responses, whereas their parent bacteria cannot
We next investigated the ability of B. fragilis OMVs to activate the intracellular receptors for peptidoglycan, NOD1 and NOD2, which detect unique components of bacterial peptidoglycan. Specifically, NOD1 detects D-glutamyl-meso-diaminopimelic acid found predominantly in Gram-negative bacteria (49), whereas NOD2 detects muramyl dipeptide found in peptidoglycan of both Gram-negative and Gram-positive bacteria (50). To investigate the immune-stimulating potential of peptidoglycan delivered by B. fragilis OMVs, HEK-Blue cells expressing human NOD1 were stimulated with an increasing dose of B. fragilis OMVs or B. fragilis bacteria (Figures 5A, B). We found that stimulation of NOD1-expressing HEK-Blue cells with an MOI of 2 × 107 OMVs per cell resulted in significant activation of NOD1 compared to non-stimulated controls (p < 0.05) (Figure 5A). Based on our quantification of peptidoglycan associated with OMVs (Figure 1D), this corresponded to approximately 12 ng of peptidoglycan associated with 2 × 107 OMVs resulting in the activation of NOD1 (Figure 5A). In comparison, we observed that NOD1 was not activated by B. fragilis bacteria at any MOI examined in this study, indicating that there are differences in the ability of B. fragilis and their OMVs to activate NOD1 (Figure 5B). We next characterized the ability of OMVs to activate NOD2, which has been previously reported to have a role in the detection of B. fragilis OMVs by bone marrow-derived dendritic cells (25, 50). However, we did not observe any significant activation of NOD2-expressing HEK-Blue cells by either B. fragilis OMVs or B. fragilis bacteria at any of MOI examined in this study (Figures 5C, D). Furthermore, heat-killed bacteria did not activate either NOD1 or NOD2, and the inability of B. fragilis bacteria to activate NOD1 signaling compared to OMVs was not due to a reduction in host cell viability (Supplementary Figures 2A–C).
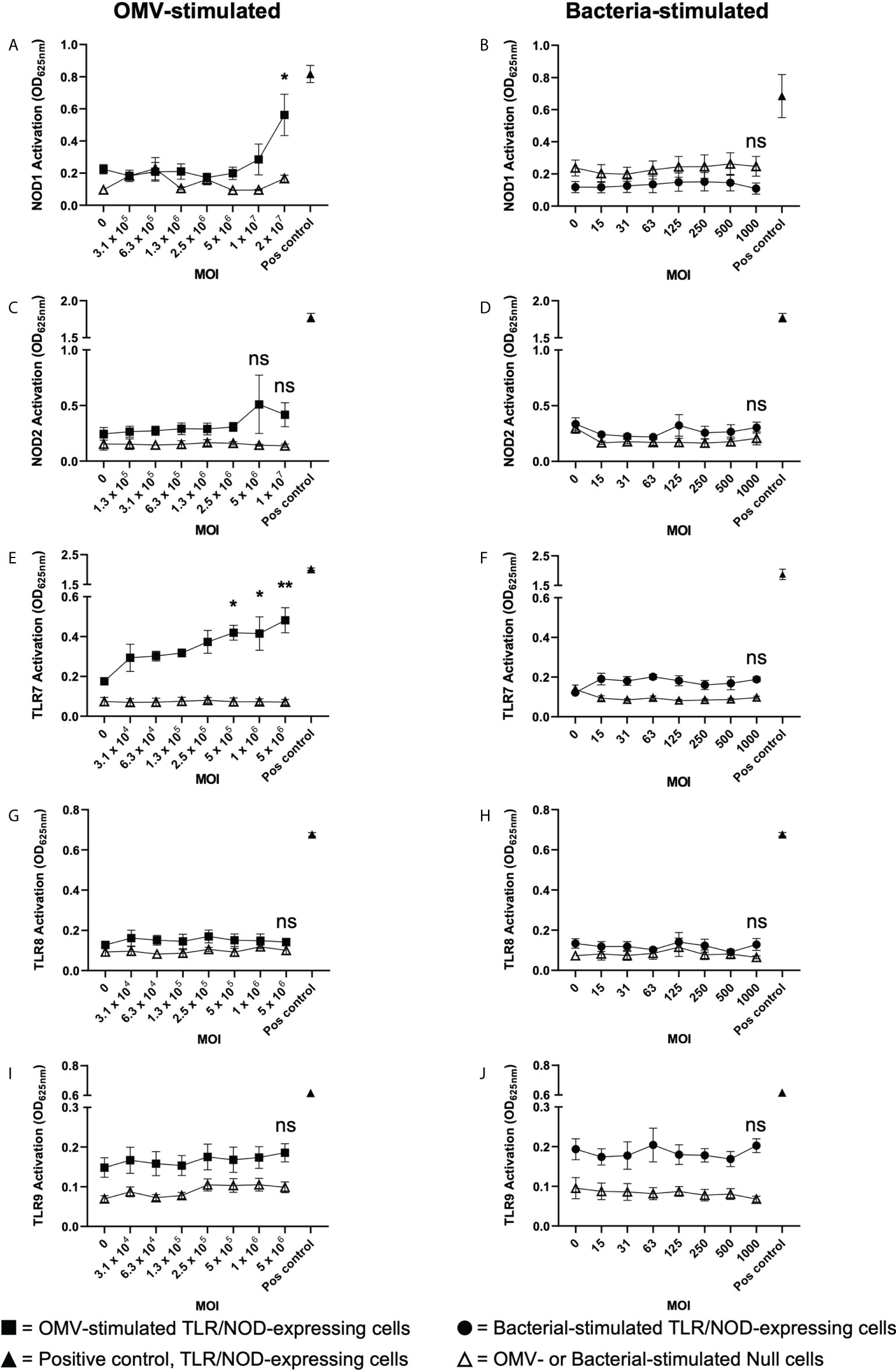
Figure 5 B fragilis OMVs activate NOD1 and TLR7 whereas B fragilis bacteria cannot. (A, B) NOD1, (C, D) NOD2, (E, F) TLR7, (G, H) TLR8 and (I, J) TLR9 expressing HEK-Blue cells were stimulated with an increasing MOI of either B. fragilis OMVs (A, C, E, G, I, squares) or B. fragilis bacteria (B, D, F, H, J, circles) for 18 hours. Open triangles indicate stimulation of the HEK-Blue null cell line with either B. fragilis OMVs or B. fragilis bacteria as a negative control in each assay. Filled triangles indicate positive controls for each respective cell line. Data represents mean ± SEM of three biological replicates. ns=not significant, *p < 0.05, **p < 0.01. (One-way ANOVA with Dunnett’s multiple comparisons test, compared to non-stimulated controls).
Having determined that B. fragilis OMVs harbor both DNA and RNA cargo (Figures 1D, E) and that RNA associated with B. fragilis OMVs can enter epithelial cells (Figure 4C), we next investigated their ability to activate the RNA receptors TLR7 and TLR8, and TLR9 that detects bacterial DNA. To address this, TLR7, TLR8 and TLR9 expressing HEK-Blue cells were stimulated with an increasing dose of B. fragilis OMVs or their parent bacteria (Figures 5E–J). Stimulation of TLR7 expressing HEK-Blue cells with an increasing dose of B. fragilis OMVs resulted in significant activation of TLR7 compared to non-stimulated controls, with a minimum MOI of approximately 5 × 105 OMVs per cell being required to induce TLR7 activation (p < 0.05) (Figure 5E). In contrast, live and heat killed B. fragilis bacteria could not activate TLR7 at any concentration examined in this study (Figure 5F, Supplementary Figure 3A). Furthermore, the inability of live or heat killed B. fragilis to activate TLR7 compared to OMVs was not due to impairing the viability of HEK-Blue cells expressing TLR7 (Supplementary Figure 3B). We also found that neither B. fragilis OMVs, nor live or heat-killed B. fragilis bacteria were able to induce the activation of TLR8 (Figures 5G, H, Supplementary Figure 3C), suggesting that B. fragilis bacteria were unable to activate either TLR7 or TLR8, and that RNA associated with B. fragilis OMVs preferentially activated TLR7 compared to TLR8 (Figure 5E). This is in contrast to what we have previously observed using S. aureus MVs, where an equivalent amount of RNA-containing S. aureus MVs could activate both TLR7 and TLR8 (6). Moreover, we found that live and heat killed B. fragilis bacteria and their OMVs could not activate TLR9 responses at all MOIs examined (Figures 5I, J, Supplementary Figure 3D). The inability of B. fragilis OMVs to activate TLR9 responses may be attributed to them having approximately ten-fold less DNA (0.311ng per 109 OMVs) than RNA (3.21ng per 109 OMVs) content (Figures 1D, E) and therefore, the amount of DNA delivered by B. fragilis OMVs may be insufficient to induce TLR9 activation. Moreover, NF-κB activity was not observed in the negative control HEK-Blue null cell line in response to stimulation with B. fragilis OMVs or bacteria, revealing that the delivery of bacterial cargo by B. fragilis OMVs is essential for their ability to activate the intracellular receptors NOD1 and TLR7 (Figure 5).
Taken together, our data identify the ability of the intestinal commensal B. fragilis to package protein, DNA, RNA, peptidoglycan and LPS into their OMVs, and that there is enrichment of immunogenic cargo in B. fragilis OMVs. Furthermore, we show the ability of enriched B. fragilis OMV-associated cargo to be delivered intracellularly to host cells, which ultimately enables B. fragilis OMVs to preferentially activate a broader range of innate immune receptors compared to their parent bacteria. Moreover, these findings identify novel mechanisms of selective immune activation mediated by B. fragilis OMVs at the host epithelial cells surface via preferential activation of TLR4, TLR7 and NOD1.
Discussion
The immunostimulatory functions of BMVs depend upon the specific cargo they contain and their ability to deliver this cargo to host cells. BMVs produced by both pathogens and commensals can package various biological cargo including nucleic acids, proteins, LPS and peptidoglycan. Furthermore, pathogen-derived BMVs can be enriched in immunostimulatory cargo, enabling them to activate host PRRs and drive immune responses in the host. However, the enrichment of immunostimulatory cargo in commensal-derived BMVs compared to their parent bacteria, and their subsequent ability to deliver this cargo and activate innate immune receptors is not equally well characterized. The findings of this study reveal that OMVs produced by the commensal B. fragilis contain protein, nucleic acids, LPS and peptidoglycan and are enriched in LPS, peptidoglycan and proteins compared to their parent bacterium (Figures 1, 2). Additionally, we show that B. fragilis OMVs can enter intestinal epithelial cells to deliver their RNA and peptidoglycan cargo intracellularly (Figure 4). Moreover, the enrichment of peptidoglycan, LPS and protein cargo into B. fragilis OMVs, in addition to their ability to deliver their cargo into host epithelial cells, enables them to activate a more diverse range of PRRs which includes TLR4, TLR7 and NOD1 compared to their parent bacteria (Figures 3, 5). Collectively, our findings identify the enrichment of select cargo into B. fragilis OMVs that subsequently results in their ability to activate a broader range of host innate immune receptors compared to their parent bacteria, revealing that B. fragilis OMVs may function to increase the potential for commensal-host crosstalk at the intestinal epithelial barrier.
In this study, we investigated the type and quantity of biological cargo associated with B. fragilis OMVs and determined their ability to activate host innate immune receptors compared to their parent bacteria. Our data revealed that B. fragilis OMVs contain a wide range of biological cargo consisting of protein, LPS, peptidoglycan, DNA and RNA (Figure 1). Interestingly, B. fragilis OMVs contained approximately ten-fold more RNA than DNA (Figures 1D, E), which is consistent with previous studies identifying that MVs derived from the Gram-positive commensals Lactobacillus reuteri (20) and Lactobacillus casei (18) also packaged significantly more RNA than DNA. While the RNA associated with pathogen-derived BMVs is becoming increasingly recognized as being able to activate innate immune receptors and to modulate cellular functions when delivered into host cells (37, 51), knowledge regarding the immunostimulatory and immunomodulatory abilities of RNA delivered by commensal-derived BMVs is limited, highlighting that future research endeavors should focus on broadening our understanding of their functions.
Our data show that B. fragilis OMVs were enriched with protein cargo compared to their parent bacterium (Figure 2A). In agreement with our findings, a previous study examining the proteome of B. fragilis OMVs found that they were enriched in acidic proteases and sugar-hydrolysing glycosidases, which facilitated the catabolism and acquisition of environmental nutrients and were thought to ameliorate the establishment and composition of the gut microbiota (15). Moreover, while selective protein packaging into pathogen-derived BMVs has been shown to promote pathogen colonisation or survival (52, 53), proteins enriched in BMVs produced by various pathogens also have important roles in facilitating bacterial pathogenesis and promoting the development of inflammatory host immune responses (12, 52–55), further supporting the notion that bacteria can regulate the proteome of their OMVs to modulate their functions.
We also observed the enrichment of LPS into B. fragilis OMVs compared to their parent bacteria (Figure 2C). Although ProQ Emerald stain used in this study cannot discriminate between different LPS isoforms at the molecular level, the LPS enriched in B. fragilis OMVs was characteristic of both larger LPS species, as well as smaller LPS, LOS or lipid A species (Figure 2C). Consequently, we observed that B. fragilis OMVs, but not their parent bacteria, induced dose-dependent activation of TLR4, the host immune receptor responsible for the detection of LPS. Therefore, our findings suggest that the enrichment of LPS into B. fragilis OMVs enhances their ability to activate TLR4 compared to their parent bacteria. In agreement with our findings, a previous study showed that long-chain LPS moieties could be enriched in Porphyromonas gingivalis OMVs compared to their parent bacteria (7). Moreover, it was also reported that OMVs produced by wild-type Neisseria meningitidis strains induced stronger TLR4 responses compared to OMVs produced by LPS-depleted strains, demonstrating that OMVs containing more LPS were more readily able to induce the activation of TLR4-mediated immune responses (56). Collectively, these studies support our findings that LPS can be enriched in B. fragilis OMVs which may contribute to their enhanced capacity to mediate TLR4 signaling compared to their parent bacteria.
In addition to examining the ability of B. fragilis OMVs to preferentially activate host cell surface expressed TLRs, we also investigated the ability of B. fragilis OMVs to enter epithelial cells, rendering their cargo accessible to intracellular PRRs, and their subsequent ability to activate these cytoplasmic PRRs. Using previously validated methods to label BMVs and their associated peptidoglycan and RNA cargo, we demonstrated that B. fragilis OMVs entered intestinal epithelial cells and delivered their fluorescently-labelled peptidoglycan and RNA cargo intracellularly (Figure 4). Considering that B. fragilis bacteria are non-invasive and do not readily secrete immunostimulatory effector molecules via a known secretion system (57, 58), OMVs are emerging as a novel secretion mechanism utilized by B. fragilis to deliver immunostimulatory cargo into the cytoplasm of host epithelial cells. Consistent with our findings, a previous study reported the ability of OMVs produced by the closely related Bacteroides thetaiotaomicron to enter intestinal epithelial cells using both Caco-2 cells and small intestinal organoid models of OMV entry (59). The entry of B. fragilis OMVs into intestinal epithelial cells has not been well described, however previous studies have reported the uptake of B. fragilis OMVs by host dendritic cells ex vivo, which was thought to facilitate the activation of the cytoplasmic NOD2 immune receptor (22, 25). Additionally, OMVs produced by commensal and probiotic strains of Escherichia coli were also found to enter intestinal epithelial cells, whereby fluorescent labelling of their peptidoglycan cargo demonstrated the intracellular delivery of peptidoglycan and the subsequent activation of NOD1-dependent immune responses (21). In contrast to the limited studies reporting the delivery of commensal-derived peptidoglycan into host cells via BMVs, there are numerous studies reporting the ability of BMVs produced by pathogens including H. pylori, Vibrio cholerae and Aggregatibacter actinomycetemcomitans to contain peptidoglycan and induce the activation of NOD1- or NOD2-dependent immune responses upon their entry into host epithelial cells (5, 42, 60–62). The BMV-mediated delivery of bacterial RNA to host cells has been also observed for pathogen-derived BMVs (37, 44, 48), resulting in the activation of intracellular detectors of microbial RNA, TLR7 and TLR8 (6, 40), but this has not been characterized in the context of commensal-derived BMVs. Therefore, our findings identify that B. fragilis can deliver bacterial RNA to host epithelial cells via OMVs and suggests the possibility that other commensal organisms may also be capable of potentially delivering bacterial-derived RNA into host cells via this mechanism, and this forms the basis of future studies. Collectively, these findings demonstrate that B. fragilis OMVs enter intestinal epithelial cells to deliver peptidoglycan and RNA cargo intracellularly, resulting in the activation of their respective cytoplasmic PRRs.
Having shown that B. fragilis OMVs are enriched in peptidoglycan (Figure 2), as well as their ability to deliver this cargo intracellularly (Figure 4), we demonstrated that B. fragilis OMVs can activate NOD1, the intracellular receptor for Gram-negative peptidoglycan (Figure 5A). However, in contrast to a previous study reporting that B. fragilis OMVs activated NOD2 following phagocytosis by dendritic cells (25), B. fragilis OMVs did not activate NOD2 in our epithelial HEK-Blue cell model of PRR activation. This may be explained by the differences in OMV entry between phagocytic dendritic cells compared to non-phagocytic epithelial cells (63), in addition to the increased expression of NOD2 by cells of myeloid origin (64). Furthermore, B. fragilis OMVs activated the intracellular RNA receptor, TLR7, but did not activate TLR8 which can also detect microbial RNA. Whilst human TLR7 and TLR8 are both responsible for the detection of single-stranded RNA compounds, and can be activated by RNA delivered by S. aureus MVs into host epithelial cells (6), evidence suggests that TLR7 may have greater ligand sensitivity than TLR8 (65), thus providing a potential explanation as to why we did not see TLR8 activation by B. fragilis OMVs in our study. In addition, we did not observe TLR9 activation in response to stimulation with B. fragilis OMVs at any MOI examined in this study, which suggests that the amount of DNA delivered by B. fragilis OMVs may not have been sufficient to mediate TLR9 activation (35). We have previously shown that BMVs produced by different strains of a bacterial species vary in their amount of DNA and RNA cargo and therefore differ in their ability to activate their respective TLRs (35). Therefore, although we did not see activation of TLR8 and TLR9 by B. fragilis OMVs in our study, we cannot exclude the possibility that stimulation of these cells with an increased amount of OMVs, or with OMVs produced by a different B. fragilis strain that harbor a greater concentration of DNA and RNA, may activate these TLRs.
Most importantly, the findings of our study revealed that B. fragilis bacteria did not induce the activation of any intracellular receptor tested in this study, which may be due to the bacterium being unable to directly deliver their biological cargo intracellularly. Therefore, the ability of the commensal B. fragilis to produce OMVs that mediate the activation of intracellular receptors NOD1 and TLR7 enables B. fragilis to activate a broader range of immune receptors at the epithelium. Collectively these findings suggest that OMV secretion by B. fragilis is a novel mechanism used by this bacterium to increase their potential to mediate immune crosstalk at the intestinal epithelium.
Overall, our findings identify that B. fragilis OMVs are enriched in immunostimulatory cargo and can transport this cargo directly into host epithelial cells to preferentially activate host PRRs compared to their parent bacteria. Furthermore, whilst previous studies have recognized the immunomodulatory properties of commensal bacteria or their BMVs, this study compares and provides evidence of key differences in the abilities of B. fragilis OMVs and B. fragilis bacteria to activate host TLRs and NODs. Therefore, OMVs emerge as a novel secretion mechanism used by B. fragilis and potentially other non-invasive commensal bacteria to mediate TLR and NOD activation in epithelial cells. In this way, commensal-derived BMVs may directly contribute to immune activation or modulation at the intestinal mucosal surface. Further research elucidating the composition and ability of other commensal and microbiota-derived BMVs to selectively deliver immunogenic cargo, and to activate and signal via host innate immune receptors is needed to improve our understanding of their contribution to maintaining homeostasis in the gastrointestinal niche.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
All authors performed the research. WG, EJ, AFH and MK-L wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by an Australian Research Council Discovery Grant (MK-L and AFH, Grant number DP190101655) and by the LIMS Bioimaging Platform (La Trobe University, Australia). MK-L is supported by a veski Inspiring Women Fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.970725/full#supplementary-material
Supplementary Figure 1 | Live and heat-killed B. fragilis bacteria activate TLR2, but not TLR4, without reducing host cell viability. HEK-Blue (A) null cells as well as (B) TLR2 and (C) TLR4 expressing HEK-Blue cells were stimulated with an increasing dose of either live (closed circles) or heat-killed (open circles) B. fragilis bacteria for 18 hours. Triangles represent positive controls for each respective cell line. Data represents mean ± SEM of three biological replicates. ns = not significant, *p < 0.05 (Unpaired t-test). (D) HEK-Blue null cells as well as (E) TLR2 and (F) TLR4 expressing HEK-Blue cells were stimulated with either live or heat-killed B. fragilis bacteria (MOI 1,000) for 18 hours, and cell viability was measured using MTT Assay. Non-stimulated cells (NS) were used as a control. Data represents mean ± SEM of at least three biological replicates. ns = not significant (One-way ANOVA with Tukey’s multiple comparisons test).
Supplementary Figure 2 | Live and heat-killed B. fragilis bacteria do not activate NOD1 or NOD2, and do not decrease the viability of NOD1-expressing cells. HEK-Blue cells expressing (A) NOD1 or (B) NOD2 were stimulated with an increasing dose of either live (closed circles) or heat-killed (open circles) B. fragilis bacteria for 18 hours. Triangles represent positive controls for each respective cell line. Data represents mean ± SEM of three biological replicates. ns = not significant (Unpaired t-test). (C) The viability of NOD1-expressing HEK-Blue cells following 18 hours stimulation with either live or heat-killed B. fragilis bacteria (MOI 1,000) was measured by MTT Assay. Non-stimulated cells (NS) were used as a control. Data represents mean ± SEM of four biological replicates. ns = not significant (One-way ANOVA with Tukey’s multiple comparisons test).
Supplementary Figure 3 | Live and heat-killed B. fragilis bacteria do not activate TLR7, TLR8 or TLR9, and do not decrease the viability of TLR7-expressing cells. HEK-Blue cells expressing (A) TLR7, (C) TLR8 or (D) TLR9 were stimulated with an increasing dose of either live (closed circles) or heat-killed (open circles) B. fragilis bacteria for 18 hours. Triangles represent positive controls for each respective cell line. Data represents mean ± SEM of three biological replicates. ns = not significant, (Unpaired t-test). (B) The viability of TLR7-expressing HEK-Blue cells following 18 hours stimulation with either live or heat-killed B. fragilis bacteria (MOI 1,000) was measured by MTT Assay. Non-stimulated cells (NS) were used as a control. Data represents mean ± SEM of four biological replicates. ns = not significant (One-way ANOVA with Tukey’s multiple comparisons test).
References
1. Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem (2000) 275(17):12489–96. doi: 10.1074/jbc.275.17.12489
2. Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol (1995) 177(14):3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
3. Sjöström AE, Sandblad L, Uhlin BE, Wai SN. Membrane vesicle-mediated release of bacterial RNA. Sci Rep (2015) 5:15329. doi: 10.1038/srep15329
4. Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, et al. A novel mechanism for the biogenesis of outer membrane vesicles in gram-negative bacteria. Nat Commun (2016) 7:10515. doi: 10.1038/ncomms10515
5. Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol (2010) 12(3):372–85. doi: 10.1111/j.1462-5822.2009.01404.x
6. Bitto NJ, Cheng L, Johnston EL, Pathirana R, Phan TK, Poon IKH, et al. Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy. J Extracell Vesicles (2021) 10(6):e12080. doi: 10.1002/jev2.12080
7. Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem (2011) 286(2):1269–76. doi: 10.1074/jbc.M110.185744
8. Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol (2015) 15(6):375–87. doi: 10.1038/nri3837
9. Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog (2002) 32(1):1–13. doi: 10.1006/mpat.2001.0474
10. Bitto NJ, Chapman R, Pidot S, Costin A, Lo C, Choi J, et al. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci Rep (2017) 7(1):7072. doi: 10.1038/s41598-017-07288-4
11. Resch U, Tsatsaronis JA, Le Rhun A, Stübiger G, Rohde M, Kasvandik S, et al. A two-component regulatory system impacts extracellular membrane-derived vesicle production in group a Streptococcus. mBio (2016) 7(6):e00207-16. doi: 10.1128/mBio.00207-16
12. Zavan L, Bitto NJ, Johnston EL, Greening DW, Kaparakis-Liaskos M. Helicobacter pylori growth stage determines the size, protein composition, and preferential cargo packaging of outer membrane vesicles. Proteomics (2019) 19(1-2):e1800209. doi: 10.1002/pmic.201800209
13. Díaz-Garrido N, Badia J, Baldomà L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J Extracell Vesicles (2021) 10(13):e12161. doi: 10.1002/jev2.12161
14. Aguilera L, Toloza L, Giménez R, Odena A, Oliveira E, Aguilar J, et al. Proteomic analysis of outer membrane vesicles from the probiotic strain. Escherichia coli Nissle 1917. Proteomics (2014) 14(2-3):222–9. doi: 10.1002/pmic.201300328
15. Elhenawy W, Debelyy MO, Feldman MF. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio (2014) 5(2):e00909-14. doi: 10.1128/mBio.00909-14
16. Zakharzhevskaya NB, Vanyushkina AA, Altukhov IA, Shavarda AL, Butenko IO, Rakitina DV, et al. Outer membrane vesicles secreted by pathogenic and nonpathogenic Bacteroides fragilis represent different metabolic activities. Sci Rep (2017) 7(1):5008. doi: 10.1038/s41598-017-05264-6
17. Hong J, Dauros-Singorenko P, Whitcombe A, Payne L, Blenkiron C, Phillips A, et al. Analysis of the Escherichia coli extracellular vesicle proteome identifies markers of purity and culture conditions. J Extracell Vesicles (2019) 8(1):1632099. doi: 10.1080/20013078.2019.1632099
18. Domínguez Rubio AP, Martínez JH, Martínez Casillas DC, Coluccio Leskow F, Piuri M, Pérez OE. Lactobacillus casei BL23 produces microvesicles carrying proteins that have been associated with its probiotic effect. Front Microbiol (2017) 8:1783. doi: 10.3389/fmicb.2017.01783
19. Li M, Lee K, Hsu M, Nau G, Mylonakis E, Ramratnam B. Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol (2017) 17(1):66. doi: 10.1186/s12866-017-0977-7
20. Hu R, Lin H, Wang M, Zhao Y, Liu H, Min Y, et al. Lactobacillus reuteri-derived extracellular vesicles maintain intestinal immune homeostasis against lipopolysaccharide-induced inflammatory responses in broilers. J Anim Sci Biotechnol (2021) 12(1):25. doi: 10.1186/s40104-020-00532-4
21. Cañas MA, Fábrega MJ, Giménez R, Badia J, Baldomà L. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front Microbiol (2018) 9:498. doi: 10.3389/fmicb.2018.00498
22. Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe (2012) 12(4):509–20. doi: 10.1016/j.chom.2012.08.004
23. Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One (2013) 8(10):e76520. doi: 10.1371/journal.pone.0076520
24. Al-Nedawi K, Mian MF, Hossain N, Karimi K, Mao YK, Forsythe P, et al. Gut commensal microvesicles reproduce parent bacterial signals to host immune and enteric nervous systems. FASEB J (2015) 29(2):684–95. doi: 10.1096/fj.14-259721
25. Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science (2016) 352(6289):1116–20. doi: 10.1126/science.aad9948
26. Seo MK, Park EJ, Ko SY, Choi EW, Kim S. Therapeutic effects of kefir grain Lactobacillus-derived extracellular vesicles in mice with 2,4,6-trinitrobenzene sulfonic acid-induced inflammatory bowel disease. J Dairy Sci (2018) 101(10):8662–71. doi: 10.3168/jds.2018-15014
27. Maerz JK, Steimle A, Lange A, Bender A, Fehrenbacher B, Frick JS. Outer membrane vesicles blebbing contributes to B. vulgatus mpk-mediated immune response silencing. Gut Microbes (2018) 9(1):1–12. doi: 10.1080/19490976.2017.1344810
28. Diaz-Garrido N, Fábrega M-J, Vera R, Giménez R, Badia J, Baldomà L. Membrane vesicles from the probiotic nissle 1917 and gut resident Escherichia coli strains distinctly modulate human dendritic cells and subsequent T cell responses. J Funct Foods (2019) 61:103495. doi: 10.1016/j.jff.2019.103495
29. Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest (2014) 124(10):4166–72. doi: 10.1172/jci72334
30. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA (2010) 107(27):12204–9. doi: 10.1073/pnas.0909122107
31. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature (2008) 453(7195):620–5. doi: 10.1038/nature07008
32. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science (2011) 332(6032):974–7. doi: 10.1126/science.1206095
33. Stentz R, Horn N, Cross K, Salt L, Brearley C, Livermore DM, et al. Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β-lactam antibiotics. J Antimicrob Chemother (2015) 70(3):701–9. doi: 10.1093/jac/dku466
34. Turner L, Bitto NJ, Steer DL, Lo C, D'Costa K, Ramm G, et al. Helicobacter pylori outer membrane vesicle size determines their mechanisms of host cell entry and protein content. Front Immunol (2018) 9:1466. doi: 10.3389/fimmu.2018.01466
35. Bitto NJ, Zavan L, Johnston EL, Stinear TP, Hill AF, Kaparakis-Liaskos M. Considerations for the analysis of bacterial membrane vesicles: Methods of vesicle production and quantification can influence biological and experimental outcomes. Microbiol Spectr. (2021) 9(3):e0127321. doi: 10.1128/Spectrum.01273-21
36. Bitto NJ, Kaparakis-Liaskos M. Methods of bacterial membrane vesicle production, purification, quantification, and examination of their immunogenic functions. Methods Mol Biol (2022) 2523:43–61. doi: 10.1007/978-1-0716-2449-4_4
37. Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, et al. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog (2016) 12(6):e1005672. doi: 10.1371/journal.ppat.1005672
38. Hadzija O. A simple method for the quantitative determination of muramic acid. Anal Biochem (1974) 60(2):512–7. doi: 10.1016/0003-2697(74)90261-9
39. Cañas MA, Giménez R, Fábrega MJ, Toloza L, Baldomà L, Badia J. Outer membrane vesicles from the probiotic Escherichia coli Nissle 1917 and the commensal ECOR12 enter intestinal epithelial cells via clathrin-dependent endocytosis and elicit differential effects on DNA damage. PLoS One (2016) 11(8):e0160374. doi: 10.1371/journal.pone.0160374
40. Cecil JD, O'Brien-Simpson NM, Lenzo JC, Holden JA, Chen YY, Singleton W, et al. Differential responses of pattern recognition receptors to outer membrane vesicles of three periodontal pathogens. PLoS One (2016) 11(4):e0151967. doi: 10.1371/journal.pone.0151967
41. Alhawi M, Stewart J, Erridge C, Patrick S, Poxton IR. Bacteroides fragilis signals through toll-like receptor (TLR) 2 and not through TLR4. J Med Microbiol (2009) 58(Pt 8):1015–22. doi: 10.1099/jmm.0.009936-0
42. Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, et al. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe (2014) 15(5):623–35. doi: 10.1016/j.chom.2014.04.001
43. Choi JW, Kim SC, Hong SH, Lee HJ. Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J Dent Res (2017) 96(4):458–66. doi: 10.1177/0022034516685071
44. Rodriguez BV, Kuehn MJ. Staphylococcus aureus secretes immunomodulatory RNA and DNA via membrane vesicles. Sci Rep (2020) 10(1):18293. doi: 10.1038/s41598-020-75108-3
45. Nicola AM, Frases S, Casadevall A. Lipophilic dye staining of Cryptococcus neoformans extracellular vesicles and capsule. Eukaryot Cell (2009) 8(9):1373–80. doi: 10.1128/ec.00044-09
46. Nakayama-Imaohji H, Hirota K, Yamasaki H, Yoneda S, Nariya H, Suzuki M, et al. DNA Inversion regulates outer membrane vesicle production in. Bacteroides fragilis. PLoS One (2016) 11(2):e0148887. doi: 10.1371/journal.pone.0148887
47. Ha JY, Choi SY, Lee JH, Hong SH, Lee HJ. Delivery of periodontopathogenic extracellular vesicles to brain monocytes and microglial IL-6 promotion by RNA cargo. Front Mol Biosci (2020) 7:596366. doi: 10.3389/fmolb.2020.596366
48. Zhang H, Zhang Y, Song Z, Li R, Ruan H, Liu Q, et al. sncRNAs packaged by Helicobacter pylori outer membrane vesicles attenuate IL-8 secretion in human cells. Int J Med Microbiol (2020) 310(1):151356. doi: 10.1016/j.ijmm.2019.151356
49. Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science (2003) 300(5625):1584–7. doi: 10.1126/science.1084677
50. Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem (2003) 278(11):8869–72. doi: 10.1074/jbc.C200651200
51. Dauros-Singorenko P, Hong J, Swift S, Phillips A, Blenkiron C. Effect of the extracellular vesicle RNA cargo from uropathogenic Escherichia coli on bladder cells. Front Mol Biosci (2020) 7:580913. doi: 10.3389/fmolb.2020.580913
52. Jang KS, Sweredoski MJ, Graham RL, Hess S, Clemons WM Jr. Comprehensive proteomic profiling of outer membrane vesicles from Campylobacter jejuni. J Proteomics (2014) 98:90–8. doi: 10.1016/j.jprot.2013.12.014
53. Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics (2007) 7(17):3143–53. doi: 10.1002/pmic.200700196
54. Deo P, Chow SH, Hay ID, Kleifeld O, Costin A, Elgass KD, et al. Outer membrane vesicles from Neisseria gonorrhoeae target PorB to mitochondria and induce apoptosis. PLoS Pathog (2018) 14(3):e1006945. doi: 10.1371/journal.ppat.1006945
55. Kwon SO, Gho YS, Lee JC, Kim SI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett (2009) 297(2):150–6. doi: 10.1111/j.1574-6968.2009.01669.x
56. Zariri A, Beskers J, van de Waterbeemd B, Hamstra HJ, Bindels TH, van Riet E, et al. Meningococcal outer membrane vesicle composition-dependent activation of the innate immune response. Infect Immun (2016) 84(10):3024–33. doi: 10.1128/iai.00635-16
57. Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut bacteroidales species. Proc Natl Acad Sci USA (2016) 113(13):3627–32. doi: 10.1073/pnas.1522510113
58. Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics (2016) 17:58. doi: 10.1186/s12864-016-2377-z
59. Jones EJ, Booth C, Fonseca S, Parker A, Cross K, Miquel-Clopés A, et al. The uptake, trafficking, and biodistribution of Bacteroides thetaiotaomicron generated outer membrane vesicles. Front Microbiol (2020) 11:57. doi: 10.3389/fmicb.2020.00057
60. Thay B, Damm A, Kufer TA, Wai SN, Oscarsson J. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-κB activation. Infect Immun (2014) 82(10):4034–46. doi: 10.1128/iai.01980-14
61. Bielig H, Rompikuntal PK, Dongre M, Zurek B, Lindmark B, Ramstedt M, et al. NOD-like receptor activation by outer membrane vesicles from Vibrio cholerae non-O1 non-O139 strains is modulated by the quorum-sensing regulator HapR. Infect Immun (2011) 79(4):1418–27. doi: 10.1128/iai.00754-10
62. Johnston EL, Heras B, Kufer TA, Kaparakis-Liaskos M. Detection of bacterial membrane vesicles by NOD-like receptors. Int J Mol Sci (2021) 22(3):1005. doi: 10.3390/ijms22031005
63. O'Donoghue EJ, Krachler AM. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol (2016) 18(11):1508–17. doi: 10.1111/cmi.12655
64. Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem (2001) 276(7):4812–8. doi: 10.1074/jbc.M008072200
Keywords: bacterial membrane vesicles, outer membrane vesicles (OMVs), Bacteroides fragilis, TLRs, NOD1, innate immunity, epithelial cells, Commensals
Citation: Gilmore WJ, Johnston EL, Bitto NJ, Zavan L, O'Brien-Simpson N, Hill AF and Kaparakis-Liaskos M (2022) Bacteroides fragilis outer membrane vesicles preferentially activate innate immune receptors compared to their parent bacteria. Front. Immunol. 13:970725. doi: 10.3389/fimmu.2022.970725
Received: 16 June 2022; Accepted: 22 August 2022;
Published: 20 September 2022.
Edited by:
Paola Massari, Tufts University, United StatesReviewed by:
Sara Ahmadi Badi, Pasteur Institute of Iran, IranQiong Liu, Nanchang University, China
Mariola J. Edelmann, University of Florida, United States
Copyright © 2022 Gilmore, Johnston, Bitto, Zavan, O'Brien-Simpson, Hill and Kaparakis-Liaskos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Kaparakis-Liaskos, bS5saWFza29zQGxhdHJvYmUuZWR1LmF1
 William J. Gilmore
William J. Gilmore Ella L. Johnston
Ella L. Johnston Natalie J. Bitto
Natalie J. Bitto Lauren Zavan
Lauren Zavan Neil O'Brien-Simpson
Neil O'Brien-Simpson Andrew F. Hill2,4,5
Andrew F. Hill2,4,5 Maria Kaparakis-Liaskos
Maria Kaparakis-Liaskos