- 1The Roslin Institute and Royal (Dick) School of Veterinary Studies, University of Edinburgh, Edinburgh, United Kingdom
- 2Division of Biomedical and Life Sciences, Faculty of Health and Medicine, Lancaster University, Lancaster, United Kingdom
Toxoplasma gondii, a zoonotic apicomplexan that infects over a billion people worldwide, can cause early death in immunocompromised individuals and defects in foetal brain development. Toxoplasma is also a major cause of abortion in small ruminants. When Toxoplasma encounters host cells, several outcomes are possible. For example, the parasite can enter the host cell or can inject its effector proteins into the cell without entering. These heterogenous outcomes occur simultaneously in the same host and likely determine disease pathogenesis. Yet, current knowledge of host-Toxoplasma interactions is largely based on averaged responses in bulk cell populations. Here, we employed single cell RNA (scRNA) and bulk RNA sequencing to investigate the transcriptional profiles that underpin heterogenous host-Toxoplasma interaction in human peripheral blood mononuclear cells. We observed that Toxoplasma preferentially infects and elicits transcriptional responses in dendritic cells in human blood. Additionally, we observed that monocytes adopt a dendritic cell-like transcriptional profile over the course of infection. Using genes expressed in sorted host cell populations representative of the different heterogenous host-Toxoplasma interaction outcomes as a reference panel, we show that genes expressed in cells infected via phagocytosis are largely expressed in dendritic cells. Thus, by integrating scRNA and bulk RNA sequencing, our study unveils the transcriptional profiles of diverse Toxoplasma-host cell interaction outcomes, providing novel avenues for targeted investigations into host gene functions during Toxoplasma infections.
1 Introduction
Toxoplasma gondii is a protozoan that infects virtually all warm-blooded animals (1, 2), including over a billion people worldwide (3, 4). When Toxoplasma interacts with phagocytic host cells, there are several possible outcomes. For example, the parasite can infect the cells via active invasion to form a parasitophorous vacuole (PV) or via phagocytosis to temporarily live in a phagosome (5–7). Besides infecting host cells, Toxoplasma can also inject its effector proteins into cells it does not invade (uninfected-injected host cells) (8). Thus, the outcome of Toxoplasma infection is a consequence of several heterogenous host-pathogen interaction outcomes that occur simultaneously within a host. For example, previous experiments showed that only human monocytes that phagocytose Toxoplasma secrete IL12 (7), which is needed to induce interferon gamma that is indispensable to effective host responses to Toxoplasma. However, other studies suggest that monocytes largely express CCL2 in response to the parasite (9). We propose that these variable observations are due to averaging heterogenous host responses in bulk cell populations and can be resolved by investigating host-Toxoplasma interactions at the single cell level.
Single cell RNA sequencing (scRNA-seq) is used to transcriptionally profile individual cells in composite cell populations (10). Previously, we used dual scRNA-seq to capture the transcriptome of both the host cell and Toxoplasma to reveal transcriptional segregation of human monocytes exposed to Toxoplasma for one hour (11), indicating that the transcriptional response to the parasite is driven by a subset of exposed cells. Nevertheless, the study was limited in scope as it involved only monocytes and a single infection time point. Here, we extended our previous study to understand the transcriptional programs that underpin heterogenous Toxoplasma interactions with human peripheral blood mononuclear cells (PBMCs) using a combination of dual scRNA-seq and bulk RNA-seq. We show that Toxoplasma preferentially infects dendritic cells (DCs) in human blood, triggering distinct transcriptional responses that vary not only between infected and bystander DCs but also among different DC subclusters. These findings suggest that Toxoplasma infection orchestrates a complex transcriptional modulation in DCs, influencing cellular responses and immunological signalling within the microenvironment. Additionally, our data show that monocytes transition to a DC-like transcriptional profile in response to infection, highlighting the potential adaptive changes in cell populations. By integrating scRNA and bulk RNA-seq, our study reveals divergent transcriptional responses within infected cells, and diverse roles of DCs during Toxoplasma infection.
2 Materials and methods
2.1 Isolation of human peripheral blood mononuclear cells
Whole blood was collected with written informed consent, according to the guidelines of, and with approval from, the Roslin Institute’s Health and Safety committee from healthy adult donors seronegative for Toxoplasma. PBMCs were isolated from whole blood using a standard Ficoll gradient protocol.
2.2 Parasites and infection
Tachyzoites of a type 1 Toxoplasma gondii (RH strain) were maintained by serial passage on human foreskin fibroblasts (HFFs). Parasites were grown in RPMI (Life Technologies) supplemented with 10% foetal bovine serum (FBS; Omega Scientific), 2 mM glutamine (Sigma), 10 mM HEPES (pH 7.5; Sigma), and 20 μg/ml gentamicin at 37°C in 5% CO2. For all infections, parasites were prepared by scraping heavily vacuolated HFFs, followed by syringe lysis. The released parasites were pelleted by centrifugation at 572 x g for 7 min, washed in phosphate-buffered saline (PBS; Life Technologies), filtered through a 5μm membrane to exclude host cell debris, and counted. The parasites were added to 2 x 106 PBMCs in 6-well tissue culture plates at an intended multiplicity of infection (MOI) of 1, briefly centrifuged to bring the cells and parasites into contact and incubated at 37°C in 5% CO2 for 1h to allow infection. The cells were then rinsed twice in PBS to remove extracellular parasites and incubated in fresh RPMI media supplemented with 10% FBS at 37°C in 5% CO2 for 12h and 24h before processing for single cell RNA sequencing. Naïve uninfected cells were used as controls at each time point.
2.3 Single cell RNA library preparation and sequencing
Naïve (control), 12h, and 24h Toxoplasma-exposed human peripheral blood mononuclear cells from three healthy adult donors (n = 3 per group) were separately processed for 10X scRNA sequencing according to the manufacturer’s recommendation (10X Genomics 3’ version 3). Due to a lack of sufficient live cells, we were unable to sequence one of the 12h replicates. All scRNA-seq library preparations and sequencing were performed at the Institute of Genetics and Cancer (IGC) and the Wellcome Clinical Research Facility, respectively, at the University of Edinburgh.
2.4 scRNA-seq analysis
Raw scRNA-seq read counts were obtained using Cell Ranger v7.0.0 (12) based on the human (GRCh38, GENCODE) and Toxoplasma (ToxoDB-61) genomes and downstream analyses were performed in R 4.4.1 using Seurat v3 (13). Cells expressing less than 1000 genes and 1000 transcripts or more than 10% mitochondrial genes were excluded. Cell cycle scores (G2M and S scores) were estimated using canonical cell cycle marker genes obtained from biomaRt R package (14).
Normalisation was performed using SCTransform (15) by regressing out the effects of mitochondrial, ribosomal, and cell cycle genes before integrating the datasets. The number of principal components (PC) was determined using an elbow plot by plotting the PC standard deviation. Dimensionality reduction was performed with Uniform Manifold Approximation and Projection (UMAP) using the 20 PC and a resolution of 0.2.
2.5 scRNA-seq cluster annotation
The naive samples were subset and the markers for each cluster identified using the FindAllMarkers function in Seurat (Supplementary Table 1). Cell types were determined by combining supervised and unsupervised cell annotation approaches. For the unsupervised method, R packages such as scType (16), Azimuth (13), and SingleR (17) were used based on their built-in cell type reference gene database. We used ‘Tissue = Immune system’ reference for scType, the ‘Human PBMC’ reference for Azimuth, and the following datasets for SingleR: HumanPrimaryCellAtlasData, BluePrintEncodeData, DatabaseImmuneCellExpressionData, NovershternHematopoieticData, and MonacoImmuneData.
DCs were subset and the FindVariableFeatures, RunUMAP, FindNeighbors, and FindClusters features in Seurat used to identify variable features and to re-cluster the cells. We then performed pathway analysis using Metascape (18) to determine biological pathways enriched in different scRNA-seq cell cluster. Pseudotime trajectory analysis was performed in Monocle3 (19) using default parameters.
2.6 Bulk RNA library preparation
Due to the difficulty to fluorescently sort sufficient PBMCs representing the different host cell-Toxoplasma interaction outcomes (e.g., actively invaded or uninfected), we used the human monocytic cell line THP-1, which has been shown to mimic primary monocyte response to Toxoplasma (9). THP-1 cells were exposed for 12h to GFP-expressing RH parasites pre-labelled with pHrodo Red, a dye that fluoresces red in acidic conditions. 105 cells representing each of the three populations: uninfected cells (GFP- pHrodo-), cells infected through active invasion (GFP+), and cells infected via phagocytosis (GFP+ pHrodo+) were then sorted in cytometry. Toxoplasma can inject its effector proteins into host cells without infecting them, in a process known to rely on the contact between the parasite and host cell (8). Therefore, we also used naïve cells separated by a transwell membrane from cells exposed to Toxoplasma to generate true bystander cells that are neither infected nor injected with parasite effector proteins but still exposed to the host cell or parasite effectors secreted into the cell culture media. RNAs from naïve (unexposed), transwell, and sorted cell populations were separately extracted using RNA extraction kits (Qiagen) and processed for bulk RNA sequencing.
2.7 Bulk RNA-sequencing analysis
Transcript level quantification was performed by pseudoaligning the reads to both the human (release 42, GRCh38.p13, GENCODE) and Toxoplasma (TgondiiGT1, ToxoDB-61) transcripts using kallisto v0.44.0 (20). The transcripts were summed to gene-level counts using only the human gene annotation file (gtf). Approximately ~261,000 transcripts and ~ 63,000 genes were analysed. Differential gene expression was performed using DESeq2 v1.44.0 R package (21) by comparing each cell sub-population (infection outcome) relative to all other sub-populations. Genes with an adjusted p-value < 0.05 and log2FC > 2 were considered significant. Multiple testing correction was done using the Benjamini-Hochberg method. Pathway analysis was performed in Metascape (Supplementary Table 7) (18).
3 Results
3.1 Cells expressing dendritic cell markers significantly increase in Toxoplasma-exposed PBMCs
PBMCs can provide a window into the complexity of the immune system and can be used to access the effect of different pathogens on the immune system. Thus, we characterised the transcriptional responses to infection in an heterogenous cell population by performing single cell RNA sequencing (scRNA-seq) analysis of human PBMCs exposed to Toxoplasma. To do this, freshly isolated PBMCs from three Toxoplasma-naïve donors were separately exposed to a clonal type I Toxoplasma strain (RH) at an intended MOI of 1 for 12h and 24h then processed for scRNA-seq analysis. Naïve (unexposed) cells were used as controls. Quality control, data normalisation, and doublets exclusion in Seurat v4.2.0 (Materials and Methods), yielded a total of 31016 high quality cells from the three donors, of which 10887, 6438, and 13691 were from the control, 12h, and 24h samples, respectively (Supplementary Table 2). Control, 12h and 24h cells had a median of 2595, 2558, and 2349 genes per cell, respectively (Supplementary Table 2). Unsupervised dimensionality reduction, clustering, and visualisation with uniform manifold approximation and projection (UMAP) identified six clusters (Figure 1A). Among these three clusters represented a heterogenous mix of monocytes (MS4A7, S100A9, CD36, CSF3R, CD81), including classical monocytes (CD14) and non-classical monocytes (FCGR3A) (Figures 1A–C). The remaining three clusters were identified as dendritic cells (CD86, CLEC4A, CD81), T cells (CCL5, NKG7), and B cells (IGHM, CD79A), based on the expression of known cell type specific markers (Figures 1A, C). All clusters contained cells from each donor, indicative of an absence of significant donor effects (Supplementary Figures S1, S2, Supplementary Table 3). However, the cell type compositions, particularly dendritic cells (DCs), changed upon Toxoplasma exposure (Figure 1D). Cell expressing DC gene markers increased from ~5% in control (unexposed) to ~30% and ~33% in 12h- and 24h-Toxoplasma-exposed cells, respectively (Figure 1D, Supplementary Table 4). The increase in cells expressing DC markers was accompanied by a decrease in cells expressing monocyte markers from ~92% in control (unexposed) to ~67% and ~65% at 12h- and 24h post-exposure cells, respectively. No significant changes in cell population were observed for the other cell types identified in the scRNA-seq data. This suggests that monocytes might acquire a DC-like marker expression phenotype in the course of Toxoplasma infection of human blood.
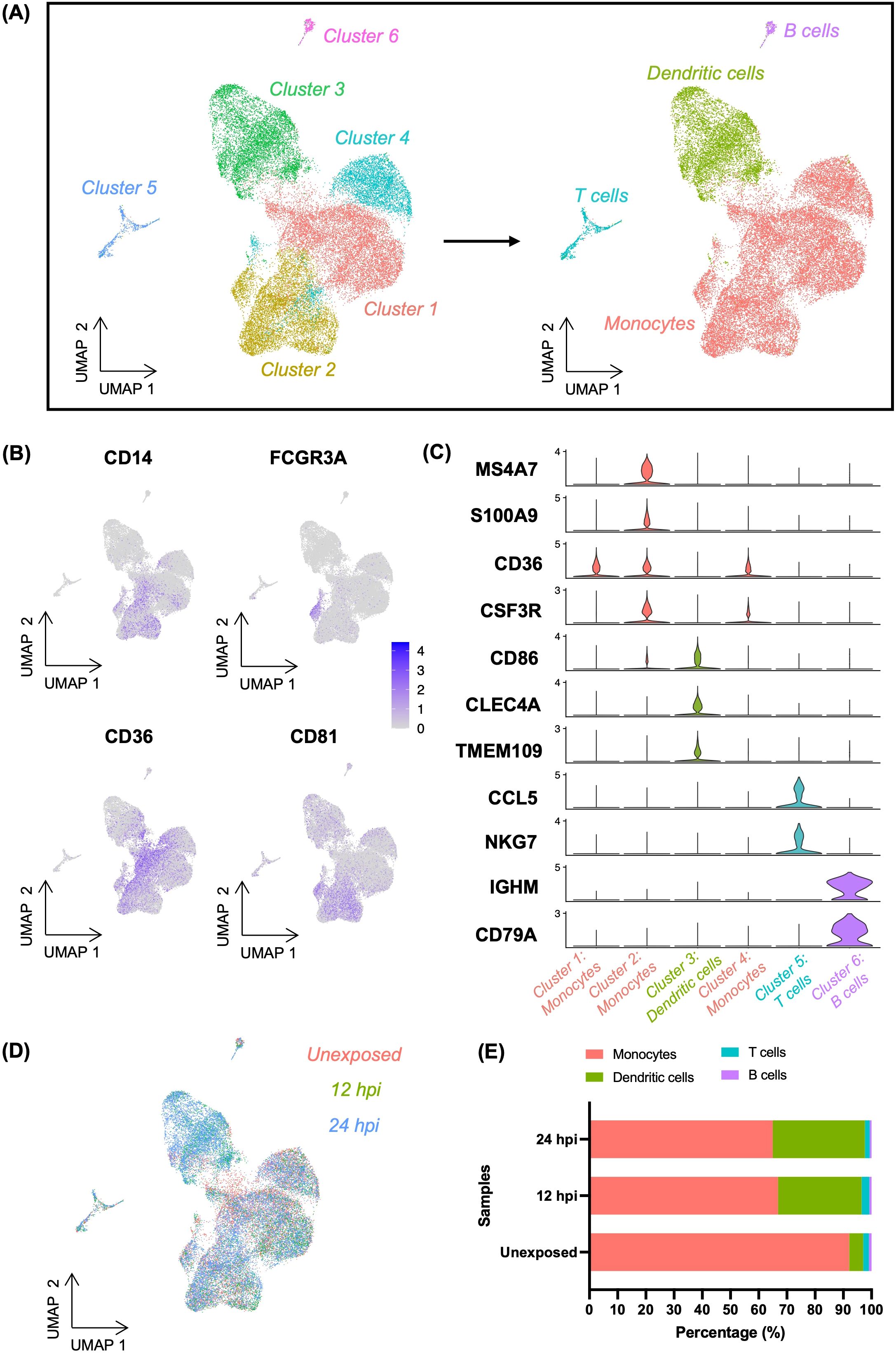
Figure 1. Classification of cells in Toxoplasma-exposed and unexposed human PBMCs. (A) UMAP projection of combined PBMCs at resolution 0.2, identifying six distinct cell clusters (left). Among these, three clusters were labelled as monocytes, which were further resolved into four distinct cell types (right). (B) Feature plots showing the expression of CD14 (classical monocyte marker) and FCGR3A (non-classical monocyte marker) as well as CD36 and CD81 (canonical monocyte markers) within the heterogenous monocytes cluster. (C) Violin plots showing the expression levels of cell type-specific gene markers across the four identified cell types. (D) UMAP depicting the distribution of cells categorised by experiment samples (Unexposed, 12hpi, and 24hpi) (E) Bar plot representing the percentage composition of each cell type across experimental samples.
3.2 Heterogenous transcriptional responses and Toxoplasma burden across dendritic cell subclusters
Previously, we reported that a subset of CD16- monocytes drives the transcriptional response in monocytes exposed to Toxoplasma for 1h (11). Here, we sought to determine whether, in the presence of other myeloid cell types, monocytes are still the dominant cell type regulating transcriptional response to the parasite over time. Through an integrative analysis of the exposed and unexposed samples, followed by clustering and visualisation in UMAP, we identified DCs as the major cell type transcriptionally distinguishing Toxoplasma-exposed and unexposed cells (Figures 2A, B, Supplementary Figure 3). Previously, DC were reported to be the major myeloid cell type responding to Toxoplasma, based on the secretion of IL12, in human blood (7). Thus, we subset the exposure-distinguishing DC population and performed dimensionality reduction to investigate if the transcriptional response of DCs to infection was homogenous. We identified four transcriptionally distinct DC subclusters, DC1-4 (Figure 2C, Supplementary Tables 5, 6), suggesting heterogenous transcriptional response within the infection-distinguishing DC subset. Interestingly, we observed significant differences in the proportion of Toxoplasma transcripts across DC subclusters. At 12h post-exposure DC4 exhibited significantly lower parasite transcript levels compared to DC1-3, suggesting differential susceptibility to infection or parasite transcriptional activity in cells of the same cell type (Figure 2D). By 24hpi, all DC subclusters showed significant differences in parasite transcript abundance, indicating dynamic changes in infection burden or parasite transcriptional activity over time (Figure 2D).
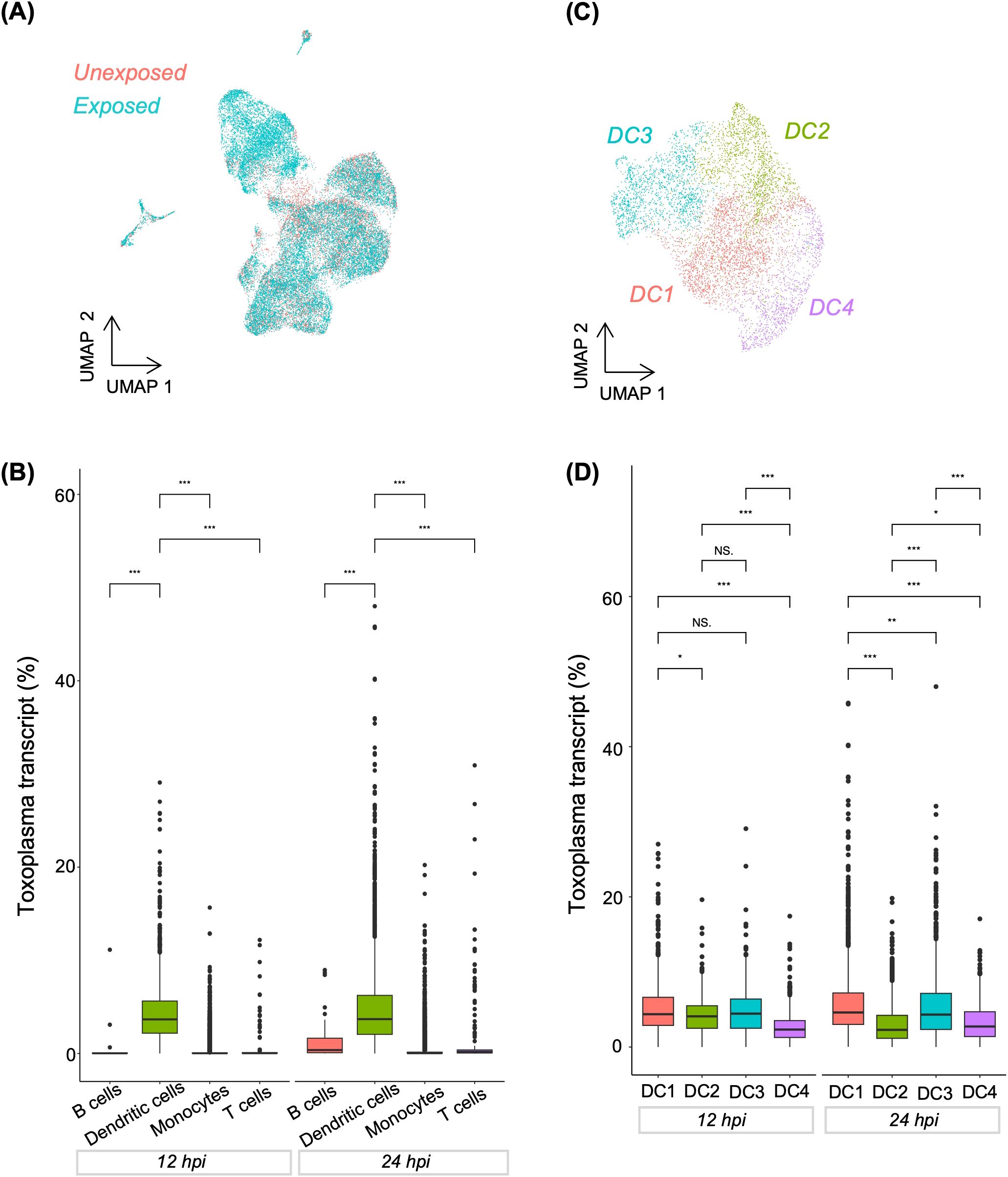
Figure 2. Distribution of Toxoplasma-infected cells in combined PBMCs and dendritic cell sub-clusters. (A) UMAP showing the distribution of cells categorised by experimental group (Unexposed and exposed). (B) Boxplot showing Toxoplasma transcript abundance across major immune cell types at 12 and 24hpi. (C) UMAP highlighting four sub-clusters within dendritic cell population. (D) Boxplot showing Toxoplasma transcript abundance across dendritic cell sub-clusters at 12 and 24hpi. (B, D) A Kruskal-Wallis test was used to assess differences in Toxoplasma transcript within each exposure group. Pairwise comparisons between cell types at each time were performed using the Wilcoxon rank-sum test. Asterisks denote statistical significance: ***p < 0.001, **p < 0.01, *p<0.05, and NS, not significant.
Response to infection can be driven by infected and bystander cells. Thus, we investigated whether the transcriptional heterogeneity in Toxoplasma-exposed DCs was determined by “infected” or “bystander” status. To distinguish infected and bystander DCs, we used similarity in cell barcodes to match each infected cells with its cognate parasite RNA. We arbitrarily considered cells whose total RNA counts consisted of more than 1% Toxoplasma RNA transcripts as infected (Figure 3A). Pathway analysis performed for exposed DCs using genes expressed in infected and bystander cells revealed that both bystander and infected DCs shared enrichment in fundamental cellular processes, such as metabolism and cell cycle regulation (Figure 3B).
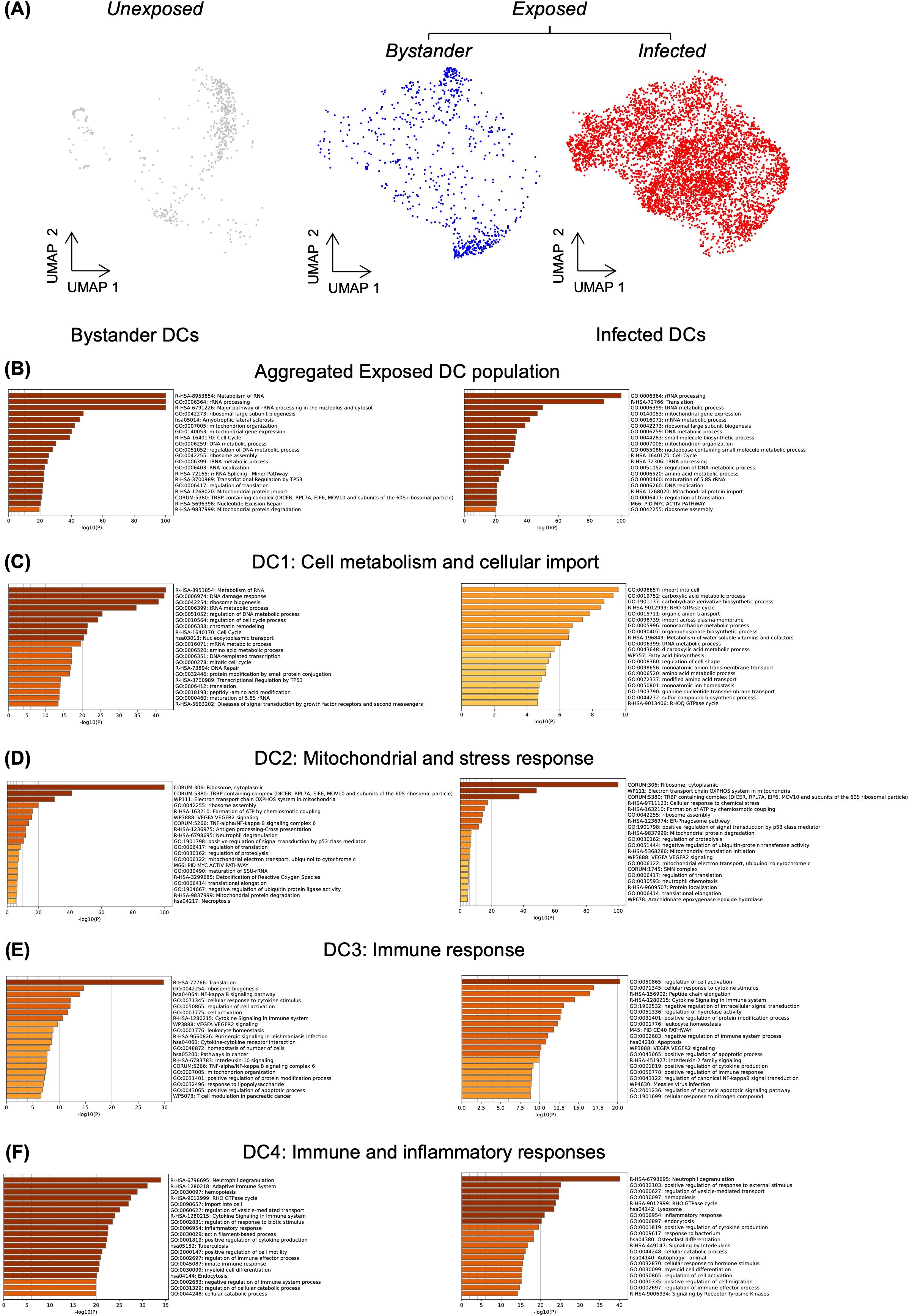
Figure 3. Differential biological pathway enrichment between dendritic cell sub-clusters. (A) UMAP showing unexposed and Toxoplasma-exposed dendritic cells. Within the exposed population, cells with less than 1% Toxoplasma RNA transcripts are classified as bystander cells, while those with more than 1% Toxoplasma RNA transcripts are classified as infected cells. Pathway enrichment analysis was performed exclusively on exposed cells using gene markers identified for bystander (left) and infected (right) dendritic cells (B) Analysis of aggregated exposed DC population. (C-F) Analysis of individual DC sub-populations (DC1-DC4), with pathway enrichment performed using Metascape.
Differential pathway enrichment was assessed between infected and bystander DCs within each subcluster. In DC1, both bystander and infected cells showed similar enrichments, suggesting potential involvement in homeostasis and metabolic pathways (Figure 3C). In DC2, bystander and infected cells demonstrated strong overlap, with both groups enriched for mitochondrial functions and ribosome assembly pathways (Figure 3D). The shared enrichment may reflect a general stress response to Toxoplasma exposure. In DC3, both bystander and infected cells showed enrichment in cytokine-related and immune regulation pathways (Figure 3E). For DC4, bystander cells were enriched in pathways related to the adaptive immune response and cytokine signalling (Figure 3F). However, infected DC4 cells showed enrichment in lysosome and endocytosis pathways (Figure 3F), which may suggest an involvement in phagocytic activities. The differential responses observed across DC subclusters indicate complex transcriptional changes following Toxoplasma infection. These changes may reflect both immune activation and responses to infection. However, more target experiments are necessary to establish the functional relevance of these findings.
3.3 Monocytes shift to a dendritic cell-like transcriptional response during Toxoplasma infection
We observed a decline in cells expressing monocyte markers, which was accompanied by an increase in cells expressing DC markers. Thus, to investigate whether the transcriptional response of monocytes changes to DC-like response during long term exposure to Toxoplasma, we conducted a pseudotime analysis using Monocle3. Identifying this transcriptional shift could offer insight into how the immune landscape adapts during Toxoplasma infection. To establish the starting point of the pseudotime trajectory, we selected nodes representing classical monocytes, which serve as progenitors for various immune cell subtypes (22). We examined cells within the monocytes cluster to identify those expressing classical monocytes markers - CD14+ and FCGR3A- (Figure 1B). These CD14+FCGR3A- cells were designated as the root nodes, establishing an origin point for tracking differentiation trajectories in the pseudotime analysis. Anchoring on these classical monocytes allowed us to map lineage-specific trajectories from monocytes to potential DC fates. The UMAP trajectory plot indicated a clear pathway from monocytes to DCs, suggesting that DCs could arise from the monocyte cluster over pseudotime (Figure 4A). To further validate this trajectory, we analysed the expression of monocyte- and DC-specific markers across pseudotime. Monocyte markers such as CD14, LYZ, and S100A9 exhibited high expression at early pseudotime, predominantly localised within the monocyte cluster (Figure 4B). However, as cells progressed along the pseudotime trajectory towards DC fates, the expression of these markers gradually declined (Figure 4B). In contrast, DC markers, such as TXN and CLEC4, showed a progressive increase in expression along the trajectory, with their highest levels observed in differentiated DCs (Figure 4B). Overall, this pseudotime analysis supports a model where Toxoplasma infection induces a shift in monocyte transcriptional response towards a DC-like transcriptional profile. These DCs likely play a critical role in antigen presentation and pathogen clearance, thus, potentially serving as pivotal cells in shaping the host’s adaptive immune response to the infection.
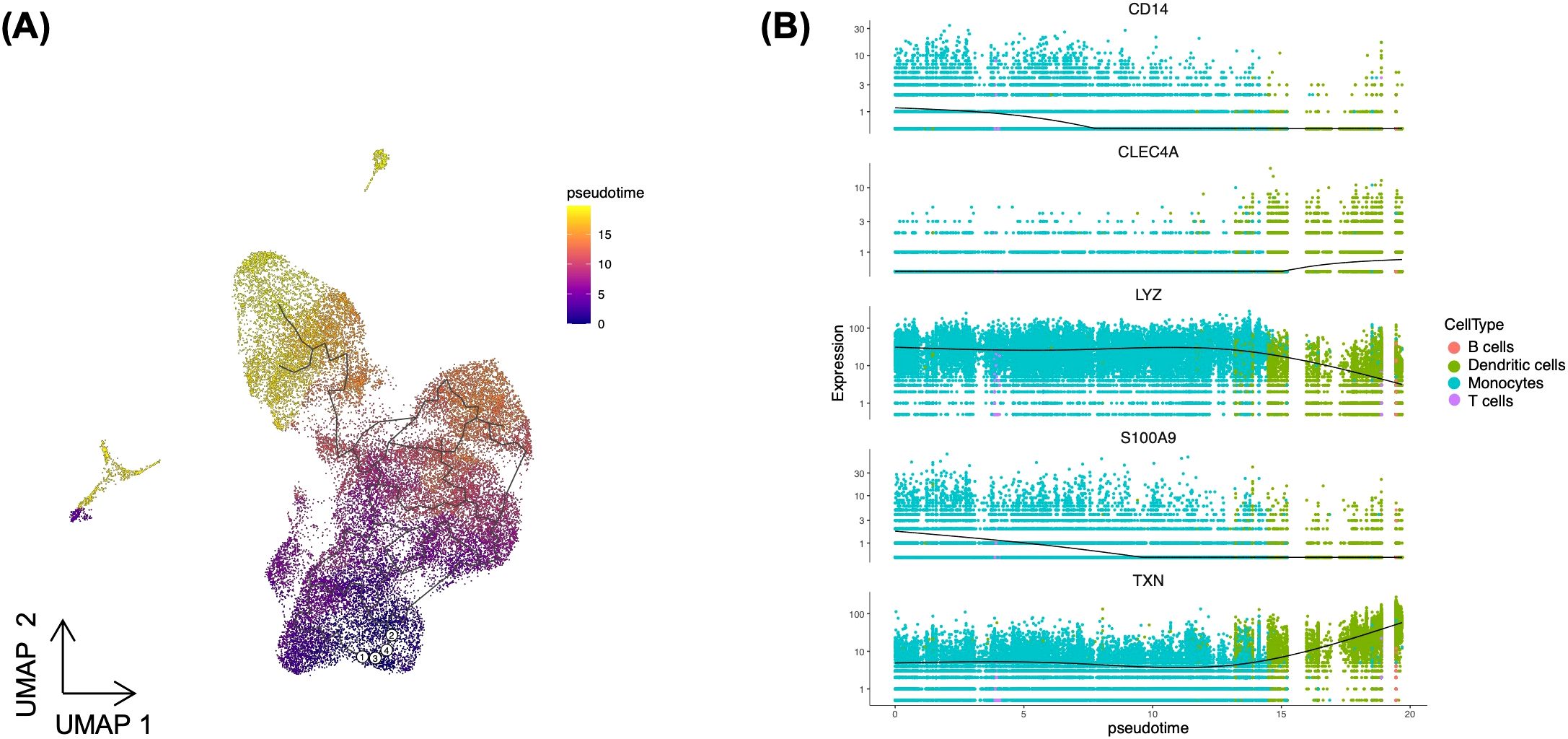
Figure 4. Pseudotime analysis illustrating dendritic cells differentiation from monocytes. (A) UMAP displaying the pseudotime trajectory of combined PBMCs. White circles (1-4) indicate root nodes representing classical monocytes, while black line depicts the trajectory. (B) Expression dynamics of dendritic cell markers (CLEC4A and TXN) and monocyte markers (CD14, LYZ, and S100A9) along the pseudotime trajectory. The black line shows smoothed average expression across pseudotime.
3.4 Transcriptional heterogeneity is observed in Toxoplasma-exposed PBMCs
Toxoplasma interactions with phagocytic host cells can result in cells that are: (i) infected via active parasite invasion; (ii) infected via phagocytic uptake of parasite; (iii) uninfected but contact-dependently injected with parasite effectors (rhoptry proteins) (“uninfected-injected”); and (iv) neither infected nor injected (“uninfected-uninjected”). These heterogenous host-pathogen interaction outcomes, which occur simultaneously within a host, are potentially underpinned by distinct transcriptional programs. Although we were able to analyse individual host cell transcriptomes from the scRNA-seq, it was not possible to determine whether the transcriptional separation at the single cell level is driven by the heterogeneous host-parasite interactions. Thus, in an attempt at differentiating these heterogenous infection outcomes in the scRNA-seq, we separately sorted cells representing the different infection outcomes and performed bulk RNA-sequencing to obtain the corresponding reference genes. Briefly, THP-1, a human monocytic cell line that has previously been shown to mimic primary monocyte response to Toxoplasma (23, 24), was infected with type I (RH) Toxoplasma that constitutively express GFP and pre-labelled with pHrodo. Through FAC sorting, we obtained uninfected cells (GFP- pHrodo-), cells infected via active invasion (GFP+), or phagocytosis (GFP+ pHrodo+) (Materials and methods). Because GFP- pHrodo- (uninfected) cells can consist of both “uninfected-injected” and “uninfected-uninjected” cells, we used cells in a transwell, which abrogates contact between THP-1 and Toxoplasma, to generate the “uninfected-uninjected” cells. Total RNAs from naïve (unexposed), transwell, and the different sorted subpopulation cells were extracted and separately processed for bulk RNA-sequencing.
To identify a panel of reference genes for each sorted population, we performed a differential expression analysis (DEA) by comparing one sorted subpopulation against all other subpopulations. Based on this analysis, three genes (RNU2-58P, TCAM1P, UQCRHP1) were upregulated, and one (ENSG00000289901) was downregulated in the actively invaded group (GFP+) relative to the other infection outcomes: phagocytosed (GFP+ pHrodo+), uninfected group (GFP- pHrodo-) and “uninfected-uninjected” group (transwell) (Figure 5A). Similarly, three (CCL22, KIF1A, RAET1K) genes were upregulated, while MIR8485 was downregulated in the phagocytosed (GFP+ pHrodo+) population (Figure 5B). Rudzki et al. showed that secretion of CCL22, an immunomodulatory chemokine, is induced by Toxoplasma GRA28, which has potential involvement in parasite dissemination. KIF1A, a kinesin-like protein, is typically associated with neurological disorder (25). RAETIK, a long non-coding RNA, lacks a known function during infection. In contrast, four (ULBP1, EGR3, RAET1K, EGR2) genes were downregulated in uninfected cells (GFP- pHrodo-) relative to the other three infection outcomes (Figure 5C). ULBP1, an NK cell receptor activator, is crucial for immunomodulation (26), while EGR2/3 are early growth response genes essential for immune homeostasis (27–29). Additionally, five genes (CCL26, MIR8485, ULBP1, CCL13, CISH) and one unannotated gene (ENSG00000289505) were downregulated in the transwell relative to the other infection groups (Figure 5D). CCL26 and CCL13 are chemokines potentially involved in triggering immune responses. CISH is known to control cytokine signals in phagosomes during bacterial infection (30, 31).
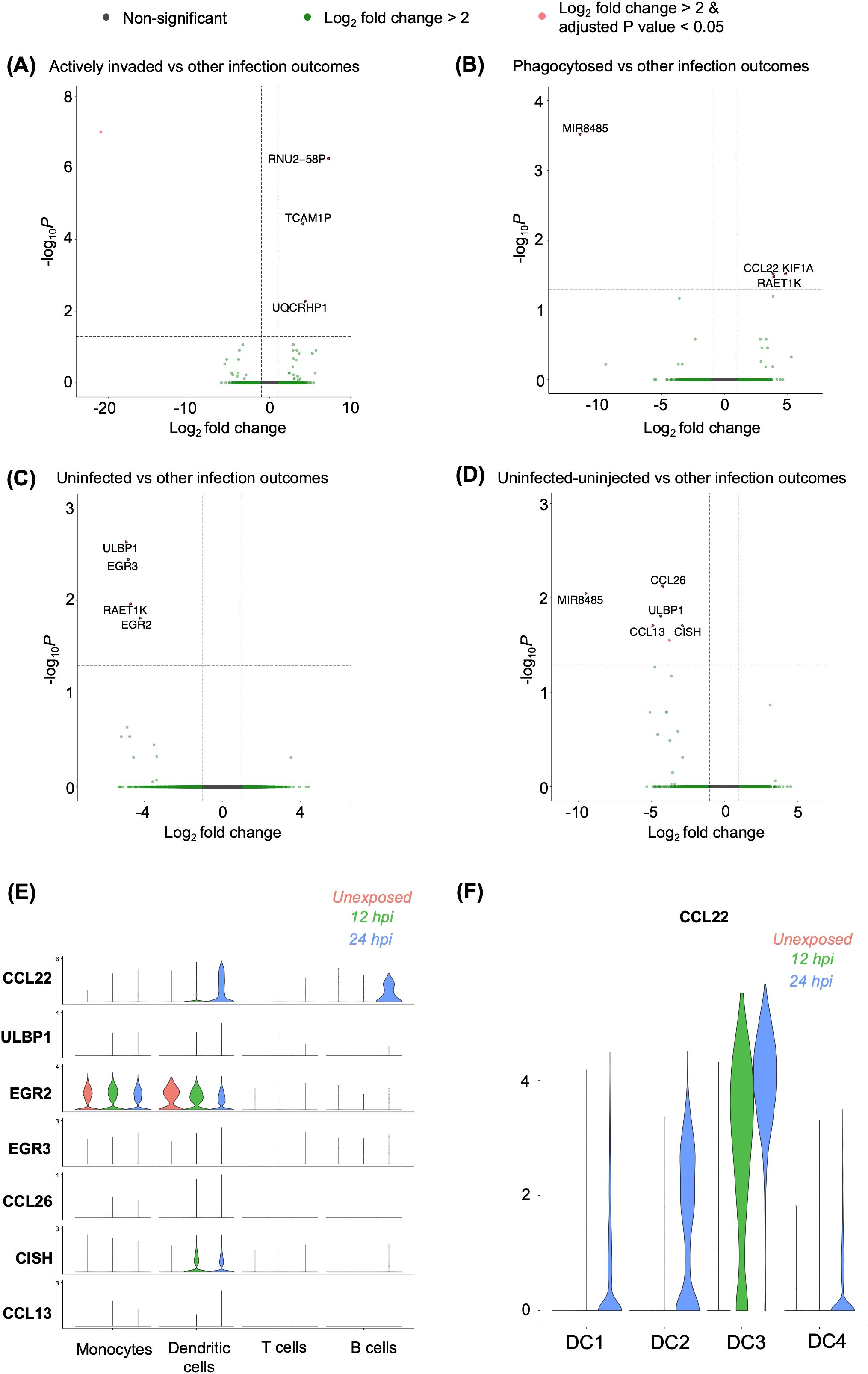
Figure 5. Differential gene expressions among heterogenous infection outcomes. (A-D) Differentially expressed genes identified in (A) actively invaded, (B) phagocytosed, (C) uninfected cells, and (D) uninfected-uninjected THP-1 cells compared to other infection outcomes, based on bulk RNA sequencing. Differential expression was determined using the criteria |Log2 fold change| > 2, and an adjusted p-value of <0.05. (E) Validation of differentially expressed genes identified in bulk RNA sequencing, shown across cell types in a single-cell RNA sequencing. (F) Expression of CCL22 within dendritic cell sub-populations.
To investigate whether different infection outcomes were overrepresented in individual cell clusters as defined by the scRNA-seq data, we analysed the expression of the differentially expressed genes from the sorted population in each cell cluster. None of the genes differentially expressed in the actively invaded group were expressed in the scRNA datasets (Figure 5A). One gene, CCL22, which is upregulated in phagocytosed group, was notably expressed in exposed DCs and B cells 24h post-infection (Figures 5B, E). Further analysis of the DC subclusters revealed that CCL22 expression was predominantly observed in infected cells in the DC3 sub-cluster compared to the other subclusters (Figure 5F). Among the downregulated genes in uninfected group (Figure 5C), we noted that EGR2 was expressed in both naïve and Toxoplasma-exposed monocytes and DCs (Figure 5E), whereas ULBP1 and EGR3 were lowly expressed in any cluster. Moreover, four genes (ULBP1, CISH, CCL26, CCL13) out of six downregulated genes in transwell were found to be lowly expressed in scRNA (Figures 5D, E). Thus, gene expression heterogeneity in Toxoplasma exposed PBMCs is largely not influenced by the heterogenous host-parasite interaction outcomes.
4 Discussion
Infection is driven by interaction between pathogens and individual host cells, resulting in different infection outcomes such as some cells becoming infected while others remain uninfected within the same individual. As such host-pathogen interactions are best studied at the single cell level. Here, we utilised a combination of dual scRNA-seq and bulk RNA-seq to (i) investigate the transcriptional profile of individual leukocytes during Toxoplasma infection, (ii) understand the transcriptional differences between bystander and infected myeloid cells, (iii) explore the origin and expansion of DCs in response to Toxoplasma exposure, and (iv) determine the transcriptional profile underpinning heterogenous Toxoplasma-immune cell interactions. Our data highlight the role of myeloid cells, especially DCs, in host response to acute Toxoplasma infection.
ScRNA-seq analyses revealed that DCs drive the transcriptional response to Toxoplasma in human blood and that monocytes adopt DC-like transcriptional profile during the course of Toxoplasma infection. These findings align with previous studies highlighting the critical role of DCs in controlling Toxoplasma infection in human blood (32–35). Previous studies have implicated monocyte-derived DCs in the pathogenesis of other parasitic infection. For instance, monocyte-derived DCs were postulated to induce T-helper cell responses to Leishmania infection (36). Similarly, monocyte-derived DCs are widely associated with inflammatory responses (37), where they are thought to contribute to both T-cell activation and innate immune responses (38). In this study, we observed increase in the proportions of cells expressing DC markers. This was accompanied by a decrease in the proportion of cells expressing monocyte markers. Subsequent pseudotime analysis showed the shift in expression of monocyte to DC markers during Toxoplasma infection. This monocyte-to-DC differentiation may represent a critical mechanism in mounting an effective immune response to the parasite. By sub-setting DCs, we did observed different infection patterns in DC subclusters. Our transcriptomic analysis further revealed that dendritic cell subclusters exhibit varying transcriptional profiles, with pathway enrichment analysis suggesting involvement in different biological processes across subclusters. This underscores the different responses of DCs to Toxoplasma infection, highlighting the complexity of host-pathogen interactions at the cellular level.
Although Toxoplasma-host cell interactions are known to simultaneously produce disparate outcomes, such as infected and uninfected-injected cells, the transcriptional programs underpinning these outcomes are equivocal. We identified differentially expressed genes in cell populations representing these different outcomes, including uninfected (cells that may have come into contact with the parasite but are not infected) and uninfected-uninjected (cells that are separated from the parasite by a transwell), highlighting potential transcriptional differences that could be missed when averaging responses across all exposed cells. Interestingly, only five differentially expressed genes in the different bulk cell populations were identified in the scRNA-seq data. This could be due to differences between THP-1, human monocytic cell line used in bulk RNA, and PBMCs used in scRNA-seq (39, 40). THP-1 contains only monocytes (40), which are professional phagocytic cells, while PBMCs have an assortment of immune cell types, including non-professional phagocytes. Additionally, while the transcriptional response in bulk cell populations is entirely based on THP-1 responses, the response in PBMCs is driven mostly by DCs. Several differentially expressed genes in both the bulk cell populations and single-cell clusters are known to modulate response to Toxoplasma infection. For example, the chemokine CCL22 is known to be induced in cells that phagocytose Toxoplasma (41, 42). Interestingly, not all DC subclusters showed increased CCL22 expression. Among the subclusters, only exposed cells within the DC3 subcluster exhibited increased CCL22 expression. Within this subcluster, both infected and bystander cells showed enrichment in immune-related pathways. This, coupled with the increased CCL22 expression, suggests that although the DC3 cluster is a dendritic cell sub-cluster, it may contain cells that phagocytose the parasite or that closely mimic the transcriptional response of monocytes that phagocytose Toxoplasma. Additionally, studies have demonstrated that EGR2 is rapidly upregulated in infected cells in response to Toxoplasma secreted factors (43). ULBP1 has been reported to control bacterial and viral infections (44, 45), suggesting its potential involvement in controlling Toxoplasma infection.
In conclusion, our scRNA-seq analysis provides insights into the diverse transcriptional responses of PBMCs during Toxoplasma infection. Dendritic cells appear to play a prominent role in the early immune response, with individual subpopulations exhibiting distinct transcriptional profiles. While comparisons with THP-1 cells offer some context, we acknowledge the limitations of comparing cell lines with primary cells. These findings underscore the importance of studying the host immune responses within diverse immune cell types. However, further experimental validation is needed to substantiate and refine the interpretations drawn from these computational analyses.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: GEO accession number GSE295224 and Zenodo DOI https://doi.org/10.5281/zenodo.15240681.
Ethics statement
The studies involving humans were approved by Roslin Institute Health and Safety Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
PC: Formal Analysis, Methodology, Writing – original draft, Writing – review & editing. BF: Methodology, Writing – review & editing. BS: Formal Analysis, Methodology, Writing – review & editing. MH: Conceptualization, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded in part by an Academy of Medical Sciences Springboard fellowship (SBF005\1127 8601884_8601897) awarded to MH, and strategic funding to the Roslin Institute by the Biotechnology and Biological Sciences Research Council (BBS/E/D/20002173). BS was partially funded by a BBSRC Core Capability Grant BB/CCG1780/1 awarded to The Roslin Institute. PC is supported by a Wellcome Hosts, Pathogens, and Global Health doctoral fellowship awarded to the University of Edinburgh.
Acknowledgments
The authors thank the Wellcome Clinical Research Facility at University of Edinburgh for providing RNA-seq services. We thank Lizzie Freyer and Angela Ingram for making the scRNA-seq libraries, Jessica Powell and Marzuq Ungogo for critical review of the manuscript drafts.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1582645/full#supplementary-material
Supplementary Figure 1 | UMAP showing cell distribution by donor across experimental groups. Cells are coloured by donor to illustrate the distribution across the unexposed, 12 and 24hpi groups.
Supplementary Figure 2 | Cell type composition across donors and experimental groups. Bar plot showing the percentage of each immune cell type for individual donors within the unexposed, 12, and 24hpi groups.
Supplementary Figure 3 | UMAP depicting Toxoplasma gondii transcript abundance. The intensity of the colour indicates the percentage of Toxoplasma transcript detected, highlighting the distribution of infected cells.
References
1. Tenter AM, Heckeroth AR, and Weiss LM. Toxoplasma gondii: from animals to humans. Int J Parasitol. 30(12-13):1217–58.
2. Dubey JP, Lindsay DS, and Speer CA. Structures of toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts †. Clin Microbiol Rev J. (1998) 11(2):267–99. doi: 10.1128/CMR.11.2.267
3. Hoffmann S, Batz MB, and Morris JG. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J Food Prot. (2012) 75:1292–302. doi: 10.4315/0362-028X.JFP-11-417
4. Montazeri M, Mikaeili Galeh T, Moosazadeh M, Sarvi S, Dodangeh S, Javidnia J, et al. The global serological prevalence of Toxoplasma gondii in felids during the last five decades, (1967-2017): A systematic review and meta-analysis. Parasites Vectors. (2020) 13(1):82. doi: 10.1186/s13071-020-3954-1
5. Dupont CD, Christian DA, Selleck EM, Pepper M, Leney-Greene M, Harms Pritchard G, et al. Parasite fate and involvement of infected cells in the induction of CD4+ and CD8+ T cell responses to toxoplasma gondii. PloS Pathog. (2014) 10:1–17. doi: 10.1371/journal.ppat.1004047
6. Zhao Y, Marple AH, Ferguson DJP, Bzik DJ, and Yap GS. Avirulent strains of Toxoplasma gondii infect macrophages by active invasion from the phagosome. Proc Natl Acad Sci United States America. (2014) 111:6437–42. doi: 10.1073/pnas.1316841111
7. Tosh KW, Mittereder L, Bonne-Annee S, Hieny S, Nutman TB, Singer SM, et al. The IL-12 response of primary human dendritic cells and monocytes to toxoplasma gondii is stimulated by phagocytosis of live parasites rather than host cell invasion. J Immunol. (2016) 196:345–56. doi: 10.4049/jimmunol.1501558
8. Koshy AA, Dietrich HK, Christian DA, Melehani JH, Shastri AJ, Hunter CA, et al. Toxoplasma co-opts host cells it does not invade. PloS Pathog. (2012) 8:18. doi: 10.1371/journal.ppat.1002825
9. Safronova A, Araujo A, Camanzo ET, Moon TJ, Elliott MR, Beiting DP, et al. Alarmin S100A11 initiates a chemokine response to the human pathogen Toxoplasma gondii. Nat Immunol. (2019) 20:64–72. doi: 10.1038/s41590-018-0250-8
10. Hwang B, Lee JH, and Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. (2018) 50:1–14. doi: 10.1038/s12276-018-0071-8
11. Patir A, Gossner A, Ramachandran P, Alves J, Freeman TC, Henderson NC, et al. Single-cell RNA-seq reveals CD16- monocytes as key regulators of human monocyte transcriptional response to Toxoplasma. Sci Rep. (2020) 10:1–10. doi: 10.1038/s41598-020-78250-0
12. Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. (2017) 8:1–12. doi: 10.1038/ncomms14049
13. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. (2021) 184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048
14. Durinck S, Spellman PT, Birney E, and Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. (2009) 4:1184–91. doi: 10.1038/nprot.2009.97
15. Hafemeister C and Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. (2019) 20:1–15. doi: 10.1186/s13059-019-1874-1
16. Ianevski A, Giri AK, and Aittokallio T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat Commun. (2022) 13:1–10. doi: 10.1038/s41467-022-28803-w
17. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. (2019) 20:163–72. doi: 10.1038/s41590-018-0276-y
18. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. (2019) 10:1–10. doi: 10.1038/s41467-019-09234-6
19. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. (2014) 32:381–6. doi: 10.1038/nbt.2859
20. Bray NL, Pimentel H, Melsted P, and Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. (2016) 34:525–7. doi: 10.1038/nbt.3519
21. Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:1–21. doi: 10.1186/s13059-014-0550-8
22. Kapellos TS, Bonaguro L, Gemünd I, Reusch N, Saglam A, Hinkley ER, et al. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front Immunol. (2019) 10(2035):1–13. doi: 10.3389/fimmu.2019.02035
23. Daigneault M, Preston JA, Marriott HM, Whyte MKB, and Dockrell DH. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PloS One. (2010) 5:1–10. doi: 10.1371/journal.pone.0008668
24. Chanput W, Mes JJ, and Wichers HJ. THP-1 cell line: An in vitro cell model for immune modulation approach. Int Immunopharmacol. (2014) 23(1):37–45. doi: 10.1016/j.intimp.2014.08.002
25. Nair A, Greeny A, Rajendran R, Abdelgawad MA, Ghoneim MM, Raghavan RP, et al. KIF1A-associated neurological disorder: an overview of a rare mutational disease. Pharmaceuticals. (2023) 16(2):147. doi: 10.3390/ph16020147
26. Cosman D, Jü Rgen Mü, Sutherland CL, Chin W, Armitage R, Fanslow W, et al. ULBPs, Novel MHC Class I-Related Molecules, Bind to CMV Glycoprotein UL16 and Stimulate NK Cytotoxicity through the NKG2D Receptor tions of most of the nonessential viral glycoproteins remain unknown. NK cells are also known to play an important role in controlling CMV infection. Patients with defective NK cell function have problems with herpesvirus infections cell activation and cytotoxicity that can override the. Immunity. (2001). 14(2):123–33. doi: 10.1016/S1074-7613(01)00095-4
27. Harris JE, Bishop KD, Phillips NE, Mordes JP, Greiner DL, Rossini AA, et al. Early growth response gene-2, a zinc-finger transcription factor, is required for full induction of clonal anergy in CD4+ T cells. J Immunol. (2004) 173:7331–8. doi: 10.4049/jimmunol.173.12.7331
28. Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat Immunol. (2005) 6:472–80. doi: 10.1038/ni1193
29. Miao T, Symonds ALJ, Singh R, Symonds JD, Ogbe A, Omodho B, et al. Egr2 and 3 control adaptive immune responses by temporally uncoupling expansion from T cell differentiation. J Exp Med. (2017) 214:1787–808. doi: 10.1084/jem.20160553
30. Carow B, Gao Y, Terán G, Yang XO, Dong C, Yoshimura A, et al. CISH controls bacterial burden early after infection with Mycobacterium tuberculosis in mice. Tuberculosis. (2017) 107:175–80. doi: 10.1016/j.tube.2017.09.007
31. Queval CJ, Song OR, Carralot JP, Saliou JM, Bongiovanni A, Deloison G, et al. Mycobacterium tuberculosis controls phagosomal acidification by targeting CISH-mediated signaling. Cell Rep. (2017) 20:3188–98. doi: 10.1016/j.celrep.2017.08.101
32. Caetano Reis Sousa B, Hieny S, Scharton-Kersten T, Jankovic D, Charest H, Germain RN, et al. In Vivo Microbial Stimulation Induces Rapid CD40 Ligand-independent Production of Interleukin 12 by Dendritic Cells and their Redistribution to T Cell Areas. J Exp Med. (1997). http://www.jem.org.
33. Channon JY, Seguin RM, and Kasper LH. Differential infectivity and division of toxoplasma gondii in human peripheral blood leukocytes. Infect And Immun. (2000) 68(8):e0159121. doi: 10.1128/IAI.68.8.4822-4826.2000
34. Diana J, Persat F, Staquet MJ, Assossou O, Ferrandiz J, Gariazzo MJ, et al. Migration and maturation of human dendritic cells infected with Toxoplasma gondii depend on parasite strain type. FEMS Immunol Med Microbiol. (2004) 42:321–31. doi: 10.1016/j.femsim.2004.06.021
35. Liu C-H, Fan Y, Dias A, Esper L, Corn RA, Bafica A, et al. Cutting Edge: Dendritic Cells Are Essential for In Vivo IL-12 Production and Development of Resistance against Toxoplasma gondii Infection in Mice. J Immunol. (2006) 177:31–5. doi: 10.4049/jimmunol.177.1.31
36. León B, López-Bravo M, and Ardavín C. Monocyte-Derived Dendritic Cells Formed at the Infection Site Control the Induction of Protective T Helper 1 Responses against Leishmania. Immunity. (2007) 26:519–31. doi: 10.1016/j.immuni.2007.01.017
37. Randolph GJ, Inaba K, Robbiani DF, Steinman RM, and Muller WA. Differentiation of Phagocytic Monocytes into Lymph Node Dendritic Cells In Vivo differentiate into DCs in an in vitro model of transendo-thelial trafficking without addition of exogenous cyto-kines (Randolph et al., 1998), supporting the idea that most efficiently when they receive a phagocytic stimulus during their transient residence in subendothelial matrix. Immunity. (1999) 42(10):2535–43. doi: 10.1016/S1074-7613(00)80149-1
38. Hespel C and Moser M. Role of inflammatory dendritic cells in innate and adaptive immunity. Eur J Immunol. (2012) 42(10):2535–43. doi: 10.1002/eji.201242480
39. Schildberger A, Rossmanith E, Eichhorn T, Strassl K, and Weber V. Monocytes, peripheral blood mononuclear cells, and THP-1 cells exhibit different cytokine expression patterns following stimulation with lipopolysaccharide. Mediators Inflammation. (2013) 2013:1–10. doi: 10.1155/2013/697972
40. Shiratori H, Feinweber C, Luckhardt S, Linke B, Resch E, Geisslinger G, et al. THP-1 and human peripheral blood mononuclear cell-derived macrophages differ in their capacity to polarize in vitro. Molecular Immunology. (2017) 88:58–68. doi: 10.1016/j.molimm.2017.05.027
41. Rudzki EN, Ander SE, Coombs RS, Alrubaye HS, Cabo LF, Blank ML, et al. Toxoplasma gondii GRA28 is required for placenta-specific induction of the regulatory chemokine CCL22 in human and mouse. (2021) 11(2). doi: 10.1128/mBio
42. ten Hoeve AL, Braun L, Rodriguez ME, Olivera GC, Bougdour A, Belmudes L, et al. The Toxoplasma effector GRA28 promotes parasite dissemination by inducing dendritic cell-like migratory properties in infected macrophages. Cell Host Microbe. (2022) 30:1570–1588.e7. doi: 10.1016/j.chom.2022.10.001
43. Phelps ED, Sweeney KR, and Blader IJ. Toxoplasma gondii rhoptry discharge correlates with activation of the early growth response 2 host cell transcription factor. Infect Immun. (2008) 76:4703–12. doi: 10.1128/IAI.01447-07
44. Agnone A, Torina A, Vesco G, Villari S, Vitale F, Caracappa S, et al. Antigen-specific T cells and cytokines detection as useful tool for understanding immunity against zoonotic infections. Clin Dev Immunol. (2012). doi: 10.1155/2012/768789
Keywords: Toxoplasma gondii, host-pathogen interactions, PBMCs, monocytes, dendritic cells, single-cell RNA sequencing
Citation: Chandrasegaran P, Faydaci B, Shih B and Hassan MA (2025) Dual single-cell and bulk RNA sequencing reveal transcriptional profiles underlying heterogenous host-parasite interactions in human peripheral blood mononuclear cells. Front. Immunol. 16:1582645. doi: 10.3389/fimmu.2025.1582645
Received: 24 February 2025; Accepted: 28 May 2025;
Published: 24 June 2025.
Edited by:
Florent Ginhoux, Singapore Immunology Network (A*STAR), SingaporeReviewed by:
Sabrina Marion, Institut Pasteur de Lille, FranceOndrej Stepanek, Institute of Molecular Genetics (ASCR), Czechia
Copyright © 2025 Chandrasegaran, Faydaci, Shih and Hassan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Musa A. Hassan, bXVzYS5oYXNzYW5Acm9zbGluLmVkLmFjLnVr
 Praveena Chandrasegaran
Praveena Chandrasegaran Bekir Faydaci1
Bekir Faydaci1 Musa A. Hassan
Musa A. Hassan