- Department of Gynecology and Obstetrics, The Second Affiliated Hospital of Fujian Medical University, Quanzhou, Fujian, China
Recurrent spontaneous abortion (RSA) seriously affects women’s reproductive health, and its pathogenesis is complex and varied. The aim of this study is to identify key molecular markers closely associated with RSA to rapidly and effectively predict the RSA, and to provide simple and practical indicators for clinical diagnosis and treatment.
Method: We obtained mRNA expression profiles from the GSE26787 and GSE165004 datasets of the Gene Expression Omnibus (GEO) database, immune-related genes (IRGs) from the ImmPort database (https://www.immport.org), and genes related to inflammatory response from the Molecular Signatures database. Different Inflammation- and immunity-related genes (DIIRGs) were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Protein-protein interaction (PPI) networks were utilized to explore the connections between various DIIRGs. The candidate DIIRGs were analyzed by the least absolute shrinkage and selection operator (LASSO) and the multiple support vector machine recursive feature elimination (mSVM-RFE). The diagnostic ability of the candidate genes was verified using receiver operating characteristic (ROC) curves. The performance of the predictive model was evaluated using a Nomo plot. We further confirmed the expression levels and diagnostic value of key genes by performing immunohistochemistry (IHC) in clinical tissue samples. The compositional patterns of the infiltration of 22 immune cell types in RSA were analyzed via the CIBERSORT algorithm.
Result: We identified 403 differentially expressed genes (DEGs) and 7 DIIRGs between RSA endometrium and Non-RSA endometrium. GO analysis showed that DIIRGs were significantly enriched in positive regulation of cell-cell adhesion, inflammatory response to antigenic stimulus, and protein tyrosine kinase activity. KEGG enrichment analyses were performed mainly on Epithelial cell signaling in Helicobacter pylori infection, NOD-like receptor signaling pathway, and Ras signaling pathway. A predictive and diagnostic model composed of three genes (CYBB, LYN, and MET). The CYBB, LYN, and MET genes were identified as diagnostic biomarkers of RSA (AUC = 0.747, AUC = 0.751, AUC=0.703), and reduced levels of CYBB and LYN expression were found to correlate with RSA in clinical samples. In addition, immune microenvironment analysis showed that CYBB and MET were positively correlated with naïve B cells and negatively correlated with CD8 T cells, LYN and MET were positively correlated with M2 macrophages and negatively correlated with eosinophils, respectively (P < 0.05).
Conclusion: Inflammation-immunity is a key factor in the pathogenesis of RSA. CYBB and LYN are regarded as the crucial genes that constitute a model and contribute to inflammation-immunity throughout the occurrence and progression of RSA. These findings provide a new perspective on the diagnosis and pathogenesis of RSA.
Introduction
Recurrent spontaneous abortion (RSA) shows up prevalently and challengingly within the scope of obstetrics and gynecology (1). The Royal College of Obstetricians and Gynecologists (RCOG) sets the definition of RSA as the happening of three or more natural miscarriages prior to the 24th week of pregnancy. However, the American Society for Reproductive Medicine’s criteria states that it is two or more natural miscarriages (2). In China at present, RSA is defined as the occurrence of three or more natural miscarriages within 28 weeks of pregnancy with the same partner (3). Not only does this condition augment psychological pressure and diminish self-esteem, resulting in psychological issues such as anxiety and depression, but also frequent abortions tend to induce endometrial abnormalities. It is shown by epidemiological survey data that a history of RSA can be found in around 1%–5% of women during their reproductive years (4). Considerable proof points to the fact that RSA is correlated with immunologic factor abnormalities, genetic defects, abnormal genital structure, endocrine disorders, inflammation, and other elements (5–7). Considering the existing absence of RSA screening, it is of great significance to identify innovative diagnostic biomarkers, which can help in the earlier detection of potential RSA, as well as to locate therapeutic targets to better the prognoses for RSA.
With the ongoing advancement of reproductive immunology in recent times, an increasing quantity of research evidence points to the fact that the occurrence of RSA is closely related to the imbalance within the maternal-fetal immune tolerance mechanisms. Two types of immune cells exist: the innate immune cells [neutrophils (8), Natural killer (NK) cells (9), myeloid-derived suppressor cells (MDSCs) (10) and macrophages (11)] and the adaptive immune cells (B (12) and T cells (13)). The disproportion of these immune cells may pose a threat to the success of pregnancy. The dysregulation of the immune system is likely to trigger immune cells to launch attacks on the embryo or the placenta (14).
Once human pregnancy initiates, the blastocyst which has hatched from the zona pellucida adheres to and is implanted within the endometrium of the maternal uterus. A sequence of crucial events, namely embryo implantation, decidualization, placentation, and parturition, is essential for the success of human pregnancy. In pregnancy, the physiological events are inflammatory processes. What is needed for the remodeling of intrauterine tissue, the feto-placental growth, and the parturition during the whole gestation is a balance of pro- and anti-inflammatory factors (15). NK cells play a vital role in initiating and resolving inflammation (16). Also, their existence is detected in each and every phase of pregnancy (17, 18).
This study aimed to uncover new DIIRGs related to RSA samples with the intention of pinpointing diagnostic Inflammation and immunity-related biomarkers by employing bioinformatics methods. Subsequently, we took further steps to confirm the DIIRGs within endometrium samples from both the normal pregnancy and RSA groups. Moreover, we explored the possible connection between the newly discovered DIIRGs and immune cells to promote further investigations into the origin and development of RSA in this particular area.
Materials and methods
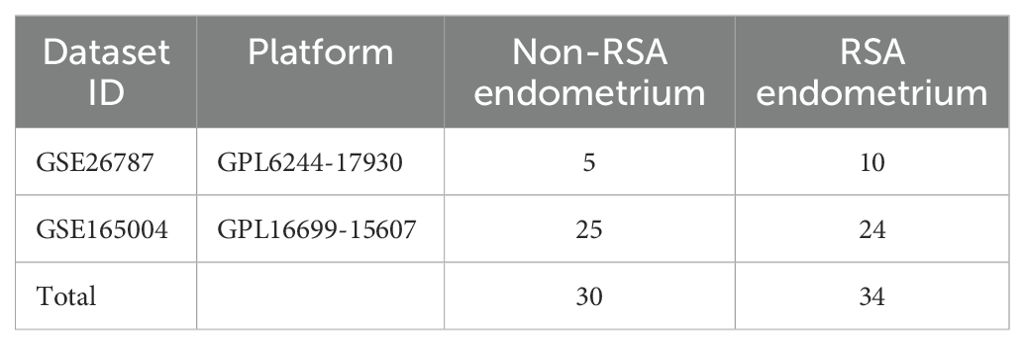
Collecting and processing microarray data
We retrieved the relevant original PE datasets, namely GSE26787 and GSE165004, from the GEO database (https://www.ncbi.nlm.nih.gov/gds). It consisted of 30 Non-RSA and 34 RSA samples, as presented in Table 1. Based on the probe annotation files, in each dataset, probes have corresponded to gene symbols. When there are multiple probes corresponding to a single gene, the average expression value of those probes is taken as the expression level of that gene. The raw data underwent background correction and normalization by using the limma package of R (http://www.bioconductor.org/). Genes with |log fold change (FC)| = 0.585 and adjusted p < 0.05 were regarded as DEGs. Download the data of IRGs from the Import database (https://www.immport.org/shared/) (Supplementary Table S1), and download the genes related to inflammatory responses from the Molecular Signatures database (https://www.gsea-msigdb.org/gsea/msigdb) (Supplementary Table S2). Subsequently, the intersection operation was performed between the IRGs, inflammation-related genes, and DEGs, leading to the derivation of DIIRGs in RSA.
Analysis of the function annotations and pathway enrichment for DIIRGs
The DIIRGs were subjected to GO and KEGG pathway enrichment analyses using the R packages “clusterProfiler”, “enrichplot”, “org.Hs.eg.db”, and “DOSE” with the aim of identifying the enriched GO terms within three distinct categories, namely cellular components, biological processes, and molecular functions, as well as the KEGG pathways. For visualizing the enrichment results, the “ggplot2” package in R was employed. A p-value less than 0.05 was regarded as the criterion for significant enrichment.
Construction of protein−protein interaction
In order to obtain a PPI network, we made use of the STRING website (https://string-db.org/) (19), where 7 DIIRGs were entered into the “multiple proteins” module, and “Homo sapiens” was chosen for the organism module. The gene symbols were extracted from protein IDs and PPIs without corresponding gene names were filtered out. Subsequently, Cytoscape 3.10.0 was applied to build the PPI network. Additionally, the cytoHubba plugin assisted in spotting the hub genes.
Constructing a diagnostic prediction model of RSA with DIIRGs
By means of Spearman correlation analysis to select DIIRGs possessing a |logFC| value of 0.585, the significant diagnostic biomarkers in RSA were pinpointed. The employment of the LASSO algorithm together with the mSVM-RFE algorithm facilitated the discovery of such biomarkers.
To conduct LASSO, which is an algorithm in regression analysis for variable selection to avoid overfitting, the “glmnet” package was applied. In addition, the mSVM-RFE algorithm was implemented using the “e1071” R package. This algorithm employs resampling methods in each iteration to stabilize feature rankings and identify the most pertinent features by removing the feature vectors generated by the SVM via supervised machine learning techniques (20). As mSVM-RFE has a lower risk of overfitting compared with SVM-RFE, we combined LASSO and mSVM-RFE to screen for overlapping genes, and then these genes were validated within the training set.
ROC curves were finally produced by means of the “pROC” R package. The ability of biomarkers to distinguish RSA from Non-RSA endometrium samples diagnostically was evaluated via the AUC of the ROC curve.
A nomogram model for the diagnostic capability of RSA was built and verified.
For the purpose of predicting RSA occurrence, the “rms” and “rmda” packages were employed to build a nomogram model. Wherein, each factor had its score marked as a “point,” and the sum of all factor scores was defined as “total points.” Then, calibration curves were produced to examine the nomogram model’s predictive capacity.
Patients and tissue specimens
Evelen paraffin-embedded samples, which consisted of five Non-RSA endometrium and six RSA endometrium samples, were collected from the Second Affiliated Hospital of Fujian Medical University (Fujian, China). The approval for this study had been obtained from the Research Ethics Committee of the aforementioned hospital prior to its initiation.
Immunohistochemistry
The IHC staining was carried out according to the procedures described previously (21). The primary antibodies, namely anti-LYN (18135-1-AP, Proteintech, USA), anti-CYBB (NOX2) (19013-1-AP, Proteintech, USA), and anti-cMET (MET) (AF6128, Affinity, England) were utilized. The staining intensity ratios of LYN, CYBB, and MET were classified into four levels: negative (scored as 0 points), light yellow (1 point), brownish yellow (2 points), and tan (3 points). Regarding the number of stained cells, it was evaluated based on the following proportions: if less than one-third, 1 point was assigned; if between one-third and two-thirds, 2 points; and if more than two-thirds, 3 points. The final expression scores of LYT, CYBB, and MET were calculated by multiplying the two aforementioned ratings. Subsequently, the slide samples were divided into two groups, namely the low-expression group and the high-expression group, which were defined by scores of less than 6 and greater than or equal to 6 respectively. The histopathological diagnoses of the patients were validated by two pathologists who specialized in obstetrics and gynecology.
The distribution of immune cells in RSA and biomarkers
The quantification of the proportion of infiltrating immune cells from the gene expression profiles in RSA was achieved by employing the CIBERSORT algorithm (http://cibersORT.stanford.edu/). The LM22 gene signature matrix, downloaded from the CIBERSORT webpage, was utilized to estimate the putative abundance of immune cells (22). The “corrplot” R package was utilized for conducting the correlation and visualization of LM22. Pearson’s correlation analysis was adopted to explore the relationships of the screened diagnostic biomarkers with the levels of infiltrating immune cells, with the results being visualized by means of the “ggplot2” R package.
Statistical analysis
Using R software (v.4.1.3), all the statistical analyses were performed. With the Mann–Whitney U test, comparisons between the Non-RSA and RSA groups were made. LASSO regression, the SVM-RFE algorithm, ROC analysis, Pearson’s correlation, and unpaired t-tests were involved in the analyses. Throughout these analyses, statistical significance was determined as p < 0.05.
Results
Study procedure
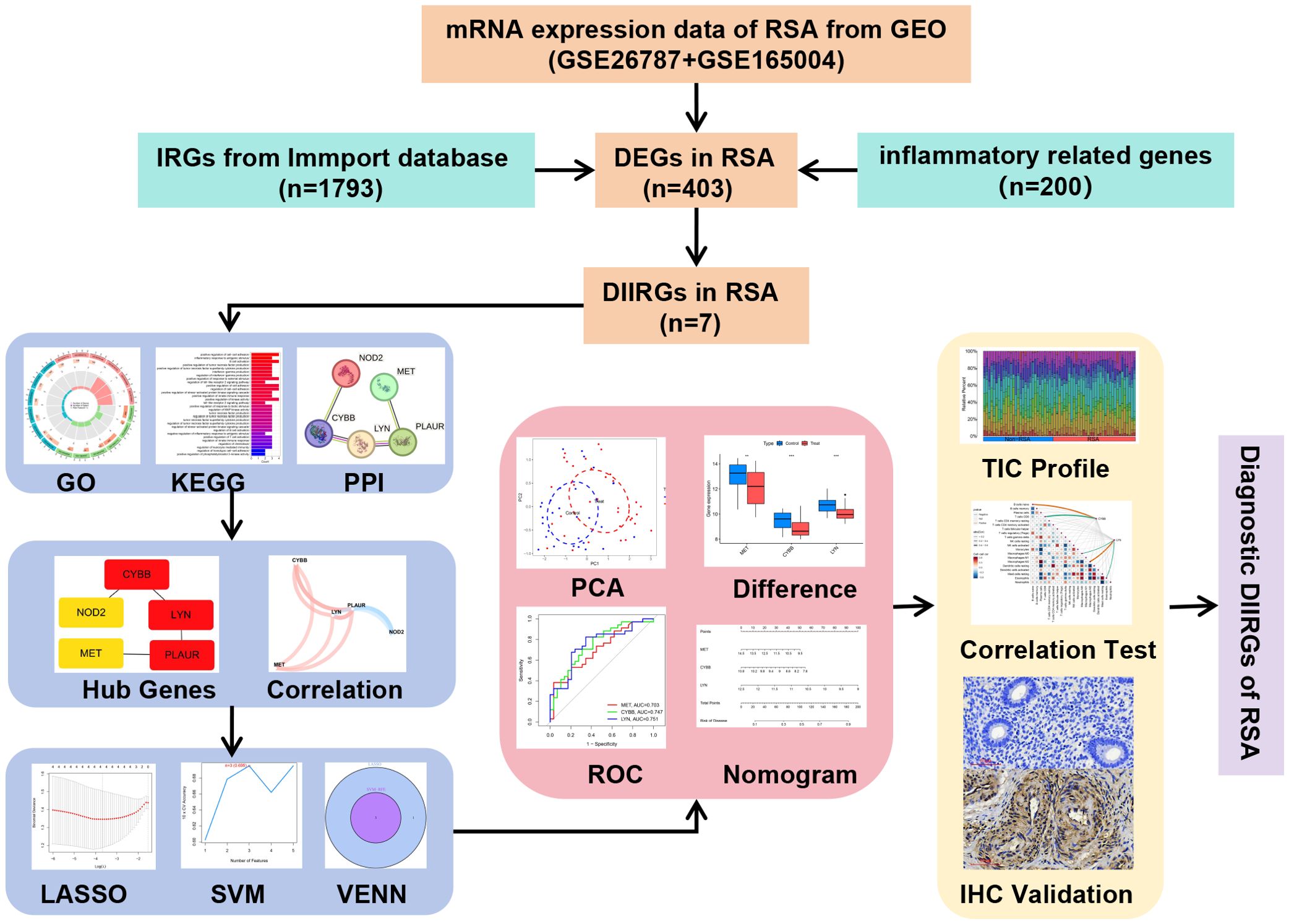
In the current study, the analytical procedure depicted in Figure 1 was adopted. The transcriptome RNA-seq data were obtained from the GEO database. The screening of the overlapping DIIRGs between the Non-RSA and RSA groups was accomplished by utilizing conjugated DEGs inflammation-related genes and immune-related genes. Following this, GO, KEGG, PPI, and hub gene network analyses were performed on the identified DIIRGs. The overlapping DIIRGs were selected via the conjugated LASSO and SVM-RFE methods; ROC curves were generated to assess the predictive power of these biomarkers and were further validated with IHC. The compositional patterns of LM22 in PE were determined by the CIBERSORT algorithm. Additionally, a correlation analysis was conducted between the diagnostic immune-related biomarkers and the infiltrating immune cells.
Identification of DIIRGs in RSA
With the utilization of the “limma” package in R, 403 DEGs between RSA patients and Non-RSA pregnancy within the GSE26787 and GSE165004 datasets were detected. The identification was based on the criteria of logFCfilter = 0.585 and adj.P.Val.Filter < 0.05 (Figure 2A). Out of these genes, 163 were notably downregulated and 240 were significantly upregulated (Figure 2B) (Supplementary Table S3). As depicted in Figure 2C, 7 DIIRGs were found through the intersection of the DEGs, IRGs, and the Inflammatory genes (Supplementary Table S4). Among the 7 DIIRGs, the expression of NOD2 was higher while the other 6 DIIRGs were remarkably downregulated in the RSA group.
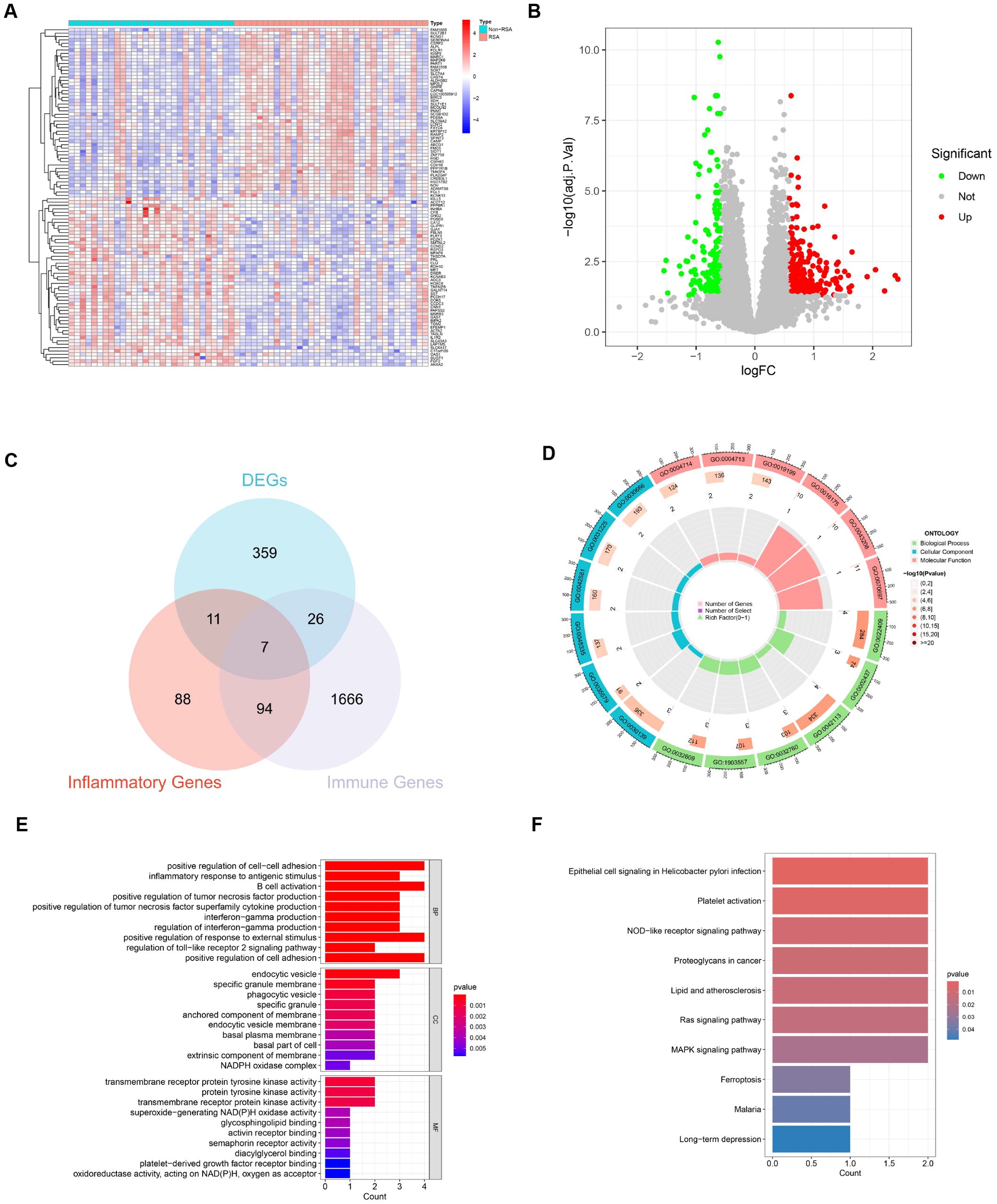
Figure 2. Identification and function of DIIRGs. (A) The expression levels of the first 50 DEGs between RSA tissue and Non-RSA samples from the GEO database are visualized through heatmaps. The genes are named in the row annotations, and the column annotations, which are the sample IDs, are not shown in the plots. The color gradient, which goes from red to blue, represents the expression levels from high to low in the heatmaps. (B) 403 DEGs between RSA and Non-RSA are illustrated by the volcano plots. In these plots, genes that are upregulated are indicated by red dots, genes that are downregulated are denoted by green dots, and genes without differential expression are represented by black dots. (C) 7 DIIRGs are contained in the Venn diagram of the intersection of differential genes, inflammatory genes, and immune genes. (D) A circle plot for GO analysis of 7 DIIRGs is presented. (E) A bar graph for GO analysis of 7 DIIRGs is shown. (F) The DIIRGs are annotated by KEGG.
GO and KEGG functional enrichment
In order to gain a more in-depth understanding of the functions and enrichment pathways of the 7 DIIRGs, the “ClusterProfiler” package in R was employed to conduct functional enrichment analysis. The results showed that when it came to genetic biological processes (BPs), the focus was predominantly on positive regulation of cell-cell adhesion and inflammatory response to antigenic stimulus. The cellular components (CCs) of these genes were mainly located in the endocytic vesicle, specific granule membrane, and phagocytic vesicle. Regarding the molecular functions (MFs), they were primarily associated with transmembrane receptor protein tyrosine kinase activity, activin receptor binding, and superoxide-generating NADPH oxidase activity, with a significance level of P < 0.05 (Figures 2D, E, Supplementary S1). Additionally, KEGG enrichment analysis suggested that the 7 DIIRGs were mainly engaged in Epithelial cell signaling in Helicobacter pylori infection, NOD-like receptor signaling pathway, and Ras signaling pathway (P <0.05, Figure 2F, Supplementary S2). All these findings point to a significant correlation between RSA and inflammation-immunity.
Construction of the PPI and hub genes network
The interactions of 7 DIIRGs were investigated by using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/). Through this, PPI networks with concealed disconnected nodes were produced, and the remaining five genes (LYN, CYBB, PLAUR, MET, NOD2) have a connection (Figure 3A). After that, the cytoHubba plugin in Cytoscape software was utilized for clustering the genes in the network. Consequently, five nodes associated with the MCC were identified and clustered (Figure 3B). Notably, four of the genes (LYN, CYBB, PLAUR, MET) showed decreased expression levels, while NOD2 showed elevated expression in RSA group (Figures 3C, D).
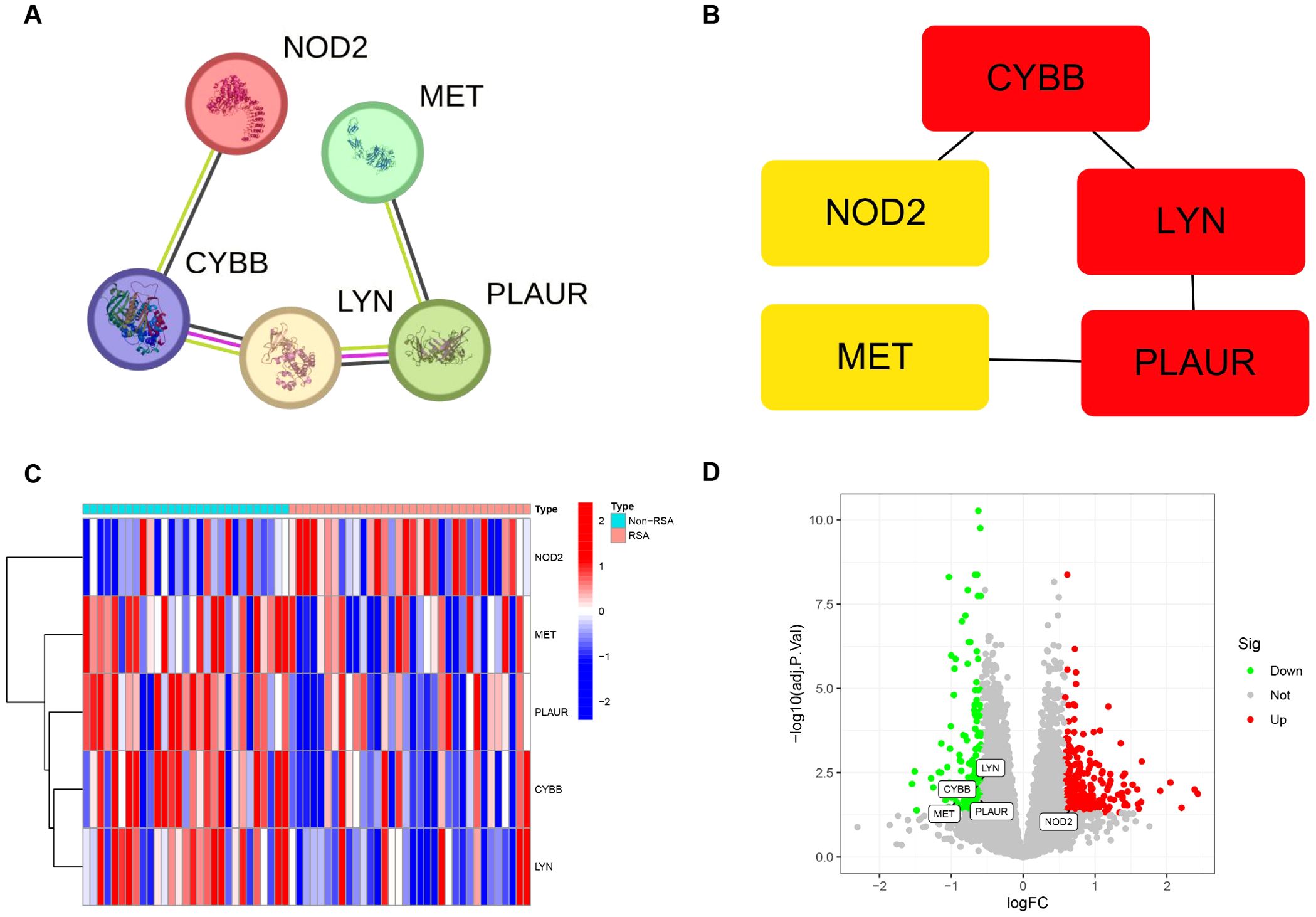
Figure 3. Association between DIIRGs and hub genes. (A) The PPI networks exploring five DIIRGs binding protein interactions are constructed by using the STRING tool. (B) Five hub genes are obtained through the MCC algorithm. (C) The expression levels of five hub genes between RSA tissue and Non-RSA samples are visualized through heatmaps. (D) Five hub genes between RSA tissue and Non-RSA samples are illustrated by the volcano plots.
The linkages among the expressing magnitudes of five hubgenes in RSA
To explore the correlations between the expression levels of the five DIIRGs, we carried out a correlation analysis with a cutoff of 0.5. By employing the “tidyverse” and “corrr” packages in R, we were able to visualize the results, thereby producing a coexpression network map (Figure 4A) and a correlation plot (Figure 4B). Moreover, scatter plots for the five gene groups with the highest correlations, where the cutoff was set at 0.6 and P<0.05 in RSA, were also generated (Figure 4C). It was found that PLAUR is positively correlated with LYN and CYBB (RLYN = 0.66; RCYBB = 0.65), while PLAUR shows a negative correlation with NOD2(RNOD2 = 0.66).
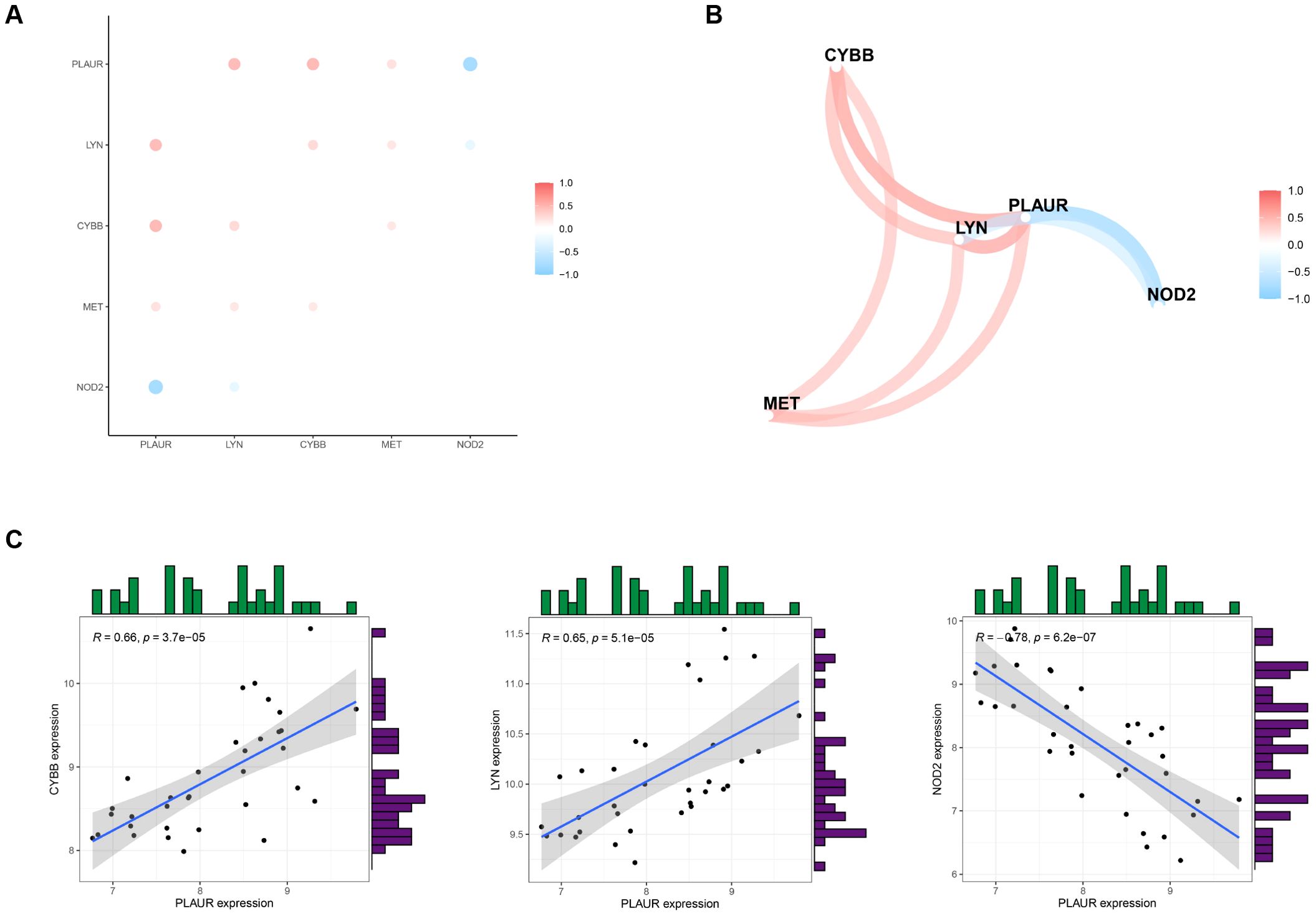
Figure 4. Analyzing the correlation of DIIRGs. (A) DIIRGs’ co-expression network map is depicted. (B) The correlation of DIIRGs is plotted. (C) A scatter plot for some highly correlated DIIRGs is provided.
A prediction model for RSA is under development
To accurately identify the key prognostic biomarkers in RSA, we utilized the LASSO and mSVM-RFE algorithms. Through LASSO analysis, four DIIRGs, namely MET, CYBB, NOD2, and LYN, were singled out from a group of five DIIRGs relevant to RSA, as shown in Figures 5A, B. Meanwhile, with the application of the mSVM-RFE model, this collection of five DIIRGs was further narrowed down to three DIIRGs, which included CYBB, LYN, and MET, as depicted in Figures 5C, D. When comparing the results of the two algorithms, it was found that the final selection consisted of three overlapping candidate genes: CYBB, LYN, and MET, as illustrated in Figure 5E.
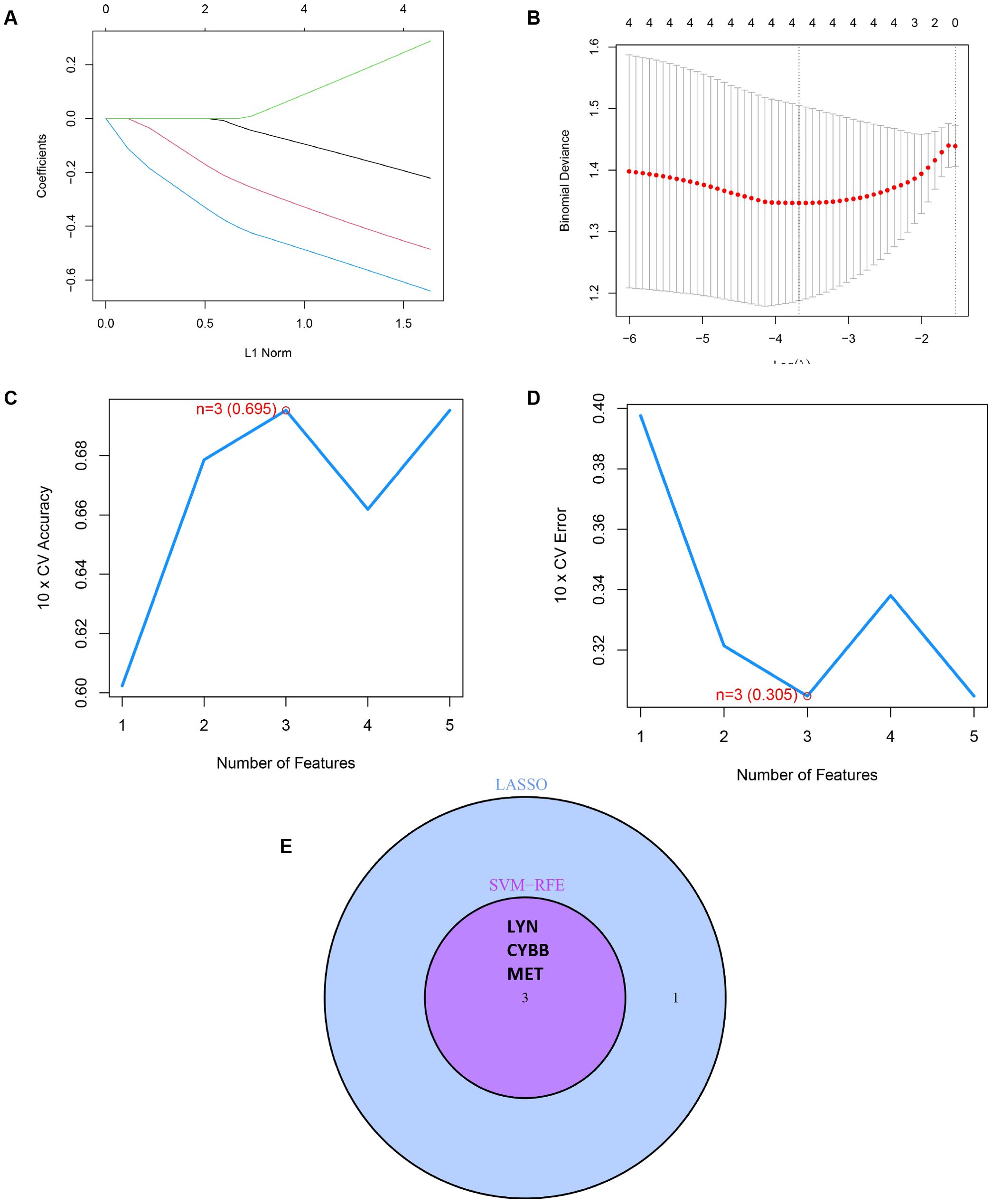
Figure 5. Construction of a prediction model for RSA. (A) A curve in the LASSO regression coefficient profiles of the 5 DIIRGs illustrates the changing course of each DIIRG. (B) The LASSO Cox regression model was employed to draw a plot of partial likelihood deviance versus log (l). (C) When k = 3, the curve of the total within the sum of the squared error curve under the corresponding cluster number k arrives at the “elbow point”. (D) At k = 3, the curve representing the average silhouette width for the corresponding cluster number k reaches its peak. (E) The Venn diagram shows the 3 diagnostic markers shared by the LASSO and SVM-RFE algorithms.
Further investigation into the three crucial DIIRGs
Three candidate DIIRGs, namely CYBB, LYN, and MET, which emerged as the common elements found through both the LASSO regression model and the mSVM-RFE model, were picked out for further probing. In Figure 6A, the chromosomal locations of CYBB, LYN, and MET are presented. The findings from the principal component analysis imply that these three candidate genes are highly effective in discriminating between pregnancies with RSA and those of Non-RSA, hinting at a potentially crucial part they play in diagnosing RSA, as shown in Figure 6B. Additionally, the predictive effectiveness of these three genes was assessed, in contrast to Non-RSA pregnancies, CYBB, LYN, and MET exhibited downregulated expression in pregnancies affected by RSA (Figure 6C). The AUC values of the ROC curves for these three DIIRGs (AUCCYBB = 0.747; AUCLYN = 0.751; AUCMET = 0.703) demonstrated a superior predictive power for RSA compared to Non-RSA pregnancies, as seen in Figure 6D. Subsequently, a risk-associated nomogram for RSA was devised (Figures 6E, F), which could function as a means to predict the capacity of the risk score to tell apart RSA from Non-RSA pregnancies.
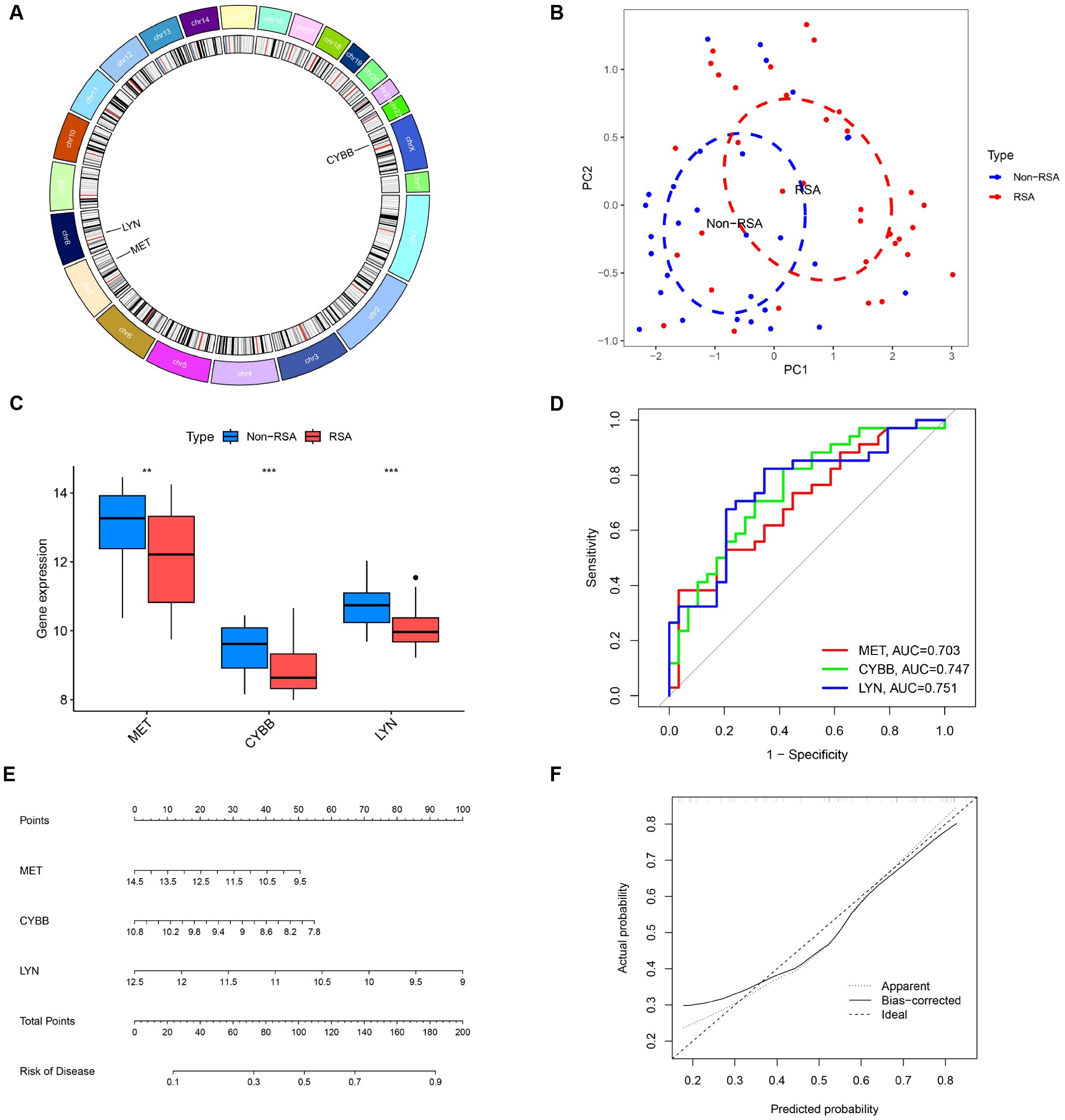
Figure 6. Additional analysis of three key DIIRGs. (A) The locations on the chromosome of three key DIIRGs. (B) RSA and Non-RSA can be clearly distinguished by principal component analysis using the three mentioned genes. (C) The relative expression levels of three key DIIRGs between RSA and Non-RSA are shown by the GSE26787+ GSE165004 datasets. (D) The performances of three key DIIRGs for predicting RSA in GSE26787+ GSE165004 datasets were verified by ROC curves. (E) Diagnostic Nomo plot related to three key DIIRGs. (F) Calibration curve of a model consisting of three key genes. **P < 0.01, ***P < 0.001.
Verification of the three crucial DIIRGs
For the sake of further clinical verification, we gauged the expression of CYBB, LYN, and MET by means of immunohistochemistry (IHC). The outcome disclosed that a low expression level of CYBB and LYN were correlated with RSA, both LYN and CYBB are expressed in the cytoplasm (Figures 7A, B; p < 0.05), the expression of MET was not significantly different between RSA tissues and Non-RSA tissues, it is also expressed in the cytoplasm (Figure 7C). Collectively, the aforementioned results imply that the CYBB and LYN genes exhibit stronger diagnostic potential.
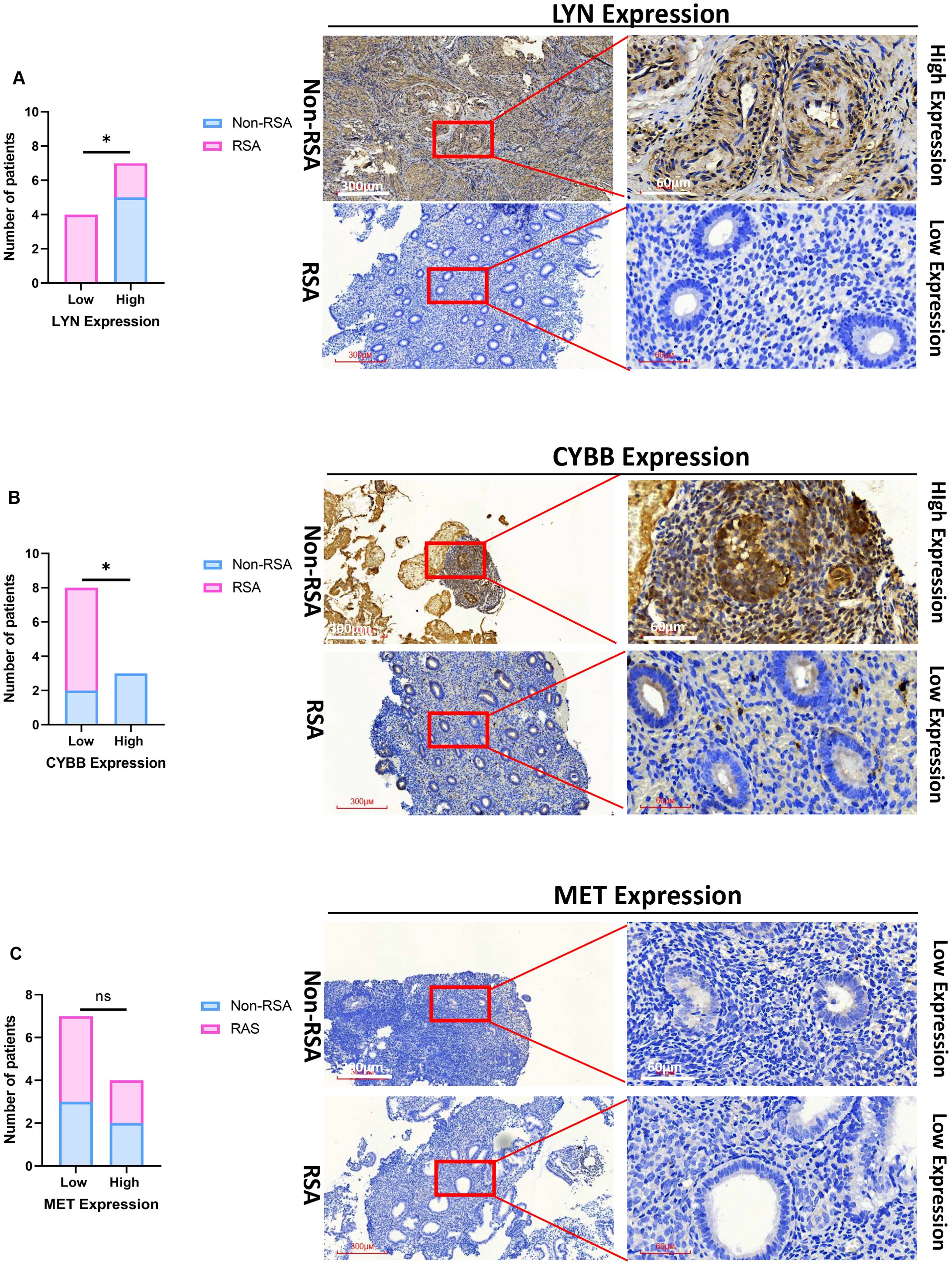
Figure 7. Validation of the three essential DIIRGs. (A) Significantly low expression of LYN was found in RSA tissues when compared with Non-RSA specimens (Non-RSA specimens = 5; RSA = 6). (B) Significantly low expression of CYBB was found in RSA. (C) No Significant expression of MET was found between RSA and Non-RSA tissues. Representative images (×40 and ×200) of IHC staining for LYN, CYBB, and MET in 6 RSA and 5 Non-RSA specimens (high expression versus low expression); *P < 0.05. ns, no significance.
The distribution of immune cells is related to CYBB, LYN, and MET
In order to obtain a more profound understanding of the connection between immune cell infiltration and RSA, the CIBERSORT algorithm was employed to ascertain the relative proportions of 22 kinds of immune cells in both the control and RSA samples, as shown in Figure 8A. Subsequently, our exploration into the correlation between the CYBB, LYN, and MET genes and infiltrating immune cells yielded the following results. CYBB and MET were positively correlated with naïve B cells and negatively correlated with CD8 T cells, LYN and MET were positively correlated with M2 macrophages and negatively correlated with eosinophils, respectively (P < 0.05), as illustrated in Figure 8B. These findings in relation to immune activity bolster the influence of the CYBB, LYN, and MET genes.
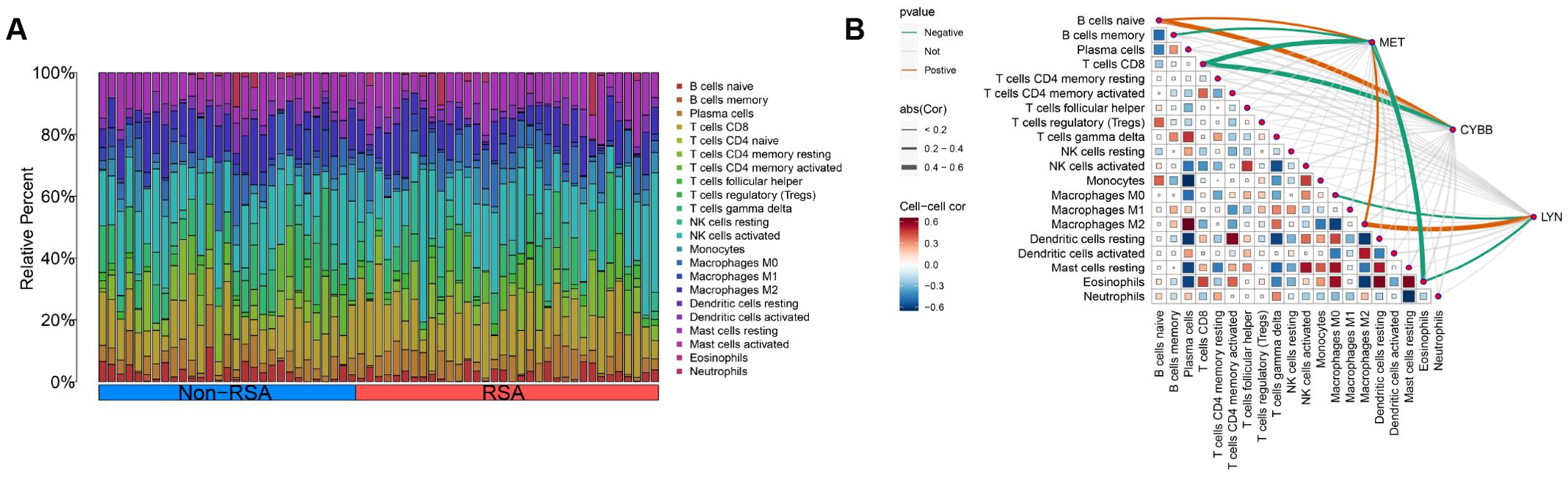
Figure 8. Distribution of immune cells is related to CYBB, LYN, and MET. (A) The distribution of 22 immune cell subtype proportions between RSA tissue and Non-RSA samples is illustrated by a bar plot. (B) Infiltrating immune cells in RSA are correlated with LYN, CYBB, and MET, and this correlation is analyzed.
Discussion
RSA is a severe reproductive disease during pregnancy. It not only poses a great threat to the physical health of women of childbearing age but also brings heavy psychological burdens. Previous studies have shown that immunodeficiency accounts for the vast majority of the multiple factors leading to unexpected recurrent spontaneous abortions (23). Alloimmune RSA is usually referred to as unexplained RSA (URSA). Current research believes that it is related to abnormal immune cells, high histocompatibility of human lymphocyte antigens, and the lack of blocking antibodies (BA) (4). For a successful pregnancy, from the implantation of the embryo, the attachment of the placenta, the growth, and development of the fetus, to the delivery process of the fetus, a specific immune microenvironment is required, and the successful establishment of maternal-fetal immune tolerance is necessary.
GO enrichment analysis revealed that inflammatory response to antigenic stimulus and B cell activation in RSA. The results show that there is a close and non-negligible association between RSA and immune inflammation. Immune factors account for roughly 50% of the pathogenetic mechanisms in RSA (24). There exist a variety of immune cells including NK cells, macrophages, and DC cells at the maternal-fetal interface (25). In both maternal and fetal tissues, NK cells stand out as the immune cells with the highest abundance (26). Meanwhile, some immunotherapeutic approaches directed at NK cells have reaped promising achievements (27). The research suggested that the occurrence of RSA might be attributed to immune inflammation and oxidative stress brought about by metabolic dysregulation (28). Daher et al. (29) pointed out that for patients suffering from RSA, NK cell activity had increased and the levels of Th1-type cytokines including IFN-r and TNF-a had gone up. And they connected these observations to immune inflammation. Elevated levels of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and Th17/Treg ratios can be detected in pregnant women with URSA. Such elevations may give rise to an immune imbalance at the maternal-fetal interface and consequently increase the likelihood of miscarriages (30). This finding reveals that the occurrence of RSA is not caused by a single factor, but is rather a complex pathological process involving multiple cell types and a variety of internal and external factors working together.
KEGG enrichment analysis mainly involves NOD-like receptor signaling pathway and Ras signaling. Nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) play a central part in both innate and adaptive immunity and they are intracellular proteins. Given that NLRs are members of pattern recognition receptors (PRRs), they can sense particular pathogen-associated molecular patterns. Subsequently, they initiate a multitude of signaling pathways, culminating in the secretion of diverse cytokines (31). It has been reported that the loss of NOD1 can heighten the inflammatory responses and modulate the pro-inflammatory reactions triggered by H. pylori infection (32). Jamontt et al. conducted a study and discovered that in IL-10-deficient mice, NOD2 signaling served to exacerbate colitis. Moreover, upon the deletion of NOD2 in IL-10/mice, a remarkable improvement in the symptoms of colitis was observed (33). It can be concluded from the above studies that this pathway is related to both inflammation and immunity. Currently, there is very little research on the NOD-like receptor (NLR) signaling pathway in RSA. NLRP7 is a member of the NLR family and a gene related to the NOD-like receptor (NLR) signaling pathway, serving as an immune sensor in innate immunity. In an in vitro decidualization model, the expression level of NLRP7 was elevated and it was translocated into the nuclei of endometrial stromal cells. When NLRP7 was overexpressed, the expression of IGFBP-1 was enhanced and the activation of the PR reporter was promoted (34). These findings indicate that NLRP7 plays a role in the in vitro decidualization process of endometrial stromal cells. The Ras signaling is one of the core pathways that regulate cell proliferation, differentiation, survival, and metabolism, and it is widely involved in biological processes such as development, immunity, and cancer. One study determines that alterations in the RAS pathway drive transcriptional remodeling of hematopoietic stem and progenitor cells (HSPCs) and the monocytic populations derived from them (35). Inflammatory signals, both intrinsic and extrinsic to the cells, act in concert with these mutations, ultimately resulting in the dysfunction of immune cells (35). Intracellular CD59 regulates the subcellular compartmentalization of Ras, which confers spatial selectivity to T - cell activation and presents a potential immunotherapeutic approach mediated by T cells (36). Up to now, there has been still limited research on the Ras signaling pathway in recurrent miscarriage. URSA is caused by abnormal H3K27 histone methylation of the RASA1 gene, which regulates the Ras-MAPK pathway in trophoblast cells (23). Cell cycle arrest and morphological changes in mouse embryonic fibroblasts deficient in RAS proteins are prevented by active R-RAS2/TC21 (37).
Based on the 7 DIIRGs that exhibited the most significant differences between the RSA and normal control groups, the LASSO regression and mSVM-RFE models were employed to screen out three genes, namely CYBB, LYN, and MET. Subsequent validation verified that the CYBB, LYN, and MET genes are correlated with RSA, as demonstrated by an area under the curve (AUC) greater than 0.7. CYBB, which can be found on the X chromosome, is precisely the gene that encodes the sizable gp91phox subunit of cytochrome b558. It should be emphasized that this cytochrome b558 constitutes the transmembrane moiety of the NADPH oxidase complex (38). It has been verified by research that chronic granulomatous disease (CGD) is the earliest discovered ailment related to CYBB. Specifically, CGD represents a primary immunodeficiency concerning phagocyte function, which stems from a deficiency in NADPH oxidase (phox) (39). Moreover, on account of mutations in CYBB, 70% of the cases show an X-linked pattern, consequently giving rise to a defect in the production of gp91PHOX (40). It has recently been demonstrated that CYBB/NOX2 within cDCs is instrumental in enhancing antigen presentation, thereby activating CD4+ T cells and leading to tissue damage mediated by TH cells (41). In a model concerning hyperhomocysteinemia-induced renal harm, the activation of NLRP3 inflammasome, which was instigated by NADPH oxidase-driven redox signaling, led to the recruitment of immune cell infiltration. Eventually, this cascade of events gave rise to glomerular injury and sclerosis (42). These results gain additional backing from our study.
LYN, which belongs to the src family of non-receptor protein tyrosine kinases, is mainly expressed in hematopoietic tissues. It is believed, similar to other members of the src family, that Lyn takes part in signal transduction from cell surface receptors without intrinsic tyrosine kinase activity (43). It is the signaling pathway of Lyn that has drawn extensive attention in the research on inflammation (44), immunity (45), allergy (46), and tumor (47). Given that Lyn participates simultaneously in both positive- and negative-regulatory pathways, a comprehensive and clear mechanistic map of its function remains mostly absent. This is because the effects it exerts are not only complex but also vary depending on cell type. In the sepsis-associated acute kidney injury (SA-AKI) model, the deletion of the LYN gene exacerbates tubular injury. Its protective mechanism involves the inhibition of STAT3 phosphorylation and cell apoptosis. The over-activation of STAT3 promotes the inflammatory response, while LYN alleviates inflammatory damage by negatively regulating the STAT3 pathway (48). In the Alzheimer’s disease (AD) model, LYN directly binds to TLR4 and regulates the inflammatory and phagocytic functions of microglia. LYN deficiency enhances the TLR4-induced neuroinflammatory response and weakens the phagocytic ability of Aβ, leading to aggravated neuronal damage (49). In systemic autoinflammation (such as LE and Ptpn6me - v/me - v mouse models), LYN deficiency or functional abnormalities (such as the pTyr508His mutation) lead to the over-activation of the NF - κB pathway, triggering severe systemic inflammatory responses (such as fever, chronic urticaria, and hypercytokinemia) (50). In B cells, LYN maintains immune tolerance by regulating BCR signaling and CSK-mediated phosphorylation (51). Low LYN expression leads to the over-activation of B cells, promotes the release of pro-inflammatory factors (such as TNF-α), and exacerbates the inflammatory response (52). The low expression of the LYN gene usually leads to reduced inhibition of inflammatory signaling pathways (such as TAT3, NF-κB, and TLR4), thereby promoting the inflammatory response and tissue damage. Our research findings indicate that LYN is lowly expressed in RSA, the finding is similar to the research mentioned above. ICAM1 activation alone can trigger eructophagy in macrophages via Lyn kinase (53). However, another study has indicated that Lyn plays a minimal part in the macrophage response to TLR4 activation (54). It appears that Lyn plays a more prominent positive part in neutrophil migration and trafficking. At the site of infection, peroxides are produced and they oxidize the cysteines within Lyn. This oxidation process triggers Lyn activation, subsequently guiding neutrophil migration to follow the peroxide gradient and move toward the infection site (55). To sum up, the LYN gene plays a dual role (either pro-inflammatory or anti-inflammatory) in the inflammatory response, the immune response of macrophages, and disease progression by regulating pathways such as TAT3, TLR4, and metabolic reprogramming. The specific effects depend on the microenvironmental signals and the types of diseases. There has been no report on LYN in RSA so far, and the mechanism of the role of LYN in RSA still needs to be further investigated.
CYBB encodes the gp91 phox subunit of the phagocyte NADPH oxidase, which is highly expressed in all phagocytes and lowly expressed in B cells (56). The CYBB gene is located on the X chromosome, and its mutation rate accounts for approximately 60-80% (57). The low expression of the CYBB gene is closely related to the occurrence and development of inflammation, and its mechanism of action involves the abnormal functions of various immune cells and the dysregulation of inflammatory signaling pathways. In the model of bronchopulmonary dysplasia (BPD), the expression of CYBB in alveolar M1 macrophages decreases, leading to the impairment of their pro-inflammatory functions. At the same time, it promotes the abnormal infiltration of macrophages in lung tissues, exacerbating inflammatory lung injury (58). In the microenvironment of pancreatic cancer, tumor cells induce the downregulation of CYBB in macrophages (such as U937 cells), promoting the polarization of macrophages towards the M2 phenotype (an anti-inflammatory/tumor-promoting phenotype). This state of low CYBB expression is associated with enhanced tumor-related inflammation and poor prognosis in patients (59). In the model of bladder infection, mice with NOX2 (CYBB) deficiency are unable to effectively inhibit the NF-κB signaling pathway due to insufficient production of reactive oxygen species (ROS). This leads to excessive infiltration of neutrophils and the loss of control of bladder inflammation (60). In colitis, CYBB cooperates with TLR4 to regulate the activation of the NLRP3 inflammasome. Low expression of CYBB may disrupt the inflammatory balance and exacerbate the damage to the intestinal mucosa (61). The inhibition of CYBB in zebrafish embryos leads to lipid metabolism disorders and excessive activation of the TLR4/NF-κB pathway, triggering liver injury and immune-inflammatory responses (62). The low expression of the CYBB gene weakens the activity of NADPH oxidase, alters the polarization state of immune cells, and activates inflammatory pathways such as NF-κB/NLRP3. Ultimately, this leads to the loss of control of the inflammatory response and tissue damage. This mechanism plays an important role in various pathological processes, including infectious diseases, autoimmune diseases, and the tumor microenvironment. Our research findings indicate that CYBB is lowly expressed in RSA, the finding is similar to the research mentioned above. It suggests that restoring the expression of CYBB or targeting its regulatory network may become a new strategy for treating inflammation-related diseases.
The MET gene, which is located on the long arm of human chromosome 7, region 2, band 1 (7q21 - 31), has 21 exons. Its encoded protein product is the hepatocyte growth factor receptor (HGFR). MET, which is a heterodimeric transmembrane tyrosine kinase receptor, is encoded by the mesenchymal-epithelial transition factor (MET) proto-oncogene and is mainly expressed in epithelial and endothelial cells (63, 64). Aberrant MET activation, which has been found in a broad range of solid tumors, facilitates uncontrolled cell proliferation, migration, and survival, leading to highly aggressive tumors (65). What’s more, several studies have demonstrated that the HGF/MET axis is necessary for the development and severity of different inflammatory and immune-mediated diseases such as colitis (66), COVID-19 (67), multiple sclerosis (68), and rheumatoid arthritis (69). An upregulation of colonic MET+ neutrophils during DSS colitis was demonstrated. The severity of DSS-induced colitis was reduced by the genetic ablation of MET in neutrophils. Concomitantly, the number of TH17 cells was decreased, which could be due to a decreased production of IL-1β by MET-deficient neutrophils (66). Intriguingly, a dual function appears to be fulfilled by the HGF/MET pathway in macrophage operations. The maintenance of a pro-inflammatory state associated with disease severity can be directly facilitated by the HGF/MET pathway (70, 71). However, it was also reported that the transition of macrophages towards an anti-inflammatory state promoting muscle repair is enhanced by HGF/MET (72). In our study, there is no significant difference in the expression of MET between RSA and normal pregnancy. MET was positively correlated with macrophages. This may weaken the anti-inflammatory effect of macrophages. Although there was no statistical difference in the verification of MET by IHC, we can increase the sample size in the future. Further investigation of MET on macrophages is needed in RSA.
Our research also has some drawbacks. Firstly, the study focused only on endometrial tissue, excluding other relevant maternal-fetal interface tissues (decidua or placenta) that could provide a more comprehensive understanding of RSA. After that, limited clinical heterogeneity (age, hormonal status and autoimmune background) were not incorporated into the analysis, which could affect the expression of immune and inflammatory genes. Due to the limited sample size, the nomogram model might need further verification prior to its clinical application. Subsequently,
limitations regarding one algorithmic approach for immune cell inference. Lastly, although the expression levels of CYBB, LYN, and MET have been confirmed by IHC, flow cytometry is still required to delve deeper into the action mechanisms of these molecules.
Conclusion
To sum up, through the application of machine-learning methods, we probed into the possible association between Inflammation-immunity and the development of RSA. Our study uncovered a remarkable relationship between them. A novel predictive and diagnostic model related to inflammation-immunity composed of CYBB and LYN genes was discovered to have low expression levels in RSA. Additionally, this model is correlated with both immune cells and inflammatory cells. These findings may expand our understanding of the inflammatory response and immune regulation in patients with RSA, providing new insights into the diagnosis and treatment of RSA.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the Research Ethics Committee of The Second Affiliated Hospital of Fujian Medical University prior to the study. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
ZW: Conceptualization, Formal Analysis, Funding acquisition, Writing – original draft. QL: Formal Analysis, Writing – review & editing. ZZ: Data curation, Writing – review & editing. YX: Data curation, Writing – review & editing. LH: Data curation, Writing – review & editing. LS: Investigation, Writing – review & editing. QS: Supervision, Writing – review & editing. YK: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Second Affiliated Hospital of Fujian Medical University Doctoral Miaopu Project (BS202401), the innovation of science and technology, Fujian province(2024Y9412), the Fujian Provincial Health Technology Project (2024GGA044), and Joint funds for the innovation of science and technology, Fujian province (2023Y9234).
Acknowledgments
The authors acknowledge the GEO database for providing data on RAS available, the Import database for providing data on IRGs, and the Molecular Signatures database for providing the genes related to inflammatory responses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1568536/full#supplementary-material
Supplementary Table 1 | The immune-related genes (IRGs) from the Import database.
Supplementary Table 2 | The genes related to inflammatory responses from the Molecular Signatures database
Supplementary Table 3 | 403 differentially expressed genes (DEGs) between RSA patients and Non-RSA patients within the GSE26787 and GSE165004 datasets
Supplementary Table 4 | 7 DIIRGs were found by intersecting the DEGs, IRGs, and the Inflammatory genes.
Supplementary S1 | The result of GO enrichment
Supplementary S2 | The result of KEGG pathway enrichment
References
1. Bender Atik R, Christiansen OB, Elson J, Kolte AM, Lewis S, Middeldorp S, et al. ESHRE guideline: recurrent pregnancy loss: an update in 2022. Hum Reprod Open. (2023) 2023:hoad002. doi: 10.1093/hropen/hoad002
2. Youssef A, Vermeulen N, Lashley E, Goddijn M, and van der Hoorn MLP. Comparison and appraisal of (inter)national recurrent pregnancy loss guidelines. Reprod biomedicine Online. (2019) 39:497–503. doi: 10.1016/j.rbmo.2019.04.008
3. Wu M, Liu P, and Cheng L. Galectin-1 reduction and changes in T regulatory cells may play crucial roles in patients with unexplained recurrent spontaneous abortion. Int J Clin Exp Pathol. (2015) 8:1973–8.
4. Deng T, Liao X, and Zhu. Recent advances in treatment of recurrent spontaneous abortion. Obstetrical gynecological survey. (2022) 77:355–66. doi: 10.1097/OGX.0000000000001033
5. Ndjapa-Ndamkou C, Govender L, and Chauke L. Role of genetic factors in recurrent miscarriages - A review. Afr J Reprod Health. (2022) 26:72–82. doi: 10.29063/ajrh2022/v26i10.9
6. Dimitriadis E, Menkhorst E, Saito S, Kutteh WH, and Brosens JJ. Recurrent pregnancy loss. Nat Rev Dis Primers. (2020) 6:98. doi: 10.1038/s41572-020-00228-z
7. Daumová M, Hadravská Š, and Putzová M. Spontaneous abortion in the first trimester of pregnancy. Ceskoslovenska patologie. (2023) 59:60–3.
8. Jiang S, He F, Gao R, Chen C, Zhong X, Li X, et al. Neutrophil and neutrophil-to-lymphocyte ratio as clinically predictive risk markers for recurrent pregnancy loss. Reprod Sci (Thousand Oaks Calif.). (2021) 28:1101–11. doi: 10.1007/s43032-020-00388-z
9. Woon EV, Nikolaou D, MacLaran K, Norman-Taylor J, Bhagwat P, Cuff AO, et al. Uterine NK cells underexpress KIR2DL1/S1 and LILRB1 in reproductive failure. Front Immunol. (2022) 13. doi: 10.3389/fimmu.2022.1108163
10. Li C, Zhang X, Kang X, Chen C, Guo F, Wang Q, et al. Upregulated TRAIL and reduced dcR2 mediate apoptosis of decidual PMN-MDSC in unexplained recurrent pregnancy loss. Front Immunol. (2020) 11. doi: 10.3389/fimmu.2020.01345
11. Ye HX, Liao GN, Dong YJ, Li L, Wang XM, Shu J, et al. miR-146a-5p enhances embryo survival in unexplained recurrent spontaneous abortion by promoting M2 polarization of decidual macrophages. Int Immunopharmacol. (2022) 110:108930. doi: 10.1016/j.intimp.2022.108930
12. Sung N, Byeon HJ, Garcia MDS, Skariah A, Wu L, Dambaeva S, et al. Deficiency in memory B cell compartment in a patient with infertility and recurrent pregnancy losses. J Reprod Immunol. (2016) 118:70–5. doi: 10.1016/j.jri.2016.09.003
13. Ander E, Diamond M, and Coyne CB. Immune responses at the maternal-fetal interface. Sci Immunol. (2019) 4. doi: 10.1126/sciimmunol.aat6114
14. Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. (2013) 31:387–411. doi: 10.1146/annurev-immunol-032712-100003
15. Yockey LJ and Iwasaki A. Interferons and proinflammatory cytokines in pregnancy and fetal development. Immunity. (2018) 49:397–412. doi: 10.1016/j.immuni.2018.07.017
16. Zitti B and Bryceson YT. Natural killer cells in inflammation and autoimmunity. Cytokine Growth factor Rev. (2018) 42:37–46. doi: 10.1016/j.cytogfr.2018.08.001
17. Zhang J, Dunk CE, Kwan M, Jones RL, Harris LK, Keating S, et al. Human dNK cell function is differentially regulated by extrinsic cellular engagement and intrinsic activating receptors in first and second trimester pregnancy. Cell Mol Immunol. (2017) 14:203–13. doi: 10.1038/cmi.2015.66
18. de Mendonça Vieira R, Meagher A, Crespo ÂC, Kshirsagar SK, Iyer V, Norwitz ER, et al. Human term pregnancy decidual NK cells generate distinct cytotoxic responses. J Immunol (Baltimore Md.: 1950). (2020) 204:3149–59. doi: 10.4049/jimmunol.1901435
19. von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, and Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. (2003) 31:258–61. doi: 10.1093/nar/gkg034
20. Zhou X and Tuck DP. MSVM-RFE: extensions of SVM-RFE for multiclass gene selection on DNA microarray data. Bioinf (Oxford England). (2007) 23:1106–14. doi: 10.1093/bioinformatics/btm036
21. Chen H, Wang J, Yang H, Chen D, and Li P. Association between FOXM1 and hedgehog signaling pathway in human cervical carcinoma by tissue microarray analysis. Oncol Lett. (2016) 12:2664–73. doi: 10.3892/ol.2016.4932
22. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. (2015) 12:453–7. doi: 10.1038/nmeth.3337
23. Zhang J, Liu X, and Gao Y. Abnormal H3K27 histone methylation of RASA1 gene leads to unexplained recurrent spontaneous abortion by regulating Ras-MAPK pathway in trophoblast cells. Mol Biol Rep. (2021) 48:5109–19. doi: 10.1007/s11033-021-06507-6
24. Liu H, Lin XX, Huang XB, Huang DH, Song S, Chen YJ, et al. Systemic characterization of novel immune cell phenotypes in recurrent pregnancy loss. Front Immunol. (2021) 12:657552. doi: 10.3389/fimmu.2021.657552
25. Yang Y, Song S, Gu S, Gu Y, Zhao P, Li D, et al. Kisspeptin prevents pregnancy loss by modulating the immune microenvironment at the maternal-fetal interface in recurrent spontaneous abortion. Am J Reprod Immunol (New York N.Y.: 1989). (2024) 91:e13818. doi: 10.1111/aji.13818
26. Mori M, Bogdan A, Balassa T, Csabai T, and Szekeres-Bartho J. The decidua-the maternal bed embracing the embryo-maintains the pregnancy. Semin immunopathology. (2016) 38:635–49. doi: 10.1007/s00281-016-0574-0
27. Moffett A and Shreeve N. First do no harm: uterine natural killer (NK) cells in assisted reproduction. Hum Reprod (Oxford England). (2015) 30:1519–25. doi: 10.1093/humrep/dev098
28. Azizi R, Soltani-Zangbar MS, Sheikhansari G, Pourmoghadam Z, Mehdizadeh A, Mahdipour M, et al. Metabolic syndrome mediates inflammatory and oxidative stress responses in patients with recurrent pregnancy loss. J Reprod Immunol. (2019) 133:18–26. doi: 10.1016/j.jri.2019.05.001
29. Saini V, Arora S, Yadav A, and Bhattacharjee J. Cytokines in recurrent pregnancy loss. Clinica chimica acta; Int J Clin Chem. (2011) 412:702–8. doi: 10.1016/j.cca.2011.01.002
30. Qian J, Zhang N, Lin J, Wang C, Pan X, Chen L, et al. Distinct pattern of Th17/Treg cells in pregnant women with a history of unexplained recurrent spontaneous abortion. Bioscience Trends. (2018) 12:157–67. doi: 10.5582/bst.2018.01012
31. Zhou Y, Yu, and Zhang W. NOD-like receptor signaling pathway in gastrointestinal inflammatory diseases and cancers. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms241914511
32. Suarez G, Romero-Gallo J, Piazuelo MB, Sierra JC, Delgado AG, Washington MK, et al. Nod1 Imprints Inflammatory and Carcinogenic Responses toward the Gastric Pathogen Helicobacter pylori. Cancer Res. (2019) 79:1600–11. doi: 10.1158/0008-5472.CAN-18-2651
33. Jamontt J, Petit S, Clark N, Parkinson SJ, and Smith P. Nucleotide-binding oligomerization domain 2 signaling promotes hyperresponsive macrophages and colitis in IL-10-deficient mice. J Immunol (Baltimore Md.: 1950). (2013) 190:2948–58. doi: 10.4049/jimmunol.1201332
34. Huang JY, Yu PH, Li YC, and Kuo PL. NLRP7 contributes to in vitro decidualization of endometrial stromal cells. Reprod Biol endocrinology: RB&E. (2017) 15:66. doi: 10.1186/s12958-017-0286-x
35. Montalban-Bravo G, Thongon N, Rodriguez-Sevilla JJ, Ma F, Ganan-Gomez I, Yang H, et al. Targeting MCL1-driven anti-apoptotic pathways overcomes blast progression after hypomethylating agent failure in chronic myelomonocytic leukemia. Cell Rep Med. (2024) 5:101585. doi: 10.1016/j.xcrm.2024.101585
36. Li L, Ding P, Lv X, Xie S, Li L, Chen J, et al. CD59-regulated ras compartmentalization orchestrates antitumor T-cell immunity. Cancer Immunol Res. (2022) 10:1475–89. doi: 10.1158/2326-6066.CIR-21-1072
37. Fernández-Pisonero I, Lorenzo-Martín LF, Drosten M, Santos E, Barbacid M, Alarcón B, et al. Active R-RAS2/TC21 prevents cell cycle arrest and morphological alterations in mouse embryonic fibroblasts lacking RAS proteins. Oncogene. (2025). doi: 10.1038/s41388-025-03367-3
38. Mortaz E, Azempour E, Mansouri D, Tabarsi P, Ghazi M, Koenderman L, et al. Common infections and target organs associated with chronic granulomatous disease in Iran. Int Arch Allergy Immunol. (2019) 179:62–73. doi: 10.1159/000496181
39. Yu HH, Yang YH, and Chiang BL. Chronic granulomatous disease: a comprehensive review. Clin Rev Allergy Immunol. (2021) 61:101–13. doi: 10.1007/s12016-020-08800-x
40. Battersby AC, Cale AM, Goldblatt D, and Gennery AR. Clinical manifestations of disease in X-linked carriers of chronic granulomatous disease. J Clin Immunol. (2013) 33:1276–84. doi: 10.1007/s10875-013-9939-5
41. Keller CW, Kotur MB, Mundt S, Dokalis N, Ligeon LA, Shah AM, et al. CYBB/NOX2 in conventional DCs controls T cell encephalitogenicity during neuroinflammation. Autophagy. (2021) 17:1244–58. doi: 10.1080/15548627.2020.1756678
42. Abais JM, Zhang C, Xia M, Liu Q, Gehr TW, Boini KM, et al. NADPH oxidase-mediated triggering of inflammasome activation in mouse podocytes and glomeruli during hyperhomocysteinemia. Antioxidants Redox Signaling. (2013) 18:1537–48. doi: 10.1089/ars.2012.4666
43. Brian BFT and Freedman T. The src-family kinase lyn in immunoreceptor signaling. Endocrinology. (2021) 162. doi: 10.1210/endocr/bqab152
44. Ding Y, Li C, Zhang Y, Ma P, Zhao T, Che D, et al. Quercetin as a Lyn kinase inhibitor inhibits IgE-mediated allergic conjunctivitis. Food Chem toxicology: an Int J published Br Ind Biol Res Assoc. (2020) 135:110924. doi: 10.1016/j.fct.2019.110924
45. Brodie EJ, Infantino S, Low MSY, and Tarlinton DM. Lyn, lupus, and (B) lymphocytes, a lesson on the critical balance of kinase signaling in immunity. Front Immunol. (2018) 9. doi: 10.3389/fimmu.2018.00401
46. Lefaudeux D, De Meulder B, Loza MJ, Peffer N, Rowe A, Baribaud F, et al. U-BIOPRED clinical adult asthma clusters linked to a subset of sputum omics. J Allergy Clin Immunol. (2017) 139:1797–807. doi: 10.1016/j.jaci.2016.08.048
47. Fattet L, Jung HY, Matsumoto MW, Aubol BE, Kumar A, Adams JA, et al. Matrix rigidity controls epithelial-mesenchymal plasticity and tumor metastasis via a mechanoresponsive EPHA2/LYN complex. Dev Cell. (2020) 54:302–316.e7. doi: 10.1016/j.devcel.2020.05.031
48. Li N, Lin G, Zhang H, Sun J, Gui M, Liu Y, et al. Lyn attenuates sepsis-associated acute kidney injury by inhibition of phospho-STAT3 and apoptosis. Biochem Pharmacol. (2023) 211:115523. doi: 10.1016/j.bcp.2023.115523
49. Islam R, Choudhary HH, Zhang F, Mehta H, Yoshida J, Thomas AJ, et al. Microglial TLR4-Lyn kinase is a critical regulator of neuroinflammation, Aβ phagocytosis, neuronal damage, and cell survival in Alzheimer’s disease. Sci Rep. (2025) 15:11368. doi: 10.1038/s41598-025-96456-y
50. Louvrier C, El Khouri E, Grall Lerosey M, Quartier P, Guerrot AM, Bader Meunier B, et al. De novo gain-of-function variations in LYN associated with an early-onset systemic autoinflammatory disorder. Arthritis Rheumatol (Hoboken N.J.). (2023) 75:468–74 doi: 10.1002/art.42354.
51. Kashiwakura JI, Kawahara S, Inagaki I, Inui K, Saitoh K, Kagohashi K, et al. STAP-2 negatively regulates BCR-mediated B cell activation by recruiting tyrosine-protein kinase CSK to LYN. FEBS Lett. (2023) 597:2433–45. doi: 10.1002/1873-3468.14730
52. L’Estrange-Stranieri E, Gottschalk TA, Wright MD, and Hibbs ML. The dualistic role of Lyn tyrosine kinase in immune cell signaling: implications for systemic lupus erythematosus. Front Immunol. (2024) 15:1395427. doi: 10.3389/fimmu.2024.1395427
53. Nguyen JA, Orsetti TL, Vernon P, Greene CJ, McKenna N, and Yates RM. Direct neutrophil and T cell contact with macrophages induces release of phagosomally processed PAMPs via eructophagy. J Cell Sci. (2025) 138. doi: 10.1242/jcs.263731
54. Ma J, Abram CL, Hu Y, and Lowell CA. CARD9 mediates dendritic cell-induced development of Lyn deficiency-associated autoimmune and inflammatory diseases. Sci Signaling. (2019) 12. doi: 10.1126/scisignal.aao3829
55. Yoo SK, Starnes TW, Deng Q, and Huttenlocher A. Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature. (2011) 480:109–12. doi: 10.1038/nature10632
56. Wu UI and Holland M. Host susceptibility to non-tuberculous mycobacterial infections. Lancet Infect Dis. (2015) 15:968–80. doi: 10.1016/S1473-3099(15)00089-4
57. Pozo-Beltrán CF, Suárez-Gutiérrez MA, Yamazaki-Nakashimada MA, Medina-Vera I, Saracho-Weber F, Macías-Robles AP, et al. B subset cells in patients with chronic granulomatous disease in a Mexican population. Allergologia immunopathologia. (2019) 47:372–7. doi: 10.1016/j.aller.2019.03.005
58. He Y, Li D, Zhang M, and Li F. Bioinformatic analysis reveals the relationship between macrophage infiltration and Cybb downregulation in hyperoxia-induced bronchopulmonary dysplasia. Sci Rep. (2024) 14:20089. doi: 10.1038/s41598-024-70877-7
59. Chen CJ, Wang HC, Hou YC, Wu YY, Shieh CC, and Shan YS. Blocking M2-like macrophage polarization using decoy oligodeoxynucleotide-based gene therapy prevents immune evasion for pancreatic cancer treatment. Mol Cancer Ther. (2024) 23:1431–45. doi: 10.1158/1535-7163.MCT-23-0767
60. Cotzomi-Ortega I, Rosowski EE, Wang X, Sanchez-Zamora YI, Lopez-Torres JM, Sanchez-Orellana G, et al. Neutrophil NADPH oxidase promotes bacterial eradication and regulates NF-κB-Mediated inflammation via NRF2 signaling during urinary tract infections. Mucosal Immunol. (2025) 18:402–17. doi: 10.1016/j.mucimm.2024.12.010
61. Kim JS, Kim HK, Kim M, Jang S, Cho E, Mun SJ, et al. Colon-targeted eNAMPT-specific peptide systems for treatment of DSS-induced acute and chronic colitis in mouse. Antioxidants (Basel Switzerland). (2022) 11. doi: 10.3390/antiox11122376
62. Xiong G, Hu H, Zhang H, Zhang J, Cao Z, Lu H, et al. Cyhalofop-butyl exposure induces the severe hepatotoxicity and immunotoxicity in zebrafish embryos. Fish shellfish Immunol. (2023) 134:108644. doi: 10.1016/j.fsi.2023.108644
63. Finisguerra V, Prenen H, and Mazzone M. Preclinical and clinical evaluation of MET functions in cancer cells and in the tumor stroma. Oncogene. (2016) 35:5457–67. doi: 10.1038/onc.2016.36
64. Bradley CA, Salto-Tellez M, Laurent-Puig P, Bardelli A, Rolfo C, Tabernero J, et al. Targeting c-MET in gastrointestinal tumours: rationale, opportunities and challenges. Nat Rev Clin Oncol. (2018) 15:150. doi: 10.1038/nrclinonc.2018.13
65. Guo R, Luo J, Chang J, Rekhtman N, Arcila M, and Drilon A. MET-dependent solid tumours - molecular diagnosis and targeted therapy. Nat Rev Clin Oncol. (2020) 17:569–87. doi: 10.1038/s41571-020-0377-z
66. Stakenborg M, Verstockt B, Meroni E, Goverse G, De Simone V, Verstockt S, et al. Neutrophilic HGF-MET signalling exacerbates intestinal inflammation. J Crohn’s colitis. (2020) 14:1748–58. doi: 10.1093/ecco-jcc/jjaa121
67. Guo J, Wang S, Xia H, Shi D, Chen Y, Zheng S, et al. Cytokine signature associated with disease severity in COVID-19. Front Immunol. (2021) 12. doi: 10.3389/fimmu.2021.681516
68. Benkhoucha M, Tran NL, Breville G, Senoner I, Bradfield PF, Papayannopoulou T, et al. CD4(+)c-Met(+)Itgα4(+) T cell subset promotes murine neuroinflammation. J Neuroinflamm. (2022) 19:103. doi: 10.1186/s12974-022-02461-7
69. Hosonuma M, Sakai N, Furuya H, Kurotaki Y, Sato Y, Handa K, et al. Inhibition of hepatocyte growth factor/c-Met signalling abrogates joint destruction by suppressing monocyte migration in rheumatoid arthritis. Rheumatol (Oxford England). (2021) 60:408–19. doi: 10.1093/rheumatology/keaa310
70. Moransard M, Sawitzky M, Fontana A, and Suter T. Expression of the HGF receptor c-met by macrophages in experimental autoimmune encephalomyelitis. Glia. (2010) 58:559–71. doi: 10.1002/glia.20945
71. Lorenc VE, Lima ESR, Hackett SF, Fortmann SD, Liu Y, Campochiaro PA, et al. Hepatocyte growth factor is upregulated in ischemic retina and contributes to retinal vascular leakage and neovascularization. FASEB bioAdvances. (2020) 2:219–33. doi: 10.1096/fba.2019-00074
Keywords: recurrent spontaneous abortion, cybb, lyn, inflammation, immunity
Citation: Wu Z, Lin Q, Zhou Z, Xie Y, Huang L, Sheng L, Shi Q and Ke Y (2025) LYN and CYBB are pivotal immune and inflammatory genes as diagnostic biomarkers in recurrent spontaneous abortion. Front. Immunol. 16:1568536. doi: 10.3389/fimmu.2025.1568536
Received: 06 February 2025; Accepted: 03 June 2025;
Published: 07 July 2025.
Edited by:
Roberta Bulla, University of Trieste, ItalyReviewed by:
Fernando Gómez-Chávez, National Polytechnic Institute (IPN), MexicoMihaela Andreescu, Colentina Clinical Hospital, Romania
Copyright © 2025 Wu, Lin, Zhou, Xie, Huang, Sheng, Shi and Ke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yumin Ke, Mzk4MDMxMzEzQHFxLmNvbQ==; Qirong Shi, d3NxeTIxNEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Zhuna Wu
Zhuna Wu Qiuya Lin†
Qiuya Lin† Liying Sheng
Liying Sheng Qirong Shi
Qirong Shi Yumin Ke
Yumin Ke